Abstract
Key message
Genetic resources contributes to the sustainable protein production in soybean.
Abstract
Soybean is an important crop for food, oil, and forage and is the main source of edible vegetable oil and vegetable protein. It plays an important role in maintaining balanced dietary nutrients for human health. The soybean protein content is a quantitative trait mainly controlled by gene additive effects and is usually negatively correlated with agronomic traits such as the oil content and yield. The selection of soybean varieties with high protein content and high yield to secure sustainable protein production is one of the difficulties in soybean breeding. The abundant genetic variation of soybean germplasm resources is the basis for overcoming the obstacles in breeding for soybean varieties with high yield and high protein content. Soybean has been cultivated for more than 5000 years and has spread from China to other parts of the world. The rich genetic resources play an important role in promoting the sustainable production of soybean protein worldwide. In this paper, the origin and spread of soybean and the current status of soybean production are reviewed; the genetic characteristics of soybean protein and the distribution of resources are expounded based on phenotypes; the discovery of soybean seed protein-related genes as well as transcriptomic, metabolomic, and proteomic studies in soybean are elaborated; the creation and utilization of high-protein germplasm resources are introduced; and the prospect of high-protein soybean breeding is described.
Introduction
Soybean (Glycine max L.) originated in China and was known as “Shu” (Shu means legumes) in ancient China. It has a cultivation history longer than 5000 years. Having the largest planting area in the world among legume crops, soybean is the main source of edible vegetable oil and high-quality vegetable protein and provides a high-quality raw material for producing forage for livestock and aquatic animals (Kim et al. 2021a; Harada and Kaga 2019). Soybean protein is one of the high-quality vegetable proteins that have important health benefits. It is rich in eight essential amino acids, vitamins, flavonoids, and polysaccharides required by the human body (He and Chen 2013). Compared with cereal crops such as rice and maize, soybean protein has better features such as a high protein content, balanced amino acids, and excellent performance in emulsifying action and oil absorption, thereby being widely used in food processing, medicine, forage, and the chemical industry (Singh et al. 2007). Soybean-based products have become a daily nutritional solution for vegetarians and an important protein source for people on vegetarian and low-fat diets (Mortensen et al. 2009). In 1999, the important health benefits of soybean protein were recognized by the United States Food and Drug Administration (FDA) (Erdman 2000). In 2000, the United States Department of Agriculture approved access to soybean foods on university campuses, and two years later, tofu and soy yogurt were further approved as substitutes for animal meat and milk in school meal plans (He and Chen 2013), which strengthened the role of soybean products among the mainstream foods used in Western countries. The multiple demands for soybean products in the production of food, medicine, forage, and chemicals have greatly promoted the continuous increase in the global needs for soybean and the rapid development of the soybean industry. The yield and global production of soybean have continued to increase. The global production of soybean in 2021 was 382 million tons, an increase of 7.75% compared with that in 2020 and double the production (177 million tons) in 2000. The soybean protein produced in 2021 was approximately 153 million tons (https://baijiahao.baidu.com/).
Studies conducted by the World Health Organization (WHO)/Food and Agriculture Organization (FAO) (1985) have shown that soybean protein can provide all the essential amino acids for the balanced nutrition for human. Soybean protein is considered to be the high biological value protein among the plant-based proteins (García et al. 1998). The quality of soybean protein is comparable to animal proteins from meat, milk, and eggs (Millward 2012; Kudełka et al. 2021). Therefore, soybean protein is high-quality protein. With the increasing global population and the continuous improvement in per capita-based living standards, people have a more comprehensive understanding of the nutritional value and health benefits of soybean protein. In particular, under the impact of the COVID-19 pandemic, the role of soybean has become more prominent in securing the supply of food and dietary nutrients. The acceptance and need for high-quality soybean products in many countries and regions have increased significantly. According to an estimation with incomplete data, more than 12,000 food recipes worldwide require soybean protein as the raw material, which largely meets people's needs for vegetable protein. In addition, soybean meal has become the important source of high-quality protein for feed industry (Banaszkiewicz 2011; He and Chen 2013). Therefore, effectively securing the sustainable supply of soybean protein has become a major focus in soybean production and research, which can be achieved through breeding for high-yield and high-protein varieties using the rich genetic resources of soybean. This paper systematically reviews the origin, production, and development of soybean as well as the utilization of genetic resources in the breeding of high-protein soybean varieties.
Nutritional and economic value of soybean protein
Soybean protein is a complete vegetable protein
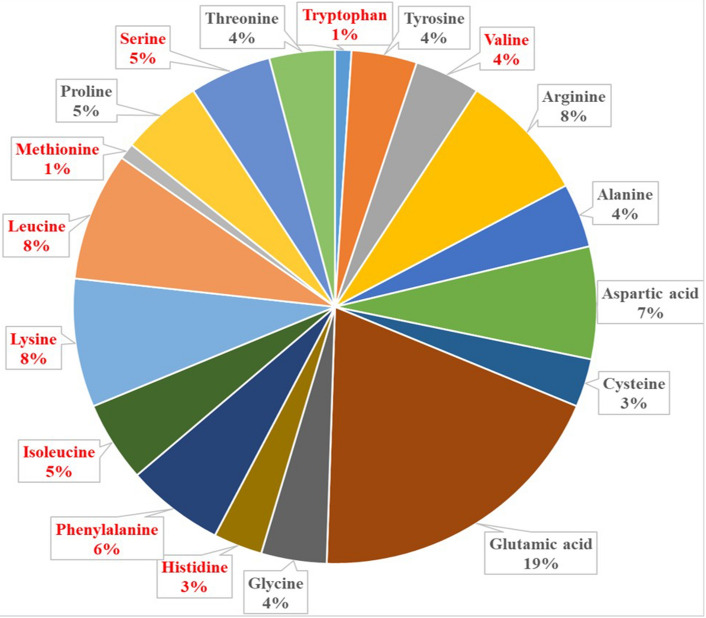
Oil and protein are the important commercial interest in soybean. Soybean seeds have the high protein content of about 40% and oil content of about 20%, respectively (Banaszkiewicz 2011). Based on 2018–2019 data from the United States Department of Agriculture (USDA), 87%, 6% and 7% of the global soybean output are used for soy oil and soy cake, foods for human consumption, and whole-bean animal feed, respectively (https://apps.fas.usda.gov/psdonline/app/index.html#/app/advQuery). Soybean-based food products are a source of high-quality vegetable protein, such as tofu, soy milk, and yuba (Rizzo and Baroni 2018). Globulin is largely responsible for the nutritional value of soybean meal, accounting for about 70% of the total protein in soybean seed (Krishnan et al. 2000; Kudełka et al. 2021). The amino acid composition of soybean protein is similar to that of animal protein (Kudełka et al. 2021). Soybean protein is also rich in eight essential amino acids required by the human body and is recognized as a complete protein source that also contains histidine, which is not produced by infants (Chen et al. 2012; Kim et al. 2021a). The percent rates of nine amino acids contents are displayed in Fig. 1, including histidine, phenylalanine, methionine, serine, valine, isoleucine, leucine, tryptophan, and lysine. According to the Protein digestibility-corrected amino acid scores (PDCAAS), soybean protein ranks first among vegetable proteins and is comparable to that of milk and egg proteins (https://foodproteins.globalfoodforums.com/food-protein-articles/soy-protein-delivers-on-nutrition-quality-sustainability/). Compared to cow’s milk, soy milk is free of lactose and cholesterol (He and Chen 2013).
Fig. 1.
The amino acid composition of soybean protein. The red grid represents essential amino acid, while the black grid represents non-essential amino acid
Soybean protein also benefits human health and prevents multiple diseases (Asif and Acharya 2013). In 1999, the FDA pointed out that the daily intake of 25 g of soybean protein may reduce the risk of heart disease (Erdman 2000). Soybean protein consumption may significantly reduce total cholesterol, LDL (low-density lipoprotein) cholesterol, and triglyceride concentrations in serum, thereby reducing the risk of coronary heart disease (Anderson et al. 1995; Zhang et al. 2003). Multiple studies have shown that the consumption of soybean protein may reduce the risk of breast and prostate cancer, benefit the kidneys, alleviate diabetes, prevent osteoporosis, lower blood pressure, relieve depression, and improve menopausal symptoms (Singh et al. 2007; Jayachandran and Xu 2019). The use of soybean protein may also inhibit fat accumulation, increase fat metabolism, and effectively regulate the expression of appetite suppressors, thereby contributing to weight reduction and obesity prevention (Messina 2016; Ramdath et al. 2017).
Soybean protein plays an important role in the food, forage, and chemical industries
Soybean is used to produce food products, food additives and industrial additives (Modgil and Kumar 2021; Kudełka et al. 2021). Soybean has been used as a basic ingredient in traditional dishes since ancient times and is the main source of high-quality vegetable protein for humans (Kim and Kwon 2001). Fermented and non-fermented foods are two types of processed foods based on soybean. Non-fermented soybean foods mainly include edamame, soybean sprouts, dehydrated soybeans, soybean flour, soy milk, tofu, bean skin, and yuba. Fermented soybean foods include soy sauce, fermented soybeans, natto, miso, soybean paste, fermented bean curd, and fermented soy milk (Kada et al. 2008; He and Chen 2013; Harada and Kaga 2019; Jayachandran and Xu 2019). In addition to traditional processing, soybean is also widely used in the production of functional foods, such as infant formula soybean milk powder, protein powder, artificial meat, dairy-free milk alternatives, and plant-based foods (Andres et al. 2012; Rizzo and Baroni 2018). Soybean-based products are also used as food additives to improve taste, increase the elasticity and oil- and water-holding capacity of foods, and enhance the storability of foods (Singh et al. 2007).
In the field of forage processing, soybean meal is the high-quality vegetable protein source considering its quantity and quality (Banaszkiewicz 2011). According to European Feed Manufacturers Federation (FEFAC) in 2007, about 18.6% edible oil and 78.7% protein-rich meal are produced in soybean after oilseed processing. The protein content in soybean meal is as high as 40–49%, and the soybean meal is mainly used for animal nutrition on the feed market (Banaszkiewicz 2011). 85% of soybean meal produced globally is used for forage production for non-ruminant animals such as poultry and pigs (Mili et al. 2012). The global production of soybean meal continues to increase. In 2019, the global production of soybean meal was 243 million tons (USDA, https://www.fas.usda.gov/), and in 2021, it increased to 251 million tons. It is predicted that the global production of soybean meal in 2022 will reach a new record of 256 million tons (https://www.feedtrade.com.cn/sbm/stat/2160577.html). The protein in soybean meal has become the most important source of protein not only in livestock forage but also in aquaculture feed. Except for methionine, the amino acid composition of soybean meal is similar to that of fish protein powder and can fully meet the amino acids needs of fish farming (INRA 2004). Due to its high quality and low price, soybean meal has gradually replaced expensive fish protein powder and is playing an increasingly important role in aquaculture (Yun et al. 2018). Soybean protein concentrate (SPC) can be used to largely replace skimmed milk powder and milk for feeding calves and can also be used as a pre-fermented feed to replace skimmed milk powder, whey protein, and fish protein powder for piglets (Dei. 2011). In the chemical industry, the widespread use of polyethylene, polypropylene, and polystyrene produced from petroleum causes serious white pollution and energy crises. Soybean protein can be sustainably reproduced at a low cost and can be an effective substitute for petroleum in some areas of the chemical industry (Tian et al. 2018a). In recent years, the problem of environmental pollution has attracted increasing attention, and the regulations have become more and more strict. How to effectively solve the problem of white pollution is a serious concern of the public. Soybean protein-based biodegradable materials are widely used in the chemical industry and play an increasingly important role in the production of plastics, foams, edible films, nanofibers, adhesives, novel composite materials, and biomedical substances, which in turn increase of the output and economic value of soybean and soybean protein (Liu et al. 2013; Luo et al. 2015; Thakur et al. 2015; Tian et al. 2018b).
The origin of soybean and current situation of soybean production
The origin and spread of soybean
It is believed that soybean has a long history of domestication in China. Cultivated soybean (Glycine max (L.) Merr.) and wild soybean (Glycine soja Sieb. et Zucc.) are two distinct species in soybean. Wild soybean is the original ancestral species of cultivated soybean and is an important germplasm resource for high-protein breeding due to its high protein content. Wild soybean occurs mainly in China, North Korea, Japan, and Russia and has the widest distribution in China (Li 1988; Kuroda et al. 2008). Cultivated soybean originated from the long-term artificial domestication and selection of wild soybean (Tian and Gai 2001) and has a cultivation history longer than 5000 years. Soybean was first introduced from China to neighboring countries Korea and Japan. In 1737, soybean was introduced into France and then into Europe. In 1765, soybean was introduced into the USA. In 1882, soybean was introduced into Argentina and began to spread in South America. In 1898, soybean was introduced from northeastern China into central and northern Russia. In 1950, Brazilians began to grow soybean to improve soil fertility, and the planting area in Brazil gradually expanded thereafter. Brazil became the world's largest soybean producer in 2021. Soybean is currently planted worldwide. Numerous soybean varieties have been developed to adapt to different ecological conditions and meet the diverse needs of multi-dietary culture (Chang 1989; Abe et al. 2003; Hymowitz and Shurtleff 2005; Patil et al. 2017).
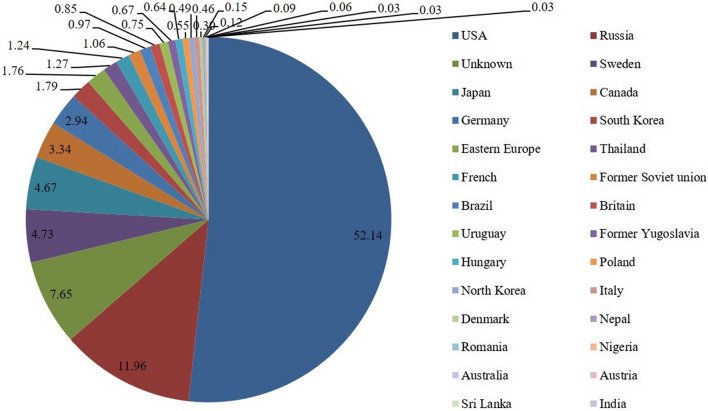
Soybean originated in China where there are very rich soybean germplasm resources with useful characteristics. The Chinese Crop Germplasm Resources Bank has collected and preserves more than 33,000 soybean germplasms from different sources (Fig. 2). The soybean germplasm resources that spread from China have promoted the breeding of new soybean varieties around the world, making important contributions to the development of the soybean industry and the sustainable production of protein worldwide. The early soybean varietal resources in the USA were mainly collected from China and its surrounding areas. Dorsett collected 1,500 soybean resources from northeastern China in 1924–1926, subsequently, Dorseet and Morse collected about 4,500 soybean resources from China, Japan, and the Korean peninsula from 1929 to 1931 (Chang 1989; Singh and Hymowitz 1999) and screened varieties such as Blackeye (introduced from Heilongjiang Province, China), Cayuga (introduced from Heilongjiang Province, China), Aksarben (introduced from Liaoning Province, China), and Barchet (introduced from Hebei Province, China) for soybean production in North America (Bernard et al. 1988). In the middle of the twentieth century, the outbreak of cyst nematode disease seriously threatened the soybean industry of the United States. The cyst nematode-resistant germplasm Peking screened from Chinese soybean resources was used in soybean breeding in the USA, and soybean varieties such as Pickett, Dyer and Bedford were successfully bred. These varieties effectively helped control the occurrence and spread of cyst nematode disease after they were used in production, and as a result, they rescued the soybean industry of the United States. Peking is still one of the main resources of resistance genes to cyst nematode among the varieties used in soybean production in the USA (Qiu et al. 2006). In the past few decades, soybean genetic resources collected from northern China have effectively promoted the breeding of new soybean varieties and the development of the soybean industry in the northern United States and Canada. The soybean genetic resources collected from southern China became the ancestral species of the varieties bred in the south of the United States (Chang 1989; Bernard et al. 1988). North American soybean varieties retain 72% of the sequence diversity of Asian landraces but have lost 79% of rare alleles and have a serious population bottleneck (Hyten et al. 2006), which is due to the few germplasms introductions from China and its surrounding areas when the breeding was conducted. Bandillo et al. (2015) conducted a microarray analysis of 14,000 soybean germplasm accessions collected by the USDA and found that 59% of the soybean resources in the USA have a Chinese origin; 49% of the American soybean ancestors have homology with Chinese soybean genes, verifying the fact that soybean originated in China and has benefited the world.
Fig. 2.
The number and ratio of soybean germplasm collected from different sources in the Chinese Crop Germplasm Resources Bank
Wysmierski and Vello (2013) identified 60 ancestral parents from 444 Brazilian soybean varieties, of which 55.3% of the genetic basis was contributed by CNS (PI 548445), S-100 (PI 548488), Roanoke (PI 548485), and Tokyo (PI 548493). Among them, CNS, S-100, and Roanoke were from China. Zhou et al. (2000) identified 74 ancestral parents among 86 soybean varieties registered in Japan from 1950 to 1988, of which 76% of the genetic basis was contributed by Japanese ancestral parents, and 2%, 5%, and 2% were contributed by foreign ancestral parents from North America (the United States and Canada), China, and Korea, respectively. The long history of soybean breeding and the geographical isolation of Japan account for the diverse genetic basis of Japanese soybean varieties. The 1,300 soybean varieties bred in China from 1923 to 2005 were derived from 670 ancestral parents, of which 113 were core parents with a large contribution to breeding and 20 were foreign resources that accounted for 17.7% of the parents (Xiong et al. 2007). In recent years, soybean varieties with high protein content and high quality such as Huachun 6 (45.80%), Huaxia 4 (46.15%), and Tongnong 10 (46.22%) bred in China include the genes from elite foreign germplasms such as Brazil 8 and Tokachi nagaha (Ouyang et al. 2009; Cui et al. 1999; https://www.chinaseed114.com/). In particular, Tokachi nagaha is one of the most widely used imported resources in soybean breeding in China. As of 2005, 195 soybean varieties had been bred, which are important backbone parents for spring soybean breeding in northern and northeastern China and have a genetic contribution rate of 0.78–50% to the new varieties (Guo et al. 2007). These studies show that the use of foreign soybean resources to broaden the genetic basis of cultivated soybean is one of the common characteristics of soybean breeding worldwide. The spread of Chinese soybean germplasm to the rest of the world has promoted the sustainable production of protein on a global scale, and the use of foreign soybean germplasm resources has effectively secured the supply of edible vegetable protein in China.
Current status of global soybean production
Before the 1950s, China was the largest soybean producer in the world. After World War II, the USA first strengthened the development and utilization of soybean varieties, and in 1954, soybean production in the USA began to surpass that in China and ranked first in the world. In the USA, soybean output has increased from 18.47 million tons in 1961 to 120 million tons in 2021, i.e., an increase of 6.5 times. Brazil is an emerging market in global soybean development and an important engine for pushing the continuous growth of global soybean production. The Brazilian soybean industry has the characteristics of a late start, rapid development, and great growth potential. Since 1974, the total annual soybean output in Brazil has surpassed that in China, and its global output share has increased sharply from 1.01% in 1961 to 36.74% in 2020. In 2021, the soybean output in Brazil was about 144 million tons, which surpassed the 120 million tons in the United States, and Brazil became the largest soybean producer in the world (https://www.fas.usda.gov/). Argentina is the third largest soybean producer and its soybean output has also grown rapidly. Soybean production in Argentina has increased from 1000 tons in 1961 to 50 million tons in 2020, accounting for 13.81% of the market share. The total annual soybean output in Argentina in 2021 was estimated to be 49.5 million tons (https://www.fas.usda.gov/). China is the fourth largest soybean producer in the world, but the average annual output is limited. The total annual output increased from 6.21 million tons in 1961 to 19.6 million tons in 2020, which is an increase of 3.16 times. However, the proportion of global output decreased from 23.1% in 1961 to 5.41% in 2020, and now China has become the world's largest soybean importer because of the large population, and the limited arable land needs to be preferentially used for producing rice and other main food crops (Hou et al. 2010; Gu et al. 2014; Kim et al. 2021b; Qiu et al. 2006; USDA, https://www.fas.usda.gov/).
