Highlights
-
•
The evolutionary analysis revealed the purifying selection on synonymous substitutions during SARS-CoV-2′s evolution.
-
•
Synonymous substitutions have caused optimized codon usage towards human genome over time, while nonsynonymous substitutions have caused a slight deoptimization.
-
•
Synonymous substitutions faced selection pressure from maintaining RNA secondary structure.
-
•
Inconsecutive evolutionary traces of SARS-CoV-2 may cause by alternating lineages.
Keywords: SARS-CoV-2, Natural selection, Synonymous substitution, Codon usage, RNA structure
Abstract
From the first emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) till now, multiple mutations that caused synonymous and nonsynonymous substitutions have accumulated. Among them, synonymous substitutions were regarded as “silent” mutations that received less attention than nonsynonymous substitutions that cause amino acid variations. However, the importance of synonymous substitutions can not be neglected. This research focuses on synonymous substitutions on SARS-CoV-2 and proves that synonymous substitutions were under purifying selection in its evolution. The evidence of purifying selection is provided by comparing the mutation number per site in coding and non-coding regions. We then study the two forces of purifying selection: synonymous codon usage and RNA secondary structure. Results show that the codon usage optimization leads to an adapted codon usage towards humans. Furthermore, our results show that the maintenance of RNA secondary structure causes the purifying of synonymous substitutions in the structural region. These results explain the selection pressure on synonymous substitutions during the evolution of SARS-CoV-2.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a member of the Coronaviridae family that was initially discovered in December 2019 in the city of Wuhan, China. To date (May 10, 2022), it has caused 515,192,979 confirmed cases of SARS-CoV-2 infection and 6254,140 deaths (https://covid19.who.int/). SARS-CoV-2 is a single-stranded RNA virus, with the genomic length of its genome is approximately 29,000 nucleotides.
Since its first appearance, it has accumulated multiple mutations on SARS-CoV-2′s genome, causing synonymous and nonsynonymous substitutions in coding areas. Nonsynonymous substitutions, or mutations at the protein level drew the most attention because amino acid mutations could produce functional or structural alterations in the viral protein (Berrio et al., 2020; Bhattacharya et al., 2022; Cheng et al., 2021; Johnson et al., 2022; Liu et al., 2022; Miller et al., 2022; Resende et al., 2021; Shah and Woo, 2022). Mutations that cause synonymous substitutions received less attention than nonsynonymous substitutions, as they do not affect the amino acid and are therefore considered “silent” mutations. However, such mutations may have a considerable impact on gene functions through mechanisms such as mRNA splicing, folding, and translation efficiency and accuracy (Shabalina et al., 2013).
Translation efficiency is an important driving force on synonymous codon usage. Synonymous codons with abundant corresponding tRNA could increase the translation efficiency, shaping biased codon usage in a genome, especially in highly expressed genes (Bulmer, 1991; Comeron, 2004; Duret, 2000; Kanaya et al., 2001). For viruses that utilize their host's translation machinery, often showed a biased codon usage toward their host (Ata et al., 2021; Cristina et al., 2016; Sharp and Li, 1987), showing adaptive evolution in synonymous codon usage for improved translation efficiency. There have been various studies on the evolution of synonymous codon usage of SARS-CoV-2. Hussain et al. found a decrease in codon adaptation index (CAI) over time (Hussain et al., 2021), while Ramazzotti et al. found an increase in the resemblance of viral codon usage to humans (Ramazzotti et al., 2022).
RNA structure has important functions in many processes related to RNA transcription and protein translation (Faure et al., 2016; Martin and Ephrussi, 2009; Mustoe et al., 2018; Sharp, 2009). In the case of the RNA virus, RNA structure may influence its replication efficiency in hosts (de Borba et al., 2015; Diviney et al., 2008), and play a role in translation regulation (Huang et al., 2012), particularly during translation initiation (Nicholson and White, 2011; Treder et al., 2008). Such forces of maintaining functional RNA structures will exert selection pressure on synonymous substitutions that lie on it.
This study focuses on synonymous substitutions in the process of SARS-CoV-2′s evolution. We firstly found some evidence of purifying synonymous substitutions in the evolution of SARS-CoV-2. And then explained the mechanism of purifying selection from two aspects: synonymous codon usage and RNA secondary structure. Based on this study, we uncovered a corner of SARS-CoV-2′s evolution: the mechanism of natural selection on “silent” mutations.
2. Materials and methods
2.1. Genome data collection
A total of 14,299 complete genomes of SARS-CoV-2 were retrieved from the GISAID database (https://www.gisaid.org/), starting from the first appearance of the virus till April 15, 2022. The dataset contains sufficient strains (about 500) for each month to reduce the possible sampling bias. Genomes containing ambiguous bases (base symbol “N” for example) could lead to inaccurate results, and thus were removed; a total of 6483 genomes were reserved after filtering. The reference genome (NC_045512.2) was downloaded from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). After genome retrieval, the multiple sequence alignment (MSA) was built by MAFFT (Katoh and Standley, 2013).
2.2. Calculation of dN/dS
Yn00 and codeml from PAML (Yang, 2007) were used to calculate the ratio between nonsynonymous substitution number per nonsynonymous site to synonymous substitution number per synonymous site (dN/dS, or ω). Several different methods were used to estimate ω, with slightly different results. Nevertheless, this does not affect the conclusions in the following analyses. Thus, the following analysis only takes the result from the Nei-Gojobori method (yn00) as an example. Results from other methods can be found in Table S1. The overall ω estimated by the Nei-Gojobori method (yn00) was calculated as the mean of pairwise ω (from each strain to the reference). Samples with no synonymous substitutions can not be used to calculate the value of ω, thus were not included in this analysis. Samples with no synonymous substitutions can not be used to calculate the value of ω, thus were excluded.
2.3. Mutation number per site
Mutation number per site in the non-coding region was used as the indicator of neutral selection. The selection pressure on coding sequences can be weighed by comparing the mutation number per site in coding and non-coding regions. The mutation number per site was calculated as the observed mutation number of a region divided by its length. Perl script was developed to perform this analysis.
