Abstract
The taxon Elasmobranchii (sharks and rays) contains one of the long-established evolutionary lineages of vertebrates with a tantalizing collection of species occupying critical aquatic habitats. To overcome the current limitation in molecular resources, we launched the Squalomix Consortium in 2020 to promote a genome-wide array of molecular approaches, specifically targeting shark and ray species. Among the various bottlenecks in working with elasmobranchs are their elusiveness and low fecundity as well as the large and highly repetitive genomes. Their peculiar body fluid composition has also hindered the establishment of methods to perform routine cell culturing required for their karyotyping. In the Squalomix consortium, these obstacles are expected to be solved through a combination of in-house cytological techniques including karyotyping of cultured cells, chromatin preparation for Hi-C data acquisition, and high fidelity long-read sequencing. The resources and products obtained in this consortium, including genome and transcriptome sequences, a genome browser powered by JBrowse2 to visualize sequence alignments, and comprehensive matrices of gene expression profiles for selected species are accessible through https://github.com/Squalomix/info.
Keywords: Shark, ray, chimaera, biodiversity genomics, whole genome sequencing, karyotype
Introduction
Although usually recognized as a kind of ‘fish’ like actinopterygian fishes, cartilaginous fishes (chondrichthyans) form a distinct class of vertebrates with more than 1,200 species, known mostly as sharks and rays ( Figure 1; Nelson et al., 2016). This taxonomic class has the longest evolutionary history among vertebrates of about 400 million years, in terms of the divergence of extant members ( Naylor et al., 2012). Whereas its diversity might not be widely recognized, species in this taxon are characterized by several unique traits including electromagnetic sensing (all cartilaginous fishes), electricity generation (electric rays), diverse morphology sometimes with a flattened body (angelsharks and most rays) and/or a toothed rostrum (sawsharks and sawfishes). The highlight of their biological enigmas is in their reproductive modes with high plasticity between oviparity and viviparity, and occasionally parthenogenesis and intersexuality ( Penfold and Wyffels, 2019). Mainly because of overfishing, many cartilaginous fish populations are declining ( Pacoureau et al., 2021), and evidence-based resource management would greatly benefit from the establishment of genomic platforms.
Figure 1. Chondrichthyan phylogeny and taxon sampling in the Squalomix Consortium.
This figure includes some chondrichthyan species selected to represent the individual taxonomic orders that reflect the local fauna of Japan and are/will be analyzed by the consortium by genome or transcriptome sequencing (as of April 10, 2022). The full list of species and current status can be found in https://github.com/Squalomix/info.
Despite these outstanding evolutionary and biological importance, modern genomic approaches have only recently been applied to cartilaginous fishes (reviewed in Kuraku, 2021). The only exception is the effort commenced before 2010 on the elephant fish Callorhinchus milii ( Venkatesh et al., 2014), a member of the Holocephali (chimaeras and ratfishes), the more species-poor chondrichthyan lineage, with a relatively small genome size of about 1.9 giga basepairs (Gbp). In contrast, most elasmobranchs have genomes of more than 3 Gbp plagued with abundant repetitive elements.
Squalomix: consortium scope and organization
The Squalomix Consortium ( Figure 2A) was launched in 2020 aiming to provide the genome sequence and other genome-wide data for chondrichthyan species including transcriptomes and epigenomes. Sample processing and data production is conducted by the Molecular Life History Laboratory at the National Institute of Genetics, Mishima, Japan, and the Laboratory for Phyloinformatics in RIKEN Kobe, Japan, which harbors a DNA Analysis Facility. The consortium is funded by academic agencies as of May 2022 and is seeking additional funding sources, especially from industrial groups oriented toward the conservation of biodiversity and marine environments. In November 2020, the Squalomix Consortium became affiliated with Earth BioGenome Project (EBP), the global initiative to promote biodiversity genomics ( Lewin et al., 2022). The collaborative network at the Squalomix Consortium includes an extensive range of expertise and worldwide distribution.
Figure 2. Squalomix Consortium.
A, Consortium logo. B, One of the main study species, the red stingray Hemitrygon akajei. Photo credit: Itsuki Kiyatake.
Versatile sample collection featuring the local fauna
In Squalomix, sample collection is performed cautiously to minimize the sacrifice of wildlife—especially those with an endangered status. The collection focuses mainly on the rich marine fauna in Japan’s neighboring temperate waters, with occasional sources from death stranding for elusive species. The project collaborates closely with local aquariums oriented toward academic science. Their contributions play indispensable roles in relaying offshore sampling and enable sustainable sampling of embryos and blood from live individuals, although the latter approach is limited to species that can be bred in captivity and are amenable to husbandry.
