Abstract
Background
To screen efficacious neoantigens for the development of LIHC mRNA vaccines, construct LIHC immune clusters, and therefore select patients who might benefit from vaccination.
Methods
RNA-seq data and clinical information of 371 TCGA-LIHC and 231 ICGC-LIHC cohorts were downloaded. Differentially expressed genes and their associations with prognosis were analyzed by GEPIA, genetic alterations were examined in the cBioPortal portal, and the association between genes and immune infiltrating cells was explored by TIMER. The immune clusters were constructed by consistency clustering, and the immune landscape was described using CIBERSORT.
Results
POLR3C and KPNA2 were identified as LIHC tumor neoantigens related to inferior prognosis and antigen-presenting cell infiltration. In addition, three immune clusters (IC1, IC2 and IC3) with significant differences in molecular, immune cytological, and clinical features were identified in both the TCGA and ICGC LIHC cohorts. Immune “hot” phenotype IC3 displayed a better survival than IC2, and immune “cold” phenotype IC1 exhibited a high tumor mutation burden.
Conclusion
In conclusion, for the development of anti-LIHC mRNA vaccines, we identified efficacious neoantigens POLR3C and KPNA2, profiled the tumor microenvironment of LIHC, and identified IC1 patients as the subgroup who might not most benefit from vaccination.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00432-022-04285-7.
Keywords: mRNA vaccine, Immunotype, Hepatocellular carcinoma, Tumor immune microenvironment, Immune landscape
Introduction
Liver cancer is one of the most common cancers worldwide, and its incidence is rising in Western countries (Ferlay et al. 2019). Hepatocellular carcinoma (HCC) accounts for approximately 85% of primary liver cancer and is most frequently associated with chronic hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, alcohol abuse, metabolic syndrome, or exposure to dietary toxins such as aflatoxins (Llovet et al. 2016). Locoregional treatments, including liver transplantation, resection, percutaneous ablation, transcatheter arterial chemoembolization (TACE) and radioembolization, are applied for resectable HCCs (European Association for the Study of the Liver 2018). However, the prognosis remains poor due to the strong and widespread resistance of HCC to cytotoxic chemotherapy.
In recent years, immunotherapy holds great promise to patients with HCC, particularly in unresectable tumors. Immunotherapy includes immunomodulators, immune checkpoint inhibitors (ICIs) represented by anti-programmed cell death 1 (PD-1) antibody therapy, tumor vaccines, and chimeric antigen receptor T cell (CAR-T) therapy, which specifically kill tumor cells by inhibiting the tumor-expressed extracellular ligands that suppress the intrinsic immune response (Rosenberg 2014; Disis 2014; Patel and Kurzrock 2015). Immunotherapy has proven to be effective and safe in treating a plethora of solid tumors, extending OS with a tolerable toxicity profile (Herzberg et al. 2017; Schizas et al. 2020a, b). While anti-PD-1 antibody therapy causes severe side effects in most patients and benefits a limited group of patients, CAR-T therapy has low efficacy in solid tumors (Inarrairaegui et al. 2018; Guo and Tang 2021). These limitations restrict the widespread clinical use of immunotherapy against HCC, and new therapeutic strategies are needed to overcome these barriers (Chen et al. 2021).
Currently, the major tumor vaccines used for HCC treatment and research include nucleic acid vaccines, peptide vaccines, lysovirus vaccines and DC vaccines. The antitumor functions such as overcoming tumor immunosuppression, enhancing the immunogenicity of tumor cells, and activating the immune system of patients were achieved by introducing tumor antigens into patients in various forms. More research was performed on peptide vaccines, such as the alpha-fetoprotein (AFP) peptide vaccine and glycosylated protein 3 (GPC3) peptide vaccine, that showed a survival benefit and promising antitumor activity in the treatment of HCC (Butterfield et al. 2014; Nakagawa et al. 2017). AFP is one of the most common serum markers for the diagnosis of HCC, and its high expression in HCC cells makes it a promising target for vaccine-based therapy (Liu et al. 2013). Although many tumor antigens have been discovered in HCC, only vaccines against AFP, GPC3, and resistance-associated protein 3 (MRP3) showed good tolerability and safety and their clinical potentials are limited. Oncolytic viruses induce an antitumor neoantigen-specific CTL response by directly lysing tumor cells, which leads to the release of soluble cancer antigens accompanied by the adverse influenza-like symptoms (Heo et al. 2013). Although DC vaccines can provoke Th and CTL cell reactions, they are time- and resource-consuming to develop and often ineffective in application (Bouzid et al. 2020). Moreover, there is a risk of inserting mutations into the DNA sequence that could ultimately negate the effectiveness of the therapy (Pardi et al. 2018). Messenger RNA (mRNA) cannot be integrated into the genome and is degradable by cellular RNases and therefore has a short half-life in vivo, which constitutes its safe and reliable profile (Pardi et al. 2018; Sayour et al. 2018). Additionally, the progress in sequence modification and vector strategies has improved the in vivo stability, cytosolic uptake and mRNA expression (McNamara et al. 2015; Sayour et al. 2015). mRNA sequences can be rapidly designed for clinical application, modified and transcribed in vitro to encode antigens of any pathology; these characteristics make mRNA vaccines cost-effective and efficient, which is important during pandemics or for individualized treatment (Luo et al. 2020). As a result, the messenger RNA vaccine can be easily adapted to target tumor-specific antigens, and several clinical trials have been launched (Pardi et al. 2018; Sayour et al. 2018). However, few studies identified HCC patient subpopulations suitable for vaccination as a part of their effort to develop tumor antigen-specific mRNA vaccines (Kole et al. 2020).
In this study, we aimed to identify LIHC neoantigens for the development of mRNA vaccines and to determine the LIHC immune profile to facilitate the selection of appropriate patients for vaccination. Two overexpressed and mutated genes associated with inferior survival and favorable antigen presenting cell (APC) infiltration status were identified in LIHC. Meanwhile, three powerful immune clusters and eight functional modules were carefully established by clustering immune-associated genes in the TCGA cohort, which were validated in an independent ICGC cohort. The immune clusters correspond to different clinical, molecular and immune cellular profiles.
Subsequently, based on the distribution of correlative genetic features, the LIHC immune landscape was defined in individuals. The results suggest that the construction of a complex tumor immune microenvironment (TIME) for each LIHC patient can help provide a theoretical basis for the development of mRNA vaccines and the screening of appropriate patients for vaccination.
