Abstract
A variety of single-gene human diseases are caused by haploinsufficiency, a genetic condition by which mutational inactivation of one allele leads to reduced protein levels and functional impairment. Translational enhancement of the spare allele could exert a therapeutic effect. Here we developed BOOST, a novel gene-editing approach to rescue haploinsufficiency loci by the change of specific single nucleotides in the Kozak sequence, which controls translation by regulating start codon recognition. We evaluated for translational strength 230 Kozak sequences of annotated human haploinsufficient genes and 4621 derived variants, which can be installed by base editing, by a high-throughput reporter assay. Of these variants, 149 increased the translation of 47 Kozak sequences, demonstrating that a substantial proportion of haploinsufficient genes are controlled by suboptimal Kozak sequences. Validation of 18 variants for 8 genes produced an average enhancement in an expression window compatible with the rescue of the genetic imbalance. Base editing of the NCF1 gene, whose monoallelic loss causes chronic granulomatous disease, resulted in the desired increase of NCF1 (p47phox) protein levels in a relevant cell model. We propose BOOST as a fine-tuned approach to modulate translation, applicable to the correction of dozens of haploinsufficient monogenic disorders independently of the causing mutation.
INTRODUCTION
Haploinsufficiency is one of the major causes of mendelian dominant diseases. It is a pathogenetic mechanism in which the mutational inactivation of one protein-coding allele reduces the expression of the functional protein to a level that is not sufficient to sustain its physiological role (1). Approximately 300 genes in the human genome have been annotated as haploinsufficient (HI) with a disease phenotype (2–4). Those genes are responsible for many single-gene disorders, including susceptibility to cancer, growth retardation, and developmental, neurological, or metabolic syndromes. Moreover, a recent large-scale analysis of human genetic variation identified more than 3000 loss-of-function intolerant genes, including all the previous ones; >70% of them are still unassigned for a human disease phenotype (5). Therefore, it is likely that the number of genes whose hemiallelic state confers a non-trivial survival or reproductive disadvantage, despite not an evident disease or disease susceptibility, will grow in the next future.
Translation is a key layer of gene expression regulation. In eukaryotes, it is fundamental to control the spatial distribution of proteins in different cells and tissues or to trigger fast and reversible responses to environmental changes (6,7). Most eukaryotic mRNAs are translated via the so-called mechanism of cap-dependent translation, in which the 43S preinitiation complex attaches to the 5’ capped end of the mRNA and scans linearly until it encounters the first AUG starting codon; recognition of the initiation codon is followed by the recruitment of the large ribosomal subunit (8). This process is recognized as an essential control step for translational efficiency, which is modulated by cis-elements present in the 5’UTR of mRNAs, such as internal ribosome entry sites (IRESs) or upstream open reading frames (uORFs) (9,10). An additional cis-acting control of translational efficiency was discovered forty years ago by Marilyn Kozak, who showed that the sequences flanking the AUG starting codon directly impact translational efficiency (11). She described a sequence, the Kozak sequence, as the optimal nucleotide context around the AUG starting codon: GCCRCCaugG, in which the purine (R) in position –3 and the G in position +1 with respect to the AUG starting codon are the most important bases to allow efficient translation (11–13). Since then, the strength of Kozak sequence has been extensively investigated also in other organisms, such as yeast (14) or zebrafish (15).
Translational modulation operated by the Kozak sequence in mammals is demonstrated by Mendelian and complex diseases in which translation efficiency is affected because of point variations near the AUG starting codon. For instance, a C-to-T mutation in position -1 with respect to the AUG codon within the Kozak sequence (–1C > T) of the α-tocopherol transfer protein gene reduces protein levels and causes the AVED (ataxia with vitamin E deficiency) monogenic disorder (16,17). As another example, a T/C polymorphism in the same –1 position of the CD40 gene (−1T > C, rs1883832) is associated with increased CD40 translation (18,19), therefore predisposing to Graves’ disease (20) and coronary heart disease (18,19). Despite the apparently strict pattern of the Kozak consensus motif, sequence variability is present around the AUG starting codon in the human genome and in the genomes of other vertebrates, and more recent efforts aimed at measuring the strength of different AUG starting codons as a function of the surrounding bases have found substantial variance (21–25). These results open to the possibility that several genes are translationally controlled by suboptimal Kozak sequences.
High-throughput approaches to select strong Kozak sequences have been recently exploited with the aim of improving the yield and product quality of bispecific antibodies (bsAbs) in CHO cells (26) or the production of natural compounds in yeast (27). However, no effort has been made to manipulate the Kozak sequence for direct therapeutic purposes.
Base editors are a recently developed CRISPR-Cas genome-editing tool able to generate point variations in genomic DNA without the need for donor DNA and without creating double-strand breaks. They were originally composed of the fusion of a Cas9 nickase with a base modification enzyme (deaminase) which performs single nucleotide substitutions in the DNA (28). Two main classes of base editors have been developed: cytosine base editors, converting C–G into T–A base pairs, and adenine base editors, substituting A–T to G–C base pairs (29). Base editors enable the implementation of gene therapy protocols while avoiding the safety issues associated with the DNA double-strand breaks caused by CRISPR-Cas9 (30) but retaining its high precision. Recent in vivo successes (31–33) prove their potential. In a previous example, base editors have been used to introduce an AUG starting codon upstream of a reporter protein to precisely report on editing efficiency (34).
We conceived that the availability of base editors and the possible presence of suboptimal Kozak sequences in the human genome could provide an elegant approach to modulate translational efficiency for the molecular compensation of some HI disorders. Moreover, such an approach would be independent of the type of alteration inactivating the defective allele and, therefore, highly suitable for gene therapy protocols.
Here, to find base conversions upregulating translational efficiency, we systematically screened 4621 variants of Kozak sequences for translational strength. We designed these variants in the main AUG context of 230 previously annotated haploinsufficient genes (2). Analysis of our library of Kozak variants, bearing the conversions that base editors can reproduce, identified 47 genes in which at least one variant was significantly stronger than the wild-type Kozak. This finding proved that weak Kozak sequences are present at about a 20% frequency in our gene sample. We validated the variants of eight genes and base-edited one of them in a cell model, providing proof of principle of the approach, which we called BOOST (Base editing cOrrection of haplOinSufficiency by Translational enhancement).
MATERIALS AND METHODS
Plasmids
Guide RNAs were cloned inside pUC19 (Addgene plasmid #50005) using BbsI (Thermo Fisher Scientific, #ER1011) restriction sites as previously described (35). The base editors used were purchased from Addgene: pCMV_ABE7.10 (#102919); pCMV_ABEmax (#112095); pCMV_AncBE4max (#112094). Guide RNAs sequences are listed in Supplementary Table S4, along with a list of in silico predicted off-target effects, as analysed by the Cas-OFFinder tool (36). sg-1 targets the EGFP Kozak sequence using a GGG PAM in the antisense strand that places the target T in position 5 of the base editor window of action. sgNCF1 targets the NCF1 Kozak sequence using a GGG PAM in the antisense strand that places the target nucleotides in positions 10–11–12 of the base editor window of action.
pWPT-/mEGFP-1T-IRES-mCherry (EGFP-1T, Addgene plasmid #190190) was obtained by designing a mutated Kozak sequence as an oligonucleotide and cloning it in pWPT-/GCCACC-mEGFP-IRES-mCherry (Addgene plasmid #49235) using EcoRI (New England Biolabs, #R3101) and XhoI (New England Biolabs, #R0146) restriction sites. In the same oligonucleotide, a PAM sequence was added to allow base editing of the Kozak sequence.
pWPT-mCherry (Addgene plasmid #190605) was obtained by cloning five stop codons as an oligonucleotide in place of the Kozak sequence in pWPT-/GCCACC-mEGFP-IRES-mCherry using EcoRI (New England Biolabs, #R3101) and XhoI (New England Biolabs, #R0146) restriction sites.
pWPT-mEGFP (Addgene plasmid #190606) was obtained by digesting pWPT-/GCCACC-mEGFP-IRES-mCherry with PstI (New England Biolabs, #R0140) and XmaI (New England Biolabs, #R0180) restriction enzymes, creating a 367 bases deletion inside the mCherry coding sequence. Blunt ends were generated with DNA Polymerase I, Large (Klenow) Fragment (New England Biolabs, #M0210).
The Kozak sequence variants were purchased as oligonucleotides and cloned in pWPT-/GCCACC-mEGFP-IRES-mCherry to validate the hits that emerged from the screening EcoRI (New England Biolabs, #R3101) and XhoI (New England Biolabs, #R0146) restriction sites. All the oligos used are listed in Supplementary Table S1.
Cell cultures
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Life Technologies); U2OS and Raji cells were cultured in Roswell Park Memorial Institute 1640 medium (RPMI, Life Technologies). All media were supplemented with 10% fetal bovine serum (FBS, Life Technologies), 1% l-glutamine and 100U/ml antibiotics (PenStrep, Life Technologies). The cells were maintained at 37°C in a 5% CO2 humidified atmosphere.
HEK293T transfection
For FACS analysis experiments, 105 HEK293T cells/well were seeded into 24-well plates (Corning). After 1 day, cells were transfected with 4 μl polyethyleneimine (PEI) per well using 500 ng of pWPT-/GCCACC-mEGFP-IRES-mCherry (EGFP-1C) or pWPT-/mEGFP-1T-IRES-mCherry (EGFP-1T). Cells were cultured for three days before cell detachment and analysis at FACS Canto.