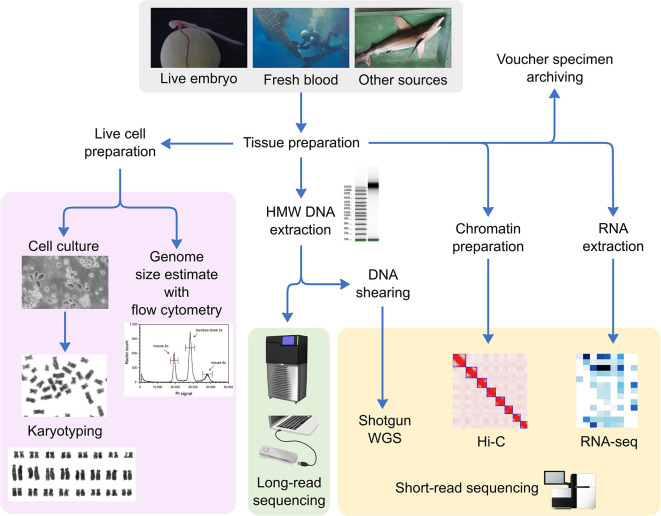
Another strength of the Squalomix Consortium is its expertise in laboratory solutions that are not confined to DNA sequencing, but additionally explore post-genome approaches to decipher the molecular basis of chondrichthyan phenotypic evolution. Access to fresh tissues from local aquaria facilitates embryological analysis, genome size quantification with flow cytometry, and karyotyping from cell cultures ( Figure 3). Remarkably, cell culture in cartilaginous fishes, which was long thought difficult because of their high body fluid osmolarity, was enabled by modifying the culture medium with balancing osmolytes ( Uno et al., 2020). Our cytological expertise also allowed various epigenomic analyses that benefit from whole genome sequencing, on transcription factor binding with ChIP-seq ( Hara et al., 2018) and chromatin openness with ATAC-seq, in addition to long-range DNA interactions with Hi-C ( Kadota et al., 2020; Onimaru et al., 2021). These techniques contributed to biological analyses based on the draft genome sequences of three shark species ( Hara et al., 2018), which launched the Squalomix Consortium.
Figure 3. Typical work flow in the Squalomix Consortium.
Whole genome sequencing (WGS) is mainly performed with the Sequel II/IIe platform (Pacific Biosciences, Inc.) to obtain high-fidelity (HiFi) long reads, which is supplemented by short-read sequencing. Extraction of high molecular weight (HMW) genomic DNA is mainly performed using the NucleoBond columns (Macherey-Nagel, Inc.) and the extracted DNA is controlled with Agilent TapeStation systems (Agilent Technologies, Inc.) as well as conventional pulse-field gel electrophoresis. Flow cytometry for genome size estimation employs the Ploidy Analyser platform (Sysmex Inc.). Hi-C sample preparation employs the iconHi-C protocol ( Kadota et al., 2020) that was optimized in-house based on several existing protocols.
Sequencing strategy and recent progress
The sequencing strategy in the Squalomix Consortium is designed to accommodate genomic characteristics of cartilaginous fishes, mostly with large, repetitive genomes. In the standard protocol formulated in January 2021 ( Figure 3), we start by estimating genome size using flow cytometry and karyotyping as well as by ‘survey’ sequencing of transcriptomes, which serves for species identity verification with an assembled mitochondrial DNA sequence. These initial steps ensure sample authenticity and quality. We then proceed to genome sequencing, which employs both short-read and long-read high-fidelity (‘HiFi’) sequencing platforms, together with Hi-C data production for chromosome-scale scaffolding based on three-dimensional DNA interactions. The long-read data are obtained using the Sequel II or IIe platforms (Pacific Biosciences, Inc.) with a minimum sequencing depth of 20x. The assembly outputs are evaluated with reference to their coverage of protein-coding gene space, as well as transcriptome data, genome size, and karyotypic organization obtained separately. These validations allow us to scrutinize the inclusion of those genomic regions that are difficult to sequence and assemble, such as the Hox C genes that were previously thought to be missing in elasmobranchs but were retrieved by elaborate annotation ( Hara et al., 2018; reviewed in Kuraku, 2021). Complete genome assemblies are critical to validate gene loss and variations in gene repertoires via synteny/phylogeny comparisons, previously suggested for visual opsins and conventional olfactory receptors ( Hara et al., 2018). The standard procedure outlined above ( Figure 3) has been applied to several study species, including the red stingray Hemitrygon akajei ( Figure 2B) for which a draft genome assembly has been made available for BLAST searches at the Squalomix sequence archive ( Figure 4A; https://transcriptome.riken.jp/squalomix/).
