Abstract
Drug-induced liver injury (DILI) and cardiotoxicity (DICT) are major adverse effects triggered by many clinically important drugs. To provide an alternative to in vivo toxicity testing, the U.S. Tox21 consortium has screened a collection of ~10K compounds, including drugs in clinical use, against >70 cell-based assays in a quantitative high-throughput screening (qHTS) format. In this study, we compiled reference compound lists for DILI and DICT and compared the potential of Tox21 assay data with chemical structure information in building prediction models for human in vivo hepatotoxicity and cardiotoxicity. Models were built with four different machine learning algorithms (e.g., Random Forest, Naïve Bayes, eXtreme Gradient Boosting, and Support Vector Machines) and model performance was evaluated by calculating the area under the receiver operating characteristic curve (AUC-ROC). Chemical structure-based models showed reasonable predictive power for DILI (best AUC-ROC = 0.75±0.03) and DICT (best AUC-ROC = 0.83±0.03), while Tox21 assay data alone only showed better than random performance. DILI and DICT prediction models built using a combination of assay data and chemical structure information did not have a positive impact on model performance. The suboptimal predictive performance of the assay data is likely due to insufficient coverage of an adequately predictive number of toxicity mechanisms. The Tox21 consortium is currently expanding coverage of biological response space with additional assays that probe toxicologically important targets and under-represented pathways that may improve the prediction of in vivo toxicity such as DILI and DICT.
Keywords: Liver injury, Hepatotoxicity, Cardiotoxicity, In vitro assay, High-throughput screening, Tox21
Introduction
Modern drug development is aimed at producing compounds that maximize therapeutic benefits while also minimizing adverse effects. Despite promising results from preclinical studies in animal and cell models, more than 30% of small molecules fail in clinical trials because they are found to be harmful to human health.1 Furthermore, 80% of drug candidates fail in human clinical trials due to unmanageable toxicity or lack of clinical efficacy.1 Drug-induced liver injury (DILI) and cardiotoxicity (DICT) are major concerns during the safety profile evaluation of current drugs and development of novel therapeutics. The liver is responsible for a wide range of functions, including xenobiotic detoxification, protein synthesis, storage and synthesis of glucose, production of the bile necessary for digestion, and regulation of blood cholesterol and triglycerides. Due to its central role in biotransformation and excretion of foreign compounds, the liver represents a primary target for adverse drug reactions. Many drugs and environmental chemicals can evoke some degree of liver injury,2,3 making DILI the single most common adverse indication, thus leading to drug candidate failure and/or withdrawal from the consumer market. However, DILI is largely unpredictable due to complex factors that give rise to liver damage, hence making prevention difficult.4 Moreover, current in vivo toxicological studies are insufficient to assess the hepatotoxic potential of compounds early in the drug development process, thereby presenting an urgent need for alternative DILI prediction strategies.5
Likewise, DICT is another major safety concern and common cause of drug withdrawals from the consumer market.6 The NIH National Cancer Institute (NCI) broadly defines cardiotoxicity as “toxicity that affects the heart”,7 and major cardiovascular adverse effects may include tachycardia, hypertension, and electrocardiographic abnormalities such as prolonged QT interval. Many antineoplastic agents have been linked to cardiovascular toxicity with extended use.8 In addition, cardiotoxicity has been observed in a diverse range of drug classes such as antipsychotics, antidepressants, and antibiotics.9 Like DILI, the mechanisms that underly DICT are not fully understood, and effective predictive approaches are critical for reducing chemical-induced cardiotoxicity.
Quantitative structure-activity relationship (QSAR) modeling and machine learning methods have become increasingly popular for predicting compound properties such as toxicity.10 To date, QSAR modeling has been widely used in liver toxicity research to study hepatotoxicity and predict DILI. While a number of hepatotoxicity prediction models have recently been developed, they often suffer from imbalanced and/or limited training/testing datasets that produce unsatisfactory predictive performance.11 Consequently, the applicability of these models is often limited due to their insufficient coverage of the chemical space. The etiopathogenesis of DILI is complex and involves the interaction of several factors, so the limited DILI data available in humans is a major drawback for QSAR modeling.12 Similarly, QSAR approaches have been conducted to predict DICT compounds. However, the performance from these cardiotoxicity models varies widely because they are heavily influenced by the type of molecules and techniques used for model building.13 Most previously published models use in-house data or proprietary data obtained from industry that are not publicly available, while a limited number of other studies that use data extracted from public databases often suffer from a certain amount of experimental uncertainty.14 Collectively, these factors compromise the practical use of prior models for reliable assessment of cardiotoxicity. Small model training datasets and lack of proof of validation are additional factors that limit the usefulness of prior models.
To improve toxicity testing and prediction, the Toxicology in the 21st Century (Tox21) consortium was established as a federal collaboration between the National Center for Advancing Translational Sciences (NCATS), the National Toxicology Program (NTP) of the National Institute of Environmental Health Sciences (NIEHS), the Environmental Protection Agency (EPA), and the Food and Drug Administration (FDA).15–17 The aim of Tox21 is to develop alternative toxicity assessment methods to quickly and efficiently predict potential adverse effects of chemicals on human health. Tox21 employs quantitative high-throughput screening (qHTS), an automated robotic process in which each compound of a large chemical library is tested at multiple concentrations, to test large collections of chemicals in multiple cell-based assays. A Tox21 screening library comprised of approximately 10,000 chemical samples, also known as the “Tox21 10K compound library”,18 has been screened for potential biological pathway disruptions that may result in toxicity.19,20 The Tox21 10K compound library contains industrial and consumer products, food additives, drugs, and chemical mixtures, and also includes the NCATS Pharmaceutical Collection (NPC),21,22 a collection of approximately 3,000 small molecule drugs approved for clinical use or investigational purposes by the U.S., European, Japanese, Australian, and Canadian authorities. The Tox21 10K compound library has been screened in more than 70 cell-based assays in qHTS format related to nuclear receptor and stress signaling pathways, and also a smaller number of assays that probe for genotoxicity, developmental toxicity, G-protein coupled receptors, and cell-death signaling. To date, the screening has generated nearly 102 million data points that are publicly available to the scientific community.23,24
In this study, we aim to build machine learning models to predict DILI and DICT. To develop robust models for DILI and DICT prediction, a comprehensive set of human in vivo toxicity data is essential for model training and testing. We compiled reference lists of DILI and DICT compounds by incorporating in vivo toxicity data integrated from a diverse set of literature sources. Both chemical structure and assay data were used as descriptors to build the DILI and DICT prediction models. These two types of descriptors were tested both independently and in combination, and the resulting model performances were assessed. Accordingly, we identified assay targets and pathways as well as chemical features that contributed the most to model performance. The chemical structural features and targets that are significantly associated with liver toxicity and/or cardiotoxicity can be further investigated as potential indicators to help us better understand the underlying pathways and mechanisms of these toxicities.
Materials and methods
In vivo toxicity data
To compile a comprehensive list of reference compounds for DILI and DICT, data were integrated from six sources including ChemIDplus (DILI and DICT), Pharmapendium® (DILI and DICT), U.S. Food and Drug Administration (FDA)’s Center for Drug Evaluation and Research (CDER) (DICT) and National Center for Toxicological Research (NCTR) (DILI and DICT),25 Enzo Life Sciences cardiotoxicity library (DICT), and the Side Effect Resource (SIDER) database26 via the European Molecular Biology Laboratory (EMBL) (DICT). ChemIDplus is a database produced by the U.S. National Library of Medicine (NLM) that collects compound records (chemical nomenclature, properties, toxicity, and structures) from more than 100 sources. Pharmapendium® is a curated database containing extensive information on adverse drug effects extracted from the FDA and European Medicines Agency (EMA) drug approval documents. The Enzo Life Sciences cardiotoxicity library is a collection of 130 diverse compounds with known cardiotoxicity, including ion channel blockage, mitochondrial toxicity, arrhythmia, and fibrosis. SIDER contains information on marketed medicines and the associated adverse drug reactions that were extracted from public documents and package inserts. A total of 474 drugs that did not have any cardiotoxicity reports (244 overlapped with the Tox21 10K compound library) were collected from the SIDER database, and these compounds were included as negative controls in our DICT reference list.
In vivo toxicity data scoring and cutoff for DILI and DICT
For the reference lists collected for DILI and DICT, each compound was assigned a toxicity score according to the frequency in which it was identified as “toxic” in the literature. First, within each data source, the compounds were each assigned a value between 0 and 1 as specified in the following. For compounds retrieved from Pharmapendium® and ChemIDplus, different types (e.g., via different mechanisms or manifestations of toxicity) of DILI and/or DICT were normally reported. Compounds with 10 or more such reports were assigned a value of 1 and others were assigned the value N/10, where N is the number of toxicity reports. For the DILI and DICT lists provided by NCTR, compounds with the most toxicity concern were assigned a value of 1, compounds with less toxicity concern were assigned a value of 0.5, and compounds with no toxicity concern were assigned a value of 0. For compounds from the other data sources, compounds listed as toxic were assigned a value of 1 and 0 otherwise. Finally, the values from all data sources were averaged for each compound to yield the final DILI or DICT score, such that a larger score indicates a higher likelihood of toxicity and 0 means non-toxic.
A total of 1,407 compounds were collected from three different sources for the DILI reference list (Supplementary Table 1). The best model performance for DILI was obtained when using 0.4 as the cutoff value to separate the data into toxic and non-toxic binary classes, i.e., a compound was considered toxic if its DILI score was > 0.4. Other compounds on the list were considered non-toxic. A total of 1,160 compounds were collected from six sources for the DICT reference list (Supplementary Table 2). Different DICT score cutoffs were tested, and the following conditions produced the best model performance for DICT prediction. Compounds with a DICT score of 0 were considered non-toxic, while compounds with a score ≥ 0.5 were considered toxic. Any compounds with a DICT score between 0 and 0.5 were considered inconclusive and removed from the modeling set.