Materials and methods
Data sources and pre-processing
The normalized RNA sequencing (RNA-seq) data and corresponding clinical information of 371 HCC patients and 50 normal samples were downloaded from the TCGA portal (https://portal.gdc.cancer.gov/repository), and another 231 HCC patient data were obtained from the ICGC portal (https://dcc.icgc.org/projects/LIRI-JP).
A total of 2917 immune-related genes were extracted by combining immune signature genes from five sources (leukocyte signature matrix 22 or LM22 (Newman et al. 2015), leukocyte signature matrix 7 or LM7 (Tosolini et al. 2017), 11 immune cell gene signatures or ImSig (Nirmal et al. 2018), the NanoString immune signature panel (https://www.nanostring.com), and ImmPort (https://www.immport.org/shared/home) and listed in Supplementary Table S1. Ninety-four immune checkpoint genes (ICGs) (Supplementary Table S2), 26 immunogenic cell death (ICD) genes (Supplementary Table S3), and 28 immune cell genes were identified by reviewing the literature (Afrache et al. 2012; Pardoll 2012; Hu et al. 2021; Campbell and Purdy 2011; Charoentong et al. 2017).
Exploration of the neoantigens
Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia2.cancer-pku.cn, version 2) was applied to integrate differential gene expression (|log2FC|> 1 and q-values < 0.01) and patient survival data using ANOVA (Tang et al. 2019). The Kaplan–Meier curve analysis was performed to investigate overall survival (OS) and relapse-free survival (RFS) with the median cutoff values, and the p value was obtained using the log rank test. The hazard ratio was calculated using the Cox proportional hazards regression model, and p values < 0.05 were considered statistically significant. The raw expression data from TCGA, ICGC and other databases were combined, and the genetic alterations in TCGA LIHC project (TCGA, PanCancer Atlas) were compared using cBio Cancer Genomics Portal (cBioPortal, http://www.cbioportal.org). The association between the abundance of tumor immune infiltrating cells (TIICs) was analyzed and visualized using the Tumor Immune Estimation Resource (Afrache et al. 2012) (TIMER, https://cistrome.shinyapps.io/timer/).
Identification and validation of the immune clusters
The gene expression data were clustered based on the expression levels of 2716 immune-related genes, and a consistency matrix was constructed to evaluate relevant immune clusters using the R package ‘ConsensusClusterPlus’ and gene modules using the R package “WGCNA”. The agglomerative hierarchical clustering algorithm was applied for the consensus matrix with 80% item resampling (pItem), 100% gene resampling (pFeature), k values varying from 2 to 6, and 1000 resamplings (reps). The optimal k value was defined by the consistency cumulative distribution function (CDF) and delta area. The immune clustering of the ICGC cohort were performed with the same settings. The prognostic values of immune clusters for OS were estimated using univariate and multivariate Cox regression. The KEGG functional annotation of each gene module was obtained by the DAVID program.
Statistical analysis
Statistical analysis was performed using R (https://www.r-project.org/). The categorical variables were analyzed by Fisher’s exact test and visualized by R package “vcd”. The Mann Whitney test was applied for continuous variables between two groups, and the he Kruskal–Wallis test and post hoc using Dunn’s multiple comparisons test to assess significant differences when comparing more than two groups. Kaplan–Meier curves and the log-rank test were applied for survival analysis by R package “survival”. A two-tailed p value < 0.05 was regarded as statistically significant.
Results
Identification of LIHC efficacious neoantigens
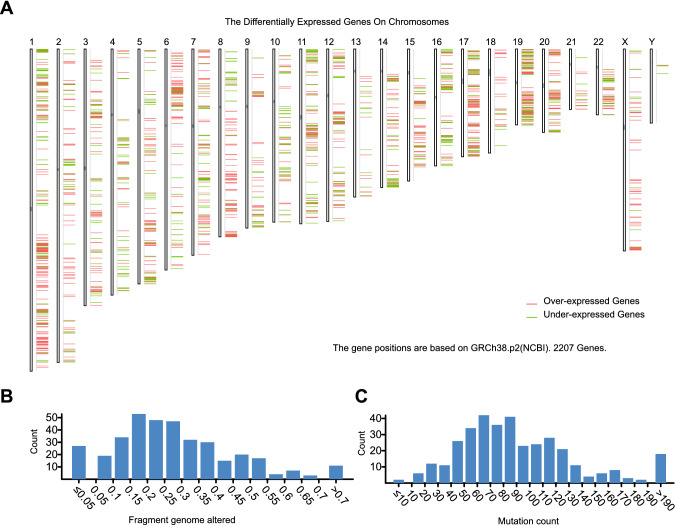
To identify effective neoantigens for LIHC, 1482 differentially overexpressed genes (Fig. 1A) (Supplementary Table S4) and 8014 mutant genes potentially encoding tumor-associated neoantigens (Fig. 1B and C) were identified. Analysis of the mutation profiles suggested that, mutations were more likely to be responsible for encoding tumor-associated antigens than genome structural variants in HCC (Supplementary Table S5), due to the altered genome fraction was rare (Supplementary Table S6). A total of 344 genes that were overexpressed and frequently mutated were identified, of which TP53, CTNNB1 and MUC16 were the most frequently mutated genes according to mutation counts (Supplementary Table S5).
Fig. 1.
