Abstract
The RNA-binding protein tristetraprolin (TTP) is a potent activator of mRNA decay, specifically for transcripts bearing AU-rich elements (AREs) in their 3′-untranslated regions. TTP functions as a mediator for mRNA decay by interacting with the decay machinery and recruiting it to the target ARE-mRNA. In this study, we report a weak, but direct interaction between TTP and the human decapping enzyme DCP2, which impacts the stability of ARE transcripts. The TTP–DCP2 interaction is unusual as it involves intrinsically disordered regions (IDRs) of both binding partners. We show that the IDR of DCP2 has a propensity for oligomerization and liquid–liquid phase separation in vitro. Binding of TTP to DCP2 leads to its partitioning into phase-separated droplets formed by DCP2, suggesting that molecular crowding might facilitate the weak interaction between the two proteins and enable assembly of a decapping-competent mRNA–protein complex on TTP-bound transcripts in cells. Our studies underline the role of weak interactions in the cellular interaction network and their contribution towards cellular functionality.
INTRODUCTION
Degradation of mRNA serves as an important mechanism for achieving rapid changes in the gene expression profile of a cell in response to external stimuli. Half-lives of mRNA can vary drastically in eukaryotes: transcripts encoding cytokines, proto-oncogenes and transcription factors are highly labile and are degraded within minutes, while those of housekeeping genes are stable over hours (1). The intrinsic stability of an mRNA transcript is often determined by specific sequences or structural elements that are usually located in the 3′-untranslated region (3′-UTR) of the mRNA. These cis-acting elements are recognized by distinct trans-acting protein factors, which in turn recruit the mRNA degradation machinery to the transcript to ensure its timely decay (2). A predominant cis-acting element in higher eukaryotes is the AU-rich element (ARE), a nonameric sequence motif rich in adenines and uridines found in multiple copies in the 3′-UTR, which is recognized with high specificity and affinity by AU-binding proteins (AUBPs) (3).
Of the AUBPs competent in triggering mRNA decay, the most extensively studied are the protein tristetraprolin (abbreviated as TTP and also known as Tis11/ZFP36) and its paralogues BRF1 (Tis11b/ZFP36L1) and BRF2 (Tis11d/ZFP36L2) (4). TTP and its paralogues are characterized by the presence of two CCCH zinc finger motifs arranged into a tandem zinc finger (TZF) domain, which is flanked at the N- and C-termini by long stretches of low complexity sequences (Figure 1A; Supplementary Figure S2D). The central TZF domain binds RNA with high affinity and is fairly well conserved among the paralogues (5), while the flanking regions diverge considerably in length and sequence (Supplementary Figure S1A). The N- and C-terminal stretches are referred to as ‘activation domains’ for their role in recruiting components of the mRNA degradation machinery to activate mRNA decay (6). Co-immunoprecipitation studies indicate that TTP and BRF1 are capable of recruiting the CCR4–NOT deadenylase complex and components of the RNA exosome, as well as the decapping machinery (DCP1, DCP2 and EDC3) and the 5′–3′ exonuclease Xrn1 (6–9). The N-terminal activation domain of TTP was shown to be important for decapping of an ARE substrate, while the C-terminal domain engages the CNOT1 protein to recruit the CCR4–NOT complex (7,10,11). Recruitment of the CCR4–NOT complex to an RNA substrate establishes a vast interaction network, involving factors that mediate decapping and translational repression, in addition to deadenylation and subsequent 3′–5′ exonucleolytic decay (12). Therefore, a direct interaction of TTP with CNOT1 is an efficient means of triggering rapid mRNA decay. The NOT1-binding region of TTP was mapped to a conserved sequence motif at its very C-terminus (denoted as NIM in Figure 1A and Supplementary Figure S1A) (10). Interestingly, a TTP construct lacking this motif was still capable of mediating decay of an ARE substrate, suggesting that additional motifs in TTP engage with the degradation machinery independently of CNOT1 to facilitate mRNA decay, and ensure redundancy in this process (10,11).
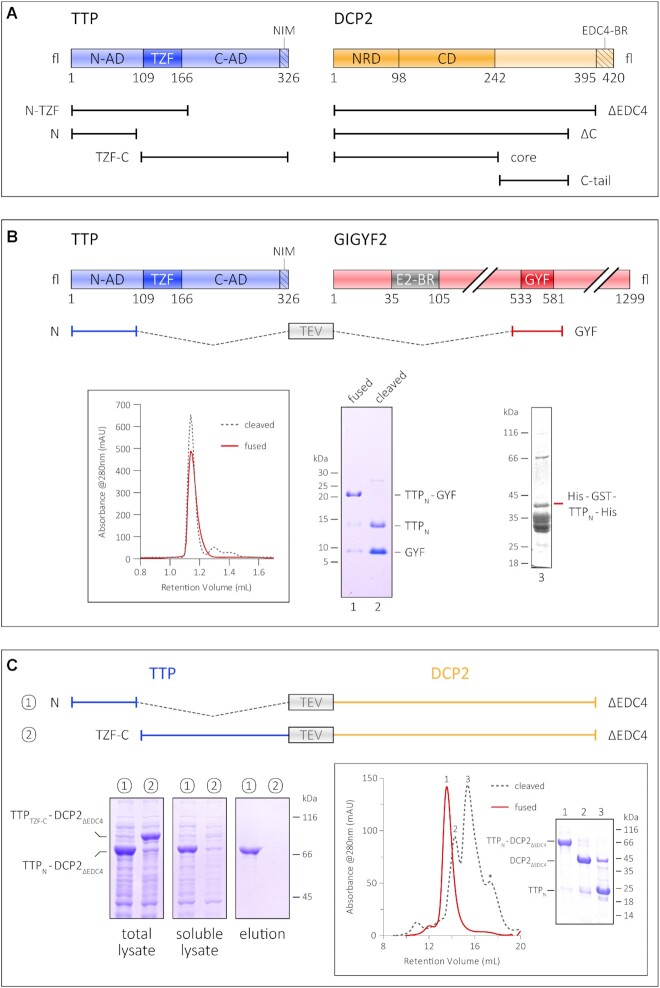
Figure 1.
Interaction of the TTP N-terminal activation domain with GIGYF2 and DCP2. (A) Schematic representation of the domain organization of TTP and DCP2 and the constructs used in this study. The central tandem zinc finger domain of TTP (dark blue) is flanked by the unstructured N- and C-terminal activation domains (N-AD and C-AD, respectively) on either side (light blue). The NOT1-interacting motif (NIM) at the very C-terminus of TTP is indicated by the hatched box. The structured core of DCP2 (dark orange) comprises its N-terminal regulatory domain (NRD) and its catalytic Nudix domain (CD), and is followed by a long, unstructured C-terminal region (light orange), the last 25 residues of which harbour a binding motif for EDC4 (EDC4-BR, indicated by the hatched box). (B) The GYF domain of GIGYF2 restores proteolytic stability of the TTP N-terminal activation domain (TTPN). A schematic representation of GIGYF2 indicates the position of the GYF domain (dark red) in the primary structure of the protein, with respect to other binding motifs, such as the eIF4E2-binding motif (in grey, denoted by E2-BR). The fusion polypeptide of TTPN and GYF, separated by a linker sequence bearing a TEV protease cleavage site, is shown. Analytical SEC and the corresponding SDS–PAGE analysis of peak fractions depict the stability of TTPN in complex with GYF. The retention volume of the single polypeptide (fused) is identical to that of non-covalently linked complex (cleaved, referring to cleavage by TEV protease), indicating formation of a stable complex. TTPN in complex with GYF is stable even upon TEV cleavage of the fusion polypeptide. In contrast, fusion of a non-interacting protein such as GST does not rescue the proteolytic instability of TTPN. (C) Schematic representation of the TTP–DCP2 fusion polypeptides, designed similarly to the TTPN–GYF fusion described above, and comparison of the stability of TTPN–DCP2ΔEDC4 with that of TTPTZF–C–DCP2ΔEDC4 by Ni2+-affinity pulldowns. Both fusion proteins are well expressed, but only TTPN–DCP2ΔEDC4 can be isolated by Ni2+-affinity purification, indicating that fusion of DCP2 rescues the behaviour of TTPN but not that of TTPTZF-C. The left, middle and right panels indicate total lysate, soluble lysate and elution, respectively. Total lysate and soluble lysate refer to samples collected before and after centrifugation of the cell suspension after sonication. Inset: analytical SEC and the corresponding SDS–PAGE analysis of peak fractions of the intact TTPN–DCP2ΔEDC4 fusion and of the protein cleaved with TEV protease. Cleavage of the fusion yields proteolytically stable TTPN, although it is not stably associated with DCP2ΔEDC4 in SEC.
To understand the contribution of the N-terminal activation domain towards TTP-mediated decay, we set out to investigate the interaction between TTP and the decapping enzyme DCP2. DCP2 is a bilobed protein consisting of an N-terminal regulatory domain (NRD) and a catalytic Nudix domain (CD) that mediates hydrolysis of the 7-methyl guanosine cap of eukaryotic mRNA (13–16). The structured core is followed by an unstructured C-terminal extension that harbours motifs for anchoring proteins such as the enhancer of decapping 4 (EDC4, also known as Hedls), which is a scaffold for assembly of decapping factors in metazoans (Figure 1A) (17). We found a weak, but direct interaction between the TTP N-terminal activation domain and DCP2 that is mediated by intrinsically disordered regions (IDRs) of both proteins and show that decay activation by the TTP N-terminal domain is dependent on DCP2. We propose a mechanism for how molecular crowding in cells might promote the weak interaction between TTP and DCP2 and facilitate assembly of decay-competent messenger ribonucleoprotein complexes (mRNPs) on ARE transcripts.