In vitro assay data
qHTS data used for modeling in this study were generated by screening the Tox21 10K compound library, including 70 assays with 203 readouts. All in vitro assay data and detailed assay descriptions used in this study are publicly available on the NCATS website (https://tripod.nih.gov/tox21/pubdata/) and PubChem.27,28 The following cell lines were used for the qHTS assays: human cell lines (80.88%), murine embryo fibroblast (7.35%), Chinese hamster ovary cell lines (5.88%), and other (5.88%). The majority of these assays cover several pathways related to nuclear receptor signaling (NR, 55.90%), stress response (SR, 11.80%), cytotoxicity (8.80%), and other targets/pathways related to toxicity (23.50%). Curve rank is a value between −9 and 9 that was used as a measure of compound activity, such that a positive number represents activation while a negative number represents inhibition.29 For modeling purposes, compounds with absolute curve rank > 0.5 were labeled as active (1), and inactive (0) otherwise.
Structure data
To build classification models in this study, two structure-based fingerprint sets were used: ToxPrint and ECFP4. Publicly available Toxprint chemotypes (v2.0_r711, https://toxprint.org/) generated within the associated ChemoTyper application (https://chemotyper.org/) consists of 729 uniquely defined chemical features.30 Extended-Connectivity Fingerprints (ECFP4) is a 1024-bit fingerprint set that was generated using the CDK package in KNIME v.4.0.2.25.31 Each bit is representative of a structural feature and was designated a value of 1 for the presence of a particular feature and 0 otherwise.
Feature selection
To identify assays significantly related to DILI or DICT, each assay was used to predict hepatotoxicity and cardiotoxicity, and these assays can be viewed as single-descriptor models. ToxPrint chemotypes were used to identify chemical structural features significantly enriched in DILI and DICT compounds. Statistical significance was measured by the Fisher’s exact test with p < 0.05 considered as significant.
Feature selection using the Fisher’s exact test was used to further optimize model performance and determine features (chemical features or assays) significant for DILI and DICT prediction. Ten independent p-values ranging from 0.01 to 0.1 were used to select features at different p-value cutoffs to train models. The best performing feature sets were selected to build the final model. The top feature sets were further combined to rebuild the model to determine if the combined feature set achieved better model performance than that of the structure or assay feature set alone.
Prediction modeling
In this study, a total of 1,407 unique compounds were used to build the DILI prediction models, including 831 non-toxic compounds and 578 toxic compounds, for chemical structure-based modeling (ToxPrint or ECFP4, Supplementary Table 1). To obtain a balanced modeling set, a subset of toxic compounds with a size roughly equal to the set of non-toxic compounds was randomly selected from the original dataset. The modeling set was further randomly split into two sets, 70% for training and 30% for model validation, and this process was repeated 100 times. For assay activity-based models (assay-only), compounds with complete activity profiles in the 70 assays (213 readouts), including 663 non-toxic compounds and 466 toxic compounds, were used for model training and validation. The same dataset and modeling procedure were repeated for the chemical structure (structure-only) and assay activity combined (structure + assay) models for DILI.
A total of 646 unique compounds were used to build DICT prediction models, including 244 non-toxic compounds and 402 toxic compounds, for chemical structure-based modeling (ToxPrint or ECFP4, Supplementary Table 2). For assay activity-based models, compounds with complete activity profiles in the 70 assays (213 readouts), including 209 non-toxic compounds and 335 toxic compounds, were used for model training and validation. The same dataset and procedure were repeated for the structure + assay combined models for DICT.
Four different machine learning classification algorithms were applied: Random Forest (RF), Naïve Bayes (NB), eXtreme Gradient Boosting (XGBoost), and Support Vector Machines (SVM). Models were built and tested using R version 4.1.2, with the “Random Forest” package for the RF classifier, the “e1071” package for the NB and SVM classifiers, and the “xgboost” package for the XGBoost classifier. The implementation of the NB classifier was adapted with the settings of Laplace smoothing, and the Gaussian Radial Basis Function kernel was used for the SVM classifier. In addition, the optimal parameters for the SVM and RF classifiers were selected using the “e1071” package. The R codes used in this study are publicly available on the GitHub repository at https://github.com/TX-2017/machine-learning.
Model performance was evaluated by calculating the area under the receiver operating characteristic curve (AUC-ROC). The ROC curve is a graphical plot that illustrates the predictive ability of a binary classification model across different thresholds. The ROC curve is created by plotting true positive rates against false positive rates at various thresholds. The area under the curve (AUC) provides an aggregate measure of model performance. A larger AUC value indicates better classifier performance. A perfect predictive model would have an AUC score of 1, while an AUC score of 0.5 indicates a random classifier. For feature selection, assays with AUC-ROC scores greater than 0.5 were considered to be predictive of toxicity and retained for further analysis.
To assess the applicability domain (AD) of the structure-based models, the closest structural neighbor in the training set was found for each of the compounds to be predicted, e.g., the Tox21 10K library. Structural similarity was determined by calculating the Tanimoto coefficient using ECFP4 fingerprints, which measures the similarity between two compounds based on the number of shared structural features. It is calculated by taking the number of structural features in common by both compounds and dividing it by the total number of structural features in either compound. The Taminoto coefficient ranges from 0, where the compounds have no chemotypes in common, to 1, when the compounds are identical. A compound that has a close structural neighbor in the training set with a Tanimoto similarity (Tmax) ≥ 0.4 was considered to be within the model’s AD.
Results
Model performance
The modeling results for both DILI and DICT are summarized in Table 1.
Table 1.
Performance of four classification models (RF, NB, XGBoost, and SVM) for each toxicity endpoint (DILI and DICT) with feature selection.
| In vivo Endpoints | Descriptor | RF | NB | XGBoost | SVM |
|---|---|---|---|---|---|
| DILI | ToxPrint | 0.68±0.03 | 0.66±0.03 | 0.67±0.03 | 0.65±0.03 |
| Assay | 0.59±0.02 | 0.61±0.04 | 0.59±0.03 | 0.59±0.03 | |
| ToxPrint + Assay | 0.67±0.02 | 0.63±0.03 | 0.66±0.03 | 0.65±0.02 | |
| ECFP4 | 0.75±0.03 | 0.74±0.02 | 0.71±0.03 | 0.72±0.03 | |
| ECFP4 + Assay | 0.72±0.03 | 0.72±0.02 | 0.68±0.03 | 0.70±0.03 | |
| DICT | ToxPrint | 0.72±0.03 | 0.75±0.02 | 0.70±0.04 | 0.71±0.04 |
| Assay | 0.58±0.04 | 0.58±0.04 | 0.57±0.04 | 0.56±0.04 | |
| ToxPrint + Assay | 0.69±0.04 | 0.66±0.04 | 0.67±0.04 | 0.68±0.04 | |
| ECFP4 | 0.79±0.04 | 0.83±0.03 | 0.73±0.04 | 0.78±0.03 | |
| ECFP4 + Assay | 0.73±0.03 | 0.71±0.06 | 0.71±0.03 | 0.74±0.04 |
The DILI modeling results showed that liver toxicity could be reasonably predicted by chemical structure, with ECFP4 demonstrating the best predictive performance. While the AUC-ROC scores of the ToxPrint-based models ranged from 0.65±0.03 to 0.68±0.03, ECFP4-based models performed better as shown by the scores ranging from 0.71±0.03 to 0.75±0.03. The AUC-ROC scores of the assay-based models ranged from 0.59±0.02 to 0.61±0.04, which were much lower than both chemical structure-based models. The AUC-ROC scores (0.63±0.03 to 0.72±0.03) of the models built on both chemical structure (Toxprint or ECFP4) and assay data showed that the addition of assay data did not improve model performance compared to chemical structure-based models alone.
Likewise, the application of chemical structure could also be used to predict cardiotoxicity as shown by the modeling results from DICT prediction. The AUC-ROC score of the ECFP4-based model ranged from 0.73±0.04 to 0.83±0.03, while the AUC-ROC score of the ToxPrint-based model ranged from 0.70±0.04 to 0.75±0.02. Similarly, the AUC-ROC scores of the assay-based models were relatively low, scoring within the range of 0.56±0.04 to 0.58±0.04. The AUC-ROC of the models built using both chemical structure (Toxprint or ECFP4) and assay data revealed that the addition of assay data did not improve the DICT model performance, suggesting that chemical structure alone was the better predictor of hepatotoxicity and cardiotoxicity. In both DICT and DILI models, ECFP4 fingerprints produced better model performance than ToxPrint fingerprints.
Feature selection was used to optimize model performance. Ten different p-value cutoffs between the range of 0.01 and 0.1 were used for feature selection, and the best cutoff value was selected for each method. Comparing the model performance before (Supplementary Table 3) and after (Table 1) application of feature selection, we observed that the impact of feature selection on model performance varied across the different descriptor types and machine learning methods. For DILI prediction, feature selection for NB with ECFP4 exhibited the greatest AUC-ROC score increase of 0.15, improving from 0.60±0.03 to 0.75±0.03. Likewise, feature selection for NB with ECFP4 substantially improved DICT prediction by an AUC-ROC score by 0.13, increasing from 0.70±0.03 to 0.83±0.03 (Table 1; Supplementary Table 3). Overall, feature selection helped to boost model performance for both DILI and DICT prediction. We observed modest improvements for RF after application of feature selection, which is likely due to the built-in feature selection already implemented in this method.
Toxic structural feature identification for DILI and DICT compounds
The ToxPrint chemotypes were used to identify chemical features that are significantly (Fisher’s exact test) enriched in toxic compounds. Several distinct features were present in hepatotoxic compounds (Figure 1), particularly aromatic rings such as bond:N(=O)_nitro_aromatic (4.1×10−2), ring:hetero_[6]_N_piperidine (4.95×10−6), and ring:hetero_[4]_N_beta_lactam (1.60×10−3).
Figure 1.
