Summary
Liver damage and an exacerbated inflammatory response are hallmarks of Ebola virus (EBOV) infection. Little is known about the intrinsic response to infection in human hepatocytes and their contribution to inflammation. Here, we present an induced pluripotent stem cell (iPSC)-derived hepatocyte-like cell (HLC) platform to define the hepato-intrinsic response to EBOV infection. We used this platform to show robust EBOV infection, with characteristic ultrastructural changes and evidence for viral replication. Transcriptomics analysis revealed a delayed response with minimal early transcriptomic changes, followed by a general downregulation of hepatic function and upregulation of interferon signaling, providing a potential mechanism by which hepatocytes participate in disease severity and liver damage. Using RNA-fluorescence in situ hybridization (FISH), we showed that IFNB1 and CXCL10 were mainly expressed in non-infected bystander cells. We did not observe an inflammatory signature during infection. In conclusion, iPSC-HLCs are an immune competent platform to study responses to EBOV infection.
Key words: iPSC-hepatocytes, Ebola virus infection, Ebola virus host response, downregulation key liver genes
Highlights
-
•
iPSC-derived HLCs model Ebola virus infection of primary human hepatocytes
-
•
Liver function genes are downregulated in Ebola virus-infected iPSC-HCLs
-
•
Delayed type I IFN response observed in non-infected bystander cells
-
•
No induction of an inflammatory response in Ebola virus-infected iPSC-HLCs
Here, Scoon and colleagues present an iPSC-based model for investigating Ebola virus infection of human primary-like hepatocytes. EBOV infection led to a general downregulation of liver function genes and a delayed interferon response in non-infected bystander cells with no inflammatory signature. This model can be used to further explore host response dynamics in a critical cell type of EBOV pathogenesis.
Introduction
Ebola virus (EBOV) infection causes a severe disease in humans with case fatality rates ranging from 40% to 90%. Severe liver damage, coagulopathy, and thrombocytopenia are hallmark clinical manifestations of EBOV disease (EVD) (Martines et al., 2015). Histopathological changes in liver autopsies of fatal EBOV cases include hepatocellular necrosis, Councilman bodies, Kupffer cell hyperplasia, and the presence of EBOV inclusions in infected cells (Martines et al., 2015; Zaki and Goldsmith, 1999). Liver damage in EVD patients and experimentally infected non-human primates (NHPs) is indicated by elevated serum levels of liver enzymes, including aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase, and γ-glutamyltransferase, and decreasing levels of albumin and calcium (de Wit et al., 2016; Geisbert et al., 2003d; Lanini et al., 2018; Rollin et al., 2007; Uyeki et al., 2016). Decreasing serum protein C levels and increasing fibrinogen levels indicate dysregulation of clotting and coagulopathy (Geisbert et al., 2003c; Liu et al., 2017; Smither et al., 2019).
In addition to organ damage, there is evidence that a dysregulated proinflammatory response is a hallmark of severe EVD (Liu et al., 2017). Monocytes and macrophages are activated upon EBOV infection (reviewed in McElroy et al., 2018), and this proinflammatory environment may affect liver damage during EBOV infection. While hepatocytes constitute over 80% of the liver volume, their immune response to EBOV has yet to be characterized. Most previous reports that have examined the intrinsic hepatic response to EBOV infection utilized immortalized hepatocarcinoma cell lines (Hartman et al., 2008; Holzer et al., 2016; Kash et al., 2006; Kuzmin et al., 2017), which have an impaired antiviral response compared with primary human hepatocytes (PHHs) (Li et al., 2005; Marozin et al., 2008). While PHHs represent a more reliable platform, limited accessibility, donor-to-donor variability, and complex in vitro conditions required for long-term culture have restricted their use (Gural et al., 2018; Khetani and Bhatia, 2008; March et al., 2015; Michailidis et al., 2020).
Here, we have employed human induced pluripotent stem cell (iPSC)-derived hepatocyte-like cells (HLCs) as a model for EBOV infection. These cells can be terminally differentiated and maintained in a monoculture for approximately 10 days and have previously been shown to effectively model viral infections (Lang et al., 2016; Sakurai et al., 2017; Schwartz et al., 2012; Shlomai et al., 2014; Tricot et al., 2018; Wilson et al., 2015; Zhou et al., 2014). We demonstrate that iPSC-HLCs recapitulate aspects of the innate human host response to EBOV infection that cannot be modeled in immortalized cell lines. Our data show that EBOV infection leads to a delayed type I interferon (IFN) response and the downregulation of genes associated with liver function in iPSC-HLCs, illuminating a mechanism by which hepatocytes participate in EVD severity and liver damage.
Results
iPSC-derived HLCs are physiologically more similar to primary hepatocytes than immortalized Huh7 cells
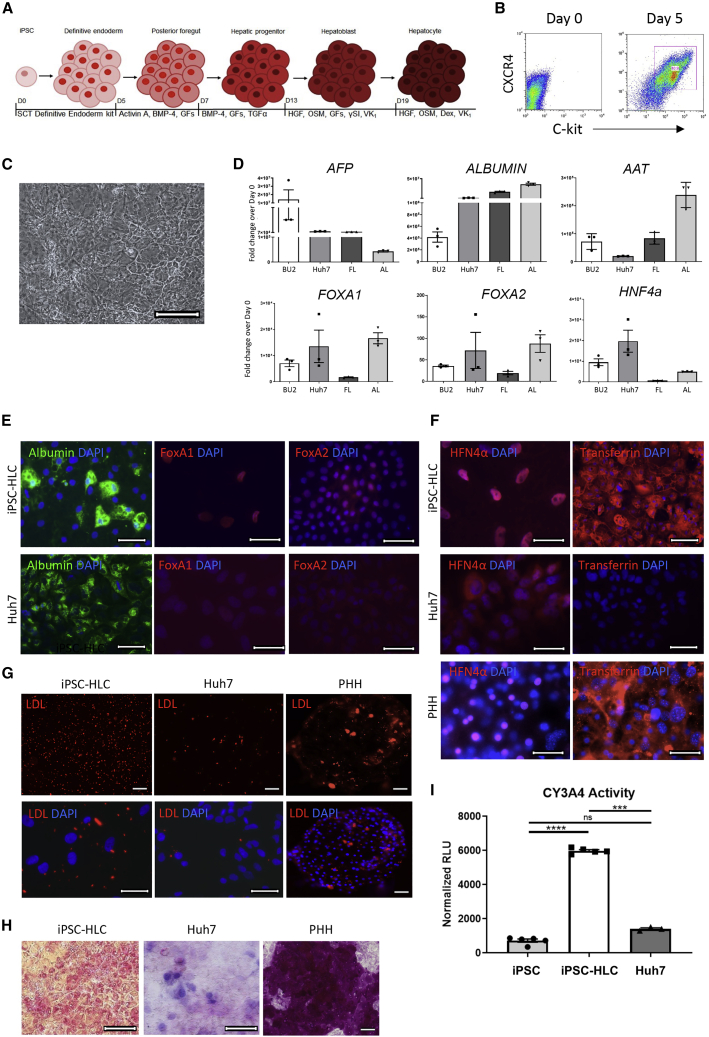
To generate human HLCs from iPSCs, we used a previously published stepwise directed differentiation protocol that follows normal hepatic development as a roadmap for iPSC lineage specification (Figure 1A) (Wilson et al., 2015). At day 5 (D5), approximately 80%–90% of the cells were CXCR4/cKit double positive, indicating robust differentiation into definitive endoderm (Figure 1B). By D20 of differentiation, most cells in culture adopted a cuboidal, highly vacuolated morphology, typical of primary hepatocytes (Figure 1C). A molecular, histological, and functional characterization of the iPSC-HLCs in comparison with either immortalized hepatocarcinoma Huh7 cells or fetal and adult liver tissue showed that the iPSC-HLCs retained expression of the fetal liver marker alpha feto protein (AFP), likely reflecting the embryonic-like characteristics of the cells in this culture system (Figure 1D). As expected, iPSC-HLCs expressed many hepatic markers, including albumin, AAT, and the transcription factors FOXA1, FOXA2, and HNF4a. The mRNA levels of these hepatic markers were generally higher in Huh7 cells compared with iPSC-HLCs. However, immunofluorescence analysis revealed striking differences in the protein levels and localization of the hepatic transcription factors in both cell types (Figure 1E). While FOXA1 and FOXA2 were translocated into the nuclei of iPSC-HLCs, they were expressed at low protein levels in Huh7 cells and homogenously distributed throughout the cytoplasm. Albumin expression was observed in both cell types (Figure 1E). We also compared the expression levels and localization of HNF4α and transferrin in iPSC-HLCs, PHHs, and Huh7 cells. Similar to FOXA1 and FOXA2, the transcription factor HNF4α was expressed at low levels and distributed homogenously in the cytoplasm of Huh7 cells, whereas it was localized in the nuclei of iPSC-HLCs and PHHs (Figure 1F). Transferrin, a clotting factor and carrier protein produced exclusively by hepatocytes in the liver, was robustly expressed in iPSC-HLCs and PHHs, but not in Huh7 cells (Figure 1F), further emphasizing the differences between the primary-like and immortalized cell platforms. Functional analysis of iPSC-HLCs and PHHs compared with Huh7 cells revealed major deficiencies in the Huh7 cells. One major function of hepatocytes in vivo is to store and metabolize lipoproteins, and an inability to bind or retain lipoproteins indicates a deficiency in hepatic function (Kwiterovich, 2000). While the majority of iPSC-HLCs and PHHs were positive in the low-density lipoprotein (LDL) uptake assay, LDL uptake was impaired in Huh7 cells (Figure 1G). Similarly, periodic acid-Schiff (PAS) staining, used to detect glycogen storage, resulted in the typical magenta staining in PHHs, whereas the Huh7 cells showed weak and diffuse staining (Figure 1H). PAS staining in iPSC-HLCs revealed bright pink-magenta staining, also indicating positive glycogen storage. Finally, we tested whether the iPSC-HLCs have proper cytochrome P450 function. Hepatocytes produce cytochrome P450 enzymes, which are involved in the metabolism of over 80% of all prescribed therapeutics (Ingelman-Sundberg et al., 2007). Immortalized hepatocytes express low levels of these enzymes unless specifically cultured in confluent conditions (Donato et al., 2013). To measure the enzymatic activity of the predominant cytochrome P450 enzyme, CY3A4, in iPSC-HLCs and Huh7 cells, we used a luminescent-based assay. In contrast to Huh7 cells that had no CY3A4 activity, iPSC-HLCs showed a strong luminescent signal indicative of CY3A4 enzymatic activity (Figure 1I).