2.4. Codon usage adaptation
Two approaches were used to calculate codon usage adaptation: codon adaptation index (CAI) and relative codon usage (RCU) adaptation.
CAI was used to quantify codon adaptation from the relative synonymous codon usage aspect. The CAI range from 0 to 1, and a higher value of CAI represents a higher codon adaptation to the host (human) (Sharp and Li, 1987). The CAI was calculated by the local version of CAIcal (Puigbo et al., 2008).
RCU was calculated by comparing the human codon usage of a variant to the human codon usage of the reference (Ramazzotti et al., 2022). For a variant, its RCU was calculated as follows:
Where n is the total number of codons for a sequence, is the human codon usage of i th codon in the sequence, and is the human codon usage of the corresponding codon in the reference sequence. The value of RCU higher than 1 means substitution has displayed more adapted codon usage toward humans. Specifically, relative codon usage adaptation for synonymous substitutions () and relative codon usage adaptation for nonsynonymous substitutions () were used, where only codons that have undergone synonymous substitutions or nonsynonymous substitutions were considered. The calculation was performed by Perl script.
The codon usage table for humans was calculated by a dataset containing highly expressed genes, which were identified according to their protein abundance. Protein abundance data were downloaded from PAXdb (Wang et al., 2015). Full coding sequence data of humans (GRCh38) were downloaded from Ensembl (https://www.ensembl.org/index.html).
The collection date of a strain was at least accurate to the month in this analysis. When calculating the number of days after the first discovery, the first day was set to be December 1, 2019, and the number of days for each month was set to be approximately 30. If the specific collection date was missing, we regarded it as the 15th of that month.
2.5. Expected RCU of randomly mutated genome
A collection of random sequences was constructed further to prove the selection pressure on the codon usage pattern. The sequences were randomly mutated from the reference genome (NC_045512), with the mutation number of 100 in the coding region, and the ratio of transitions to transversions was estimated by yn00 (Yang, 2007). The procedure for calculating RCU for randomly mutated sequences is the same as described in the previous section. Assume that the selection force has been driven by codon usage optimization, the higher RCU value from the target dataset than that from the randomly mutated dataset can be observed.
2.6. Effective number of codons and GC content at the third codon position
Effective number of codons (ENC) and GC content at the third codon position (GC3s) were calculated by codonW (http://codonw.sourceforge.net). ENC is calculated as:
(k = 2, 3, 4 and 6) is the mean of values for the k-fold degenerate amino acids, which is calculated as:
Where n is the observed number of the codons for an amino acid and is the total number of the i th codon for that amino acid (Wright, 1990).
The relationship between expected ENC and GC3s can be approximated by:
Where GC3s is the proportion of G + C in the third codon position (Met-and Trp-excluded) (Wright, 1990).
2.7. RNA structure detection
We used raw data (SRA format) from GSE158052 (Morandi et al., 2021) in Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds/) to detect RNA structure. Reads were mapped by bowtie2 (Langmead and Salzberg, 2012), then sorted the mapping results and summarized mismatch numbers by samtools v1.9 (Danecek et al., 2021). The mismatch ratio of the dimethyl sulfate (DMS) signal was calculated as mismatch numbers divided by the sequencing depth. Next, we calculated the Gini coefficient with the window that contains 50 A and C (adenine and cytosine), and used the Gini coefficient as the strength of RNA structure. Details of the algorithm and the visualization of RNA structure data can be found in RSVdb (Yu et al., 2021).
2.8. Codon substitution density
Synonymous and nonsynonymous codon substitution densities were both calculated in the study. The codon substitution density comprises two parts: how many frequently substituted codons are in the region, and how to define frequently substituted codons.
A process was carried out on defining a frequently substituted codon: there could be some newly occurred substitutions, or the substitution has been wiped out very quickly. In most cases, such substitution only has a few observed counts in the MSA position. These substitutions could not provide the evidence of neutral selection. Thus, codons containing too few substitutions (less than 0.4%) were regarded as no substitution occurred.
The codon substitution density is the number of frequently substituted codons divided by the length of the area. Codon substitution density was calculated by sliding window, parameters according to those when calculating the Gini coefficient.
3. Results
3.1. Natural selection on synonymous substitutions
3.1.1. Estimation of selection pressure (dN/dS)
The ratio of nonsynonymous substitution number per nonsynonymous site to synonymous substitution number per synonymous site (dN/dS, or ω) was commonly used to calculate the selection pressure on a protein-coding sequence. On the genome level, the average ω of all strains compared to reference is 0.8446 (Nei-Gojobori method by yn00, results from other methods can be find in Table S1), indicating an overall purifying selection of SARS-CoV-2. The value of ω also indicated that approximately 15.54% of nonsynonymous substitutions were purified by natural selection if synonymous substitutions were completely under neutral selection.
3.1.2. Evidence of purifying the synonymous substitution
Selection pressure involved in the translation processes is more likely to influence the coding sequences. To verify the selection pressure on synonymous mutations, an indicator of neutral selection is required, such as, spontaneous mutation rate. However, estimating the mutation rate accurately on a dataset with a high number of samples might be complex (De Maio et al., 2021; Desai et al., 2021; Sanjuan et al., 2010). Thus, the mutation number per site in non-coding was used as the indicator of neutral selection. The selection pressure on synonymous mutation will cause fewer observed mutation numbers in coding regions. Comparing the observed mutation number between the coding sequence and non-coding regions enables us to weigh the selection pressure on coding regions.
The first evidence of purifying synonymous substitutions came from comparing the observed mutation number per site between coding sequences and non-coding regions (details of coding sequences showed in Table 1). The observed mutation number per site in the coding sequence (length of 29,264 nt) is 6.203, and in non-coding regions (length of 643 nt) is 18.95. The mutation number per site in non-coding regions is much higher (p < 0.001, U test), indicating that coding regions are under overall purifying selection.