The major soybean producing countries mainly focused in the USA, Brazil, Argentina, China and India, accounting for about 85% of the world’s soybean production (Gupta and Manjaya 2022). Considering their total soybean output, these countries will provide some good prospects for increasing sustainable soybean production. According to statistics, the United States accounts for about 40% of the annual exports to China (http://www.amis-outlook.org/news/detail/en/c/1144134/). With the increasing expansion of soybean export, soybean becomes the greatest cultivated crop than wheat and corn in the United States (Nosowitz 2017). In 2017, Brazil (USD 26.1 billion), the United States (USD 22.8 billion), and Argentina (USD 3 billion) were the largest soybean-exporting countries by total value, but China (USD 38.1 billion) was the largest importing countries (https://resourcetrade.earth/). Nevertheless, the global trade flow is largely reshaped in soybean. The current United States–China trade war has a direct effect on soybean industry that soybean export from the United States will likely be turned to other markets, and Brazil, Argentina and Ukraine will become more attractive markets for soybean purchases in China (http://www.amis-outlook.org/news/detail/en/c/1144134/). In 2019, China accounts for around 80% of Brazil’s soybean exports (https://www.hellenicshippingnews.com/brazil-soybeans-lose-protein-china-sales-at-risk/). The soybean production has not been interrupted by the impact of COVID-19 in the main producing countries (https://ussoy.org/u-s-soy-industry-strives-to-maintain-export-channels-supply-chain-of-high-quality-soy-amidst-covid-19-concerns/). Moreover, climate change also has a strong effect on soybean production. The World Bank estimates that soybean yields in Brazil could drop by 30% or more by 2050, but less drop in Argentina (Fernandes et al. 2012). According to the published report from the EU Commission, the soybean planting area in the 27 EU countries has more than doubled over the past 10 years and was almost 958,000 hectares in 2021 (https://www.biobased-diesel.com/post/soybean-area-in-eu-is-growing). In 2021, Italy (286,000 hectares), France (172,000 hectares), Romanian (160,000 hectares), Croatia (85,000 hectares) and Austria (76,000 hectares) were the five largest EU soybean producer. Europe produced about 10 million tons of traditional soybean per year, while genetically modified soybean imports accounted for more than 80% of plant protein, and 90–95% of that was used for animal feed (https://insights.figlobal.com/plant-based/european-soy-quest-protein-self-sufficiency).
Meanwhile, the versatile consumption demand for soybean is the major factor in maintaining the soybean production growth. In addition, the continuous intensification of the contradiction between population expansion and resource shortages as well as the increase in people's living standard have led to an increase in the global demand for soybean, which has further promoted the rapid development of the soybean industry and has caused a change in the market share of soybean producers. Soybean varieties with high yields and high protein contents will play a very important role in maintaining human health and sustainable social development.
Phenomics of soybean protein
Genetic characteristics of soybean protein
Soybean seed protein content is a quantitative trait determined mainly by the additive effects of multiple genes that interact with each other. This trait has a complex genetic mechanism and can be affected by the interaction between genotype and the environment (Pathan et al. 2013). The protein content of soybean seed is often negatively correlated with the oil content because the synthesis of protein and that of oil compete for limited carbon and energy supplies. The soybean seed protein content is also negatively correlated with agronomic traits including the grain yield, plant height, number of nodes and branches on the main stem, the flowering time, and maturity date (Dhungana et al. 2017; Chung et al. 2003; Cober and Voldeng 2000; Phansak et al. 2016). These negative correlations are main obstacles to the breeding of soybean varieties with high yield, high oil content and high protein content. In addition, the protein content of soybean seeds varies significantly among genotypes and can be affected by environmental conditions, planting location, sowing date, water and fertilizer conditions, and temperature (Li et al. 2004a, b). The database of soybean germplasm resources showed that seed protein contents of wild soybean and cultivated soybean are 35.5–56.9% and 31.7–57.9%, respectively (https://npgsweb.ars-grin.gov/). These wide variations will provide great potentiality for genetic improvement of protein content (Wang et al. 2021a).
Distribution of wild soybean germplasm resources in terms of the protein content
Soybean originated in China where there are the most abundant wild soybean resources. More than 7000 wild soybean resources were collected and are preserved in the China National Crop Gene Bank, accounting for about 90% of the wild soybean resources worldwide (Dong 2008). Wild soybean has rich genetic diversity and resistance to diseases and stresses as well as a higher protein content (46–48%) than that (38–42%) of cultivated soybean (Patil et al. 2018). Therefore, wild soybean can be used in the genetic improvement of soybean, especially the breeding of varieties with high protein content (Wang et al. 1998; Hu et al. 2011; Xue et al. 2014; Li et al. 2017). Wee et al. (2018) analyzed 334 Japanese wild soybeans and found that their average protein content was 54% (48.6–57.0%). Li (1993) found that the average protein content of more than 5200 wild soybean accessions collected in China was 44.99%, and their protein content showed a distribution pattern of high in the south and low in the north. In addition, Wang et al. (1998) determined the protein content of 6115 wild soybean resources and found that their average content was 45.36%, which is 1.05% and 2.06% higher than that of cultivated soybean collected from China and foreign countries, respectively. These studies further show that wild soybeans are characterized by a high protein content.
Distribution of cultivated soybean germplasm resources in terms of the protein content
There is an inverse relationship between the protein content of cultivated soybean and latitude; the lower the latitude, the higher the protein content (Wang et al. 1998, Rotundo et al. 2017). The protein content of a particular variety varies significantly under different growing conditions. Cv. Williams was planted in 186 locations in 76 countries and regions around the world, and its average, lowest, and highest protein contents were 42.53%, 32.8%, and 48.7%, respectively (Li et al. 2004a, b). The protein content of soybean grown in the southern United States is generally higher than that grown in the northern US (Rotundo et al. 2017). In China, the protein content of soybean grown in the Huanghuai region and southern multiple cropping areas is higher than that grown in Northeast China, which is the main soybean producing area (Liu et al. 2017). Wang et al. (1998) reported that the protein content of cultivated soybean collected and preserved in China was an average of 2.06% higher than that of soybean introduced from abroad (Wang et al. 1998). The sowing season and sowing date also significantly affect the soybean protein content. Spring sowing is favorable for oil biosynthesis, while autumn sowing and short-day conditions are favorable for protein accumulation. In addition, delayed sowing may result in a decrease in the soybean protein content.
There are abundant genetic variations in the protein content among diverse soybean germplasm resources. An analysis of soybean germplasm collected and preserved by the Chinese Crop Germplasm Resource Bank showed that the average protein content of Chinese soybean germplasm was 42.47%, whereas that of foreign soybean germplasm was 41.23% (http://crop.agridata.cn). Grieshop and Fahey (2001) analyzed the protein content of 48 Brazilian soybean accessions, 49 Chinese soybean accessions, and 36 American soybean accessions and found that the protein content of Chinese soybean (42.14%) was significantly higher than that of Brazilian (40.86%) and American (41.58%) soybeans. A study of soybean germplasm encompassing maturity groups from 00 to VIII in the USA indicated that the protein content was higher in the south (41.1%) than in the north (40.7%) (Yaklich et al. 2002). Shi et al. (2010) analyzed 105 soybean accessions from the United States, Japan, and Korea and found that their average protein content was 42.9% and that of United States accessions was 41.3%. In addition, Lee et al. (2021) found that the average protein content of soybeans from South Korea, North Korea, Japan, the USA, and Russia was 39.7%, 39.2%, 38.8%, 38.0% and 37.2%, respectively. In Europe, the soybean varieties are mainly from MG 0 to MG II in Italy, while the soybean varieties are mainly from MG 000 to MG II in France (Rüdelsheim and Smets 2012). Soybean cultivation had great potential for European agriculture, but adaptation was the central issue for soybean sustainable development in Europe (Kurasch et al. 2017a). Kurasch et al. (2017a) revealed that desirable varieties by breeding improvement might be the prerequisite for expansion of soybean cultivation in Europe, and the protein content of 1008 RILs from an early European soybean variety is about 39–42% or above 42%. In China, the soybean varieties are mainly bred for food; therefore, the protein content of Chinese soybean germplasm is higher than that of foreign counterparts. Wang et al. (1998) analyzed the protein content of 21,050 soybean accessions collected from several provinces in China and found that their average protein content was 44.31%, and the average protein content of the accessions collected from individual provinces was 42.89% (Heilongjiang), 42.24% (Jilin), 43.30% (Liaoning), 45.05% (Jiangsu), 44.35% (Anhui), 47.10% (Jiangxi), 46.03% (Hunan), 46.17% (Guizhou), and 45.27% (Fujian). These results showed that soybean protein content display the geographic distribution variation in seed protein content.
The variability of seed composition such as protein, oil and fatty acid concentrations may be influenced by genotype, environment, management practices and their interactions (Bellaloui et al. 2011). Different latitude regions show environmental fluctuations especially in photoperiod and temperature. Soybean is a short-day plant, and its photo- and thermo-sensitivities seriously restrict the growing season and geographic distribution (Szczerba et al. 2021). Elevated temperature (> 28 °C) and shortened day length may contribute to maintaining high protein content by increasing the rate of nitrogen translocation to the seed and seed growth rate (Cure et al. 1982; Dornbos and Mullen 1992; Gibson and Mullen 1996). In 2003–2004, soybean seed showed significant differences on the content of crude protein, essential amino acids and non-essential amino acids in different Brazilian states as the diversity of growing conditions, including climatic changes, topography, and soil fertility (Goldflus et al. 2006). Due to the warmer weather and longer days, the same genetically modified soybean had higher protein content in Brazil (about 37%) than United States (about 14.1%) in 2017 (https://geneticliteracyproject.org/2018/01/30/brazils-higher-protein-gmo-soybeans-hurting-us-exports-china/). Rotundo et al. (2009) reported that high protein varieties have greater assimilate availability per seed during seed-filling period, and higher protein levels in high yielding varieties can be achieved with greater sucrose and nitrogen assimilate supply per seed. Additionally, maintaining adequate soil moisture during the reproductive stage is critical for the accumulation of protein content in soybean seed (Wijewardana et al. 2019). Moreover, UV-B radiation can cause significant alteration that protein content decline linearly with higher doses of UV-B radiation (Reddy et al. 2016). Therefore, different environmental conditions during pod-filling period may have a considerable effect on soybean protein content.
Soybean protein-related genomics
Soybean protein content-related genes
A high protein content is an important goal of soybean quality breeding. The cloning of soybean protein content-related genes/quantitative trait loci (QTL) is helpful for breeding new soybean varieties with high protein content and high yield using molecular technology. Several researchers have used a variety of materials and methods to study the genes/loci related to the soybean protein content. Existing studies have identified the QTLs related to the soybean protein content on 20 soybean chromosomes, and related new QTLs are still being reported (Bandillo et al. 2015; Kim et al. 2016; Lee et al. 2019; Wang et al. 2020a). For example, QTLs cqProt-001 and cqProt-003 were first identified on chromosomes 15 and 20 (Diers et al. 1992), QTL Satt127 and Iasu-A144H-1 were detected in wild soybean to have an association with the protein content (Sebolt et al. 2000), QTLs have been mapped on the B1 and L linkage groups (Chapman et al. 2003), the protein content-related QTLs Seed protein 34–10, Seed protein 36–31, and Seed protein 36–32 were identified on chromosome 19 (Lu et al. 2013; Mao et al. 2013), Kim et al. (2016) identified a high protein and low oil content QTL on chromosome 15 in PI 407788A, and Warrington et al. (2015) identified a major QTL associated with the protein and amino acid contents on chromosome 20. According to data in the Soybase database, 249 QTLs related to the soybean protein content have been identified (https://soybase.org/), mostly in the linkage group I (chromosome 20), explaining 7–65% of the observed phenotypic variations, followed by those detected in linkage group E (chromosome 15), linkage group C2 (chromosome 6), and linkage group G (chromosome 18) (Table 1).
Table 1.
Information of QTLs for protein content in soybean
| Chr | Linkage Group | Numbers of QTL | Genetic contribution (%) |
|---|---|---|---|
| 1 | D1a | 7 | 4.7–27.6 |
| 2 | D1b | 8 | 5.16–18.0 |
| 3 | N | 12 | 5.6–17.9 |
| 4 | C1 | 15 | 6.8–31.0 |
| 5 | A1 | 10 | 4.6–23.0 |
| 6 | C2 | 19 | 4.8–27.6 |
| 7 | M | 14 | 4.0–27.1 |
| 8 | A2 | 10 | 5.0–12.1 |
| 9 | K | 17 | 2.35–24.4 |
| 10 | O | 9 | 6.0–21.0 |
| 11 | B1 | 9 | 3.0–15.7 |
| 12 | H | 8 | 3.0–32.0 |
| 13 | F | 14 | 6.0–18.1 |
| 14 | B2 | 12 | 5.0–19.0 |
| 15 | E | 18 | 5.65–24.0 |
| 16 | J | 3 | 7.6 |
| 17 | D2 | 10 | 6.64–26.0 |
| 18 | G | 16 | 2.89–20.1 |
| 19 | L | 13 | 4.0–27.0 |
| 20 | I | 25 | 7.0–65.0 |
Lestari et al. (2013) aligned repeated genomic regions associated with the soybean protein content and identified 35 and 19 candidate genes on chromosomes 10 and 20, respectively. Yang et al. (2019) mapped a protein content-related gene within a 329-kb region on chromosome 15 and predicted that the Glyma.15g049200 gene might be related to the soybean protein content. In addition. Bolon et al. (2010) mapped a protein content-related QTL in an 8.4 Mb interval between Sat_174 (24.54 Mb) and ssrqtl_38 (32.92 Mb) on chromosome 20, and Hwang et al. (2014) further narrowed the region to 2.4 Mb between 27.6 and 30.0 Mb through association analysis and screened six candidate genes (Glyma20g19680, Glyma20g21030, Glyma20g21080, Glyma20g19620, Glyma20g19630, Glyma20g21040). Valliyodan et al. (2016) analyzed 106 soybean lines through re-sequencing and found three gene clusters related to the protein content in a 2.4 MB interval on chromosome 20, in which Glyma20g19680, Glyma20g21030, and Glyma20g21080 were further identified as the candidate genes. Vaughn et al. (2014) also identified a protein content-related QTL located about 1 Mb downstream from the region identified by Hwang, and the location of the most significant SNP site was 31,972,955 bp. A total of 13 candidate genes associated with seed protein were detected, 8 of which were highly expressed in mature soybean seeds (Huang et al. 2018). Fliege et al. (2022) conducted fine mapping of chromosome 20 and cloned the major gene Glyma.20g85100 that controls the protein content, which provides a detailed insights into the haplotype analysis of Glyma.20G085100 for variations in protein content. Analysis of transgenic plants indicated that the regulatory effect of the cqSeed protein-003 QTL on the protein content was caused by a transposon insertion in the CCT domain encoded by the Glyma.20G85100 gene. Qin et al. (2022) reported that seven candidate genes associated with seed protein content were identified in a 471-kb haplotype block from Chr6_18844283 to Chr6_19315351, including polynucleotidyl transferase (Glyma.06G202900 and Glyma.06G203100), polygalacturonase activity (Glyma.06G202600 and Glyma.06G203000), ATP synthase (Glyma.06G203200), and genes without annotation (Glyma.06G202700 and Glyma.06G202800). The discovery of more soybean protein content genes is an urgent need for promoting the breeding of new soybean varieties with high protein contents.
Genes related to the oil content may also be involved in protein synthesis and metabolism. For example, phosphoenolpyruvate carboxylase (PEPC) is involved in the dual regulation of both protein and fatty acid synthesis in seeds. Inhibition of the expression of the endogenous GmPEPC gene in soybean promoted the accumulation of oil, while the increase in PEPC activity favored the synthesis of protein (Zhang et al. 2017; Zhao et al. 2006). Transcription factors GmDof4 and GmDof11 increased the oil content in grains by activating the expression of fatty acid synthesis-related genes such as acetyl-CoA carboxylase gene and by inhibiting the expression of protein synthesis-related genes (Wang et al. 2007; Sun et al. 2018). The ABI3 (Abscisic acid insensitive 3) gene is involved in regulating the synthetic pathways of both protein and oil (Lazarova et al. 2002). Transgenic soybean lines expressing the Arabidopsis QQS (Qua-Quine Starch) gene showed 8–10% increase in seed protein content by regulating metabolic processes affecting the partitioning of carbon and nitrogen among proteins and carbohydrates (Li et al. 2015). Furthermore, some soybean sugar transporters such as GmSWEET10a, GmSWEET10b and GmSWEET39 had pleiotropic effect on seed protein and oil content (Miao et al.2020; Wang et al. 2020b; Zhang et al. 2020). POWR1 (Protein, Oil, Weight, Regulator 1) gene plays a critical role in controlling the seed quality and yield traits by regulating likely nutrient transport and lipid metabolism, and transgenic study demonstrated that the high-protein POWR1 allele may be employed to meet the worldwide requirement for high-protein soybean in the breeding process (Goettel et al. 2022).
Genes related to soybean seed protein components
So far, 625 seed proteins have been identified in soybean seeds and they are classified into 11 groups based on their functions, of which 197 storage proteins are the main components of the proteins in mature soybean seeds (Krishnan et al. 2009; Zhang et al. 2016). Soybean seed storage proteins can be classified into four basic types based on their differences in solubility: albumins, globulins, prolamins, and glutelins (Natarajan et al. 2006). The proportion of each storage protein relative to total protein is affected by both genotype and the environment. The storage proteins of cultivated soybean seeds are mainly composed of 2S, 7S, 11S, and 15S globulin complexes, of which 7S and 11S globulins account for 60–80% of the total seed protein (Singh et al. 2015). 7S globulin (also known as β-conglycinin), which accounts for about 30% of soybean seed protein, is a heterotrimeric glycoprotein that consists of α, α’, and β subunits and has a molecular weight of 126–170 kDa (Hirano 2021; Singh et al. 2015). 11S globulin is a hexameric protein of 320–375 kDa, of which each subunit consists of an acidic (A) and a basic (B) polypeptide chain, accounting for about 40–60% of total seed protein (Tandang-Silvas et al. 2010; Zhao et al. 2021). The proportions of 7S and 11S globulin in soybean seeds are negatively correlated, and the ratio of 11S to 7S (11S/7S) globulin in soybean seed was reported to directly affect the nutritional quality and functional properties of soybean seed protein, thereby affecting the application value of soybean protein (Singh et al. 2015). The content of sulfur-containing amino acids in 11S globulin is 5–6 times that in 7S globulin. A high content of 11S globulin results in higher nutritional quality of soybean protein. Therefore, breeding soybean varieties with high content of 11S globulin can improve the nutritional value of soybean protein and related products (Achouri et al. 2005; Liu et al. 2006; Magni et al. 2018). Furthermore, compared with 11S globulin, 7S globulin has fewer disulfide bonds and a higher lysine content, thus having better emulsifying properties (Fujiwara et al. 2014; Kagawa et al. 1987). Compared to animal proteins, the soybean protein has lower content of sulfur amino acids (Henkel 2000). The sulfur-containing amino acid content in soybean is an important evaluation index of protein quality, however, the limitation of protein quality is the deficiency in sulfur-containing amino acids such as methionine (Met) and cysteine (Cys) (Krishnan et al. 2000). The FAO recommended that the requirement of Cys + Met contents are 3.5% of the total protein (George and de Lumen 1991). In regular soybean varieties, the Met + Cys contents are approximately 2.4% of the total protein (Panthee et al. 2006). Moreover, the value of soybean protein is limited by the Met + Cys contents (2.9g/16gN) (Banaszkiewicz 2011). According to Liebig’s law of the minimum and the concept of the well-known Liebig barrel, Met and Cys are the first and the second limiting amino acid, respectively (Lemme et al. 2020). Therefore, the levels of sulfur-containing amino acids (Met + Cys) are extremely important for soybean. A total of 113 genes encoding for sulfur-containing amino acid enzymes were identified in soybean (https://www.genome.jp/kegg/pathway.html). Panthee et al (2006) identified seven QTLs governing the content of sulfur-containing amino acids using a population of F6-derived recombinant inbred lines (RILs), including four (Satt235, Satt252, Satt427 and Satt436) for Cys and three (Satt252, Satt564 and Satt590) for Met concentration. Two QTLs and nine QTLs associated with both Cys and Met content were found in F5:9-derived progeny of RILs and 137 Canadian short‑season soybean lines, respectively (Fallen et al. 2013; Malle et al. 2020). The synthesis of sulfur-containing amino acids was regulated by soybean cysteine β-lyase gene GmCBL1 and its homologous gene GmCBL2, and overexpression of GmCBL1 and GmCBL2 significantly increased the contents of Met+Cys in transgenic hairy roots. These promising molecular markers and candidate genes may be useful for genetic selection for elevated soybean protein quality. Soybean antigen protein, also known as soybean allergen, may cause allergic reactions in piglets, calves, and fish, which damage the intestinal tract and impede the growth and development of the animals (Sun et al. 2005; Qiang et al. 2018; Li et al. 2021). The allergen database AllergenOnline (http://www.allergenonline.org/) includes 43 soybean allergens, i.e., glycinin, β-conglycinin, Gly m Bd 28 K, Gly m Bd 30 K, soybean hydrophobin (Gly m 1), soybean hull protein, soybean inhibitory protein, and Kunitz trypsin inhibitor (KTI). Glycinin (11S) and β-conglycinin (7S) are the main substances that cause allergy in young animals, and the acidic subunit of 11S globulin can cause allergy in both humans and animals (Adachi et al. 2003). 7S globulin includes lectin, β-amylase, Glym Bd 28 K, Glym Bd 30 K, lipoxygenase, and β-conglycinin. Β-conglycinin is a trimer that consists of α, α’, and β subunits, of which the α and α’ subunits have the highest antigenicity (Fang and Qiu 2005; Amigo-Benavent et al. 2011). 2S globulin also includes a Kunitz trypsin inhibitor, cytochrome c urease, and 2S globulin (Tay et al. 2006). Therefore, determining the contents of 7S and 11S globulin and the proportion of each component in diverse soybean genetic resources and screening elite germplasm resources are of great significance to the improvement of the nutritional and processing quality of soybean protein and the sustainable production and application of protein.