MATERIALS AND METHODS
Protein expression and purification
All protein sequences used in this study are of human origin.
TTPN
Residues 1–99 of TTP, corresponding to the N-terminal activation domain (TTPN), were cloned into a modified pET28a vector bearing an N-terminal 6×His-MBP (maltose-binding protein) tag, followed by a Tobacco etch virus (TEV) protease cleavage site. The protein was expressed in Escherichia coli BL21(DE3) gold pLysS cells grown in Terrific broth (TB). Protein expression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 18°C. The culture was harvested 20 h post-induction and the pellet was resuspended in lysis buffer (50 mM Tris pH 7.5, 1000 mM NaCl, 10 mM imidazole, 10% glycerol, 5 mM β-mercaptoethanol) supplemented with 1 mM phenylmethylsulphonyl fluoride (PMSF) and 0.5 mg of DNase I. Cells were lysed by sonication, clarified by centrifugation and filtration, and subjected to Ni2+-affinity chromatography (IMAC) to isolate the target protein. His-MBP–TTPN was eluted from the Ni2+-affinity resin (Machery-Nagel #745400.100) in elution buffer (lysis buffer supplemented with 250 mM imidazole). To cleave the His-MBP tag, TEV protease was added to the eluted protein to a final ratio of 1:50 (w/w). The protein mixture was dialysed to remove imidazole and subjected to a reverse Ni2+-affinity purification (the flowthrough containing primarily TTPN and small amounts of His-MBP was collected). Following removal of His-MBP contaminants by incubation with dextrin resin, the TTPN protein was concentrated and loaded on a size-exclusion chromatography (SEC) column (HiLoad 26/60 Superdex 75 pg, GE Healthcare). Peak fractions from the first SEC step, performed in SEC buffer A (20 mM Tris pH 7.5, 1000 mM NaCl, 5% glycerol), were pooled and reloaded onto the SEC column to remove any traces of His-MBP contaminants. Peak fractions from the second SEC step, performed in SEC buffer B (50 mM potassium phosphate pH 7.0, 50 mM NaCl), were pooled, concentrated and flash-frozen in liquid nitrogen. In this and all other purifications, protein purity was monitored by sodium dodecylsulphate–polyacrylamide gel electrophoresis (SDS–PAGE) analyses. The purification of TTPN for Förster resonance energy transfer (FRET) and liquid–liquid phase separation (LLPS) is described in detail in the Supplementary Methods.
DCP2 proteins and GYF
DCP2ΔC and DCP2core were cloned into a modified pET28a vector with an N-terminal 6×His-thioredoxin (Trx) tag, whereas DCP2C-tail and GYF were cloned into pET28a with an N-terminal 6×His tag. Proteins were expressed in E. coli BL21(DE3) Star pRare as described above for TTPN. All proteins were purified using a combination of Ni2+-affinity chromatography and SEC, employing lysis buffer A and SEC buffer A. An intermediate heparin-affinity chromatography step (Hitrap Heparin column 5 ml) was additionally performed for DCP2ΔC. The affinity tags (6×His or 6×His-Trx) were removed by cleavage with human rhinovirus (HRV) 3C protease prior to SEC.
GST-tagged proteins
GYF and DCP2 proteins were cloned into a modified pET vector bearing an N-terminal 6×His-glutathione S-transferase (GST) tag. The protease cleavage site between the GST tag and the protein of interest was replaced by a seven residue long glycine–serine–alanine-rich linker, resulting in an uncleavable His-GST tag in all constructs. The proteins were expressed and purified by Ni2+-affinity chromatography, as described above for His-MBP–TTPN, followed by SEC (Hiload Superdex 200 16/600 for His-GST–DCP2ΔC/DCP2core and Hiload Superdex 75 16/600 for His-GST2–DCP2C-tail/GYF) in SEC buffer A. As with His-Trx–DCP2ΔC, an intermediate heparin-affinity chromatography step was performed for His-GST–DCP2ΔC. Peak fractions were pooled and flash-frozen as above.
GST–SMG6TPR (residues 520–1186) was a gift from Elena Conti (Max Planck Institute of Biochemistry, Martinsried, Germany).
Fluorescent labelling of proteins
DCP2C-tail was labelled with Cy3 (Cytiva Amersham Cy3 Maleimide Mono-Reactive Dye, product no. PA23031) and TTPN with Cy5 (Cytiva Amersham Cy5 Maleimide Mono-Reactive Dye, product no. PA25031) for FRET experiments. Both constructs contained a single cysteine which was coupled to the dye via a maleimide reactive group. Each protein (200 nmol) diluted in labelling buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 100 μM TCEP) was pre-incubated at 37°C for 30 min before adding one vial of fluorescent dye resuspended in 50 μl of dimethylsulphoxide (DMSO). The reaction (total volume of 2 ml) was incubated at 25°C for 2 h, followed by an overnight incubation at 4°C. The excess dye was separated from the labelled protein by SEC (Superdex 75 10/300 GL, GE Healthcare) performed in SEC buffer D (20 mM HEPES pH 7.5, 100 mM NaCl). The SEC run was monitored at 280 nm to detect the protein as well as at 550 nm (for Cy3) or 650 nm (for Cy5) to track the fluorescent label. Peak fractions containing the labelled protein were pooled and concentrated. DCP2 proteins for fluorescence microscopy studies were labelled with maleimide-reactive XFD488 dye (AAT Bioquest #1878, identical to Alexafluor488) using a similar protocol as for Cy3 labelling of the protein. All buffers used for XFD488 labelling of DCP2 contained 150 mM NaCl. Fluorescent dye at 100 nmol was added to 100 nmol (DCP2C-tail) or 50 nmol (DCP2core and DCP2ΔC) of protein in a total volume of 1 ml. The SEC run was monitored at 280 nm and 499 nm.
FRET measurements
The experimental setup and analysis of FRET were performed according to (18). Briefly, DCP2C-tail was used as the FRET donor and TTPN as the FRET acceptor in our setup. DCP2 was maintained at a fixed final concentration of 5 μM, while TTP was titrated in increasing concentrations, from 0 to 50 μM. In addition to the FRET reaction containing both labelled proteins, a donor bleed-through control (Cy3-DCP2C-tail and unlabelled TTPN) and an acceptor direct excitation control (unlabelled DCP2C-tail and Cy5-TTPN) were set up for each titration point. Proteins were mixed in a total volume of 35 μl in SEC buffer C (20 mM HEPES pH 7.5, 150 mM NaCl). Following overnight incubation at 4°C, samples were transferred to a black PerkinElmer OptiPlate 384-F and measured with a Tecan Spark plate reader at 20°C (20 flashes, band width of 10 nm) in three different settings: FRET excitation at 520 nm and detection at 667 nm; donor bleed-through excitation at 520 nm and detection at 565 nm; and acceptor direct excitation at 640 nm and detection at 667 nm. FRET values were corrected for donor bleed-through and acceptor direct excitation, after being adjusted against the buffer background. Values within a replicate series were normalized against the 35 μM TTP data point and fitted to a one site-specific binding model with Hill slope using GraphPad Prism 5.
GST pulldowns
A 5–8 μg aliquot of GST-tagged bait proteins and 25 μg of prey protein were mixed in GST pulldown buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 10% glycerol, 0.1% NP-40) to a total volume of 40 μl. Approximately 15% of the reaction was used as an input for the SDS–PAGE analysis. The reaction mixture was incubated at 4°C for 6 h, following which 12 μl of a 50% slurry of glutathione Sepharose resin (GE Healthcare) was added. The mixture was further supplemented with 200 μl of GST pulldown buffer and incubated at 4°C for 1 h. The beads were extensively washed with GST pulldown buffer. Bound proteins were eluted in GST pulldown buffer supplemented with 30 mM reduced glutathione and analysed by SDS–PAGE and Coomassie or silver staining.
Ni2+-affinity pulldowns
Fusions of TTP and DCP2 connected via a TEV protease-cleavable linker were cloned into a modified pET28a vector bearing an N-terminal 6×His-Trx tag. Escherichia coli BL21(DE3) gold pLysS cells transformed with these plasmids were grown in 40 ml of TB cultures at 37°C to an optical density at 600 nm (OD600 nm) of 2.5. Protein expression was induced with 0.5 mM IPTG at 18°C and cultures were harvested 20 h post-induction. Pellets derived from a culture volume equivalent to a 1 ml culture at an OD600 nm of 20 were resuspended in 1.4 ml of lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 10 mM imidazole) and lysed by sonication. Lysates were cleared by centrifugation, and supernatants were mixed with 40 μl of equilibrated Ni2+-bead slurry (Machery-Nagel #745400.100) and incubated for 1 h at 4°C. Beads were washed three times with lysis buffer prior to addition of 40 μl of elution buffer (50 mM Tris pH 7.5, 150 mM NaCl, 500 mM imidazole). Samples were incubated with elution buffer for 15 min at 30°C. Samples of total lysate, cleared (soluble) lysate and elution were analysed by SDS–PAGE.
Analytical size-exclusion chromatography
All samples were prepared in a total volume of 40 μl in SEC buffer A supplemented with 2 mM dithiothreitol (DTT) and analysed on a Superdex 75 3.2/300 Increase column (GE Healthcare). For the TTP–DCP2 interaction, TTPN and DCP2C-tail were mixed in equimolar ratios (ranging from 25 to 100 μM) and incubated overnight at 4°C. The individual proteins were also analysed by SEC.
