Summary
Mesenchymal stromal cells (MSCs) transplantation could enhance bone repair. However, the cell fate of transplanted MSCs, in terms of their local distribution and spatial associations with other types of cells were poorly understood. Here, we developed a single-cell 3D spatial correlation (sc3DSC) method to track transplanted MSCs based on deep tissue microscopy of fluorescent nanoparticles (fNPs) and immunofluorescence of key proteins. Locally delivered fNP-labeled MSCs enhanced tibial defect repair, increased the number of stem cells and vascular maturity in mice. fNP-MSCs persisted in the defect throughout repair. But only a small portion of transplanted cells underwent osteogenic differentiation (OSX+); a significant portion has maintained their expression of mesenchymal stem cell and skeletal stem cell markers (SCA-1 and PRRX1). Our results contribute to the optimization of MSC-based therapies. The sc3DSC method may be useful in studying cell-based therapies for the regeneration of other tissue types or disease models.
Keywords: mesenchymal stromal cells, bone repair, blood vessels, fluorescent nanoparticle, stem cell fate, spatial analysis
Highlights
-
•
Transplanted marrow stromal cells associated with vessels during bone defect repair
-
•
Small proportion of transplanted cells differentiated into osteogenic cells
-
•
A proportion of transplanted cells maintained expressions of stem cell markers
Mesenchymal stromal cell (MSC) transplantation could enhance bone repair, but their cell fate is poorly understood. In this article, Liu and colleagues utilize a single-cell 3D spatial correlation (sc3DSC) method to reveal that a large portion of transplanted MSCs maintain stemness, localize around blood vessels, increase blood vessel formation, and induce invasion of host progenitors into the bone defect.
Introduction
Cell therapies based on mesenchymal stromal cells (MSCs) have shown promise as a therapeutic strategy to promote bone repair based on their capacity for self-renewal and multipotent differentiation (Oryan et al., 2017). Although multiple studies have demonstrated that transplantation of MSCs promoted bone regeneration in preclinical models (Dupont et al., 2010; Seebach et al., 2015), MSC-based therapies still face some major challenges, such as low survival rates (Mei et al., 2014). The beneficial behaviors of transplanted stromal cells, including migration, differentiation, and cell-cell interactions, during bone repair are becoming increasingly clear (Featherall et al., 2018). Marrow MSCs could participate in bone regeneration not only by osteogenic differentiation but also by supporting the local microenvironment, such as regulating angiogenesis (Green et al., 2018). In addition, transplanted MSCs could also recruit resident pericytes and promote vascular maturation in a spinal cord injury model (Menezes et al., 2020). However, the local spatial distribution of transplanted MSCs and their cell fate in a bone defect remains unclear. Therefore, MSC-based therapies for bone regeneration could be vastly improved by understanding the spatiotemporal distributions of transplanted MSCs.
Recent development of fluorescence probes has enabled the tracking of implanted cells without genetic manipulations (Chen et al., 2018). Fluorescent nanoparticles (fNPs) with aggregation-induced emission and the cell-penetrating peptide Tat have enabled long-term tracking of implanted MSCs (Ding et al., 2014). This class of fNPs has low toxicity, is photophysically stable, and is retained in small animal models of skin injury, limb ischemia, and liver regeneration (Cao et al., 2019). However, the effectiveness of this fNP in tracking implanted MSCs within a healing bone defect has not been investigated.
Recent advances in deep tissue microscopy have enabled the observation of single cells that spans up to hundreds of micrometers in the skeletal system (Acar et al., 2015; Ramasamy et al., 2016; Sivaraj et al., 2021). However, this method also created a vast amount of data. The torturous 3D structures of blood vessels and blood volume pose additional challenges to quantify their relationship to surrounding cells. In our previous work, an automated approach was developed to quantify the 3D spatial correlation of one cell type and blood vessels in a bone repair model (Liu et al., 2019). Here, we improved this methodology to study transplanted fNP-labeled MSCs and multiple other cell types involved in bone repair, as well as blood vessel and blood volume, using a deep learning algorithm based on U-Net (Zhou et al., 2018).
MSC transplantation as a therapeutic has great potential in improving bone regeneration. However, the spatiotemporal progression in terms of cell fates and cell-cell interactions still need to be elucidated. In this study, we hypothesized that, in addition to osteogenic differentiation, a portion of transplanted MSCs retain stem-like phenotype, and promote bone regeneration by recruiting and regulating multiple cell types that are essential for bone regeneration. In this effort, we utilized a custom-made fNP to label marrow-derived MSCs. Using immunofluorescent deep-tissue microscopy, this study aims to reveal the spatiotemporal behavior of transplanted MSCs in a mouse monocortical tibial defect (MTD) model. We were able to quantify the proportions of transplanted cells and their osteogenic progenies, and their effects on blood vessels, mesenchymal stem cells, skeletal stem cells, and more mature osteoblasts.
Results
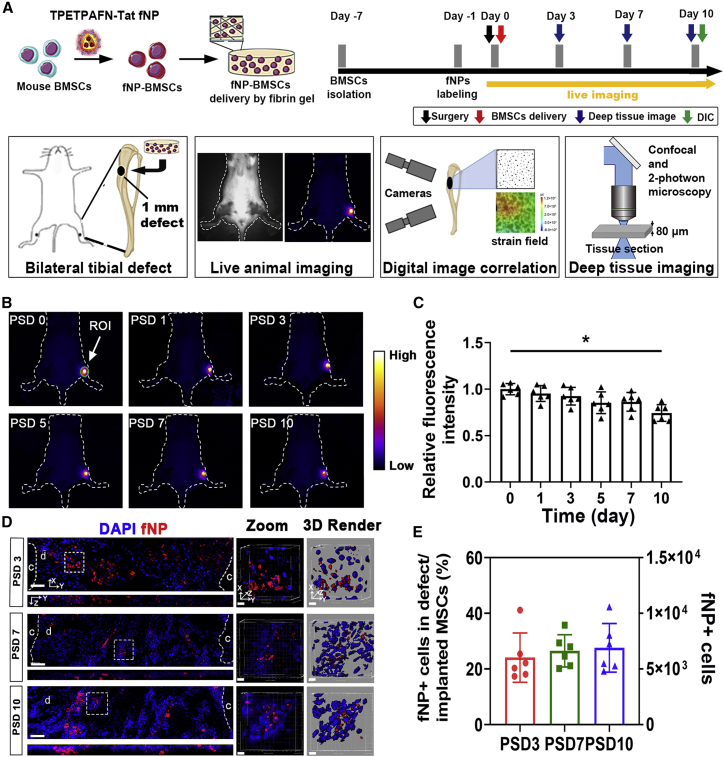
The survival and retention of locally delivered MSCs in tibial defect
Fluorescent signals of fNPs were localized in the cytoplasm of MSCs after 12 h of fNP incubation in vitro (Figure S1). Over 80% of fNP-labeled MSCs expressed SCA-1; less than 10% of which expressed OSX and F4/80 (Figure S1). The fNP-labeled MSCs were capable of differentiating into osteogenic lineage cells in vitro, with no significant difference to non-labeled MSCs (Figure S2). MSCs labeled with second near infrared (NIR-II) fNPs (Li et al., 2021) were locally delivered to tibial defects (Figure 1A). The NIR-II signals were observed in the defect area throughout the bone repair process (Figure 1B). NIR-II signals were not observed in the contralateral tibia without MSC transplantation. Compared with post surgery day 0 (PSD 0), the fluorescence intensity did not show significant decrease on PSD 7, but was reduced by 28% ± 12% on PSD 10 (Figure 1C). The fNP signals were observed in the cytoplasm of transplanted cells and confirmed their penetration into the defect (Figure 1D). Out of total transplanted MSCs, 23% ± 8% migrated into the defect (Figure 1E).
Figure 1.
Long-term tracking of implanted MSCs in tibial defects during bone repair
(A) Schematic of fNP labeling and MSC transplantation.
(B) Long-term tracking of implanted MSCs in the tibial defect site with NIR-II imaging, quantification was performed in the region of interest (ROI).
(C) Fluorescence intensity of the ROI over the defect area; n = 6 mice; ∗p < 0.05.
(D) Confocal microscopy of implanted cells (fNP) within the defect on PSD 3, 7, and 10. Zoomed-in images of regions enclosed by white dotted lines are shown to the right, followed by 3D rendered surfaces; c, cortical bone; d, defect area. Scale bars, 80 μm (in the left pane) and 5 μm (in zoomed-in images).
(E) The number of MSCs from (C) at each time point; n = 6 mice.