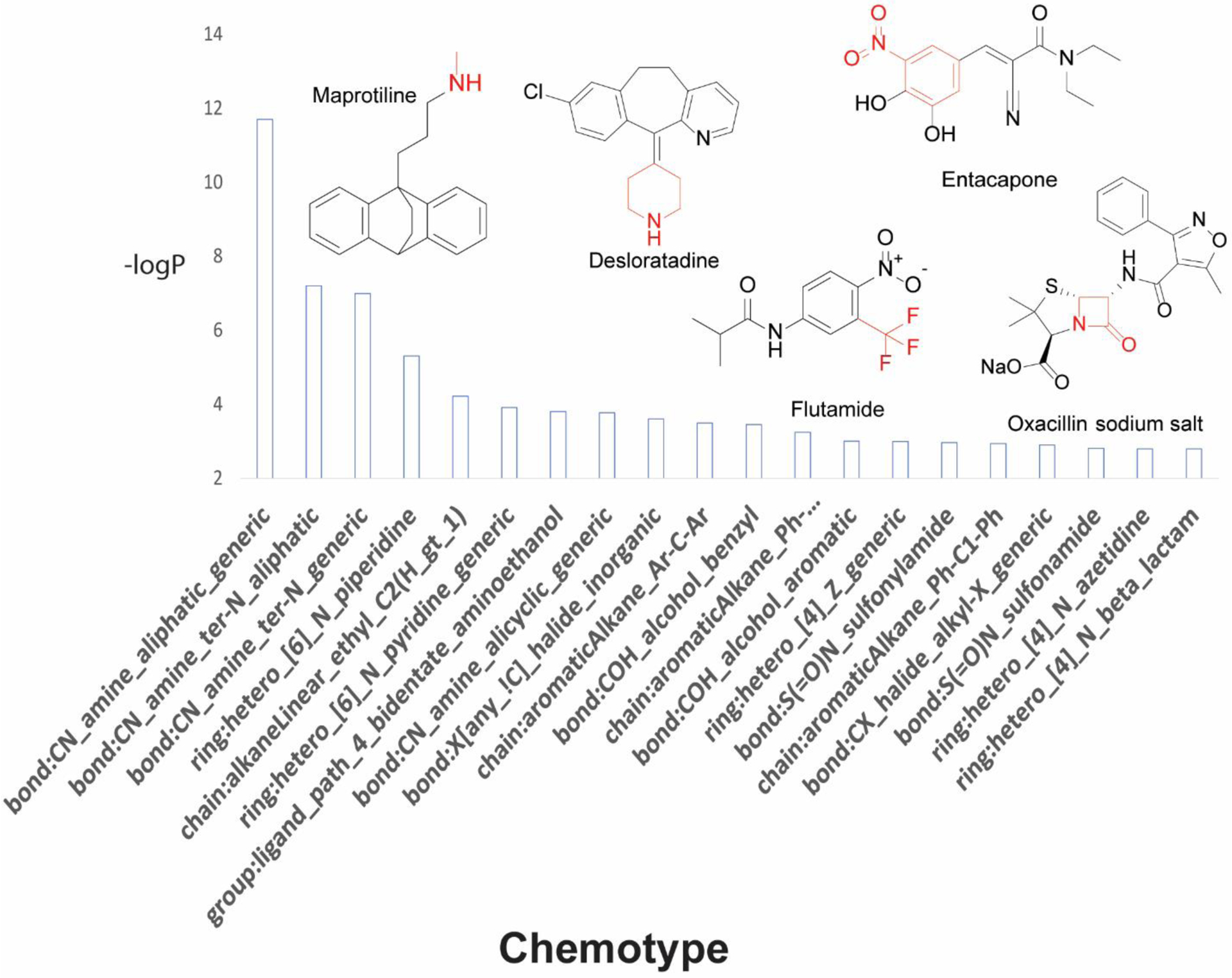
Chemotypes significantly enriched in compounds active for DILI. Chemotypes are sorted by significance based on p-values. Representative drugs that contain these structural fragments are displayed with significant chemotypes highlighted in red.
Moreover, small chemical fragments such as bond:CX_halide_alkyl-X_trihalo_(1_1_1-) (6.22×10−3) and bond:CN_amine_aliphatic_generic (2.00×10−12) were found enriched in DILI compounds. For cardiotoxic compounds (Figure 2), we also observed the presence of six-membered rings including ring:hetero_[6]_Z_1- (4.35×10−6), ring:hetero_[6]_N_piperazine (3.49×10−4), ring:hetero_[6]_N_pyridine (2.60×10−2), and bond:CC(=O)C_quinone_1_4-benzo (2.75×10−2).
Figure 2.
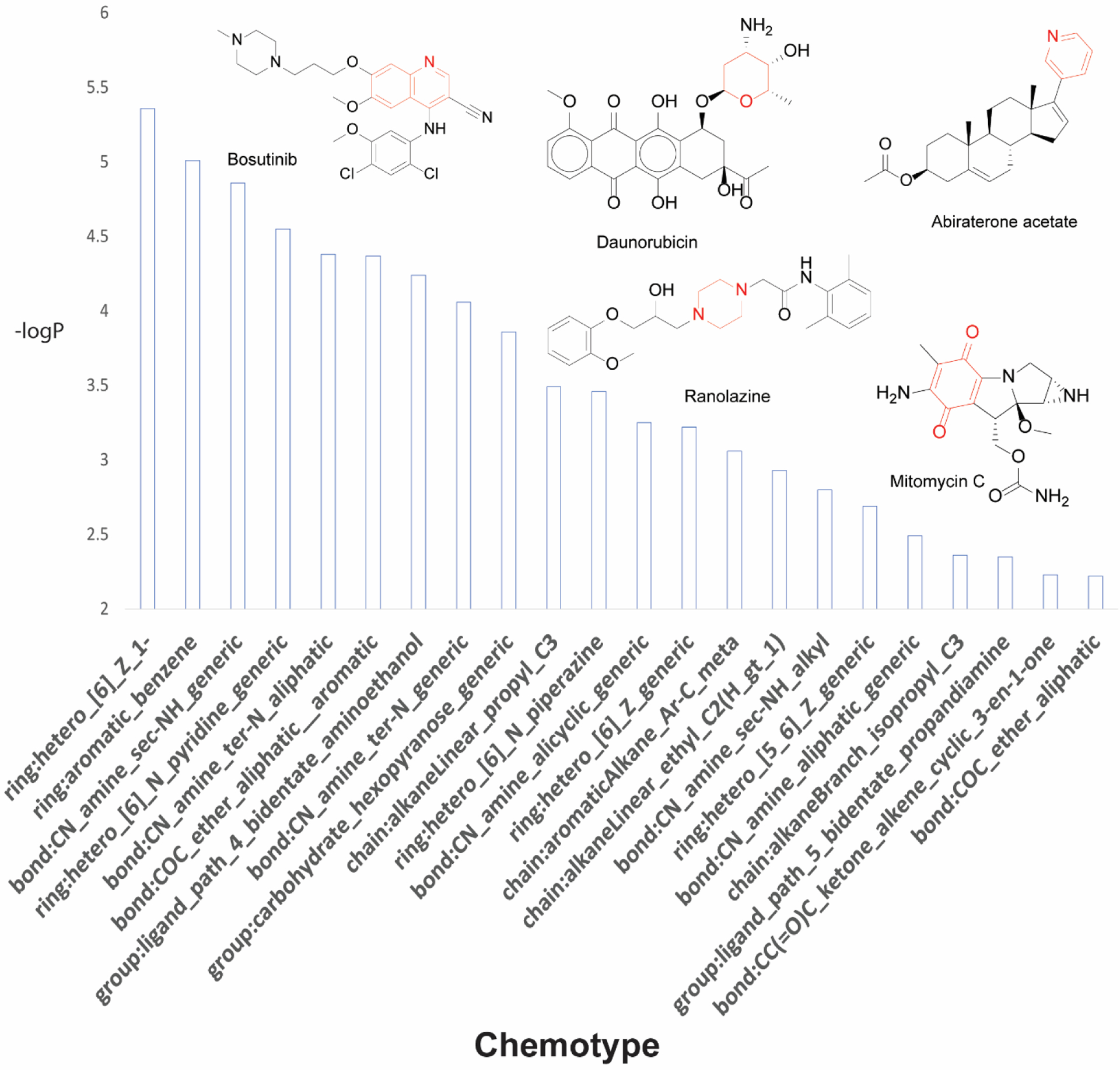
Chemotypes significantly enriched in compounds active for DICT. Chemotypes are sorted by significance based on p-values. Representative drugs that contain these structural fragments are displayed with significant chemotypes highlighted in red.
In addition, the double-ring structure ring:hetero_[6_6]_N_quinoline (1.62×10−2) was identified in cardiotoxic compounds. The most significant chemotypes identified for both hepatotoxic and cardiotoxic compounds are presented in Supplementary Tables 4 and 5, respectively.
Assays predictive of DILI and DICT
We examined the performance of each assay within the Tox21 assay panel in predicting DILI and DICT. The top five assays that are the most predictive of DILI measured by their AUC-ROC scores, in descending order of significance, are: tox21-p450-2c9-p1_ratio (0.60), tox21-ror-cho-antagonist-p1_ratio (0.58), tox21-ar-bla-agonist-p1_ratio (0.57), tox21-gr-hela-bla-antagonist-p1_ratio (0.57), and tox21-er-bla-antagonist-p1_ch2 (0.57). Likewise, the top five assays for predicting DICT and their corresponding AUC-ROC scores, in descending order of significance, are as follows: tox21-pparg-bla-agonist-p1_ratio (0.60), tox21-casp3-cho-p1_viability (0.60), tox21-ahr-p1_viability (0.59), tox21-erb-bla-antagonist-p1_ratio (0.59), and tox21-casp3-hepg2-p1_viability (0.59). A full list of the top 20 most predictive assays for DILI and DICT are shown in Tables 2 and 3, respectively.
Table 2.
Top 20 Tox21 assays that are predictive of DILI.
| # | DILI Assay | Target | AUC |
|---|---|---|---|
| 1 | tox21-er-bla-antagonist-p1_ch1 | ER-BLA antagonist viability | 0.57 |
| 2 | tox21-pxr-p1_ratio | PXR agonist | 0.57 |
| 3 | tox21-err-p1_agonist | ERR | 0.56 |
| 4 | tox21-mitotox-p1_rhodamine | Mitochondria toxicity | 0.56 |
| 5 | tox21-err-p1_ratio | ERR | 0.56 |
| 6 | tox21-are-bla-p1_ratio | ARE | 0.55 |
| 7 | tox21-casp3-cho-p1_viability | Caspase-3/7 cytotoxicity counter screen | 0.55 |
| 8 | tox21-mitotox-p1_ratio | Mitochondria toxicity | 0.55 |
| 9 | tox21-pr-bla-antagonist-p1_ch2 | PR-BLA antagonist | 0.55 |
| 10 | tox21-are-bla-p1_ch2 | ARE | 0.55 |
| 11 | tox21-p450-2c9-p1_ratio | CYP2C9 | 0.55 |
| 12 | tox21-rxr-bla-agonist-p1_ch1 | RXR-BLA control viability | 0.55 |
| 13 | tox21-pgc-err-p1_ratio | PGC-ERR | 0.54 |
| 14 | tox21-cre-antagonist-p1_ch2 | CRE antagonist | 0.54 |
| 15 | tox21-ar-bla-antagonist-p1_ratio | AR-BLA antagonist | 0.54 |
| 16 | tox21-car-antagonist-p1_ratio | CAR antagonist | 0.54 |
| 17 | tox21-pparg-bla-agonist-p1_ratio | PPARγ agonist | 0.54 |
| 18 | tox21-ahr-p1_ratio | AhR | 0.54 |
| 19 | tox21-cre-agonist-p1_ch1 | CRE control viability | 0.54 |
| 20 | tox21-cre-antagonist-p1_ratio | CRE antagonist | 0.54 |
Table 3.
