Abstract
Single-strand selective uracil–DNA glycosylase 1 (SMUG1) initiates base excision repair (BER) of uracil and oxidized pyrimidines. SMUG1 status has been associated with cancer risk and therapeutic response in breast carcinomas and other cancer types. However, SMUG1 is a multifunctional protein involved, not only, in BER but also in RNA quality control, and its function in cancer cells is unclear. Here we identify several novel SMUG1 interaction partners that functions in many biological processes relevant for cancer development and treatment response. Based on this, we hypothesized that the dominating function of SMUG1 in cancer might be ascribed to functions other than BER. We define a bad prognosis signature for SMUG1 by mapping out the SMUG1 interaction network and found that high expression of genes in the bad prognosis network correlated with lower survival probability in ER+ breast cancer. Interestingly, we identified hsa-let-7b-5p microRNA as an upstream regulator of the SMUG1 interactome. Expression of SMUG1 and hsa-let-7b-5p were negatively correlated in breast cancer and we found an inhibitory auto-regulatory loop between SMUG1 and hsa-let-7b-5p in the MCF7 breast cancer cells. We conclude that SMUG1 functions in a gene regulatory network that influence the survival and treatment response in several cancers.
INTRODUCTION
Single-strand selective uracil–DNA glycosylase 1 (SMUG1) forms the Family 3 of uracil–DNA glycosylases and is present in vertebrates, insects and some eubacteria (1,2). Although structurally related to two other uracil–DNA glycosylases, uracil–DNA glycosylase (UNG) and thymine–DNA glycosylase (TDG), the SMUG1 amino acid sequence diverges substantially. SMUG1 initiates repair of DNA base damage via the base excision repair (BER) pathway, removing uracil, both from single-stranded DNA as well as U:G mismatches and U:A pairs (3), and several pyrimidine oxidation products (e.g. 5-formyluracil (4,5), 5-carboxyuracil (6) and 5-hydroxymethyl uracil (5-hmU) (5,7,8)). SMUG1 has pan-nuclear localization with some enrichment in subnuclear structures, like nucleoli and Cajal bodies (9). We recently showed that UNG and SMUG1 act synergistically with respect to uracil repair in mouse (10), which shows that SMUG1 is important for uracil repair in vivo.
Depending on their origin, SMUG1 substrates may have high mutagenic potential: uracil arising from cytosine deamination gives mutagenic U:G pairs which give rise to C-to-T transitions upon replication. A small, but additive mutagenic effect was found upon suppressing SMUG1 expression in Ung−/− mouse embryonic fibroblasts (MEFs) (10). Whole genome sequencing of thymic lymphomas arising in Ung/Smug1-deficient mice showed that loss of uracil BER led to both an expected accumulation of C-to-T transition mutations and a noticeable increase in transversion mutations at A:T base pairs likely contributed by mismatch repair (10). This suggests that SMUG1 has a role in protecting the genome from spontaneous mutations, but the impact on mutation accumulation is rather modest.
When present in U:A pairs, uracil is read by replicative polymerases as a cognate T:A pair. Similarly, when 5-hmU is present in a 5-hmU:A base pair after direct oxidation of thymine it will likely not be mutagenic (11), but it might affect the binding of proteins such as transcription factors (12,13). The observation that C-to-T mutations in Ung/Smug1-deficient mice was found primarily in CpG dinucleotides suggests that SMUG1 might, indirectly, affect gene regulation. Similarly, the SMUG1 substrate 5-hmU, which might form as an intermediate of oxidative demethylation of 5-methylcytosine (5-mC) (14), might influence gene expression through epigenetic regulation. Thus, SMUG1 might influence cancer risk directly through an anti-mutagenic function and indirectly through gene regulation.
Interestingly, several studies indicate that SMUG1 status is associated with modified cancer risk and response to therapy (15–17). Although SMUG1 appears to be constitutively regulated during the cell cycle (15), it is up-regulated in breast cancer and in breast cancer cell lines (overexpressed in 167 of 210 analyzed cancer cell lines, EMBL-EBI Gene expression Atlas, http://www.ebi.ac.uk/gxa/). Previous antibody based staining of breast cancer tissue arrays, suggested that low SMUG1 expression was correlated with aggressive breast cancer (16) and served, both, as an independent prognostic biomarker in ER+ breast cancers and as predictive marker for response to adjuvant chemotherapy (17). Potentially regulatory single-nucleotide polymorphisms within the SMUG1 gene were identified as independent prognostic factors that predicted poor survival in colon cancer (18,19) and as risk modifiers in bladder (20) and cervical carcinoma (21). Recently, analyses of tumor sequencing data indicated that mutations in the SMUG1 promoter correlate with an increase in C-to-T mutations in breast and colorectal cancer as well as melanoma (22). Hence, there are indications that SMUG1 expression might affect risk of cancer development and response to adjuvant therapy. The mechanisms behind these observations remains unknown, as current evidence suggests that the impact of SMUG1 on spontaneous mutagenesis is modest. Evidence from our laboratory indicates that SMUG1 might have a general function in RNA metabolism (9,23). Thus, SMUG1 appears as a multifunctional protein, potentially affecting many cellular functions including carcinogenesis, cancer pathogenesis and evolution.
The present work aimed at exploring the association of SMUG1 expression with tumor progression and chemoresistance. We define a bad prognosis signature in different cancer types by mapping out a SMUG1 interaction network and identify the existence of an auto-regulatory loop between SMUG1 and let-7b-5p in breast cancer. As let-7-5p is an established risk modulator in breast cancer, it is likely that SMUG1 also modulates cancer risk and response to therapy through the regulation of gene expression.
MATERIALS AND METHODS
Cell lines
MCF7, MDA-MB-231, BT-474 and HeLa cell lines were grown in Dulbecco's modified Eagle's medium, GlutaMAX (DMEM, Life Technologies) supplemented with 10% (vol/vol) fetal bovine serum (FBS, Lonza) and 1× penicillin–streptomycin (Life Technologies) whereas ZR-751 cells were cultured in Roswell Park Memorial Institute 1640 medium (RPMI1640, Life Technologies) containing 10% (vol/vol) FBS and 1× penicillin/streptomycin. Cells were grown at 37°C with 5% CO2.
CRISPR-Cas9-mediated generation of stable SMUG1 knockout (KO) cells
The stable SMUG1 knockout (KO) cell lines (MCF7 and MDA-MB-231) were generated using CRISPR-Cas9 technology. Alt-R® CRISPR-Cas9 system (Integrated DNA Technologies) was used according to the manufacturers’ instructions. Briefly, the gRNA targeting the exon 3 of SMUG1 was created by annealing a tracrRNA (Integrated DNA Technologies) with SMUG1 specific crRNA (Integrated DNA Technologies). The gRNA was then incubated with HiFidelity Cas9 protein (Integrated DNA Technologies) to form individual ribonucleotide protein complexes (RNPs). The cell lines were electroporated with RNPs and seeded for colony formation. Electroporation was carried out as per manufacturers’ instructions (Neon® Transfection system, Thermo Fisher Scientific). After 10–15 days, colonies were picked and plated into 96-well plates. Clones emerging were collected and validated by sequencing (Eurofins Genomics) and by western blotting. The sequence of crRNA used for generating SMUG1 KO cells is provided below:-SMUG1 crRNA: 5′-GGGCATCATCTACAATCCCGGTTTTAGAGCTATGCT-3′
DNA, miRNA and siRNA transfections
For overexpression and siRNA experiments, cells were seeded onto 6-well plates and transfected 24 h later. The construct used for SMUG1 overexpression was pcDNA3.1-SMUG1-Flag (Genscript) or pEYFP-N1-SMUG1 (Clontech); an empty vector was used in control transfections. For the rescue experiments, cells were electroporated with the Neon Transfection system (Life Technologies) following the manufacturers’ instructions; the constructs used were pHH25-SMUG1, pHH25-SMUG1 E29/31R or pHH25-SMUG1 H239L (23). For siRNA experiments, either a scrambled control or SMUG1 specific siRNAs were used (Ambion). For miRNA transfections, hsa-let-7b-5p mimic or inhibitor and negative control miRNAs were used (Life Technologies). FuGENE® 6 (Roche), Lipofectamine™ 3000 and Lipofectamine™ RNAiMAX (Invitrogen) transfection reagents were used as per manufacturer's indications. Transfected cells were harvested 24 or 72 h after transfection (siRNA treated).
Yeast-two hybrid (Y2H) screen
ULTImate Y2H screening was performed at Hybrigenics Services per their standard protocols. Briefly, the coding sequence for full-length human SMUG1 (NM_001243787.1; aa 1–270) was PCR-amplified and cloned into pB29 plasmid (N-bait-LexA-C fusion) as a carboxy (C)-terminal fusion to LexA (LexA-SMUG1). The pB29-LexA-SMUG1 construct was used to screen a random primed human breast tumor epithelial cell cDNA library (RP1) cloned into the pB43 plasmid (N-bait-GAL4-C fusion). Each screen was performed to ensure a minimum of 50 million interactions tested.
Co-immunoprecipitation
Co-immuprecipitation experiments were carried out as previously described (9). Briefly, 2 × 107 cells were harvested and washed twice with ice-cold PBS. Cells were suspended in 500 μl lysis buffer (20 mM Tris–HCl pH 7.5, 400 mM KCl, 20% glycerol, 1 mM DTT, 1× Complete EDTA-free protease inhibitor cocktail) and incubated on ice for 10 min before three freeze-thaw cycles in liquid N2 and ice. Cellular debris were discarded by centrifugation at 15 000 × g for 15 min at 4°C and the whole cell extract was dialysed overnight at 4°C against 1 l dialysis buffer (25 mM Tris–HCl pH 7.5, 5 mM MgCl2, 100 mM KCl, 10% glycerol, 1 mM DTT, 0.5 mM PMSF). Approximately 1 mg cell lysate was used per immunoprecipitation. The cell lysates were preliminary treated with 160 U DNase I-RNase free for 30 min at 30°C. The cell lysates were then incubated with 50 μl anti-GFP antibody (Roche) overnight at 4°C. The following day, 40 μl Protein G Sepharose™ 4 Fast Flow (GE Healthcare) pre-equilibrated with IP buffer (25 mM Tris–HCl pH 7.9, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 1 mM DTT, 1× Complete EDTA-free protease inhibitor cocktail) containing 100 mM KCl was added and gently mixed for 2 h at 4°C. Immune complexes were washed three times with IP buffer containing 150 mM KCl and two times with IP buffer containing 100 mM KCl, each for 5 min at 4°C. Immunoprecipitates were boiled in SDS-sample buffer (25 mM Tris–HCl pH 6.8, 2% SDS, 10% glycerol, 0.05% bromophenol blue, 5% 2-mercaptoethanol) for 5 min, separated by SDS-polyacrylamide gel electrophoresis (PAGE), stained in colloidal Coomassie blue and subject to mass spectrometry.
Liquid chromatography–mass spectrometry (LC–MS) and protein identification
Immunopurified proteins from total cell extracts from HeLa cell clones expressing the ectopic SMUG1-eGFP-tagged protein or transfected with the empty vector were processed in parallel. After staining with colloidal Coomassie blue, Coomassie G-250 stained gel pieces were in-gel digested with 0.2 μg trypsin (Promega) for 16 h at 37°C. The digestion was stopped by adding 5 μl 50% formic acid and the generated peptides were purified using a ZipTip C18 (Millipore), and dried using a Speed Vac concentrator (Concentrator Plus, Eppendorf). The tryptic peptides were dissolved in 10 μl 0.1% formic acid/2% acetonitrile and 5 μl analyzed using an Ultimate 3000 nano-HPLC system connected to a LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source. For liquid chromatography separation, an Acclaim PepMap 100 column (C18, 3 μm beads, 100 Å, 75 μm inner diameter, 15 cm length) (Dionex) was used. A flow rate of 300 nL/min was employed with a solvent gradient of 4–35% B in 47 min, to 50% B in 20 min and then to 80% B in 2 min. Solvent A was 0.1% formic acid and solvent B was 0.1% formic acid/90% acetonitrile. The mass spectrometers were operated in the data-dependent mode to automatically switch between MS1 and MS2 acquisition. Survey full scan MS spectra (from m/z 300 to 2000) were acquired with the resolution R = 60 000 at m/z 400 (LTQ-Orbitrap XL), after accumulation to a target of 1e6. The maximum allowed ion accumulation times were 60 msec. The method used allowed sequential isolation of up to six most intense ions, depending on signal intensity (intensity threshold: 1.7e4), for fragmentation using collision induced dissociation (CID) at a target value of 10 000 charges in the linear ion trap of the LTQ-Orbitrap XL. Target ions already selected for MS2 were dynamically excluded for 60 s. For accurate mass measurements, the lock mass option was enabled in MS mode.
