SUMMARY
The diversity of visual input processed by the mammalian visual system requires the generation of many distinct retinal ganglion cell (RGC) types, each tuned to a particular feature. The molecular code needed to generate this cell type diversity is poorly understood. Here we focus on the molecules needed to specify one type of retinal cell: the upward-preferring ON direction-selective ganglion cell (up-oDSGC) of the mouse visual system. Single-cell transcriptomic profiling of up- and down-oDSGCs shows that the transcription factor Tbx5 is selectively expressed in up-oDSGCs. Loss of Tbx5 in up-oDSGCs results in a selective defect in the formation of up-oDSGCs and a corresponding inability to detect vertical motion. A downstream effector of Tbx5, Sfrp1, is also critical for vertical motion detection but not up-oDSGC formation. These results advance our understanding of the molecular mechanisms that specify a rare retinal cell type and show how disrupting this specification leads to a corresponding defect in neural circuitry and behavior.
eTOC BLURB
Little is known about the molecular code needed to specify different retinal ganglion cell types. Al-Khindi et al. show that the transcription factor Tbx5 is expressed by upward-preferring ON DSGCs and is required for their initial formation and the proper detection of vertical motion.
INTRODUCTION
Processing the diverse array of information in the environment requires the nervous system to generate a correspondingly diverse array of cell types. This is particularly evident in the mammalian retina, where ~40-45 types of retinal ganglion cells (RGCs) each process a specific feature of the visual scene, such as spatial contrast, color information, detection of looming stimuli, and image motion.1-3 How the nervous system generates this RGC diversity at a molecular level is poorly understood.
Prior work provides important insight into the molecular network required for the specification of RGCs as a class.4 Atoh7 (also termed Math5) is a transcription factor expressed by retinal progenitor cells that is necessary for their commitment to the RGC fate and their survival.5-7 Pou4f2 (also termed Brn3b), a transcription factor positively regulated by Atoh7, is required for the specification of a majority of RGCs (>70%).8,9 Downstream of Pou4f2 lies Pou4f1 (also termed Brn3a), a transcription factor that regulates the differentiation of monostratified RGCs.8 Importantly, neither Atoh7 nor any Pou4f family member is specific for the differentiation of any one RGC subtype. Recent work has begun to shed light on genes that specify particular RGC subtypes. One gene, Eomes (also termed Tbr2), commits newly-born RGCs to become intrinsically photosensitive RGCs.10,11 Another gene, Tbr1, is required for the formation and survival of 2 types of OFF-sustained RGCs.12,13 The molecular code required to specify other RGC types, however, remains unclear.
The retinal direction-selective (DS) circuit plays a critical role in vision and provides a unique opportunity to study the molecular mechanisms responsible for generating RGC diversity. Direction-selective ganglion cells (DSGCs) function as motion detectors, only firing action potentials when light moves in a particular direction across their receptive fields.14,15 Some DSGCs, termed ON DSGCs (oDSGCs), respond to slow motion and are essential for stabilizing images on the retina during head movement through the optokinetic reflex (OKR).14,16 Classically, oDSGCs can be divided into three subtypes, each tuned to a direction parallel to one of the three semicircular canals,17 though more recent work categorizes oDSGCs into four subtypes with each tuned to one of the four cardinal directions.18 Upon detecting a moving stimulus, oDSGCs activate the nuclei of the accessory optic system (AOS) in the midbrain: the medial terminal nucleus (MTN), lateral terminal nucleus (LTN), dorsal terminal nucleus (DTN), and the nucleus of the optic tract (NOT).17,19 These AOS nuclei, in turn, coordinate compensatory eye movements, in the form of the OKR, to minimize image motion on the retina.
Certain oDSGC types are very similar, differing in one fundamental functional property. oDSGCs that respond to upward or downward motion, for example, have different directional preferences, but otherwise have similar dendrite morphologies, similar ON-sustained responses to light stimuli, and similar axon projections to the MTN.20 Little is known about the molecular programs needed for the initial specification of these, or any, closely related RGC types.
Here, we use single-cell RNA sequencing (scRNAseq) to transcriptionally profile the upward- and downward-preferring oDSGCs of the AOS. We describe genetic differences between these two closely related cell types and show that the transcription factor Tbx5 is necessary for the specification of upward-preferring oDSGCs and the corresponding ability to detect vertical motion. We also identify one downstream effector of Tbx5, Sfrp1, that is critical for vertical motion detection. This work advances our understanding of the molecules needed to specify a particular retinal cell type and shows how disruption of this specification leads to a selective defect in both neural circuitry and behavior.
RESULTS
ON DSGCs tuned to upward or downward motion have unique transcriptional signatures
To determine transcriptional differences between DSGCs, we focused on two DSGC subtypes of the AOS: the upward-tuned ON DSGCs (up-oDSGCs) and downward-tuned ON DSGCs (down-oDSGCs) that project axons to the MTN. Up-oDSGCs were selectively labeled using the Spig1-GFP knock-in mouse line.21 To label down-oDSGCs, a retrogradely transported fluorescent dye, Cholera Toxin Subunit B-Alexa647 (CTB-A647), was stereotactically injected into the MTN of postnatal day 3 (P3) Spig1-GFP mice (Figure 1A). Brains were examined after injection to confirm the presence of CTB-A647 in the MTN and the absence of dye in adjacent regions such as the superior colliculus (SC) (Figures S1A-S1F). Examining retinas from these mice revealed both GFP+CTB+ up-oDSGCs and GFP−CTB+ down-oDSGCs, often occurring in pairs, as previously described20,21 (Figure 1B). DSGCs acquire their directional preference during a narrow time window between ~P7-P8.22,23 Therefore, we harvested retinas before (P4, P5), during (P7, P8), and after (P10, P12) the acquisition of DS (Figure 1C). As noted previously,20,21 Spig1-GFP mice express GFP nonspecifically in the dorsotemporal retina, so we removed the dorsotemporal retina under fluorescent guidance prior to tissue dissociation. Single GFP+CTB+ up-oDSGCs and GFP−CTB+ down-oDSGCs were isolated using fluorescence activated cell sorting (FACS) and subjected to full-length single-cell RNA sequencing (scRNAseq) at high read depth (1.5 x 106 ± 0.4 x 106[M ± SD] reads/cell, 89 ± 4% alignment to genome) using a modified Smart-seq2 protocol.24,25 Our decision to use FACS-enriched Smart-seq2 (high read depth, low sample size) over unbiased droplet-based 3’ end-tagging methods1,26 (low read depth, high sample size) was guided by three reasons. First, given that up-oDSGCs and down-oDSGCs are closely related DSGC subtypes that share many properties, including dendrite morphology and axon projections, we hypothesized that these two cell types may have high transcriptional similarity. A sequencing approach characterized by high read depth, which provides more information about lower abundance transcripts, could be better at detecting subtle transcriptional differences that would distinguish these two highly similar cell types. Second, given that up- and down-oDSGCs are rare,21,27 we speculated that collecting up- and down-oDSGCs in a targeted manner with FACS would provide a more certain yield than collecting RGCs in an agnostic fashion using an unbiased droplet-based technology. Third, given that down-oDSGCs are completely uncharacterized at a transcriptional level, we would have no way to identify which cells correspond to down-oDSGCs if RGCs were collected in an agnostic manner using droplet-based approaches.
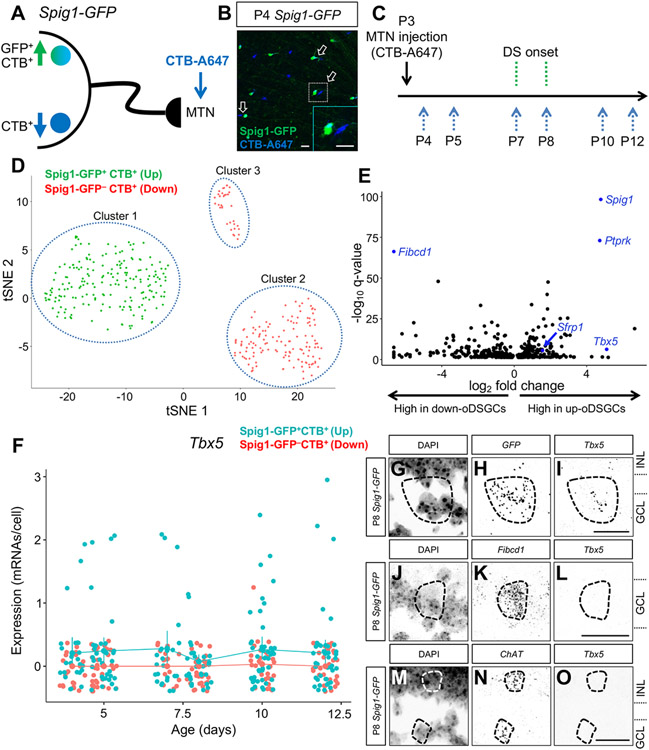
Figure 1. Transcriptomic profiling of MTN-projecting ON DSGCs.
(A) Upward-preferring Spig1-GFP+ and downward-preferring Spig1-GFP− ON direction-selective ganglion cells (oDSGCs) were retrogradely labeled by stereotactic injection of CTB-A647 into the medial terminal nucleus (MTN). (B) Whole-mount retina at P4 shows GFP+CTB+ up and GFP−CTB+ down oDSGCs after successful MTN targeting at P3. Note the couplets of retrogradely labeled RGCS (arrows) that are differentially labeled (inset). (C) Experimental design for single-cell RNA sequencing. MTN injections were done at P3. Cells were collected before (P4, P5), during (P7, P8), and after (P10, P12) DS onset. (D) tSNE plot of single-cell RNA sequencing data reveals 3 RGC populations: 2 groups of MTN-projecting oDSGCs (Spig1-GFP+CTB+ cells–cluster 1 and Spig1-GFP− CTB+ cells–cluster 2) and 1 group that likely corresponds to ipRGCs (Spig1-GFP−CTB+ cells–cluster 3). (E) Volcano plot showing q-values and effect sizes for genes that are significantly differentially expressed between up- and down-oDSGCs (all genes shown have q<0.05). Spig1, Ptprk, Sfrp1, and Tbx5 were differentially expressed in up-oDSGCs, whereas Fibcd1 was differentially expressed in down-oDSGCs. (F) Tbx5 expression in up-oDSGCs was constant from P4 through P12 and was absent in GFP− CTB+ down-oDSGCs. Note the extremely low Tbx5 transcript number per cell and the small fraction of Spig1-GFP+CTB+ RGCs that express Tbx5 at any given time point. Data presented as mean ± 95% confidence intervals. (G-O) RNAscope in situ hybridization in P8 Spig1-GFP retinas shows Tbx5 expression in GFP+ up-oDSGCs (G-I), absent Tbx5 expression in Fibcd1+ down-oDSGCs (J-L), and absent Tbx5 expression in ChAT+ SACs (M-O). GCL, ganglion cell layer. INL, inner nuclear layer. Scale bars, 25 um. See also Figure S1.