Figure 1.
iPSC-HLCs display molecular hepatic identity and retain hepatic functions
(A) Outline of the directed differentiation protocol used to generate iPSC-HLCs. SCT, STEMCELL Technologies; GFs, growth factors (fibroblast growth factor 2 [FGF-2], vascular endothelial growth factor [VEGF], epidermal growth factor [EGF]; Dex, dexamethasone; VK1, vitamin K; OSM, oncostatin M.
(B) Representative flow cytometry analysis of five replicates of BU2 D0 and D5 iPSC differentiation into endoderm stained for definitive endoderm markers CXCR4 and c-kit (n = 5 independent replicates).
(C) Brightfield image of BU2 iPSC-HLCs at D23. Scale bar represents 200 μm.
(D) TaqMan qRT-PCR analysis of triplicate samples of three independent differentiations of BU2 iPSC-HLCs at D23, Huh7 cells, fetal liver, and adult liver (n = 3 technical replicates of three independent experiments for BU2; n = 3 technical replicates for all other cell types). Expression of each marker is normalized to β-actin and visualized as the fold change over D0 BU2 iPSCs. AFP, α-fetoprotein; AAT, alpha-1 antitrypsin protein; FOXA1, forkhead box A1; FOXA2, forkhead box A2; HNF4α, hepatocyte nuclear factor 4 alpha. Error bars represent SEM.
(E) Expression of albumin, FOXA1, and FOXA2, determined by immunofluorescence analysis in iPSC-HLCs (top row) and Huh7 cells (bottom row). Representative images of BU1 and BU2 and Huh7 samples (n = 1 from each donor for a total of two independent replicates per cell type). Images taken at 40× magnification or 63× magnification with oil, scale bars represent 20 μm. Albumin, BU2; FOXA1, BU1; FOXA2, BU1.
(F) Expression of HNF4α and transferrin, determined by duplicate immunofluorescence analysis of BU1 and BU2 iPSC-HLCs (top row), Huh7 cells (middle row), and donor A primary human hepatocytes PHHs (bottom row) (n = 1 technical replicate from each iPSC-HLC donor and n = 2 independent replicates for Huh7 cell samples and PHH donor A samples for a total of two independent replicates per cell type). Images taken at 40× magnification, 60× magnification, or 63× magnification with oil, scale bars represent 20 μm HNF4α, BU2; transferrin, BU2.
(G) Representative images of two replicates of low-density lipoprotein (LDL)-uptake assay for BU1 and BU2 iPSC-HLCs, Huh7 cells, and donor A PHHs (n = 2 technical replicates for each iPSC-HLC donor and cell type). LDL analog fluoresces red when endocytosed into the cell. Images taken at 10× (top row and bottom row, right image, scale bar represents 100 μm) or 40× magnification (bottom row, left and middle images, scale bar represents 20 μm). Cell nuclei were stained with DAPI; 10× magnification, BU2; 40× magnification, BU1.
(H) Representative PAS staining of three replicates of BU2 iPSC-HLCs, Huh7 cells, and donor A PHHs (n = 3 independent replicates per cell type). Images taken at 20× and 40× magnification, scale bar represents 100 μm.
(I) Detection of active CY3A4 in triplicate wells of a singular differentiation of BU2 iPSCs, BU2 iPSC-HLCs, and Huh7 cells using a P450-Glo assay (n = 3 independent replicates per cell type). Relative luciferase units (RLU) for each sample were normalized to RLU from supernatants of untreated cells. Error bars represent SEM of three technical replicates. ns, not significant; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001.
It was previously reported that infection with vesicular stomatitis virus (VSV), a strong inducer of the type I IFN response, leads to type I IFN expression in PHHs but not in Huh7 cells (Marozin et al., 2008). To examine if the iPSC-HLCs were able to adequately respond to an acute viral infection, iPSC-HLCs and Huh7 cells were infected with VSV and IFNB1 expression was analyzed by qRT-PCR. In contrast to Huh7 cells, IFNB1 expression was strongly upregulated in infected iPSC-HLCs (Figure S1). This indicates that, similar to PHHs, iPSC-HLCs are IFN competent and able to respond to viral stimuli. It also confirms that Huh7 cells may not represent a suitable platform to analyze antiviral responses to viral infections.
Overall, these results indicate that iPSC-HLCs recapitulate functional and molecular hepatocyte features more faithfully than Huh7 cells do and therefore provide a better model to study EBOV hepatic infection.
iPSC-HLCs and PHHs support robust EBOV infection and replication
Next, we determined if iPSC-HLCs were permissive to EBOV infection. As a comparison, we also infected PHHs using a previously established platform based on cryopreserved adult primary hepatocytes collected from donors, allowing for the maintenance of primary hepatocytes in culture for up to several weeks (Khetani and Bhatia, 2008; March et al., 2015). We initially used micropatterned co-cultures (MPCCs) that consist of PHH islands supported by J2-3T3 murine embryonic fibroblasts. The MPCC system was successfully used to for hepatitis C and hepatitis B virus infection studies (March et al., 2015). However, despite robust EBOV infection of the PHHs in this co-culture system (Figures S2A and S2B), the MPCCs were less suitable for EBOV studies because the mouse fibroblasts were infected in addition to the PHHs, albeit at low infection rates (Figures S2A and S2B). Since the goal of this study was to dissect the host response of human hepatocytes to EBOV infection, infected mouse fibroblasts could potentially confound the results. Therefore, we took advantage of a controlled apoptosis system to remove the 3T3-J2s fibroblasts from the MPCCs prior to infection (Chen et al., 2020).
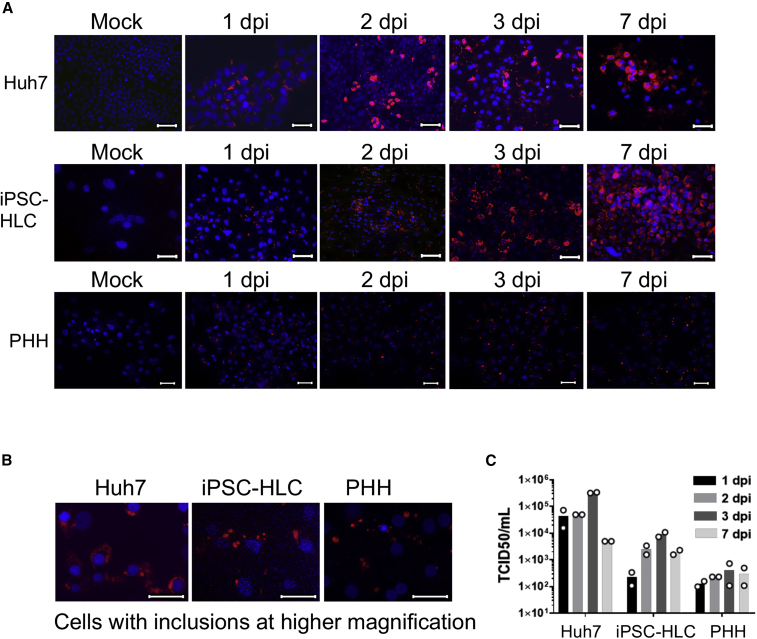
iPSC-HLCs from three different donors (BU1–3), PHHs from two donors in fibroblast-depleted MPCCs, and Huh7 cells were infected with EBOV over a time course of 7 days and analyzed by immunofluorescence staining. At 1 day post infection (dpi), about 50% of iPSC-HLCs and PHHs were infected and about 90% of the Huh7 cells (Figure 2A). Viral inclusion formation, a hallmark of EBOV infection, was observed in all three cell types (Figure 2B). In parallel, we collected supernatants from the infected cells to determine viral titers over the course of infection. Viral titers peaked at 3 dpi, with the highest titers produced in Huh7 cells (5 × 105) followed by iPSC-HLCs (2 × 104) (Figure 2C). The increase in viral titers correlated with an increase in the percentage of infected cells in culture, which consistently showed 80%–90% infection by 3 dpi (Figure 2A). Infected PHHs showed a modest increase in viral production and PHHs over time with overall lower titers (1 × 103) compared with the iPSC-HLCs. In summary, these data show that iPSC-HLCs and PHHs are permissive to EBOV infection and that iPSC-HLCs support EBOV replication more efficiently than PHHs do.
Figure 2.
iPSC-HLCs and PHH are susceptible to EBOV infection
(A) Immunofluorescence analysis of EBOV-infected Huh7 cells, BU2 iPSC-HLCs, and donor A PHHs. Cells were mock infected or infected with EBOV at an MOI of 10, fixed at the indicated time points, and stained with an antibody directed against EBOV nucleoprotein (red). Cell nuclei were stained with DAPI. Images are representative of triplicate replicates (n = 3 technical replicates per cell type). Images of iPSC-HLCs and Huh7 cells taken at 40× magnification. Images of PHHs taken at 20× magnification. Scale bar represents 20 μm.
(B) Higher magnification to visualize viral inclusions. Images taken at 63× magnification. Scale bar represents 20 μm.
