Abstract
African swine fever virus (ASFV) is a highly pathogenic double-stranded DNA virus. It affects various breeds of pigs, causing serious economic losses and health threats because of its rapid spread and high pathogenicity and infectivity. This situation is not helped by the lack of a validated vaccine or effective therapies. Since the 1960s, different strains of ASFV have been subjected to serial passage in a variety of cell lines. The attenuated ASFV strains obtained through serial passage are not only candidates for ASF vaccine research, but also are useful to study the molecular genetic characteristics and pathogenic mechanism of the virus. This review summarizes related studies on the attenuated strains of ASFV acquired through cell passage over the last 60 years, with the aim of providing inspiration for the rational design of vaccines in future.
Keywords: African swine fever virus, Attenuation, Cell-adapted strain, Vaccine
Introduction
African swine fever (ASF) is a contagious disease of swine caused by African swine fever virus (ASFV), which is a large enveloped virus containing a double-stranded DNA genome of approximately 170–190 kbp. ASFV infection of domestic pigs and wild boars can induce a spectrum of disease severity, from subclinical to highly lethal, depending on host characteristics and the particular circulating virus strain, whereas African wild boar (warthogs, bush pigs, and giant forest hogs) infection is generally inapparent and acts as an ASFV reservoir host [1, 2].
ASF was first reported in Kenya in 1921. It spread widely on the African continent in the first and middle parts of the twentieth century. It was then introduced into Portugal from Angola in 1957 and began to spread in Europe, causing huge losses to the pig industry in many European countries. However, except for Sardinia, European countries succeeded in eradicating the disease in 1995 by utilizing stringent disease control programs (e.g., eradication and improved biosecurity measures) [3]. Scientists have been researching ASFV since Portugal lost nearly 20,000 pigs in 1957 because of an outbreak of ASF. With the advent of cell culture techniques in the 1960s, the era of vaccine candidate development began, and many attenuated ASFV strains were developed. However, until now, the development of a safe and effective ASF vaccine is still a formidable challenge. Early studies found that after continuous passage in cells, virulent strains no longer caused typical clinical signs after infecting pigs and the inoculated pigs could resist attack by homologous viruses [4, 5]. Meanwhile, some studies also found that after serial passage in cell lines, some strains, although displaying attenuated virulence, could not provide subsequent protection against homologous viruses [6]. In addition, the passage results of same ASFV strains in different cell lines varied in vivo [7]. However, no comprehensive studies have been carried out to illustrate the development of attenuated ASFV through serial passage in cell culture during 60 years of research. Therefore, in the present review, we summarize the current knowledge of the attenuated ASFVs obtained through serial passage in cell culture worldwide and analyze the features of attenuated strains and the rules of the passage, providing a reference for research on cell-adapted strains of ASFV and the development of new vaccines.
The attenuation of ASFV through passaging in primary cell culture
Research on ASFV cell-adapted strains first began in the 1960s. In 1960, Malmquist and Hay successfully isolated ASFV from swine bone marrow (BM) and buffy-coat (BC) culture and discovered the hemadsorption (HAD) reaction in infected pig leukocyte (PL) cultures [8]. Since then, scientists have tried to attenuate or modify ASFV by passage in cultures of spontaneously susceptible cells or by adaptation and passage in a variety of kidney cell lines [9]. In 1961, Ribeiro et al. passaged the Portuguese isolate 1455 through primary bone marrow cells (PBMCs) and showed that the virus gradually became attenuated [10]. They noted that the virus obtained within the 60th passage still induced unspecified locomotive disorders, as well as the typical signs of ASF in pigs; however, the locomotive disorders were much reduced by the 70th passage. In addition, animals immunized with this attenuated strain were protected from challenge with the ASFV strains circulating in both Portugal and Spain. Plowright and Ferris succeeded in passaging the Hinde strain of ASFV nine times in PBMCs [4, 11], but did not describe the process in detail. In 1963, Botija passaged five strains of ASF isolated from Spanish pigs using primary pig kidney cells (PPKs) and PLs and confirmed that attenuation of virulence occurred [12]. Indeed, one of the strains completely lost its pathogenicity to pigs after 60 passages. These preliminary studies demonstrated that ASFV could be attenuated through serial passage in cell culture, suggesting the practical application of this technique for vaccine development (Table 1).
Table 1.
Passages of African swine fever virus through primary cell culture
| Parental virus | Cell culture | Passages | Virulence evaluation | References | ||
|---|---|---|---|---|---|---|
| Strain | Virulence | Safety (No. surviving) | Protection | |||
| Lisbon 60 | Virulent | PBMCa | 89 | / | / | [9] |
| Portuguese 1455 | Virulent | PBMC | 70 | Attenuated | + | [10] |
| Hinde | Virulent | PPKb | 9 | / | / | [4] |
| Spencer | Virulent | PPK | 44 | Attenuated (4/4) | + | [4] |
| Portuguese | Virulent | PPK | 34 | Attenuated (2/2) | + | [4] |
| Gasson | Virulent | PPK | 23 | Attenuated (1/2) | −* | [4] |
| Hinde WH II | Virulent | BCc | 60 | Attenuated (102/111)e | + (40–59%) | [15] |
| Ugandan | Virulent | BC | 65 | Attenuated (2/2) | + | [16] |
| CV | Virulent | PLd and PBMC | 44 | Over-Attenuated (2/2) | − | [17] |
| Congo K49 | Virulent | PBMC | 262 | Attenuated (10/11) | + | [18, 19] |
| France F-32 | Virulent | PBMC | 135 | Attenuated (1/5) | − | [18, 19] |
| Lisbon-57 | Virulent | PBMC and PL | / | Attenuated | + (50–70%) | [20] |
| Katanga-78 | Virulent | PBMC | / | Attenuatedf | + (50–80%) | [20] |
| Congo-49 | Virulent | PBMC | / | Attenuatedg | + (75%) | [20] |
| Mozambique-78 | Virulent | PBMC | / | Attenuatedh | + (90%) | [20] |
| France-32 | Virulent | PBMC | / | Attenuated | + | [20] |
| Rhodesia | Virulent | PBMC and PL | / | Attenuatedi | + | [20] |
aPBMC primary bone marrow cell
bPPK primary pig kidney cell
cBC buffy-coat culture
dPL pig leukocyte
eSeveral pigs developed a mild febrile reaction and slight swellings over their joints
f42% of the pigs had a slight temperature reaction
gThe vaccinated pigs developed depression, high-temperature reaction (fever), and hemorrhages in parenchymal organs
h20% of pigs showed a high temperature after vaccination
iViremia (up to 105.0 hemagglutinating unit (HAU)50/mL) was detected in vaccinated pigs two weeks after infection
*The vaccinated pig remained normal until 30 days after challenge when it suddenly showed a high temperature and signs suggestive of ASF infection
/ unknown; + provided protection; − provided no protection
The Portuguese ASF vaccine strain, comprising the 81st passage of the Lisbon 60 virus through PBMCs, was used in animal experiments in 1967 [13]. It was inoculated intramuscularly (IM) into pig No. 114 and then heparinized blood drawn from this pig at the 3rd day, the peak of its febrile response, was inoculated IM in 1.0 mL amounts into four pigs (Nos. 115, 116, 117, and 118). After vaccination with the attenuated virus, there was only one day of clinical response (an increase in temperature). On the 71st day after vaccination, the four vaccinated pigs were given in feed comprising blood and tissue containing the virulent Portuguese strain. This failed to elicit a thermal response or to induce clinical infection. To test the resistance of vaccinates further, on the 15th day after being orally exposed to virulent ASFV, two of the four pigs (Nos. 115 and 117) were challenged by IM injection (3.0 mL of blood containing the challenge strain). The second challenge (IM) of No. 117 failed to elicit a thermal reaction after this inoculation, and the pig showed disease resistance. On the 108th day (the 22nd day after the second challenge), the temperature of pig No. 115 began to fluctuate for four rounds and gradually increased until it reached the highest temperature (41.3 ℃), at which point, the pig was sacrificed.