Data were acquired using Xcalibur v2.5.5 and raw files were processed to generate peak list in Mascot generic format (*.mgf) using ProteoWizard. Database searches were performed using Mascot in-house version 2.2.0 to search the SwissProt database (Human, 20 411 proteins) assuming the digestion enzyme trypsin, at maximum one missed cleavage site, fragment ion mass tolerance of 0.6 Da, parent ion tolerance of 10 ppm, oxidation of methionines, acetylation of the protein N-terminus, pyroglutamate formation of N-terminal peptides with glutamine, and propionamide formation of cysteines as variable modifications.
Proximity ligation assay (PLA)
After fixation with PFA 4% for 20 min at RT, Flag-tagged SMUG1 cells were permeabilized for 5 min in PBS 0.25% (vol/vol) Triton X-100. Cells were incubated in blocking solution (FBS 10% in TBS 0.1% [vol/vol] Tween-20) for 1 h at RT. Incubation with primary antibodies (anti-Flag (Sigma-Aldrich) and anti-SFPQ (Abcam), anti-MATR3 (Abcam), anti-RPLP0 (Abcam), anti-NPM1 (Abcam) or anti-DNA Ligase I (Abcam) diluted 1:200 in blocking solution) was carried out overnight at 4°C. After three washes in PLA Washing buffer A, PLA was performed following the manufacturer's protocol. Briefly, PLUS and MINUS PLA probes were diluted 1:5 in Duolink® Antibody diluent and added to the coverslips for 1 h at 37°C. Cells were washed twice in PLA Washing buffer A and the ligation step (ligase diluted 1:40 in Ligation buffer 1×) was carried out for 30 min at 37°C followed by amplification (Polymerase diluted 1:80 in Amplification buffer 1×) for 100 min at 37°C. Coverslips were washed twice in PLA Washing buffer B for 10 min each, in PLA Washing buffer A for 1 min and counterstained for SFPQ, MATR3, RPLP0 or DNA Ligase I, incubating the cells with Alexa Fluor 488 conjugated anti-rabbit (Life Technologies) for 2 hr at RT. Cells were washed twice in PLA Washing buffer A for 2 min, rinsed with PLA Washing buffer B 0.01× and mounted with Prolong Diamond Antifade mounting medium. Technical control, represented by the omission of the anti-Flag antibody, resulted in loss of PLA signal.
Antibodies for immunofluorescence and immunoblotting
Primary antibodies used for immunofluorescence were SFPQ (1:200, Abcam), MATR3 (1:200, Abcam), RPLP0 (1:500, Abcam), DNA Ligase I (1:200, Abcam), NPM1 (1:500, Abcam) and Flag (1:200, Sigma-Aldrich). Secondary antibody for immunofluorescence (Alexa Fluor 488 conjugated goat-anti-rabbit) was purchased from Life Technologies. Immunoblotting was carried out using the following antibodies: SMUG1 (1:2000, Abcam) and α-Tubulin (1:3000, Sigma-Aldrich) or GAPDH (1:2000, Cell Signaling Technology) as loading controls.
Western blot
Whole-cell lysates were prepared in RIPA buffer [10 mM Tris–HCl (pH 8.0), 140 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA, 0.1% SDS (wt/vol), 0.1% sodium deoxycholate (wt/vol) and 1% Triton X-100 (vol/vol)] containing protease inhibitors (Sigma-Aldrich). Protein extracts were run on any kD Mini-PROTEAN TGX precast gel (Bio-Rad) and blotted on nitrocellulose membrane. Blots were blocked in 5% non-fat milk dissolved in 1x PBS, 0.1% Tween-20 (blocking solution). After the incubation with the specific primary antibody, secondary antibody incubation was carried out for 1 h (1:3000 in blocking solution) at RT. Blots were developed with SuperSignal West Pico Chemiluminescent substrate (Thermo Scientific). The chemiluminescent signals were detected with a ChemiDoc Imaging system (BioRad).
RNA isolation and qPCR
Total RNA was isolated with miRNeasy mini kit (Qiagen) following the manufacturer's instructions. Reverse transcription was performed using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative PCR was carried out on a QuantStudio 7 Flex detection system (Applied Biosystems) with the Power SYBR green PCR master mix (Applied Biosystems). Each sample was analysed in triplicate. Primer sequences are provided in Supplementary Table S1.
microRNA specific qPCR assay (miQPCR)
microRNA specific qPCR assay was performed as described previously (24) with minor modifications. Briefly, following treatment of MCF7 with siRNAs or miRNAs, cells were rinsed in PBS, scraped from the wells and lysed in 700 μl of Qiazol. Cellular RNAs were isolated using the miRNeasy mini kit (Qiagen) by following manufacturer's instructions. For elongating miRNAs, 1 μg of total RNA was diluted in 4 μl of nuclease free water, mixed with 4 μl of Elongation mix (1× T4 Rnl2 Buffer (New England Biolabs), 5 μM MgCl2, 15% PEG 8000, 1.5 μM miLINKER (Integrated DNA Technologies), 4U RNase inhibitor (Life Technologies) and 40U Rnl2tr K227Q (New England Biolabs)) and incubated for 2 h at 25°C. At the end of the incubation, 12 μl of cDNA Synthesis mix (1× RT buffer, 2 μl mQ-RT primer (Life technologies), 1× dNTPs, 1 μl MultiScribe™ Reverse Transcriptase and 6 μl of nuclease free water) were added to each sample and reverse transcription was carried out as per manufacturer's instructions. cDNAs were then diluted 200 times and qPCR/ddPCR assays were performed. Primer sequences are provided in Supplementary Table S2.
Metabolic labeling
MCF7 cells silenced for SMUG1 or treated with control siRNAs (72 h) were pulsed for three hours with 4-thiouridine (4sU, Sigma) at a final concentration of 150 μM. After 3 h, media was changed in normal DMEM High Glucose and cell pellets collected at different time points (up to 12 h).
microRNA half-life measurement
Total RNA was isolated using the miRNeasy kit (Qiagen) following the manufacturer's procedure. The protocol followed for the half-life measurement was performed as described previously (25) with few modifications. Briefly, 100 μg 4sU-labeled RNA were used for the biotinylation reaction. Biotinylation reactions were carried in labeling buffer (10 mM Tris pH 7.4, 1 mM EDTA pH 8.0) and 0.2 mg/ml EZ-Link Biotin-HPDP (Pierce) for 2 h at 25°C. Unbound Biotin-HPDP was removed by chloroform/isoamylalcohol (24:1) extraction using MaXtract (high density) tubes (Qiagen), following the kit procedure. RNA was precipitated adding an equal volume of isopropanol and 1:10 volume of 5M NaCl; samples were then centrifuged 17 000 × g for 20 min. The pellet was washed with an equal volume of 75% ethanol and precipitated again at 17 000 × g for 10 min. The pellet was resuspended in 100 μl RNase-free water. Biotinylated RNA was captured using Dynabeads MyOne Streptavidin T1 beads (Invitrogen). Biotinylated RNA was incubated with 100 μl Dynabeads with rotation for 15 min at 25°C. Beads were magnetically fixed and washed with 1 × Dynabeads washing buffer (5 mM Tris pH 7.5, 0.5 mM EDTA pH 8.0 and 1M NaCl). cDNA synthesis of the captured RNA-4sU was performed on beads using the SuperScript VILO cDNA synthesis kit (Life technologies), adding in the mastermix the RT probe for TaqMan® Assay for hsa-let-7b-5p (Life Technologies).
digital droplet PCR (ddPCR)
The cDNA was assayed via ddPCR using Droplet Digital PCR QX system (Bio-Rad). Briefly, the cDNA was added to a 20 μl PCR mixture containing 10 μl 2x QX200 ddPCR Supermix for Probes (No dUTP) (Bio-Rad) and 1 μl of TaqMan® Assay for hsa-let-7b-5p (Life Technologies). 20 μl of PCR mixture and 70 μl Droplet generation oil for Probe (Bio-Rad) were mixed. Droplets were generated using a QX100 Droplet Generator (Bio-Rad). The following PCR conditions were used: after enzyme activation at 95°C for 10 min, 40 cycles at 94°C for 30 s and 60°C for 1 min were followed by 1 step at 98°C for 10 min. Reactions were read in the QX200 Droplet Reader (Bio-Rad).
RNA co-immunoprecipitation assay
RNA immunoprecipitation assay was performed as described previously (9) with minor modifications. Briefly, cells were washed twice in PBS and cross-linked in 1% formaldehyde–1× PBS for 10 min at room temperature. Glycine (pH 2.5) was added to 0.2 M in order to quench the reaction before washing the cells twice with ice-cold PBS. Cells were lysed in lysis buffer A [50 mM HEPES (pH 7.8), 1 mM EDTA (pH 8.0), 1% Triton X-100 (vol/vol), EDTA-free protease inhibitor complex] and sonicated for 10 cycles with 30 s ON and 30 s OFF per cycle using a Bioruptor (Diagenode). The sonicated lysate was diluted in 1 volume of lysis buffer B [50 mM HEPES (pH 7.8), 1 mM EDTA (pH 8.0), 50 mM MgCl2, 10 mM CaCl2, 0.4 U/μl RNaseOUT recombinant ribonuclease inhibitor (Invitrogen)]. DNA was digested with DNAse I RNAse free (Life Technologies) at 37°C for 15 min and digestion was stopped adding 20 mM EDTA (pH 8.0). The lysate was centrifuged at 4°C for 5 min at 20 000 × g. The supernatant was incubated with SMUG1 and normal rabbit IgG covalently coupled to Protein G dynabeads (Invitrogen). The RIP was performed at 4°C overnight. Beads were washed once with Binding buffer [50 mM HEPES (pH 7.8), 20 mM EDTA (pH 8.0), 0.5% Triton X-100 (vol/vol), 25 mM MgCl2, 5 mM CaCl2], FA500 buffer [50 mM HEPES (pH 7.8), 1 mM EDTA (pH 8.0), 1% Triton X-100 (vol/vol), 500 mM NaCl, 0.1% Na-deoxycholate (wt/vol)], LiCl buffer [10 mM Tris–HCl (pH 7.5), 1% Triton X-100 (vol/vol), 1 mM EDTA (pH 8.0), 250 mM LiCl, 0.5% Na-deoxycholate (wt/vol)] and TES buffer [10 mM Tris–HCl (pH 7.5), 1 mM EDTA (pH 8.0), 10 mM NaCl]. RNA–protein complexes were eluted twice with 2.5 bead volumes of Elution buffer [100 mM Tris–HCl (pH 7.8), 10 mM EDTA (pH 8.0), 1% SDS (wt/vol)] for 10 min at 37°C. RNA-protein complexes and input samples were reverse-crosslinked with 200 mM NaCl for 1 h at 65°C and incubated at 42°C for 1 h after adding 20 μg proteinase K. The RNA was extracted with Trizol solution (Invitrogen) and analyzed by qPCR as percentage of input.
Colony formation assay
MCF7 cells silenced for SMUG1, treated with hsa-let-7b-5p mimic miRNA (24 hr), or SMUG1 KO cell lines (MCF7 and MDA-MB-231) were harvested and seeded as single-cell suspension in six multi-well plates (1000 cells/well for MCF7 and 400 cells/well for MDA-MB-231). Cells were incubated at 37°C with 5% CO2 for twelve days. Cells were then fixed and stained with 0.3% crystal violet (Sigma-Aldrich) in 70% ethanol for 30 min at RT. The number of colonies were counted. Six wells were set up per each condition.
Wound healing assay
MCF7 cells silenced for SMUG1, treated with hsa-let-7b-5p mimic miRNA (48 h), or MCF7 SMUG1 KO cell lines were serum starved for 16 h. A scratched area was created using a sterile 200 μl pipette tip and cells were incubated in complete medium for 48 h. The migration capacity of the cells into the wound area was monitored by acquiring images with an inverted microscope at different time points (0, 6, 24, 36 and 48 h). Wound closure was quantified by measuring the wound area at the different time points using ImageJ software and presented as the percentage relative to the initial scratched area.