For base editing experiments in HEK293T cells, 105 cells/well were seeded into 24-well plates (Corning) and transfected with 4 μl PEI per well using 750 ng of base editor plasmid, 250 ng of sgRNA plasmid and 100 ng of pWPT-/mEGFP-1T-IRES-mCherry (EGFP-1T). Cells were cultured for five days before DNA extraction.
For High Content Screening System Operetta (PerkinElmer) analysis, 105 HEK293T cells/well were seeded into 24-well plates (Corning) and transfected with 4 μl PEI per well using 100 ng of pWPT-mEGFP-IRES-mCherry bearing either the wild type or a variant of the target Kozak sequences emerged from the high-throughput screening. Twenty-four hours post-transfection, cells were detached and plated in a 96-well plate (Corning) (8000 cells/well). Seventy-two hours post-transfection, cells were analysed at the High Content Screening System Operetta (PerkinElmer). At the same time point, cells were collected for protein extraction and western blot analysis.
Electroporation of Raji cells
For base editing experiments, cells were transfected using the Neon transfection system (MPK5000) according to the manufacturer's instructions. Briefly, 7 × 105 Raji cells/condition were harvested and washed in PBS (Invitrogen). Cells were then resuspended in 100 μl of R buffer and electroporated with 1500 ng of AncBE4max base editor and 250 ng of sgRNA plasmid with the following conditions: 1350 V, 30 s, one pulse. After electroporation, cells were immediately transferred to a 12-well plate (Corning) containing a pre-warmed antibiotic-free medium. Cells were cultured for five days before DNA and protein extraction. To increase base editing efficiency, five days post electroporation Raji edited cells were serially diluted to obtain single clones. Single-cell clones were picked, base editing efficiency was analysed (Supplementary Figure S4B) and the best-edited clones (Var 2 and Var 4) were selected for further experiments.
Analysis of the efficiency of base editing
Genomic DNA was extracted using QuickExtract DNA Extraction Solution (Epicentre, #QE09050). The target region was PCR-amplified using MyTaq HS RedMix 2X (Meridian Bioscience, #BIO-25047). The oligos used are listed in Supplementary Table S1. PCR products were purified using NucleoSpin Gel and PCR clean-up (Macherey-Nagel, REF 740609.50), and Sanger sequenced and analysed by EditR software to evaluate base editing efficiency (37). Editing efficiencies are shown compared to the percentage of editing obtained with the scrambled sgRNA (sgCTRL).
Kozak sequences library construction
The Kozak sequence variants were synthesised as oligonucleotides on a custom Agilent 244K microarray designed for this purpose. Two libraries were synthesised with two different synthesis processes. Library B was synthesised with a process that reduces the error rate from 1/250–1/500 bases to 1/600–1/1200 bases. The libraries were purchased as pooled unamplified lyophilised ssDNA oligonucleotides. The oligonucleotides designed were 98 nt long. Eleven central nucleotides in each oligo (four before and four after the ATG) represent the Kozak sequence and the variable part of each oligonucleotide. The remaining nucleotides represent the homology arms with the final reporter vector and the restriction sites of the desired enzymes for cloning (XhoI and EcoRI). The oligo design is described in Supplementary Table S1.
The library was cloned inside pWPT-mCherry, to avoid background EGFP signal in case of reconstitution of the empty vector during Gibson assembly or by inefficient digestion of the destination vector.
For the plasmid library, pWPT-mCherry was digested with EcoRI (New England Biolabs, #R3101) and XhoI (New England Biolabs, #R0146). The oligonucleotides library was cloned in the linearised vector using NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs, #E2621), compatible with ligation of ssDNA oligos and dsDNA assembly. In particular, 1pmol of resuspended library oligos was ligated with 100 ng of digested purified vector following the manufacturer's instructions. TOP10 Escherichia coli were transformed with the ligation product. The colonies were scraped, and DNA was purified through a Midiprep purification (Qiagen, #12143).
Lentiviral transduction
Lentiviral particles of the Kozak variants library were produced by seeding 10 × 106 HEK293T cells into 15 cm dishes. The day after, the plates were transfected with 25 μg of the vector and 16.25 μg psPax2 (Addgene, #12260) packaging vector and 8.75 μg pMD2.G (Addgene, #12259) using PEI. After 6 h of incubation, the medium was replaced with fresh complete DMEM. 48h later, the supernatant containing the viral particles was collected, spun down at 250 g for 5 min, and filtered through a 0.45 μm PES filter. Lentiviral particles were concentrated by ultracentrifugation for 2 h at 150000 g at 4°C with a 20% sucrose cushion. The titres of the lentiviral vectors (reverse transcriptase units, RTU) were measured using the product enhanced reverse transcriptase (SG-PERT) assay as previously described (38).
FACS-seq experiments were carried out on HEK293T cells. To ensure 1000× coverage of the library, viral particles were added to 25 × 106 HEK293T cells. The titre of viral particles was calculated to obtain 25% of infection frequency (MOI = 0.3), as validated by flow cytometry three days post-transduction, ensuring that transduced cells received a single copy of the vector.
For U2OS transduction, lentiviral particles of the single Kozak variants were produced as described above. 5 × 104 cells/well were seeded into 24-well plates (Corning) and the day after were transduced with 3 RTU of lentiviral vectors. Twenty-four hours post-transduction, cells were detached and plated in a 96-well plate (Corning) (4000 cells/well). Seventy-two hours post-transduction, cells were analysed at High Content Screening System Operetta (PerkinElmer). At the same time point, cells were collected for protein extraction and western blot analysis.
Fluorescence-activated cell sorting (FACS)
HEK293T cells transduced with the Kozak variants library were sorted into four gates according to the EGFP/mCherry ratio as a measure of Kozak strength. All sortings were performed using the FACS Aria IIIu (Becton Dickinson, BD Biosciences) using the GFP channel (488 nm excitation laser, 500 nm splitter, 530/30 nm emission filter) and the mCherry channel (561 nm excitation laser, splitter 600 nm, 610/20 nm emission filter) and FACS Diva Software (BD Biosciences version 8.0.2). 561 nm laser (Yellow-Green) allows optimal mCherry excitation. Cells were resuspended in PBS without Ca2+ Mg2+ complemented with 2% BSA, 1% Pen-Strep and 1,5mM EDTA and filtered through a 35 μm filter (Becton Dickinson). Cells were sorted at low pressure (20–25 psi) with the 100 μm nozzle.
The sorting was divided into two rounds. In the first round, three days after lentiviral particle transduction, 5 × 106 HEK293T cells positive for mCherry expression were sorted to ensure 1000× coverage of the library. This first round was needed to enrich the transduced cell population. The sorted cells were expanded for three days, and mCherry expression was evaluated before proceeding with the sorting. In the second round of sorting (day 8 post-transduction), mCherry-positive cells were sorted in 4 bins (gates) according to their EGFP/mCherry ratio, so that 25% of the population fell in each gate. To maintain 1000× coverage, 1.25 × 106 cells were collected from each bin. Immediately after the sorting, a small sample of cells from each bin was re-run to check for purity.
The cell sorter FACS Aria IIIu (Becton Dickinson, BD Biosciences) was also used to analyse the EGFP/mCherry ratio of the target Kozak sequences and respective variants individually selected through the screening (3 days post transient transfection). Data were analysed with FlowJo software (v. 10.7.1).
Deep sequencing
The library of Kozak sequences was deep sequenced before the high-throughput screening to check for proper representation of all the variants. After cell sorting, the Kozak sequences from the four subpopulations were PCR amplified and deep sequenced. Briefly, genomic DNA was extracted from the subpopulations using the DNeasy Blood & Tissue Kit (Qiagen, #69504). The DNA from each population was loaded in PCR reactions with 400 ng input each. A second PCR was performed for the ligation of standard Illumina adapters. The PCR products were purified with Ampure XP beads (Beckman Coulter), mixed in equimolar ratios, and sequenced with the Illumina MiSeq on an SR250 v2 flow cell (Illumina, San Diego, CA, USA).
Reads were quality checked with FASTQC (v0.11.4) (39) and filtered to retain only those sequences that were not shifted (starting codon: CCA/CNA). Furthermore, only reads with no mismatch in the translation starting codon (ATG/CTG) were retained. Following the extraction of the 11 bases Kozak sequences from the reads, the occurrences of expected sequences (n = 4838 unique sequences) were calculated, and the unexpected sequences were removed.
Sequence read counts were converted in counts per million mapped reads (CPM) and filtered to retain only sequences with at least five CPM considering all four gates (Supplementary Figure S1C, F). Subsequently, the Gini index was calculated with the DescTools R package (v0.00.39) on each sequence's normalised counts, and a cut-off of 0.25 was applied. During the data analysis, the variants were always considered related to their wild-type; once a wild-type sequence was filtered out at each filtering step, the derived variant sequences and only those were filtered out too.
Considering the expression distribution of the wild-type sequences in the four gates as the expected one and the corresponding variant sequences distribution as the observed ones, a Chi-square goodness of fit test was performed between each pair of wild-type and variant sequences. Benjamini-Hochberg (BH) multiple testing correction was applied to all P-values, and a 0.01 cut-off was set on the adjusted P-values. Finally, the expected value (EV) was computed for each sequence on the percentage of normalised counts in each gate. The EV of each wild-type sequence was subtracted from the EV of the respective variant sequences. By considering weak the wild-type sequences with an EV ≦2 (22.5% of the wild-type analysed in the screening), only variants sequences with positive values were retained as possible hits to be validated.
Library A and Library B were intersected, considering the variant and respective wild-type Kozak sequences which satisfy the CPM, adjusted P-value, and EV difference thresholds defined before.
A 0.5 cutoff on the difference between the EV of the variants and EV of the wild-type was then applied to have a more robust identification of the variants that significantly increased translation.
