Summary
The DNA sensor cyclic GMP-AMP synthase (cGAS) is important for antiviral and anti-tumor immunity. cGAS generates cyclic GMP-AMP (cGAMP), a diffusible cyclic dinucleotide that activates the antiviral response through the adaptor protein Stimulator of Interferon Genes (STING). cGAMP cannot passively cross cell membranes, but recent advances have established a role for extracellular cGAMP as an “immunotransmitter” that can be imported into cells. However, the mechanism by which cGAMP exits cells remains unknown. Here, we identifed ABCC1 as a direct, ATP-dependent cGAMP exporter in mouse and human cells. We show that ABCC1 overexpression enhanced cGAMP export and limited STING signaling, and that loss of ABCC1 reduced cGAMP export and potentiated STING signaling. We demonstrate that ABCC1 deficiency exacerbated cGAS-dependent autoimmunity in the Trex1−/− mouse model of Aicardi-Goutières syndrome. Thus, ABCC1-mediated cGAMP export is a key regulatory mechanism that limits cell intrinsic activation of STING and ameliorates STING-dependent autoimmune disease.
Keywords: ABCC1, MRP1, cGAS, cGAMP, STING, Aicardi-Goutières Syndrome, Interferons
eTOC:
cyclic GMP-AMP (cGAMP) is an “immunotransmitter” that can be imported into cells to activate antiviral responses, but how cGAMP exits cells is currently unknown. Maltbaek et al identify ABCC1 as an active cGAMP exporter that modulates STING-dependent immunity.
Introduction
The cGAS-STING DNA sensing pathway has emerged as a key innate immune response that is important for antiviral immunity (Goubau et al., 2013), contributes to autoimmune diseases (Crowl et al., 2017), and mediates aspects of anti-tumor immunity (Li and Chen, 2018). Cyclic GMP-AMP synthase (cGAS) is a nucleotidyltransferase that binds to double-stranded DNA and catalyzes the formation of cyclic GMP-AMP (cGAMP; Sun et al., 2013; Wu et al., 2013), a diffusible cyclic dinucleotide (CDN) that activates the adaptor protein stimulator of interferon genes (STING) on the endoplasmic reticulum (Ishikawa et al., 2009). Activated STING then serves as a platform for the inducible recruitment of TANK-binding kinase 1 (TBK1), which phosphorylates and activates the transcription factor interferon regulatory factor 3 (IRF3), leading to the induction of the type I interferon (IFN)-mediated antiviral response (Liu et al., 2015).
The formation of cGAMP by cGAS has its roots in ancient nucleotidyltransferases that regulate signaling and metabolism in prokaryotes (Kranzusch et al., 2015; Woodward et al., 2010). Like all RNA nucleotide-based second messengers that exist across all kingdoms of life (Nelson and Breaker, 2017), cGAMP is unable to passively enter or exit cells. The only known enzyme that degrades cGAMP in metazoans is an extracellular phosphodiesterase called ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1; Li et al., 2014), which hydrolyzes cGAMP into GMP and AMP (Kato et al., 2018). cGAMP can avoid ENPP1-mediated degradation via movement through gap junctions (Ablasser et al., 2013) and by packaging into enveloped viral particles (Bridgeman et al., 2015; Gentili et al., 2015). However, tumor cells have been shown to release soluble cGAMP (Marcus et al., 2018), and this pool of extracellular cGAMP is subject to potent degradation by ENPP1 (Carozza et al., 2020). Moreover, extracellular cGAMP can be imported into cells through at least three distinct transmembrane transporters to activate STING signaling (Cordova et al., 2021; Lahey et al., 2020; Luteijn et al., 2019; Ritchie et al., 2019; Zhou et al., 2020a; Zhou et al., 2020b).
Based on these recent insights, a paradigm in cGAMP biology is emerging in which extracellular cGAMP serves as an “immunotransmitter” that contributes to STING-dependent immune responses (Ritchie et al., 2022). Central to this paradigm is the notion that the cells that produce cGAMP and the cells that respond to cGAMP can be distinct. A fundamental question remains: how does cGAMP exit the cells that produce it? One possibility is that cGAMP is passively released from cells upon loss of cell membrane integrity that accompanies cell death (Marcus et al., 2018). Another possibility, suggested in a recent study, is that there are transport mechanisms that mediate cGAMP release from live cells (Carozza et al., 2020). However, the molecular mechanisms of cGAMP export remain unknown. Here, we identify the ATP-binding cassette transporter ABCC1, also known as multidrug resistance protein 1 (MRP1), as a transporter that mediates direct, ATP-dependent cGAMP export. We demonstrate that ABCC1-mediated cGAMP export limited cell intrinsic STING responses and restrained cGAS-dependent autoimmunity in vivo. These findings reveal a key molecular site of action for cGAMP export and establish a regulatory mechanism that controls STING signaling.
Results
cGAMP is exported from live cells
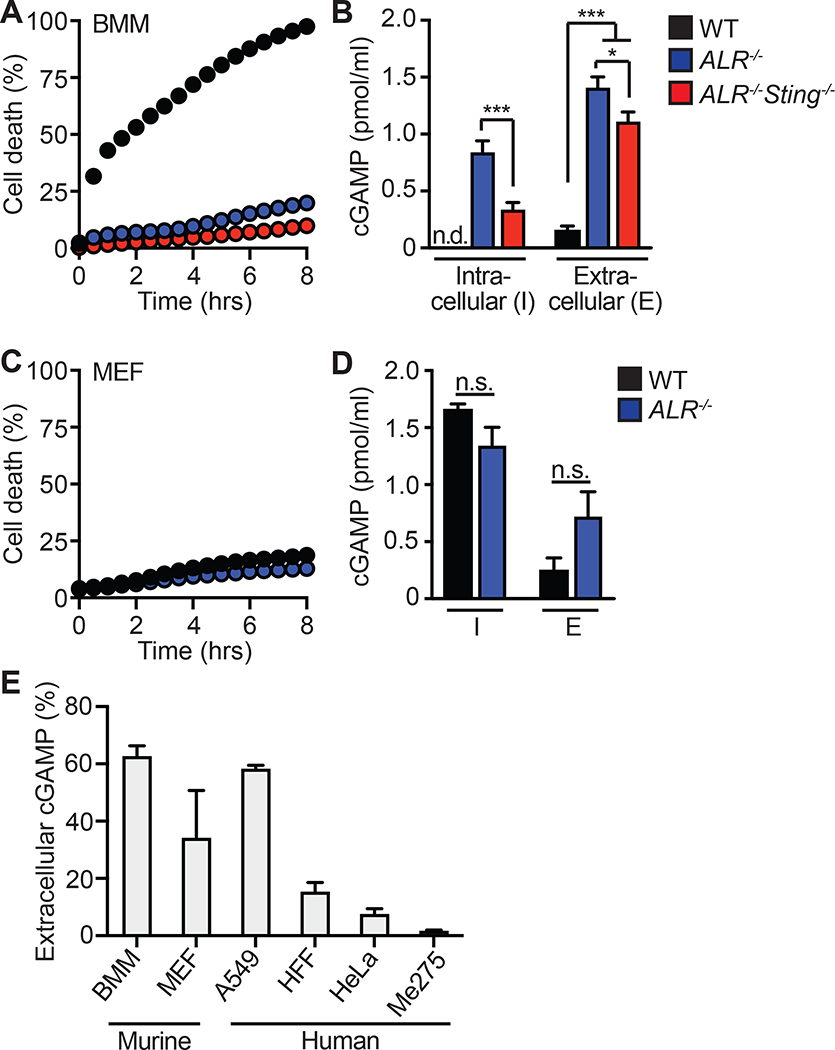
Intracellular DNA detection activates both the cGAS-cGAMP-STING pathway to trigger IFN-mediated antiviral responses and the AIM2 inflammasome to activate rapid, inflammatory cell death known as pyroptosis (Fink and Cookson, 2005; Hornung et al., 2009). This AIM2-dependent cell death pathway is particularly potent in myeloid cells, although it can function in non-myeloid cell types (Maelfait et al., 2020). To study cGAMP dynamics in the absence of the AIM2 inflammasome, we prepared primary bone marrow derived macrophages (BMMs) from mice lacking all 13 mouse AIM2-like receptors (ALRs; Gray et al., 2016). As previously reported, we found that DNA-activated rapid pyroptosis was almost completely abolished in ALR−/− BMMs (Figure 1A). We then measured intracellular and extracellular concentrations of cGAMP 8 hours after transfection with calf thymus (CT) DNA using a sensitive cGAMP ELISA (Burleigh et al., 2020; Volkman et al., 2019). We normalized the input volumes of cell extracts and extracellular media to allow direct comparison of absolute cGAMP amounts between these two compartments. We found that ALR−/− BMMs produced significantly more total cGAMP after DNA transfection than WT control BMMs, consistent the enhanced IFN response that has been observed in cells from AIM2- and ALR-deficient mice (Figure 1B) (Gray et al., 2016; Hornung et al., 2009). However, we recovered the majority of this cGAMP from the extracellular media, not from inside the cells (Figure 1B). We next tested whether STING signaling was required for the appearance of extracellular cGAMP. We found that ALR−/−Sting−/− BMMs exported similar amounts of cGAMP compared to ALR−/− BMMs (Figure 1B). The ALR−/−Sting−/− BMMs retained significantly less intracellular cGAMP than the ALR−/− BMMs (Figure 1B), perhaps reflecting the absence of STING protein, which is a high affinity cGAMP receptor. These data indicate that cGAMP export occurs independently of STING activation.
Figure 1. cGAMP is exported from live cells following cGAS activation.
(A) Cell death of BMMs from WT, ALR−/−, ALR−/−Sting−/− mice transfected with calf thymus DNA (CT DNA) quantified using an IncuCyte imaging system.
(B) cGAMP quantification using ELISA from cell lysates and supernatants in (A) 8 hours after transfection.
(C) Cell death of MEFs from WT or ALR−/− mice transfected with calf thymus DNA (CT-DNA) quantified using an IncuCyte imaging system.
(D) cGAMP quantification using ELISA from cell lysates and supernatants in (C) 8 hours after transfection.
(E) Percent extracellular cGAMP of indicated cell types 8 hours after CT DNA transfection. cGAMP ELISA was used to determine relative extracellular and intracellular concentrations before calculating the percent extracellular. Error bars represent mean ± SD of three biological replicates per group. *p<0.05, ***p<0.001. All data are shown are derived from a single experiment. Comparative results were obtained across three independent experiments. See also Figure S1.
We next studied primary murine embryonic fibroblasts (MEFs), which do not undergo ALR-dependent pyroptosis in response to DNA detection (Figure 1C) (Gray et al., 2016). We found that MEFs exported significant amounts of cGAMP in the absence of cell death (Figure 1D), but not as much as BMMs in terms of relative extracellular versus intracellular cGAMP amounts (Figure 1B). We extended these findings to immortalized human foreskin fibroblasts (HFF) and human tumor cell lines, including A549 lung adenocarcinoma cells (Giard et al., 1973), HeLa cervical carcinoma cells (Gey et al., 1952), and Me275 melanoma cells (Valmori et al., 1998). We verified that none of these cells expressed detectable Enpp1 mRNA or ENPP1 protein that could otherwise degrade extracellular cGAMP (Figure S1A, S1B). We found that the amount of extracellular cGAMP was low or nearly absent in HFF, HeLa, and Me275 cells, even though these cells all produced detectable intracellular cGAMP in response to DNA transfection (Figure S1C). A549 cells were the only human cell type tested that exported cGAMP with similar “efficiency” compared to mouse BMMs (Figure 1E). In all of these cell types, we found that passing the supernatants through a 10 kilodalton filter did not affect recovery of extracellular cGAMP (Figure S1D), suggesting that membrane-bound exosomes do not contribute to export of endogenous cGAMP from these cells (Bridgeman et al., 2015; Gentili et al., 2015). Thus, we have confirmed that live cells export soluble cGAMP (Carozza et al., 2020), and we have identified a nearly 60-fold range of export “efficiency” among diverse mouse and human cell types (Figure 1E).