Cell cycle analysis
0.5 × 106 SMUG1 knock-down cells (MCF7) or SMUG1 KO cell lines (MCF7 and MDA-MB-231) were collected, washed once in cold PBS and fixed in ice-cold 70% ethanol. Cells were stored overnight at –20°C. The cells were centrifuged, washed twice in cold PBS and stained with a solution containing 0.04 mg/ml propidium iodide (PI, Sigma-Aldrich), 0.1 mg/ml ribonuclease A (Sigma-Aldrich) in PBS. Samples were incubated at 25°C for 30 min in the dark. Cell cycle distribution was determined by flow cytometry analysis (FACSCanto™ II, BD Biosciences). Cells were analyzed using FlowJO™ v10.8 software (BD Biosciences). Forward scatter (FSC-A) and side scatter (SSC-A) were used to identify cell population while PI fluorescence pulse area (PI-A) and PI fluorescence pulse width (PI-W) were used to identify single cells. Cell cycle distribution were analyzed in PI histogram plots. Data is available in FlowRepository under the following accession codes: FR-FCM-Z5F8 (Cell cycle analyses on MCF7 siSMUG1), FR-FCM-Z5FA (Cell cycle analyses on MCF7 overexpressing hsa-let-7b-5p), FR-FCM-Z5FC (Cell cycle analyses on MCF7 SMUG1 KO), and FR-FCM-Z5FD (Cell cycle analyses on MDA-MB-231 SMUG1 KO).
Transwell migration assay
WT and SMUG1 KO MDA-MB-231 cells were serum starved for 16 h, trypsinized and resuspended in serum-free medium. Cells were seeded 5 × 104 cells/well in the upper chamber of a 24-well insert with 8-μm membrane (Corning). Growth medium supplemented with 15% FBS was used as an attractant in the lower chamber. After 24-h incubation, cell migrated through the membrane were fixed and stained with 0.3% crystal violet (Sigma) in 70% ethanol for 30 min at RT. Images were captured by microscope using a 10× magnification and six random fields were counted.
Analysis of cell displacement
MCF7 cells (WT and SMUG1 KO clones) were seeded at sub-confluent density (2 × 104 cells/well) in collagen IV-coated 96-well glass bottom plates (Merck). Coating was carried out by treatment of each well with 20 μg/ml of collagen IV (Merck) in PBS at 4°C for 20 h. After seeding, the plates were placed in a CO2 incubator for 6 h to allow cells to attach to the collagen-coated glass surface. Subsequently, cells were placed in an ImageXpress Micro Confocal high-content microscope controlled by the MetaXpress 6 software and equipped with an environmental chamber maintaining 5% CO2 and 37°C (Molecular Devices). Time lapse series of phase contrast images were acquired using a 20 × 0.45 NA Ph1 air objective, camera binning = 2, a frame rate of 3 min between frames and a total imaging period of 12 h. Time lapse series were analyzed by particle tracking using the TrackMate plugin in Fiji ImageJ (26,27) in combination with an in-house Python-based script (Python 3.7.6). Average displacement speed was calculated as the average of all tracked cell displacements within a 12 h period of imaging.
SMUG1-PPI network construction
The list of SMUG1-interacting partners was used to construct the corresponding PPI network by defining the interactions between the partners using the InWeb_InBioMap tool, applying the suggested parameters (28). The SMUG1-PPI network was represented as an undirected graph (i.e. nodes and edges symbolize proteins and interactions between them, respectively), and it was visualized via Cytoscape (v3.6.1) (29). The network enrichment analysis was performed using the ClueGO tool, using standard parameters (30). The hubs of the network were obtained by using the Cytohubba tool based on the global metric, betweenness centrality (31).
Tumor datasets and differential gene expression analysis
The differential gene expression results from TCGA and normal datasets (GTEX data) for the genes encoding the proteins in the SMUG1-PPI network were obtained via the GDC data portal hub (https://portal.gdc.cancer.gov/, last accessed July 2018). In order to better estimate the differentially expressed genes between the tumor and the corresponding normal datasets, we obtained ‘in-silico empirical’ negative controls, i.e. the least significantly DE genes based on a first-pass DE analysis performed prior to RUVg normalization (32). Pearson correlations were calculated between the gene expression profiles of SMUG1 and SMUG1-PPI in cancer patients or in the control groups using the stats package inside the R/Bioconductor environment.
Pathway enrichment analysis for SMUG1 correlated genes in TCGA–BRCA
The list of genes positively and negatively correlated with SMUG1 in breast cancer RNA-seq data (TCGA–BRCA, n = 1034; SCAN-B/GSE96058, n = 3273; SCAN-B/GSE81538, n = 405, for a total patients n = 4712) was obtained using the online correlation module of Breast Cancer Gene Expression Miner dataset v4.3 (http://bcgenex.ico.unicancer.fr/BC-GEM/GEM-Requete.php) (33). Pathway enrichment analysis was done using g:Profiler (34) and visualized with the Cytoscape Enrichment Map application (35) as described in (36). Clusters of nodes were labelled using the AutoAnnotate Cytoscape application (37).
SMUG1 mRNA–hsa-let-7b-5p/hsa-let-7c-5p correlations in TCGA
Normalized mRNA and miRNA expression data, together with clinical data (PAM50 subtypes and estrogen receptor (ER) status) for the TCGA cohort, were downloaded from the Xena browser (https://xenabrowser.net/datapages/). Altogether, 747 tumor samples had matching mRNA and miRNA data. Spearman's rank correlation was used to compute the correlation between SMUG1 mRNA and miRNA expression. Spearman's rank correlations between SMUG1 and hsa-let-7b-5p/hsa-let-7c-5p expression were computed in R and visualized as dot plots (https://www.R-project.org/).
Survival analysis
For the TCGA–BRCA dataset, differentially expressed genes (multiple correction adjustment using the Benjamini–Hochberg method, Padj < 0.05; absolute log fold change difference ≥ 1) corresponding to SMUG1 interacting partners and SMUG1 median expression were used to perform survival analyses. Kaplan–Meier plots were drawn using the RTCGA Bioconductor package, which uses maximally selected rank statistics (maxstat) to determine the optimal cutpoint for continuous variables. Samples stratification was done within the 30–70% percentile range of gene expression by the optimal cutpoint value. The Benjamini-Hochberg method was used for p-value correction of Kaplan–Meier plots.
miRNA regulators analysis
miRNAs targeting the gene sets for selected cancer-specific PPI sub-modules were retrieved by miRWalk (38). Only experimentally validated miRNAs (from Mirtarbase) having miRWalk score higher than 0.95 were selected. The functional enrichment analysis of miRNAs was achieved by using mirPath v3.0 from the DIANA Tools (39).
Breast cancer cohort for SMUG1 and hsa-let-7b-5p expression
Formalin-fixed, paraffin-embedded breast tumor samples from 66 breast cancer patients were collected at Akershus University Hospital, Norway. Individual data and information on primary and advanced disease were collected from electronic health records and treated anonymously according to strict privacy standards. Ethical permission for this study was approved by the regional ethical committee of south-east Norway (No. 2014-895).
Chromogenic in situ Hybridization (CISH)
Chromogenic in situ Hybridization (CISH) for hsa-let-7b-5p was carried out using miRNAscope™ HD Assay Red (ACD). Briefly, 4 μm-thick FFPE tissue sections were baked in dry oven for 1 h at 60°C. Sections were deparaffinized in xylene, and post-fixed in 10% neutral buffered formalin overnight, followed by RNAscope® hydrogen peroxide incubation for 10 min. Heated target retrieval was perfomed in RNAscope® 1× Target Retrieval Reagent for 15 min. Sections were then transferred into ACD EZ-Batch™ slide holder and incubated with RNAscope® Protease Plus at 40°C for 30 min within HybEZ™ Humidifying system. Hsa-let-7b-5p probe (ACD) were then added for 2 h at 40°C. miRNAscope™ Positive Control Probe-SR-RNU6-S1 and miRNAscope™ Negative Control Probe-SR-Scramble-S1 were added on separate FFPE control slides for 2 h at 40°C. Signal amplification Amp1-6 were applied sequentially and incubated 15 or 30 min according to the manufacturer's instruction. Fast red working solution was added for 10 min to detect the red signal. The samples were then counterstained with hematoxylin and mounted with EcoMount. Whole slide images were scanned at 40× using Aperio ScanScope AT. Tissue sections were examined under a standard bright field microscope at 40× magnification and scored by semi-quantitative scoring guideline utilizing the estimated number of punctate dots present within each cell boundary (score 0, no staining or less than 1 dot/cell; score 1, 2–10 dots/cell; score 2, 11–20 dots/cell and score 3, >20 dots/cell).
Immunohistochemistry (IHC) for SMUG1
Briefly, antigen retrieval was performed with EnVision™ FLEX Target Retrieval Solution, low pH (Dako) for 20 min at 97°C in PT-Link station (Dako); endogenous peroxidase activity was quenched by incubating the slides in EnVision™ FLEX peroxidase blocking reagent (Dako) for 10 min; miRNAscope™ Negative Control Probe-SR-Scramble-S1 was added for 2 h at 40°C; Protein block (Histolab) was incubated for 10 min; then SMUG1 (Origene) was diluted 1:200 in 5% bovine serum albumin (BSA, Merk) and slides were incubated for 1 h at 40°C; donkey anti-goat IgG-HRP secondary antibody (Santa Cruz) was diluted 1:1000 in 5% BSA and added on slides for 1 h at RT; at last, sections were reacted with 3,30‐diamino‐benzidine tetrahydrochloride (DAB) solution (Dako) for 10 min and counterstained with hematoxylin (Dako) for 10 min. The total scores of SMUG1 were calculated by multiplying the staining intensity for the individual scores of the positive cells (40). Scores of positive cells were defined as: 0 (≤5%); 1 (5–24%); 2 (25–49%); 3 (50–74%) and 4 (>75%). Staining intensity scores were defined as follows: weak (1 point); medium (2 points); and strong (3 points). The total score is divided into the following levels: −, 0 points; +, 1–4 points, ++, 5–8 points; +++, ≥9 points where ‘−’ and ‘+’ are considered low expressions, and ‘++’ and ‘+++’ high expressions. The IHC results were evaluated by two independent pathologists blinded of clinical information.
Quantification and statistical analysis
All quantified data are presented as mean ± s.e.m., mean ± s.d. and fold change unless stated otherwise (refer to figure legends for detailed information). Student t-test or one-way ANOVA were used to assess the statistical significance in GraphPad Prism 7 (GraphPad software). A P <0.05 was considered as statistical significant. P values were indicated with asterisks. Replicates, statistical tests carried out and statistical significances are reported in the corresponding figure legends.
The Kaplan–Meier estimator and log-rank tests were performed using the functions Surv, survfit, and survdiff (R package survival v2.42–3). In the box-and-whisker plots, the line within each box represents the median. Upper and lower edges of each box represent 75th and 25th percentile, respectively. The whiskers represent the lowest datum still within [1.5 × (75th − 25th percentile)] of the lower quartile and the highest datum still within [1.5 × (75th − 25th percentile)] of the upper quartile.
RESULTS
SMUG1 expression is increased in ER positive breast cancer
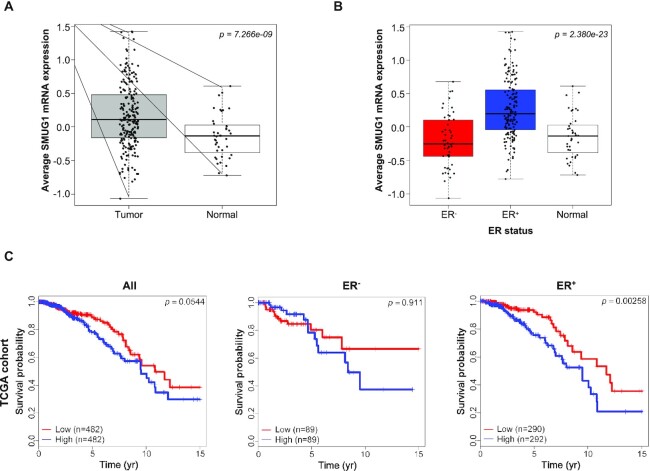
Reduced levels of SMUG1 mRNA and protein were previously correlated with increased aggressiveness and poor prognosis in primary breast cancers, pointing at SMUG1 as a possible negative prognostic marker for adjuvant therapy in breast tumors (16). However, previous studies showed a very modest increase in spontaneous mutagenesis when SMUG1 expression was suppressed in MEFs (41). This suggested to us that the negative correlation between SMUG1 and poor prognosis, if causal, might be ascribed to functions of SMUG1 other than its anti-mutagenesis properties. To further explore the possible underlying mechanisms, we first analyzed SMUG1 expression levels in breast tumor vs adjacent normal tissues in the TCGA–BRCA cohort (Figure 1). When considering all breast tumor samples as one group we found increased SMUG1 mRNA levels compared to normal breast tissue (Figure 1A). As estrogen receptor (ER) positive breast tumors have a very different biology than ER negative tumors, we further stratified SMUG1 expression according to ER status and found that ER+ tumors showed significant increase of SMUG1 mRNA levels when compared to both ER− and normal adjacent tissue (Figure 1B and Supplementary Figure S1). Breast cancer-specific mortality stratified by high or low SMUG1 expression levels was assessed using Kaplan–Meier plots (Figure 1C and Supplementary Figure S2). Surprisingly, for the breast cancer cohorts we analyzed (with at least 100 patients included), low SMUG1 mRNA expression levels were associated with better survival compared to patients with high levels of SMUG1. Although not always significant, the same trend was observed in all 7 cohorts analyzed (Figure 1C and Supplementary Figure S2). Interestingly, improved survival with low SMUG1 expression was more pronounced for ER+ samples (Figure 1C and Supplementary Figure S2). While it is clear that patients with better prognosis tended to have a lower SMUG1 expression, it also appeared that SMUG1 expression alone cannot be used as an independent predictor of survival for breast cancer as, in contrast to previously reported (16), the multivariate cox regression analyses were not significant. If SMUG1 has an indirect role in cancer development, we would expect its expression to be correlated with gene expression programs associated with oncogenesis. Genes positively and negatively correlated with SMUG1 in breast cancer RNA-seq data were identified using the online correlation module of Breast Cancer Gene Expression Miner dataset v4.3 (http://bcgenex.ico.unicancer.fr/BC-GEM/GEM-Requete.php) (Supplementary Table SI, worksheet ‘Gene_correlation_table’) (33). Gene ontology (GO) analyses of these genes did not show any enrichment for classical pathways associated with oncogenesis, pointing instead to mitochondrial electron transport and respiratory processes as main pathways associated with genes co-expressed with SMUG1 in breast cancer (Supplementary Figure S3). In addition, also regulation of T cells, immune response and fatty acid transport were enriched (Supplementary Table SI, worksheet ‘GO_enrichment_table’).