In 1967, Greig et al. serially passaged several different ASFV strains through PPK cells and challenged them with a homologous virus [4]. They found that there was no temperature response after pigs infected with the 35th passage of the Spencer strain (originating from Johannesburg, South Africa, in 1951) and the evidence showed that pigs died because of a bacterial infection rather than ASFV. The 39th passage had no virulence, and the inoculated pigs had no clinical signs and were able to resist challenge with the homologous virus; the 34th passage of the Portuguese challenge virus (originating from Portugal) induced a non-pathogenic temperature increase in pigs and provided protection of inoculated pigs against the homologous strain. The 23rd passage of the Gasson virus (isolated in 1950 in Kenya) was attenuated but could not provide protection (in two inoculated pigs: one died and the other returned to normal after an increasing body temperature but failed to resist the challenge with the homologous strain).
In 1968, Stone et al. used the attenuated Lisbon 60 strain (modified by Ribeiro et al. [10]) to immunize 22 pigs via IM injection (106 median tissue culture infectious dose (TCID50)/mL) and challenged them with the homologous strain on the 117th day after immunization (using a spleen homogenate diluted to 104 50% lethal dose (LD50) from the sick pig after being challenged by the virus) [14]. All inoculated pigs remained alive and exhibited no clinical signs of ASF. By contrast, 17 of 18 pigs that were challenged with the virulent Lisbon 60 ASFV had a thermal response and 4 pigs died. On the 23rd day after challenge, 14 pigs were killed to prepare a lung and spleen-mixed homogenate, which was inoculated into two healthy non-immunized pigs: both pigs died. Coggins’ team passaged the Hinde WH II virus [15] (isolated from Kenya) through BC culture for 60 serial passages and then the virus was passaged rapidly another 60 times, followed by a further 49 passages using a terminal dilution technique to produce Hinde WH II-TD49). The results of the challenge with virulent ASFV showed that the Hinde WH II-TD49 virus conferred partial resistance to virulent ASFV. In addition, viruses recovered from the two pigs that died following inoculation with the Hinde WH II-TD49 virus were fully virulent in susceptible pigs thereafter. Another study used the Ugandan strain of ASFV that had been passaged in porcine BC cultures [16] and found that some hemadsorption (HAD) reacting attenuated virus changed into non-hemadsorping (NHD) viruses; however, the NHD viruses segregated in vitro sometimes re-acquired the HAD characteristic after being passed back to pigs. Serial passage of a virulent virus in vitro often results in loss of virulence in an original animal host, while serial passage of a cell culture-adapted virulent virus in vivo often gains virulence in an animal host.
In 1979, Thomson et al. inoculated two pigs with an attenuated CV strain (the virulent CV strain isolated from South Africa in 1962 was attenuated by 43 passages in PLs and 1 passage in PBMCs) and challenged them with virulent virus [17]. The results showed that the inoculated pigs were not protected from lethal infection with the virulent homologous virus.
In a comparative analysis of ASFV genotypes and serogroups, the All-Russian Research Institute for Veterinary Virology and Microbiology (VNIIVViM) analyzed multiple ASFV strains that had been attenuated by cell passage, such as Congo KK-262 (an attenuated K49 derivative produced by 50 passages in PPK cell lines and 262 passages in PBMCs) and France FK-32/135 (obtained by 135 passages of F-32 in PBMCs) [18]. Animals immunized with the KK-262 virus showed a stable level of protection when challenged with virulent Congo K49. Most of the immunized animals survived and no clinical signs of ASF and viremia were detected. However, animals immunized with the France-attenuated (FK-32/135) virus were not protected from challenge with virulent Congo K49 [19]. In addition, the main results from the FRCVM (previously known as VNIIVViM) for the further analysis of various seroimmunotype-attenuated ASFV strains showed that most of them were attenuated by cell passage (mainly in PBMCs), showing varying degrees of protection against challenge with virulent ASFV. Comprehensive analysis of all attenuated strains indicated that FK-32/135 showed the best potential, but its inoculation route still needs further research and exploration [20].
The attenuation of ASFV via passage in continuous cell culture
In addition to the attenuation of ASFV through primary cell lines, scientists have also conducted many experiments on the passage and propagation of ASFV in continuous cell lines, which are mostly used for the study of viral gene function, virulence-associated genes, pathogenic mechanisms, and diagnosis (https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.09.01_ASF.pdf). Primary cells have disadvantages, including low reproducibility, high batch-to-batch variation, long time consumption, expensive cell extraction, and animal welfare considerations [21]; therefore, it is important to identify passaged cell lines that support the efficient replication of ASFV for biological research, virus purification, and the production of live-attenuated vaccines (Table 2).
Table 2.
