Abstract
Chlamydomonas reinhardtii transitions from mitotically dividing vegetative cells to sexually competent gametes of two distinct mating types following nutrient deprivation. Gametes of opposite mating type interact via their cilia, initiating an intraciliary signaling cascade and ultimately fuse forming diploid zygotes. The process of gametogenesis is genetically encode, and a previous study revealed numerous significant changes in mRNA abundance during this life-cycle transition. Here we describe a proteomic analysis of cilia derived from vegetative and gametic cells of both mating types in an effort to assess the global changes that occur within the organelle during this process. We identify numerous membrane- and/or matrix-associated proteins in gametic cilia that were not detected in cilia from vegetative cells. This includes the pro-protein from which the GATI-amide gametic chemotactic modulator derives, as well as receptors, a dynamin-related protein, ammonium transporters, two proteins potentially involved in the intraciliary signaling cascade-driven increase in cAMP, and multiple proteins with a variety of interaction domains. These changes in ciliary composition likely directly affect the functional properties of this organelle as the cell transitions between life-cycle stages.
INTRODUCTION
Cilia are highly conserved microtubule-based cellular extensions that function as motile, sensory, and secretory organelles. These structures are of ancient origin, dating to before the divergence from the last eukaryotic common ancestor (Satir et al., 2008), and are found in a very broad array of extant organisms (Carvalho-Santos et al., 2011; Kumar et al., 2019). Cilia are extremely complex, and upward of 1000 different types of proteins are estimated to be involved in their formation and function (van Dam et al., 2019); this represents about 5% of the coding capacity of the human genome. In addition, cilia have a membrane lipid content distinct from that of the plasma membrane (Garcia et al., 2018). In mammals, defects in ciliary formation, motility, and/or signaling lead to numerous developmental defects and complex syndromes (ciliopathies) that may involve multiple organs, and have phenotypes ranging from infertility and polycystic kidneys to skeletal and neurological malformations, epilepsy, and insulin resistance (Reiter and Leroux, 2017).
The unicellular green alga Chlamydomonas reinhardtii has proven to be an exceptional system in which to study the organization and function of cilia, as it is readily amenable to genetic, biochemical, and physiological analysis (see various chapters in Witman, 2009). This organism grows as a haploid, with vegetative cells dividing mitotically. Following nutrient deprivation, these cells undergo a developmental process that results in formation of pregametes of two different genetically determined mating types (termed minus and plus); pregametes become sexually competent following exposure to blue light, which is detected by phototropin (Huang and Beck, 2003). These gametes exhibit opposite chemotactic responses to an amidated peptidergic signal; the peptide attracts minus gametes but repels plus gametes (Luxmi et al., 2019). As chemotaxing gametes retain their cell walls, it is quite possible that the receptor(s) responsible for signaling alterations in dynein-driven motility reside in the ciliary membrane. Gametes of opposite mating type initially interact via their cilia, which leads to complex intraciliary signaling and an increase in cAMP (reviewed in Snell and Goodenough, 2009). Ultimately, the gametes undergo cell fusion and form a quadriciliate cell that further develops into a diploid zygote (Figure 1a). Once nutrients are again available, zygotes pass through meiosis and hatch releasing haploid progeny (Sasso et al., 2018).
FIGURE 1:
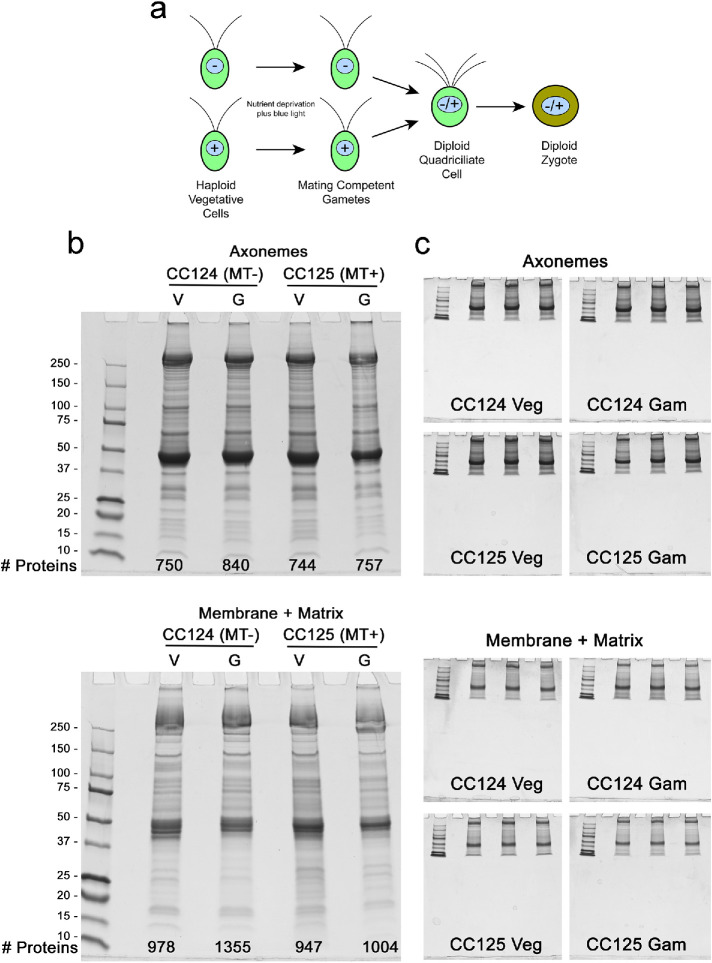
Cilia samples used for mass spectrometry. a) Schematic illustrating the transition from vegetative to gametic Chlamydomonas cells, and formation of a diploid quadriciliate cell that subsequently develops into a zygote. Mating type is indicated by – and + signs within the nucleus. b) Axoneme and detergent-soluble membrane plus matrix samples from vegetative (Veg) and gametic (Gam) cilia of wild-type Chlamydomonas of both mating types (CC124 and CC125) were electrophoresed in 4–15% gradient gels and stained with Coomassie blue; in combination, the amounts loaded for membrane plus matrix and axoneme fractions of each sample were 50 μg cilia. The number of proteins identified in each sample is indicated at the bottom of each gel. c) Additional aliquots of the samples shown in panel b were run in triplicate using a short gel format and stained with Coomassie blue. Following imaging, the protein-containing gel segments were excised, trypsinized, and subjected to mass spectral analysis.