The small molecule inhibitor MK-571 blocks cGAMP export
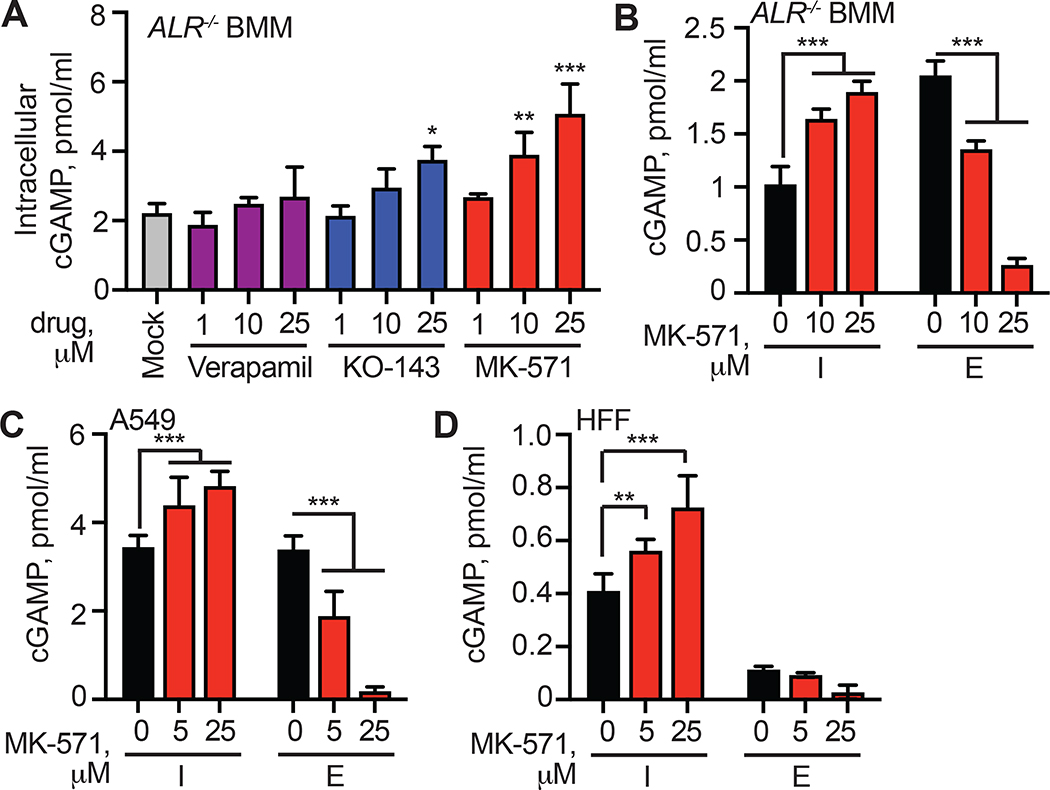
Having ruled out cell death and extracellular vesicles as potential mechanisms of cGAMP export, we focused on transmembrane transporters as potential cGAMP exporters. There are two broad classes of such proteins: 1) transporters that passively export substrates based on electrochemical gradients, and 2) transporters that use the energy of ATP hydrolysis to move substrates across membranes, sometimes against a concentration gradient. Influenced by the prior identification of energy-dependent transporters that mediate efflux of cyclic dinucleotides in bacteria (Woodward et al., 2010), we focused on active transport mechanisms that are accomplished in eukaryotes by the ATP-binding cassette (ABC) family of transmembrane proteins (Dean et al., 2001). ABC transporters are named because of a shared intracellular domain that binds ATP and translates the energy of ATP hydrolysis into conformational changes that move substrates from the cytosol to extracellular space or into membrane-bound intracellular compartments. The human genome encodes 49 ABC transporters that move a broad spectrum of small molecules with diverse masses and chemical properties (Vasiliou et al., 2009). Many ABC transporters have been scrutinized because of their ability to export chemotherapeutic drugs (Robey et al., 2018; Sun et al., 2012), which has resulted in the development of several pharmacological inhibitors of entire families of ABC proteins. We used three such inhibitors of distinct classes of ABC transporters and measured their effect on cGAMP export from ALR−/− BMMs: verapamil, which inhibits the ABCB1 transporter (Safa, 1988); KO-143, which blocks the ABCG2 transporter (Allen et al., 2002); and MK-571, a drug that has well characterized inhibitory activity against ABCC1 (Chee et al., 2018; Cole, 2014; Dean and Annilo, 2005; Gallman et al., 2021). We found that verapamil did not influence cGAMP export, but pretreatment with MK-571 resulted in a significant, dose-dependent increase in intracellular cGAMP after DNA transfection (Figure 2A), consistent with blockade of cGAMP export. The highest dose of KO-143 also significantly increased intracellular cGAMP, but it was previously reported that KO-143 has an inhibitory effect on the transport activity of both ABCB1 and ABCC1 at this concentration (Weidner et al., 2015). Thus, we focused on the dose-dependent blockade of cGAMP export by MK-571. MK-571 pretreatment not only resulted in enhanced retention but also reduced export of cGAMP from mouse BMMs (Figure 2B). We extended these findings to human cells. As with mouse cells, pre-treatment with MK-571 resulted in enhanced retention of intracellular cGAMP after DNA transfection in A549 cells (Figure 2C) and HFFs (Figure 2D). Furthermore, MK-571-treated A549 cells had reduced extracellular cGAMP (Figure 2C), and the low amount of extracellular cGAMP in HFFs was further reduced by MK-571 treatment (Figure 2D). These data reveal that MK-571 blocks cGAMP export and causes intracellular cGAMP accumulation in a dose dependent manner in both mouse and human cells.
Figure 2. MK-571 blocks cGAMP export in mouse and human cells.
(A) ALR−/− BMMs were treated with 1, 10, or 25 μM of indicated inhibitor or mock, followed by CT DNA transfection. 8 hours later, intracellular cGAMP was quantified in cell lysates using ELISA.
(B) ALR−/− BMMs were treated with 10 or 25 μM MK-571 or mock, followed by CT DNA transfection. 8 hours later, cell lysates and supernatants were harvested and cGAMP was quantified using ELISA (I: intracellular; E: extracellular).
(C, D) The indicated human cell types were treated with MK-571 (0–25 μM) followed by CT DNA transfection. 8 hours later, cell lysates and supernatants were harvested and cGAMP was quantified using ELISA. Statistical analysis was performed using a one-way ANOVA comparing all drug treatments to mock (A) or a two-way ANOVA comparing drug treatments to mock-treated conditions in the intracellular or extracellular compartment (B-D). Tests corrected for multiple comparisons using the Holm-Sidak method. Error bars represent mean ± SD of three biological replicates per group. **p<0.01, ***p<0.001. All data shown are derived from a single representative experiment. Comparative results were obtained across three independent experiments.
ABCC1 is a cGAMP exporter
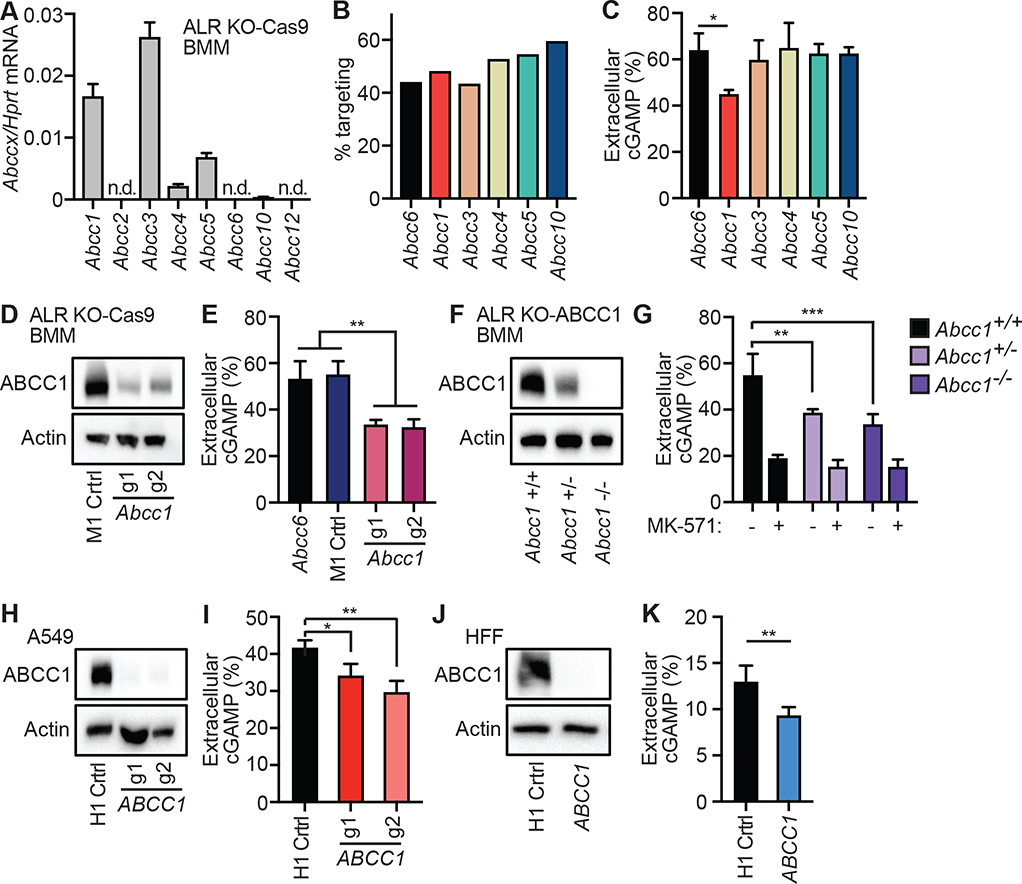
MK-571 inhibits ABCC1, also known as multidrug resistance protein 1 (MRP1) (Cole et al., 1992; Leier et al., 1994). In the three decades since its identification, ABCC1 has been demonstrated to mediate the export of a large range of endogenous and exogenous macromolecules, including cysteinyl leukotriene C4 (LTC4), the conjugated estrogen E217βG, glutathione (GSH), anthracycline chemotherapeutic drugs, antibiotics, statins (Cole, 2014), and sphingosine-1-phosphate (Mitra et al., 2006). ABCC1 is a member of a family that includes several related transporters, and it is now known that MK-571 blocks additional members of the ABCC family, including ABCC2, ABCC4, and ABCC5 (Barrington et al., 2015; Reid et al., 2003). Because of the potential confounding effects of MK-571 promiscuity on interpretation of our pharmacological data, we sought genetic evidence to explain the potent inhibition of cGAMP export by MK-571. We therefore undertook a lentiCRISPR-based genetic screen in primary mouse BMMS. We generated ALR−/− mice that constitutively express Cas9 (Platt et al., 2014), and we developed quantitative RT-PCR assays to measure the expression of each member of the murine ABCC family. We found that ALR−/−Cas9+ BMMs expressed detectable mRNA transcripts for five of the eight murine ABCC family members (Figure 3A). We designed guide RNAs (gRNAs) to target each of these five expressed ABCC channels, cloned them into a lentiCRISPR vector, prepared lentivirus particles, and transduced primary BMMs followed by selection in puromycin (Gray et al., 2016). We included an Abcc6 gRNA as a negative control that targets a member of the Abcc family that is not expressed in these cells. After three days of selection, we observed robust but incomplete disruption of the specific target genes using TIDE analysis (Tracking of Indels by Decomposition) (Brinkman et al., 2014) (Figure 3B). We transfected each population of targeted BMMs with CT DNA for 8 hours, measured intracellular and extracellular cGAMP concentrations, and then calculated the percent of extracellular cGAMP. We found that targeting of Abcc1, but not any of the other expressed Abcc genes, resulted in a significant decrease in the percent of extracellular cGAMP recovered following DNA transfection (Figure 3C, Figure S2A). We repeated this experiment, including an additional M1 non-targeting control gRNA (Sanjana et al., 2014), together with a second gRNA targeting a distinct site in the Abcc1 gene. Both Abcc1 gRNAs depleted ABCC1 protein as measured by immunoblot (Figure 3D) and resulted in a significant decrease in extracellular cGAMP (Figure 3E, Figure S2B). To assess whether Abcc1 was responsible for all cGAMP export in BMMs, we crossed ABCC1-deficient mice to our ALR−/− mice. Heterozygous Abcc1+/−ALR−/− BMMs had reduced ABCC1 protein, whereas homozygous Abcc1−/−ALR−/− BMMs lacked ABCC1 protein expression (Figure 3F). We found a significant reduction in extracellular cGAMP after DNA stimulation in both Abcc1+/− and Abcc1−/− BMMs compared to controls (Figure 3G, Figure S2C). In addition, pretreatment with MK-571 further reduced cGAMP export, even in Abcc1+/− and Abcc1−/− cells (Figure 3G, Figure S2C). Together, these data provide genetic evidence that ABCC1 is responsible for a significant fraction of cGAMP export in BMMs. Moreover, because MK-571 inhibits residual cGAMP export in cells lacking ABCC1, our findings suggest the existence of additional, non-redundant, MK-571-sensitive export mechanisms.