Top 20 Tox21 assays that are predictive of DICT.
| # | DICT Assay | Target | AUC |
|---|---|---|---|
| 1 | tox21-pparg-bla-agonist-p1_ratio | PPARγ agonist | 0.60 |
| 2 | tox21-casp3-cho-p1_viability | Caspase-3/7 cytotoxicity counter screen | 0.60 |
| 3 | tox21-ahr-p1_viability | AhR cytotoxicity counter screen | 0.59 |
| 4 | tox21-erb-bla-antagonist-p1_ratio | ER-β antagonist | 0.59 |
| 5 | tox21-casp3-hepg2-p1_viability | Caspase-3/7 cytotoxicity counter screen | 0.59 |
| 6 | tox21-err-p1_viability | ERR cytotoxicity counter screen | 0.58 |
| 7 | tox21-elg1-luc-agonist-p1_viability | ATAD5 cytotoxicity counter screen | 0.58 |
| 8 | tox21-pparg-bla-antagonist-p1_ratio | PPARγ antagonist | 0.58 |
| 9 | tox21-gh3-tre-antagonist-p1_viability | TR-β antagonist cytotoxicity counter screen | 0.58 |
| 10 | tox21-dt40-p1_100 | Cell viability | 0.58 |
| 11 | tox21-pxr-p1_viability | PXR agonist cytotoxicity counter screen | 0.58 |
| 12 | tox21-shh-3t3-gli3-agonist-p1_viability | Hedgehog agonist cytotoxicity counter screen | 0.58 |
| 13 | tox21-shh-3t3-gli3-antagonist-p1_viability | Hedgehog antagonist cytotoxicity counter screen | 0.57 |
| 14 | tox21-aromatase-p1_viability | Aromatase cytotoxicity counter screen | 0.57 |
| 15 | tox21-mitotox-p1_rhodamine | Mitochondria toxicity | 0.57 |
| 16 | tox21-pr-bla-antagonist-p1_ratio | PR-BLA antagonist | 0.57 |
| 17 | tox21-vdr-bla-antagonist-p1_ratio | VDR-BLA antagonist | 0.57 |
| 18 | tox21-ppard-bla-agonist-p1_viability | PPAR-δ-BLA agonist cytotoxicity counter screen | 0.57 |
| 19 | tox21-pgc-err-p1_viability | PGC-ERR cytotoxicity counter screen | 0.57 |
| 20 | tox21-pparg-bla-antagonist-p1_ch1 | PPAR-δ-BLA antagonist control viability | 0.57 |
Predicting the DILI and DICT potential of the Tox21 10K compound library
We applied the ECFP-based RF, NB, XGBoost, and SVM models, which achieved good performance during model training and testing, to predict the DILI and DICT potential of the compounds in the Tox21 10K compound library (Supplementary Table 6). Each compound was assigned a toxicity probability based on model predictions, and the majority of compounds in the 10K library were found to be non-toxic for both DILI and DICT. The ROC curve was used to determine the optimal probability cutoff for toxicity. With respect to hepatotoxicity, the RF model predicted 2,807 compounds (35%) as toxic. We observed a similar trend using the NB model, which identified 3,745 compounds (47%) as potentially toxic. The XGBoost model predicted 2,384 compounds (30%) as toxic, while the SVM model predicted 2,065 hepatotoxic compounds (26%). With respect to cardiotoxicity, the RF model predicted 4,420 compounds (55%) as toxic. Similarly, the NB model predicted 3,075 toxic compounds (38%). The XGBoost model predicted 2,532 compounds (32%) as toxic, while the SVM model identified 2,163 potentially cardiotoxic compounds (27%). All four methods predicted a consensus 1,374 hepatotoxic compounds (17%) and 1,469 cardiotoxic compounds (18%), resulting in smaller numbers of predicted toxic compounds.
The top 10 toxic compounds for DILI and their average predicted probabilities, in order of descending probability, are: floxacillin sodium salt (0.91), floxacillin sodium hydrate (0.91), dicloxacillin sodium salt monohydrate (0.91), cloxacillin sodium (0.91), oxacillin sodium salt (0.91), nafcillin sodium monohydrate (0.90), piperacillin sodium salt (0.89), cefixime (0.89), azlocillin sodium (0.89), and methicillin sodium hydrate (0.89). All compounds are reported as toxic according to our DILI reference list (Supplementary Table 1), and this finding validates our models’ robustness to detect known liver toxicants.
The top 10 toxic compounds for DICT that fell within the model AD (Tmax ≥ 0.4) ranked in descending order by the average of their predicted probabilities are: cisapride (0.87), orphenadrine dihydrogen citrate (0.85), roxithromycin (0.85), tamoxifen citrate (0.84), everolimus (0.83), dirithromycin (0.82), azithromycin (0.81), erythromycin ethylsuccinate (0.81), sirolimus (0.81), and doxepin hydrochloride (0.81). Six out of these ten compounds (cisapride, orphenadrine dihydrogen citrate, roxithromycin, tamoxifen citrate, sirolimus, and doxepin hydrochloride) were known toxicants in our DICT reference list (Supplementary Table 2). The remaining four compounds (everolimus, dirithromycin, azithromycin, and erythromycin ethylsuccinate) have literature evidence potentially linking them to cardiotoxicity as detailed below, so these compounds may warrant further investigation.
Everolimus (CAS# 159351-69-6) is an antineoplastic agent used to treat various types of cancer including kidney, pancreas, breast, and brain cancer. Everolimus is an mTOR inhibitor that has been reported to have a direct cardiotoxic effect by blocking signaling for blood vessel formation which may contribute to coronary artery disease (CAD) risk factors such as hyperglycemia, hyperlipidemia, and hypertension.32 Dirithromycin (CAS# 62013-04-1), azithromycin (CAS# 83905-01-5), and erythromycin ethylsuccinate (CAS# 1264-62-6) are all antibiotics used to treat several different types of bacterial infections. Specifically, they belong to the macrolides class of antibiotics, which are commonly used to treat both acute and chronic infections. Macrolides are also known to cause QT prolongation and cardiac arrhythmias. Dirithromycin was discontinued in the United States; however, it is still available in many European countries.33 In 2013, the FDA warned azithromycin to be linked to arrhythmia-related adverse cardiac events especially in patients who are at high risk for cardiovascular events.34 Despite these cautions, azithromycin continues to be a popular broad-spectrum antibiotic. In a previous study, azithromycin was found to have low hERG liability based on in vitro assay results.35 Erythromycin ethylsuccinate is an antibiotic that carries the highest risk of cardiotoxicity among the more commonly used macrolides.36 Specifically, it is known to cause significant prolongation of the QT interval and potentially a fatal ventricular tachyarrhythmia called Torsade de Pointes (TdP).
Overall, the RF, NB, XGBoost, and SVM models predicted that the majority of compounds in the 10K library were not likely to be toxic. The DILI and DICT prediction results using RF, NB, XGBoost, SVM, and consensus models are summarized in Table 4.
Table 4.
Distribution of model-predicted DILI and DICT compounds in the Tox21 10K library with and without the application of the applicability domain (AD) using Random Forest (RF), Naïve Bayes (NB), eXtreme Gradient Boosting (XGBoost), Support Vector Machines (SVM), and a consensus of all four methods.
| RF | NB | XGBoost | SVM | Consensus | ||
|---|---|---|---|---|---|---|
| DILI | without AD | 2,807 (35%) | 3,745 (47%) | 2,384 (30%) | 2,065 (26%) | 1,374 (17%) |
| with AD * | 1,536 (19%) | 1,838 (23%) | 1,389 (17%) | 1,268 (16%) | 845 (11%) | |
| DICT | without AD | 4,420 (55%) | 3,075 (38%) | 2,532 (32%) | 2,163 (27%) | 1,469 (18%) |
| with AD * | 2,357 (29%) | 1,698 (21%) | 1,502 (19%) | 1,357 (17%) | 953 (12%) | |
Tanimoto coefficient ≥ 0.4
Furthermore, we presented the distributions of the predicted toxicity probabilities of compounds in the Tox21 10K library generated from the four machine learning methods (RF, NB, XGBoost, and SVM) in Supplementary Figure 2. These results reiterate our findings that the majority of compounds in the Tox21 10K library were non-toxic regarding both DILI and DICT.
The reliability of the predictions obtained from structure-based models is often limited by the model AD. We assessed the similarity of each compound in the prediction set to the compounds in the model training set (see Methods for details). When considering only compounds that fall within the model AD (Tmax ≥ 0.4), we observed an overall reduction in the predicted number of toxic compounds. For DILI models, the NB method predicted the most toxic compounds (1,838; 23%), while SVM identified the least number of compounds (1,268; 16%) as toxic. For DICT models, the RF method picked up the most toxic compounds (2,357; 29%). Similarly, SVM predicted the least number of cardiotoxic compounds (1,357; 17%). The consensus of all four methods further reduced the total number of predicted toxic compounds for both DILI and DICT, resulting in the prediction of 845 (11%) hepatotoxic compounds and 953 (12%) cardiotoxic compounds.
As observed in Table 4, the use of the AD narrowed the predicted number of toxic compounds by removing compounds that are not structurally similar to the model training set. Defining the AD greatly reduced the number of both hepatotoxic and cardiotoxic compound predictions in the Tox21 10K compound library, with predicted compounds decreasing by at least 10% using RF, NB, XGBoost, and SVM methods separately. The compounds in our training set mostly consists of drugs while the Tox21 10K compound library contains many environmental chemicals that do not have drug-like structures, thus they may fall outside of the model AD. Our QSAR models were limited by the chemical space of the training set that was used to build the models. Nonetheless, similar modeling approaches to predict hepatotoxicity and/or cardiotoxicity have been utilized in previous studies to fill in data gaps related to human organ toxicity.37,38 The applicability of these models could be improved by expanding the training dataset to cover a wider range of chemical structural space beyond drug-like molecules.