Although cilia on both vegetative and gametic cells are motile and power cell locomotion, they exhibit different responses to the chemotactic GATI-amide peptide (Luxmi et al., 2019), differential localization of adhesion molecules (Goodenough and Heuser, 1999), and altered intraciliary signaling (Pan and Snell, 2000). Thus, ciliary composition likely changes at various life-cycle stages. Although a comprehensive whole-cell transcriptomic analysis of mRNA abundance during gametogenesis in Chlamydomonas is available (Ning et al., 2013), a corresponding examination of proteins present specifically in gametic cilia, where release of the amidated ectosome-associated chemotactic modulator (Luxmi et al., 2019) and the initial steps of cell–cell contact and signaling occur during mating (Pan et al., 2003), has not been reported. Here we analyze the composition of cilia isolated from vegetative and gametic cells of both mating types to assess developmentally driven changes in ciliary composition. Our data reveal numerous alterations, especially in detergent-soluble components that likely directly affect ciliary activity, thereby altering organellar functions and responses at these life stages.
RESULTS AND DISCUSSION
Global analysis of vegetative and gametic cilia proteomes
To assess compositional differences that occur during gametogenesis, cilia were isolated from vegetative and gametic Chlamydomonas cells of both mating types and fractionated into a detergent-soluble membrane plus matrix fraction and an axonemal fraction; protein concentrations and sample volumes were carefully adjusted so that the same total cilia mass was analyzed for each preparation. These samples were then electrophoresed, stained with Coomassie blue, trypsinized, and subjected to LC-MS/MS analysis. The protein content of each sample is illustrated in Figure 1b, and the actual triplicate “short gel” samples used for mass spectrometry are shown in Figure 1c. A global analysis of the proteins identified and how many were unique to a particular fraction is provided in Supplemental Table S1. The complete dataset indicating the normalized total spectral counts for every protein identified in each replicate is provided in Supplemental Table S2.
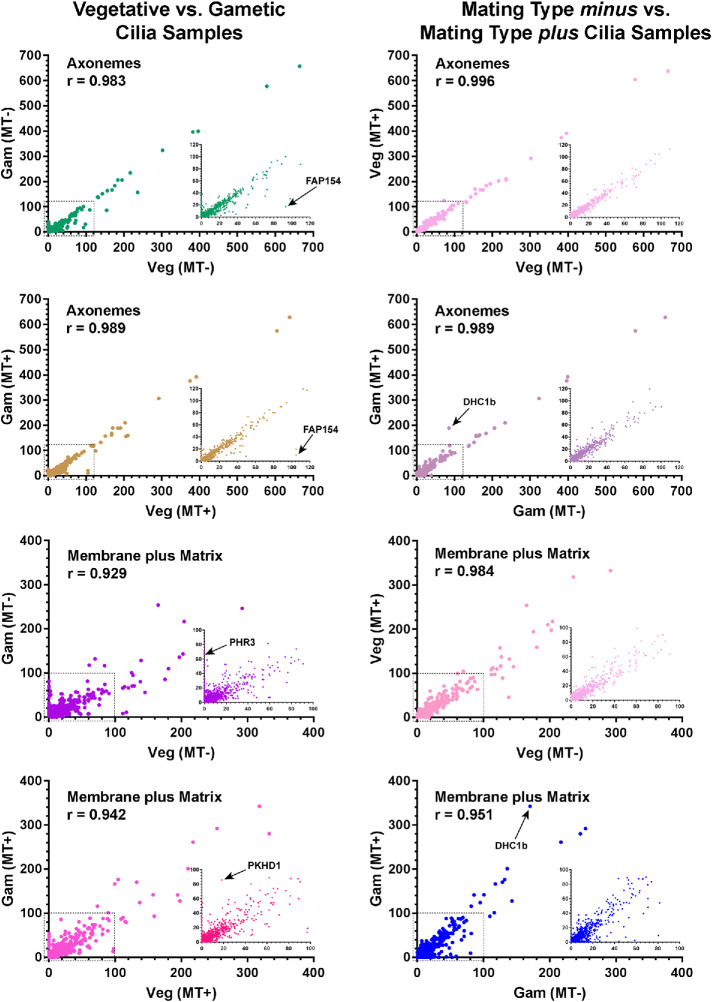
A global comparison of the abundance of every identified protein in vegetative and gametic cilia from both mating types and the Pearson correlation coefficients (r) between vegetative/gametic and mating type minus/plus datasets are shown in Figure 2. Axonemal samples show a very high degree of consistency, with few proteins deviating from an approximate 1:1 ratio. This provides a direct measure of the equivalence of cilia amounts between the samples. In contrast, considerably more variation is evident in the detergent extracts.