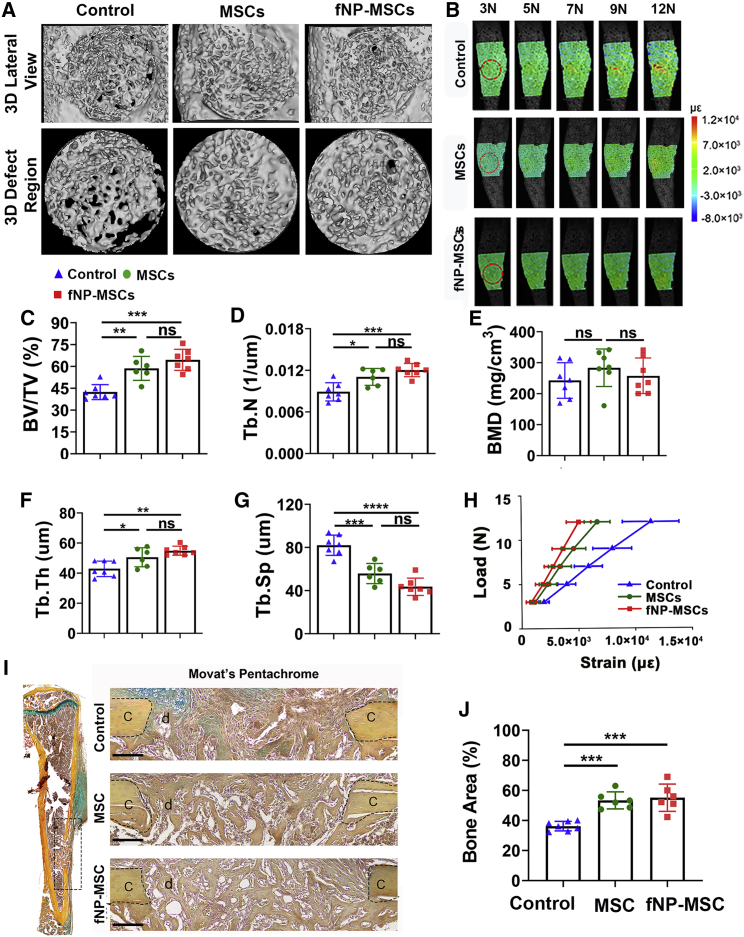
Locally delivered MSCs with fNP labeling improved structure and function of bone defect tissue
Microcomputed tomography (micro-CT) data showed that, compared with control, transplantation of either unlabeled MSCs or fNP-labeled MSCs (fNP-MSCs) significantly increased new bone volume (BV) on PSD 10 (Figure 2A), BV fraction (BV/total tissue volume [TV]) by 47% ± 7% and 50% ± 5%, trabecular number (Tb.N) by 39% ± 4% and 45% ± 6%, trabecular thickness (Tb.Th) by 24% ± 8% and 32% ± 6%, and decreased trabecular separation (Tb.Sp) by 36% ± 4% and 45% ± 6%, respectively (Figures 2C–2G). Bone mineral density was not affected (Figure 2E). Movat’s pentachrome staining showed that new bone area has increased in both MSC and fNP-MSC transplantation groups on PSD 10, compared with the control group (Figures 2I and 2J).
Figure 2.
Locally delivered MSCs improved structure and function of regenerating tissue in bone defects; this improvement was not affected by fNP labeling
(A) Micro-CT of the defect on PSD 10.
(B) Strain fields on tibia surface with defects under 3–12 N of axial loading, measured by DIC; red dotted line, defect region.
(C–G) Structural properties and bone mineral density (BMD) of defect regions on PSD 10; n = 6 mice; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(H) Axial strain on defect region under 3–12 N of loading; n = 6 mice. ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(I) Movat’s pentachrome staining of a tibia with MTD (left) and zoom-in defect regions (right). Scale bars, 80 μm; c, cortical bone; d, defect area.
(J) Bone area from Movat’s pentachrome staining; n = 6 mice; ∗∗∗p < 0.001.
Using digital image correlation (DIC), elevation of axial strain across the defect area was correlated with increased load (Figure 2B). MSC and fNP-MSC transplantation reduced axial strain at the defect area by 43% ± 7% and 50% ± 9%, respectively, under 12 N of load (Figure 2H). MSC transplantation also reduced axial strain on the surrounding uninjured areas of the tibia (Figure S3).
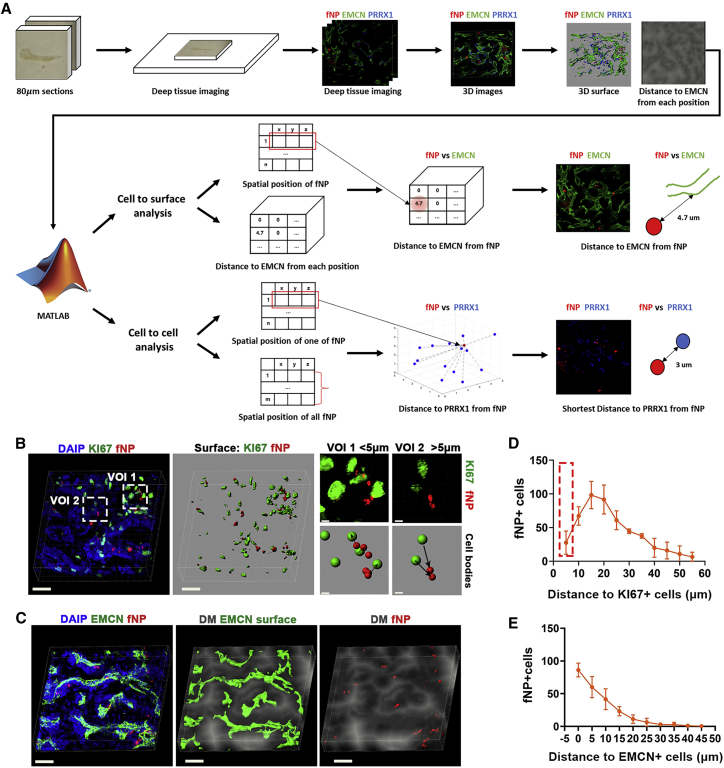
Quantitative spatial analysis of transplanted cells in defect via sc3DSC
The sc3DSC method was developed to quantify (1) the spatial distribution of transplanted MSCs, (2) phenotype markers and secreted proteins of all cells within a bone defect, and (3) the spatial relationship between different cell types during bone repair. The cell-to-cell method was utilized to quantify the distance between transplanted fNP+ cells and other cell types (Figures 3B and 3D). Fluorescent signals with a distance less than 5 μm from the center of each other were considered to be co-localized in the same cell (Figure 3B). The cell-to-surface method was utilized to quantify biological marker proteins, fNPs, and their spatial correlation with the blood vessels (Figures 3C and 3E).
Figure 3.
Spatial correlation analysis of transplanted MSCs during bone repair through sc3DSC
(A) The workflow of sc3DSC analysis.
(B) Cell-to-cell method; points from different channels with less than 5 μm in distance were considered to be in the same cell (right). Scale bars, 20 μm (in the left and middle panels) and 3 μm (in the right panel).
(C) Cell-microvessel/protein correlation analysis via the cell-to-surface method; distance mask depicts the distance from every voxel in extravascular spaces to the closest voxel containing vascular endothelial cells (EMCN, green); the center points of transplanted cells (fNP, red) were correlated with the distance mask with respect to EMCN. Scale bar, 25 μm.
(D and E) Spatial statistical analysis between transplanted cells (fNP) versus proliferating cells (KI67) and microvessels (EMCN).
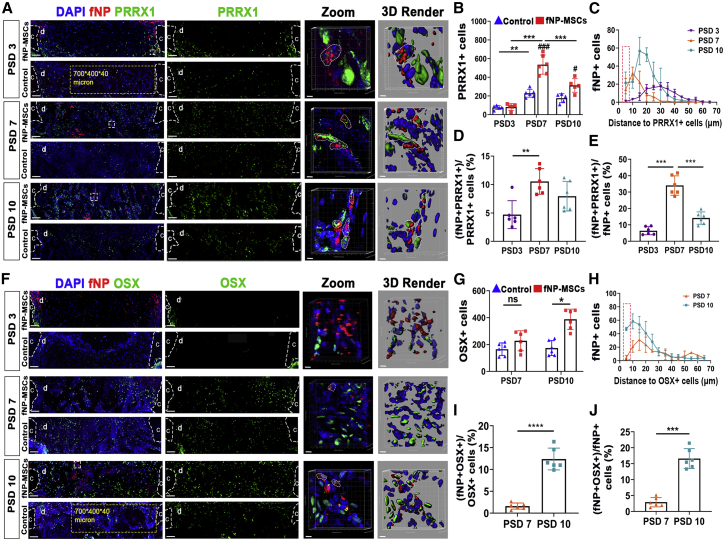
Locally delivered MSCs increased osteoblastic cell numbers in the bone defect, but few underwent osteogenic differentiation
Having observed the enhanced bone regeneration and the presence of locally delivered MSCs, we proceeded to examine the cell fate of transplanted MSCs. Using sc3DSC, we analyzed DAPI, PRRX1, and fNP signals in 3D space and found that transplanted MSCs differentiated into skeletal stem cells (PRRX1+) (Wilk et al., 2017) (Figure 4A). MSC transplantation resulted in higher number of PRRX1+ cells (131% ± 13% on PSD 7, and 58% ± 6% on PSD 10), but not on PSD 3 (Figure 4B). PRRX1+ cells were in closer proximity of implanted cells on PSD 7 and PSD 10 compared with PSD 3 (Figure 4C). The proportion of fNP+ PRRX1+ cells in the defect was 5% ± 3%, 10% ± 4%, and 7% ± 3% on PSD 3, 7, and 10, respectively (Figure 4D). Furthermore, the proportions of fNP+ PRRX1+ cells out of the total number of fNP+ cells were 6% ± 2%, 33% ± 8%, and 15% ± 5% on PSD 3, 7, and 10, respectively (Figure 4E). Some transplanted MSCs differentiated toward the osteogenic phenotype, showing both fNP and OSX signals (Figure 4F). MSC transplantation increased the number of OSX+ cells by 103% ± 7% on PSD 10, but had no effect on PSD 7 (Figure 4G). Compared with PSD 7, more OSX+ cells were in proximity of transplanted cells on PSD 10 (Figure 4H). The proportion of fNP+ OSX+ cells over total OSX+ cells was 2% ± 1% and 13% ± 4% on PSD 7 and PSD 10, respectively (Figure 4I); the proportion of fNP+ OSX+ cells over fNP+ cells in the defect area was 3% ± 1% and 17% ± 4% on PSD 7 and PSD 10, respectively (Figure 4J). Expressions of osteogenic genes (Ocn, Opn, Runx2, Alp, and Col1) were significantly higher in the MSC implantation group on PSD 10 (Figure S4). Transplantation of MSCs increased collagen fiber volume by 110% ± 11% in defect region on PSD 10, but had no obvious effect on PSD 7 (Figure S5).
