Abstract
BACKGROUND
Hyperpolarized 13C MRI quantitatively measures enzyme-catalyzed metabolism in cancer and metabolic diseases. Whole-abdomen imaging will permit dynamic metabolic imaging of several abdominal organs simultaneously in healthy and diseased subjects.
PURPOSE
Image hyperpolarized [1-13C]pyruvate and products in the abdomens of healthy volunteers, overcoming challenges of motion, magnetic field variations, and spatial coverage. Compare hyperpolarized [1-13C]pyruvate metabolism across abdominal organs of healthy volunteers.
STUDY TYPE
Prospective technical development.
SUBJECTS
13 healthy volunteers (8 male), 21–64 years (median 36).
FIELD STRENGTH/SEQUENCE
3T. Proton: T1-weighted spoiled gradient echo, T2-weighted single-shot fast spin echo, multi-echo fat/water imaging. Carbon-13: echo-planar spectroscopic imaging, metabolite-specific echo-planar imaging.
ASSESSMENT
Transmit magnetic field was measured. Variations in main magnetic field (ΔB0) determined using multi-echo proton acquisitions were compared to carbon-13 acquisitions. Changes in ΔB0 were measured after localized shimming. Improvements in metabolite signal-to-noise ratio were calculated. Whole-organ regions of interests were drawn over the liver, spleen, pancreas, and kidneys by a single investigator. Metabolite signals, time-to-peak, decay times, and mean first-order rate constants for pyruvate-to-lactate (kPL) and alanine (kPA) conversion were measured in each organ.
STATISTICAL TESTS
Linear regression, one-sample Kolmogorov-Smirnov tests, paired t-tests, one-way ANOVA, Tukey’s multiple comparisons tests. P≤0.05 considered statistically significant.
RESULTS
Proton ΔB0 maps correlated with carbon-13 ΔB0 maps (slope=0.93, y-intercept=−2.88, R2=0.73). Localized shimming resulted in mean frequency offset within ±25 Hz for all organs. Metabolite SNR significantly increased after denoising. Mean kPL and kPA were highest in liver, followed by pancreas, spleen, and kidneys (all comparisons with liver were significant).
DATA CONCLUSION
Whole-abdomen coverage with hyperpolarized carbon-13 MRI was feasible despite technical challenges. Multi-echo gradient echo 1H acquisitions accurately predicted chemical shifts observed using carbon-13 spectroscopy. Carbon-13 acquisitions benefited from local shimming. Metabolite energetics in the abdomen compiled for healthy volunteers can be used to design larger clinical trials in patients with metabolic diseases.
Keywords: hyperpolarized MR, [1-13C]pyruvate, abdomen, liver, metabolic imaging
INTRODUCTION
Hyperpolarized (HP) carbon-13 (13C) MRI permits rapid, dynamic, quantitative measurements of in vivo enzyme-catalyzed metabolism in humans (1). In recent years, it has been applied in patients with cardiac diseases, cancers, and other metabolic diseases, with the goal of improved patient care and outcomes (2–6). HP 13C MRI is enabled by dynamic nuclear polarization of 13C-labeled compounds, such as [1-13C]pyruvate (7). Upon dissolution, HP compounds can be safely injected into human subjects while retaining an observable signal that is ≥10,000 times higher than thermal values (1).
HP [1-13C]pyruvate is of particular clinical relevance because pyruvate sits at the intersection of multiple major metabolic pathways that are metabolically reprogrammed in cancers and other diseases (8–10). As the product of glycolysis, pyruvate is either exchanged into lactate or else into acetyl-CoA and bicarbonate prior to the tricarboxylic acid cycle. Pyruvate can also be converted to alanine via alanine amino transferase. This process leads to three potentially observable HP 13C products from [1-13C]pyruvate: [1-13C]lactate, 13C-bicarbonate, and [1-13C]alanine. Analysis and kinetic modeling can quantify the enzymatic conversion rate of pyruvate to lactate through lactate dehydrogenase (LDH) (1, 11, 12). The apparent first-order rate constant, kPL, is a potential biomarker that is elevated in cancer and reflects treatment response in animal models and in humans (1–3, 13–15). Hence, HP [1-13C]pyruvate MRI has seen much success in characterizing metabolism in the prostate, brain, heart, the kidneys, and in quantifying response to therapy in multiple cancers (2–5, 16, 17). The apparent first-order rate constant of pyruvate conversion to alanine, kPA, can also be measured and quantified.
The liver is a common site of tumors and metabolic diseases including non-alcoholic fatty liver disease (NAFLD). NAFLD, which is characterized by the ectopic accumulation of fat in the liver, can progress to non-alcoholic steatohepatitis (NASH), with the risk of cirrhosis, portal hypertension, and hepatocellular carcinoma (18). The current gold standard for distinguishing between simple steatosis and NASH is with a biopsy. New improved methods are needed to better distinguish between simple steatosis and NASH noninvasively. Pre-clinical models of liver injury, diabetes, and fatty liver have revealed that HP 13C MRI is responsive to changes in liver metabolism that occur in these diseases (19–22).