(C) Viral titers of each EBOV-infected hepatocyte platform at the indicated time points. Bars represent the average of two technical replicates (n = 2 technical replicates per cell type). iPSC-HLCs, BU2; PHH, donor A.
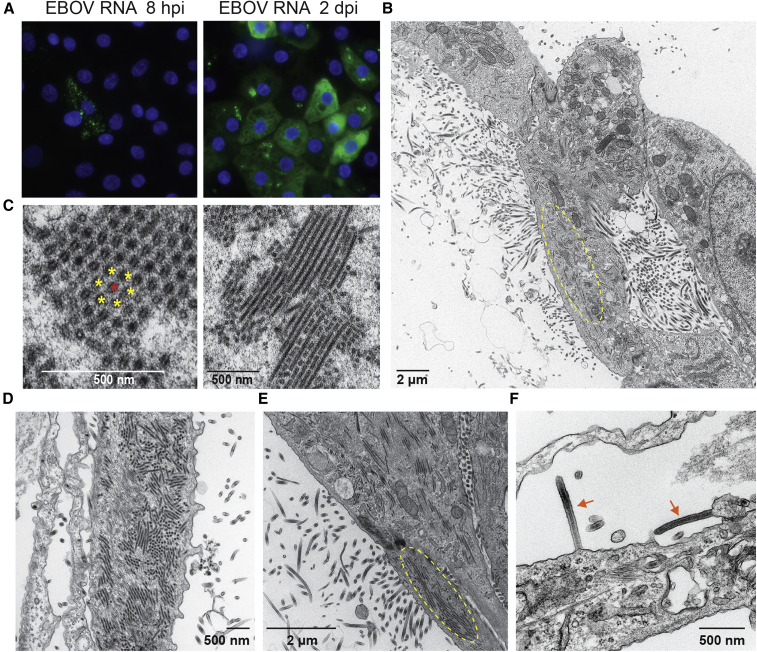
Since the iPSC-HLCs showed robust EBOV infection, we focused on this cell type for a more phenotypical characterization. To visualize spatial and temporal viral RNA production in infected cells at early and late time points, we performed single-cell, single-molecule fluorescent RNA in situ hybridization (RNA-FISH) using RNAscope technology. The used probes are directed against EBOV VP35 positive-sense RNA, which includes VP35 mRNA and antigenomic RNA. Robust viral mRNA production was observed at 8 h post infection (hpi) with a punctate staining pattern throughout the cytoplasm, each small dot likely representing a single mRNA molecule, whereas larger bright puncta may represent RNA aggregates (Figure 3A). At 2 dpi, the viral RNA was homogenously distributed throughout the cytoplasm and aggregated in clusters that co-localize with viral inclusions.
Figure 3.
Productive EBOV infection of iPSC-HLCs
(A) RNA-FISH analysis of BU2 EBOV-infected iPSC-HLCs at the indicated time points using probes directed against EBOV VP35 mRNA. Scale bar represents 20 μm. Images are representative of BU1, BU2, and BU3 samples (n = 1 technical replicate per iPSC donor per time point for a total of three independent replicates per time point).
(B–F) Transmission electron microscopy of BU2 EBOV-infected iPSC-HLCs at 1 and 2 dpi. Cells were infected with an MOI of 3. Image is representative of infections in BU1, BU2, and BU3 (n = 1 replicates per iPSC donor per time point for a total of three independent replicates per time point). (B) Overview of BU2 EBOV-infected cell at 2 dpi. Circled area indicates the accumulation of filamentous viral nucleocapsids into inclusions. (C) Higher magnification of cross-sectioned (left) and longitudinal sectioned (right) viral inclusions. Single nucleocapsids are marked with yellow and red asterisks, respectively, to visualize the hexagonal pattern. (D and E) Release of viral particles at 1 dpi (D) and 2 dpi (E). Circled area in (E) indicates viral inclusions. (F) Arrows indicate mature, budding Ebola virions. Cells were fixed at 2 dpi. Images were taken from cells derived from different donors: (A–D) donor BU2, (E) donor BU1, (F) donor BU3.
Electron microscopy (EM) analysis of EBOV-infected iPSC-HLCs confirmed the formation of EBOV nucleocapsids in the cytoplasm of the infected cells and the release of viral particles (Figure 3B). Newly synthesized EBOV nucleocapsids accumulated in viral inclusions and were arranged in parallel tubes in a hexagonal pattern in which each nucleocapsid is surrounded by six neighboring nucleocapsids, in line with previously reported observations (Figure 3C) (Kolesnikova et al., 2017). The accumulation of viral inclusions and the release of viral particles was observed at 1 and 2 dpi for all three donors (Figures 3D–3F).
To follow the course of EBOV infection in iPSC-HLCs by live cell imaging, we used a recombinant EBOV clone expressing green fluorescence protein (GFP) from an additional transcription unit (Towner et al., 2005). Although the cells were infected with a high MOI of 10, we observed peak infection only at 3 dpi (Figure S3). At 7 dpi, the infected cells showed a pronounced cytopathic effect (CPE), leading to reduced GFP expression. Overall, these data indicate that iPSC-HLCs can be productively infected with EBOV and that the infection leads to cell damage at late time points.
EBOV infection induces a delayed type I IFN response in human hepatocytes
To analyze the transcriptional profile induced by EBOV infection across the three infection platforms, iPSC-HLCs, PHHs, and Huh7 cells were infected with purified Ebola virions, and the cellular RNA was harvested 24 hpi for bulk RNA sequencing analysis. The chosen time point was based on the transcriptional response to EBOV infection in human monocyte-derived macrophages, which display a strong transcriptional signature at 24 hpi (Olejnik et al., 2017). We calculated the log fold change (logFC) in expression for each aligned transcript between the EBOV-infected sample and mock control to determine which transcripts were differentially expressed for each condition and ranked these genes based on absolute logFC. However, our results indicated that there was no significant differential gene expression in infected compared with non-infected cells in either cell type (Figure S4).
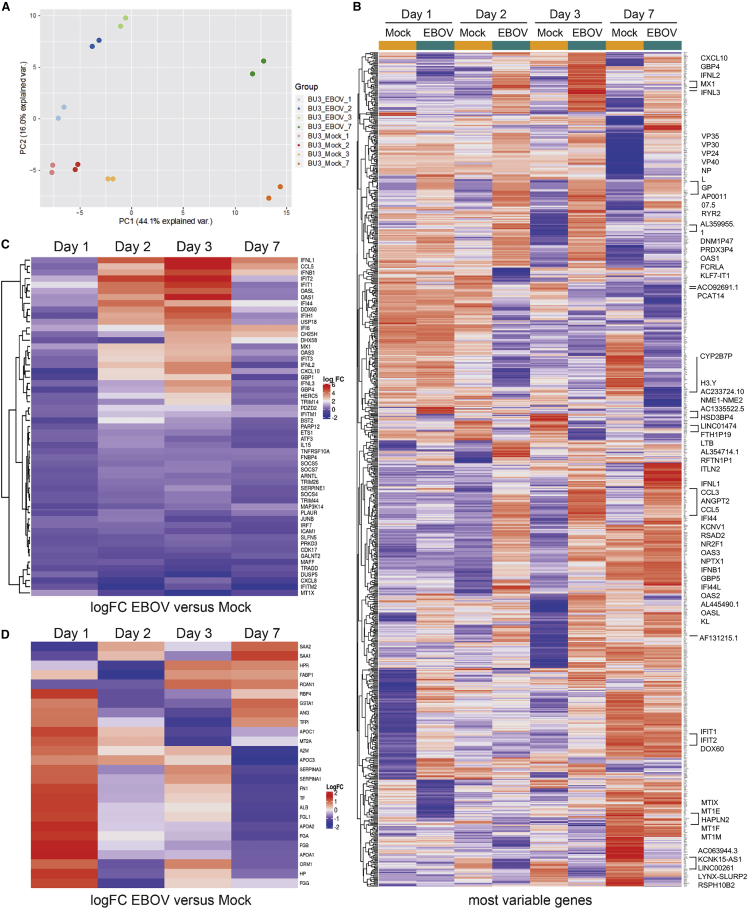
Since we observed peak infection of EBOV-infected iPSC-HLCs as late as 3 dpi (Figure S3), this inspired us to explore the transcriptional response to EBOV infection in liver cells at later time points. iPSC-HLCs were infected with EBOV and cellular RNA was isolated at 1, 2, 3, and 7 dpi for bulk transcriptomics analysis. Principal component analysis (PCA) of mock-infected compared with EBOV-infected samples at all time points showed clear clustering of the replicates, separating the non-infected from the infected samples at each time point, particularly at 2, 3, and 7 dpi (Figure 4A). We observed minimal gene expression changes at 1 dpi, confirming our previous RNA sequencing data. When we ranked the differentially expressed genes based on the false discovery rate (FDR)-corrected p value, there were only 20 upregulated genes and 14 downregulated genes with an adjusted p value ≤ 0.01 at 1 dpi (Table S1). We then applied the same analysis across all time points in mock-infected and EBOV-infected iPSC-HLCs over time and plotted the 250 most variable genes as a heatmap (Figure 4B). Clear gene expression signatures related to EBOV infection emerge at 2 and 3 dpi, converging into a different signature by 7 dpi. Starting 2 dpi, we observed a pronounced type I and III IFN signature, specifically upregulation of IFNB1, IFNL1-3, and a set of IFN-stimulated genes (ISGs). There was a strong overlap of the top 30 differentially expressed genes (DEGs) at 2 and 3 dpi (20 out of 30), including 13 ISGs (Figures 4B, 4C, and S5A). To further assess the induction of a type I/III IFN response in EBOV-infected cells, we plotted the fold change of a panel of type I/III IFNs and 50 ISGs for each time point, confirming the upregulation of ISGs at 2 dpi and, even more pronounced, 3 dpi, including IFNL1-3, IFNB1, chemokine ligand 5 (CCL5), 2′-5′-oligoadenylate synthetase like (OASL), OAS1, interferon-induced protein with tetratricopeptide repeats 1 (IFIT1), IFIT2, and the poorly characterized interferon-induced protein 44 (IFI44), which was shown to have antiviral activity (Busse et al., 2020; Carlton-Smith and Elliott, 2012; Power et al., 2015) (Figure 4C). By 7 dpi, the IFN response signature was no longer present (Figure 4C). To confirm that we observe similar antiviral responses within 2–3 dpi across multiple donors, we assessed the expression of CXCL10, CCL5, and ISG15 by qRT-PCR at each time point in EBOV-infected HLCs from multiple donors (Figure S5C). While we observed variations in gene expression for each donor, the expression of each of these ISGs peaked between 2 and 3 dpi for each donor, confirming that this time point is critical for the initiation of antiviral responses in EBOV-infected HLCs (Figure S5C).