The TTP–GYF interaction was analysed by mixing together 6.3 nmol TTPN and 7.5 nmol GYF (1:1.2 molar ratio), followed by a 2 h incubation at 4°C prior to SEC (Superdex 75 3.2/300 Increase column). As above, single proteins were also analysed by SEC (6.3 nmol each protein). Furthermore, the TTPN–GYF and the TTPN–DCP2ΔEDC4 fusion constructs, in both their single polypeptide form (intact fusion) and protease-cleaved form (resulting in two polypeptides), were analysed by analytical and semi-analytical SEC, respectively. A total of 80 μg of TTPN–GYF was analysed in each analytical SEC run on a Superdex 75 3.2/300 Increase column. Approximately 2.5 mg of TTPN–DCP2ΔEDC4 (fusion or cleaved) was analysed by semi-analytical SEC using a Superdex 200 10/300 column.
Luciferase and RT–qPCR assays
A total of 0.3 × 106 human embryo kidney (HEK)293 cells in Dulbecco's modified Eagle's medium (DMEM; Gibco #31966-021) supplemented with 10% foetal bovine serum were seeded in each well of 12-well plates. Replicates were set up for protein and RNA extraction for luciferase assays and quantitative reverse transcription–polymerase chain reaction (RT–qPCR) to determine the small interfering RNA (siRNA) knockdown efficiency of DCP2.
Cells were transfected with 20 pmol siRNA against DCP2 (or a control siRNA, siScrambled) using ROTIFect (Carl Roth #P001.3), according to the manufacturer's instructions. At 24 h post-siRNA transfection, cells were transfected with 100 ng of either TTP or green fluorescent protein (GFP) expression plasmids and 500 ng of luciferase plasmids [Renilla luciferase reporters containing or lacking the tumour necrosis factor α (TNFα) ARE in the 3′-UTR, and firefly luciferase as a control] using polyethyleneimine (PEI; Polysciences #24765). Cells were harvested 24 h post-plasmid transfection in 200 μl of lysis buffer (60 mM Tris pH 7.5, 30 mM NaCl, 1 mM EDTA, 1% Triton X-100) for protein extraction or 500 μl of Trizol (Bio&Sell) for RNA extraction.
Luciferase assays were performed in duplicate for all biological replicates. Renilla and firefly luciferase assays were performed using commercially available reagents (Renilla-Juice Luciferase Assay #102531 and Beetle-Juice Luciferase assay Firefly #102511 from PJK GmbH) and activities were measured in a plate reader (Tecan GENios).
To determine levels of TNFα mRNA, HEK293 cells were seeded and transfected with siRNAs and TTP plasmids as described above. Wherever mentioned, cells were treated with 5 μg/ml actinomycin D (or DMSO as a control) for 8 h before harvesting.
A 200 ng aliquot of DNA-free total RNA extracted from replicates was used for gene-specific cDNA synthesis by MLV-reverse transcriptase (Qiagen). qPCRs were performed using the PowerUp SYBR Green Master Mix (ThermoFisher Scientific) on a Stratagene Mx3005P instrument. The data represent the mean of duplicates of one representative analysis. Primers used for reverse transcription and qPCR were: DCP2 fwd, GCAGCAGAATTCTTTGATGAAGTG and DCP2 rev, GCTGTCCCTCAGCATGTTCT (used for reverse transcription of DCP2 cDNA); TNFα fwd, CGAGTGACAAGCCTGTAGCCC and TNFα rev, CTCAGCTCCACGCCATTGGC (used for reverse transcription of TNFα cDNA); and GAPDH fwd, CTTCGCTCTCTGCTCCTCCTGTTCG and GAPDH rev, ACCAGGCGCCCAATACGACCAAAT (used for reverse transcription of glyceraldehyde phosphate dehydrogenase)
Liquid–liquid phase separation microscopy
Microscopy was performed with unlabelled proteins for phase contrast microscopy, and Cy5-TTPN and XFD488-DCP2 proteins for fluorescent microscopy. Individual proteins or protein mixtures were diluted to twice the concentrations indicated in the figures, incubated overnight at 4°C and were mixed with a crowding solution [30% polyethylene glycol (PEG) 8000, 100 mM sodium citrate pH 7.0] in a 1:1 ratio immediately before the microscopy analysis.
Phase contrast microscopy
TTPN and DCP2 constructs (DCP2ΔC, DCP2core and DCP2C-tail) were diluted in SEC buffer C (20 mM HEPES pH 7.5, 150 mM NaCl) individually in different concentrations ranging from 495 to 20 μM or in equimolar mixtures of TTP and DCP2 (concentrations of 250, 100, 70, 50 and 20 μM of each protein). A Zeiss Primo Star microscope with a Plan-Achromat ×40/0.65 Ph2 lens (with Ph2 condenser for phase contrast) was used to visualize formation of phase-separated droplets. The Supplementary Video depicting fusion of phase-separated droplets is of DCP2C-tail at 125 μM in the presence of 15% PEG8000 (final concentrations).
Fluorescence microscopy
Fluorescence microscopy was performed as above, using fluorescently labelled TTPN and DCP2 proteins. Proteins were diluted and mixed together in a 1:1 ratio with 180 μM of each protein and incubated overnight at 4°C. The mixtures were further diluted 1:1 in the crowding solution (see above) immediately prior to analysis. A Zeiss Axio Observer 7 ApoTome microscope with a Plan-Apochromat ×63/1.4 DIC oil immersion lens was used to visualize droplets containing the two proteins. DCP2 was detected via the X-channel (475 nm filter) and TTP was detected via the Y-channel (630 nm filter). Optical sectioning was performed using the Zeiss Apotome to improve the contrast of the fluorescent droplets by eliminating out-of-focus light and reducing the background from multiple focal planes.
RESULTS
The N-terminal activation domain of TTP associates with the decapping enzyme DCP2
In vitro biochemical and biophysical studies on human TTP have largely been marred by the proteolytically unstable nature of the full-length protein as well as shorter constructs spanning the N- and C-terminal activation domains. Previous structural and biochemical studies have relied on short synthetic peptides corresponding to binding motifs and fusions with MBP to stabilize the protein, although other commonly used stability tags such as GST, GFP and Trx do not have a similar effect (Figure 1B; Supplementary Figure S1B). Previous reports present evidence for improving the solubility and stability of a protein by linking it covalently to its interaction partner (19,20). We hypothesized that binding of an interaction partner to TTP would minimize its proteolytic degradation and stabilize the protein. The translational repressor GIGYF2 is a known interactor of TTP, with its GYF domain directly binding the TTP-PPPGϕ motifs (21). Therefore, we designed a construct where the N-terminal activation domain of TTP (TTPN) containing a PPPGF motif was fused to the GIGYF2-GYF domain (hereafter referred to as GYF) to generate a single polypeptide, with the two interacting partners connected by a short linker bearing a TEV protease cleavage site (Figure 1B). The resultant fusion construct is remarkably stable and yields large amounts of a homogenous species. Interestingly, TTPN remains stable even after cleavage of the polypeptide with TEV protease to yield a non-covalently associated TTP–GYF complex, indicating that most of the proteolytic degradation of TTP occurs during its expression (Figure 1B, lane 2). The behaviour of the TTP–GYF fusion is especially striking in comparison with the negative control, His-GST–TTP, suggesting that the stability of TTPN is the result of fusion to an interaction partner and not merely to another polypeptide (Figure 1B, lane 3). Encouraged by this observation, we proceeded to generate fusion constructs of the TTP N- and C-terminal activation domains with DCP2 (Figure 1C). Full-length DCP2 is also proteolytically unstable when expressed in E. coli (Supplementary Figure S1C) as the activators/enhancers of decapping that stabilize DCP2 upon interaction in mammalian cells, such as DCP1, EDC1, EDC3 and EDC4, are not available in E. coli. Therefore, a C-terminal truncation of DCP2 lacking the 26 residue EDC4-binding region (DCP2ΔEDC4) was used instead (Figure 1A). Fusion of DCP2ΔEDC4, like GYF, conferred stability on TTPN and led to the isolation of highly pure fusion protein, but did not have a stabilizing effect on the TTP C-terminal activation domain (Figure 1C, left panels). The TTPC-TZF–DCP2ΔEDC4 fusion protein is expressed in an amount comparable with TTPN–DCP2ΔEDC4, but little to no protein was observed in the soluble lysate and the elution of the small-scale Ni2+-affinity pulldown, suggesting that the protein is unstable in solution. As with the GYF fusion, cleavage of the TTPN–DCP2ΔEDC4 chimera with TEV protease also resulted in stable TTPN, despite the inability of the two proteins to form a stable complex at the low concentrations used in SEC (Figure 1C, inset). Our results suggest a direct interaction between TTPN and DCP2, consistent with previous reports of the N-terminal activation domain of TTP engaging the decapping machinery.