Table. 1.
Information of coding sequences of SARS-CoV-2.
| Product | Start | Stop | Length |
|---|---|---|---|
| ORF1a/ORF1ab polyprotein | 266 | 21,555 | 7096 |
| Surface glycoprotein | 21,563 | 25,384 | 1273 |
| ORF3a protein | 25,393 | 26,220 | 275 |
| Envelope protein | 26,245 | 26,472 | 75 |
| Membrane glycoprotein | 26,523 | 27,191 | 222 |
| ORF6 protein | 27,202 | 27,387 | 61 |
| ORF7a protein | 27,394 | 27,759 | 121 |
| ORF7b | 27,756 | 27,887 | 43 |
| ORF8 protein | 27,894 | 28,259 | 121 |
| Nucleocapsid phosphoprotein | 28,274 | 29,533 | 419 |
| ORF10 protein | 29,558 | 29,674 | 38 |
We raise the null hypothesis that synonymous substitutions were under neutral selection. Under this condition, the ω is 0.8446, which means approximately 15.54% of nonsynonymous substitutions were purified by natural selection. The ratio of nonsynonymous to synonymous site is roughly 3.384 in SARS-CoV-2′s genome, indicating that 77.19% of substitutions caused by random mutations are nonsynonymous. Thus, the expected mutation number per site in coding regions without the pressure of purifying nonsynonymous substitutions is 7.049, still lower than the observed mutation number per site in non-coding regions (p < 0.001, U test). Based on this finding, we have evidence to refute the null hypothesis, indicating that synonymous substitution was subjected to purifying selection rather than neutral selection during the evolution of SARS-CoV-2.
Non-coding regions, especially 5′ and 3′ UTR, may have some evolutionary bias caused by other mechanisms. Thus, we provide the second evidence that comes from ORF8. In some strains of SARS-CoV-2, ORF8 is truncated to 26 aa due to premature termination that CAA change into TAA at the 27th codon, leading to a silent segment with 285 nt lengths. As this segment is no longer translated, it is free from the selection pressure associated with the translation process and protein function, making it an ideal material for comparing the selection pressure. Within the strains that contain the premature termination variation, the observed mutation number per site is 1.018 in the coding sequence, and 8.231 in the silent segment. Followed the processes above, we were able to determine that mutation in coding regions was under purifying selection (p < 0.001, U test), and synonymous substitution was also under purifying selection (p < 0.001, U test). Evidence from the silent segment of ORF8 further proved the purifying selection on synonymous substitutions.
Evidence from non-coding regions, and the silent segment from ORF8 both proved that synonymous substitution was not likely under neutral selection, but under purifying selection with the evolution of SARS-CoV-2. The force of purifying selection is essential to gaining a better understanding of SARS-CoV-2′s evolution.
3.2. Synonymous substitution was driven by codon usage optimization
3.2.1. Compositional effect and selective effect on codon usage
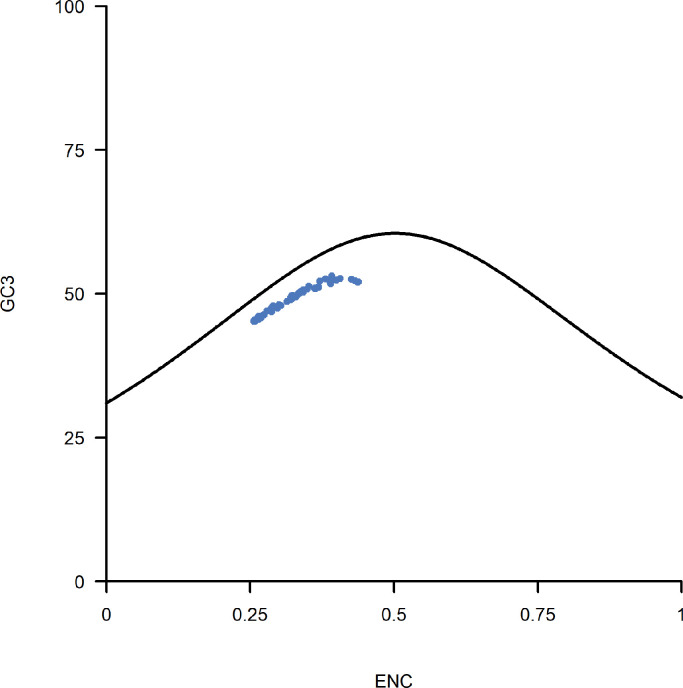
Nucleotide composition is the primary factor of codon usage for most RNA viruses, and coronavirus makes no exception (Berkhout and van Hemert, 2015; van Hemert et al., 2016). ENC-GC3s plot was carried out to investigate how much dose G + C composition affects SARS-CoV-2′s codon usage. If a gene is only subject to G + C compositional constraint, it will lie on or just below the theoretical ENC curve (Wright, 1990). Otherwise, the gene should be under selection pressure. The result is shown in Fig. 1. Plots of all strains sit not far from the theoretical ENC curve. This result indicates that G + C composition is the dominant factor of SARS-CoV-2′s codon usage, but nature selection also affects SARS-CoV-2′s codon usage. Natural selection may play more important roles in some genes than the genome level (S gene, for example) (Posani et al., 2022; Tyagi et al., 2022). The average codon adaptation index (CAI) of all strains at the genome level is 0.7200. This result also indicates the influence of natural selection on shaping the codon usage of SARS-CoV-2.
Fig. 1.
ENC (Effective number of codons)-GC3s (GC content at the third codon position) plot. The black curve represents the theoretical ENC value with no nature selection. If a gene is only subject to G + C compositional constraint, it will lie on or just below the theoretical ENC curve.
