Abstract
Background:
The SOMAscan assay has an advantage over immunoassay-based methods because it measures a large number of proteins in a cost-effective manner. However, the performance of this technology compared to the routinely used immunoassay techniques needs to be evaluated.
Objective:
We performed comparative analyses of SOMAscan and immunoassay-based protein measurements for five cerebrospinal fluid (CSF) proteins associated with Alzheimer’s disease (AD) and neurodegeration: NfL, Neurogranin, sTREM2, VILIP-1 and SNAP-25.
Methods:
We compared biomarkers measured in ADNI (N=689), Knight-ADRC (N=870), DIAN (N=115), and Barcelona-1 (N=92) cohorts. Raw protein values were transformed using z-score in order to combine measures from the different studies. sTREM2 and VILIP-1 had more than one analyte in SOMAscan; all available analytes were evaluated. Pearson’s correlation coefficients between SOMAscan and immunoassays were calculated. Receiver operating characteristic curve and area under the curve were used to compare prediction accuracy of these biomarkers between the two platforms.
Results:
Neurogranin, VILIP-1 and NfL showed high correlation between SOMAscan and immunoassay measures (r > 0.9). sTREM2 had a fair correlation (r > 0.6), whereas SNAP-25 showed weak correlation (r = 0.06). Measures in both platforms provided similar predicted performance for all biomarkers except SNAP-25 and one of the sTREM2 analytes. sTREM2 showed higher AUC for SOMAscan based measures.
Conclusion:
Our data indicate that SOMAscan performs as well as immunoassay approaches for NfL, Neurogranin, VILIP-1 and sTREM2. Our study shows promise for using SOMAscan as an alternative to traditional immunoassay-based measures. Follow-up investigation will be required for SNAP-25 and additional established biomarkers.
Keywords: Alzheimer’s Disease, CSF biomarkers, Assays, SOMAscan, Correlation
Introduction
Immunoassays have been at the forefront of protein measurement with their use in research being reported as early as the 1950s [1]. The introduction of enzyme linked immunosorbent assay (ELISA) in the 1971 manuscript by Engvall and Perlmann helped further the popularity of immunoassays in protein measurements [2]. The basic principle of immunoassays relies on a binding reaction between a molecule and a highly specific antibody labeled with a quantifiable molecule. After several washing steps, the read out is based quantifiable molecule, i.e., an indirect measure of the analyte. The popularity of immunoassays, particularly ELISAs and related tests, can be attributed to their ease of use and high degree of sensitivity and specificity [3,4]. However, one limitation of this technique is that it can only measure a limited number of proteins at once, usually one protein, due to cross-reactivity issues [5]. This becomes a major hindrance, particularly in the case of complex traits where multiple proteins need to be evaluated. Thus, there has been an increased interest in high throughput, large scale proteomic measurement platforms in recent years [6].
SOMAscan, a multiplexed, modified aptamer-based protein measurement platform developed by SomaLogic, Inc., is one such promising technique [7]. The current version (v4.1) of the assay is capable of simultaneously measuring ~7,000 protein targets from a single 150 μl sample [8]. Although the volume of sample required for measurement is similar to traditional immunoassays, the fact that the traditional platforms require multiple aliquots to measure a comparable number of proteins adds to the overall cost of processing as well as the total sample volume required. Furthermore, Slow Offrate Modified Aptamers (SOMAmer), single stranded deoxy oligonucleotides used by SOMAscan, have been shown to have high specificity and reproducibility [7,9,10]. The scalable nature of these aptamers also allows for addition of new targets as more human proteins are discovered [7]. Despite these obvious advantages of SOMAscan over traditional immunoassay-based approaches, the novel nature of this technology warrants the need to evaluate its performance in comparison to that of more established approaches.
Efforts have been made to assess the inter-platform relation between the two technologies. Raffield et al., (2020) reported Spearman correlation coefficients from −0.13 to 0.97 (median ~ 0.5) for 63 proteins measured by SOMAscan and multiplex immunoassays [6]. Another study, based on plasma and urine biomarkers of acute kidney injury in 54 cardiac surgery patients, found correlation of around 0.75 between the two platforms [11]. While these findings provide an encouraging backdrop, these comparisons have been mostly focused on plasma and serum-based samples and we cannot directly infer the agreement of the protein measures between the platforms in the context of cerebrospinal fluid (CSF) biomarkers or in neurologic cohorts. Given the importance of CSF biomarkers in neurodegenerative diseases, it is essential that we assess the similarity between CSF biomarker measurements from the two platforms. In addition, the variation in strength of correlation observed for different biomarkers also raises questions regarding the generalization of these findings in the settings of other disease conditions.
In this study, we compare the performance of SOMAscan and immunoassay-based platforms in five CSF proteins associated with AD and neurodegeneration in a multi-cohort setting. The five biomarkers of interest are neurofilament light chain protein (NfL), neurogranin, visinin like protein 1 (VILIP-1), soluble triggering receptor expressed on myeloid cells 2 (sTREM2) and synaptosomal-associated protein 25 (SNAP-25). These biomarkers were chosen based on their importance in AD and neurodegeneration as well as their availability in both SOMAscan and an immunoassay panel after quality control (QC) measures. Other well validated biomarkers such as ptau, Aβ40, or Aβ42 and emerging biomarkers such as YKL-40 could not be included in these analyses either because they were not present in the SOMAscan panel or did not pass QC (YKL-40; Supplementary Figure 1).
CSF, NfL, and neurogranin have been proposed as markers of neurodegeneration, a hallmark of AD pathology and other neurodegenerative diseases [12]. Decreased levels of sTREM2, a marker of microglial activation and inflammatory changes, have been associated with increased rate of AD progression and is reported to be an endophenotype for AD [13–16]. Similarly, SNAP-25, a member of the soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) complex, has been proposed to be associated with decline in cognitive function resulting from synapse degradation, whereas variants associated with VILIP-1 have also been implicated in AD pathology [17–20]. In light of the importance of these biomarkers in AD, it is our goal to evaluate the concordance between the inter-platform measurements of these proteins.
Method and Materials
Ethics Statement
The Institutional Review Board of Washington University School of Medicine in St. Louis approved the study and research was performed in accordance with the approved protocols.
Cohorts:
This study examined 1,766 unique subjects with proteins measured by immunoassays and SOMAscan from four different cohorts: Alzheimer’s Disease Neuroimaging Initiative (ADNI), Dominantly Inherited Alzheimer Network (DIAN), Charles F. and Joanne Knight Alzheimer Disease Research Center (Knight ADRC) and Barcelona-1. The common samples between the platforms were identified based on subject ID and CSF draw date match.
ADNI
Data used in the analyses performed in this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). For up-to-date information, see www.adni-info.org.
Knight ADRC
Charles F. and Joanne Knight Alzheimer Disease Research Center (Knight ADRC), housed at Washington University in St. Louis, is one of 30 ADRCs funded by NIH. The goal of this collaborative research effort is to advance AD research with the ultimate goal of treatment or prevention of AD. The subjects included in this study are from the Memory and Aging Project (MAP) supported by Knight ADRC. As part of the project, subjects undergo annual psychometric testing and interviews along with biennial or triennial PET, MRI and CSF collection. Further details on Knight ADRC and MAP can be found at https://knightadrc.wustl.edu/
DIAN
The Dominantly Inherited Alzheimer Network (DIAN), led by Washington University School of Medicine in St. Louis, is focused on the study of Autosomal Dominant AD (ADAD). It is a family-based long-term observational study with standardized clinical and cognitive testing, brain imaging, and biological fluid collection (blood, cerebrospinal fluid) from subjects with the intent of identifying changes in pre-symptomatic and symptomatic gene carriers who are expected to develop AD. Since the focus of this study is on ADAD, which has an early age of onset compared to sporadic AD, the subjects in this cohort are younger on average compared to other cohorts. The data used in this study are from data freeze 15 (DF15). Additional details on DIAN can be found at https://dian.wustl.edu/.
Barcelona-1
Barcelona -1 is a longitudinal observational study consisting of ~300 subjects at baseline carried out in the Memory and Disorder unit at the University Hospital Mutua de Terrassa, Terrassa, Barcelona, Spain. Cases include subjects diagnosed with AD dementia (ADD), non-AD dementias (non-ADD), mild cognitive impairment (MCI), or subjective memory complaints (SMC). Clinical information was collected at baseline as well as longitudinally and lumbar puncture (LP) and amyloid PET were performed if subjects had diagnosis of MCI, early-onset dementia (<65 years), or dementia with atypical clinical features [21].
Sample Collection and Biomarker measurement
CSF samples were collected after an overnight fast, processed, and stored at −80 °C for both SOMAscan and immunoassay-based measurement. In total, 689 samples from ADNI, 870 samples from Knight ADRC, 115 Samples from DIAN and 92 samples from Barcelona-1 were included.