Only cells that expressed the neuronal marker Tubb3 and the RGC marker RBPMS were included in our analyses (Figures S1G and S1H). RGCs passing quality control expressed an average of 7324 ± 959 genes (M ± SD). oDSGCs were rare; up-oDSGCs and down-oDSGCs each accounted for 0.002-0.005% of all retinal cells isolated by FACS between P4-P12. Our observed oDSGC frequency was similar to the expected frequency based on prior estimates (up- and down-oDSGCs each account for ~0.005% of all retinal cells21,27). In total, between 24-45 GFP+CTB+ up-oDSGCs and 18-40 GFP−CTB+ down-oDSGCs were isolated at each developmental time point.
Unsupervised and semi-supervised clustering analysis (based on Spig1 expression; see STAR Methods) revealed 3 RGC types: one GFP+CTB+ cluster and two GFP−CTB+ clusters (one large and one small) (Figure 1D), with no overlap among these clusters at any postnatal developmental time point. Each cluster expressed its own set of unique marker genes. The single GFP+CTB+ cluster specifically expressed protein tyrosine phosphatase receptor kappa (Ptprk) and, as expected, Spig1; the larger GFP−CTB+ cluster specifically expressed fibrinogen C domain-containing protein 1 (Fibcd1); and the smaller GFP−CTB+ cluster expressed calbindin 2 (Calb2) (Figures S1I-S1L). The smaller GFP−CTB+ cluster also expressed several markers of intrinsically photosensitive retinal ganglion cells (ipRGCs), such as eomesodermin (Eomes) and melanopsin (Opn4) (Figures S1M and S1N). The presence of two GFP−CTB+ clusters was unexpected; prior work, using retrograde CTB labeling from the MTN, only identified a single type of down-oDSGC based on morphology and electrophysiological properties.20,21 The smaller cluster of GFP−CTB+ RGCs likely represents ipRGC contamination, given expression of the ipRGC marker genes Opn4 and Eomes. Contamination by ipRGCs is plausible given the close spatial proximity of the suprachiasmatic nucleus (the primary target of ipRGC axons) and the MTN on the ventral surface of the neonatal mouse brain. Therefore, we removed the small cluster of GFP−CTB+ RGCs from our subsequent data analyses and do not consider it further in this study.
To gain insight into genes that might underlie oDSGC directional tuning, we identified the genes differentially expressed between up- and down-oDSGCs averaged across all developmental time points (Figure 1E). As expected, Spig1 transcripts were highly enriched in the GFP+CTB+ up-oDSGC cluster. Several genes showed striking differential expression (DE) patterns, such as Ptprk, T-box transcription factor 5 (Tbx5), and secreted frizzled-related protein 1 (Sfrp1) in up-oDSGCs, and Fibcd1 in the larger cluster of down-oDSGCs. These results demonstrate that two rare RGC subtypes differing primarily in their tuning to upward or downward motion can be unambiguously distinguished by DE gene expression profiles.
Our ability to transcriptionally separate the GFP+CTB+ up-oDSGC cluster and the GFP−CTB+ down-oDSGC cluster was notable since recent transcriptomic analyses of mouse RGCs were not able to resolve up- and down-oDSGCs into two separate cell types. For example, 10X Genomics 3’ end-tagging technology was used to sequence 6225 P5 mouse RGCs at 100,000 reads/cell.1 In this data set, RGC cluster 32 strongly expresses Spig1 (also known as Fstl4) and Ptprk, suggesting that cluster 32 corresponds to up-oDSGCs. However, RGCs in cluster 32 also strongly express Fibcd1, a marker of down-oDSGCs in our data set. Similarly, 10X Genomics 3’ end-tagging technology used to sequence 35,699 P56 mouse RGCs2 identified RGC cluster 10 as being most similar to RGC cluster 32 from the P5 data set described above.1 In this data set, RGC cluster 10 expresses both the up-oDSGC marker Ptprk and the down-oDSGC marker Fibcd1. Moreover, Gpr88, a marker of cluster 10, is expressed by both up- and down-oDSGCs in our data set (Figure S1O). These droplet-based scRNAseq methods are characterized by higher sample size but lower read depth (i.e., fewer mRNAs/unique molecular identifiers [UMIs] sampled per cell), in contrast to the approach described here, which has lower sample size but higher read depth. Droplet-based high-sample-size, low-depth scRNAseq approaches provide an excellent avenue for global characterization of cell type number within tissues; however, they may yield insufficient transcriptional information to resolve very similar cell types such as up- and down-oDSGCs. Our ability to transcriptionally differentiate up- and down-oDSGCs shows that high-depth, low-sample-size scRNAseq approaches can complement global droplet-based approaches to profile rare populations of transcriptionally similar cell types.
Onset of direction selectivity at P7-P8 does not correlate with a stepwise change in DSGC gene expression
DSGCs acquire their directional preference during a critical interval between P7-P8.22,23 One possible mechanism for this phenomenon is a sudden, stepwise change in gene expression in oDSGCs around the P7-P8 time window. We tested this hypothesis by examining the temporal changes in gene expression between P4 (before the onset of DS) and P12 (after the onset of DS) by ordering the cells along a pseudotime trajectory.28 Between P4 and P12, oDSGCS showed dramatic changes in gene expression, with some genes increasing over time and others decreasing. Up- and down-oDSGCs showed age-related changes in 2035 and 1624 genes, respectively. Of these, 976 genes were common to both up- and down-oDSGCs, 1059 were unique to up-oDSGCs, and 648 were unique to down-oDSGCs (Figure S1P). Examining the temporal changes in (1) the top 200 shared genes, (2) top 200 up-oDSGC-specific genes, and (3) top 200 down-oDSGC-specific genes revealed no genes whose expression changed in a stepwise, non-linear fashion around the P7-P8 time window. Rather, genes that changed over time tended to have a smooth, linear change in their temporal kinetics between P4 and P12 (Figures S1Q-S1T). These results suggest that DS acquisition is unlikely to be explained by a sudden, non-linear change in gene expression intrinsic to oDSGCs.
The transcription factor Tbx5 is selectively expressed in up-oDSGCs
One gene that showed selective enrichment in up-oDSGCs was Tbx5, which encodes a member of the T-box transcription factor family. Tbx5 is expressed in an asymmetric fashion in many tissues, including the upper limbs,29 the developing heart,30 and the embryonic retina as early as E10.5.31,32 Given its asymmetric expression patterns in many tissues, we asked whether Tbx5 regulates DSGC directional preference, perhaps by regulating asymmetric inhibitory synaptic connectivity between up-oDSGCs and starburst amacrine cells (SACs)22,23 or by influencing the development of up-oDSGCs.
Tbx5 is expressed at constant levels in up-oDSGCs between P4 and P12, though only at very low levels (1-2 detected mRNAs/cell) and only detected in ~14% of up-oDSGCs (Figure 1F). Tbx5 expression, however, was absent in down-oDSGCs. In situ hybridization of P8 Spig1-GFP retinal tissue showed exclusive Tbx5 expression in GFP+ up-oDSGCs (Figures 1G-1I) and no expression in Fibcd1+ down-oDSGCs (Figures 1J-1L), ChAT+ SACs (Figures 1M-1O), or elsewhere in the retina. Querying Tbx5 expression in existing RGC scRNAseq databases at P5 and P56 RGCs reveals high and relatively specific expression in clusters that include up-oDGSCs, among other RGC subtypes (data not shown).1,2 These results show that Tbx5 is selectively expressed by up-oDSGCs both before and after the onset of DS.
Tbx5 is necessary for the vertical optokinetic reflex
To assess Tbx5 function in up-oDSGCs, we took advantage of the Pcdh9-Cre mouse line,33 which labels a type of MTN-projecting oDSGC that prefers vertical motion.34 Intraocular injections of AAV9 FLEX-tdTomato into the eyes of P2 Spig1-GFP/+;Pcdh9-Cre/+ transheterozygous mice showed almost exclusive co-labeling between Spig1-GFP+ and tdTomato+ RGCs at P10, suggesting that Spig1-GFP and Pcdh9-Cre label the same class of upward-preferring ON DSGC (Figures S2A-S2E). Additionally, single-cell transcriptomic profiling of P12 Pcdh9-Cre RGCs showed overlap between these cells and Spig1-GFP RGCs on the tSNE plot (Figure S2F), further indicating that Spig1-GFP and Pcdh9-Cre both label the same up-oDSGC population.
We measured the optokinetic reflex (OKR), a behavioral readout of oDSGC and AOS function,35,36 in Pcdh9-Cre;Tbx5flox/flox mice, which lack Tbx5 in up-oDSGCs. In response to continuously-rotating stimuli, Pcdh9-Cre;Tbx5flox/flox mice showed normal horizontal OKR performance, but these mice had no vertical OKR in either the ventrodorsal or dorsoventral directions (Figures 2A-2D).
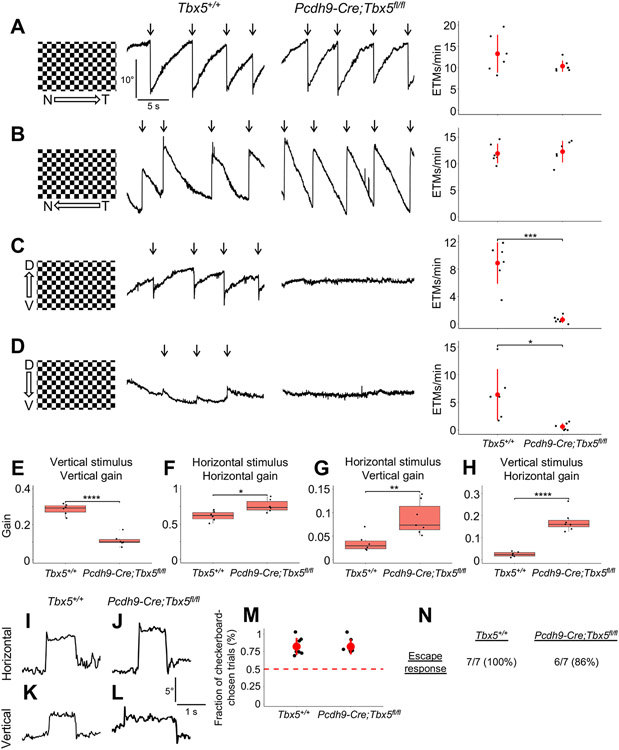
Figure 2. Pcdh9-Cre;Tbx5fl/fl mice show selective defects in vertical motion detection.
(A-D) Optokinetic reflex (OKR) results in adult Tbx5+/+ and Pcdh9-Cre;Tbx5fl/fl mice in response to nasotemporal (A), temporonasal (B), ventrodorsal (C), or dorsoventral (D) continuous motion, measured by scoring eye tracking movements (ETMs) per minute (arrows). Pcdh9-Cre;Tbx5fl/fl mice exhibited absent OKRs in both the ventrodorsal and dorsoventral directions. Data presented as mean ± SD. (E-H) Pcdh9-Cre;Tbx5fl/fl animals showed reduced gain in response to vertical sinusoidal stimuli (E) and enhanced gain in response to horizontal sinusoidal stimuli (F). Pcdh9-Cre;Tbx5fl/fl mice demonstrated enhanced vertical gain in response to horizontal stimuli (G) and enhanced horizontal gain in response to vertical stimuli (H). Data presented as mean ± IQR with whiskers representing the range. (I-L) Tbx5+/+ and Pcdh9-Cre;Tbx5fl/fl mice both performed voluntary horizontal and vertical saccades. (M) Pcdh9-Cre;Tbx5fl/fl mice showed normal performance on the visual cliff task, a measure of depth perception (dashed line represents chance performance). Data presented as mean ± SD. (N) Pcdh9-Cre;Tbx5fl/fl mice showed normal escape behaviors on the looming task, a measure of the ability to detect overhead looming stimuli. N, nasal. T, temporal. D, dorsal. V, ventral. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figure S2.