Figure 4.
Upregulation of ISGs and downregulation of genes involved in liver function in EBOV-infected iPSC-HLCs at 2 and 3 dpi
(A) Principal component analysis (PCA) of transcriptional responses of EBOV- and mock-infected BU3 iPSC-HLCs at each time point (n = 2 technical replicates per infection condition per time point). PCA1 represents the majority of variance and is associated with the day of differentiation. PCA2 represents the second largest contributor of variance and is associated with infection status.
(B) Heatmap displaying the 250 most variable genes at each time point. Highlighted are the top 30 upregulated and downregulated DEG at 3 dpi. Key for the value of the logFC to the right of the plot.
(C) Heatmap displaying the logFC for a list of ISGs (right of the plot) of EBOV-infected iPSC-HLCs compared with mock-infected cells at the indicated time points. Key logFC change in expression shown to the right.
(D) Heatmap displaying the logFC for a list of genes involved in liver function (right of the plot) of mock- and EBOV-infected iPSC-HLCs at 3 dpi. Shown are the technical replicates for each condition. Key logFC change in expression shown to the right.
A striking feature of the host response signature in the infected iPSC-HLCs was a general downregulation of genes at later time points, many of them associated with characteristic liver functions. For instance, several of the downregulated genes were associated with aminotransferases and ion transport (9 of 30 genes) (Figure S5B). AGXT, cytochrome P450 enzyme CYP3A7, the bile acid transporter nuclear receptor subfamily 1 group H member 4 (NRIH4), SERPINA7, ITIH1, and ITIH3 were all significantly downregulated at 7 dpi (Figure S5B). Genes involved in plasma formation and clotting, including ALB, insulin-like growth factor 2 (IGF2), podocan (PODN), and n-acetyltransferase 16 (NAT16), were also downregulated at this time point (4 of 30) (Figure S5B). We further confirmed the significant downregulation of liver-associated genes in EBOV-infected iPSC-HLCs 3 dpi by using a curated list of liver-specific genes for comparison (Wilson et al., 2015), including albumin (ALB), serpina genes (SERPINA 1 and SERPINA 3), fibrinogen genes (FGA, FGB, and FGG), apolipoprotein genes (APOA1, APOA2, and APOC1), and angiopoietin (ANG) (Figure 4D). To determine if the downregulation of these genes at later time points was due to EBOV-induced apoptosis, we analyzed cells for caspase 3 activity using a luminescent assay (Figure S7). There was minimal CPE in EBOV-infected HLCs and Huh7 cells and minimal caspase activity compared with staurosporine-treated control cells (Figures S7A and S7B), indicating that EBOV did not induce apoptosis in the infected cells. Overall, our data suggest that EBOV infection leads to severe liver cell damage associated with loss of hepatic functions.
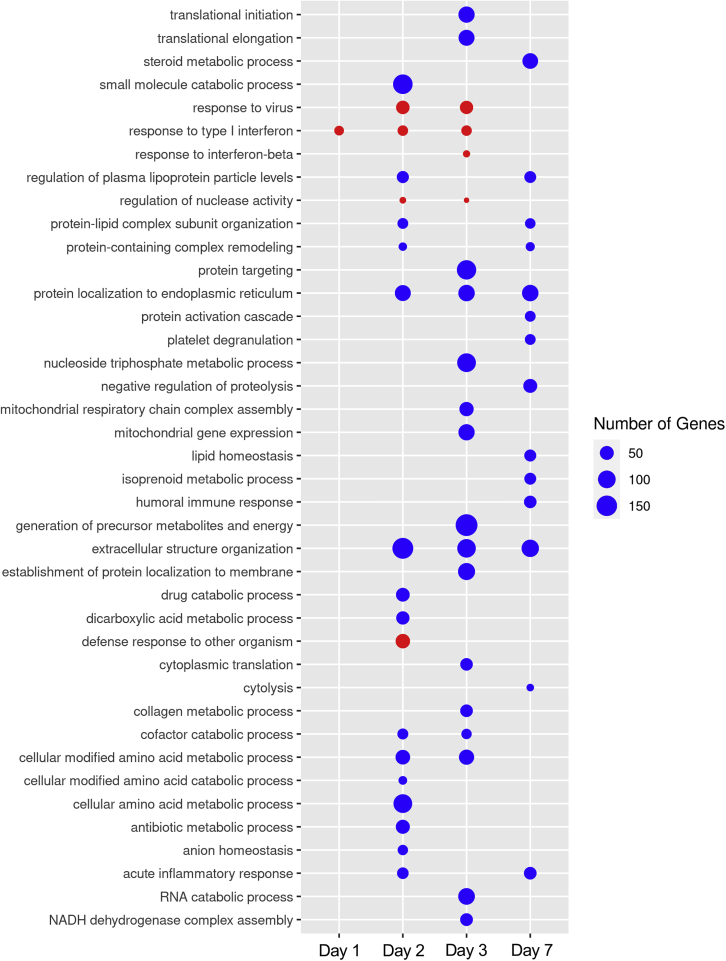
To identify pathways that were perturbed during EBOV infection, we performed gene set enrichment analysis (GSEA) on the DEGs in EBOV-infected cells compared with mock-infected controls, using WebGestalt (Liao et al., 2019). Gene ontology (GO) analysis showed moderate upregulation of IFN response pathways in EBOV-infected cells at 2 and 3 dpi (Figure 5). Hallmark pathway analysis confirmed these results, highlighting that the IFNα response pathway is upregulated at 2 and 3 dpi (Table S2). Importantly, there was no upregulation of inflammatory response pathways, suggesting that EBOV infection does not trigger a proinflammatory response in primary-like hepatocytes. While there were only few moderately upregulated pathways in EBOV-infected cells, pathway downregulation was more pronounced over the course of infection, which we observed in the DEG profile. This includes pathways related to metabolic processing, protein synthesis, protein transport, protein membrane localization, and extracellular structure organization, indicating cellular shutdown (Figure 5). By hallmark pathway analysis, there were no upregulated pathways at 7 dpi (Table S2). By GO analysis, responses to type I IFN and response to virus were slightly upregulated at 7 dpi, which we hypothesize is driven by the low expression of IFNB1 and IFNL1-3 at this time point (Figures 4 and 5). Hallmark pathway analysis suggested a downregulation of coagulation and xenobiotic metabolism pathways, indicating that EBOV-infected hepatocytes respond with a reduced capacity to produce clotting factors or metabolize drugs, which are suggested implications of liver damage during EBOV infection of NHPs (Table S2) (Geisbert et al., 2003c, 2003d). This analysis highlights the changing dynamics in the host response of EBOV-infected iPSC-HLCs.
Figure 5.
EBOV infection of iPSC-HLCs leads to a general downregulation of critical pathways with the exception of viral response pathways
Gene Ontology (GO) analysis of the DEGs in EBOV- compared with mock-infected BU3 iPSC-HLCs for each time point (n = 2 technical replicates per infection condition per time point). FDR ≤0.01. Time point labeled on the bottom of the plot, GO pathways labeled on the left of the plot, key located on the right of the plot. Red bubbles mark pathways that are upregulated at each time point, and blue bubbles denote pathways that are downregulated at each time point. The GO analysis by WebGestalt determined that all p values = 0, indicating that the p value is <0.001 for each pathway.
