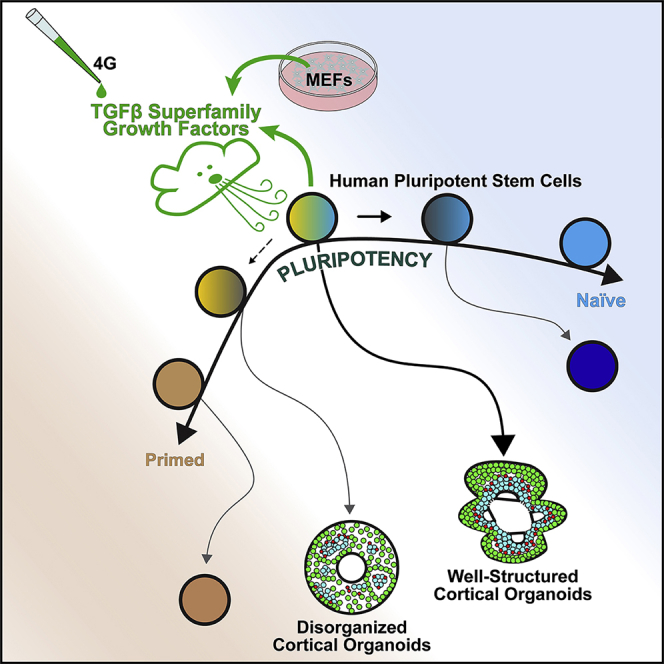
Summary
Telencephalic organoids generated from human pluripotent stem cells (hPSCs) are a promising system for studying the distinct features of the developing human brain and the underlying causes of many neurological disorders. While organoid technology is steadily advancing, many challenges remain, including potential batch-to-batch and cell-line-to-cell-line variability, and structural inconsistency. Here, we demonstrate that a major contributor to cortical organoid quality is the way hPSCs are maintained prior to differentiation. Optimal results were achieved using particular fibroblast-feeder-supported hPSCs rather than feeder-independent cells, differences that were reflected in their transcriptomic states at the outset. Feeder-supported hPSCs displayed activation of diverse transforming growth factor β (TGFβ) superfamily signaling pathways and increased expression of genes connected to naive pluripotency. We further identified combinations of TGFβ-related growth factors that are necessary and together sufficient to impart broad telencephalic organoid competency to feeder-free hPSCs and enhance the formation of well-structured brain tissues suitable for disease modeling.
Keywords: brain organoid, neurogenesis, neural stem cell, embryonic stem cell, pluripotent stem cell, differentiation, neural development, stem cell heterogeneity, pluripotency, cerebral cortex, ganglionic eminence, hippocampus, choroid plexus
Graphical abstract
Highlights
-
•
hPSC maintenance conditions influence the quality of cortical organoid formation
-
•
Identification of an intermediate pluripotency state optimal for cortical organoids
-
•
MEF feeder support enhances activation of diverse TGFβ signaling pathways
-
•
The organoid-promoting effects of MEFs can be mimicked by a TGFβ factor mixture
Watanabe et al. show that hPSCs maintained under different conditions exhibit variabilities in the expression and function of TGFβ superfamily factors, which tracks with their state of pluripotency along the naive-to-primed spectrum and affects their capacity to form well-structured cortical organoids. These findings illustrate an important source of variability in organoid studies and outline a strategy for rectification.
Introduction
The emergence of methods to direct the formation of neurons from human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs)—collectively hPSCs—provides unprecedented opportunities for investigating mechanisms behind healthy human brain development and neurological disease. Attention has most recently turned to the creation of tridimensional structures termed organoids or spheroids, which display spatial organization of neural progenitors and diverse populations of neurons and glial cells that better approximate the features of the developing human brain in vivo than two-dimensional cultures in vitro (Chiaradia and Lancaster, 2020; Velasco et al., 2020). Thus far, organoids have been used to model a variety of disorders impacting brain growth such as microcephaly and lissencephaly, and efforts to model more complex neurological disorders that principally impact neural circuit formation and function such as schizophrenia, autism spectrum disorders, and degenerative conditions are under way (Amin and Pasca, 2018; Samarasinghe et al., 2021; Zhang et al., 2021). Success in these endeavors, however, depends on the reproducible creation of well-structured organoids in which neural networks can be assembled.
Several studies have suggested that organoid reproducibility is best accomplished using protocols that direct the formation of specific brain regions such as the cerebral cortex and basal ganglia (Watanabe et al., 2017; Qian et al., 2018; Sloan et al., 2018; Xiang et al., 2018) reflected by a high degree of organoid consistency seen at the transcriptomic level (Qian et al., 2016; Watanabe et al., 2017; Velasco et al., 2019; Yoon et al., 2019). However, the cytoarchitecture of the organoids, particularly the laminar organization of the cortex, can still markedly vary (Bhaduri et al., 2020a, 2020b). Some of these differences could arise from variabilities inherent to the PSC lines used in a given experiment (Ortmann and Vallier, 2017) and the lack of standardization in protocols used for neuronal differentiation (Anderson et al., 2021). The importance of organoid quality has been further raised by recent studies suggesting that in vitro culture can impart metabolic stress and thereby negatively impact progenitor maturation, cell type specification, and developmental trajectories (Bhaduri et al., 2020b). Given the importance of anatomical structure for neural circuit formation and function in vivo, organoids with irregular structure could serve as poor models for studying the mechanisms behind healthy human brain development and disease.
Previously, we established efficient and reproducible cerebral organoid methods that exhibited improved structural features, including marked expansion of the outer subventricular zone (SVZ), abundant formation of basal progenitors and astrocytes, and enhanced production of upper-layer neurons (Watanabe et al., 2017). We further documented that the developmental trajectory of these organoids was strikingly like that of the human fetal brain in vivo. Our methods also allowed use of developmental patterning signals to create distinct forebrain regions such as the ganglionic eminences (GE), permitting the formation of cortex-GE fusion organoids in which excitatory and inhibitory neurons can intermix to form functional neural networks that mimic activities seen in the human fetal cortex (Samarasinghe et al., 2021). However, in conducting these experiments, we observed that optimal results were achieved when hPSCs were maintained under certain culture conditions, suggesting that even with a well-established organoid protocol, variability could still arise from changes inherent to the starting stem cell population.
While there are many potential contributors to PSC variance, one feature that has been gaining recognition is their state of pluripotency: naive, primed, or a transition phase termed formative (Smith, 2017; Kinoshita et al., 2020). Naive and primed states reflect the developmental progression of pre- to post-implantation epiblast populations in vivo. Formative PSCs have intermediate properties of the two extremes, exhibiting features of the early post-implantation embryonic stage in vivo in which lineage-specific gene expression is minimal and cells are still capable of adopting early developmental fates such as germline stem cells (Kalkan et al., 2017; Smith, 2017; Rostovskaya et al., 2019; Kinoshita et al., 2020). Naive and primed PSC states are interconvertible in vitro (Weinberger et al., 2016); thus, the different ways in which hPSCs are maintained could potentially alter their capacity to effectively form brain organoids.
Here, we define variances in the transcriptional state of hPSCs maintained under different conditions including certain mouse embryonic fibroblast (MEF) feeder-supported and feeder-free (FF) conditions, which were associated with markedly different differentiation outcomes with respect to the efficiency and structural quality of cerebral organoids. We further show that some of the major differences are the expression and activity of transforming growth factor β (TGFβ) superfamily signals derived from MEFs and the hPSCs themselves, which modulate the state of hPSC pluripotency along the naive-to-primed trajectory. The quality of organoids produced by MEF-supported hPSCs was reduced by inhibition of TGFβ signaling, while outcomes from FF hPSCs was improved by pre-treatment of these cells with a mixture of four TGFβ growth factors. Collectively, these studies identify the state of hPSC pluripotency influenced by TGFβ superfamily signaling as a major source of variability that can impact brain organoid formation and illustrate a strategy for modulating this state to help ensure reliability and consistency in organoids to support developmental studies and disease research.
Results
hPSC growth conditions influence their capacity to form well-structured cortical organoids
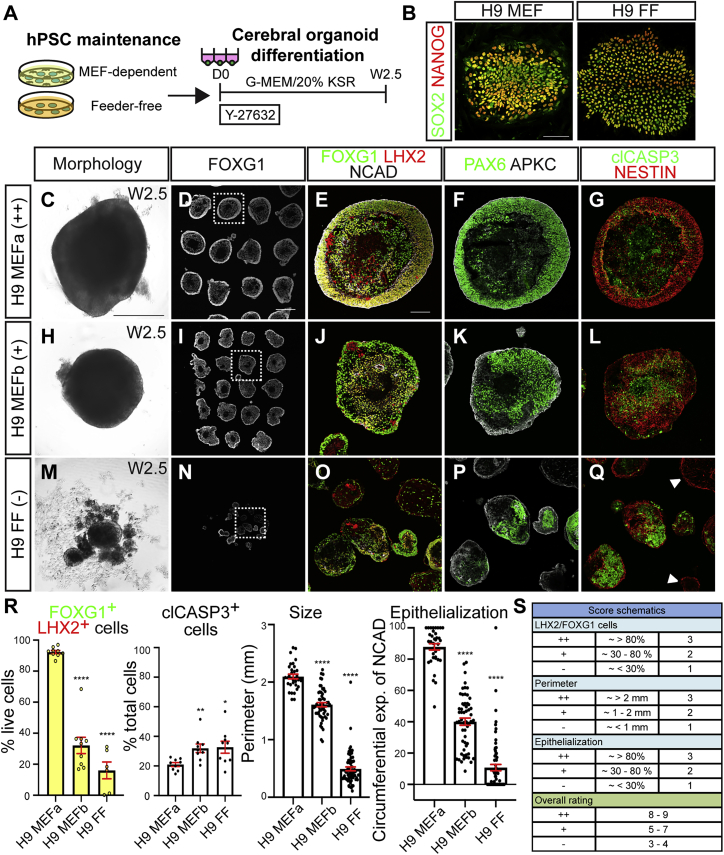
To examine how hPSC maintenance conditions influence cortical organoid formation, we first compared differentiation outcomes of H9 hESCs cultured on certain batches of MEFs and different lot numbers of KnockOut Serum Replacement (KSR) media supplement (hereafter referred to as MEFa and MEFb conditions), or under mTeSR1-based FF conditions (Figures 1A and 1B). While hPSC maintenance conditions were varied, organoid differentiation procedures in this and all subsequent experiments were held constant unless otherwise indicated (Figure S1A). When the hESCs reached 70%–80% confluence, cells were dissociated and plated to form one uniformly sized aggregate in each well of a 96-well V-bottom plate (day 0) (D0 in Figures 1A and S1A; see also Watanabe et al., 2017). After 2.5 weeks in vitro (W2.5), nearly all organoids generated from MEFa-supported H9 exhibited spherical and epithelialized morphologies with >80% of the cells expressing canonical forebrain and cortical progenitor markers such as FOXG1, LHX2, PAX6, and NESTIN (Figures 1C–1G and 1R). Expression of N-CADHERIN (NCAD) and atypical protein kinase C (aPKC; PKCζ) was continuous around the periphery of the aggregate, indicating consistency in the apicobasal polarization of the neural progenitors. Comparable results were seen with cortical organoid formation from a second hESC line, UCLA1, and three hiPSC lines: XFiPSC, Rett syndrome patient wild type and MECP2 mutant iPSC, and Hips2, maintained using the same MEFa conditions (Figures S1B–S1F and S1W–S1Y‴).