Figure 3. Identification of ABCC1 as a cGAMP exporter.
(A) Quantification of ABCC family member mRNA transcript expression in ALR−/−Cas9+ BMMs by RT-qPCR.
(B) ALR−/−Cas9+ BMMs were transduced with lentiCRISPR encoding the indicated ABCC family-specific gRNAs, selected for 3 days, and then percent genomic targeting was calculated using Sanger sequencing and Tracking of Indels by DEcomposition (TIDE) analysis.
(C) Cells from (B) were transfected with CT DNA and then 8 hours later cGAMP was quantified in supernatants and cell lysates using ELISA.
(D) ALR−/−Cas9+ BMMs were transduced with lentiCRISPR encoding Abcc1 or M1 control-specific gRNAs as described in (B) followed by immuno blot analysis for ABCC1 protein.
(E) Cells from (D) were transfected with CT DNA and then 8 hours later cGAMP was quantified in supernatants and cell lysates using ELISA.
(F) BMMs were harvested from ALR−/− BMMs mice that were crossed to different ABCC1 genotypes and then evaluated by immuno blot for ABCC1 protein.
(G) Cells from (F) were transfected with CT DNA and then 8 hours later cGAMP was quantified in supernatants and cell lysates using ELISA.
(H) A549 cells were transduced with lentiCRISPR encoding ABCC1- or H1 control-specific gRNAs as described in (B) and ABCC1 protein expression was evaluated by immuno blot.
(I) Cells from (H) were transfected with CT DNA and then 8 hours later cGAMP was quantified in supernatants and cell lysates using ELISA.
(J) HFFs were transduced with lentiCRISPR encoding ABCC1- or H1 control-specific gRNAs as described in (B) and ABCC1 protein was assessed by immuno blot.
(K) Cells from (J) were transfected with CT DNA and then 8 hours later cGAMP was quantified in supernatants and cell lysates using ELISA. Statistical analysis was performed using a one-way ANOVA comparing targeted lines to relevant controls and corrected for multiple comparisons using the Holm-Sidak method. Error bars represent mean ± SD of three biological replicates per group. *p<0.05, **p<0.01, ***p<0.001. All data shown are derived from a single representative experiment. Comparative results were obtained across two (A-C, F, G) or three (D, E, H-K) independent experiments. See also Figure S2.
Next, we tested whether disruption of ABCC1 reduces cGAMP export in human cells. We designed two distinct gRNAs to target human ABCC1 with lentiCRISPR as described above. We first tested these gRNAs in A549s and found that they depleted human ABCC1 protein (Figure 3H). We assessed functional loss of ABCC1 transport activity following targeting by staining cells with Fluo-3, a calcium-binding fluorescent compound that is a known substrate of ABCC1-dependent export (Keppler et al., 1999; Prechtl et al., 2000). We used measurement of Fluo-3 fluorescence as a proxy for ABCC1 activity: cells with high ABCC1 activity were dimmer than cells with low ABCC1 activity (Figure S2D). Flow cytometry analysis showed that ABCC1 targeted A549 cells had a significant increase in Fluo-3 mean fluorescence intensity (MFI) compared to H1 control targeted cells, demonstrating that these gRNAs reduce ABCC1 transport activity (Figure S2E, S2F). ABCC1 targeted A549 cells had a significant decrease in percentage of extracellular cGAMP compared to control H1 targeted cells (Figure 3I, Figure S2G). We then targeted ABCC1 in HFFs (Figure 3J), and we found a significant decrease in the percent of extracellular cGAMP in the ABCC1-targeted cells compared to H1 control targeted cells (Figure 3K, Figure S2H). Taken together, these data demonstrate that disruption of human ABCC1 reduces cGAMP export. Moreover, and like Abcc1−/− mouse cells (Figure 3G), the more complete inhibition of cGAMP export by MK-571 treatment compared to ABCC1 disruption in A549 cells and HFF implies the existence of additional cGAMP export mechanism(s) in human cells that are sensitive to MK-571 (Figure 2C, Figure 2D).
ABCC1 is a direct, ATP-dependent cGAMP exporter
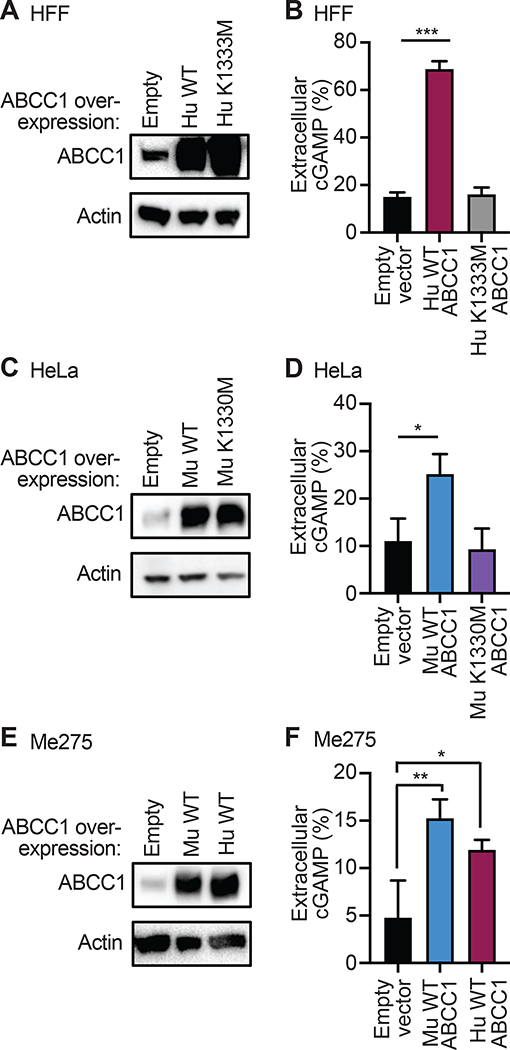
We found that ABCC1 protein expression correlated with the diverse efficiencies of cGAMP export that we identified among several mouse and human cell lines (Figure S3A, S3B). We therefore tested whether ABCC1 overexpression could increase cGAMP export in cells with lower endogenous ABCC1 expression. To do this, we transduced HFFs, which had relatively low ABCC1 expression and poor cGAMP export efficiency, with lentivirus encoding human ABCC1. To test the requirement for ATP hydrolysis in cGAMP export, we introduced a K1333M point mutation into the second nucleotide binding domain (NBD) of ABCC1 that is known to reduce ABCC1-mediated LTC4 transport by approximately 70% (Gao et al., 2000). After selection of transduced cells, we observed increased ABCC1 protein expression (Figure 4A) and decreased Fluo-3 retention in the ABCC1-overexpressing HFFs relative to those transduced with control lentivirus (Figure S3C, S3D), demonstrating increased ABCC1 function. HFFs expressing K1333M mutant ABCC1 also had a significantly lower Fluo-3 MFI than control HFFs, but not as low as HFFs overexpressing WT ABCC1 (Figure S3C, S3D). To our knowledge, Fluo-3 fluorescence has never been studied in the context of the K1333M mutation, so it is possible that Fluo-3 export by ABCC1 is partially ATP-independent. We next tested these cells for cGAMP export after DNA transfection. We found that HFFs overexpressing WT ABCC1 exported more than triple the percent of extracellular cGAMP compared to controls (Figure 4F, Figure S3E). In contrast, the K1333M mutant failed to enhance cGAMP export (Figure 4F, Figure S3E).
Figure 4. ABCC1 is an ATP-dependent cGAMP exporter.
(A) HFFs were transduced with lentivirus encoding human WT or K1333M mutant ABCC1 or control (empty vector) and selected for 5 days in hygromycin. Cells were evaluated for ABCC1 protein expression by immuno blot.
(B) Cells from (A) were transfected with CT DNA and then 8 hours later cGAMP was quantified in supernatants and cell lysates using ELISA.
(C) HeLa cells were transduced with lentivirus encoding murine WT or K1330M mutant ABCC1 or control (empty vector) and selected for 5 days in hygromycin. Cells were evaluated for ABCC1 protein expression by immuno blot.
(D) Cells from (C) were transfected with CT DNA and then 8 hours later cGAMP was quantified in supernatants and cell lysates using ELISA.
(E) Me275 cells were transduced with lentivirus encoding human or murine WT ABCC1 or control (empty vector) and selected for 5 days in hygromycin. Cells were evaluated for ABCC1 protein expression by immuno blot.
(F) Cells from (E) were transfected with CT DNA and then 8 hours later cGAMP was quantified in supernatants and cell lysates using ELISA. Statistical analysis was performed using a one-way ANOVA comparing WT or mutant ABCC1-overexpression cells to empty vector control (B, D, F) and corrected for multiple comparisons using the Holm-Sidak method. Error bars represent mean ± SD of three biological replicates per group. *p<0.05, **p<0.01, ***p<0.001. All data shown are derived from a single representative experiment. Comparative results were obtained across three independent experiments. See also Figure S3.
To evaluate functional orthology between human and mouse ABCC1, we cloned the murine ABCC1 open reading frame into a lentiviral vector, as well as the corresponding K1330M NBD mutation (Gao et al., 2000). We transduced HeLa cells, which, like HFFs, have relatively low endogenous ABCC1 expression and low efficiency of cGAMP export. After selection of transduced cells, we observed increased ABCC1 protein in HeLa cells expressing both the murine WT ABCC1 and the K1330M mutant relative to those transduced with control lentivirus (Figure 4C). We transfected these cells with CT DNA and found that HeLa cells overexpressing WT murine ABCC1, but not K1330M ABCC1, had a significant increase in the percent of extracellular cGAMP (Figure 4D, Figure S3F). Finally, we found that overexpression of either human or mouse ABCC1 significantly enhanced cGAMP export in Me275 human melanoma cells (Figure 4E, 4F, Figure S3G). Together, these data indicate that both human and mouse ABCC1 mediate ATP-dependent cGAMP export.