FIGURE 2:
Relative abundance of proteins in vegetative and gametic cilia. The abundance of each protein (normalized total spectral count; average of n = 3) identified in vegetative and gametic cilia from both mating type minus and plus cells is represented in scatterplots; the Pearson correlation coefficients (r) for each comparative analysis are shown. The insets show expanded views of the regions indicated by the dashed boxes. The abundance of nearly all axonemal proteins is highly correlated between samples. Several proteins that show highly distinct abundance changes are indicated: FAP154 (Cre08.g362100) contains a PAS domain; PHR3 (Cre12.g532850) is a multipass transmembrane protein in the ionotropic glutamate receptor family; and PKHD1 (Cre07.g340450) is a signal peptide–containing transmembrane protein with pectin lyase, G8, and immunoglobulin-like folds. Intriguingly, the dynein heavy chain (DHC1b) that powers retrograde intraflagellar transport shows a significant increase in mating type plus gamete cilia. The major membrane glycoprotein FMG1 is not shown on the membrane plus matrix plots, as it is present at >threefold the amount of any other protein, and thus its inclusion greatly distorts the data by forcing all other points much closer to the ordinate. A single data point in the mating type minus vegetative vs. gametic membrane comparison is obscured by the inset.
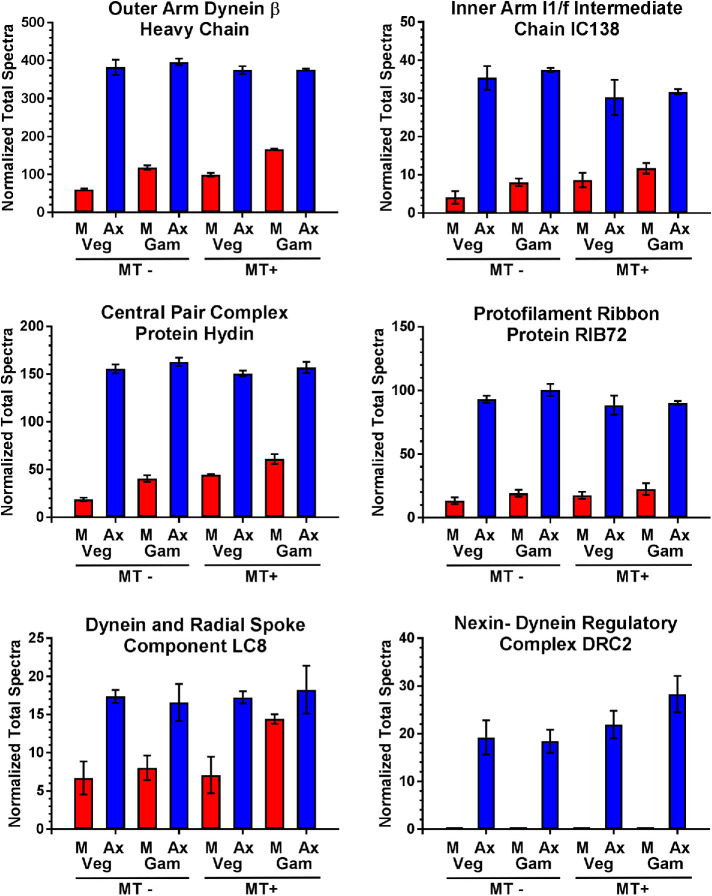
To assess intersample consistency of the numeric data, we analyzed the abundance of multiple core axonemal components required for normal ciliary motility that would not be expected to vary either between vegetative and gametic cilia or as a function of mating type. These include components of the inner and outer dynein arms, radial spokes, protofilament ribbons, nexin–dynein regulatory complex, and central pair microtubule complex (Figure 3). These proteins show highly consistent abundance levels between the cilia preparations and their replicates with small standard deviations. One exception is LC8, which is part of multiple axonemal complexes and shows enhanced levels in the mating type plus gametic cilia detergent extract; this correlates well with an increase in the dynein heavy chain that powers retrograde intraflagellar transport in the same fraction (Figure 2). Furthermore, although both α- and β-tubulins (Cre04.g216850 and Cre12.g549550) were present in large and consistent amounts, only two peptides were identified in a single sample for ε-tubulin (Cre03.g172650) and none for either γ- (Cre06.g299300) or δ-tubulin (Cre03.g187350); these three proteins are known to be in the basal body, and in the case of γ-tubulin also the transition zone, but generally absent from the cilium proper e.g. (Dutcher and Trabuco, 1998; Silflow et al., 1999; Dutcher et al., 2002).
FIGURE 3:
Spectral count consistency between cilia samples. The total normalized spectral counts for six components of core axonemal substructures identified in membrane plus matrix (M) and axonemal (Ax) fractions of cilia from CC124 (MT-) and CC125 (MT+) vegetative and gametic cells are shown. The data are highly consistent between ciliary samples, except for an increase in LC8 in the plus gamete cilia detergent extract samples. Note that no peptides for DRC2 were identified in any membrane plus matrix sample. Plots show mean ± SD (n = 3).
Multiple proteins are exclusively present in gametic cilia
We next asked what proteins were exclusively found in gametic and not vegetative cell cilia, as this might highlight key signaling and/or regulatory pathways. The initial cutoff criterion used was stringent, requiring that zero peptides for a protein be found in any of the 12 vegetative cilia samples examined (i.e., membrane/matrix and axonemal samples from both mating types analyzed in triplicate). A total of 33 proteins met this criterion (Table 1), including a dynamin-related protein, which contains the dynamin central and effector domains plus a plekstrin homology region but lacks the GTPase motif, two annotated ammonium transporters, and several putative receptors. Importantly, only gametic samples contained the GATI-amide chemotactic modulator (Cre03.g204500), which is present in gametic cilia (Luxmi et al., 2019) and essentially not expressed in vegetative cells (Ning et al., 2013). Proteins present in both vegetative and gametic cilia but whose abundance changed ≥5-fold between these samples are indicated in Supplemental Tables S3 and S4; these include a dynamin-related GTPase, a RAN GTPase activating protein, multiple subunits of the T-complex chaperonin, ion channels, and putative receptors.
TABLE 1:
Proteins identified only in gametic ciliaa.