Identification of potential tumor antigens of LIHC. A Identification of potential tumor-associated neoantigens of LIHC. Chromosomal distribution of up- and downregulated genes in LIHC is presented. The histogram shows the interval distribution of the samples in the B altered genomic fraction and C mutation count
Assessment of tumor neoantigens related to LIHC prognosis and APCs
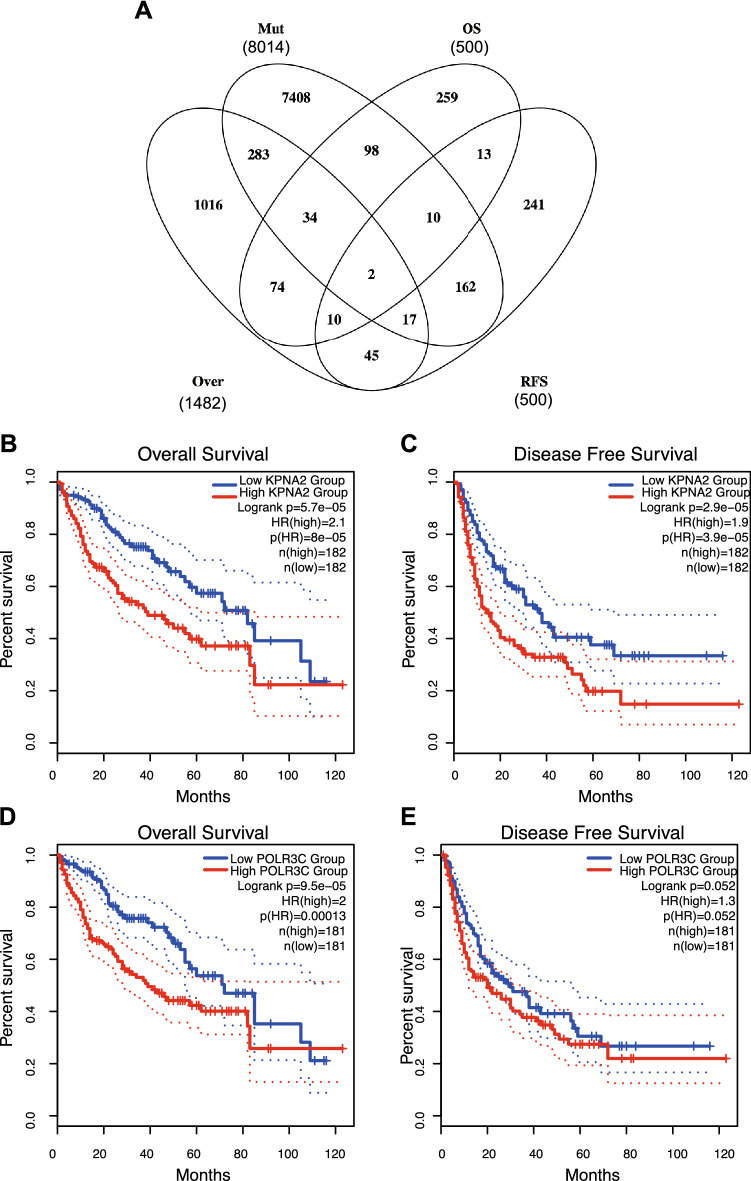
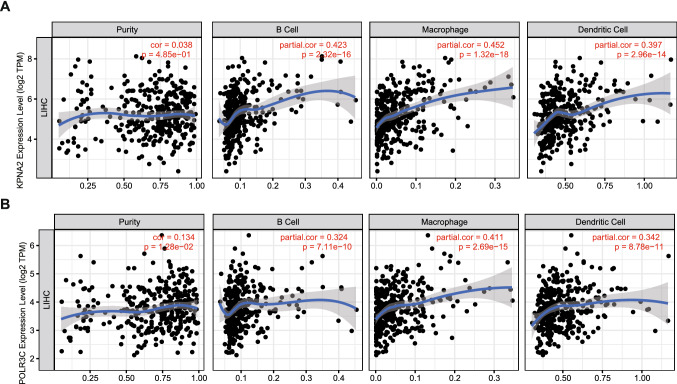
Next, the prognosis-associated tumor-specific neoantigens were identified from the above genes as potential targets for mRNA vaccine development. Thirty-eight genes were significantly related to the OS of LIHC patients, of which two genes showed a close association with RFS (Fig. 2A). LIHC patients with KPNA2 overexpression in primary tumor tissues had significantly worse survival than those in the KPNA2low group, as shown in Fig. 2B and C. Meanwhile, patients with tumors expressing high levels of POLR3C were also associated with inferior prognosis (Fig. 2D and E). Overall, 2 candidate genes were considered to be essential for LIHC development and progression. Moreover, higher expression levels of KPNA2 and POLR3C were significantly related to increased infiltration of B cells, macrophages, and DCs into the tumor (Fig. 3A and B). The results indicate that prognosis-related neoantigens can be directly processed and presented to T cells by APCs and then recognized by B cells to initiate an immune response, and these antigens can be used for the development of mRNA vaccines against LIHC.
Fig. 2.
Identification of LIHC prognosis-related tumor neoantigens. A Venn diagram showing the candidate neoantigens associated with high expression, mutations, OS and RFS (2 candidates in total). B–E Kaplan–Meier curves showing the OS of LIHC patients stratified on the basis of B POLR3C and D KPNA2 expression levels and RFS according to C POLR3C and E KPNA2 expression levels
Fig. 3.
Identification of tumor antigens associated with APCs. Correlation between the expression levels of A KPNA2 and B POLR3C and the infiltration of macrophages, dendritic cells and B cells in LIHC tumors
Discrimination of potential LIHC immune clusters
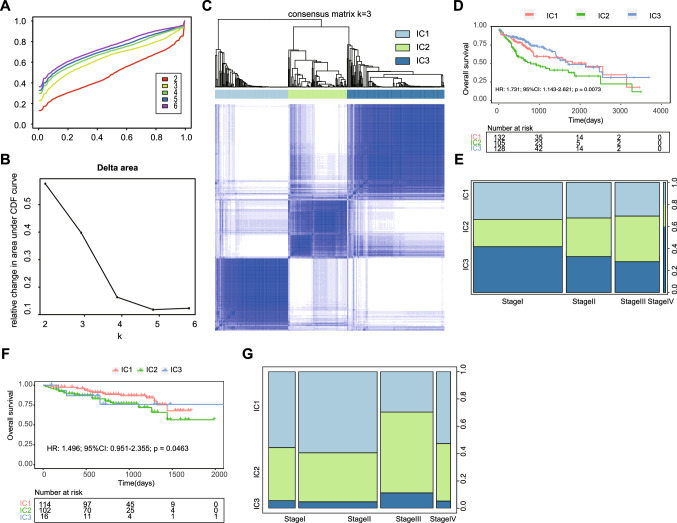
Immunotypes could be used to represent the immune status of the tumors and their microenvironment, thus helping to identify patients who might benefit from mRNA vaccination. Therefore, we analyzed the expression profiles of 2917 immune-associated genes in 377 LIHC samples from the TCGA database and constructed consensus clusters. We then selected immune-associated genes present in stable clusters with k = 3 (Fig. 4A and B) and constructed 3 immune clusters, IC1, IC2 and IC3 (Fig. 4C), based on their cumulative distribution functions and functional delta areas. IC2 was associated with the worst prognosis, while IC1 and IC3 had better survival probabilities (HR 1.731; 95% CI 1.143–2.621; log-rank p = 0.0073) (Fig. 4D). The cluster distribution at different tumor stages showed that patients diagnosed at differential stages were irregularly clustered (Fig. 4E). Similar to the results obtained in the TCGA cohort, immune subtypes were also prognostically relevant (HR 1.496; 95% CI 0.951–2.355; log-rank p = 0.0463) in the ICGC cohort (Fig. 4F) and significantly altered at different stages (Fig. 4G). In conclusion, immunophenotyping can be used to predict the prognosis of patients with LIHC in different cohorts.
