Abstract
Purpose:
Perineural invasion (PNI), a common occurrence in oral squamous cell carcinomas, is associated with poor survival. Consequently, these tumors are treated aggressively. However, diagnostic criteria of PNI vary and its role as an independent predictor of prognosis has not been established. To address these knowledge gaps, we investigated spatial and transcriptomic profiles of PNI-positive and PNI-negative nerves.
Experimental Design:
Tissue sections from 142 patients were stained with S100 and cytokeratin antibodies. Nerves were identified in two distinct areas: tumor bulk and margin. Nerve diameter and nerve-to-tumor distance were assessed; survival analyses were performed. Spatial transcriptomic analysis of nerves at varying distances from tumor was performed with NanoString GeoMx Digital Spatial Profiler Transcriptomic Atlas.
Results:
PNI is an independent predictor of poor prognosis among patients with metastasis-free lymph nodes. Patients with close nerve-tumor distance have poor outcomes even if diagnosed as PNI negative using current criteria. Patients with large nerve(s) in the tumor bulk survive poorly, suggesting that even PNI-negative nerves facilitate tumor progression. Diagnostic criteria were supported by spatial transcriptomic analyses of >18,000 genes; nerves in proximity to cancer exhibit stress and growth response changes that diminish with increasing nerve-tumor distance. These findings were validated in vitro and in human tissue.
Conclusions:
This is the first study in human cancer with high-throughput gene expression analysis in nerves with striking correlations between transcriptomic profile and clinical outcomes. Our work illuminates nerve-cancer interactions suggesting that cancer-induced injury modulates neuritogenesis, and supports reclassification of PNI based on nerve-tumor distance rather than current subjective criteria.
Translational Relevance.
Perineural invasion (PNI) occurs frequently in oral squamous cell carcinoma. However, diagnostic criteria of PNI vary and its role as an independent predictor of prognosis has not been established. We reveal that PNI is an important predictor of prognosis among patients with no lymph node metastasis. Moreover, patients with nerves closer to tumor have poor outcomes even if diagnosed as PNI negative using current criteria. Consistent with clinical findings, spatial transcriptomic analysis of nerves in sections of human tumors shows a gradient of gene expression that is dependent on nerve-tumor distance. Together, these findings support broadening the definition of PNI to make it more clinically relevant. Moreover, the transcriptomic data reveal an injury response to the tumor; mechanisms involved in response to stress and growth are upregulated in nerves proximal to cancer. Understanding how these mechanisms contribute to clinical outcomes is crucial for developing new treatment strategies that reduce tumor recurrence.
Introduction
Investigation of the dynamic interaction between cancer and nerves is the focus of the emerging field of cancer neuroscience (1). The impact of these studies has not been translated into clinical practice. For example, perineural invasion (PNI) is an adverse pathologic feature in multiple cancers including prostate, gastric, and oral squamous cell carcinoma (OSCC; refs. 2–4). Consequently, diagnosis of PNI has serious implications for treatment planning and prognosis. However, the diagnostic criteria are archaic; molecular changes between what are currently classified as PNI-positive and PNI-negative nerves have not been investigated in human cancer. In this study, applying spatial and molecular investigations of nerves in human cancer, we provide evidence for reclassification of PNI based on current knowledge of cancer neuroscience.
PNI is defined as “tumor in close proximity to nerve and involving at least 33% of its circumference or tumor cells within any of the three layers of the nerve sheath” (5). However, in mechanistic studies, nerves modulate tumor progression prior to physical contact with cancer cells (6–10). In an in vivo model in which neural ganglia and oral cancer cells were grafted adjacent to each other, nerves were attracted toward cancer cells overexpressing galanin receptor 2 (8). Furthermore, nerves have effects that vary with taxonomy and tumor location. For example, adrenergic nerves promote cancer cell survival in early-stage tumor development, while cholinergic nerves regulate invasion, migration, and metastasis enhancing tumor dissemination in advanced-stage prostate cancer (6). In OSCC, p53 deficiency modulates neuronal responses in the tumor microenvironment, and drives sensory nerves to adrenergic differentiation via a miRNA-regulated mechanism (10). Interestingly, surgical denervation of sensory nerves inhibited the presence of tumor-related adrenergic nerves and, consequently, tumor growth in mice (10). Similarly, surgical and chemical denervation of the stomach inhibited tumor formation and progression in mouse models of gastric cancer through inhibition of muscarinic acetylcholine M3 receptor (7). Together, these findings show that nerves impact tumor progression even in the absence of direct physical contact.
On the basis of evidence supporting nerve-tumor crosstalk, our group investigated PNI and other nerve parameters in 71 patients with OSCC. These results suggested that nerves influence tumor behavior in the absence of PNI (4). In the current study in human OSCC, we investigated spatial transcriptomic changes in nerves at varying distance from cancer cells. These findings were supported by in vitro studies. Furthermore, using an independent cohort of patients, we validated our previous translational work on the association of PNI and other nerve-related parameters with clinical outcome in OSCC. Our findings support that PNI should be redefined on the basis of nerve-tumor distance rather than the extent to which nerves are surrounded by cancer cells.
Materials and Methods
Patient population
Deidentified tissue sections of oral cavity OSCC were obtained from the Tissue Core of the University of Michigan Head and Neck Cancer Specialized Program of Research Excellence (HNSPORE)/Head and Neck Oncology Program (HNOP). Institutional Review Board approval and patient consent were obtained by the HNSPORE/HNOP prior to specimen collection. The study population consisted of 142 patients (Fig. 1A). Of these, 71 were from a previously published cohort (4) here named cohort 1, and a new population of 71 patients, designated cohort 2. Tissue sections for the first cohort were from pretreatment biopsies, and the second cohort were from pretreatment biopsies or a single-tissue block of the excision specimen. Cohort 1 data were collected between November 2008 and June 2012, while cohort 2 data were collected between June 2012 and September 2014. Follow-up data for cohort 1 were updated since publication in 2018; cohort 2 and updated cohort 1 data are shown in Supplementary Table S1.
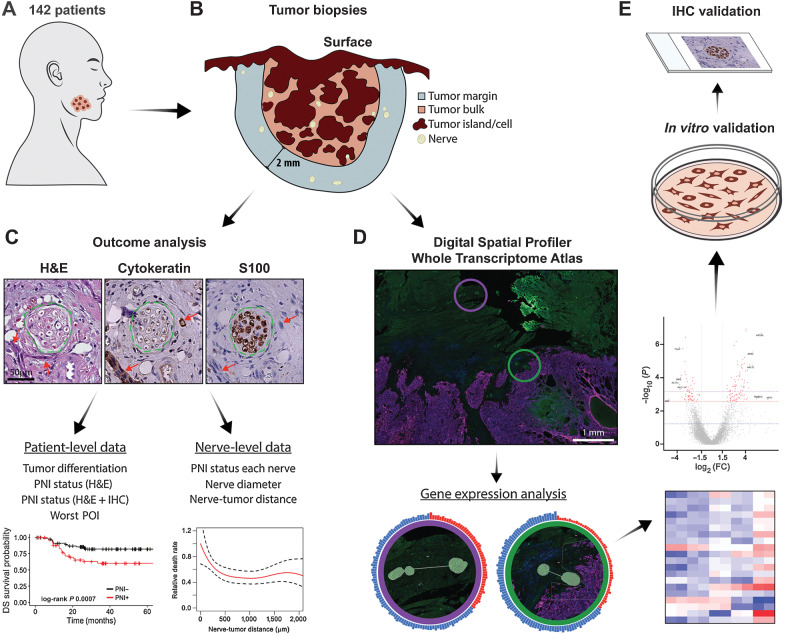
Figure 1.
Schematic summary of study design. A, Cohort of 142 patients with OSCC. B, Tumor biopsy samples were serially sectioned. All nerves in the tumor bulk and in a 2 mm margin around the tumor bulk were assessed. C, H&E, cytokeratin, and S100 stains were used to locate tumor cells and nerves. Green dashed lines show the nerve outline and red arrows indicate tumor cells. Scale bar, 50 μm. Analysis of serial sections generated patient-level and nerve-level data, used for outcome analysis. D, A subset of samples (n = 8) was selected for spatial transcriptomic analysis. Morphology markers for cytokeratin and S100 were used to guide identification of nerve areas in relation to tumor. AOIs for a nerve close to tumor (green circle) and a nerve far from tumor (purple circle) are overlaid in the immunofluorescence image and enlarged below. Tissues were probed for over 18,000 genes; differential gene expression analysis was performed comparing nerves in different areas of the tumor specimen. E,In vitro validation was performed in neuronal and Schwann cells, and rat DRG; IHC validation was performed in patient tissue specimens.
The combined study population of 142 patients (73 males, 69 females) had a mean age of 62.8 years and median follow-up time of 43.9 months. Most patients were treated with surgery; 58 (40.8%) patients received surgery alone, 49 (34.5%) received surgery plus adjuvant radiotherapy, and 35 (24.6%) received surgery plus adjuvant chemoradiation. OSCC recurred in 41 patients, 35 of whom died of disease. Table 1 summarizes demographic and disease-related characteristics of each cohort and the combined population. For comparison, Supplementary Fig. S1 shows survival data for cohort 2 using the same analyses published previously for cohort 1 (4).
Table 1.
Demographics and disease-related characteristics of the sample.