We next examined compounds by consumer product use categories39,40 in the Tox21 10K compound library that were predicted to be hepatotoxic or cardiotoxic. The distribution of DILI and DICT compounds in each use category was calculated and the significance of the enrichment was determined using the Fisher’s exact test. The top ten categories enriched with predicted hepatotoxic compounds (in order of descending significance) are: “drug” (95.5%; 1.00×10−20), “discontinued” (36.4%; 1.00×10−20), “antibiotic” (9.2%; 6.55×10−20), “food” (3.5%; 6.75×10−20), “orphan” (1.5%; 1.63×10−17), “pediatric” (5.1%; 1.20×10−9), “diuretic” (3.7%; 1.57×10−9), “anticancer” (6.3%; 4.14×10−9), “anti-inflammatory” (5.5%; 6.13×10−9), and “antihypertensive” (4.1%; 2.86×10−7). The categories “antibiotic”, “diuretic”, “anticancer”, “anti-inflammatory”, and “antihypertensive” refer to pharmaceutical-related compounds. “Drug” refers to any drug product or compound related to the manufacturing of drugs, whereas “food” applies to products designated for human consumption, excluding food additives, as well as manufacture and facilities contaminants related to food.
Furthermore, the top ten categories enriched with potential cardiotoxic compounds along with the corresponding statistical significance p-value (in order of descending significance) are: “drug” (89.9%; 1.00×10−20), “discontinued” (23.7%; 4.05×10−13), “food” (31.8%; 1.51×10−12), “pediatric” (5.6%; 2.00×10−9), “anti-HIV” (4.3%; 1.35×10−8), “antipsychotic” (3.3%; 2.45×10−7), “orphan” (9.6%; 6.82×10−6), “biomarker” (3.5%; 7.39×10−5), “dopamine antagonist” (1.8%; 7.21×10−4), and “angiotensin” (1.3%; 0.0011). The following categories refer to pharmaceutical-related compounds: “drug”, “discontinued”, “pediatric”, “anti-HIV”, “antipsychotic”, “orphan”, “biomarker”, “dopamine antagonist”, and “angiotensin”. Compounds in the “food” category can potentially induce cardiotoxicity through food consumption and/or contaminants. For instance, sorbitan monooleate (CAS # 1338-43-8) is a food additive with emulsifying properties. More recently, the European Food Safety Authority (EFSA) Panel on Food Additives and Nutrient Sources Added to Food (ANS) re-evaluated the safety of various sorbitan esters (including sorbitan monooleate) as food additives and concluded that while there is no safety concern at the reported uses and use levels, their recommendation is that the authorities consider lowering the current limits for sorbitan esters to ensure that these compounds will not be a significant source of toxic elements (arsenic, cadmium, lead, and mercury) in food. Another compound albendazole (CAS # 54965-21-8) belongs to an anthelmintic class of drugs that is widely used for the treatment of gastrointestinal, parasitic infections in animals. It has been approved for farmed ruminants and recently considered as a regulatory treatment against parasites for fish in aquaculture.41 Sucralose (CAS # 56038-13-2) is an artificial sweetener and sugar substitute. In recent decades, there has been increased attention in the safety concerns of low/no-calorie sweeteners such as sucralose. A recent rat study revealed that a 10-week consumption of sucralose mixture resulted in a significant vascular endothelial dysfunction.42 Xylazine (CAS # 7361-61-7) is an adrenergic agonist used in veterinary medication for sedation, analgesia, and muscle relaxation in animals. Food safety concerns may arise from xylazine and metabolite residue in animal-derived food products.43 Five out of ten use categories (“drug”, “discontinued”, “food”, “pediatric”, and “orphan”) overlapped between DILI and DICT compounds, suggesting that these product use categories contain compounds that may contribute to both hepatotoxicity and cardiotoxicity. The majority of the compounds identified by our models are categorized as pharmaceutical-related compounds. Since the training data consisted mostly of drugs, application of our structure-based models is likely to identify other drug-like compounds that are potentially toxic for human consumption.
Discussion
To improve the prediction of DILI and DICT, we compiled a comprehensive dataset that incorporates human in vivo hepatotoxicity and cardiotoxicity data from a diverse set of data sources. The resulting DILI and DICT reference compound lists were used to develop and train the models in this study for the prediction of hepatotoxicity and cardiotoxicity. In addition, assays and chemical structure features that contributed the most to DILI and DICT were identified. The top 20 DILI predictive assays are listed in Table 2. Some of these assays measure targets or pathways that have known connections to hepatotoxicity such as nuclear factor kappa B (NF-κB)44, pregnane X receptor (PXR)45, Nrf2/ARE46, P45047, and mitochondria toxicity48. These assays can serve as a validation for our methods’ utility in identifying hepatotoxicity-related targets and pathways.
NF-κB represents a family of transcription factors that is involved in several aspects of the immune and stress/inflammatory responses. Basal NF- κB activity is essential for cell survival at different stages of development, particularly for protection of the liver against apoptosis. Inactivation of the NF- κB pathway can lead to adverse effects, including liver failure and apoptosis induction. Moreover, PXR is a member of the orphan nuclear receptor family that is highly expressed in the liver where it serves as a master xenobiotic sensor and plays a critical role in drug metabolism.49 PXR tightly regulates the gene expression involved in the detoxification and elimination of drugs in the liver (hepatic drug-clearance system); however, its undesired activation plays an integral role in DILI and has been reported to induce liver injury by increasing the expression of drug metabolizing enzymes.50
The nuclear factor erythroid 2-related factor 2 (Nrf2) is a transcription factor that binds to the antioxidant response element (ARE), and activation of the Nrf2/ARE pathway induces cellular defense against oxidative stress and inflammation. Loss of Nrf2 broadly increases the sensitivity of hepatotoxic chemicals, suggesting that Nrf2 is a pleiotropic transcriptional factor that may serve as a modifier in the development and progression of chronic diseases, including liver injury. The tox21-are-bla-p1 assay detects compounds that can induce the Nrf2/ARE pathway resulting in toxicity. In addition, cytochrome P450 is a superfamily of enzymes that contributes to the metabolism of a wide range of xenobiotic and endogenous compounds. CYP-mediated metabolism of drugs to toxic reactive metabolites leads to the pathogenesis of DILI.51 The Tox21 P450 assays identify CYP inhibitors and substrates which could interfere with the CYP activities producing toxic metabolites. Lastly, some drugs can cause liver injury through mitochondrial toxicity. Drug-induced mitochondria dysfunction can interfere with drug metabolism and renal excretion, ultimately leading to hepatotoxicity.52,53 The tox21-mitotox-p1 assay detects compounds that can disrupt the mitochondria membrane potential resulting in mitochondria dysfunction. Other assay targets identified in our analysis to be predictive of DILI can improve our understanding of the underlying pathways and mechanisms that may lead to hepatotoxicity.
Furthermore, the top 20 predictive assays for DICT based on our analysis are listed in Table 3. Some assay targets were well linked to cardiotoxicity, while others may begin to bridge the gap between understanding cardiotoxicity mechanisms and the first stages of drug development. For example, Tox21 assays that measure the peroxisome proliferator-activated receptor gamma (PPARγ), estrogen receptor beta (ER-β), and vitamin D receptor (VDR) signaling pathways as well as mitochondrial membrane potential, which is an indicator of mitochondrial toxicity, were found to be predictive of cardiotoxicity. PPARs are lipid-activated transcription factors that are critically involved in the regulation of lipid and glucose homeostasis. PPARs are highly expressed in cardiac cells, so activation of PPARγ can exhibit cardiovascular risks through both PPARγ-dependent (on-target) and -independent (off-target) mechanisms. The Tox21 PPARγ assays run in agonist and antagonist modes detect PPARγ agonists and antagonists, respectively. Both modes were found to be predictive of DICT based on our analysis (Table 3). PPARγ agonists such as thiazolidinediones (TZDs), are commonly used as antidiabetic drugs, with two widely used TZDs – pioglitazone and rosiglitazone – being reported to increase the risk of heart failure and other cardiovascular events in diabetic patients.54,55 Nonetheless, the role of PPARγ and the heart remains elusive, so there exists a continual need to better understand the role of PPAR in the physiology and pathology of cardiovascular-related diseases.
ER-β is a receptor that is expressed on cardiomyocytes, where it exerts a protective effect on cardiac function by initiating signaling pathways essential for the growth, repair, and survival of cardiomyocytes.56 Modulation of estrogen receptors by potential therapeutic agents is currently being considered for the prevention and treatment of various pathological conditions, including cancer and cardiovascular diseases.57 For breast cancer patients, however, cardiotoxicity is one of the most concerning side effects of treatments which inhibit ER-β. There is a strong association between the drug tamoxifen, an antineoplastic agent that is also an ER antagonist, and an increased risk for cardiotoxicity manifested in several forms (e.g., thrombotic/thromboembolic events and pulmonary embolism).58 The tox21-erb-bla-antagonist-p1 assay that identifies ER-β antagonists is among the top 20 predictive assays of DICT.
VDR, the molecular mediator for Vitamin D signaling is present in almost all tissues, including vascular smooth muscle cells, cardiomyocytes, and endothelial cells. High doses of vitamin D in humans have been strongly associated with extensive arterial calcium phosphate deposits, with both activation and inhibition of VDR promoting different types of calcification. The tox21-vdr-bla-antagonist-p1 assay detects compounds that inhibit VDR activity and is among the top 20 DICT predictive assays. Vascular calcification is a disorder commonly linked with cardiovascular mortality in which calcium mineralization occurs along vascular walls, often leading to vessel stiffening and reduced compliance.59 Vascular calcification is complex, and the exact impact of VDR signaling on this process is not well understood.