Fig. 4.
Identification of potential immune clusters of LIHC. A Cumulative distribution function curve and B delta area of immune-related genes in the TCGA cohort. C Sample clustering heatmap. D Kaplan–Meier curves showing the OS of LIHC immune clusters in the TCGA cohort. E Distribution of IC1-IC3 across LIHC stages in the TCGA cohort. F Kaplan–Meier curves showing the OS of LIHC immune clusters in the ICGC cohort. G Distribution ratio of IC1-IC3 across LIHC stages in the ICGC cohort
The mutational characteristics of LIHC immune clusters
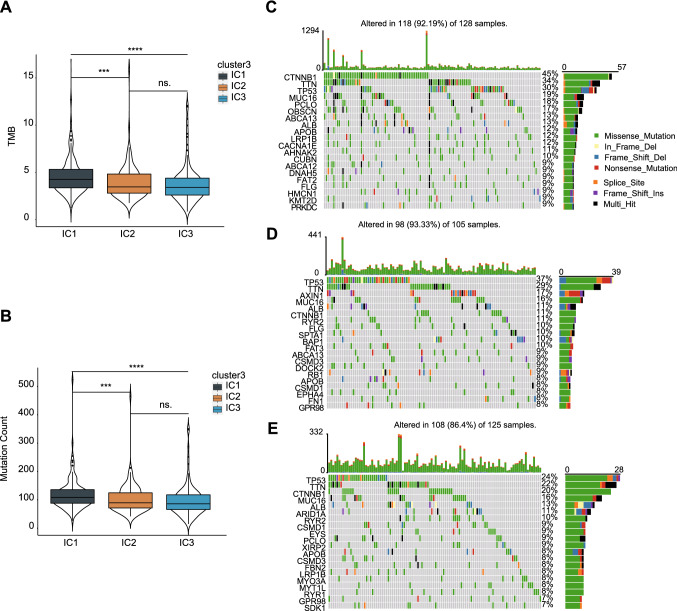
Tumor mutation burden (TMB) and somatic mutation counts are positively correlated with anticancer immunity (Rooney et al. 2015). Therefore, to explore the antitumor immunity of immune clusters, we calculated the TMB and mutation counts for each LIHC patient in the TCGA cohort. IC1 showed a significantly higher TMB than IC2 and IC3 (Kruskal–Wallis test, p < 0.0001) (Dunn’s multiple comparisons test, IC1 vs. IC2, p = 0.001; IC1 vs. IC3, p < 0.0001; IC2 vs. IC3, p > 0.9999), as shown in Fig. 5A. Similar trends were observed for the mutation count (Kruskal–Wallis test, p < 0.0001) (Dunn’s multiple comparisons test, IC1 vs. IC2, p = 0.002; IC1 vs. IC3, p < 0.0001; IC2 vs. IC3, p > 0.9999) (Fig. 5B). Furthermore, 20 genes, including TP53, CTNNB1 and MUC16, were most frequently mutated in all clusters (Fig. 5C–E). These findings suggest that the immune cluster can predict TMB and somatic mutation counts in LIHC patients and that patients in IC1 may respond positively to the mRNA vaccine.
Fig. 5.
Association between immune clusters and TMB and mutation count. A TMB and B mutation count in LIHC clusters IC1-IC3. C–E Twenty highly mutated genes in immune clusters C IC1, D IC2 and E IC3. *p < 0.05, **p < 0.01, ***p < 0.001
The immune regulators in LIHC immune clusters
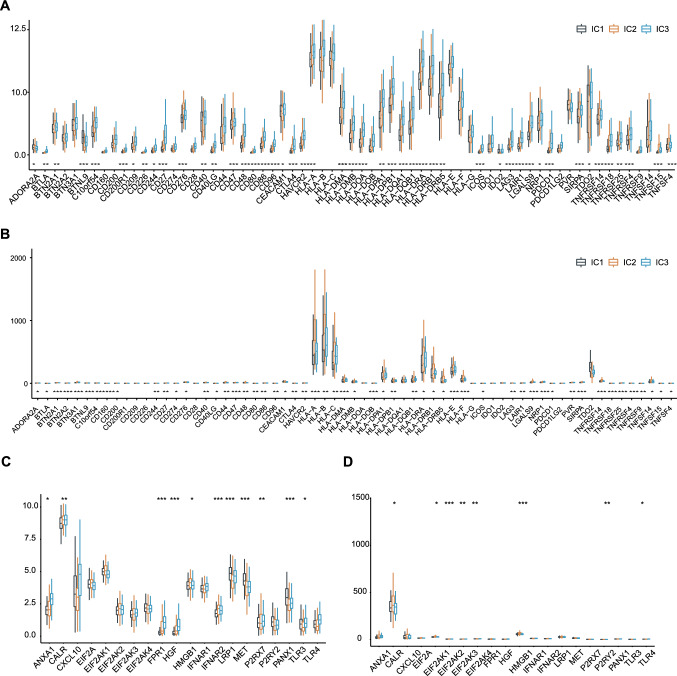
The expression levels of immune checkpoint (ICP) and immunogenic cell death (ICD) regulators in each immune cluster were analyzed due to their importance in cancer immunity. Ninety-four ICP-related genes were detected in both TCGA and ICGC cohorts, and out of these 94 genes, the expressed genes of 81 (87.23%) in the TCGA cohort (Fig. 6A) and 55 (58.51%) in the ICGC cohort (Fig. 6B) were significantly altered among the immune clusters. For example, BTLA, C10orf54, CD244, CD27, CD276, CD40LG, CD47, CD48, CD80, CD86, CD96, CTLA4, HAVCR2, HLA-A, HLA-DMA, HLA-DMB, HLA-DPA1, HLA-DPB1, HLA-DQB1, HLA-DRA, HLA-DRB1, HLA-DRB5, ICOS, LAG3, LAIR1, LGALS9, NRP1, PDCD1, SIRPA, TNFRSF14, TNFRSF18, TNFRSF25, TNFRSF4, TNFSF15, and TNFSF4 were significantly upregulated in IC2 tumors in both the TCGA and ICGC cohorts, while CEACAM1 and TDO2 were overexpressed in IC1 tumors in both the TCGA and ICGC cohorts. Among the 26 ICD genes, 20 were detected in the ICGC cohort and the expression of 8 (40%) gene showed significantly altered expression in the immune clusters (Fig. 6D). Accordingly, 26 ICD genes were detected in the TCGA cohort, of which 11 (42.3%) exhibited significant differences across the clusters (Fig. 6C). For example, CALR, EIF2A, EIF2AK1, EIF2AK2, EIF2AK3, HMGB1, and P2RY2 were statistically upregulated in IC2 tumors in the ICGC cohort, while ANXA1, CALR, FPR1, HGF, HMGB1, and IFNAR2 high expressed in IC2 tumors in the TCGA cohort. These findings suggested that immune clusters can reflect the expression levels of ICP and ICD regulators and therefore be used to predict response to mRNA vaccines.