The IDRs of TTP and DCP2 mediate a direct but weak interaction between the proteins
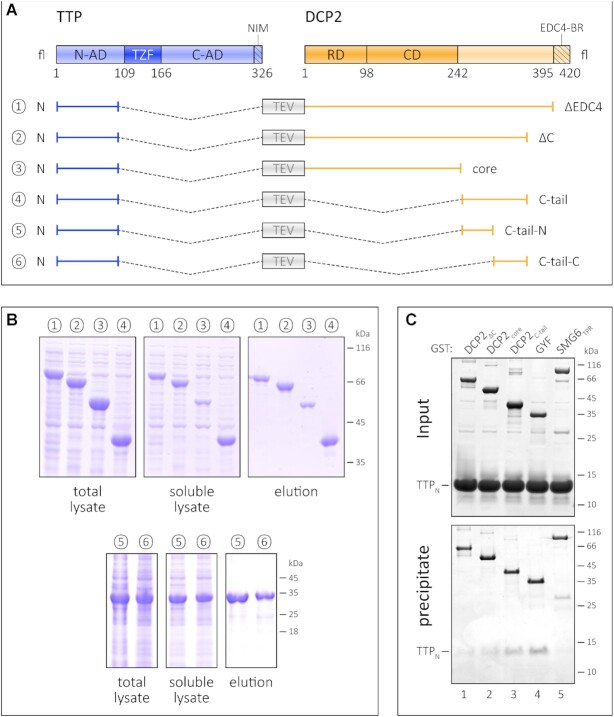
In order to determine the region or domain of DCP2 that mediates stabilization of TTP, we designed a series of constructs where TTPN is linked to different DCP2 constructs (Figure 2A). As mentioned above, fusion of TTPN to DCP2ΔEDC4 results in a stable protein (Figures 1C and 2B). Truncation of an additional 40 residues from the C-terminus of the DCP2ΔEDC4 construct (DCP2ΔC) does not mitigate its ability to stabilize TTP (Figure 2B, compare lanes 1 and 2). The DCP2 NRD–Nudix core does not stabilize TTPN to the same extent as the other constructs, as the amount of TTPN–DCP2core protein obtained in the soluble lysate and elution of the Ni2+-affinity pulldown is significantly lower than for the other fusion constructs (Figure 2B, compare lane 3 with lanes 1, 2 and 4). Moreover, the TTPN–DCP2core appears to degrade during subsequent purification steps (Supplementary Figure S2A) and cannot be purified by SEC, unlike fusions encompassing the DCP2 C-tail which remain stable over time. Taken together, our observations indicate that the binding motif for TTP resides in the C-terminal extension of DCP2 that is proximal to its NRD–Nudix core (referred to as the C-tail). Consistently, fusion of the DCP2 C-tail is sufficient to confer stability on TTPN (Figure 2B, lane 4).
Figure 2.
The TTP-binding site resides in the C-terminal IDR of DCP2. (A) Schematic representation of the fusion constructs comprising TTPN and the different regions of DCP2. As in Figure 1, all fusion constructs (1–6) contain a linker with a TEV protease cleavage site separating the TTP and DCP2 sequences. (B) Ni2+-affinity pulldowns to compare the effect of the DCP2 NRD–CD core and the unstructured C-terminus on the stability of TTPN. The left, middle and right panels for each set of constructs (1–4 and 5–6) indicate total lysate, soluble lysate and elution, respectively. Fusion of DCP2core to TTPN has the least impact on its proteolytic stability (see also Supplementary Figure S2A), while all DCP2 constructs bearing the C-tail have a remarkable stabilizing effect. The truncated DCP2 C-tail constructs, lacking the N- or the C-terminal half of the C-tail, are also capable of stabilizing TTPN upon fusion. The numbers 1–6 for the fusion constructs correspond to those labelled in (A) above. (C) GST pulldown assays of isolated TTPN with GST–DCP2 proteins. GST–GYF and GST–SMG6TPR serve as positive and negative controls, respectively. The top and the bottom panels indicate input and precipitate, respectively. In this experiment, proteins have been visualized by silver staining. GST–DCP2C-tail co-precipitates TTPN to a similar extent as GST–GYF, while the amount of TTPN co-precipitated with GST–DCP2core is negligible. Despite containing the C-tail, GST–DCP2ΔC fails to co-precipitate TTPN, possibly due to folding back of the C-tail on the NRD–CD core (see also Supplementary Figure S2B).
To further investigate the association of TTP and DCP2 in vitro, we performed GST pulldowns using GST-tagged DCP2 proteins and untagged TTPN (Figure 2C). The NMD endonuclease SMG6TPR and GYF were used as negative and positive controls for this assay, respectively. As anticipated, TTPN co-precipitated with GST–DCP2C-tail, but did not bind GST–DCP2core as much (Figure 2C, compare lanes 3 and 2). Surprisingly, the DCP2ΔC construct, which was capable of stabilizing TTP upon fusion, did not co-precipitate any TTPN protein. We attribute this to an interaction between the DCP2 core and its C-tail, which in an intramolecular context (in DCP2ΔC) takes precedence over the intermolecular TTPN–DCP2C-tail interaction (Supplementary Figure S2B). Indeed, the inability of DCP2ΔC to mediate stable interactions with TTPNin trans allowed us to isolate large amounts of homogenous TTPN from the parent fusion protein for our biochemical and biophysical studies (Supplementary Figure S2C; Supplementary Methods). The interaction of TTPN with DCP2C-tail presents an unusual mode of interaction, where the binding regions of both interacting partners are predicted to be disordered (Supplementary Figure S2D). Co-immunoprecipitation studies using HEK293 cells exogenously expressing a tagged TTP construct encompassing the N-terminal activation domain and the RNA-binding domain (TTPN-TZF) and full-length DCP2 revealed that HA-DCP2fl could be co-precipitated with Flag-HA-TTPN-TZF (Supplementary Figure S2E, left panel). However, due to low expression of HA-DCP2C-tail, its interaction with TTP in HEK293 cells could not be investigated (Supplementary Figure S2E, right panel).
To narrow down the TTPN-binding region within the DCP2 C-tail IDR, we generated two new fusion constructs where either the N- (residues 249–299) or the C-terminal (residues 299–353) half of the DCP2 C-tail was fused to TTPN (Figure 2A, constructs 5 and 6). Both constructs appear to stabilize TTPN to the same extent, as assessed by Ni2+-affinity pulldown (Figure 2B, samples 5 and 6). This suggests that the TTP-binding sites are dispersed throughout the DCP2 C-tail instead of being localized to a particular stretch of amino acids. To corroborate our observations, we performed cross-linking mass spectrometry (XLMS) on mixtures of TTPN and DCP2ΔC, using the lysine–cysteine cross-linker succinimidyl 4-(p-maleimidophenyl) butyrate (SMPB). XLMS was carried out on TTP–DCP2 mixtures that were pre-incubated at two concentrations, 0.5 and 2 mg/ml. TTPN has no lysines, and a single cysteine residue which cross-links to lysines in DCP2. Consistent with our biochemical data, we found a high density of cross-links clustered at the DCP2 C-tail (Supplementary Figure S3; Supplementary Table S1). Inter-links were also detected between the DCP2 core and TTP, though not all available lysines participated in cross-linking. We observe that the numbers of cross-linked spectral matches of TTP to the C-terminal part of DCP2ΔC (amino acid positions 250–353) do not change, and even increase, when the protein concentration is lowered, while the numbers of cross-linked spectral matches between TTP and DCP2ΔC from amino acid positions 1 to 250 generally decrease. These results argue against non-specific oligomerization or aggregation, but rather suggest a preferred interaction of TTP with the C-terminal region of DCP2ΔC, i.e. amino acids 250–353. A closer look at the intra-links for DCP2ΔC reveals a large number of cross-links between the DCP2 C-tail and the core domain, reminiscent of the interaction between the DCP2 C-tail and the core observed in GST pulldown assays (Supplementary Figures S2B and S3).
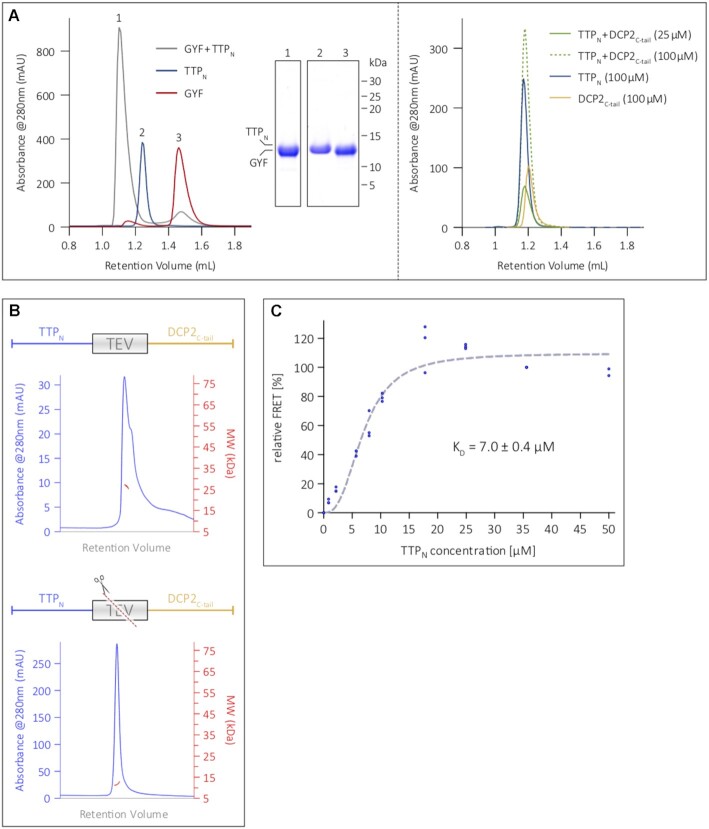
We next performed analytical SEC assays to compare the interaction of TTPN with GYF and DCP2C-tail. Incubation of TTPN and GYF in a 1:1.2 molar ratio for 16 h at 4°C resulted in formation of a stable complex, as indicated by a shift in peak retention volume in comparison with the individual proteins (Figure 3A, left panel). In contrast, TTPN did not form a stable complex with DCP2C-tail, even at high concentrations of 100 μM, suggesting a weaker interaction between the two proteins (Figure 3A, right panel; see also Figure 1C). The molecular weight of the TTPN–DCP2C-tail fusion, as determined by SEC coupled with multi-angle light scattering (SEC-MALS), is ∼26 kDa, and is reduced to ∼11 kDa upon cleavage with TEV protease (Figure 3B). This indicates that unlike GYF, DCP2C-tail does not remain associated with TTPN upon cleavage of the fusion polypeptide, which is consistent with a weak interaction between the two proteins.