Figure 1.
High SMUG1 expression levels associate with poor overall survival in breast cancer cohort. (A, B) Boxplot representation of SMUG1 expression in TCGA–BRCA cohort in normal adjacent versus tumor samples (A); in ER+, ER−versus normal adjacent samples (B). Kruskal Wallis p-value is denoted. (C) Kaplan–Meier curves showing overall survival rates in TCGA–BRCA cohort with low (red) or high (blue) SMUG1 expression in all, ER− and ER+ patients. SMUG1 median expression was used for patients stratification. Log rank test P-value is denoted.
Proteomic analysis of SMUG1 interactors using mass spectrometry and yeast two-hybrid data
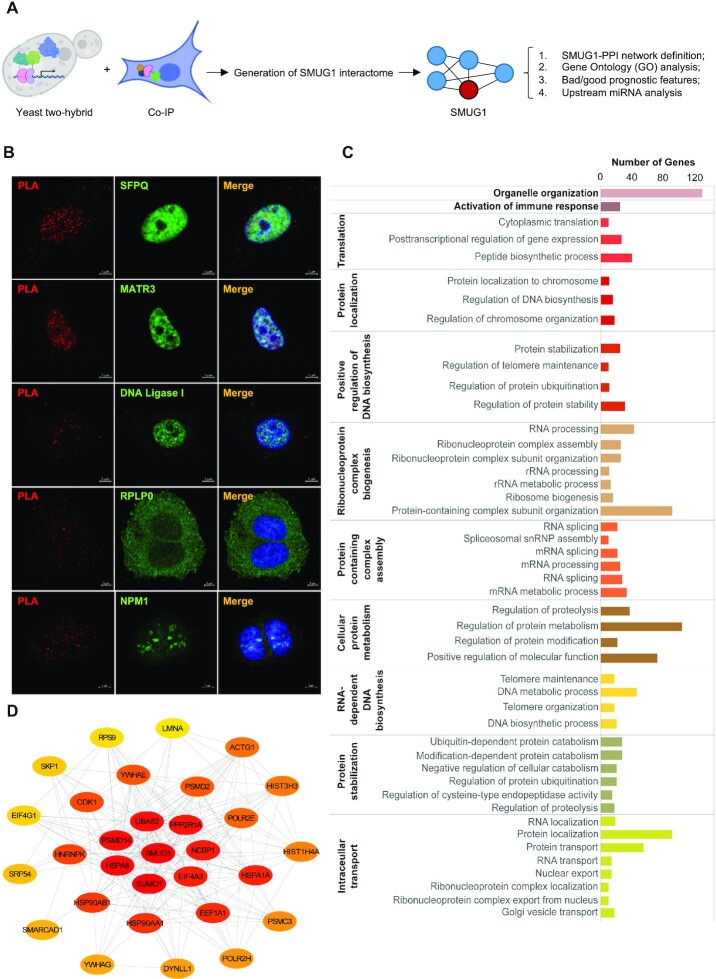
SMUG1 is a multifunctional protein involved in RNA quality control as well as in DNA repair (9,23,42). We therefore investigated whether the functional impact of SMUG1 in breast cancer might be defined, not only through SMUG1 expression, but also through the expression and activity of the complex network of SMUG1 involving its interacting partners. Although a few protein and RNA partners of SMUG1 are known (9,23), the SMUG1 interactome is still poorly defined, in particular with respect to protein-protein interactions. Thus, to gain a deeper understanding of the possible role of SMUG1 in breast cancer, we identified novel SMUG1 interactors using yeast two-hybrid (Y2H) and co-immunoprecipitation followed by mass spectrometry approaches (Figure 2A). First, we identified binary in vivo protein-protein interactions (PPI) taking advantage of a high throughput Y2H screen using a breast cancer library as bait. To expand the dataset of SMUG1 PPIs, the list of positive hits from the Y2H screen (ELAVL1, EXOC6 and GPN3) was complemented with SMUG1-associated proteins identified through mass spectrometric analyses following co-immunoprecipitation of eGFP-SMUG1 in HeLa cells. In sum, a list of 534 potential SMUG1 interactors were identified (Supplementary Table SII, worksheet ‘SMUG1_PPI_HeLa’). Direct protein-protein interaction with SMUG1 was confirmed via Proximity Ligation assay (PLA) for selected interactors involved in RNA metabolism (MATR3, RPLP0, NPM1 and SFPQ) and DNA repair (DNA Ligase I) (Figure 2B and Supplementary Figure S4).
Figure 2.
SMUG1 interacts with proteins involved in RNA metabolism and gene transcription. (A) Schematic representation of the workflow used for determining the SMUG1 interacting proteins in connection with analysis of TCGA expression data. (B) Proximity Ligation Assay (PLA) showing Flag-tagged SMUG1 (red) and SFPQ, MATR3, DNA Ligase I, NPM1 and RPLP0 (green) interaction in MCF7 cells. Scale bars, 1 μm. (C) Top 30 hubs of the SMUG1-PPI network, based on global metric, betweenness centrality. Color shades represent the significance of the hub, with red color as the most significant and yellow color as the least. (D) Functional enrichment analysis of SMUG1-PPI Network based on GO-Biological Processes (P < 0.05).
The list of interactors was, next, used to establish a SMUG1-PPI network (Supplementary Table SII, worksheet ‘SMUG1_PPI_total’). Direct and/or indirect interactions between these molecules were retrieved by the InWeb_InBioMap web tool, giving rise to undirected PPI network with 525 nodes and 5685 edges (data not shown). Gene ontology analyses for functional gene enrichment based on biological process (BP) identify 389 unique IDs. The enriched BP are consistent with functions of SMUG1 in DNA and RNA metabolism, but also, suggested a role for SMUG1 in protein metabolism/stability (Figure 2C and Supplementary Table SIII). This complex PPI-network was then further analyzed focusing on its most critical elements, by performing a hub analysis based on the betweenness centrality metric, a measure of how often a node occurs on all shortest paths among pairs of nodes in a network, meaning the importance of each node/protein for the connectivity of the network (43). The resulting top 30 hub nodes were extracted from the main SMUG1-PPI network as hub-subnetworks (Figure 2D and Supplementary Table SIV). This hub module was then analyzed with ClueGO to categorize these genes/IDs in the GO biological processes (Supplementary Table SIV). As shown in Figure 2C, ribonucleoprotein complex biogenesis, RNA processing/splicing and regulation of cellular protein metabolism were the most enriched processes, for the global network. Taken together, this analysis revealed a prominent representation of RNA and protein metabolism within the SMUG1-PPI network, suggesting that SMUG1 may act as a central hub connecting the different subnetworks with diverse functions.
Definition of a prognostic SMUG1 signature in cancers
In order to evaluate whether the association with poor/good prognosis improved when considering SMUG1-interactome in breast and other cancer types, we interrogated TCGA cancer cohorts. The genes that were significantly differentially expressed (Padj < 0.05, absolute log fold change > 1) and significantly correlated (P < 0.05, absolute Pearson correlation > 0.6) with the expression profile of SMUG1 were calculated through the analysis of 33 TCGA datasets (Supplementary Table SV and Table S3). To assess whether the SMUG1 interactome correlated more consistently with clinical outcomes than SMUG1 expression alone (Figure 1C), Kaplan–Meier plots were obtained for each gene in each dataset, allowing us to define good and bad prognosis gene signatures on a per cancer basis. The distribution of the differentially expressed genes with respect to good or bad prognosis (P < 0.05) signature per cancer datasets are summarized in Supplementary Table SVI. Six datasets, having different ratios in the number of bad-good prognosis genes (higher number of bad prognosis genes: hepatocellular carcinoma [LIHC] and human skin cutaneous melanoma [SKCM]; similar number of genes between bad and good prognosis: acute myeloid leukemia [LAML] and lower grade glioma [LGG]; lower number of bad prognosis genes: breast cancer [BRCA] and glioblastoma [GBM]) were analyzed further. In general, the SMUG1 interactome did not show any defined prognostic signature in the majority of the datasets analyzed: although a few datasets (i.e. LIHC and SKCM) showed a clear bad prognosis signature (Supplementary Figure S5A and Supplementary Tables SV and VI). This suggests that additional regulators might affect the prognosis. In fact, although the number of genes for good and bad prognosis are similar in the TCGA–BRCA dataset, Kaplan–Meier analysis using the survival outcomes of patients having high/low expression of the bad prognosis genes (n = 85) showed that high expression of the bad prognosis genes was significantly associated with lower survival probability (P < 0.0001) (Supplementary Figure S5B and Table SV, worksheets ‘BRCA’; ‘Report’).
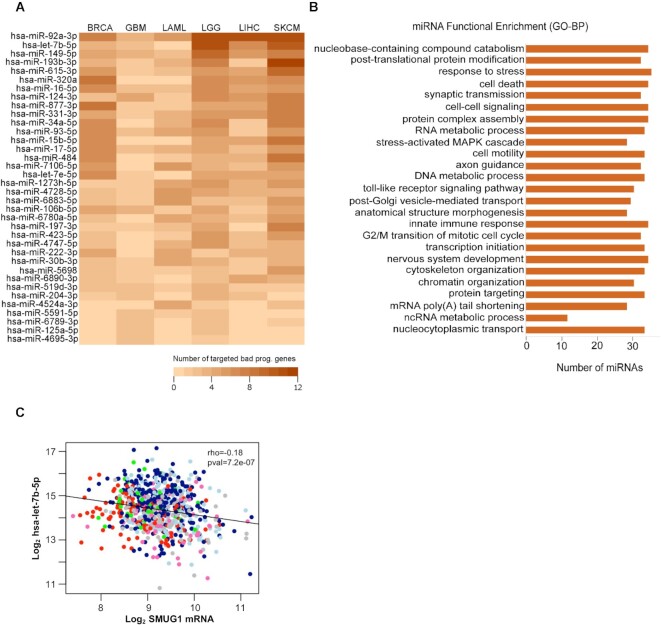
To assess the possibility these gene sets have common upstream regulators, we used the miRWalk tool to identify common miRNA regulators. We found that 36 miRNAs target the highest number of genes in each bad prognostic network (Figure 3A and Supplementary Table SV, worksheet ‘Heatmap’). Among the top 10 miRNAs of all datasets, miR-92a-3p, hsa-let-7b-5p, miR-149-5p, miR-193b-3p, miR-615-3p and miR-320a target the highest number of genes in these networks (Figure 3A). Gene Ontology analyses indicate the involvement of these miRNAs in several biological processes relevant for cancer biology, such as epigenetic regulation of gene expression and maintenance of differentiation (Figure 3B and Supplementary Table SVII).
Figure 3.