Clinical translation of these methods to benefit patients requires reliable whole-abdomen imaging methods. Whole-abdomen coverage using HP 13C MRI is challenging for the following reasons. First, the large field of view requires adequate transmitter and receiver coil coverage. Second, physiologic motion such as respiration, cardiac activity, and bowel peristalsis requires a fast, motion-insensitive imaging sequence. Finally, the presence of air-filled structures such as the lungs and the bowel can cause non-uniformities in the magnetic field (B0) that could confound metabolite-specific imaging methods.
The first aim of this study was to develop a specialized approach combining several important imaging tools to overcome these limitations for performing HP 13C MRI throughout the whole abdomen in healthy volunteers. The second aim of this study was to obtain preliminary estimates of normal HP [1-13C]pyruvate metabolism and its variation in solid abdominal organs of healthy volunteers.
MATERIALS AND METHODS
This study was approved by the local Institutional Review Board (IRB), with written informed consent under a protocol approved by the US Food and Drug Administration Investigational New Drug (FDA IND) application.
Radiofrequency Coil Array
HP 13C data were acquired using a volumetric transmitter and an 8-channel flexible receiver array (QTAR, Clinical MR Solutions, Brookfield, WI) that covered the abdomen. Figure 1A and 1B show the internal coil arrays of the transmitter and receiver components, respectively. The transmitter coil was 33.0 × 22.9 cm and each receiver coil was 21.5 × 15.0 cm.
Figure 1.
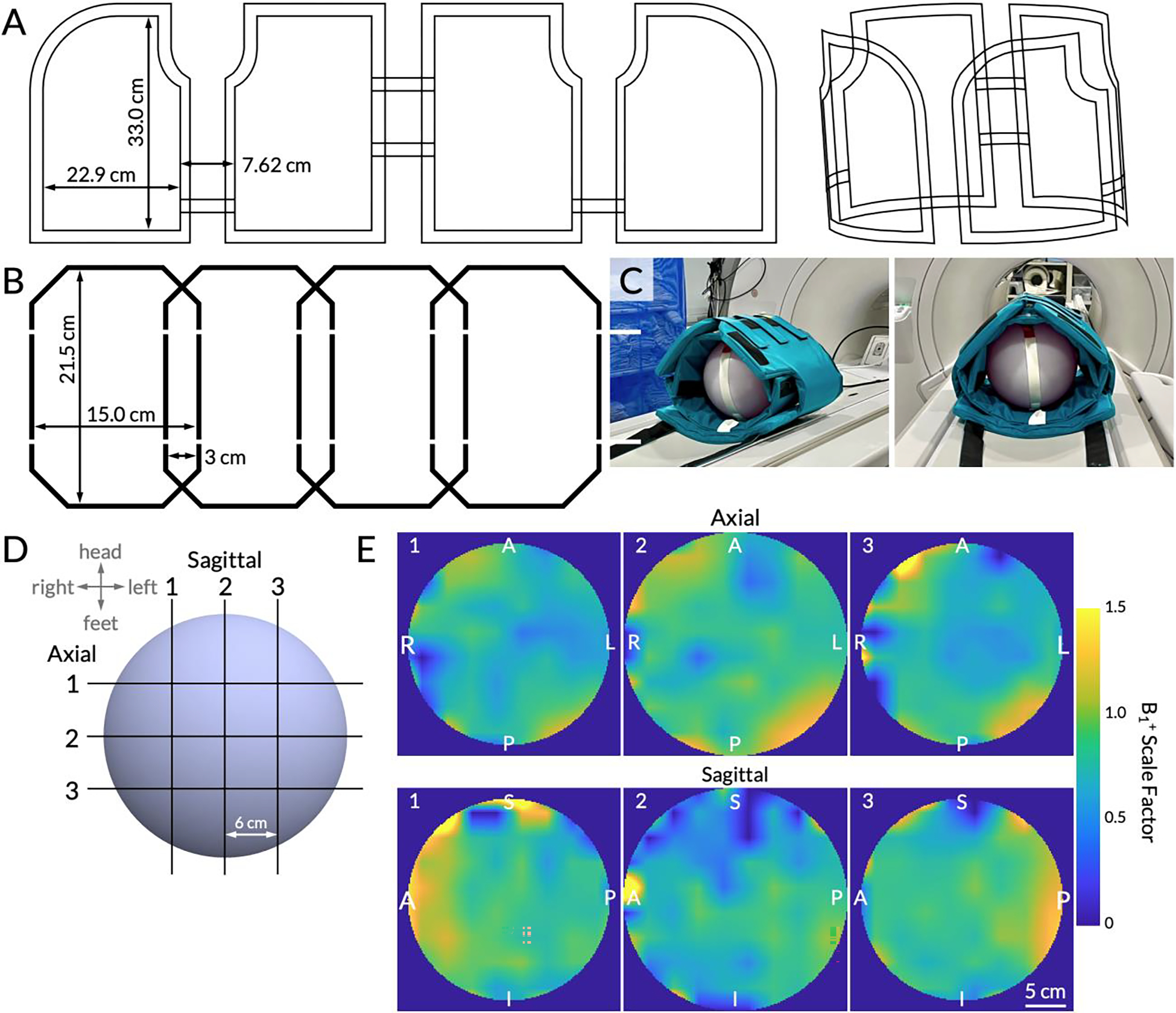
(A) Schematic conductor paths of the 13C volumetric transmitter, shown flat and in 3D. (B) Schematic conductor paths of the 13C receiver array. One of two identical 4-channel arrays is shown. (C) The coil setup using a 27.8 cm wide spherical dimethyl silicon phantom for Bloch-Siegert B1+ mapping. The inner vest is the 8-channel receiver array and the outer vest is the transmitter. (D) A schematic of the multi-slice B1+ maps. Three slices were acquired in the sagittal and axial planes with a 6 cm center-to-center separation. The slice numbers correspond to the B1+ maps (interpolated for display) shown in (E). Right/Left, Anterior/Posterior, and Superior/Inferior labels are displayed on the edges. The adjacent color bar shows the B1+ scale factor ranging from 0 to 1.5, with 1.0 indicating transmit power that matches the prescribed value.