Figure 4.
Locally delivered MSC increased osteoblastic lineage cell numbers in the defect, but few underwent osteogenic differentiation
(A) Fluorescent microscopy showing cells expressing the skeletal stem cell marker PRRX1 and implanted cells (fNP). Scale bars, 80 μm; c, cortical bone; d, defect area; zoomed-in images of regions enclosed by white dotted line are shown to the right, followed by 3D rendered surfaces. Scale bars, 5 μm; yellow dotted line, volume of interest (VOI); white dotted lines in the zoomed-in images, fNP+ PRRX1+ cells.
(B) Number of PRRX1+ cells; ∗∗p < 0.01, ∗∗∗p < 0.001, #p < 0.05, ###p < 0.001; n = 6 mice.
(C) Distances between implanted cells (fNP+) to PRRX1+ cells in defect at each time point; red dotted lines, fNP+ PRRX1+ cells (distance < 5 μm).
(D) Proportion of fNP+ PRRX1+ cells over total PRRX1+ cells in the VOI from (A); ∗∗p < 0.01; n = 6 mice.
(E) Proportion of fNP+ PRRX1+ cells over fNP+ cells in the VOI from (A); ∗∗∗p < 0.001; n = 6 mice.
(F) Fluorescent microscopy showing osteoblasts (OSX) and implanted cells (fNP). Scale bars, 80 μm; c, cortical bone; d, defect area; zoomed-in images of regions enclosed by white dotted line, followed by 3D rendered surfaces. Scale bars, 5 μm; the white dotted lines indicate fNP+ OSX+ cells.
(G) Number of OSX+ cells in the VOI from (F); ∗p < 0.05; n = 6 mice.
(H) Distances between implanted cells (fNP+) to osteoblast (OSX+) in defect at each time point; red dotted lines, fNP+ OSX+ cells (distance < 5 μm).
(I and J) Proportion of fNP+ OSX+ cells over total OSX+ cells and fNP+ cells, respectively, in the VOI from (F); ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; n = 6 mice.
Locally delivered MSCs increased SCA-1+ cells population during bone defect repair
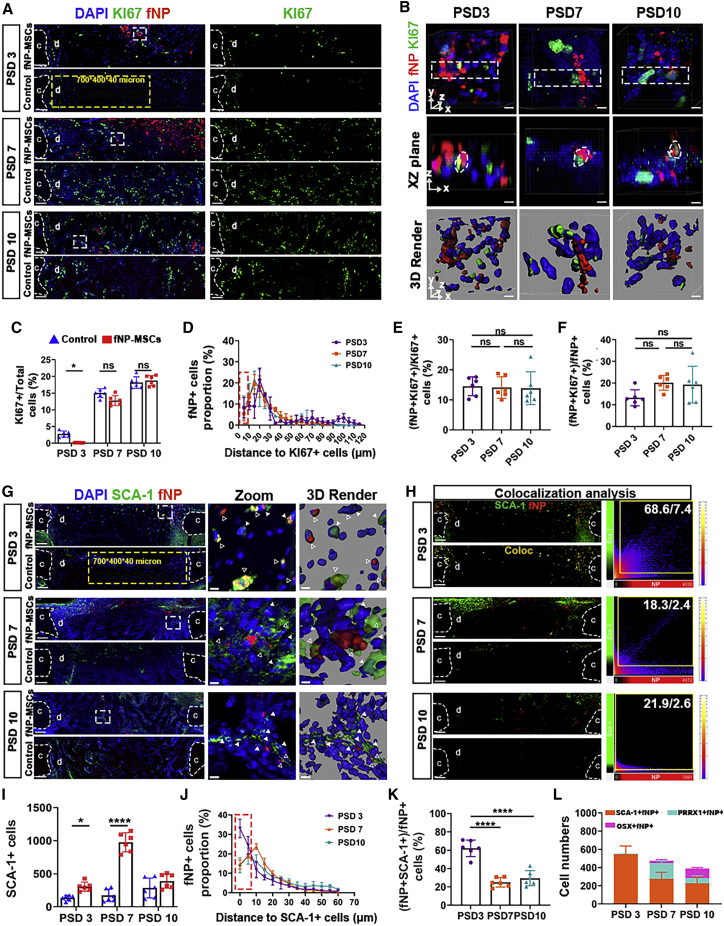
We found that implanted MSCs had proliferative activity at all time points (Figures 5A and 5B). MSC transplantation decreased the proportion of KI67+ proliferating cells by 216% ± 3% on PSD 3, but had no effect on PSD 7 and 10 (Figure 5C). The distance between KI67+ cells and fNP+ cells did not change at these time points (Figure 5D). Out of all transplanted cells, 14% ± 3% were proliferative on PSD 3; this proportion was maintained to PSD 10 (Figure 5F). The proportion of proliferating implanted cells (fNP+ KI67+) over total proliferating cells (KI67+) was 12% ± 3% on PSD3, and showed no change on PSD 7 and PSD 10 (Figure 5E).
Figure 5.
Locally delivered MSCs maintained their SCA-1 expression and increased SCA-1+ cell population during bone defect repair
(A) Fluorescent microscopy showing cells with proliferative activity (KI67) and implanted cells (fNP); c, cortical bone; d, defect area. Scale bars, 80 μm; yellow dotted line, VOI.
(B) XY (top) and XZ plane (middle) of zoomed-in regions enclosed by white dotted lines in (A), followed by 3D rendered surfaces (bottom). Scale bars, 5 μm; white dotted lines, fNP+ KI67+ cells.
(C) Proportion of KI67+ cells in VOI from (A); ∗p < 0.05; n = 6 mice.
(D) Distance distribution of fNP+ cells to KI67+ cells; region enclosed by red dotted line, fNP+ KI67+ cells (distance < 5 μm).
(E and F) Proportion of fNP+ KI67+ cells over total fNP+ cells and total KI67+ cells, respectively, at each time point. n = 6 mice.
(G) Fluorescent microscopy showing SCA-1+ cells and fNP+ implanted cells; c, cortical bone; d, defect area. Scale bars, 80 μm; yellow dotted lines, VOI; zoomed-in images of regions enclosed by white dotted line are shown to the right, followed by 3D rendered surfaces; white triangles, SCA-1+ fNP– cells; empty triangles, SCA-1+ fNP+ cells. Scale bars, 5 μm.
(H) Co-expression analysis of SCA-1 and fNP. Scale bars, 80 μm.
(I) Number of SCA-1+ cells in VOI from (G); ∗p < 0.05, ∗∗∗∗p < 0.0001; n = 6.
(J) Distance distribution of fNP+ cells to SCA-1+ cells; red dotted line, fNP+ SCA-1+ cells (distance < 5μm).
(K) Proportion of fNP+ SCA-1+ cells over total fNP+ cells; ∗∗∗∗p < 0.0001; n = 6.
(L) Qualitative summary of implanted cells with stem cell marker (SCA-1) or osteogenic lineage cell marker (PRRX1, OSX) expression in defect area at each time point.
Cells with fNP SCA-1 signals were present in defects and the periosteum throughout bone repair (Figure 5G). Higher numbers of host stem cells (fNP- SCA-1+) were in the MSC implantation group on PSD 3 and PSD 7; and they were spatially adjacent to the implanted cells (fNP+) (Figure 5G). Voxel-wise co-localization analysis revealed that 68.6% of fNP+ voxels were SCA-1+ on PSD 3, but this decreased to 26.9% and 15.3% on PSD 7 and PSD 10, respectively (Figure 5H). Lower proportions of SCA-1+ voxels were fNP+ (7.4% on PSD 3, 2.6% on PSD7, and 2.4% on PSD 10) (Figure 5H). MSC transplantation significantly increased the number of SCA-1+ cells in the defect by 182% ± 10% and 419% ± 18% on PSD 3 and PSD 7, respectively, but had no effect on PSD 10 (Figure 5I). sc3DSC analysis showed that more SCA-1+ cells were adjacent (<10 μm) to implanted fNP+ cells on PSD 3 and PSD 7, compared with PSD 10 (Figure 5J). We observed 61% ± 8% of fNP+ SCA-1+ cells over total fNP+ cells on PSD 3; but the percentage decreased significantly on PSD 3 to PSD 10 (Figure 5K). The proportion of implanted cells expressing mesenchymal stem cell and skeletal stem cell markers (SCA-1 and PRRX1) was higher than the proportion of implanted cells expressing more mature osteogenic markers (OSX) on PSD 3, PSD7, and PSD 10 (Figure 5L).