For the consensus sequences of the gates, the distribution of EVs of both variants and wild-type sequences was subdivided into quartiles, each representing one of the four distribution-derived gates. Sequences for each gate were extracted, and a consensus sequence representing the nucleotide frequency in each position of the Kozak sequence was generated using the seqLogo R package (v1.52).
For targeted deep sequencing for indels generation and base editing analysis, the locus of interest was PCR-amplified from genomic DNA extracted from HEK293T cells and Raji cells five days after transient transfection or electroporation, respectively, of the base editor and sgNCF1 or sgCTRL. Amplicons were indexed by PCR using Nextera indexes (Illumina), purified with Ampure XP beads (Beckman Coulter), quantified with the Qubit dsDNA High Sensitivity Assay kit (Invitrogen), mixed in equimolar ratios, and sequenced with the Illumina MiSeq on an SR250 v2 flow cell (Illumina, San Diego, CA, USA). The primers used to generate the amplicons are reported in Supplementary Table S1. Raw sequencing data (FASTQ files) were analysed using CRISPResso online tool (40).
High content screening
Microplates with seeded and transfected cells (Corning 96-well plate) were imaged on the High Content Screening System Operetta™ (PerkinElmer). In each well, images were acquired in 9 preselected fields with LWD 10x objective over four channels: brightfield, digital phase contrast (DPC) based on brightfield images, with excitation filters 460–490 and 520–550 nm and emission filters 500–550 and 560–630 nm for GFP and mCherry reporters, respectively. For the feature extraction, the images were analysed by Harmony software version 4.1 (PerkinElmer). Briefly, individual cell nuclei were segmented in the DPC channel. Nucleus morphology, GFP, and mCherry mean intensity were quantified in the cell nuclei population. Single-cell object features were extracted from each sample well. To discriminate between GFP/mCherry negative and positive cells, a threshold was determined based on the GFP/mCherry intensity frequency distribution of all samples for each experiment.
Western blotting
For protein extraction, cells were homogenised in RIPA buffer with a complete protease inhibitor (PI) cocktail (Roche) and quantified with a BCA (bicinchoninic acid) assay. For protein extraction from the single sucrose fractions, 10% Trichloroacetic acid (TCA) was added and mixed. After incubation overnight at –20°C, samples were centrifuged at 14000 rpm for 10 min at 4°C. Samples were washed three times with 1 ml ice cold Acetone and centrifuged at 14000 rpm for 5 min. Pellets were resuspended in 30 μl of RIPA buffer + PI. Protein lysates were resolved on SDS-PAGE and transferred to the PVDF membrane. Membrane blocking was performed with 5% milk (BioRad)-TBS-T for one hour. Incubation with the primary antibodies was performed overnight at 4°C. Incubation with the secondary antibodies was performed for 1 hour at room temperature. The following antibodies were used: mouse anti-EGFP (sc-9996, Santa Cruz); rabbit anti-Cherry (PA5-34974, Thermo Fisher Scientific); mouse anti-beta tubulin (sc-53140, Santa Cruz); mouse anti-alpha-actinin (sc-17829, Santa Cruz); goat anti-p47phox (PA1-9073, Thermo Fisher Scientific); rabbit anti-RPS6 (5G10, Cell Signaling); rabbit anti-RPL26 (ab59567, Abcam); secondary anti-mouse IgG HRP (sc-2005, Santa Cruz); secondary anti-rabbit IgG HRP (#31460, Thermo Fisher Scientific); secondary anti-goat IgG HRP (ab97100, Abcam). Blots were imaged with the Uvitec Alliance Mini imaging system (UVITEC, Cambridge, UK) after incubation with ECL Prime or Select detection reagent (GE Healthcare, Buckinghamshire, UK). The intensity of the bands was quantified by densitometry using the ImageJ analysis program.
Polysome profiling
Polysomal profiling was performed according to previously described protocols (41). Briefly, the cells were treated with cycloheximide and then lysed in 700 μl of cold lysis buffer. The lysate was centrifuged at 13000g for 10 min at 4°C to pellet cell debris and loaded on a linear 15–50% [w/v] sucrose gradient. The lysate was then centrifuged in an SW41Ti rotor (Beckman) at 40000 rpm for 1 h 40 min at 4°C in a Beckman Optima Optima XPN-100 Ultracentrifuge. Fractions of 1 ml of volume were then collected, monitoring the absorbance at 254 nm with the UA-6 UV/VIS detector (Teledyne Isco).
Total RNA extraction
Raji cells were pelleted and lysed in 1 ml of Trizol (Thermo Fisher) per 5 × 106 cells. For polysomal RNA extraction, sucrose fractions corresponding to polysomes and total RNA were pooled together and lysed in 1 ml of Trizol (Thermo Fisher).
Chloroform was added corresponding to ⅕ of the total volume after 15min incubation at RT. Samples were centrifuged at 12000g for 15 min at 4°C. The formed aqueous phase was transferred to a new tube, and 1 ml of isopropanol was added. After 1h incubation at –80°C, samples were centrifuged at 12000 g for 10 min at 4°C, the supernatant was removed, and pellets were washed with 1 ml of 70% ethanol. Finally, samples were centrifuged at 5000 g for 10 min at 4°C, the supernatant was removed and the pellet was air-dried for 5–10 min before being dissolved in 20 μl DEPC-treated water.
Quantitative real-time PCR
For NCF1 qPCR, 1 μg of total RNA was reverse transcribed using the RevertAid RT kit (ThermoFisher, K1619) following manufacturers’ instructions. Sybr-green qPCR was performed as follows: 20 ng template cDNA, ExcelTaq™ 1X Q-PCR Master Mix (SYBR, NO ROX, SMOBIO, #TQ1100), and 0.4 μM of each primer in a reaction volume of 15 μl. The qPCR reaction was performed on a CFX96 real-time PCR Detection System (Bio-Rad Laboratories) with the following cycling conditions: 95°C for 2 min, followed by 40 cycles at 95°C for 15 s, 60°C for 60 s. HPRT1 expression was used as a reference. The ΔΔCq method was used to calculate the relative mRNA levels of each gene.
For polysome fractionation analysis, the gene-specific translation efficiency (TE) was calculated as the ratio between the fold change at the polysomal level and the fold change at the total level of the gene of interest, as described before (41).
Statistical analyses
For high Content Screening System (Operetta) analysis, the violin plots report the data distribution from at least three biological replicates. The dashed line indicates the median of the population. For FACS analysis quantification, qPCR, and base editing efficiency analysis, the data were normalised over the wild type of each respective gene and are reported as mean ± SD (standard deviation) of at least three biological replicates. Statistical significance was determined by an unpaired two-tailed t-test (comparing each variant to the corresponding wild-type), as indicated in the figure legends (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001).
RESULTS
Base editing-mediated Kozak optimisation enhances protein levels in a reporter system
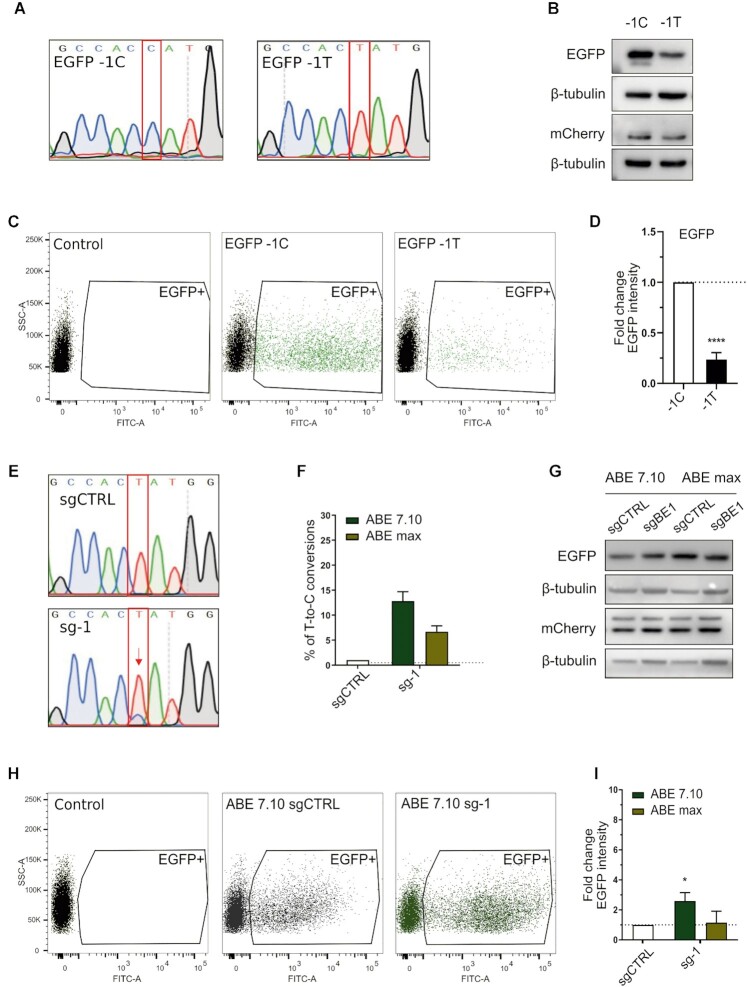
To evaluate the feasibility of BOOST, and the ability to control translational regulation through CRISPR-Cas base editing, we first performed an experiment in a reporter system. We designed this initial model inspired by a mutation causing AVED (ataxia with vitamin E deficiency), where a T instead of a C in position –1 with respect to the AUG decreases translational efficiency (16).