B1+ Mapping
In order to characterize the spatial uniformity of the transmitter coil, transmit field (B1+) mapping was performed on a 27.8 cm-diameter spherical dimethyl silicon phantom using a Bloch-Siegert acquisition scheme on a clinical 3 T scanner (MR750, GE Healthcare, Milwaukee, WI) with commercial software (RTHawk, HeartVista, Inc., Los Altos, CA) (Figure 1C) (23). The imaging sequence followed previously published methods beginning with a single-band spectral-spatial radiofrequency (RF) pulse with a 130 Hz full width at half maximum (FWHM) passband and an 868 Hz stopband (all subsequently stated frequencies in text and in figures have been scaled to reflect 13C imaging) (23, 24). The excitation pulse was followed by an off-resonance Fermi pulse (duration = 12 ms, frequency offset = ±4.5 kHz, and phase shift constant, KBS, of 6.76 rad/G2) and a single-shot spiral readout (23, 25). Other scan parameters were: repetition time (TR) = 300 ms, echo time (TE) = 21 ms, acquisition matrix = 30 × 30 voxels, in-plane spatial resolution = 30 × 30 mm, flip angle = 75°, acquisition bandwidth = 250 kHz, and signal averages = 200.
Three 3.0 cm-thick slices were acquired in sagittal and axial planes, each separated by 6.0 cm (Figure 1D shows a schematic of these slices). Data were reconstructed using MATLAB 2021a (The MathWorks, Inc., Natick, MA) following previously published methods (23). The complex conjugate of the positive phase map was multiplied with the negative phase map. After taking the sum in the coil dimension, the phase, ϕBS, of the resulting matrix was obtained. The B1+ map was calculated using the equation:
where B1,ref is a reference maximum power of the Fermi pulse (0.3 G) (23, 25–27). The resulting B1+ values were normalized such that a value of 1.0 indicated delivery of the prescribed flip angle. Images were interpolated to match the resolution of proton (1H) images with a final matrix size of 576 × 576. Regions outside the phantom were masked as determined by the phantom’s size and the image voxel size. The mean and standard deviation of the B1+ scale factors were calculated over a circular 10 cm region of interest (ROI) placed in the center of the middle slice and the four quadrants.
Study Eligibility
Inclusion criteria for this pilot study were age ≥18 years at the time of study. Subjects were excluded if they had 1) poorly controlled hypertension (systolic blood pressure >160 mmHg or diastolic blood pressure >100 mmHg); 2) congestive heart failure (New York Heart Association status ≥2); 3) pregnant or nursing; or 4) any contraindications to MRI.
1H MRI Scans
Proton imaging was performed with standard clinical imaging sequences and the full-volume body coil for both transmit and receive (Table 1). T1-weighted spoiled gradient echo and T2-weighted single-shot fast spin echo images were obtained for scan planning and anatomic registration of 13C data. In addition, multi-echo gradient echo acquisition (Iterative Decomposition of water and fat with Echo Asymmetry and Least square estimation (IDEAL IQ), GE Healthcare) was used to generate fat fraction, T2*, and ΔB0 maps. All images were acquired at end-inspiration to match the positioning of the 13C images.
Table 1.
Scan parameters for all proton (1H) and carbon-13 (13C) human scans.