Identification of upstream miRNA regulators of SMUG1 interacting proteins in TCGA datasets. (A) Heatmap of the miRNAs (n = 36) regulating the highest number of SMUG1 interacting proteins in BRCA, GBM, LAML, LGG, LIHC and SKCM datasets. Heatmap is ordered, from highest to lowest, according to the total number of proteins in each dataset which is regulated by corresponding miRNAs on y-axis. Color scale is showing the number of genes targeted by those miRNAs in each dataset, lightest as lowest and darkest as highest number of genes. (B) Functional annotation of top 10 miRNAs of all datasets (n = 36) presented in (A) based on Gene Ontology—Biological Process terms (P < 0.05). (C) Scatterplot showing SMUG1 mRNA expression (x-axis) versus hsa-let-7b-5p expression (y-axis) for breast tumor samples of the TCGA–BRCA cohort (n = 747). Tumors are color-coded according to the PAM50 molecular subtype; dark blue = luminal A (LumA; n = 267), light blue = luminal B (LumB; n = 49), red = basal-like (basal; n = 115), pink = Her2-enriched (HER2; n = 62), green = normal-like (n = 49), gray = subtype not available (n = 106). Spearman′s rho and the associated P-value are indicated.
SMUG1 and hsa-let-7b-5p correlation in the TCGA–BRCA dataset
A clear role for the let-7 miRNA family as tumor suppressors has been demonstrated (44) and patients with breast cancer showed dysregulated levels of hsa-let-7b-5p and hsa-let-7c-5p (44–47). For this reason, we interrogated the TCGA breast cancer dataset with respect to a possible correlation existing between SMUG1 levels and these two members of the let-7 family, hsa-let-7b-5p and hsa-let-7c-5p, hereafter named let-7b-5p and let-7c-5p. Interestingly, only let-7b-5p had a weak but significant negative correlation with SMUG1 levels (Spearman's rho = –0.18, correlation P-value = 7.2e−7; Figure 3C), in particular in Luminal A and B datasets (Supplementary Figure S6) that also have the highest SMUG1 expression level (Supplementary Figure S1). This suggests a potential co-regulatory mechanism between SMUG1 and let-7b-5p.
SMUG1 affects let-7b-5p levels in breast cancer cells
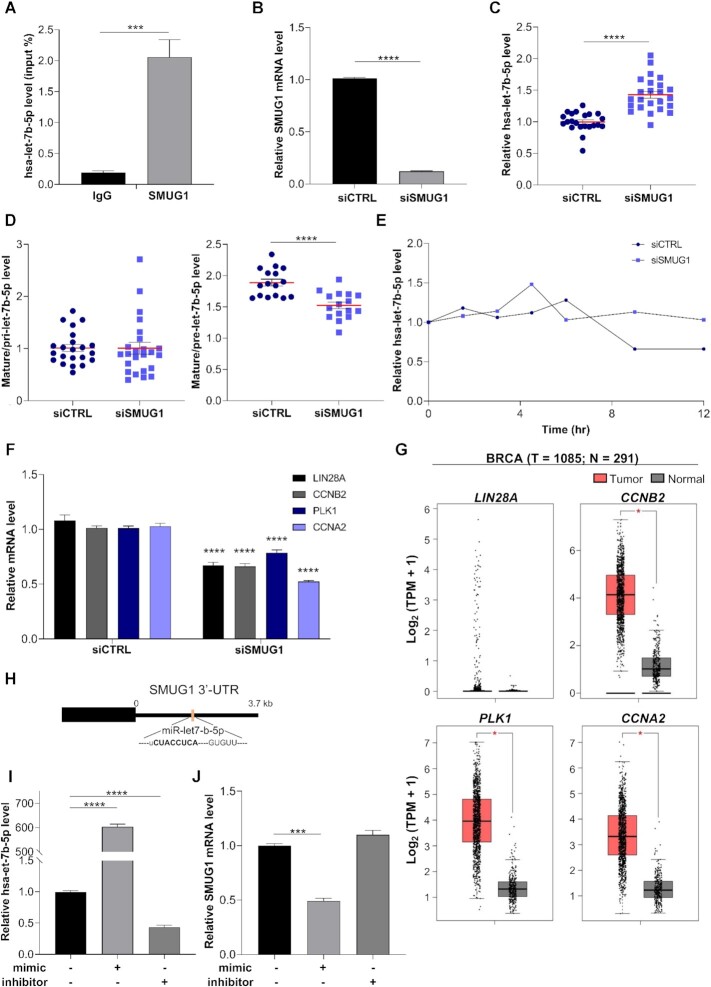
Prompted by the negative association existing between SMUG1 and let-7b-5p, as observed in the TCGA breast cancer data, we studied the possible co-regulation of SMUG1 and let-7b-5p in MCF7 cells, a well-established breast adenocarcinoma cell line expressing both estrogens and progesterone receptors. As SMUG1 binds RNA molecules (9,23), we asked if SMUG1 physically interacts with let-7b-5p. In RNA-immunoprecipitation experiments (RNA-IP), we detected statistically significant enrichment of let-7b-5p using an anti-SMUG1 antibody compared to the immunoglobulin G (IgG) control (Figure 4A). Interestingly, SMUG1 also binds miR-92a-3p in vivo, the only common upstream miRNA in all the TCGA datasets considered (Figure 3A), as expected due to its overexpression in malignant tumors (48), and other members of the let-7 microRNA family (Supplementary Figure S7A). We then checked the expression levels of let-7b-5p and of the other miRNAs bound by SMUG1 in MCF7 cells transiently silenced for SMUG1 (Figure 4B and C and Supplementary Figure S7B and C). As seen in Figure 4C, the let-7b-5p expression increased in SMUG1 knock-down cells. Increased expression of let-7b-5p in SMUG1 knock-down cells, was also confirmed in three other breast cancer cell lines (MDA-MB-231, BT-474 and ZR-751), confirming a role for SMUG1 in let-7b-5p regulation (Supplementary Figure S8). Interestingly, the same behavior was also observed for the other miRNAs bound by SMUG1 (Supplementary Figure S7C), suggesting that SMUG1 may have a role in miRNA processing or degradation. Considering the function of SMUG1 in regulating hTERC maturation and processing (23), we followed the let-7b-5p maturation process from pri-/pre-let-7b to the mature form (Figure 4D). We measured the levels of mature let-7b-5p and the two immature forms, pri-let-7b and pre-let-7b (Figure 4D). Transient silencing of SMUG1 did not affect the ratio between mature and pri-let-7b (Figure 4D, left), but a significant reduction in mature vs pre-let-7b ratio was observed in several cell lines (Figure 4D, right, and Supplementary Figure S8G–I). Taken together, these data suggest that the precursor transcripts, in the form of pre-let-7b, accumulate in SMUG1-depleted cells, pointing to a role for SMUG1 during miRNA processing from pre-miRNA to the mature form. In vivo metabolic labeling experiments showed an increased stability of let-7b-5p in SMUG1 knock-down cells further confirming not only an increase of let-7b-5p expression but also activity (Figure 4E), and indicating that post-transcriptional processing might be responsible for the expression levels and stability of let-7b-5p in the absence of SMUG1. To test whether SMUG1 substrates might be present in let-7b-5p molecules, we used a previously described assay based on reduced amplification of transcripts containing modified bases after SMUG1 digestion (23). No reduction in amplification for let-7b-5p was observed between siCTRL and siSMUG1 cells, indicating the absence of SMUG1 substrates in let-7b-5p (data not shown).
Figure 4.
SMUG1 regulates the let-7b-5p miRNA in MCF7 cells. (A) RNA immunoprecipitation (RIP) of let-7b-5p by SMUG1 in MCF7 cells using an antibody against SMUG1, quantified by qPCR. Data are presented as percent of input RNA. IgG, negative control. (B) SMUG1 silencing efficiency in MCF7 cells as quantified by qPCR. (C) Relative let-7b-5p levels in siCTRL and siSMUG1 cells measured by miQPCR. (D) Relative mature let-7b-5p to pri-let-7b (left) and to pre-let-7b (right) ratios in MCF7 cells silenced for SMUG1 expression as measured by miQPCR and ddPCR. (E) Half-life of let-7b-5p in siCTRL and siSMUG1 cells as measured by ddPCR. (F) Relative let-7b-5p target mRNA levels in siCTRL and siSMUG1 cells measured by qPCR. (G) Gene expression levels of LIN28A, CCNB2, PLK1 and CCNA2 in TCGA–BRCA and normal samples. Tumor data are matched with TCGA–BRCA normal and GTEx data. T, tumor (red); N, normal (grey). (H) Schematic representation of the positioning of the let-7b-5p binding site within the human SMUG1 3’ untranslated region (UTR). (I, J) Relative let-7b-5p (I) and SMUG1 mRNA (J) levels in MCF7 cells treated with the let-7b-5p mimic, the let-7b-5p inhibitor or the negative control miRNAs, quantified by miQPCR and qPCR, respectively. (A) Data represent means ± s.d., n = 3. (B–E, I, J) Data represent means ± s.e.m., n = 24. (A–G, I, J) * P≤ 0.05, * * P≤ 0.01, * * * P≤ 0.001, * * * * P≤ 0.0001 (two-tailed Student's t-test).
In order to test the functional impact on the let-7b-5p-mRNA regulatory axis, we selected some known let-7b-5p target mRNAs associated with pluripotency and proliferation (LIN28A, CCNB2, PLK1 and CCNA2) (49) and analyzed their expression profile via qRT-PCR in siCTRL and siSMUG1 cells. As expected, increased let-7b-5p miRNA levels corresponded to significantly decreased mRNA levels in siSMUG1 cells, indicating that let-7b-5p negatively regulated the expression of LIN28A, CCNB2, PLK1 and CCNA2 (Figure 4F). Considering the negative correlation between SMUG1 and let-7b-5p and the increased levels of SMUG1 in breast cancer samples, the mRNA of genes regulated by let-7b-5p should be upregulated. We therefore investigated the mRNA expression levels of these genes (LIN28A, CCNB2, PLK1 and CCNA2) in normal and BRCA cancer specimens by GEPIA (Figure 4G). All the genes showed a higher expression in BRCA samples than controls (Figure 4G). Interestingly, correlation analyses of SMUG1 mRNA expression and the mRNAs of these downstream targets of let-7b-5p showed significant positive (although weak) correlations in ER+ tumors (Supplementary Figure S9). Hence, SMUG1 affects let-7b-5p levels and their expression is negatively correlated in breast cancer (Figures 3C and 4C).
Let-7b-5p negatively regulates SMUG1 expression in breast cancer cells
Prompted by our previous results, we suspected that SMUG1 mRNA itself might be a let-7b-5p target. Indeed, TargetScan predicted the presence of a let-7b-5p target sequence 1.75 kb into the SMUG1 3’UTR (Figure 4H). Validation of SMUG1 as a let-7b-5p target was also confirmed in other tissues as kidney, bone marrow, mammary gland and cervix using the online DIANA-TarBase v8 tool (50). Partial inhibition of let-7b-5p did not affect SMUG1 expression, probably due to the amount of endogenous let-7b-5p left (Figure 4I, J and Supplementary Figure S10A). However, overexpression of let-7b-5p mimic in MCF7 cells led to significant reduction of SMUG1 mRNA and protein levels (Figure 4I, J and Supplementary Figure S10B). Together, these data support a negative regulation loop between let-7b-5p and SMUG1 in breast cancer samples.
Increased let-7b-5p levels reduces cell proliferation and migration in breast cancer cells
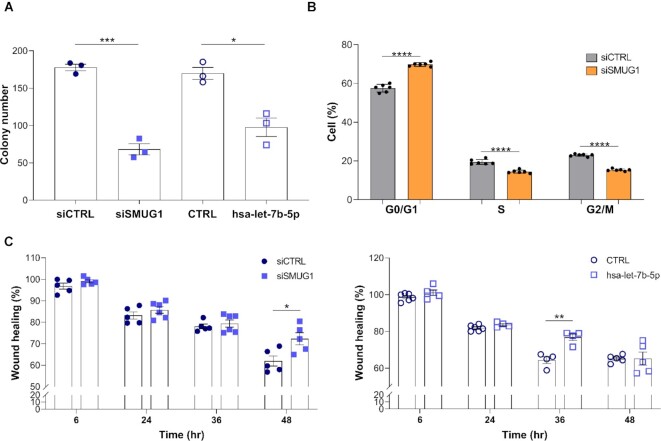
Previous studies suggested a role for let-7b-5p as tumor suppressor in breast cancer development and progression (51–53). Thus, we tested the impact on SMUG1 knockdown, and its consequent increased level of let-7b-5p, on cell proliferation and migration (Figure 5). We first looked at cell proliferation in MCF7 transiently silenced for SMUG1 and overexpressing let-7b-5p. Increased levels of the microRNA caused a reduced number of colonies in both conditions (Figure 5A and Supplementary Figure S11A). Cell cycle analyses indicated a mild, but significant, accumulation of cells in the G1 phase (Figure 5B and Supplementary Figure S11B), in concordance with the cell proliferation data. Tumor suppressors can control tumorigenesis not only via their anti-proliferative activities, but also through modulation of cell migration, a property critical for tumor invasion and metastasis. The cell spreading capacity of MCF7 cells silenced for SMUG1 or overexpressing let-7b-5p was tested using the wound healing assay. As shown in Figure 5C and Supplementary Figure S11C, cells with elevated let-7b-5p levels presented a reduced migration capacity compared to the controls. Thus, in combination these results confirmed the role of let-7b-5p as tumor suppressor affecting both cell proliferation and migration in breast cancer. This suppressor activity of let-7b-5p might explain the observed association of low SMUG1 levels with increased survival in ER+ breast cancers (Figure 1 and Supplementary Figure S2), suggesting an indirect effect of SMUG1 in cancer development.