Figure 1.
The quality of cortical organoid formation can markedly vary depending on hPSC maintenance conditions
(A) hPSC were maintained using different media conditions, dissociated, and used to create cerebral organoids with the same protocol.
(B) H9 hESCs maintained under either MEF-supported or FF conditions express canonical pluripotency markers such as SOX2 and NANOG.
(C–Q) Comparison of the features and quality of week 2.5 (W2.5) organoids derived from H9 cells maintained with the support of two different batches of MEF and KSR lots (collectively termed MEFa and MEFb conditions) versus mTeSR1-based FF conditions (FF).
(R) Quantification of the percentage of FOXG1+LHX2+ or clCASP3+ cells out of total live cells per organoid (n = 9 organoids per condition, over 4,400 cells counted for each condition, from three different batches of organoids; three organoids per batch), organoid perimeter (n ≥ 32 organoids per condition, from three different batches of organoids, 10–11 organoids per batch), and circumferential expression of NCAD (n ≥ 35 organoids per condition, from three different batches of organoids, 11–12 organoids per batch). Data are presented as mean ± SEM.
(S) Organoid scoring rubric used to classify cortical organoid formation at W2.5 as high quality (++), intermediate quality (+), or poor quality (−). Scores are based upon the three criteria plotted in (R) and indicated on panel labels in many subsequent figures.
Scale bars, 100 μm (B and E) and 500 μm (C and D). See also Figures S1 and S2.
By contrast, H9 cultured under the MEFb conditions formed organoids of smaller size and mixed composition, with irregular shape, reduced coexpression of FOXG1 and LHX2, patchy PAX6 expression, and reduced neuroepithelial characteristics (Figures 1H–1L and 1R). The quality of cortical organoids generated from H9 cells maintained using mTeSR1-based FF conditions appeared even worse (Figures 1M–1R). Poor outcomes were similarly seen with XFiPSC and other iPSC lines maintained under the same FF and MEFb conditions (Hips2, E9, and CHD2 indel mutant; Figures 2D–2G, S1Z–S1BB‴, and S5J).
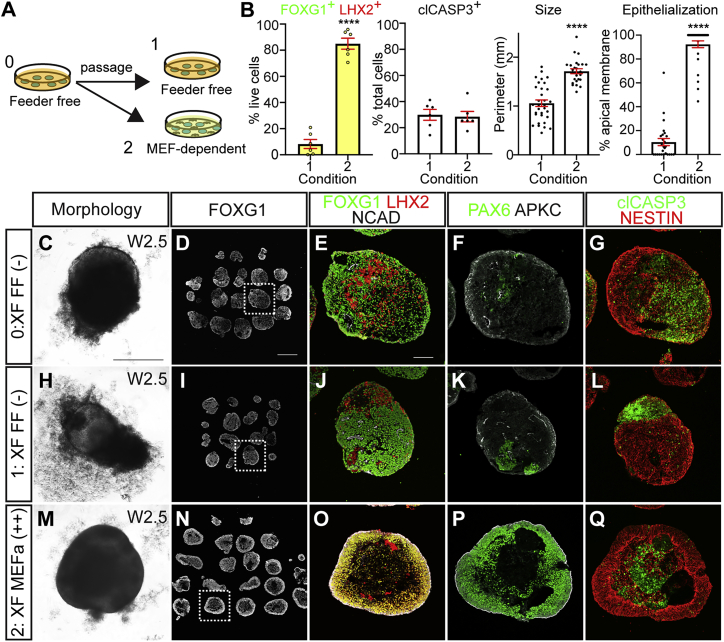
Figure 2.
Feeder-free hPSCs regain cortical organoid competency when passaged under certain MEF-supported conditions
(A) Schematic depicting how FF hPSCs (condition 0) were either maintained in the same FF culture condition (condition 1) or adapted to MEFa-supported conditions (condition 2).
(B) Quantification of the percentage of FOXG1+LHX2+ or clCASP3+ cells out of total live cells per organoid (n = 6 organoids per condition; >6,500 cells counted per condition, from three batches of organoids, two organoids per batch), organoid perimeter (n ≥ 25 organoids per condition, from three batches of organoids, 8–9 organoids per batch), and circumferential expression of NCAD (n ≥ 26 organoids per condition, from three batches of organoids, 8–9 organoids per batch). Data are presented as mean ± SEM.
(C–Q) Morphological and immunohistochemical analyses of cortical organoid formation at W2.5. Scale bars, 500 μm (C and D) and 100 μm (E).
See also Figure S1.
Importantly, we observed that the quality of organoids at the W2.5 time point predicted their developmental trajectory. Most organoids that showed consistent expression of cortical progenitor markers and distinct neuroepithelial architecture at W2.5 displayed robust expression of later cortical developmental markers and exhibited well-defined layered organization at week 8 (W8) and later time points (Figures S1W–S1Y‴, S1CC, and S1DD‴; see also Watanabe et al., 2017). We also examined the expression of genes that have been singled out as hallmarks of metabolic stress such as ARCN1, GORASP2, and PGK1 (Bhaduri et al., 2020b) and found that their expression across development in organoids derived from MEFa-supported hESCs was similar to that seen in the developing human brain in vivo (Figures S2A–S2C). These findings were further confirmed by immunostaining for ARCN1 (Figures S2D–S2G′). Conversely, organoids that appeared dysmorphic at W2.5 did not improve with continued culture, and by W8 showed patchy expression of cortical markers and inconsistent histological organization (Figures S1Z–S1BB‴ and S1EE–S1FF‴). For these reasons, we focused much of our attention toward identifying the factors that influenced organoid quality at W2.5.
To encapsulate the structural quality of organoid formation across these and other experiments, we developed a 9-point and three-tier rating scale considering three criteria: perimeter size, cortical marker expression, and extent of epithelialization. High-, mixed-, and low-quality organoid batches were respectively assigned ratings of ++ (8–9 points), + (5–7 points), and − (≤4 points) (Figures 1R and 1S; Table S1). We hereafter refer to the hPSCs yielding these three outcomes as organoid-competent, semi-competent, and non-competent.
hPSCs yielding lower-quality organoids can be improved by modifying their maintenance conditions
We next examined whether the non-competent hPSCs grown under FF conditions were forevermore fated to yield poor-quality organoids or whether their capacity to form well-structured organoids could be restored by adapting the cells to the MEFa conditions (Figure 2A). XFiPSCs cultured under FF conditions were accordingly split and passaged in parallel under FF mTeSR1 or MEFa conditions, then simultaneously differentiated into cortical organoids. In as little as a single passage, XFiPSCs regained organoid competency in stark contrast to the cells that were maintained under FF conditions (Figures 2B–2Q). The capacity of non-competent FF hPSCs to transform into competent hPSCs by the MEFa conditions was further confirmed using H9 hESCs (Figures S1L–S1V). Together, these results demonstrate that states of hPSC organoid competency are malleable and can be readily altered by changes in hPSC maintenance conditions.