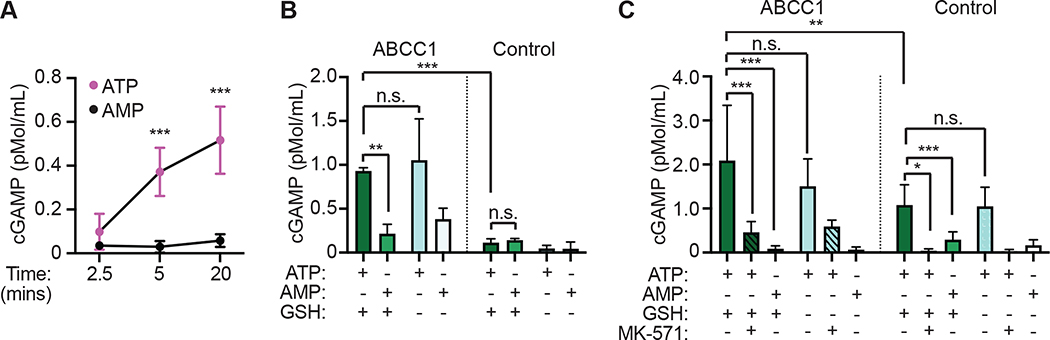
One potential caveat of our functional studies of cGAMP export in cells is the possibility that cGAMP is not a direct substrate of ABCC1, and that cGAMP export is instead stimulated indirectly by the movement of a distinct, unrelated ABCC1 substrate. We therefore performed biochemical assays to evaluate cGAMP transport using commercially available “inside out” vesicles prepared from Spodoptera frugiperda (fall armyworm Sf9) insect cells. These inside out vesicles are a “gold standard” in the biochemistry of transporters and have been used to define three broad classes of ABCC1 substrates: 1) molecules that are transported on their own, 2) molecules that are transported as covalent conjugates with glutathione, and 3) molecules that are co-transported with glutathione in the absence of covalent conjugation (Cole, 2014). Using vesicles derived from Sf9 cells overexpressing human ABCC1, we found robust, ATP-dependent cGAMP transport that increased over time (Figure 5A). We then focused on the 5-minute and 20-minute time points for more detailed analysis. At 5 minutes of incubation, we found that ABCC1-expressing vesicles transported cGAMP in the presence of ATP but not AMP, whereas control vesicles did not (Figure 5B). In addition, we observed that ABCC1-expressing vesicles transported similar amounts of cGAMP in the presence or absence of glutathione (Figure 5B). At the 20-minute time point, we observed cGAMP transport into the control Sf9-derived vesicles that did not express human ABCC1 (Figure 5C). The amount of cGAMP transported into the control vesicles was significantly less than that transported into the ABCC1-expressing vesicles. However, this endogenous cGAMP transport was similar to the transport observed in ABCC1-expressing vesicles in that it required ATP and was glutathione-independent (Figure 5C). We found that MK-571 inhibited cGAMP transport into both the ABCC1-expressing vesicles and the control vesicles (Figure 5C). These data provide biochemical evidence that ABCC1 mediates direct, ATP-dependent, glutathione-independent transport of cGAMP. Moreover, the discovery of pharmacologically similar cGAMP transport in plain Sf9 cell-derived vesicles suggests that ATP-dependent cGAMP export is a conserved mechanism that exists in both arthropods and chordates.
Figure 5. Identification of a conserved mechanism of direct, ATP-dependent cGAMP export.
(A) Time course of cGAMP transport using Sf9 cell-derived vesicles expressing human ABCC1 in the presence of ATP or AMP.
(B) 5-minute vesicle transport assays using Sf9 insect cell-derived vesicles expressing human ABCC1 or control vesicles with cGAMP in the presence of ATP or AMP, and with or without glutathione (GSH).
(C) 20-minute vesicle transport assays using Sf9 insect cell-derived vesicles expressing human ABCC1 or control vesicles with cGAMP in the presence of ATP or AMP, with or without glutathione (GSH), and with or without MK-571 (25 mM).
Statistical analysis was performed using one-way ANOVA comparing within time points (A) or comparing the mean of each group to every other group (B, C), and corrected for multiple comparisons using the Holm-Sidak method. Error bars represent the mean ± SD of 3–8 biological replicates per group. *p<0.05, **p<0.01, ***p<0.001. All data shown are derived from a single experiment. Comparative results were obtained across three (A, B) or two (C) independent experiments.
cGAMP export controls cell-intrinsic STING signaling
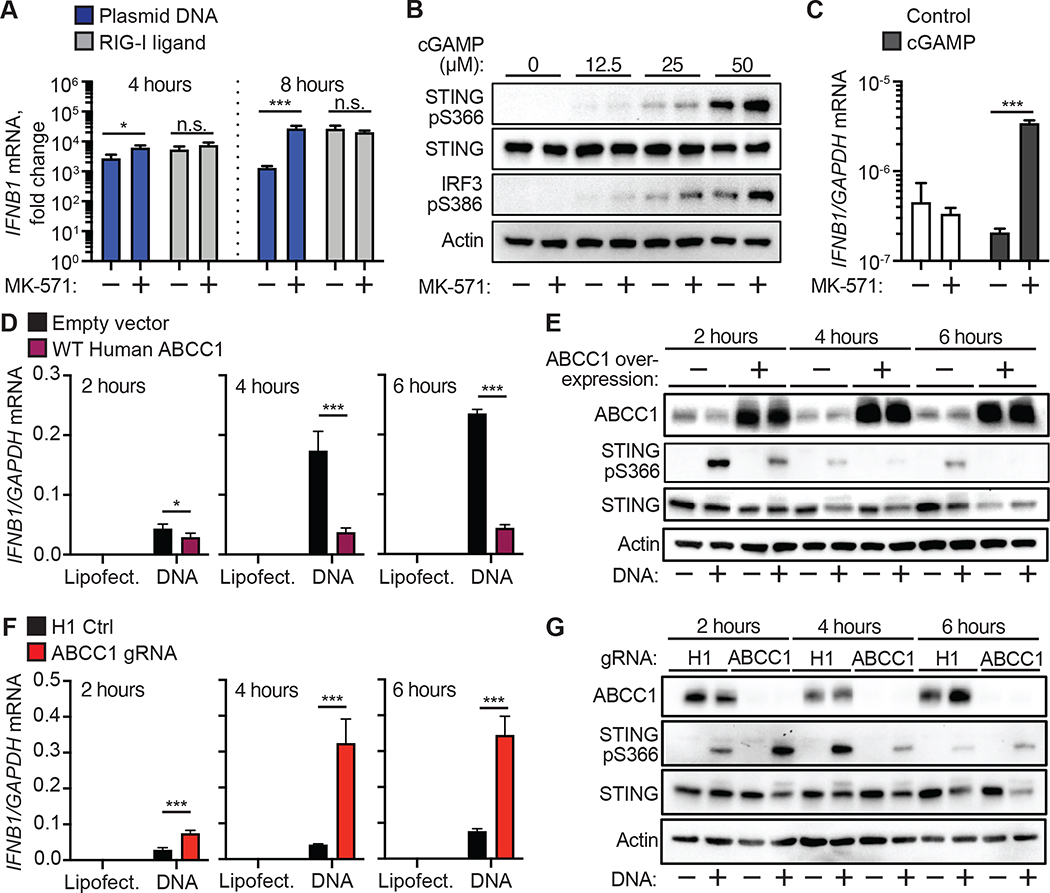
cGAMP binding to STING activates TBK1- and IRF3-dependent transcription of type I IFN genes (Liu et al., 2015). We hypothesized that reduction of intracellular cGAMP concentrations through ABCC1-dependent export would similarly reduce cell-intrinsic STING signaling. Consistent with this hypothesis, we found that pretreatment of HFFs with MK-571 potently enhanced DNA-activated IFNB1 transcription but not RNA-activated IFNB1 transcription (Figure 6A). Similarly, MK-571 pretreatment enhanced the response in cells treated with extracellular cGAMP, as evidenced by enhanced phospho-STING and phospho-IRF3 protein (Figure 6B), as well as increased IFNB1 transcription (Figure 6C). These data suggest that cGAMP export limits cell intrinsic STING signaling in response to DNA and imported cGAMP.
Figure 6. cGAMP export controls cell-intrinsic STING signaling.
(A) Quantification of IFNB1 induction by RT-qPCR in HFFs that were treated with 25 μM MK571 or mock followed by transfection with plasmid DNA or RIG-I ligand for 4 or 8 hrs.
(B) HFFs were treated with 25 μM MK-571 or mock, and then cGAMP (0–50 μM) was added to the extracellular media. Immuno blot analysis was performed 4 hours after cGAMP addition for phosphorylated STING, STING, and phosphorylated IRF3.
(C) HFFs were treated with 25 μM MK-571 or mock, and then cGAMP (0 or 50 μM) was added to the extracellular media. INFB1 induction was quantified by RT-qPCR 4 hours after cGAMP addition.
(D) Quantification of IFNB1 induction by RT-qPCR in HFFs overexpressing ABCC1 or empty vector control following transfection with CT DNA. Cells were harvested at the indicated time points.
(E) Immuno blot analysis of cells from (D) for ABCC1, phosphorylated STING, and STING protein expression.
(F) Quantification of IFNB1 induction by RT-qPCR in ABCC1- or H1 control-targeted HFFs following transfection with CT DNA. Cells were harvested at the indicated time points.
(G) Immuno blot analysis of cells from (F) for ABCC1, phosphorylated STING, and STING protein expression. Statistical analysis was performed using a two-way ANOVA comparing mock or MK-571 treatment within each transfected ligand group (A, C) or comparing control to ABCC1-modulated cells within each transfected ligand group (D, F). All tests were corrected for multiple comparisons using the Holm-Sidak method. Error bars represent mean ± SD of three biological replicates per group. *p<0.05, ***p<0.001. All data shown are derived from a single representative experiment. Comparative results were obtained across three independent experiments.
Given our findings that pharmacological blockade of cGAMP export enhanced STING signaling, we hypothesized that modulation of ABCC1 expression alters type I IFN production. To test this, we stimulated our ABCC1-overexpressing HFFs with CT DNA and found that these cells had significantly lower IFNB1 mRNA induction compared to control cells throughout a 6-hour time course (Figure 6D). Within this same experiment, we observed decreased phosphor-STING at each time point in ABCC1-overexpressing cells, consistent with the reduction in IFNB1 transcription (Figure 6E). Conversely, we found that targeting ABCC1 in HFFs using lentiCRISPR (Figure 3J) resulted in strongly enhanced IFNB1 activation and accelerated kinetics of phospho-STING after DNA transfection when compared to H1 non-targeting control cells (Figure 6F, 6G). These findings indicate that modulation of ABCC1 expression alters the antiviral response to DNA and suggest that ABCC1 acts as a potent negative regulator of cell intrinsic STING signaling.