The 7S and 11S storage proteins are encoded by a multigene family (Fischer and Goldberg 1982; Scallon et al. 1985). The 7S globulin gene family has at least 15 members encompassing CG-1 to CG-15 (Krishnan et al. 2000). CG-1 encodes the α´ subunit, and CG-4 encodes the β subunit (Harada et al. 1989). Genetic analysis of plant material completely lacking the 7S globulin indicated that this was controlled by a dominant locus, Scg-1 (Teraishi et al. 2001). The Scg-1 locus contains the α-subunit gene and β-subunit gene, which are involved in regulating the synthesis of soybean 7S globulin. Silencing both of the genes induced the complete lack of 7S globulin and an increase in the 11S globulin content in soybean (Tsubokura et al. 2012). Overexpression of OASS simultaneously increased the content of 7S and 11S globulin, thus increasing the total protein content and contributing to the significant increase in the cysteine and methionine contents (Alaswad et al. 2021). A total of seven genes were found to encode soybean 11S globulin, Gy1–Gy7. The Gy1–Gy5 genes consist of four exons and three introns (Nielsen et al. 1989; Beilinson et al. 2002). Gy1, Gy2, and Gy3 encode A1aB2, A2B1a, and A1bB1b subunits, respectively, which mainly consist of sulfur-containing amino acids, cysteine, and methionine. Gy4 and Gy5 encode the A5A4B3 and A3B4 subunits, respectively (Nielsen et al. 1989; Beilinson et al. 2002). The Gy1 and Gy2 genes are located on chromosome 3, and they are 3 kb apart and closely linked. Gy3 is located on chromosome 19 (Nielsen et al. 1989). Gy4 and Gy5 are located on chromosomes 10 and 13, respectively (Diers et al. 1992). In addition to the five genes described above, Beilinson et al. also identified two new genes (Gy6 and Gy7). Gy6 is located downstream of Gy2 on chromosome 3 and is a pseudogene that cannot encode functional proteins. Gy7 is in tandem with the 3´ end of Gy3 on chromosome 19 and has very low expression. Therefore, it is a functional but weakly expressed gene (Beilinson et al. 2002; Nielsen et al. 1989). There are 29 QTLs related to the 7S and 11S globulin contents of soybean storage proteins in the soybean database (www.soybase.org). Satt461, Satt292, and Satt156 are in linkage group D2, I, and L, respectively, and are associated with the content of 11S globulin. Satt461 and Satt249 are in linkage group D2 and J, respectively, and are associated with the content of 7S globulin (Panthee et al. 2004). Liu et al. (2009) mapped 28 QTLs related to the content of 11S and 7S globulins using a population of recombinant inbred lines. Jian et al. (2013) found 14 SSR sites associated with 11S and 7S globulins through association analysis. Mutagenesis is widely used to induce mutations in the subunits of soybean protein. Zhang et al. (2018) screened six mutants with an obvious mutation in the subunit genes and 10 mutants that had an 11S/7S ratio higher than 3 from EMS-induced mutant lines. Hayashi et al. (2001, 2009) obtained a mutant lacking 7S globulin by means of γ-ray irradiation, and its regulatory genes were mapped on chromosome 19 between markers Satt523 and Sat_388 (3.39 cM) (Hayashi et al. 2001, 2009). From the abundant soybean germplasm resources, studies have screened a variety of elite mutants that lack α, α’, A3, (α’ + A4), or (α’ + α) (Liu et al. 2010; Song et al. 2013; Tuo et al. 2014; Zhang et al. 2015). Patil et al. (2017) developed a CGY-2-NIL near-isogenic line with a mutation in the CGY-2 gene. The creation and identification of these genetic materials, markers, and loci are of great significance in the discovery of the genes that control the quality of the soybean protein and for the genetic improvement of soybean quality and provide a basis for the isolation of soybean genes controlling the 7S and 11S globulin contents and the further application of these genes in soybean breeding and production.
Genes related to the synthesis and transport of soybean seed protein
Understanding how the molecular mechanisms of synthesis, transport, and storage of seed proteins has great significance for seed quality improvement in soybean (The et al. 2020; Yang et al. 2020). The anabolism of soybean seed protein mainly involves two processes: amino acid synthesis (ammonia assimilation and transport) and the transcription, translation, and processing of storage protein genes (Warsame et al. 2018). The anabolism of soybean seed protein begins with the fixation and conversion of nitrogen (Fabre and Planchon 2000). Soybean converts atmospheric nitrogen into NO3− by the action of nitrogenase in rhizobia in a symbiotic nitrogen fixation system formed in soybean roots, which is then converted into NO2− by the action of nitrate reductase (NR). Catalyzed by nitrite reductase (NiR), NO2− is reduced to NH4+ (Ohyama et al. 2017). NH4+ is immediately transported to the cytoplasm of root nodule cells where it is used to synthesize adenine and guanine that are then oxidized and decomposed into ureide. After being transported from roots to hulls, ureide is degraded into glyoxylic acid and ammonium ions (Cabanos et al. 2021). Ammonium ions are converted into glutamine and glutamate through the glutamine synthase–glutamate synthase pathway, and after being activated, they are transported by tRNA to mRNA on the rough surface of the plasma reticulum to synthesize 11S and 7S globulins that are then transported by Golgi receptors to the vacuole for processing and modification. Finally, all storage proteins are combined in a protein body for the synthesis and accumulation of soybean seed protein (Cabanos et al. 2021). Therefore, the seed protein content is derived from the transport of nitrogen compounds (such as amino acids, ureides, peptides) from roots, nodules, and mature leaves (Tegeder and Masclaux-Daubresse 2018).
Some related genes involved in protein endoplasmic reticulum synthesis pathway may cause the significant variations in protein content between Jidou 12 and Ji HJ117 (Guo et al. 2019). Amino acid transporters (AATs) family such as Cationic Amino Acid Transporters (CATs), Amino Acid Permeases (AAPs), and organic nitrogen compounds transporters (NCTs) were identified in recent studies (Cheng et al. 2016; Joaquim et al. 2022). Joaquim et al. (2022) reported that some NCT-related genes AAP7, AVT3, CAT9, UMAMIT25 and UPS2 were highly expressed in the high protein content seed, indicating that genetic manipulation of these NCT genes may contribute to elevated protein content in soybean seed. The putative amino acid permease transporter gene (AAP8, Glyma.08G113400) and seed storage 2S albumin protein gene (Glyma.08G112300) may be responsible for the high water-soluble protein content in soybean. (Zhang et al. 2017).
The synthesis and transport of storage proteins in soybean seed
During soybean seed development, both 7S and 11S glycinin are initially synthesized on the endoplasmic reticulum (ER) as precursors, and then transported to protein storage vacuoles (PSVs) via the vesicles (Hohl et al. 1996; Vitale and Raikhel 1999). In PSV, most of them are processed into mature subunits and deposited (Müntz 1998). Such as 11S glycinin of soybean, it is first synthesized on the ER as proglycinin, which contains a short signal peptide that directs the precursor transferred to the lumen of the ER, then, the signal sequence is removed and the resultant proglycinin assembles into trimers (Müntz 1998). Proglycinins are sorted to the PSV via the dense vesicle (DV)-mediated post-Golgi trafficking pathway, where they are processed into mature hexameric structure by specific posttranslational cleavage that occurs between Asn and Gly residues (Müntz 1998; Robinson et al. 1998). In addition to the typical “ER-Golgi-PSV” pathway, ER-derived vesicles were observed in β-conglycinin-inhibited transgenic soybean seed, which resembled precursor accumulating vesicles of pumpkin seeds or the protein bodies accumulated in cereal seeds (Hara-Nishimura et al. 1998). Glycinin is a major component of these ER-derived vesicles, thus these ER-derived novel vesicles are called Protein Bodies (PBs) (Kinney et al. 2001). Moreover, the ER-derived precursor-accumulating (PAC) vesicles also involve in the trafficking of Glycinin during the early stage of soybean cotyledon development (Mori et al. 2004). In summary, storage proteins of soybean are transported to the PSV by three pathways: (1) ER-Golgi-PSV, storage proteins are synthesized on the ER as precursors and transported to the PSV through the Golgi by DVs (Robinson et al. 1998). (2) ER-PBs, storage proteins are synthesized on the ER and directly bud from ER to form PBs (Kinney et al. 2001). (3) ER-PAC-PSV, storage proteins are synthesized on the ER and transported to the PSV bypassing the through the Golgi via PAC vesicles (Mori et al. 2004).
In maturing seed cells, vacuolar sorting determinants (VSDs) are required to direct proteins into transport vesicles destined for the PSV. Three kinds of VSD have been identified so far: sequence-specific VSD (ssVSD), C-terminal VSD (ctVSD), and physical structure VSD (psVSD) (Matsuoka and Neuhaus 1999; Vitale and Raikhel 1999). The ssVSDs contain conserved amino acid sequences, such as the NPIR-like motif, which are necessary for recognition by a vacuolar sorting receptor (Koide et al. 1997; Nishizawa et al. 2003). The ssVSDs can be located in N-terminal (e.g., sporamin in sweet potato), C-terminal (e.g., 2S albumin in Brazil nut), or within mature proteins (e.g., 11S glycinin in soybean) (Kirsch et al. 1996; Nishizawa et al. 2006; Saalbach et al. 1996; Vitale and Hinz 2005). ctVSDs are present in C-terminal regions of polypeptides and have highly variable sequence, they are often enriched in hydrophobic amino acids (Neuhaus and Rogers 1998; Nishizawa et al. 2003). In contrast to the ssVSD and ctVSD, there is limited information on the psVSD, which is postulated to depend on the integrity of long internal sequence stretches (Neuhaus and Rogers 1998). A transient expression assay in developing soybean cotyledons demonstrated that the C-terminal 10 residues of the β-conglycinin α′- and β-subunits (PLSSILRAFY; PFPSILGALY; CT10) were shown to act as a necessary and sufficient vacuolar targeting signals in soybean cotyledon cells (Nishizawa et al. 2003). In contrast to β-conglycinin, three types of VSD may coexist in 11S glycinin of soybean (Nishizawa et al. 2006), suggesting that plants have evolved complicated sorting mechanism.
Study of the soybean seed protein content
Transcriptomics
High-throughput sequencing-based transcriptomics and proteomics are effective methods to analyze changes in genes and their products in the biological processes of organisms (Lan et al. 2012). Studies show that the soybean seed protein content-related genes are transcriptionally activated and then repressed during embryogenesis, while genes encoding mRNA are transcribed at a similar rate. In the absence of DNA methylation, both transcription and post-transcription processes regulate the mRNA levels of genes that encode soybean seed protein, providing information for improving the soybean seed protein content (Walling et al. 1986). Song et al. (2016) found a large number of differentially expressed genes between a near-isogenic line (cgy-2-NIL) lacking the allergenic α subunit of β-conglycinin and its recurrent parent. The cgy-2 allele is derived from a functional allele that is closely related to the amino acid quality. The β subunit of soybean seed storage protein is of great importance to the balance of sulfur-containing amino acids and their processing quality. Zhang et al. (2019) found that sulfur-containing amino acids (Cys + Met) were significantly increased (31.5%) in soybean seeds with a low content of the β subunit, implying a close relationship between the β subunit and sulfur assimilation, which work together to coordinate the synthesis of soybean seed protein. In production, the protein content of variety Ji HJ117 (52.99%) was higher than that of the control variety Jidou 12 (46.48%), and the variation may be caused by the difference in protein synthetic pathways that take place in the endoplasmic reticulum during seed development (Guo et al. 2019).
Metabolomics
Metabolomics is an important method to elucidate the regulatory mechanism of metabolism related to soybean seed growth and the synthesis and degradation of proteins. Lin et al. (2014) identified 169 metabolites in the seeds of 29 conventional soybean varieties and found that the level of 104 metabolites varied significantly among the varieties. At the same time, metabolite markers that can be used to distinguish genetically related soybean varieties were identified, and the results provide a genetic basis for further analysis of soybean seed metabolites. Schmidt et al. (2011) systematically analyzed the soybean SP2 mutant (seed storage protein knockout mutant) using proteomic, metabolomic, and transcriptomic approaches and found that the rebalancing of the seed protein content was largely due to the selective accumulation of a small number of proteins, while the rebalancing of protein components was accompanied by only slight transcriptomic and metabolomic changes.
Proteomics
Proteins are the final products of gene expression and the specific executors of gene functions. Changes at the proteomic level can directly reflect changes in the execution of gene functions during the growth and development of organisms (Li et al. 2019a). Although the genomic differences between wild and cultivated soybeans have been extensively studied and gradually clarified, differences in their seed protein expression have not been determined. Li et al. (2007) analyzed the differences in seed storage proteins between three wild soybeans and three cultivated soybeans, which provided important information for identifying important specific genes in wild and cultivated soybeans. Hashiguchi et al. (2020) identified 65 proteins that were differentially expressed between wild and cultivated soybean seeds, which may promote the use of wild relatives in transgenic breeding to improve soybean protein and other agronomic traits. Glycinin subunits are believed to have an important role in soybean breeding and the improvement of the biochemical properties of soybean proteins. Cho et al. (2014) identified 10 glycinins from the cotyledon tissue of the soybean seed coat, which provides a theoretical basis for clarifying the genetic regulation of glycinin expression in seeds. Min et al. (2015) found that the urea cycle might be involved in the accumulation of glycinin and β-conglycinin subunits (SSPs), thereby increasing the protein content of soybean seeds. The protein content of a soybean mutant induced by fast neutron (FN) irradiation was increased by 15%. Islam et al. (2020) compared the difference in seed protein expression between the wild-type and FN-induced mutant and found that the level of basic 7S globulin in the mutant was fourfold higher than that in wild-type soybean seeds. This study provides a valuable germplasm resource for clarifying the molecular mechanism regulating the synthesis and degradation of soybean protein. The protein profiles of the high-oil variety Jiyu 73 (JY73) and high-protein variety Zhonghuang 13 (ZH13) were compared, and it was found that the high protein content of ZH13 was mainly due to the high expression of major storage proteins and the proteins related to nitrogen and carbon metabolism (Xu et al. 2015).
Creation of soybean germplasm with high protein content and the breeding of new varieties
Integration of multiple breeding approaches accelerates the discovery and utilization of soybean genetic resources with high protein content
There are two major ways to genetically improve soybean protein: one is the use of conventional breeding methods to improve the soybean protein content or protein quality (Tian et al. 2021; Hao et al. 2021; Zhang et al. 2021); the other way is to modify specific genes to achieve the targeted improvement of soybean protein (Lin et al. 2010; Wu et al. 2021; Zhang et al. 2021). Hybridization and mutation breeding are the two major techniques methods used in the breeding of soybean varieties with high protein content, and hybridization is the most commonly used technique to generate variation. According to the genetic characteristics of soybean protein traits, parents with high protein content are selected for hybridization with high-yielding parents to combine the desired traits of both parents, and then new soybean varieties with high protein content and high yield are selected among the progenies. This is the most conventional approach used in the breeding of soybean varieties with high protein content. Mutation breeding for improving soybean quality began in the mid-1970s. The improvement of soybean protein content and quality through physical and chemical mutagenesis is an effective approach for improving soybean quality, which mainly includes chemical, physical, and spatial mutagenesis. The soybean variety Yumeminori with null α´, α subunits and low allergenicity was isolated from the γ-ray irradiation progeny of Kari-kei 434 (Takahashi et al. 1994, 2004), and a four bp insertion mutation in the exon of CG-2 gene was responsible for the absence of α subunit in Yumeminori (Ishikawa et al. 2006). Guo et al. (2005) used physical mutagens such as 60Co-γ rays and thermal neutron irradiation as well as chemical mutagens such as EMS and sodium azide to treat soybean and successively bred the vegetable soybean variety Huaihadou 1 (protein content of 44.93%), the high-protein mutant lines 903,525 (47.60%), 903,526 (47.02%), and 903,527 (47.53%), which have also high resistance to viral diseases and gray spot disease, and two high-protein stable mutants, 923,725 (45.38%) and 923,738 (45.24%). Wei et al. (2019) screened nine stable germplasms (m1–m9) with high protein content among the mutant lines of EMS-treated Zhongpin 661 in the M7 generation, and their average protein content was 48.17%, which is 16.94% higher than the protein content (41.19%) of the wild type. Besides, Wang et al. (2021b) screened a new soybean line Nanxiadou 25 from the 60Coγ radiation-induced mutant lines of Rongxiandongdou, and Nanxiadou 25 was high protein (50.1%), good shading tolerance, and strong lodging tolerance soybean variety widely cultivated in southwest China. A range of high-protein materials obtained through mutagenesis has enriched the soybean gene pool. Recurrent selection is an effective method to synergistically improve the protein content and yield. Without selective backcrossing for the protein content, the protein content of the backcrossed population generally decreases, and selective backcrossing using material with high protein content can effectively increase the protein content of the population (Zhao et al. 2006).
Combining molecular biological technology with conventional breeding approaches may effectively shorten the breeding time and improve the efficiency of selection in crop quality improvement (Jun et al. 2007). The development of markers related to the soybean protein content such as SSR marker satt182, satt419, satt239, and satt598 has made selection more efficient (Li et al. 2020). The development of high-throughput and low-cost sequencing technologies, chips, and genome-wide selection have improved the selection efficiency in the genetic improvement of the soybean protein content. Increasing the training set size and improving the relationship between the training and validation sets were reported to improve the efficiency in selecting varieties with high protein content, and the predictive ability reached R2 = 0.81 (Stewart-Brown et al. 2019). The selection of a representative and diverse training population increased the predictive ability related to the protein content to 0.92 (Jarquin et al. 2016). For a training population with fewer than 350 individuals, the predictive ability of the protein content increased with the training population size, and the prediction accuracy increased significantly with the number of markers when fewer than 1000 markers were used in the model (Beche et al. 2021). The use of intragenic markers in predicting the protein content increased the predictive ability by 0.02 compared with the use of intergenic markers, and the addition of progeny lines to the training population established with germplasm resources increased the predictive ability from 0.47 to 0.65 (Sun et al. 2022). In addition, the inclusion of epistatic effects in the model also improved the accuracy of predicting the protein content (Duhnen et al. 2017). With the advancement of sequencing technology and an in-depth understanding of the factors that contribute to the prediction accuracy, genome-wide selection is expected to display superiority in the selection of multi-gene-controlled complex traits including the protein content. The cloning of protein content-related genes is conducive to the targeted modification of specific genes and the improvement of soybean protein. The content of sulfur-containing amino acids in soybean is affected by genes, the environment, and the interaction between genes and the environment. Traditional breeding approaches have low selection efficiency and slow progress. The use of molecular biological methods is conducive to improving the efficiency of selecting for the amino acid content. Dinkins et al. (2001) transformed a 15 kDa δ-zein gene into soybean using particle bombardment, and in the transgenic plants, the content of methionine was increased by 12–20% and the content of cysteine was increased by 15–35%. In the same way, Li et al. (2005) obtained transgenic soybean plants with a 27 kDa γ-zein gene, in which the content of methionine was increased by 15.49–18.57% and the content of cysteine was increased by 26.97–29.33% compared with control plants. A proglycinin gene Gy1 (A1aB1b) with a synthetic DNA encoding four continuous methionines (V3-1) was connected between the hpt gene and the modified green fluorescent protein sGFP (S65T) gene, and a resultant plasmid was introduced into soybean plants using particle bombardment. The result showed that compared to control plants, transgenic soybean plants accumulated higher levels of glycinin (El-Shemy et al. 2007). The soybean resources with high protein content and high-quality proteins have greatly benefited sustainable protein production.