We thus created two versions of the bicistronic reporter vector pWPT-EGFP-IRES-mCherry (42): EGFP wild-type Kozak sequence (C in position –1, EGFP-1C) or a suboptimal motif having a T in position -1 (EGFP-1T) (Figure 1A). We found that this single base change reduced EGFP translation by 4–5-fold, as observed by western blot (Figure 1B) and FACS analysis (Figure 1C, D).
Figure 1.
Boosting of a suboptimal Kozak sequence by base editing. (A) Sanger sequencing chromatograms representing the wild-type (EGFP-1C) and the mutated EGFP version (EGFP-1T), with a single variation in position –1 of the Kozak sequence. (B) Western blot analysis of EGFP and mCherry expression in HEK293T cells transiently transfected with EGFP-1C or EGFP-1T plasmids. (C) Representative FACS dot plots of HEK293T cells three days after transient transfection. (D) FACS analysis of HEK293T cells transiently transfected with the respective plasmids. The average EGFP intensity of EGFP-1T is normalised over EGFP-1C. Data are reported as mean ± SD of n = 3 biological replicates. Statistically significant differences were calculated by unpaired t-test. (E) Representative Sanger sequencing chromatograms of HEK293T cells edited with the ABE7.10 base editor and sg-1, compared with ABE7.10 combined with a scrambled sgRNA (sgCTRL). (F) Percentage of correct T-to-C conversion analysed with the EditR software. (G) Western blot analysis of EGFP and mCherry expression in HEK293T cells edited with ABE 7.10 or ABEmax combined with sg-1 or sgCTRL. (H) Representative FACS dot plots of cells edited with ABE7.10 and sg-1, compared with ABE7.10 combined with a scrambled sgRNA (sgCTRL) 3 days after transfection. (I). FACS analysis of EGFP expression in cells transfected with the base editors (ABE7.10 and ABEmax) and sgCTRL or sg-1. The average EGFP intensity of sg-1 is normalised over sgCTRL. Data are means ± SD from n = 3 biological replicates. Statistically significant differences were calculated by unpaired t-test (P value = 0,0483).
The EGFP -1T > C Kozak model was designed to be a target for the sg-1 sgRNA coupled to the adenine base editors ABE7.10 (43) and ABEmax (44) to produce a T > C variation and recreate the optimal Kozak sequence (the target T is in position 5 of the base editor window of action counting the PAM as positions 21–23). After the co-transfection of EGFP-1T, the base editor, and the sgRNA in HEK293T cells, about 13% of base editing was achieved with ABE7.10 (Figure 1E, F), resulting in a 2-fold increase in EGFP translation (Figure 1G–I). These data confirm that variation of the Kozak sequence in the 5’UTR region just upstream of the AUG impacts translational efficiency, and even a single substitution can alter EGFP expression up to 4-fold. Moreover, they demonstrate the possibility of modulating protein levels by editing the Kozak sequence with CRISPR-Cas base editors.
Design and generation of the library of actionable Kozak variants
The BOOST gene-editing strategy would be ideal to be applied where translational enhancement could be a viable therapeutic approach. Thus, we aimed at performing a high throughput screening of Kozak variants of haploinsufficient genes to find variants up-regulating gene expression. In particular, we screened wild-type (WT) Kozak sequences of annotated HI genes and compared them with respective variants to identify the specific set of actionable changes up-regulating the translation efficiency of each WT Kozak sequence. The variants were designed according to the modifications that can be reproduced by base editors (i.e. transitions). We created a non-biased library of Kozak mutants, ignoring every previous knowledge of the most performing Kozak sequence (GCCRCCaugG). To build our library, we took into account the HI genes present in the most recent literature annotation (2), together with some genes recently described as having a high HI prediction (HiPred score (45), Supplementary Table S2). We discarded the genes associated with complex diseases, including cancer, as our approach of translational enhancement is more suited for monogenic disorders. We obtained a list of 230 HI genes, from which we built a library of Kozak sequence variants based on the following principles:
We defined the Kozak width from nucleotide –4 to nucleotide + 7 (the A of the starting codon ATG being base + 1, e.g. NNNN_ATG_NNNN);
We rationally created variants bearing conversions reproducible with the initially developed base editors (i.e. transitions) starting from each wild-type Kozak present in the library in such a way that the same transition could be present one or multiple times, so as to mimic the tendency of base editors to introduce multiple transitions in a given window (e.g. WT: AAAA_ATG_AAAA; V1: GAAA_ATG_AAAA; V2: GGAA_ATG_AAAA; V3: GGGA_ATG_AAAA… and so on);
Each variant bears one kind of transition at a time, meaning that no variant simultaneously bears two types of transitions.
This led to a library of 5539 sequences, 4838 of which are unique.
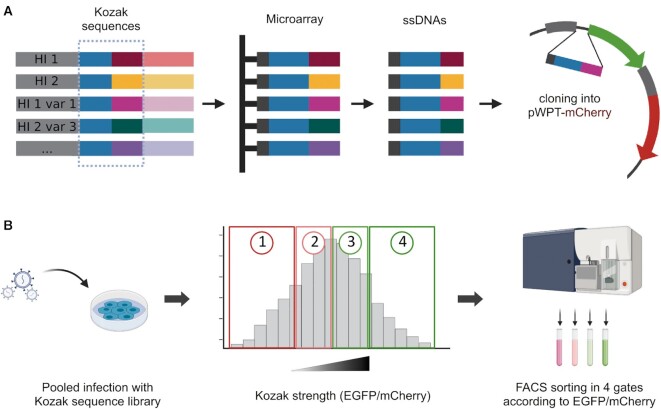
The Kozak sequence variants were synthesised as oligonucleotides on a custom Agilent 244K microarray designed for this purpose. As the destination vector, we chose the lentiviral bicistronic reporter previously described. To avoid the EGFP background signal caused by random reconstitution of the digested vector during Gibson assembly or by inefficient digestion of the destination vector, we created an alternative plasmid by replacing the EGFP Kozak sequence of pWPT-EGFP-IRES-mCherry with five stop codons, creating a pWPT-mCherry (Supplementary Figure S1A). After assembly, the library of wild-type and variant Kozak sequences would substitute the EGFP Kozak sequence, directing EGFP expression (Figure 2A, Supplementary Figure S1B). In the reporter vector, mCherry expression is regulated by an IRES. Therefore, it is translated by the same transcript and could be used as a normalising signal. The resulting vector bearing the Kozak library was deep-sequenced to check for a good representation of all the variants (Supplementary Figure S1C). The obtained reporter bearing the library was used to transduce HEK293T cells, which were then cell sorted in four gates according to the normalised EGFP translational efficiency (EGFP/mCherry) (Figure 2B).
Figure 2.
Schematic workflow of library generation and selection screening for Kozak strength. (A) The Kozak variants were designed as oligonucleotides bearing the overhangs to be cloned in the destination vector. The oligos were synthesised on a custom microarray. The library was cloned in place of the EGFP Kozak sequence in a bicistronic reporter vector. (B) The Kozak sequence library was used to transduce HEK293T cells. Transduced cells were sorted according to their EGFP/mCherry ratio as a measure of Kozak strength. The four gates were drawn so that each gate contained 25% of the total population.
Evaluation of protein levels from the wild-type and variant Kozak sequences
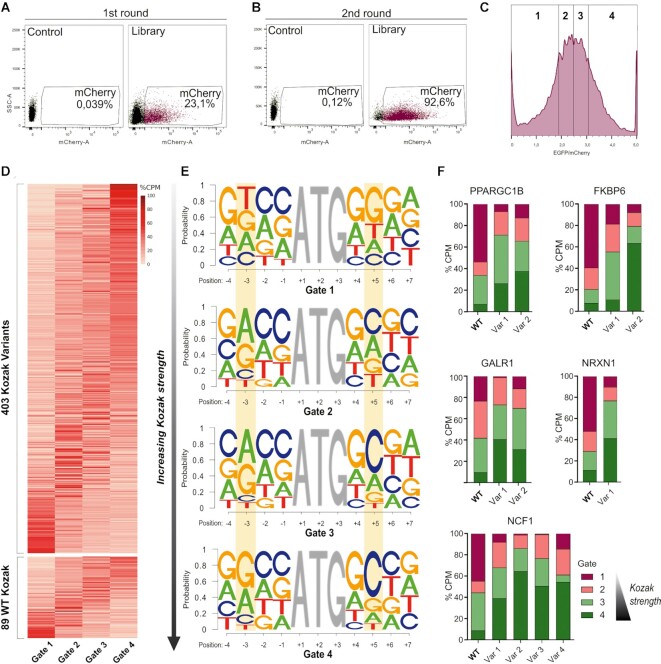
To quantify the translational efficiency of the wild-type and variant Kozak sequences of the HI genes, we modified the previously described FACS-seq procedure (21). The screening was carried out in two rounds of cell sorting. In the first round, to enrich the transduced cells expressing the library, 5 × 106 mCherry positive cells were sorted to ensure 1000X library coverage (Figure 3A, Supplementary Figure S1D); in the second round, the resulting mCherry positive cells were sorted according to their EGFP/mCherry ratio in 4 bins of different fluorescence intensity ratios (Figure 3B, C, Supplementary Figure S1E). Three days post-transduction of the Kozak variant library, we analysed the expression of the fluorescent proteins (Figure 3A, Supplementary Figure S1D). FACS analysis assessed 23.1% of mCherry positive cells (the reporter internal control) (Figure 3A). mCherry-positive cells were sorted and seeded to achieve expansion and complete recovery. Forty-eight hours later (5 days post-transduction), EGFP and mCherry expression were assessed again (Figure 3B, Supplementary Figure S1E). 92.6% of the sorted cells were mCherry positive, confirming the validity of the first sorting step and allowing us to proceed with the second round. To measure the strength of the Kozak sequence variants, we divided the population of mCherry-positive cells into four gates according to the ratio between the EGFP and mCherry emissions (EGFP/mCherry). The gates were created so that each bin contained 25% of the total population of interest (Figure 3C). 1.25 × 106 cells were sorted for each gate to maintain 1000× library coverage (see Materials and Methods). After sorting, a sample from each sorted bin was rerun to check for purity post sorting (see Materials and Methods).