In addition to continuously-rotating stimuli, we measured the OKR using sinusoidally-moving stimuli, which allows us to compute eye movement gain (eye velocity divided by stimulation velocity).16 Pcdh9-Cre;Tbx5flox/flox mice showed reduced vertical OKR gain in response to sinusoidal vertical stimuli (Figure 2E), as expected based on their responses to continuous vertical stimuli. However, when presented with horizontal sinusoidal stimuli, Pcdh9-Cre;Tbx5flox/flox animals showed an enhanced horizontal OKR gain compared to wild-type controls (Figure 2F). Surprisingly, Pcdh9-Cre;Tbx5flox/flox mice also showed increased vertical gain in response to horizontal sinusoidal motion and, reciprocally, increased horizontal gain in response to vertical sinusoidal motion (Figures 2G and 2H) – a phenomenon termed “cross-coupling”, which has been previously observed in humans with ocular pathologies such as strabismus and idiopathic infantile nystagmus.37-39 The visual defects we observed were specific to the OKR; voluntary horizontal and vertical saccadic eye movements (Figures 2I-2L), depth perception (Figure 2M), and looming stimulus detection (Figure 2N) were all normal in Pcdh9-Cre;Tbx5flox/flox mice. Taken together, these results show that deletion of Tbx5 in up-oDSGCs causes a selective defect in both the upward and downward OKR. Additionally, deletion of Tbx5 in up-oDSGCs unmasks a cross-coupling response in which vertical or horizontal stimuli induce eye movements in the orthogonal direction.
Tbx5 is required for the initial formation of up-oDSGCs
To investigate the neural basis underlying the vertical OKR defect seen in Pcdh9-Cre;Tbx5flox/flox mice, we examined retinas from Spig1-GFP;Pcdh9-Cre;Tbx5+/+ and Spig1-GFP;Pcdh9-Cre;Tbx5flox/flox mice. At embryonic day 13.5 (E13.5) – approximately the time of RGC birth – there were no Spig1-GFP+ up-oDSGCs in Spig1-GFP;Pcdh9-Cre;Tbx5flox/flox retinas (Figures 3A and 3B). By P0, Spig1-GFP+ up-oDSGCs were completely absent from Spig1-GFP;Pcdh9-Cre;Tbx5flox/flox retinas, and this absence persisted to later ages (e.g., P12) (Figures 3C-3F). The absence of up-oDSGCs in Spig1-GFP;Pcdh9-Cre;Tbx5flox/flox mice is unlikely to be explained by apoptosis given that up-oDSGCs were still absent even after deletion of Bax, a pro-apoptotic gene (Figures S3A and S3B). Additionally, the absence of retinal neurons in Spig1-GFP;Pcdh9-Cre;Tbx5flox/flox mice was restricted to up-oDSGCs and did not apply more generally to SACs or RGCs (Figures S3C-S3F). Overall retinal architecture was also intact in Pcdh9-Cre;Tbx5flox/flox mice: throughout the retina, ChAT labeling revealed normal SAC dendrite stratification in the ON and OFF IPL sublaminae and calbindin labeling showed normal IPL lamination patterns (Figures S3G-S3L).
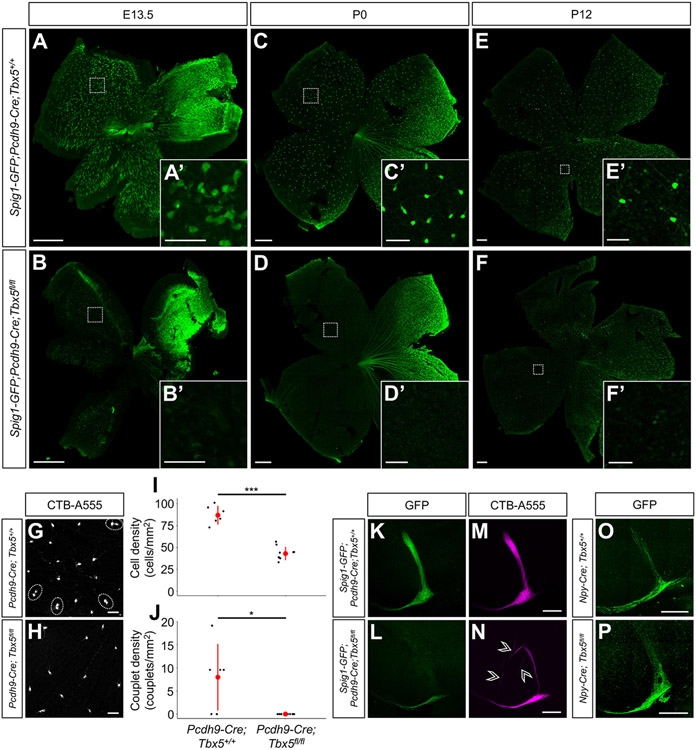
Figure 3. Up-oDSGCs are absent in Spig1-GFP;Pcdh9-Cre;Tbx5fl/fl mice.
(A-F) Spig1-GFP+ up-oDSGCs were absent in Spig1-GFP;Pcdh9-Cre;Tbx5fl/fl mice at E13.5 (A and D), P0 (B and E), and P12 (C and F). (G-H) Whole-mount retinas of P6 Pcdh9-Cre;Tbx5+/+ (G) and Pcdh9-Cre;Tbx5fl/fl (H) retinas are shown following retrograde injection of CTB-555 into the MTN at P4. RGC couplets, corresponding to 1 up- and 1 down-oDSGC, were observed in Pcdh9-Cre;Tbx5+/+ retinas (dashed circles in G), but were absent in Pcdh9-Cre;Tbx5fl/fl retinas. (I-J) P6 Pcdh9-Cre;Tbx5fl/fl mice showed a ~50% reduction in MTN-projecting RGC density (I) and absence of up- and down-oDSGC couplets (J). Data presented as mean ± SD. (K-N) Following intraocular CTB-A555 injections at P10, the dorsal MTN was hypoinnervated in P12 Spig1-GFP;Pcdh9-Cre;Tbx5fl/fl mice, while the ventral MTN was normally innervated. Mistargeted CTB+ axons were observed in the proximity of the MTN in Spig1-GFP;Pcdh9-Cre;Tbx5fl/fl mice (arrowheads in N). (O-P) The MTN was appropriately innervated in adult Npy-Cre;Tbx5fl/fl mice following intraocular AAV2 FLEX-GFP injection. Scale bars, 250 um in (A-F) and (K-P); 50 um in (A’-F’) and (G-H). *p < 0.05; ***p < 0.001. See also Figure S3.
To exclude the possibility that Spig1-GFP no longer reliably labels up-oDSGCs in Tbx5 mutant mice, retinas from P6 Pcdh9-Cre;Tbx5flox/flox mice were examined after retrograde CTB-A555 injection into the MTN at P4. Compared to controls, the density of MTN-projecting RGCs was reduced by ~50% in Pcdh9-Cre;Tbx5flox/flox mice at P6 (Figures 3G-3I). Additionally, though MTN-projecting CTB-labeled RGCs in wild-type mice often were observed in couplets (one RGC preferring upward motion and the other downward20), no retrogradely labeled RGC couplets were ever observed in Pcdh9-Cre;Tbx5flox/flox retinas (Figures 3G, 3H, and 3J).
To further assess the fidelity of Spig1-GFP as a marker of up-oDSGCs in Tbx5 mutant mice, RGC axon central projections were examined after intraocular CTB-A555 injections. In P12 Spig1-GFP;Pcdh9-Cre;Tbx5flox/flox animals, GFP+ axons were absent from the dorsal MTN, the target of up-oDSGCs, but present in the ventral MTN, the target of down-oDSGCs20 (Figures 3K and 3L). The presence of few Spig1-GFP+ axons in the ventral MTN likely arises from non-specific GFP expression in down-oDSGCs from the dorsotemporal retina, as described previously.20 CTB labeling of all RGC axons showed hypoinnervation of the dorsal MTN, with preserved innervation of the ventral MTN (Figures 3M and 3N). Occasional mistargeted CTB+GFP− axons were observed in the vicinity of the dorsal MTN (Figure 3N, arrows). This axon targeting defect was specific to the MTN; other retinorecipient targets, such as the dorsal terminal nucleus (DTN) and the nucleus of the optic tract (NOT), which mediate the horizontal OKR,17 and the SC showed normal innervation (Figures S3M-S3P).
To confirm that Tbx5 regulates the initial formation of up-oDSGCs rather than up-oDSGC survival and maintenance, we examined brains from Npy-Cre;Tbx5flox/flox mice. Our scRNAseq data set showed that Npy is expressed in both up- and down-oDSGCs after P4 (Figure S3Q). Intraocular injections of AAV2 FLEX-GFP in P3 Npy-Cre mice revealed RGC axon projections to both the dorsal and ventral MTN, as well as the DTN and NOT (responsible for horizontal image stabilization), at P18 (Figures S3X-S3Z). Few, if any, axon projections were observed in other retinorecipient targets such as the suprachiasmatic nucleus, lateral geniculate nucleus, and SC (Figures S3AA-S3AC). These data indicate that, beyond P17, Npy-Cre labels all AOS-projecting DSGCs. Surprisingly, adult Npy-Cre;Tbx5flox/flox mice showed normal MTN innervation, suggesting that both up- and down-oDSGCs were still present (Figures 3O-3P). Npy-Cre;Tbx5flox/flox mice also showed normal OKR performance (data not shown). Npy-Cre expression, however, is not present in up-oDSGCs until well after their initial formation since retinas from P0 Npy-Cre;Spig1-GFP;LSL-tdTomato mice showed mutual exclusivity between Spig1-GFP+ up-oDSGCs and Npy-Cre+tdTomato+ RGCs (Figures S3R-S3T), though Spig1-GFP+ up-oDSGCs became tdTomato+ by P8 (Figures S3U-S3W). These data indicate that Npy-Cre expression in up-oDSGCs does not begin until after the P1-P4 time window. The use of Npy-Cre;Tbx5flox/flox mice thus represents a way to remove Tbx5 from up-oDSGCs postnatally. The presence of normal MTN innervation and normal OKR behavior in Npy-Cre;Tbx5flox/flox mice suggests that Tbx5 is not required for the survival or maintenance of up-oDSGCs. Taken together, these results show that Tbx5 is necessary for the initial formation of up-oDSGCs, but not their subsequent maintenance or survival.
Sfrp1 is a downstream effector of Tbx5 in the AOS
What does Tbx5, a transcription factor, regulate that affects the development and function of oDSGCs? To begin to provide insight into this question, we cross-referenced our scRNAseq data with bulk RNAseq data from Tbx5 wild-type and null E9.5 mouse heart tissue.40 We examined the overlap between genes differentially expressed between Tbx5+ up-oDSGCs and Tbx5− down-oDSGCs, and genes differentially expressed between E9.5 Tbx5+/+ and Tbx5−/− heart tissue; any genes common to both data sets were considered potential downstream effectors of Tbx5. Figure 4A displays, in rank order, the genes potentially upregulated and downregulated by Tbx5 in oDSGCs, based on genes regulated by Tbx5 in heart tissue.