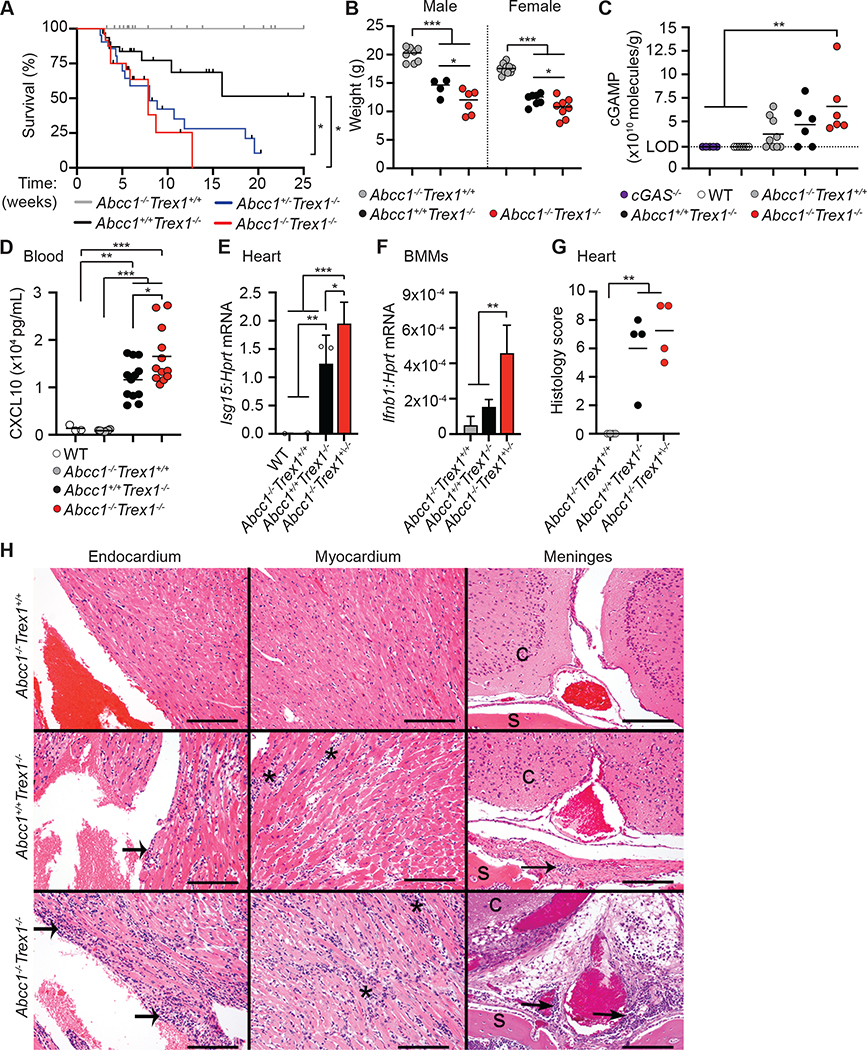
ABCC1 deficiency exacerbates cGAS-dependent autoimmunity in Trex1−/− mice
Mutations in the human gene that encodes the TREX1 DNA exonuclease cause a rare and severe autoimmune disease called Aicardi-Goutières Syndrome (AGS) (Crow et al., 2006). We have previously shown that TREX1 is an essential and specific negative regulator of the cGAS-STING DNA sensing pathway (Stetson et al., 2008). Trex1−/− mice have a median life span of around 110 days and develop severe autoimmunity that requires cGAS (Gao et al., 2015; Gray et al., 2015), STING (Gall et al., 2012), type I IFNs (Stetson et al., 2008), and lymphocytes (Gall et al., 2012; Stetson et al., 2008). Whereas Trex1−/− mice have been valuable to identify signaling pathways and immune cells that are important for causing disease, no genetic modifiers that limit or restrict disease have yet been identified. We utilized Trex1−/− mice to explore the contribution of cGAMP export in a clinically relevant mouse model of human autoimmunity. Since ABCC1 expression correlated negatively with type I IFN production after DNA stimulation (Figure 6), we hypothesized that loss of ABCC1 in Trex1−/− mice would lead to accelerated mortality from enhanced IFN-mediated disease. We intercrossed Trex1−/− and Abcc1−/− mice and monitored survival. We found that both Abcc1−/−Trex1−/− and Abcc1+/−Trex1−/− mice exhibited significantly accelerated mortality compared to Abcc1+/+Trex1−/− mice (Figure 7A). The accelerated mortality of Trex1−/− mice on both Abcc1+/− and Abcc1−/− backgrounds was consistent with the similarly reduced cGAMP export in both Abcc1−/− and Abcc1+/− BMMs (Figure 3G), suggesting that haploinsufficiency for Abcc1 is sufficient to modify the course of disease. At 35 days of age, Abcc1−/−Trex1−/− mice were more severely runted than Abcc1+/+Trex1−/− mice (Figure 7B). We measured tissue cGAMP and found that hearts from over half of the Abcc1−/− mice that we sampled had detectable cGAMP, whereas we could not detect cGAMP in heart tissue from any Abcc1+/+ (WT) mice or from Cgas−/− mice that served as a negative control for the assay (Figure 7C). This increase was consistent with previously reported steady state accumulation of glutathione (a distinct ABCC1 substrate) in the hearts of Abcc1−/− mice (Lorico et al., 1997). Heart cGAMP was detectable in most Abcc1+/+Trex1−/− mice and significantly increased in Abcc1−/−Trex1−/− mice relative to control mice (Figure 7C). Despite the limit of detection of the cGAMP ELISA assay, these data suggest that ABCC1 deficiency results in increased tissue cGAMP in both steady state and during cGAS-dependent disease.
Figure 7. ABCC1 deficiency enhances cGAS-dependent autoimmunity in Trex1−/− mice.
(A) Survival of Abcc1−/−Trex1+/+ (n= 33), Abcc1+/+Trex1−/− (n= 31), Abcc1+/−Trex1−/− (n= 21), and Abcc1−/−Trex1−/− (n= 24) mice.
(B) Weights of mice at 35 days of age; Abcc1+/+Trex1−/− (n= 8 M, 10 F), Abcc1+/+Trex1−/− (n= 4 M, 7 F), and Abcc1−/−Trex1−/− (n= 6 M, 8 F).
(C) Quantification of intracellular cGAMP recovered from heart tissue by ELISA; Cgas−/− (n= 5), WT (n= 7), Abcc1−/−Trex1+/+ (n= 9), Abcc1+/+Trex1−/− (n= 6), Abcc1−/−Trex1−/− (n= 6). Values are normalized to individual heart weights (LOD = limit of detection at 2.3×1010 molecules/g).
(D) Quantification of Cxcl10 protein in serum measured by ELISA of the indicated genotypes; WT (n= 3), Abcc1−/− (n= 8), Abcc1+/+Trex1−/− (n= 12), Abcc1−/−Trex1−/− (n= 12).
(E) Quantification of Isg15 mRNA transcripts by RT-qPCR in heart tissue of the indicated genotypes; n= 3 mice per group.
(F) Quantification of Ifnb1 mRNA transcripts by RT-qPCR in BMMs of the indicated genotypes; n= 3 or 4 mice per group.
(G) Histological score in heart tissue measured at 40 days of age; n= 4 mice per group.
(H) Representative (of 4 of each genotype) H&E-stained heart (endocardium and myocardium) and brain at the level of cerebrum (c) with meninges, periosteum, and skull (s) tissue sections from Abcc1−/−Trex1+/+, Abcc1+/+Trex1−/−, and Abcc1−/−Trex1−/− mice. Abcc1+/+Trex1−/− and Abcc1−/−Trex1−/− mice have inflammation extending along the endocardial surface (wide arrows), which appears more pronounced in Abcc1−/−Trex1−/− mice. Abcc1+/+Trex1−/− and Abcc1−/−Trex1−/− mice both have myocardial inflammation (asterisks) with variably severe myocardial degeneration. Abcc1+/+Trex1−/− and Abcc1−/−Trex1−/− mice have perivascular lymphocytic infiltrates (narrow arrows) in the meninges and periosteum of the skull which is variably severe and not present in all mice, but which is generally minimal in the Abcc1+/+Trex1−/− mice. The scale bars in each panel indicate 100 μm. Statistical analysis for the survival curves was calculated with a log-rank (Mantel-Cox) test (A). Statistical analysis for all other experiments was performed using a one-way ANOVA comparing each group to every other group (C, D), comparing transcripts of each gene between genotypes (E, F), or comparing Abcc1−/−Trex1+/+ to Abcc1+/+Trex1−/− and Abcc1−/−Trex1−/− mice in (G). All ANOVA tests were corrected for multiple comparisons using the Holm-Sidak method. Error bars represent mean ± SD. The ROUT method was used to detect potential outliers and no outliers were detected across the data shown. *p<0.05, **p<0.01, ***p<0.001.
We evaluated the IFN-dependent chemokine CXCL10 in serum as an additional measure of the chronic antiviral response and found that CXCL10 was elevated in Abcc1−/−Trex1−/− mice compared to Abcc1+/+Trex1−/− mice (Figure 7D). Similarly, Isg15 mRNA expression in heart tissue was higher in Abcc1−/−Trex1−/− mice compared to Abcc1+/+Trex1−/− mice, consistent with enhanced IFN signaling (Figure 7E). We next prepared BMMs from age- and sex-matched mice and found that expression of Ifnb1 mRNA was increased in Abcc1−/−Trex1−/− BMMs compared to Abcc1+/+Trex1−/− BMMs (Figure 7F), suggesting that loss of ABCC1 enhances the spontaneous type I IFN response caused by Trex1 deficiency.
Lastly, we performed a blinded histological analysis of affected tissues, comparing Abcc1−/−Trex1−/− mice and Abcc1+/+Trex1−/− mice to Abcc1−/− controls. While we observed a trend towards more severe heart pathology in Abcc1−/−Trex1−/− mice compared to Abcc1+/+Trex1−/− mice, this did not reach statistical significance in the context of our scoring criteria, likely because of the severe inflammation that has been well documented in the Trex1−/− hearts (Figure 7G, 7H). We also noted the presence of focal to multifocal, generally mild, perivascular lymphoid aggregates (with lower numbers of other inflammatory cells) associated with the meninges and periosteum of the skull in 3/4 Abcc1−/−Trex1−/− mice compared to 1/4 Abcc1+/+Trex1−/− mice (Figure 7H). Taken together, these data suggest that ABCC1 is a genetic modifier and negative regulator of the cGAS-cGAMP-STING pathway in vivo in a model of chronic cGAS activation.
Discussion
In this study, we have provided pharmacological, genetic, functional, biochemical, and in vivo evidence identifying ABCC1 as an important molecular site of action for cGAMP export. We have shown that loss of ABCC1 resulted in increased intracellular cGAMP concentrations and enhanced STING signaling. Conversely, overexpression of ABCC1 reduced intracellular cGAMP concentrations and downregulated STING signaling. We demonstrated that ABCC1 mediated direct, ATP-dependent transport of cGAMP and have shown that cGAMP export is conserved in insect cells. Finally, we have demonstrated that ABCC1 is a genetic modifier that limits cGAS-dependent autoimmunity in vivo in the well-characterized Trex1−/− mouse model of human AGS.
Genetic disruption of ABCC1 reduced but did not completely ablate cGAMP export in vitro, suggesting that alternative cGAMP export mechanisms exist. Consistent with the recent identification of multiple transporters that import cGAMP into cells (Cordova et al., 2021; Lahey et al., 2020; Luteijn et al., 2019; Ritchie et al., 2019; Zhou et al., 2020a; Zhou et al., 2020b), our findings demonstrate the existence of multiple cGAMP exporters. It was recently proposed that the LRRC8-containing volume regulated anion channel (VRAC) might serve as a “passive” cGAMP exporter, but these studies clearly demonstrated that LRRC8 can only export cGAMP under artificial hypotonic conditions that activate the channel through cell swelling (Lahey et al., 2020; Zhou et al., 2020a). While our CRISPR screen did not implicate other ABCC family members in cGAMP export, our finding that MK-571 further inhibitted cGAMP export in Abcc1−/− BMMs and the fact that other nucleotide derivates such as cyclic AMP and cyclic GMP can be moved by ABCC family proteins (Guo et al., 2003; Jedlitschky et al., 2000; Reid et al., 2003) suggest that cGAMP might be a substrate of multiple, related ABC transporters. To identify additional cGAMP exporters, our ALR-deficient, Cas9-expressing mice remain a valuable tool for CRISPR-based screens in primary cells. Regardless of the potential for multiple cGAMP export mechanisms, we emphasize that ABCC1 plays a nonredundant role in cGAMP export and regulation of cGAS-STING signaling, both in vitro and in vivo.
Using inside out vesicles derived from Sf9 insect cells, we provided biochemical evidence that ABCC1 is a direct, ATP-dependent, glutathione-independent cGAMP transporter. These data suggest that cGAMP belongs to the class of ABCC1 substrates that are neither conjugated to glutathione nor co-transported with glutathione (Cole, 2014). Recent structural studies have identified a bipartite substrate binding site in ABCC1 comprised of a positively charged pocket that can bind anions like glutathione, together with a large hydrophobic pocket that can accommodate lipid moieties of glutathione-conjugated substrates like LTC4 (Johnson and Chen, 2017). We hypothesize that cGAMP engages the same positively charged pocket as gluthathione itself using its negatively charged phosphate groups, which would explain why cGAMP export does not require glutathione as a cofactor. Further biochemical and structural studies will provide additional insight into the cGAMP export mechanism by ABCC1.