Competent and non-competent hPSCs exist in distinct transcriptomic states related to TGFβ superfamily signaling and expression of genes connected to naive pluripotency
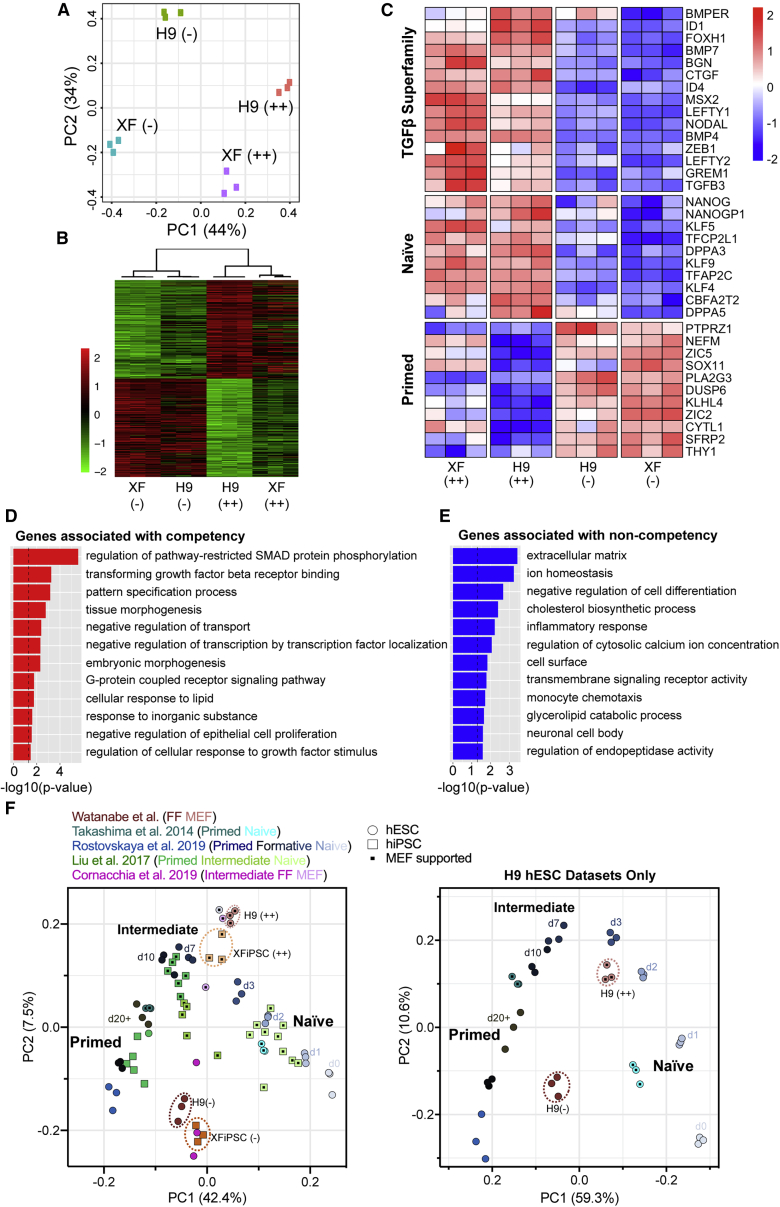
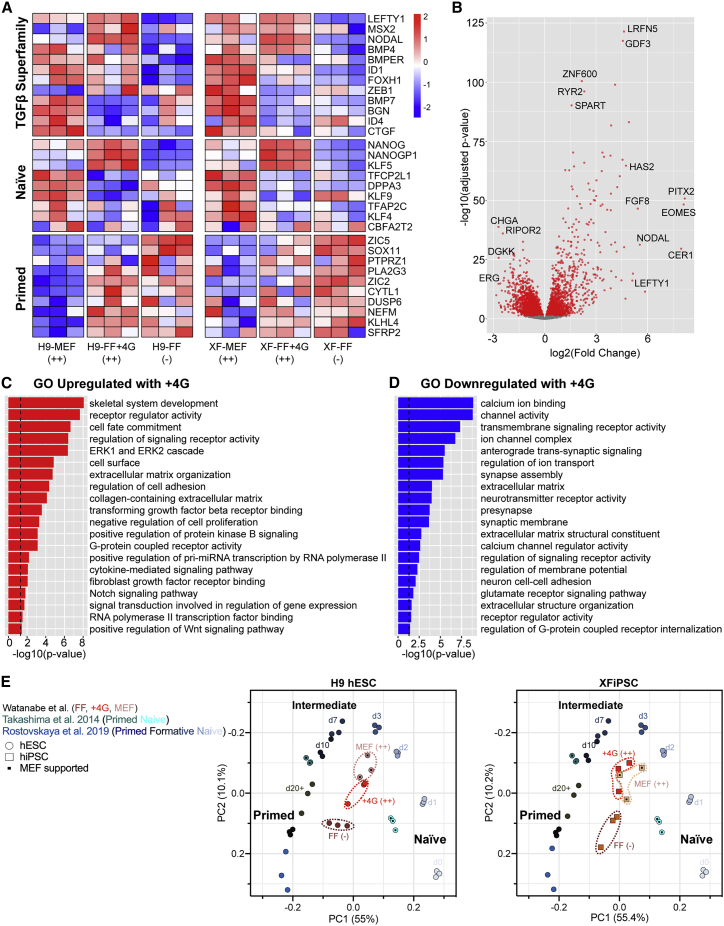
Given the capacity of hPSCs to switch between states of organoid competency, we sought to define the transcriptomic signatures associated with this difference. We conducted bulk RNA-sequencing analysis on triplicate samples of both H9 hESCs and XFiPSCs grown under the MEFa and FF conditions. Principal component analyses based on either mRNA transcripts or long non-coding RNAs showed that each hPSC replicate group was separated by both cell line origin and cell culture conditions (Figures 3A and S3A). Hierarchical clustering using the top 300 most differentially expressed genes revealed that the influences of cell culture conditions predominated over cell line origin (Figure 3B). Gene ontology (GO) analysis revealed that genes associated with growth factor signaling, SMAD protein phosphorylation, and developmental processes were highly upregulated in the organoid-competent feeder-supported hPSCs (Figure 3D). By contrast, genes associated with metabolic pathways such as cholesterol biosynthesis were enriched in FF hPSCs, as has previously been reported (Figure 3E; Zhang et al., 2016; Cornacchia et al., 2019). Extending these GO results, we identified two overt signatures among the genes most highly expressed by organoid-competent hPSCs: TGFβ superfamily signaling and connection to naive pluripotency. By contrast, non-competent hPSCs showed enrichment for genes associated with primed pluripotency (Figures 3C and S3B; Tables S2 and S3). Using Disease Association Protein-Protein Link Evaluator (DAPPLE), a permutation algorithm that looks for significant connectivity in genes associated with a disease state or condition based on known protein-protein interaction data (Rossin et al., 2011), we found that certain bone morphogenetic protein (BMP)-, TGFβ-, LEFTY-, and NODAL-associated molecules were highly connected in competent hPSCs (Figure S3C).
Figure 3.
Cortical organoid competence is associated with a distinct transcriptomic signature defined by elevated TGFβ superfamily signaling and expression of genes connected to naive pluripotency
(A) Principal component (PC) analysis based on RNA transcripts expressed by undifferentiated MEFa-supported versus FF H9 hESCs and XFiPSCs (XF) prior to organoid formation. ++ and − indicate the observed quality of organoids formed from these cells. Three independent experimental replicates per hPSC line and condition are displayed.
(B) Hierarchical clustering of the top 300 differentially expressed genes between the samples.
(C) Heatmap showing the expression of genes associated with TGFβ superfamily signaling, naive pluripotency, and primed pluripotency. The color of the tiles indicates scaled regressed gene expression data.
(D and E) Gene ontology analysis of the upregulated and downregulated transcripts.
(F) PC analysis relating the global transcriptome of our H9 and XFiPSC samples (annotated with dashed circles) to previously published datasets for naive, formative/intermediate, and primed hPSCs. Right plot displays the same information for H9 datasets alone. The x axis (PC1) was reversed to best match the left plot. Days of formative transition from naive to primed pluripotency (day 0 to day 20+) are indicated according to Rostovskaya et al. (2019).
See also Figures S3 and S4; Table S4.
As organoid-competent hPSCs were prominently associated with MEF support, we performed mass spectrometry analysis of secreted proteins present in MEF-conditioned culture media (Figure S3D). Among the top 18 proteins with spectral counts greater than 20, the sixth most abundant was Inhibin beta A (INHBA), another TGFβ superfamily member whose homodimers constitute ACTIVIN A, a growth factor used in the 5i/L/A method for naive hPSC conversion (Theunissen et al., 2014). Thus, the strong signature of TGFβ superfamily signaling associated with MEF-supported hPSCs likely reflects both the paracrine actions of growth factors produced by MEFs such as ACTIVIN A on top of the autocrine actions of signals produced by the hPSCs themselves.
We next used cell marker enrichment analyses to compare the transcriptomes of both organoid-competent and non-competent hPSCs to reference datasets for naive and primed hPSCs, along with a variety of cell lineages. Non-competent hPSCs were most enriched for genes associated with primed pluripotency as well as multi-lineage differentiation (Figures S3B–S3E and Table S3). While competent hPSCs showed increased expression of some genes connected to naive pluripotency, they did not strongly align with naive hPSCs in this analysis (Figures 3D and S3B). At the same time, competent hPSCs showed reduced association with multi-lineage differentiation compared with non-competent hPSCs (Figures S3E), suggesting that they reflect a state of pluripotency which is neither primed nor naive.
To further assess how the observed states of organoid competency relate to hPSC pluripotency, we compared global transcriptomes of our undifferentiated hPSCs with published datasets representing both naive and primed groups, as well as intermediate states representing the formative stages of pluripotency that accompany the developmental transition from pre- to post-implantation blastocysts (Takashima et al., 2014; Liu et al., 2017; Cornacchia et al., 2019; Rostovskaya et al., 2019). Principal component analysis using all genes showed pronounced separation of naive and primed hPSC samples, splitting into two groups along the principal component 1 (PC1) axis (Figures 3F, S3F, and S3J; Table S4). hPSCs representing early to middle stages in formative transition formed clusters in between the naive and primed states. When overlaid onto this map, our non-competent FF hPSCs clustered adjacent to the primed hPSC datasets whereas the competent MEF-supported hPSCs formed groups that were closer to, yet distinct from, naive hPSCs. The reference datasets that appeared closest to our organoid-competent hPSCs were MEF-supported cells reported by Cornacchia et al. (2019) and those representing days 3 and 7 of formative transition as reported by Rostovskaya et al. (2019).
We further examined the status of semi-competent hPSCs associated with maintenance of cells under the suboptimal MEFb condition. Principal component analysis of the global transcriptome showed that organoid-competent hPSCs, non-competent hPSCs, and semi-competent hPSCs formed distinct groupings, with semi-competent hPSCs positioned between competent and non-competent hPSCs along the PC1 axis that segregated cells along the naive-to-primed pluripotency spectrum (Figure S3H). Consistent with this finding, many genes associated with primed pluripotency showed medium expression levels in semi-competent hPSCs when compared with competent hPSCs and non-competent hPSCs (Figure S3I). While we anticipated that both TGFβ superfamily molecules and genes associated with naive pluripotency would similarly exhibit medium expression levels relative to organoid-competent and non-competent hPSCs, this was not the case. Rather, TGFβ molecules and naive-pluripotency-connected factors were instead upregulated in semi-competent hPSCs compared with competent hPSCs. These results raise the possibility that TGFβ superfamily signaling levels might have to be finely tuned to achieve optimal cortical organoid formation.
Additional GO analysis showed that genes that were highly upregulated in organoid-competent hPSCs compared with semi-competent hPSCs were associated with extracellular matrix, negative regulation of differentiation, and positive regulation of MAPK signaling (Figure S3G). One of the most enriched GO terms was “negative regulation of insulin-like growth factor receptor signaling pathway,” which stood out as potentially relevant given that one of the key components of the hPSC culture media known to show variability across lots, KSR, contains high amounts of insulin (Wataya et al., 2008). By contrast, genes enriched in semi-competent hPSCs were associated with the breakdown of small molecules and natural killer (NK) cell-mediated cytotoxicity.