Figure 3.
High-throughput determination of protein levels from Kozak sequence variants. (A) mCherry expression of the transduced cells in FACS-seq first round of sorting. 5 × 106 mCherry-positive cells (23.1% of the total) were sorted. (B) FACS-seq second round of sorting. (C) mCherry-positive cells from the gate drawn in (B) were divided into four gates according to EGFP/mCherry expression, defined in such a way that each bin contains 25% of the total population of interest. (D) The heatmap represents the distribution of the percentage of the count per million reads (CPM) in the four gates of the candidate HI genes and variants which passed the statistical analysis. In the upper panel, the Kozak variants are represented. The WT Kozak sequences of the HI genes are shown in the lower panel. Each column corresponds to one of the four gates, while each row stands for one of the Kozak sequences. Rows are ordered by the expected value (EV) of the corresponding sequence. (E) Logo representation of the Kozak sequences extracted from each of the four gates. In each panel, the positions along the Kozak sequence (with A of ATG being position +1) are represented on the x-axis, and the probability of occurrence of each base is shown on the y-axis. Gate 1 (upper panel) represents the lowest translational efficiency, while gate 4 (lower panel) corresponds to the most performing Kozak sequences. Relevant positions (–3 and +5) are highlighted in yellow. (F) Percentage of the count per million reads (CPM) in the four gates of the wild-type (WT) and the respective variants (Var) of the five selected genes.
The Kozak sequence region from the cells collected in each bin was PCR-amplified. Deep sequencing of all fractions allowed us to compare the strength of each HI wild-type Kozak to its variants. Eighty-nine wild-type sequences and 403 variant sequences passed the statistical analysis (Figure 3D). The heatmap shows that each sequence (row) is significantly present in one gate (column) and decreases progressively in the adjacent ones, as expected. As shown in the heatmap, a significant number of wild-type sequences are present in the lower gates (22.5% of the selected wild-type genes, see Materials and Methods), meaning that the corresponding genes are regulated by a relatively weak Kozak sequence and can thus be potentially up-regulated (Figure 3D, lower panel).
We then generated a motif for each of the four gates, representing the nucleotide frequency at each position of the Kozak sequence (Figure 3E). We analysed the motifs and compared them with the optimal Kozak sequence described in the literature for mammals, GCCRCCaugGCG. Here, a purine in position -3 is considered the most important for strong translational efficiency, while a pyrimidine is associated with evident leaky scanning (46). We noticed that in Gate 1 (Figure 3E, upper panel), thymine was overall the most represented nucleotide in position -3. This is in agreement with previous findings since Gate 1 includes the least performing Kozak sequences. Interestingly, moving onwards with the gates, adenine or guanine become the predominant nucleotides in that position, indicating increased Kozak strength. Moreover, a G-stretch can be observed in the consensus of Gate 1 after the AUG. This stretch gradually disappears with the increasing translational efficiency until cytosine becomes predominant in position +5, a feature of high-performing Kozak sequences in mammals, as previously documented (47).
We also investigated the relationship between the reporter translational efficiency levels and specific conversions, by mapping each wild-type-to-variant nucleotide transition for each position in the Kozak sequence. However, this analysis did not show evidently favoured conversions (data not shown).
Aiming at selecting Kozak variants up-regulating the corresponding WT, we first calculated the Gini Index, which measures the statistical dispersion of the variants' expression distribution, to retrieve sequences with an unequal distribution and thus with a greater representation in one of the four gates (see Methods). Secondly, we calculated for each Kozak sequence an Expected Value (EV) (see Methods), and by subtracting the EV of each wild-type sequence from the EV of the corresponding variants, we selected only the variants with maximal distance from the respective WT. We obtained 47 wild-type and 149 variant sequences (Supplementary Table S3). From this list, we selected the five HI genes and their corresponding variants with the best overall scores (Gini index and EV value) for validation. These genes were PPARGC1B, FKBP6, GALR1, NRXN1 and NCF1 (Figure 3F).
We also repeated the high-throughput screening with a second oligonucleotide library (Library B, see Materials and Methods) synthesised and cloned independently from the first to corroborate our findings (Supplementary Figure S1F, G). We checked for sequence representation and processed the sequencing data as described above. We intersected the results obtained with the two libraries, and found that 50,3% of the Kozak sequences in Library A and 54,2% in Library B were present in their intersection. Interestingly, the five hit genes selected from the first screening passed the statistical analysis again. Moreover, all the variants identified in the first replicate confirmed their trend to up-regulate the translational efficiency compared to their corresponding wild-type Kozak sequence (Supplementary Figure S1G).
Validation of protein up-regulation by selected hit Kozak sequence variants
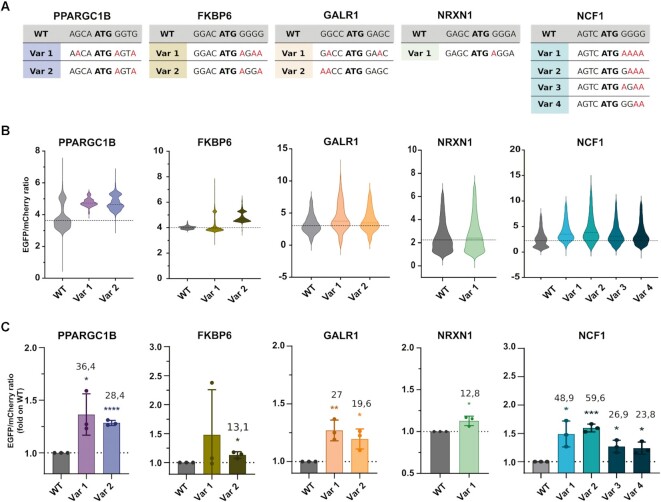
To validate the selected hits, we cloned each of the Kozak sequences (the wild-type and hit variants of the five selected genes) in place of the plasmid EGFP Kozak sequence in our reporter vector, creating one new plasmid for each sequence. We transiently transfected HEK293T cells with the respective wild-type and hit Kozak variants and measured the fluorescence by high content image analysis three days after transfection (Figure 4). These analyses confirmed that 10 of the 11 tested Kozak variants increase the translational efficiency compared to their respective wild-type sequence (Figure 4B, C). Among the five targets, PPARGC1B variants increased translation on average by 30%, as measured by high content image analysis. One of the two variants tested for FKBP6 resulted in a significant translational up-regulation, about a 15% increase in fluorescence. GALR1 variants showed a similar trend, with Var 1 being more efficient than Var 2 (27% increase). NRXN1 variant induced 12.8% up-regulation of the fluorescence. Finally, all four variants selected for NCF1 enhanced translation (20–60% over the wild-type) (Figure 4C). Three additional genes were selected and validated from the screening, and all the tested variants increased the translational efficiency from the respective wild-type (Supplementary Figure S2). Overall, these data supported the validity of our HI genes Kozak screening and data analysis approach and provided for all the singularly tested wild-type sequences at least one Kozak sequence variant that can significantly enhance protein production.
Figure 4.
Validation of actionable hit variants. (A) Wild-type (WT) and variants (Var) Kozak sequences of the selected hit genes. (B) Translational enhancement analysed as EGFP/mCherry expression by high content image analysis. The violin plots report the data distribution from n = 3 biological replicates. The dashed line indicates the population median. (C) The histogram represents the mean of the populations analysed by high content image analysis. Data are means ± SD from n = 3 biological replicates. The numbers indicate the percentage of mean increase of the variants over the WT. Statistically significant differences were calculated using the unpaired t-test of each variant versus the corresponding WT.
We decided to validate the BOOST approach for the NCF1 Kozak sequence since it was the case for which we reached the highest translational up-regulation and with more Kozak variants. To further confirm this enhancement, we produced lentiviral particles of each NCF1 Kozak variant-bearing reporter and transduced HEK293T cells at low MOI (reproducing the protocol used for the library transduction). We then analysed the EGFP/mCherry ratio three days post-transduction with flow cytometry (Supplementary Figure S3A, B). The results confirmed the observations of the transient transfection, with Var 2 and Var 4 being the best-performing variants. We then confirmed translation enhancement by the NCF1 Kozak variants by transduction at low MOI in U2OS cells. (Supplementary Figure S3C, D). Here, Var 2 enhanced translation by 30.2% and Var 4 by 44.7% in high content image analysis.