Figure 3.
The N-terminal activation domain of TTP engages DCP2 in a low-affinity interaction. (A) Analytical SEC analyses of equimolar mixtures of TTPN and GYF (left panel), and TTPN and DCP2C-tail (right panel). SEC runs of the individual proteins (TTPN, DCP2C-tail and GYF) are shown for comparison. TTP and DCP2 do not form a stable complex, even at reasonably high concentrations of 100 μM, indicating weak binding, while the interaction of TTP with GYF is considerably stronger and results in a complex (see corresponding SDS–PAGE analysis of the peak fractions). (B) SEC-MALS to analyse the TTPN–DCP2C-tail fusion polypeptide in its intact (top) and TEV protease-cleaved (bottom) states. The intact fusion has a molecular weight of 26.5 kDa (in good agreement with the theoretical molecular weight of 23.2 kDa). Cleavage of the fusion by TEV protease results in a molecular weight of 11.6 kDa, which does not match that of the complex, but approximates the molecular weights of isolated TTPN (11.1 kDa) and DCP2C-tail (12.1 kDa). (C) Intermolecular FRET assay using Cy3-DCP2C-tail (donor) and Cy5-TTPN (acceptor) to determine the equilibrium dissociation constant (KD) of the interaction. The mean relative FRET percentage calculated from three independent experiments (only two measurements for the 50 μM data point) were fitted to a one-site specific binding curve with Hill slope. The resultant KD of 7 μM indicates a low-affinity interaction. All data points are shown in the figure. Plotting of the data points and curve-fitting were performed using Graphpad Prism 5.0.
To quantify the strength of the interaction between TTP and DCP2 in solution, we used FRET, which allows measurements over a wide range of binding affinities. DCP2C-tail and TTPN were labelled at a single position with the fluorescent dyes Cy3 and Cy5, respectively (Supplementary Figure S4A). An increasing amount of the acceptor, Cy5-TTPN, was titrated into a constant amount of the Cy3-DCP2C-tail donor, with the unlabelled TTP and DCP2 proteins serving as controls in the FRET setup. The tendency of DCP2C-tail to oligomerize at high concentrations under non-reducing conditions, as observed in SEC-MALS (Supplementary Figure S4B), precluded its use as an acceptor at high concentrations in FRET. Saturation of relative FRET efficiency with increasing concentrations of TTPN suggests a specific binding to DCP2C-tail with an estimated KD of 7 μM (Figure 3C).
Functional impact of the N-terminal domain of TTP
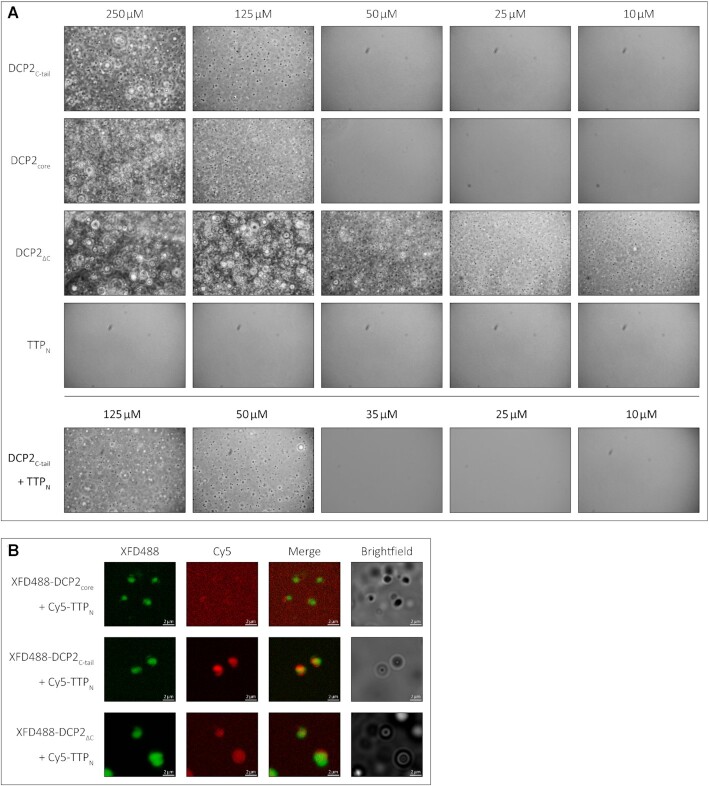
The modest affinity of the interaction between TTP and DCP2 necessitates that these proteins are present in sufficiently high amounts in cells at a given point of time to ensure binding when the need arises. However, a quantitative study of protein abundance in HEK293 cells suggests that DCP2 is present in relatively low amounts, in comparison with other decapping and decay factors such as DCP1, EDC3, EDC4 and CNOT1 (22). TTP and its paralogues are highly unstable and present in low amounts in uninduced cells (23). A possible way to circumvent the low cellular concentration of a protein is to increase its local concentration by sequestering it into a cellular compartment (24). Two cytoplasmic compartments associated with mRNA processing (decay, storage or translational repression) are processing bodies (P-bodies) and stress granules (25,26). These membrane-less compartments are assembled through liquid-liquid phase-separation (LLPS), usually induced by multivalent, low-affinity protein–protein/RNA interactions (27,28). These compartments are dynamic in nature and enable rapid recruitment and release of their components. DCP2 is a core component of P-bodies and is also present in stress granules (29–33). Interestingly, the N- and C-terminal activation domains of TTP were also shown to be capable of localizing to P-bodies, while TTP was also found to be a component of stress granules (34). We set out to investigate if the weak intra- and intermolecular interactions mediated by the IDRs of DCP2 and TTP have an impact on their localization to membrane-less compartments. To this end, we first analysed the tendency of DCP2ΔC, DCP2core, DCP2C-tail and TTPN to phase-separate into liquid droplets in vitro by phase-contrast microscopy. Consistent with IDRs playing a role in phase separation and the tendency of the DCP2C-tail protein to form higher order oligomers, we observed that DCP2C-tail formed droplets upon addition of a crowding reagent (PEG8000), at concentrations of ≥125 μM but not at 50 μM, indicating that the critical concentration lies within this broad range. The droplets showed typical characteristics of LLPS, such as growth, merging and reversible deformation (Supplementary Video). The DCP2 NRD–CD core also showed phase separation under these conditions, although it does not contain long stretches of IDRs. Not surprisingly, DCP2ΔC, which contains both the core and the C-tail, displayed the strongest propensity to form droplets, even at a relatively low protein concentration (Figure 4A, top three panels). However, TTPN, which is predicted to be disordered, did not undergo phase separation in vitro (Figure 4A, fourth panel). Interestingly, addition of TTPN to DCP2C-tail lowers the critical concentration for phase separation (between 35 and 50 μM), suggesting that the presence of TTP modulates the network of interactions driving LLPS (Figure 4A, bottom panel). Although the two short DCP2 C-tail constructs, DCP2C-tail-N and DCP2C-tail-C, interact with TTPNin vitro, they do not form phase-separated droplets on their own or in the presence of TTPN (Supplementary Figure S5). The inability of the truncated DCP2 C-tail constructs to undergo LLPS implies that the short IDRs lack sufficient binding sites and therefore cannot build up the multivalent interaction network that drives phase separation.
Figure 4.
The low-affinity interaction of TTP with DCP2 induces LLPS of TTPNin vitro. (A) Representative images of in vitro LLPS of the DCP2 proteins and TTPN in the presence of a molecular crowding reagent, PEG8000. A range of protein concentrations from 250 to 10 μM were analysed. All DCP2 proteins undergo LLPS at high concentrations, while DCP2ΔC, which has the strongest tendency to undergo phase separation, also does so at the lowest concentration investigated. TTPN does not form phase-separated droplets in any of the tested conditions. Mixtures of TTPN and DCP2C-tail show phase separation at a lower concentration than DCP2C-tail alone. Images were acquired by phase-contrast microscopy. (B) Representative images of in vitro phase separation of equimolar (90 μM) mixtures of Cy5-labelled TTPN and XFD488-labelled DCP2 in the presence of a molecular crowding reagent, PEG8000. Overlap of Cy5 and XFD488 signals suggests co-localization of the two proteins. TTPN partitions into phase-separated droplets in the presence of DCP2C-tail and DCP2ΔC, but not in the presence of DCP2core. Brightfield images show additional phase-separated droplets in different focal planes that are not visible in the fluorescent images due to optical sectioning by the Zeiss Apotome. The imperfect overlap between the XFD488 and Cy5 fluorescent images stems from moving droplets and a time lag in image acquisition. Scale bars, 2 μm.
We next tested if the weak interaction between TTPN and DCP2C-tail in solution leads to recruitment of TTP to phase-separated droplets. In order to track both proteins, we labelled TTPN and the DCP2 proteins with the fluorophores Cy3 and XFD488, respectively, and mixed them in equimolar ratios (final concentration of 90 μM). As expected, all DCP2 proteins formed phase-separated droplets (Figure 4B). Interestingly, TTPN partitioned into phase-separated droplets formed by DCP2ΔC and DCP2C-tail (as indicated by an overlap between the Cy3 and XFD488 signals), but not into those formed by DCP2core (Figure 4B). Our observations suggest that the previously reported localization of the TTP N-terminal activation domain to P-bodies in cells might be a consequence of its weak interaction with DCP2.