Passages of African swine fever virus through continuous cell culture
| Parental virus | Cell culture | Passage | Virulence evaluation | References | ||
|---|---|---|---|---|---|---|
| Strain | Virulence | Safety (No. surviving) | Protection | |||
| L'60BM89 | Attenuated | Vero | 15 | / | / | [22] |
| Hinde | Virulent | PK-2a | 104 | Attenuated (8/8)m | / | [23] |
| Tengani | Virulent | BHK21 | 137 | / | / | [25] |
| BA71 | Virulent | Vero | 100 | Attenuated | / | [28] |
| E75 | Virulent | CV-1 | 4 | Attenuated (8/12)n | + (100%) | [29, 30] |
| ASFV-G | Virulent | Vero | 110 | Over-Attenuated | − | [6] |
| E70 | Virulent | MSj | 44 | Attenuated (9/9) | / | [31, 32] |
| Tengani | Virulent | Vero | 27 | Attenuated (6/6)o | + | [33] |
| Stavropol | Virulent | A4C2/9k | 33 | Attenuatedp | − | [7] |
| Stavropol | Virulent | CV-1 | 20 | Over-Attenuated | − | [7] |
| Stavropol | Virulent | PSGK-60k | 20 | Un-attenuated | − | [7] |
| TSP-80 | Virulent | PPK-66b/17l, CV, and PBMC | / | Attenuated | + (80–100%) | [20] |
| TS-7 | Virulent | PBMC and PPK-66b | / | Attenuated | + | [20] |
| Uganda | Virulent | PBMC and PPK-66b | 50 | Attenuatedq | + | [20] |
| ASFV-HLJ/18 | Virulent | HEK293T | 121 | / | / | [34] |
| Odintsovo 02/14 | Virulent | CV-1 | 30 | Attenuated (5/6) | + § | [35] |
| Odintsovo 02/14 | Virulent | CV-1 | 50 | Attenuated | / | [36] |
| OURT88/3 | Natural Attenuated | ZMAC-4 | 12 | / | + (100%) | [37] |
jMS: MS monkey kidney cell
kPSGK-60: continuous pig kidney cell
lPPK-66b: continuous pig kidney cell
mStrains after 75 passages and above produced a very mild or no noticeable response
nThe best inoculation dose was i.m.i. (intramuscular injection) with 104 hemagglutinating unit (HAU), the survival rate of infected pigs was 100%, and there was no viremia or only limited viremia and fever
oA pig had a diphasic pyrexia within a 12-day clinical course
pVariants of strain Stavropol of passage 14 retained their virulent properties. Passages 24 and 33 lost pathogenicity
qIt caused a temperature reaction in 9% of the inoculated pigs
§A group of weak animals showed swollen joints, bleeding, and necrotic lesions on their skin
In 1961, Ribeiro et al. passaged L'60BM89 virus (Lisbon 60 virus serially passaged 89 times through PBMCs) 15 times through Vero cells [22], proving that ASFV could also be propagated through a continuous cell line. Since then, this technique has been studied intensively by several research groups. In 1962, Malmquist et al. passaged the Hinde strain of ASFV (isolated from Kenya) in PK-2a cells (porcine kidney cells adapted to continuous cultivation) and found that increased passage of the Hinde strain in PK-2a cells decreased its virulence. The pigs inoculated with virus passaged 75 or more (90\103\104) times in cells survived completely [23]. Pigs infected with the 90th passage of Hinde virus in PK-2a cells all died after challenge with approximately 105 TCID50 of the homologous virulent virus. However, the pig immunized with the 103rd passage of Hinde virus and then inoculated with the 50th passage virus could resist attack by the homologous virulent virus [5], which proved that the virus strain attenuated by cell passage can confer imperfect protection.
In 1965, Hess et al. successively passaged several different strains of ASFV (formerly adapted to PLs or chick embryo cell cultures) in PK-2a cells. After many passages, the virulence of all strains changed [4, 24]. In 1974, Tessler et al. attenuated the Tengani strain of ASFV by 26 passages in swine BC cultures and 137 passages in a line of baby hamster kidney cells (BHK21) to test the virus titration method [25]. In 1976, Enjuanes et al. passaged the BA71 strain (isolated from the spleen of an affected pig in Badajoz, Spain in 1971 and passaged 36 times in swine monocytes) in Vero cells 100 times and obtained a non-pathogenic virus strain (BA71V) that was adapted to Vero cells [26–28]. In 1981, Gonzalvo et al. passaged the virulent E75 strain (isolated from Spain in 1975) in CV-1 cells (green monkey kidney fibroblasts) four times to obtain E75CV1 [29] and confirmed that it was partially effective in resisting the challenge with the homogenous virus. In recent research, the protective potential of E75CV1 against challenge with the homologous E75 virus was confirmed; however, it showed poor cross‑protection against the heterologous strain BA71 [30].
In 1987, the Spain 70 strain (isolated in Pontevedra in 1970, E70) was passaged 6 times in PLs (E70 L6) and then passaged again 14 (E70 MS14) and 44 (E70 MS44) times in the MS monkey kidney cell line (MS). Tabares et al. compared the restriction enzyme cleavage maps of ASFV grown in PLs (E70 L6) with that adapted in MS cells (E70 MS14) [31]. The results showed that after adaption to growth in MS cells, the size of DNA from ASFV strain E70 L6 decreased, mainly because of deletion and addition in the terminal region of the genome. To gain a better understanding of the genes involved in attenuation and those associated with the observed changes in phenotype with specific genetic variations, Zsak’s team constructed recombinant NL-S gene-deletion mutants from the E70 monkey cell culture-adapted virus, E70 MS44. The results indicated that the NL-S gene of ASFV, while not essential for growth in swine macrophages in vitro, is a significant viral virulence factor and might function as a host range gene [32].
In a study of ASFV subpopulations in 1991, Pan et al. successively passaged the Tengani virus (TG, isolated in Malawi, which is highly infectious and highly virulent), followed by cloning the TG strain in BC and Vero cells, and tested them in pigs [33]. It was found that the virulence and infectivity of TG decreased significantly after 27 passages in Vero cells.
The ASFV-G virus was passaged serially 110 times in Vero cells by Krug et al. in 2014. The adaptability of Vero cells reduced its ability to replicate in primary pig macrophages [6]. The virulence of ASFV gradually became attenuated through successive passages. In pigs inoculated with 102 HAD50 (50% hemadsorbing dose) of ASFV-G/VP30 (passaged 30 times through Vero cells), the signs of ASF were milder and the manifestation of the disease was rather transient compared with inoculation with the same dose of parental ASFV-G. In addition, IM inoculation of swine with 104 HAD50 of ASFV-G/VP110 induced no signs of ASF; however, it was unable to confer any protection against the challenge with the virulent virus. The animals that survived infection with low-dose (102 HAD50) ASFV-G/VP30, VP80, and VP110 were also challenged, but none of them obtained protection. The authors analyzed the genotypes and phenotypes of the viruses obtained in different passages (30/80/110) and found that deletions gradually accumulated in specific areas at the right and left variable ends of the genome.