We discovered fortuitously that Sf9 insect cells have an endogenous mechanism of cGAMP export that is pharmacologically similar to ABCC1-mediated cGAMP export. Although the existence of this endogenous cGAMP transport mechanism in Sf9 cells makes it difficult to perform rigorous kinetic analyses of ABCC1-mediated cGAMP transport, this finding suggests that cGAMP export is conserved in both arthropods and chordates. In light of the recent identification of cGAS-like receptors in Drosophila that produce 2’,3’ cGAMP and 3’,2’ cGAMP (Holleufer et al., 2021; Slavik et al., 2021), our discovery of cGAMP export in insect cells raises new questions about the immunotransmitter functions of cGAMP in arthropod immunity. Identification and characterization of the orthologous, MK-571-sensitive cGAMP exporters in insect cells will be an important future goal.
Our findings have important implications for our understanding of the regulatory mechanisms involved in cGAS-STING signaling. We demonstrated that genetic ablation or pharmacological blockade of ABCC1 enhanced cell-intrinsic STING signaling, whereas overexpression of ABCC1 reduced STING signaling. Thus, by limiting intracellular cGAMP concentrations, ABCC1 negatively regulates the cGAS-STING pathway and provides a mechanism to expose extracellular cGAMP to ENPP1-mediated degradation (Li et al., 2014). In further support of this, loss of Abcc1 in the Trex1−/− mouse model led to accelerated and exacerbated disease. Thus, ABCC1 is a genetic modifier that limits cGAS-dependent immune disease. There are numerous polymorphisms in the human ABCC1 gene that have been associated with the varied responses to anthracycline-based chemotherapies in cancer patients (Cole, 2014). It will be interesting to determine whether these or other ABCC1 polymorphisms influence the efficiency of cGAMP export and the strength of cGAS-STING signaling. We speculate that hypomorphic ABCC1 alleles might be evolutionarily advantageous because they provide superior protection against certain virus infections, but these same alleles might predispose to cGAS-dependent autoimmune disease. Because haploinsufficiency for Abcc1 is sufficient to exacerbate disease in Trex1−/− mice, we propose that subtle differences in expression and function of ABCC1 might have a significant impact on STING-dependent immunity.
Our studies did not directly test the immunotransmitter function of cGAMP signaling that is additionally regulated by cGAMP degradation and cGAMP import. Ultimately, the influence of cGAMP export on STING signaling could be context and disease dependent, and it is possible that ABCC1-mediated export can play a positive regulatory role in propagating STING signaling to bystander cells. For example, we identified highly variable cGAMP export efficiencies across multiple human cancer cell lines and found that ABCC1 overexpression converted poor cGAMP exporters into more efficient exporters. Prior studies highlighted an essential role for tumor-derived extracellular cGAMP in priming antitumor immune responses (Carozza et al., 2020; Marcus et al., 2018). Our findings raise the possibility that high expression of ABCC1 on tumor cells enhances antitumor immune responses through increased export of cGAMP in the tumor microenvironment. However, it is known that overexpression of ABCC1 by tumor cells renders them resistant to certain chemotherapeutic drugs that have been previously defined as export substrates of ABCC1 (Cole, 2014). Thus, the potential clinical utility of ABCC1 inhibitors in cancer patients to limit efflux of anti-cancer chemotherapeutics might be offset by their potential to reduce the export of an important innate immune signal that primes anti-tumor immunity. Therefore, our finding that cGAMP is a substrate of ABCC1 warrants further study of the consequences of ABCC1 blockade in cancer.
Finally, there have been numerous recent advances in our understanding of the ancient evolutionary origins of the cGAS-STING pathway. These include the discoveries of diverse cGAS orthologs, the generation of 2’−5’-linked oligonucleotides, and the functional conservation of STING in prokaryotic antiviral immunity (Burroughs et al., 2015; Gui et al., 2019; Lowey et al., 2020; Morehouse et al., 2020; Whiteley et al., 2019). Our identification of ABCC1 as a cGAMP exporter and our discovery of conserved cGAMP export in insect cells raises comparisons to the regulation of intracellular CDN concentrations in prokaryotes. The ABC transporter superfamily exists not only in eukaryotes but also prokaryotes and archaea (Higgins, 1992), and the regulation of intracellular CDN concentrations through energy-dependent efflux is documented in bacteria (Woodward et al., 2010). We postulate that cGAMP export by ABCC1 fits into this framework in which the central regulatory components involved in animal cGAS-STING signaling all evolved from ancient prokaryotic mechanisms of defense against bacteriophages. A detailed evolutionary analysis is called for to explore this possibility. The possibility that ABCC1 exports other CDNs, such as those of microbial origin, also remains to be determined.
In summary, we have identified ABCC1 as an important molecular site of action for cGAMP export. Our discovery highlights ABCC1 as a negative regulator of cell-intrinsic STING signaling and completes the cycle of cGAMP production, export, and import, further rationalizing the existence of an extracellular cGAMP degradation mechanism. Further investigation into ABCC1-mediated cGAMP export has the potential to inform therapeutic approaches to enhance the protective functions of type I IFN in human diseases.
Limitations of Study
One limitation of our study is that we have not yet identified all of the mechanisms that account for cGAMP export. A full characterization of how cGAMP export contributes to regulation of the cGAS-STING pathway will require the identification of the residual cGAMP export mechanisms that operate in the absence of ABCC1.
METHODS
Resource availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Daniel Stetson (stetson@uw.edu).
Materials availability
Plasmids and cell lines generated in this study will be made available to research labs, accompanied by a standard material transfer agreement for non-commercial use.
Data and code availability
This paper does not report original code
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
ALR−/−, Sting (Tmem173)−/−, and Trex1−/− mice have been described previously (Gall et al., 2012; Gray et al., 2016; Stetson et al., 2008). C57BL/6J wild-type (stock #000664) and Abcc1−/− (stock #028129) mice were purchased from Jackson Laboratories. All mice used in this study were C57BL/6J and housed in a specific pathogen-free facility at the University of Washington with approval of the University of Washington Institutional Animal Care and Use Committee.
Cell lines and tissue culture
Primary mouse BMMs and MEFs were derived and cultured as previously described (Brunette et al., 2012). HeLa, A549, and HepG2 were purchased from ATCC. Me275 cells, established at the Ludwig Institute for Cancer Research (Valmori et al., 1998), were provided by A. Rongvaux (Fred Hutchinson Cancer Research Center). Telomerase-immortalized human foreskin fibroblasts (HFFs) were provided by D. Galloway (Fred Hutchinson Cancer Research Center). All adherent cell lines were cultured in DMEM supplemented with 10% FCS, L-glutamine, penicillin/streptomycin, sodium pyruvate, and HEPES.
METHOD DETAILS
Quantification of cell death
BMMs and MEFs were seeded at a density of 2×104 and 1×104, respectively, in a 24-well plate. Cell death was assayed with a 2-color IncuCyte Zoom in-incubator imaging system (Essen Biosciences, Ann Arbor, MI, USA) and analyzed as described (Orozco et al., 2014). SytoxGreen and SytoGreen (25 nM, Life Technologies) were used to calculate the frequency of dead cells.
cGAMP measurements
For in vitro measurements, extracellular cGAMP was measured directly from cell supernatants that were maintained at a volume of 200 μL. To quantify intracellular cGAMP, cells were washed once with PBS and then lysed in 200 μL RIPA buffer (150 mM NaCl, 1% Triton-X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0) for 15 minutes on ice. Lysates were then cleared of insoluble material before being used for cGAMP measurement. Relative cGAMP measurements were quantified using Direct 2’3’-Cyclic GAMP ELISA Kit (Arbor Assays).
To quantify cGAMP in mouse heart tissues, hearts were cleaned and weighed, minced, and then digested in dissociation buffer (2.7 mg/mL Collagenase A (Sigma), 23 U/mL DNase I (Sigma), 2 mM CaCl2 in PBS) for 1 hour at 37° C. Digestion was terminated with termination buffer (2% FBS, 5 mM EDTA in PBS). Samples were strained through 70 μm mesh strainers, centrifuged, and then subjected to red blood cell lysis in buffer (150 mM NH4CL, 10 mM KHCO3, 0.1 mM Na EDTA in water). Remaining cells were washed twice and suspended in 200 μL RIPA buffer for 15 minutes of lysis, then centrifuged to clear of insoluble material before direct measurement in cGAMP ELISA Kit. Limit of detection (LOD) was calculated by averaging the weights of all control (cGAS−/− and WT) hearts to use in calculation of molecules per gram of tissue within the given limit of detection of the ELISA Kit (0.04 pMol/mL).
Cell treatments and stimulations
For all in vitro experiments, cells were seeded in triplicate at a density of 1×105 (A549, HeLa, Me275), or 2.5×104 (HFFs) cells per well in 24-well plates. For nucleic acid transfections, calf thymus genomic DNA (Sigma) was diluted in water and used at 4 μg/mL (for human cells) or 1 μg/mL (for mouse cells); RIG-I ligand was synthesized in vitro as previously described using HiScribe T7 High Yield RNA Synthesis Kit (Saito et al., 2008) and used at 1 μg/mL; midiprepped pcDNA3 was used for plasmid stimulations at 4 μg/mL. For all transfections, nucleic acids were complexed with Lipofectamine 2000 (Invitrogen) at a ratio of 1 μg nucleic acid to 1 μL lipid. 2’3’ cGAMP (Invivogen) was diluted in water and directly added to cell culture media at 12.5 – 50 μM. MK-571, Verapamil, and KO-143 (Sigma) were suspended in DMSO and used to treat cells at concentrations from 1–50 μM for 1 hour prior to stimulation. Mock-treated cells received the same amounts of plain DMSO.
Immuno blotting and antibodies
Cells were harvested in RIPA lysis buffer supplemented with phosphatase and protease inhibitors (Pierce). Lysates were vortexed and incubated on ice for 15 minutes followed by clearing by centrifugation for 15 minutes. Cleared samples were then resuspended in running buffer with a final concentration of 25 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 0.0012% bromophenol blue. We discovered that ABCC1 aggregated if samples were boiled, so for ABCC1 immuno blots, we heated the samples at 37C for 7 minutes. All other samples were boiled for 7 minutes. Proteins were separated on 4–12% Bis-Tris SDS-PAGE gel (Life Technologies) and transferred to Immobilon-P PVDF membrane (Millipore). Blots were blocked in 5% BSA/TBST or 5% non-fat dry milk depending on primary antibody used. Membranes were probed overnight at 4° C with the following primary antibodies: anti-mouse/human ABCC1 (Abcam ab260038), anti-mouse/human Actin (Cell Signaling #3700), anti-human phospho-STING (Cell Signaling #19781), anti-mouse phospho-STING (Cell Signaling #72971), anti-mouse/human STING (Cell Signaling #13647), anti-human phospho-IRF3 (Abcam ab196035). Membranes were then probed with goat anti-rabbit-HRP or goat anti-mouse-HRP (Jackson ImmunoResearch) and developed by ECL 2 chemiluminescence (Pierce).
Fluo-3 AM staining and flow cytometry
Single cell suspensions were washed in PBS followed by staining with 2.5 uM Fluo-3 AM (Sigma) in FACS buffer (5% FBS in HBSS) and allowed to incubate for 20 mins on ice. 4 volumes of FACS buffer were then added and samples were incubated for an additional 40 mins at 37 C without CO2. After washing, samples were acquired on the Canto-II (BD Bioscience). Raw data were analyzed with FlowJo v10.