Somalogic
The SOMAscan panel used for this study measured ~7500 aptamers mapping to approximately 6600 unique protein targets. Protein measurements are reported in relative fluorescence unit (RFU). Initial data normalization procedures for SOMAscan protein measurements were performed by SomaLogic. Briefly, Hybridization normalization was performed at the sample level. Aptamers were then divided into three normalization groups: S1, S2 and S3; based on the observed signal to noise ratio in technical replicates and samples. This division was done to avoid combining features with different level of protein signal for additional normalization steps. Median normalization was then performed to remove other assay biases such as protein concentration, pipetting variation, variation in reagent concentrations, and assay timing among others [10]. Finally, normalization to a reference was performed on individual samples to account for additional technical variance as well as biological variance. This normalization step was performed using iterative Adaptive Normalization by Maximum Likelihood (ANML), a modification of median normalization, until convergence is reached. Additional details on normalization procedures are documented in Somalogic’s technical note [22].
Quality control was performed on the normalized data provided by Somalogic using an in-house pipeline, where aptamers were removed if they met any of the following three criteria: (A) had maximum absolute difference between calibration scale factor and median scale factor, calculated for each plate, greater than 0.5, (B) had median coefficient of variation (CV) more than 0.15, or (C) fell outside 1.5-fold of interquartile range (IQR), on either end, in more than 85% of samples. IQR was calculated using log10 transformed protein levels. QC steps were independent of the three-normalization group defined by Somalogic. The QC steps (A) and (B) are adaptation of metrics used by Somalogic that have been found to be effective previously [10]. The cut-offs used were also derived based on Somalogic’s recommendation for previous versions of the panel.
Subject level QC was then performed wherein a subject was flagged as an outlier if log10 transformed RFU levels for that subject fell outside the 1.5-fold of IQR in more than 85% of aptamers. If an analyte was found to be shared by ≈ 70 % of these subject outliers, these aptamers were excluded from the matrix in addition to those removed by the three criteria mentioned above. Subject outliers were recalled following the shared analyte removal, and a second IQR check was done, at the end of which remaining sample outliers were removed from the matrix. An additional limit of detection criteria (LOD) was also used to flag aptamers independently. Somalogic defines LOD, for their SOMAscan platform, as the lowest concentration of analyte that can be consistently detected [23]. For our analysis, if an aptamer had LOD greater than average RFU level in buffer + 2SD in more than 15% of total samples, it was flagged. All five biomarkers of interest passed QC.
Immunoassays
Details on Immunoassay based biomarker measurement within each cohort and for each biomarker have been described previously. Briefly, NfL was measured using a commercially available immunoassay kit (UmanDiagnostics AB, Umeå, Sweden) in both Knight ADRC and ADNI [24–26]. Neurogranin measurements were available for samples from Knight ADRC, ADNI and Barcelona-1. Measurements within Knight ADRC and ADNI were performed using the Erenna® immunoassay system [27,28]. In samples from the Barcelona-1, EUROIMMUN enzyme-linked immunosorbent assay kits were utilized to measure Neurogranin levels [21,29]. Similar to Neurogranin, VILIP-1 and SNAP-25 were also measured using the Erenna® immunoassay system-based sandwich immunoassay in ADNI [28]. Although, both these biomarkers were measured using the same Erenna system, those measurements were done independently at different lab and batches, so they were treated as independent measurements. within Knight ADRC as well, variation existed in that SNAP-25 levels were measured using a single molecule counting system in Knight ADRC (Supplementary Table 1) [20,30]. sTREM2 levels in both Knight ADRC and ADNI were measured using the in-house immunoassay developed at Washington University in St. Louis [13,31,32]. For samples in DIAN, sTREM2 measurements were obtained from a Meso Scale Discovery (MSD) platform-based immunoassay as described previously [33]
Since immunoassay-based data were collected retrospectively from in-house and publicly available data, QC was performed for each biomarker within each cohort separately due to the technical variation among cohorts and within biomarkers (Supplementary Table 1). Duplicates were removed if multiple measurements for a biomarker were available for the same subject at the same date of CSF draw. Outliers were defined using the 1.5-fold IQR criteria, as described for SOMAscan based measurement. Any measurements detected as outlier were removed.
Statistical analysis
Statistical analysis was performed using available data from all cohorts together as well as in each cohort individually. To compare the protein levels for the different biomarkers across platforms (SOMAscan vs. Immunoassays), Pearson’s correlation coefficient was used. First, raw protein measurements of each biomarker were log10 transformed and scaled with mean 0 and variance 1 to get a Z-score value for analysis. Comparison of SOMAscan-based and immunoassays measurements within each cohort was performed using the Z-scores as well as the raw values. For analyses combining data from multiple cohorts Z-scores, instead of raw values, were used to account for the difference in units of protein measurement reported by the two platforms and the variation in immunoassay techniques used between the cohorts. For immunoassay-based measures, Z-scores were calculated within each cohort separately. For SOMAscan, Z- scores were calculated prior to stratification by cohort because the protein measures were obtained at the same time using a common panel for all cohorts. Second, samples with protein measurement for both platforms were kept. SOMAscan panel had more than one aptamer targeting some proteins: two aptamers targeting VILIP-1 and three targeting sTREM2. The two analytes targeting VILIP-1 had a correlation of 0.95 and the correlation for the three analytes targeting TREM2 ranged from 0.92 – 0.94 (Supplementary Figure 2). All available aptamers were included in analysis for both cohort specific and combined analysis. Cutoffs for the strength of correlation were used as reported by previously published literature [34].
To compare the predictive accuracy of the biomarkers between platforms, receiver operating characteristic (ROC) curve and area under the curve (AUC) values were assessed. Univariate logistic regression was performed with clinical status (AD cases vs. controls) at CSF draw as the response variable and standardized protein measure as the predictor variable. Multivariate logistic models with standard scores as predictor variables and CSF draw and gender included as covariates were also evaluated to confirm the findings from the univariate models. A subset of common samples with reported AD disease or control status at the time of CSF draw were used to compute ROC. P<0.05 was considered as significance threshold. All statistical analyses were performed using R statistical software version 3.5.2.
Results
Cohort demographics
A total of 870 CSF samples collected from the Knight-ADRC were included, of which 175 have AD and 3 have ADAD. 626 samples were cognitively normal controls and 66 had other types of neurogenerative diseases (referred to as “Others” hereafter). The average age of sample at CSF draw date was 70.88 years (±8.69). NfL measurements were obtained from 53 AD cases, 155 controls and 13 Others. Neurogranin was measured in samples from 171 AD cases, 606 controls and 64 Others. SNAP-25 measurement was available for 52 AD cases, 109 controls and 12 Others. sTREM2 and VILIP-1 were measured in samples from 79 AD cases, 226 controls, 18 Others and samples from 53 AD, 113 controls and 12 Others respectively. For the DIAN study 115 CSF samples were included for analysis; 72 of these samples were carriers of autosomal dominant pathogenic variants and 36 were non-carriers. In contrast to other cohorts, the cases in DIAN were ADAD cases with average age at CSF of draw of 41.12 years (±10.18). ADNI included a total of 689 unique subjects with an average age of 73.49 years (±7.56) at CSF draw; 519 of them had a clinical diagnosis of AD and 149 were diagnosed as cognitive normal individuals. An additional 21 samples had mild cognitive impairment (MCI). NfL was measured in 106 AD cases, 39 controls and 7 MCI samples. Neurogranin and VILIP-1 were both measured in CSF samples from 34 cases, 17 controls and 4 MCI, whereas SNAP-25 was measured in samples from 37 cases, 16 controls and 4 MCI at time of lumbar puncture. sTREM2 measurements were available for 512 cases, 149 controls and 20 MCI. Barcelona-1 included data from 23 AD cases, 1 control and 68 subjects with other diagnosis resulting a total of 92 unique subjects. The average age at CSF draw for the samples was 70.03 (±7.51). Of the five biomarkers of interest, only Neurogranin was measured using immunoassay (Table 1).
Table 1:
Sample characteristics by cohort.
| Cohort | Samples (N) | Average Age (±SD) |
|
Disease Status | Proteins |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Male (%) | Cases (%) | Control (%) | Others/MCI (%) | NfL | NG | sTREM2 | VILIP-1 | SNAP-25 | |||
|
| |||||||||||
| ADNI | 689 | 73.49 (±7.56) | 57.33 | 75.33 | 21.63 | 3.05 | 152 | 55 | 684 | 58 | 57 |
| Knight ADRC | 870 | 70.88 (±8.69) | 45.98 | 20.46 | 71.95 | 7.59 | 221 | 841 | 323 | 179 | 173 |
| DIAN | 115 | 41.12 (±10.18) | 49.57 | 62.61 | 31.30 | 6.09 | NA | NA | 113 | NA | NA |
| Barcelona -1 | 92 | 70.03 (±7.51) | 57.61 | 25.00 | 1.08 | 73.91 | NA | 92 | NA | NA | NA |
Characteristics of samples with both SOMAscan and immunoassay-based protein measurements, matched based on subject ID and cerebrospinal fluid (CSF) draw date, between the platforms within each cohort. Age and disease status correspond to age and status recorded at CSF draw date. Age is reported in years (mean± SD). Cases represent sporadic AD cases for ADNI, Knight-ADRC and Barcelona 1 and autosomal dominant AD cases for DIAN. Subjects with disease status other than AD or control were grouped under “Others” category. NA values denote that no information was available for the protein within that cohort. Sample number differed between aptamers targeting the same biomarker. The highest number of samples used is reported if applicable.