| Sequence | Repetition time (TR); echo time (TE) | In-plane spatial resolution | Matrix size; FOV | # Slices; slice thickness; slice gap | Flip angle | Pixel Bandwidth | Acquisition time; temporal resolution |
|---|---|---|---|---|---|---|---|
| T1-weighted spoiled gradient echo (1H anatomical images) | 3.73–4.12 ms; 1.67–1.88 ms | 0.352 × 0.352 to 0.938 × 0.938 mm | 512 × 512 to 1024 × 1024; 36 × 36 to 48 × 48 cm | 44–152; 4.4–10 mm; 0 mm | 8° | 391–558 Hz | 30 s; N/A |
| T2-weighted single-shot fast spin echo (axial 1H anatomical images) | 545–2200 ms; 48.6–83.8 ms | 0.703 × 0.703 to 1.563 × 1.563 mm | 256 × 256 to 512 × 512; 30 × 30 to 40 × 40 cm | 40–92; 5–6 mm; 0 mm | 90° | 195–326 Hz | < 60 s total scan time divided into ≥ 4 acquisitions; N/A |
| T2-weighted single-shot fast spin echo (coronal 1H anatomical images) | 1690–4400 ms; 9.70–86.4 ms | 0.664 × 0.664 to 0.781 × 0. 781 mm | 512 × 512; 340 × 340 to 400 × 400 cm | 10–36; 5–14 mm; 0–1 mm | 90° | 195–244 Hz | < 60 s total scan time divided into ≥ 4 acquisitions; N/A |
| Multi-echo gradient echo (ΔB0 maps) | 6.73–7.70 ms; 3.11–3.592 ms; 7 echoes; 0.86 ms echo spacing | 1.406 × 1.406 to 1.875 × 1.875 mm | 256 × 256; 36 × 36 to 48 × 48 cm | 16–46; 7–10 mm; 0 mm | 3–4° | 710 Hz | < 35 s; N/A |
| Echo-planar spectroscopic imaging (EPSI) (13C spectroscopic images) | 130 ms; 3.5–5.2 ms; 53 echoes; 1.83 ms echo spacing (545 Hz spectral bandwidth) | 20 × 20 to 22 × 22 mm | 16 × 18; 32 × 36 cm to 35.2 × 39.6 cm | 1; 20–30 mm; N/A | pyruvate: 10°; lactate: 20°; alanine: 15° (constant flip angles) | Slice-selective multiband spectral-spatial; | 60 s; 3 s |
| Echo-planar imaging (EPI) (13C metabolite images) | 64–72 ms; 22 ms | 20 × 20 mm | 16 × 16; 32 × 32 cm | 7–14; 20 mm; 20 mm | pyruvate: 30°; lactate: 60°; alanine: 60° (constant flip angles) | Slice-selective single-band spectral-spatial | 60 s; 3 s |
Hyperpolarized 13C MRI Scans
Hyperpolarization of [1-13C]pyruvate
A mixture of 1.46 g Good Manufacturing Practice grade [1-13C]pyruvic acid (ISOTEC Stable Isotope Division, MilliporeSigma, Miamisburg, OH) and 15 mM trityl radical (GE Healthcare, Milwaukee, WI) was prepared and loaded in pharmacy kits (GE Healthcare, Milwaukee, WI) according to the IRB- and FDA IND-approved manufacturing processes. Following polarization for at least two hours in a 5 T SPINlab clinical-research polarizer (GE Healthcare, Milwaukee, WI), the sample was rapidly warmed, dissolved, and neutralized in 18.25 g of buffer composed of 333 mM Tris, 600 mM sodium hydroxide, and 333 mg/L disodium EDTA. Injection was performed following approval of a staff clinical pharmacist. The injection rate was 5 mL/s followed by a 20 mL saline flush. The injection volume was determined using the weight-based dose of 0.43 mL/kg (total body weight) with a maximum of 40 mL.
Radiofrequency Power Calibration
The RF pulse was calibrated using a spherical dimethyl silicon phantom and a 2 ms hard pulse. The transmit gain for a 90° flip was recorded and confirmed using a 30° flip which resulted in a signal with half the maximum amplitude.
Subject and Coil Positioning
After calibration, the subject was placed on the scanner bed and the QTAR receiver array was first fitted around the abdomen followed by the transmitter. The positioning of the coil was iteratively adjusted using embedded fiducial markers and 1H anatomical scans to ensure coil coverage from the top of the liver to the bottom of the kidneys.
13C Center Frequency Determination
An initial estimate of the center frequency for the HP [1-13C]pyruvate acquisition was determined using the center frequency of the 1H scans and a conversion factor empirically derived from prior acquisitions (28). For a subset of the subjects receiving two HP 13C scans, further adjustments were made according to the procedure described below.
Shimming
For all subjects, shimming was initially determined based on the scanner’s built-in auto-shimming algorithm that was performed at the time of the IDEAL IQ acquisition. For a subset of scans, additional shimming was performed using a localized shim box to reduce B0 inhomogeneities within the target volume. The targeted shim region was centered on the right hepatic lobe and kept below the diaphragm to avoid air-tissue interfaces. The size of the local shim box varied depending on the size of the liver and measured 10 cm Left/Right, 8–12 cm Anterior/Posterior, and 11–15 cm Superior/Inferior. The z-shim was further adjusted manually to reduce residual B0 inhomogeneities in the through-plane direction.
Quantification of the final state of B0 homogeneity within each organ was performed post-injection using whole organ 3D ROIs drawn in MATLAB by an investigator (PML, 4 years). Frequency variations without and with local shimming were visualized with box plots. Outliers were defined as values more than 1.5 times the interquartile range from the top or bottom of the box. Upper and lower quartile bounds were used to compare not only frequency variance but also to evaluate whether the majority of the voxels in each organ were within the FWHM, or 50% excitation, of the excitation pulse’s passband.
Hyperpolarized 13C Acquisition Timing
The serial dynamic HP 13C MRI acquisition was started 5 s after the saline injection completed. Each subject was asked to hold their breath as long as easily tolerated, followed by free breathing for the rest of the 60 s acquisition.
Echo-planar Spectroscopic Imaging
The 2D dynamic echo-planar spectroscopic imaging (EPSI) pulse sequence consisted of a slice-selective multiband spectral-spatial pulse followed by an EPSI readout similar to prior prostate cancer studies (3, 29, 30). The scan parameters were (Table 1): TR = 130 ms, TE = 3.5–5.2 ms, number of echoes = 53, echo spacing = 1.83 ms, spectral bandwidth = 545 Hz, in-plane spatial resolution = 20 × 20 to 22 × 22 mm, matrix size = 16 phase encode × 18 readout voxels, slice thickness = 20–30 mm, constant flip angles = 10° for pyruvate, 20° for lactate, and 15° for alanine, acquisition time = 60 s, and temporal resolution = 3 s.