Figure 5.
Increased levels of let-7b-5p in SMUG1 knock-down cells negatively affect cell proliferation and migration. (A) Colony formation assay in MCF7 cells transiently silenced for SMUG1 and control (siSMUG1 and siCTRL) siRNAs or treated with let-7b-5p mimic and the negative control (CTRL) miRNAs. Bar graph showing the colony number after culturing the cells for 12 days. Data represent means ± s.e.m, n = 3. (B) Cell cycle analyses of MCF7 cells transiently silenced with SMUG1 and control siRNAs. Data represent means ± s.e.m., n = 6. (C) Cell migration in MCF7 cells transiently silenced for SMUG1 and control siRNAs or treated with let-7b-5p mimic and the negative control miRNAs as measured by wound-healing assay. Bar graphs showing the percentage of migration at the different time points relative to time 0. Data represent means ± s.e.m., n = 4. (A–C) * P≤ 0.05, ** P≤ 0.001, *** P≤ 0.001, **** P≤ 0.0001 (two-tailed Student's t-test and one-way ANOVA).
SMUG1 knock-out alters let-7b-5p maturation, ultimately reducing cell proliferation and migration
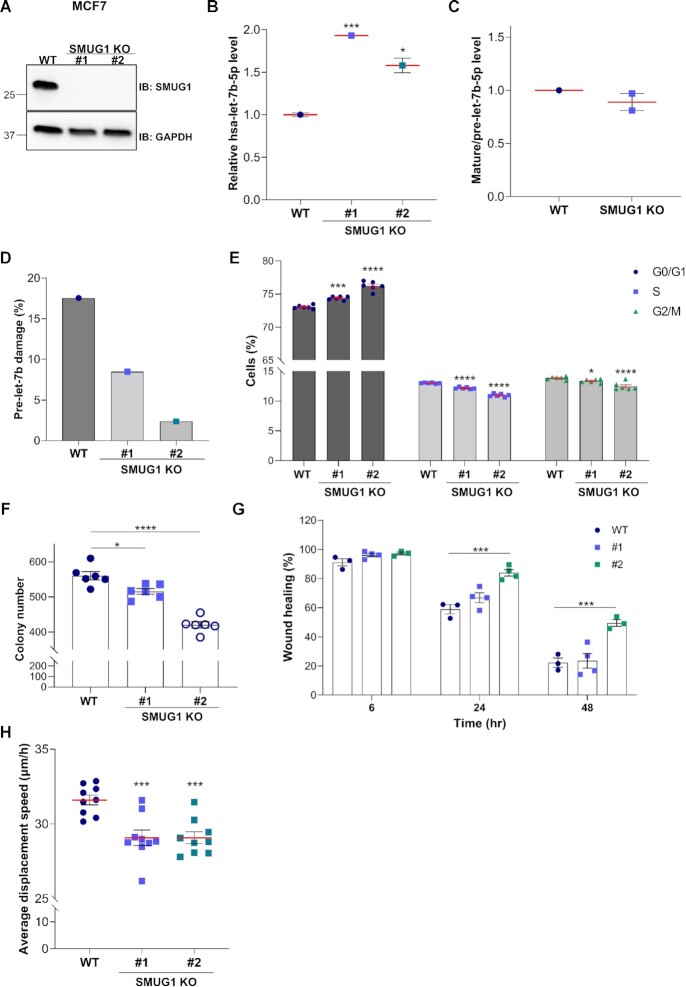
In order to confirm the data obtained in MCF7 transiently silenced for SMUG1, we generated SMUG1 knock-out (KO) clones using CRISPR/Cas9 technology in two breast cancer cell lines, MCF7 and MDA-MB-231. As expected SMUG1 transcription was not affected (data not shown), but no protein was detected using an antibody directed towards the N-terminal domain of SMUG1 (Figure 6A and Supplementary Figure S12A). Real-time PCR for let-7b-5p levels revealed a ∼ 2-fold increase of this microRNA in two independent clones of each cell line (Figure 6B and Supplementary Figure S12B). We observed a reduced mature-to-pre-let-7b ratio in both SMUG1 KO MCF7 and MDA-MB-231 cells (Figure 6C and Supplementary Figure S12C), supporting a possible role for SMUG1 in the microRNA maturation process. Next, we tested the presence of SMUG1 substrates on pre-let-7b and we saw a reduced number of possible substrates in both the clones (Figure 6D and Supplementary Figure S12D), suggesting that the accumulation of pre-let-7b in SMUG1 KO and the increased levels of the mature form might be the consequence of an inefficient RNA processing. As in the knock-down system, both cell proliferation and cell migration were negatively affected by the elevated levels of let-7b-5p observed in the SMUG1 KO clones (Figure 6E–H, Supplementary Figures S12E–G and S13). The observed accumulation of cells in G1 phase (Figure 6E and Supplementary Figure S12E) was again accompanied by reduced cellular proliferation, as measured by the colony formation assay (Figure 6F, Supplementary Figure S12F and Figure S13A), as well as cellular migration, measured by wound healing and by high content imaging for MCF7 cells (Figure 6G, H and Supplementary Figure S13B) and by the transwell migration assay for the MDA-MB-231 clones (Supplementary Figure S12G).
Figure 6.
SMUG1 KO cells present increased levels of let-7b-5p and show negative effects on cell proliferation in MCF7 cells. (A) Representative western blotting showing SMUG1 protein levels in in two independent SMUG1 KO clones in MCF7 cells. GAPDH was used as loading control. (B) Relative let-7b-5p levels in WT and SMUG1 KO cells measured by QPCR. (C) Relative mature hsa-let-7b-5p to pre-let-7b ratios in SMUG1 KO cells as measured by ddPCR. (D) Pre-let-7b RNA damage in two independent SMUG1 KO clones as measured by ddPCR. (E) Cell cycle distribution of SMUG1 KO cells. Data represent means ± s.e.m., n = 6. (F) Colony formation assay in SMUG1 KO cells. Bar graph showing the colony number after culturing the cells for 12 days. Data represent means ± s.e.m., n = 3. (G) Cell migration in SMUG1 KO clones as measured by wound-healing assay. Bar graphs showing the percentage of migration at the different time points relative to time 0. Data represent means ± s.e.m., n = 3. (H) Average displacement speed calculated based on particle tracking data. Box plots showing the average cell displacement speed per each clone over a time period of 12 h, n = 9 separate microscopic fields of view. (B, E–H) * P ≤ 0.05, *** P ≤ 0.001, **** P ≤ 0.0001 (two-tailed Student's t-test and one-way ANOVA).
Finally, to better characterize what SMUG1 function is required for let-7b-5p regulation, we performed complementation assays (Supplementary Figure S14). Let-7b-5p levels were reduced when the expression of SMUG1-WT and of DKC1-binding mutant (SMUG1 E29/31R) was restored in SMUG1 KO cells. Interestingly, the expression of a SMUG1 mutant with significantly hampered damage-excision activity due to inefficient binding to the substrate (SMUG1 H239L) failed to complement the microRNA levels, pointing at SMUG1 substrate affinity and RNA binding as critical features for let-7b-5p regulation in breast cancer cells possibly through facilitating the recruitment of critical processing factors.
SMUG1 and let-7b-5p show anti-correlative expression levels in breast cancer tissues
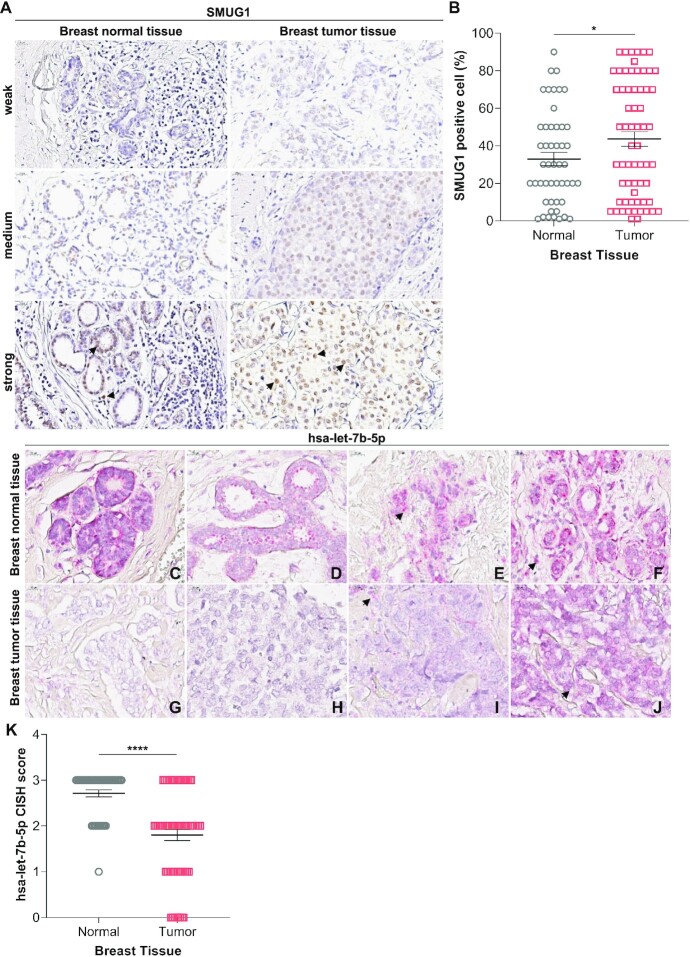
To assess whether the negative correlation between SMUG1 and let-7b-5p expression found in RNA-seq datasets (Figure 3C) was confirmed in tumor tissue, we checked SMUG1 and let-7b-5p expression and subcellular localization in serial sections (Supplementary Figure SS15) from tissues from a cohort of 66 breast cancer samples comprising Luminal A (LumA, 21 samples), Luminal B (LumB, 12 samples), Triple negative (TNBC, 17 samples), and HER2-positive (HER2, 16 samples) subtypes. The clinico-pathological characteristics are summarized in Supplementary Table SVIII. Expression levels and subcellular localization were analyzed for both SMUG1 and let-7b-5p, using IHC and CISH, respectively (Figure 7). SMUG1 presented a general nuclear staining with positive nucleoli in all tissue types (Figure 7A), consistent with its known direct protein interaction with DKC1 and function in ribosomal RNA biogenesis (9,23). Interestingly, a positive correlation between SMUG1 and its interacting protein expression could be observed as well in breast cancer tissues, confirming the bioinformatic analyses (Supplementary Figure SS16). An increased percentage of SMUG1-positive cells was observed in tumor samples, where 41% of the samples presented high SMUG1 expression level versus 25% of the normal ones (Figure 7B), in accordance with SMUG1 mRNA data (Figure 1A). We then focused on let-7b-5p levels. As shown in Figure 7C-J, let-7b-5p displayed a general cytoplasmic localization in both tissues (Figure 7C–J). Interestingly, decreased let-7b-5p levels were seen in the tumor when compared to the adjacent normal tissue (Figure 7K). In conclusion, the negative correlation between SMUG1 and let-7b-5p was confirmed in tumor tissue.
Figure 7.
SMUG1 and let-7b-5p show inverse expression levels in breast cancer tissues. (A) Representative images of IHC for SMUG1-positive in normal (left panels) and tumor (right panels) breast tissue, with intensity scores as weak, medium and strong, respectively. Magnification ×400. Arrow head, SMUG1 positive cells. (B) Scatterplot showing percentage of SMUG1 positive cells in normal versus tumor breast samples, as measured by IHC. (C–J) Representative images of CISH for miRNA let7b-5p in breast cancer cohort. Arrow head, let-7b-5p positive cells. (C–F) miRNA let-7b-5p – positive adjacent normal breast tissue from the same resection specimens corresponding to G-J, respectively. Arrow head, let-7b-5p positive cells. (G–J) miRNA let-7b-5p – positive breast tumor, Score 0, 1, 2, and 3, respectively. Cells with red dots were scored as positive. Magnification ×400. IHC and CISH were performed in consecutive tissue sections. (K) Scatterplot showing let-7b-5p levels in normal vs tumor breast samples, as measured by CISH. (B, K) * P≤ 0.05, **** P≤ 0.0001 (two-tailed Student's t-test) compared to the normal samples.