Utilization of wild soybean in the breeding of varieties with high protein content
The use of elite germplasm or genes of wild soybean to broaden the genetic basis of cultivated soybean is conducive to the breeding of new soybean varieties with high yield, high quality, and high protein content. Using cultivated soybean Heinong 35 and wild soybean ZYD 355 and ZYD665 as parents, Lai et al. (2005) bred a new soybean line, Longpin 8807, with protein content higher than 48.29%. Du and Yu (2010) crossed the cultivated soybean N23674 (protein content 42.49%) with wild soybean BB52 (protein content 40.09%) that grew naturally in a tidal flat area and screened the F4:5 line 4035 that had a distinctly superior quality: a high protein content (49.13%); increases of 0.03–0.76% in the contents of the essential amino acids threonine, lysine, histidine, phenylalanine, leucine, isoleucine, and valine compared with the two parents; and the sulfur-containing amino acid content was the same as that of the wild parent and 0.28% higher than that of the cultivated soybean parent. Eickholt et al. (2019) crossed wild soybean PI 366122 with cultivated soybean variety N7103 and selected eight new breeding lines with high protein content. The protein content of the lines was on average 0.4–4.0% higher than that of N7103 (43.7%), and the highest was 47.7%. The availability of varieties and lines with high protein content developed using wild soybeans has facilitated the breeding of new soybean varieties with high quality, high yield, and a high protein content and has promoted the development of the soybean industry to effectively secure the sustainable and stable supply of protein.
Utilization of cultivated soybean in the breeding of varieties with high protein content
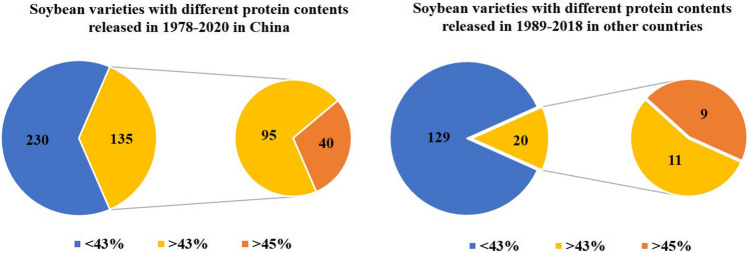
Many efforts have been made to genetically improve the protein content in soybean. The choice of the most suitable parents is crucial in improving the soybean protein content using conventional breeding approaches, and the principle of parental matching depends on the heritability of the soybean protein content and gene action. The protein content of hybrid progenies is closely correlated with the protein content of the parents (Liu et al. 2016). The protein content of hybrid progenies tends to be stable in the F4 generation, and at this time, the selection efficiency is higher. The protein content of hybrid progenies can be directly affected by that of their male parent if they have a female parent with high protein content (Wang et al. 2013). Elevated seed protein level in high yielding varieties should not be at the expense of seed numbers (Rotundo et al. 2009). In the breeding of soybean varieties with high protein content, the correlation between the protein content and other agronomic traits in hybrid progenies needs to be considered. In particular, the improvement of the protein content, 100-grain weight, and yield needs to be synergistic, which forms the basis for the breeding of new soybean varieties with high yield and high protein content (Chen et al. 2002; Wang et al. 2021a, b). Moreover, protein content needs to be prioritized over yield and other agronomic traits, and breeding strategies also need to be further adjusted to meet protein requirements in harvested soybeans (https://germination.ca/is-high-protein-high-yielding-soy-possible/). High protein soybean means the increased protein amount for animal feed, and a single percentage point increase in the protein content will represent millions of tons of additional protein (https://www.syngenta.ca/market-news/new-research-could-increase-soybean-protein-content). The Bayer breeding company have attempted to balance protein, yield, herbicide tolerance and disease resistance for global food security, and the objective of its soybean program project was to develop elite commercial soybean products with an increase in seed protein while maintaining yield and agronomic traits (https://germination.ca/is-high-protein-high-yielding-soy-possible/). In Canadian specific food-grade soybean program, more efforts were made to develop the leading high-protein varieties with higher soybean protein and minimal impacts on yield for the soybean markets (https://germination.ca/is-high-protein-high-yielding-soy-possible/). From 1978 to 2020, a total of 135 soybean varieties had the protein content higher than 43% in China, of which 40 soybean varieties had higher protein content (> 45%) (Fig. 3). From 2003 to 2016, a total of 251 soybean varieties were approved in China, of which 17.27% had a protein content higher than 43% (Liu et al. 2017). From 1989 to 2018, a total of 20 soybean varieties had the protein content higher than 43% in other countries, while only 9 soybean varieties had higher protein content (> 45%) (Fig. 3). A series of diversified soybean germplasms with higher protein content (> 45%) were bred through hybridization in the world, such as X3585-116-3-B (50.1%), Nandou12 (51.79%), NC 104 (50.7%), BARC-9 (52.9%), D90-7256 (50.5%), Hipro (53.9%) and Saedanbaek (48.2%) (Table 2). Many soybean varieties had the significant higher content for seed protein in China, such as 903,525 (47.60%), 903,526 (47.02%), Guichundou108 (47.86%), Zhechun 4 (47.96%), Fudou 234 (47.88%) and Nandou12 (51.79%) (Table 2). For the United States, conventional breeding had been used to generate some high protein soybean varieties, including NC 104 (50.70%), BARC-8 (52.80%), BARC-9 (52.90%), D90-7256 (50.50%) in 1986–2020 (Table 2). Besides, Lee (PI548656) and Harosoy (PI548573) were important breeding materials in United States (https://soybase.org/uniformtrial/index.php?filter=+Essex+&page=lines&test=ALL). AC Proteus (52.10%), AC Proteina (49.80%), X3585-116-3-B (50.10%), HS-151 (46.70%), HS-161 (46.20%) and HS-162 (48.10%) were the high protein varieties that were identified and reported in Canada (Table 2). Ontario was the main soybean-producing province in Canada, and the Ontario soybean variety trials in 2005–2021 were conducted by the Ontario Soybean and Canola Committee (www.Gosoy.ca). In 2021, three soybean varieties Primo (48.4%), BAFFIN (48.5%) and Atiron (48.7%) exhibited higher protein content than most soybean varieties in the Ontario soybean variety trial. Besides, more soybean varieties with high protein content are accessible in the Canadian food-grade soybean variety database (https://buycanadiansoybeans.ca/food-grade soybeans/). Greya (46.30%), Saedanbaek (48.20%), and Hipro (53.90%) were the high protein varieties in Russia, Korea, and Republic of Korea, respectively (Table 2).
Fig. 3.
The contribution of soybean varieties with different protein contents released in the world
Table 2.
List of high-protein (> 45%) soybean germplasms developed by conventional breeding
| Country | Germplasm | Protein content (%) | Pedigree | Reference |
|---|---|---|---|---|
| China | 903,525 | 47.60 | Hefeng 22/PI407788A | Guo et al. (2005) |
| China | 903,526 | 47.02 | Hefeng 22/PI407788A | Guo et al. (2005) |
| China | Xingdou 5 | 45.90 | Kefeng 6/Yudou 28 | Zhu et al. (2008) |
| China | Huaxia 4 | 46.15 | Guizao 1/Brazil 8 | Ouyang et al. (2009) |
| China | Huachun 6 | 45.80 | Guizao 1/Brazil 8 | Li (2010) |
| China | Gongqiudou 5 | 45.46 | Gongxuan 1/Gongqiudou 3 | Fan et al. (2016) |
| China | Meng 1301 | 45.26 | Hedou 3/Fudou 9 | Zhang et al. (2019) |
| China | Guichundou108 | 47.86 | Quandou 937/Guizao 1 | Yang et al. (2020) |
| China | Mudou 15 | 45.08 | Heinong 48/Longpin8807 | Wang et al. (2020c) |
| China | Xudou 25 | 45.49 | Xudou 9/Yushandadou | Zhou et al. (2021) |
| China | Zhonglongdou 106 | 45.96 | Heinong48/Wuxing 4 | Liu et al. (2021) |
| China | Nannong 49 | 46.84 | Xiangchundou 26/Taiwan 292 | https://www.chinaseed114.com/ |
| China | Huaxia 14 | 45.40 | Guixiadou 2/Nandou 12 | https://www.chinaseed114.com/ |
| China | Xudou 23 | 45.22 | Xudou 9/Zheng 90,007 | https://www.chinaseed114.com/ |
| China | Huachun 11 | 46.73 | Huachun 3/Fudou 310 | https://www.chinaseed114.com/ |
| China | Shengdou 32 | 45.69 | Xudou 18/Hedou 12 | https://www.chinaseed114.com/ |
| China | Nandou 12 | 51.79 | Chengdou 4/Gongdou 6 | https://www.chinaseed114.com/ |
| China | Quqiu 6 | 46.00 | Zheqiudou 3/Maoqiupeng | https://www.chinaseed114.com/ |
| China | Shengdou 40 | 45.61 | Huachun 3/Fudou 234 | https://www.chinaseed114.com/ |
| China | Wandou 39 | 45.81 | Hedou 3/Jidou 12 | https://www.chinaseed114.com/ |
| China | Zhechun 4 | 47.96 | Wuxing-5/Zhechun3 | https://www.chinaseed114.com/ |
| China | Fudou 234 | 47.88 | Pudou8008/Huangshadou | https://www.chinaseed114.com/ |
| America | NC 104 | 50.70 | D55-4110/N56–4071 | Carter et al. (1986) |
| America | NC 105 | 48.70 | D55-4110/N56–4071 | Carter et al. (1986) |
| America | NC 106 | 50.40 | D55-4110/N56–4071 | Carter et al. (1986) |
| America | BARC-7 | 49.10 | CX797-21/D80-6931 | Leffel (1992) |
| America | BARC-8 | 52.80 | CX797-21/NC-2–62 | Leffel (1992) |
| America | BARC-9 | 52.90 | CX797-21/NC-2–62 | Leffel (1992) |
| America | D90-7256 | 50.50 | Forrest/D76-8070 | Hartwig (1996) |
| America | Prolina | 46.10 | Complex hybridization | Burton and Wilson (1999) |
| America | DMK93-9048 | 46.20 | D86-3429/Braxton | Kenty et al. (2001) |
| America | R95-1705 | 46.70 | Hutcheson/BARC-7 | Chen et al. (2008) |
| America | N6202 | 45.70 | N6201/N95-7390 | Carter et al. (2010) |
| America | R05-1415 | 46.90 | MFS-591/V96-4486 | Chen et al. (2011) |
| America | R05-1772 | 46.10 | R95-1705/V96-4181 | Chen et al. (2011) |
| America | UA 5814HP | 45.40 | R95-1705/S00-9980–22 | Chen et al. (2017) |
| America | X3144-48–1-B | 46.90 | AC Proteus/Maple Glen | Samanfar et al. (2019) |
| America | TN15-4009 | 46.00 | TN09-016/S05-11,482 | Pantalone and Wyman (2020) |
| Canada | AC Proteus | 52.10 | Complex hybridization | Voldeng et al. (1996a) |
| Canada | Maple Glen | 45.70 | Complex hybridization | Voldeng et al. (1996b) |
| Canada | AC Proteina | 49.80 | Complex hybridization | Cober and Voldeng (2000) |
| Canada | X3585-116–3-B | 50.10 | Maple Glen/AC Proteus | Cober and Voldeng (2000) |
| Canada | HS-151 | 46.70 | Complex hybridization | Yu et al. (2016a) |
| Canada | HS-161 | 46.20 | Complex hybridization | Yu et al. (2016b) |
| Canada | HS-162 | 48.10 | Complex hybridization | Yu et al. (2016c) |
| Canada | HS-182 | 45.70 | Complex hybridization | Yu et al. (2019a) |
| Canada | AAC Wigle | 45.80 | RCAT0606SCN/SG01-0217EMMM-1 | Yu et al. (2019b) |
| Russia | Greya | 46.30 | Vilana/Valenta | Zelentsov et al. (2020) |
| Korea | Saedanbaek | 48.20 | MD87L/SS92414 | Kim et al. (2014) |
| Republic of Korea | Hipro | 53.90 | Saedanbaek/Daepung | Kim et al. (2021c) |
Mutagenesis is an effective means to increase the protein content of soybean. After the ethyl methanesulfonate (EMS) treatment of Jidou 1 and Jidou 6 seeds, three mutant lines with high protein content were selected in the M4 generation (Yu et al. 1995). A batch of soybean varieties with a high protein content such as Heinong 41 (protein content 45.23%) and 90-3527 (protein content 47.53%) was successively bred using γ-ray and thermal neutron irradiation as well as chemical mutagens such as EMS and sodium azide (NaN3) (Wang et al. 2000). Wei et al. (2019) reported that continuous selection of plants with a high protein content in the M2 generation resulted in an overall decrease in the protein content. In contrast, continuous selection of plants with a low protein content in the M2 generation resulted in distinct genetic gain. Gene silencing (Herman et al. 2003) and gene editing technologies (Sugano et al. 2020) have been successfully used to reduce soybean allergenic proteins. Soybean varieties with a high protein content and high quality such as Qihuang 34 and Kexin 3 were bred through the combination of mutation and hybridization techniques. Soybean varieties and mutant lines were also developed by means of 60Co γ-ray and thermal neutron irradiation and treatment using the chemical mutagens EMS and sodium azide, which include vegetable soybean variety Huaihadou 1 (protein content 44.93%), the three high-protein mutant lines 903,525 (47.60%), 903,526 (47.02%), and 903,527 (47.53%) that mature early and have high resistance to viral diseases and gray spot, and two high-protein stable mutants, 923,725 (45.38%) and 923,738 (45.24%) (Guo et al. 2005). Wei et al. (2019) screened nine stable germplasms (m1–m9) from the M7 generation of EMS-treated Zhongpin 661, and they had an average protein content of 48.17%, which is 6.98% higher than that of the wild type (41.19%). Those high-protein resources have greatly contributed to the sustainable production of protein.
With regard to the productive efficiency, protein yield per unit area should be taken seriously in practical breeding process. Protein yield is calculated by multiplying yield and protein content. The high positive correlation relationship was found between protein yield and yield, however, the low correlation relationship was found between protein yield and protein content (Kurasch et al. 2017b). Besides, the dense-tolerant trait is a good agronomic trait for the improvement of yield and further protein yield in soybean. Dense-tolerant planting genotype can be introduced to achieve the more self-sufficient in protein yield. Europe depended heavily on soybean imports to meet the growing demand for plant protein, especially for animal feed (https://insights.figlobal.com/plant-based/european-soy-quest-protein-self-sufficiency). There is an urgent need to solve the soybean protein shortage and improve the protein self-sufficiency in Europe. Food-grade soybean is generally used for production of soy milk, tofu and other soy-based food (Jegadeesan and Yu 2020). Kurasch et al. (2017b) revealed that high-yield soybean variety appears to be the powerful strategy for achieving the high protein yield for animal feed, while high protein soybean variety appears to be beneficial for food-grade soybean production in Europe. Coupling high yields with high protein is the Holy Grail in practical breeding process (https://germination.ca/is-high-protein-high-yielding-soy-possible/). Nevertheless, how to balance it well is of great importance for soybean production.
Breeding soybean varieties with high-quality protein for forage
Soybean is an important source of protein for forage, which is not only rich in nutrients but also contains allergenic proteins and protease inhibitors that prevent the animals from digesting and absorbing proteins. Screening germplasm resources that lack allergenic proteins and utilizing conventional hybridization integrated with modern molecular technology are important strategies to bread high-quality soybean varieties for forage. About 80% of Japanese soybeans lack the allergenic protein Gly m Bd 28 K (Ogawa et al. 2000). Guan et al. (2004) found one accession lacking the β subunit and 77 accessions lacking the allergenic protein 28 K among 175 accessions of Chinese soybean germplasm resources. Soybean Kunitz trypsin inhibitor is a common anti-nutritional factor. Jiang et al. (2020) used conventional hybridization aided by molecular marker-assisted selection employing non-denaturing polyacrylamide gel electrophoresis and selected a new soybean line HZ8009 without the Kunitz trypsin inhibitor. Phytic acid (phytate) benefits the normal growth and development of soybean plants but is an anti-nutritional factor and cannot be digested and utilized by humans and non-ruminants. Myo-inositol phosphate synthase (MIPS) is involved in the synthesis of phytic acid. Hize et al. (2002) and Yuan et al. (2007) silenced the gene using mutagenesis or a molecular biological approach and developed LR33 and Gm-lpa-TW-1 that have a greatly reduced phytic acid content in the seeds; the seed phytic acid content of Gm-lpa-TW-1 decreased by more than 50% compared with that of the wild type parent. A molecular marker for the LPA trait of Gm-lpa-TW-1 was developed (Yuan et al. 2010). These studies and the creation of genetic materials provide references for the improvement of soybean protein quality and the breeding of soybean varieties for forage utilizing modern biological technologies.
Breeding multifunctional soybean varieties with different seed storage protein components
Since 7S and 11S globulins had a great impact on the nutrition and quality of soybean products, studies had been carried out to develop soybean varieties with different subunit compositions. An inverse correlation was found between the 7S and 11S globulin contents, and the main allergen was included in the subunit of 7S globulin (Ogawa et al. 1989, 1995). At the early stage, soybean breeding was mainly focused on low content of 7S globulin and high content of 11S globulin in soybean seed. Yumeminori was regarded as a low level of 7S globulin (null α´, α subunits), high level of 11S globulin, high content of sulfur amino acids and low allergenicity soybean variety, which was developed by using the γ-ray irradiation method (Takahashi et al. 2004). Another new soybean variety Nagomimaru with α and α' subunits deficiency was developed from the progeny of cross between Kariko 0542 and Tachinagaha, and Nagomimaru was suitable for making soy milk in Japan (Hajika et al. 2009). Soybean seed with high protein content (> 45%), high 11S/7S ratio and suitable subunit composition of 11S is desirable for tofu production and texture (Jegadeesan and Yu 2020). Moreover, the quality and stability of soymilk is also affected by the subunit compositions (Nik et al. 2009). Poysa et al. (2006) showed that α´ subunit of 7S globulin deficiency, A4 subunit of 11S globulin deficiency and A3 subunit of 11S globulin played major role in contributing to tofu quality. IWATE-3 introduced from Japan, which lacks the α′ subunit of 7S glycinin and all the 11S glycinin, was used for elite soybean germplasm in Canadian soybean breeding (Zarkadas et al. 2007; Yu et al. 2016b, c). Additionally, a wild soybean lacking all three α, α', and β subunits of 7S globulin was a valuable genetic resource for soybean breeding (Hajika et al. 1996, 1998). Two high protein soybean varieties HS-161 lacking the α' subunit of 7S globulin and A3 subunit of 11S globulin and HS-162 lacking the α' subunit of 7S globulin and A4 subunit of 11S globulin are suitable for soybean food production (Yu et al. 2016b, c). Particularly, HS-162 had excellent processing quality for tofu and soy milk, and HS-162 made firmer tofu than the check tofu-type soybean variety Harovinton (44.90% of seed-protein content) (Buzzell et al. 1991; Yu et al. 2016c). It was reported that 7S globulin can improve lipid metabolism (Mochizuki et al. 2009). The soybean variety Nanahomare contains approximately 1.8-fold 7S globulin than ordinary varieties, but lacks all the 11S globulin (Yagasaki et al. 2010). Nishimura et al. (2016) revealed that 7S globulin-rich soybean variety Nanahomare played a crucial role in decreasing the serum triglyceride level. Nanahomare might be used as a potential soybean resource for 7S globulin-rich soybean breeding and provide the required amounts of 7S globulin for prevention of lifestyle-related diseases such as high serum triglyceride. These soybean varieties with modified subunits can be good genetic resources for improvement of nutrient content and functional components.
Future expectations
Clarification of the genetic basis for the formation of protein traits
Soybean protein is a complex quantitative trait. The soybean protein content is closely related to the climate. High-rainfall and high-temperature regions such as Tonghua, Jilin Province (Northeast China), Zhengzhou, Henan Province (Huanghuaihai region), and southern China have mostly favorable conditions for the production of a high protein content. The soybean growing regions were established by the Department of Plant Industry Management of the Ministry of Agriculture and Rural Affairs in 2003, and the set standard for high-protein soybean is 43% for northern regions and 45% for Huanghuaihai and the southern regions. The soybean protein content is negatively correlated with other traits such as yield and the oil content, which cause difficulties in the breeding of high-protein soybean. In order to break the linkage between the protein content and other traits including yield and the oil content, it is necessary to identify the genes related to the formation of soybean protein on a genome-wide scale. In particular, it is necessary to discover the genes that cause the stable expression of a high protein content. For example, soybean variety Jinghe 1 has a high protein content (higher than 48%) in Heihe city, Heilongjiang Province, China. It has extraordinary environmental stability and a high protein content in high latitude regions. Therefore, it is an important resource for discovering genes related to a high protein content. The genetic loci associated with the soybean protein content, key genes and regulatory mechanisms involved in the synergistic regulation of the soybean protein content and yield, as well as the genetic basis for the formation of soybean protein and the key molecular modules or regulatory networks for design breeding can be systematically and precisely elucidated based on the available germplasm resources, large amount of data, as well as technical means in population genetics, genomics, systems biology, and bioinformatics.