EPSI-derived ΔB0 Maps
EPSI data were processed according to previously published methods, including tensor rank truncation, phase corrections, and baseline correction, but not B0 (this would nullify the desired ΔB0 maps) or B1+ corrections (31). For each voxel, spectra were summed through time and the highest value within the chemical shift range of 171.13 to 174.10 ppm was taken to be the pyruvate peak. An empirical pyruvate peak reference value of 172.58 ppm was used to compute the ΔB0 for each voxel, resulting in EPSI-derived ΔB0 maps (subsequently referred to as 13C ΔB0 maps).
These 13C ΔB0 maps were compared against those acquired using the 1H multi-echo gradient echo acquisitions (subsequently referred to as 1H ΔB0 maps). First, the 1H ΔB0 maps were scaled to account for the different gyromagnetic ratio of 13C and then an ROI was drawn around the abdomen by an investigator (PML, 4 years). The 1H ΔB0 maps were then down-sampled to match the spatial resolution of the 13C ΔB0 maps (Figure 2A). An empirical signal-to-noise ratio (SNR) threshold of 40.0 for pyruvate was used to filter out voxels in both ΔB0 maps. In addition to taking their difference, the two ΔB0 maps were plotted against each other and linear regressions were computed to quantify correlations. To investigate biases, the differences in frequencies between the two ΔB0 maps were plotted against the 1H ΔB0 map.
Figure 2.
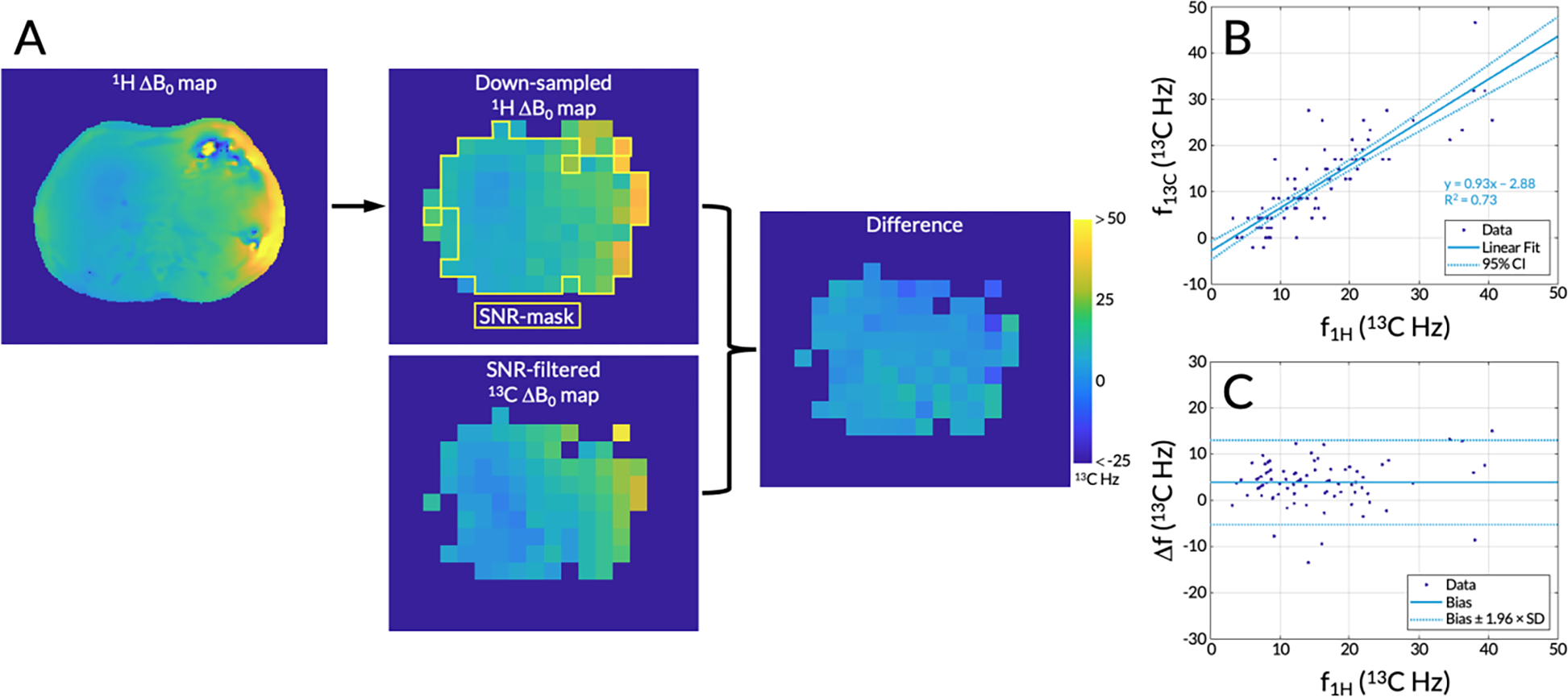
(A) A representative 1H ΔB0 map, which was down-sampled to match the spatial resolution of the 13C ΔB0 map. A signal-to-noise ratio (SNR) filter using empirical thresholds based on the echo-planar spectroscopic imaging (EPSI) data was applied to both ΔB0 maps. Voxels in the 1H ΔB0 map that passed this SNR filter are highlighted with a yellow outline. Both ΔB0 maps were scaled using the 13C gyromagnetic ratio and the difference between the two maps was taken. The same color bar is applicable to all images. (B) A correlation plot of the frequencies from each separate ΔB0 map plotted against each other with 95% confidence interval (CI) shown. (C) A Bland-Altman plot showing the bias between the two ΔB0 maps with dotted lines indicating ±1.96 × the standard deviation, or 95% of the differences having tested for normality.