IFNB1 and CXCL10 expression in EBOV-infected iPSC-HLCs is delayed and mainly observed in non-infected bystander cells
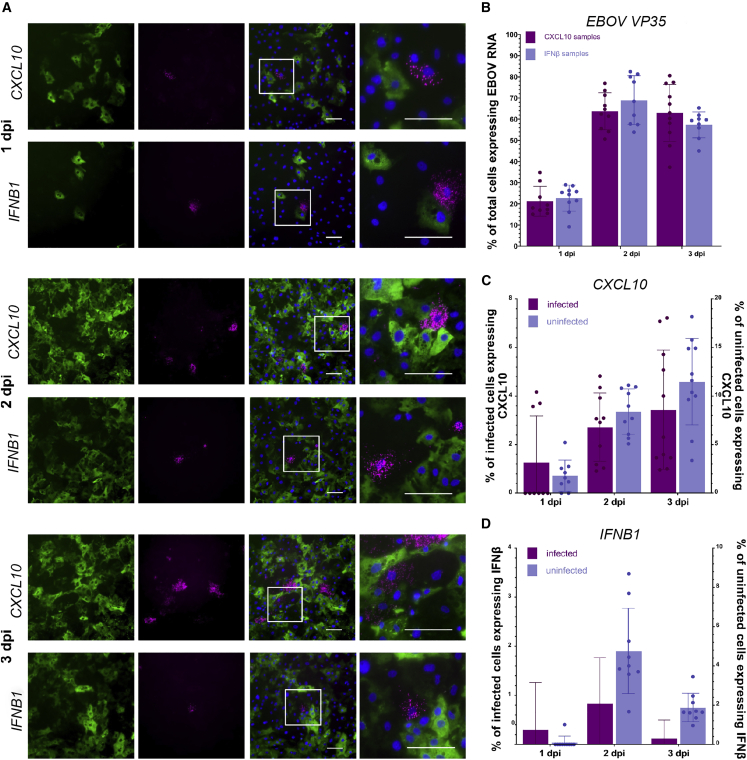
Bulk RNA analysis only provides the average gene expression levels of a diverse cell population and does not allow us to discriminate between the host response signatures in infected and non-infected cells. Given the clear but moderate type I/III IFN response in EBOV-infected iPSC-HLCs, we hypothesized that there might be significant heterogeneity regarding the IFN response in this cell population. To assess the IFN and inflammatory response at single-cell level, we performed RNA-FISH analysis with infected iPSC-HLCs using probes targeting IFNB1, CXCL10, and IL6 mRNA in combination with EBOV-specific probes targeting VP35 mRNA. The number of the cells with detectable VP35 expression increased from about 20% at 1 dpi to 60%–70% at 2 and 3 dpi (Figures 6A and 6B). Consistent with the RNA sequencing data, less than 2% of the cells expressed CXCL10 (1.6%) or IFNB1 (0.2%) at 1 dpi (Figures 6A, 6C, and 6D). We observed an increase in both CXCL10- and IFNB1-expressing cells at 2 and 3 dpi with up to 2% of IFNB1 and 6% of CXCL10-expressing cells. Luminex analysis confirmed the secretion of CXCL10 in the supernatants of infected cells (Figure S6D). Of note, CXCL10 and IFNB1 were mainly expressed in uninfected bystander cells. The ratio of uninfected to infected IFNB1-expressing cells increased from 50% at day 1 to 92% at day 3 (Figure S6A). The signal intensity of EBOV mRNA was weak in cells expressing IFNB1 or CXCL10, suggesting that these cells represent early stages of infection with low expression rates of VP35 and VP24, the known EBOV IFN antagonists. There was no detectable IL6 expression in infected iPSC-HLCs at any time point post infection, confirming the absence of an inflammatory response in these cells (Figure S6B). In contrast to the iPSC-HLCs, we did not observe IFNB1 or CXCL10 expression in Huh7 cells (Figure S6C). This is in line with previous studies using immortalized hepatocytes that did not observe a delayed IFN response upon EBOV infection (Hartman et al., 2008; Kuzmin et al., 2017). Overall, the RNA-FISH results corroborate the findings from the transcriptomic analysis and suggest that EBOV infection of iPSC-HLCs induces a delayed and moderate type I IFN response that is not observed in infected Huh7 cells.
Figure 6.
RNA-FISH analysis reveals expression of IFNB1 and CXCL10 in uninfected bystander cells and cells with weak EBOV mRNA signal intensity
(A) EBOV-infected BU1 and BU2 iPSC-HLCs were fixed at the indicated times and analyzed by RNA-FISH using a combination of probes against EBOV VP35 mRNA (green) and CXCL10 (magenta, top row), or IFNB1 (IFNβ, magenta, bottom row) mRNA. Shown are representative images from both donors at each time point at 20× magnification, and right column shows inset region of merged images (n = 2 technical replicates per iPSC donor per time point for a total of four replicates per time point; two technical replicates of two different donors). Scale bars represent 100 μm.
(B–D) Quantification of gene expression from images using at least 1,200 cells and eight fields of view per time point and probe combination. (B) Percentage of total cells positive for EBOV VP35 mRNA at each time point. Error bars represent SD. (C) Percentage of infected cells expressing CXCL10 mRNA, left axis, and percentage of uninfected cells expressing CXCL10 mRNA, right axis. Error bars represent SD. (D) Percentage of infected cells expressing IFNB1 mRNA, left axis, and percentage of uninfected cells expressing IFNB1 mRNA, right axis. Error bars represent SD. (A) Upper, BU2; lower, BU2. (B) Upper, BU1; lower, BU1. (C) Upper, BU1; lower, BU1.
EBOV can be transmitted from infected macrophages to iPSC-HLCs
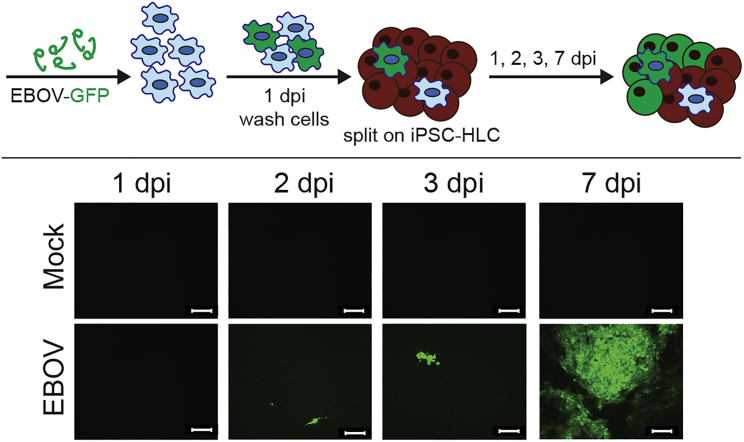
Since there is evidence that migrating monocytes or tissue-resident macrophages are early target cells in the liver (Geisbert et al., 2003b; Greenberg et al., 2020), we explored whether EBOV can be transmitted from infected macrophages to iPSC-HLCs in a co-culture model. Donor-matched human primary monocyte-derived macrophages (MDMs) were infected with recombinant EBOV expressing GFP at an MOI of 10 to ensure that all MDMs were EBOV infected. At 1 dpi, the infected MDMs were detached, rigorously washed, and seeded onto iPSC-HLCs. Spread of infection was visualized by GFP expression (Figure 7). While only single green cells were observed at 2 dpi, likely representing the infected MDMs, the infection spread throughout the entire iPSC-HLC monolayer over the time course of 7 days, indicating that EBOV infection of hepatocytes can be initiated by infected macrophages.
Figure 7.
Transmission of EBOV-GFP from monocyte-derived macrophages (MDMs) to iPSC-HLCs
Top, infection scheme and workflow. Bottom, live fluorescent imaging of BU2 MDM and BU2 iPSC-HLC co-cultures; 10× magnification. Images taken from one well over the period of 1–7 dpi. Scale bar represents 20 μm. Images are representative of two independent experiments performed with BU1, BU2, and BU3 (n = 1 per iPSC donor at each time point for a total of three independent replicates per time point for each infection. This infection time course was performed twice for a total of six independent replicates per time point).
Discussion
Although the liver plays a crucial role in EBOV disease, the intrinsic response of human hepatocytes to EBOV infection is not well defined. In vitro hepatocyte platforms are limited in their capacity to recapitulate human immune responses. Immortalized hepatocytes like Huh7 cells have impaired IRF3 signaling (Marozin et al., 2008), which our study highlights as an important signaling cascade in the EBOV-induced hepatic host response. Additionally, immortalized cell lines can vary significantly from lot to lot, preventing an accurate comparison of results across studies, even within the same cell line (Ben-David et al., 2018). PHHs are difficult to culture and require intricately engineered culture systems to support in vitro survival (Khetani and Bhatia, 2008; March et al., 2015; Xiang et al., 2019). In the co-culture system presented in this study, PHHs destabilize 2 to 3 days after the removal of the fibroblast stroma, which challenges the use of this PHH culture system for longer infection times (Chen et al., 2020). There are recent studies that utilize a combination of small molecules to maintain stable PHHs in culture for up to 4 weeks, which could allow for longitudinal EBOV infections to be performed on a primary hepatocyte (Xiang et al., 2019). However, these culture systems still require delicate engineering to maintain their function. Additionally, all PHH systems are dependent on donor cell availability, which can limit the genetic backgrounds or diversity of samples used in immune response studies. For these reasons, we used iPSC-HLCs as an alternative to traditional immortalized and primary culture models. We have demonstrated that iPSC-HLCs are terminally differentiated hepatocytes and primary-like in their functional capacity. iPSC-HLCs are also differentiated as a monoculture that can be sustained in a functional state for 10 days, allowing for longer infection studies than PHH culture systems. iPSC-based platforms also offer additional advantages, such as the ability to differentiate into multiple cell types from one donor to create organ-specific co-culture systems within one genetic background. This advantage is applicable to EBOV pathogenesis studies since EBOV infects many different cell types and organs, which may contribute to disease progression.
Using the iPSC-based platform, we began to define the intrinsic hepatic response to EBOV infection. The most prominent transcriptional changes are defined by the downregulation of pathways involved in critical cellular processes later in infection, such as protein production and transport, and extracellular structure organization. This general shutdown of cell function is contrasted by a moderate upregulation of type I and type III IFN responses dominated by the expression of IFNB1, IFNL1, and select ISGs beginning 2 dpi and increasing at 3 dpi. At single-cell level, IFNB1 and CXCL10 expression was observed in uninfected bystander cells and cells with low expression levels of EBOV mRNA. This is in line with RNA-FISH analysis on liver samples obtained from EBOV-infected NHPs, showing ISG15 expression predominantly in uninfected bystander cells (Greenberg et al., 2020). In addition, single-cell analysis of blood from EBOV-infected NHPs and infected human peripheral blood mononuclear cells revealed that ISG expression is mainly elicited in uninfected bystander monocytes or monocytes with low amounts of viral RNA, corroborating our results. Although EBOV generally suppresses type I IFN induction in the infected cells (Kotliar et al., 2020; McElroy et al., 2018), there is a strong proinflammatory response in EVD patients and infected animals associated with specialized immune cells, including macrophages and monocytes (Caballero et al., 2016; Greenberg et al., 2020; Kotliar et al., 2020; Liu et al., 2017; McElroy et al., 2016). In contrast to monocytes and macrophages, there was no induction of a proinflammatory response in EBOV-infected human iPSC-HLCs, indicating that, indeed, immune cells are the main drivers of EBOV-induced inflammation. Of note, iPSC-HLCs have been shown to be able to respond to exogenous stimuli by mounting an inflammatory signature, indicating that the lack of an inflammatory response in EBOV-infected cells is not due to the inability of the cells to induce inflammatory cytokines (Fischer et al., 2020).