Collectively, these data illustrate the wide diversity in the states of pluripotency exhibited by hPSCs maintained under different culture conditions, some of which are favorable or unfavorable for cortical organoid formation. hPSC states under MEF-dependent conditions also appeared to be variable across different laboratories (Figure 3F), which may reflect the cell culture practices of individual groups but also considerable variance in the batches of MEF and media components used in each case. Importantly, while our best results were achieved with MEFa-supported hPSCs that displayed increased expression of some naive pluripotency genes, naive hPSCs themselves yielded poor-quality organoids (Figures S1G–S1K and S1V).
TGFβ superfamily signaling in hPSCs is required for effective cortical organoid formation
Since multiple genes associated with TGFβ superfamily signaling were upregulated in competent hPSCs and ACTIVIN A was being secreted by MEFs, we tested whether TGFβ superfamily signaling was required for high-quality cortical organoid formation. Five days before the start of organoid differentiation, we pre-treated MEF-supported hPSCs with either SB-431542, a selective small-molecular inhibitor of TGFβ/ACTIVIN receptors, or LDN-193189, which inhibits BMP receptors at low doses and both BMP and TGFβ/ACTIVIN receptors when used at higher concentrations (Figure S4A; Inman et al., 2002; Yu et al., 2008). MEF-supported hPSCs exposed to either low or high doses of these inhibitors displayed markedly reduced cortical organoid quality and size (Figures S4B–S4D). Transcriptomic and GO analyses confirmed that the expression and activity of many TGFβ signaling molecules were downregulated after SB-431542 application along with factors associated with naive pluripotency, while genes associated with primed pluripotency demonstrated intermediate changes (Figures S4E–S4H). SB-431542-treated hPSCs were also enriched for genes associated with sensory organ and tissue morphogenesis, developmental growth, synapse assembly, and forebrain development, although the expression of differentiated cell lineage markers was minimal. Thus, while SB-431542 changed the transcriptional profile of the hPSCs, it did not appear to trigger premature differentiation.
Next, we examined whether the addition of TGFβ superfamily molecules to non-competent FF hPSC cultures could improve their capacity to form well-structured organoids. As several TGFβ superfamily molecules such as LEFTYA, BMP4, TGFβ1, TGFβ3, NODAL, and ACTIVIN A were either enriched in competent hPSCs or produced by the supporting MEFs, we tested the effects of pre-treating FF hPSCs with each of these growth factors alone or in pairwise combinations. Supplementation of the cultures with most of these factors, apart from LEFTYA, improved the overall quality of cortical organoid formation with some additive effects (Figure S5 and Table S1). The combination of BMP4 and TGFβ1 displayed the most potency but was inconsistent (Figure S5E and Table S1). Moreover, even when there appeared to be favorable outcomes, the overall organoid quality scored below that routinely achieved with MEF-supported competent hPSCs. We also observed dose-dependent effects and the potential for overshoot. For example, 0.1 ng/mL BMP4 improved cortical organoid formation, but this benefit was lost when 0.15 ng/mL was used (Figure S5C). Taken together, these results demonstrate that BMP and TGFβ superfamily signaling in undifferentiated hPSCs is required for effective cortical organoid production, but individual or pairs of these growth factors are not alone sufficient to convey competence.
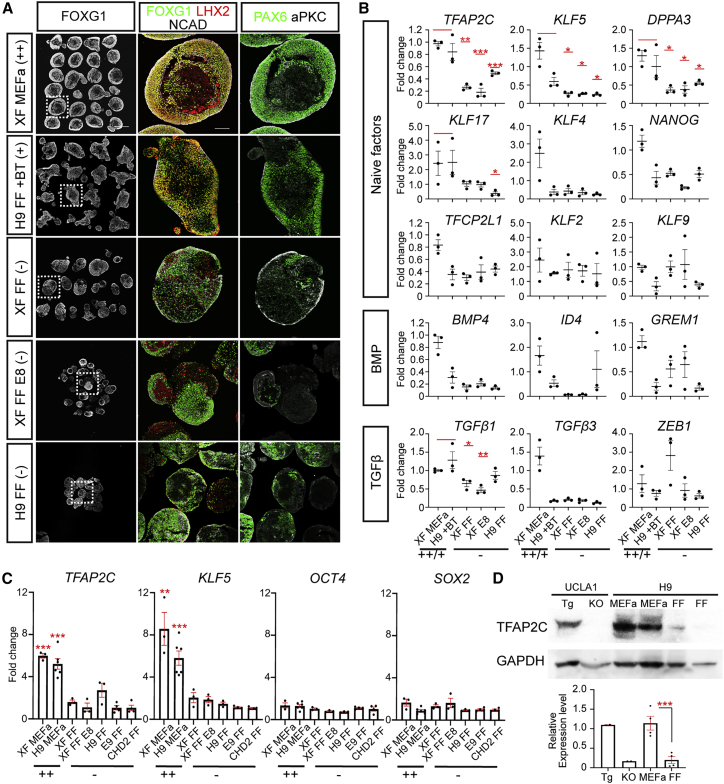
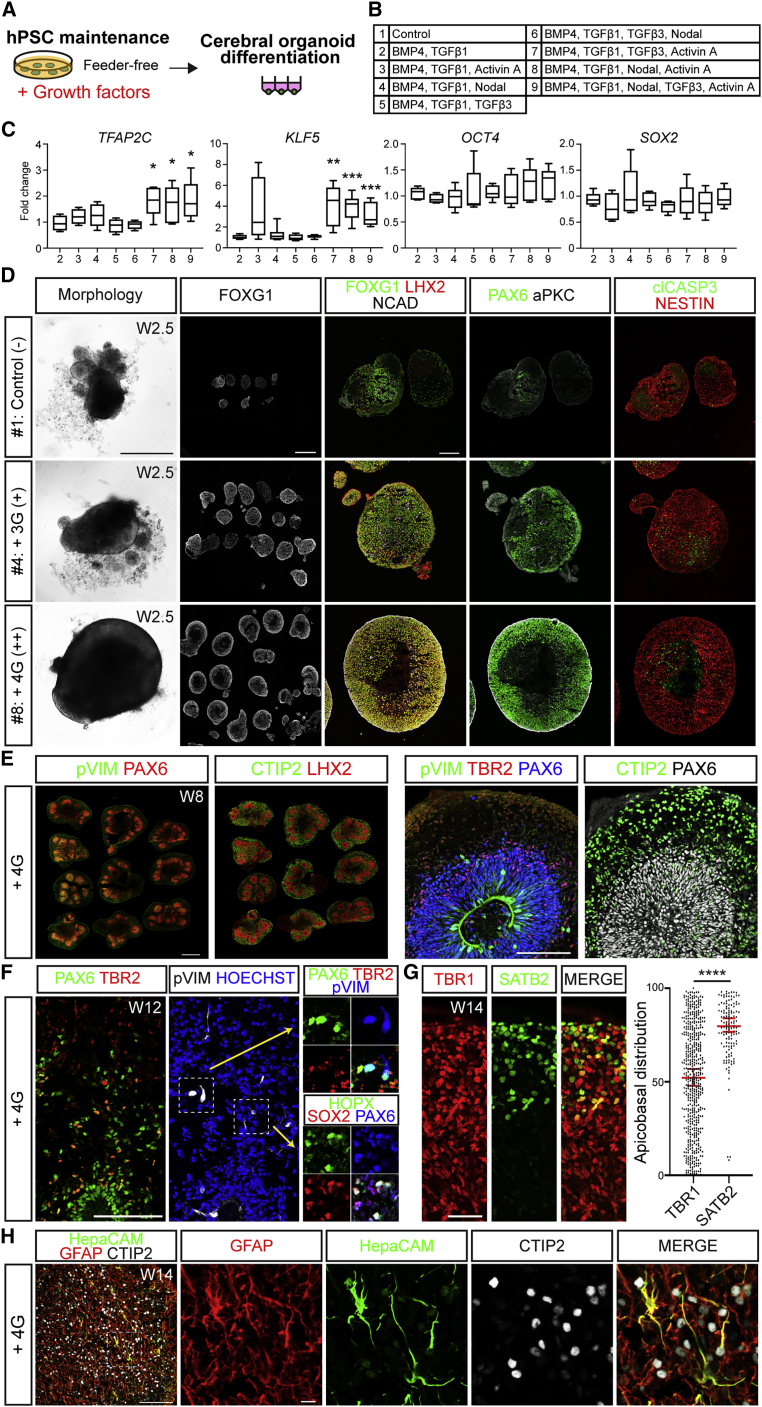
TFAP2C and KLF5 can serve as indicators of hPSC competency to form well-structured cortical organoids
Given that multiple TGFβ superfamily growth factors showed partial benefit to cortical formation, we sought to assess the effects of more complex combinations. However, as our primary readout was quality of organoid formation after several weeks of differentiation, we sought a more efficient screening strategy. We reasoned that the transcriptional signature of TGFβ superfamily signaling and naive pluripotency genes associated with competent hPSCs could be used to predict their capacity to produce high-quality organoids. We thus collected RNA samples from undifferentiated hPSCs cultured under five different conditions: MEFa-dependent XFiPSCs (representing fully competent [++] cells), FF H9 supplemented with BMP4 and TGFβ1 (semi-competent [+] cells), XFiPSCs and H9 hESCs grown under mTeSR1-based FF conditions (non-competent [−] cells), and XFiPSCs grown under E8-based FF conditions (non-competent [−] cells), and then scored their organoid production quality at W2.5 (Figure 4A). Using qRT-PCR analyses, we found that elevated expression of certain naive pluripotency-associated genes in the hPSCs, TFAP2C, KLF5, and DPPA3, predicted well-structured organoid formation more so than genes associated with TGFβ superfamily signaling such as BMP4, TGFβ1 ID4, GREM1, and ZEB1 (Figure 4B). Moreover, TFAP2C, KLF5, and DPPA3 showed very low expression in the hPSCs that yielded the poorest-quality organoids. We further confirmed these results for TFAP2C and KLF5 with additional replicates, cell lines, and cell culture conditions (Figure 4C). While TFAP2C and KLF5 levels were variable, other pluripotency genes such as OCT4 and SOX2 were unchanged. We also observed elevated expression of TFAP2C protein in MEFa-supported H9 hESCs compared with FF H9 hESCs (Figure 4D). Based on these lines of evidence, we concluded that TFAP2C and KLF5 levels could serve as prospective indicators of hPSC competency to create well-structured organoids.