Figure 4. Overview of the Squalomix data sharing platform.
A, Sequence similarity search (BLAST) in elasmobranch genome and transcriptome sequences. B, Molecular phylogeny inference facilitated by the existing combination of aLeaves (that hosts products of Squalomix) and MAFFT webservers ( Kuraku et al., 2013). C, Interactive genome browser employing JBrowse2 version 1.6.9 ( Buels et al., 2016) for the zebra shark Stegostoma tigrinum (or S. fasciatum) based on its first genome assembly sSteFas1.1 (NCBI Genome ID, GCA_022316705.1). The websites providing these functions are found through the main consortium gateway ( https://github.com/Squalomix/info).
Cooperation toward the global goals
The Squalomix Consortium aims not only to sequence and analyze the genomes but also to tightly interact with other research groups whose target species list contains cartilaginous fishes including other EBP-affiliated projects (see below). To maximize mutual benefit among those projects, some animal samples from our collection could be provided for genome sequencing at other sites. The Squalomix Consortium offers laboratory experiments for genome size quantification or karyotype analysis for species listed by other consortia, provided that fresh cells are available. The sample transfer will be processed in accordance with the Nagoya Protocol and other relevant regulations. Inclusive cooperation respecting complementary expertise is expected to overcome the long-standing difficulty in studying elasmobranchs sustainably and contribute to disentangling the marine ecosystems for effective conservation.
Data sharing platforms
Once produced, genome assemblies pass rigid quality controls and are deposited in the NCBI Genome under the NCBI BioProject ID PRJNA707598 and made available as database for BLAST searches at our Squalomix sequence archive ( https://transcriptome.riken.jp/squalomix/). This archive also has a link to the up-to-date listing of the species for which genome sequences are available, filed by the GenomeSync database ( http://genomesync.org/). The archive website also hosts a gateway to genome browsers powered by JBrowse2 that allow users to visualize specific genomic regions and load additional tracks including base composition, gene models, repetitive elements, and aligned RNA-seq reads ( Figure 4C). We also provide comprehensive matrices of expression profiles for predicted genes of the brownbanded bamboo shark Chiloscyllium punctatum and the cloudy catshark Scyliorhinus torazame that were already quantified and normalized based on RNA-seq data of various tissues for our past publication ( Hara et al., 2018).
Other pioneering efforts tackling elasmobranch genomes
Some elasmobranch genomes have already been sequenced by other pioneering working groups ( https://www.ncbi.nlm.nih.gov/data-hub/genome/?taxon=7777&reference_only=true). This includes the Vertebrate Genomes Project (VGP), whose data production format employs a suite of modern promising solutions including optical mapping and Hi-C scaffolding as well as long-read and short-read sequencing, to cover all vertebrate species ( Rhie et al., 2021). The initial VGP progress report released the genome sequences of the thorny skate Amblyraja radiata (NCBI Genome ID, GCA_010909765.2). The Darwin Tree of Life (DToL) Project partly links with VGP and aims to sequence all eukaryotic species in Britain and Ireland. DToL’s first chondrichthyan genome is that of the small-spotted catshark Scyliorhinus canicula, the egg-laying species most widely studied in developmental biology and endocrinology (NCBI Genome ID, GCA_902713615.1). The recently launched European Reference Genome Atlas (ERGA) also plans to produce reference chromosome anchored genomes of multiple species from this geography including cartilaginous fish aiming to empower conservation efforts ( Formenti et al., 2022). Researchers in China launched the Fish10K project that partially targets cartilaginous fishes ( Fan, et al., 2020). In addition, the DNA Zoo project puts special emphasis on Hi-C scaffolding ( Rao et al., 2014), often using available genome assemblies already released by other groups as input and performing chromosome-scale genome scaffolding using Hi-C data even in the presence of intra-specific genomic variations. So far, the DNA Zoo effort produced the chromosome-scale genome assemblies of the brownbanded bamboo shark C. punctatum and the whale shark Rhincodon typus, each of which was produced using samples from multiple individuals ( Hoencamp et al., 2021). All the above efforts are expected to be coordinated under the overarching EBP initiative, in order to play complementary roles towards the global aim of generating high-quality genomic resources.