3.2.2. Increasing trend of synonymous codon usage over time
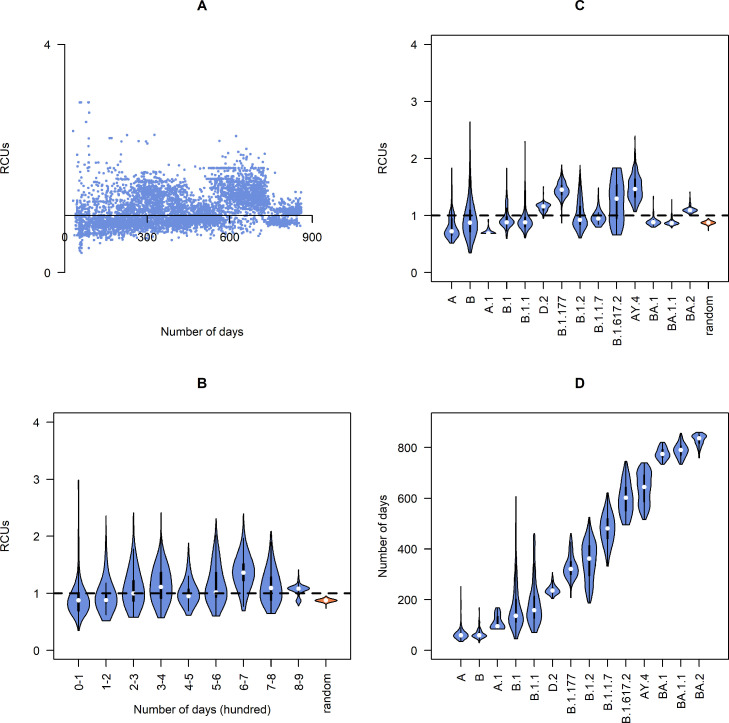
As mentioned in the introduction, the use of synonymous codons might affect translation efficiency. Viruses with higher translation efficiency are more likely to survive from natural selection, leading to the preservation of codons frequently used by humans, and the purifying of codons that humans rarely use. This selection pressure should increase the value of RCU (relative codon usage) within synonymous substituted codons. The trend of RCU changes over time is shown in Fig. 2A and B. In the early months, only a few synonymous substitutions occurred, which caused a high variance of RCU value. The variance decreased over time with the increasing number of synonymous substitutions. Another apparent trend is the increasing RCU value for most stages.
Fig. 2.
RCU (relative codon usage) value for synonymous substitutions and randomly mutated sequences toward the human. A: RCU value for each sample, plot by its collection date. B: Distribution of RCU value for different periods, the orange box shows the distribution of RCU for randomly mutated sequences. C: Distribution of RCU value for some lineages, the orange box shows the distribution of RCU for randomly mutated sequences. D: Distribution of collection date for lineages.
3.2.3. Synonymous codon usage pattern in different lineages
While the above results alone are not solid enough to prove the selection pressure from codon usage optimization, the analysis has two primary limits. The first limitation is that random mutation could cause increasing or decreasing RCU; we need to calculate the RCU for randomly mutated sequences to verify that this trend of increasing RCU was not the result of random mutation. The second limitation is that several dominant lineages occupied the different pandemic stages; the overall tendency could be highly affected by the codon usage bias of these lineages, so each lineage along is necessary to be tested by comparing to randomly mutated sequences.
To eliminate these limitations, RCU for some lineages and randomly mutated sequences were calculated (Fig. 2B and C). The RCU value for most stages was significantly higher than random (Fig. 2B), which proved that the increasing trend was not caused by random mutation. Most lineages have higher RCU than random, and a weak correlation between RCU and the collection date in different lineages was observed (Fig. 2C and D). These results showed the selection pressure from synonymous codon usage optimization on most lineages, especially for lineages in the later stage of the pandemic.
3.3. Synonymous substitution was driven by RNA secondary structure maintaining
RNA secondary structure is another force that could exert selection pressure on synonymous substitutions. Any mutations could potentially break the RNA secondary structure, but synonymous substitutions are more likely affected by the force of maintaining RNA structure. Because a mutation that caused nonsynonymous substitution could be linked with important protein functions, making it harder to be wiped out.
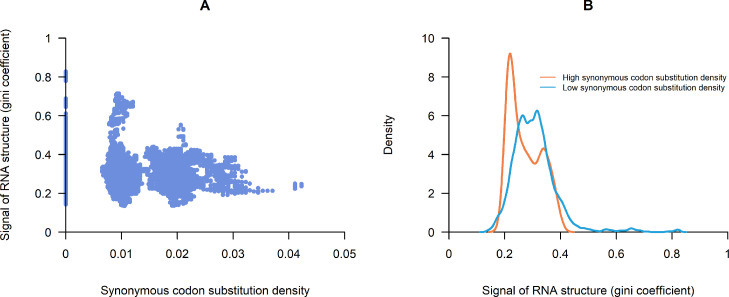
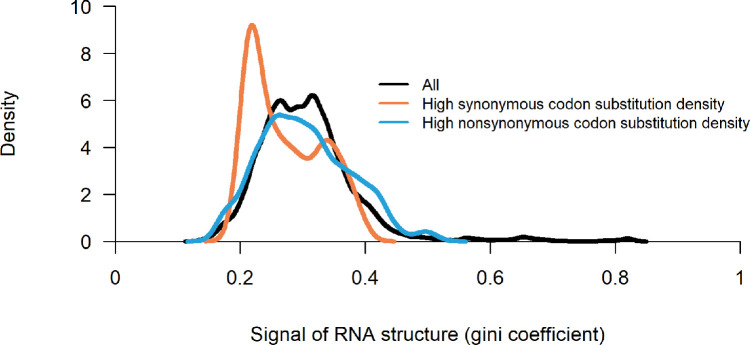
As a single mutation can hardly reflect its impact on RNA secondary structure in a region, the codon substitution density was calculated to better demonstrate the structural effect of mutations. Fig. 3 depicts the relationship between synonymous codon substitution density and RNA structure strengths. The strength of RNA structure showed a weak negative correlation to synonymous codon substitution density (Fig. 3A). However, most mutations sited on areas with low RNA structure strength, they were not likely under purifying selection from RNA structure. So that the overall correlation could conceal the selection pressure on minority mutations in areas with strong RNA structure, making it a defective indicator of the selection pressure from RNA structure. Thus, we split these areas into two groups according to the threshold of the synonymous codon substitution density of 0.025. Synonymous mutation densities above 0.025 were regarded as regions with frequent synonymous substitutions, and vice versa. The result showed that RNA structure strength is significantly lower in regions with frequent synonymous substitutions (p < 0.001, T-test) (Fig. 3B).