| Cohort 1 (N = 71)a | Cohort 2 (N = 71) | Combined (N = 142) | |
|---|---|---|---|
| Patient characteristics | n (%)b | n (%)b | n (%)b |
| Age | 60.2 (SD = 12.9) | 65.4 (SD = 12.3) | 62.8 (SD = 12.8) |
| Gender | |||
| Male | 41 (57.7) | 32 (45.1) | 73 (51.4) |
| Female | 30 (42.2) | 39 (54.9) | 69 (48.6) |
| Smoking | |||
| Never | 15 (21.1) | 20 (28.2) | 35 (24.6) |
| Current | 34 (47.9) | 26 (36.6) | 60 (42.3) |
| Former | 22 (31.0) | 21 (29.6) | 43 (30.3) |
| Missing | 0 (0) | 4 (5.6) | 4 (2.8) |
| ACE-27 comorbidities | |||
| None | 22 (31.0) | 12 (16.9) | 34 (23.9) |
| Mild | 32 (45.1) | 38 (53.5) | 70 (49.3) |
| Moderate | 10 (14.1) | 15 (21.1) | 25 (17.6) |
| Severe | 7 (9.8) | 6 (8.5) | 13 (9.1) |
| Tumor characteristics | n (%)b or mean | n (%)b or mean | n (%)b or mean |
|---|---|---|---|
| Oral cavity subsite | |||
| Tongue | 34 (47.8) | 48 (67.6) | 82 (57.7) |
| Gingiva | 15 (21.1) | 4 (5.6) | 19 (13.4) |
| Floor of mouth | 10 (14) | 7 (9.9) | 17 (12.0) |
| Retromolar area | 9 (12.6) | 2 (2.8) | 11 (7.7) |
| Other | 3 (4.2) | 5 (7.0) | 8 (5.6) |
| Missing | 0 (0) | 5 (7.0) | 5 (3.5) |
| T stage | |||
| 1 | 12 (16.9) | 11 (15.5) | 23 (16.2) |
| 2 | 21 (29.5) | 31 (43.7) | 52 (36.6) |
| 3 | 12 (16.9) | 5 (7.0) | 17 (12.0) |
| 4 | 26 (36.6) | 24 (33.8) | 50 (35.2) |
| N stage | |||
| 0 | 41 (57.7) | 53 (74.6) | 94 (66.2) |
| 1 | 8 (11.3) | 7 (9.9) | 15 (10.6) |
| 2 | 1 (1.4) | 2 (2.8) | 3 (2.1) |
| 2a | 2 (2.8) | 0 (0.0) | 2 (1.4) |
| 2b | 17 (23.9) | 4 (5.6) | 21 (14.8) |
| 2c | 2 (2.8) | 5 (7.0) | 7 (4.9) |
| AJCC Clinical Stage (7th Edition)c | |||
| I | 10 (14.1) | 11 (15.5) | 21 (14.8) |
| II | 14 (19.7) | 26 (36.6) | 40 (28.2) |
| III | 12 (16.9) | 6 (8.5) | 18 (12.7) |
| IV | 35 (49.3) | 28 (39.4) | 63 (44.3) |
| Histopathologic characteristics | n (%)b or mean | n (%)b or mean | n (%)b or mean |
|---|---|---|---|
| Differentiation | |||
| Poor | 11 (15.5) | 3 (4.2) | 14 (9.9) |
| Moderate | 27 (38.0) | 33 (46.5) | 60 (42.2) |
| Well | 33 (46.5) | 35 (49.3) | 68 (47.9) |
| Worst pattern of invasiond | |||
| POI 1 | 5 (7.0) | 0 (0.0) | 5 (3.5) |
| POI 2 | 19 (26.8) | 4 (5.6) | 23 (16.2) |
| POI 3 | 16 (22.5) | 6 (8.5) | 22 (15.5) |
| POI 4 | 26 (36.6) | 48 (67.6) | 74 (52.1) |
| POI 5 | 5 (7.0) | 12 (16.9) | 17 (12.0) |
| Missing | 0 (0) | 1 (1.4) | 1 (0.7) |
| PNI (H&E) | |||
| No | 55 (77.5) | 50 (70.4) | 105 (73.9) |
| Yes | 16 (22.5) | 21 (29.6) | 37 (26.1) |
| PNI (H&E + IHC)e | |||
| No | 47 (66.2) | 34 (47.9) | 81 (57.0) |
| Yes | 24 (33.8) | 37 (52.1) | 61 (43.0) |
| Expanded N stage (H&E + IHC)e | |||
| N0, PNI negative | 30 (42.2) | 26 (36.6) | 56 (39.4) |
| N0, PNI positive | 11 (15.5) | 27 (38.0) | 38 (26.8) |
| N+ | 30 (42.2) | 18 (25.4) | 48 (33.8) |
| Number of nerves with PNI | |||
| None | 47 (66.2) | 34 (47.9) | 81 (57.0) |
| One (unifocal) | 6 (8.5) | 6 (8.5) | 12 (8.5) |
| Two or more (multifocal) | 18 (25.3) | 31 (43.7) | 49 (34.5) |
| Depth of invasion (mm)f | 13.21 | 12.57 | 12.9 |
| Missing | 2 (2.8) | 2 (2.8) | 4 (2.8) |
IHC
For cohort 1, IHC stains were described previously (4). For cohort 2, 5 μm serial sections were stained with hematoxylin and eosin (H&E) or the following antibodies: S100 to highlight nerves, cytokeratin AE1/AE3 to highlight epithelial cells, and myelin basic protein (MBP) for validation of spatial transcriptomic data. Details are in Supplementary Materials and Methods.
Data collection
Slides were digitally scanned with iScan Coreo (Ventana Medical Systems, Roche Diagnostics International) and images were analyzed with Ventana Image Viewer software v.3.1.4 for cohort 1, and Halo Image analysis platform v3.1 (IndicaLabs) for cohort 2. Both programs have the same tools for manually annotating nerves and tumor areas. Two distinct adjacent areas were analyzed in all 142 specimens using Halo: (i) tumor bulk containing tumor islands and tumor stroma, and (ii) tumor margin, defined as a 2 mm margin around the tumor bulk (Fig. 1B).
Tumor differentiation and PNI (positive or negative) were scored by a board-certified Oral and Maxillofacial Pathologist using H&E sections and current criteria; a PNI-positive nerve is invaded by or has at least 33% of its circumference surrounded by tumor cells (5). Using the same criteria (5) for both cohorts, other investigator(s) scored PNI independently for each nerve using H&E and IHC sections. This resulted in two independent analyses, PNI (H&E) and PNI (H&E+IHC), respectively, for each patient. The first cohort was analyzed by one investigator (L.B. Schmitd). The second cohort was analyzed by L.B. Schmitd and a second investigator trained and supervised by L.B. Schmitd. Using S100-stained sections, nerves were located and measured for area and diameter, which was defined as the smallest axis of the nerve cross-section. Nerves smaller than approximately 10 μm in diameter and further than approximately 2 mm from tumor bulk were excluded. Cytokeratin-stained sections were used to determine worst pattern of invasion (POI; ref. 11), measure distance between each nerve and the nearest tumor island, and locate tumor in association with nerves to score PNI (Fig. 1C). Because specimens were deidentified, data were collected without knowledge of demographics, treatment, or clinical outcome.
NanoString GeoMx Digital Spatial Profiler
Eight of the 142 patients were selected for transcriptome spatial analysis using the NanoString GeoMx Digital Spatial Profiler (DSP)/Human NGS Whole Transcriptome Atlas (Fig. 1D). Cases were selected on the basis of location (tongue) and presence of nerves in tissue sections. Each slide with sections from 2 patients was stained with fluorescence-labeled antibodies to allow identification of tissue: pan-cytokeratin (AE1/AE3, Novus Biologicals, #NBP2-33200AF647) for epithelial cells and S100 (Dako Omnis GA50461-2, dilution 1:2) for nerves. Nerve areas of interest (AOI) were selected for analysis based on nerve-tumor proximity as follows: (i) NC (nerves close): within 100 μm from tumor with at least 50% surrounded by tumor cells (n = 6); (ii) N100: within 100 μm from tumor but excluded from NC (n = 8); (iii) N1000: 100 to 1,000 μm from tumor (n = 16); (iv) NF (nerves far): beyond 1 mm from any tumor cell (n = 16). AOIs were drawn manually around nerves. Only nerves with at least 16,000 μm2 and at least 50 cells were selected to ensure proper RNA probe counts.
Cell culture and qPCR
Peripheral neuronal (50B11; ref. 12) and Schwann (S16, ATCC, #CRL-2941, RRID:CVCL_B072) cell lines, and ex vivo rat dorsal root ganglia (DRG) were used for mRNA expression validation experiments (Fig. 1E). Experiments using rats were done in accordance with Institutional Animal Care and Use Committee rules from our institution. Briefly, both cell lines were treated with conditioned medium from an OSCC cell line (UM-SCC-29) for 0 to 48 hours. DRGs were co-cultured in the presence or absence of UM-SCC-29 for up to 72 hours. Cells or DRGs were then lysed, RNA extracted, and qPCR perfomed to validate spatial transcriptomic data. Details are in Supplementary Materials and Methods. Primers are listed in Supplementary Table S2.
Statistical analysis
Data from clinical samples were submitted for analysis to cancer biostatisticians from the HNOP. Data were stratified as patient-level and nerve-level, the first including tumor characteristics for each patient, and the second including information derived from IHC for each nerve as described in Fig. 1C.
Descriptive statistics were calculated for patient-level and nerve-level characteristics. Kaplan–Meier methods were used to estimate overall survival (OS), disease-specific survival (DSS), and disease-free survival (DFS) probabilities within groups, defined by patient-level characteristics. Time of diagnosis was used as baseline when defining OS, DSS, and DFS. Regression tree methods based on the log-rank statistic were used to identify potential thresholds of patient-level characteristics related to differences in survival probabilities. Cox regression modeling was used to examine the relationship between continuous patient-level characteristics and survival. Logistic regression was used to investigate the relationship between PNI and other variables.
Weighted Cox regression modeling was used to examine the association between nerve-level characteristics and survival, where nerves from each subject were weighted by the inverse of the number of nerves for that subject. A Cox generalized additive model was fitted using the penalized splines method to explore the shape of the relationship between OS and DSS and the distance between the nerve and nearest tumor island, as well as the diameter of nerves in the tumor bulk. All analyses were conducted using R package, version 3.3.0.
Data obtained with the NanoString GeoMx DSP platform were analyzed using dedicated software (GeoMx_NGS_Pipeline_2.2.0.2 and GeoMx Analysis Suite 2.3). AOI quality control (QC) detected raw reads above 1,000 for all AOIs, and good alignment rate and sequencing saturation. Probe QC detected 24 local and 0 global outliers. All the local outliers were negative control probes, and were removed from each AOI individually. AOIs with deduplicated probe counts Q3 (upper quartile) values lower than 2 were excluded (n = 7). Two additional AOIs were removed because they had less than 1% of genes with read counts higher than limit of quantification (LOQ). LOQ was defined as geomean(NegProbe) × geoSD(NegProbe)2 for each AOI. Target filtering was applied to retain 8,162 gene targets (of 18,814) with read counts above LOQ in at least 10% of the AOIs. Q3 normalization was then applied on the filtered AOIs and gene targets. Differential gene expression across groups was analyzed using linear mixed effect models; differentially expressed genes (DEG) were defined as fold change (FC) >1.5 and FDR <0.1 (13, 14). iPathwayGuide analysis (https://ipathwayguide.advaitabio.com) used all filtered gene targets as background.
Data availability
Raw data for this study were generated at the University of Michigan Advanced Genomics core facility (https://brcf.medicine.umich.edu/cores/advanced-genomics/). Derived data supporting the findings of this study are available from the corresponding author upon request.
Results
We profiled all nerves in both tumor bulk and a 2 mm margin from the tumor in 142 OSCC biopsies (Fig. 1). IHC analysis shows that PNI is present in 43% of patients, although PNI is observed in only 26.1% of patients by H&E alone; that is, assessment with H&E+IHC increases PNI detection (Table 1). Subsequent results reflect PNI assessment by H&E+IHC. The demographic, tumor, and histopathologic characteristics are in Table 1; descriptive survival statistics are in Supplementary Table S1. The odds of being PNI positive are increased by having American Joint Committee on Cancer (AJCC; 7th edition) stages III [OR, 5.31; 95% confidence interval, CI (1.27–22.24)] and IV [OR, 3.86; 95% CI (1.17–12.78)], larger tumors [T3 OR, 5.14; 95% CI (1.29–20.52) and T4 OR, 3.32; 95% CI (1.07–10.34)], and worst POI grades 4–5 [OR, 2.81; 95% CI (1.09–7.26); Supplementary Table S3].