DISCUSSION
In the present study, we identified a regulatory loop between SMUG1 and let-7b-5p miRNA in breast cancer cells. Functional relevance of this regulatory loop is supported by a negative correlation between the expression of SMUG1 and let-7b-5p in breast cancer datasets (Figures 3C and 7). On the mechanistic level, SMUG1 physically interacts with let-7b-5p and other let-7 microRNAs in vivo and regulates let-7b-5p expression. Consequently, when SMUG1 expression was down-regulated or constitutively knock-out in breast cancer cells, we observed increased levels of let-7b-5p and accumulation of its precursor, pointing at miRNA misprocessing events during the maturation steps in SMUG1 knock-down and knock-out cells (Figures 4 and 6, Supplementary Figures S8 and S12). The same negative correlation between SMUG1 and let-7b-5p expression levels was also confirmed in vivo by IHC and CISH on a breast cancer cohort (Figure 7). While the let-7b-5p target sequence does not contain bases that could be converted to 5-hmU, e.g. by direct oxidation or AID/APOBEC deamination of 5-hydroxymethylcytosine (5-hmC) derived from 5-mC, the precursor form contains several Cs in the hairpin-loop region that could be converted into SMUG1 substrates (Figure 6 and Supplementary Figure S12). Our data suggests that in absence of SMUG1 the equilibrium between distinct pre-let-7b populations (modified vs unmodified) shifts preferentially towards the unmodified/moderately modified, possibly due to a change in the stability. Interestingly, SMUG1 is also a downstream target of let-7b-5p, having a let-7b-5p response element in its 3′-UTR region, suggesting a co-regulatory mechanism between SMUG1 and let-7b-5p (Figure 4).
microRNA expression is often altered in malignancies and although some have been implicated and well-studied in breast cancer, for example let-7a, little is known about the specific relationship of let-7b, breast cancer subtypes and clinical outcomes (51,54,55). A few independent studies observed deregulation of let-7b during early breast cancer progression and showed that its expression is downregulated during epithelial-mesenchymal transition and associated with less aggressive breast cancer (54–58). It has been suggested that let-7b has tumor suppressor properties in breast cancer development and progression. Accordingly, SMUG1 KO breast cancer cells with elevated let-7b-5p levels showed accumulation in the G1 phase, consistent with reduced proliferation. With respect to cell mobility, we observed reduced cell migration for the clones analyzed, albeit with some clonal variation in the magnitude of the effect (Figures 5 and 6, Supplementary Figure S12). We also found a significant negative correlation between let-7b expression and patient overall survival, relapse-free survival and tumor lymph node metastasis in breast cancer cohorts (51–53). Even though let-7b levels are generally reduced in tumor tissues, its expression varies within the different subtypes and as a function of the grade, with its level decreasing as the grade increases. In less differentiated and more aggressive subtypes, as basal-like and HER2+, let-7b expression is reduced; on the contrary, relative increased levels are described for luminal A, luminal B and normal-like tumors (51,59). Bioinformatic analyses of SMUG1 mRNA expression in different breast cancer cohorts showed increased SMUG1 levels in tumor compared to control samples (Figure 1). Even though ER+ datasets showed increased SMUG1 mRNA levels when compared to ER− tumors and normal tissue, reduced levels of SMUG1 were associated with better survival, especially in the ER+ dataset of the cohorts analyzed (Figure 1 and Supplementary Figure S2). Thus, the higher survival observed for low SMUG1 in ER+ cohorts could be associated with the increased levels of let-7b-5p and its tumor suppressor activities rather than a direct role of SMUG1 in tumorigenesis. A previous study reported that low SMUG1 expression was associated with worse prognosis in the UPSA breast cancer cohort, and this effect was predominant in ER+ tumors. On the contrary, in ER− tumors that received chemotherapy and in gastric cancers low SMUG1 expression showed better survival, suggesting a complex role for SMUG1 in carcinogenesis (16). It is difficult to compare our results to those previously observed, because the final Kaplan–Meier result could be influenced by several factors such as the normalization method of the microarray data and the stratification of the SMUG1 groups. These differences led us to look in more detail into the UPSA data and we generated Kaplan–Meier curves by splitting the cohorts according to the expression of genes with known association with survival, i.e.ESR1, ERBB2 and MKI67, and found the expected results, such as higher ERBB2 expression associates with worse survival or high ESR1 with better outcome. These conflicting results suggest that SMUG1 mRNA expression alone cannot be used as a robust prognostic marker for breast cancer nor its response to adjuvant therapy.
SMUG1 protein-protein interactors or its associated RNAs form a complex network that affects SMUG1 functions and influence the surrounding cellular environment. Since SMUG1 is a multifunctional protein, analyzing its PPI could reveal how its different functions are working together to determine its impact on breast cancer survival, where the dominating effects may be ascribed to functions other than DNA repair, as previously described for Apurinic/apyrimidinic Endonuclease 1, another key DNA repair protein in the BER pathway (60). We characterized the SMUG1 protein–protein interactome via a combined approach of Y2H and immunoprecipitation. We analyzed the impact of SMUG1 interactors, defined as bad prognosis genes in TCGA datasets, on breast cancer cohort showing lower survival probability in patients having these genes highly expressed (Figure 3 and Supplementary Figure S5). Taken together our results suggest that SMUG1 role in breast cancer is associated with RNA-related functions and its protein-protein network rather than only its DNA repair activity. Inefficient DNA repair and defective DNA damage response increase the mutation frequency, leading to genomic instability and cancer development (61). Even though SMUG1 was suspected to have an anti-tumor role via preventing the accumulation of mutations arising from deamination of cytosine residues and from several pyrimidine oxidation products (i.e. 5-hmU, 5-fU and 5-caU) (5–8,10,42), previous studies showed a modest increase in the mutagenesis ratio in MEFs knock-out for SMUG1 (41). Apart from its role in DNA repair, SMUG1 is involved in RNA metabolism and RNA quality control (9,23). In fact, increased SMUG1 protein levels significantly protect against killing 5-fluorouracil (FU)-treated cells, increasing cell and drug resistance in tumors (62). All drugs used in adjuvant chemotherapy induce ribosome biogenesis defects (63) suggesting that the role of SMUG1 in RNA metabolism may affect the treatment response. A role for SMUG1 in RNA metabolism may also be consistent with the observation that SMUG1 contributes to the cellular response to recovery from FU (4).
We conclude that the correlation of SMUG1 alone with survival does probably not reflect a direct role of SMUG1 in cancer development and it should not be used as a prognostic marker. However, the present analysis suggests that SMUG1 is part of a gene regulatory network (possibly regulated by miRNAs) that influence survival and treatment response in several cancers.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (64) partner repository with the dataset identifier PXD025929. Data associated with FACS analysis is available in FlowRepository under accession FR-FCM-Z5F8, FR-FCM-Z5FA, FR-FCM-Z5FC and FR-FCM-Z5FD. All other datasets presented in this manuscript are available online.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Christian Köhler and Bernd Thiede for mass spectrometry analyses, Anna Lång and Stig Ove Bøe for microscopy analyses, Francisco José Naranjo Galindo for flow cytometry assistance, and Kulbhushan Sharma for sharing reagents and discussion on gene editing. High-content imaging was performed at the UiO/OUS Core facility for Advanced Light Microscopy (the Gaustad node). Mass spectrometry-based proteomic analyses were performed by the Proteomics Core Facility, Department of Biosciences, University of Oslo. This facility is a member of the National Network of Advanced Proteomics Infrastructure (NAPI), which is funded by the Research Council of Norway INFRASTRUKTUR-program (project number: 295910).
Notes
Present address: Dilara Ayyildiz, Princess Máxima Center for Pediatric Oncology Heidelberglaan 25, 3584 CS Utrecht, The Netherlands.
Present address: Laure Jobert, LifeTechnologies AS, Ullernschauseen 52, 0379 Oslo, Norway.
Contributor Information
Lisa Lirussi, Institute of Clinical Medicine, Department of Clinical Molecular Biology, University of Oslo, N-0318 Oslo, Norway; Section of Clinical Molecular Biology, Akershus University Hospital (AHUS), 1478 Lørenskog, Norway.
Dilara Ayyildiz, Laboratory of Molecular Biology and DNA repair, Department of Medicine, University of Udine, p.le M. Kolbe 4, 33100 Udine, Italy.
Yan Liu, Section of Clinical Molecular Biology, Akershus University Hospital (AHUS), 1478 Lørenskog, Norway.
Nicola P Montaldo, Institute of Clinical Medicine, Department of Clinical Molecular Biology, University of Oslo, N-0318 Oslo, Norway.
Sergio Carracedo, Institute of Clinical Medicine, Department of Clinical Molecular Biology, University of Oslo, N-0318 Oslo, Norway; Section of Clinical Molecular Biology, Akershus University Hospital (AHUS), 1478 Lørenskog, Norway.
Miriam R Aure, Department of Medical Genetics, Institute of Clinical Medicine, Faculty of Medicine, University of Oslo and Oslo University Hospital, 0450 Oslo, Norway.
Laure Jobert, Institute of Clinical Medicine, Department of Clinical Molecular Biology, University of Oslo, N-0318 Oslo, Norway.
Xavier Tekpli, Department of Medical Genetics, Institute of Clinical Medicine, Faculty of Medicine, University of Oslo and Oslo University Hospital, 0450 Oslo, Norway.
Joel Touma, Department of Breast and Endocrine Surgery, Akershus University Hospital (AHUS), 1478 Lørenskog, Norway; Institute of Clinical Medicine, University of Oslo, Campus AHUS, 1478 Lørenskog, Norway.
Torill Sauer, Institute of Clinical Medicine, University of Oslo, Campus AHUS, 1478 Lørenskog, Norway; Department of Pathology, Akershus University Hospital, 1478 Lørenskog, Norway.
Emiliano Dalla, Laboratory of Molecular Biology and DNA repair, Department of Medicine, University of Udine, p.le M. Kolbe 4, 33100 Udine, Italy.
Vessela N Kristensen, Department of Medical Genetics, Institute of Clinical Medicine, Faculty of Medicine, University of Oslo and Oslo University Hospital, 0450 Oslo, Norway; Department of Pathology, Akershus University Hospital, 1478 Lørenskog, Norway.
Jürgen Geisler, Institute of Clinical Medicine, University of Oslo, Campus AHUS, 1478 Lørenskog, Norway; Department of Oncology, Akershus University Hospital (AHUS), 1478 Lørenskog, Norway.
Silvano Piazza, Bioinformatics Core Facility, Centre for Integrative Biology (CIBIO), University of Trento, via Sommarive 18, 38123, Povo (Trento), Italy.
Gianluca Tell, Laboratory of Molecular Biology and DNA repair, Department of Medicine, University of Udine, p.le M. Kolbe 4, 33100 Udine, Italy.
Hilde Nilsen, Institute of Clinical Medicine, Department of Clinical Molecular Biology, University of Oslo, N-0318 Oslo, Norway; Section of Clinical Molecular Biology, Akershus University Hospital (AHUS), 1478 Lørenskog, Norway; Department of Microbiology, Oslo University Hospital, N-0424 Oslo, Norway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Council of Norway [229633]; South East Regional Health Authority [274901 to H.N.]; Norwegian Cancer Society [4501723-2015 to L.L.]. Funding for open access charge: Norwegian Cancer Association. The research leading to these results has received funding from AIRC under IG 2017 [ID. 19862 project to GT].
Conflict of interest statement. None declared.