Fig. 3.
Relationship between synonymous codon substitution density and RNA structure in “replicate 1″ of data GSE158052. A: Negative correlation between synonymous codon substitution density and the signal of RNA structure (defined by Gini coefficient). B: Distribution of signal of the RNA structure, with high synonymous codon substitution density, and low synonymous codon substitution density, respectively, the threshold of synonymous codon substitution density is 0.025. “Replicate 2″ of data GSE158052 has the same trend, which showed in fig. S1.
3.4. Nonsynonymous substitution affects codon usage and RNA structure
Nonsynonymous substitutions could also be under the selection pressure from codon usage optimization and maintaining RNA structures. However, the trail of these forces could be covered by major selection pressure such as antigenic shift and receptor binding enhancement.
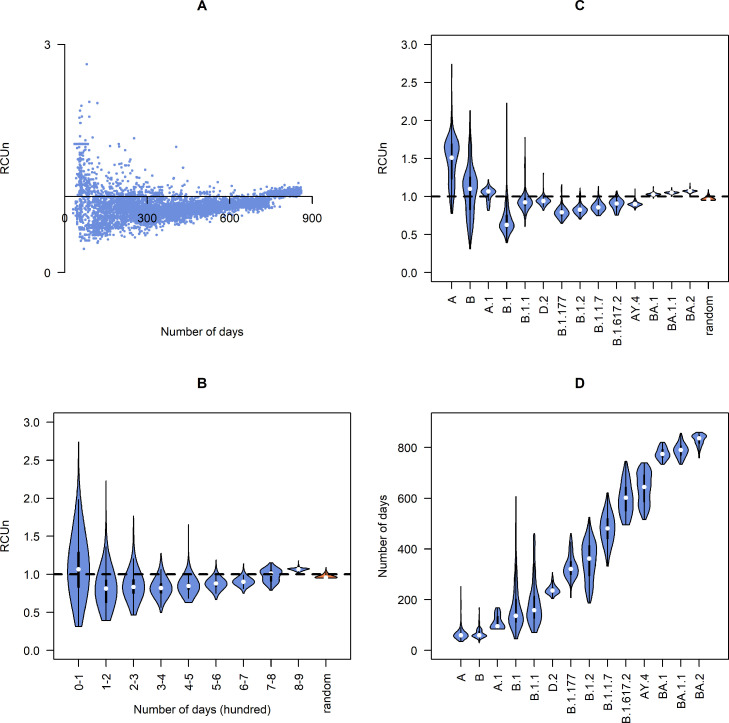
Our results showed that nonsynonymous substitutions could greatly affect codon usage patterns. Nonsynonymous substitutions resulted in a decrease in RCU in the early months (Fig. 4A and B), indicating that nonsynonymous substitutions cause deoptimization of codon usage in the early stages. After that, the value of RCU gradually increased, possibly because of a weak pressure on nonsynonymous substitutions from codon usage.
Fig. 4.
RCU (relative codon usage) value for nonsynonymous substitutions and randomly mutated sequences toward the human. A: RCU value for each sample, plot by its collection date. B: Distribution of RCU value for different periods, the orange box shows the distribution of RCU for randomly mutated sequences. C: Distribution of RCU value for some lineages, the orange box shows the distribution of RCU for randomly mutated sequences. D: Distribution of collection date for lineages.
RNA structure also could bring selection pressures on nonsynonymous substitutions. It showed an inconspicuous trend of purifying nonsynonymous substitution in regions with stable RNA secondary structure (Gini coefficient higher than 0.6) (Fig. 5). The selection pressure is significantly lower on nonsynonymous substitutions than that on synonymous substitutions (Fig. 5). This result confirmed the suggestions above: nonsynonymous substitutions have the selection pressure from maintaining the RNA secondary structure, but there are other forces to keep some nonsynonymous substitutions with important functions that could weaken the effect of this pressure.
Fig. 5.
Distribution of the signal of the RNA structure (defined by Gini coefficient) in “replicate 1″ of data GSE158052. Distributions were drawn with all datasets (black), high synonymous codon substitution density (orange), and high nonsynonymous codon substitution density (blue), respectively. The threshold of synonymous and nonsynonymous codon substitution density is 0.025. “Replicate 2″ of data GSE158052 has the same trend, which showed in fig. S2.
4. Discussions
4.1. Natural selection in non-coding regions
Mutations that caused synonymous substitutions were commonly used as the marker of neutral selection, while a mass of evidence has proved that the selection of synonymous substitution was not neutral (Ngandu et al., 2008; Rahman et al., 2021; Shabalina et al., 2013; Wynn and Christensen, 2015). This study provided evidence from non-coding regions and the silent segment from ORF8, showing that synonymous substitutions in SARS-CoV-2 were under purifying selection. However, non-coding regions could also be under some selection pressure rather than completely under neutral selection.
RNA secondary structure, for example, could exert selection pressure on non-coding regions since it serves crucial functions in these regions (Melidis et al., 2021; Miao et al., 2021; Zhao et al., 2020). As a result, the higher purifying selection pressure in coding regions relative to non-coding regions may not be due to a force on RNA secondary structure maintenance. Thus, more precisely speaking, our evidence from non-coding regions, and the silent segment from ORF8 can only show a more intense purifying selection in coding regions than in non-coding regions.