Figure 4.
TFAP2C and KLF5 are prospective indicators of hPSCs that can effectively form high-quality cortical organoids
(A) Representative examples of W2.5 cortical organoids derived from hPSCs maintained under different culture conditions associated with a range of organoid quality outcomes: XFiPSCs grown under MEFa-supported conditions (++ cells), H9 grown under FF conditions supplemented with BMP4 and TGFβ1 (+ cells, see Figure S5E), and XFiPSCs or H9 cells grown under either mTeSR1- or E8-based FF conditions (− cells). All conditions shown have three independent experimental replicates per hPSC line, ≥12 organoids per replicate from three different batches of organoids, and four organoids per batch. See also Table S1.
(B) qRT-PCR analyses. Expression levels displayed are normalized to XF cultured under the MEFa-supported condition. Data are presented as mean ± SEM, n ≥ 3 independent experimental replicates. Statistical analysis compares the combination of XF MEFa (++) and H9 FF + BT (+) against each FF condition.
(C) qRT-PCR analysis of gene expression in additional hPSC lines and culture conditions. Expression levels were compared with E9 FF hiPSC samples, and data represent mean ± SEM. n ≥ 3 independent experimental replicates.
(D) Western blot analysis showing high TFAP2C protein expression in MEFa-supported or FF H9 hESCs. Lysates from UCLA1 hESCs with a doxycycline-inducible TFAP2C transgene (Tg: + 1 μg/mL doxycycline) or gene knockout (KO) were run as controls. Expression levels relative to GAPDH are plotted. Data are presented as mean ± SEM, n ≥ 4 independent experimental replicates.
See also Figures S5.
Competency to form well-structured cortical organoids can be conferred onto non-competent FF hPSCs by a mixture of four TGFβ superfamily molecules
Using TFAP2C and KLF5 expression as a readout, we systematically tested the impact of pre-treating non-competent FF hPSCs with various combinations and concentrations of the five TGFβ superfamily growth factors that showed positive benefits in our previous experiments. We included BMP4 and TGFβ1 in all combinations, since these two factors had shown the most promising organoid-promoting activities (Figure S5). Eight different combinations of BMP4, TGFβ1, TGFβ3, NODAL, and ACTIVIN A were tested on FF H9 hESCs and subjected to qRT-PCR analyses 3 days later (Figures 5A–5C). Significant increases in TFAP2C and KLF5 without changes in OCT4 and SOX2 were observed with three conditions: #7, BMP4/TGFβ1/ACTIVIN A/TGFβ3; #8, BMP4/TGFβ1/ACTIVIN A/NODAL; and #9, all five growth factors together (Figures 5B and 5C). We then carried out cortical organoid formation from FF H9 cells pre-conditioned with growth factor combinations #4, #7, #8, and #9.
Figure 5.
Identification of combinations of TGFβ superfamily growth factors that can enhance cortical organoid production from feeder-free hPSCs
(A) FF H9 hESCs were supplemented with various combinations of TGFβ superfamily growth factors for 3 days prior to the start of organoid formation. Organoid differentiation conditions were held constant.
(B) List of the combinations of growth factors tested.
(C) qRT-PCR analyses identifying the combinations of TGFβ superfamily growth factors that upregulated TFAP2C and KLF5, but not OCT4 or SOX2, in undifferentiated cells. Expression levels displayed are normalized to FF H9 without any growth factor supplementation. Data are presented as mean ± SEM, n ≥ 4 independent experimental replicates.
(D) Representative images of cortical organoids formed from FF H9 supplemented with or without growth factor combinations #4 or #8.
(E) Immunohistochemical analysis of 4G-derived cortical organoids at W8. See Figure S1 for comparable analysis of W8 organoids grown under MEF-supported and FF conditions without 4G.
(F) At W12, 4G-derived cortical organoids typically exhibit an expanded SVZ with abundant formation of SOX2+, PAX6+, HOPX2+, and pVIM+ basal radial glial cells.
(G) By W14, the cortical organoids display formation of both deep (TBR1+) and superficial-layer (SATB2+) neurons.
(H) W12 and older organoids also contain many GFAP+ and HepaCAM+ astrocytes.
Scale bars, 500 μm (D), 100 μm (E, F, first panel in H), 50 μm (G), and 10 μm (second panel in H). See also Figures S5 and S6.
Without any growth factor supplementation, FF H9 cells yielded poor-quality cortical organoids (Figure 5D and Table S1). By contrast, FF H9 treated with growth factor combinations #7, #8, and #9 all showed dramatic improvement in organoid formation with quality scores above 7 (Figures 5C and 5D; Table S1). Among these three conditions, the addition of four growth factors (hereafter referred to as “4G,” #7 and #8) produced the best results with little to no difference betwen these two mixtures (Table S1). FF H9 supplemented with the combination of BMP4/TGFβ1/NODAL (#4), which did not significantly upregulate TFAP2C and KLF5, showed partial rescue of organoid quality (Figure 5D and Table S1), illustrating the value of these genes in predicting organoid outcomes. We confirmed the capacity of the 4G condition #7 (BMP4, TGFβ1, TGFβ3, and ACTIVIN A) to increase TFAP2C and KLF5 expression and enhance cortical organoid formation in FF XFiPSCs and two additional FF non-competent hiPSC lines (Figures S5J and S5K), suggesting that the 4G mixture may be of general benefit when using FF cells.
We next examined how the overall transcriptomic state of hPSCs was altered by 4G #7 addition (Figure 6). Many genes encoding TGFβ superfamily signaling molecules that were characteristic of MEF-supported hPSCs were upregulated following 4G addition to FF hPSCs (Figures 6A and 6B). We also found that subsets of genes connected to naive pluripotency were similarly increased by the 4G mixture, while genes associated with primed pluripotency were downregulated. GO analyses of the genes that were upregulated by 4G treatment were enriched for terms associated with TGFβ and fibroblast growth factor receptor binding, ERK signaling, and both Notch and WNT pathways (Figure 6C). In comparison, genes that were suppressed by 4G were principally enriched for terms associated with calcium channel signaling, extracellular matrix, and transmembrane signaling (Figure 6D). Principal component analysis of all genes lastly showed that 4G-treated hPSCs were shifted away from non-competent FF hPSCs along the naive-to-primed pluripotency spectrum and clustered close to MEFa-supported competent hPSCs (Figure 6E). Thus, 4G supplementation appears to be sufficient to alter the global state of hPSC pluripotency toward a condition that is more favorable for cortical organoid formation.
Figure 6.
4G supplementation shifts the transcriptome of hPSCs to a more naive state
(A) Heatmap showing the effects of different hPSC culture conditions on the expression of genes associated with TGFβ superfamily signaling, naive pluripotency, and primed pluripotency. The color of the tiles indicates scaled regressed gene expression data.
(B) Volcano plot demonstrating the fold change of gene expression (x axis) and adjusted p value (y axis).
(C and D) Gene ontology analysis of the upregulated (C) and downregulated (D) transcripts in 4G-treated hPSCs.
(E) Principal component (PC) analysis showing that FF hPSCs treated with 4G become transcriptionally similar to MEFa-supported cells. In both plots, the y axis (PC2) was reversed to facilitate comparisons with the plots presented Figures 3 and S3.
Cortical organoids derived from 4G-treated FF hPSCs reproducibly exhibit well-defined laminar architecture
We next examined whether organoids created from non-competent hPSCs using the 4G supplementation method exhibited the histological organization and expression of molecular markers of cortical development seen in our previous studies using MEFa-supported hPSCs (Watanabe et al., 2017; Samarasinghe et al., 2021). In our protocol, we add leukemia inhibitory factors (LIF) at W5 to activate STAT3 signaling and thereby stimulate basal radial glial cell (bRGC) expansion and, subsequently, astrogliogenesis (Watanabe et al., 2017). At W8, 4G-associated organoids exhibited prominent layered organization with a well-defined ventricular zone containing LHX2+PAX6+ apical radial glial cells, SVZ containing TBR2+ intermediate progenitors (IP), and subplate/cortical plate compartment with many CTIP2+ neurons (Figure 5E). The layered organization was also reproducible across individual organoids within the group. By contrast, organoids derived from FF H9 without 4G showed patchy expression of PAX6 and LHX2, reduced numbers of TBR2+ IP and correspondingly thinned SVZ, and a poorly defined subplate/cortical plate with relatively few CTIP2+ neurons (Figures S1FF–S1FF‴).
With continued culture, the 4G-associated organoids came to exhibit a well-defined outer SVZ with abundant formation of dispersed bRGCs expressing canonical markers such as SOX2, PAX6, HOPX, and pVIM yet negative for the IP marker TBR2 (Figure 5F). In our previous publication, LIF addition also improved upper-layer neuronal production and laminar organization of the organoids (Watanabe et al., 2017). The same appeared true with W14 4G-associated organoids, which displayed abundant formation of SATB2+ upper-layer neurons that were localized outward in the cortical plate (Figure 5G), reminiscent of the distribution of deep- and upper-layer neurons seen in organoids derived from MEFa-supported hPSCs and gestational week 15.5 human cortices (Watanabe et al., 2017). GFAP+ astrocytes with characteristic star-shaped morphologies, along with a smaller number of HepaCAM+ cells, were also produced (Figure 5H). Together, these experiments illustrate how the addition of a mixture of diverse TGFβ superfamily molecules to undifferentiated non-competent hPSCs can alter their developmental trajectory and promote the reproducible formation of high-quality cortical organoids that exhibit key cytoarchitectural features of the developing human cortex in vivo.