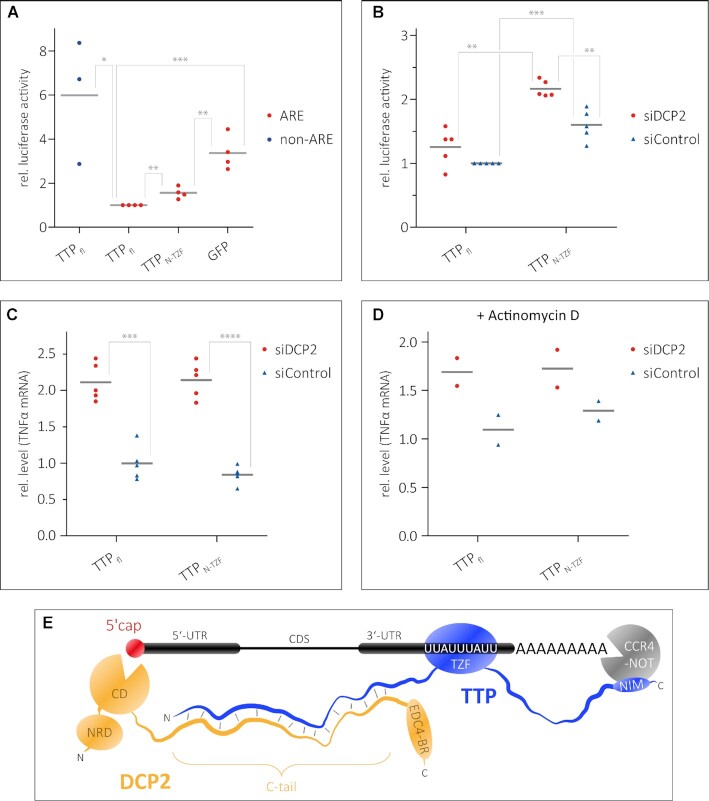
The observed low-affinity interaction between TTPN and the DCP2C-tail also raises the question of whether the N-terminal activation domain of TTP plays a role in ARE-mediated decay, independent of the C-terminal NIM region. To assess this, we used a Renilla luciferase (R-Luc) reporter containing two copies of the ARE motif of TNFα in its 3′-UTR, followed by the 3′ end of the non-coding RNA MALAT1 (35). The levels of R-Luc activity in HEK293 cells in the presence of different TTP constructs were measured as a readout of reporter mRNA levels. A firefly luciferase (F-Luc) reporter was used as a transfection control, while an R-Luc reporter lacking the ARE motifs was used as a control for ARE-specific decay (Figure 5A). The R-Luc activity of the R-Luc-ARE reporter in the presence of TTPN-TZF is ∼1.5-fold higher than in the presence of full-length TTP, but considerably lower than in the presence of GFP, which served as a negative control (Figure 5A). This suggests that the TTP N-terminal activation domain is capable of mediating decay of ARE-RNA, independent of the C-terminal activation domain and the NIM sequence therein. A TTP construct lacking the N-terminal activation domain was not tested in these assays as the nuclear export signal of TTP resides within its N-terminus (36,37), and its absence would render the protein incapable of localizing to the cytoplasm. Since TTPN interacts with the DCP2C-tail, we next proceeded to test if the ability of the TTP N-terminal domain to activate decay is dependent on DCP2. To this end, we transfected HEK293 cells with an siRNA against DCP2 or with a control siRNA (consists of a scrambled sequence that does not specifically target any transcript) 24 h prior to transfecting the TTP and reporter plasmids. RT–qPCR assays showed that siRNA-mediated knockdown of DCP2 is effective and reduces DCP2 transcript levels to ∼2% of its normal level (Supplementary Figure S6A). Reduction of DCP2 levels leads to an increase in R-Luc activity of the R-Luc-ARE reporter in HEK293 cells transfected with TTPN-TZF, but has no significant effect in cells expressing full-length TTP (Figure 5B). Since luciferase activity is an indirect readout of mRNA stability, we measured mRNA levels of the endogenous TTP target TNFα, the AREs of which are in the R-Luc-ARE reporter used above. TNFα transcript levels were measured by RT–qPCR in DCP2-depleted HEK293 cells (or control siRNA-treated cells) which were transfected either with full-length TTP or with TTPN-TZF. Knockdown of DCP2 leads to an increase of TNFα mRNA levels by 2-fold, in the presence of both full-length TTP and TTPN-TZF (Figure 5C). This effect persists in cells where transcription has been shut off by treatment with actinomycin D, although the differences are less pronounced (Figure 5D). Taken together, our observations show that DCP2 has a significant impact on TTP-mediated decay of ARE-mRNA. While TTP mediates direct interactions with DCP2, it does not stimulate decapping activity in vitro (Supplementary Figure S6B), suggesting that its predominant function is to facilitate the assembly of a decay-competent mRNP by recruiting bulk decay factors to the TTP-bound mRNA via DCP2. Indeed, a plethora of factors/activators such as DCP1, EDC1, EDC3 and EDC4 are required to stimulate decapping activity of human DCP2 as no single protein is capable of modulating the conformation of DCP2 to induce a switch from an inactive to an active conformation (38).
Figure 5.
Recruitment of DCP2 by TTPN contributes to decay of an ARE-mRNA reporter. (A and B) Luciferase reporter assays as a readout of the level of an mRNA transcript containing AREs in the 3′-UTR. HEK293 cells were transfected with RLuc-ARE-A90-MALAT1 (ARE) reporter and plasmids expressing full-length TTP or TTPN-TZF. An FLuc–GFP reporter was used as a transfection control. An RLuc-A90-MALAT1 lacking the AREs (non-ARE) and GFP were used as negative controls in (A). siDCP2 refers to siRNA-mediated knockdown of DCP2, while siControl indicates treatment with a scrambled siRNA sequence that does not target any specific transcript in cells (B). Grey bars represent the mean of n independent experiments (n = 4 for ARE reporter samples and n = 3 for non-ARE reporter in A, n = 5 in B). Individual data points are shown in all cases. The RLuc activity is normalized to FLuc; all RLuc/FLuc activities are expressed relative to that observed for full-length TTP (siControl-treated sample for B), which was set to 1. Asterisks indicate the significance of the observed difference in each case as determined from P-values derived from unpaired t-tests. *P <0.05 (significant), **P <0.01 (very significant), ***P <0.001 (extremely significant). (A) The N-terminal activation domain of TTP is capable of mediating decay of the ARE reporter. A reporter lacking the ARE sequence (non-ARE) is not degraded by full-length TTP, and GFP does not degrade the ARE reporter, reaffirming that the observed degradation of the ARE reporter by the TTP proteins is a specific effect. (B) A significant effect of DCP2 knockdown on ARE reporter levels is observed in cells expressing TTPN-TZF but not in cells expressing full-length TTP. (C and D) Detection of transcript levels of an endogenous TTP target, TNFα, by RT–qPCR as a direct measure of mRNA stability. The levels of TNFα mRNA were measured in DCP2-depleted (siDCP2) or control (siControl) HEK293 cells expressing either full-length TTP or TTPN-TZF. Depletion of DCP2 leads to increased levels of the TNFα transcript in both cases, suggesting that mRNA decay by TTP is impeded in the absence of DCP2. Asterisks are as described above; ****P <0.0001 (extremely significant). The effects of reduction of cellular DCP2 levels are less pronounced in cells treated with the transcription inhibitor actinomycin D (D). (E) A model representing the modular interactions mediated by TTP that lead to the assembly of a decay-competent mRNP on an ARE-mRNA. The TZF domain of TTP binds RNA, while the N- and C-terminal activation domains engage DCP2 (and possibly the decapping machinery) and the CCR4–NOT deadenylation complex, respectively. Bridging of events at the 5′ and 3′ end by additional protein factors not shown in the figure (such as the LSm complex and PATL1) might further stabilize the weak TTP–DCP2 interactions.
DISCUSSION
The function of TTP in mediating rapid mRNA turnover hinges on its ability to recruit components of the mRNA degradation machinery to the target mRNA transcript. This is enabled by the modular domain organization of TTP, comprising its central RNA-binding TZF domain and the flanking N- and C-terminal activation domains. Although the interactions of the TZF domain with an ARE-RNA as well as that of the C-terminal activation domain with the CCR4–NOT complex are fairly well characterized, the interactions mediated by the N-terminal activation domain have not been studied in as much molecular detail. Here we investigate the interaction of the TTP N-terminal activation domain (TTPN) with the decapping enzyme DCP2 in vitro. We show, using a combination of biochemical and biophysical methods, that TTPN mediates a direct, but weak interaction with DCP2. This interaction is an unusual one as it involves IDRs of both interacting partners. This is in contrast to the typical examples of protein–protein interactions, where the binding interface involves folded domains on both interaction partners or where an IDR of one protein interacts with a structured domain of its binding partner to form a well-ordered interface. A recent report on the binding of the linker histone (H1) to the nuclear protein prothymosin α (ProTα) highlighted the interaction between disordered regions of the two proteins, where both proteins retain their structural disorder in the complex (39). The interaction between two IDRs represents a new mode of biomolecular binding that has largely remained unexplored in vitro due to the focus of structural biology methods on protein order and structurally defined binding sites.