4.2. Nonsynonymous substitution has effects on codon usage and RNA structure
We put forward 2 possible forces of purifying the synonymous substitution: synonymous substitution codon usage and RNA secondary structure. However, nonsynonymous substitutions could also include constraints by these forces. Nonsynonymous substitutions that cause amino acid variations always lead to the functional change of a viral protein, hence directly facing the selection forces driven by protein function (such as antigenic shift and receptor binding enhancement). Such functional changing will result in a high selection advantage that will outweigh the effects of other selection forces and erase the effect of these forces. Conversely, synonymous substitutions that are not constrained by protein function may allow us to quantify these minor forces on SARS-CoV-2′s evolution.
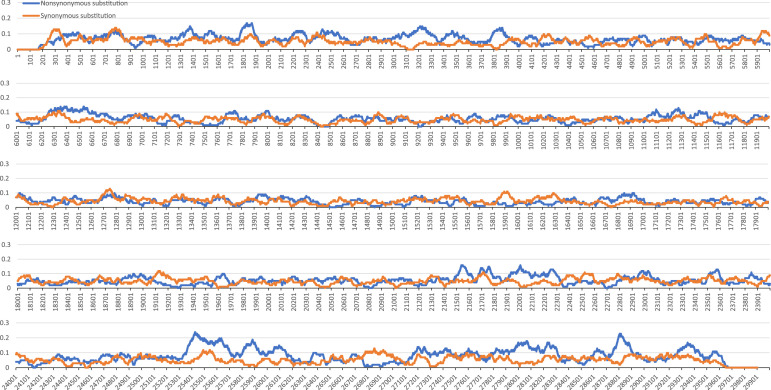
These differences can be reflected by the location of synonymous and nonsynonymous substitutions (Fig. 6). Synonymous substitutions are evenly distributed throughout the genome, indicating that synonymous substitutions were subjected to minimal selection or that these selection pressures are insensitive to the genome location. However, nonsynonymous substitutions tend to be gathered in some areas, indicating the strong force of natural selection on nonsynonymous substitutions. The different localization of synonymous and nonsynonymous substitutions proved one aspect of the outcome of different natural selections.
Fig. 6.
Location of synonymous substitutions and nonsynonymous substitutions. Value in the figure is represented by the ratio of substituted codons with the window of 100 nt.
The effect of nonsynonymous substitution can also be revealed by the different results between Hussain et al. (Hussain et al., 2021) and Ramazzotti et al. (Ramazzotti et al., 2022). Hussain et al. used the entire coding sequence and found a decreased CAI over time. The decreased CAI may result from nonsynonymous substitutions, as our results showed that nonsynonymous substitutions had caused deoptimization in the early months. However, Ramazzotti et al. only considered mutations that cause synonymous substitutions and found an increased similarity of viral codons toward humans. This discussion highlighted the importance of the effect of nonsynonymous substitutions on lesser selection forces.
4.3. Multiple effects of natural selection
Most processes involved in evolution can bring multiple effects on natural selection. Codon usage adaptation, for example, is composed of two parts: retaining preferred codons and purifying undesired codons, which includes both positive and purifying selection. Both parts increase the RCU value, but purifying selection will lead to fewer synonymous substitution numbers. Ramazzotti et al. (Ramazzotti et al., 2022) showed that favorable codons have more chances to reach fixation, proving the positive selection from codon usage optimization.
RNA secondary structure also has a dual effect on natural selection. Despite the fact that RNA structure is linked to a variety of important roles, it has been discovered that it generally reduces translation efficiency (Kozak, 1986; Kramer and Gregory, 2018; Pelletier and Sonenberg, 1985). Breaking an RNA secondary structure sometimes led to increasing fitness, which could lead to positive selection. It is hard to distinguish different forces driven by RNA secondary structure. However, our findings revealed that the synonymous substitutions in RNA secondary structures tend to be wiped out, indicating that SARS-CoV-2 trends to maintaining its RNA secondary structure in evolution. So, the main force from RNA secondary structure is more likely to be purifying selection in SARS-CoV-2′s evolution.
4.4. Distinct lineages caused inconsecutive evolutionary traces
The pandemic of SARS-CoV-2 can be decomposed into the pandemic of some main lineages (B.1.1.7 and B.1.617.2, for example) with distinct codon usage patterns. An apparent consistency can be observed between the trend of RCU for different times (Figs. 2B and 4B) and the trend of RCU for different lineages (Figs. 2C and 4C). Although the overall trend of increasing RCU in Fig. 2 was observed, the alternating lineages created the fluctuation in the value of RCU, causing a relaxed correlation between RCU and collection date. Increasing codon usage adaptation may be more evident in longer evolutionary histories that demand continuous attention.
This observation led to an essential issue on SARS-CoV-2′s evolution. Different lineages have distinct evolutionary paths; some lineages diverged in the early months of the pandemic and have independently evolved for a long time. The direction of selection for these lineages may not be the same, and with the hitchhiking effect and genetic drift, together, they made the distinct genomic characters for different lineages. With the alternating pandemic of different lineages, the inconsecutive evolutionary traces of SARS-CoV-2 were observed. Researchers should pay attention to this because the inconsecutive evolutionary traces could lead to some false conclusions. Such diverged evolutionary route also warns us of the complicated disease we could be facing.
5. Conclusion
This study focused on "silent" mutations rather than nonsynonymous substitutions, which revealed an interesting evolutionary aspect of SARS-CoV-2. We first found the evidence of purifying selection on synonymous substitutions, then studied the two forces that could cause the purifying selection. Codon usage analysis showed that synonymous substitutions in SARS-CoV-2 had shaped a biased codon usage patterns towards the human's genome over time. Maintaining RNA secondary structure also caused selection pressure on synonymous substitutions, making them more likely to be observed in areas with weak RNA structures.
Even though SARS-CoV-2 has received a lot of attention, the epidemic remains uncontrollable and continues to affect our daily lives. To get over this, thorough data and more in-depth research on SARS-CoV-2 are still urgently needed.
Supporting information
Fig. S1: Synonymous codon substitution density and RNA structure in “replicate 2″ of data GSE158052.
Fig. S2: RNA structure of synonymous and nonsynonymous substitution in “replicate 2″ of data GSE158052.