4G supplementation can enhance the quality of cortical organoids generated from FF hPSCs using other protocols
Given the benefits of 4G treatment for FF hPSCs with our cortical organoid protocol, we sought to test whether it could also improve differentiation outcomes from FF cells using other approaches such as the widely used spheroid method (Yoon et al., 2019). We accordingly generated cortical spheroids from H9 hESCs grown under two FF media conditions: Spheroid Essential 8 (SE8) and mTeSR1 (SmTeSR), with or without 4G addition. 4G treatment appeared to have a positive impact, as spheroids derived from 4G-treated cells showed greater numbers of FOXG1+LHX2+ cortical progenitors and enhanced epithelialization at W2.5 compared with the untreated controls, without any change to overall size of the spheroids, abundance of NESTIN+ neural progenitors, or extent of cell death (Figures S6A–S6Q). The appearance of the spheroids at W2.5 again seemed to predict outcomes at later time points, as W8 organoids derived from 4G-treated hPSCs showed marked improvements in the expression of cortical markers such as PAX6, TBR2, pVIM, and CTIP2, which tended to be patchy in spheroids made from untreated cells (Figures S6R–S6Y). Collectively, these data suggest that 4G administration may be of general benefit for creating cortical organoids/spheroids starting from FF hPSCs.
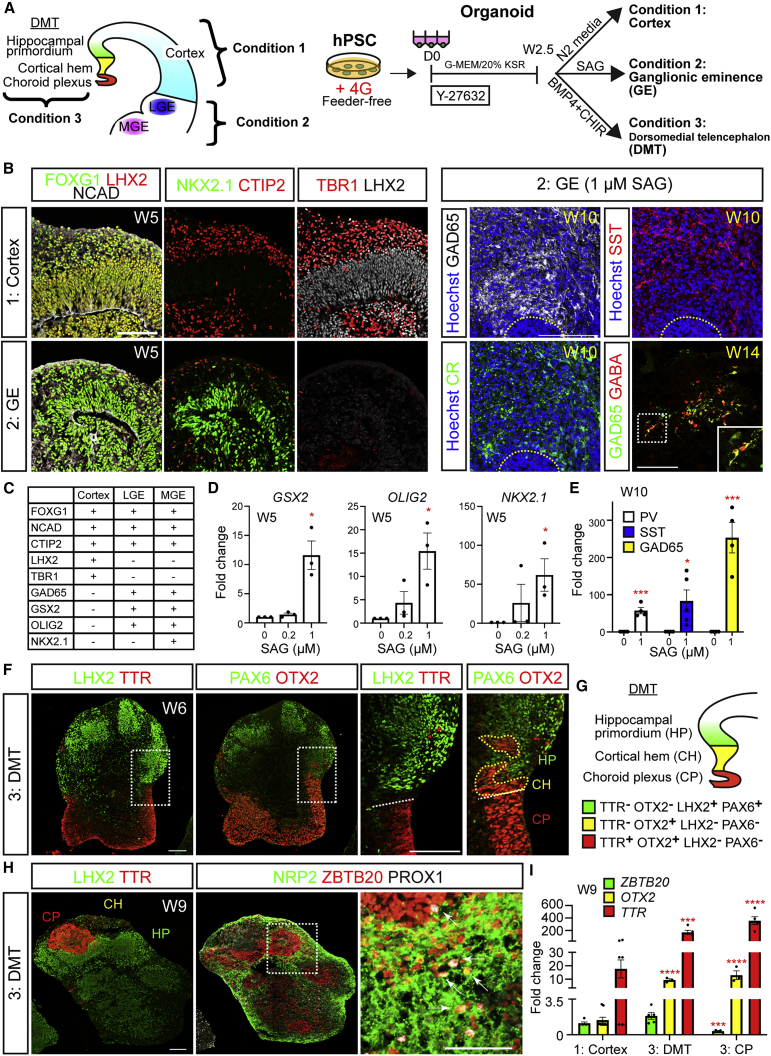
4G-treated hPSCs display a broad capacity to form cerebral, ganglionic eminence, and hippocampal organoids
We lastly tested whether hPSCs exposed to the 4G mixture are biased toward forming cortical organoid structures or broadly capable of forming different telencephalic regions. During development in vivo, organizers secrete patterning molecules to form signaling gradients that specify positional identity (Monuki, 2007). For example, high levels of Sonic hedgehog (SHH) provide ventral patterning to form the GE whereas high concentrations of BMPs/WNTs provide dorsal patterning to form the dorsomedial telencephalon (DMT), which includes the choroid plexus, cortical hem, and hippocampal primordium. Neuroepithelial cells that are not exposed to any of these signals default to forming cerebral cortex. To mimic the ventralizing actions of SHH in vivo, we exposed organoids to varying concentrations of Smoothened agonist (SAG) at W2.5 (Figure 7A) as previously described (Kadoshima et al., 2013; Watanabe et al., 2017; Samarasinghe et al., 2021). In the absence of SAG, W5 organoids exhibited staining for many cortical markers, including FOXG1, NCAD, LHX2, CTIP2, and TBR1 (Figures 7B and 7C). Following the addition of 1 μM SAG, GE markers such as FOXG1, NKX2.1, OLIG2, GSX2, and CTIP2, were highly expressed while cortex-specific markers (LHX2 and TBR1) were suppressed (Figures 7B–7D). By W10, SAG-treated organoids expressed multiple markers of differentiated interneurons including GAD65, SOMATOSTATIN (SST), CALRETININ, and PARVALBUMIN, which contrasted with the untreated organoids that lacked these markers and instead exhibited cortical features (Figures 7B–7E). At W14, the mature inhibitory neuron marker GABA was expressed throughout these ventralized organoids. Hence, forebrain organoids derived from 4G-treated hPSCs can be readily differentiated into GE-like organoids upon SHH pathway activation.
Figure 7.
hPSCs cultured under 4G conditions can form multiple telencephalic organoid subtypes: cortex, ganglionic eminence, and dorsomedial telencephalon
(A) Schematics showing the organoid protocols used to create cortex, ganglionic eminence (GE), and dorsomedial telencephalon (DMT) structures. The DMT includes choroid plexus (CP), cortical hem (CH), and hippocampal primordium (HP).
(B and C) Representative examples of immunostaining of cortical and GE organoids (B) and summary of marker expression (C).
(D and E) qRT-PCR analyses of GE progenitor and inhibitory neuron gene expression. Expression levels are normalized to organoids without SAG application. Data are presented as mean ± SEM. n ≥ 3 independent experimental replicates are plotted, about 5–10 organoids per replicate.
(F and G) DMT induction with the application of BMP4 and CHIR. CP regions are distinguished by TTR and OTX2 staining adjacent to OTX2+TTR− CH cells. HP, distinguished by LHX2 and PAX6 costaining, also formed adjacent to the CH as is seen in vivo (schematized in G).
(H) HP was distinguished by staining for ZBTB20 and NEUROPILIN2 (NRP2), as well as the dentate gyrus granule cell marker PROX1.
(I) qRT-PCR analyses of DMT markers. Expression levels are normalized to organoids without BMP4 and CHIR application. Data are presented as mean ± SEM. n ≥ 3 independent experimental replicates are plotted, about 5–10 organoids per replicate.
All scale bars, 100 μm. See also Figure S7.
We next dorsalized early forebrain organoids through a combination of BMP and WNT pathway stimulation at W2.5 (6.5 ng/mL BMP4 and 3 μM CHIR 99021), as previously described (Sakaguchi et al., 2015). At W6, we observed formation of distinct DMT compartments within the organoids including choroid plexus-like regions positive for TTR and OTX2 adjacent to a cortical hem-like region that expressed OTX2 alone (Figures 7F and 7G). Next to these hem-like regions were cells with the characteristics of hippocampal primordium including PAX6, LEF1, NEUROPILIN2 (NRP2), and ZBTB20 expression (Figure S7A). By W9, ZBTB20 and NRP2 levels increased, and additional hippocampal markers such as the dentate gyrus (DG) granule cell marker PROX1 appeared adjacent to the choroid plexus and cortical hem-like regions (Figures 7H and S7B–S7F). With additional time, expression of the kainate receptor GLUK1, a characteristic feature of CA3 neurons, also became evident in these organoids (Figures S7F and S7G). The GLUK1+ cells notably segregated away from PROX1+ cells, reminiscent of the separation of DG and CA3 compartments in vivo. Compared with untreated samples, BMP4- and CHIR-treated organoids, as well as choroid plexus-like structures that were isolated from these organoids, showed upregulation of DMT markers including OTX2, TTR, LMX1A, and MSX1 by qRT-PCR analyses (Figures 7I and S7H). ZBTB20 and LHX2 are both expressed in the neocortex and hippocampal primordium but not in the choroid plexus in vivo (Mangale et al., 2008; Sakaguchi et al., 2015). Consistent with in vivo expression patterns, we observed reduced expression of ZBTB20 and LHX2 in the isolated choroid plexus-like structures compared with untreated samples and intact BMP4- and CHIR-treated organoids. Together, these results demonstrate that non-competent FF hPSCs pre-treated with the 4G mixture of TGFβ-related growth factors possess broad developmental competence and can be directed to form diverse telencephalic structures.
Discussion
hPSC-derived brain organoids offer unprecedented opportunities for studying the mechanisms of human brain development and neurological disease. Success in these endeavors critically depends on the reproducible creation of organoids that approximate the human brain in vivo both in gene expression patterns and histological structure. Our studies show that the state of hPSC pluripotency is a key variable that needs to be considered, as the structural features and consistency of organoids can markedly differ depending on how hPSCs are maintained prior to differentiation.
Pluripotency in vitro has been primarily described in the context of three major stages: naive, formative/intermediate, and primed, referring to pre-, early post-, and later post-implantation epiblasts in vivo. Here, we show that hPSCs maintained under different culture conditions can exhibit marked variance in their transcriptional states spanning the spectrum of naive to primed pluripotency. Notably, cortical organoids formed from cells that exhibit a particular intermediate state defined by the expression of certain genes associated with TGFβ superfamily signaling and naive pluripotency displayed better structural organization than organoids generated from hPSCs at the more extreme ends of the pluripotency spectrum. We further observed that a subset of genes associated with naive pluripotency, exemplified by TFAP2C and KLF5, can serve as predictive indicators of the quality of cortical organoids that the cells can produce. Moreover, activation of TGFβ superfamily signaling in non-competent hPSCs can alter their transcriptional profile and boost their capacity to form organoids with desired cytoarchitectural features. These findings together suggest means for both identifying and modulating PSC states to help ensure the overall quality and consistency of organoids across experiments.