PNI is a strong driver of survival among node-negative patients
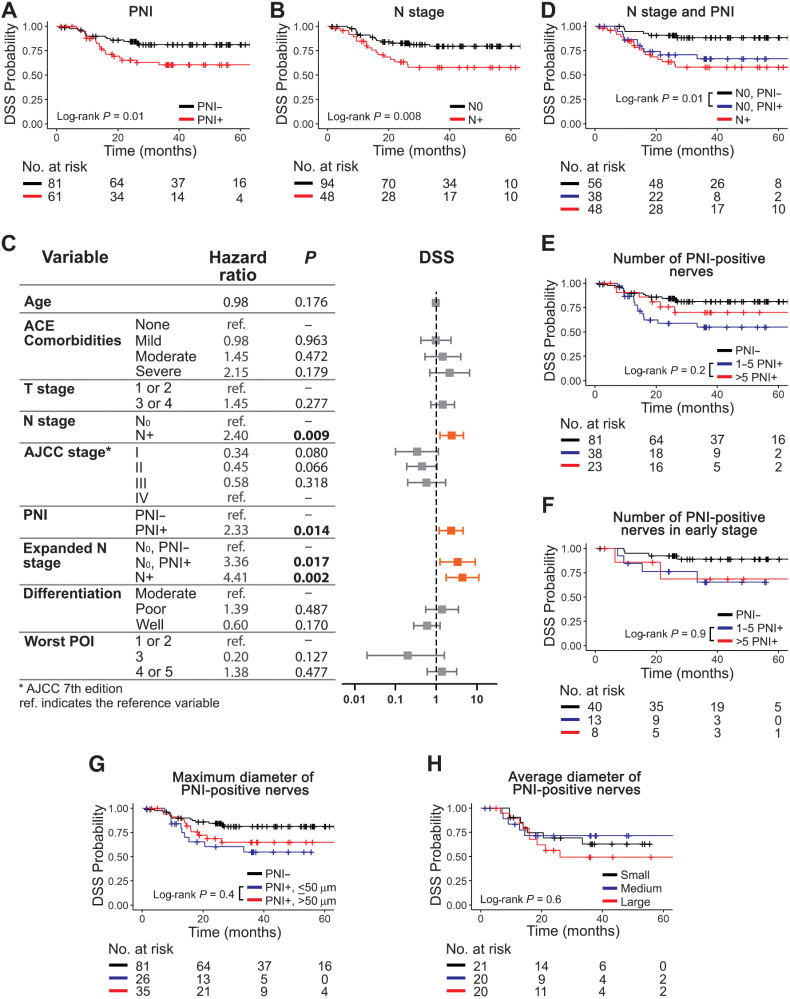
PNI is often associated with lymph node metastasis (15) and may be used as a predictor of poor prognosis among patients with early-stage disease, especially clinically N0 patients (16). Lymph node metastasis is a strong predictor of poor survival; patients typically receive adjuvant therapy when positive nodes are detected (17). Our sample has 66.2% clinically N0 and 43% early-stage patients (AJCC 7th Edition stages I and II; Table 1). Survival analyses show that PNI (Fig. 2A; Supplementary Fig. S2A and S2B) and lymph node involvement (Fig. 2B; Supplementary Fig. S2C and S2D) associate with worse survival. Univariate Cox modeling (Fig. 2C; Supplementary Table S4) shows that PNI associates with poor DSS [HR, 2.33; 95% CI (1.18–4.58)], OS [HR, 1.93; 95% CI (1.13–3.29)], and DFS [HR, 1.92; 95% CI (1.04–3.56)]. Similarly, there is significant association between node positivity and poor DSS [HR, 2.40; 95% CI (1.24–4.66)] and OS [HR, 1.81; 95% CI (1.06–3.08)].
Figure 2.
PNI is associated with poor DSS among node-negative patients. Number and diameter of PNI-positive nerves has no correlation with survival. PNI-positive (A) and node-positive (B) patients survive poorly. C, Univariate Cox modeling of patient-level data. Significant HRs are shown in bold and depicted in orange in the plot. D, Among node-negative patients, detection of PNI is significantly associated with poor DSS. The number of PNI-positive nerves is not associated with DSS (E), even among patients with early-stage OSCC (F). Maximum (G) or average (H) diameter of PNI-positive nerves do not associate with DSS. Tertiles were used for nerve diameter: small (< 29.2 μm), medium (29.2–47.5 μm), and large (> 47.5 μm). Kaplan–Meier survival curves using patient-level data are shown in A, B, and D through H. Log-rank P values are displayed in each plot and the number of patients at risk for each timepoint is shown below each survival plot. PNI was assessed using H&E + IHC.
To investigate the relationship between PNI and N stage, we split N0 patients into PNI positive and PNI negative. Among N0 patients, there is a strong relationship between PNI positivity and worse DSS (Fig. 2D), OS, and DFS (Supplementary Fig. S2E and S2F). N0, PNI-positive patients show significantly higher HRs compared with N0, PNI-negative patients [Fig. 2C; Supplementary Table S4; DSS, HR, 3.36; 95% CI (1.24–9.09); OS, HR, 2.45; 95% CI (1.19–5.06); DFS, HR, 2.36; 95% CI (1.03–5.38)], despite a higher rate of adjuvant radiotherapy for the N0, PNI-positive patients (data not shown). Tumor differentiation and worst POI do not correlate with any of the outcomes (Fig. 2C; Supplementary Table S4). However, we addressed them because higher grades/scores of these histopathologic features have been associated with worse patient survival and tumor aggressiveness (18–21). Overall, these data indicate that PNI is a valuable prognostic marker for survival among N0 patients.
More and larger PNI-positive nerves do not associate with worse survival
To clarify the value of PNI in predicting prognosis, the association between number of PNI-positive nerves, nerve diameter, and survival were analyzed. These parameters have been assessed in multiple studies with discordant results (16, 22–24).
PNI was scored as multifocal (>one) versus unifocal (only one); no difference in patient survival is observed in this cohort (Supplementary Fig. S3A–S3C). PNI-positive biopsies have one to 42 PNI-positive nerves; six or more PNI-positive nerves in the tissue sample do not associate with worse DSS (Fig. 2E), OS, or DFS (Supplementary Fig. S3D and S3E), compared with ≤5 PNI-positive nerves. These data suggest no predictive value for the accumulation of PNI-positive nerves.
PNI is more likely to occur in late-stage tumors (Supplementary Table S3); however, it is also a strong driver of survival among N0 patients (Fig. 2D). Consequently, we assessed the correlation between number of PNI-positive nerves and survival in early-stage patients (AJCC stages I–II). DSS is very similar between groups with one to five, and more than five PNI-positive nerves (Fig. 2F). Patients with early-stage disease (n = 61) with fewer PNI-positive nerves have a higher chance of receiving surgery alone (69%) compared to patients with six or more PNI-positive nerves (25%) who received more aggressive therapy. The lack of differences in DSS could be explained by adjuvant therapy being given more often to patients with a higher number of PNI-positive nerves.
Previous studies (11, 22) showed that survival of patients with PNI-positive nerves larger than 1 mm in diameter is poorer than patients with smaller PNI-positive nerves. Nerves ≥1 mm are rare in our sample. Among 9,137 nerves, only 7 are larger than 1 mm, none of which are PNI positive. PNI-positive nerve diameter ranges from 10 to 888 μm. Using regression tree analysis, we split PNI-positive patients based on maximum nerve diameter of 50 μm. There is no correlation between the largest PNI-positive nerve in each patient and DSS, OS, or DFS (Fig. 2G; Supplementary Fig. S3F and S3G). Furthermore, we evaluated the average nerve diameter of PNI-positive nerves dividing patients into groups based on tertiles: small (<29.2 μm), medium (29.2–47.5 μm), and large (>47.5 μm). There is no association between large average diameter of PNI-positive nerves and survival (Fig. 2H; Supplementary Fig. S3H and S3I), suggesting that large PNI-positive nerves are not meaningful for survival.
Our data support that although PNI is an important predictor of poor prognosis, the quantity and size of PNI-positive nerves is not critical for patient survival.
Distance between nerves and tumor predicts outcome
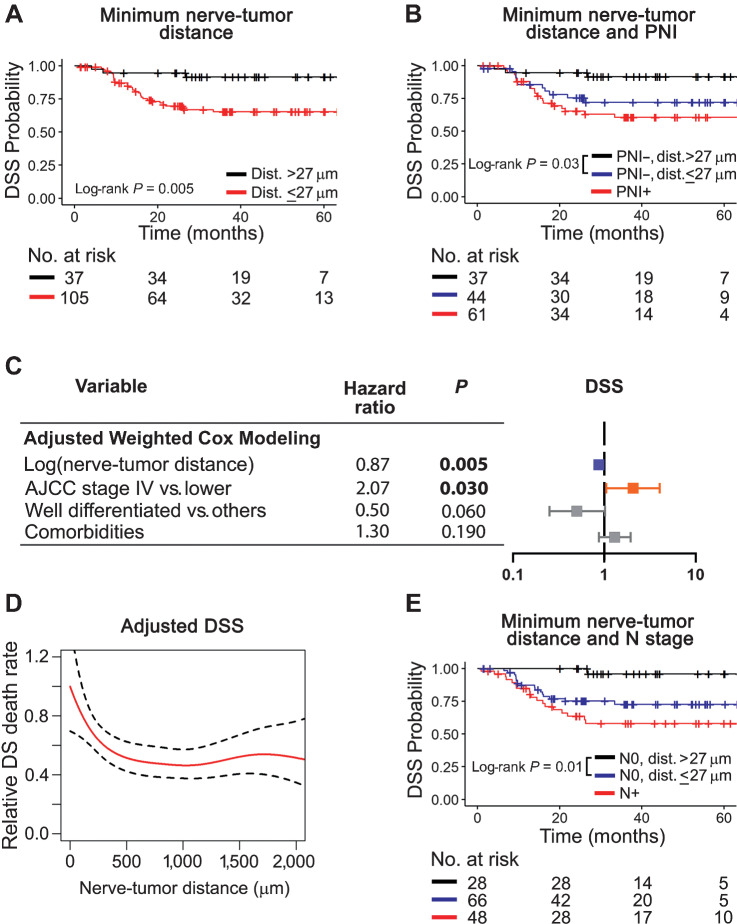
Although 43% of our OSCC population has PNI-positive nerve(s), the relative number of PNI-positive nerves is very low (4.5% of all nerves; Supplementary Table S5). Given that most nerves in the tumor microenvironment are PNI negative, we investigated the association of other nerve parameters with survival. Previously, we showed that the minimum distance between nerve and tumor is relevant for survival; PNI-negative patients with close nerve-tumor proximity have low survival (4). In the combined cohort of 142 patients with analysis of 9,137 nerves, nerve-tumor distance was assessed at the patient-level and as individual nerves across patients. Nerve-level data are in Supplementary Table S5 and nerve-related characteristics of patients are in Supplementary Table S6. For patient-level analysis, because each nerve has a unique nerve-tumor distance, the minimum nerve-tumor distance within each patient was selected as a parameter for survival analyses. Regression tree methods split the subjects into two groups based on greatest difference with respect to DSS; a split was obtained at 27 μm for minimum nerve-tumor distance. With this cutoff, there is significant association between shorter nerve-tumor distances and poor DSS (Fig. 3A), OS, and DFS (Supplementary Fig. S4A and S4B). Not surprisingly, univariate analysis (Supplementary Table S4) verifies that smaller HRs are significantly associated with large nerve-tumor distances [DSS, HR, 0.56; 95% CI (0.36–0.85); OS, HR, 0.67; 95% CI (0.49–0.92); DFS, HR, 0.66; 95% CI (0.46–0.95)] when minimum nerve-tumor distance is considered for each patient. In other words, smaller nerve-tumor distances significantly associate with poor survival. However, because the current definition of PNI implies a nerve-tumor distance equal or close to zero but no criteria for “close” currently exist, OSCCs that were PNI-negative were split using the same 27 μm cutoff. Significantly poorer DSS for close nerve-tumor distances is evident in the PNI-negative group (Fig. 3B); a similar trend is observed for OS and DFS (Supplementary Fig. S4C and S4D). This prompted investigation of correlations between nerve-tumor distances using all 9,114 nerves with available nerve-tumor distance information. Adjusted Cox regression model weighted by the inverse of number of nerves within each subject (Fig. 3C; Supplementary Table S7) shows that greater nerve-tumor distances are significantly associated with improved DSS [HR, 0.87; 95% CI (0.79–0.96)] and OS [HR, 0.88; 95% CI (0.81–0.95)].