Similar Standard Score distribution between the platforms
We performed Z-score normalization in order to compare the protein levels between the platforms and cohorts (Figure 1). The normalized levels for NfL, Neurogranin, VILIP-1 and SNAP-25 had significantly higher levels in cases than controls (Figure 1), as expected. These findings were consistent across both platforms, and no significant differences were found in the z-scores for a specific analyte across cohorts. Significance was determined using a two tailed, two sample t-Test with assumption of unequal variance. No significant difference in sTREM2 level were observed between cases and controls when using SOMAscan or immunoassays.
Figure 1: Z-score distribution across SOMAscan and immunoassay platforms.
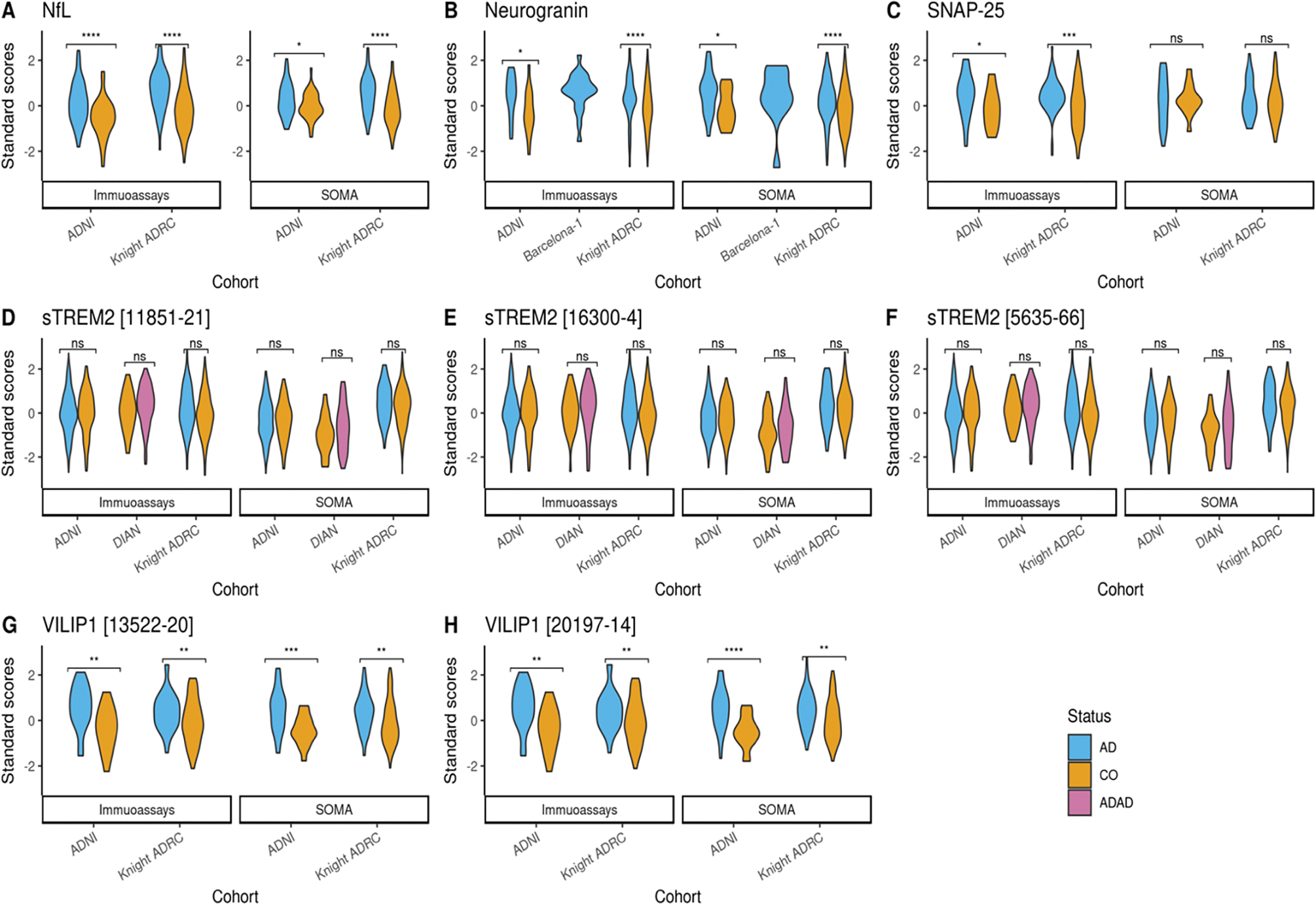
(A) Violin plot showing the standard score distribution for NfL protein across ADNI and Knight ADRC (B)Violin plot showing the standard score distribution for neurogranin across ADNI, Knight ADRC and Barcelona-1 (C) Violin plot showing the standard score distribution for SNAP-25 protein across ADNI and Knight ADRC (D-F) Violin plot showing the standard score distribution for different aptamers targeting sTREM2 protein across ADNI, Knight ADRC and DIAN (G-H) Violin plot showing the standard score distribution for different aptamers targeting VILIP-1 protein across ADNI and Knight ADRC. ns= Not significant. Z-scores were calculated using log10 transformed raw protein levels. Colors represent disease status at CSF draw date. Score distributions were plotted for samples with disease status reported as sporadic Alzheimer’s disease (AD), Autosomal Dominant Alzheimer’s disease (ADAD) or control (CO) only. X-axis shows cohorts that had information on the measured protein for each platform. Y-axis shows the distribution of standard scores. P-values shows the significance differences between cases and controls, stratified by cohort and the platforms. If the SOMAscan panel had more than one analyte targeting a protein, multiple plots were generated corresponding to each.
Correlation between SOMAscan and immunoassay measurements
We tested for correlation between biomarker measurements between immunoassay vs SOMAscan on different aliquots from the same individual and CSF draw date. Pearson’s correlation coefficient was calculated using all common samples. We found significant positive correlation for NfL, neurogranin and VILIP-1 measurements (r > 0.9). For VILIP-1, the SOMAscan 7K panel included two different aptamers, and a significant and similar correlation was found for both (r = 0.91; p-value = 5.28 ×10−89; n = 236 and 0.92; p-value = 2.07 ×10−96; n=237 respectively; Figure 2 and Table 2). The correlation of the three sTREM2 SOMAscan aptamers with immunoassays was also significant but showed lower correlation coefficients than VILIP-1, neurogranin and NfL (r ranging from 0.64 to 0.66 [p value < 10−140; n =1107–1120]). SNAP-25 was the only biomarker that showed no significant correlation between the two platforms (r = 0.06, P-value >0.05; n = 230).
Figure 2: Correlations between SOMAscan and immunoassay platforms for different proteins across cohorts.