Systematic identification of genetic variation in genes related to the protein content
The essence of breeding is to utilize the association between genotype and phenotype to screen useful genes or allelic variations to obtain varieties with desired comprehensive traits. The soybean protein content is mainly regulated by multiple genes rather than a single gene. Pyramiding multiple desired genes or allelic loci is the strategy used in soybean breeding for a high protein content. Therefore, examining the genetic variation of genes related to the soybean protein content in diverse germplasm resources and their correlation with phenotypes and the screening of germplasm with excellent allelic variation are the keys to improving the efficiency of breeding soybean varieties with high protein content. The availability of cultivated and wild soybean reference genomes as well as functional chips such as SoySNP618K (Li et al. 2022) and ZDX1 (Sun et al. 2022) facilitate the re-sequencing or microarray-based sequence analysis of germplasm resources. Large-scale processing of massive amounts of data obtained from sequencing or microarray analysis improves the efficiency and accuracy of detecting genetic variation, which can be combined with the analysis of phenotypic data of the soybean protein content obtained from various growing conditions to identify useful alleles and establish linear or non-linear models predicting the relationship between the genetic variation and protein content, so as to understand the regularity of the interaction between genes related to the protein content and the environment and to discover desirable alleles that adapt to specific environments.
Molecular design boosts high-protein soybean breeding
The breeding of breakthrough varieties often depends on the utilization of rare and desired resources (Wehrmann et al. 1987). The availability of the soybean reference genome (Schmutz et al. 2010), wild soybean genome (Kim et al. 2010), pan-genome (Li et al. 2014), and graph-based pan-genome (Liu and Tian 2020) is conducive to the discovery of genes related to the protein content of soybean. The utilization of genes related to the protein content of wild soybean can improve the protein content of cultivated soybean. The discovery of QTLs and genes related to the protein content of cultivated soybean can facilitate the breeding of soybean varieties with high protein content by means of transformation and gene-editing technologies (Wu et al. 2021; Valliyodan et al. 2016). With the establishment and improvement of massive amounts of data, the comprehensive use of modern breeding technologies on the basis of bioinformatics and CRISPR/Cas9 has become an important method for plant improvement and germplasm creation (Gao et al. 2021). Li et al. (2019b) designed sgRNAs for nine different main storage protein genes and used CRISPR/Cas9 technology to edit the soybean seed storage protein gene family. The mutations in three storage protein genes were detected in soybean hairy roots, and the mutation frequency ranged between 3.8 and 43.7%. These studies laid a basis for the use of molecular design to boost the breeding of new soybean varieties with high protein content.
It is expected that genes related to the soybean protein content and their genetic variation on a whole-genome scale will be systematically elucidated. The key molecular modules and regulatory networks of genes related to the protein content and the genetic mechanisms of their interactions with yield and the environment will be clarified. In addition, new technologies such as gene editing, gene transformation, synthetic biology, and artificial intelligence are maturing, and under the background of the advancement of above-mentioned technologies, molecular design breeding or intelligent breeding will be applied in the targeted breeding of soybeans with high protein content, high quality, and high yield (Fig. 4), which will benefit the sustainable production of protein and secure the supply of food and dietary nutrients on a global scale.
Fig. 4.
Strategy for breeding of high-protein soybean
Acknowledgements
This work was supported by the Agricultural Science and Technology Innovation Program (ASTIP) of Chinese Academy of Agricultural Sciences, National Natural Science Foundation of China (31960408), Double Thousand Plan of Jiangxi Province (jxsq 2019201073).
Author contributions statement
LQ provided the key ideas of the review; BG, LS, SJ, HR, RS, ZW, HH, JW, XW, XL, DX jointly wrote the manuscript; BG, LS, SJ, XW prepared the tables and figures; LQ, BG, LS, SJ, CG, DX, WL revised and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Agricultural Science and Technology Innovation Program (ASTIP) of Chinese Academy of Agricultural Sciences, National Natural Science Foundation of China (31960408), and Double Thousand Plan of Jiangxi Province (jxsq 2019201073).
Declaration
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bingfu Guo, Liping Sun and Siqi Jiang have contributed equally to this work.
Change history
11/24/2022
A Correction to this paper has been published: 10.1007/s00122-022-04241-6
References
- Abe J, Xu DH, Suzuki Y, Kanazawa A, Shimamoto Y. Soybean germplasm pools in Asia revealed by nuclear SSRs. Theor Appl Genet. 2003;106:445–453. doi: 10.1007/s00122-002-1073-3. [DOI] [PubMed] [Google Scholar]
- Achouri A, Boye JI, Yaylayan VA, Yeboah FK. Functional properties of glycated soy 11S glycinin. J Food Sci. 2005;70:269–274. doi: 10.1111/j.1365-2621.2005.tb07172.x. [DOI] [Google Scholar]
- Adachi M, Kanamori J, Masuda T, Yagasaki K, Kitamura K, Mikami B, Utsumi S. Crystal structure of soybean 11S globulin: glycinin A3B4 homohexamer. Proc Natl Acad Sci USA. 2003;100:7395–7400. doi: 10.1073/pnas.0832158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaswad AA, Song B, Oehrle NW, Wiebold WJ, Mawhinney TP, Krishnan HB. Development of soybean experimental lines with enhanced protein and sulfur amino acid content. Plant Sci. 2021;308:110912. doi: 10.1016/j.plantsci.2021.110912. [DOI] [PubMed] [Google Scholar]
- Amigo-Benavent M, Clemente A, Ferranti P, Caira S, del Castillo MD. Digestibility and immunoreactivity of soybean β-conglycinin and its deglycosylated form. Food Chem. 2011;129:1598–1605. doi: 10.1016/j.foodchem.2011.06.015. [DOI] [Google Scholar]
- Anderson JW, Johnstone BM, Cook-Newell ME. Meta-analysis of the effects of soy protein intake on serum lipids. N Engl J Med. 1995;333:276–282. doi: 10.1056/NEJM199508033330502. [DOI] [PubMed] [Google Scholar]
- Andres A, Cleves MA, Bellando JB, Pivik RT, Casey PH, Badger TM. Developmental status of 1-year-old infants fed breast milk, cow’s milk formula, or soy formula. Pediatr. 2012;129:1134–1140. doi: 10.1542/peds.2011-3121. [DOI] [PubMed] [Google Scholar]
- Asif M, Acharya M. Phytochemicals and nutritional health benefits of soy plant. Int J Nutr Pharmacol Neur Dis. 2013;3:64–70. doi: 10.4103/2231-0738.106998. [DOI] [Google Scholar]
- Banaszkiewicz T. Nutritional value of soybean meal. Soybean Nutr. 2011;12:1–20. [Google Scholar]
- Bandillo N, Jarquin D, Song Q, Nelson R, Cregan P, Specht J, Lorenz A. A population structure and genome-wide association analysis on the USDA soybean germplasm collection. Plant Genome. 2015;8:4–24. doi: 10.3835/plantgenome2015.04.0024. [DOI] [PubMed] [Google Scholar]
- Beche E, Gillman JD, Song Q, Nelson R, Beissinger T, Decker J, Shannon G, Scaboo AM. Genomic prediction using training population design in interspecific soybean populations. Mol Breeding. 2021;41:1–15. doi: 10.1007/s11032-021-01203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilinson V, Chen Z, Shoemaker RC, Fischer RL, Goldberg RB, Nielsen NC. Genomic organization of glycinin genes in soybean. Theor Appl Genet. 2002;104:1132–1140. doi: 10.1007/s00122-002-0884-6. [DOI] [PubMed] [Google Scholar]
- Bellaloui N, Reddy KN, Bruns HA, Gillen AM, Mengistu A, Zobiole LH, Fisher DK, Abbas HK, Zablotowicz RM, Kremer RJ. Soybean seed composition and quality: Interactions of environment, genotype, and management practices. In: Gillen A, editor. Soybeans: cultivation, uses and nutrition. 1. New York: Nova Science Publishers, Inc.; 2011. pp. 1–42. [Google Scholar]
- Bernard RL, Juvik GA, Hartwig EE, Edwards CJ. Origins and pedigrees of public soybean varieties in the United States and Canada. Techl Bull U S Depart Agric. 1988;1976:61. [Google Scholar]
- Bolon YT, Joseph B, Cannon SB, Graham MA, Diers BW, Farmer AD, May GD, Muehlbauer GJ, Specht JE, Tu ZJ, Weeks N, Xu WW, Shoemaker RC, Vance CP. Complementary genetic and genomic approaches help characterize the linkage group I seed protein QTL in soybean. BMC Plant Biol. 2010;10:1–24. doi: 10.1186/1471-2229-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JW, WILSON R, Registration of Prolina Soybean. Crop Sci. 1999;39:294–295. doi: 10.2135/cropsci1999.0011183X003900010066x. [DOI] [Google Scholar]
- Buzzell RI, Anderson TR, Hamill AS, Welacky TW. Harovinton soybean. Can J Plant Sci. 1991;71:525–526. doi: 10.4141/cjps91-075. [DOI] [Google Scholar]
- Cabanos C, Matsuoka Y, Maruyama N. Soybean proteins/peptides: a review on their importance, biosynthesis, vacuolar sorting, and accumulation in seeds. Peptides. 2021;143:170598. doi: 10.1016/j.peptides.2021.170598. [DOI] [PubMed] [Google Scholar]
- Carter TE, Jr, Burton JW, Brim CA. Registration of NC 101 to NC 112 soybean germplasm lines contrasting in percent seed protein. Crop Sci. 1986;26:841–842. doi: 10.2135/cropsci1986.0011183X002600040057x. [DOI] [Google Scholar]
- Carter TE, Jr, Rzewnicki PE, Burton JW, Villagarcia MR, Bowman DT, Taliercio E, Kwanyuen P. Registration of N6202 soybean germplasm with high protein, favorable yield potential, large seed, and diverse pedigree. J Plant Regist. 2010;4:73–79. doi: 10.3198/jpr2009.08.0462.crg. [DOI] [Google Scholar]
- Chang RZ (1989) Utilization of Chinese soybean genetic resources abroad. World Agric 20–21
- Chapman A, Pantalone VR, Ustun A, Allen FL, Landau-Ellis D, Trigiano RN, Gresshoff PM. Quantitative trait loci for agronomic and seed quality traits in an F2 and F4:6 soybean population. Euphytica. 2003;129:387–393. doi: 10.1023/A:1022282726117. [DOI] [Google Scholar]
- Chen LH, Li J, Liu LJ, Zu W, Ma XF. The relationship between protein accumulation regulation and yield formation in soybean. J Northeast Agric Univ. 2002;33:116–124. [Google Scholar]
- Chen P, Sneller CH, Ishibashi T, Cornelious B. Registration of high-protein soybean germplasm line R95–1705. J Plant Regist. 2008;2:58–59. doi: 10.3198/jpr2007.03.0135crg. [DOI] [Google Scholar]
- Chen P, Ishibashi T, Dombek DG, Rupe JC. Registration of R05–1415 and R05–1772 high-protein soybean germplasm lines. J Plant Regist. 2011;5:410–413. doi: 10.3198/jpr2010.05.0304crg. [DOI] [Google Scholar]
- Chen KI, Erh MH, Su NW, Liu WH, Chou CC, Cheng KC. Soyfoods and soybean products: from traditional use to modern applications. Appl Microbiol Biotechnol. 2012;96:9–22. doi: 10.1007/s00253-012-4330-7. [DOI] [PubMed] [Google Scholar]
- Chen P, Florez-Palacios L, Orazaly M, Manjarrez-Sandoval P, Wu C, Rupe JC, Dombek DG, Kirkpatrick T, Robbins RT. Registration of ‘UA 5814HP’Soybean with high yield and high seed-protein content. J Plant Regist. 2017;11:116–120. doi: 10.3198/jpr2016.09.0046crc. [DOI] [Google Scholar]
- Cheng L, Yuan H, Ren R, Zhao S, Han Y, Zhou Q, Ke D, Wang Y, Wang L. Genome-wide identification, classification, and expression analysis of amino acid transporter gene family in Glycine max. Front Plant Sci. 2016;7:515. doi: 10.3389/fpls.2016.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kwon SJ, Roy SK, Kim HS, Lee CW, Woo SH. A systematic proteome study of seed storage proteins from two soybean genotypes. Korean J Crop Sci. 2014;59:359–363. doi: 10.7740/kjcs.2014.59.3.359. [DOI] [Google Scholar]
- Chung J, Babka HL, Graef GL, Staswick PE, Lee DJ, Gregan PB, Shoemaker RC, Specht JE. The seed protein, oil, and yield QTL on soybean linkage group I. Crop Sci. 2003;43:1053–1067. doi: 10.2135/cropsci2003.1053. [DOI] [Google Scholar]
- Cober ER, Voldeng HD. Developing high-protein, high-yield soybean populations and lines. Crop Sci. 2000;40:39–42. doi: 10.2135/cropsci2000.40139x. [DOI] [Google Scholar]
- Cui YS, Li GF, Zhang J, Xie LS, Zheng LJ, Ren WJ, Pan RC. Pedigree analysis of high protein soybean varieties. Chin Agric Sci Bull. 1999;15:43–45. [Google Scholar]
- Cure JD, Patterson RP, Raper JCD, Jackson WA. Assimilate distribution in soybeans as affected by photoperiod during seed development. Crop Sci. 1982;22:1245–1250. doi: 10.2135/cropsci1982.0011183X002200060039x. [DOI] [Google Scholar]
- Dei HK. Soybean as a feed ingredient for livestock and poultry. London: IntechOpen; 2011. [Google Scholar]
- Dhungana SK, Kulkarni KP, Kim M, Ha BK, Kang S, Song JT, Shin DH, Lee JD. Environmental stability and correlation of soybean seed starch with protein and oil contents. Plant Breed Biotech. 2017;5:293–303. doi: 10.9787/PBB.2017.5.4.293. [DOI] [Google Scholar]
- Diers BW, Keim P, Fehr WR, Shoemaker RC. RFLP analysis of soybean seed protein and oil content. Theor Appl Genet. 1992;83:608–612. doi: 10.1007/BF00226905. [DOI] [PubMed] [Google Scholar]
- Dinkins RD, Reddy MSS, Meurer CA, Yan B, Trick H, Thibaud-Nissen FO, Finer JJ, Parrott WA, Collins GB. Increased sulfur amino acids in soybean plants overexpressing the maize 15 kDa zein protein. In Vitro Cell Dev Plant. 2001;37:742–747. doi: 10.1007/s11627-001-0123-x. [DOI] [Google Scholar]
- Dong YS. Advances of research on wild soybean in China. J Jilin Agric Univ. 2008;30:394–400. [Google Scholar]
- Dornbos DL, Mullen RE. Soybean seed protein and oil contents and fatty acid composition adjustments by drought and temperature. J Am Oil Chem Soc. 1992;69:228–231. doi: 10.1007/BF02635891. [DOI] [Google Scholar]
- Du LL, Yu BJ. Analysis of salt tolerance, agronomic traits and seed quality of Glycine max, saltborn Glycine soja and their hybrids. Chin J Oil Crop Sci. 2010;32:77–82. [Google Scholar]
- Duhnen A, Gras A, Teyssèdre S, Romestant M, Claustres B, Daydé J, Mangin B. Genomic selection for yield and seed protein content in soybean: a study of breeding program data and assessment of prediction accuracy. Crop Sci. 2017;57:1325–1337. doi: 10.2135/cropsci2016.06.0496. [DOI] [Google Scholar]
- Eickholt D, Carter TE, Taliercio E, Dickey D, Dean LO, Delheimer J, Li Z. Registration of USDA-max 9 soja Core Set-1: recovering 99% of wild soybean genome from PI 366122 in 17 agronomic interspecific germplasm lines. J Plant Regist. 2019;13:217–236. doi: 10.3198/jpr2017.09.0059crg. [DOI] [Google Scholar]
- El-Shemy HA, Khalafalla MM, Fujita K, Ishimoto M. Improvement of protein quality in transgenic soybean plants. Biol Plant. 2007;51:277–284. doi: 10.1007/s10535-007-0055-3. [DOI] [Google Scholar]
- World Health Organization/Food and Agriculture Organization/United Nations University (1985) Energy and protein requirements Report of a Joint FAO/WHO/UNU Expert Consultation. In: WHO technical report series, no. 724. WHO, Geneva [PubMed]
- Erdman JW. Soy protein and cardiovascular disease a statement for healthcare professionals from the nutrition committee of the AHA. Circulation. 2000;102:2555–2559. doi: 10.1161/01.CIR.102.20.2555. [DOI] [PubMed] [Google Scholar]
- Fabre F, Planchon C. Nitrogen nutrition, yield and protein content in soybean. Plant Sci. 2000;152:51–58. doi: 10.1016/S0168-9452(99)00221-6. [DOI] [Google Scholar]
- Fallen BD, Hatcher CN, Allen FL, Kopsell DA, Saxton AM, Chen P, Kantartzi SK, Cregan PB, Hyten DL, Pantalone VR. Soybean seed amino acid content QTL detected using the universal soy linkage panel 1.0 with 1,536 SNPs. J Plant Genome Sci. 2013;1:68–79. [Google Scholar]
- Fan S, Li L, Xiang S, Yang H. Breeding and cultivation technology of a new summer soybean variety Gongqiudou 5 with high-yield and high-protein. Soybean Sci Technol. 2016;4:3. [Google Scholar]
- Fang XQ, Qiu LJ (2005) Progress on characterization and genetic improvement of allergic proteins in soybean. Natl Sym Crop Biotech Mutage Abs :37–38
- Fernandes ECM, Soliman A, Donatelli M, Tubiello FN. Climate change and agriculture in Latin America, 2020–2050. Washington: World Bank; 2012. [Google Scholar]
- Fischer RL, Goldberg RB. Structure and flanking regions of soybean seed protein genes. Cell. 1982;29:651–660. doi: 10.1016/0092-8674(82)90181-7. [DOI] [PubMed] [Google Scholar]
- Fliege CE, Ward RA, Vogel P, Nguyen H, Quach T, Guo M, Viana JPG, Santos LB, Specht JE, Clemente TE, Matthew H, Diers BW. Fine mapping and cloning of the major seed protein quantitative trait loci on soybean chromosome 20. Plant J. 2022;110:114–128. doi: 10.1111/tpj.15658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Cabanos C, Toyota K, Kobayashi Y, Maruyama N. Differential expression and elution behavior of basic 7S globulin among cultivars under hot water treatment of soybean seeds. J Biosci Bioeng. 2014;117:742–748. doi: 10.1016/j.jbiosc.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Gao ZK, Jiang J, Han ZQ, Huang ZP, Xiong FQ, Tang XM, Wu HN, Zhong RC, Liu J, Tang RH, He LQ. CRISPR/Cas9 system and its research progress in grain and oil crop genetic improvement. Chin Agric Sci Bull. 2021;37:26–34. [Google Scholar]
- García MC, Marina ML, Laborda F, Torre M. Chemical characterization of commercial soybean products. Food Chem. 1998;62:325–331. doi: 10.1016/S0308-8146(97)00231-8. [DOI] [Google Scholar]
- George AA, de Lumen BO. A novel methionine-rich protein in soybean seed: Identification, amino acid composition, and n-terminal sequence. J Agric Food Chem. 1991;39:224–227. doi: 10.1021/jf00001a046. [DOI] [Google Scholar]