We analysed the sequences of the IDRs of DCP2 and TTP to find sequence motifs that might be involved in protein–protein interactions. The DCP2 C-tail sequence revealed several lysine residues interspersed with multiple glutamine and serine residues. However, no sequence motif could be identified in this stretch. Similarly, TTPN contains several proline, glycine and serine residues and, apart from the tetraproline motif (PPPPGF), contains no identifiable sequence motif. Therefore, specific interaction motifs could not be identified and validated by mutational analyses. The interaction between TTPN and DCP2C-tail is a fairly weak one, of a modest affinity of ∼7 μM. Although the two proteins can be co-precipitated in GST pulldown assays, they do not form a stable complex in SEC, indicating rapid dissociation of the interacting partners from each other. Studies of protein–protein interactions, especially from a structural biochemistry perspective, have typically been dominated by strong, stoichiometric interactions that result in the formation of stable complexes. However, a high-throughput study of the human interactome revealed that weak, substoichiometric interactions in fact dominate the interactome and drive functionality in cells (40). While strong interactions define core complexes that exist in isolation, weak interactions in vitro translate to transient, dynamic interactions in cells that can be regulated and remodelled easily. Assembly of decay-competent mRNPs on mRNA transcripts destined for targeted decay entail multiple weak interactions among components of the complex, which are then readily disassembled after decay of the mRNA (41). Using ARE reporters, we show that TTPN-TZF, which lacks the C-terminal NIM, is still capable of mediating mRNA decay, though not as efficiently as the full-length protein. A reduction of cellular DCP2 levels by siRNA knockdown significantly hampers the ability of TTPN-TZF to mediate decay. This effect is mirrored in the endogenous TTP target, TNFα, where depletion of DCP2 was found to stabilize the TNFα transcript in cells expressing full-length TTP or its N-terminal activation domain. These observations suggest that the weak interaction between TTPN and DCP2C-tail is sufficient to induce degradation of a target transcript even in the absence of the stronger interaction of TTP-NIM with the CCR4–NOT deadenylation complex. To test the contribution of the C-terminal activation domain of TTP alone, it would be necessary to fuse an exogenous nuclear export signal from a different protein to this construct and ensure that TTPC is localized to the cytoplasm where bulk mRNA decay takes place. The interaction with DCP2 also incorporates an element of redundancy into TTP-mediated ARE-mRNA decay, which stringently regulates the levels of several transcripts involved in innate immunity (such as TNFα) (1). Interestingly, the eIF4E transporter protein 4E-T, which was shown to interact with TTP in a proximity-dependent biotin identification (BioID) assay, bridges the 3′-end degradation machinery with the decapping machinery at the 5′ end, suggesting that TTP can assemble a decay-competent mRNP on its target transcript in multiple ways (12).
Weak multivalent interactions between proteins and/or RNA often lead to LLPS in vitro and partitioning of proteins into membraneless organelles in cells. An example of such organelles in cells are cytoplasmic P-bodies which are rich in mRNA decay factors (such as DCP1/2, EDC4, 4E-T and DDX6), but are now increasingly believed to be sites of mRNA storage for translationally repressed mRNA, rather than sites of decay (26,29,42). The N- and C-terminal activation domains of TTP were reported to localize to P-bodies and recruit their mRNA cargo to these compartments (34). We show that TTPN does not undergo phase separation on its own at high concentrations, even in the presence of a molecular crowding reagent. However, DCP2, which is a core component of P-bodies, has a strong tendency to undergo LLPS in vitro. The DCP2ΔC construct encompassing the catalytic core and C-tail forms LLPS droplets at relatively low protein concentrations. Both the core and the unstructured C-tail of DCP2 also have a tendency to partition into droplets, albeit at higher concentrations than the DCP2ΔC protein. The tendency of DCP2 to form LLPS droplets might stem from intramolecular interactions of the DCP2 C-tail with itself or the catalytic core (Supplementary Figures S2B, S3 and S4B). In the presence of equimolar amounts of DCP2ΔC and DCP2C-tail, TTPN also undergoes LLPS in vitro, an effect not observed with DCP2core, which lacks the TTP-binding region. These observations suggest that DCP2 is the ‘scaffold’ for condensation and TTP is the ‘client’, the addition of which lowers the critical concentration needed for phase separation of the scaffold (43). The DCP2-driven phase separation of TTP might lead to its recruitment to P-bodies in cells, although it should be noted that TTP also interacts with other core components of P-bodies (29). The sequestering of TTP and DCP2 in phase-separated droplets in vitro or in P-bodies in cells also serves to increase their local concentration, which in turn would augment binding of the two proteins. Interestingly, low complexity IDRs shown to mediate phase separation of proteins such as FUS, hnRNPA1 and the C-terminal domain of RNA polymerase II are often enriched in residues that have high potential for engaging in π–π interactions. Exposed peptide backbones of residues, such as proline, glycine and serine, have π bonds that can mediate π–π interactions, as do side chain aromatic, guanidinium and amide groups, such as tyrosine, tryptophan, arginine, asparagine and glutamine. π–π contacts are a relatively overlooked protein feature that can mediate phase separation (44). Since the DCP2 C-tail is rich in glutamine residues and the N-terminal activation domain of TTP has several prolines and glycine, we speculate that the nature of interactions driving LLPS of DCP2 and TTP are indeed π–π interactions.
In summary, we show that TTP uses its intrinsically disordered N-terminal activation domain to engage the unstructured C-tail of the decapping enzyme DCP2 in a weak, fuzzy-type interaction to mediate decay of its target ARE-mRNA (Figure 5E). In contrast to Dcp2 in fungi, the human DCP2 has a relatively short disordered C-terminal region (170 residues in humans as opposed to 486 in Schizosaccharomyces pombe Dcp2). As a result, human DCP2 contains relatively fewer short linear interaction motifs (SLIMs) than yeast Dcp2 proteins. Interestingly, the DCP1 proteins in humans are much longer than in yeast (582 residues in human DCP1a, of which 456 are in an unstructured tail, compared with 127 residues in S. pombe). A general notion is that many of the SLIMs found in yeast Dcp2 are transferred to metazoan Dcp1, preserving the interaction network within the decapping assembly (45). Therefore, it would be worthwhile to test if the human DCP1 proteins, and specifically their IDRs, modulate binding of TTP to DCP2. The interaction with DCP2 is probably one of the many interactions mediated by TTP to assemble a transient decay-competent mRNP on the target transcript, and probably represents a common event in other targeted mRNA decay pathways. A very recent study by Tibble and co-authors show that decapping can be both activated and repressed in membraneless compartments, depending on protein–protein interactions that drive phase separation and induce conformational changes of S. pombe Dcp1/Dcp2 (46). Although TTP and DCP2 can partition in phase-separated droplets and have been found to be present in P-bodies in cells, it remains unclear as to where decay of the target mRNA eventually takes place and what releases TTP from P-bodies in cells. Our studies highlight the importance of investigating weak interactions, particularly between intrinsically disordered proteins, to understand the dynamic assembly and disassembly of macromolecular complexes in mediating cellular functions.
DATA AVAILABILITY
All original data will be made available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jens Lykke-Andersen for the gift of the human DCP2 plasmid, and Christian Freund for the human GIGYF2 plasmid. The luciferase reporter plasmids were a generous gift from Catia Igreja and the former Izaurralde group. We would like to acknowledge the assistance of the Core Facility BioSupraMol (Freie Universität Berlin) supported by the Deutsche Forschungsgemeinschaft (DFG). We thank Tanja Matković-Rachid and Francesca Bottanelli for sharing their expertise in optical microscopy experiments, and Eric Simko and Eugene Valkov for the in vitro decapping assay. We are grateful to Janosch Henning and Cecilia Perez-Borrajero as well as members of our laboratory for helpful discussions and comments on the manuscript.
Author contributions: V.D.M. purified the proteins used in this study with assistance from N.M., M.M. and S.C. Biochemical assays, RT–qPCR assays and microscopy experiments were performed by V.D.M. M.M. assisted in biochemical assays. FRET assays and SEC-MALS were performed by N.M. and V.D.M. Cross-linking mass spectrometry was performed by J.S. and H.U. Luciferase assays were carried out by T.D., S.C. and V.D.M. The study was designed by S.C. and V.D.M.; the manuscript was written by S.C. and V.D.M. with input from all authors.
Contributor Information
Vincent D Maciej, Institute of Chemistry and Biochemistry, Freie Universität Berlin, Takustrasse 6, D-14195 Berlin, Germany.
Nevena Mateva, Institute of Chemistry and Biochemistry, Freie Universität Berlin, Takustrasse 6, D-14195 Berlin, Germany.
Juliane Schwarz, Max Planck Institute for Multidisciplinary Sciences, Bioanalytical Mass Spectrometry Group, Am Fassberg 11, D-37077 Goettingen, Germany; University Medical Center Goettingen, Bioanalytics, Institute for Clinical Chemistry, Robert Koch Strasse 40, D-37075 Goettingen, Germany.
Theresa Dittmers, Institute of Chemistry and Biochemistry, Freie Universität Berlin, Takustrasse 6, D-14195 Berlin, Germany.
Megha Mallick, Institute of Chemistry and Biochemistry, Freie Universität Berlin, Takustrasse 6, D-14195 Berlin, Germany.
Henning Urlaub, Max Planck Institute for Multidisciplinary Sciences, Bioanalytical Mass Spectrometry Group, Am Fassberg 11, D-37077 Goettingen, Germany; University Medical Center Goettingen, Bioanalytics, Institute for Clinical Chemistry, Robert Koch Strasse 40, D-37075 Goettingen, Germany.