To study the immunobiological properties of attenuated variants of ASFV adapted to continuous cell cultures, Balysheva et al. passaged the virulent ASFV strain Stavropol 01/08 (Stavropol) through several different cell lines [7]. The results showed that the variants of Stavropol strain at the 14th passage in A4C2/9k cells (a hybrid cell line SPEV TK with pig lymphocytes) and at the 20th passage in PSGK-60 cells (pig kidney cells) retained their virulence. At the 24th and 33rd passage in A4C2/9k cells and the 20th passage in African green monkey cells (CV-1) the virus had lost its pathogenicity for pigs but did not protect them from challenge with the virulent virus.
In recently published research, Tao Wang tested different cell lines to study cell-adaptive ASFV strains. They continuously passaged ASFV-WT in HEK293T (Human embryonic kidney 293T) cells for 121 passages [34]. After continuous passaging, ASFV adapted to H293T cells showed reduced infectivity toward primary porcine alveolar macrophages. The discovery of deletions of some genes associated with viral pathogenesis and virulence in pigs prompted the authors to conclude that the virulence of ASFV-VP121 to pigs might have been attenuated. Additionally, specialists of the FGBI “ARRIAH” in Russia prepared an ASFV strain (ASF/ARRIAH/CV-1) [35] that was adapted to growth in CV-1 cells. The mortality of pigs inoculated with the 30th passage the virus was 16.7%, and all the surviving pigs acquired resistance to repeated inoculation with the virulent isolate, Arm07. In a follow-up study, they prepared the ASFV/ARRIAH/CV-1/50 strain [36], which was passaged 50 times in CV-1 cells, for complete genome sequence analysis and comparison. The results showed that the adaptation of the ASFV to growth in CV-1 continuous cell culture led to the appearance of a large deletion in the 3′ variable region of the genome.
In 2020, in the process of studying alternatives to primary cells for ASFV replication, Portugal et al. passaged the attenuated strain of ASFV, OURT88/3, in Zuckermann macrophage-4 (ZMAC-4) cells for 12 generations. The results showed that it did not reduce the ability of this virus to induce protection against challenge with the virulent virus [37]. However, the adaptation of OURT88/3 viral strain to ZMAC-4 cell line needs to be further verified by virus DNA sequencing and other means.
Gene analysis of cell-adapted strains of African swine fever virus
Many studies have confirmed that the virus might change its pathogenicity and/or virulence to animals during cell passage. The attenuated ASFV strains were obtained by serial passaging; therefore, initially, it was not known which changes were introduced in the viral genome. Hence, efforts were made to determine the changes responsible for loss of virulence. Importantly, in 1982, Wesley and Pan first showed that, during the process of adapting the wild-type ASFV to cell culture, major changes in restriction endonuclease cleavage sites occurred [38]. Similarly, restriction enzyme cleavage maps for the fragments for ASFV DNA after adaptation to growth in MS cells (strain E70MS14) revealed that before adaptation to growth in MS cells, the size of the DNA from ASFV strain E70 L6 was 173 kbp and after adaptation it was only 156 kbp. The decrease in size was produced by deletions (15.2 kbp located near the left terminus and 2.4 kbp located near the right terminal fragment) and additions mainly in the terminal regions of the genome. In addition, it was found that the DNA of the ASFV E70 strain was homologous during the first 14 passages of the virus in monkey kidney cells; however, after 44 passages (E70MS44), the DNA was heterogeneous [31].
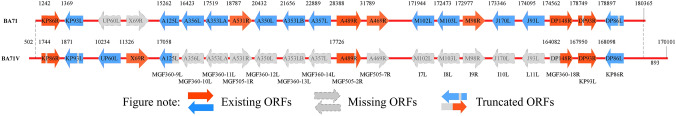
In 1995, the complete genome sequence of BA71V was reported. A comparison showed that the identity of the complete gene sequence between the attenuated BA71V and the Malawi strain was about 92%. The loss of gene colinearity between the two strains was mainly caused by deletions in the BA71V isolate associated with the gene families at the ends of the genome [28]. Furthermore, compared with BA71, BA71V has acquired two genes (UP60L and X69R). More importantly, large gene deletions occurred between MGF360-9L to MGF505-7R and I7L to MGF360-18R, which might be an important reason for the attenuation of BA71V (Fig. 1, Table 3). Another study showed that there were four large deletions in the left and right variable regions, three of which occurred in locations where genetic changes are frequently observed between strains [39].
Fig. 1.
Differential genes of BA71 and BA71V. Compared with BA71, BA71V has acquired two genes (UP60L and X69R), and the expression levels of six genes were different, including KP86R, KP93L, A125L (multigene family (MGF)360-9L), A489R (MGF505-2R), DP93R, and DP86L. More importantly, large gene deletions occurred between MGF360-9L to MGF505-7R and I7L to MGF360-18R, which might be an important reason for the attenuation of BA71V
Table 3.
Comparison of MGF deletions detected in attenuated African swine fever viruses
| Parental strain | Adapted strain | Parental strain | Adapted strain | Parental strain | Adapted strain | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BA71 | BA71V | ASFV-G | ASFV-G-VP30 | ASFV-G-VP60 | ASFV-G-VP80 | ASFV-G-VP110 | ASFV-G-ΔI177L/ΔLVR | Pig/HLJ/2018 | ASFV-P31 | ASFV-P61 | ASFV-P101 | ||
| MGF 100 | 1L | + | + | + | − | − | + | + | + | + | + | + | + |
| 1R | − | − | + | + | + | + | + | + | + | + | − | − | |
| 2L | + | + | + | + | + | + | + | + | + | + | + | + | |
| 3L | + | + | T | T | T | T | + | T | T | T | T | T | |
| MGF 110 | 1L | + | + | T | T | T | T | + | T | T | T | T | T |
| 2L | + | + | + | + | + | + | + | + | + | + | + | + | |
| 3L | F | F | + | + | + | + | + | + | + | + | + | + | |
| 4L | + | + | + | + | + | + | + | + | + | + | + | + | |
| 5L | + | + | F | F | F | F | F | F | F | F | F | − | |
| 6L | + | + | F | F | F | F | F | F | F | F | F | − | |
| 7L | T | T | + | + | + | + | + | + | + | + | + | − | |
| 8L | − | − | − | − | − | − | + | − | − | − | − | − | |