Figure 3.
Close nerve-tumor distances associate with poor survival. A, Patients with a nerve-tumor distance ≤27 μm survive poorly. B, Among PNI-negative patients, those with nerve tumor distance ≤27 μm survive poorly and similarly to PNI-positive patients. C, Adjusted Cox modeling of nerve-level data. Data are weighted by the number of nerves per patient and adjusted for variables shown. Significant HRs are shown in bold and depicted in blue for low and orange for high. D, Adjusted Cox additive modeling for relative DSS as a function of nerve-tumor distance using nerve-level data; a decrease in relative DSS death rate is observed as nerve-tumor distance increases. The model is adjusted for AJCC 7th edition stage and tumor differentiation. E, Among node-negative patients, nerve-tumor distance ≤27 μm associates with poor DSS. Kaplan–Meier survival curves using patient-level data are shown in A, B, and E. Log-rank P values are displayed in each plot and the number of patients at risk for each timepoint is shown below each survival plot. PNI was assessed using H&E + IHC.
PNI has been associated with increased tumor depth (25). Because AJCC 7th edition classification does not take into consideration depth of invasion (DOI), we updated patient data according to AJCC 8th edition which incorporates depth. Two patients without available information were excluded; updated staging is shown in Supplementary Fig. S5A–S5C. When using AJCC 8th edition to adjust the Cox regression model (Supplementary Fig. S5D), nerve-tumor distance continues to show a significant HR, supporting that shorter distance is independently predictive of DSS regardless of AJCC classification.
Nerve-tumor distance is relevant for patient survival. To investigate whether there is a threshold above which nerve-tumor distance is no longer related to survival, nonlinear models were used. A Cox generalized additive model explores the relationship between nerve-tumor distance of individual nerves to death rate for both DSS (Fig. 3D) and OS (Supplementary Fig. S4E). Nerves are weighted by the inverse of the number of nerves per patient and the analysis is adjusted by AJCC 7th edition stage and differentiation for DSS. For OS, adjusted analysis is done for AJCC stage, differentiation, and age. The estimated relative death rate decreases as the nerve-tumor distance increases; the hazard of death is clearly high for short nerve-tumor distances and gradually decreases, stabilizing at a distance of approximately 500 μm, suggesting that distances shorter than 500 μm are more relevant to survival. Adjusting the analysis using AJCC 8th edition stage shows similar results (Supplementary Fig. S5E).
Our results (Fig. 2), show that PNI is associated with poor DSS among the N0 population. To understand the predictive value of nerve-tumor distance among N0 patients, we split the N0 population based on the 27 μm cutoff for minimum nerve-tumor distance. Similar to that shown for PNI, shorter nerve-tumor distances associate with poor DSS (Fig. 3E), OS, and DFS, (Supplementary Fig. S4F–S4G) in N0 patients, suggesting that nerve-tumor distance is a driver of survival among patients without lymph node metastasis. Interestingly, using nerve-tumor distance instead of PNI status to score N0 patients for survival, the number of patients at risk increases from 38 (N0, PNI positive) to 66 (N0, ≤27 μm).
Diameter of nerves within tumor bulk associates with patient survival
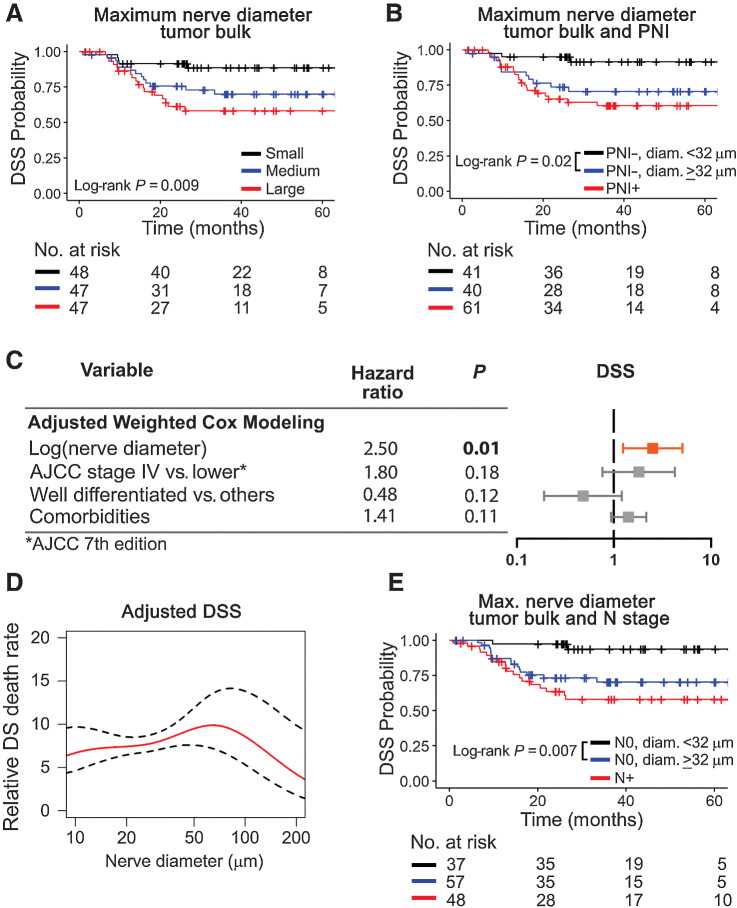
Because nerves closer to tumor associate with poor survival (Fig. 3), we hypothesized that having larger nerves in proximity to tumor also suggest poor outcome. To investigate this association, we restricted the analysis to nerves present in the tumor bulk (n = 2,975 nerves), of which 96% are within 500 μm of a tumor cell. Splitting patients by maximum nerve diameter in the tumor bulk into tertiles, survival is poor when nerve diameter is high (DSS, Fig. 4A; OS and DFS; Supplementary Fig. S6A anad S6B). We calculated maximum nerve diameter that best separates the PNI-negative patients using regression tree methods. PNI-negative patients with a maximum nerve diameter ≥32 μm survive poorly compared with PNI negative with smaller nerves in the tumor bulk (DSS; Fig. 4B); PNI-negative patients with nerve diameter ≥32 μm behave similarly to PNI-positive patients (Fig. 4B; Supplementary Fig. S6C and S6D). The association between larger nerve diameter in the tumor bulk and poor survival is significant even when adjusting for AJCC stage and comorbidities [DSS, HR, 3.73; 95% CI (1.01–13.80); Supplementary Table S8]. Multivariate Cox modeling using nerve-level data also reveals an increased HR for larger nerve diameter in the tumor bulk (Fig. 4C; Supplementary Table S7). Adjusting for AJCC 8th edition does not change the significance of these results (Supplementary Fig. S5F). To investigate the threshold of nerve diameter that is most relevant for patient survival, we used additive models for DSS (Fig. 4D; Supplementary Fig. S5G) and OS (Supplementary Fig. S6E) using all nerves in the tumor bulk from 142 patients. The relative patient death rate increases as nerve diameter increases, regardless of the AJCC classification used, and the effect is lost above a diameter of approximately 100 μm. Similar to what happens for nerve-tumor distance, larger nerve diameter in the tumor bulk associates with poor DSS (Fig. 4E), OS, and DFS (Supplementary Fig. S6F and S6G) in N0 patients.
Figure 4.
Large nerve diameter in tumor bulk associates with poor DSS. A, Patients are divided in tertiles for maximum nerve diameter in tumor bulk into low (≤32.28 μm), medium (33.73–88.13 μm), and large (≥ 89.61 μm). Patients with large nerves in the tumor bulk survive poorly. B, Among PNI-negative patients, nerve diameter ≥32 μm in the tumor bulk significantly associates with poor DSS. C, Adjusted Cox modeling of nerve-level data. Data are weighted by the number of nerves per patient and adjusted for variables shown in the table. Significant HRs are shown in bold and depicted in orange for high. D, Adjusted Cox additive modeling for relative DSS as a function of nerve diameter in the tumor bulk using nerve-level data; an increase in relative DSS death rate is observed as nerve diameter increases. The model is adjusted for AJCC 7th edition stage and tumor differentiation. E, Among node-negative patients, a nerve diameter ≥32 μm associates with poor DSS. Kaplan–Meier survival curves using patient-level data are shown in A, B, and E. Log-rank P values are displayed in each plot and the number of patients at risk for each timepoint is shown below each survival plot. PNI was assessed using H&E + IHC.
To evaluate whether nerve-related parameters are influenced by tumor DOI, we did linear regression analyses (Supplementary Fig. S7A–S7C). Nerve-tumor distance and nerve diameter do not correlate with DOI. Even the number of nerves per patient does not increase with DOI (Supplementary Fig. S7C). As expected, large tumors have increased DOI (Supplementary Fig. S7D). PNI-positive and PNI-negative patients have similar overall depth (Supplementary Fig. S7E). However, splitting in tiers of 5 mm depth, PNI positivity is more frequent with tumor DOI >10 mm (Supplementary Fig. S7F). These data support that DOI has a weak influence on PNI, nerve-tumor distance, and nerve diameter in our sample.
Overall, our spatial morphometric analyses of nerves in oral cancer reveal that nerves close to tumor associate with poor patient outcome. In addition, among nerves close to tumor, diameter is likely relevant for survival.