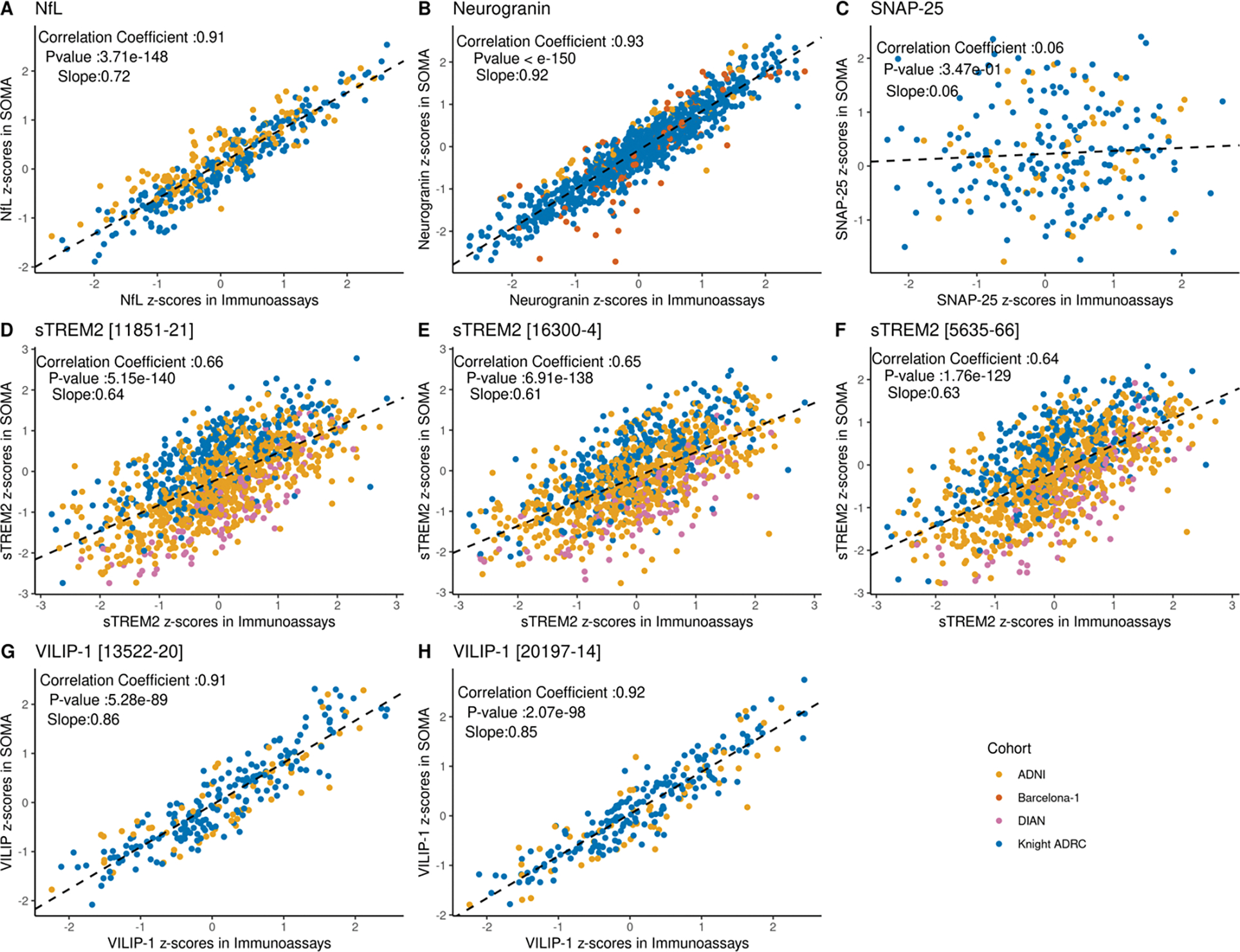
For each protein, Pearson’s correlation coefficient was calculated using the Z-scores from all common samples between the two platforms across all available cohorts. Significance was determined based on a p-value threshold of 0.05. Data points are color coded to show the cohort source. Slope in each plot corresponds to a regression line fitted using Immunoassay score as independent variable and SOMA score as dependent variable. (A) Scatter plot showing the correlation of standard scores for NfL protein (r = 0.91 and p-value = 3.71 × 10−148) (B) Scatter plot showing the correlation of standard scores for neurogranin protein (r = 0.93 and p-value <10−150) (C) Scatter plot showing the correlation of standard scores for SNAP-25 protein (r = 0.06 and p value = 0.35) (D-F) Scatter plot showing the correlation of standard scores for sTREM2. Three aptamers targeting sTREM2 proteins were available in the SOMAscan panel. Correlation was evaluated for all three aptamers. (r = 0.64 to 0.66; p-value = 5.15 × 10−140 to 1.76× 10−129) (G-H) Scatter plots showing the correlation of standard scores for two different aptamers targeting VILIP-1 protein (r = 0.91; p-value = 5.28 × 10−89 and 0.92; p-value = 2.07 × 10−98 respectively).
Table 2:
Pearson’s correlation coefficient of proteins measured by SOMAscan and immunoassay platforms.
| Analyte Soma ID | Correlation Coefficient | R-Squared | P-value | Slope | |
|---|---|---|---|---|---|
|
| |||||
| NfL | 10082–251 | 0.91 | 0.84 | 3.71 ×10−148 | 0.72 |
| Neurogranin | 18303–39 | 0.93 | 0.86 | <10−150 | 0.92 |
| SNAP-25 | 13105–7 | 0.06 | 0.004 | 0.34 | 0.06 |
| sTREM2 | 11851–21 | 0.66 | 0.44 | 5.15 ×10−140 | 0.64 |
| 16300–4 | 0.65 | 0.43 | 6.91 ×10−138 | 0.61 | |
| 5635–66 | 0.64 | 0.41 | 1.76 ×10−129 | 0.63 | |
| VILIP-1 | 13522–20 | 0.91 | 0.82 | 5.28 ×10−89 | 0.86 |
| 20197–14 | 0.92 | 0.85 | 2.07 ×10−98 | 0.85 | |
Standard scores were used to calculate the correlation coefficient. Significance was determined based on a p-value threshold of 0.05. Slope corresponds to a regression line fitted using immunoassay score as independent variable and SOMA score as dependent variable. Samples from all available cohorts were used for correlation calculation.
Cohort specific analyses
Correlation of biomarkers within each cohort using Z-scores and raw values were additionally compared (Supplementary Figure 3–10; Supplementary Table 2–3). In ADNI, NfL, neurogranin and VILIP-1 showed strong correlations between the platforms (r > 0.8; Supplementary Figure 3). NfL showed the highest correlation with r = 0.89 (p-value = 7.32 ×10−53; n = 152).
In Knight ADRC, neurogranin showed the highest correlation with a coefficient of 0.95 (p-value < 10−150; n = 841) followed by NfL with r = 0.94 (p-value = 6.81×10−107; n= 221; Supplementary Figure 4). VILIP-1 also showed a significant correlation with r > 0.9 in both aptamers targeting it.
Consistent with our findings from the combined analyses, SNAP-25 did not show a significant correlation in either ADNI or Knight ADRC (P-value > 0.05; n = 57 and n = 221 respectively; Supplementary Figure 3–4).
For DIAN, only the sTREM2 immunoassay measurement was available at the time of these analyses. However, the correlation for sTREM2 was highest in DIAN among all cohorts (Supplementary Figure 5). DIAN used an MSD immunoassay platform whereas both ADNI and Knight ADRC used an in-house assay to measure sTREM2. This finding highlights the variation in correlation between SOMAscan and immunoassay platform based on the technical nuances involved. We observed r value as high as 0.85 (Analyte 11851–21; p-value = 1.92×10−31; n = 109) for the protein denoting a strong correlation between sTREM2 measurement in DIAN (Supplementary Figure 5).
Finally, correlation was also evaluated within Barcelona-1for neurogranin (Supplementary Figure 6). Findings for neurogranin were consistent with the other cohorts, with it showing a strong correlation (r = 0.84, p-value = 1.67×10−25; n = 92).
The consistently strong correlations observed between the platforms for Nfl, neurogranin, VILIP-1 and sTREM2 show that SOMAscan based protein measures are in accordance with immunoassay-based measures. As made evident in the case of sTREM2, variation in correlations seen across cohort could have been due to the difference in sample characteristics and immunoassay technique and utilized by each cohort.
The correlations found for each analyte in each cohort were not significantly different when using Z-scores or raw values, confirming that using Z-scores does not lead to artifactual findings.
SOMAscan and immunoassay-based measurements lead to similar prediction performance
Next, we compared the prediction accuracy of the biomarkers using the Z-scores obtained for the two platforms. Our goal was to evaluate if the measures from the two platforms show any difference in their ability to differentiate cases and controls for the same group of subjects. A univariate model, using only the standardized protein measures, was evaluated for both combined as well as cohort specific data. A second multivariate model using age at CSF draw and gender as covariates was also analyzed.
For the univariate model, we did not find any significant difference in the AUCs for NfL, neurogranin, VILIP-1 or two of the three aptamers targeting sTREM2 (Figure 3, Table 3). For NfL, we observed an AUC of 0.66 in both SOMAscan (AUCSOMA) and in immunoassay (AUCIMMUNO; p-value = 0.68; n = 353). We observed similar trends in neurogranin and VILIP-1. Both AUCSOMA and AUCIMMUNO for neurogranin were 0.64 (p-value =0.56; n = 849). VILIP-1 showed AUCsoma = 0.68 for both aptamers and an AUCIMMUNO = 0.66 and 0.67 (p-value = 0.29 and p-value = 0.56; n = 221 and 220 respectively). A significant difference in ROCs was observed in one of the aptamers for sTREM2 (SOMA id: 5635–66), where SOMAscan (AUCsoma = 0.58) performed significantly better than the immunoassays (AUCIMMUNO = 0.508 (p-value =0.04; n = 962). The other two sTREM2 aptamers (SOMA id: 16300–4 and 11851–21) also showed higher AUCs for SOMAscan than the immunoassay values, but the difference was not statistically significant (p- value 0.12 and 0.06; n = 966 and 968, respectively). Consistent with the poor correlation observed for the SNAP-25 measures between the two platform, there was a significant difference in ROCs (p-value = 0.01, AUCsoma = 0.53 and AUCIMMUNOASSAY = 0.66; n = 214), with the immunoassay-based measure presenting better prediction power.