Sutapa Chakrabarti, Institute of Chemistry and Biochemistry, Freie Universität Berlin, Takustrasse 6, D-14195 Berlin, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This study was supported by the Deutsche Forschungsgemeinschaft (DFG) [CH1245/3-1, CH1245/3-2 and CH1245/5-1 to S.C.] and the Max Planck Gesellschaft (to H.U). S.C. is supported by the Heisenberg Program of the DFG. S.C. and H.U. gratefully acknowledge support from the DFG Priority Programme SPP 1935. The authors declare that there are no conflicts of interest. Funding for open access charges: DFG [CH1245/3-2 to S.C.] and Freie Universität Berlin.
Conflict of interest statement. None declared.
REFERENCES
- 1. Khabar K.S. The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J. Interferon Cytokine Res. 2005; 25:1–10. [DOI] [PubMed] [Google Scholar]
- 2. Mayr C. Regulation by 3′-untranslated regions. Annu. Rev. Genet. 2017; 51:171–194. [DOI] [PubMed] [Google Scholar]
- 3. Khabar K.S. Hallmarks of cancer and AU-rich elements. Wiley Interdiscip. Rev. RNA. 2017; 8:e1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wells M.L., Perera L., Blackshear P.J.. An ancient family of RNA-binding proteins: still important!. Trends Biochem. Sci. 2017; 42:285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hudson B.P., Martinez-Yamout M.A., Dyson H.J., Wright P.E.. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 2004; 11:257–264. [DOI] [PubMed] [Google Scholar]
- 6. Lykke-Andersen J., Wagner E.. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005; 19:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fenger-Gron M., Fillman C., Norrild B., Lykke-Andersen J.. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005; 20:905–915. [DOI] [PubMed] [Google Scholar]
- 8. Hau H.H., Walsh R.J., Ogilvie R.L., Williams D.A., Reilly C.S., Bohjanen P.R.. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J. Cell. Biochem. 2007; 100:1477–1492. [DOI] [PubMed] [Google Scholar]
- 9. Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M.. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001; 107:451–464. [DOI] [PubMed] [Google Scholar]
- 10. Fabian M.R., Frank F., Rouya C., Siddiqui N., Lai W.S., Karetnikov A., Blackshear P.J., Nagar B., Sonenberg N.. Structural basis for the recruitment of the human CCR4–NOT deadenylase complex by tristetraprolin. Nat. Struct. Mol. Biol. 2013; 20:735–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sandler H., Kreth J., Timmers H.T., Stoecklin G.. Not1 mediates recruitment of the deadenylase caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011; 39:4373–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nishimura T., Padamsi Z., Fakim H., Milette S., Dunham W.H., Gingras A.C., Fabian M.R.. The eIF4E-binding protein 4E-T is a component of the mRNA decay machinery that bridges the 5′ and 3′ termini of target mRNAs. Cell Rep. 2015; 11:1425–1436. [DOI] [PubMed] [Google Scholar]
- 13. Mugridge J.S., Ziemniak M., Jemielity J., Gross J.D.. Structural basis of mRNA-cap recognition by Dcp1–Dcp2. Nat. Struct. Mol. Biol. 2016; 23:987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. She M., Decker C.J., Svergun D.I., Round A., Chen N., Muhlrad D., Parker R., Song H.. Structural basis of Dcp2 recognition and activation by Dcp1. Mol. Cell. 2008; 29:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valkov E., Muthukumar S., Chang C.T., Jonas S., Weichenrieder O., Izaurralde E.. Structure of the Dcp2–Dcp1 mRNA-decapping complex in the activated conformation. Nat. Struct. Mol. Biol. 2016; 23:574–579. [DOI] [PubMed] [Google Scholar]
- 16. Wurm J.P., Holdermann I., Overbeck J.H., Mayer P.H.O., Sprangers R.. Changes in conformational equilibria regulate the activity of the Dcp2 decapping enzyme. Proc. Natl Acad. Sci. USA. 2017; 114:6034–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang C.T., Bercovich N., Loh B., Jonas S., Izaurralde E.. The activation of the decapping enzyme DCP2 by DCP1 occurs on the EDC4 scaffold and involves a conserved loop in DCP1. Nucleic Acids Res. 2014; 42:5217–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mattiroli F., Gu Y., Luger K.. FRET-based stoichiometry measurements of protein complexes in vitro. Bio Protoc. 2018; 7:e2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gadd M.S., Bhati M., Jeffries C.M., Langley D.B., Trewhella J., Guss J.M., Matthews J.M.. Structural basis for partial redundancy in a class of transcription factors, the LIM homeodomain proteins, in neural cell type specification. J. Biol. Chem. 2011; 286:42971–42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kozono H., White J., Clements J., Marrack P., Kappler J.. Production of soluble MHC class II proteins with covalently bound single peptides. Nature. 1994; 369:151–154. [DOI] [PubMed] [Google Scholar]
- 21. Fu R., Olsen M.T., Webb K., Bennett E.J., Lykke-Andersen J.. Recruitment of the 4EHP–GYF2 cap-binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU-rich elements. RNA. 2016; 22:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cho N.H., Cheveralls K.C., Brunner A.D., Kim K., Michaelis A.C., Raghavan P., Kobayashi H., Savy L., Li J.Y., Canaj H.et al.. OpenCell: proteome-scale endogenous tagging enables the cartography of human cellular organization. Science. 2022; 375:eabi6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanduja S., Blanco F.F., Dixon D.A.. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip. Rev. RNA. 2011; 2:42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alberti S. The wisdom of crowds: regulating cell function through condensed states of living matter. J. Cell Sci. 2017; 130:2789–2796. [DOI] [PubMed] [Google Scholar]
- 25. Decker C.J., Parker R.. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012; 4:a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aizer A., Kalo A., Kafri P., Shraga A., Ben-Yishay R., Jacob A., Kinor N., Shav-Tal Y.. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J. Cell Sci. 2014; 127:4443–4456. [DOI] [PubMed] [Google Scholar]
- 27. Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D.et al.. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015; 57:936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J., Choi J.M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D.et al.. A molecular grammar governing the driving forces for phase separation of Prion-like RNA binding proteins. Cell. 2018; 174:688–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hubstenberger A., Courel M., Benard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.B., Munier A., Fradet M.et al.. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017; 68:144–157. [DOI] [PubMed] [Google Scholar]
- 30. Sheth U., Parker R.. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003; 300:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Teixeira D., Parker R.. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007; 18:2274–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Youn J.Y., Dunham W.H., Hong S.J., Knight J.D.R., Bashkurov M., Chen G.I., Bagci H., Rathod B., MacLeod G., Eng S.W.M.et al.. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell. 2018; 69:517–532. [DOI] [PubMed] [Google Scholar]
- 33. Youn J.Y., Dyakov B.J.A., Zhang J., Knight J.D.R., Vernon R.M., Forman-Kay J.D., Gingras A.C.. Properties of stress granule and P-body proteomes. Mol. Cell. 2019; 76:286–294. [DOI] [PubMed] [Google Scholar]
- 34. Franks T.M., Lykke-Andersen J.. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007; 21:719–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peter D., Weber R., Sandmeir F., Wohlbold L., Helms S., Bawankar P., Valkov E., Igreja C., Izaurralde E.. GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression. Genes Dev. 2017; 31:1147–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lai W.S., Wells M.L., Perera L., Blackshear P.J.. The tandem zinc finger RNA binding domain of members of the tristetraprolin protein family. Wiley Interdiscip. Rev. RNA. 2019; 10:e1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Phillips R.S., Ramos S.B., Blackshear P.J.. Members of the tristetraprolin family of tandem CCCH zinc finger proteins exhibit CRM1-dependent nucleocytoplasmic shuttling. J. Biol. Chem. 2002; 277:11606–11613. [DOI] [PubMed] [Google Scholar]
- 38. Mugridge J.S., Tibble R.W., Ziemniak M., Jemielity J., Gross J.D.. Structure of the activated Edc1–Dcp1–Dcp2–Edc3 mRNA decapping complex with substrate analog poised for catalysis. Nat. Commun. 2018; 9:1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Borgia A., Borgia M.B., Bugge K., Kissling V.M., Heidarsson P.O., Fernandes C.B., Sottini A., Soranno A., Buholzer K.J., Nettels D.et al.. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018; 555:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F.et al.. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015; 163:712–723. [DOI] [PubMed] [Google Scholar]
- 41. Gehring N.H., Wahle E., Fischer U.. Deciphering the mRNP code: RNA-bound determinants of post-transcriptional gene regulation. Trends Biochem. Sci. 2017; 42:369–382. [DOI] [PubMed] [Google Scholar]
- 42. Kamenska A., Simpson C., Vindry C., Broomhead H., Benard M., Ernoult-Lange M., Lee B.P., Harries L.W., Weil D., Standart N.. The DDX6–4E-T interaction mediates translational repression and P-body assembly. Nucleic Acids Res. 2016; 44:6318–6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vernon R.M., Chong P.A., Tsang B., Kim T.H., Bah A., Farber P., Lin H., Forman-Kay J.D.. Pi–Pi contacts are an overlooked protein feature relevant to phase separation. Elife. 2018; 7:e31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fromm S.A., Truffault V., Kamenz J., Braun J.E., Hoffmann N.A., Izaurralde E., Sprangers R.. The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex. EMBO J. 2012; 31:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tibble R.W., Depaix A., Kowalska J., Jemielity J., Gross J.D.. Biomolecular condensates amplify mRNA decapping by biasing enzyme conformation. Nat. Chem. Biol. 2021; 17:615–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original data will be made available upon request.