Metabolite-specific Echo-planar Imaging
[1-13C]pyruvate, [1-13C]lactate and [1-13C]alanine signals were acquired using a multi-slice metabolite-specific echo-planar imaging (EPI) acquisition (32). The scan parameters were (Table 1): TR (time per metabolite per slice) = 64–72 ms, TE = 22 ms, in-plane spatial resolution = 20 × 20 mm, matrix size = 16 × 16 voxels, number of slices = 7–14, slice thickness = 20 mm, constant flip angles = 30° for pyruvate and 60° for lactate and alanine, acquisition time = 60 s, and temporal resolution = 3 s. A variable number of slices was used in order to cover the top of the liver to the bottom of the kidneys.
When the EPI scan was the sole 13C acquisition, the center frequency was determined using the center frequency of the 1H scans and the empirical conversion factor discussed earlier. When the EPI scan was preceded by an EPSI scan, the initial estimate of the center frequency was adjusted based on the acquired spectra.
Data Processing and Kinetic Modeling
Kinetic modeling was performed based on data from metabolite-specific EPI acquisitions. Data were phase corrected, Fourier transformed, pre-whitened, and coil-combined using the pyruvate signals to estimate the coil weights in MATLAB, as previously described (33). Data were denoised using a patch-based higher-order singular value decomposition (HOSVD) with the following previously-published parameters: patch size = 5 × 5, search window size = 11, step length = 2, kglobal = 0.4 and klocal = 0.8 (34).
Data were linearly interpolated in the slice dimension by a factor of 4 and then interpolated in the x and y dimensions by zero-padding k-space also by a factor of 4. A 2D radially symmetric Fermi filter was computed using the following Fermi-Dirac distribution:
where x and y are the number of voxels in the image’s two dimensions, , and , with N being the number of voxels in the image’s longest dimension. The zero-padded k-space data were multiplied by the Fermi filter and subsequently 2D inverse Fourier transformed.
An ROI was drawn around the entire abdomen by an investigator (PML, 4 years) to generate a mask to remove voxels outside of the abdomen. The SNR of each voxel was computed as the AUC 13C signal divided by the standard deviation of background noise (calculated using test scans acquired prior to injection). Empirical SNR thresholds (pyruvate = 50.0, lactate = 20.0, and alanine = 10.0) were then used to create SNR masks to filter out noise voxels. Quantification of SNR improvement was measured by the percent of voxels within the abdominal ROI that passed these metabolite SNR thresholds before and after denoising. Additionally, the mean SNR of the voxels within the abdominal ROI was computed for each metabolite over the 20 timepoints and compared against the SNR thresholds (divided by time due to data being dynamic) to evaluate improvements in SNR after denoising.
For each subject with an EPI scan, the pyruvate dynamic curves of every voxel in five organs of interest (the liver, right and left kidneys, pancreas, and spleen) were characterized by the metrics time-to-peak and decay time. Time-to-peak was defined as the timepoint when the pyruvate signal reached its highest value after the start of acquisition. Timepoints greater than 50 s were rejected.
Decay time was defined as the interval between the peak timepoint and the following half-maximum timepoint. To calculate the decay time, the two timepoints before and after the signal crossed the half-maximum value were obtained. If neither of the two timepoints were greater than 50 s, the signal was linearly interpolated to estimate the half-maximum timepoint. The half-maximum timepoint was subtracted by the time-to-peak to give the decay time. Finally, both metrics in all organs were normalized by the mean values of the right and left kidneys.
Metabolite signals were summed over the entire time course to produce metabolite area under the curve (AUC) maps. First-order conversion rates of pyruvate to lactate (kPL) and of pyruvate to alanine (kPA) were computed using an inputless two-site exchange model (11). Fitting was performed across a range of flip angles centered around the nominal values and the flip angles that resulted in the lowest lactate and alanine root-mean-square error were chosen. In addition to the flip angles, other parameters given to the model were: number of phase encodes per timepoint = 1, number of timepoints = 20, TR = 3, T1 of pyruvate = 30 s, T1 of lactate = 25 s, and T1 of alanine = 25 s. Initial estimates of kPL and kPA of 0.02 s−1 and 0.01 s−1, respectively, were used as starting conditions for the fits.
Both 1H and 13C MR images were imported into ITK-SNAP (35). Five 3D ROIs were drawn on the 1H images for the liver, right and left kidneys, pancreas, and spleen by an investigator (PML, 4 years). From these 3D ROIs, mean and standard deviation values of pyruvate time-to-peak and decay time, pyruvate, lactate, and alanine AUC signals, and kPL and kPA values were computed for each organ. Pyruvate time-to-peak, decay time, kPL, and kPA values for each organ were also averaged across EPI scans.