A crucial question that still needs to be addressed from our observations is that hepatocytes are consistently silent 1 dpi, despite significant infection at this time point. This delayed antiviral response might provide an environment that favors undisturbed viral propagation and open a window for early therapeutic intervention through stimulating an antiviral response by inducing IFN production in uninfected bystander cells. Indeed, US Food and Drug Administration (FDA)-approved therapeutics that stimulate antiviral responses have been shown to inhibit EBOV infection in immortalized cell culture systems and could be tested using the primary-like platform described in this study (Jasenosky et al., 2019).
Another critical component of the hepatic response to EBOV infection is the downregulation of coagulation and acute-phase reactant pathways beginning 2 dpi. Coagulopathy is a well-established clinical indicator of severe EBOV infection, although the exact impact of consumptive coagulopathy on the pathogenesis of EVD is poorly understood (Feldmann and Geisbert, 2011). In our study, iPSC-HLCs downregulated the coagulation cascade 2–7 dpi. We did not observe an increase in acute-phase reactants that has been observed in NHPs (Ward et al., 2019). Our data indicate downregulation of critical clotting factors and plasma proteins, including ALB; fibrinogen chains alpha, beta, and gamma (FGA, FGB, and FGG); protein c (PROC); and F3 as early as 2 dpi. This discrepancy could be evidence of a compensatory feedback loop designed to prevent coagulation dysfunction or consumptive coagulopathy. Fibrinogen levels are lower in EBOV patients than healthy controls and increase in patients who survive, and low levels of serum ALB is a hallmark indicator of EBOV-induced liver damage in NHPs (Geisbert et al., 2003a; Smither et al., 2019). This is emphasized by the network analysis of the 7 dpi DEG that highlighted HNF1A as a critical regulator of the gene expression profile. In EBOV-infected NHPs, another hepatic nuclear factor homeobox gene (HNF4A) was significantly downregulated in the liver, and the authors attributed the downregulation of this transcription factor with the associated downregulation of genes involved in cholesterol synthesis, coagulation, and hepatic metabolism (Jankeel et al., 2020). HNF1A is known to regulate the expression of HNF4A, which may highlight overlap in the transcriptional signatures between our EBOV-infected iPSC-HLCs and bulk NHP liver tissue (Boj et al., 2001; Jankeel et al., 2020). Of the HNF4A targets listed in this study as downregulated transcripts in the liver, we observed significant downregulation in 11 of 17 genes (PCK2, PDK4, APOB, CYP3A7, CYP2A6, F7, TF, F3, TTR, and SERPINC1) (Jankeel et al., 2020). Clearly, the expression of coagulation factors in hepatocytes is affected during EBOV infection, but the role that this may play in pathogenesis, organ failure, or patient outcome is poorly understood. This platform allows for further investigation of the hepatic responses to EBOV infection that can help elucidate the complex mechanisms that underlie liver pathology infection.
The use of co-culture systems, such as the one highlighted in this study, can help determine which cell types in the liver are responsible for specific host responses, helping to define a cell-intrinsic disease signature that is difficult to discern in the whole-organ environment. The data presented here corroborate that genes associated with liver function are perturbed in EBOV-infected hepatocytes. Future studies using this platform can help identify how EBOV impairs liver function during disease progression and inform the development of therapeutic interventions that may universally prevent EBOV fatality.
Experimental procedures
iPSC culture and directed differentiation into hepatocytes
iPSC lines used in this study were BU1, BU2, and BU3. iPSCs were generated and characterized as previously described (Park et al., 2017). Briefly, reprogramming of peripheral blood was done with the STEMCCA reprogramming vector (Somers et al., 2010) and, upon reprogramming, the reprogramming cassette was excised.
iPSCs were maintained on Matrigel-coated six-well tissue culture plates in 2 mL of mTeSR 1 feeder-free medium (STEMCELL Technologies) with 200 ng/mL Primocin (Invitrogen). The directed differentiation from iPSCs into hepatocytes was performed as previously described (Wilson et al., 2015). iPSCs were seeded onto Matrigel-coated six-well tissue culture plates with 2 mL of mTeSR feeder-free medium. For D1–D5 of the differentiation protocol, the STEMdiff Definitive Endoderm kit (STEMCELL Technologies) was used according to manufacturer’s instructions. At D5, iPSC-derived endoderm cells were split at a 1:4 or 1:5 ratio, depending on the iPSC line, and differentiated into HLCs following the schedule and cytokine concentrations outlined in Table S4. The culture medium was changed on D5 and D6. From D7 until the start of each experiment, the culture medium was changed every other day. At D19, the cells were terminally differentiated, and cells were used for experiments between D19 and D26. From D0 until the day of infection, the cells were incubated in a hypoxic incubator at 37°C. During infection, cells were incubated in normoxic conditions.
Primary hepatocyte cell culture
The PHH co-culture with murine embryonic fibroblasts 3T3-J2s were generated by seeding cryopreserved primary human hepatocytes purchased from BioIVT on collagen-patterned 96-well plates as described previously (Khetani and Bhatia, 2008; March et al., 2015). 3T3-J2s were a kind gift provided by Howard Green, Harvard Medical School. Fibroblast-depleted micropatterned PHH cultures were obtained by incorporation of 3T3-J2s expressing an inducible apoptosis switch (inducible caspase-9 [Casp9]). 3T3-J2s were lentivirally transduced with the pMSCV-F-del Casp9.IRES.GFP plasmid (15567, Addgene) followed by FACS purification (top 15%, FACSAria II, BD Biosciences) (Chen et al., 2020). On the day of infection, PHH co-cultures were dosed with a chemical inducer of dimerization (0.5 μM AP20187, Takarabio) to trigger fibroblast removal via apoptosis. PHH co-cultures were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Corning) supplemented with 10% fetal bovine serum (Gibco), 1% insulin/transferrin/selenous acid and linoleic acid (ITS; BD Biosciences), 7 ng/mL glucagon (Sigma-Aldrich), 40 ng/mL dexamethasone (Sigma-Aldrich), 15 mM HEPES (Gibco), and 100 μg/mL penicillin-streptomycin (Corning). Cells were kept at 37C with 5% CO2. Donors utilized in this study are labeled as donor A and donor B.
Virus infection and purification
Work with infectious EBOV was performed at Boston University’s National Emerging Infectious Diseases Laboratories, Boston, MA and at the NIH/NIAID Integrated Research Facility, Fort Detrick, MD. EBOV isolate Mayinga (GenBank: AF086833.2; kindly provided by H. Feldmann, NIH NIAID Rocky Mountain Laboratories, Hamilton, MT) and recombinant EBOV-GFP (Mayinga isolate; kindly provided by the World Reference Center for Emerging Viruses and Arboviruses, University of Texas Medical Branch, Galveston, TX) expressing GFP from an additional transcription unit (Towner et al., 2005) were propagated in Vero E6 cells as described before (Olejnik et al., 2017). Cell supernatants were centrifuged at 1,000 × g for 10 min at 4 °C to remove cellular debris. Clarified supernatants were purified over a 20% sucrose cushion by centrifugation at 25,000 rpm for 2 h at 4 °C and viral pellets were resuspended in PBS. Viral titers were determined by tissue culture infectious dose 50 (TCID50) assays and focus forming unit (FFU) assays as previously described (Olejnik et al., 2017). All EBOV infection studies were performed with purified virions. If not stated otherwise, cells were infected at an MOI of 10. After addition of the inoculum, virions were allowed to adsorb for 20 min to 1 h at 37°C before cells were washed and fresh cell culture medium was added.
qRT-PCR and RNA sequencing
Cells were harvested for RNA analysis by using either the RNeasy Kit (Qiagen) or TRIzol Reagent (Thermo Fisher) according to manufacturer’s instructions. Detailed descriptions of qRT-PCR analysis, Illumina sequencing, and SmartSeq2 sequencing, including data analysis, are provided in the supplemental methods.
Immunofluorescence analysis
Cells were fixed with either 4% PFA or 10% formalin and permeabilized with acetone-methanol. Primary antibody names and dilutions are listed in Table S3. A detailed description of the procedures is provided in the supplemental methods.
Fluorescence in situ hybridization assay
Fluorescence in situ hybridization (RNA-FISH) was performed using the Advanced Cell Diagnostics RNAscope v2 kit according to manufacturer’s instructions (ACD Bio) and as previously described (Hume et al., 2022). A detailed protocol is provided in the supplemental methods. The following probes (all from ACD Bio) were used: ebolavirus Zaire VP35-C1, human IL6-C2, human IFNβ-C3, and human CXCL10-C3.
EM
For conventional thin-section microscopic evaluation, 4 × 105 EBOV-infected BU1, BU2, and BU3 iPSC-HLCs were inactivated and preserved in 2.5% glutaraldehyde (E.M. Sciences, Warrington, PA), in Millonig’s sodium phosphate buffer (Tousimis Research, Rockville, MD), for 24 h (n = 1 for each donor at each time point for a total of three samples per time point). After fixation was complete, the cells were washed in Millonig’s buffer, and incubated for 2 h in 1.0% osmium tetroxide, in the same buffer. Following rinsing steps in ultrapure water and en bloc staining with 2.0% uranyl acetate, the samples were dehydrated in a series of graded ethanol washes and infiltrated and embedded in Spurr’s plastic resin (E.M. Sciences). Embedded blocks were sectioned using a Leica UC7 Ultramicrotome, and 70- to 80-nm sections were collected on 200 mesh copper grids, and post-stained with Reynold’s lead citrate. Samples were examined in an FEI Tecnai Spirit Twin transmission electron microscope, operating at 80 kV.