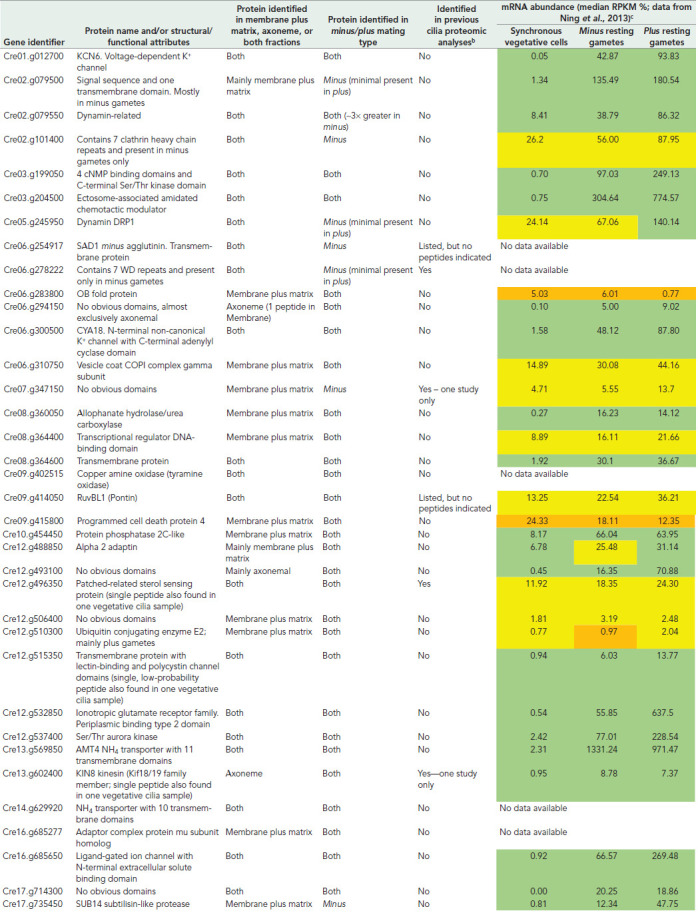
|
aAlso included in this table are three intriguing proteins (Dynamin DRP1, a Patched-related receptor, and the Kin8 kinesin) for which a single peptide was found in one of 12 vegetative cilia samples.
bThe results from previous proteomic analyses of Chlamydomonas cilia published by Pazour et al. (2005), Wang et al. (2017), Jordan et al. (2018), Picariello et al. (2019), Zhao et al. (2019), and Dai et al. (2020) are tabulated at http://chlamyfp.org/.
cThese data are from Ning et al. (2013) and were obtained using the 21gr (mating type plus) and 6145C (mating type minus) wild-type strains. The color code is green = >fourfold abundance increase; yellow = abundance increase, but <four-fold; orange = no significant increase.
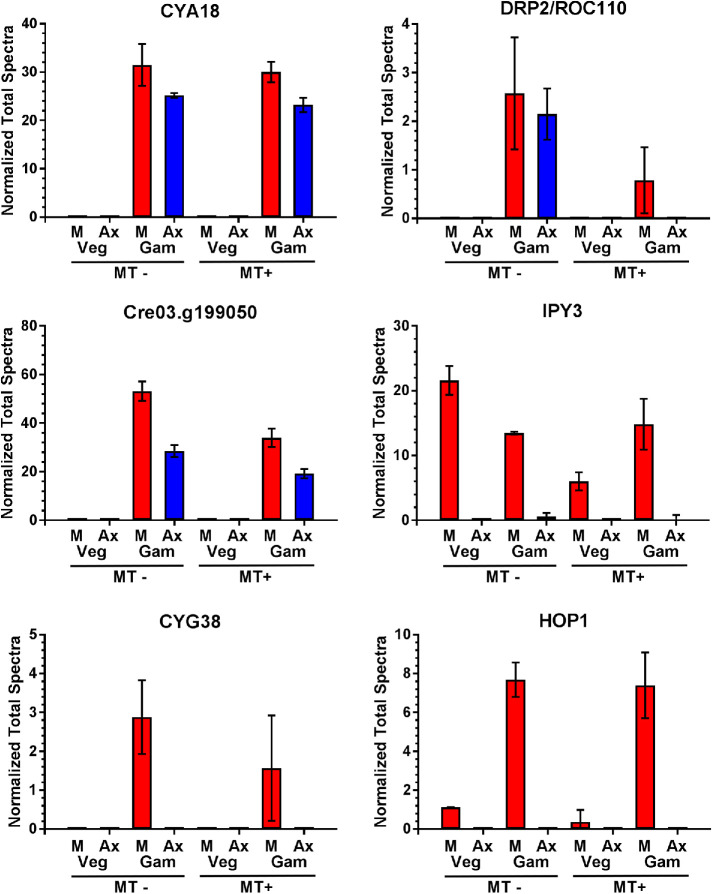
Also identified only in gametic cilia were two proteins with domains suggesting a potential role in the signaling pathway, where ciliary adhesion leads to an intraciliary kinase cascade and ultimately an increase in cellular cAMP that presages cell fusion and zygote formation (Figure 4 and Table 1). One (Cre03.g199050) contains four N-terminal cNMP binding domains and a C-terminal Ser/Thr kinase module and thus might be a downstream effector following cAMP increase. The second is CYA18 (Cre06.g300500), which consists of a noncanonical K+ channel followed by an adenylyl cyclase domain. A similar protein was previously identified in ciliates and Plasmodium and demonstrated to exhibit both ion channel properties and cyclase activity (Schultz et al., 1992; Weber et al., 2004); BLAST searches also reveal CYA18 orthologs in haptophytes, cryptophytes, and stramenopiles, but not opisthokonts or excavates. CYA18 is the most abundant channel and cyclase in gametic cilia and may represent a cyclase activated during mating-induced signaling. The presence of an N-terminal channel domain in CYA18 indicates a potentially direct connection between cAMP formation and K+ flux that might play an important role in gametic ciliary signaling. This concept can be tested readily, as inward K+ flux is easily controlled by external application of specific channel blocking agents. An additional (putative guanylate) cyclase (CYG38) was also present in low abundance only in gametic cilia.