Fig. 6.
Association between immune clusters and ICPs and ICD regulators. A, B Differential expression of ICP genes among the LIHC immune clusters in the A TCGA and B ICGC cohorts. C, D Differential expression of ICD regulators genes among the LIHC immune clusters in the C TCGA and D ICGC cohorts. *p < 0.05, **p < 0.01, ***p < 0.001
The serological biomarkers in LIHC immune clusters
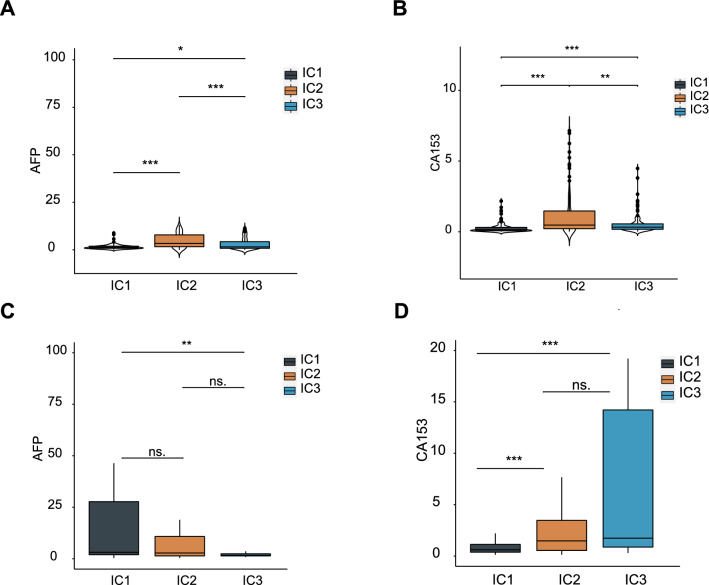
AFP and CA153 are diagnostic and prognostic serological biomarkers of LIHC, and higher levels of both indicate vigorous cancer progression, adverse prognosis or relapse. In this study, levels of both AFP and CA153 in different immune clusters showed significant differences in both ICGC and TCGA cohorts. For instance, IC2 showed higher AFP and CA153 levels (Fig. 7A and B), whereas IC1 and IC3 showed moderate AFP and CA153 levels in the TCGA cohort, respectively. Importantly, these results were consistent with the better prognosis observed in IC1 and IC3 patients. In addition, frequencies of differential levels of AFP and CA153, i.e., patients expressing AFP but not CA153 (Fig. 7C and D), were relatively consistent in both groups, suggesting that CA153 levels were highly susceptible to biological or environmental factors.
Fig. 7.
Association between immune clusters and AFP and CA153. A, B AFP (A) and CA153 (B) expression in LIHC immune clusters in TCGA cohorts. C, D AFP (C) and CA153 (D) expression in LIHC immune clusters in ICGC cohorts
The biological properties of LIHC immune clusters
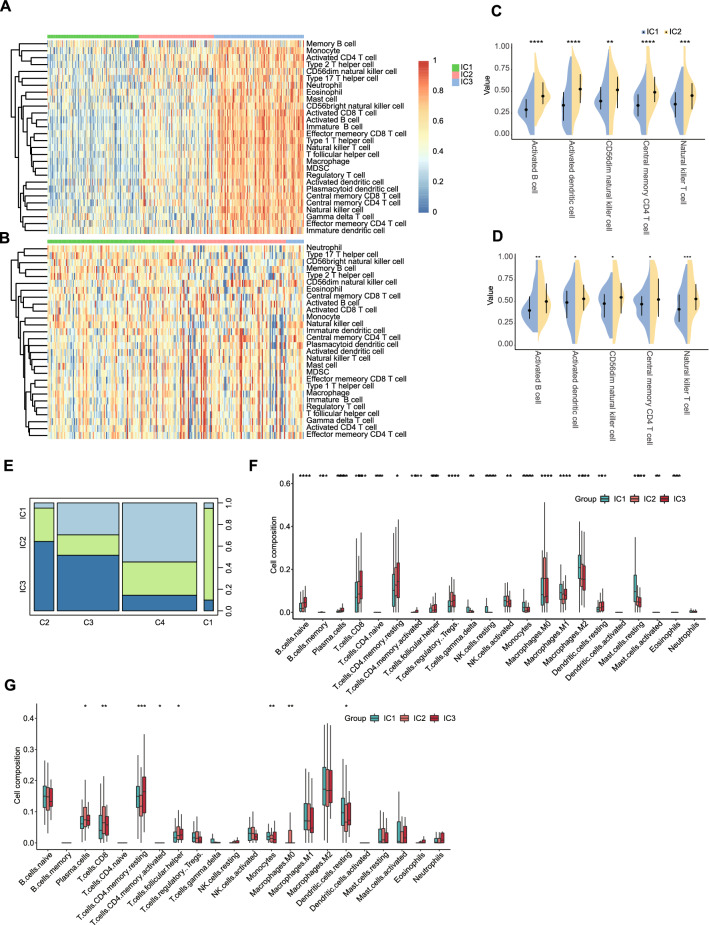
The tumor immune status of an individual determines the response to mRNA vaccines. Therefore, to characterize the immune cell components in the three immune clusters, we performed ssGSEA scoring of 28 previously reported signature genes in the TCGA and ICGC cohorts. As shown in Fig. 8A, the immune cell components varied among IC1, IC2 and IC3, and the immune cell components in IC1 were overall lower than those in IC2 and IC3 in the TCGA cohort. Meanwhile, in the ICGC cohort, IC2 and IC3 showed similar immune cell components, and the immune cell composition of the IC1 cluster was quite different (Fig. 8B). Based on the above findings, we analyzed the immune cell components of IC1 and IC2 patients. Shown in Fig. 8C and D are immune cells that differed in both TCGA and ICGC cohorts such as activated B cells, activated dendritic cells, CD56dim natural killer cells, central memory CD4 T cells, and natural killer T cells.
Fig. 8.