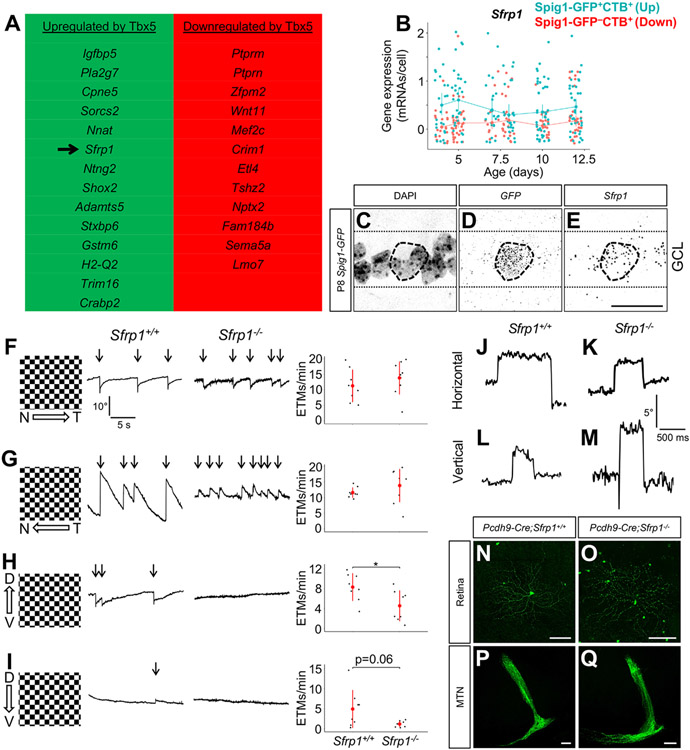
Figure 4. Sfrp1 is a potential downstream effector of Tbx5.
(A) Cross-referencing bulk RNAseq data from Tbx5+/+ and Tbx5−/− cardiac tissue40 with our oDSGC single-cell RNAseq data yielded a list of genes potentially upregulated or downregulated by Tbx5 (rank-ordered by q-value). (B) Sfrp1 was expressed at slightly higher levels in up-oDSGCs compared to down-oDSGCs between P4 and P12. Data presented as mean ± 95% confidence intervals. (C-E) RNAscope in situ hybridization in P8 Spig1-GFP retinas shows Sfrp1 expression in GFP+ up-oDSGCs within the ganglion cell layer (GCL). (F-I) Optokinetic reflex (OKR) measurements in adult Sfrp1+/+ and Sfrpt−/− mice showed comparable performance in response to nasotemporal (F) and temporonasal (G) motion. Sfrpt−/− mice showed impaired OKR performance in response to ventrodorsal motion (H) and a trend to statistically significant impairment in response to dorsoventral motion (I). Data presented as mean ± SD. (J-M) Voluntary horizontal (J-K) and vertical (L-M) saccades were intact in Sfrp1−/− mice. (N-Q) Adult up-oDSGCs, labeled with intraocular AAV2 FLEX-GFP in Pcdh9-Cre;Sfrp1+/+ and Pcdh9-Cre;Sfrp1−/− mice, appeared grossly normal with regards to dendrite morphology (N-O) and axon projections to the MTN (P-Q). Scale bars, 25 um in (C-E); 100 um in (N-Q). *p < 0.05. ETM, eye-tracking movement. See also Figure S4.
One of these genes, Sfrp1, is notable because it is a member of the Wnt/Frizzled signaling pathway, which controls tissue polarity and asymmetry in many contexts.41 Sfrp1 encodes a secreted protein that binds to Wnt proteins in the extracellular space, thereby inhibiting Wnt signaling.42,43 In heart tissue and cardiomyocyte-differentiating induced pluripotent stem cells, Tbx5 positively regulates Sfrp1 at the level of RNA expression.40,44 ChIP-seq experiments performed in cultured cardiomyocytes show Tbx5 binding sites within and around the Sfrp1 locus,45 lending further support to a Tbx5-Sfrp1 transcriptional regulatory network. Therefore, we investigated the role of Sfrp1 in vertical oDSGC AOS circuit function. Between P4-P12, Sfrp1 expression was significantly higher in up-oDSGCs than down-oDSGCs (q=2.6 x 10−6; Figure 4B). In situ hybridization of P8 Spig1-GFP retinal tissue confirmed Sfrp1 expression in GFP+ up-oDSGCs (Figures 4C-4E). We examined the OKR in Sfrp1−/− mice46 and observed a selective defect in response to upward motion (p<0.05), a possible mild defect in response to downward motion (p=0.06), and normal OKR performance in response to horizontal motion (Figures 4F-4I). In wild-type mice, the downward OKR response is weak at baseline,47 which may explain the borderline downward defect seen in Sfrp1−/− mice. Voluntary vertical and horizontal saccades were intact in Sfrp1−/− mice (Figures 4J-4M), as was depth perception (Figure S4A) and performance on the looming task (Figure S4B). Intraocular injections of AAV2 FLEX-GFP in Pcdh9-Cre;Sfrp1−/− mice showed no gross defects in up-oDSGC morphology (Figures 4N and 4O) and intact MTN innervation (Figures 4P and 4Q). Overall retinal architecture was intact in adult Sfrp1−/− mice: ChAT labeling showed normal SAC dendrite stratification in the ON and OFF IPL sublaminae and calbindin labeling showed normal IPL lamination patterns (Figures S4C-S4H). Taken together, these results show that Sfrp1, a known downstream target of Tbx5 expressed in up-oDSGCs, is required for the normal vertical OKR response.
DISCUSSION
In this study, we sought to determine the molecular mechanisms that direct the development of AOS image stabilization circuits, focusing on the initial specification of upward-preferring oDSGCs. Using single-cell RNAseq, we transcriptionally profiled up- and down-oDSGCs at various postnatal developmental time points and showed that these oDSGCs have distinct transcriptional profiles, even before the acquisition of direction selectivity at P7-P8.22,23 Comparing the transcriptional signatures of up- and down-oDSGCs revealed that the transcription factor Tbx5 is selectively expressed by up-oDSGCs. Deleting Tbx5 in up-oDSGCs caused a selective defect in the initial formation of up-oDSGCs, with resulting aberrant innervation of the MTN and a behavioral defect in vertical image stabilization, as measured by the OKR. We also observe an interesting connection between Tbx5 function in cardiac development and the AOS, since a downstream target of Tbx5 in the developing heart, Sfrp1, is expressed in up-oDSGCs and is also necessary for a normal vertical OKR response. These observations provide insight into the molecular mechanisms needed to generate a specific and rare RGC subtype and demonstrate how interfering with the initial differentiation of this RGC subtype leads to profound and specific visual-behavioral deficits.
Over the past several years, droplet-based scRNAseq approaches have provided invaluable insight into cell type diversity within the retina.1,2,26,48 These high-sample-size, low-depth droplet-based methods allow for an unbiased survey of cell types, including types that were not previously known to exist.26 Our data show how high-depth, low-sample-size scRNAseq approaches can complement droplet-based approaches to distinguish transcriptional differences between closely-related cell types. Up- and down-oDSGCs are so transcriptionally similar that droplet-based scRNAseq sequencing methods, even using upwards of 35,000 RGCs, are not able to distinguish them.1,2 These results suggest that the shallower read depth characteristic of 3’ end-tagging, droplet-based scRNAseq may not sufficiently capture subtle differences in gene expression between cell types as similar as up- and down-oDSGCs. Taken together, these results suggest that the current estimate for the number of mouse RGC types – 401 to 462 – is likely an underestimate.
The mutual exclusivity between up- and down-oDSGC clusters (Figure 1D) informs our understanding of DSGC fate choice. The absence of cluster overlap, even at early developmental time points prior to the acquisition of DS, indicates that up- and down-oDSGCs are fated to prefer upward, or downward, motion early in their development, well before they form synapses with SACs. Consistent with bromodeoxyuridine birthdating data,49 the developmental events that determine DSGC directional preference appear to be transcriptionally determined early in development, with minimal SAC contribution. During DS acquisition at P7-P8, DSGCs and SACs sculpt their synapses to become congruent with the DSGC’s pre-established gene expression program.
Analyzing gene expression across development informs our understanding of models for how DSGCs acquire a directional preference. One idea is that DSGCs express (or repress) a gene at P7-P8 that induces the formation of asymmetric SAC input. This model, however, is not supported by our data since we did not observe any genes with a sudden, non-linear onset or offset of expression at the P7-P8 junction. Though unlikely, it is possible that such a gene exists but was not captured by our scRNAseq methodology. Based on our results, it is unlikely that the onset of DS at P7-P8 arises from a transcriptional change intrinsic to the DSGC. Other models, however, remain possible. The DSGC could, for instance, express a DS-instructive molecule both before and after DS onset, with DS acquisition being dictated by a post-transcriptional change in the DSGC or by altered expression of a molecule in SACs. Although the precise molecular events that trigger DS onset at P7-P8 remain a mystery, future analyses of differentially expressed genes in up- and down-oDSGCs may shed light on this issue.
In Pcdh9-Cre;Tbx5flox/flox mice, the downward component of the OKR was abolished despite the presence of intact down-oDSGCs. This result was unexpected and implies that both up- and down-oDSGCs are necessary for a normal downward OKR. The vertical OKR, however, is a complex behavior that involves both eyes: during a rotational stimulus, presentation of downward motion – and activation of down-oDSGCs – in one eye occurs simultaneously with presentation of upward motion – and activation of up-oDSGCs – in the other eye. Signals from both eyes are integrated in the cerebellar flocculus, which then initiates optokinetic nystagmus via the oculomotor, trochlear, and abducens nuclei.16 Given the binocular integration that occurs in the cerebellum, the necessity of both up- and down-oDSGCs for a normal downward OKR may not be surprising. Down-oDSGCs are present in Pcdh9-Cre;Tbx5flox/flox mice, and future work will determine whether down-oDSGCs do, in fact, show normal DS responses in Pcdh9-Cre;Tbx5flox/flox mice.
Although Pcdh9-Cre;Tbx5flox/flox mice showed reduced OKR gain in response to vertical stimuli, they showed enhanced OKR gain in response to horizontal stimuli. The reason for this enhancement in horizontal OKR gain may relate to reciprocal GABAergic inhibitory connections between the MTN, responsible for the vertical OKR, and the NOT, responsible for the horizontal OKR.50,51 With up-oDSGCs absent in Pcdh9-Cre;Tbx5flox/flox mice, the MTN may no longer be able to inhibit the NOT, resulting in a compensatory increase in NOT activity and horizontal OKR gain. Altered MTN-NOT connectivity may also explain the increased horizontal gain in response to vertical stimuli, and increased vertical gain in response to horizontal stimuli, seen in Pcdh9-Cre;Tbx5flox/flox animals, a phenomenon termed cross-coupling.37,38,52 Since up-oDSGCs are absent, the ability of the MTN to inhibit the NOT may be altered. oDSGCs preferring horizontal motion, though not tuned to vertical motion, still respond mildly to vertical stimuli, resulting in NOT activation.17 In wild-type animals, this NOT activation is suppressed by strong inhibition from the MTN. This MTN inhibition, however, may not occur in Pcdh9-Cre;Tbx5flox/flox mice, resulting in horizontal eye motion following vertical stimuli. Increased vertical gain in response to horizontal stimuli, meanwhile, may be attributed to residual functionality in the surviving down-oDSGCs. Down-oDSGCs, though not tuned to horizontal motion, may still respond slightly to horizontal stimuli, leading to increased vertical gain following horizontal stimuli. This “unmasking” of the horizontal, or vertical, OKR in response to motion in the orthogonal direction may also arise from dysfunction in OKR integration centers downstream of the NOT and MTN, such as the inferior olive and cerebellar flocculus.16 Future studies should assess functionality in these downstream areas to pinpoint the region responsible for this cross-coupling OKR phenomenon.