FIGURE 4:
Proteins with varying ciliary localization following gametogenesis. The total normalized spectral counts are plotted for proteins identified in the various samples. Examples of several proteins present exclusively in gametic cilia (CYA18, CYG38, DRP2/ROC110, and Cre03.g199050), and of proteins mostly present in gametic cilia (HOP1) or found in both vegetative and gametic cilia (the IPY3 pyrophosphatase) are shown. Plots show mean ± SD (n = 3).
Generation of 3′–5′ cyclic nucleotides from NTPs releases pyrophosphate, which must be hydrolyzed to generate a sufficiently negative change in ΔG’ so that the back reaction does not occur. Intriguingly, a single pyrophosphatase (IPY3) was present in the membrane/matrix fractions of both vegetative and gametic cilia samples; this implies that there may be multiple pathways (e.g., ubiquitin activation) that lead to ciliary generation of pyrophosphate that must be hydrolyzed. Although not quite making the zero peptide cutoff, abundance of the cochaperone Sti1p/HOP1 that interacts with and regulates HSP70/HSP90 also increased dramatically in gametic cilia (Figure 4).
Several proteins were mainly or exclusively present in gametic samples of a single mating type, such as the minus agglutinin (Cre06.g254917) that localizes to the ciliary membrane and mediates direct associations with agglutinin molecules on the surface of plus gamete cilia (Ferris et al., 2005). Intriguingly, the plus agglutinin was not identified in any sample, which may reflect low sequence complexity (it is 12% proline, 18% alanine, and 12% glycine) and/or very different abundance levels in resting gametes; indeed, only a few peptides for this protein were identified previously in ectosomes from actively mating gametes (Luxmi et al., 2019). Several other mating type–specific proteins are more enigmatic. Cre02.g079500 encodes a protein with a canonical signal sequence placing it in the secretory pathway and a C-terminal transmembrane segment. It is essentially Chlamydomonas-specific and only weak homology (∼35% identity) was found even in volvocine relatives. Cre06.g278222 consists of seven WD repeats and exhibits 76% identity to the vertebrate receptor for activated protein kinase C 1-like (RACK1) adaptor that is involved in protein kinase C signaling. In addition, a clathrin heavy chain (Cre02.g101400) was identified that is part of a coexpression cluster associated with mating activation induced in minus gametes (Molla-Herman et al., 2010); clathrin has been reported to be present at the ciliary pocket (Clement et al., 2013) and to affect ciliary assembly. In contrast, an E2 ubiquitin-conjugating enzyme UBC21 encoded at Cre12.g510300 was present in plus gamete cilia but apparently absent from vegetative cell cilia.
Previous proteomic studies of Chlamydomonas vegetative cell cilia have defined the core components of these organelles; see for example (Pazour et al., 2005) and http://chlamyfp.org. With a few exceptions, the proteins identified here only in gametic cilia have not been found previously in Chlamydomonas cilia (Table 1). There are also examples of proteins present in vegetative cell cilia that are missing following gametogenesis. One intriguing example is UMM7 (Cre08.g380000), which contains two enzymatic cores—an N-terminal 5-histidylcysteine sulfoxide synthase and a C-terminal S-adenosylmethionine-dependent methyltransferase. Predictably, this enzyme likely synthesizes an ovothiol, such as 1-N-methyl-4-mercaptohistidine or a closely related compound. Ovothiols are strongly reducing antioxidants found in trypanosomes, diatoms, euglenoids, and the eggs of marine invertebrates, where they scavenge hydrogen peroxide and potentially form mixed disulfides with exposed cysteine residues of proteins (Castellano and Seebeck, 2018). As dynein-driven ciliary motility and the sign of phototactic behavior are both controlled by changes in redox poise (Wakabayashi and King, 2006; Wakabayashi et al., 2011), defining how ovothiols affect the redox state of the cilium may provide essential insight into this regulatory process.
Numerous flagellar-associated proteins exhibit altered abundance in gametic versus vegetative cell cilia
The original Chlamydomonas ciliary proteome identified numerous proteins of unknown function that were given the generic designation FAP (flagellar-associated protein; Pazour et al., 2005). We find that several show dramatic decreases in abundance following gametogenesis, while others increase considerably. These proteins were solubilized, either partially or completely, by detergent treatment, indicating that they are components of the membrane and/or the soluble ciliary matrix and not integral axonemal proteins. This supports the concept that the ciliary membrane undergoes multiple alterations in functional specialization as cells transition to mating competency.
Four of these proteins appear to be membrane-associated. FAP24 contains a signal sequence placing it on the external face of the ciliary membrane; levels are reduced by ∼50% in gametic cilia. It also has a TroA-like domain that usually function as ferric siderophores and/or are involved in binding transition metals such as Mn2+, Cu2+, and Zn2+. FAP102 exhibits a more complex abundance pattern. Although present and essentially unchanging in vegetative and gametic plus mating type cilia, FAP102 is almost completely absent from mating type minus vegetative cilia and increases more than 40-fold during gametogenesis. FAP102 contains a signal sequence but no obvious structural domains and is redox active (Wakabayashi and King, 2006). Two transmembrane proteins, FAP154 and FAP49, which are almost identical (except at the extreme termini) and encoded by adjacent genes, are abundant in vegetative cell cilia and decrease by ∼5-fold following gametogenesis. Both have seven N-terminal transmembrane domains, a central PAS domain, and a C-terminal region containing at least four additional transmembrane segments; PAS domains act as environmental sensors, often of light, oxygen, or other small molecules.