Cellular and molecular characteristics of immune clusters. A Differential enrichment scores of 28 immune cell signatures among LIHC immune clusters in the A TCGA and B ICGC cohorts. C, D Common differential immune cell signatures in the C TCGA and D ICGC cohorts. E Overlap of LIHC immune clusters with 6 pan-cancer immune clusters. F, G Differential enrichment scores of 22 immune signatures among LIHC immune clusters in TCGA (F) and ICGC (G) cohorts. *p < 0.05, **p < 0.01, ***p < 0.001
To demonstrate the reliability of this immunophenotype, we first explored the relationship between three immune clusters and six previously reported pan-cancer immune clusters (C1–C6), where LIHC was mainly clustered into C4 (Thorsson et al. 2018). As shown in Fig. 8E, IC1 and IC2 mainly overlap with C4, and IC3 overlaps with C3. C3 is associated with better prognosis. Thus, IC3 was associated with an overall favorable immune-activated phenotype and characterized by a highly diverse immune signature. These results were consistent with the relatively favorable OS of patients in IC3 compared to those in the IC2 cluster. In contrast, IC1 patients with better prognosis and IC2 patients with the poorest survival largely overlapped with C3. The finding not only demonstrated the reliability of our immunotyping method but also argued for previous classification. In addition, we also speculate that it is possible that the favorable OS of patients in IC1 may be due to the tumor mutational load rather than the immune microenvironment. Additionally, we also applied CIBERSORT to quantify the immune infiltration into tumors, and a similar trend in the levels of immune cell infiltration was seen in both the TCGA and ICGC cohorts (Fig. 8F and G). The infiltration and activation of T cells and other antitumor immune cells in immune clusters, as well as the suppression of immunosuppressive cells, largely determine the therapeutic potential of mRNA vaccines. Therefore, IC2 and IC3 were immunologically “hot” while IC1 was immunologically “cold” phenotype. These results suggest that the immune clusters reflect the immune status of LIHC and allow the identification of patients suitable for mRNA vaccination. In addition, patients in immunologically “hot” IC2 and IC3 clusters exhibited an immunosuppressive microenvironment that might potentially benefit from mRNA vaccination.
The LIHC immune gene co-expression modules and hub genes
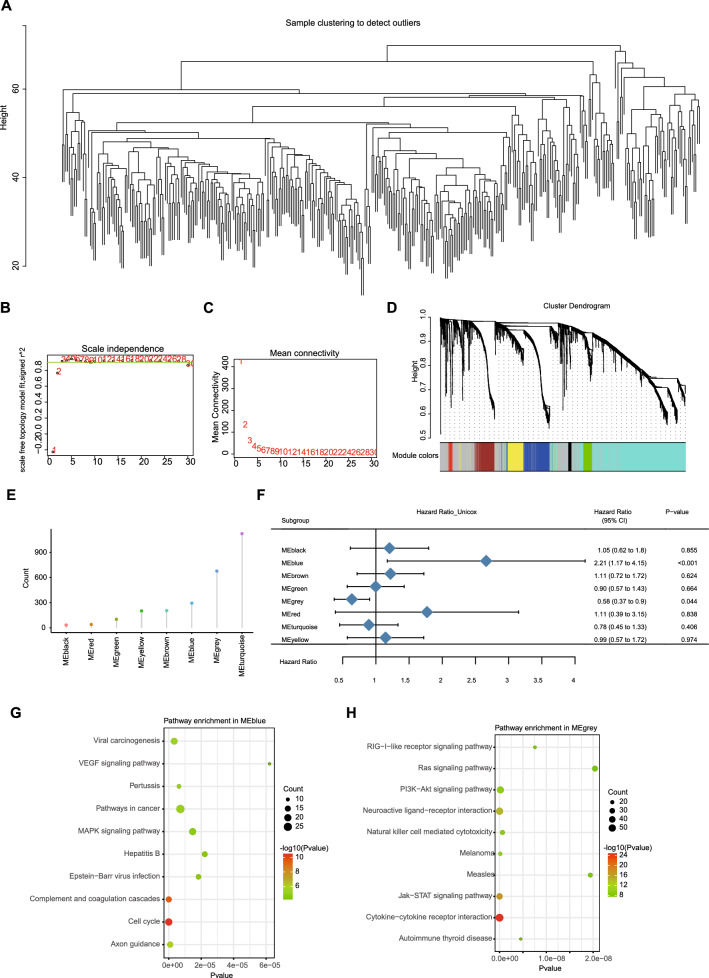
Samples were clustered using WGCNA (Fig. 9A) and a soft threshold of 3 was defined for the scale-free network (Fig. 9B and C) to deploy the LIHC immune gene co-expression module. The expression matrix was then converted to a topology matrix after conversion to an adjacency matrix. An average linkage hierarchy clustering approach with at least 30 genes was applied to each network according to the standard hybrid dynamic shear tree criteria. For each module, Eigengenes were computed and nearby modules were combined into a one module with height = 0.25, depth split = 4, and minimum module size = 30. Eight co-expression modules with 2716 transcripts were obtained (Fig. 9D), and the count of highly related genes in the eight modules were further analyzed (Fig. 9E) (Supplementary Table S7). The MEblue and MEgrey modules were strongly related to prognosis of LIHC, as shown by prognostic correlation analysis (Fig. 9F).
Fig. 9.
Identification of immune gene co-expression modules of LIHC. A Sample clustering. B Scale-free fit index for various soft-thresholding powers (β). C Mean connectivity for various soft-thresholding powers. D Dendrogram of all differentially expressed genes clustered based on a dissimilarity measure (1-TOM). E Gene counts in each module. F Forest maps of single factor survival analysis of 8 modules of LIHC. G, H Dot plot showing the top 10 KEGG terms in the MEblue module (G) and the MEgrey module (H). The dot size and color intensity represent the gene count and enrichment level, respectively
Furthermore, genes in the MEblue module were enriched in cancer pathways, but genes in the MEgrey module were enriched in KEGG pathways related to immunity, such as cytokine − cytokine receptor interaction, Jak − STAT signaling pathway, and neuroactive ligand − receptor interaction (Fig. 9G and H). The analysis of module-relevant genes of the MEblue module was consistent with the above findings that patients in the MEblue module had a worse prognosis. This makes mRNA vaccines potentially applicable to patients with highly expressed genes clustered into the MEblue module rather than the MEgrey module. Therefore, five hub genes, including TOP2A, CDK1, CENPF, CCNB2 and MCM2, were identified as potential for mRNA vaccine development with > 90% correlation in the MEblue module.