Figure 3: ROC curve showing sensitivity and specificity for different proteins in SOMAscan and immunoassay platforms.
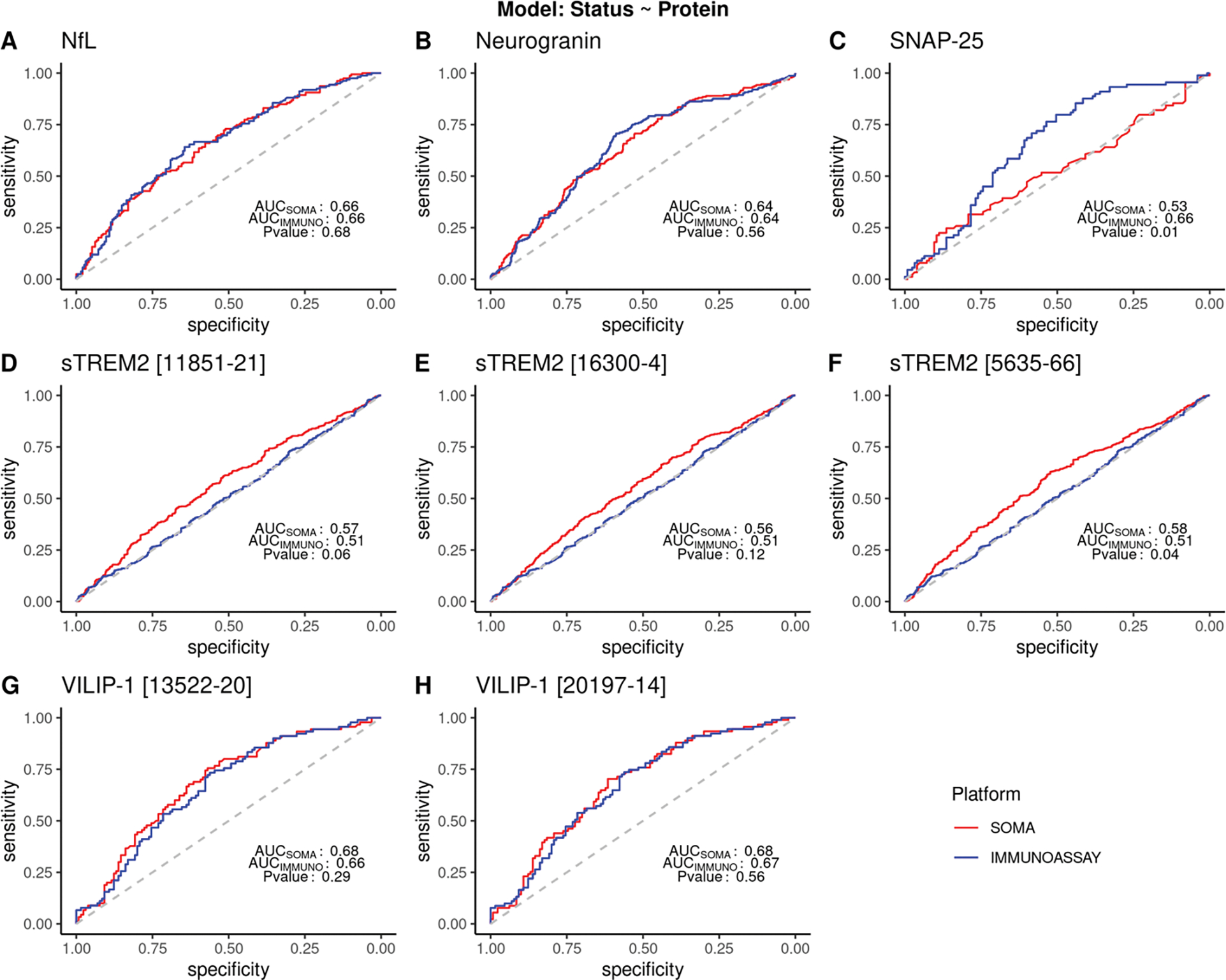
ROC and AUC were calculated using a logistic model with protein standard scores as the predictor variable and disease status as the response variable. Clinical disease status reported at the time of CSF draw was used. Common samples from all cohorts were combined and analyzed. sTREM2 analyses included samples from Knight ADRC and ADNI only. Samples reported to have status other than cases or controls were excluded from the analysis. P<0.05 denotes significant difference in the ROC between the platforms. (A) ROC curve for NfL (AUC= 0.66 for SOMAscan and immunoassay; p= 0.68). (B) ROC curve for neurogranin (AUC 0.64 for SOMAscan and immunoassay; p value = 0.56) (C) ROC curve for SNAP-25 (AUC for SOMAscan = 0.53 and for immunoassay = 0.66 respectively; with p-value of 0.01) (D-F) ROC curve for three aptamers from the SOMAscan panel for sTREM2. (AUC for SOMAscan = 0.56 −0.58; AUC for immunoassay= 0.51; p -value = 0.04 −0.12) (G-H) ROC curve for two aptamers from the SOMAscan panel targeting VILIP-1 protein (AUC values: 0.68 for SOMAscan; 0.67 and 0.66 for immunoassay; p- value = 0.56 and 0.29) respectively.
Table 3:
Area Under Curves for different proteins between SOMA and immunoassay platforms.
| Protein | Analyte | Samples | AUC | |||
|---|---|---|---|---|---|---|
| Cases | Controls | SOMA | Immunoassay | P-value | ||
|
| ||||||
| NfL | 10082–251 | 159 | 194 | 0.66 | 0.66 | 0.68 |
| Neurogranin | 18303–39 | 225 | 624 | 0.64 | 0.64 | 0.56 |
| SNAP-25 | 13105–7 | 89 | 125 | 0.53 | 0.66 | 0.01 |
| 11851.21 | 591 | 375 | 0.57 | 0.51 | 0.06 | |
| sTREM2 | 16300.4 | 593 | 375 | 0.56 | 0.51 | 0.12 |
| 5635.66 | 589 | 373 | 0.58 | 0.51 | 0.04 | |
| VILIP-1 | 13522.20 | 90 | 130 | 0.68 | 0.66 | 0.56 |
| 20197.14 | 91 | 130 | 0.68 | 0.67 | 0.29 | |
The samples used for receiver operating curve (ROC) include common samples across all available cohorts for that protein. Samples were excluded if they were reported to have status other than AD cases or controls. sTREM2 analyses included samples from ADNI and Knight ADRC only. P-value denotes the test of significance for a difference between the ROCs between the two platforms.
We re-calculated ROCs and AUC by adjusting the model for age at CSF draw and gender (Supplementary Figure 11; Supplementary Table 4). Improvements in AUCs were observed for all the aptamers except NfL, which did not show any change. Trends of the AUCs in both platforms were similar to the univariate model with no significant difference in AUCs in case of NfL, VILIP-1, or neurogranin and a significant difference in SNAP-25. However, sTREM2 [analyte 11851.21 and 5635.66] showed significant difference in the multivariate model with higher AUC for SOMAscan platform (AUCSOMA = 0.62 and AUCIMMUNO =0.59; p -value=0.02 for both). sTREM2 analyte 16300–4 also showed higher AUC for SOMAscan platform although the difference was not significant (AUCSOMA = 0.61 and AUCIMMUNO =0.59; p -value=0.07).
Performance of both univariate and multivariate models were also evaluated in individual cohorts (Supplementary Figure 12–20; Supplementary Table 5–7). Analyses using Barcelona-1 were not performed because it did not have sufficient control samples. For the univariate model, results consistent with our findings in combined analysis were observed. We observed similar and not significantly different AUCs for neurogranin and VILIP-1 in both Knight ADRC and ADNI (ADNI: neurogranin p-value = 0.63; VILIP p-value = 0.60 and 0.12; Knight ADRC: neurogranin p-value =0.18; VILIP-1 p-value = 0.08 and 0.32). However, we observed significant differences for NfL in both cohorts (Supplementary Figure 12–13; Supplementary Table 5). The NfL immunoassay measures showed significantly better prediction ability than SOMAscan within ADNI (AUCSOMA = 0.62; AUCIMMUNO = 0.71; p-value =0.001; n= 145) and Knight ADRC (AUCSOMA = 0.71 and AUCIMMUNO = 0.75; p-value =0.02; n = 208). On the other hand, SNAP-25, which had a significant difference in ROCs between SOMAscan and immunoassays in the combined analysis, did not show any significant difference in either cohort when evaluated individually. This observation is likely due to the smaller sample size in the individual cohorts and lower statistical power as a result. In ADNI, SNAP-25 showed AUCSOMA = 0.50 and AUCIMMUNO = 0.68 (p-value =0.12; n = 53). Similarly, in Knight ADRC, SNAP-25 AUCSOMA was 0.54 and AUCIMMUNO was 0.65 (p-value =0.06; n = 161). None of the three aptamers targeting sTREM2 showed any significant difference in prediction accuracy between SOMAscan and immunoassay in the three cohorts (Supplementary Figure 12–14; Supplementary Table 5). No changes were observed when the models were tested using raw protein values (Supplementary Figure 15–17; Supplementary Table 6).