Nerves closer to tumor have a unique transcriptomic signature
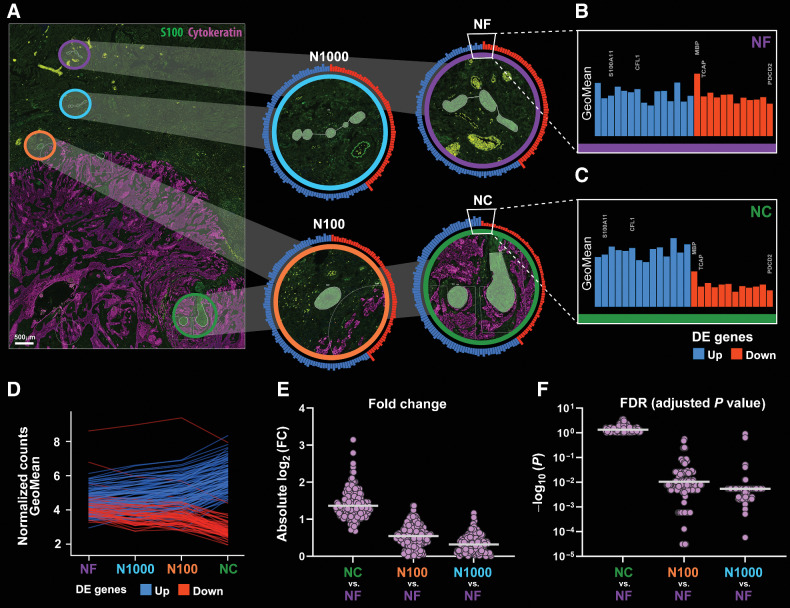
Because nerves closer to tumor are meaningful for clinical outcomes in OSCC, we hypothesized that these nerves have a different molecular profile compared with nerves that are not embedded within tumor, that is, within the tumor margin. To explore differences between nerves close to and at a distance from tumor, we used the NanoString GeoMx DSP Whole Transcriptome Atlas platform in 8 of the 142 patients. Nerve AOIs were categorized on the basis of proximity to tumor as shown in Fig. 5A. NC designates nerves within 100 μm of tumor and at least 50% surrounded by tumor cells. Gradual increase in distance between tumor and nerve is represented in the AOIs N100 (nerve within 100 μm from tumor but excluding NC) and N1000 (100 μm to 1 mm from tumor). NF refers to nerves beyond 1 mm from tumor.
Figure 5.
Transcriptomic profile of nerves varies with nerve-tumor distance. A, Biopsy sections were stained with morphology markers (cytokeratin in purple and S100 in green) for identification of AOIs. AOIs are overlaid in the fluorescence image and enlarged in panels at right: NC (within 100 μm from tumor with at least 50% surrounded by tumor cells) is identified in green, N100 (within 100 μm from tumor cells but excluded from NC) in orange, N1000 (between 100 and 1,000 μm from tumor) in light blue, and NF (beyond 1,000 μm from tumor) in purple. Circular bar graphs around each enlarged AOI represent log2-transformed normalized gene counts (GeoMeans) of 159 genes that were differentially expressed between NC and NF. Blue bars are upregulated and red bars are downregulated in NC. Enlarged GeoMean bar graphs of NF (B) and NC (C) show differences in gene expression. D, GeoMeans for 159 genes as a function of the type of AOI. Upregulated genes go gradually up whereas downregulated genes go gradually down from NF to NC. Gradual differences are observed for the log2(FC; E) and adjusted P values (FDR; F) for 159 genes as a function of nerve-tumor distance. NC versus NF has the largest log2(FC) and the smallest FDR values, while N1000 versus NF are most similar in terms of gene expression. Scale bar (A), 500 μm.
Overall, 8,162 genes are detected in all AOIs. There are 159 DEGs for the NC versus NF comparison; 95 are upregulated while 64 are downregulated in NC. Log2-transformed normalized gene counts (GeoMeans) are depicted in the circle plots in all four types of AOIs (Fig. 5A) and partially zoomed in as linear plots (Fig. 5B and C). Comparing GeoMeans between NF and NC, a gradient effect in gene expression is observed as a function of nerve-tumor distance. Gradual changes in expression of all 159 genes are shown in Fig. 5D, with genes upregulated in NC in blue and downregulated in NC in red. To further explore this gradient effect, we plotted both adjusted P values and log2FC (fold change) values for all DEGs across groups. As expected, absolute logFC values decrease gradually from the NC versus NF comparison with the N1000 versus NF comparison (Fig. 5E). Conversely, adjusted P values increase (Fig. 5F), confirming that dissimilarities found in the NC versus NF comparison are decreased as the distance between tumor and nerve increases.
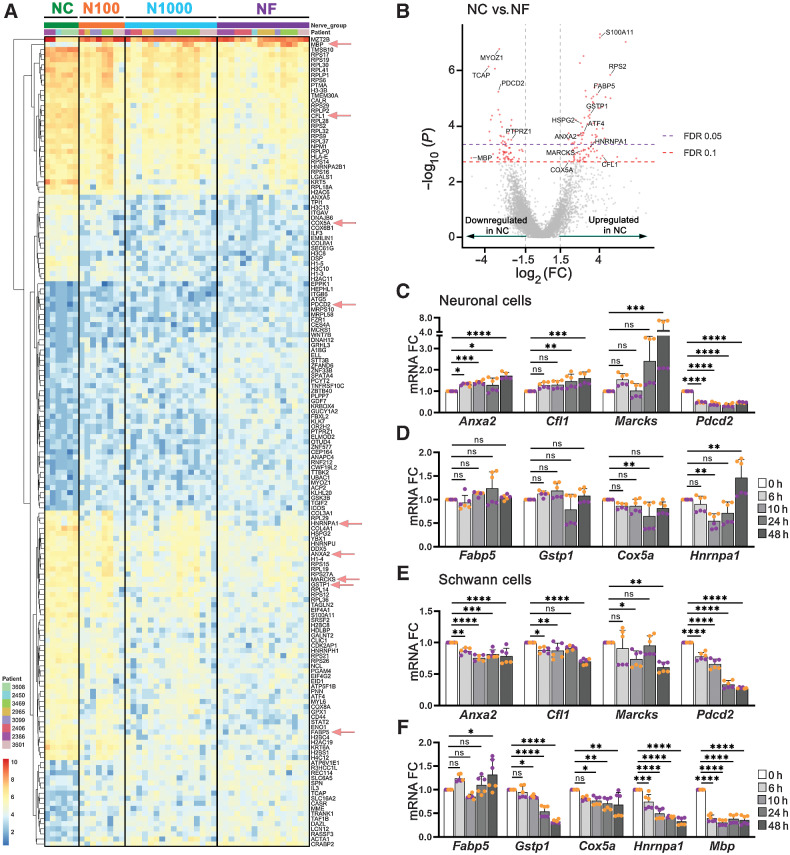
DEGs from the NC versus NF comparison are shown in Fig. 6A and B. The top 50 upregulated and downregulated genes are listed in Supplementary Table S9. Exploring DEGs further, ribosome (KEGG: 03010) is the most well-represented pathway, with 26 upregulated and 1 downregulated gene in NC, suggesting that nerves close to tumor have increased protein translation. Alcoholism (KEGG: 05034) is also significantly represented by upregulation of 12 genes. Of interest, pathways of neurodegeneration multiple diseases (KEGG: 05022) and amyotrophic lateral sclerosis (ALS, KEGG: 05014) were among the six most well-represented pathways, with 10 and nine genes, respectively. ALS is a neurodegenerative disease; suggested mechanisms include impaired proteostasis, aberrant RNA processing, mitochondrial dysfunction and oxidative stress, microglia activation, and axonal dysfunction (26). The top six well-represented pathways and top 10 gene ontology terms for molecular function, biological process, and cellular component are listed in Supplementary Fig. S8.
Figure 6.
DEGs in nerves close to and far from tumor. A, Heatmap of 46 AOIs across 8 patients, showing 159 significant (FDR ≤ 0.1) differentially expressed genes in NC versus NF. Each row of the heatmap represents the transformed log2 values of one DEG across all samples (blue, low expression; red, high expression). AOI types are identified on top. Genes selected for in vitro validation (C–F) are highlighted with red arrows. B, Volcano plot of differentially expressed genes, identified in red. FDR ≤ 0.05 and ≤ 0.10 are identified by dashed lines. C and D, 50B11 neuronal cells were treated with conditioned medium from cancer cells (UM-SCC-29) for up to 48 hours and mRNA expression of selected genes was assessed. E and F, S16 Schwann cells were treated as described in C and D. Each color represents an independent experiment in C, D, E, and F. Timepoints are depicted in shades of gray. (One-way ANOVA P values: * < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001).
Overall, these data suggest that nerves in proximity to tumor undergo a degenerative process and react to tumor stress factors in a distance-dependent manner.
Nerves in proximity to tumor have changes in myelin that reflect injury or degeneration
Among the 159 DEGs, two genes are constituents of the peripheral nervous system (PNS) myelin sheath (AnxA2, Mbp; ref. 27). Myelin, produced by Schwann cells, surrounds neuronal axon projections to facilitate propagation of electrical impulses during neurotransmission. Annexin A2 (ANXA2) is upregulated in NC and accumulates in Schwann cells upon injury or stress and causes cytoskeleton remodeling for nerve growth (28). MBP is among the most abundant myelin proteins, approximately 8% of all myelin proteins (27). Our data show downregulation of MBP in NC. Downregulation of MBP also occurs in sensory nerves undergoing myelin degeneration in oral mucosa (29). MBP is posttranscriptionally downregulated by DDX5 (30). Interestingly, our data show upregulation of DDX5 and downregulation of MBP in NC.
PTPRZ1, downregulated in NC, is expressed exclusively in the nervous system. In the central nervous system, it suppresses oligodendrocyte differentiation and remyelination (31). Little is known about its function in the PNS. This gene encodes for protein receptor-type tyrosine-protein phosphatase zeta (R-PTP-zeta), a putative receptor for heterophilic NCAM (neural cell adhesion molecule) in Schwann cells that may be involved in neuron-Schwann cell communication and enhancing Schwann cell migration (32). Because PTPRZ1 suppresses myelination, downregulation of this gene in NC may represent an effort to increase myelination necessary for nerve regeneration.
Axonogenesis and stress response genes are upregulated in nerves near tumor cells
Nerves close to tumor show upregulation of genes implicated in cytoskeletal organization such as CFL1 and MARCKS. Cofilin1 (CFL1) is a known regeneration-associated gene (33, 34), involved in actin and microtubule organization necessary for filopodia and axon elongation. It is involved in neurite growth, axon guidance and synaptogenesis (35); dysregulation of Cofilin activity is present in neurodegenerative diseases such as Alzheimer's (36). MARCKS (myristoylated alanine-rich C-kinase substrate) is critical for neurite initiation, branching, and neuronal migration (37). These findings indicate a growth/regenerative response in nerves proximal to cancer cells. Another upregulated gene in NC, HSPG2, encodes the protein Perlecan, an extracellular matrix and basement membrane component crucial for cell adhesion and migration during nervous system development (38).
Upregulated genes also show an important response to stress in nerves close to tumor. These genes are involved in endoplasmic reticulum stress (ATF4), cell detoxification (FABP5, GSTP1), neuronal survival via inhibition of necrosis (PTMA), and reduction of apoptosis after glucose deprivation (COX5A) and ischemia (HNRNPA1). Interestingly, PDCD2, a gene related to induction of apoptosis (39, 40), is downregulated in NC. In injured mouse dorsal ganglia, Pdcd2 is upregulated, but only in the non-peptidergic nociceptor population of neurons (33). Overall, our data suggest nerve reprogramming to survival mode that in turn, supports axon growth.