Multiple FAPs that are reduced and/or absent in gametic cilia contain protein–protein interaction motifs including ankyrin repeats (FAP26 and FAP79), armadillo/β-catenin-like repeats (FAP28), tetratricopeptide repeats (FAP185), and a prefoldin-like domain (FAP88). In addition, FAP280 is reduced ∼10-fold in gametes. This membrane/matrix-associated protein contains a multiprotein bridging factor domain found in a wide range of eukaryotes and is potentially involved in transcriptional coactivation. Finally, two additional proteins (FAP177 and FAP181) with no obvious domains are both reduced following gametogenesis.
Several proteins were found almost exclusively in gametic axonemes. For example, Cre10.g423600 encodes a protein containing an N-terminal NTPase domain, 3 EF-hands that are predicted to bind Ca2+, and 11 C-terminal WD repeats. This increased ∼5-fold in minus gamete cilia samples and over 10-fold in plus gamete samples. Similarly, the Cre06.g294150 protein, which has no obvious domains and multiple long regions of low complexity, was not found in any vegetative cilia sample.
Changes in ciliary receptors following gametogenesis
Chlamydomonas contains almost 150 genes annotated as encoding various receptors including members of the scavenger, ionotropic glutamate, seven-transmembrane domain (listed as G protein–coupled receptor-related), patched-related, Toll-like, and lectin-binding receptor families, as well as the blue light receptor phototropin (Huang et al., 2004; Merchant et al., 2007; Luxmi et al., 2019).
We identified a patched-related receptor (Cre12.g496350) almost exclusively in gametic cilia; however, several previous analyses also found evidence for this protein in vegetative cilia (Jordan et al., 2018; Picariello et al., 2019; Zhao et al., 2019), and its mRNA expression levels only change by ∼2-fold during gametogenesis (Ning et al., 2013; see Table 1). The abundance of a “GPCR-related” protein (Cre13.g604050) with extracellular pectate lyase repeats likely involved in carbohydrate binding showed a similar gamete-specific pattern with only a single peptide found in one vegetative cilia sample; this putative receptor has not been identified in cilia previously. Importantly, although the topology of this protein is superficially similar to that of GPCRs, Chlamydomonas lacks canonical Gαβγ subunits (Merchant et al., 2007; Urano et al., 2012). An additional transmembrane protein (Cre12.g532850) in the ionotropic glutamate receptor family and containing a type 2 periplasmic binding fold often present in small molecule sensors was present only in gametic cilia; this correlates well with the large increase in mRNA expression seen in gametic samples (Ning et al., 2013 and Table 1). In contrast, peptides from a scavenger receptor (Cre05.g240700) and the blue light receptor phototropin required for the acquisition of mating competency decreased in gametic samples.
mRNA and protein abundance changes following gametogenesis
Whole-cell transcriptomic analyses are often used as a proxy for proteomic studies; if the mRNA increases the general assumption is that protein levels follow, although the comparative magnitude and timing of that increase is not always clear. Our analysis of the ciliary proteomics of gametogenesis combined with the detailed transcriptomic study of Ning et al. (2013) provides an opportunity to make a broad assessment of the correlation between these measures at the organelle level. The transcriptomic analysis measured whole cell mRNA levels in both asynchronous and synchronized vegetative cells and resting gametes, as well as gametes treated with lysin to remove the cell walls or activated with dibutyryl cAMP (Ning et al., 2013). One important caveat for this comparison is that the wild-type strains used in the mRNA analysis are different from those used here for proteomics, which may introduce strain-specific anomalies. Even so, in general there is a strong correlation between increased mRNA levels and the appearance of a protein in cilia following gametogenesis (Table 1). In a few cases, this correlation fails, which might result from experimental issues or incorrect identification of a protein as gamete-specific or be due to vegetative cells maintaining a store of the protein product in the cytoplasm and only relocating it to cilia following a gametogenic signal.
In conclusion, this comparative analysis has identified numerous developmental changes in ciliary composition and found both vegetative cell– and gamete-specific cilia components. Furthermore, it provides a resource for understanding functional specializations that occur in cilia from Chlamydomonas cells of different mating types following life-cycle stage transitions.
METHODS
Chlamydomonas culture
Wild-type Chlamydomonas reinhardtii strains CC124 (mating type minus) and CC125 (mating type plus) were grown in 2 × 1 l cultures to a density of ∼5 × 106 cells/ml in R medium containing acetate on a 12 h:12 h light/dark cycle and aerated with a 5:1 air to CO2 mixture. One culture of each strain was processed as described below to provide vegetative cell cilia samples. The second culture was harvested by centrifugation (Fiberlite F10 rotor, 1100 × g, 7 mins, 20°C), resuspended in M-N/5 medium and incubated overnight to induce gametogenesis. Successful gamete formation was assessed by mating and formation of quadriciliate cells. This second culture was then processed to provide the gametic cilia samples.
Cilia isolation and fractionation
Cilia isolation was performed following (Craige et al., 2013) with modifications. Cells were harvested by low speed centrifugation (Fiberlite F10 rotor, 1,100 × g, 7 min, 20°C),and washed three times for vegetative cells and twice for gametic cells with 10 mM HEPES pH 7.5. Cells were then resuspended in 30 mM HEPES pH 7.5, 5 mM MgSO4, 4% sucrose (HMS; 10 ml per tube) on ice; all solutions contained protease inhibitor cocktail (Sigma-Aldrich, P9599) and 1 mM DTT hereafter. Subsequently, 100 μl of 5.3% (wt./vol) CaCl2 was added to each tube followed by 2 ml of 25 mM dibucaine.HCl to induce deciliation, which was assessed by phase contrast microscopy. Deciliated cell bodies were collected by low speed centrifugation (1800 ×g, 5 min in a Sorvall ST8 centrifuge with a swing-out rotor).