Findings for the multivariate models were in line with our observations in the univariate models within each cohort (Supplementary Figure 18–20, Supplementary Table 7). NfL was the only biomarker with a significant difference in AUCs between SOMAscan and immunoassays (ADNI: p-value = 0.01, n = 145; Knight ADRC: p-value = 0.03, n = 208).
The lack of significant differences in AUCs between NfL, neurogranin and VILIP-1 observed between the two platforms in the combined analyses further provides evidence supporting the concordance of protein measures between SOMAscan and classic immunoassays. The variation in the prediction power within each cohort can be attributed to the lack of statistical power owing to decreased sample sizes.
Discussion
The need for a high-throughput protein quantification method has driven the search for alternatives to traditional immunoassay-based protein techniques. SOMAscan, developed by SomaLogic Inc. (Boulder, CO), is a viable option and has already been used in large-scale proteomic studies [35–37]. However, before any novel technology becomes mainstream, comparing its performance to well-established measures can provide evidence for or against its adoption. In this study, we compared the performance of SOMAscan with immunoassay-based platforms for several CSF proteins that have been proposed as important markers for AD and other neurodegenerative diseases (NfL, neurogranin, VILIP-1, sTREM2 and SNAP-25]). The choice of proteins for inclusion in this comparative study was based on availability of their measures in both the SOMAscan and immunoassay platforms. Well validated biomarkers such as ptau, Aβ40, or Aβ42 are not covered on the Somalogic 7K panel. YKL-40 is included but did not pass QC in our analyses (Supplementary Figure 1). GFAP, an emerging biomarker, was included in the panel and passed QC, but we did not have access to immunoassay-based measure for GFAP at the time of the analyses for any of the cohorts. When more than one analyte targeting the same biomarker weas present in the SOMAscan panel (Supplementary Figure 2), analyses were performed for all of them.
We leveraged data from four different cohorts (ADNI, Knight ADRC, DIAN, and Barcelona-1) and compared the performance of the platforms using the correlation between the measures and their prediction accuracy using harmonized protein values. We observed significant correlation in four of the five AD biomarkers. Nfl, neurogranin and VILIP-1 all had r > 0.9. Only one biomarker, SNAP-25, had a poor correlation which failed to pass the significance threshold of p-value < 0.05. The results were consistent across combined and cohort-specific analyses. In univariate models tested using data from all cohorts, NfL, neurogranin, VILIP-1 and two aptamers targeting sTREM2 did not have any significant difference between the ROCs among the platforms, suggesting similar discriminating power between them. However, a significant difference was observed in one of three aptamers in the SOMAscan panel targeting sTREM2, with higher AUC in SOMA than in immunoassays, suggesting that the SOMAscan based measurements for sTREM2 are better than those for immunoassays. SNAP-25 also exhibited a significant difference, but immunoassay-based measures showed higher AUC. Similar findings were found within-cohort using both Z-scores and raw values. Overall good agreement was found between protein measures from SOMAscan and immunoassays. Apart from the SOMAscan discovery panel spanning over 6000 proteins, custom, disease specific panels, that can be used in clinical trial or disease monitoring, are also available. These biomarkers could be part of such AD specific diagnostic tool in future.
The findings from our study are consistent with what has been previously reported in literature comparing SOMAscan and other immunoassay platforms. A multi cohort study from the TOPMed consortium compared protein measurements from SOMAscan with other immunoassay platforms for cardiovascular and smoking related diseases reported a similar wide range of correlations between the protein measures [6]. Among the 63 proteins assessed in plasma or serum samples, 13 (20%) had a low correlation (r < 0.3), 33 (52%) had moderate correlation (r = 0.3–0.7) and 17 (27%) had high correlation (r > 0.7) [6]. We observed a similar range of correlations for our biomarkers. Of the five biomarkers analyzed in our study, strong correlation was observed in four, a higher percentage (80%) than the 27% reported here.
Our study is one of the very few that has compared SOMAscan to immunoassays for CSF AD and neurogenerative biomarkers. Given the fact that immunoassays are the most widely used platforms for protein measurement, it is necessary that they be used as a reference to evaluate this novel platform [38]. Previous studies have compared SOMAscan with immunoassays, but mostly in plasma or urine samples [6,11,38]. CSF biomarkers are considered more reliable when it comes to the neurodegenerative diseases [39]. Particularly in diseases like AD, the ability to monitor a wide range of the CSF proteome is a major feat in terms of early disease detection and diagnosis, as changes in CSF proteome can precede clinical symptoms [40]. Also, a multi cohort approach, as used in our study, gives us the opportunity to make comparisons across various immunoassay techniques. For neurogranin and sTREM2, we had more than 800 samples from various cohorts. To our knowledge, this is the largest number of CSF based samples ever used for comparison across these two platforms.
Despite the several strengths of our study, our results should be considered with caution. First, SOMAscan reports protein levels in relative fluorescence units unlike immunoassay-based methods that report absolute values of protein quantification. Thus, we cannot directly comment on the interchangeability of raw measures reported by the two platforms or perform comparisons at the raw value level. This can, however, be addressed through alternative approaches like comparison of the performance of data driven dichotomization cut-offs, based on linear mixed models and the expectation-maximization algorithm, to define biomarker positive vs negative status. We have used this data-driven approach in several studies demonstrating that this method leads to similar cut-offs as those calculated experimentally [41,42]. This approach has several advantages as it can be used in Z-scores, can be implemented in studies using different platforms or sample composition, and does not require a-priori knowledge to perform the dichotomization. Although we observed high correlations among most of our biomarkers the same cannot be said about SNAP-25. Follow-up studies will be needed with larger sample sizes to provide enough statistical power in order to reach a conclusion for this analyte.
Additionally, the finding for these five CSF biomarkers cannot be generalized to other AD biomarkers. Several classic AD biomarkers such as GFAP, amyloid beta and pTau could not be included in the analysis because they were missing in one of the two platforms. YKL-40, although present in both the SOMAscan and Immunoassay panels, was excluded because it failed our QC criteria in SOMAscan. Given the importance of these biomarkers in AD diagnosis, follow up studies will be needed as more data becomes available. The field of AD biomarker study is moving towards blood-based biomarker detection. Since the findings from one tissue-based biomarker cannot be directly translated into another, similar comparisons are required to comment on the performance of SOMAscan with blood-based AD biomarkers.
To conclude, the findings from our study show the potential of the SOMAscan assay as an alternative to traditional immunoassay- based approaches for CSF NfL, neurogranin, VILIP-1 and sTREM2. However, we cannot conclude the same about SNAP-25. The variation in results across cohorts also highlights the need of an additional orthogonal approach to confirm the interchangeability of protein measures between the platforms. Further, we only compared CSF based biomarkers, so additional studies will be needed to confirm that these findings replicate in plasma or using other tissue-based biomarkers.
Supplementary Material
ACKNOWLEDGMENTS:
We thank all the participants and their families, as well as the many involved institutions and their staff.
The recruitment and clinical characterization of research participants at Washington University were supported by NIH P30AG066444 (JCM), P01AG03991(JCM), and P01AG026276(JCM). O.H. is an Archer Foundation Research Scientist.
This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, the Neurogenomics and Informatics Center (NGI: https://neurogenomics.wustl.edu/) and the Departments of Neurology and Psychiatry at Washington University School of Medicine.
ADNI resources: Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12–2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutica l Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
DIAN resources: “Data collection and sharing for this project was supported by The Dominantly Inherited Alzheimer Network (DIAN, U19AG032438) funded by the National Institute on Aging (NIA),the Alzheimer’s Association (SG-20–690363-DIAN), the German Center for Neurodegenerative Diseases (DZNE), Raul Carrea Institute for Neurological Research (FLENI), Partial support by the Research and Development Grants for Dementia from Japan Agency for Medical Research and Development, AMED, and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), Spanish Institute of Health Carlos III (ISCIII), Canadian Institutes of Health Research (CIHR), Canadian Consortium of Neurodegeneration and Aging, Brain Canada Foundation, and Fonds de Recherche du Québec – Santé. This manuscript has been reviewed by DIAN Study investigators for scientific content and consistency of data interpretation with previous DIAN Study publications. We acknowledge the altruism of the participants and their families and contributions of the DIAN research and support staff at each of the participating sites for their contributions to this study “
Funding:
This work was supported by grants from the National Institutes of Health (R01AG044546 (CC), P01AG003991(CC, JCM), RF1AG053303 (CC), RF1AG058501 (CC), U01AG058922 (CC), and R01AG057777 (OH)), and the Chuck Zuckerberg Initiative (CZI).