Co-culture of iPSC-HLCs and macrophages
On the day prior to infection, MDMs from BU1, BU2, and BU3 seeded in a T75 flask were split by incubating cells with 0.1% Trypsin-EDTA solution for 15 min at 37°C and 5% CO2 (n = 1 flask per donor). Cells were centrifuged at 300 × g for 5 min at 4°C and resuspended in 1 mL of MDM medium. Cells were counted and 1 × 106 cells were seeded into six-well low-attachment plates (Corning). On the day of infection, macrophages were infected with EBOV-GFP at an MOI of 10. At 1 dpi, MDMs were incubated with 1 mL of Gentle Cell Dissociation buffer (GCD; Gibco) for 2 min at 37°C and 5%CO2. Cells were centrifuged at 300 × g for 5 min at 4°C. GCD was removed, and cell pellets were washed three times in 1 mL of PBS. After the third wash, cells were resuspended in 1 mL of the appropriate medium and seeded in six- or 12-well tissue culture plates alone or onto D20 iPSC-HLCs at a 1:1 ratio (n = 1 per donor per time point for a total of three samples per time point).
Author contributions
W.A.S. differentiated iPSC-HLCs, performed EBOV infection studies and most of the post-infection experiments, was involved in data analysis, and wrote the first draft of the manuscript. L.M.S. and S.M.R. generated primary hepatocyte cultures. C.V.M., J.V.L., and F.W. performed bioinformatics analyses. J.G.B. performed the EM analysis. S.M., R.F.J., and A.J.H. performed BSL-4 infection studies. E.L.S. performed FISH analysis, imaging, and image quantification. J.O. performed RNA and Luminex analysis. A.M. performed RNA analysis. E.Y.F. performed functional assays of the PHHs. S.N.B. was involved in the design of the primary hepatocyte experiments. E.M. and G.M. designed and directed the study, performed data analysis and interpretation, and edited the manuscript.
Acknowledgments
We would like to thank H. Feldmann, J. Connor, and the World Reference Center for Emerging Viruses and Arboviruses for providing virus stocks; J. Alonso and H. Green for providing cells; and G. Olinger and the Defense Biological Product Assurance Office (DBPAO) through BEI for providing antibodies. This work was supported by NIH grants R21AI137793 (to G.M. and E.M.), N0175N92020C00005 (to G.M.), R21AI135912 (to E.M.), R01AI133486 (to E.M.), and NIH training grants T32HL007035 (to E.L.S.) and TL1TR001410 (to W.A.S.). S.N.B. is a Howard Hughes Medical Institute Investigator.
Conflicts of interest
The authors declare no competing interests.
Published: September 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.08.003.
Contributor Information
Elke Mühlberger, Email: muehlber@bu.edu.
Gustavo Mostoslavsky, Email: gmostosl@bu.edu.
Supplemental information
Data and code availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplemental information. RNA sequencing data have been deposited to Gene Expression Omnibus (GEO: GSE184640). Pluripotent stem cell lines used in this study, along with maintenance standard operating procedures and directed differentiation protocols, are available from the CReM iPSC Repository at Boston University and Boston Medical Center and can be found at http://crem.bu.edu.
References
- Ben-David U., Siranosian B., Ha G., Tang H., Oren Y., Hinohara K., Strathdee C.A., Dempster J., Lyons N.J., Burns R., et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560:325–330. doi: 10.1038/s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj S.F., Parrizas M., Maestro M.A., Ferrer J. A transcription factor regulatory circuit in differentiated pancreatic cells. Proc. Natl. Acad. Sci. USA. 2001;98:14481–14486. doi: 10.1073/pnas.241349398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse D.C., Habgood-Coote D., Clare S., Brandt C., Bassano I., Kaforou M., Herberg J., Levin M., Eléouët J.F., Kellam P., Tregoning J.S. Interferon-induced protein 44 and interferon-induced protein 44-like restrict replication of respiratory syncytial virus. J. Virol. 2020;94:e00297-20. doi: 10.1128/JVI.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero I.S., Honko A.N., Gire S.K., Winnicki S.M., Melé M., Gerhardinger C., Lin A.E., Rinn J.L., Sabeti P.C., Hensley L.E., Connor J.H. In vivo Ebola virus infection leads to a strong innate response in circulating immune cells. BMC Genom. 2016;17:707. doi: 10.1186/s12864-016-3060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton-Smith C., Elliott R.M. Viperin, MTAP44, and protein kinase R contribute to the interferon-induced inhibition of Bunyamwera Orthobunyavirus replication. J. Virol. 2012;86:11548–11557. doi: 10.1128/JVI.01773-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.X., Chhabra A., Song H.H.G., Fleming H.E., Chen C.S., Bhatia S.N. Controlled apoptosis of stromal cells to engineer human microlivers. Adv. Funct. Mater. 2020;30:1910442. doi: 10.1002/adfm.201910442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Rosenke K., Fischer R.J., Marzi A., Prescott J., Bushmaker T., van Doremalen N., Emery S.L., Falzarano D., Feldmann F., et al. Ebola laboratory response at the eternal love winning africa campus, monrovia, Liberia, 2014-2015. J. Infect. Dis. 2016;214:S169–S176. doi: 10.1093/infdis/jiw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato M.T., Jover R., Gómez-Lechón M.J. Hepatic cell lines for drug hepatotoxicity testing: limitations and strategies to upgrade their metabolic competence by gene engineering. Curr. Drug Metab. 2013;14:946–968. doi: 10.2174/1389200211314090002. [DOI] [PubMed] [Google Scholar]
- Feldmann H., Geisbert T.W. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer L., Lucendo-Villarin B., Hay D.C., O'Farrelly C. Human PSC-derived hepatocytes express low levels of viral pathogen recognition receptors, but are capable of mounting an effective innate immune response. Int. J. Mol. Sci. 2020;21:E3831. doi: 10.3390/ijms21113831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert T.W., Hensley L.E., Jahrling P.B., Larsen T., Geisbert J.B., Paragas J., Young H.A., Fredeking T.M., Rote W.E., Vlasuk G.P. Treatment of Ebola virus infection with a recombinant inhibitor of factor VIIa/tissue factor: a study in rhesus monkeys. Lancet. 2003;362:1953–1958. doi: 10.1016/S0140-6736(03)15012-X. [DOI] [PubMed] [Google Scholar]
- Geisbert T.W., Hensley L.E., Larsen T., Young H.A., Reed D.S., Geisbert J.B., Scott D.P., Kagan E., Jahrling P.B., Davis K.J. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 2003;163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert T.W., Young H.A., Jahrling P.B., Davis K.J., Kagan E., Hensley L.E. Mechanisms underlying coagulation abnormalities in ebola hemorrhagic fever: overexpression of tissue factor in primate monocytes/macrophages is a key event. J. Infect. Dis. 2003;188:1618–1629. doi: 10.1086/379724. [DOI] [PubMed] [Google Scholar]
- Geisbert T.W., Young H.A., Jahrling P.B., Davis K.J., Larsen T., Kagan E., Hensley L.E. Pathogenesis of Ebola hemorrhagic fever in primate models: evidence that hemorrhage is not a direct effect of virus-induced cytolysis of endothelial cells. Am. J. Pathol. 2003;163:2371–2382. doi: 10.1016/S0002-9440(10)63592-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg A., Huber B.R., Liu D.X., Logue J.P., Hischak A.M.W., Hart R.J., Abbott M., Isic N., Hisada Y.M., Mackman N., et al. Quantification of viral and host biomarkers in the liver of rhesus macaques: a longitudinal study of Zaire ebolavirus strain kikwit (EBOV/Kik) Am. J. Pathol. 2020;190:1449–1460. doi: 10.1016/j.ajpath.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gural N., Mancio-Silva L., Miller A.B., Galstian A., Butty V.L., Levine S.S., Patrapuvich R., Desai S.P., Mikolajczak S.A., Kappe S.H.I., et al. In vitro culture, drug sensitivity, and transcriptome of plasmodium vivax hypnozoites. Cell Host Microbe. 2018;23:395–406.e4. doi: 10.1016/j.chom.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A.L., Ling L., Nichol S.T., Hibberd M.L. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J. Virol. 2008;82:5348–5358. doi: 10.1128/JVI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzer M., Krähling V., Amman F., Barth E., Bernhart S.H., Carmelo V.A.O., Collatz M., Doose G., Eggenhofer F., Ewald J., et al. Differential transcriptional responses to Ebola and Marburg virus infection in bat and human cells. Sci. Rep. 2016;6:34589. doi: 10.1038/srep34589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume A.J., Heiden B., Olejnik J., Suder E.L., Ross S., Scoon W.A., Bullitt E., Ericsson M., White M.R., Turcinovic J., et al. Recombinant Lloviu virus as a tool to study viral replication and host responses. PLoS Pathog. 2022;18:e1010268. doi: 10.1371/journal.ppat.1010268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelman-Sundberg M., Sim S.C., Gomez A., Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Jankeel A., Menicucci A.R., Woolsey C., Fenton K.A., Mendoza N., Versteeg K., Cross R.W., Geisbert T.W., Messaoudi I. Early transcriptional changes within liver, adrenal gland, and lymphoid tissues significantly contribute to ebola virus pathogenesis in cynomolgus macaques. J. Virol. 2020;94:e00250-20. doi: 10.1128/JVI.00250-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasenosky L.D., Cadena C., Mire C.E., Borisevich V., Haridas V., Ranjbar S., Nambu A., Bavari S., Soloveva V., Sadukhan S., et al. The FDA-approved oral drug nitazoxanide amplifies host antiviral responses and inhibits ebola virus. iScience. 2019;19:1279–1290. doi: 10.1016/j.isci.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash J.C., Mühlberger E., Carter V., Grosch M., Perwitasari O., Proll S.C., Thomas M.J., Weber F., Klenk H.D., Katze M.G. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J. Virol. 2006;80:3009–3020. doi: 10.1128/JVI.80.6.3009-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani S.R., Bhatia S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- Kolesnikova L., Nanbo A., Becker S., Kawaoka Y. Inside the cell: assembly of filoviruses. Curr. Top. Microbiol. Immunol. 2017;411:353–380. doi: 10.1007/82_2017_15. [DOI] [PubMed] [Google Scholar]