Taken together, the gene profile of nerves within cancer reveals a response that promotes growth and survival to overcome damage. The gradient of gene expression dependent on nerve-tumor distance, suggests the injury to nerves is molecular rather than physical.
Studies in neuronal and Schwann cells, DRG, and human tissue verify spatial transcriptomic data
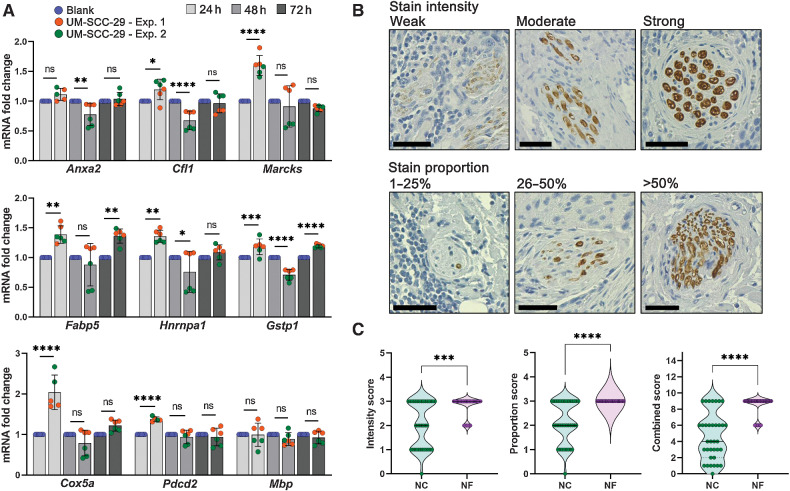
To explore whether gene expression changes in nerves close to tumor are due to tumor-secreted factors, we performed in vitro experiments using neuronal (50B11) and Schwann (S16) cells, and ex vivo DRGs. Cells were cultured in conditioned medium from cancer cells for up to 48 hours and expression of Anxa2, Cfl1, Marcks, Fabp5, Gstp1, Cox5a, Hnrnpa1, Pdcd2, and Mbp, was evaluated by qPCR (Fig. 6C–F). Neuronal cells significantly upregulate Anxa2, Cfl1, Marcks, and Hnrnpa1, and downregulate Pdcd2, consistent with patient transcriptomic data. Schwann cells upregulate Fabp5 and downregulate Mbp and Pdcd2, also consistent with spatial transcriptomic data. Cox5a and Gstp1 changes in both cell lines did not validate the spatial transcriptomic findings. Overall, most tested targets were changed in the expected direction in at least neuronal or Schwann cells. DRGs were co-cultured with UM-SCC-29 for up to 72 hours; expression of the same genes was assessed by qPCR. Similarly to cell-based experiments, changes in DRGs mostly replicate spatial transcriptomic data (Fig. 7A).
Figure 7.
Validation of DEGs in nerves close to cancer. A, DRG from rat were co-cultured for the designated timepoints in the presence or absence of UM-SCC-29 cells; mRNA expression of selected genes was measured. Timepoints are depicted in shades of gray. (One-way ANOVA P values: * < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001). B and C, IHC for MBP; nerves close to (NC, n = 35) and far from tumor (NF, n = 40) were scored for intensity and proportion of stain, and a combined score (intensity × proportion) was generated. Scale bars = 50 μm; n = 8 patients. Quantification is shown in C (Mann–Whitney test P values: *** < 0.001; **** < 0.0001).
To investigate protein expression in nerves close to cancer, we performed IHC for MBP in patient biopsies. Nerves close and far from tumor were scored for intensity and proportion of stain (Fig. 7B). A significant reduction of MBP protein expression is observed in nerves close to tumor (Fig. 7C), similar to MBP gene expression from spatial transcriptomic data.
Discussion
This study shows the first unbiased spatial gene expression profile of nerves in human cancer. Our clinical and transcriptomic findings elucidate cancer-nerve interactions associated with poor survival. We identify parameters beyond the current clinical definition of PNI that are associated with poor survival. These may be especially pertinent among patients without lymph node metastasis, where adjuvant therapy is not routinely recommended after surgery. Considering both PNI-positive and PNI-negative nerves in cancer, we show that shorter nerve-tumor distances predict poor survival. In support of these clinical findings, spatial transcriptome analysis shows that nerves close to tumor cells have an injury-response program. Importantly, gene expression changes were gradual based on nerve-tumor cell distance. Together, these findings support redefining PNI and expanding nerve-related criteria used to predict patient outcome and select treatment.
Advanced tumor size, DOI, and lymph node metastasis, currently recognized as important predictors of poor prognosis in OSCC, are incorporated into disease staging (41). Adjuvant radiotherapy is routinely recommended for clinical stage of at least T3 and/or node-positive patients (41). However, up to 50% of patients with stage I or II OSCC develop recurrence despite the early stage (42–45); these patients often do not have indications for adjuvant therapy (44, 45). Other parameters such as PNI could help guide treatment selection (11, 16). In fact, the National Comprehensive Cancer Network Guidelines (46) for OSCC recommend postoperative chemoradiation or radiation for PNI, considering it a strong adverse feature. Our study shows that among N0 patients, detection of PNI in biopsies helps stratify patients into a group with more aggressive disease. OS, DSS, and DFS rates for patients with N0, PNI-positive OSCC mirrors the poor survival of N-positive patients (Fig. 2; Supplementary Fig. S2). Similarly, among N0 patients, shorter nerve-tumor distance and large nerve diameter in the tumor bulk associate with poor outcomes (Fig. 3 and 4; Supplementary Figs. S4 and S6). Detection of PNI and our expanded criteria may increase patients eligible for adjuvant therapy beyond current recommendations, potentially improving clinical outcomes.
PNI is notoriously variable in definition, generating poor concordance among pathologists (15, 47, 48). This has led to great variability in clinical studies focused on PNI (15). To overcome this limitation, some studies investigated parameters associated with PNI to enhance its significance as a predictor of prognosis (11, 16, 22–24, 49–52). These include number (16, 22, 23, 52), location (51, 52), and diameter of PNI-positive nerves (11, 22, 49, 51, 53).
The number of PNI-positive foci has been variably linked to survival. PNI was defined as focal (one), moderate (two to five), or extensive (more than five), with no association between number of PNI-positive nerves and outcome (16). Conversely, more than five PNI-positive nerves were associated with worse DSS (24). Tarsitano and colleagues (23) reported shorter time to local or regional recurrence associated with extensive PNI. Other studies defining high PNI as multifocal (more than one; refs. 22, 49, 52) likely because few patients presented with >5 PNI-positive nerves (52), reported increased local failure (22) and worse DSS (49, 52). In contrast, our results show that multifocal does not decrease survival compared with unifocal PNI. Because patients with PNI are more likely to have higher tumor stage and receive adjuvant treatment, potentially improving survival, we evaluated the effect of PNI foci among early-stage patients. Accumulation of PNI-positive nerves has no contribution to poor DSS, OS, and DFS, even in early-stage OSCC.
PNI-positive nerves ≥1 mm in diameter correlated with locoregional failure (11, 22, 23). However, PNI-positive nerves larger than 1 mm are not common on incisional biopsies of OSCC; our study has zero PNI-positive nerves larger than 1 mm. In a sample of 105 patients, only 10 had PNI-positive nerves larger than 0.5 mm (49). Previous findings suggest that PNI-positive nerves <1 mm associate with worse OS, DSS, and local recurrence (4, 49, 53). Here, we analyze PNI-positive nerve diameter using both maximum and average diameter for each patient; no associations between OS, DSS, or DFS and large PNI-positive nerves are found. Discordance across studies could be due to variability in defining PNI, and the use of H&E alone for detection of PNI.
Although most recent studies use the same diagnostic criteria (5), the definition of PNI is subjective due to the term “close proximity” between tumor and nerve. Furthermore, only a few studies used IHC to confirm PNI (4, 54, 55). The current study uses a standardized definition of PNI (5) and IHC to highlight both nerves and tumor, increasing detection of PNI by 17%. We and others reported that IHC increases PNI detection because it helps identify small nerves and isolated tumor cells or small tumor islands (4, 54). Given the association of nerve-tumor parameters with outcome, our findings support the use of IHC in routine pathologic examination of carcinomas.
To provide objective criteria for nerve proximity to cancer (PNI-positive), we comprehensively analyzed all nerves in biopsies, measuring distance between nerve and the nearest tumor island/or cell. Strikingly, our results show that nerve-tumor distance helps predict poor survival, even among patients scored as PNI-negative. PNI-negative patients with any nerve-tumor distance less than 27 μm survived poorly compared with PNI-negative patients with larger distances. Using the 27 μm cutoff instead of PNI to split N0 patients, survival is also poor. This represents an increase of 28 N0 patients (PNI-negative but with distance less than 27 μm) who survive poorly and could perhaps benefit from adjuvant therapy. However, which distance to consider PNI is not defined; previously, using 71 biopsies and the same criteria, we showed similar results with a different cutoff (4). There is a gradual decrease in strength of association of distance with clinical outcomes as distance increases; nerves beyond 500 μm are not relevant. Importantly, our results advocate for a more inclusive definition of PNI considering nerves in proximity to tumor as PNI positive, regardless of extent of nerve involvement.
Within the tumor bulk, large nerve diameter is associated with poor survival; PNI-negative patients with nerves larger than 32 μm have worse survival than those with smaller nerves. Importantly, nerve diameter and nerve-tumor distance are independently associated with poor DSS, even when adjusting for AJCC 8th edition stage, which incorporates DOI. While nerve diameter in the tumor bulk associates with poor survival, diameter of PNI-positive nerves is not important. Similarly, patients with pancreatic cancer with larger nerves detected in tissue microarrays have poor overall survival (56). We believe that both nerve-tumor distance and nerve diameter reflect the biology of tumor-nerve interactions; nerve diameter may be a proxy for nerve activity.
The exchange between peripheral nerves and the local microenvironment are selectively regulated by the blood-nerve barrier, mainly comprised of endothelial cells. In physiologic conditions, this semipermeable barrier maintains homeostasis and integrity of nerves, regulating passage of nutrients and macromolecules that modulate the surrounding tissues (57). However, in pathologic conditions such as nerve injury, inhibition of tight junctions of the blood-nerve barrier increases permeability, modulating leukocyte translocation and immunosurveillance (58). Spatial transcriptomic analysis in our study supports a neurodegenerative/repair phenotype in response to nearby tumor that may trigger neuroplasticity toward axonogenesis. Nerves close to tumor show increased protein synthesis machinery, survival, stress response, and growth, including genes associated with cytoskeletal changes and axon regeneration. Given the injury phenotype, nerves that approximate cancer would be more affected by tumor-secreted factors and are more likely to influence tumor behavior. This is consistent with clinical data showing that relative death rate decreases as a function of larger nerve-tumor distances.