The supernatant containing detached cilia was laid over a 25% sucrose solution made in 30 mM HEPES pH 7.5, 5 mM MgSO4. The tubes were then spun in a swing-out rotor (2400 × g, 10 min in a Sorvall ST8 centrifuge) to pellet any remaining cell bodies through the sucrose under layer. The top sucrose layer and the 4%/25% sucrose interface (the latter contains most of the cilia) were transferred into fresh tubes. Cilia were harvested by centrifugation in a Fiberlite F21S 8 × 50y rotor (30,000 × g, 20 min, 4°C). Pellets were resuspended in 0.5 ml of HMEK buffer (30 mM HEPES pH 7.5, 5 mM MgSO4, 0.5 mM EDTA, 25 mM KCl) and spun in a Fiberlite F21S 8 × 50y rotor (30,000 × g, 20 min, 4°C). Isolated cilia were resuspended in appropriate volumes of HMEK to adjust the concentration to 4 mg/ml. Protein concentration was determined with the BCA assay using BSA as standard. IGEPAL-CA630 detergent (Sigma-Aldrich, I3021) was then added to a final concentration of 1% (vol/vol) and extraction allowed to occur on ice for 30 mins with mixing every 10 mins. Following centrifugation (Fiberlite F21S 8 × 50y rotor, 30,000 × g, 20 mins, 4°C), the supernatant was removed and used as the membrane plus matrix sample, while the extracted axoneme pellet was resuspended in gel sample buffer directly.
Gel samples derived from equal total cilia mass equivalents (50 μg) were denatured at 80°C for 10 min and fractionated in 4–15% SDS gradient MINI-PROTEAN TGX polyacrylamide gels (Biorad, Hercules, CA). For mass spectrometry, samples were separated using a short gel protocol in which the sample entered only about 2 cm into the gel. Each sample was run in triplicate in a different electrophoresis unit to ensure crossover contamination did not occur. Following staining with newly prepared Coomassie blue, each lane was excised, subject to in-gel tryptic digestion and the peptide products analyzed by mass spectrometry (see below).
Mass spectrometry
Trypsinized cilia samples were spiked with 2 pmol trypsin-digested yeast alcohol dehydrogenase (ADH) for quality control and normalization. Mass spectral (LC-MS/MS) analysis was performed at the University of Massachusetts medical school mass spectrometry facility. Each sample was injected into a Thermo Scientific Q exactive quadrupole-Orbitrap hybrid mass spectrometer with Waters NanoAcquity ultra-performance liquid chromatography. Six ADH peptides were used to evaluate the acquisitions and yielded a mass error of ∼2 ppm. Data were searched against the Chlamydomonas genome v.5.5 with Mascot in Proteome Discoverer 2.1.1.21 using a fragment tolerance of 0.050 and a parent ion tolerance of 10 ppm; a maximum of two missed cleavages were allowed. Modifications assessed were carbamidomethyl on Cys, C-terminal Gly-loss plus amide, N-terminal pyroglutamylation on Gln, oxidation on Met, and N-terminal acetylation and phosphorylation on Ser, Thr, and Tyr. Protein threshold was set to 99.0% minimum with a two-peptide minimum; data were analyzed using Scaffold ver. 5.0.1.
Total normalized spectral count data were plotted using GraphPad Prism ver.7. For bar charts, mean ± SD is shown; for scatterplots of vegetative versus gametic cilia samples, the average spectral counts from n = 3 samples were used. Pearson correlation coefficients (r) were calculated using GraphPad Prism.
Domain Analysis
Protein domain organization was analyzed using SMART (http://smart.embl-heidelberg.de/), Prosite (https://prosite.expasy.org/), and cDART (https://www.ncbi.nlm.nih.gov/Structure/lexington/docs/ cdart_about.html). SignalP was employed to assess the presence of potential signal sequences (https://services.healthtech.dtu.dk/ service.php?SignalP-5.0), and TOPCONS used to predict membrane protein topology (https://topcons.net/; Tsirigos et al., 2015).
Supplementary Material
Acknowledgments
We thank Crysten Blaby-Haas (Brookhaven National Laboratory) for assistance with bioinformatics and Betty Eipper (University of Connecticut Health Center) for insightful comments. This study was supported by Grants RO1-GM051293 and R35-GM140631 from the National Institutes of Health.
Abbreviations used:
- FAP
flagellar-associated protein
- GPCR
G protein-coupled receptor
- LC
light chain
- PAS
Per-Arnt-Sim domain
Footnotes
This article was published online ahead of print in MBoC in Press (http://www .molbiolcell.org/cgi/doi/10.1091/mbc.E22-02-0033) on April 7, 2022.