Statistical Analysis
Statistical analysis was conducted in GraphPad Prism 9 (GraphPad Software, San Diego, CA) and MATLAB. Linear regression and Bland-Altman analysis were conducted to investigate correlations between the 1H and 13C ΔB0 maps. One-sample Kolmogorov-Smirnov tests were used to test for normality (P ≥ 0.05 to accept the null hypothesis that data come from a normal distribution). Upon confirming normality, paired t-tests were used to evaluate effects of denoising and an ordinary one-way ANOVA was conducted on the averaged kPL and kPA values. Tukey’s multiple comparisons tests were also used to investigate differences in kPL and kPA between organs. P ≤ 0.05 was considered to be statistically significant.
RESULTS
B1+ Characterization of QTAR
Multi-slice B1+ maps of the QTAR coil were acquired with a center-to-center separation of 60 mm (Figure 1D). B1+ maps acquired using a Bloch-Siegert acquisition revealed higher-than-nominal transmit power in the right-anterior regions (Figure 1E, axial images #1–3 and sagittal image #1). Higher transmit power was also observed in the left-posterior regions, similarly reflected in the corresponding sagittal image (Figure 1E, axial images #1–3 and sagittal image #3).
The mean B1+ scale factor in a central, circular 10 cm region was 0.80±0.10 (Figure 1E, axial image #2). The maximum value was 1.03 and the minimum was 0.36. In the right-anterior region of the central axial slice (where the liver would be), the mean B1+ scale factor was 0.90±0.16. The left-posterior region had a mean B1+ scale factor of 0.95±0.13. Lower mean B1+ scale factors were observed in the right-posterior and the left-anterior regions, which had mean B1+ scale factors of 0.80±0.16 and 0.75±0.13, respectively.
Healthy Volunteer Scans
A total of thirteen healthy volunteers were prospectively recruited for this study. A summary of all HP 13C acquisitions is shown in Table S1. Eight subjects were male and five were female. Subjects were between 21 and 64 years old with a median age of 36 years. Of the thirteen subjects, subject #6 was unable to be imaged due to hardware failure. Due to the iterative nature of this pilot study, two subjects were scanned exclusively with a different pulse sequence or with a different coil than the other subjects. Therefore, data from these subjects were not considered in the final analysis. Of the remaining ten subjects, three had an EPSI acquisition alone, five had an EPI acquisition alone, and two had both.
Pre-injection quality control results were as follows: sterile pyruvate concentration of 245±13 mM, pH of 7.6±0.3, temperature of 32.4±1.1 °C, and a polarization of 32.7±8.5%. Time from dissolution to injection was 51.2±3.3 s.
1H vs. 13C ΔB0 Maps using Pyruvate’s Frequency Offsets
In one representative case, 1H and 13C ΔB0 maps correlated significantly with a linear regression slope of 0.93±0.12 (95% confidence interval), a y-intercept of −2.88±1.99, and an R2 of 0.74 (Figure 2B). The bias between the 1H and 13C ΔB0 maps was 3.90±9.18 Hz and differences ranged from −13.51 to 15.07 Hz (Figure 2C). A one-sample Kolmogorov-Smirnov test accepted the null hypothesis that the differences between the two maps came from a normal distribution (P = 0.34). Hence, 95% of the differences fall between ±1.96 × the standard deviation, that is, −5.27 and 13.08 Hz (Figure 2C).
B0 Inhomogeneity Corrections
Representative axial 1H ΔB0 maps without and with local shimming are shown in Figure 3A. The frequency offsets of various regions of interest in the liver and kidneys are highlighted. The location of the shim box is shown on top of T2-weighted anatomical references. Box plots of the frequency variation within each organ of interest are shown in Figure 3B, both without and with local shimming. Outliers are indicated with X’s. The dashed lines at ±50 Hz correspond to the FWHM, or 50% excitation, of the single-band spectral-spatial pulse’s passband used in the metabolite-specific EPI sequence (36).
Figure 3.