LentiCRISPR targeting
VSV-G pseudotyped, self-inactivating lentivirus was prepared by transfecting a 60–80% confluent 10-cm plate of HEK 293T cells with 1.5 μg of pVSV-G expression vector, 3 μg of pMDLg/pRRE, 3 μg pRSV-Rev and 6 μg of pRRL lentiCRISPR vectors using Poly(ethyleneimine) (PEI; Sigma). Media was replaced 24 hours post-transfection and harvested 24 hours later for filtration with a 0.45 μm filter (SteriFlip, Millipore). Approximately 1 million cells were transduced with 10 mL filtered virus. Following 3–5 days of selection in appropriate antibiotic, successful targeting was verified through population-level Sanger sequencing and analysis using Tracking of Indels by DEcomposition (TIDE) (Brinkman et al., 2014).
For CRISPR/Cas9 gene targeting, we generated pRRL lentiviral vectors in which a U6 promoter drives expression of a gRNA, and an MND promoter drives expression of Cas9, a T2A peptide, and either puromycin or hygromycin resistance (Gray et al., 2016). Primary mouse cells targeted in Figure 3 received lentivirus that did not encode Cas9, as the cells came from mice that have constitutive Cas9 expression. gRNA sequences were designed using Benchling. gRNA sequences are as follows, where the (G) denotes a nucleotide added to enable robust transcription off the U6 promoter. Guide RNA sequences are listed in Table S1.
Cloning of ABCC1
PCR and In-Fusion cloning (Takara Clontech) were used to generate ABCC1 constructs. WT and K1333M mutant human ABCC1 constructs were generated using a human ABCC1 cDNA clone (Transomic, Clone ID TOH6003) as template. Mouse WT and K13330 mutant ABCC1 constructs were generated using a mouse ABCC1 cDNA clone (Transomic, Clone ID BC090617) as template.
Vesicle transport assays
We used inside-out plasma membrane vesicles derived from Sf9 cells (SBVT04 for hMRP1 and SBCT03 for control; Sigma Aldrich) to determine whether cGAMP is a direct substrate of ABCC1/MRP1. All reagents were prepared in incubation buffer (10 mM Tris-HCl, 10 mM MgCl2, 250 mM sucrose, pH 7.0) and incubated for 3 minutes at 37° C prior to use in the reactions. Reactions were initiated by adding ATP or AMP (for a final concentration of 5 mM) to 50 μg vesicles with or without 2’3’ cGAMP (final concentration 5 μM). Some reactions also contained 1 mM glutathione (GSH) and/or 25 μM MK-571. Reactions were incubated in 96-well plates for 2.5, 5, 10, or 20 minutes at 37° C. Reactions were terminated by putting plates on ice and adding 200 μl pre-chilled wash buffer per well (10 mM Tris-HCl, 100 mM NaCl, 250 mM sucrose) and transferred to a prewet 96-well filtration plate (EMD Millipore; MSFBN6B10). Each well was washed 5 times with 200 μl wash buffer under vacuum pressure. The trapped vesicles were lysed by addition of 100 μl RIPA lysis buffer and eluted into collection plates. Samples were subsequently assayed for cGAMP by ELISA, (Arbor Assays; K067). We used three distinct lots of ABCC1 vesicles and two distinct lots of control vesicles with similar results.
Quantitative RT-PCR
For quantitative RT-PCR analysis, triplicate cell samples or 50 uL fresh mouse blood were harvested into Trizol reagent before purification via Direct-zol RNA miniprep (Zymo Research) per manufacturer’s instructions with an additional dry spin after disposing of the final wash to prevent carryover. cDNA was generated using EcoDry premix (Takara Bio). Samples were assayed in duplicate and transcript expression was measured using iTaq Universal SYBR green supermix (BioRad) on the Bio-Rad CFX96 Real-Time system. Sequences of PCR primers are listed in Table S2.
CXCL10 ELISA
Mouse blood was collected into Eppendorf tubes and allowed to clot for 45 minutes at room temperature, followed by two rounds of centrifugation and collection of top serum layer. Serum samples were assayed directly with Mouse IP-10 (CXCL10) ELISA Kit (Abcam) and analyzed per manufacturer’s instructions.
Histology and Pathology
Tissues were fixed in 10% neutral buffered formalin and routinely paraffin embedded. Tissue sections (5 μm) were stained with hematoxylin and eosin and histological scores were assigned in a blinded manner as previously described (Gray et al., 2016). Images were captured from glass slides using NIS-Elements BR 3.2 64-bit and plated in Adobe Photoshop Elements. Image white balance, lighting, and contrast were adjusted using auto corrections applied to the entire image.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Quantitative data were visualized and analyzed using GraphPad Prism software, ImageJ, and FlowJo. Specific statistical tests and experimental replicate numbers are noted in the figure legends.
Supplementary Material
Table S2: Related to STAR Methods. Sequences of quantitative RT-PCR primers used in this study
Table S1: Related to STAR Methods. Sequences of guide RNAs used in this study
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse/human ABCC1 | Abcam | Cat # ab260038 |
| anti-mouse/human Actin | Cell Signaling | Cat # 3700 |
| anti-human phospho-STING | Cell Signaling | Cat # 19781 |
| anti-mouse phospho-STING | Cell Signaling | Cat # 72971 |
| anti-mouse/human STING | Cell Signaling | Cat # 13647 |
| anti-human phospho-IRF3 | Abcam | Cat # ab196035 |
| goat anti-rabbit-HRP | Jackson ImmunoResearch | Cat # 111-035-003 |
| goat anti-mouse-HRP | Jackson ImmunoResearch | Cat # 115-035-146 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SytoxGreen | Life Technologies | Cat # S7020 |
| SytoGreen | Life Technologies | Cat # S7575 |
| DNase I | Sigma | Cat # D4263 |
| Collagenase A | Sigma | Cat # 11088793001 |
| Fluo-3 AM solution | Sigma-Aldrich | Cat # 39294 |
| calf thymus genomic DNA | Invitrogen | Cat # 15633019 |
| 2’3’ cGAMP | Invivogen | Cat # tlrl-nacga23 |
| Lipofectamine 2000 | Invitrogen | Cat # 11668019 |
| MK-571 | Sigma-Aldrich | Cat # M7571 |
| Verapamil | Sigma-Aldrich | Cat # V4629 |
| KO-143 | Sigma-Aldrich | Cat # K2144 |
| Critical Commercial Assays | ||
| Direct 2’3’-Cyclic GAMP ELISA | Arbor Assays | Cat # K067 |
| HiScribe T7 High Yield RNA Synthesis Kit | New England Biolabs | Cat # E2040S |
| Mouse IP-10 ELISA Kit | Abcam | Cat # ab260067 |
| Direct-zol RNA miniprep | Zymo Research | Cat # R2072 |
| Experimental Models: Cell Lines | ||
| C57BL/6 primary MEFs, (mouse embryonic fibroblasts) | Gray EE et al, J Immunol 2015 195:1939 | N/A |
| C57BL/6 primary BMMs, (bone marrow macrophages) | Gray EE et al, J Immunol 2015 195:1939 | N/A |
| HEK 293T cells | ATCC | CRL-3216 |
| A549 cells | ATCC | CCL-185 |
| HeLa cells | ATCC | CCL-2 |
| HepG2 | ATCC | HB-8056 |
| Me275 | Valmori D et al, J Immunol 1998 160(4):1750-8 | N/A |
| Tert-HFFs (tert-immortalized human foreskin fibroblasts) | Gift from D. Galloway; Fred Hutchinson Cancer Center | N/A |
| Experimental Models: Organisms/Strains | ||
| mouse: ALR−/−: C57BL/6J | Gray et al. Immunity. 2016 Aug 16;45(2):255–66 | N/A |
| mouse: Abcc1−/−: C57BL/6J | Jackson Laboratories | Cat # 028129 |
| mouse: Sting−/−: C57BL/6J | Gray et al. Immunity. 2016 Aug 16;45(2):255–66 | N/A |
| mouse: Trex1−/−: C57BL/6J | Stetson et al. Cell 2008 Aug 22; 134(4): 587–98 | N/A |
| Oligonucleotides | ||
| See Table S1 for qPCR primer sequences | See table S1 | N/A |
| See Table S2 for guide RNA sequences | See table S2 | N/A |
| Recombinant DNA | ||
| pRRL CRISPR vectors (guide sequences in Table S2) | Gray et al. Immunity. 2016 Aug 16;45(2):255–66 | N/A |
| Plasmid: pRRL-Hu-WT-ABCC1 | This paper | N/A |
| Plasmid: pRRL-Hu-K1333M-ABCC1 | This paper | N/A |
| Plasmid: pRRL-Mu-WT-ABCC1 | This paper | N/A |
| Plasmid: pRRL-Mu-K1330M-ABCC1 | This paper | N/A |
| Software and Algorithms | ||
| Prism v 9.0 | GraphPad | N/A |
| Adobe Photoshop Elements 2020 | Adobe | N/A |
| FlowJo | BD Biosciences | N/A |
| ImageLab | Bio-Rad | N/A |
| Tracking of Indels by DEcomposition (TIDE) | Brinkman et al. Nucleic Acids Res. 2014 Dec 16; 42(22): e168. | N/A |
Highlights:
The immunostimulatory molecule cGAMP is actively exported from live cells
ABCC1 mediates active, ATP-dependent cGAMP export
ABCC1-mediated cGAMP export limits activation of the STING-interferon pathway
ABCC1 limits cGAS-dependent autoimmunity in vivo
Acknowledgements
We thank N. Mausolf and S. Miller for expert mouse colony management, B. Johnson (University of Washington Histology and Imaging Core) for histology preparation, and all the members of the Stetson lab for helpful discussions. This work was supported by NIH T32-GM007270-44A1 (J.H.M.); NIH R21-AI159037 (D.B.S.); and Howard Hughes Medical Institute Faculty Scholar Award 55108572 (D.B.S.).