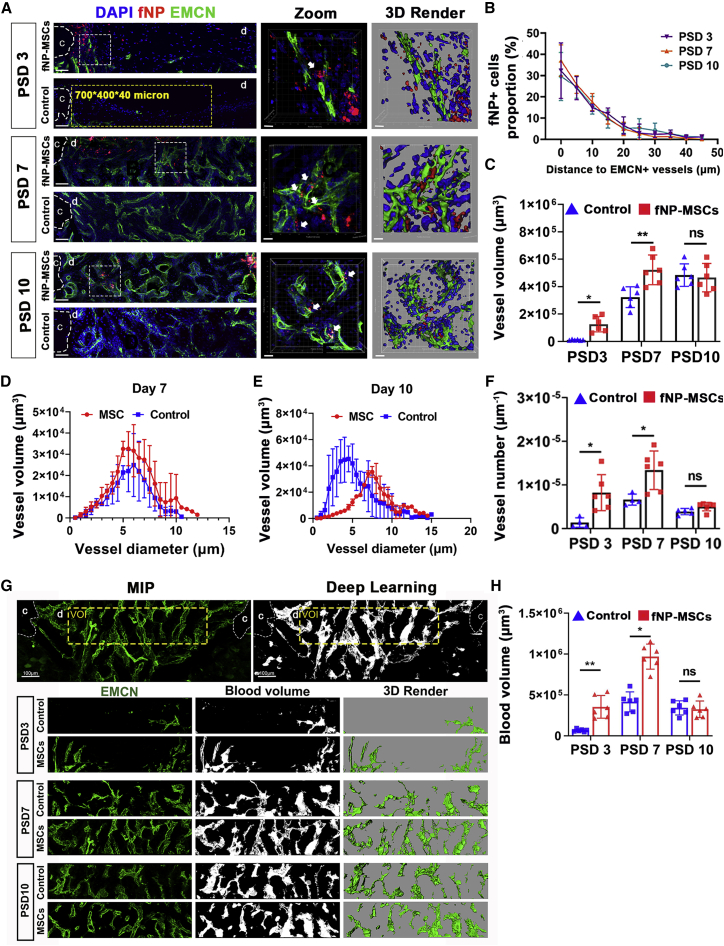
Locally delivered MSCs were in close proximity to newly formed vessels and improved neovascularization in bone defects
To elucidate the relationship between transplanted MSCs and newly formed vessels, we applied sc3DSC analysis for EMCN+ vessels and fNP+ implanted cells at the defect during the vessel invasion (PSD 3–7) and vessel remodeling phase (PSD 7–10). Implanted cells were in close proximity to newly formed vessels throughout tibial defect repair (Figure 6A). The distance between fNP+ cells and EMCN+ vessels did not change significantly at each time point (Figure 6B). MSC transplantation increased vessel volume and vessel number on PSD 3 and PSD 7 (Figures 6C and 6F). MSC transplantation produced thicker vessels on PSD 10 (Figure 6E), but had no effect on the distribution of vessel diameters on PSD 7 (Figure 6D). The blood volume, which was generated by deep learning, followed closely to the vascular structure (Figures 6G and S6). Compared with control, MSC transplantation increased blood volume by 361% ± 19% and 116% ± 14% on PSD 3 and PSD 7, respectively (Figure 6H).
Figure 6.
Locally delivered MSCs had peri-vascular localization and promoted angiogenesis
(A) Fluorescent microscopy showing vessels (EMCN) and implanted cells (fNP). Scale bars, 80 μm; yellow dotted line, VOI for quantitative analysis; c, cortical bone; d, defect area; zoomed-in images of regions enclosed by white dotted line are shown to the right, followed by 3D rendered surfaces. Scale bars, 10 μm; white arrows, fNP+ cells adjacent to newly formed vessels.
(B) The distance distribution of implanted cells (fNP) to EMCN+ vessels.
(C–F) Vessel volume, the distribution of vessel diameter versus volume, and vessel number at each time point; ∗p < 0.05, ∗∗p < 0.01; n = 6 mice.
(G) Fluorescent microscopy showing newly formed vessels (green) and blood volume generated with neural network (white) within the defect. Scale bars, 100 and 50 μm for up and down planes, respectively; c, cortical bone; d, defect area.
(H) Blood volume quantification; ∗p < 0.05, ∗∗p < 0.01; n = 6.
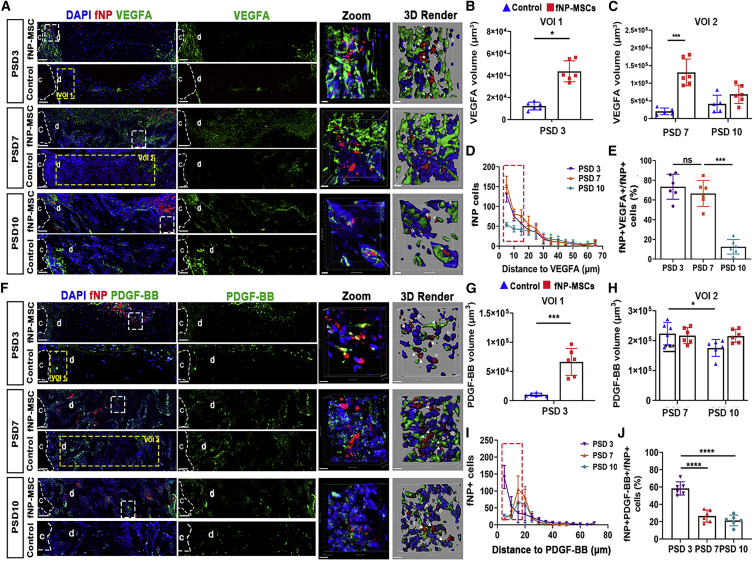
Locally delivered MSCs secreted VEGFA and PDGF-BB during the early phase of bone repair
MSC transplantation increased the relative mRNA expression level of VEGFA, HIF-1α, and platelet-derived growth factor B (PDGF-BB) by 353% ± 9%, 476% ± 11%, and 661% ± 25%, respectively (Figure S4). Transplanted MSCs secreted VEGFA and PDGF-BB proteins within the defect (Figure 7A and 7F). MSC transplantation increased VEGFA volume by 171% ± 6% and 417% ± 19% on PSD 3 and PSD 7, respectively, but had no effect on PSD 10 (Figures 7B and 7C); and increased PDGF-BB volume by 261% ± 6% only on PSD 3 (Figures 7G and 7H). We observed fewer fNP+ VEGFA+ cells on PSD 10 compared with PSD 3 and 7 (Figure 7D). Out of total transplanted cells, 71% ± 6% and 70% ± 8% expressed VEGFA (fNP+ VEGFA+/fNP+ cells) on PSD 3 and 7, respectively (Figure 7E). The proportion of fNP+ PDGF-BB+ cells over fNP+ cells was 58% ± 9% on PSD 3, and decreased to 22% ± 4% and 19% ± 6% on PSD 7 and 10 (Figure 7J).
Figure 7.
Transplanted MSCs secreted VEGFA and PDGF-BB in the bone defect
(A) Fluorescent microscopy showing VEGFA and implanted cells (fNP) within the defect; c, cortical bone; d, defect area. Scale bars, 80 μm; regions enclosed by yellow dotted lines: VOI1 was selected as quantitative regions with 200, 400, and 40 μm on X, Y, and Z on PSD 3; VOI1 was adjacent to cortical bone; VOI2 was selected as quantitative regions with 700, 400, and 40 μm on X, Y, and Z on PSD 7 and 10, the middle portion of the defect is selected; zoomed-in images of regions enclosed by white dotted lines are enlarged to right panels, followed by 3D rendered surfaces; white arrows indicate fNP+ VEGFA+ cells; empty arrows indicate fNP+ VEGFA– cells. Scale bars, 10 μm.
(B and C) Quantification of VEGFA; ∗p < 0.05, ∗∗∗p < 0.001; n = 6.
(D) The distance distribution of fNP+ cells to the VEGFA surface.
(E) The proportion of fNP+ VEGFA+ cells relative to fNP+ cells in defect area; ∗∗∗p < 0.001; n = 6.
(F) Representative confocal microscopy images of PDGF-BB (green) and implanted cells (fNP, red) within the defect. Scale bars, 80 μm; zoomed-in images of regions enclosed by white dotted lines are enlarged to right panel of respective groups, followed by 3D rendered surface; white arrows indicate fNP+ PDGF-BB+ cells; empty arrows indicate fNP+ PDGF-BB– cells. Scale bars, 10 μm.
(G and H) Quantification assessment for PDGF-BB volumes. ∗p < 0.05, ∗∗∗p < 0.001; n = 6.
(I) The distance distribution of fNP+ cells to the PDGF-BB surface.
(J) The proportion of fNP+ PDGF-BB+ cells relative to fNP+ cells; ∗∗∗∗p < 0.0001; n = 6.
A small portion of fNP-labeled cells showed co-localization with F4/80+ macrophages (fNP+ F4/80+) on PSD 3, 7, and 10 (Figures S7A–S7C). The proportion of fNP+ F4/80+ voxels was 8% ± 2% of total cells on PSD 3, and decreased to only 1.1% ± 0.3% on PSD 7, and 2.3% ± 0.1% on PSD 10 (Figure S7E).