REFERENCES
- [1].Yalow RS, Berson SA IMMUNOASSAY OF ENDOGENOUS PLASMA INSULIN IN MAN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Engvall E, Perlmann P (1971) Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry 8, 871–874. [DOI] [PubMed] [Google Scholar]
- [3].Darwish IA (2006) Immunoassay Methods and their Applications in Pharmaceutical Analysis: Basic Methodology and Recent Advances. International Journal of Biomedical Science : IJBS 2, 217. [PMC free article] [PubMed] [Google Scholar]
- [4].Sakamoto S, Putalun W, Vimolmangkang S, Phoolcharoen W, Shoyama Y, Tanaka H, Morimoto S (2018) Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. Journal of Natural Medicines 72, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hoofnagle AN, Wener MH (2009) The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J Immunol Methods 347, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Raffield LM, Dang H, Pratte KA, Jacobson S, Gillenwater LA, Ampleford E, Barjaktarevic I, Basta P, Clish CB, Comellas AP, Cornell E, Curtis JL, Doerschuk C, Durda P, Emson C, Freeman CM, Guo X, Hastie AT, Hawkins GA, Herrera J, Johnson WC, Labaki WW, Liu Y, Masters B, Miller M, Ortega VE, Papanicolaou G, Peters S, Taylor KD, Rich SS, Rotter JI, Auer P, Reiner AP, Tracy RP, Ngo D, Gerszten RE, O’Neal WK, Bowler RP (2020) Comparison of Proteomic Assessment Methods in Multiple Cohort Studies. Proteomics 20, e1900278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, Flather D, Forbes A, Foreman T, Fowler C, Gawande B, Goss M, Gunn M, Gupta S, Halladay D, Heil J, Heilig J, Hicke B, Husar G, Janjic N, Jarvis T, Jennings S, Katilius E, Keeney TR, Kim N, Koch TH, Kraemer S, Kroiss L, Le N, Levine D, Lindsey W, Lollo B, Mayfield W, Mehan M, Mehler R, Nelson SK, Nelson M, Nieuwlandt D, Nikrad M, Ochsner U, Ostroff RM, Otis M, Parker T, Pietrasiewicz S, Resnicow DI, Rohloff J, Sanders G, Sattin S, Schneider D, Singer B, Stanton M, Sterkel A, Stewart A, Stratford S, Vaught JD, Vrkljan M, Walker JJ, Watrobka M, Waugh S, Weiss A, Wilcox SK, Wolfson A, Wolk SK, Zhang C, Zichi D (2010) Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLOS ONE 5, e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].(2021) SomaScan® Assay v4.1 SomaScan Assay v4.1.
- [9].Tin A, Yu B, Ma J, Masushita K, Daya N, Hoogeveen RC, Ballantyne CM, Couper D, Rebholz CM, Grams ME, Alonso A, Mosley T, Heiss G, Ganz P, Selvin E, Boerwinkle E, Coresh J (2018) Reproducibility and Variability of Protein Analytes Measured Using a Multiplexed Modified Aptamer Assay. [DOI] [PMC free article] [PubMed]
- [10].Candia J, Cheung F, Kotliarov Y, Fantoni G, Sellers B, Griesman T, Huang J, Stuccio S, Zingone A, Ryan BM, Tsang JS, Biancotto A Assessment of Variability in the SOMAscan Assay OPEN. [DOI] [PMC free article] [PubMed]
- [11].Kukova LZ, Mansour SG, Coca SG, de Fontnouvelle CA, Thiessen-Philbrook HR, Shlipak MG, El-Khoury JM, Parikh CR (2019) Comparison of Urine and Plasma Biomarker Concentrations Measured by Aptamer-Based versus Immunoassay Methods in Cardiac Surgery Patients. [DOI] [PubMed]
- [12].Mattsson N, Insel PS, Palmqvist S, Portelius E, Zetterberg H, Weiner M, Blennow K, Hansson O (2016) Cerebrospinal fluid tau, neurogranin, and neurofilament light in Alzheimer’s disease. EMBO Mol Med 8, 1184–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Piccio L, Buonsanti C, Cella M, Tassi I, Schmidt RE, Fenoglio C, Rinker J, Naismith RT, Panina-Bordignon P, Passini N, Galimberti D, Scarpini E, Colonna M, Cross AH (2008) Identification of soluble TREM-2 in the cerebrospinal fluid and its association with multiple sclerosis and CNS inflammation. Brain 131, 3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ewers M, Biechele G, Suárez-Calvet M, Sacher C, Blume T, Morenas-Rodriguez E, Deming Y, Piccio L, Cruchaga C, Kleinberger G, Shaw L, Trojanowski JQ, Herms J, Dichgans M, Brendel M, Haass C, Franzmeier N (2020) Higher CSF sTREM2 and microglia activation are associated with slower rates of beta-amyloid accumulation. EMBO Mol Med 12,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ewers M, Franzmeier N, Suárez-Calvet M, Morenas-Rodriguez E, Caballero MAA, Kleinberger G, Piccio L, Cruchaga C, Deming Y, Dichgans M, Trojanowski JQ, Shaw LM, Weiner MW, Haass C (2019) Increased soluble TREM2 in cerebrospinal fluid is associated with reduced cognitive and clinical decline in Alzheimer’s disease. Sci Transl Med 11,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Deming Y, Filipello F, Cignarella F, Cantoni C, Hsu S, Mikesell R, Li Z, Del-Aguila JL, Dube U, Farias FG, Bradley J, Budde J, Ibanez L, Fernandez MV, Blennow K, Zetterberg H, Heslegrave A, Johansson PM, Svensson J, Nellgård B, Lleo A, Alcolea D, Clarimon J, Rami L, Molinuevo JL, Suárez-Calvet M, Morenas-Rodríguez E, Kleinberger G, Ewers M, Harari O, Haass C, Brett TJ, Benitez BA, Karch CM, Piccio L, Cruchaga C (2019) The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer’s disease risk. Science Translational Medicine 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Brinkmalm A, Brinkmalm G, Honer WG, Frölich L, Hausner L, Minthon L, Hansson O, Wallin A, Zetterberg H, Blennow K, Öhrfelt A (2014) SNAP-25 is a promising novel cerebrospinal fluid biomarker for synapse degeneration in Alzheimer’s disease. Mol Neurodegener 9, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cao L, Wang F, Yang QG, Jiang W, Wang C, Chen YP, Chen GH (2012) Reduced thyroid hormones with increased hippocampal SNAP-25 and Munc18–1 might involve cognitive impairment during aging. Behavioural brain research 229, 131–137. [DOI] [PubMed] [Google Scholar]
- [19].Tarawneh R, D’Angelo G, MacY E, Xiong C, Carter D, Cairns NJ, Fagan AM, Head D, Mintun MA, Ladenson JH, Lee JM, Morris JC, Holtzman DM (2011) VISININ-LIKE PROTEIN-1: DIAGNOSTIC AND PROGNOSTIC BIOMARKER IN ALZHEIMER DISEASE. Ann Neurol 70, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kester MI, Teunissen CE, Sutphen C, Herries EM, Ladenson JH, Xiong C, Scheltens P, van der Flier WM, Morris JC, Holtzman DM, Fagan AM (2015) Cerebrospinal fluid VILIP-1 and YKL-40, candidate biomarkers to diagnose, predict and monitor Alzheimer’s disease in a memory clinic cohort. Alzheimers Res Ther 7,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Álvarez I, Diez-Fairen M, Aguilar M, González JM, Ysamat M, Tartari JP, Carcel M, Alonso A, Brix B, Arendt P, Pastor P (2021) Added value of cerebrospinal fluid multimarker analysis in diagnosis and progression of dementia. Eur J Neurol 28, 1142–1152. [DOI] [PubMed] [Google Scholar]
- [22].(2012) SOMAscan® v4 Data Standardization and File Specification Technical Note.