- Gibson LR, Mullen RE. Soybean seed composition under high day and night growth temperatures. J Am Oil Chem Soc. 1996;73:733–737. doi: 10.1007/BF02517949. [DOI] [Google Scholar]
- Goettel W, Zhang H, Li Y, Qiao Z, Jiang H, Hou D, Song Q, Pantalone VR, Song BH, Yu D, An YQC. POWR1 is a domestication gene pleiotropically regulating seed quality and yield in soybean. Nat Commun. 2022;13:1–11. doi: 10.1038/s41467-022-30314-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldflus F, Ceccantini M, Santos W. Amino acid content of soybean samples collected in different Brazilian states: Harvest 2003/2004. Braz J Poultry Sci. 2006;8:105–111. doi: 10.1590/S1516-635X2006000200006. [DOI] [Google Scholar]
- Grieshop CM, Fahey GC. Comparison of quality characteristics of soybeans from Brazil, China, and the United States. J Agric Food Chem. 2001;49:2669–2673. doi: 10.1021/jf0014009. [DOI] [PubMed] [Google Scholar]
- Gu QP, Zhou J, Du JD. Development and enlightenment of soybean industry in USA, Brazil and Argentina. Southern Rural. 2014;000:36–40. [Google Scholar]
- Guan RX, Chang RZ, Qiu LJ, Liu ZX, Guo ST. Analysis of protein subunit7S/11S constitution and allergen lacking of soybean [Glycine max (L.) Merrill] cultivars. Acta Agron Sin. 2004;30:1076–1079. [Google Scholar]
- Guo YH, Wang PY, Xu DC, Meng LF, Li FM, Dong DJ, Lin WG, Wu JJ, Zhang L, Zhong P, Han L. Study on mutagenesis and improvement of soy protein content. Soy Bull. 2005;6:12–13. [Google Scholar]
- Guo JJ, Chang RZ, Zhang JX, Zhang JS, Guan RX, Qiu LJ. Genetic contribution of Japanese soybean germplasm Tokachi nagaha to Chinese soybean cultivar. Soybean Sci. 2007;26:807–812. [Google Scholar]
- Guo JW, Shi XL, Liu Q, Zhao QS, Di R, Liu BQ, Yan L, Wang FM, Zhang MC, Zhao BH, Yang CY. Transcriptome analysis of protein synthesis related genes in soybean. N China Agric J. 2019;34:61–73. [Google Scholar]
- Gupta SK, Manjaya JG. Advances in improvement of soybean seed composition traits using genetic, genomic and biotechnological approaches. Euphytica. 2022;218:1–33. doi: 10.1007/s10681-022-03046-4. [DOI] [Google Scholar]
- Hajika M, Takahashi M, Sakai S, Igita K. A new genotype of 7 S globulin (β-conglycinin) detected in wild soybean (Glycine soja Sieb. et Zucc.) Breed Sci. 1996;46:385–386. [Google Scholar]
- Hajika M, Sakai S, Matsunaga R. Dominant inheritance of a trait lacking β-conglycinin detected in a wild soybean line. Breed Sci. 1998;48:383–386. [Google Scholar]
- Hajika M, Takahashi K, Yamada T, Komaki K, Takada Y, Shimada H, Sakai T, Shimada S, Adachi T, Tabuchi K, Kikuchi A, Yumoto S, Nakamura S, Ito M. Development of a new soybean cultivar for soymilk," Nagomimaru". Bull Natl Inst Crop Sci. 2009;10:1–20. [Google Scholar]
- Hao QN, Wang AA, Long ZF, Chen HF, Shan ZH, Chen SL, Deng JB, Zhou XA. Effects of da- 6 on the characteristics, yield and quality of soybean varieties in south China. Soybean Sci. 2021;40:799–804. [Google Scholar]
- Hara-Nishimura I, Shimada T, Hatano K, Takeuchi Y, Nishimura M (1998) Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10:825–836 [DOI] [PMC free article] [PubMed]
- Harada K, Kaga A. Recent genetic research on Japanese soybeans in response to the escalation of food use worldwide. Euphytica. 2019;215:1–27. doi: 10.1007/s10681-019-2396-3. [DOI] [Google Scholar]
- Hartwig EE. Registration of soybean germplasm line D90–7256 having high seed protein and low oligosaccharides. Crop Sci. 1996;36:212. [Google Scholar]
- Hashiguchi T, Hashiguchi M, Tanaka H, Gondo T, Akashi R. Comparative analysis of seed proteome of Glycine max and Glycine soja. Crop Sci. 2020;60:1530–1540. doi: 10.1002/csc2.20131. [DOI] [Google Scholar]
- Hayashi M, Miyahara A, Sato S, Kato T, Yoshikawa M, Taketa M, Hayashi M, Pedrosa A, Onda R, Imaizumi-Anraku H. Construction of a genetic linkage map of the model legume lotus japonicus using an intraspecific F2 population. DNA Res. 2001;8:301–310. doi: 10.1093/dnares/8.6.301. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Kitamura K, Harada K. Genetic mapping of Cgdef gene controlling accumulation of 7S globulin (beta-conglycinin) subunits in soybean seeds. J Hered. 2009;100:802–806. doi: 10.1093/jhered/esp046. [DOI] [PubMed] [Google Scholar]
- He F, Chen J. Consumption of soybean, soy foods, soy isoflavones and breast cancer incidence: differences between Chinese women and women in Western countries and possible mechanisms. Food Sci Hum Well. 2013;2:146–161. doi: 10.1016/j.fshw.2013.08.002. [DOI] [Google Scholar]
- Henkel J. Soy: health claims for soy protein, questions about other components. FDA Consumer Mag. 2000;34:13–18. [PubMed] [Google Scholar]
- Herman EM, Helm RM, Jung R, Kinney AJ. Genetic modification removes an immunodominant allergen from soybean. Plant Physiol. 2003;132:36–43. doi: 10.1104/pp.103.021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H. Basic 7S globulin in plants. J Proteomics. 2021;240:104209. doi: 10.1016/j.jprot.2021.104209. [DOI] [PubMed] [Google Scholar]
- Hize WD, Carlson TJ, Kerr PS, Sebastian SA. Biochemical and molecular characterization of a mutation that confers a decreased raffinosaccharide and phytic acid phenotype on soybean seeds. Plant Physiol. 2002;128:650–660. doi: 10.1104/pp.010585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl I, Robinson DG, Chrispeels MJ, Hinz G. Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. J Cell Sci. 1996;109:2539–2550. doi: 10.1242/jcs.109.10.2539. [DOI] [PubMed] [Google Scholar]
- Hou SW, Hu JH, Li MZ, Sun XM. Development status and trend of soybean production in the world. Agric Technol. 2010;30:1–2. [Google Scholar]
- Hu XM, Zhang BX, Zhu YM, Lai YC, Li W, Li W, Bi YD, Xiao JL, Qi N, Lin H, Liu GY, Yang XF, Liu LY, Zhang LL. The studies and utilization of wild soybean (Glycine soja) J Anhui Agric Sci. 2011;39:13311–13313. [Google Scholar]
- Huang S, Yu J, Li Y, Wang J, Wang X, Qi H, Xu M, Qin H, Yin Z, Mei H, Chang H, Gao H, Liu S, Zhang Z, Zhang S, Zhu R, Liu C, Wu X, Jiang H, Zhen Hu, Xin D, Chen Q, Qi Z. Identification of soybean genes related to soybean seed protein content based on quantitative trait loci collinearity analysis. J Agric Food Chem. 2018;67:258–274. doi: 10.1021/acs.jafc.8b04602. [DOI] [PubMed] [Google Scholar]
- Hwang EY, Song QJ, Jia GF, Specht J, Hyten DL, Costa J, Cregan PB. A genome-wide association study of seed protein and oil content in soybean. BMC Genomics. 2014;15:1–12. doi: 10.1186/1471-2164-15-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymowitz T, Shurtleff WR. Debunking soybean myths and legends in the historical and popular literature. Crop Sci. 2005;45:473–476. doi: 10.2135/cropsci2005.0473. [DOI] [Google Scholar]
- Hyten DL, Song QJ, Zhu YL, Choi IY, Nelson RL, Costa JM, Specht JE. Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA. 2006;103:16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INRA (Institut Scientifique de Recherche Agronomique) (2004) Tables of composition and nutritional value of feed materials, 2 ed. In: Sauvant D, Perez JM, Tran G (eds) Wageningen Academic Publishers, Netherlands, p 186
- Ishikawa G, Takada Y, Nakamura T. A PCR-based method to test for the presence or absence of β-conglycinin α′- and α-subunits in soybean. Mol Breed. 2006;17:365–374. doi: 10.1007/s11032-006-9010-9. [DOI] [Google Scholar]
- Islam N, Krishnan HB, Natarajan S. Proteomic profiling of fast neutron-induced soybean mutant unveiled pathways associated with increased seed protein content. Proteome Res. 2020;19:3936–3944. doi: 10.1021/acs.jproteome.0c00160. [DOI] [PubMed] [Google Scholar]
- Jarquin D, Specht J, Lorenz A. Prospects of genomic prediction in the USDA soybean germplasm collection: historical data creates robust models for enhancing selection of accessions. G3-Genes Genom Genet. 2016;6:2329–2341. doi: 10.1534/g3.116.031443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran M, Xu B. An insight into the health benefits of fermented soy products. Food Chem. 2019;271:362–371. doi: 10.1016/j.foodchem.2018.07.158. [DOI] [PubMed] [Google Scholar]
- Jegadeesan S, Yu K (2020) Food grade soybean breeding, current status and future directions. In: Hasanuzzaman (ed) Legume crops: prospects, production and uses, 4rd edn
- Jian S, Wen Z, Li H, Yuan D, Li J, Zhang H, Ye Y, Lu W. Identification of QTLs for glycinin (11S) and β-conglycinin (7S) fractions of seed storage protein in soybean by association mapping. Acta Agron Sin. 2013;38:820–828. doi: 10.3724/SP.J.1006.2012.00820. [DOI] [Google Scholar]
- Jiang Y, Xue EY, Lu WC, Cui GW, Li YM, Han TF, Wang SD. Breeding and feeding quality and analysis of a new soybean strain deficient in Kunitz trypsin inhibitor. Acta Pratacul Sin. 2020;29:91–98. [Google Scholar]
- Joaquim PI, Molinari MD, Marin SR, Barbosa DA, Viana AJC, Rech EL, Henning FA, Nepomuceno AL, Mertz-Henning LM. Nitrogen compounds transporters: candidates to increase the protein content in soybean seeds. J Plant Interact. 2022;17:309–318. doi: 10.1080/17429145.2022.2039791. [DOI] [Google Scholar]
- Jun TH, Van K, Kim MY, Lee SH, Walker DR. Association analysis using SSR markers to find QTL for seed protein content in soybean. Euphytica. 2007;162:179–191. doi: 10.1007/s10681-007-9491-6. [DOI] [Google Scholar]
- Kada S, Yabusaki M, Kaga T, Ashida H, Yoshida K. Identification of two major ammonia-releasing reactions involved in secondary natto fermentation. Biosci Biotech Biochem. 2008;72:1869–1876. doi: 10.1271/bbb.80129. [DOI] [PubMed] [Google Scholar]
- Kagawa H, Yamauchi F, Hirano H. Soybean basic 7S globulin represents a protein widely distributed in legume species. FEBS Lett. 1987;226:145–149. doi: 10.1016/0014-5793(87)80568-9. [DOI] [Google Scholar]
- Kenty MM, Young LD, Kilen TC. Registration of DMK93-9048 soybean germplasm with resistance to foliar feeding insects and stem canker and possessing high protein. Crop Sci. 2001;41:603–603. doi: 10.2135/cropsci2001.412603x. [DOI] [Google Scholar]
- Kim JS, Kwon CS. Estimated dietary isoflavone intake of Korean population based on National Nutrition Survey. Nutr Res. 2001;21:947–953. doi: 10.1016/S0271-5317(01)00310-4. [DOI] [PubMed] [Google Scholar]
- Kim MY, Lee S, Van K, Kim TH, Jeong SC, Choi IY, et al. Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc Natl Acad Sci USA. 2010;107:22032–22037. doi: 10.1073/pnas.1009526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Ko JM, Baek IY, Jeon MK, Han WY, Park KY, Lee BW, Lee YH, Jung CS, Oh KW, Ha TJ, Moon JK, Yun HT, Lee JH, Choi JK, Jung JH, Lee SS, Jang YJ, Son CK, Kang DS. Soybean cultivar for tofu, 'Saedanbaek' with disease resistance, and high protein content. Korean J Breed Sci. 2014;46:295–301. doi: 10.9787/KJBS.2014.46.3.295. [DOI] [Google Scholar]
- Kim M, Schultz S, Nelson RL, Diers BW. Identification and fine mapping of a soybean seed protein QTL from PI 407788A on chromosome 15. Crop Sci. 2016;56:219–225. doi: 10.2135/cropsci2015.06.0340. [DOI] [Google Scholar]
- Kim IS, Kim CH, Yang WS. Physiologically active molecules and functional properties of soybeans in human health-A current perspective. Int J Mol Sci. 2021;22:4054. doi: 10.3390/ijms22084054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Lozano R, Kim JH, Bae DN, Kim ST, Park JH, Choi MS, Kim J, Ok HC, Park SK, Gore MA, Moon JK, Jeong SC. The patterns of deleterious mutations during the domestication of soybean. Nat Commun. 2021;12:1–14. doi: 10.1038/s41467-020-20337-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Shin I, Park SK, Choi MS, Lee JD, Ha BK, Lee J, Kang YJ, Jeong S, Moon J, Kang S. Soybean cultivar ‘Hipro’ for tofu and soymilk with high seed protein content and pod shattering resistance. Korean Soc Breed Sci. 2021;53:60–68. doi: 10.9787/KJBS.2021.53.1.60. [DOI] [Google Scholar]
- Kinney AJ, Jung R, Herman EM. Cosuppression of the α subunits of β-conglycinin in transgenic soybean seeds induces the formation of endoplasmic reticulum–derived protein bodies. Plant Cell. 2001;13:1165–1178. doi: 10.1105/tpc.13.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch T, Saalbach G, Raikhel NV, Beevers L. Interaction of a potential vacuolar targeting receptor with amino- and carboxyl-terminal targeting determinants. Plant Physiol. 1996;111:469–474. doi: 10.1104/pp.111.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide Y, Hirano H, Matsuoka K, Nakamura K. The N-terminal propeptide of the precursor to sporamin acts as a vacuole-targeting signal even at the C-terminus of the mature part in tobacco cells. Plant Physiol. 1997;114:863–870. doi: 10.1104/pp.114.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HB, Jiang G, Krishnan AH, Wiebold WJ. Seed storage protein composition of non-nodulating soybean (Glycine max (L.) Merr.) and its influence on protein quality. Plant Sci. 2000;157:191–199. doi: 10.1016/S0168-9452(00)00283-1. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Oehrle NW, Natarajan SS. A rapid and simple procedure for the depletion of abundant storage proteins from legume seeds to advance proteome analysis: a case study using Glycine max. Proteomics. 2009;9:3174–3188. doi: 10.1002/pmic.200800875. [DOI] [PubMed] [Google Scholar]
- Kudełka W, Kowalska M, Popis M. Quality of soybean products in terms of essential amino acids composition. Molecules. 2021;26:5071. doi: 10.3390/molecules26165071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasch AK, Hahn V, Leiser WL, Vollmann J, Schori A, Bétrix CA, Mayr B, Winkler J, Mechtler K, Jonas A, Sudaric A, Pejic I, Sarcevic H, Jeanson P, Balko C, Signor M, Miceli F, Strijk P, Rietman H, Muresanu E, Djordjevic V, Pospišil A, Barion G, Weigold P, Streng S, Krön M, Würschum T. Identification of mega-environments in Europe and effect of allelic variation at maturity E loci on adaptation of European soybean. Plant Cell Environ. 2017;40:765–778. doi: 10.1111/pce.12896. [DOI] [PubMed] [Google Scholar]
- Kurasch AK, Hahn V, Leiser WL, Starck N, Würschum T. Phenotypic analysis of major agronomic traits in 1008 RILs from a diallel of early European soybean varieties. Crop Sci. 2017;57:726–738. doi: 10.2135/cropsci2016.05.0318. [DOI] [Google Scholar]
- Kuroda Y, Kaga A, Tomooka N, Vaughan DA. Gene flow and genetic structure of wild soybean (Glycine soja) in Japan. Crop Sci. 2008;48:1071–1079. doi: 10.2135/cropsci2007.09.0496. [DOI] [Google Scholar]
- Lai YC, Lin H, Fang WC, Yao ZC, Qi N, Wang QX, Yang XF, Li H. Research on the excellent resource of wild soybean screen appraise and utilization in Heilongjiang. Chin Agric Sci Bull. 2005;21:379–382. [Google Scholar]
- Lan P, Li W, Schmidt W. Complementary proteome and transcriptome profiling in phosphate- deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol Cell Proteomics. 2012;11:1156–1166. doi: 10.1074/mcp.M112.020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarova G, Zeng Y, Kermode AR. Cloning and expression of an ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene homologue of yellow-cedar (Chamaecyparis nootkatensis) J Exp Bot. 2002;53:1219–1221. doi: 10.1093/jexbot/53.371.1219. [DOI] [PubMed] [Google Scholar]
- Lee S, Van K, Sung M, Nelson R, Lamantia J, McHale LK, Mian MAR. Genome-wide association study of seed protein, oil and amino acid contents in soybean from maturity groups I to IV. Theor Appl Genet. 2019;132:1639–1659. doi: 10.1007/s00122-019-03304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kim HS, Hwang TY. Variation in protein and isoflavone contents of collected domestic and foreign soybean (Glycine max (L) Merrill) Germplasms in Korea. Agriculture. 2021;11:735. doi: 10.3390/agriculture11080735. [DOI] [Google Scholar]
- Leffel RC. Registration of high-protein soybean germplasm lines BARC-6, BARC-7, BARC-8, and BARC-9. Crop Sci. 1992;32:502. doi: 10.2135/cropsci1992.0011183X003200020054x. [DOI] [Google Scholar]
- Lemme A, Naranjo V, de Paula Dorigam JC. Utilization of methionine sources for growth and Met+Cys deposition in broilers. Animals. 2020;10:2240. doi: 10.3390/ani10122240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestari P, Van K, Lee J, Kang YJ, Lee SH. Gene divergence of homeologous regions associated with a major seed protein content QTL in soybean. Front Plant Sci. 2013;4:176. doi: 10.3389/fpls.2013.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Studies on the ecological and geographical distribution of the chinese resources of wild soybean (G. soja) Sci Agric Sinica. 1993;26:47–55. [Google Scholar]
- Li FS. Utilization and prospect of wild soybean genetic resources. Chin Seed Ind. 1988;1:16–22. [Google Scholar]
- Li B. High yield cultivation technique of new soybean variety Huachun 6. Crop Sci. 2010;24:2. [Google Scholar]
- Li WD, Lu WG, Liang HZ, Wang SF, Yuan BJ, Geng Z, Wang SG. Effects of eco-physiological factors on soybean protein content. Acta Agron Sin. 2004;30:1065–1068. [Google Scholar]
- Li WX, Zhu ZH, Liu SC, Liu F, Zhang XF, Li Y, Wang SM. Quality characters of Chinese soybean (Glycine max) varieties and germplasm resources. J Plant Genet Resour. 2004;5:185–192. [Google Scholar]
- Li ZW, Meyer S, Essig JS, Liu Y, Schapaugh MA, Muthukrishnan S, Hainline BE, Trick HN. High-level expression of maize γ-zein protein in transgenic soybean (Glycine max) Mol Breed. 2005;16:11–20. doi: 10.1007/s11032-004-7658-6. [DOI] [Google Scholar]
- Li CM, Yang SP, Gai JY, Yu DY. Comparative proteomic analysis of wild (Glycine soja) and cultivated (Glycine max) soybean seeds. Prog Biochem Biophys. 2007;34:1296–1302. [Google Scholar]
- Li YH, Zhou GY, Ma JX, Jiang WK, Jin LG, Zhang ZH, Guo Y, Zhang JB, Sui Y, Zheng LT, Zhang SS, Zuo QY, Shi XH, Li YF, Zhang WK, Hu YY, Kong GY, Hong HL, Tan B, Ji S, Liu ZX, Wang YS, Ruan H, Yeung CKL, Liu J, Wang H, Zhang LJ, Guan RX, Wang KJ, Li WB, Chen SY, Chang RZ, Jiang Z, Jackson SA, Li RQ, Qiu LJ. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat Biotechnol. 2014;32:1045–1052. doi: 10.1038/nbt.2979. [DOI] [PubMed] [Google Scholar]