REFERENCES
- 1. Haushalter K.A., Todd Stukenberg M.W., Kirschner M.W., Verdine G.L.. Identification of a new uracil–DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol. 1999; 9:174–185. [DOI] [PubMed] [Google Scholar]
- 2. Mi R., Dong L., Kaulgud T., Hackett K.W., Dominy B.N., Cao W.. Insights from xanthine and uracil DNA glycosylase activities of bacterial and human SMUG1: switching SMUG1 to UDG. J. Mol. Biol. 2009; 385:761–778. [DOI] [PubMed] [Google Scholar]
- 3. Kavli B., Sundheim O., Akbari M., Otterlei M., Nilsen H., Skorpen F., Aas P.A., Hagen L., Krokan H.E., Slupphaug G.. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002; 277:39926–39936. [DOI] [PubMed] [Google Scholar]
- 4. Nagaria P., Svilar D., Brown A.R., Wang X.H., Sobol R.W., Wyatt M.D.. SMUG1 but not UNG DNA glycosylase contributes to the cellular response to recovery from 5-fluorouracil induced replication stress. Mutat. Res. 2013; 743-744:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Masaoka A., Matsubara M., Hasegawa R., Tanaka T., Kurisu S., Terato H., Ohyama Y., Karino N., Matsuda A., Ide H.. Mammalian 5-formyluracil–DNA glycosylase. 2. Role of SMUG1 uracil–DNA glycosylase in repair of 5-formyluracil and other oxidized and deaminated base lesions. Biochemistry. 2003; 42:5003–5012. [DOI] [PubMed] [Google Scholar]
- 6. Darwanto A., Theruvathu J.A., Sowers J.L., Rogstad D.K., Pascal T., Goddard W., Sowers L.C.. Mechanisms of base selection by human single-stranded selective monofunctional uracil–DNA glycosylase. J. Biol. Chem. 2009; 284:15835–15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boorstein R.J., Cummings A., Marenstein D.R., Chan M.K., Ma Y., Neubert T.A., Brown S.M., Teebor G.W. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem. 2001; 276:41991–41997. [DOI] [PubMed] [Google Scholar]
- 8. Kemmerich K., Dingler F.A., Rada C., Neuberger M.S.. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung-/-Msh2-/- mice. Nucleic Acids Res. 2012; 40:6016–6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jobert L., Skjeldam H.K., Dalhus B., Galashevskaya A., Vagbo C.B., Bjoras M., Nilsen H.. The human base excision repair enzyme SMUG1 directly interacts with DKC1 and contributes to RNA quality control. Mol. Cell. 2013; 49:339–345. [DOI] [PubMed] [Google Scholar]
- 10. Alsoe L., Sarno A., Carracedo S., Domanska D., Dingler F., Lirussi L., SenGupta T., Tekin N.B., Jobert L., Alexandrov L.B.et al.. Uracil accumulation and mutagenesis dominated by cytosine deamination in CpG dinucleotides in mice lacking UNG and SMUG1. Sci. Rep. 2017; 7:7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pfaffeneder T., Spada F., Wagner M., Brandmayr C., Laube S.K., Eisen D., Truss M., Steinbacher J., Hackner B., Kotljarova O.et al.. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014; 10:574–581. [DOI] [PubMed] [Google Scholar]
- 12. Theruvathu J.A., Darwanto A., Hsu C.W., Sowers L.C.. The effect of pot1 binding on the repair of thymine analogs in a telomeric DNA sequence. Nucleic Acids Res. 2014; 42:9063–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cui J., Gizzi A., Stivers J.T.. Deoxyuridine in DNA has an inhibitory and promutagenic effect on RNA transcription by diverse RNA polymerases. Nucleic Acids Res. 2019; 47:4153–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schuermann D., Weber A.R., Schar P.. Active DNA demethylation by DNA repair: facts and uncertainties. DNA Repair (Amst.). 2016; 44:92–102. [DOI] [PubMed] [Google Scholar]
- 15. Kavli B., Otterlei M., Slupphaug G., Krokan H.E.. Uracil in DNA–general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst.). 2007; 6:505–516. [DOI] [PubMed] [Google Scholar]
- 16. Abdel-Fatah T.M., Albarakati N., Bowell L., Agarwal D., Moseley P., Hawkes C., Ball G., Chan S., Ellis I.O., Madhusudan S.. Single-strand selective monofunctional uracil–DNA glycosylase (SMUG1) deficiency is linked to aggressive breast cancer and predicts response to adjuvant therapy. Breast Cancer Res. Treat. 2013; 142:515–527. [DOI] [PubMed] [Google Scholar]
- 17. Abdel-Fatah T.M., Perry C., Arora A., Thompson N., Doherty R., Moseley P.M., Green A.R., Chan S.Y., Ellis I.O., Madhusudan S.. Is there a role for base excision repair in estrogen/estrogen receptor-driven breast cancers?. Antioxid. Redox Signal. 2014; 21:2262–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oliveira D.M., Laudanna C., Migliozzi S., Zoppoli P., Santamaria G., Grillone K., Elia L., Mignogna C., Biamonte F., Sacco R.et al.. Identification of different mutational profiles in cancers arising in specific colon segments by next generation sequencing. OncoTargets Ther. 2018; 9:23960–23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Broderick P., Bagratuni T., Vijayakrishnan J., Lubbe S., Chandler I., Houlston R.S.. Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer. 2006; 6:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xie H., Gong Y., Dai J., Wu X., Gu J.. Genetic variations in base excision repair pathway and risk of bladder cancer: a case-control study in the united states. Mol. Carcinog. 2015; 54:50–57. [DOI] [PubMed] [Google Scholar]
- 21. Ye F., Wang H., Liu J., Cheng Q., Chen X., Chen H.. Association of SMUG1 SNPs in intron region and linkage disequilibrium with occurrence of cervical carcinoma and HPV infection in chinese population. J. Cancer. 2019; 10:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Juul M., Bertl J., Guo Q., Nielsen M.M., Switnicki M., Hornshoj H., Madsen T., Hobolth A., Pedersen J.S.. Non-coding cancer driver candidates identified with a sample- and position-specific model of the somatic mutation rate. Elife. 2017; 6:e21778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kroustallaki P., Lirussi L., Carracedo S., You P., Esbensen Q.Y., Gotz A., Jobert L., Alsoe L., Saetrom P., Gagos S.et al.. SMUG1 promotes telomere maintenance through telomerase RNA processing. Cell Rep. 2019; 28:1690–1702. [DOI] [PubMed] [Google Scholar]
- 24. Benes V., Collier P., Kordes C., Stolte J., Rausch T., Muckentaler M.U., Haussinger D., Castoldi M.. Identification of cytokine-induced modulation of microRNA expression and secretion as measured by a novel microRNA specific qPCR assay. Sci. Rep. 2015; 5:11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rabani M., Levin J.Z., Fan L., Adiconis X., Raychowdhury R., Garber M., Gnirke A., Nusbaum C., Hacohen N., Friedman N.et al.. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011; 29:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W.. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017; 115:80–90. [DOI] [PubMed] [Google Scholar]
- 27. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li T., Wernersson R., Hansen R.B., Horn H., Mercer J., Slodkowicz G., Workman C.T., Rigina O., Rapacki K., Staerfeldt H.H.et al.. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat. Methods. 2017; 14:61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pages F., Trajanoski Z., Galon J.. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009; 25:1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y.. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014; 8(Suppl. 4):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Risso D., Ngai J., Speed T.P., Dudoit S.. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014; 32:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jezequel P., Frenel J.S., Campion L., Guerin-Charbonnel C., Gouraud W., Ricolleau G., Campone M.. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database (Oxford). 2013; 2013:bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J.. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019; 47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Merico D., Isserlin R., Stueker O., Emili A., Bader G.D.. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010; 5:e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., Wadi L., Meyer M., Wong J., Xu C.et al.. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, cytoscape and enrichmentmap. Nat. Protoc. 2019; 14:482–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kucera M., Isserlin R., Arkhangorodsky A., Bader G.D.. AutoAnnotate: a cytoscape app for summarizing networks with semantic annotations. F1000Res. 2016; 5:1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sticht C., De La Torre C., Parveen A., Gretz N.. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018; 13:e0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G.. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015; 43:W460–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeng Xiao, Y., Zhu J., Peng C., Liang W., Lin H. Knockdown of nucleophosmin 1 suppresses proliferation of triple-negative breast cancer cells through activating CDH1/Skp2/p27kip1 pathway. Cancer Manag Res. 2019; 11:143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. An Q., Robins P., Lindahl T., Barnes D.E.. C → t mutagenesis and gamma-radiation sensitivity due to deficiency in the smug1 and ung DNA glycosylases. EMBO J. 2005; 24:2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nilsen H., Haushalter K.A., Robins P., Barnes D.E., Verdine G.L., Lindahl T.. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil–DNA glycosylase. EMBO J. 2001; 20:4278–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Unnithan S.K.R., Kannan B., Jathavedan M.. Betweenness centrality in some classes of graphs. Int. J. Combinatorics. 2014; 2014:12. [Google Scholar]
- 44. Chirshev E., Oberg K.C., Ioffe Y.J., Unternaehrer J.J.. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019; 8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Joosse S.A., Muller V., Steinbach B., Pantel K., Schwarzenbach H.. Circulating cell-free cancer-testis MAGE-A RNA, BORIS RNA, let-7b and miR-202 in the blood of patients with breast cancer and benign breast diseases. Br. J. Cancer. 2014; 111:909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Qattan A., Intabli H., Alkhayal W., Eltabache C., Tweigieri T., Amer S.B.. Robust expression of tumor suppressor miRNA’s let-7 and miR-195 detected in plasma of Saudi female breast cancer patients. BMC Cancer. 2017; 17:799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li X.X., Gao S.Y., Wang P.Y., Zhou X., Li Y.J., Yu Y., Yan Y.F., Zhang H.H., Lv C.J., Zhou H.H.et al.. Reduced expression levels of let-7c in human breast cancer patients. Oncol. Lett. 2015; 9:1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li M., Guan X., Sun Y., Mi J., Shu X., Liu F., Li C.. miR-92a family and their target genes in tumorigenesis and metastasis. Exp. Cell. Res. 2014; 323:1–6. [DOI] [PubMed] [Google Scholar]
- 49. Xi X., Chu Y., Liu N., Wang Q., Yin Z., Lu Y., Chen Y.. Joint bioinformatics analysis of underlying potential functions of hsa-let-7b-5p and core genes in human glioma. J. Transl. Med. 2019; 17:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karagkouni D., Paraskevopoulou M.D., Chatzopoulos S., Vlachos I.S., Tastsoglou S., Kanellos I., Papadimitriou D., Kavakiotis I., Maniou S., Skoufos G.et al.. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018; 46:D239–D245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bozgeyik E. Bioinformatic analysis and in vitro validation of Let-7b and Let-7c in breast cancer. Comput. Biol. Chem. 2020; 84:107191. [DOI] [PubMed] [Google Scholar]
- 52. Ma L., Li G.Z., Wu Z.S., Meng G.. Prognostic significance of let-7b expression in breast cancer and correlation to its target gene of BSG expression. Med. Oncol. 2014; 31:773. [DOI] [PubMed] [Google Scholar]
- 53. Wang H., Ren Y., Qian C., Liu J., Li G., Li Z.. Over-expression of CDX2 alleviates breast cancer by up-regulating microRNA let-7b and inhibiting COL11A1 expression. Cancer Cell Int. 2020; 20:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Quesne J.L., Jones J., Warren J., Dawson S.J., Ali H.R., Bardwell H., Blows F., Pharoah P., Caldas C.. Biological and prognostic associations of miR-205 and let-7b in breast cancer revealed by in situ hybridization analysis of micro-RNA expression in arrays of archival tumour tissue. J. Pathol. 2012; 227:306–314. [DOI] [PubMed] [Google Scholar]
- 55. van Schooneveld E., Wildiers H., Vergote I., Vermeulen P.B., Dirix L.Y., Van Laere S.J.. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res. 2015; 17:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J.et al.. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007; 131:1109–1123. [DOI] [PubMed] [Google Scholar]
- 57. Dangi-Garimella S., Yun J., Eves E.M., Newman M., Erkeland S.J., Hammond S.M., Minn A.J., Rosner M.R.. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009; 28:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sempere L.F., Christensen M., Silahtaroglu A., Bak M., Heath C.V., Schwartz G., Wells W., Kauppinen S., Cole C.N.. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007; 67:11612–11620. [DOI] [PubMed] [Google Scholar]
- 59. Blenkiron C., Goldstein L.D., Thorne N.P., Spiteri I., Chin S.F., Dunning M.J., Barbosa-Morais N.L., Teschendorff A.E., Green A.R., Ellis I.O.et al.. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007; 8:R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ayyildiz D., Antoniali G., D’Ambrosio C., Mangiapane G., Dalla E., Scaloni A., Tell G., Piazza S.. Architecture of the human ape1 interactome defines novel cancers signatures. Sci. Rep. 2020; 10:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Alhmoud J.F., Woolley J.F., Al Moustafa A.E., Malki M.I. DNA damage/repair management in cancers. Cancers (Basel). 2020; 12:1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. An Q., Robins P., Lindahl T., Barnes D.E.. 5-Fluorouracil incorporated into DNA is excised by the smug1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007; 67:940–945. [DOI] [PubMed] [Google Scholar]
- 63. Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I.. The nucleolus under stress. Mol. Cell. 2010; 40:216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M.et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (64) partner repository with the dataset identifier PXD025929. Data associated with FACS analysis is available in FlowRepository under accession FR-FCM-Z5F8, FR-FCM-Z5FA, FR-FCM-Z5FC and FR-FCM-Z5FD. All other datasets presented in this manuscript are available online.