Our findings support that both nerve degeneration and repair are stimulated by proximity to cancer. A previous study suggested that Schwann cells associated with ganglioneuromas have a repair-specific phenotype similar to injured nerves (59). In contrast to epithelial or mesenchymal tumors, ganglioneuromas, which are derived from neuronal/glial tissue have a large population of glial cells. In addition, transcriptomic changes in the margin of melanomas suggest neuronal and glia-associated genes involved in nerve repair, while changes in tumor bulk suggest degeneration (60). However, both studies used bulk RNA sequencing and nerves were not profiled separately from the stroma. As a confounding issue, tumor and stromal cells such as fibroblasts and the immune infiltrate may express nerve growth factors, neuropeptides, and response to stress and repair genes, similar to cells in nerves.
Transcriptomic analysis of peripheral nerves represents mainly Schwann cells and neurons. We anticipated that changes in spatial data would represent Schwann cells more than neurons. Surprisingly, in vitro validation shows DEGs more related to neuronal cells. Because neuronal soma, where the nucleus is located, lies at great distances from nerves in cancer, transcripts captured from neuronal cells would be limited to axons. Indeed, neuronal cells have functional protein production machinery in axons, dendrites, and synaptic areas (61), both in developing and mature neurons (62). Because axons can extend for more than a meter from the soma to the site of innervation, neurons are highly compartmentalized and express transcripts in axons to facilitate rapid responses to the local environment (63, 64). For example, transcript levels are regulated by extracellular cues and lead to rapid translation of proteins at the injury site (65, 66).
Our study is the first to correlate spatial transcriptomic changes in nerves in human cancer with clinical outcomes. Using data from The Cancer Genome Atlas (67) and from c-Bioportal (68), others have studied the transcriptomic changes in tumors with and without PNI. Genes associated with epithelial-to-mesenchymal transition, metastasis, and invasion were upregulated in PNI-positive tumors, suggesting that PNI is an invasive process (67). Neurogenesis and axon guidance genes were also upregulated (68). However, bulk RNA samples do not elucidate spatial distribution of transcripts because tumor stroma and parenchyma are pooled.
Here we show that nerve-cancer cell proximity is important for prognosis in OSCC, and modulates the nerve transcriptome. These findings have implications for tumor biology and more importantly, open avenues of investigation leading to better treatment selection. In fact, these investigations could focus only on nerves close to tumor, which our study suggests are important for survival.
Limitations
IHC for the two cohorts was performed several years apart; however, all slides were digitized shortly after staining, preventing loss of quality. Although sections from the two cohorts were analyzed using different software, both digital pathology programs have similar tools for distance and area assessment.
Measuring all nerves in a large tumor sample is time consuming and a limitation for clinical use. However, our data suggest that a pathologist would need to measure only nerves close to the tumor, which are relevant for survival.
For spatial transcriptomic analysis, an important limitation is the area of tissue required to profile nerves both close and far from tumor cells; only two tumors per slide were possible because of the large tissue area required for each. Even in tumors with several nerves, not all nerves could be analyzed, due to low nuclear count or small size. Nerves smaller than 100 μm in diameter do not generate enough expression data for analysis and were excluded preventing transcriptomic analysis as a function of diameter. Another important aspect of the GeoMx analysis is the ability to select different data filters to remove unreliable data. Depending on the stringency of the analysis, DEGs may vary. Therefore, as recommendations for analysis evolve, transcriptomes detected will change. Furthermore, this study is exploratory in nature; interpretation of the function of DEGs requires mechanistic studies in the context of nerve-tumor interactions and may be tumor type specific.
Supplementary Material
Acknowledgments
The authors thank the many investigators in the University of Michigan HNSPORE/ HNOP for their contributions to patient recruitment, assistance in data collection, and encouragement including Carol R. Bradford, MD, Thomas E. Carey, PhD, Douglas B. Chepeha, MD, Sonia Duffy, PhD, Avraham Eisbruch, MD, Joseph Helman, DDS, Kelly M. Malloy, MD, Jonathan McHugh, MD, Scott A. McLean, MD, Tamara H. Miller, RN, Jeff Moyer, MD, Mark E. Prince, MD, Nancy Rogers, RN, Matthew E. Spector, MD, Nancy E. Wallace, RN, Heather Walline, PhD, Brent Ward, DDS, and Francis Worden, MD. We greatly thank patients and their families who tirelessly participated in survey and specimen collections in the University of Michigan HNSPORE/ HNOP. The authors also thank Lisa Peterson, MPH of the University of Michigan HNSPORE/ HNOP, Ms. Theresa Cody for excellent tissue sectioning, Margareth Hogan and Olivia Koues from the University of Michigan Advanced Genomics Core, and people from NanoString for help with conducting the spatial transcriptomic experiment and with data analysis. Support was provided by NIH/NIDCR DE027551 grant and a clinical research grant from the department of Periodontics and Oral Medicine (N.J. D'Silva), University of Michigan CEW+ Riecker Graduate Student Research Fund (L.B. Schmitd), and Rogel Cancer Center Core Grant CA46592 (E.L. Bellile and J.M.G. Taylor). The transcriptomic analysis work was funded and supported by the University of Michigan Biosciences Initiative Single-Cell Spatial Analysis Program.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 3403
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
L.B. Schmitd reports funding by NIH/NIDCR DE027551 (NJD) and the University of Michigan CEW+ Riecker Graduate Student Research Fund during the conduct of the study. C. Perez-Pacheco reports funding by NIH/NIDCR DE027551 (NJD) during the conduct of the study. N.J. D'Silva reports grants from NIH/NIDCR (DE027551) and NIH/NCI (CA 250214) during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
L.B. Schmitd: Conceptualization, data curation, formal analysis, investigation, writing–original draft, writing–review and editing. C. Perez-Pacheco: Data curation, investigation, writing–original draft, writing–review and editing. E.L. Bellile: Formal analysis, writing–original draft, writing–review and editing. W. Wu: Conceptualization, data curation, formal analysis, writing–original draft, writing–review and editing. K. Casper: Data curation, writing–original draft, writing–review and editing. M. Mierzwa: Data curation, writing–review and editing. L.S. Rozek: Resources, writing–review and editing. G.T. Wolf: Resources, writing–review and editing. J.M.G. Taylor: Formal analysis, writing–review and editing. N.J. D'Silva: Conceptualization, resources, supervision, funding acquisition, visualization, writing–original draft, writing–review and editing.
References
- 1. Monje M, Borniger JC, D'Silva NJ, Deneen B, Dirks PB, Fattahi F, et al. Roadmap for the emerging field of cancer neuroscience. Cell 2020;181:219–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao B, Lv W, Mei D, Luo R, Bao S, Huang B, et al. Perineural invasion as a predictive factor for survival outcome in gastric cancer patients: a systematic review and meta-analysis. J Clin Pathol 2020;73:544–51. [DOI] [PubMed] [Google Scholar]
- 3. Zareba P, Flavin R, Isikbay M, Rider JR, Gerke TA, Finn S, et al. Perineural invasion and risk of lethal prostate cancer. Cancer Epidemiol Biomarkers Prev 2017;26:719–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmitd LB, Beesley LJ, Russo N, Bellile EL, Inglehart RC, Liu M, et al. Redefining perineural invasion: integration of biology with clinical outcome. Neoplasia 2018;20:657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liebig C, Ayala G, Wilks JA, Berger DH, Albo D. Perineural invasion in cancer: a review of the literature. Cancer 2009;115:3379–91. [DOI] [PubMed] [Google Scholar]
- 6. Magnon C, Hall SJ, Lin J, Xue X, Gerber L, Freedland SJ, et al. Autonomic nerve development contributes to prostate cancer progression. Science 2013;341:1236361. [DOI] [PubMed] [Google Scholar]
- 7. Zhao CM, Hayakawa Y, Kodama Y, Muthupalani S, Westphalen CB, Andersen GT, et al. Denervation suppresses gastric tumorigenesis. Sci Transl Med 2014;6:250ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scanlon CS, Banerjee R, Inglehart RC, Liu M, Russo N, Hariharan A, et al. Galanin modulates the neural niche to favour perineural invasion in head and neck cancer. Nat Commun 2015;6:6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deborde S, Omelchenko T, Lyubchik A, Zhou Y, He S, McNamara WF, et al. Schwann cells induce cancer cell dispersion and invasion. J Clin Invest 2016;126:1538–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amit M, Takahashi H, Dragomir MP, Lindemann A, Gleber-Netto FO, Pickering CR, et al. Loss of p53 drives neuron reprogramming in head and neck cancer. Nature 2020;578:449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brandwein-Gensler M, Teixeira MS, Lewis CM, Lee B, Rolnitzky L, Hille JJ, et al. Oral squamous cell carcinoma: histologic risk assessment, but not margin status, is strongly predictive of local disease-free and overall survival. Am J Surg Pathol 2005;29:167–78. [DOI] [PubMed] [Google Scholar]
- 12. Chen W, Mi R, Haughey N, Oz M, Höke A. Immortalization and characterization of a nociceptive dorsal root ganglion sensory neuronal line. J Peripher Nerv Syst 2007;12:121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brady L, Kriner M, Coleman I, Morrissey C, Roudier M, True LD, et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat Commun 2021;12:1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bergholtz H, Carter JM, Cesano A, Cheang MCU, Church SE, Divakar P, et al. Best practices for spatial profiling for breast cancer research with the GeoMx(®) digital spatial profiler. Cancers 2021;13:4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmitd LB, Scanlon CS, D'Silva NJ. Perineural invasion in head and neck cancer. J Dent Res 2018;97:742–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chinn SB, Spector ME, Bellile EL, McHugh JB, Gernon TJ, Bradford CR, et al. Impact of perineural invasion in the pathologically N0 neck in oral cavity squamous cell carcinoma. Otolaryngol Head Neck Surg 2013;149:893–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pfister DG, Spencer S, Adelstein D, Adkins D, Anzai Y, Brizel DM, et al. Head and neck cancers, version 2.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2020;18:873–98. [DOI] [PubMed] [Google Scholar]
- 18. Thomas B, Stedman M, Davies L. Grade as a prognostic factor in oral squamous cell carcinoma: a population-based analysis of the data. Laryngoscope 2014;124:688–94. [DOI] [PubMed] [Google Scholar]
- 19. Subramaniam N, Balasubramanian D, Murthy S, Kumar N, Vidhyadharan S, Vijayan SN, et al. Predictors of locoregional control in stage I/II oral squamous cell carcinoma classified by AJCC 8th edition. Eur J Surg Oncol 2019;45:2126–30. [DOI] [PubMed] [Google Scholar]