| 9L | − | − | + | + | + | + | + | + | + | + | − | − | |
| 11L | F | F | T | T | T | T | + | T | T | T | − | − | |
| 12L | F | F | + | + | + | + | + | + | + | + | − | − | |
| 13L | F | F | + | + | + | + | + | + | + | + | − | − | |
| 14L | F | F | F | F | F | F | + | F | F | F | − | − | |
| MGF 300 | 1L | + | + | + | + | + | + | + | − | + | − | − | − |
| 2R | + | + | + | + | + | + | + | − | + | − | − | − | |
| 4L | + | + | + | + | + | + | + | − | + | − | − | − | |
| MGF 360 | 1L | + | + | + | + | + | + | + | + | + | + | + | + |
| 2L | + | + | + | + | + | + | + | + | + | + | + | + | |
| 3L | + | + | + | + | + | + | + | + | + | + | + | + | |
| 4L | F | F | + | + | + | + | + | F | + | + | − | − | |
| 5L | F | F | − | − | − | − | + | − | − | − | − | − | |
| 6L | F | F | + | + | + | + | + | − | + | + | − | − | |
| 8L | + | + | + | + | + | + | + | − | + | − | − | − | |
| 9L | + | T | + | + | + | + | + | − | + | − | − | − | |
| 10L | + | − | + | + | + | + | + | − | + | − | − | − | |
| 11L | + | − | + | + | + | + | + | F | + | − | − | − | |
| 12L | + | − | + | + | + | + | + | + | + | + | + | − | |
| 13L | + | − | + | + | T | T | T | + | + | + | + | − | |
| 14L | + | − | + | + | − | − | − | + | + | + | + | − | |
| 15R | + | + | + | + | + | + | + | + | + | + | + | + | |
| 16R | + | + | + | + | + | + | + | + | + | + | + | + | |
| 18R | + | T | + | T | T | + | + | + | + | + | + | + | |
| 19R | + | + | + | + | + | + | + | + | + | + | + | + | |
| 21R | T | T | + | + | + | + | + | + | + | + | + | + | |
| 22R | − | − | − | − | − | − | − | − | − | − | − | − | |
| MGF 505 | 1R | − | − | + | + | + | + | − | + | + | + | + | − |
| 2R | + | T | + | + | − | − | − | + | + | + | + | + | |
| 3R | + | + | + | + | − | − | − | + | + | + | + | + | |
| 4R | + | + | + | + | − | − | − | + | + | + | + | + | |
| 5R | + | + | + | + | − | T | T | + | + | + | + | + | |
| 6R | F | F | + | + | T | + | + | + | + | + | + | + | |
| 7R | + | F | + | + | + | + | + | + | + | + | + | + | |
| 9R | + | + | + | + | + | + | + | + | + | + | + | + | |
| 10R | + | + | + | + | + | + | + | + | + | + | + | + | |
| 11L | + | + | + | T | T | + | + | + | + | + | + | + | |
MGF multigene family; T truncated protein; F fusion with another protein
During the process of passaging ASFV-G through Vero cells, two major deletions that mapped to specific regions within the left and right variable ends of the viral genome were observed [6]. The deletion at the right end of the genome first occurred at passage 30 and the deletion of the variable end on the left side of the genome were first observed at passage 60. Single mutations occurred in nine different genes in passage 110 relative to the parental ASFV-G genome (Table 3). Deletions at the right variable end of the genome were similar to the deletions in cell-adapted BA71V. The deletion of a multigene family at the left variable end of the genome is common in many attenuated ASFV strains (OURT88/3 [40], Pr4△35 [41], and E70△NL [42]). This deletion often leads to attenuated virulence and affects the ability of ASFV to grow in macrophage cell cultures. Therefore, it is inferred that the attenuation of ASFV-G/VP110 might be caused by the deletion of the MGF360 and MGF505 genes on the left side of the ASFV genome.
After the passage of ASFV through H293T cells, stepwise losses and cumulative mutations at the left variable end of MGF genes were discovered [34]. The 121st passage of ASFV through H293T cells had a total of about 25 kbp of deleted gene sequences, including 22 MGF genes (Table 3) and two non-MGF genes (X96R and 285L). Through comparison, it was found that the deleted MGF genes in ASFV-P121 were very similar to the MGF genes in the BA71V strain, and these genes were related to virus replication in HEK293T and Vero cells. They also found that the 3′-terminal mutations in the genome were mainly concentrated in the I7L, I8L, I9L, I10L, and MGF100-3L genes. However, further research is needed on the function of these genes and the mechanism of the replication of ASFV in H293T cells.
Russian scholars conducted complete genome sequence analysis of two ASFV strains adapted to grow in CV-1 cell culture for 30 and 50 passages and compared them with the parental virus. The results indicated a large deletion within the genomes of the highly passaged strains [36]. Similar to the genome of cell-adapted strain BA71V [39], the genomes of ASFV/ARRIAH/CV-1/30 and ASFV/ARRIAH/CV-1/50 also had deletions of large fragments in the right variable region.
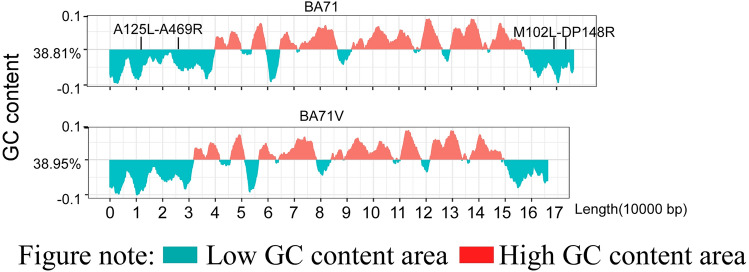
The above description showed that gene loss of attenuated ASFV strains mainly occurs in the MGF region, whether at the 5′ or 3′ end. This might be related to the low content of GC nucleotides in this region, which is more obvious at the 3′ end in the region from MA102L to DP148R at the 3′ end of BA71V (Fig. 2) for the average cost of GC-rich amino acids is lower than that of AT-rich amino acids [43]. Some genes in MGF360 and MGF505 are virulence genes and the deletion of the MGF360/505 gene will lead to an increase in the type I interferon mRNA level, showing that the MGF family can regulate innate immunity [44–46], which was supported by a later study on MGF505-7R [47]. In addition, it is reported that the deletion of MGF110-9L will reduce the virulence of ASFV wild virus and provide homologous protection [48], but does not affect the proliferation of the virus. However, the ASFV cell adaptation strains ASFV-G-ΔI177L/ΔLVR [49] and ASFV-P101 [34] showed varying degrees of deletion in the MGF region (Table.3), further suggesting that the loss of MGF plays a role in virus adaption to cells. However, the functions of many genes and members of the ASFV multigene family are not fully understood. Analysis of the genetic structure changes of ASFV in the continuous cell line adaptation process and comparing the biological characteristics of the virus strains will help to further understand the influence of the genome structure on the phenotypic characteristics, which will inspire the construction of better ASFV gene-deletion strains and the development of safe and reliable vaccines.
Fig. 2.