REFERENCES
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M (2011). Tracing the origins of centrioles, cilia, and flagella. J Cell Biol 195, 341–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano I, Seebeck FP (2018). On ovothiol biosynthesis and biological roles: from life in the ocean to therapeutic potential. Natural Product Reports 35, 1241–1250. [DOI] [PubMed] [Google Scholar]
- Clement CA, Ajbro KD, Koefoed K, Vestergaard ML, Veland IR, Henriques de Jesus MPR, Pedersen LB, Benmerah A, Andersen CY, Larsen LA, Christensen ST (2013). TGF-β signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Reports 3, 1806–1814. [DOI] [PubMed] [Google Scholar]
- Craige B, Brown JM, Witman GB (2013). Isolation of Chlamydomonas flagella. Curr Protoc Cell Biol, Unit-3.41.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Ichikawa M, Peri K, Rebinsky R, Huy Bui K (2020). Identification and mapping of central pair proteins by proteomic analysis. Biophysics Physicobiol 17, 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK, Morrissette NS, Preble AM, Rackley C, Stanga J (2002). Epsilon-tubulin is an essential component of the centriole. Mol Biol Cell 13, 3859–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK, Trabuco EC (1998). The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol Biol Cell 9, 1293–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Waffenschmidt S, Umen JG, Lin H, Lee J-H, Ishida K, Kubo T, Lau J, Goodenough UW (2005). Plus and minus sexual agglutinins from Chlamydomonas reinhardtii. Plant Cell 17, 597–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G III, Raleigh DR, Reiter JF (2018). How the ciliary membrane Is organized inside-out to communicate outside-in. Curr Biol 28, R421–R434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE (1999). Deep-etch analysis of adhesion complexes between gametic flagellar membranes of Chlamydomonas reinhardtii (Chlorophyceae). J Phycol 35, 756–767. [Google Scholar]
- Huang K, Beck CF (2003). Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 100, 6269–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Kunkel T, Beck CF (2004). Localization of the blue-light receptor phototropin to the flagella of the green alga Chlamydomonas reinhardtii. Mol Biol Cell 15, 3605–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MA, Diener DR, Stepanek L, Pigino G (2018). The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat Cell Biol 20, 1250–1255. [DOI] [PubMed] [Google Scholar]
- Kumar D, Mains RE, Eipper BA, King SM (2019). Ciliary and cytoskeletal functions of an ancient monooxygenase essential for bioactive amidated peptide synthesis. Cell Mol Life Sci 76, 2329–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxmi R, Kumar D, Mains RE, King SM, Eipper BA (2019). Cilia-based peptidergic signaling. PLoS Biol 17, e3000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla-Herman A, Ghossoub R, Blisnick T, Meunier A, Serres C, Silbermann F, Emmerson C, Romeo K, Bourdoncle P, Schmitt A, et al. (2010). The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J Cell Sci 123, 1785–1795. [DOI] [PubMed] [Google Scholar]
- Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, et al. (2013). Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev 27, 1198–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Misamore MJ, Wang Q, Snell WJ (2003). Protein transport and signal transduction during fertilization in Chlamydomonas. Traffic 4, 452–459. [DOI] [PubMed] [Google Scholar]
- Pan JM, Snell WJ (2000). Signal transduction during fertilization in the unicellular green alga, Chlamydomonas. Curr Opin Microbiol 36, 596–602. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB (2005). Proteomic analysis of a eukaryotic cilium. J Cell Biol 170, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picariello T, Brown JM, Hou Y, Swank G, Cochran DA, King OD, Lechtreck K, Pazour GJ, Witman GB (2019). A global analysis of IFT-A function reveals specialization for transport of membrane-associated proteins into cilia. J Cell Sci 132, jcs.220749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR (2017). Genes and molecular pathways underpinning ciliopathies. Nature Rev Mol Cell Biol 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso S, Stibor H, Mittag M, Grossman AR (2018). From molecular manipulation of domesticated Chlamydomonas reinhardtii to survival in nature. Elife 7, e39233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P, Mitchell DR, Jékely G (2008). How did the cilium evolve? Curr Topics Dev Biol, 85, 63–82. [DOI] [PubMed] [Google Scholar]
- Schultz JE, Klumpp S, Benz R, Schürhoff-Goeters WJ, Schmid A (1992). Regulation of adenylyl cyclase from Paramecium by an intrinsic potassium conductance. Science 255, 600–603. [DOI] [PubMed] [Google Scholar]
- Silflow CD, Liu B, LaVoie M, Richardson EA, Palevitz BA (1999). γ-tubulin in Chlamydomonas: characterization of the gene and localization of the gene product in cells. Cell Motil 42, 285–297. [DOI] [PubMed] [Google Scholar]
- Snell WJ, Goodenough UW (2009). Flagellar adhesion, flagellar-generated signaling and gamete fusion during mating. In: The Chlamydomonas Sourcebook. Cell Motility and Behavior, vol. 3, ed. Witman GB. San Diego, CA: Academic Press, 369–394. [Google Scholar]
- Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A (2015). The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic acids research 43, W401–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones JC, Wang H, Matthews M, Bradford W, Bennetzen JL, Jones AM (2012). G protein activation without a GEF in the plant kingdom. PLoS Genetics 8, e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam TJP, Kennedy J, van der Lee R, de Vrieze E, Wunderlich KA, Rix S, Dougherty GW, Lambacher NJ, Li C, Jensen VL, et al. (2019). CiliaCarta: an integrated and validated compendium of ciliary genes. PLoS One 14, e0216705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, King SM (2006). Modulation of Chlamydomonas reinhardtii flagellar motility by redox poise. J Cell Biol 173, 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K-I, Misawa Y, Mochiji S, Kamiya R (2011). Reduction–oxidation poise regulates the sign of phototaxis in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 108, 11280–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gu L, Meng D, Wu Q, Deng H, Pan J (2017). Comparative proteomics reveals timely transport into cilia of regulators or effectors as a mechanism underlying ciliary disassembly. J Proteome Res 16, 2410–2418. [DOI] [PubMed] [Google Scholar]
- Weber JH, Vishnyakov A, Hambach K, Schultz A, Schultz JE, Linder JU (2004). Adenylyl cyclases from Plasmodium, Paramecium and Tetrahymena are novel ion channel/enzyme fusion proteins. Cellular Signalling 16, 115–125. [DOI] [PubMed] [Google Scholar]
- Witman GB (2009). The Chlamydomonas Sourcebook. Volume 3—Cell Motility and Behavior. Academic Press. [Google Scholar]
- Zhao L, Hou Y, Picariello T, Craige B, Witman GB (2019). Proteome of the central apparatus of a ciliary axoneme. J Cell Biol 218, 2051–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.