- [23].Armbruster DA, Pry T (2008) Limit of Blank, Limit of Detection and Limit of Quantitation. The Clinical Biochemist Reviews 29, S49. [PMC free article] [PubMed] [Google Scholar]
- [24].Meeker KL, Butt OH, Gordon BA, Fagan AM, Schindler SE, Morris JC, Benzinger TLS, Ances BM (2022) Cerebrospinal fluid neurofilament light chain is a marker of aging and white matter damage. Neurobiol Dis 166,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Henson RL, Doran E, Christian BT, Handen BL, Klunk WE, Lai F, Lee JH, Diana Rosas H, Schupf N, Zaman SH, Lott IT, Fagan AM, Aizenstein HJ, Ances BM, Andrews HF, Bell K, Birn RM, Brickman AM, Bulova P, Cheema A, Chen K, Clare I, Clark L, Cohen AD, Constantino JN, Doran EW, Feingold E, Foroud TM, Hartley SL, Head E, Hom C, Honig L, Ikonomovic MD, Johnson SC, Jordan C, Ilyas Kamboh M (2020) Cerebrospinal fluid biomarkers of Alzheimer’s disease in a cohort of adults with Down syndrome. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring 12, e12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zetterberg H, Skillbäck T, Mattsson N, Trojanowski JQ, Portelius E, Shaw LM, Weiner MW, Blennow K (2016) Association of Cerebrospinal Fluid Neurofilament Light Concentration With Alzheimer Disease Progression. JAMA Neurol 73, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kester MI, Teunissen CE, Crimmins DL, Herries EM, Ladenson JH, Scheltens P, van der Flier WM, Morris JC, Holtzman DM, Fagan AM, Author C (2015) Neurogranin as a Cerebrospinal Fluid Biomarker for Synaptic Loss in Symptomatic Alzheimer Disease Original Investigation. JAMA Neurol 72, 1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Longitudinal Analysis of Cerebrospinal Fluid Visinin-like protein-1, chitinase-3 like-1, synaptosomal-associated protein 25 and neurogranin.
- [29].Kirsebom B-E, Nordengen K, Waterloo K, Torsetnes B, Islad Ottir BG, Brix B, Vanmechelen E, Br Athen G, Hessen E, Aarsland D, Fladby T Cerebrospinal fluid neurogranin/b-site APP-cleaving enzyme 1 predicts cognitive decline in preclinical Alzheimer’s disease. [DOI] [PMC free article] [PubMed]
- [30].Butt OH, Long JM, Henson RL, Herries E, Sutphen CL, Fagan AM, Cruchaga C, Ladenson JH, Holtzman DM, Morris JC, Ances BM, Schindler SE (2021) Cognitively normal APOE ε4 carriers have specific elevation of CSF SNAP-25. Neurobiol Aging 102, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schindler SE, Cruchaga C, Joseph A, McCue L, Farias FHG, Wilkins CH, Deming Y, Henson RL, Mikesell RJ, Piccio L, Llibre-Guerra JJ, Moulder KL, Fagan AM, Ances BM, Benzinger TLS, Xiong C, Holtzman DM, Morris JC (2021) African Americans Have Differences in CSF Soluble TREM2 and Associated Genetic Variants. Neurology Genetics 7, e571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Deming Y, Filipello F, Cignarella F, Cantoni C, Hsu S, Mikesell R, Li Z, Del-Aguila JL, Dube U, Farias FG, Bradley J, Budde J, Ibanez L, Fernandez MV, Blennow K, Zetterberg H, Heslegrave A, Johansson PM, Svensson J, Nellgård B, Lleo A, Alcolea D, Clarimon J, Rami L, Molinuevo JL, Suárez-Calvet M, Morenas-Rodríguez E, Kleinberger G, Ewers M, Harari O, Haass C, Brett TJ, Benitez BA, Karch CM, Piccio L, Cruchaga C (2019) The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer’s disease risk. Sci Transl Med 11,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Morenas-Rodríguez E, Li Y, Nuscher B, Franzmeier N, Xiong C, Suárez-Calvet M, Fagan AM, Schultz S, Gordon BA, Benzinger TLS, Hassenstab J, McDade E, Feederle R, Karch CM, Schlepckow K, Morris JC, Kleinberger G, Nellgard B, Vöglein J, Blennow K, Zetterberg H, Ewers M, Jucker M, Levin J, Bateman RJ, Haass C, Adams S, Allegri R, Araki A, Barthelemy N, Bechara J, Berman S, Bodge C, Brandon S, Bill Brooks W, Brosch J, Buck J, Buckles V, Carter K, Cash L, Chen C, Chhatwal J, Chrem P, Chua J, Chui H, Cruchaga C, Day GS, de La Cruz C, Denner D, Diffenbacher A, Dincer A, Donahue T, Douglas J, Duong D, Egido N, Esposito B, Farlow M, Feldman B, Fitzpatrick C, Flores S, Fox N, Franklin E, Friedrichsen N, Fujii H, Gardener S, Ghetti B, Goate A, Goldberg S, Goldman J, Gonzalez A, Gräber-Sultan S, Graff-Radford N, Graham M, Gray J, Gremminger E, Grilo M, Groves A, Häsler L, Hellm C, Herries E, Hoechst-Swisher L, Hofmann A, Holtzman D, Hornbeck R, Igor Y, Ihara R, Ikeuchi T, Ikonomovic S, Ishii K, Jack C, Jerome G, Johnson E, Käser S, Kasuga K, Keefe S, Bill Klunk W, Koeppe R, [Google Scholar]; Koudelis D, Kuder-Buletta E, Laske C, Levey A, Lopez O, Marsh J, Martinez R, Martins R, Mason NS, Masters C, Mawuenyega K, McCullough A, Mejia A, MountzMD J, Mummery C, Nadkarni N, Nagamatsu A, Neimeyer K, Niimi Y, Noble J, Norton J, O’Connor A, Obermüller U, Patira R, Perrin R, Ping L, Preische O, Renton A, Ringman J, Salloway S, Schofield P, Senda M, Seyfried N, Shady K, Shimada H, Sigurdson W, Smith J, Smith L, Snitz B, Sohrabi H, Stephens S, Taddei K, Thompson S, Wang P, Wang Q, Weamer E, Xu J, Xu X (2022) Soluble TREM2 in CSF and its association with other biomarkers and cognition in autosomal-dominant Alzheimer’s disease: a longitudinal observational study. The Lancet Neurology 21, 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mukaka MM (2012) A guide to appropriate use of Correlation coefficient in medical research. Malawi Medical Journal : The Journal of Medical Association of Malawi 24, 69. [PMC free article] [PubMed] [Google Scholar]
- [35].Yang C, Farias FG, Ibanez L, Sadler B, Fernandez MV, Wang F, Bradley JL, Eiffert B, Bahena JA, Budde JP, Li Z, Dube U, Sung YJ, Mihindukulasuriya KA, Morris JC, Fagan A, Perrin RJ, Benitez B, Rhinn H, Harari O, Cruchaga C Genomic and multi-tissue proteomic integration for understanding the biology of disease and other complex traits. [Google Scholar]
- [36].Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS (2018) Genomic atlas of the human plasma proteome. Nature 558, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, Sarwath H, Thareja G, Wahl A, Delisle RK, Gold L, Pezer M, Lauc G, Selim MAED, Mook-Kanamori DO, Al-Dous EK, Mohamoud YA, Malek J, Strauch K, Grallert H, Peters A, Kastenmüller G, Gieger C, Graumann J (2017) Connecting genetic risk to disease end points through the human blood plasma proteome. Nature Communications 2017 8:1 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu RX, Thiessen-Philbrook HR, Vasan RS, Coresh J, Ganz P, Bonventre J v., Kimmel PL, Parikh CR (2021) Comparison of proteomic methods in evaluating biomarker-AKI associations in cardiac surgery patients. Translational Research 238, 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Begcevic I, Brinc D, Drabovich AP, Batruch I, Diamandis EP (2016) Identification of brain-enriched proteins in the cerebrospinal fluid proteome by LC-MS/MS profiling and mining of the Human Protein Atlas. Clinical Proteomics 13, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kroksveen AC, Opsahl JA, Aye TT, Ulvik RJ, Berven FS (2011) Proteomics of human cerebrospinal fluid: Discovery and verification of biomarker candidates in neurodegenerative diseases using quantitative proteomics. Journal of Proteomics 74, 371–388. [DOI] [PubMed] [Google Scholar]
- [41].Deming Y, Li Z, Kapoor M, Harari O, Del-Aguila JL, Black K, Carrell D, Cai Y, Fernandez MV, Budde J, Ma S, Saef B, Howells B, lin Huang K, Bertelsen S, Fagan AM, Holtzman DM, Morris JC, Kim S, Saykin AJ, de Jager PL, Albert M, Moghekar A, O’Brien R, Riemenschneider M, Petersen RC, Blennow K, Zetterberg H, Minthon L, van Deerlin VM, Lee VMY, Shaw LM, Trojanowski JQ, Schellenberg G, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Peskind ER, Li G, di Narzo AF, Kauwe JSK, Goate AM, Cruchaga C (2017) Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta Neuropathol 133, 839–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ali M, Sung YJ, Wang F, Fernández M v., Morris JC, Fagan AM, Blennow K, Zetterberg H, Heslegrave A, Johansson PM, Svensson J, Nellgård B, Lleo A, Alcolea D, Clarimon J, Rami L, Molinuevo JL, Suárez-Calvet M, Morenas-Rodríguez E, Kleinberger G, Haass C, Ewers M, (ADNI) the ADNI, Cruchaga C (2021) Leveraging large multi-center cohorts of Alzheimer Disease endophenotypes to understand the role of Klotho heterozygosity on disease risk. medRxiv 16, 2021.10.07.21264646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