Table. S1: Result with different ω estimation methods.
Funding
This work was supported by the National Natural Science Foundation of China (31771474).
Author statement
Conceptualization: HB; data curation: HB and QS; formal analysis: HB and GA; investigation: HB; visualization: HB; funding acquisition: ST; writing: HB; review & editing: ST, and SUR.
Data availability
All data in this paper are available in GenBank, GISAID, PAXdb or Ensembl databases.
Perl scripts were developed to perform analyses in the research. All codes are available upon request.
CRediT authorship contribution statement
Haoxiang Bai: Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing – review & editing. Galal Ata: Formal analysis. Qing Sun: Data curation. Siddiq Ur Rahman: Writing – review & editing. Shiheng Tao: Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank all the researchers who have shared SARS-CoV-2 data on the GISAID database.
We also thank Hao Wang and Yi Zhang for advices.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198966.
Appendix. Supplementary materials
Reference
- Ata G., Wang H., Bai H.X., Yao X.T., Tao S.H. Edging on mutational bias, induced natural selection from host and natural reservoirs predominates codon usage evolution in hantaan virus. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.699788. ARTN 699788 10.3389/fmicb.2021.699788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., van Hemert F. On the biased nucleotide composition of the human coronavirus RNA genome. Virus Res. 2015;202:41–47. doi: 10.1016/j.virusres.2014.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrio A., Gartner V., Wray G.A. Positive selection within the genomes of SARS-CoV-2 and other Coronaviruses independent of impact on protein function. PeerJ. 2020;8 doi: 10.7717/peerj.10234. ARTN e10234 10.7717/peerj.10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Dhama K., Agoramoorthy G., Chakraborty C. Omicron variant (B.1.1.529) of SARS-CoV-2: understanding mutations in the genome, S-glycoprotein, and antibody-binding regions. Geroscience. 2022 doi: 10.1007/s11357-022-00532-4. 10.1007/s11357-022-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Han X.D., Zhu Z.J., Qi C.L., Wang P., Zhang X. Functional alterations caused by mutations reflect evolutionary trends of SARS-CoV-2. Brief. Bioinform. 2021;22:1442–1450. doi: 10.1093/bib/bbab042. 10.1093/bib/bbab042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J.M. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics. 2004;167:1293–1304. doi: 10.1534/genetics.104.026351. 10.1534/genetics.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristina J., Fajardo A., Sonora M., Moratorio G., Musto H. A detailed comparative analysis of codon usage bias in Zika virus. Virus Res. 2016;223:147–152. doi: 10.1016/j.virusres.2016.06.022. 10.1016/j.virusres.2016.06.022. [DOI] [PubMed] [Google Scholar]
- Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10 doi: 10.1093/gigascience/giab008. ARTN giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Borba L., Villordo S.M., Iglesias N.G., Filomatori C.V., Gebhard L.G., Gamarnik A.V. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J. Virol. 2015;89:3430–3437. doi: 10.1128/Jvi.02677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio N., Walker C.R., Turakhia Y., Lanfear R., Corbett-Detig R., Goldman N. Mutation rates and selection on synonymous mutations in SARS-CoV-2. Genome Biol. Evol. 2021;13 doi: 10.1093/gbe/evab087. ARTN evab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S., Rashmi S., Rane A., Dharavath B., Sawant A., Dutt A. An integrated approach to determine the abundance, mutation rate and phylogeny of the SARS-CoV-2 genome. Brief Bioinform. 2021;22:1065–1075. doi: 10.1093/bib/bbaa437. 10.1093/bib/bbaa437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diviney S., Tuplin A., Struthers M., Armstrong V., Elliott R.M., Simmonds P., Evans D.J. Hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J. Virol. 2008;82:9008–9022. doi: 10.1128/JVI.02326-07. 10.1128/Jvi.02326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L. tRNA gene number and codon usage in the C-elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–289. doi: 10.1016/S0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- Faure G., Ogurtsov A.Y., Shabalina S.A., Koonin E.V. Role of mRNA structure in the control of protein folding. Nucleic. Acids. Res. 2016;44:10898–10911. doi: 10.1093/nar/gkw671. 10.1093/nar/gkw671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.W., Chan M.Y., Hsu W.L., Huang C.C., Tsai C.H. The 3 '-terminal hexamer sequence of classical swine fever virus RNA plays a role in negatively regulating the IRES-Mediated translation. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033764. ARTN e33764 10.1371/journal.pone.0033764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Rasool S.T., Pottathil S. The evolution of severe acute respiratory syndrome coronavirus-2 during pandemic and adaptation to the host. J. Mol. Evol. 2021;89:341–356. doi: 10.1007/s00239-021-10008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, B.A., Zhou, Y., Lokugamage, K.G., Vu, M.N., Bopp, N., Crocquet-Valdes, P.A., Kalveram, B., Schindewolf, C., Liu, Y., Scharton, D., et al. (2022). Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. bioRxiv. 10.1101/2021.10.14.464390. [DOI] [PMC free article] [PubMed]
- Kanaya S., Yamada Y., Kinouchi M., Kudo Y., Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 2001;53:290–298. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30 doi: 10.1093/molbev/mst010. 772-780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U. S. A. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M.C., Gregory B.D. Does RNA secondary structure drive translation or vice versa? Nat. Struct. Mol. Biol. 2018;25:641–643. doi: 10.1038/s41594-018-0100-2. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–U354. doi: 10.1038/Nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.H., Iketani S., Guo Y.C., Chan J.F.W., Wang M., Liu L.Y., Luo Y., Chu H., Huang Y.M., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602 doi: 10.1038/s41586-021-04388-0. 676-+. 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Martin K.C., Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melidis L., Hill H.J., Coltman N.J., Davies S.P., Winczura K., Chauhan T., Craig J.S., Garai A., Hooper C.A.J., Egan R.T., et al. Supramolecular cylinders target bulge structures in the 5 ' UTR of the RNA genome of SARS-CoV-2 and inhibit viral replication**. Angew. Chem. Int. Edit. 2021;60:18144–18151. doi: 10.1002/anie.202104179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z.C., Tidu A., Eriani G., Martin F. Secondary structure of the SARS-CoV-2 5′-UTR. Rna Biol. 2021;18:447–456. doi: 10.1080/15476286.2020.1814556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, N.L., Clark, T., Raman, R., and Sasisekharan, R. (2022). A structural dynamic explanation for observed escape of SARS-CoV-2 BA.2 variant mutation S371L/F. bioRxiv. 10.1101/2022.02.25.481957.