Discussion
This is the first study, to our knowledge, to screen LIHC antigens for mRNA vaccine development. An atlas of differentially expressed and mutated genes in LIHC was constructed and a range of novel antigens with prognostic relevance were identified, among which POLR3C and KPNA2 were promising mRNA vaccine candidates. Their upregulation was positively correlated with inferior prognosis and high infiltration of B cell, macrophage cell and dendritic cell. It follows that these neoantigens have a crucial function in the evolution and advancement of LIHC, and can be immediately processed and presented to CD8 + T cells in the setting of sufficient lymphocyte infiltration for triggering immune attack. While the function of these neoantigens remains to be validated, their potency as targets for mRNA vaccine development is bolstered by previous studies. For example, KPNA2 is an adaptor protein in the classical nuclear protein import machinery that mediates the import of intranuclear signaling factors and the export of response molecules to the cytoplasm (Leung et al. 2003). More specifically, KPNA2 is a major adapter for the nuclear localization of NBS1, which is a key regulator of the MRE11/RAD50/NBS1 complex (Tseng et al. 2005). The complex plays an important role in the early processing of double-strand breaks (DSBs). NBS1 has two important roles according to its subcellular localization, with KPNA2 being the major determinant of the subcellular localization of NBS1. In the nucleus, NBS1 acts as a tumor suppressor involved in DNA DSB repair and maintains genomic stability. However, in the cytoplasm, NBS1 plays an oncogenic role by binding and activating the phosphatidylinositol 3-kinase/AKT pathway to promote tumorigenesis (Teng et al. 2006). KPNA2 is overexpressed in most advanced breast cancers and other tumors with poor survival (Dressman et al. 2006), making KPNA2 a potential diagnostic marker. Sortiriou et al. improved the accuracy of tissue tumor grading by reclassifying grade 2 tumors into two groups of high and low risks of recurrence based on KPNA2 gene expression, attesting to its prognostic value. In prostate cancer, nuclear KPNA2 overexpression was significantly associated with shorter OS and RFS, as shown by immunohistochemical analysis. In several independent study cohorts, nuclear KPNA2 expression was a very important, independent, and unfavorable risk factor for OS (Dahl et al. 2006; Dankof et al. 2007; Gluz et al. 2008). POLR3C is a subunit of RNA polymerase III, which is involved in cancer progression through ribosomal RNA (rRNA) and transfer RNA (tRNA) biosynthesis (Vannini and Cramer 2012).
Furthermore, the effectiveness of the vaccine would be successfully improved when patients with high levels of intrinsic immune resistance are identified. Therefore, considering the limited number of known tumor antigens and paucity of information on the immune environment of HCC, there is an urgent need to identify more meaningful tumor-specific antigens and immune clusters. To distinguish those most likely to benefit from mRNA vaccination, three immune clusters with distinct molecular, cytological and clinical profiles, were divided from LIHC patients based on immune gene expression profiles. In the TCGA and ICGC cohorts, patients in the IC1 and IC3 clusters displayed a better prognosis than those in the IC2 cluster. It indicated that the prognosis of LIHC patients could be predicted by immunophenotyping, and its reliability was proved by traditional predictive markers such as AFP, CA153 and clinical staging. Additionally, immunophenotypes can also predict individual responses to mRNA vaccines. For example, patients classified in highly immuno-infiltrated IC3 cohort probably responsed well to mRNA vaccines. In the TCGA and ICGC cohorts, high expression of ICP in the IC2 cluster indicated an immunosuppressive tumor microenvironment of IC2, which might be suppressed by the effective immune response triggered by the mRNA vaccine. While the IC3 cluster had elevated ICD regulator expression in both TCGA and ICGC cohorts, indicating a higher immune response to mRNA vaccines. Additionally, the heterogeneous immune profile of individual patients within the immune cluster suggests the necessary development of personalized mRNA vaccines, to reduce the impact of immune components. Hub genes as identified in the MEblue module, including TOP2A, CDK1, CENPF, CCNB2 and MCM2, were potential biomarkers for personalized mRNA vaccines.
To further investigate the influence of mRNA vaccine efficacy, we explored the immune cell composition in tumor immune clusters. IC2 and IC3, which were immunologically “hot” phenotypes, had significantly higher scores for activated B cells, activated dendritic cells, CD56dim natural killer cells, central memory CD4 T cells, and natural killer T cells compared to IC1, which was immunologically “cold” phenotype. Consistent molecular features and immune patterns further demonstrated that patients at various immune clusters had different degrees of immune reactions to mRNA vaccines. Pan-cancers were classified into C1-C6 clusters according to previous immunophenotyping studies. Most LIHC samples were grouped into the C4 cluster. C3 was related to superior prognosis, C1 and C2 to intermediate prognosis, and C6 to inferior prognosis. In this study, LIHC patients were categorized into IC1-3 clusters. IC1 and IC2 overlapped mainly with C4, whereas IC3 overlapped with C3. This was consistent with the superior prognosis of IC3 and the inferior prognosis of IC2, which validated the reliability of our immunotyping approach. Although the potential neoantigens screened in this study should be validated in future studies.
Conclusions
In this study, for the development of an mRNA vaccine, we identified potential LIHC neoantigens POLR3C and KPNA2, constructed tumor immunophenotyping, and found that patients in immune cluster IC3 had the potential to benefit from vaccination. This provides a theoretical basis for the development of anti-LIHC mRNA vaccines, prognostic prediction, and selection of suitable patients for vaccination.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
YFL, QQH and LL contributed to the study conception and design. Material preparation, data collection and analysis were performed by YFL, QQH, SZ, XYC, and MT. The first draft of the manuscript was written by YFL and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by CAMS Innovation Fund for Medical Sciences (2021-I2M-1-012), National High Level Hospital Clinical Research Funding (BJ-2022-151) and the Fundamental Research Funds for the Central Universities (3332019120).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaoyan Chen, Email: chenxiaoyan0206@163.com.
Min Tang, Email: tangmin3911@bjhmoh.cn.
Lin Li, Email: lilin_51@hotmail.com.