- Li L, Zheng W, Zhu Y, Ye H, Tang B, Arendsee ZW, Jones D, Li R, Ortiz D, Zhao X, Du C, Nettleton D, Scott MP, Salas-Fernandez MG, Yin Y, Wurtele ES. QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc Natl Acad Sci USA. 2015;112:14734–14739. doi: 10.1073/pnas.1514670112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Guo R, Jiao Y, Jin XF, Zhang HY, Shi LX. Comparison of salt tolerance in soja based on metabolomics of seedling roots. Front Plant Sci. 2017;8:1101. doi: 10.3389/fpls.2017.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Nadeem M, Sun GL, Wang XB, Qiu LJ. Male sterility in soybean: Occurrence, molecular basis and utilization. Plant Breed. 2019;138:659–676. doi: 10.1111/pbr.12751. [DOI] [Google Scholar]
- Li CL, Nguyen V, Liu J, Fu WQ, Chen C, Yu KF, Cui YH. Mutagenesis of seed storage protein genes in soybean using CRISPR/Cas9. BMC Res Not. 2019;12:1–7. doi: 10.1186/s13104-019-4207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Liu Q, Yang QC, Shu WT, Li JH, Chang SH, Zhang DH, Zhang BL, Zhang LC, Geng Z. Application of molecular marker assisted breeding of soybean high protein gene. J Shanxi Agric Sci. 2020;48:1192–1197. [Google Scholar]
- Li DL, Yang ZY, Zhu R, Li L, Yu Z, Wu LF. Effects of glycinin on intestinal health of aquatic animals and its improvement measures. Soybean Sci. 2021;40:420–425. [Google Scholar]
- Li YF, Li YH, Su SS, Reif JC, Qi ZM, Wang XB, Wang X, Tian Y, Li DL, Sun RJ, Liu ZX, Xu ZJ, Fu GH, Ji YL, Chen QS, Liu JQ, Qiu LJ. SoySNP618K array: a high-resolution single nucleotide polymorphism platform as a valuable genomic resource for soybean genetics and breeding. J Integr Plant Biol. 2022;64:632–648. doi: 10.1111/jipb.13202. [DOI] [PubMed] [Google Scholar]
- Lin YH, Zang LJ, Li W, Zhang Lf XuR. QTLs mapping related protein content of soybean. Soybean Sci. 2010;29:207–209. [Google Scholar]
- Lin H, Rao J, Shi JX, Hu CY, Cheng F, Wilson ZA, Zhang DB, Quan S. Seed metabolomic study reveals significant metabolite variations and correlations among different soybean cultivars. J Integr Plant Biol. 2014;56:826–836. doi: 10.1111/jipb.12228. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tian Z. From one linear genome to a graph-based pan-genome: a new era for genomics. Sci China Life Sci. 2020;63:1938–1941. doi: 10.1007/s11427-020-1808-0. [DOI] [PubMed] [Google Scholar]
- Liu S, Ohta K, Dong C, Thanh VC, Ishimoto M, Qin Z, Hirata Y. Genetic diversity of soybean (Glycine max (L.) Merrill) 7S globulin protein subunits. Genet Resour Crop Ev. 2006;53:1209–1219. doi: 10.1007/s10722-005-2434-y. [DOI] [Google Scholar]
- Liu SH, Zhou RB, Yu DY, Chen SY, Gai JY. QTL mapping of protein related traits in soybean (Glycine max (L.) Merr.) Acta Agron Sin. 2009;35:2139–2149. doi: 10.3724/SP.J.1006.2009.02139. [DOI] [Google Scholar]
- Liu S, Teng W, Jiang Z, Zhang B, Ge Y, Diao G, Zheng T, Zeng R, Wu S, Li W. Development of soybean germplasm lacking of 7S globulin α-subunit. Acta Agron Sin. 2010;36:1409–1413. doi: 10.3724/SP.J.1006.2010.01409. [DOI] [Google Scholar]
- Liu D, Zhu C, Peng K, Guo Y, Chang PR, Cao X. Facile preparation of soy protein/poly (vinyl alcohol) blend fibers with high mechanical performance by wet-spinning. Ind Eng ChemRes. 2013;52:6177–6181. doi: 10.1021/ie400521a. [DOI] [Google Scholar]
- Liu CH, Zhang CS, Ding HL, Zhu HD, Fei ZH, Zhang AH, Hu SY, Wu YH, Feng LJ. Effects of different parent combinations on protein content of soybean progeny. Soybean Sci Technol. 2016;5:4–8. [Google Scholar]
- Liu J, Xu RX, Shi L, Wang M, Xu YY, Jiang HF, Jiang SC, Xing DY. Variation trend of major traits of national authorized soybean cultivars from 2003 to 2016. Anhui Agric Sci Bull. 2017;23(11):60–66. [Google Scholar]
- Liu M, Lai Y, Bi Y, Luan X, Li W, Di S, Fan C, Wang Y. Breeding and cultivation technology of a high protein soybean cultivar Zhonglongdou106. Heilongjiang Agric Sci. 2021;12:137–140. [Google Scholar]
- Lu W, Wen Z, Li H, Yuan D, Li J, Zhang H, Huang Z, Cui S, Du W. Identification of the quantitative trait loci (QTL) underlying water soluble protein content in soybean. Theor Appl Genet. 2013;126:425–433. doi: 10.1007/s00122-012-1990-8. [DOI] [PubMed] [Google Scholar]
- Luo J, Luo J, Yuan C, Zhang W, Li J, Gao Q, Chen H. An eco-friendly wood adhesive from soy protein and lignin: performance properties. RSC Adv. 2015;5:100849–100855. doi: 10.1039/C5RA19232C. [DOI] [Google Scholar]
- Magni C, Sessa F, Capraro J, Duranti M, Maffioli E, Scarafoni A. Structural and functional insights into the basic globulin 7S of soybean seeds by using trypsin as a molecular probe. Biochem Biophys Res Commun. 2018;496:89–94. doi: 10.1016/j.bbrc.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Malle S, Eskandari M, Morrison M, Belzile F. Genome-wide association identifies several QTLs controlling cysteine and methionine content in soybean seed including some promising candidate genes. Sci Rep. 2020;10:1–14. doi: 10.1038/s41598-020-78907-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao T, Jiang Z, Han Y, Teng W, Zhao X, Li W, Morris B. Identification of quantitative trait loci underlying seed protein and oil contents of soybean across multi-genetic backgrounds and environments. Plant Breed. 2013;132:630–641. doi: 10.1111/pbr.12091. [DOI] [Google Scholar]
- Matsuoka K, Neuhaus JM. Cis-elements of protein transport to the plant vacuoles. J Exp Bot. 1999;50:165–174. doi: 10.1093/jxb/50.331.165. [DOI] [Google Scholar]
- Messina M. Soy and health update: evaluation of the clinical and epidemiologic literature. Nutrients. 2016;8:754. doi: 10.3390/nu8120754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Yang S, Zhang K, He J, Wu C, Ren Y, Gai J, Li Y. Natural variation and selection in GmSWEET39 affect soybean seed oil content. New Phytol. 2020;225:1651–1666. doi: 10.1111/nph.16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili D, Stanaûev V, Marjanoviû-Jeromela A, Stanaûev V, Puvaýa N, Zari S (2012) Effect of feed on the basis of soybean in pig nutrition. Sci Pap 67–72
- Millward DJ. Amino acid scoring patterns for protein quality assessment. Brit J Nutr. 2012;108:31–34. doi: 10.1017/S0007114512002462. [DOI] [PubMed] [Google Scholar]
- Min CW, Gupta R, Kim SW, Lee SE, Kim YC, Bae DW, Han YW, Lee BW, Ko JM, Agrawal GK, Rakwal R, Kim ST. Comparative biochemical and proteomic analyses of soybean seed cultivars differing in protein and oil content. J Agric Food Chem. 2015;63:7134–7142. doi: 10.1021/acs.jafc.5b03196. [DOI] [PubMed] [Google Scholar]
- Mochizuki Y, Maebuchi M, Hirotsuka M, Wadahara H, Moriyama T, Kawada T, Urade R. Changes in lipid metabolism by soy β-conglycinin-derived peptides in HepG2 cells. J Agric Food Chem. 2009;57:1473–1480. doi: 10.1021/jf8031793. [DOI] [PubMed] [Google Scholar]
- Modgil R, Kumar V. Soybean (Glycine max) In: Tanwar B, Ankit G, editors. Oilseeds: health attributes and food applications. 1. Singapore: Springer; 2021. pp. 1–46. [Google Scholar]
- Mori T, Maruyama N, Nishizawa K, Higasa T, Yagasaki K, Ishimoto M, Utsumi S. The composition of newly synthesized proteins in the endoplasmic reticulum determines the transport pathways of soybean seed storage proteins. Plant J. 2004;40:238–249. doi: 10.1111/j.1365-313X.2004.02204.x. [DOI] [PubMed] [Google Scholar]
- Mortensen A, Kulling SE, Schwartz H, Rowland I, Ruefer CE, Rimbach G, Cassidy A, Magee P, Millar J, Hall WL, Kramer BF, Sorensen IK, Sontag G. Analytical and compositional aspects of isoflavones in food and their biological effects. Mol Nutr Food Res. 2009;53:266–309. doi: 10.1002/mnfr.200800478. [DOI] [PubMed] [Google Scholar]
- Müntz K. Deposition of storage proteins. Plant Mol Biol. 1998;38:77–99. doi: 10.1023/A:1006020208380. [DOI] [PubMed] [Google Scholar]
- Natarajan SS, Xu C, Bae H, Caperna TJ, Garrett WM. Characterization of storage proteins in wild (Glycine soja) and cultivated (Glycine max) soybean seeds using proteomic analysis. J Agric Food Chem. 2006;54:3114–3120. doi: 10.1021/jf052954k. [DOI] [PubMed] [Google Scholar]
- Neuhaus JM, Rogers JC. Sorting of proteins to vacuoles in plant cells. Plant Mol Biol. 1998;38:127–144. doi: 10.1023/A:1006032627036. [DOI] [PubMed] [Google Scholar]
- Nielsen NC, Dickinson CD, Cho TJ, Thanh VH, Scallon BJ, Fischer RL, Sims TL, Drews GN, Goldberg RB. Characterization of the glycinin gene family in soybean. Plant Cell. 1989;1:313–328. doi: 10.1105/tpc.1.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik AM, Tosh SM, Woodrow L, Poysa V, Corredig M. Effect of soy protein subunit composition and processing conditions on stability and particle size distribution of soymilk. LWT-Food Sci Technol. 2009;42:1245–1252. doi: 10.1016/j.lwt.2009.03.001. [DOI] [Google Scholar]
- Nishimura M, Ohkawara T, Sato Y, Satoh H, Takahashi Y, Hajika M, Nishihira J. Improvement of triglyceride levels through the intake of enriched-β-conglycinin soybean (Nanahomare) revealed in a randomized, double-blind, placebo-controlled study. Nutrients. 2016;8:491. doi: 10.3390/nu8080491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa K, Maruyama N, Satoh R, Fuchikami Y, Higasa T, Utsumi S. A C-terminal sequence of soybean β-conglycinin α′ subunit acts as a vacuolar sorting determinant in seed cells. Plant J. 2003;34:647–659. doi: 10.1046/j.1365-313X.2003.01754.x. [DOI] [PubMed] [Google Scholar]
- Nishizawa K, Maruyama N, Utsumi S. The C-terminal region of α′ subunit of soybean β-conglycinin contains two types of vacuolar sorting determinants. Plant Mol Biol. 2006;62:111–125. doi: 10.1007/s11103-006-9007-0. [DOI] [PubMed] [Google Scholar]
- Nosowitz D (2017) Soy is set to become our biggest crop by acreage. But what are we doing with this soy? Modern Farmer Media. https://modernfarmer.com/2017/12/soy-set-become-biggest-crop-acreage-soy/. Accessed 04 Dec 2017
- Ogawa T, Tayama E, Kitamura K, Kaizuma N. Genetic improvement of seed storage proteins using three variant alleles of 7S globulin subunits in soybean (Glycine max L.) Jpn J Breed. 1989;39:137–147. doi: 10.1270/jsbbs1951.39.137. [DOI] [Google Scholar]
- Ogawa T, Bando N, Tsuji H, Nishikawa K, Kitamura K. α-Subunit of β-conglycinin, an allergenic protein recognized by IgE antibodies of soybean-sensitive patients with atopic dermatitis. Biosci Biotechnol Biochem. 1995;59:831–833. doi: 10.1271/bbb.59.831. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Samoto M, Takahashi K. Soybean allergens and hypoallergenic soybean products. J Nutr Sci Vitaminol. 2000;46:271–279. doi: 10.3177/jnsv.46.271. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Ohtake N, Sueyoshi K, Ono Y, Tsutsumi K, Ueno M, Tanabata S, Sato T, Takahashi Y (2017) Amino acid metabolism and transport in soybean plants. In: Asao T, Asaduzzaman MD (eds) Amino acid, new insights and roles in plant and animal. pp 171–196
- Ouyang ZC, Cheng YB, Cao YQ, Nian H. High efficiency cultivation technique regulation of the new soybean variety Huaxia 4. Crops. 2009;25:100–100. [Google Scholar]
- Pantalone V, Wyman C. Registration of TN15-4009 soybean germplasm with resistance to soybean cyst nematode, southern root knot nematode, and peanut root knot nematode. J Plant Regist. 2020;14:77–81. doi: 10.1002/plr2.20007. [DOI] [Google Scholar]
- Panthee DR, Kwanyuen P, Sams CE, West DR, Saxton AM, Pantalone VR. Quantitative trait loci for β-conglycinin (7S) and glycinin (11S) fractions of soybean storage protein. J Am Oil Chem Soc. 2004;81:1005–1012. doi: 10.1007/s11746-004-1014-4. [DOI] [Google Scholar]
- Panthee DR, Pantalone VR, Sams CE, Saxton AM, West DR, Orf JH, Killam AS. Quantitative trait loci controlling sulfur containing amino acids, methionine and cysteine, in soybean seeds. Theor Appl Genet. 2006;112:546–553. doi: 10.1007/s00122-005-0161-6. [DOI] [PubMed] [Google Scholar]
- Pathan SM, Vuong T, Clark K, Lee J, Shannon JG, Roberts CA, Ellersieck MR, Burton JW, Cregan PB, Hyten DL, Nguyen HT, Sleper DA. Genetic mapping and confirmation of quantitative trait loci for seed protein and oil contents and seed weight in soybean. Crop Sci. 2013;53:765–774. doi: 10.2135/cropsci2012.03.0153. [DOI] [Google Scholar]
- Patil G, Mian R, Vuong T, Pantalone V, Song Q, Chen P, Shannon GJ, Carter TC, Nguyen HT. Molecular mapping and genomics of soybean seed protein: a review and perspective for the future. Theor Appl Genet. 2017;130:1975–1991. doi: 10.1007/s00122-017-2955-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil G, Vuong TD, Kale S, Valliyodan B, Deshmukh R, Zhu C, Wu X, Bai Y, Yungbluth D, Lu F, Kumpatla S, Grover Shannon J, Varshney RK, Nguyen HT. Dissecting genomic hotspots underlying seed protein, oil, and sucrose content in an interspecific mapping population of soybean using high-density linkage mapping. Plant Biotechnol J. 2018;16:1939–1953. doi: 10.1111/pbi.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phansak P, Soonsuwon W, Hyten DL, Song Q, Cregan PB, Graef GL, Specht JE. Multi-population selective genotyping to identify soybean [Glycine max (L.) Merr.] seed protein and oil QTLs. G3-Genes Genom Genet. 2016;6:1635–1648. doi: 10.1534/g3.116.027656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poysa V, Woodrow L, Yu K. Effect of soy protein subunit composition on tofu quality. Food Res Int. 2006;39:309–317. doi: 10.1016/j.foodres.2005.08.003. [DOI] [Google Scholar]
- Qiang JN, Zhao Y, Zhao WD, Li P, Na RGA, Chang MN, Wang X. Study on gut allergenic responses of soybean major antigen protein. Soybean Sci. 2018;37:322–325. [Google Scholar]
- Qin J, Wang F, Zhao Q, Shi A, Zhao T, Song Q, Ravelombola W, An H, Yan L, Yang C, Zhang M. Identification of candidate genes and genomic selection for seed protein in soybean breeding pipeline. Front Plant Sci. 2022;13:882732. doi: 10.3389/fpls.2022.882732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu LJ, Chang RJ, Yuan CP, Guan RX, Zhang Y, Li YH. Prospect and present statue of gene discovery and utilization for introduced soybean germplasm. J Plant Genet Resour. 2006;7:1–16. [Google Scholar]
- Ramdath DD, Padhi EM, Sarfaraz S, Renwick S, Duncan AM. Beyond the cholesterol-lowering effect of soy protein: a review of the effects of dietary soy and its constituents on risk factors for cardiovascular disease. Nutrients. 2017;9:324. doi: 10.3390/nu9040324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KR, Patro H, Lokhande S, Bellaloui N, Gao W. Ultraviolet-B radiation alters soybean growth and seed quality. Food Nutr Sci. 2016;7:55–66. [Google Scholar]
- Rizzo G, Baroni L. Soy, soy foods and their role in vegetarian diets. Nutrients. 2018;10:43. doi: 10.3390/nu10010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Bäumer M, Hinz G, Hohl I. Vesicle transfer of storage proteins to the vacuole: the role of the Golgi apparatus and multivesicular bodies. J Plant Physiol. 1998;152:659–667. doi: 10.1016/S0176-1617(98)80027-8. [DOI] [Google Scholar]
- Rotundo JL, Borrás L, Westgate ME, Orf JH. Relationship between assimilate supply per seed during seed filling and soybean seed composition. Field Crop Res. 2009;112:90–96. doi: 10.1016/j.fcr.2009.02.004. [DOI] [Google Scholar]
- Rotundo JL, Miller-Garvin JE, Naeve SL. Regional and temporal variation in soybean seed protein and oil across the United States. Crop Sci. 2017;56:797–808. doi: 10.2135/cropsci2015.06.0394. [DOI] [Google Scholar]
- Rüdelsheim PLJ, Smets G (2012) Baseline information on agricultural practices in the EU Soybean (Glycine max (L.) Merr.). Perseus BVBA 42
- Saalbach G, Rosso M, Schumann U. The vacuolar targeting signal of the 2S albumin from Brazil nut resides at the C terminus and involves the C-terminal propeptide as an essential element. Plant Physiol. 1996;112:975–985. doi: 10.1104/pp.112.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanfar B, Cober ER, Charette M, Tan LH, Bekele WA, Morrison MJ, Kilian A, Belzile F, Molnar SJ. Genetic analysis of high protein content in ‘AC Proteus’ related soybean populations using SSR, SNP, DArT and DArTseq markers. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-55862-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallon B, Thanh VH, Floener LA, Nielsen NC. Identification and characterization of DNA clones encoding group-II glycinin subunits. Theor Appl Genet. 1985;70:510–519. doi: 10.1007/BF00305984. [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Barbazuk WB, Sandford M, May G, Song ZH, Zhou WX, Nikolau BJ, Herman EM. Silencing of soybean seed storage proteins results in a rebalanced protein composition preserving seed protein content without major collateral changes in the metabolome and transcriptome. Plant Physiol. 2011;156:330–345. doi: 10.1104/pp.111.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Sebolt AM, Shoemaker RC, Diers BW. Analysis of a quantitative trait locus allele from wild soybean that increases seed protein concentration in soybean. Crop Sci. 2000;40:1438–1444. doi: 10.2135/cropsci2000.4051438x. [DOI] [Google Scholar]
- Shi A, Chen P, Zhang B, Hou A. Genetic diversity and association analysis of protein and oil content in food-grade soybeans from Asia and the United States. Plant Breeding. 2010;129:250–256. doi: 10.1111/j.1439-0523.2010.01766.x. [DOI] [Google Scholar]
- Singh RJ, Hymowitz T. Soybean genetic resources and crop improvement. Genome. 1999;42:605–616. doi: 10.1139/g99-039. [DOI] [Google Scholar]
- Singh P, Kumar R, Sabapathy S, Bawa A. Functional and edible uses of soy protein products. Compr Rev Food Sci F. 2007;7:14–28. doi: 10.1111/j.1541-4337.2007.00025.x. [DOI] [Google Scholar]