Prior studies have indicated a role for Tbx5 in dorsal-ventral patterning in the retina,31,32,53 raising the question of whether there exists a relationship between Tbx5’s role in dorsal-ventral patterning and its role in regulating up-oDSGC development. We suspect that Tbx5 has, at least, two roles in the retina that occur at different developmental times involving different spatial domains: 1) dorsal-ventral patterning early in embryonic development before RGCs are born (e.g., E10.5), with high Tbx5 expression in the dorsal retina;31,53 and 2) up-oDSGC specification later in development (e.g., E13.5), both in the dorsal and ventral retina. The absence of up-oDGSCs in both the dorsal and ventral retina in Spig1-GFP;Pcdh9-Cre;Tbx5fl/fl mice indicates that Tbx5’s role in up-oDSGC specification is not restricted to a particular spatial domain but, rather, generalizes to both the dorsal and ventral retina. Future studies will examine whether Tbx5’s role in dorsal-ventral patterning and its role in up-oDSGC development are truly independent.
The behavioral and histologic phenotypes seen in Pcdh9-Cre;Tbx5fl/fl mice resemble those previously seen in Sema6a−/− mice,54 raising the question of whether Tbx5 regulates Sema6a expression. Currently, we do not have any evidence for a direct regulatory relationship between Tbx5 and Sema6a, and several lines of evidence argue that the similar effects of Tbx5 and Sema6a deletion on up-oDSGC biology and OKR behavior are independent. Our single-cell RNAseq data shows that both up- and down-oDSGCs express Sema6a at comparable levels (there is no significant difference in Sema6a expression between up- and down-oDSGCs; data not shown). However, only up-oDSGCs express Tbx5. This dissociation between Tbx5 and Sema6a expression makes it unlikely that Tbx5 regulates Sema6a. Bulk RNAseq data from Tbx5+/+ and Tbx5−/− heart tissue also shows no regulatory relationship between Tbx5 and Sema6a.40 The Tbx5 and Sema6a deletion phenotypes also differ: in Pcdh9-Cre;Tbx5fl/fl mice, up-oDSGCs do not form initially, whereas in Sema6a−/− mice, up-oDSGCs initially develop but then subsequently die over the course of development after failing to establish connections with their targets in the MTN.54 Additionally, both dorsal and ventral MTN innervation by up-oDSGCs and down-oDSGCs, respectively, is compromised in Sema6a mutants, further suggesting that Sema6a expression is not under direct control of Tbx5. Taken together, these data indicate that Tbx5 likely does not regulate Sema6a.
Cross-referencing our scRNAseq data with existing RNAseq data from Tbx5+/+ and Tbx5−/− heart tissue40 revealed a set of potential effector genes downstream of Tbx5. One of these potential downstream effectors, Sfrp1, was expressed by up-oDSGCs and was required for a normal upward OKR response. How, on a cellular and molecular level, Sfrp1 regulates the upward OKR is unclear. Since Sfrp1 is a secreted protein, it may be functioning in a cell non-autonomous manner, affecting either down-oDSGCs or other cells in the DS circuit. Indeed, Sfrp1 is unlikely to be exerting its effects on up-oDSGCs given the normal dendrite morphology and axonal MTN projections seen in Pcdh9-Cre;Sfrp1−/− mice. Future studies will investigate how Sfrp1 and other potential downstream Tbx5 effectors regulate the OKR and DSGC biology.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alex L. Kolodkin (kolodkin@jhmi.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Github and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | AVES |
Cat # GFP-1020;
RRID: AB_10000240 |
| Goat polyclonal anti-choline acetyltransferase | Millipore |
Cat # AB144P;
RRID: AB_11214092 |
| Rabbit polyclonal anti-DsRed | Living Colors |
Cat # 632496;
RRID: AB_10013483 |
| Guinea pig polyclonal anti-RBPMS | PhosphoSolutions |
Cat# 1832-RBPMS;
RRID: AB_2492226 |
| Rabbit polyclonal anti-calbindin D-28k | Swant |
Cat # CB38;
RRID: AB_10000340 |
| Brilliant Violet 421 rat anti-CD11b | BioLegend |
Cat # 101251;
RRID:AB_2562904 |
| Bacterial and virus strains | ||
| AAV9 FLEX-tdTomato (6.8E+12) | UNC Vector Core | N/A |
| AAV9 FLEX-GFP (3.7E+12) | UNC Vector Core | N/A |
| AAV2 FLEX-GFP (3.7E+12) | UNC Vector Core | N/A |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Papain | Worthington | Cat # LS003126 |
| DAPI (4’, 6-Diamidino-2-Phenylindole, Dihydrochloride) | Life Technologies |
Cat # D1306;
RRID: AB_2629482 |
| Cholera Toxin Subunit B (Recombinant), Alexa Fluor 555 Conjugate | Thermo Fisher | Cat # C34776 |
| Cholera Toxin Subunit B (Recombinant), Alexa Fluor 647 Conjugate | Thermo Fisher | Cat # C34778 |
| Propidium iodide | Thermo Fisher | Cat # P1304MP |
| Critical commercial assays | ||
| Smart-seq2 reagents | Picelli et al. 50 | N/A |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat # FC-131-1024 |
| RNAscope Fluorescent Multiplex Kit | Advanced Cell Diagnostics | Cat # 320850 |
| RNAscope Probe – Mm-Sfrp1 | Advanced Cell Diagnostics | Cat # 404981 |
| RNAscope Probe – eGFP | Advanced Cell Diagnostics | Cat # 400281 |
| RNAscope Probe – GFP-C2 | Advanced Cell Diagnostics | Cat # 409011-C2 |
| RNAscope Probe – Mm-Tbx5-C2 | Advanced Cell Diagnostics | Cat # 519581-C2 |
| RNAscope Probe – Mm-Chat-C3 | Advanced Cell Diagnostics | Cat # 410071-C3 |
| RNAscope Probe – Mm-Fibcd1-C3 | Advanced Cell Diagnostics | Cat # 524021-C3 |
| RNAscope 3-plex Negative Control Probe | Advanced Cell Diagnostics | Cat # 320871 |
| RNAscope 3-plex Positive Control Probe Mm | Advanced Cell Diagnostics | Cat # 320881 |
| Reagent or resource | ||
| Superscript III First-Strand Synthesis SuperMix | Thermo Fisher | Cat # 18080400 |
| Deposited data | ||
| Raw data files for scRNA-sequencing | This study | |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Mouse: Spig1-GFP | Yonehara et al. 19 | N/A |
| Mouse: Tg(Pcdh9-cre)NP276Gsat/Mmucd | MMRRC |
MMRRC Stock # 036084-UCD;
RRID: MMRRC_036084-UCD |
| Mouse: Tbx5 flox/+ |
Gift from B.
Bruneau |
N/A |
| Mouse: B6.Cg-Npy tm1(cre)Zman /J | Jackson Laboratory |
Jackson Laboratory Stock # 027851;
RRID: IMSR_JAX:027851 |
| Mouse: B6.Cg-Gt(ROSA)26Sor tm14(CAG-tdTomato)Hze /J | Jackson Laboratory |
Jackson Laboratory Stock # 007914;
RRID: IMSR_JAX:007914 |
| Mouse: Sfrp1 +/− | Gift from J. Nathans | N/A |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and algorithms | ||
| Tophat | Trapnell et al. 53 | RRID: SCR_013035 |
| Cufflinks | Trapnell et al. 53 |
http://cole-traonell-lab.github.io/cufflinks/; RRID: SCR_014597 |
| Cuffdiff | Trapnell et al. 53 |
http://cole-traonell-lab.github.io/cufflinks/; RRID: SCR_001647 |
| R version 3.5.1 | The R project | https://www.r-project.org/ |
| ImageJ | NIH |
https://imagej.nih.gov/ij/; RRID: SCR_003070 |
| Igor Pro version 6.37 | WaveMetrics | RRID: SCR_000325 |
| MATLAB_R2012a version 7.14.0.739 | MathWorks | RRID: SCR_001622 |
| LabVIEW 2012 version 12.0f3 |
National
Instruments |
RRID: SCR_014325 |
| Other | ||
| Illumina Nextseq 500 | Illumina | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at The Johns Hopkins University School of Medicine. The day of birth was designated as postnatal day 0 (P0). Mice of either sex ranging in age from P3 to P112 were used. Spig1-GFP mice were a gift from M. Noda and were described previously.20,21 Pcdh9-Cre mice were obtained from the Mutant Mouse Resource and Research Center (MMRRC). Tbx5flox/+ mice were a gift from B. Bruneau and were described previously.55 Npy-Cre mice (Stock #027851) were obtained from the Jackson Laboratory. Sfrp1−/− mice were a gift from J. Nathans and were described previously.46 All mice were maintained on a C57BL/6J background. Animals were housed in a 12-hour light-dark cycle. Behavioral tests were performed at consistent hours during the light cycle.
METHOD DETAILS
Stereotactic surgery and MTN injections
P3 Spig1-GFP mice were anesthetized using 2% isoflurane and placed into a stereotactic apparatus. Alexa 555- or Alexa 647-conjugated cholera toxin B (CTB) (1 mg/ml, ThermoFisher Scientific) was injected into the medial terminal nucleus (200 nl at 10 nl/s) using a Hamilton Neuros syringe coupled to a computerized microsyringe pump controller (World Precision Instruments). Five minutes elapsed between the injection and removal of the needle to allow diffusion of CTB around the injection site. The MTN coordinates at P3 are as follows: anterior/posterior −1.534 mm, medial/lateral ±0.91 mm, dorsal/ventral −4.916 mm, needle tilted 52° anteriorly (coordinates are relative to the intersection of the superior sagittal sinus and the inferior cerebral vein).
Intraocular AAV Injections
Mice were anaesthetized with 2% isoflurane. A hole was made at the corneal limbus with a 30 G needle and 2 ul of AAV was injected intravitreally using a Hamilton syringe.
The following viruses were used in this study: AAV9 CAG-FLEX-tdTomato (6.8 × 1012 GC/ml; UNC Vector Core), AAV9 CAG-FLEX-GFP (3.7 × 1012GC/ml; UNC Vector Core), AAV2 CAG-FLEX-GFP (3.7 × 1012 GC/ml; UNC Vector Core).