Enhancement of NCF1 translation by base editing of its Kozak sequence
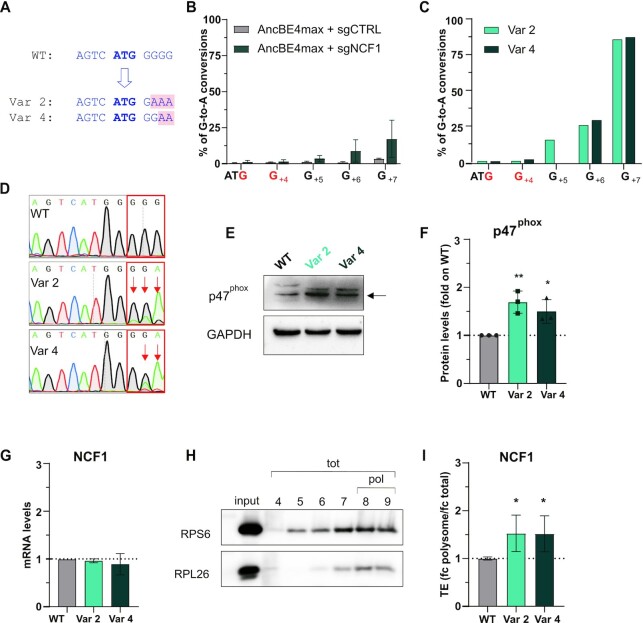
Being assured of the reporter effect of NCF1 Kozak Var 2 and Var 4 in different cell lines, we base-edited the NCF1 endogenous locus. The base conversions to reproduce Var 2 and Var 4 are both G > A, a substitution that cytosine base editors can insert (CBE) targeting C nucleotides complementary to the targeted G. Thus, sgNCF1, the sgRNA targeting the NCF1 Kozak, was selected aiming at recreating the strong NCF1 Kozak variants 2 and 4. The target G nucleotides are in a G-stretch that comprises the G of the starting codon (Figure 5A). Therefore sgNCF1 was designed so that the three target guanines are at the limit of the base editor window of action (positions 10–12 counting the PAM as positions 21–23), to avoid undesired bystander effects (Supplementary Table S4). No other G was in the base editing range inside the NCF1 open reading frame.
Figure 5.
BOOST of the NCF1 Kozak sequence. (A) Schematic representation of the NCF1 wild-type (WT), variant 2 (Var 2) and variant 4 (Var 4) Kozak sequences. The starting codon is bold blue; the base changes in the variants are highlighted in pink. (B) Editing efficiency in the Raji bulk population at target and bystander (in red) guanines analysed with the EditR software five days post-electroporation of AncBE4max and sgNCF1 or sgCTRL. The percentage of corrected G-to-A conversions (y-axis) is shown for each position in the NCF1 Kozak sequence (x-axis, with the A of ATG being position + 1). Data are means ± SD from n = 3 independent experiments. (C). Editing efficiency in the two clones isolated from the bulk population (Var 2 and Var 4 cells) at target and bystander (in red) guanines. (D) Sanger sequencing chromatograms of NCF1 Kozak sequence in Raji WT, Var 2 and Var 4 cells. (E) Western blot analysis of the p47phox protein in Raji cells (WT, Var 2, and Var 4). One representative blot result is shown. The arrow indicates the 47KDa band corresponding to p47phox. (F) Western blot quantification. p47phox levels were normalised on the housekeeping protein, and the fold change with respect to the WT levels is shown, n = 3 biological replicates. (G) qPCR of NCF1 on WT, Var 2 or Var 4 Raji cells. Data are means ± SD from n = 3 independent experiments. (H). Representative western blot of two polysomal markers (RPS6 and RPL26) in the fractions isolated by sucrose gradient centrifugation. The input is the cellular cytoplasmic lysate loaded on the sucrose gradient. tot= fractions corresponding to the total RNA; pol= fractions selected as polysomes and used in (I). (I) Translational efficiency (TE) quantification of NCF1 in Var 2 and Var 4 cells with respect to the WT cells. TE is the ratio between polysomal (fractions 8–9) and total (fractions 4–9) mRNA levels (fold change polysome/fold change total) measured by qPCR. Data are means ± SD from n = 3 independent experiments. Statistically significant differences were calculated by unpaired t-test of each variant versus the WT.
We first validated sgNCF1 in HEK293T cells, to ensure base editing efficiency and the absence of bystander effects (Supplementary Figure S4A). Next, we performed base editing of the NCF1 Kozak sequence in Raji cells, a B lymphocyte cell line derived from Burkitt's lymphoma that constitutively expresses the gene of interest. We electroporated Raji cells with AncBE4max (44) and sgNCF1 plasmids, obtaining an editing efficiency in the bulk population lower than 30% in the best-edited position, as analysed by Sanger sequencing five days after electroporation (Figure 5B). Of note, human cells have six copies of the sgNCF1 target region, two from the NCF1 gene and four from a pair of NCF1 pseudogenes (NCF1B and NCF1C) (48). To improve the readout of the editing, we then decided to isolate cells clones, and we found and expanded clones having the desired base editor-mediated nucleotide changes equivalent to the Kozak NCF1 variants 2 and 4 (Var 2 and Var 4 clones) (Supplementary Figure S4B, Figure 5C, D). In both clones, G+7 was largely (∼84%) converted to A, editing efficiency in G+6 was ∼32%, while G+5 was partially edited (∼16%) for Var 2 but left unedited for Var 4, reproducing the desired variants (Figure 5A, D). The editing levels were also confirmed by deep sequencing of the target locus (Supplementary Figure S4C) and were compatible with the expected total NCF1 copy number of at least 6 (NCF1, and the pseudogenes NCF1B and NCF1C). No bystander editing was observed (Supplementary Figure S4B, Figure 5C, positions in red). Additionally, deep sequencing showed very low indel formation (<0,2% in all samples) following base editing of NCF1, both in the bulk population of edited cells and in the expanded clones (Supplementary Figure S4D). Next, we evaluated the expression of p47phox, the protein encoded by NCF1, in the edited cells. Western blot analysis revealed increased p47phox expression with both variants compared to the wild-type (Figure 5E, F). In particular, Var 2 had increased protein levels by 69.2% and Var 4 by 49.7% (Figure 5F). We also analysed the NCF1 mRNA level in the wild-type cells and the clones finding that they were unchanged. These results strongly support the idea that the increase in gene expression results from an enhancement in the translation of NCF1 due to the Kozak sequence editing (Figure 5G). To confirm this, we performed a sucrose gradient fractionation in Raji wild-type, Var 2, and Var 4 clones (Figure 5H, I). First, we identified the fractions containing the polysomes (representing the actively translating ribosomes) by western blot analysis of two polysome markers: RPS6 (40S ribosomal protein S6), and RPL26 (60S ribosomal protein L26) (Figure 5H). We then measured the translational efficiency (TE) (41) of NCF1 by qPCR by calculating the ratio between the fold change of the NCF1 mRNA in the polysomal RNA (fractions 8–9) and in the total RNA (fractions 4–9) (Figure 5I). This experiment showed that the increase in protein levels corresponds to the increased loading of mRNA on the polysomes.
Collectively, these results showed that BOOST is a new gene editing approach targeting the Kozak sequence of a gene. It introduces suitable variants triggering the translational up-regulation of the target gene through base editing.
DISCUSSION
Although HI is responsible for hundreds of human diseases and disease susceptibilities, the attempts made until now to rescue haploinsufficient genes’ expression have essentially been based on gene augmentation approaches that are identical to the gene replacement strategies addressed to cope with recessive disorders (49). In both cases, the insufficient or absent expression of the gene, respectively, is compensated by the delivery, primarily viral, of an intronless additional copy of the gene, transcriptionally controlled by an additional promoter. Several FDA-approved gene replacement procedures are in the clinic, and more are in trials. An expected critical issue when applying the paradigm to HI diseases, in which residual gene expression is present in the affected tissues, often reaching 50% of the normal levels, is tight control of the increment in these levels. The reason for this caution derives from the several documented examples of diseases in which naturally occurring or induced overexpression of a gene leads to detrimental effects as much as its lack of expression does. For instance, haploinsufficiency of the PMP22 gene causes hereditary neuropathy with liability to pressure palsies. In contrast, duplication of the same gene causes Charcot-Marie-Tooth disease type 1A, two inherited neuropathies revealing a strict PMP22 dosage sensitivity (50). Moreover, loss-of-function mutations in MeCP2 cause the X-linked Rett syndrome, but also, a mild overexpression of MeCP2 (2-fold) in mice can cause cell death and a Rett-like phenotype (51).
Genome surgery methods could represent an exciting alternative for the therapeutic rescue of HI genes. A CRISPR-Cas-based genome editing approach aimed at enhancing gene expression at the transcriptional level has been proposed to upregulate the transcription of the haploinsufficient SCN1A gene in Dravet syndrome (52). The authors used the CRISPR-a approach, in which a catalytically inactive Cas9 is fused to transcriptional activators to induce overexpression of target genes (53). CRISPR-a resulted in the increased transcription of the SCN1A gene, demonstrating the feasibility of HI rescue by increasing the expression of the spare functional allele. An obvious limitation of this approach is that CRISPR-a requires permanent overexpression of the transcriptional activator and does not install irreversible editing in the genome. Therefore, its use in therapy would require multiple rounds of treatment or permanent genomic instalment of the CRISPR-a, both significant obstacles for a clinical application.
Here, we propose BOOST, an innovative gene therapy approach designed to rescue haploinsufficiency, that relies on CRISPR-Cas base editors to insert nucleotide variations in the Kozak sequence. Applying this approach, the Kozak sequence of the HI gene is made translationally stronger, engaging more ribosomes. How many HI genes could be in principle amenable to this transformation? We devised a way to explore this experimentally for 230 HI genes whose hemiallelic loss causes disease, by designing for each of them a number of actionable (i.e. which could be obtained by base editing) variants and systematically weighing their strength by an EGFP-based reporter system (21). We identified 149 promising variants to upregulate the translation of 47 HI Kozak sequences, about 20% of the total. Considering also that purposely we did not explore all the possible range of variations but only those compatible with the originally developed base editors, we confirm that a significant number of HI genes are controlled by suboptimal Kozak sequences, or at least can be enhanced by Kozak sequence variants. This number could also be extended, for at least two reasons. First, we selected the screening hits by setting strict thresholds to maximise the difference between variants and wild-type sequences. Second, the fast pace at which novel base editors, such as the near PAMless CBE and ABE (54), and other genome editing systems, such as the prime editing (55), are being developed will likely make any Kozak modification possible. These latest advancements are directly relevant to the selection of variant Kozak candidates because in our proof of principle screen, due to the constraint of introducing only transitions, we also mutagenised four base positions downstream of the AUG to allow for enough variability, which has the disadvantage of changing the second or third protein amino acid in some variants. Unbiased mutagenesis upstream the AUG of 4 bases, as it would be allowed by the application of prime editing, would generate 256 variants, probably enough to identify some upregulating Kozak variant for virtually any HI gene of interest.