- Kotliar D., Lin A.E., Logue J., Hughes T.K., Khoury N.M., Raju S.S., Wadsworth M.H., 2nd, Chen H., Kurtz J.R., Dighero-Kemp B., et al. Single-cell profiling of ebola virus disease in vivo reveals viral and host dynamics. Cell. 2020;183:1383–1401.e19. doi: 10.1016/j.cell.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin I.V., Schwarz T.M., Ilinykh P.A., Jordan I., Ksiazek T.G., Sachidanandam R., Basler C.F., Bukreyev A. Innate immune responses of bat and human cells to filoviruses: commonalities and distinctions. J. Virol. 2017;91:e02471-16. doi: 10.1128/JVI.02471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiterovich P.O., Jr. The metabolic pathways of high-density lipoprotein, low-density lipoprotein, and triglycerides: a current review. Am. J. Cardiol. 2000;86:5L–10L. doi: 10.1016/s0002-9149(00)01461-2. [DOI] [PubMed] [Google Scholar]
- Lang J., Vera D., Cheng Y., Tang H. Modeling dengue virus-hepatic cell interactions using human pluripotent stem cell-derived hepatocyte-like cells. Stem Cell Rep. 2016;7:341–354. doi: 10.1016/j.stemcr.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanini S., Portella G., Vairo F., Kobinger G.P., Pesenti A., Langer M., Kabia S., Brogiato G., Amone J., Castilletti C., et al. Relationship between viremia and specific organ damage in ebola patients: a cohort study. Clin. Infect. Dis. 2018;66:36–44. doi: 10.1093/cid/cix704. [DOI] [PubMed] [Google Scholar]
- Li K., Chen Z., Kato N., Gale M., Jr., Lemon S.M. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem. 2005;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Speranza E., Muñoz-Fontela C., Haldenby S., Rickett N.Y., Garcia-Dorival I., Fang Y., Hall Y., Zekeng E.G., Lüdtke A., et al. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol. 2017;18:4. doi: 10.1186/s13059-016-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March S., Ramanan V., Trehan K., Ng S., Galstian A., Gural N., Scull M.A., Shlomai A., Mota M.M., Fleming H.E., et al. Micropatterned coculture of primary human hepatocytes and supportive cells for the study of hepatotropic pathogens. Nat. Protoc. 2015;10:2027–2053. doi: 10.1038/nprot.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozin S., Altomonte J., Stadler F., Thasler W.E., Schmid R.M., Ebert O. Inhibition of the IFN-beta response in hepatocellular carcinoma by alternative spliced isoform of IFN regulatory factor-3. Mol. Ther. 2008;16:1789–1797. doi: 10.1038/mt.2008.201. [DOI] [PubMed] [Google Scholar]
- Martines R.B., Ng D.L., Greer P.W., Rollin P.E., Zaki S.R. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J. Pathol. 2015;235:153–174. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- McElroy A.K., Harmon J.R., Flietstra T.D., Campbell S., Mehta A.K., Kraft C.S., Lyon M.G., Varkey J.B., Ribner B.S., Kratochvil C.J., et al. Kinetic analysis of biomarkers in a cohort of US patients with ebola virus disease. Clin. Infect. Dis. 2016;63:460–467. doi: 10.1093/cid/ciw334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy A.K., Mühlberger E., Muñoz-Fontela C. Immune barriers of Ebola virus infection. Curr. Opin. Virol. 2018;28:152–160. doi: 10.1016/j.coviro.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidis E., Vercauteren K., Mancio-Silva L., Andrus L., Jahan C., Ricardo-Lax I., Zou C., Kabbani M., Park P., Quirk C., et al. Expansion, in vivo-ex vivo cycling, and genetic manipulation of primary human hepatocytes. Proc. Natl. Acad. Sci. USA. 2020;117:1678–1688. doi: 10.1073/pnas.1919035117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejnik J., Forero A., Deflubé L.R., Hume A.J., Manhart W.A., Nishida A., Marzi A., Katze M.G., Ebihara H., Rasmussen A.L., Mühlberger E. Ebolaviruses associated with differential pathogenicity induce distinct host responses in human macrophages. J. Virol. 2017;91:e00179-17. doi: 10.1128/JVI.00179-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Gianotti-Sommer A., Molina-Estevez F.J., Vanuytsel K., Skvir N., Leung A., Rozelle S.S., Shaikho E.M., Weir I., Jiang Z., et al. A comprehensive, ethnically diverse library of sickle cell disease-specific induced pluripotent stem cells. Stem Cell Rep. 2017;8:1076–1085. doi: 10.1016/j.stemcr.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power D., Santoso N., Dieringer M., Yu J., Huang H., Simpson S., Seth I., Miao H., Zhu J. IFI44 suppresses HIV-1 LTR promoter activity and facilitates its latency. Virology. 2015;481:142–150. doi: 10.1016/j.virol.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollin P.E., Bausch D.G., Sanchez A. Blood chemistry measurements and D-Dimer levels associated with fatal and nonfatal outcomes in humans infected with Sudan Ebola virus. J. Infect. Dis. 2007;196:S364–S371. doi: 10.1086/520613. [DOI] [PubMed] [Google Scholar]
- Sakurai F., Mitani S., Yamamoto T., Takayama K., Tachibana M., Watashi K., Wakita T., Iijima S., Tanaka Y., Mizuguchi H. Human induced-pluripotent stem cell-derived hepatocyte-like cells as an in vitro model of human hepatitis B virus infection. Sci. Rep. 2017;7:45698. doi: 10.1038/srep45698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R.E., Trehan K., Andrus L., Sheahan T.P., Ploss A., Duncan S.A., Rice C.M., Bhatia S.N. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 2012;109:2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai A., Schwartz R.E., Ramanan V., Bhatta A., de Jong Y.P., Bhatia S.N., Rice C.M. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc. Natl. Acad. Sci. USA. 2014;111:12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smither S.J., O'Brien L.M., Eastaugh L., Woolley T., Lever S., Fletcher T., Parmar K., Hunt B.J., Watts S., Kirkman E. Haemostatic changes in five patients infected with ebola virus. Viruses. 2019;11 doi: 10.3390/v11070647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers A., Jean J.C., Sommer C.A., Omari A., Ford C.C., Mills J.A., Ying L., Sommer A.G., Jean J.M., Smith B.W., et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cell. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J.S., Paragas J., Dover J.E., Gupta M., Goldsmith C.S., Huggins J.W., Nichol S.T. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- Tricot T., Helsen N., Kaptein S.J.F., Neyts J., Verfaillie C.M. Human stem cell-derived hepatocyte-like cells support Zika virus replication and provide a relevant model to assess the efficacy of potential antivirals. PLoS One. 2018;13:e0209097. doi: 10.1371/journal.pone.0209097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeki T.M., Mehta A.K., Davey R.T., Jr., Liddell A.M., Wolf T., Vetter P., Schmiedel S., Grünewald T., Jacobs M., Arribas J.R., et al. Clinical management of ebola virus disease in the United States and europe. N. Engl. J. Med. 2016;374:636–646. doi: 10.1056/NEJMoa1504874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M.D., Brueggemann E.E., Kenny T., Reitstetter R.E., Mahone C.R., Trevino S., Wetzel K., Donnelly G.C., Retterer C., Norgren R.B., Jr., et al. Characterization of the plasma proteome of nonhuman primates during Ebola virus disease or melioidosis: a host response comparison. Clin. Proteomics. 2019;16:7. doi: 10.1186/s12014-019-9227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.A., Ying L., Liesa M., Segeritz C.P., Mills J.A., Shen S.S., Jean J., Lonza G.C., Liberti D.C., Lang A.H., et al. Emergence of a stage-dependent human liver disease signature with directed differentiation of alpha-1 antitrypsin-deficient iPS cells. Stem Cell Rep. 2015;4:873–885. doi: 10.1016/j.stemcr.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C., Du Y., Meng G., Soon Yi L., Sun S., Song N., Zhang X., Xiao Y., Wang J., Yi Z., et al. Long-term functional maintenance of primary human hepatocytes in vitro. Science. 2019;364:399–402. doi: 10.1126/science.aau7307. [DOI] [PubMed] [Google Scholar]
- Zaki S.R., Goldsmith C.S. Pathologic features of filovirus infections in humans. Curr. Top. Microbiol. Immunol. 1999;235:97–116. doi: 10.1007/978-3-642-59949-1_7. [DOI] [PubMed] [Google Scholar]
- Zhou X., Sun P., Lucendo-Villarin B., Angus A.G.N., Szkolnicka D., Cameron K., Farnworth S.L., Patel A.H., Hay D.C. Modulating innate immunity improves hepatitis C virus infection and replication in stem cell-derived hepatocytes. Stem Cell Rep. 2014;3:204–214. doi: 10.1016/j.stemcr.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplemental information. RNA sequencing data have been deposited to Gene Expression Omnibus (GEO: GSE184640). Pluripotent stem cell lines used in this study, along with maintenance standard operating procedures and directed differentiation protocols, are available from the CReM iPSC Repository at Boston University and Boston Medical Center and can be found at http://crem.bu.edu.