References
- Afrache H, Gouret P, Ainouche S, Pontarotti P, Olive D. The butyrophilin (BTN) gene family: from milk fat to the regulation of the immune response. Immunogenetics. 2012;64:781–794. doi: 10.1007/s00251-012-0619-z. [DOI] [PubMed] [Google Scholar]
- Bouzid R, Peppelenbosch M, Buschow SI. Opportunities for conventional and in situ cancer vaccine strategies and combination with immunotherapy for gastrointestinal cancers, a review. Cancers. 2020;12:1121. doi: 10.3390/cancers12051121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield LH, Economou JS, Clark Gamblin T, Geller DA. Alpha fetoprotein DNA prime and adenovirus boost immunization of two hepatocellular cancer patients. J Transl Med. 2014;12:86. doi: 10.1186/1479-5876-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology. 2011;132:315–325. doi: 10.1111/j.1365-2567.2010.03398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoentong P, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- Chen P, Fang QX, Chen DB, Chen HS. Neoantigen vaccine: an emerging immunotherapy for hepatocellular carcinoma. World J Gastrointest Oncol. 2021;13:673–683. doi: 10.4251/wjgo.v13.i7.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl E, et al. Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin alpha2 as a potential novel prognostic marker in breast cancer. Clin Cancer Res. 2006;12:3950–3960. doi: 10.1158/1078-0432.CCR-05-2090. [DOI] [PubMed] [Google Scholar]
- Dankof A, et al. KPNA2 protein expression in invasive breast carcinoma and matched peritumoral ductal carcinoma in situ. Virchows Arch. 2007;451:877–881. doi: 10.1007/s00428-007-0513-5. [DOI] [PubMed] [Google Scholar]
- Disis ML. Mechanism of action of immunotherapy. Semin Oncol. 2014;41(Suppl 5):3–13. doi: 10.1053/j.seminoncol.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Dressman HK, et al. Gene expression profiles of multiple breast cancer phenotypes and response to neoadjuvant chemotherapy. Clin Cancer Res. 2006;12:819–826. doi: 10.1158/1078-0432.CCR-05-1447. [DOI] [PubMed] [Google Scholar]
- European Association for the Study of the Liver Electronic address, e. e. e. & European Association for the Study of the, L. EASL Clinical Practice Guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- Ferlay J, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- Gluz O, et al. Nuclear karyopherin alpha2 expression predicts poor survival in patients with advanced breast cancer irrespective of treatment intensity. Int J Cancer. 2008;123:1433–1438. doi: 10.1002/ijc.23628. [DOI] [PubMed] [Google Scholar]
- Guo J, Tang Q. Recent updates on chimeric antigen receptor T cell therapy for hepatocellular carcinoma. Cancer Gene Ther. 2021 doi: 10.1038/s41417-020-00259-4. [DOI] [PubMed] [Google Scholar]
- Heo J, et al. Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat Med. 2013;19:329–336. doi: 10.1038/nm.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg B, Campo MJ, Gainor JF. Immune checkpoint inhibitors in non-small cell lung cancer. Oncologist. 2017;22:81–88. doi: 10.1634/theoncologist.2016-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu FF, Liu CJ, Liu LL, Zhang Q, Guo AY. Expression profile of immune checkpoint genes and their roles in predicting immunotherapy response. Brief Bioinform. 2021 doi: 10.1093/bib/bbaa176. [DOI] [PubMed] [Google Scholar]
- Inarrairaegui M, Melero I, Sangro B. Immunotherapy of hepatocellular carcinoma: facts and hopes. Clin Cancer Res. 2018;24:1518–1524. doi: 10.1158/1078-0432.CCR-17-0289. [DOI] [PubMed] [Google Scholar]
- Kole C, et al. Immunotherapy for hepatocellular carcinoma: a 2021 update. Cancers. 2020;12:2859. doi: 10.3390/cancers12102859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung SW, Harreman MT, Hodel MR, Hodel AE, Corbett AH. Dissection of the karyopherin alpha nuclear localization signal (NLS)-binding groove: functional requirements for NLS binding. J Biol Chem. 2003;278:41947–41953. doi: 10.1074/jbc.M307162200. [DOI] [PubMed] [Google Scholar]
- Liu C, et al. Value of alpha-fetoprotein in association with clinicopathological features of hepatocellular carcinoma. World J Gastroenterol. 2013;19:1811–1819. doi: 10.3748/wjg.v19.i11.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;14:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- Luo W, et al. Novel therapeutic strategies and perspectives for metastatic pancreatic cancer: vaccine therapy is more than just a theory. Cancer Cell Int. 2020;20:66. doi: 10.1186/s12935-020-1147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara MA, Nair SK, Holl EK. RNA-based vaccines in cancer immunotherapy. J Immunol Res. 2015;2015:794528. doi: 10.1155/2015/794528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, et al. Association between high-avidity T-cell receptors, induced by alpha-fetoprotein-derived peptides, and anti-tumor effects in patients with hepatocellular carcinoma. Gastroenterology. 2017;152:1395–1406 e1310. doi: 10.1053/j.gastro.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirmal AJ, et al. Immune cell gene signatures for profiling the microenvironment of solid tumors. Cancer Immunol Res. 2018;6:1388–1400. doi: 10.1158/2326-6066.CIR-18-0342. [DOI] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SA. Decade in review-cancer immunotherapy: entering the mainstream of cancer treatment. Nat Rev Clin Oncol. 2014;11:630–632. doi: 10.1038/nrclinonc.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayour EJ, Sanchez-Perez L, Flores C, Mitchell DA. Bridging infectious disease vaccines with cancer immunotherapy: a role for targeted RNA based immunotherapeutics. J Immunother Cancer. 2015;3:13. doi: 10.1186/s40425-015-0058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayour EJ, Mendez-Gomez HR, Mitchell DA. Cancer vaccine immunotherapy with RNA-loaded liposomes. Int J Mol Sci. 2018;19:2890. doi: 10.3390/ijms19102890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizas D, Charalampakis N, Kole C, Mylonas KS, Katsaros I, Zhao M, Ajani JA. Immunotherapy for esophageal cancer: a 2019 update. Immunotherapy. 2020;12:203–218. doi: 10.2217/imt-2019-0153. [DOI] [PubMed] [Google Scholar]
- Schizas D, et al. Immunotherapy for pancreatic cancer: a 2020 update. Cancer Treat Rev. 2020;86:102016. doi: 10.1016/j.ctrv.2020.102016. [DOI] [PubMed] [Google Scholar]
- Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:556–560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Wu KJ, Tseng SF, Wong CW, Kao L. Importin KPNA2, NBS1, DNA repair and tumorigenesis. J Mol Histol. 2006;37:293–299. doi: 10.1007/s10735-006-9032-y. [DOI] [PubMed] [Google Scholar]
- Thorsson V, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosolini M, et al. Assessment of tumor-infiltrating TCRVgamma9Vdelta2 gammadelta lymphocyte abundance by deconvolution of human cancers microarrays. Oncoimmunology. 2017;6:e1284723. doi: 10.1080/2162402X.2017.1284723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SF, Chang CY, Wu KJ, Teng SC. Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J Biol Chem. 2005;280:39594–39600. doi: 10.1074/jbc.M508425200. [DOI] [PubMed] [Google Scholar]
- Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.