The reproduction of the two candidates Kozak variants validated for NCF1 on the endogenous NCF1 locus by base editing increased NCF1 (p47phox) protein levels by 50–60%, with a similar trend to that observed in the reporter system (Figures 4 and 5E, F). The increase for the 18 validated variants was instead 20–50% (Figure 4, Supplementary Figure S2). Considering the need for HI genes, but probably also for all the dosage-sensitive disease genes, of a limited enhancement of expression, we got sufficient proof of the narrow window of tunability by Kozak variations, which should keep safe from cytotoxic overexpression. On the other hand, small increases in translation should be, in principle, sufficient to rescue HI. For example, the − 1T > C polymorphism in the Kozak sequence of the CD40 gene predisposing to Graves’ disease leads to an increase in protein levels of ∼15–30%, proving that modest changes in translational efficiency provided by Kozak variations have biological relevance (18). From another perspective, the treatment with nonsense suppressor drugs of patients affected by insufficient protein production due to nonsense mutations, such as cases of cystic fibrosis (56) and Duchenne's muscular dystrophy (57), demonstrated that a translational increase of 25% correlated with improvement in functional studies (58).
A further feature of the BOOST approach is the independence of the Kozak sequence variant developed for each HI gene from the mutation affecting the lost allele in every single patient. These mutations can be very heterogeneous for a single disease (59,60); therefore, differently from what happens in corrective gene editing, our method shares with the conventional gene augmentation therapy the advantage to develop a single clinical candidate for each disease. We see several opportunities in the application of BOOST to pathogenetic HI genes: the instalment of a permanent conversion in a cis sequence directly pushing protein production, dampening noise in the system; the limited span of the increase in translational efficiency obtainable, ideally sufficient to rescue the HI phenotype but not large enough to create imbalances; the flexibility assured by the several different Kozak variants that can be tested for each gene.
BOOST provides a number of Kozak variants with the capacity of increasing translation compared to the respective wild-type Kozak sequences. Despite the screening allowed us to identify a substantial number of them, it did not provide any usable variant for the great majority of haploinsufficiency genes, which could be due to the fact that their Kozak sequences are no further optimizable, or that the screening was not sufficiently sensitive, or both. Our validation of the improved variants resulted always successful, but provided in some cases increases in the range of 10–20% which, added to the inefficiency of delivery, could result insufficient to provide a substantial effect in in vivo applications. Moreover, even if in very few instances, the sequence variations installed by base editing could disrupt a splice site or, in case of editing downstream the ATG, likely produce variation of the second or third protein amino acid, which in some cases may affect protein stability (61). Finally, despite the Kozak sequence is a universal controller of translation efficiency, the different availability of translation initiation factors in different cell types or cell states can modulate the impact of Kozak optimization. For instance, we know that the availability of eIF1, eIF1A, and eIF5, the three factors involved in efficient start codon recognition, can shift translation to alternative translation initiation sites in certain cell states (62–64) and so potentially affect Kozak optimization. These limitations suggest that in order to map the extent of BOOST applicability to haploinsufficiency disease gene therapy a careful evaluation at the endogenous locus will be required, preferably on patient-induced pluripotent stem cells differentiated toward the tissues most affected by the diseases.
The many advancements in the efficiency and specificity of base editing, together with constantly improved delivery methods, are opening concrete possibilities for therapeutic approaches (65,66). The data presented here will be relevant to promote applications for potentially a large fraction of the known HI diseases, and also for those recessive diseases where some residual wild-type transcript is translated. For instance, in Friedreich Ataxia a trinucleotide expansion leads to a dramatic decrease in FXN protein production (67). Since low levels of wild-type mature mRNA are still produced (68), the BOOST approach could be applied. Finally, our work takes advantage of CRISPR-Cas base editors to fine-tune translation of single transcripts, thus expanding genome editing applications to a novel key control level of gene expression.
DATA AVAILABILITY
All sequencing data were deposited to the Gene Expression Omnibus (GEO) with GSE210035 accession number. The following plasmids were deposited to Addgene: pWPT-/mEGFP-1T-IRES-mCherry (#190190); pWPT-mCherry (#190605); pWPT-mEGFP (#190606); pUC19-sgNCF1 (#190193); puC19-sg-1 (#190607).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the following Core Facilities at the Department CIBIO for their technical support: HTS and Validation, NGS Facility.
Notes
Present address: Gianluca Petris, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Cambridge CB10 1SA, UK.
Contributor Information
Chiara Ambrosini, Laboratory of Translational Genomics, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
Eliana Destefanis, Laboratory of Translational Genomics, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
Eyemen Kheir, Laboratory of Molecular Virology, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
Francesca Broso, Laboratory of Translational Genomics, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
Federica Alessandrini, Laboratory of Translational Genomics, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
Sara Longhi, Laboratory of Translational Genomics, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
Nicolò Battisti, Laboratory of Translational Genomics, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
Isabella Pesce, Cell Analysis and Separation Core Facility, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
Erik Dassi, Laboratory of RNA Regulatory Networks, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
Gianluca Petris, Medical Research Council Laboratory of Molecular Biology (MRC LMB), Cambridge CB2 0QH, UK.
Anna Cereseto, Laboratory of Molecular Virology, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
Alessandro Quattrone, Laboratory of Translational Genomics, Department of Cellular, Computational and Integrative Biology - CIBIO, University of Trento, Trento 38123, Italy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione ‘Ogni giorno per Emma’ and Associazione ‘Sorriso di Ilaria di Montebruno’ [to A.Q.]; donation by Enrico and Ivana Zobele [to A.Q.]; European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement RE-GENESis [897663] [to G.P.]. The Core Facilities at the Department CIBIO are supported by the European Regional Development Fund (ERDF) 2014–2020. Funding for open access charge: overhead project funds.
Conflict of interest statement. The authors C.A., E.D., G.P., A.C. and A.Q. have filed a provisional application for a patent regarding this technology. The remaining authors declare no competing financial interests.
REFERENCES
- 1. Torgerson T., Ochs H.. Genetics of primary immune deficiencies. Stiehm's Immune Defic. 2014; 2014:73–81. [Google Scholar]
- 2. Han X., Chen S., Flynn E., Wu S., Wintner D., Shen Y.. Distinct epigenomic patterns are associated with haploinsufficiency and predict risk genes of developmental disorders. Nat. Commun. 2018; 9:2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang N., Lee I., Marcotte E.M., Hurles M.E.. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010; 6:e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dang V.T., Kassahn K.S., Marcos A.E., Ragan M.A.. Identification of human haploinsufficient genes and their genomic proximity to segmental duplications. Eur. J. Hum. Genet. 2008; 16:1350–1357. [DOI] [PubMed] [Google Scholar]
- 5. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B.et al.. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016; 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hershey J.W.B., Sonenberg N., Mathews M.B.. Principles of translational control. Cold Spring Harb. Perspect. Biol. 2019; 11:a032607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hernández G., Osnaya V.G., Pérez-Martínez X.. Conservation and variability of the AUG initiation codon context in eukaryotes. Trends Biochem. Sci. 2019; 44:1009–1021. [DOI] [PubMed] [Google Scholar]
- 8. Hinnebusch A.G. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci. 2017; 42:589–611. [DOI] [PubMed] [Google Scholar]
- 9. Ambrosini C., Garilli F., Quattrone A.. Reprogramming translation for gene therapy. Prog. Mol. Biol. Transl. Sci. 2021; 182:439–476. [DOI] [PubMed] [Google Scholar]
- 10. Barbosa C., Peixeiro I., Romão L.. Gene expression regulation by upstream open reading frames and human disease. PLos Genet. 2013; 9:e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic. Acids. Res. 1981; 9:5233–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:2850–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kozak M. Emerging links between initiation of translation and human diseases. Mamm. Genome. 2002; 13:401–410. [DOI] [PubMed] [Google Scholar]
- 14. Li J., Liang Q., Song W., Marchisio M.A.. Nucleotides upstream of the kozak sequence strongly influence gene expression in the yeast s. cerevisiae. J. Biol. Eng. 2017; 11:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grzegorski S.J., Chiari E.F., Robbins A., Kish P.E., Kahana A.. Natural variability of kozak sequences correlates with function in a zebrafish model. PLoS One. 2014; 9:e108475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ouahchi K., Arita M., Kayden H., Hentati F., Hamida M.B., Sokol R., Arai H., Inoue K., Mandel J.-L., Koenig M.. Ataxia with isolated vitamin e deficiency is caused by mutations in the α–tocopherol transfer protein. Nat. Genet. 1995; 9:141–145. [DOI] [PubMed] [Google Scholar]
- 17. Usuki F., Maruyama K.. Ataxia caused by mutations in the alpha-tocopherol transfer protein gene. J. Neurol. Neurosurg. Psychiatry. 2000; 69:254–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jacobson E.M., Concepcion E., Oashi T., Tomer Y.. A graves’ disease-associated kozak sequence single-nucleotide polymorphism enhances the efficiency of CD40 gene translation: a case for translational pathophysiology. Endocrinology. 2005; 146:2684–2691. [DOI] [PubMed] [Google Scholar]