Discussion
The main challenge of validating and improving MSC therapies for bone regeneration is the uncertain spatial distribution of implanted cells. Previous fluorescence probes were not able to continuously track transplanted MSCs in vivo due to a combination of low resolution, poor depth penetration, and short-term retention (Ni et al., 2020). We report a series of fNPs as exogenous trackers, which allow long-term tracking of MSCs with bright fluorescence, excellent biological and photophysical stabilities, low influence on viability and differentiation, and superb retention (Ding et al., 2014; Li et al., 2013a). These fNPs outperformed commercial products of exogenous fluorescent trackers, offering great promises in investigating cell fates during regeneration. We were able to directly observe locally delivered MSCs with fNP labeling at single-cell resolution during long bone defect repair. The fluorescent signal from NIR-II fNPs could penetrate soft tissues, and allowed imaging of transplanted MSCs in living mice (Li et al., 2021). In this study, locally delivered MSCs were applied onto the periosteal side of an MTD. Using a combination of live imaging and deep tissue microscopy, we showed that locally delivered MSCs were retained around the defect site and had migrated into the defect. Micro-CT analysis showed that MSCs both with and without fNP labeling, had comparable efficacy at improving the BV and trabecular structure of regenerated bone tissue, indicating that fNP labeling would not affect the osteogenic effect of transplanted MSCs.
The complex mixture of soft and hard tissues within a regenerating bone defect has posed significant challenges with regard to measuring its mechanical properties (Ruspi et al., 2017). Previous studies successfully utilized DIC to measure the local and global strain field on large bones, such as human spine (Palanca et al., 2018) and sheep tibia (Thompson et al., 2007). However, the main drawback of these DIC strategies was resolution being in the macro scale, which limited its use for small animal models and correlating mechanical strain with cells. Recently, DIC with microscopy has been developed to measure local strain field on an irregular surface with micrometer resolution (Carrieroa et al., 2018), thus allowing the measurement of mechanical properties of mouse bones (Sztefek et al., 2010). Using this method, we found that strains on the surface of intact bone from control groups were 2,000–3,000 με in response to 9 N of force, which was consistent with previous analyses (Sztefek et al., 2010). We observed higher stiffness in bone defects after MSC transplantation as shown by reduced strain, indicating functional recovery of the defect tissue.
We investigated the spatiotemporal distribution of locally delivered MSCs, using the sc3DSC technique to quantify their progenies in relation to host cells, which revealed exogenous MSC cell fates and functions during bone formation. Previous work has shown that allogeneic MSCs could directly differentiate into osteogenic cells after transplantation (Julien et al., 2021). Our results showed that bone defects with transplanted MSCs had significantly higher numbers of PRRX1+ skeletal stem cells and OSX+ osteoblasts. However, only a small proportion of the total implanted MSCs underwent osteogenic differentiation, suggesting that implanted MSCs were enhancing bone repair through other mechanisms, such as promoting the invasion of host mesenchymal stem cells and skeletal stem cells into the defect area, and subsequent regulation of their proliferation and differentiation.
MSCs are capable of self-renewal and osteogenic differentiation, but whether implanted MSCs could maintain these characteristics during bone repair is still unclear. Immunofluorescence microscopy of mesenchymal stem cells and skeletal stem cell markers (SCA-1 and PRRX1) allowed us to track the biomarker-based stemness of implanted MSCs. Using sc3DSC, we found that 30%–40% of implanted MSCs had SCA-1 and PRRX1expression up to our longest time point (PSD 10), suggesting that they maintained stemness throughout the bone repair process. During early stages of bone repair, implanted MSCs induced higher numbers of host stem cells (fNP-SCA-1+) in the defect, most of which were located in close proximity to implanted cells, suggesting the recruitment of host stem cells. Interestingly, MSC implantation decreased the proportion of KI67+ proliferating cells on PSD 3, but not on PSD 7 and 10. The higher proportion of stem cells and progenitors due to MSC implantation may have an inhibitory effect on proliferation, while promoting osteogenic differentiation, thus accelerating bone repair. This is corroborated by higher numbers of PRRX1+ skeletal stem cells on PSD 7, and higher numbers of more mature OSX+ osteogenic cells on PSD 10. Therefore, MSC implantation could promote bone regeneration by accelerating the osteogenesis process.
Paracrine effects of MSCs plays a critical role in tissue regeneration. Regulatory factors secreted by mesenchymal stem cells could regulate immune responses (Pajarinen et al., 2019), osteogenesis (Lee et al., 2019), and angiogenesis (Li et al., 2013b), as well as supporting the bone marrow niche. Here, we found that transplanted MSCs secreted PDGF-BB, a major growth factor (Xie et al., 2014) and stem cell chemoattractant (Gao et al., 2019). Administration of recombinant PDGF-BB could increase bone formation in osteonecrosis (Su et al., 2020), osteoporosis (Chen et al., 2015; Mitlak et al., 1996), and bone defect repair (Del Rosario et al., 2015). Previous results demonstrated that PDGF-BB could promote the proliferation, migration, and osteogenic differentiation of MSCs, and enhance the expansion of endothelial progenitors, thus improving angiogenesis and osteogenesis during bone remodeling (Chen et al., 2015; Zhang et al., 2018). We found that implanted MSCs secreted and increased the amount of PDGF-BB in bone defects on PSD 3. At the same time point, MSC implantation has enhanced blood vessel volume and the number of SCA-1+ cells. These results suggest that implanted MSCs may have contributed to the recruitment of host SCA-1+ cells and promoted vessel formation through the secretion of PDGF-BB.
Work by Kusumbe et al. (2014) had shown a specific type of vessels, type H vessels, were coupled with osteogenic cells. Implanted MSCs could regulate new vessel formation in bone repair, as transplanted MSCs increased vessels volume (McDermott et al., 2019). In this study, sc3DSC analysis showed that transplanted MSCs were localized close to newly formed vessels during bone repair, similar to the localization of host MSCs. In addition, more osteogenic cells were found near blood vessels in MSC transplantation groups. Taken together, MSC transplantation could enhance bone repair by enhancing osteogenesis-angiogenesis coupling. In addition, transplanted MSCs seemed to have pro-angiogenic functions, as we have found higher vessel volume during the early phase of repair (PSD 3 and 7). Endogenous MSCs could secrete angiogenic factors, including VEGF, PDGF-BB, and bFGF (Wang et al., 2013). We examined whether transplanted MSCs secreted angiogenic factors during bone defect healing. Our data showed that over 70% of fNP+ cells were expressing the pro-angiogenic factor VEGFA within the defect during this time frame. The distribution of vessel thickness showed a higher peak in the MSC transplantation groups on PSD 10, which could be due to accelerated vascular remodeling, which is vital during bone regeneration (Boerckel et al., 2011; Einhorn and Gerstenfeld, 2015).
Blood availability could also affect bone repair, as adequate blood supply would be vital for nutrient and waste transport. However, quantification of blood volume in a bone defect has been difficult. Micro-CT angiography together with a contrast agent had been utilized to measure blood volume (McDermott et al., 2019; Sawall et al., 2020). But capillary vessels posed significant challenge to radiological contrast agents, which were limited by high viscosity and the pressure required. The best method to study skeletal blood volume involves manual filling of immunofluorescence data (Gomariz et al., 2018). Here, we utilized a U-Net-based deep learning algorithm (Zhou et al., 2018) to fill microvessels to obtain the blood volume automatically.
As the uptake of nanoparticles by endogenous macrophages is possible (Dupont et al., 2010). We found a small proportion of F4/80+ macrophages with fNP signals in the defect area on PSD 3, which was in agreement with previous data showing internalized nanoparticles in macrophages (Dupont et al., 2010; Liu et al., 2021), but this decreased to around 1% during later time points. This F4/80+ fNP+ cell population during the early phase of repair was likely due to post-transplant cell death of implanted MSCs (Swijnenburg et al., 2008). However, the contribution of macrophages was small compared with implanted MSCs, as fNP+ F4/80+ cells only consisted of around 8% of total cells on PSD 3. Then, fNP+ macrophages were quickly cleared from the defect region. Significant proportion of implanted cells maintained SCA-1 expression and small proportion of implanted cells showed osteogenic differentiation, suggesting low off-target effects on tracking transplanted MSCs. The higher proportion of cells with F4/80 and fNP co-expression on PSD 10 may be due to the apoptosis of fNP-MSC-derived osteoblasts and endocytosis of cell debris by macrophages (Jilka et al., 1998).
One limitation in our study is the lack of using multiple marker genes together to define well-known cell types. As the fNPs utilized in this study occupied a wide range of the light spectrum and limited multiplexing of immunofluorescent staining, we utilized single phenotype markers to study the behavior of skeletal stem cells and osteogenic lineage cells. However, cells could express SCA-1 and PRRX1 simultaneously during osteogenic differentiation (Takarada et al., 2016). PRRX1+ SCA-1+ cells also have multipotent and self-renewal potential, but stops expressing SCA-1 and PRRX1 when differentiated into more mature osteogenic cells, and start to express OSX (Esposito et al., 2020; Nakashima et al., 2002; Takarada et al., 2016). Note that SCA-1 and OSX should not overlap and would not affect our conclusion regarding the cell fate of transplanted cells. We hope future development in ultra-narrow bandgap probes and fluorescent markers and imaging method would allow us to integrate more markers simultaneously and investigate different stage of osteogenic differentiation of transplanted MSCs.
In conclusion, we have utilized fNP labeling, live imaging, deep tissue microscopy, and sc3DSC analysis to track and quantify cell fates and activities of transplanted MSCs during bone defect repair. Our results showed spatiotemporal distribution of transplanted MSCs, spatial correlation with other cell types, osteogenic differentiation, and paracrine activities. The primary contribution of transplanted MSCs at enhancing bone repair seemed to be the acceleration of osteogenesis by recruiting endogenous skeletal stem cells and osteogenic cells, as well as the regulation of newly established vascular network within the defect space. These findings would contribute to the discovery of new cell-based therapy candidates that promote bone repair. These newly developed methodologies may be useful for analyzing transplanted cells in other tissue types for the purpose of promoting regeneration and treating diseases.