Data availability
Products from this consortium are deposited in NCBI under the BioProject ID PRJNA707598 and are available at our Squalomix sequence archive ( https://transcriptome.riken.jp/squalomix/).
Acknowledgments
The authors representing the Squalomix Consortium thank the animal caretakers and administrative staff at the aquaria and the DNA sequencing facilities that are assisting the consortium. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
Funding Statement
The consortium is funded by intramural budgets granted by RIKEN and the National Institute of Genetics, Japan, as well as JSPS KAKENHI Grant Numbers 20H03269 and 16H06279 (PAGS).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
References
- Buels R, Eric Y, Diesh CM, et al. : JBrowse: a dynamic web platform for genome visualization and analysis. Gen. Biol. 2016;17:66. 10.1186/s13059-016-0924-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Song Y, Yang L, et al. : Initial data release and announcement of the 10,000 Fish Genomes Project (Fish10K). GigaScience. 2020;9:giaa080. 10.1093/gigascience/giaa080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formenti G, et al. : The era of reference genomes in conservation genomics. Trends Genet. 2022;37:197–202. [DOI] [PubMed] [Google Scholar]
- Hara Y, Yamaguchi K, Onimaru K, et al. : Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat. Ecol. Evol. 2018;2:1761–1771. 10.1038/s41559-018-0673-5 [DOI] [PubMed] [Google Scholar]
- Hoencamp, et al. : 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science. 2021;372:984–989. 10.1126/science.abe2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota M, Nishimura O, Miura H, et al. : Multifaceted Hi-C benchmarking: what makes a difference in chromosome-scale genome scaffolding? Gigascience. 2020;9:giz158. 10.1093/gigascience/giz158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S: Shark and ray genomics for disentangling their morphological diversity and vertebrate evolution. Dev. Biol. 2021;477:262–272. 10.1016/j.ydbio.2021.06.001 [DOI] [PubMed] [Google Scholar]
- Kuraku S, Zmasek CM, Nishimura O, et al. : aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nuc. Acids Res. 2013;41:W22–W28. 10.1093/nar/gkt389 Reference Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin, et al. : The Earth BioGenome Project 2020: Starting the clock. Proc. Natl. Acad. Sci. USA. 2022;119:e2115635118. 10.1073/pnas.2115635118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor GJP, Caira JN, Jensen K, et al. : Elasmobranch Phylogeny: A mitochondrial estimate based on 595 species. Carrier JC, Musick JA, Heithaus MR, editors. The Biology of Sharks and Their Relatives. Boca Raton: CRC Press, Taylor & Francis Group;2012; pp.31–56. 10.1201/b11867-4 [DOI] [Google Scholar]
- Nelson JS, Grande T, Wilson MVH: Fishes of the world. Fifth ed. Hoboken, New Jersey: John Wiley & Sons;2016; p.1online resource. [Google Scholar]
- Onimaru K, Tatsumi K, Tanegashima C, et al. : Developmental hourglass and heterochronic shifts in fin and limb development. elife. 2021;10:e62865. 10.7554/eLife.62865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacoureau N, Rigby CL, Kyne PM, et al. : Half a century of global decline in oceanic sharks and rays. Nature. 2021;589:567–571. 10.1038/s41586-020-03173-9 [DOI] [PubMed] [Google Scholar]
- Penfold LM, Wyffels JT: Reproductive Science in Sharks and Rays. Adv. Exp. Med. Biol. 2019;1200:465–488. 10.1007/978-3-030-23633-5_15 [DOI] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, et al. : A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, et al. : Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021;592:737–746. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Nozu R, Kiyatake I, et al. : Cell culture-based karyotyping of orectolobiform sharks for chromosome-scale genome analysis. Commun. Biol. 2020;3:652. 10.1038/s42003-020-01373-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Lee AP, Ravi V, et al. : Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. 10.1038/nature12826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Koyanagi M, Kuraku S: Visual and nonvisual opsin genes of sharks and other nonosteichthyan vertebrates: genomic exploration of underwater photoreception. J. Evol. Biol. 2020;34:968–976. 10.1111/jeb.13730 [DOI] [PubMed] [Google Scholar]