The mitochondrial membrane potential is an indicator for normal cell function that results from the electron transport and oxidative phosphorylation process, an essential component for ATP production.60 Given that the mitochondria is abundantly found in cardiac muscle cells, it plays a critical role in maintaining myocardial tissue homeostasis as well as regulating cardiac muscle contraction and heartbeat.61,62 As a result, cardiomyocytes are more susceptible to mitochondrial dysfunction and oxidative stress due to the mitochondria-rich environment and low levels of antioxidants compared to other cells.61 Many drugs, such as antineoplastic, antiangiogenic, antiviral, and antidiabetic drugs, have been linked to adverse cardiovascular effects through interference with mitochondria-related signaling pathways.62 Three common mechanisms of mitochondrial toxicity are the ROS/Redox system, calcium homeostasis system, and endoplasmic reticulum stress signaling.63
While inhibition of the hERG channel is known to elicit QT interval prolongation and potentially lead to TdP, the Tox21 hERG inhibition assay64 was not among the top 20 most predictive assays for cardiotoxicity, with an AUC-ROC score (0.52) slightly above random. To investigate the cause of this miscorrelation, we checked the activity of the compounds on the DICT reference list in the hERG assay. Of the 441 reference compounds tested in the hERG assay, 241 compounds (54.6%) did not inhibit the hERG channel but were classified as cardiotoxic (toxicity score > 0.5), 106 compounds (24.0%) inhibited the hERG channel but were not classified as cardiotoxic, and only 94 compounds (21.3%) both inhibited hERG activity and were cardiotoxic (Supplementary Figure 2). These findings demonstrate that the hERG channel inhibition is not the only mechanism for DICT and does not necessarily lead to clinical cardiotoxicity, with a notable example being the antiarrhythmic drug verapamil. This calcium-channel blocker inhibits the hERG channel but does not cause QT prolongation or TdP, which could likely be attributed to the inhibition of multiple channels that mitigates the hERG-blocking effect.65 In the Tox21 hERG assay, verapamil showed hERG inhibition with IC50 values between 4–11 μM. Additionally, verapamil was assigned a toxicity score of 0.17 on our DICT reference list, indicating that this compound was seldomly identified as cardiotoxic in the literature. Drug-induced blockage of the hERG channel can lead to a variety of cardiovascular ailments, so it crucial to recognize the hERG liabilities of compounds.35
Other compounds, such as the antihypertensive drug alfuzosin, may not inhibit hERG channels but still lead to QT prolongation through interactions with other ion channels.66 Alfuzosin is used to treat benign prostatic hyperplasia by increasing the sodium current, rather than blocking the hERG potassium current, to delay cardiac repolarization. The Tox21 qHTS data revealed that alfuzosin was inactive in the hERG assay; however, the toxicity score of 0.4 on the DICT reference list indicates that ~40% of the literature sources reported this drug as cardiotoxic.
Lastly, we found that 65% of the top 20 cardiotoxicity predictive assays are cell viability assays. The cell viability assay serves as the counter screen for cell-based pathway assays by assessing the integrity of cells in which a loss in signal indicates cell death or cytotoxicity interference. It is not surprising that cytotoxicity could lead to in vivo toxicity such as cardiotoxicity, but the exact toxicity mechanism could not be inferred from generic cytotoxicity assays. Since most of the Tox21 assays that correlated with cardiotoxicity were cell viability assays rather than target- or pathway-specific assays, it suggests that the current Tox21 assay panel has insufficient coverage of the targets/pathways that detect cardiotoxicity. To address this limitation, the Tox21 program is currently screening additional assays to improve the coverage of the toxicity-related biological response space.
In vitro assays are valuable tools for studying a compound’s mechanism of action and can be used to screen tens of thousands of compounds in a short timeframe when compared to in vivo studies. One of the limitations of in vitro assays is that they are missing some functional physiological machinery, such as drug metabolism enzymes (e.g., CYPs), which may fail to identify the chemicals that need metabolic activation. To overcome this limitation, the Tox21 partners have developed an in vitro assay that incorporates metabolic components67 to mimic real physiological conditions. This type of assays will be good candidates to be adapted for regulatory use in the future.
Our models also identified chemical features that are significantly enriched in hepatotoxic and cardiotoxic compounds. Examples of compounds that contain significant hepatotoxic chemotypes include oxacillin, flutamide, entacapone, maprotiline, and desloratadine (Figure 1; Supplementary Table 4). Oxacillin (ring:hetero_[4]_N_beta_lactam) is a penicillin antibiotic used to treat a wide variety of bacterial infections.68 It has been linked to two forms of hepatotoxicity: (1) an acute elevation of hepatic enzymes (e.g. alanine aminotransferase) and (2) a more prolonged idiosyncratic liver injury similar to the hepatotoxicity describe with other second-generation penicillins (e.g. dicloxacillin).68 Flutamide (bond:CX_halide_alkyl-X_trihalo_(1_1_1-)) belongs to a class of drugs known as anti-androgens, and it is used in the management and treatment of prostate cancer. Drug-induced elevation of serum aminotransferase levels have led to several instances of acute liver injury, thereby making flutamide a potent hepatotoxin in certain patients. Entacapone (bond:N(=O)_nitro_aromatic) is a peripheral catechol-O-methyltransferase (COMT) inhibitor often used as a beneficial adjunct to treat Parkinson’s disease. Although rare, acute liver dysfunction and low rates of serum enzyme elevations have been linked to entacapone therapy.68,69 The antidepressant drug maprotiline (bond:CN_amine_aliphatic_generic) has been reported in both clinical case reports and in vitro animal studies to cause hepatic damage after prolonged use over the course of several years. Specifically, abnormally high elevations of liver enzymes and jaundice were reported in patients.70,71 Finally, desloratadine (ring:hetero_[6]_N_piperidine) is the major metabolic derivative of loratadine, a second-generation antihistamine used widely to treat allergy symptoms. In rare cases, desloratadine has been connected to instances of clinically apparent acute liver injury.72
Moreover, compounds that contain structure features enriched in cardiotoxic compounds include daunorubicin, ranolazine, bosutinib, abiraterone acetate, and mitomycin C (Figure 2; Supplementary Table 5). Daunorubicin (ring:hetero_[6]_Z_1-) is an antibiotic that belongs to the anthracyclines class of drugs that is widely used in the treatment of a variety of malignancies. Anthracycline use has been associated with cardiotoxicities such as arrhythmia, systolic dysfunction, long QT interval, and in some cases hypertension, myocardial ischemia, and thromboembolism.73 Ranolazine (ring:hetero_[6]_N_piperazine) is an antianginal agent that is primarily indicated for the treatment of patients with coronary artery disease and chronic stable angina. It inhibits the sodium ion current in cardiac cells, thus interfering in transmembrane cardiac action potential.74 TdP has not been identified as a side effect of this drug, however, the risk for developing this condition may increase in patients that are also administered other QT-prolonging medications.75 Bosutinib (ring:hetero_[6_6]_N_quinoline), a BCR-ABL1 tyrosine kinase inhibitor (TKI), has been available for several years as a treatment for chronic-, accelerated-, and blast-phase chronic myeloid leukemia (CML) for patients with resistance or intolerance to prior therapy. While the incidence of bosutinib-induced cardiotoxicity is relatively rare, cardiovascular adverse events such as hypertension, palpitations, and even cardiac failure were found to be associated with bositinib.76 Abiraterone acetate (ring:hetero_[6]_N_pyridine) is often used in combination for the treatment of metastatic castration-resistant prostate cancer. Patients with prostate cancer that were treated with abiraterone acetate reported the highest incidence of cardiotoxic effects.77 Specifically, hypertension and cardiac disorders were prominently observed in Phase III of a placebo-controlled trial that evaluated the risk ratio of abiraterone acetate-related adverse events.78,79 Mitomycin C (bond:CC(=O)C_quinone_1_4-benzo), an anti-cancer drug exhibiting antibiotic properties, is used in chemotherapy treatment. Several studies dating back from the 1970s suggests that cardiotoxicity induced by mitomycin C is dose dependent, and these adverse effects are enhanced in patients that are also treated with doxorubicin.80,81 The chemical features collectively identified from this study can serve as structural alerts of hepatotoxicity or cardiotoxicity and should be avoided when designing chemicals for new drug development to minimize their toxicity potential.
Overall, the assays previously discussed can serve as a starting point to better understand the underlying biological mechanisms that lead to DILI and DICT. The adverse outcome pathway (AOP) is an analytical construct that organizes biological interactions over the span of multiple levels and toxicity mechanisms to promote more meaningful analysis of toxicological effects that can advance human health risk assessment of chemicals.82 AOPs are useful for linking molecular initiating events (MIEs), the initial chemical-induced disruption in a biological system, with key events (KEs), the intermediate progression of toxicity, that ultimately leads to a specific adverse outcome (AO). The Tox21 in vitro assay targets that contributed the most to DILI and/or DICT could be MIEs or KEs within an AOP, thus complementing the AOP concept. For example, the assay “tox21-pxr-p1_ratio” was found to be one of the most relevant assays to DILI (AUC-ROC = 0.57) in our study (Table 2), and pregnane X receptor (PXR) plays a critical role in the detoxification of xenobiotics and bile acid homeostasis in liver.83 PXR was recorded as the MIE in an AOP concept that is related to hepatic steatosis (https://aopwiki.org/aops/60). The assay “tox21-p450-2c9-p1_ratio” was found to be one of the top relevant assays to DILI (AUC-ROC = 0.55) in our study (Table 2), and CYP2C9 is a liver enzyme responsible for the metabolism of a wide range of clinical drugs (e.g., warfarin).84 However, there is no record of CYP2C9 being involved in an AOP framework. The assay “tox21-pparg-bla-agonist-p1_ratio” was found to be the most relevant to DICT (AUC-ROC = 0.60) in our study (Table 3), and PPARγ has been reported to be closely associated with cardiotoxic regulation.85 However, PPARγ was only documented as the MIE in a few AOP concepts that are unrelated to DICT, such as lung fibrosis (https://aopwiki.org/aops/206), sarcomas in rats, mice, and hamsters (https://aopwiki.org/aops/163), and pulmonary fibrosis (https://aopwiki.org/aops/347).