At one extreme, naive hPSCs are locked into a program of self-renewal and genomic hypomethylation from which the cells need to be released to respond to inductive cues. At the opposite extreme, primed hPSCs are associated with the expression of lineage-specific fate determinants that limit the range of cell types into which they can differentiate (Weinberger et al., 2016; Morgani et al., 2017). We found that naive hPSCs were unable to directly form cortical organoids, consistent with recent studies showing that these cells must first pass through formative/intermediate states before undergoing lineage-specific differentiation (Rostovskaya et al., 2019). Moreover, cell lineages that are established early in embryonic development, such as germline stem cells, are more effectively produced when starting from the formative intermediate state rather than the primed state. It is notable that several transcription factors expressed during the formative state, such as OTX2, OCT6, SOX2, and SOX3, are also highly expressed in the early neuroectoderm (Morgani et al., 2017; Smith, 2017), thus suggesting a predisposition for these cells to form neural structures.
When compared with reference datasets collected from hPSCs spanning the range of naive to primed pluripotency, we found that both our FF and MEF-supported hPSCs classified as transcriptionally intermediate yet were markedly distinct from each other. On a global transcriptomic level, optimal MEF-supported cells appear to be shifted more toward, yet distinct from, naive hPSCs compared with FF cultures or suboptimal MEF-supported cells. Indeed, the optimal feeder-supported cells that we classified as competent hPSCs most closely resembled formative hPSCs partly through their capacitation trajectory (Rostovskaya et al., 2019). These results strongly suggest that there are hPSC states within the spectrum of naive and primed pluripotency that are either more or less capable of effectively forming well-structured neuroectodermal organoids. These intermediate hPSC states could be neuroectoderm-specific or universal to other lineages. It then follows that as cells become primed, they may become increasingly heterogeneous with subpopulations biased or committed to specific lineage identities including mesoderm and endoderm (Mohammed et al., 2017; Nguyen et al., 2018), which are less favorable for brain organoid formation.
While our study shows that it is critical to understand how exogenous factors influence hPSC states, genetic background also matters. Many of the TGFβ superfamily growth factors that influence pluripotency and developmental trajectory are produced by the hPSCs themselves and could vary because of intrinsic differences. If the specific genetic background of a given hPSC line were to favor the maintenance of intermediate states that are more or less conducive for efficient neuroectodermal differentiation, it could potentially influence organoid formation and thereby cloud the analysis of how a given disease-associated mutation affects neural development and function. An important future endeavor will be to characterize the intrinsic states of hPSC pluripotency across genetically diverse hPSCs and use this information to achieve culture conditions that uniformly maintain organoid competence.
Many patient-derived induced PSCs have been derived under FF conditions owing to its convenience and a desire to eliminate exposure of hPSCs to animal products to enable their use in clinical applications (Amit and Itskovitz-Eldor, 2006; Seifinejad et al., 2010). While MEF support remains a benchmark standard in many labs, our findings show that similar organoid results may be achieved with FF cells by pre-conditioning the cultures with the 4G mixture of TGFβ superfamily growth factors defined in our studies. Furthermore, organoids produced from 4G-treated hPSCs display a high degree of developmental potential, as they can be readily transformed into different regions of the forebrain. It is nevertheless important to note that the intrinsic production of TGFβ growth factors could markedly vary between cell lines or FF media conditions. Thus, the concentrations of the 4G factors that we utilized in our study should be viewed as starting guidelines and may need to be adjusted for different cell lines. Indeed, our experiments show that it is possible to overshoot with certain proteins such as BMP4.
Although the transcriptional state of hPSCs can be profoundly impacted by maintenance under FF or MEF-supported conditions, other contributing influences should not be overlooked. There can still be batch variabilities in other cell culture components as well as procedural differences in hPSC culture techniques. For example, our comparative transcriptomic analysis showed that MEF-supported H9 ESCs reported by the Smith lab clustered more toward primed hPSCs compared with MEF-supported H9 grown in either the Studer lab or our own lab. These observations demonstrate how the same hPSC line maintained in different labs can show significant variability in their state of pluripotency, which could, in turn, influence performance in organoid differentiation protocols. The systematic implementation of uniform hPSC maintenance conditions across laboratories may thus be an essential prerequisite to achieving consistent organoid differentiation outcomes.
Experimental procedures
Telencephalic organoid differentiation
Cerebral and basal GE organoid formation was performed as described in Watanabe et al. (2017). Of note, KSR is known to affect differentiation efficiency, and we carried out the differentiation protocol using the same KSR lots (Invitrogen, KSR 10828010 and 10828028; lot 1670543) as in Watanabe et al. (2017). For the formation of dorsomedial telencephalic (DMT) organoids, we used a modified version of previously described methods (Sakaguchi et al., 2015). From day 18 to day 24, 6.5 ng/mL BMP4 (Invitrogen, P2026055) and 3 μM GSK3 inhibitor (CHIR 99021; Fisher Scientific, 442310) were added with DMEM/F-12 (Hyclone) supplemented with 1% N2 (Invitrogen), 1% chemically defined lipid concentrate (CDLC; Invitrogen), and 10% fetal bovine serum (FBS qualified source US region; Invitrogen, 10437028) under 40% O2 and 5% CO2 conditions at 37°C. On day 42, DMT organoids were cut in half and the base medium was changed to Neurobasal medium (Invitrogen), supplemented with 1% N2 (Invitrogen), 1% CDLC, 10% FBS, 2% B-27 supplement without vitamin A (Invitrogen), GlutaMAX (Invitrogen), and 100 μg/mL Primocin (InvivoGen). After day 42, DMT organoids were cut in half every 2 weeks. From day 56, DMT organoids were cultured in Lumox dishes (Sarstedt). Medium was subsequently changed every 2–3 days until the organoids were collected for analysis.
Tissue processing and immunohistochemistry
Brain organoids were fixed, cryoprotected, embedded, frozen, and cryosectioned as previously described (Watanabe et al., 2017). Sectioned tissues were collected onto Superfrost Plus slides (Fisher Scientific) and blocked for 30 min in PBS with 1% heat-inactivated equine serum (Hyclone), 0.1% Triton X-100, and 0.01% sodium azide, and incubated in primary antibodies (see Table S6) in the blocking solution overnight at 4°C. After three washes in PBST (0.1% Triton X-100), tissue was incubated with secondary antibodies for 1 h at room temperature. After three washes, tissue was mounted in ProLong Diamond (Invitrogen) with coverslips and stored in darkness at 4°C prior to imaging. Gestation week 13–14 human fetal brain tissue was fixed in 4% paraformaldehyde in PBS for 3–4 days at 4°C. Gestation week 7 human fetal brain tissue was fixed overnight at 4°C. Brain tissue was then washed twice in PBS and immersed in 30% sucrose in PBS overnight at 4°C. The tissue was lastly frozen in optimal cutting temperature compound and cryosectioned at 20 μm thickness.
RNA isolation and processing for RNA-sequencing analyses
Samples were lysed in QIAzol and RNA extracted following manufacturer’s instructions (miRNeasy Micro Kit; Qiagen). For RNA-sequencing analyses, hPSCs, particularly H9 hESCs and XFiPSCs, were collected with three replicates (three independent experiments, four conditions, 12 samples in total) for feeder-dependent and feeder-independent conditions. RNA integrity was confirmed with the Agilent 2200 TapeStation (RNA integrity number >8) and sent to the UCLA Neuroscience Genomic Core. cDNA libraries were generated using TruSeq with Ribo-Zero Gold (Illumina) and sequenced using an Illumina HiSeq 4000 system, yielding about 50 million reads per sample. We utilized paired-end RNA sequencing with 75 bp reads.
Additional experimental procedures and descriptions of materials can be found in the supplemental information.
Data and code availability
All RNA-sequencing data reported in our study were deposited within the Gene Expression Omnibus (GEO) repository and are accessible through the series accession number GEO: GSE140057 (https://www.ncbi.nlm.nih.gov/geo/). The proteomic analyses were deposited to PRIDE Archive (https://www.ebi.ac.uk/pride/archive/) with the accession number PXD013662.
Acknowledgments
We thank Samantha Butler, Jack Parent, Xinyu Zhou, Mirjana Maletic-Savatic, Ranmal Samarasinghe, and members of Novitch lab for invaluable discussions and comments on the manuscript, and Jack Parent for the gift of hiPSC lines. M.W. is thankful to Hideya Sakaguchi for sharing and offering advice on the hippocampal organoid protocol. This work used computational and storage services associated with the Hoffman2 Shared Cluster provided by UCLA Institute for Digital Research and Education’s Research Technology Group. This work was supported by the UCLA Jonsson Comprehensive Cancer Center and Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research (BSCRC) Ablon Scholars Program, research awards from the Rose Hills Foundation, the California Institute for Regenerative Medicine (CIRM) (DISC1-08819), and the NIH (R01NS089817, R01DA051897, R01MH130061, and P50HD103557) to B.G.N. M.W. was supported by postdoctoral training awards provided by the UCLA BSCRC, the Uehara Memorial Foundation, UCLA Brain Research Institute, and award from the NIH (K99/R00HD096105). J.E.B., N.V., F.T., A.K., and O.A.M. were supported partly by the UCLA-California State University Northridge CIRM-Bridges training program (TB1-00183). J.E.B. also acknowledges the support of the UCLA BSCRC Rose Hills Foundation Graduate Scholarship Training Program. W.G. was supported by a UCLA Dissertation Year Fellowship. A.J.C. was supported by a UCLA BSCRC postdoctoral fellowship. A.T.C. was supported by a grant from the NIH (R01HD079546). K.P. was supported by research awards from the UCLA BSCRC, the David Geffen School of Medicine, the UCLA Jonnson Comprehensive Cancer Center, the NIH (R01HD098387), and a Faculty Scholar grant from the Howard Hughes Medical Institute. M.J.G. was supported by a SFARI Bridge to Independence Award and grants from the NIH (R01MH123922 and R01MH121521). We also acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30NS062691) and both the Cells, Circuits, and Systems Analysis and Genetics and Genomics Cores of the Semel Institute of Neuroscience at UCLA supported by the NICHD (U54HD087101 and P50HD103557).