Single-cell RNA sequencing cell Isolation
Following stereotactic injections of CTB-Alexa 647 into the MTN at P3, Spig1-GFP mice were sacrificed at P4, P5, P7, P8, P10, and P12. Retinas were dissected in cold Hibernate A medium (BrainBits) lacking Ca2+, Mg2+, and phenol red. Successful MTN injection was confirmed by the presence of GFP+ CTB-Alexa 647+ and GFP− CTB-Alexa 647+ retinal cells arranged in pairs under a fluorescence dissecting microscope. The dorsotemporal quadrant of each retina, which expresses GFP non-specifically, was dissected under fluorescent guidance and removed.19 Retinas were dissociated in papain (Worthington, LS003126), 1.1 mM EDTA, and 5.5 mM Cysteine-HCl dissolved in Hibernate A (no Ca2+, Mg2+, or phenol red) for 30 min at 37°C. Cells were then incubated with Brilliant Violet 421-conjugated Anti-CD11b antibody (1:300; BioLegend, Catalog No. 101251) for 30 min at 4 C to label CD11b+ microglia, which can be CTB-Alexa 647+. Cells were incubated with propidium iodide to label dead cells.
Single GFP+ CTB-Alexa 647+ CD11b− and GFP− CTB-Alexa 647+ CD11b− cells were sorted into individual tubes containing 3500 U/ml RNase inhibitor (New England Biolabs), 140 U/ml DNase (New England Biolabs), and 0.17% v/v Triton X-100 in water using a MoFlo Legacy cell sorter (Beckman Coulter, Miami, FL). Single-cell DNA libraries were prepared according to the Smart-seq2 protocol56 and sequenced using an Illumina NextSeq 500 sequencer (75 bp paired-end reads, 400 million total reads).
Immunohistochemistry
For whole-mount retina staining, mice were transcardially perfused with phosphate buffered saline (PBS) followed by 4% paraformaldehyde (PFA). Enucleated eyeballs were fixed in 4% PFA for 1 hour at 4°C, then washed 3 times with PBS to remove residual PFA. Retinas were dissected and incubated for 2 days at room temperature with primary antibodies in PBS containing 0.5% Triton X-100, 3% donkey (or goat) serum, and 0.05% sodium azide. Retinas were washed 4 times for 1 hour at room temperature with PBS and 0.1% Triton X-100, then incubated for 2 days at 4°C with secondary antibodies in PBS containing 0.1% Triton X-100 and 3% donkey (or goat) serum. Retinas were washed 4 times for 1 hour at room temperature with PBS and 0.1% Triton X-100, then mounted and imaged with a Zeiss LSM 700 confocal microscope.
For cross-sectional retina staining, mice were transcardially perfused with PBS followed by 4% PFA. Enucleated eyeballs were fixed in 4% PFA for 1 hour at 4°C, then washed 3 times with PBS to remove residual PFA. A hole was made in the cornea and the eyeballs were cryopreserved in PBS containing 30% (w/v) sucrose overnight at 4°C. Eyeballs were frozen in Neg-50 frozen section medium (Richard-Allen Scientific, Kalamazoo, MI) and sliced using a cryostat at a thickness of 25 um. Retinal sections were blocked in PBS containing 5% (v/v) fetal bovine serum (FBS) and 0.4% Triton X-100 at room temperature for 1 hour, then incubated with primary antibodies in PBS containing 5% (v/v) FBS and 0.4% Triton X-100 overnight at 4°C. Sections were washed 6 times for 5 minutes at room temperature with PBS and 0.1% Triton X-100, then incubated with secondary antibodies in PBS containing 5% (v/v) fetal bovine serum (FBS) and 0.4% Triton X-100 at room temperature for 1 hour. Sections were washed 6 times for 5 minutes at room temperature with PBS and 0.1% Triton X-100, then mounted for imaging.
For cross-sectional brain staining, mice were transcardially perfused with PBS followed by 4% PFA. Brains were fixed in 4% PFA overnight at 4°C, then washed 3 times with PBS to remove residual PFA. Brains were embedded in PBS containing 4% (w/v) agarose and sliced using a vibratome at a thickness of 100 um. Brain sections were blocked in PBS containing 10% (v/v) donkey (or goat) serum and 0.1% Triton X-100 at room temperature for 1 hour, then incubated with primary antibodies in PBS containing 10% (v/v) donkey (or goat) serum and 0.1% Triton X-100 for 3 days at 4°C. Sections were washed 4 times for 1 hour at room temperature with PBS and 0.1% Triton X-100, then incubated with secondary antibodies in PBS containing 10% (v/v) donkey (or goat) serum and 0.1% Triton X-100 for 2 days at 4°C. Sections were washed 4 times for 1 hour at room temperature with PBS and 0.1% Triton X-100, then mounted for imaging.
Primary antibodies used in this study include: chicken anti-GFP (AVES, 1:1000), rabbit anti-DsRed (Living Colors, 1:1000), guinea pig anti-RBPMS (PhosphoSolutions, 1:500), goat anti-ChAT (Millipore, 1:200), rabbit anti-calbindin (Swant [CB38], 1:2000).
RNAscope in situ hybridization
Fluorescent in situ hybridization was performed on fresh frozen retinas from P8 Spig1-GFP mice. Animals were anesthetized using 2% isoflurane and decapitated. Enucleated eyeballs were embedded in Neg-50 frozen section medium (Richard-Allen Scientific, Kalamazoo, MI), frozen on dry ice, and sliced using a cryostat at a thickness of 14 um. In situ hybridization was performed using the RNAscope Fluorescent Multiplex system v1 (Advanced Cell Diagnostics, Newark, CA) according to the manufacturer’s instructions. Probes for GFP, Tbx5, Sfrp1, Chat, and Fibcd1 were obtained from the manufacturer (Advanced Cell Diagnostics, Newark, CA). Dotted lines were drawn around the collection of mRNA puncta for the gene of interest. In our experience, the mRNA puncta were often located outside of the DAPI+ nucleus, but were grouped in a circular shape that presumably corresponds to the cell soma. Punctate dots within cells indicated positive staining.
Headpost implantation surgery
Adult mice (≥6 weeks of age) were anesthetized using isoflurane. Four 1.00UNM × 0.120” stainless steel screws were placed into the skull on both sides of the sagittal suture between the coronal and lambdoidal sutures. The screws were incorporated into a pedestal of dental cement (Ortho-Jet, Lang Dental; Wheeling, Illinois, USA) and an acrylic headpost, composed of two M1.4 hex nuts embedded in dental cement (fabricated in-house), was placed on top of the pedestal. Mice were allowed to recover for ≥7 days before behavioral testing.
Optokinetic reflex recording
Mice with headposts were restrained in an animal holder (fabricated in-house) and were placed inside a square box, each side of which consisted of a computer monitor (12 in length × 20 in width) displaying a black-and-white checkerboard pattern (stripe width, 5° of visual angle). Mice were positioned so that their two eyes saw different pairs of monitors, allowing presentation of visual rotational stimuli around the anterior-posterior axis. A fixed infrared video camera inside the box recorded eye movements. Linear displacement of the pupil in the 2D video image (in mm) was transformed into rotational displacement (in degrees) using the video-oculography method described by Stahl et al.57 Two types of visual stimuli were used to elicit the optokinetic reflex: sinusoidal stimuli (temporal frequency, 0.1 or 0.2 Hz; sinusoid amplitude, 5° of visual angle; 10 cycles per trial) and continuously rotating stimuli (rotation speed, 5° of visual angle/s; each stimulus cycle consisted of 30 s of rotating checkerboard, followed by 30 seconds of grey screen; 10 cycles per trial). Stimuli were presented in both vertical and horizontal directions. Eye movement recording and stimulus presentation programs were built in-house using MATLAB R2012a v7.14.0.739 (MathWorks, Inc., Natick, MA) and LabVIEW v12.0f3 (National Instruments, Austin, TX) software.
Data were analyzed using Igor Pro v6.37 (WaveMetrics, Inc., Portland, OR). Saccadic eye movements were manually removed from sinusoidal stimulus data. Gain, defined as the angular velocity of the pupil relative to the angular velocity of the stimulus, was computed using a custom in-house script.16 Continuously rotating stimuli were analyzed manually in Igor Pro v6 by calculating the rate of “eye tracking movements” (ETMs) in each stimulus cycle, with 1 ETM defined as 1 slow phase plus 1 saccade.47
Visual cliff assay
The visual cliff apparatus58 consists of a table whose surface is composed of transparent plexiglass (16 in width × 16 in length × 24 in height). An opaque black-and-white checkerboard pattern covers half the table, then drops off vertically to the floor, then covers the floor beneath the transparent half, thereby creating the illusion of a checkerboard “cliff.” A camera was placed above the table to record behavior. A mouse was placed onto an opaque platform (1 in width × 3.5 in length × 1.5 in height) positioned in the center of the table. The mouse was allowed to choose a side (checkerboard or cliff) and its behavior was recorded. A mouse “chose” a side when all four paws touched the table. The test was repeated for a total of 12 trials. Every 4 trials, the table was rotated 90° to minimize the use of external visual cues for decision making. Overhead lighting intensity was identical on the checkerboard and cliff sides of the table for all trials. The slope of the table was measured to ensure that there was no incline between the checkerboard and cliff sides. On the first trial, once the mouse chose a side, the platform was removed and the mouse was allowed to explore the platform for 10 minutes during which its behavior was recorded.
Looming assay
The looming assay apparatus59 consists of a transparent plexiglass box (10 in width × 20 in length × 15 in height) with a computer monitor placed on top facing down. A plastic shelter, shaped as a triangular prism (4.5 in each side × 6.25 in long), was placed on one end of the box. A 4.5 in2 square was drawn in the middle of the box. A video camera with night vision capability was positioned to record mouse behavior. The experiment was performed in a dark room with light exclusively coming from the computer monitor on top of the box.
A mouse was dark-adapted for 1 hour and then placed in the box, where it was allowed to explore for 10 minutes. Following the 10 minute acclimation period, a looming stimulus was presented if the mouse contacted the square in the middle of the box. The looming stimulus consisted of an expanding black circle on a white background that appeared 15 times over the course of 10 s. Successful looming responses were defined as freezing or escaping to the shelter in response to the looming stimulus.
QUANTIFICATION AND STATISTICAL ANALYSIS
Graphs were generated using the ggplot2 package in R v3.5.1 (The R Foundation for Statistical Computing, Auckland, New Zealand). Student’s t-tests and Chi-squared tests were performed in R. Significance was defined as p < 0.05.
Single-cell RNA sequencing data analysis
Single-cell RNA sequencing FASTQ files were aligned to the GRCm38.p6 (mm10) mouse reference genome using HISAT2 v2.1.0.60 SAM files were converted to BAM files using SAMtools v1.3.1.61 Transcript abundance estimation and normalization were performed using the Cuffquant and Cuffnorm tools, respectively, in Cufflinks v2.2.1.62 Downstream analyses were performed using Monocle228 in R v3.5.1 (The R Foundation for Statistical Computing, Auckland, New Zealand).
Normalized FPKM values from Cuffnorm were converted to estimates of absolute mRNA counts per cell using the Monocle2 Census method.63 Cells with a log10 total mRNA count above or below 2 standard deviations of the mean log10 total mRNA count were excluded from downstream analyses.
Unsupervised clustering analysis was performed in Monocle2 using high-variance genes with a “mean_expression” value of >0.1. Principal component analysis was performed on these high-variance genes, and a subset of principal components (n = 6, by elbow plot) was used for tSNE visualization. When performing tSNE dimensionality reduction, the following variables were included as covariates: developmental age, block, plate, run, num_genes_expressed. Developmental age was included as a covariate because we aimed to determine the number of RGC clusters independent of developmental age. “Block”, “plate”, and “run” were included as covariates to regress out known batch effects from sample processing. The number of genes expressed in each cell (“num_genes_expressed”) was included as a proxy for the sensitivity of scRNAseq in each cell. Clustering was performed using the Monocle2 density peak clustering algorithm.