In this study, we found that the addition of assay data to chemical structure does not always improve the model performance; these results are not surprising and similar results have been reported in previous studies.37 Hepatotoxicity and cardiotoxicity are complex and multifactorial, whereas in vitro assay typically only represents a single toxicity mechanism that covers only a small number of compounds in the modeling set, such that assay results are often biased with much more inactives than actives. Hence, the contribution of an individual assay tends to be small in a global model (e.g., a model for a complex toxicity endpoint) and the relevance of this assay becomes negligible during the modeling process. On the other hand, assay data could play a more significant role when developing a local model (e.g., a specific toxicity pathway model). Although our model performance did not benefit from the addition of assay data, recent studies have demonstrated successful use of assay data for modeling and validating QSAR predictions.86–88 In cases where structure-based models cannot be achieved (e.g., a defined chemical structure is not available), assay data can be used to build models for toxicity prediction and elucidation of the biological mechanisms of drug-induced toxicity. QSAR models, such as the models constructed in this study, are often limited by the extent of chemical space coverage. Our models were trained using a compound collection enriched with drug-like molecules, therefore making them less suitable for making predictions on other types of chemicals (e.g., environmental chemicals). Presently, Tox21 is focusing on the expansion of pathway coverage by adding assays that probe under-represented pathways in the current portfolio to improve the predictivity of Tox21 data for adverse drug effects as observed in DILI and DICT.
Conclusion
In this study, we compiled reference compound lists for DILI and DICT to build human in vivo hepatotoxicity and cardiotoxicity prediction models. The chemical structure-based models showed good performance in identifying hepatotoxic or cardiotoxic compounds, while Tox21 assay data-based models only showed better than random performance. The optimal models were applied to predict potential hepatotoxic and cardiotoxic compounds within the Tox21 10K compound library. These models can be applied to make predictions on other large compound libraries regarding DILI and DICT. In addition, we identified significant hepatotoxic and cardiotoxic chemical structure features and assays that were most predictive of DILI and DICT. The findings uncovered in this study can be used to better understand the underlying pathways and mechanisms for drug-induced toxicity as it relates to the liver and heart as well as highlight structural features that may alert us of these toxicities. Tox21 is currently expanding the coverage of biological response space with additional assays that probe toxicologically important targets and under-represented pathways to improve the prediction of in vivo toxicity, including DILI and DICT.
Supplementary Material
Highlights.
Reference compound lists for DICT and DILI were compiled to build human in vivo hepatotoxicity and cardiotoxicity prediction models
Chemical structure-based models showed reasonable predictive power for DILI and DICT prediction
Significant chemical features and assays for predicting hepatotoxicity and cardiotoxicity were identified
Predicting the DILI and DICT potential of the Tox21 10K compound library revealed that most compounds were non-toxic
Acknowledgement
This work was supported by the Intramural Research Programs of the National Toxicology Program (Interagency agreement #Y2-ES-7020-01), National Institute of Environmental Health Sciences and the National Center for Advancing Translational Sciences, National Institutes of Health. The views expressed in this article are those of the authors and do not necessarily reflect the statements, opinions, views, conclusions, or policies of the National Center for Advancing Translational Sciences, the National Institutes of Health, the United States government, or the United States Food and Drug Administration. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.(NCATS), N. C. f. A. T. S. Predictive Efficacy and Toxicology, <https://ncats.nih.gov/translation/issues/tox> (2021).
- 2.Han D et al. Regulation of drug-induced liver injury by signal transduction pathways: critical role of mitochondria. Trends Pharmacol Sci 34, 243–253, doi: 10.1016/j.tips.2013.01.009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaeschke H, McGill MR & Ramachandran A Oxidant stress, mitochondria, and cell death mechanisms in drug-induced liver injury: lessons learned from acetaminophen hepatotoxicity. Drug Metab Rev 44, 88–106, doi: 10.3109/03602532.2011.602688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holt M & Ju C Drug-induced liver injury. Handb Exp Pharmacol, 3–27, doi: 10.1007/978-3-642-00663-0_1 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Kim E & Nam H Prediction models for drug-induced hepatotoxicity by using weighted molecular fingerprints. BMC Bioinformatics 18, 227, doi: 10.1186/s12859-017-1638-4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferri N et al. Drug attrition during pre-clinical and clinical development: understanding and managing drug-induced cardiotoxicity. Pharmacol Ther 138, 470–484, doi: 10.1016/j.pharmthera.2013.03.005 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Institute, N. N. C. Dictionary of Cancer Terms, <https://www.cancer.gov/publications/dictionaries/cancerterms/def/cardiotoxicity?redirect=true> (
- 8.Dokmanovic M, King KE, Mohan N, Endo Y & Wu WJ Cardiotoxicity of ErbB2-targeted therapies and its impact on drug development, a spotlight on trastuzumab. Expert Opin Drug Metab Toxicol 13, 755–766, doi: 10.1080/17425255.2017.1337746 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Lee HM et al. Computational determination of hERG-related cardiotoxicity of drug candidates. BMC Bioinformatics 20, 250, doi: 10.1186/s12859-019-2814-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo Piparo E et al. QSAR models for Daphnia magna toxicity prediction of benzoxazinone allelochemicals and their transformation products. J Agric Food Chem 54, 1111–1115, doi: 10.1021/jf050918f (2006). [DOI] [PubMed] [Google Scholar]
- 11.He S et al. An In Silico Model for Predicting Drug-Induced Hepatotoxicity. Int J Mol Sci 20, doi: 10.3390/ijms20081897 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekins S Progress in computational toxicology. J Pharmacol Toxicol Methods 69, 115–140, doi: 10.1016/j.vascn.2013.12.003 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Chekmarev DS et al. Shape signatures: new descriptors for predicting cardiotoxicity in silico. Chem Res Toxicol 21, 1304–1314, doi: 10.1021/tx800063r (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siramshetty VB, Chen Q, Devarakonda P & Preissner R The Catch-22 of Predicting hERG Blockade Using Publicly Accessible Bioactivity Data. J Chem Inf Model 58, 1224–1233, doi: 10.1021/acs.jcim.8b00150 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Collins FS, Gray GM & Bucher JR Toxicology. Transforming environmental health protection. Science 319, 906–907, doi:319/5865/906 [pii] 10.1126/science.1154619 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kavlock RJ, Austin CP & Tice RR Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal 29, 485–487; discussion 492–487, doi:RISK1168 [pii] 10.1111/j.1539-6924.2008.01168.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tice RR, Austin CP, Kavlock RJ & Bucher JR Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 121, 756–765, doi: 10.1289/ehp.1205784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richard AM et al. The Tox21 10K Compound Library: Collaborative Chemistry Advancing Toxicology. Chem Res Toxicol 34, 189–216, doi: 10.1021/acs.chemrestox.0c00264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang R et al. Expanding biological space coverage enhances the prediction of drug adverse effects in human using in vitro activity profiles. Sci Rep 8, 3783, doi: 10.1038/s41598-018-22046-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang R et al. Modelling the Tox21 10 K chemical profiles for in vivo toxicity prediction and mechanism characterization. Nat Commun 7, 10425, doi: 10.1038/ncomms10425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang R et al. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci Transl Med 3, 80ps16, doi:3/80/80ps16 [pii] 10.1126/scitranslmed.3001862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang R et al. The NCATS Pharmaceutical Collection: a 10-year update. Drug Discov Today 24, 2341–2349, doi: 10.1016/j.drudis.2019.09.019 (2019). [DOI] [PubMed] [Google Scholar]
- 23.NCATS. Tox21 Data Browser, <https://tripod.nih.gov/tox21/pubdata> (2021).
- 24.PubChem. Tox21 phase II data, <http://www.ncbi.nlm.nih.gov/pcassay?term=tox21> (2021).
- 25.Chen M et al. DILIrank: the largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov Today 21, 648–653, doi: 10.1016/j.drudis.2016.02.015 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Kuhn M, Letunic I, Jensen LJ & Bork P The SIDER database of drugs and side effects. Nucleic Acids Res 44, D1075–1079, doi: 10.1093/nar/gkv1075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y et al. PubChem’s BioAssay Database. Nucleic acids research 40, D400–412, doi: 10.1093/nar/gkr1132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.PubChem. Tox21 phase II data, <http://www.ncbi.nlm.nih.gov/pcassay?term=tox21> (2016).
- 29.Huang R A Quantitative High-Throughput Screening Data Analysis Pipeline for Activity Profiling. Methods Mol Biol 2474, 133–145, doi: 10.1007/978-1-0716-2213-1_13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang C et al. New publicly available chemical query language, CSRML, to support chemotype representations for application to data mining and modeling. J Chem Inf Model 55, 510–528, doi: 10.1021/ci500667v (2015). [DOI] [PubMed] [Google Scholar]
- 31.Rogers D & Hahn M Extended-connectivity fingerprints. J Chem Inf Model 50, 742–754, doi: 10.1021/ci100050t (2010). [DOI] [PubMed] [Google Scholar]
- 32.Hedhli N & Russell KS Cardiotoxicity of molecularly targeted agents. Curr Cardiol Rev 7, 221–233, doi: 10.2174/157340311799960636 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Administration, U. S. F. D. Drugs@FDA: FDA-Approved Drugs, <https://www.accessdata.fda.gov/scripts/cder/daf/> (2022).
- 34.Administration, U. S. F. a. D. FDA Drug Safety Communication: Azithromycin (Zithromax or Zmax) and the risk of potentially fatal heart rhythms, <https://www.fda.gov/drugs/drug-safety-and-availability/fda-drug-safety-communication-azithromycin-zithromax-or-zmax-and-risk-potentially-fatal-heart> (2013).
- 35.Ngan DK, Xu T, Xia M, Zheng W & Huang R Repurposing drugs as COVID-19 therapies: A toxicity evaluation. Drug Discov Today, doi: 10.1016/j.drudis.2022.04.001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farzam K NT, Quick J Erythromycin. (StatPearls Publishing, 2022). [PubMed] [Google Scholar]
- 37.Xu T et al. Predictive Models for Human Organ Toxicity Based on In Vitro Bioactivity Data and Chemical Structure. Chem Res Toxicol 33, 731–741, doi: 10.1021/acs.chemrestox.9b00305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma H et al. Deep Graph Learning with Property Augmentation for Predicting Drug-Induced Liver Injury. Chem Res Toxicol 34, 495–506, doi: 10.1021/acs.chemrestox.0c00322 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.EPA. CompTox Chemicals Dashboard, <ftp://newftp.epa.gov/COMPTOX/Sustainable_Chemistry_Data/Chemistry_Dashboard/CPDat/> (2018).