Experimental procedures
Animals and reagents
Twelve-week-old female C57BL/6J mice (Shanghai Model Organism) were housed in the Southern University of Science and Technology Animal Care Facility and had access ad libitum to standard mouse chow and water for the duration of the study. All in vivo animal protocols were approved by the Institutional Animal Care and Use Committee.
Bone marrow-derived MSC isolation, fNP labeling, and local delivery
Bone marrow-derived MSCs (BMSCs) were isolated from the femur and tibia of 8–12-week-old female C57BL/6J mice according to methods published previously (Zhang et al., 2008). In brief, bone marrow of female C57BL/6 mice (6–8 weeks old) was extracted by rinsing the bone cavity with culture medium (DMEM + 1% PS + 10% FBS). The cells were resuspended in the culture media containing basic fibroblast growth factors and cultured for 5 days. The extracted BMSCs (2 × 106 cells) were cultured in a 25 cm2 cell culture flask.
fNPs were obtained by assembling Tat and 2,3-bis(4-(phenyl-(4-(1,2,2 triphenylvinyl)phenyl)amino)phenyl)fumaronitrile. These fNPs were fabricated by utilizing DSPE-PEG2000 (1 mg) and its maleimide derivative DSPE-PEG2000-Mal (1 mg) as the encapsulation matrix, and reacted with Tat peptide (100 mM in 40 μL). The chemical structure of these nanoparticles was as reported previously (Li et al., 2013a). BMSCs were incubated with fNPs (50 μg/mL) in growth medium for 18 h to label cells before transplantation.
BMSCs were delivered locally on the periosteal side of the tibial defect mixed within a fibrin gel (Enzo Life Sciences, USA). The cultured BMSCs or fNP-labeled BMSCs were resuspended in fibrinogen at a concentration of 2.5 × 104 cells/μL immediately prior to tibial defect surgery. Implants contained 2.5 × 104 fNP-labeled MSCs, or cell-free fibrin gel was injected into the defect area.
Long-bone defect repair model and local delivery of BMSCs
MTD surgery was performed as described previously (Liu et al., 2018). In brief, under isoflurane anesthesia, a longitudinal skin incision was made over the lower limb. Using a high-speed drill, 1-mm-diameter circular defects were created on the anterior medial surface of a mouse tibia centered between the tibiofibular junction and the tibial tubercle. The defect center was 4 mm distal to the proximal articulating surfacing of the tibia. After saline irrigation, the incision was closed with 6-0 nylon sutures. Buprenorphine was administered immediately before surgery and at 24 and 48 h after surgery.
Tracking of BMSCs by NIR-II live imaging during bone defect repair
The NIR-II fluorescence imaging (Suzhou NIR-Optics) of locally delivered MSCs were performed at different time points (1, 3, 5, 7, and 10 days) post-surgical transplantation, using an 808-nm laser with a power density of 30 mW/cm2 for excitation and a 1,020 nm long-pass filter to collect the fluorescence signal with an exposure time of 50 ms (n = 6). The measurement of the light intensity of fNP probes was quantified with ImageJ.
Micro-CT
The tibias were harvested on post-surgery day 10, and scanned using a micro-CT scanner (Skyscan 1172, Bruker, USA) with the voltage of 60 kV, current of 100 μA, and a resolution of 6 μm. The CTAn software was used to analyze total BV/TV, trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular spacing (Tb.Sp). The 3D reconstruction of the intact bone and defect region was performed by Mimics 17 (Materialise, USA). The volume of interest was the circular region of the tibial defect, on the same frontal plane as the existing cortical bone. A successful repair process will result in the formation of cortical bone in this volume of interest (VOI). BV/TV was defined as the ratio of the segmented bone volume to the total volume of VOI. Tb.N, Th.Th, and Tb.Sp were defined as measure of the average number of trabeculae per unit length, mean thickness of trabeculae, and distance between trabeculae in VOI. The computation of Tb.N, Th.Th, and Tb.Sp was based on 3D calculations, namely, a sphere fitting method (Bouxsein et al., 2010), where for thickness measurement the spheres are fitted to the object and for separation the spheres are fitted to the background. The basic approach is to determine the diameter of the largest possible sphere that can be fitted through each voxel that is completely contained within the object (or background) and then to average these diameters.
Mechanical testing
The mechanical properties of the bone defect tissue were determined using DIC according to methods published previously (Thompson et al., 2007). The surface of tibias was coated with a layer of matt, water-based white acrylic paint (XF-2, Tamiya Paint, USA) then speckled with matt, water-based, black paint (0741, Haoshun, China), using a high-precision airbrush. Then painted tibias were placed in the loading cups of electromagnetic mechanical test system (ElectroForce 3200 system, TA Instruments, USA). Axial compressive mechanical load was applied at a rate of 8 N/min up to a maximum of 12 N and the strain field was determined using the 3D DIC system (XTDIC-Micro, XTOP, China).
Immunofluorescent staining of thick bone sections
Mice were sacrificed on PSD 3, 7, and 10. Thick (90 μm) longitudinal tibia frozen sections were prepared as described previously (Kusumbe et al., 2015). In brief, each tibia was stripped of soft tissues and fixed in 4% paraformaldehyde (PFA) at 4°C for 4 h, decalcified in 0.5 M EDTA at 4°C for 48 h, cryoprotected at 4°C for 48 h, and cryoembedded. Cryosections were prepared using a cryostat (Leica 3050S) with a thickness of 80 μm and stored at −80°C.
For immunostaining, the tibia sections were hydrated, permeabilized, blocked, and incubated with primary antibodies against endomucin (sc-65495, 1:100, Santa Cruz), osterix (ab22552, 1:200, Abcam), paired related homeobox 1 (PRRX1) (ab211292, 1:200, Abcam), KI67 (ab15580, 1:200, Abcam), and F4/80 (ab6640, 1:200, Abcam) overnight. Next day, the sections were incubated with anti-rabbit Alexa Fluor 488 (A32790, 1:300, Invitrogen) secondary antibody for 1 h at room temperature. Sections were washed with PBS and mounted with DAPI FluoroMount-G (0100-20, SouthernBiotech) and then sealed with coverslips.
Confocal and two-photon imaging
3D fluorescent images were acquired using a Nikon A1R confocal laser scanning microscope with a 20× objective lens. The z stacks of 80 μm in height and an x-y detect area at a size of 1,024 × 1,024 pixels, with a resolution of 0.624 μm, were taken for each slide. The 1-mm defect was imaged by tiling three z stacks, spanning 1,500 mm along the long axis of the tibia.
The second harmonic generation of collagen fibers was acquired on an Olympus FVMPE-RS Multiphoton laser scanning microscope (Japan). Images were excited with an 860-nm laser, and emissions were detected using 420–465 filters. The z stack of 40 μm height and an x-y detect area, at a size of 1,024 × 1,024 pixels, with a resolution of (0.623) μm, were taken for each sample.
Spatiotemporal analysis of transplanted MSCs by sc3DSC
The sc3DSC method was developed to quantify (1) the spatial distribution of fNP+ cells, (2) phenotype markers and secreted proteins of all cells within a bone defect, and (3) the spatial relationship between different cell types during bone repair. The workflow of sc3DSC is shown in Figure 3A. The 3D surfaces of each confocal channel was created by utilizing the surface function in Imaris (v.7.1, Oxford Instruments, Switzerland). The spatial position of cells (fNP+, PRRX1+, OSX, and KI67) were obtained by the position output in Imaris and saved as excel files. The distance distribution to selected surfaces (EMCN, VEGFA, and PDGF-BB) was generated by Distance Transformation tool in Imaris, and saved as tiff files. For cell-to-surface analysis, the position information of fNP+ cells and the distance distribution to surface were input into MATLAB script. The distance distribution of fNP+ cells versus surface was analyzed by utilizing cell-to-surface code (cell-to-surface code.m). For cell-to-cell analysis, the position information of fNP+ cells and PRRX1+ cells were input into cell-to-cell code (cell-to-cell code.m) in MATLAB, which output the minimum distance between the cells.
The fNP signals were located in the cytoplasm of transplanted cells, but some cell markers (PRRX1, OSX, and KI67) were located in cellular nucleus (Esposito et al., 2020). The diameter of mesenchymal stem cells is about 20–30 μm (Ge et al., 2014), and the diameter of osteoprogenitor is approximately 20–50 μm (Malaval et al., 1999). Thus, fluorescent signals with a distance less than 5 μm from the center of each other were considered to be co-localized in the same cell. When the distance between fNPs and VEGFA/PDGF-BB was less than 10 μm, VEGFA/PDGF-BB was determined to be secreted by fNP+ cells. Spatial correlation between implanted MSCs and newly formed vessels was analyzed using the cell-to-surface method. Co-localization between fNPs and VEGFA/SCA-1 was evaluated in Imaris using the co-localization method.
In deep tissue images of longitudinal thick sections of the MTD, rectangular VOIs of 700, 400, and 40 μm on X, Y, and Z were selected as quantitative regions. The VOIs were in the space previously occupied by cortical bone.