GC content distribution and gene loss regions of BA71 and BA71v. The above description showed that gene loss of attenuated ASFV strains mainly occurs in the MGF region, whether at the 5′ or 3′ end. This might be related to the low content of GC nucleotides in this region, which is also the cause of gene loss in the region from MA102L to DP148R at the 3′ end of BA71V
Concluding remarks
ASFV spreads rapidly and causes severe disease; therefore, the development of vaccines to prevent ASFV infections is a high priority. Sixty years ago, scientists tried to attenuate the virulent ASFV strains through serial passage of viruses in cell culture; however, soon after, they realized that achieving an optimal balance of attenuation and immunogenicity is challenging, especially attenuating virulent ASFV sufficiently to abrogate its severe disease potential while retaining its immunogenicity. Since the introduction of ASFV into Spain and Portugal in 1960, scientists have serially sub-cultured the strains isolated from the field through PBMCs and used them for swine vaccination in Spain and Portugal [10]. Among the half a million pigs vaccinated in Portugal, some of them developed ASF- or vaccination-related diseases. It was found that the pigs’ immune status has an impact on the immune effect of the attenuated virus. Later studies also confirmed that attenuated NHD variant Kc-160 (Katanga-160), inoculated at the same dose, caused only a weak or moderate clinical response in 75–80% of healthy pigs, but caused the death of 20% of pigs with lowered immune status [20].
Serial passage in primary or continuous cell culture is the classic means of attenuating ASFV. Whether in primary or continuous cell culture, through serial passage, the virulence of the virus gradually decreases; meanwhile, the virus immunogenicity also decreases, making it difficult to produce protection against subsequent challenge by the homologous virus. In addition, attempts to attenuate ASFV by additional passage in cultured cells have been accompanied by an altered expression or even complete deletion of genes encoding certain virulence or immune-evasion factors. Among the erasable regions, ASFV multigene families are the first to bear the brunt. The BA71V, ASFV-G-VP110, and ASFV-P101 strains mentioned above clearly show this tendency. Further comparison of the different outcomes between serial passage in primary and continuous cell culture appeared to show that ASFV can attenuate faster or to a greater extent within the fewer passage times in continuous cell culture, especially in a heterologous permanent cell line. For instance, the E75 strain (Genotype I) lost partial pathogenicity when adapted to continuous cell cultures of heterologous (CV-1) origin cells within four consecutive passages. Meanwhile, for primary cell culture, virulent strains, such as Gasson and Portuguese (Genotype I), need at least 23 and 34 passages, respectively, in primary pig kidney cells to become partially attenuated. Meanwhile, serial passage in continuous cell culture appears to have a higher tendency to induce sequence changes in ASFV genomes, resulting in adaptation that is best suited to growth in specific cell lines.
Besides the preparation of vaccine strains, cell-adapted strains are of great significance to understand the mutation of the virus in the process of adapting to cells, which, through complete genome sequencing analysis, will determine the function of viral genes [39, 50]. Serial passaging can produce attenuated strains, some of which might provide considerable levels of protection. With the help of recombinant DNA technology and sequencing technology, analysis of the region from which the attenuated strains lose genes through cell passage will guide the further development of recombinant live-attenuated vaccines.
Conventional ASFV isolation and culture are based on PBMCs and porcine alveolar macrophages; however, these cells need to be collected from pigs and thus they are difficult to standardize [7]. ASFV can replicate in some established cell lines only after a process involving blind serial passages [6, 31], which introduces uncertainty. To solve these problems, scientists have sought or constructed new cell lines. Currently, COS-1 [51, 52], IPAM [21, 52], MA104 [53], WSL [21], A4C2/9K [7, 54], ZMAC-4 [37], and IPKM [55] cell lines have been found to be suitable to propagate ASFV. Although some cell-adapted strains [37, 49] provided effective protection against the parental strain, practical investigations of these strains are required to determine whether they might be suitable candidates for commercial production of an attenuated vaccine strain against ASFV. Meanwhile, further in-depth research is needed to determine the genomic changes that occur in the adaptation process and their impact on virus characteristics.
Author contributions
XZ, ZW, and SG conceived the article, analyzed the literature, and drafted the manuscript. YZ, HL, and YL made significant contributions to the analysis of the literature and the production of tables and pictures. NH and YZ played an important role in revising the language logic and grammar of the article. YC, XW, and ZW critically revised the work.
Funding
This study was supported by the Shandong Province Major Science and Technology Innovation Project: Research and Application of Key Technologies for African Swine Fever Prevention and Control (Grant No. 2020CXGC010801-04) and the National Key Research and Development Program of China (Grant No. 2021YFD1801300).
Declarations
Conflict of interest
The authors declare no conflicts of interest associated with this manuscript.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaoyue Zhang, Zhenzhong Wang, and Shengqiang Ge equally contributed to this paper.
Contributor Information
Yumei Cai, Email: caiyum@163.com.
Xiaodong Wu, Email: wuxiaodong@cahec.cn.
Zhiliang Wang, Email: wangzhiliang@cahec.cn.
References
- 1.Tulman ER, Delhon GA, Ku BK, Rock DL. African swine fever virus. Curr Top Microbiol Immunol. 2009;328:43–87. doi: 10.1007/978-3-540-68618-7_2. [DOI] [PubMed] [Google Scholar]
- 2.OIE (2019) OIE Technical Disease Card: African swine fever. https://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/AFRICAN_SWINE_FEVER.pdf.
- 3.Cwynar P, Stojkov J, Wlazlak K. African Swine fever status in Europe. Viruses. 2019 doi: 10.3390/v11040310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greig AS, Boulanger P, Bannister GL. African swine fever. V. Cultivation of the virus in primary pig kidney cells. Can J Comp Med Vet Sci. 1967;31:24–31. [PMC free article] [PubMed] [Google Scholar]
- 5.Malmquist WA. Serologic and immunologic studies with African swine fever virus. Am J Vet Res. 1963;24:450–459. [PubMed] [Google Scholar]
- 6.Krug PW, Holinka LG, O'Donnell V, Reese B, Sanford B, Fernandez-Sainz I, Gladue DP, Arzt J, Rodriguez L, Risatti GR, Borca MV. The progressive adaptation of a georgian isolate of African swine fever virus to vero cells leads to a gradual attenuation of virulence in swine corresponding to major modifications of the viral genome. J Virol. 2015;89:2324–2332. doi: 10.1128/JVI.03250-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balysheva VI, Prudnikova EY, Galnbek TV, Balyshev VM. Immunological properties of attenuated variants of African swine fever virus isolated in the Russian Federation. Russ Agric Sci. 2015;41:178–182. doi: 10.3103/S1068367415020056. [DOI] [Google Scholar]
- 8.Malmquist WA, Hay D. Hemadsorption and cytopathic effect produced by African Swine Fever virus in swine bone marrow and buffy coat cultures. Am J Vet Res. 1960;21:104–108. [PubMed] [Google Scholar]
- 9.Becker Y. African swine fever. Developments in veterinary virology. Martinus Nijhoff Publishing. 1987;9:81. [Google Scholar]
- 10.Manso-Ribeiro J, Nunes-Petisca JL, Lopez-Frazao F, Sobral M. Vaccination against ASF. Bull Off Int Epizoot. 1963;60:921–937. [Google Scholar]
- 11.Plowright W, Ferris RD (1957) East African Veterinary Research Organisation. Annual Report 1956–57. East African Veterinary Research Organisation Annual Report 28.