- 40.Williams AJ et al. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. Journal of cheminformatics 9, 61, doi: 10.1186/s13321-017-0247-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busatto Z, de Franca WG, Cyrino JEP & Paschoal JAR Assessment of elimination profile of albendazole residues in fish. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 35, 77–85, doi: 10.1080/19440049.2017.1400186 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Risdon S et al. Artificial sweeteners impair endothelial vascular reactivity: Preliminary results in rodents. Nutr Metab Cardiovasc Dis 30, 843–846, doi: 10.1016/j.numecd.2020.01.014 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Zheng X, Mi X, Li S & Chen G Determination of xylazine and 2,6-xylidine in animal tissues by liquid chromatography-tandem mass spectrometry. J Food Sci 78, T955–959, doi: 10.1111/1750-3841.12144 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Korunes KL et al. A gene expression biomarker for predictive toxicology to identify chemical modulators of NF-kappaB. PLoS One 17, e0261854, doi: 10.1371/journal.pone.0261854 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lynch C et al. Characterization of human pregnane X receptor activators identified from a screening of the Tox21 compound library. Biochem Pharmacol 184, 114368, doi: 10.1016/j.bcp.2020.114368 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia M et al. Identification of Chemical Compounds that Induce HIF-1{alpha} Activity. Toxicol Sci, doi:kfp123 [pii] 10.1093/toxsci/kfp123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun H et al. Prediction of Cytochrome P450 Profiles of Environmental Chemicals with QSAR Models Built from Drug-like Molecules. Mol Inform 31, 783–792, doi: 10.1002/minf.201200065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Attene-Ramos MS et al. Profiling of the Tox21 chemical collection for mitochondrial function to identify compounds that acutely decrease mitochondrial membrane potential. Environ Health Perspect 123, 49–56, doi: 10.1289/ehp.1408642 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kliewer SA et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92, 73–82, doi: 10.1016/s0092-8674(00)80900-9 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Wang YM, Chai SC, Brewer CT & Chen T Pregnane X receptor and drug-induced liver injury. Expert Opin Drug Metab Toxicol 10, 1521–1532, doi: 10.1517/17425255.2014.963555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chalasani N & Bjornsson E Risk factors for idiosyncratic drug-induced liver injury. Gastroenterology 138, 2246–2259, doi: 10.1053/j.gastro.2010.04.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tujios S & Fontana RJ Mechanisms of drug-induced liver injury: from bedside to bench. Nat Rev Gastroenterol Hepatol 8, 202–211, doi: 10.1038/nrgastro.2011.22 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Fromenty B, Berson A & Pessayre D Microvesicular steatosis and steatohepatitis: role of mitochondrial dysfunction and lipid peroxidation. J Hepatol 26 Suppl 1, 13–22, doi: 10.1016/s0168-8278(97)82328-8 (1997). [DOI] [PubMed] [Google Scholar]
- 54.Graham DJ et al. Risk of acute myocardial infarction, stroke, heart failure, and death in elderly Medicare patients treated with rosiglitazone or pioglitazone. JAMA 304, 411–418, doi: 10.1001/jama.2010.920 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Kalliora C & Drosatos K The Glitazars Paradox: Cardiotoxicity of the Metabolically Beneficial Dual PPARalpha and PPARgamma Activation. J Cardiovasc Pharmacol 76, 514–526, doi: 10.1097/FJC.0000000000000891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanchez-Soria P & Camenisch TD ErbB signaling in cardiac development and disease. Semin Cell Dev Biol 21, 929–935, doi: 10.1016/j.semcdb.2010.09.011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paterni I, Granchi C, Katzenellenbogen JA & Minutolo F Estrogen receptors alpha (ERalpha) and beta (ERbeta): subtype-selective ligands and clinical potential. Steroids 90, 13–29, doi: 10.1016/j.steroids.2014.06.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martel S, Maurer C, Lambertini M, Ponde N & De Azambuja E Breast cancer treatment-induced cardiotoxicity. Expert Opin Drug Saf 16, 1021–1038, doi: 10.1080/14740338.2017.1351541 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Zittermann A, Schleithoff SS & Koerfer R Vitamin D and vascular calcification. Curr Opin Lipidol 18, 41–46, doi: 10.1097/MOL.0b013e328011c6fc (2007). [DOI] [PubMed] [Google Scholar]
- 60.Zorova LD et al. Mitochondrial membrane potential. Anal Biochem 552, 50–59, doi: 10.1016/j.ab.2017.07.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou B & Tian R Mitochondrial dysfunction in pathophysiology of heart failure. J Clin Invest 128, 3716–3726, doi: 10.1172/JCI120849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varga ZV, Ferdinandy P, Liaudet L & Pacher P Drug-induced mitochondrial dysfunction and cardiotoxicity. Am J Physiol Heart Circ Physiol 309, H1453–1467, doi: 10.1152/ajpheart.00554.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim CW & Choi KC Effects of anticancer drugs on the cardiac mitochondrial toxicity and their underlying mechanisms for novel cardiac protective strategies. Life Sci 277, 119607, doi: 10.1016/j.lfs.2021.119607 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Krishna S et al. High-Throughput Chemical Screening and Structure-Based Models to Predict hERG Inhibition. Biology 11, 209 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Redfern WS et al. Relationships between preclinical cardiac electrophysiology, clinical QT interval prolongation and torsade de pointes for a broad range of drugs: evidence for a provisional safety margin in drug development. Cardiovasc Res 58, 32–45, doi: 10.1016/s0008-6363(02)00846-5 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Liang P et al. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation 127, 1677–1691, doi: 10.1161/CIRCULATIONAHA.113.001883 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li S et al. Profiling the Tox21 Chemical Collection for Acetylcholinesterase Inhibition. Environ Health Perspect 129, 47008, doi: 10.1289/EHP6993 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Creton S et al. Cell transformation assays for prediction of carcinogenic potential: state of the science and future research needs. Mutagenesis 27, 93–101, doi: 10.1093/mutage/ger053ger053 [pii] (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fisher A, Croft-Baker J, Davis M, Purcell P & McLean AJ Entacapone-induced hepatotoxicity and hepatic dysfunction. Mov Disord 17, 1362–1365; discussion 1397–1400, doi: 10.1002/mds.10342 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Weinstein RP & Gosselin JY Case report of hepatotoxicity associated with maprotiline. Can J Psychiatry 33, 233–234, doi: 10.1177/070674378803300314 (1988). [DOI] [PubMed] [Google Scholar]
- 71.Voican CS, Corruble E, Naveau S & Perlemuter G Antidepressant-induced liver injury: a review for clinicians. Am J Psychiatry 171, 404–415, doi: 10.1176/appi.ajp.2013.13050709 (2014). [DOI] [PubMed] [Google Scholar]
- 72.in LiverTox: Clinical and Research Information on Drug-Induced Liver Injury (2012). [PubMed]
- 73.Truong J, Yan AT, Cramarossa G & Chan KK Chemotherapy-induced cardiotoxicity: detection, prevention, and management. Can J Cardiol 30, 869–878, doi: 10.1016/j.cjca.2014.04.029 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Banerjee K, Ghosh RK, Kamatam S, Banerjee A & Gupta A Role of Ranolazine in cardiovascular disease and diabetes: Exploring beyond angina. Int J Cardiol 227, 556–564, doi: 10.1016/j.ijcard.2016.10.102 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Reed M, Kerndt CC, Gopal S & Nicolas D in StatPearls (2021).
- 76.Brummendorf TH et al. Bosutinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukaemia: results from the 24-month follow-up of the BELA trial. Br J Haematol 168, 69–81, doi: 10.1111/bjh.13108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Santoni M et al. Incidence and risk of cardiotoxicity in cancer patients treated with targeted therapies. Cancer Treat Rev 59, 123–131, doi: 10.1016/j.ctrv.2017.07.006 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Thakur A, Roy A, Ghosh A, Chhabra M & Banerjee S Abiraterone acetate in the treatment of prostate cancer. Biomed Pharmacother 101, 211–218, doi: 10.1016/j.biopha.2018.02.067 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Roviello G, Corona SP & Generali D Low dose versus standard dose of corticosteroids in the management of adverse events of special interest from abiraterone acetate: data from a literature-based meta-analysis. Med Oncol 34, 166, doi: 10.1007/s12032-017-1028-9 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Verweij J, Funke-Kupper AJ, Teule GJ & Pinedo HM A prospective study on the dose dependency of cardiotoxicity induced by mitomycin C. Med Oncol Tumor Pharmacother 5, 159–163, doi: 10.1007/BF02986439 (1988). [DOI] [PubMed] [Google Scholar]
- 81.Villani F et al. Possible enhancement of the cardiotoxicity of doxorubicin when combined with mitomycin C. Med Oncol Tumor Pharmacother 2, 93–97, doi: 10.1007/BF02934854 (1985). [DOI] [PubMed] [Google Scholar]
- 82.Hamm J et al. Alternative approaches for identifying acute systemic toxicity: Moving from research to regulatory testing. Toxicol In Vitro 41, 245–259, doi: 10.1016/j.tiv.2017.01.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagner M et al. CAR and PXR agonists stimulate hepatic bile acid and bilirubin detoxification and elimination pathways in mice. Hepatology 42, 420–430, doi: 10.1002/hep.20784 (2005). [DOI] [PubMed] [Google Scholar]
- 84.Parikh SJ et al. Insights into the Genetic Variations of Human Cytochrome P450 2C9: Structural Analysis, Characterization and Comparison. Int J Mol Sci 22, doi: 10.3390/ijms221910206 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xi Y et al. PPAR-Mediated Toxicology and Applied Pharmacology. Cells 9, doi: 10.3390/cells9020352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ciallella HL, Russo DP, Aleksunes LM, Grimm FA & Zhu H Revealing Adverse Outcome Pathways from Public High-Throughput Screening Data to Evaluate New Toxicants by a Knowledge-Based Deep Neural Network Approach. Environ Sci Technol 55, 10875–10887, doi: 10.1021/acs.est.1c02656 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ciallella HL et al. Predicting Prenatal Developmental Toxicity Based On the Combination of Chemical Structures and Biological Data. Environ Sci Technol 56, 5984–5998, doi: 10.1021/acs.est.2c01040 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jia X, Wen X, Russo DP, Aleksunes LM & Zhu H Mechanism-driven modeling of chemical hepatotoxicity using structural alerts and an in vitro screening assay. J Hazard Mater 436, 129193, doi: 10.1016/j.jhazmat.2022.129193 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