- 19. Sultan C.S., Weitnauer M., Turinsky M., Kessler T., Brune M., Gleissner C.A., Leuschner F., Wagner A.H., Hecker M.. Functional association of a CD40 gene single-nucleotide polymorphism with the pathogenesis of coronary heart disease. Cardiovasc. Res. 2020; 116:1214–1225. [DOI] [PubMed] [Google Scholar]
- 20. Tomer Y., Concepcion E., Greenberg D.A.. A C/T single-nucleotide polymorphism in the region of the CD40 gene is associated with graves’ disease. Thyroid. 2002; 12:1129–1135. [DOI] [PubMed] [Google Scholar]
- 21. Noderer W.L., Flockhart R.J., Bhaduri A., Diaz de Arce A.J., Zhang J., Khavari P.A., Wang C.L.. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014; 10:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Diaz de Arce A.J., Noderer W.L., Wang C.L.. Complete motif analysis of sequence requirements for translation initiation at non-AUG start codons. Nucleic Acids Res. 2018; 46:985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Acevedo J.M., Hoermann B., Schlimbach T., Teleman A.A.. Changes in global translation elongation or initiation rates shape the proteome via the kozak sequence. Sci. Rep. 2018; 8:4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benitez-Cantos M.S., Yordanova M.M., O’Connor P.B.F., Zhdanov A.V., Kovalchuk S.I., Papkovsky D.B., Andreev D.E., Baranov P.V.. Translation initiation downstream from annotated start codons in human mRNAs coevolves with the kozak context. Genome Res. 2020; 30:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakagawa S., Niimura Y., Gojobori T., Tanaka H., Miura K.-I.. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008; 36:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blanco N., Williams A.J., Tang D., Zhan D., Misaghi S., Kelley R.F., Simmons L.C.. Tailoring translational strength using kozak sequence variants improves bispecific antibody assembly and reduces product-related impurities in CHO cells. Biotechnol. Bioeng. 2020; 117:1946–1960. [DOI] [PubMed] [Google Scholar]
- 27. Xu L., Liu P., Dai Z., Fan F., Zhang X.. Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different kozak variants. Microb. Cell Fact. 2021; 20:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rees H.A., Liu D.R.. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018; 19:770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rees H.A., Liu D.R.. Publisher correction: base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018; 19:801. [DOI] [PubMed] [Google Scholar]
- 31. Koblan L.W., Erdos M.R., Wilson C., Cabral W.A., Levy J.M., Xiong Z.-M., Tavarez U.L., Davison L.M., Gete Y.G., Mao X.et al.. In vivo base editing rescues hutchinson-gilford progeria syndrome in mice. Nature. 2021; 589:608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Newby G.A., Yen J.S., Woodard K.J., Mayuranathan T., Lazzarotto C.R., Li Y., Sheppard-Tillman H., Porter S.N., Yao Y., Mayberry K.et al.. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature. 2021; 595:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levy J.M., Yeh W.-H., Pendse N., Davis J.R., Hennessey E., Butcher R., Koblan L.W., Comander J., Liu Q., Liu D.R.. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng. 2020; 4:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Katti A., Foronda M., Zimmerman J., Diaz B., Zafra M.P., Goswami S., Dow L.E.. GO: a functional reporter system to identify and enrich base editing activity. Nucleic. Acids. Res. 2020; 48:2841–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G.et al.. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014; 343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bae S., Park J., Kim J.-S.. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of cas9 RNA-guided endonucleases. Bioinformatics. 2014; 30:1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S.. EditR: a method to quantify base editing from sanger sequencing. CRISPR J. 2018; 1:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pizzato M., Erlwein O., Bonsall D., Kaye S., Muir D., McClure M.O.. A one-step SYBR green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J. Virol. Methods. 2009; 156:1–7. [DOI] [PubMed] [Google Scholar]
- 39. Ewels P., Magnusson M., Lundin S., Käller M.. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016; 32:3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.-C.. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016; 34:695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tebaldi T., Zuccotti P., Peroni D., Köhn M., Gasperini L., Potrich V., Bonazza V., Dudnakova T., Rossi A., Sanguinetti G.et al.. HuD is a neural translation enhancer acting on mTORC1-Responsive genes and counteracted by the Y3 small Non-coding RNA. Mol. Cell. 2018; 71:256–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ferreira J.P., Overton K.W., Wang C.L.. Tuning gene expression with synthetic upstream open reading frames. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:11284–11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R.. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017; 551:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R.. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018; 36:843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shihab H.A., Rogers M.F., Campbell C., Gaunt T.R.. HIPred: an integrative approach to predicting haploinsufficient genes. Bioinformatics. 2017; 33:1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005; 361:13–37. [DOI] [PubMed] [Google Scholar]
- 47. Niimura Y. Comparative analysis of the base biases at the gene terminal portions in seven eukaryote genomes. Nucleic Acids Res. 2003; 31:5195–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brunson T., Wang Q., Chambers I., Song Q.. A copy number variation in human NCF1 and its pseudogenes. BMC Genet. 2010; 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun W., Zheng W., Simeonov A.. Drug discovery and development for rare genetic disorders. Am. J. Med. Genet. A. 2017; 173:2307–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Paassen B.W., van der Kooi A.J., van Spaendonck-Zwarts K.Y., Verhamme C., Baas F., de Visser M.. PMP22 related neuropathies: charcot-marie-tooth disease type 1A and hereditary neuropathy with liability to pressure palsies. Orphanet J. Rare Dis. 2014; 9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Collins A.L., Levenson J.M., Vilaythong A.P., Richman R., Armstrong D.L., Noebels J.L., David Sweatt J., Zoghbi H.Y.. Mild overexpression of mecp2 causes a progressive neurological disorder in mice. Hum. Mol. Genet. 2004; 13:2679–2689. [DOI] [PubMed] [Google Scholar]
- 52. Colasante G., Lignani G., Brusco S., Di Berardino C., Carpenter J., Giannelli S., Valassina N., Bido S., Ricci R., Castoldi V.et al.. dCas9-Based Scn1a gene activation restores inhibitory interneuron excitability and attenuates seizures in dravet syndrome mice. Mol. Ther. 2020; 28:235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M.. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013; 31:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P.. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020; 368:290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A.et al.. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019; 576:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Du M., Liu X., Welch E.M., Hirawat S., Peltz S.W., Bedwell D.M.. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:2064–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kayali R., Ku J.-M., Khitrov G., Jung M.E., Prikhodko O., Bertoni C.. Read-through compound 13 restores dystrophin expression and improves muscle function in the mdx mouse model for duchenne muscular dystrophy. Hum. Mol. Genet. 2012; 21:4007–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gregory-Evans C.Y., Wang X., Wasan K.M., Zhao J., Metcalfe A.L., Gregory-Evans K.. Postnatal manipulation of pax6 dosage reverses congenital tissue malformation defects. J. Clin. Invest. 2014; 124:111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morel Swols D., Foster J. 2nd, Tekin M.. KBG syndrome. Orphanet J. Rare Dis. 2017; 12:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Campagnoli M.F., Ramenghi U., Armiraglio M., Quarello P., Garelli E., Carando A., Avondo F., Pavesi E., Fribourg S., Gleizes P.E.et al.. RPS19 mutations in patients with diamond-blackfan anemia. Hum. Mutat. 2008; 29:911–920. [DOI] [PubMed] [Google Scholar]
- 61. Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011; 20:1298–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kearse M.G., Wilusz J.E.. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev. 2017; 31:1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fijalkowska D., Verbruggen S., Ndah E., Jonckheere V., Menschaert G., Van Damme P.. eIF1 modulates the recognition of suboptimal translation initiation sites and steers gene expression via uORFs. Nucleic Acids Res. 2017; 45:7997–8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barth-Baus D., Bhasker C.R., Zoll W., Merrick W.C.. Influence of translation factor activities on start site selection in six different mRNAs. Translation (Austin). 2013; 1:e24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Koblan L.W., Erdos M.R., Gordon L.B., Collins F.S., Brown J.D., Liu D.R.. Base editor treats progeria in mice. Nature. 2021; 589:608–614. [DOI] [PubMed] [Google Scholar]
- 66. Suh S., Choi E.H., Leinonen H., Foik A.T., Newby G.A., Yeh W.-H., Dong Z., Kiser P.D., Lyon D.C., Liu D.R.et al.. Restoration of visual function in adult mice with an inherited retinal disease via adenine base editing. Nat Biomed Eng. 2021; 5:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pallardó F.V., Pagano G., Rodríguez L.R., Gonzalez-Cabo P., Lyakhovich A., Trifuoggi M.. Friedreich ataxia: current state-of-the-art, and future prospects for mitochondrial-focused therapies. Transl. Res. 2021; 229:135–141. [DOI] [PubMed] [Google Scholar]
- 68. Pianese L., Turano M., Lo Casale M.S., De Biase I., Giacchetti M., Monticelli A., Criscuolo C., Filla A., Cocozza S.. Real time PCR quantification of frataxin mRNA in the peripheral blood leucocytes of friedreich ataxia patients and carriers. J. Neurol. Neurosurg. Psychiatry. 2004; 75:1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data were deposited to the Gene Expression Omnibus (GEO) with GSE210035 accession number. The following plasmids were deposited to Addgene: pWPT-/mEGFP-1T-IRES-mCherry (#190190); pWPT-mCherry (#190605); pWPT-mEGFP (#190606); pUC19-sgNCF1 (#190193); puC19-sg-1 (#190607).