- 12.Botija CS. Modification of African classical swine fever virus in cell culture It is helpful to understand the pathogenic effect and protective power of attenuated strains "in Spanish". Bull Off Int Epizoot. 1963;60:901–919. [Google Scholar]
- 13.Bannister GL, Gray DP, Boulanger P, Willis NG. African swine fever. I. Antiserum production. Can J Comp Med. 1967;31:91–95. [PMC free article] [PubMed] [Google Scholar]
- 14.Stone SS, DeLay PD, Sharman EC. The antibody response in pigs inoculated with attenuated African swine fever virus. Can J Comp Med. 1968;32:455–460. [PMC free article] [PubMed] [Google Scholar]
- 15.Coggins L, Moulton JE, Colgrove GS. Studies with hinde attenuated African swine fever virus. Cornell Vet. 1968;48:525–540. [PubMed] [Google Scholar]
- 16.Coggins L. Segregation of a nonhemadsorbing African swine fever virus in tissue culture. Cornell Vet. 1968;58:12–20. [PubMed] [Google Scholar]
- 17.Thomson GR, Gainaru MD, van Dellen AF. African swine fever: pathogenicity and immunogenicity of two non-haemadsorbing viruses. Onderstepoort J Vet Res. 1979;46:149–154. [PubMed] [Google Scholar]
- 18.Malogolovkin A, Burmakina G, Titov I, Sereda A, Gogin A, Baryshnikova E, Kolbasov D. Comparative analysis of African swine fever virus genotypes and serogroups. Emerg Infect Dis. 2015;21:312–315. doi: 10.3201/eid2102.140649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burmakina G, Malogolovkin A, Tulman ER, Zsak L, Delhon G, Diel DG, Shobogorov NM, Morgunov YP, Morgunov SY, Kutish GF, Kolbasov D, Rock DL. African swine fever virus serotype-specific proteins are significant protective antigens for African swine fever. J Gen Virol. 2016;97:1670–1675. doi: 10.1099/jgv.0.000490. [DOI] [PubMed] [Google Scholar]
- 20.Sereda AD, Balyshev VM, Kazakova AS, Imatdinov AR, Kolbasov DV. Protective properties of attenuated strains of African Swine fever virus belonging to seroimmunotypes I–VIII. Pathogens. 2020;9:274. doi: 10.3390/pathogens9040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez EG, Riera E, Nogal M, Gallardo C, Fernandez P, Bello-Morales R, Lopez-Guerrero JA, Chitko-McKown CG, Richt JA, Revilla Y. Phenotyping and susceptibility of established porcine cells lines to African Swine Fever Virus infection and viral production. Sci Rep. 2017;7:10369. doi: 10.1038/s41598-017-09948-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manso Ribeiro J, Rosa Azevedo JA. La peste porcine africaine au Portugal. Bull Off Int Epizoot. 1961;55:88–108. [Google Scholar]
- 23.Malmquist WA. Propagation, modification, and hemadsorption of African swine fever virus in cell cultures. Am J Vet Res. 1962;23:241–247. [PubMed] [Google Scholar]
- 24.Hess WR, Cox BF, Heuschele WP, Stone SS. Propagation and modification of African swine fever virus in cell cultures. Am J Vet Res. 1965;26:141–146. [PubMed] [Google Scholar]
- 25.Tessler J, Hess WR, Pan IC, Trautman R. Immunofluorescence plaque assay for African swine fever virus. Can J Comp Med. 1974;38:443–447. [PMC free article] [PubMed] [Google Scholar]
- 26.Enjuanes L, Carrascosa AL, Moreno MA, Vinuela E. Titration of African swine fever (ASF) virus. J Gen Virol. 1976;32:471–477. doi: 10.1099/0022-1317-32-3-471. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Barreno B, Sanz A, Nogal ML, Vinuela E, Enjuanes L. Monoclonal antibodies of African swine fever virus: antigenic differences among field virus isolates and viruses passaged in cell culture. J Virol. 1986;58:385–392. doi: 10.1128/JVI.58.2.385-392.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanez RJ, Rodriguez JM, Nogal ML, Yuste L, Enriquez C, Rodriguez JF, Vinuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalvo F, Carnero M, Bruyel V. Immunological responses of pigs to partially attenuated ASF virus and their resistance to virulent homologous and heterologous viruses. In: Wilkinson PJ, editor. African Swine fever-proceedings of a CEC/FAO research seminar. Sassari: Commission of the European Communities; 1981. pp. 206–216. [Google Scholar]
- 30.Lacasta A, Monteagudo PL, Jimenez-Marin A, Accensi F, Ballester M, Argilaguet J, Galindo-Cardiel I, Segales J, Salas ML, Dominguez J, Moreno A, Garrido JJ, Rodriguez F. Live attenuated African swine fever viruses as ideal tools to dissect the mechanisms involved in viral pathogenesis and immune protection. Vet Res. 2015;46:135. doi: 10.1186/s13567-015-0275-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabares E, Olivares I, Santurde G, Garcia MJ, Martin E, Carnero ME. African swine fever virus DNA: deletions and additions during adaptation to growth in monkey kidney cells. Arch Virol. 1987;97:333–346. doi: 10.1007/BF01314431. [DOI] [PubMed] [Google Scholar]
- 32.Zsak L, Lu Z, Kutish GF, Neilan JG, Rock DL. An African swine fever virus virulence-associated gene NL-S with similarity to the herpes simplex virus ICP34.5 gene. J Virol. 1996;70:8865–8871. doi: 10.1128/JVI.70.12.8865-8871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan IC. African swine fever virus: generation of subpopulations with altered immunogenicity and virulence following passage in cell cultures. J Vet Med Sci. 1992;54:43–52. doi: 10.1292/jvms.54.43. [DOI] [PubMed] [Google Scholar]
- 34.Wang T, Wang L, Han Y, Pan L, Yang J, Sun M, Zhou P, Sun Y, Bi Y, Qiu HJ. Adaptation of African swine fever virus to HEK293T cells. Transbound Emerg Dis. 2021;68:2853–2866. doi: 10.1111/tbed.14242. [DOI] [PubMed] [Google Scholar]
- 35.Ali Mazloum NGZ. Analysis of changes in African swine fever virus genetic structure and biological properties during adaptation to continuous cell culture. Vet Sci Today. 2018 doi: 10.29326/2304-196X-2018-4-27-21-25. [DOI] [Google Scholar]