Histological staining
The tibias with MTD were extracted on PSD 10, fixed in 4% PFA for 24 h, and decalcified in a large volume of 0.5 M EDTA at 4°C for 3 weeks before tissue processing. Then, tissue samples were dehydrated and embedded in paraffin (Leica ASP 300S, Leica). Five-micrometer paraffin sections were created using a microtome (Leica CM1950, Leica) and stained with hematoxylin and eosin (G1120, Solarbio, China) and Movat’s pentachrome (A55580, OKA, China) according to the manufacturers’ instructions.
qPCR
A 2-mm section of tibia containing the defect was collected on PSD 10 and ground into powder at −80°C using TissueLyser (Jingxin, China). The total RNA was extracted using TRIzol (15596, Thermo Scientific, USA). A RevertAid First Strand cDNA Synthesis Kit (K1622, Thermo Fisher Scientific, USA) was utilized to perform cDNA synthesis from isolated total RNA. The relative expression levels of osteogenic- and angiogenic-related genes in the defect were assessed by qPCR using GoTaq qPCR Master Mix (A6002, Promega, USA). Primer sequences are listed in Table S1. Gene expression was presented as 2−(ΔΔCT) after normalizing to 18 s.
Statistical analysis
Statistical significance between two groups were evaluated by Student’s t test. Multiple group comparison was performed with one-way and two-way ANOVA tests with Tukey’s multiple comparisons test. Experimental data were expressed as means ± SD. Results were considered statistically significant if p < 0.05. GraphPad 8.0 software (GraphPad Software, La Jolla, CA, USA) was used for all statistical analyses.
Author contributions
C.L. and K.L. conceived the project. C.L. and C. Y designed the experiments. C.Y. performed most of the experiments and wrote the manuscript. Z.L. provided the fNPs, and performed labeling of MSCs. Y.L. performed the DIC mechanical testing. Q.L. provided the deep learning algorithm. R.H. and L.F. performed blood volume analysis. M.L. implemented the sc3DSC computer codes and assisted with spatial correlation analyses. C.L. supervised the experiments and analyzed results. D.W., K.L., Q.L., and C.L. contributed to review and editing of the paper.
Acknowledgments
This work was supported by grants from the Shenzhen Science and Technology Innovation Commission (grant nos. KQTD20200820113012029 and 20201022173046002), the China Postdoctoral Science Foundation (grant no. 2021M691414), Guangdong Provincial Key Laboratory of Advanced Biomaterials (2022B1212010003), the National Natural Science Foundation of China (31870991), and the SUSTech Startup fund. We acknowledge the SUSTech Core Research Facilities for confocal microscopy and Micro-CT imaging. Live imaging was performed at the Imaging Core of the Department of Biomedical Engineering of SUSTech (Platform. “NIR-II live imaging platform”).
Conflicts of interest
The authors declare no competing interests.
Published: September 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.08.008.
Contributor Information
Kai Li, Email: lik@sustech.edu.cn.
Chao Liu, Email: liuc33@sustech.edu.cn.
Supplemental information
References
- Acar M., Kocherlakota K.S., Murphy M.M., Peyer J.G., Oguro H., Inra C.N., Jaiyeola C., Zhao Z., Luby-Phelps K., Morrison S.J. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015;526:126–130. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerckel J.D., Uhrig B.A., Willett N.J., Huebsch N., Guldberg R.E. Mechanical regulation of vascular growth and tissue regeneration in vivo. Proc. Natl. Acad. Sci. USA. 2011;108:E674–E680. doi: 10.1073/pnas.1107019108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouxsein M.L., Boyd S.K., Christiansen B.A., Guldberg R.E., Jepsen K.J., Müller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- Cao H., Yue Z., Gao H., Chen C., Cui K., Zhang K., Cheng Y., Shao G., Kong D., Li Z., et al. In vivo real-time imaging of extracellular vesicles in liver regeneration via aggregation-induced emission luminogens. ACS Nano. 2019;13:3522–3533. doi: 10.1021/acsnano.8b09776. [DOI] [PubMed] [Google Scholar]
- Carrieroa A., Pereirab A.F., Wilson A.J., Castagno S., Javaheri B., Pitsillides A.A., Marenzana M., Shefelbine S.J. Spatial relationship between bone formation and mechanical stimulus within cortical bone: combining 3D fluorochrome mapping and poroelastic finite element modelling. Bone Rep. 2018;8:72–80. doi: 10.1016/j.bonr.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Lin S., Huang D., Zhang Y., Li C., Wang M., Wang Q. Revealing the fate of transplanted stem cells in vivo with a novel optical imaging strategy. Small. 2018;14:1702679. doi: 10.1002/smll.201702679. [DOI] [PubMed] [Google Scholar]
- Chen W., Baylink D.J., Brier-Jones J., Neises A., Kiroyan J.B., Rundle C.H., Lau K.H.W., Zhang X.B. PDGFB-based stem cell gene therapy increases bone strength in the mouse. Proc. Natl. Acad. Sci. USA. 2015;112:E3893–E3900. doi: 10.1073/pnas.1501759112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rosario C., Rodríguez-Évora M., Reyes R., Delgado A., Évora C. BMP-2, PDGF-BB, and bone marrow mesenchymal cells in a macroporous beta-TCP scaffold for critical-size bone defect repair in rats. Biomed. Mater. 2015;10:045008. doi: 10.1088/1748-6041/10/4/045008. [DOI] [PubMed] [Google Scholar]
- Ding D., Mao D., Li K., Wang X., Qin W., Liu R., Chiam D.S., Tomczak N., Yang Z., Tang B.Z., et al. Precise and long-term tracking of adipose-derived stem cells and their regenerative capacity via superb bright and stable organic nanodots. ACS Nano. 2014;8:12620–12631. doi: 10.1021/nn505554y. [DOI] [PubMed] [Google Scholar]
- Dupont K.M., Sharma K., Stevens H.Y., Boerckel J.D., García A.J., Guldberg R.E. Human stem cell delivery for treatment of large segmental bone defects. Proc. Natl. Acad. Sci. USA. 2010;107:3305–3310. doi: 10.1073/pnas.0905444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einhorn T.A., Gerstenfeld L.C. Fracture healing: mechanisms and interventions. Nat. Rev. Rheumatol. 2015;11:45–54. doi: 10.1038/nrrheum.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito A., Wang L., Li T., Miranda M., Spagnoli A. Role of Prx1-expressing skeletal cells and Prx1-expression in fracture repair. Bone. 2020;139:115521. doi: 10.1016/j.bone.2020.115521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherall J., Robey P.G., Rowe D.W. Continuing challenges in advancing preclinical science in skeletal cell-based therapies and tissue regeneration. J. Bone Miner. Res. 2018;33:1721–1728. doi: 10.1002/jbmr.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S.-y., Lin R.-b., Huang S.-h., Liang Y.-j., Li X., Zhang S.-e., Ouyang D.-q., Li K., Zheng G.-s., Liao G.-q. PDGF-BB exhibited therapeutic effects on rat model of bisphosphonate-related osteonecrosis of the jaw by enhancing angiogenesis and osteogenesis. Bone. 2019;144:115117. doi: 10.1016/j.bone.2019.115117. [DOI] [PubMed] [Google Scholar]
- Ge J., Guo L., Wang S., Zhang Y., Cai T., Zhao R.C.H., Wu Y. The size of mesenchymal stem cells is a significant cause of vascular obstructions and stroke. Stem Cell Rev. Rep. 2014;10:295–303. doi: 10.1007/s12015-013-9492-x. [DOI] [PubMed] [Google Scholar]
- Gomariz A., Helbling P.M., Isringhausen S., Suessbier U., Becker A., Boss A., Nagasawa T., Paul G., Goksel O., Székely G., et al. Quantitative spatial analysis of haematopoiesis-regulating stromal cells in the bone marrow microenvironment by 3D microscopy. Nat. Commun. 2018;9:2532. doi: 10.1038/s41467-018-04770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A.C., Rudolph-Stringer V., Straszkowski L., Tjin G., Crimeen-Irwin B., Walia M., Martin T.J., Sims N.A., Purton L.E. Retinoic acid receptor gamma activity in mesenchymal stem cells regulates endochondral bone, angiogenesis, and B lymphopoiesis. J. Bone Miner. Res. 2018;33:2202–2213. doi: 10.1002/jbmr.3558. [DOI] [PubMed] [Google Scholar]
- Jilka R.L., Weinstein R.S., Bellido T., Parfitt A.M., Manolagas S.C. Osteoblast programmed cell death (apoptosis): modulation by growth factors and cytokines. J. Bone Miner. Res. 1998;13:793–802. doi: 10.1359/jbmr.1998.13.5.793. [DOI] [PubMed] [Google Scholar]
- Julien A., Kanagalingam A., Martínez-Sarrà E., Megret J., Luka M., Ménager M., Relaix F., Colnot C. Direct contribution of skeletal muscle mesenchymal progenitors to bone repair. Nat. Commun. 2021;12:2860. doi: 10.1038/s41467-021-22842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe A.P., Ramasamy S.K., Adams R.H. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature. 2014;507:323–328. doi: 10.1038/nature13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe A.P., Ramasamy S.K., Starsichova A., Adams R.H. Sample preparation for high-resolution 3D confocal imaging of mouse skeletal tissue. Nat. Protoc. 2015;10:1904–1914. doi: 10.1038/nprot.2015.125. [DOI] [PubMed] [Google Scholar]