- 20. Ross AS, Miller Whalen F, Elenitsas R, Xu X, Troxel AB, Schmults CD. Diameter of involved nerves predicts outcomes in cutaneous squamous cell carcinoma with perineural invasion: an investigator-blinded retrospective cohort study. Dermatol Surg 2009;35:1859–66. [DOI] [PubMed] [Google Scholar]
- 21. Xu B, Salama AM, Valero C, Yuan A, Khimraj A, Saliba M, et al. The prognostic role of histologic grade, worst pattern of invasion, and tumor budding in early oral tongue squamous cell carcinoma: a comparative study. Virchows Arch 2021;479:597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aivazian K, Ebrahimi A, Low TH, Gao K, Clifford A, Shannon K, et al. Perineural invasion in oral squamous cell carcinoma: quantitative subcategorisation of perineural invasion and prognostication. J Surg Oncol 2015;111:352–8. [DOI] [PubMed] [Google Scholar]
- 23. Tarsitano A, Tardio ML, Marchetti C. Impact of perineural invasion as independent prognostic factor for local and regional failure in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol 2015;119:221–8. [DOI] [PubMed] [Google Scholar]
- 24. Wei PY, Li WY, Tai SK. Discrete perineural invasion focus number in quantification for T1-T2 oral squamous cell carcinoma. Otolaryngol Head Neck Surg 2019;160:635–41. [DOI] [PubMed] [Google Scholar]
- 25. Newman M, Dziegielewski PT, Nguyen NTA, Seikaly HS, Xie M., O'Connell DA, et al. Relationship of depth of invasion to survival outcomes and patterns of recurrence for T3 oral tongue squamous cell carcinoma. Oral Oncol 2021;116:105195. [DOI] [PubMed] [Google Scholar]
- 26. Le Gall L, Anakor E, Connolly O, Vijayakumar UG, Duddy WJ, Duguez S. Molecular and cellular mechanisms affected in ALS. J Pers Med 2020;10:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Patzig J, Jahn O, Tenzer S, Wichert SP, de Monasterio-Schrader P, Rosfa S, et al. Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. J Neurosci 2011;31:16369–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Negro S, Stazi M, Marchioretto M, Tebaldi T, Rodella U, Duregotti E, et al. Hydrogen peroxide is a neuronal alarmin that triggers specific RNAs, local translation of Annexin A2, and cytoskeletal remodeling in schwann cells. RNA 2018;24:915–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duan X, Li M, Liu F, Song X, Zhang C, Zhu M, et al. Sensorimotor nerve lesion of upper airway in patients with obstructive sleep apnea. Respir Physiol Neurobiol 2021;293:103720. [DOI] [PubMed] [Google Scholar]
- 30. Hoch-Kraft P, White R, Tenzer S, Krämer-Albers EM, Trotter J, Gonsior C. Dual role of the RNA helicase DDX5 in post-transcriptional regulation of myelin basic protein in oligodendrocytes. J Cell Sci 2018;131:jcs204750. [DOI] [PubMed] [Google Scholar]
- 31. Kuboyama K, Fujikawa A, Masumura M, Suzuki R, Matsumoto M, Noda M. Protein tyrosine phosphatase receptor type z negatively regulates oligodendrocyte differentiation and myelination. PLoS One 2012;7:e48797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thomaidou D, Coquillat D, Meintanis S, Noda M, Rougon G, Matsas R. Soluble forms of NCAM and F3 neuronal cell adhesion molecules promote Schwann cell migration: identification of protein tyrosine phosphatases zeta/beta as the putative F3 receptors on Schwann cells. J Neurochem 2001;78:767–78. [DOI] [PubMed] [Google Scholar]
- 33. Hu G, Huang K, Hu Y, Du G, Xue Z, Zhu X, et al. Single-cell RNA-seq reveals distinct injury responses in different types of DRG sensory neurons. Sci Rep 2016;6:31851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frendo ME, da Silva A, Phan KD, Riche S, Butler SJ. The Cofilin/Limk1 pathway controls the growth rate of both developing and regenerating motor axons. J Neurosci 2019;39:9316–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Menon S, Gupton SL. Building blocks of functioning brain: cytoskeletal dynamics in neuronal development. Int Rev Cell Mol Biol 2016;322:183–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Q, Yuan W, Yang X, Wang Y, Li Y, Qiao H. Role of cofilin in alzheimer's disease. Front Cell Dev Biol 2020;8:584898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu XH, Deng CY, Liu Y, He M, Peng J, Wang T, et al. MARCKS regulates membrane targeting of Rab10 vesicles to promote axon development. Cell Res 2014;24:576–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Melrose J, Hayes AJ, Bix G. The CNS/PNS extracellular matrix provides instructive guidance cues to neural cells and neuroregulatory proteins in neural development and repair. Int J Mol Sci 2021;22:5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang J, Wei W, Jin HC, Ying RC, Zhu AK, Zhang FJ. Programmed cell death 2 protein induces gastric cancer cell growth arrest at the early S phase of the cell cycle and apoptosis in a p53-dependent manner. Oncol Rep 2015;33:103–10. [DOI] [PubMed] [Google Scholar]
- 40. Baron BW, Hyjek E, Gladstone B, Thirman MJ, Baron JM. PDCD2, a protein whose expression is repressed by BCL6, induces apoptosis in human cells by activation of the caspase cascade. Blood Cells Mol Dis 2010;45:169–75. [DOI] [PubMed] [Google Scholar]
- 41. Lydiatt W, O'Sullivan B, Patel S. Major changes in head and neck staging for 2018. Am Soc Clin Oncol Educ Book 2018;38:505–14. [DOI] [PubMed] [Google Scholar]
- 42. Huang TY, Hsu LP, Wen YH, Huang TT, Chou YF, Lee CF, et al. Predictors of locoregional recurrence in early stage oral cavity cancer with free surgical margins. Oral Oncol 2010;46:49–55. [DOI] [PubMed] [Google Scholar]
- 43. Liu CH, Chen HJ, Wang PC, Chen HS, Chang YL. Patterns of recurrence and second primary tumors in oral squamous cell carcinoma treated with surgery alone. Kaohsiung J Med Sci 2013;29:554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolfensberger M, Zbaeren P, Dulguerov P, Müller W, Arnoux A, Schmid S. Surgical treatment of early oral carcinoma-results of a prospective controlled multicenter study. Head Neck 2001;23:525–30. [DOI] [PubMed] [Google Scholar]
- 45. Lim YC, Choi EC. Surgery alone for squamous cell carcinoma of the oral cavity: survival rates, recurrence patterns, and salvage treatment. Acta Otolaryngol 2008;128:1132–7. [DOI] [PubMed] [Google Scholar]
- 46. National Comprehensive Cancer Network. NCCN Guidelines: Head and Neck Cancers v. 2.2022. Available from: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1437.
- 47. Chi AC, Katabi N, Chen HS, Cheng YL. Interobserver variation among pathologists in evaluating perineural invasion for oral squamous cell carcinoma. Head Neck Pathol 2016;10:451–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yan F, Cheng YL, Katabi N, Nguyen SA, Chen HS, Morgan P, et al. Interobserver variation in evaluating perineural invasion for oral squamous cell carcinoma: phase 2 survey study. Head Neck Pathol 2021;15:935–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cracchiolo JR, Xu B, Migliacci JC, Katabi N, Pfister DG, Lee NY, et al. Patterns of recurrence in oral tongue cancer with perineural invasion. Head Neck 2018;40:1287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hasmat S, Ebrahimi A, Gao K, Low TH, Palme C, Gupta R, et al. Multifocal perineural invasion is a better prognosticator than depth of invasion in oral squamous cell carcinoma. Head Neck 2019;41:3992–9. [DOI] [PubMed] [Google Scholar]
- 51. Miller ME, Palla B, Chen Q, Elashoff DA, Abemayor E., St John MA, et al. A novel classification system for perineural invasion in noncutaneous head and neck squamous cell carcinoma: histologic subcategories and patient outcomes. Am J Otolaryngol 2012;33:212–5. [DOI] [PubMed] [Google Scholar]
- 52. Caponio VCA, Troiano G, Togni L, Zhurakivska K, Santarelli A, Laino L, et al. Pattern and localization of perineural invasion predict poor survival in oral tongue carcinoma. Oral Dis 2021 May 8. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 53. Fagan JJ, Collins B, Barnes L, D'Amico F, Myers EN, Johnson JT. Perineural invasion in squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg 1998;124:637–40. [DOI] [PubMed] [Google Scholar]
- 54. Kurtz KA, Hoffman HT, Zimmerman MB, Robinson RA. Perineural and vascular invasion in oral cavity squamous carcinoma: increased incidence on re-review of slides and by using immunohistochemical enhancement. Arch Pathol Lab Med 2005;129:354–9. [DOI] [PubMed] [Google Scholar]
- 55. Shen WR, Wang YP, Chang JY, Yu SY, Chen HM, Chiang CP. Perineural invasion and expression of nerve growth factor can predict the progression and prognosis of oral tongue squamous cell carcinoma. J Oral Pathol Med 2014;43:258–64. [DOI] [PubMed] [Google Scholar]
- 56. Ferdoushi A, Griffin N, Marsland M, Xu X, Faulkner S, Gao F, et al. Tumor innervation and clinical outcome in pancreatic cancer. Sci Rep 2021;11:7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maiuolo J, Gliozzi M, Musolino V, Carresi C, Nucera S, Macrì R, et al. The role of endothelial dysfunction in peripheral blood nerve barrier: molecular mechanisms and pathophysiological implications. Int J Mol Sci 2019;20:3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kanda T, Numata Y, Mizusawa H. Chronic inflammatory demyelinating polyneuropathy: decreased claudin-5 and relocated ZO-1. J Neurol Neurosurg Psychiatry 2004;75:765–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weiss T, Taschner-Mandl S, Janker L, Bileck A, Rifatbegovic F, Kromp F, et al. Schwann cell plasticity regulates neuroblastic tumor cell differentiation via epidermal growth factor-like protein 8. Nat Commun 2021;12:1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shurin GV, Kruglov O, Ding F, Lin Y, Hao X, Keskinov AA, et al. Melanoma-induced reprogramming of schwann cell signaling aids tumor growth. Cancer Res 2019;79:2736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Agrawal M, Welshhans K. Local translation across neural development: a focus on radial glial cells, axons, and synaptogenesis. Front Mol Neurosci 2021;14:717170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, et al. Dynamic axonal translation in developing and mature visual circuits. Cell 2016;166:181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, et al. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci 2010;30:15464–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Holt CE, Martin KC, Schuman EM. Local translation in neurons: visualization and function. Nat Struct Mol Biol 2019;26:557–66. [DOI] [PubMed] [Google Scholar]
- 65. Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, et al. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol 2007;178:965–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mofatteh M. mRNA localization and local translation in neurons. AIMS Neurosci 2020;7:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Z, Liu R, Jin R, Fan Y, Li T, Shuai Y, et al. Integrating clinical and genetic analysis of perineural invasion in head and neck squamous cell carcinoma. Front Oncol 2019;9:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guo JA, Hoffman HI, Shroff SG, Chen P, Hwang PG, Kim DY, et al. Pan-cancer transcriptomic predictors of perineural invasion improve occult histopathologic detection. Clin Cancer Res 2021;27:2807–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for this study were generated at the University of Michigan Advanced Genomics core facility (https://brcf.medicine.umich.edu/cores/advanced-genomics/). Derived data supporting the findings of this study are available from the corresponding author upon request.