- 36.Mazloum A, Igolkin A, Zinyakov N, Van Schalkwyk A, Vlasova N. Changes in African swine fever virus (Asfarviridae: Asfivirus: African swine fever virus) genome associated with adaptation to reproduction in continuous cell culture. Vopr Virusol. 2021;66:211–216. doi: 10.36233/0507-4088-50. [DOI] [PubMed] [Google Scholar]
- 37.Portugal R, Goatley LC, Husmann R, Zuckermann FA, Dixon LK. A porcine macrophage cell line that supports high levels of replication of OURT88/3, an attenuated strain of African swine fever virus. Emerg Microbes Infect. 2020;9:1245–1253. doi: 10.1080/22221751.2020.1772675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wesley RD, Pan IC. African swine fever virus DNA: restriction endonuclease cleavage patterns of wild-type, Vero cell-adapted and plaque-purified virus. J Gen Virol. 1982;63:383–391. doi: 10.1099/0022-1317-63-2-383. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez JM, Moreno LT, Alejo A, Lacasta A, Rodriguez F, Salas ML. Genome sequence of African swine fever virus BA71, the virulent parental strain of the nonpathogenic and tissue-culture adapted BA71V. PLoS ONE. 2015;10:22. doi: 10.1371/journal.pone.0142889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapman DAG, Tcherepanov V, Upton C, Dixon LK. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J Gen Virol. 2008;89:397–408. doi: 10.1099/vir.0.83343-0. [DOI] [PubMed] [Google Scholar]
- 41.Zsak L, Lu Z, Burrage TG, Neilan JG, Kutish GF, Moore DM, Rock DL. African swine fever virus multigene family 360 and 530 genes are novel macrophage host range determinants. J Virol. 2001;75:3066–3076. doi: 10.1128/JVI.75.7.3066-3076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neilan JG, Zsak L, Lu Z, Kutish GF, Afonso CL, Rock DL. Novel swine virulence determinant in the left variable region of the African swine fever virus genome. J Virol. 2002;76:3095–3104. doi: 10.1128/jvi.76.7.3095-3104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du MZ, Zhang C, Wang H, Liu S, Wei W, Guo FB. The GC content as a main factor shaping the amino acid usage during bacterial evolution process. Front Microbiol. 2018;9:2948. doi: 10.3389/fmicb.2018.02948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gil S, Spagnuolo-Weaver M, Canals A, Sepúlveda N, Oliveira J, Aleixo A, Allan G, Leito A, Martins CLV. Expression at mRNA level of cytokines and A238L gene in porcine blood-derived macrophages infected in vitro with African swine fever virus (ASFV) isolates of different virulence. Arch Virol. 2003;148:2077–2097. doi: 10.1007/s00705-003-0182-x. [DOI] [PubMed] [Google Scholar]
- 45.Yang K, Xue Y, Niu H, Shi C, Cheng M, Wang J, Zou B, Wang J, Niu T, Bao M, Yang W, Zhao D, Jiang Y, Yang G, Zeng Y, Cao X, Wang C. African swine fever virus MGF360-11L negatively regulates cGAS-STING-mediated inhibition of type I interferon production. Vet Res. 2022;53:7. doi: 10.1186/s13567-022-01025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang K, Huang Q, Wang R, Zeng Y, Cheng M, Xue Y, Shi C, Ye L, Yang W, Jiang Y, Wang J, Huang H, Cao X, Yang G, Wang C. African swine fever virus MGF505-11R inhibits type I interferon production by negatively regulating the cGAS-STING-mediated signaling pathway. Vet Microbiol. 2021;263:109265. doi: 10.1016/j.vetmic.2021.109265. [DOI] [PubMed] [Google Scholar]
- 47.Li D, Zhang J, Yang W, Li P, Ru Y, Kang W, Li LL, Ran Y, Zheng H. African swine fever virus protein MGF-505–7R promotes virulence and pathogenesis by inhibiting JAK1- and JAK2-mediated signaling. J Biol Chem. 2021;297:101190. doi: 10.1016/j.jbc.2021.101190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li D, Liu Y, Qi X, Wen Y, Li P, Ma Z, Liu Y, Zheng H, Liu Z. African swine fever virus MGF-110-9L-deficient mutant has attenuated virulence in pigs. Virologica Sinica. 2021;36:187–195. doi: 10.1007/s12250-021-00350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borca MV, Rai A, Ramirez-Medina E, Silva E, Velazquez-Salinas L, Vuono E, Pruitt S, Espinoza N, Gladue DP. A cell culture-adapted vaccine virus against the current african swine fever virus pandemic strain. J Virol. 2021;95:e0012321. doi: 10.1128/jvi.00123-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santurde G, Ruiz Gonzalvo F, Carnero ME, Tabares E. Genetic stability of African swine fever virus grown in monkey kidney cells. Brief report. Arch Virol. 1988;98:117–122. doi: 10.1007/BF01321012. [DOI] [PubMed] [Google Scholar]
- 51.de León P, Bustos MJ, Carrascosa AL. Laboratory methods to study African swine fever virus. Virus Res. 2013;173:168–179. doi: 10.1016/j.virusres.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 52.Carrascosa AL, Bustos MJ, de Leon P. Methods for growing and titrating African swine fever virus: field and laboratory samples. Curr Protoc Cell Biol. 2011 doi: 10.1002/0471143030.cb2614s53. [DOI] [PubMed] [Google Scholar]
- 53.Rai A, Pruitt S, Ramirez-Medina E, Vuono EA, Silva E, Velazquez-Salinas L, Carrillo C, Borca MV, Gladue DP. Identification of a continuously stable and commercially available cell line for the identification of infectious African swine fever virus in clinical samples. Viruses. 2020;12:820. doi: 10.3390/v12080820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balysheva VI, Prudnikova EY, Galnbek TV, Balyshev VM. Continuous cell subline A4C2/9K and its application to the African swine fever virus study. Vopr Virusol. 2015;60:43–47. [PubMed] [Google Scholar]
- 55.Masujin K, Kitamura T, Kameyama K, Okadera K, Nishi T, Takenouchi T, Kitani H, Kokuho T. An immortalized porcine macrophage cell line competent for the isolation of African swine fever virus. Sci Rep. 2021;11:4759. doi: 10.1038/s41598-021-84237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]