- Morandi E., Manfredonia I., Simon L.M., Anselmi F., van Hemert M.J., Oliviero S., Incarnato D. Genome-scale deconvolution of RNA structure ensembles. Nat. Methods. 2021;18 doi: 10.1038/s41592-021-01075-w. 249-+. 10.1038/s41592-021-01075-w. [DOI] [PubMed] [Google Scholar]
- Mustoe A.M., Busan S., Rice G.M., Hajdin C.E., Peterson B.K., Ruda V.M., Kubica N., Nutiu R., Baryza J.L., Weeks K.M. Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell. 2018;173 doi: 10.1016/j.cell.2018.02.034. 181-+. 10.1016/j.cell.2018.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngandu N.K., Scheffler K., Moore P., Woodman Z., Martin D., Seoighe C. Extensive purifying selection acting on synonymous sites in HIV-1 Group M sequences. Virol. J. 2008;5 doi: 10.1186/1743-422X-5-160. Artn 160 10.1186/1743-422x-5-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson B.L., White K.A. 3 ' Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr. Opin. Virol. 2011;1:373–380. doi: 10.1016/j.coviro.2011.10.002. 10.1016/j.coviro.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Posani E., Dilucca M., Forcelloni S., Pavlopoulou A., Georgakilas A.G., Giansanti A. Temporal evolution and adaptation of SARS-CoV-2 codon usage. Front. Biosci.-Landmrk. 2022;27 doi: 10.31083/j.fbl2701013. ARTN 013 10.31083/j.fbl2701013. [DOI] [PubMed] [Google Scholar]
- Rahman S., Pond S.L.K., Webb A., Hey J. Weak selection on synonymous codons substantially inflates dN/dS estimates in bacteria. P. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2023575118. ARTN e2023575118 10.1073/pnas.2023575118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazzotti D., Angaroni F., Maspero D., Mauri M., D'Aliberti D., Fontana D., Antoniotti M., Elli E.M., Graudenzi A., Piazza R. Large-scale analysis of SARS-CoV-2 synonymous mutations reveals the adaptation to the human codon usage during the virus evolution. Virus Evol. 2022;8 doi: 10.1093/ve/veac026. ARTN veac026 10.1093/ve/veac026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P.C., Naveca F.G., Lins R.D., Dezordi F.Z., Ferraz M.V.F., Moreira E.G., Coelho D.F., Motta F.C., Paixao A.C.D., Appolinario L., et al. The ongoing evolution of variants of concern and interest of SARS-CoV-2 in Brazil revealed by convergent indels in the amino (N)-terminal domain of the spike protein. Virus Evol. 2021;7 doi: 10.1093/ve/veab069. ARTN veab069 10.1093/ve/veab069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan R., Nebot M.R., Chirico N., Mansky L.M., Belshaw R. Viral mutation rates. J. Virol. 2010;84:9733–9748. doi: 10.1128/Jvi.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina S.A., Spiridonov N.A., Kashina A. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic. Acids. Res. 2013;41:2073–2094. doi: 10.1093/nar/gks1205. 10.1093/nar/gks1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Woo H.G. Omicron: a HEAVILY mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front. Immunol. 2022;12 doi: 10.3389/fimmu.2021.830527. ARTN 830527 10.3389/fimmu.2021.830527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.A. The centrality of RNA. Cell. 2009;136 doi: 10.1016/j.cell.2009.02.007. 577-580. 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic. Acids. Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treder K., Kneller E.L.P., Allen E.M., Wang Z.H., Browning K.S., Miller W.A. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008;14:134–147. doi: 10.1261/rna.777308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi N., Sardar R., Gupta D. Natural selection plays a significant role in governing the codon usage bias in the novel SARS-CoV-2 variants of concern (VOC) PeerJ. 2022;10:e13562. doi: 10.7717/peerj.13562. 10.7717/peerj.13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert F., van der Kuyl A.C., Berkhout B. Impact of the biased nucleotide composition of viral RNA genomes on RNA structure and codon usage. J. Gen. Virol. 2016;97:2608–2619. doi: 10.1099/jgv.0.000579. 10.1099/jgv.0.000579. [DOI] [PubMed] [Google Scholar]
- Wang M.C., Herrmann C.J., Simonovic M., Szklarczyk D., von Mering C. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. The 'effective number of codons' used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Wynn E.L., Christensen A.C. Are synonymous substitutions in flowering plant mitochondria neutral? J. Mol. Evol. 2015;81:131–135. doi: 10.1007/s00239-015-9704-x. [DOI] [PubMed] [Google Scholar]
- Yang Z.H. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yu H.P., Zhang Y., Sun Q., Gao H.J., Tao S.H. RSVdb: a comprehensive database of transcriptome RNA structure. Brief Bioinform. 2021;22 doi: 10.1093/bib/bbaa071. ARTN bbaa071 10.1093/bib/bbaa071. [DOI] [PubMed] [Google Scholar]
- Zhao J.X., Qiu J.M., Aryal S., Hackett J.L., Wang J.X. The RNA architecture of the SARS-CoV-2 3 '-Untranslated region. Viruses-Basel. 2020;12 doi: 10.3390/v12121473. ARTN 1473 10.3390/v12121473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this paper are available in GenBank, GISAID, PAXdb or Ensembl databases.
Perl scripts were developed to perform analyses in the research. All codes are available upon request.