- Lee Y.C., Chan Y.H., Hsieh S.C., Lew W.Z., Feng S.W. Comparing the osteogenic potentials and bone regeneration capacities of bone marrow and dental pulp mesenchymal stem cells in a rabbit calvarial bone defect model. Int. J. Mol. Sci. 2019;20:E5015. doi: 10.3390/ijms20205015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Qin W., Ding D., Tomczak N., Geng J., Liu R., Liu J., Zhang X., Liu H., Liu B., et al. Photostable fluorescent organic dots with aggregation-induced emission (AIE dots) for noninvasive long-term cell tracing. Sci. Rep. 2013;3:1150. doi: 10.1038/srep01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li Z., Hu D., Wang S., Zha M., Lu S.B., Sheng Z., Li K. Targeted NIR-II emissive nanoprobes for tumor detection in mice and rabbits. Chem. Commun. 2021;57:6420–6423. doi: 10.1039/d1cc01804c. [DOI] [PubMed] [Google Scholar]
- Li Z., Liao W., Zhao Q., Liu M., Xia W., Yang Y., Shao N. Angiogenesis and bone regeneration by allogeneic mesenchymal stem cell intravenous transplantation in rabbit model of avascular necrotic femoral head. J. Surg. Res. 2013;183:193–203. doi: 10.1016/j.jss.2012.11.031. [DOI] [PubMed] [Google Scholar]
- Liu C., Cabahug-Zuckerman P., Stubbs C., Pendola M., Cai C., Mann K.A., Castillo A.B. Mechanical loading promotes the expansion of primitive osteoprogenitors and organizes matrix and vascular morphology in long bone defects. J. Bone Miner. Res. 2019;34:896–910. doi: 10.1002/jbmr.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Carrera R., Flamini V., Kenny L., Cabahug-Zuckerman P., George B.M., Hunter D., Liu B., Singh G., Leucht P., et al. Effects of mechanical loading on cortical defect repair using a novel mechanobiological model of bone healing. Bone. 2018;108:145–155. doi: 10.1016/j.bone.2017.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Feng G., Tang B.Z., Liu B. Recent advances of AIE light-up probes for photodynamic therapy. Chem. Sci. 2021;12:6488–6506. doi: 10.1039/d1sc00045d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaval L., Liu F., Roche P., Aubin J.E. Kinetics of osteoprogenitor proliferation and osteoblast differentiation in vitro. J. Cell. Biochem. 1999;74:616–627. [PubMed] [Google Scholar]
- McDermott A.M., Herberg S., Mason D.E., Collins J.M., Pearson H.B., Dawahare J.H., Tang R., Patwa A.N., Grinstaff M.W., Kelly D.J., et al. Recapitulating bone development through engineered mesenchymal condensations and mechanical cues for tissue regeneration. Sci. Transl. Med. 2019;11:eaav7756. doi: 10.1126/scitranslmed.aav7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J., Hong Y., Lam J.W.Y., Qin A., Tang Y., Tang B.Z. Aggregation-induced emission: the whole is more brilliant than the parts. Adv. Mater. 2014;26:5429–5479. doi: 10.1002/adma.201401356. [DOI] [PubMed] [Google Scholar]
- Menezes K., Rosa B.G., Freitas C., da Cruz A.S., de Siqueira Santos R., Nascimento M.A., Alves D.V.L., Bonamino M., Rossi M.I., Borojevic R., et al. Human mesenchymal stromal/stem cells recruit resident pericytes and induce blood vessels maturation to repair experimental spinal cord injury in rats. Sci. Rep. 2020;10:19604. doi: 10.1038/s41598-020-76290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitlak B.H., Finkelman R.D., Hill E.L., Li J., Martin B., Smith T., D'Andrea M., Antoniades H.N., Lynch S.E. The effect of systemically administered PDGF-BB on the rodent skeleton. J. Bone Miner. Res. 1996;11:238–247. doi: 10.1002/jbmr.5650110213. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Ni J.S., Li Y., Yue W., Liu B., Li K. Nanoparticle-based cell trackers for biomedical applications. Theranostics. 2020;10:1923–1947. doi: 10.7150/thno.39915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oryan A., Kamali A., Moshiri A., Baghaban Eslaminejad M. Role of mesenchymal stem cells in bone regenerative medicine: what is the evidence? Cells Tissues Organs. 2017;204:59–83. doi: 10.1159/000469704. [DOI] [PubMed] [Google Scholar]
- Pajarinen J., Lin T., Gibon E., Kohno Y., Maruyama M., Nathan K., Lu L., Yao Z., Goodman S.B. Mesenchymal stem cell-macrophage crosstalk and bone healing. Biomaterials. 2019;196:80–89. doi: 10.1016/j.biomaterials.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanca M., Marco M., Ruspi M.L., Cristofolini L. Full-field strain distribution in multi-vertebra spine segments: an in vitro application of digital image correlation. Med. Eng. Phys. 2018;52:76–83. doi: 10.1016/j.medengphy.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Ramasamy S.K., Kusumbe A.P., Schiller M., Zeuschner D., Bixel M.G., Milia C., Gamrekelashvili J., Limbourg A., Medvinsky A., Santoro M.M., et al. Blood flow controls bone vascular function and osteogenesis. Nat. Commun. 2016;7:13601. doi: 10.1038/ncomms13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruspi M.L., Palanca M., Faldini C., Cristofolini L. Full-field in vitro investigation of hard and soft tissue strain in the spine by means of Digital Image Correlation. Muscles Ligaments Tendons J. 2017;7:538–545. doi: 10.11138/mltj/2017.7.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawall S., Beckendorf J., Amato C., Maier J., Backs J., Vande Velde G., Kachelrieß M., Kuntz J. Coronary micro-computed tomography angiography in mice. Sci. Rep. 2020;10:16866. doi: 10.1038/s41598-020-73735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebach C., Henrich D., Schaible A., Relja B., Jugold M., Bönig H., Marzi I. Cell-based therapy by implanted human bone marrow-derived mononuclear cells improved bone healing of large bone defects in rats. Tissue Eng. Part A. 2015;21:1565–1578. doi: 10.1089/ten.TEA.2014.0410. [DOI] [PubMed] [Google Scholar]
- Sivaraj K.K., Jeong H.-W., Dharmalingam B., Zeuschner D., Adams S., Potente M., Adams R.H. Regional specialization and fate specification of bone stromal cells in skeletal development. Cell Rep. 2021;36:109352. doi: 10.1016/j.celrep.2021.109352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Liu G., Liu X., Zhou Y., Sun Q., Zhen G., Wang X., Hu Y., Gao P., Demehri S., et al. Angiogenesis stimulated by elevated PDGF-BB in subchondral bone contributes to osteoarthritis development. JCI Insight. 2020;5:135446. doi: 10.1172/jci.insight.135446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swijnenburg R.J., Schrepfer S., Cao F., Pearl J.I., Xie X., Connolly A.J., Robbins R.C., Wu J.C. In vivo imaging of embryonic stem cells reveals patterns of survival and immune rejection following transplantation. Stem Cells Dev. 2008;17:1023–1029. doi: 10.1089/scd.2008.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztefek P., Vanleene M., Olsson R., Collinson R., Pitsillides A.A., Shefelbine S. Using digital image correlation to determine bone surface strains during loading and after adaptation of the mouse tibia. J. Biomech. 2010;43:599–605. doi: 10.1016/j.jbiomech.2009.10.042. [DOI] [PubMed] [Google Scholar]
- Takarada T., Nakazato R., Tsuchikane A., Fujikawa K., Iezaki T., Yoneda Y., Hinoi E. Genetic analysis of Runx2 function during intramembranous ossification. Development. 2016;143:211–218. doi: 10.1242/dev.128793. [DOI] [PubMed] [Google Scholar]
- Thompson M.S., Schell H., Lienau J., Duda G.N. Digital image correlation: a technique for determining local mechanical conditions within early bone callus. Med. Eng. Phys. 2007;29:820–823. doi: 10.1016/j.medengphy.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang Y., Gou W., Lu Q., Peng J., Lu S. Role of mesenchymal stem cells in bone regeneration and fracture repair: a review. Int. Orthop. 2013;37:2491–2498. doi: 10.1007/s00264-013-2059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk K., Yeh S.-C.A., Mortensen L.J., Ghaffarigarakani S., Lombardo C.M., Bassir S.H., Aldawood Z.A., Lin C.P., Intini G. Postnatal calvarial skeletal stem cells expressing PRX1 reside exclusively in the calvarial sutures and are required for bone regeneration. Stem Cell Rep. 2017;8:933–946. doi: 10.1016/j.stemcr.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Cui Z., Wang L., Xia Z., Hu Y., Xian L., Li C., Xie L., Crane J., Wan M., et al. PDGF-BB secreted by preosteoclasts induces angiogenesis during coupling with osteogenesis. Nat. Med. 2014;20:1270–1278. doi: 10.1038/nm.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Yu W., Niibe K., Zhang W., Egusa H., Tang T., Jiang X. The effects of platelet-derived growth factor-BB on bone marrow stromal cell-mediated vascularized bone regeneration. Stem Cells Int. 2018;2018:3272098. doi: 10.1155/2018/3272098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Ou G., Hamrick M., Hill W., Borke J., Wenger K., Chutkan N., Yu J., Mi Q.S., Isales C.M., et al. Age-related changes in the osteogenic differentiation potential of mouse bone marrow stromal cells. J. Bone Miner. Res. 2008;23:1118–1128. doi: 10.1359/JBMR.080304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. UNet++: a nested U-net architecture for medical image segmentation. Deep Learn. Med. Image Anal. Multimodal. Learn. Clin. Decis. Support. 2018;11045:3–11. doi: 10.1007/978-3-030-00889-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.