Footnotes
Declaration of Interests
D.B.S. is a co-founder and shareholder of Danger Bio, LLC, and a scientific advisor for Related Sciences, LLC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, and Hornung V (2013). Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JD, van Loevezijn A, Lakhai JM, van der Valk M, van Tellingen O, Reid G, Schellens JH, Koomen GJ, and Schinkel AH (2002). Potent and specific inhibition of the breast cancer resistance protein multidrug transporter in vitro and in mouse intestine by a novel analogue of fumitremorgin C. Mol Cancer Ther 1, 417–425. [PubMed] [Google Scholar]
- Barrington RD, Needs PW, Williamson G, and Kroon PA (2015). MK571 inhibits phase-2 conjugation of flavonols by Caco-2/TC7 cells, but does not specifically inhibit their apical efflux. Biochem Pharmacol 95, 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgeman A, Maelfait J, Davenne T, Partridge T, Peng Y, Mayer A, Dong T, Kaever V, Borrow P, and Rehwinkel J (2015). Viruses transfer the antiviral second messenger cGAMP between cells. Science 349, 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, and van Steensel B (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, and Stetson DB (2012). Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med 209, 1969–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh K, Maltbaek JH, Cambier S, Green R, Gale M Jr., James RC, and Stetson DB (2020). Human DNA-PK activates a STING-independent DNA sensing pathway. Sci Immunol 5, eaba4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Zhang D, Schaffer DE, Iyer LM, and Aravind L (2015). Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res 43, 10633–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carozza JA, Bohnert V, Nguyen KC, Skariah G, Shaw KE, Brown JA, Rafat M, von Eyben R, Graves EE, Glenn JS, et al. (2020). Extracellular cGAMP is a cancer cell-produced immunotransmitter involved in radiation-induced anti-cancer immunity. Nat Cancer 1, 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee YC, Pahnke J, Bunte R, Adsool VA, Madan B, and Virshup DM (2018). Intrinsic Xenobiotic Resistance of the Intestinal Stem Cell Niche. Dev Cell 46, 681–695 e685. [DOI] [PubMed] [Google Scholar]
- Cole SP (2014). Targeting multidrug resistance protein 1 (MRP1, ABCC1): past, present, and future. Annu Rev Pharmacol Toxicol 54, 95–117. [DOI] [PubMed] [Google Scholar]
- Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, and Deeley RG (1992). Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 258, 1650–1654. [DOI] [PubMed] [Google Scholar]
- Cordova AF, Ritchie C, Bohnert V, and Li L (2021). Human SLC46A2 Is the Dominant cGAMP Importer in Extracellular cGAMP-Sensing Macrophages and Monocytes. ACS Cent Sci 7, 1073–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. (2006). Mutations in the gene encoding the 3’−5’ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet 38, 917–920. [DOI] [PubMed] [Google Scholar]
- Crowl JT, Gray EE, Pestal K, Volkman HE, and Stetson DB (2017). Intracellular Nucleic Acid Detection in Autoimmunity. Annu Rev Immunol 35, 313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, and Annilo T (2005). Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet 6, 123–142. [DOI] [PubMed] [Google Scholar]
- Dean M, Rzhetsky A, and Allikmets R (2001). The human ATP-binding cassette (ABC) transporter superfamily. Genome Res 11, 1156–1166. [DOI] [PubMed] [Google Scholar]
- Fink SL, and Cookson BT (2005). Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun 73, 1907–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M Jr., Barber GN, and Stetson DB (2012). Autoimmunity Initiates in Nonhematopoietic Cells and Progresses via Lymphocytes in an Interferon-Dependent Autoimmune Disease. Immunity 36, 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallman AE, Wolfreys FD, Nguyen DN, Sandy M, Xu Y, An J, Li Z, Marson A, Lu E, and Cyster JG (2021). Abcc1 and Ggt5 support lymphocyte guidance through export and catabolism of S-geranylgeranyl-l-glutathione. Sci Immunol 6, eabg1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, and Chen ZJ (2015). Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci U S A 112, E5699–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Cui HR, Loe DW, Grant CE, Almquist KC, Cole SP, and Deeley RG (2000). Comparison of the functional characteristics of the nucleotide binding domains of multidrug resistance protein 1. J Biol Chem 275, 13098–13108. [DOI] [PubMed] [Google Scholar]
- Gentili M, Kowal J, Tkach M, Satoh T, Lahaye X, Conrad C, Boyron M, Lombard B, Durand S, Kroemer G, et al. (2015). Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science 349, 1232–1236. [DOI] [PubMed] [Google Scholar]
- Gey GO, Coffman WD, and Kubicek MT (1952). Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Research 12, 264–265. [Google Scholar]
- Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, and Parks WP (1973). In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst 51, 1417–1423. [DOI] [PubMed] [Google Scholar]
- Goubau D, Deddouche S, and Reis ESC (2013). Cytosolic sensing of viruses. Immunity 38, 855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Treuting PM, Woodward JJ, and Stetson DB (2015). Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutieres Syndrome. J Immunol 195, 1939–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Winship D, Snyder JM, Child SJ, Geballe AP, and Stetson DB (2016). The AIM2-like Receptors Are Dispensable for the Interferon Response to Intracellular DNA. Immunity 45, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui X, Yang H, Li T, Tan X, Shi P, Li M, Du F, and Chen ZJ (2019). Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kotova E, Chen ZS, Lee K, Hopper-Borge E, Belinsky MG, and Kruh GD (2003). MRP8, ATP-binding cassette C11 (ABCC11), is a cyclic nucleotide efflux pump and a resistance factor for fluoropyrimidines 2’,3’-dideoxycytidine and 9’-(2’-phosphonylmethoxyethyl)adenine. J Biol Chem 278, 29509–29514. [DOI] [PubMed] [Google Scholar]
- Higgins CF (1992). ABC transporters: from microorganisms to man. Annu Rev Cell Biol 8, 67–113. [DOI] [PubMed] [Google Scholar]
- Holleufer A, Winther KG, Gad HH, Ai X, Chen Y, Li L, Wei Z, Deng H, Liu J, Frederiksen NA, et al. (2021). Two cGAS-like receptors induce antiviral immunity in Drosophila. Nature 597, 114–118. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, and Fitzgerald KA (2009). AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, and Barber GN (2009). STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlitschky G, Burchell B, and Keppler D (2000). The multidrug resistance protein 5 functions as an ATP-dependent export pump for cyclic nucleotides. J Biol Chem 275, 30069–30074. [DOI] [PubMed] [Google Scholar]
- Johnson ZL, and Chen J (2017). Structural Basis of Substrate Recognition by the Multidrug Resistance Protein MRP1. Cell 168, 1075–1085 e1079. [DOI] [PubMed] [Google Scholar]
- Kato K, Nishimasu H, Oikawa D, Hirano S, Hirano H, Kasuya G, Ishitani R, Tokunaga F, and Nureki O (2018). Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1. Nat Commun 9, 4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler D, Cui Y, Konig J, Leier I, and Nies A (1999). Export pumps for anionic conjugates encoded by MRP genes. Adv Enzyme Regul 39, 237–246. [DOI] [PubMed] [Google Scholar]
- Kranzusch PJ, Wilson SC, Lee AS, Berger JM, Doudna JA, and Vance RE (2015). Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2’,3’ cGAMP Signaling. Mol Cell 59, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahey LJ, Mardjuki RE, Wen X, Hess GT, Ritchie C, Carozza JA, Bohnert V, Maduke M, Bassik MC, and Li L (2020). LRRC8A:C/E Heteromeric Channels Are Ubiquitous Transporters of cGAMP. Mol Cell 80, 578–591 e575. [DOI] [PubMed] [Google Scholar]
- Leier I, Jedlitschky G, Buchholz U, Cole SP, Deeley RG, and Keppler D (1994). The MRP gene encodes an ATP-dependent export pump for leukotriene C4 and structurally related conjugates. J Biol Chem 269, 27807–27810. [PubMed] [Google Scholar]
- Li L, Yin Q, Kuss P, Maliga Z, Millan JL, Wu H, and Mitchison TJ (2014). Hydrolysis of 2’3’-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat Chem Biol 10, 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, and Chen ZJ (2018). The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 215, 1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, et al. (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630. [DOI] [PubMed] [Google Scholar]
- Lorico A, Rappa G, Finch RA, Yang D, Flavell RA, and Sartorelli AC (1997). Disruption of the murine MRP (multidrug resistance protein) gene leads to increased sensitivity to etoposide (VP-16) and increased levels of glutathione. Cancer Res 57, 5238–5242. [PubMed] [Google Scholar]
- Lowey B, Whiteley AT, Keszei AFA, Morehouse BR, Mathews IT, Antine SP, Cabrera VJ, Kashin D, Niemann P, Jain M, et al. (2020). CBASS Immunity Uses CARF-Related Effectors to Sense 3’−5’- and 2’−5’-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection. Cell 182, 38–49 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn RD, Zaver SA, Gowen BG, Wyman SK, Garelis NE, Onia L, McWhirter SM, Katibah GE, Corn JE, Woodward JJ, et al. (2019). SLC19A1 transports immunoreactive cyclic dinucleotides. Nature 573, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelfait J, Liverpool L, and Rehwinkel J (2020). Nucleic Acid Sensors and Programmed Cell Death. J Mol Biol 432, 552–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus A, Mao AJ, Lensink-Vasan M, Wang L, Vance RE, and Raulet DH (2018). Tumor-Derived cGAMP Triggers a STING-Mediated Interferon Response in Non-tumor Cells to Activate the NK Cell Response. Immunity 49, 754–763 e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P, Oskeritzian CA, Payne SG, Beaven MA, Milstien S, and Spiegel S (2006). Role of ABCC1 in export of sphingosine-1-phosphate from mast cells. Proc Natl Acad Sci U S A 103, 16394–16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morehouse BR, Govande AA, Millman A, Keszei AFA, Lowey B, Ofir G, Shao S, Sorek R, and Kranzusch PJ (2020). STING cyclic dinucleotide sensing originated in bacteria. Nature 586, 429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, and Breaker RR (2017). The lost language of the RNA World. Sci Signal 10, eaam8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco S, Yatim N, Werner MR, Tran H, Gunja SY, Tait SW, Albert ML, Green DR, and Oberst A (2014). RIPK1 both positively and negatively regulates RIPK3 oligomerization and necroptosis. Cell Death Differ 21, 1511–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prechtl S, Roellinghoff M, Scheper R, Cole SP, Deeley RG, and Lohoff M (2000). The multidrug resistance protein 1: a functionally important activation marker for murine Th1 cells. J Immunol 164, 754–761. [DOI] [PubMed] [Google Scholar]
- Reid G, Wielinga P, Zelcer N, De Haas M, Van Deemter L, Wijnholds J, Balzarini J, and Borst P (2003). Characterization of the transport of nucleoside analog drugs by the human multidrug resistance proteins MRP4 and MRP5. Mol Pharmacol 63, 1094–1103. [DOI] [PubMed] [Google Scholar]
- Ritchie C, Carozza JA, and Li L (2022). Biochemistry, Cell Biology, and Pathophysiology of the Innate Immune cGAS-cGAMP-STING Pathway. Annu Rev Biochem 91, 599–628. [DOI] [PubMed] [Google Scholar]
- Ritchie C, Cordova AF, Hess GT, Bassik MC, and Li L (2019). SLC19A1 Is an Importer of the Immunotransmitter cGAMP. Mol Cell 75, 372–381 e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, and Gottesman MM (2018). Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer 18, 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safa AR (1988). Photoaffinity labeling of the multidrug-resistance-related P-glycoprotein with photoactive analogs of verapamil. Proc Natl Acad Sci U S A 85, 7187–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, and Gale M Jr. (2008). Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavik KM, Morehouse BR, Ragucci AE, Zhou W, Ai X, Chen Y, Li L, Wei Z, Bahre H, Konig M, et al. (2021). cGAS-like receptors sense RNA and control 3’2’-cGAMP signalling in Drosophila. Nature 597, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, and Medzhitov R (2008). Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, and Chen ZJ (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YL, Patel A, Kumar P, and Chen ZS (2012). Role of ABC transporters in cancer chemotherapy. Chin J Cancer 31, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmori D, Fonteneau JF, Lizana CM, Gervois N, Lienard D, Rimoldi D, Jongeneel V, Jotereau F, Cerottini JC, and Romero P (1998). Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J Immunol 160, 1750–1758. [PubMed] [Google Scholar]
- Vasiliou V, Vasiliou K, and Nebert DW (2009). Human ATP-binding cassette (ABC) transporter family. Hum Genomics 3, 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman HE, Cambier S, Gray EE, and Stetson DB (2019). Tight nuclear tethering of cGAS is essential for preventing autoreactivity. Elife 8, e47491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner LD, Zoghbi SS, Lu S, Shukla S, Ambudkar SV, Pike VW, Mulder J, Gottesman MM, Innis RB, and Hall MD (2015). The Inhibitor Ko143 Is Not Specific for ABCG2. J Pharmacol Exp Ther 354, 384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AT, Eaglesham JB, de Oliveira Mann CC, Morehouse BR, Lowey B, Nieminen EA, Danilchanka O, King DS, Lee ASY, Mekalanos JJ, et al. (2019). Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, and Portnoy DA (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, and Chen ZJ (2013). Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Chen X, Planells-Cases R, Chu J, Wang L, Cao L, Li Z, Lopez-Cayuqueo KI, Xie Y, Ye S, et al. (2020a). Transfer of cGAMP into Bystander Cells via LRRC8 Volume-Regulated Anion Channels Augments STING-Mediated Interferon Responses and Anti-viral Immunity. Immunity 52, 767–781 e766. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Fei M, Zhang G, Liang WC, Lin W, Wu Y, Piskol R, Ridgway J, McNamara E, Huang H, et al. (2020b). Blockade of the Phagocytic Receptor MerTK on Tumor-Associated Macrophages Enhances P2X7R-Dependent STING Activation by Tumor-Derived cGAMP. Immunity 52, 357–373 e359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2: Related to STAR Methods. Sequences of quantitative RT-PCR primers used in this study
Table S1: Related to STAR Methods. Sequences of guide RNAs used in this study
Data Availability Statement
This paper does not report original code
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.