Semi-supervised clustering was performed in Monocle2 using Spig1 as a known marker gene of up-oDSGCs. The markerDiffTable function classified cells according to the presence of Spig1 expression and identified marker genes differentially expressed between Spig1-expressing and Spig1-nonexpressing cells. The specificity of these marker genes was determined by computing Jensen-Shannon distances.64 The top 500 most specific marker genes in Spig1-expressing and Spig1-nonexpressing cells were used as input for principal component analysis. A subset of principal components (n = 6, by elbow plot) was used for tSNE visualization. The following variables were included as covariates during tSNE dimensionality reduction: developmental age, block, plate, run, num_genes_expressed. Clustering was performed using the Monocle2 density peak clustering algorithm.
Differential gene expression testing between up- and down-oDSGCs was performed using the Monocle2 VGAM model comparison test.28 Up- and down-oDSGCs from P4 to P12 were included in differential gene expression testing. The following full model was fit to each expressed gene: ~cell_type + block + plate + run + num_genes_expressed, and compared to a reduced model in which “cell_type” was removed. Significantly differentially expressed genes were defined as those with a q-value <0.05.
To examine gene expression changes over time, a pseudotime analysis was performed using Monocle2. Up- and down-oDSGCs were analyzed separately. First, a differential gene expression test was performed to identify genes that change as a function of age. Only genes with a q-value of <0.01 were selected for downstream pseudotime analysis. These genes were used to order cells along a pseudotime trajectory from P4 to P12. We then performed a differential gene expression test to examine gene expression changes as a function of pseudotime. The following full model was fit to each expressed gene: ~sm.ns(Pseudotime) + block + plate + run + num_genes_expressed, and compared to a reduced model in which “sm.ns(Pseudotime)” was removed. Significantly differentially expressed genes were defined as those with a q-value <0.05.
Supplementary Material
HIGHLIGHTS.
Up- and down-oDSGCs have distinct transcriptional profiles
Tbx5 is expressed by, and required for, the development of up-oDSGCs
Loss of Tbx5 in up-oDSGCs causes a selective defect in vertical image stabilization
Sfrp1, a Tbx5downstream effector, is required for vertical image stabilization
ACKNOWLEDGMENTS
We are grateful to Masaharu Noda, Benoit Bruneau, and Jeremy Nathans for generously providing Spig1-GFP, Tbx5flox/flox, and Sfrp1−/− mice, respectively. We thank Nicole Kropkowski for technical assistance. We also thank Seth Blackshaw, Samer Hattar, Jeremy Nathans, and members of the Kolodkin laboratory for comments on the manuscript and helpful discussions. This work was supported by the NIH (R01EY032095 and R01EY027713; A.L.K.), and funding from the Howard Hughes Medical Institute (A.L.K. and T.A.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Rheaume BA, Jereen A, Bolisetty M, Sajid MS, Yang Y, Renna K, Sun L, Robson P, and Trakhtenberg EF (2018). Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun 9, 2759. 10.1038/S41467-018-05134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, Levin JZ, Lin D, Wang C, Lieber CM, Regev A, He Z, and Sanes JR (2019). Single-cell profiles of retinal ganglion cells differing in resilience to injury reveal neuroprotective genes. Neuron 104, 1039–1055.e12. 10.1016/j.neuron.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F, Li E, De L, Wu Q, and Zhang Y (2021). OFF-transient alpha RGCs mediate looming triggered innate defensive response. Curr. Biol 31, 2263–2273.e3. 10.1016/j.cub.2021.03.025. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen-Ba-Charvet KT, and Rebsam A (2020). Neurogenesis and specification of retinal ganglion cells. Int. J. Mol. Sci 21, 451. 10.3390/ijms21020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodie-Kommit J, Clark BS, Shi Q, Shiau F, Kim DW, Langel J, Sheely C, Ruzycki PA, Fries M, Javed A, Cayouette M, Schmidt T, Badea T, Glaser T, Zhao H, Singer J, Blackshaw S, and Hattar S (2021). Atoh7-independent specification of retinal ganglion cell identity. Sci. Adv 7, eabe4983. 10.1126/sciadv.abe4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown NL, Patel S, Brzezinski J, and Glaser T (2001). Math5 is required for retinal ganglion cell and optic nerve formation. Development 128, 2497–2508. 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, and Gan L (2001). Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 15, 24–29. 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badea TC, Cahill H, Ecker J, Hattar S, and Nathans J (2009). Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron 61, 852–864. 10.1016/j.neuron.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu F, Jiang H, and Xiang M (2008). A comprehensive negative regulatory program controlled by Brn3b to ensure ganglion cell specification from multipotential retinal precursors. J. Neurosci 28, 3392–3403. 10.1523/JNEUROSCI.0043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao CA, Li H, Zhang Z, Kiyama T, Panda S, Hattar S, Ribelayga CP, Mills SL, and Wang SW (2014). T-box transcription regulator Tbr2 is essential for the formation and maintenance of Opn4/melanopsin-expressing intrinsically photosensitive retinal ganglion cells. J. Neurosci 34, 13083–13095. 10.1523/JNEUROSCI.1027-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweeney NT, Tierney H, and Feldheim DA (2014). Tbr2 is required to generate a neural circuit mediating the pupillary light reflex. J. Neurosci 34, 5447–5453. 10.1523/JNEUROSCI.0035-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiyama T, Long Y, Chen CK, Whitaker CM, Shay A, Wu H, Badea TC, Mohsenin A, Parker-Thornburg J, Klein WH, Mills SL, Massey SC, and Mao CA (2019). Essential roles of Tbr1 in the formation and maintenance of the orientation-selective J-RGCs and a group of OFF-sustained RGCs in mouse. Cell Rep. 27, 900–915.e5. 10.1016/j.celrep.2019.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Reggiani J, Laboulaye MA, Pandey S, Chen B, Rubenstein J, Krishnaswamy A, and Sanes JR (2018). Tbr1 instructs laminar patterning of retinal ganglion cell dendrites. Nat. Neurosci 21, 659–670. 10.1038/s41593-018-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaney DI, Sivyer B, and Taylor WR (2012). Direction selectivity in the retina: symmetry and asymmetry in structure and function. Nat. Rev. Neurosci 13, 194–208. 10.1038/nrn3165. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton NR, Scasny AJ, and Kolodkin AL (2021). Development of the vertebrate retinal direction-selective circuit. Dev. Biol 477, 273–283. 10.1016/j.ydbio.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kodama T, and du Lac S (2016). Adaptive acceleration of visually evoked smooth eye movements in mice. J. Neurosci 36, 6836–6849. 10.1523/JNEUROSCI.0067-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhande OS, Estevez ME, Quattrochi LE, El-Danaf RN, Nguyen PL, Berson DM, and Huberman AD (2013). Genetic dissection of retinal inputs to brainstem nuclei controlling image stabilization. J. Neurosci 33, 17797–17813. 10.1523/JNEUROSCI.2778-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabbah S, Gemmer JA, Bhatia-Lin A, Manoff G, Castro G, Siegel JK, Jeffery N, and Berson DM (2017). A retinal code for motion along the gravitational and body axes. Nature 546, 492–497. 10.1038/nature22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson JI (1984). The accessory optic system. Annu. Rev. Neurosci 7, 13–41. 10.1146/annurev.ne.07.030184.000305. [DOI] [PubMed] [Google Scholar]
- 20.Yonehara K, Ishikane H, Sakuta H, Shintani T, Nakamura-Yonehara K, Kamiji NL, Usui S, and Noda M (2009). Identification of retinal ganglion cells and their projections involved in central transmission of information about upward and downward image motion. PloS One 4, e4320. 10.1371/journal.pone.0004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yonehara K, Shintani T, Suzuki R, Sakuta H, Takeuchi Y, Nakamura-Yonehara K, and Noda M (2008). Expression of SPIG1 reveals development of a retinal ganglion cell subtype projecting to the medial terminal nucleus in the mouse. PloS One 3, e1533. 10.1371/journal.pone.0001533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yonehara K, Balint K, Noda M, Nagel G, Bamberg E, and Roska B (2011). Spatially asymmetric reorganization of inhibition establishes a motion-sensitive circuit. Nature 469, 407–410. 10.1038/nature09711. [DOI] [PubMed] [Google Scholar]
- 23.Wei W, Hamby AM, Zhou K, and Feller MB (2011). Development of asymmetric inhibition underlying direction selectivity in the retina. Nature 469, 402–406. 10.1038/nature09600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, and Sandberg R (2013). Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098. 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 25.Chevée M, Robertson JJ, Cannon GH, Brown SP, and Goff LA (2018). Variation in activity state, axonal projection, and position define the transcriptional identity of individual neocortical projection neurons. Cell Rep. 22, 441–455. 10.1016/j.celrep.2017.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A, and Sanes JR (2016). Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323.e30. 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeon CJ, Strettoi E, and Masland RH (1998). The major cell populations of the mouse retina. J. Neurosci 18, 8936–8946. 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sulaiman FA, Nishimoto S, Murphy GR, Kucharska A, Butterfield NC, Newbury-Ecob R, and Logan MP (2016). Tbx5 buffers inherent left/right asymmetry ensuring symmetric forelimb formation. PLoS Genet. 12, e1006521. 10.1371/journal.pgen.1006521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, and Seidman CE (1999). Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol 211, 100–108. 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- 31.Hasegawa Y, Takata N, Okuda S, Kawada M, Eiraku M, and Sasai Y (2016). Emergence of dorsal-ventral polarity in ESC-derived retinal tissue. Development 143, 3895–3906. 10.1242/dev.134601. [DOI] [PubMed] [Google Scholar]
- 32.Koshiba-Takeuchi K, Takeuchi JK, Matsumoto K, Momose T, Uno K, Hoepker V, Ogura K, Takahashi N, Nakamura H, Yasuda K, and Ogura T (2000). Tbx5 and the retinotectum projection. Science 287, 134–137. 10.1126/science.287.5450.134. [DOI] [PubMed] [Google Scholar]