Author contributions
M.W., N.V., F.T., J.E.B., A.K., N.V.B., O.A.M., and N.D. performed all organoid culture experiments and coordinated on various analytical procedures. L.S.E. performed spheroid culture experiments, and H.I.K. provided valuable reagents and guidance in the spheroid culture experiments. J.E.B., J.R.H., L.d.l.T.-U., S.S., and M.J.G. performed bioinformatics analyses. C.A.P. provided analysis of the human brain specimens. W.G. and H.R.C. contributed mass spectrometry analyses of MEF-secreted factors. A.J.C., D.C., K.P., and A.T.C. provided valuable reagents, datasets, and guidance in the naive pluripotent stem cell experiments and analyses. M.W. and B.G.N. conceived and designed the experiments with helpful input from the other authors. M.W., J.E.B., and B.G.N. wrote the manuscript with input from the other authors.
Conflicts of interest
The authors declare no competing interests.
Published: September 29, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.08.013.
Contributor Information
Momoko Watanabe, Email: momokow@uci.edu.
Bennett G. Novitch, Email: bnovitch@ucla.edu.
Supplemental information
References
- Amin N.D., Paşca S.P. Building models of brain disorders with three-dimensional organoids. Neuron. 2018;100:389–405. doi: 10.1016/j.neuron.2018.10.007. [DOI] [PubMed] [Google Scholar]
- Amit M., Itskovitz-Eldor J. Feeder-free culture of human embryonic stem cells. Methods Enzymol. 2006;420:37–49. doi: 10.1016/S0076-6879(06)20003-X. [DOI] [PubMed] [Google Scholar]
- Anderson N.C., Chen P.F., Meganathan K., Afshar Saber W., Petersen A.J., Bhattacharyya A., Kroll K.L., Sahin M., Cross-IDDRC Human Stem Cell Working Group Balancing serendipity and reproducibility: pluripotent stem cells as experimental systems for intellectual and developmental disorders. Stem Cell Rep. 2021;16:1446–1457. doi: 10.1016/j.stemcr.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri A., Andrews M.G., Kriegstein A.R., Nowakowski T.J. Are organoids ready for prime time? Cell Stem Cell. 2020;27:361–365. doi: 10.1016/j.stem.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri A., Andrews M.G., Mancia Leon W., Jung D., Shin D., Allen D., Jung D., Schmunk G., Haeussler M., Salma J., et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–148. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaradia I., Lancaster M.A. Brain organoids for the study of human neurobiology at the interface of in vitro and in vivo. Nat. Neurosci. 2020;23:1496–1508. doi: 10.1038/s41593-020-00730-3. [DOI] [PubMed] [Google Scholar]
- Cornacchia D., Zhang C., Zimmer B., Chung S.Y., Fan Y., Soliman M.A., Tchieu J., Chambers S.M., Shah H., Paull D., et al. Lipid deprivation induces a stable, naive-to-primed intermediate state of pluripotency in human PSCs. Cell Stem Cell. 2019;25:120–136.e10. doi: 10.1016/j.stem.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman G.J., Nicolás F.J., Callahan J.F., Harling J.D., Gaster L.M., Reith A.D., Laping N.J., Hill C.S. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Kadoshima T., Sakaguchi H., Nakano T., Soen M., Ando S., Eiraku M., Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T., Olova N., Roode M., Mulas C., Lee H.J., Nett I., Marks H., Walker R., Stunnenberg H.G., Lilley K.S., et al. Tracking the embryonic stem cell transition from ground state pluripotency. Development. 2017;144:1221–1234. doi: 10.1242/dev.142711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M., Barber M., Mansfield W., Cui Y., Spindlow D., Stirparo G.G., Dietmann S., Nichols J., Smith A. Capture of mouse and human stem cells with features of formative pluripotency. Cell Stem Cell. 2021;28:2180. doi: 10.1016/j.stem.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Nefzger C.M., Rossello F.J., Chen J., Knaupp A.S., Firas J., Ford E., Pflueger J., Paynter J.M., Chy H.S., et al. Comprehensive characterization of distinct states of human naive pluripotency generated by reprogramming. Nat. Methods. 2017;14:1055–1062. doi: 10.1038/nmeth.4436. [DOI] [PubMed] [Google Scholar]
- Mangale V.S., Hirokawa K.E., Satyaki P.R.V., Gokulchandran N., Chikbire S., Subramanian L., Shetty A.S., Martynoga B., Paul J., Mai M.V., et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H., Hernando-Herraez I., Savino A., Scialdone A., Macaulay I., Mulas C., Chandra T., Voet T., Dean W., Nichols J., et al. Single-cell landscape of transcriptional heterogeneity and cell fate decisions during mouse early gastrulation. Cell Rep. 2017;20:1215–1228. doi: 10.1016/j.celrep.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monuki E.S. The morphogen signaling network in forebrain development and holoprosencephaly. J. Neuropathol. Exp. Neurol. 2007;66:566–575. doi: 10.1097/nen.0b013e3180986e1b. [DOI] [PubMed] [Google Scholar]
- Morgani S., Nichols J., Hadjantonakis A.-K. The many faces of Pluripotency: in vitro adaptations of a continuum of in vivo states. BMC Dev. Biol. 2017;17:7. doi: 10.1186/s12861-017-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Q.H., Lukowski S.W., Chiu H.S., Senabouth A., Bruxner T.J.C., Christ A.N., Palpant N.J., Powell J.E. Single-cell RNA-seq of human induced pluripotent stem cells reveals cellular heterogeneity and cell state transitions between subpopulations. Genome Res. 2018;28:1053–1066. doi: 10.1101/gr.223925.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann D., Vallier L. Variability of human pluripotent stem cell lines. Curr. Opin. Genet. Dev. 2017;46:179–185. doi: 10.1016/j.gde.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Qian X., Jacob F., Song M.M., Nguyen H.N., Song H., Ming G.L. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 2018;13:565–580. doi: 10.1038/nprot.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossin E.J., Lage K., Raychaudhuri S., Xavier R.J., Tatar D., Benita Y., International Inflammatory Bowel Disease Genetics, C. Cotsapas C., Daly M.J. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovskaya M., Stirparo G.G., Smith A. Capacitation of human naive pluripotent stem cells for multi-lineage differentiation. Development. 2019;146:dev172916. doi: 10.1242/dev.172916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi H., Kadoshima T., Soen M., Narii N., Ishida Y., Ohgushi M., Takahashi J., Eiraku M., Sasai Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarasinghe R.A., Miranda O.A., Buth J.E., Mitchell S., Ferando I., Watanabe M., Allison T.F., Kurdian A., Fotion N.N., Gandal M.J., et al. Identification of neural oscillations and epileptiform changes in human brain organoids. Nat. Neurosci. 2021;24:1488–1500. doi: 10.1038/s41593-021-00906-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifinejad A., Tabebordbar M., Baharvand H., Boyer L.A., Salekdeh G.H. Progress and promise towards safe induced pluripotent stem cells for therapy. Stem Cell Rev. Rep. 2010;6:297–306. doi: 10.1007/s12015-010-9121-x. [DOI] [PubMed] [Google Scholar]
- Sloan S.A., Andersen J., Pașca A.M., Birey F., Pașca S.P. Generation and assembly of human brain region-specific three-dimensional cultures. Nat. Protoc. 2018;13:2062–2085. doi: 10.1038/s41596-018-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. Formative pluripotency: the executive phase in a developmental continuum. Development. 2017;144:365–373. doi: 10.1242/dev.142679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W., et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L., et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:524–526. doi: 10.1016/j.stem.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A., et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S., Paulsen B., Arlotta P. 3D brain organoids: studying brain development and disease outside the embryo. Annu. Rev. Neurosci. 2020;43:375–389. doi: 10.1146/annurev-neuro-070918-050154. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Buth J.E., Vishlaghi N., de la Torre-Ubieta L., Taxidis J., Khakh B.S., Coppola G., Pearson C.A., Yamauchi K., Gong D., et al. Self-organized cerebral organoids with human-specific features predict effective drugs to combat zika virus infection. Cell Rep. 2017;21:517–532. doi: 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wataya T., Ando S., Muguruma K., Ikeda H., Watanabe K., Eiraku M., Kawada M., Takahashi J., Hashimoto N., Sasai Y. Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc. Natl. Acad. Sci. USA. 2008;105:11796–11801. doi: 10.1073/pnas.0803078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L., Ayyash M., Novershtern N., Hanna J.H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Yoshiaki T., Patterson B., Cakir B., Kim K.Y., Cho Y.S., Park I.H. Generation and fusion of human cortical and medial ganglionic eminence brain organoids. Curr. Protoc. Stem Cell Biol. 2018;47:e61. doi: 10.1002/cpsc.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.J., Elahi L.S., Pașca A.M., Marton R.M., Gordon A., Revah O., Miura Y., Walczak E.M., Holdgate G.M., Fan H.C., et al. Reliability of human cortical organoid generation. Nat. Methods. 2019;16:75–78. doi: 10.1038/s41592-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P.B., Deng D.Y., Lai C.S., Hong C.C., Cuny G.D., Bouxsein M.L., Hong D.W., McManus P.M., Katagiri T., Sachidanandan C., et al. BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat. Med. 2008;14:1363–1369. doi: 10.1038/nm.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.Y., Song H., Ming G.L. Modeling neurological disorders using brain organoids. Semin. Cell Dev. Biol. 2021;111:4–14. doi: 10.1016/j.semcdb.2020.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Badur M.G., Divakaruni A.S., Parker S.J., Jäger C., Hiller K., Murphy A.N., Metallo C.M. Distinct metabolic states can support self-renewal and lipogenesis in human pluripotent stem cells under different culture conditions. Cell Rep. 2016;16:1536–1547. doi: 10.1016/j.celrep.2016.06.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-sequencing data reported in our study were deposited within the Gene Expression Omnibus (GEO) repository and are accessible through the series accession number GEO: GSE140057 (https://www.ncbi.nlm.nih.gov/geo/). The proteomic analyses were deposited to PRIDE Archive (https://www.ebi.ac.uk/pride/archive/) with the accession number PXD013662.