- Singh A, Meena M, Kumar D, Dubey AK, Hassan MI. Structural and functional analysis of various globulin proteins from soy seed. Crit Rev Food Sci Nutr. 2015;55:1491–1502. doi: 10.1080/10408398.2012.700340. [DOI] [PubMed] [Google Scholar]
- Song B, Lan L, Tian F, Tuo Y, Bai Y, Jiang Z, Shen L, Li W, Liu S. Development of soybean lines with α’-subunit or (α’+α)-subunits deficiency in 7S globulin by backcrossing. Acta Agron Sin. 2013;38:2297–2305. doi: 10.3724/SP.J.1006.2012.02297. [DOI] [Google Scholar]
- Song B, An LX, Han YJ, Gao HX, Ren HB, Zhao X, Wei XS, Krishnan HB, Liu SS. Transcriptome profile of near-isogenic soybean lines for β-conglycinin α-subunit deficiency during seed maturation. PLOS ONE. 2016;11:e0159723. doi: 10.1371/journal.pone.0159723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Brown BB, Song QJ, Vaughn JN, Li ZL. Genomic selection for yield and seed composition traits within an applied soybean breeding program. G3-Genes Genom Genet. 2019;9:2253–2265. doi: 10.1534/g3.118.200917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Hirose A, Kanazashi YH, Adachi K, Hibara M, Itoh T, Mikami M, Endo M, Hirose S, Maruyama N, Abe J, Yamada T. Simultaneous induction of mutant alleles of two allergenic genes in soybean by using site-directed mutagenesis. BMC Plant Biol. 2020;20:1–15. doi: 10.1186/s12870-020-02708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Qin GX, Zhang QH. Effects of soybean antigen protein on growth performance, dietary nutrient digestibility and intestinal absorption capacity of calves. Chin J Anim Sci. 2005;41:30–33. [Google Scholar]
- Sun Q, Xue J, Lin L, Liu D, Wu J, Jiang J, Wang Y. Overexpression of soybean transcription factors GmDof4 and GmDof11 significantly increase the oleic acid content in seed of Brassica napus L. Agronomy. 2018;8:222. doi: 10.3390/agronomy8100222. [DOI] [Google Scholar]
- Sun R, Sun B, Tian Y, Su S, Zhang Y, Zhang W, Wang J, Yu P, Guo B, Li H (2022) The practical soybean breeding pipeline by developing high throughput functional array ZDX1. Theor Appl Gene 10.21203/rs.3.rs-837237/v1 [DOI] [PMC free article] [PubMed]
- Szczerba A, Płażek A, Pastuszak J, Kopeć P, Hornyák M, Dubert F. Effect of low temperature on germination, growth, and seed yield of four soybean (Glycine max L.) cultivars. Agronomy. 2021;11:800. doi: 10.3390/agronomy11040800. [DOI] [Google Scholar]
- Takahashi K, Banba H, Kikuchi A, Ito M, Nakamura S. An induced mutant line lacking the a-subunit of b-conglycinin in soybean (Glycine max (L.) Merrill) Breed Sci. 1994;44:65–66. [Google Scholar]
- Takahashi K, Shimada S, Shimada H, Takada Y, Sakai T, Kono Y, Adachi T, Tabuchi K, Kikuchi A, Yumoto S, Nakamura S, Ito M, Banba H, Okabe A. A new soybean cultivar ‘‘Yumeminori’’ with low allergenicity and high content of 11S globulin. Bull Natl Agric Res Cent Tohoku Reg. 2004;102:23–39. [Google Scholar]
- Tandang-Silvas MR, Fukuda T, Fukuda C, Prak K, Cabanos C, Kimura A, Itoh T, Mikami B, Utsumi S, Maruyama N. Conservation and divergence on plant seed 11S globulins based on crystal structures. Biochim Biophys Acta. 2010;1804:1432–1442. doi: 10.1016/j.bbapap.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Tay SL, Kasapis S, Perera CO, Barlow PJ. Functional and structural properties of 2S soy protein in relation to other molecular protein fractions. J Agric Food Chem. 2006;54:6046–6053. doi: 10.1021/jf060387a. [DOI] [PubMed] [Google Scholar]
- Tegeder M, Masclaux-Daubresse C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018;217:35–53. doi: 10.1111/nph.14876. [DOI] [PubMed] [Google Scholar]
- Teraishi M, Takahashi M, Hajika M, Matsunaga R, Uematsu Y, Ishimoto M. Suppression of soybean b-conglycinin genes by a dominant gene, Scg-1. Theor Appl Genet. 2001;103:1266–1272. doi: 10.1007/s001220100702. [DOI] [Google Scholar]
- Thakur MK, Thakur VK, Gupta RK, Pappu A. Synthesis and applications of biodegradable soy based graft copolymers: a review. ACS Sustain Chem Eng. 2015;4:1–17. doi: 10.1021/acssuschemeng.5b01327. [DOI] [Google Scholar]
- The SV, Snyder R, Tegeder M. Targeting nitrogen metabolism and transport processes to improve plant nitrogen use efficiency. Front Plant Sci. 2020;11:628366. doi: 10.3389/fpls.2020.628366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian QZ, Gai JY. A review on the research of soybean origination and evolution. Soy Sci. 2001;20:54–59. [Google Scholar]
- Tian H, Guo G, Fu X, Yao Y, Yuan L, Xiang A. Fabrication, properties and applications of soy-protein-based materials: a review. Int J Biol Macromol. 2018;120:475–490. doi: 10.1016/j.ijbiomac.2018.08.110. [DOI] [PubMed] [Google Scholar]
- Tian H, Wu J, Xiang A. Polyether polyol-based rigid polyurethane foams reinforced with soy protein fillers. J Vinyl Addit Technol. 2018;24:105–111. doi: 10.1002/vnl.21608. [DOI] [Google Scholar]
- Tian YX, Gao FJ, Cao PP, Gao Q, Xia WR. Effect of sowing date on agronomic characters, quality and yield of new high protein soybean varieties (lines) J Nucl Agric Sci. 2021;35:1900–1907. [Google Scholar]
- Tsubokura Y, Hajika M, Kanamori H, Xia Z, Watanabe S, Kaga A, Katayose Y, Ishimoto M, Harada K. The beta-conglycinin deficiency in wild soybean is associated with the tail-to-tail inverted repeat of the alpha-subunit genes. Plant Mol Biol. 2012;78:301–309. doi: 10.1007/s11103-011-9865-y. [DOI] [PubMed] [Google Scholar]
- Tuo Y, Huo CQ, Tian FD, Song B, Shen LW, Wei XS, Guo BW, Li WB, Liu SS. Soybean7S α-subunit deficiency lines developed by backcrossing assisted by SSR marker background selection. Chin J Oil Crop Sci. 2014;36:1–9. [Google Scholar]
- Valliyodan B, Dan Q, Patil G, Zeng P, Huang J, Dai L, Chen C, Li Y, Joshi T, Song L, Vuong TD, Musket TA, Xu D, Shannon JG, Shifeng C, Liu X, Nguyen HT. Landscape of genomic diversity and trait discovery in soybean. Sci Rep. 2016;6:1–10. doi: 10.1038/srep23598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn JN, Nelson RL, Song Q, Cregan PB, Li Z. The genetic architecture of seed composition in soybean is refined by genome-wide association scans across multiple populations. G3-Genes Genom Genet. 2014;4:2283–2294. doi: 10.1534/g3.114.013433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Raikhel NV. What do proteins need to reach different vacuoles? Trends Plant Sci. 1999;4:149–155. doi: 10.1016/S1360-1385(99)01389-8. [DOI] [PubMed] [Google Scholar]
- Vitale A, Hinz G. Sorting of proteins to storage vacuoles: how many mechanisms? Trends Plant Sci. 2005;10:316–323. doi: 10.1016/j.tplants.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Voldeng HD, Guillemette RJD, Leonard DA, Cober ER. AC Proteus soybean. Can J Plant Sci. 1996;76:153–154. doi: 10.4141/cjps96-031. [DOI] [Google Scholar]
- Voldeng HD, Guillemette RJD, Leonard DA, Cober ER. Maple Glen soybean. Can J Plant Sci. 1996;76:475–476. doi: 10.4141/cjps96-085. [DOI] [Google Scholar]
- Walling L, Drews GN, Goldberg RB. Transcriptional and post-transcriptional regulation of soybean seed protein mRNA levels. P Natl A Sci. 1986;83:2123–2127. doi: 10.1073/pnas.83.7.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WZ, Liu XY, Cao YS, Zhang M. Study on protein content of Soybean germplasm resources in China. Crop Var Res. 1998;1:35–36. [Google Scholar]
- Wang PY, Xu DC, Guo YH, Meng LF, Zhao XN. Induced mutation for soybean quality. Acta Agric Nucl Sin. 2000;14:21–23. [Google Scholar]
- Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 2007;52:716–729. doi: 10.1111/j.1365-313X.2007.03268.x. [DOI] [PubMed] [Google Scholar]
- Wang XF, Ma W, Fu J. Correlation between protein content in filial generation and yield of soybean. Soybean Sci. 2013;32:573–575. [Google Scholar]
- Wang W, Han DZ, Yan HR, Luan XY, Wang J, Qiu LJ. QTL mapping of soybean protein content from high-protien soybean Zhongyin 1106. J Plant Genet Res. 2020;21:130–138. [Google Scholar]
- Wang S, Liu S, Wang J, Yokosho K, Zhou B, Yu YC, Liu Z, Frommer WB, Ma JF, Chen LQ, Guan Y, Shou H, Tian Z. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl Sci Rev. 2020;7:1776–1786. doi: 10.1093/nsr/nwaa110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li W, Zong C, Qi Y, Sun X, Bai Y, Sun G, Wang X, Xu D, Hou G, Zhang S, Ren H. Breeding and cultivation of a new soybean variety with high yield and high protein Mudou 15. Soybean Sci Technol. 2020;1:60–62. [Google Scholar]
- Wang JL, Zong CM, Wang DL, Wang YP, Jiang HX, Yang DX, Fu MM, Wang L, Ren HX, Zhao TJ, Du WG, Gai JY. Identification, evaluation and improvement utilization of northeast China soybean germplasm population in Jiamusi. Chin J Oil Crop Sci. 2021;43:996–1005. [Google Scholar]
- Wang J, Mao L, Zeng Z, Yu X, Lian J, Feng J, Yang W, An J, Wu H, Zhang M, Liu L. Genetic mapping high protein content QTL from soybean ‘Nanxiadou 25’and candidate gene analysis. BMC Plant Biol. 2021;21:1–13. doi: 10.1186/s12870-021-03176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington CV, Abdel-Haleem H, Hyten DL, Cregan PB, Orf JH, Killam AS, Bajjalieh N, Li Z, Boerma HR. QTL for seed protein and amino acids in the Benning x Danbaekkong soybean population. Theor Appl Genet. 2015;128:839–850. doi: 10.1007/s00122-015-2474-4. [DOI] [PubMed] [Google Scholar]
- Warsame AO, O’Sullivan DM, Tosi P. Seed storage proteins of faba bean (Vicia faba L): current status and prospects for genetic improvement. J Agric Food Chem. 2018;66:12617–12626. doi: 10.1021/acs.jafc.8b04992. [DOI] [PubMed] [Google Scholar]
- Wee CD, Hashiguchi M, Ishigaki G, Muguerza M, Oba C, Abe J, Harada K, Akashi R. Evaluation of seed components of wild soybean (Glycine soja) collected in Japan using near-infrared reflectance spectroscopy. Plant Genet Resour. 2018;16:94–102. doi: 10.1017/S1479262116000472. [DOI] [Google Scholar]
- Wehrmann V, Fehr W, Cianzio S, Cavins J. Transfer of high seed protein to high-yielding soybean cultivars. Crop Sci. 1987;27:927–937. doi: 10.2135/cropsci1987.0011183X002700050020x. [DOI] [Google Scholar]
- Wei ZY, Li ZF, Liu ZX, Qiu LQ. The selection effect and germplasm innovation for high protein content on EMS mutated Zp661 progenies. J Plant Genet Resour. 2019;20:1579–1587. [Google Scholar]
- Wijewardana C, Reddy KR, Bellaloui N. Soybean seed physiology, quality, and chemical composition under soil moisture stress. Food Chem. 2019;278:92–100. doi: 10.1016/j.foodchem.2018.11.035. [DOI] [PubMed] [Google Scholar]
- Wu YC, Guo BF, Gu YZ, Luan XY, Qiu HM, Liu XL, Li HY, Qiu LJ. Mapping of a new quantitative locus qPRO-19-1 associated with seed crude protein content in soybean (Glycine max L.) J Plant Genet Resour. 2021;22:139–148. [Google Scholar]
- Wysmierski PT, Vello NA. The genetic base of Brazilian soybean cultivars: evolution over time and breeding implications. Genet Mol Biol. 2013;36:547–555. doi: 10.1590/S1415-47572013005000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong DJ, Zhao TJ, Gai JY. The core ancestors of soybean cultivars released during 1923–2005 in China. Soybean Sci. 2007;26:641–647. [Google Scholar]
- Xu XP, Liu H, Tian LH, Dong XB, Shen SH, Qu LQ. Integrated and comparative proteomics of high-oil and high-protein soybean seeds. Food Chem. 2015;172:105–116. doi: 10.1016/j.foodchem.2014.09.035. [DOI] [PubMed] [Google Scholar]
- Xue ZC, Zhao SJ, Gao HY, Sun S. The salt resistance of wild soybean (Glycine soja Sieb. et Zucc. ZYD 03262) under NaCl stress is mainly determined by Na+ distribution in the plant. Acta Physiol Plant. 2014;36:61–70. doi: 10.1007/s11738-013-1386-7. [DOI] [Google Scholar]
- Yagasaki K, Sakamoto H, Seki K, Yamada N, Takamatsu M, Taniguchi T, Takahashi K. Breeding of a new soybean cultivar “Nanahomare”. Hokuriku Crop Sci. 2010;45:61–64. [Google Scholar]
- Yaklich RW, Vinyard B, Camp M, Douglass S. Analysis of seed protein and oil from soybean northern and southern region uniform tests. Crop Sci. 2002;42:1504–1515. doi: 10.2135/cropsci2002.1504. [DOI] [Google Scholar]
- Yang H, Wang W, He Q, Xiang S, Tian D, Zhao T, Gai J. Identifying a wild allele conferring small seed size, high protein content and low oil content using chromosome segment substitution lines in soybean. Theor Appl Genet. 2019;132:2793–2807. doi: 10.1007/s00122-019-03388-z. [DOI] [PubMed] [Google Scholar]
- Yang G, Wei Q, Huang H, Xia J. Amino acid transporters in plant cells: a brief review. Plants. 2020;9:967. doi: 10.3390/plants9080967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XP, Du LE, Wei YC. EMS-induced soybean mutation can screen germplasm resources with high protein or high fat content. Crop Var Resour. 1995;1:24–26. [Google Scholar]
- Yu K, Woodrow L, Poysa V. Registration of lipoxygenase free food grade soybean Germplasm, HS-151. Can J Plant Sci. 2016;96:148–150. doi: 10.1139/cjps-2015-0167. [DOI] [Google Scholar]
- Yu K, Woodrow L, Chun Shi M, Anderson D, Poysa V. Registration of 7S β-conglycinin α′ and 11S glycinin A3 null food-grade soybean germplasm, HS-161. Can J Plant Sci. 2016;97:377–379. [Google Scholar]
- Yu K, Woodrow L, Shi CM, Anderson D, Poysa V. Registration of 7S β-conglycinin α′ and 11S glycinin A4 null food-grade soybean germplasm, HS-162. Can J Plant Sci. 2016;97:536–538. [Google Scholar]
- Yu K, Woodrow L, Shi MC, Anderson D. Registration of HS-182 and HS-183 food-grade soybean [Glycine max (L.) Merr.] germplasm. Can J Plant Sci. 2019;99:568–571. doi: 10.1139/cjps-2018-0332. [DOI] [Google Scholar]
- Yu K, Woodrow L, Shi C. AAC Wigle soybean. Can J Plant Sci. 2019;99:985–987. doi: 10.1139/cjps-2019-0165. [DOI] [Google Scholar]
- Yuan FJ, Zhao HJ, Ren XL, Zhu SL, Fu XJ, Shu QY. Generation and characterization of two novel low phytate mutations in soybean (Glycine max L. Merr.) Theor Appl Gene. 2007;115:945–957. doi: 10.1007/s00122-007-0621-2. [DOI] [PubMed] [Google Scholar]
- Yuan FJ, JiangY ZhuSL, Li BQ, Fu XJ, Zhu DH, Dong DK, Shu QY. Mapping of MIPS1 and development of CAPS marker for low phytic acid mutation in soybean. Sci Agric Sin. 2010;43:3912–3918. [Google Scholar]
- Yun A, Kim J, Jeong HS, Lee KW, Kim HS, Kim PY, Cho SH. Inclusion effect of soybean meal, fermented soybean meal, and Saccharina japonica in extruded pellet for juvenile abalone (Haliotis discus, Reeve 1846) Fish Aquat Sci. 2018;21:1–8. doi: 10.1186/s41240-018-0104-1. [DOI] [Google Scholar]
- Zarkadas CG, Gagnon C, Poysa V, Khanizadeh S, Cober E, Chang V, Gleddie S. Protein quality and identification of the storage protein subunits of tofu and null soybean genotypes, using amino acid analysis, one-and two-dimensional gel electrophoresis, and tandem mass spectrometry. Food Res Int. 2007;40:111–128. doi: 10.1016/j.foodres.2006.08.005. [DOI] [Google Scholar]
- Zelentsov SV, Moshnenko EV, Budnikov EN, Trunova MV, Bubnova LA, Saenko GM, Lukomets AV, Ramazanova SA. High-protein soybean cultivar Greya. Oil Crops. 2020;4:91–95. doi: 10.25230/2412-608X-2020-4-184-91-95. [DOI] [Google Scholar]
- Zhang X, Shu X, Gao Y, Yang G, Li Q, Li H, Jin F. Soy food consumption is associated with lower risk of coronary heart disease in hinese women. J Nutr. 2003;133:2874–2878. doi: 10.1093/jn/133.9.2874. [DOI] [PubMed] [Google Scholar]
- Zhang GM, Zhang YQ, Shu YJ, Ma H. Screening and identification of three types of soybean lines lacking different seed storage protein subunits. Soybean Sci. 2015;34:21–31. [Google Scholar]
- Zhang YQ, Lu X, Li QT, Chen SY, Zhang JS. Recent advances in identification and functional analysis of genes responsible for soybean nutritional quality. Sci Agric Sin. 2016;49:4299–4309. [Google Scholar]
- Zhang D, Lü H, Chu S, Zhang H, Zhang H, Yang Y, Li H, Yu D. The genetic architecture of water-soluble protein content and its genetic relationship to total protein content in soybean. Sci Rep. 2017;7:1–13. doi: 10.1038/s41598-017-04685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MJ, Li ZF, Yu LL, Wang J, Qiu LJ. Identification and screening of protein subunit variation germplasm from both mutants and natural population in soybean. Crops. 2018;34:44–50. [Google Scholar]
- Zhang X, Xu RX, Hua W, Wang W, Han DZ, Zhang F, Gu YZ, Guo Y, Wang J, Qiu LJ. Involvement of sulfur assimilation in the low β subunit content of soybean seed storage protein revealed by comparative transcriptome analysis. Crop J. 2019;7:504–515. doi: 10.1016/j.cj.2019.01.001. [DOI] [Google Scholar]
- Zhang HY, Goettel W, Song QJ, Jiang H, Hu ZB, Wang ML, An YC. Selection of GmSWEET39 for oil and protein improvement in soybean. PLOS Genet. 2020;16:e1009114. doi: 10.1371/journal.pgen.1009114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RP, Gao MJ, Zhang BX, Wang JJ, Han XC, Liu XL, Wang XY, Li JR. Breeding and cultivation technology of a new high protein soybean variety Heinong 511. Soybean Sci. 2021;40:851–853. [Google Scholar]
- Zhao S, Zhang M, Jiang C, Yang C, Liu B, Cui Y. Study on quality improvement effect and separate character of soybean male sterile (ms1) recurrent selection population. S Sci Agric Sin. 2006;39:2422–2427. [Google Scholar]
- Zhao G, Zhu L, Yin P, Liu J, Pan Y, Wang S, Yang L, Ma T, Liu H, Liu X. Mechanism of interactions between soyasaponins and soybean 7S/11S proteins. Food Chem. 2021;368:130857. doi: 10.1016/j.foodchem.2021.130857. [DOI] [PubMed] [Google Scholar]
- Zhou X, Carter TE, Cui Z, Miyazaki S, Burton JW. Genetic base of Japanese soybean cultivars released during 1950 to 1988. Crop Sci. 2000;40:1794–1802. doi: 10.2135/cropsci2000.4061794x. [DOI] [Google Scholar]
- Zhou C, Yan Q, Zhuang Y, Pei Y. Characteristics and high yield cultivation techniques of summer sowing soybean variety Xudou 25. Agric Sci Technol. 2021;001:299–300. [Google Scholar]
- Zhu Z, Dong H, Bai Y, Zhou C, Zhang J, Zheng H (2008) Breeding and cultivation techniques of Xingdou No.5. Inner Mongolia Agric Sci Technol 006:71