- 33.Martersteck EM, Hirokawa KE, Evarts M, Bernard A, Duan X, Li Y, Ng L, Oh SW, Ouellette B, Royall JJ, Stoecklin M, Wang Q, Zeng H, Sanes JR, and Harris JA (2017). Diverse central projection patterns of retinal ganglion cells. Cell Rep. 18, 2058–2072. 10.1016/j.celrep.2017.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lilley BN, Sabbah S, Hunyara JL, Gribble KD, Al-Khindi T, Xiong J, Wu Z, Berson DM, and Kolodkin AL (2019). Genetic access to neurons in the accessory optic system reveals a role for Sema6A in midbrain circuitry mediating motion perception. J. Comp. Neurol 527, 282–296. 10.1002/cne.24507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oyster CW, Takahashi E, and Collewijn H (1972). Direction-selective retinal ganglion cells and control of optokinetic nystagmus in the rabbit. Vision Res. 12, 183–193. 10.1016/0042-6989(72)90110-1. [DOI] [PubMed] [Google Scholar]
- 36.Sugita Y, Miura K, Araki F, Furukawa T, and Kawano K (2013). Contributions of retinal direction-selective ganglion cells to optokinetic responses in mice. Eur. J. Neurosci 38, 2823–2831. 10.1111/ejn.12284. [DOI] [PubMed] [Google Scholar]
- 37.Economides JR, Suh YW, Simmons JB, Adams DL, and Horton JC (2020). Vertical optokinetic stimulation induces diagonal eye movements in patients with idiopathic infantile nystagmus. Invest. Ophthalmol. Vis. Sci 61, 14. 10.1167/iovs.61.6.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clement G, and Berthoz A (1990). Cross-coupling between horizontal and vertical eye movements during optokinetic nystagmus and optokinetic after nystagmus elicited in microgravity. Acta Otolaryngol. 109, 179–187. 10.3109/00016489009107432. [DOI] [PubMed] [Google Scholar]
- 39.Garbutt S, Han Y, Kumar AN, Harwood M, Rahman R, and Leigh RJ (2003). Disorders of vertical optokinetic nystagmus in patients with ocular misalignment. Vision Res. 43, 347–357. 10.1016/s0042-6989(02)00387-5. [DOI] [PubMed] [Google Scholar]
- 40.Waldron L, Steimle JD, Greco TM, Gomez NC, Dorr KM, Kweon J, Temple B, Yang XH, Wilczewski CM, Davis IJ, Cristea IM, Moskowitz IP, and Conlon FL (2016). The cardiac TBX5 interactome reveals a chromatin remodeling network essential for cardiac septation. Dev. Cell 36, 262–275. 10.1016/j.devcel.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu CW, and Sokol SY (2016). Wnt proteins can direct planar cell polarity in vertebrate ectoderm. Elife 5, e16463. 10.7554/eLife.16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rattner A, Hsieh JC, Smallwood PM, Gilbert DJ, Copeland NG, Jenkins NA, and Nathans J (1997). A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc. Natl. Acad. Sci. U S A 94, 2859–2863. 10.1073/pnas.94.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Q, D'Amore PA, and Sokol SY (1998). Functional and biochemical interactions of Wnts with FrzA, a secreted Wnt antagonist. Development 125, 4767–4776. 10.1242/dev.125.23.4767. [DOI] [PubMed] [Google Scholar]
- 44.Kathiriya IS, Rao KS, Iacono G, Devine WP, Blair AP, Hota SK, Lai MH, Garay BI, Thomas R, Gong HZ, Wasson LK, Goyal P, Sukonnik T, Hu KM, Akgun GA, Bernard LD, Akerberg BN, Gu F, Li K, Speir ML, … Bruneau BG (2021). Modeling human TBX5 haploinsufficiency predicts regulatory networks for congenital heart disease. Dev. Cell 56, 292–309.e9. 10.1016/j.devcel.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He A, Kong SW, Ma Q, and Pu WT (2011). Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. U S A 108, 5632–5637. 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joesting MS, Cheever TR, Volzing KG, Yamaguchi TP, Wolf V, Naf D, Rubin JS, and Marker PC (2008). Secreted frizzled related protein 1 is a paracrine modulator of epithelial branching morphogenesis, proliferation, and secretory gene expression in the prostate. Dev. Biol 317, 161–173. 10.1016/j.ydbio.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yonehara K, Fiscella M, Drinnenberg A, Esposti F, Trenholm S, Krol J, Franke F, Scherf BG, Kusnyerik A, Müller J, Szabo A, Jüttner J, Cordoba F, Reddy AP, Németh J, Nagy ZZ, Munier F, Hierlemann A, and Roska B (2016). Congenital nystagmus gene FRMD7 is necessary for establishing a neuronal circuit asymmetry for direction selectivity. Neuron 89, 177–193. 10.1016/j.neuron.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shekhar K, Whitney IE, Butrus S, Peng YR, and Sanes JR (2022). Diversification of multipotential postmitotic mouse retinal ganglion cell precursors into discrete types. eLife 11, e73809. 10.7554/eLife.73809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De la Huerta I, Kim IJ, Voinescu PE, and Sanes JR (2012). Direction-selective retinal ganglion cells arise from molecularly specified multipotential progenitors. Proc. Natl. Acad. Sci. U S A 109, 17663–17668. 10.1073/pnas.1215806109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giolli RA, Blanks RH, and Lui F (2006). The accessory optic system: basic organization with an update on connectivity, neurochemistry, and function. Prog. Brain Res 151, 407–440. 10.1016/S0079-6123(05)51013-6. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt M, van der Togt C, Wahle P, and Hoffmann KP (1998). Characterization of a directional selective inhibitory input from the medial terminal nucleus to the pretectal nuclear complex in the rat. Eur. J. Neurosci 10, 1533–1543. 10.1046/j.1460-9568.1998.00161.x. [DOI] [PubMed] [Google Scholar]
- 52.Ghasia FF, Shaikh AG, Jacobs J, and Walker MF (2015). Cross-coupled eye movement supports neural origin of pattern strabismus. Invest. Ophthalmol. Vis. Sci 56, 2855–2866. 10.1167/iovs.15-16371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Behesti H, Holt JK, and Sowden JC (2006). The level of BMP4 signaling is critical for the regulation of distinct T-box gene expression domains and growth along the dorso-ventral axis of the optic cup. BMC Dev. Biol 6, 62. 10.1186/1471-213X-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun LO, Brady CM, Cahill H, Al-Khindi T, Sakuta H, Dhande OS, Noda M, Huberman AD, Nathans J, and Kolodkin AL (2015). Functional assembly of accessory optic system circuitry critical for compensatory eye movements. Neuron 86, 971–984. 10.1016/j.neuron.2015.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, and Seidman JG (2001). A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106, 709–721. 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 56.Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart- seq2. Nat. Protoc 9, 171–181. 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 57.Stahl JS, van Alphen AM, and De Zeeuw CI (2000). A comparison of video and magnetic search coil recordings of mouse eye movements. J. Neurosci. Methods 99, 101–110. 10.1016/s0165-0270(00)00218-1. [DOI] [PubMed] [Google Scholar]
- 58.Fox MW (1965). The visual cliff test for the study of visual depth perception in the mouse. Anim. Behav 13, 232–233. 10.1016/0003-3472(65)90040-0. [DOI] [PubMed] [Google Scholar]
- 59.Yilmaz M, and Meister M (2013). Rapid innate defensive responses of mice to looming visual stimuli. Curr. Biol 23, 2011–2015. 10.1016/j.cub.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578. 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C (2017). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315. 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, and Rinn JL (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Github and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | AVES |
Cat # GFP-1020;
RRID: AB_10000240 |
| Goat polyclonal anti-choline acetyltransferase | Millipore |
Cat # AB144P;
RRID: AB_11214092 |
| Rabbit polyclonal anti-DsRed | Living Colors |
Cat # 632496;
RRID: AB_10013483 |
| Guinea pig polyclonal anti-RBPMS | PhosphoSolutions |
Cat# 1832-RBPMS;
RRID: AB_2492226 |
| Rabbit polyclonal anti-calbindin D-28k | Swant |
Cat # CB38;
RRID: AB_10000340 |
| Brilliant Violet 421 rat anti-CD11b | BioLegend |
Cat # 101251;
RRID:AB_2562904 |
| Bacterial and virus strains | ||
| AAV9 FLEX-tdTomato (6.8E+12) | UNC Vector Core | N/A |
| AAV9 FLEX-GFP (3.7E+12) | UNC Vector Core | N/A |
| AAV2 FLEX-GFP (3.7E+12) | UNC Vector Core | N/A |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Papain | Worthington | Cat # LS003126 |
| DAPI (4’, 6-Diamidino-2-Phenylindole, Dihydrochloride) | Life Technologies |
Cat # D1306;
RRID: AB_2629482 |
| Cholera Toxin Subunit B (Recombinant), Alexa Fluor 555 Conjugate | Thermo Fisher | Cat # C34776 |
| Cholera Toxin Subunit B (Recombinant), Alexa Fluor 647 Conjugate | Thermo Fisher | Cat # C34778 |
| Propidium iodide | Thermo Fisher | Cat # P1304MP |
| Critical commercial assays | ||
| Smart-seq2 reagents | Picelli et al. 50 | N/A |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat # FC-131-1024 |
| RNAscope Fluorescent Multiplex Kit | Advanced Cell Diagnostics | Cat # 320850 |
| RNAscope Probe – Mm-Sfrp1 | Advanced Cell Diagnostics | Cat # 404981 |
| RNAscope Probe – eGFP | Advanced Cell Diagnostics | Cat # 400281 |
| RNAscope Probe – GFP-C2 | Advanced Cell Diagnostics | Cat # 409011-C2 |
| RNAscope Probe – Mm-Tbx5-C2 | Advanced Cell Diagnostics | Cat # 519581-C2 |
| RNAscope Probe – Mm-Chat-C3 | Advanced Cell Diagnostics | Cat # 410071-C3 |
| RNAscope Probe – Mm-Fibcd1-C3 | Advanced Cell Diagnostics | Cat # 524021-C3 |
| RNAscope 3-plex Negative Control Probe | Advanced Cell Diagnostics | Cat # 320871 |
| RNAscope 3-plex Positive Control Probe Mm | Advanced Cell Diagnostics | Cat # 320881 |
| Reagent or resource | ||
| Superscript III First-Strand Synthesis SuperMix | Thermo Fisher | Cat # 18080400 |
| Deposited data | ||
| Raw data files for scRNA-sequencing | This study | |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Mouse: Spig1-GFP | Yonehara et al. 19 | N/A |
| Mouse: Tg(Pcdh9-cre)NP276Gsat/Mmucd | MMRRC |
MMRRC Stock # 036084-UCD;
RRID: MMRRC_036084-UCD |
| Mouse: Tbx5 flox/+ |
Gift from B.
Bruneau |
N/A |
| Mouse: B6.Cg-Npy tm1(cre)Zman /J | Jackson Laboratory |
Jackson Laboratory Stock # 027851;
RRID: IMSR_JAX:027851 |
| Mouse: B6.Cg-Gt(ROSA)26Sor tm14(CAG-tdTomato)Hze /J | Jackson Laboratory |
Jackson Laboratory Stock # 007914;
RRID: IMSR_JAX:007914 |
| Mouse: Sfrp1 +/− | Gift from J. Nathans | N/A |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and algorithms | ||
| Tophat | Trapnell et al. 53 | RRID: SCR_013035 |
| Cufflinks | Trapnell et al. 53 |
http://cole-traonell-lab.github.io/cufflinks/; RRID: SCR_014597 |
| Cuffdiff | Trapnell et al. 53 |
http://cole-traonell-lab.github.io/cufflinks/; RRID: SCR_001647 |
| R version 3.5.1 | The R project | https://www.r-project.org/ |
| ImageJ | NIH |
https://imagej.nih.gov/ij/; RRID: SCR_003070 |
| Igor Pro version 6.37 | WaveMetrics | RRID: SCR_000325 |
| MATLAB_R2012a version 7.14.0.739 | MathWorks | RRID: SCR_001622 |
| LabVIEW 2012 version 12.0f3 |
National
Instruments |
RRID: SCR_014325 |
| Other | ||
| Illumina Nextseq 500 | Illumina | N/A |