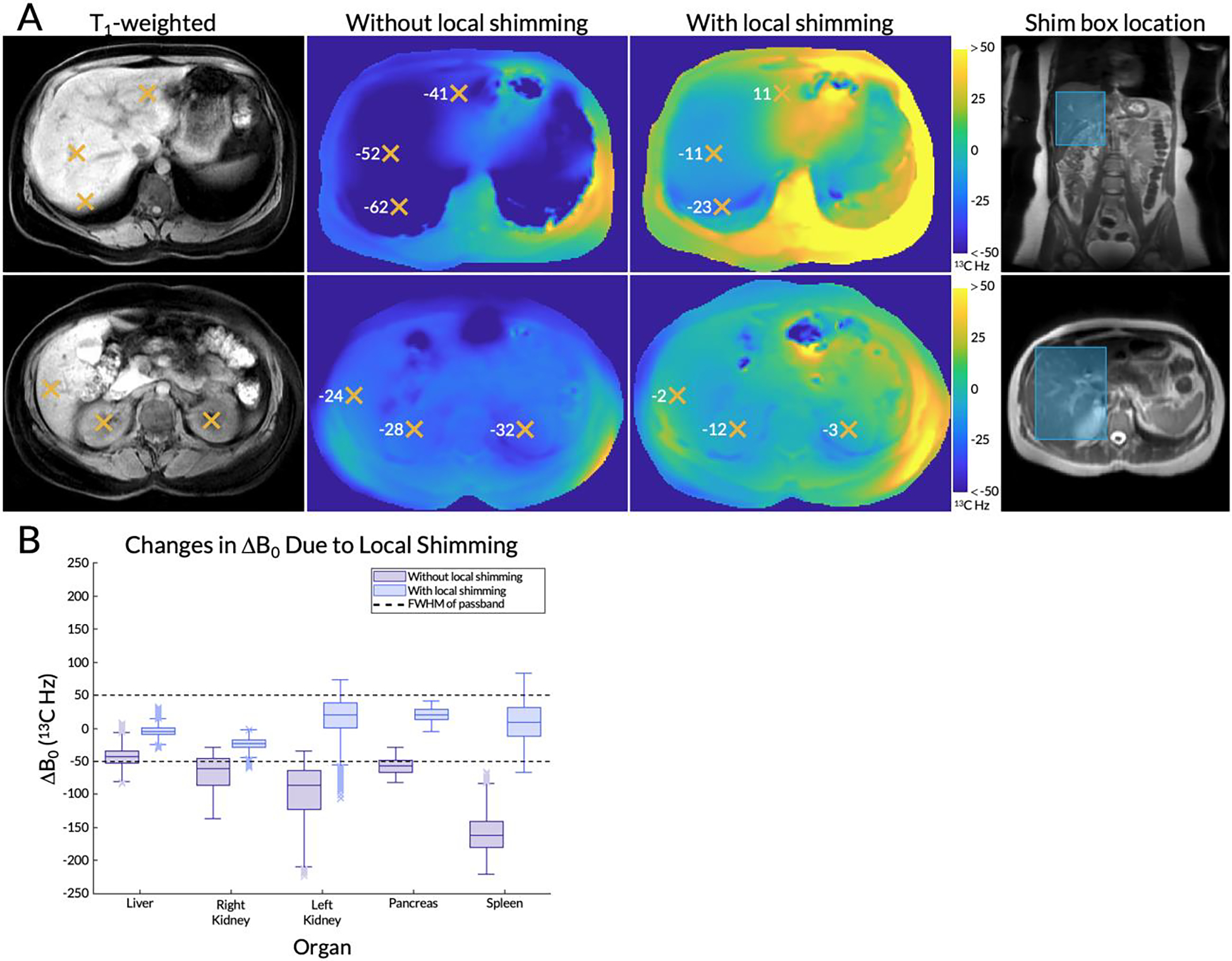
(A) Axial 1H ΔB0 maps from a representative subject for two slices of interest (T1-weighted anatomical images on the left) without and with local shimming. The frequency offsets (scaled to 13C Hz) of various regions of interest in the liver and kidneys are highlighted with orange X’s. The location of the shim box is shown on top of T2-weighted anatomical references. (B) A box and whisker plot of the frequency variation within each organ, both with and without local shimming. Outliers are indicated with X’s. The dashed lines at ±50 13C Hz correspond to the full width at half maximum (FWHM), or 50% excitation, of the single-band spectral-spatial pulse’s passband used in the metabolite-specific echo-planar imaging (EPI) sequence.
Without local shimming, frequency variations in the liver ranged from −80.74 to −6.54 Hz (excluding outliers), with a median of −43.26 Hz. The lower quartile was −53.07 Hz, at the edge of the spectral-spatial pulse’s passband. The upper quartiles of the frequency variations in the right and left kidneys were −46.03 and −64.14 Hz, respectively. Hence, the majority of their voxels was outside of the 50% excitation passband. The pancreas had an upper quartile of −49.30 Hz and the spleen’s upper whisker reached −83.51 Hz, with its entire interquartile range far beyond the passband. After local shimming in the liver, median frequency offsets and variations in B0 were both reduced in the liver as expected (median was −5.03 Hz and whiskers ranged from −24.65 to 15.09 Hz), but also in other organs. All organs had a median value between ±25 Hz and the upper and lower whiskers of the liver, right kidney, and pancreas were also within the passband. The left kidney and spleen (located further from the shim box) contained some voxels with frequencies outside of the passband.
Patch-Based Denoising
In one representative subject, EPI data denoised using HOSVD had reduced background noise across all timepoints (Figure 4A). Figure 4B shows the fifth timepoint (time = 12 s) of the central 13C slice enlarged, with arrows pointing to the head and tail of the pancreas. Quantification over data from all seven EPI scans showed that before denoising, 27±13% of all pyruvate voxels in the abdomen passed the SNR threshold of 50.0. This increased significantly to 87±15% after denoising (Figure 4C). Similar significant increases were observed for lactate (20±8.0% to 87±13%) and alanine (22±10% to 91±12%). The large variance observed after denoising is attributable principally to data from one subject (subject #9, Figure S1). Despite this outlier, tests for normality using data from all seven subjects passed with all P ≥ 0.19 in accepting the null hypothesis. For the same slice highlighted in Figure 4B, the mean SNR within the abdomen was also elevated throughout the entire time course after denoising (Figure 4D). Before denoising, the mean SNR dropped below the empirical SNR thresholds (scaled for dynamic data) at time = 30 s for pyruvate, time = 36 s for lactate, and time = 51 s for alanine. After denoising, the SNR did not drop below the thresholds over the 60 second time course.
Figure 4.
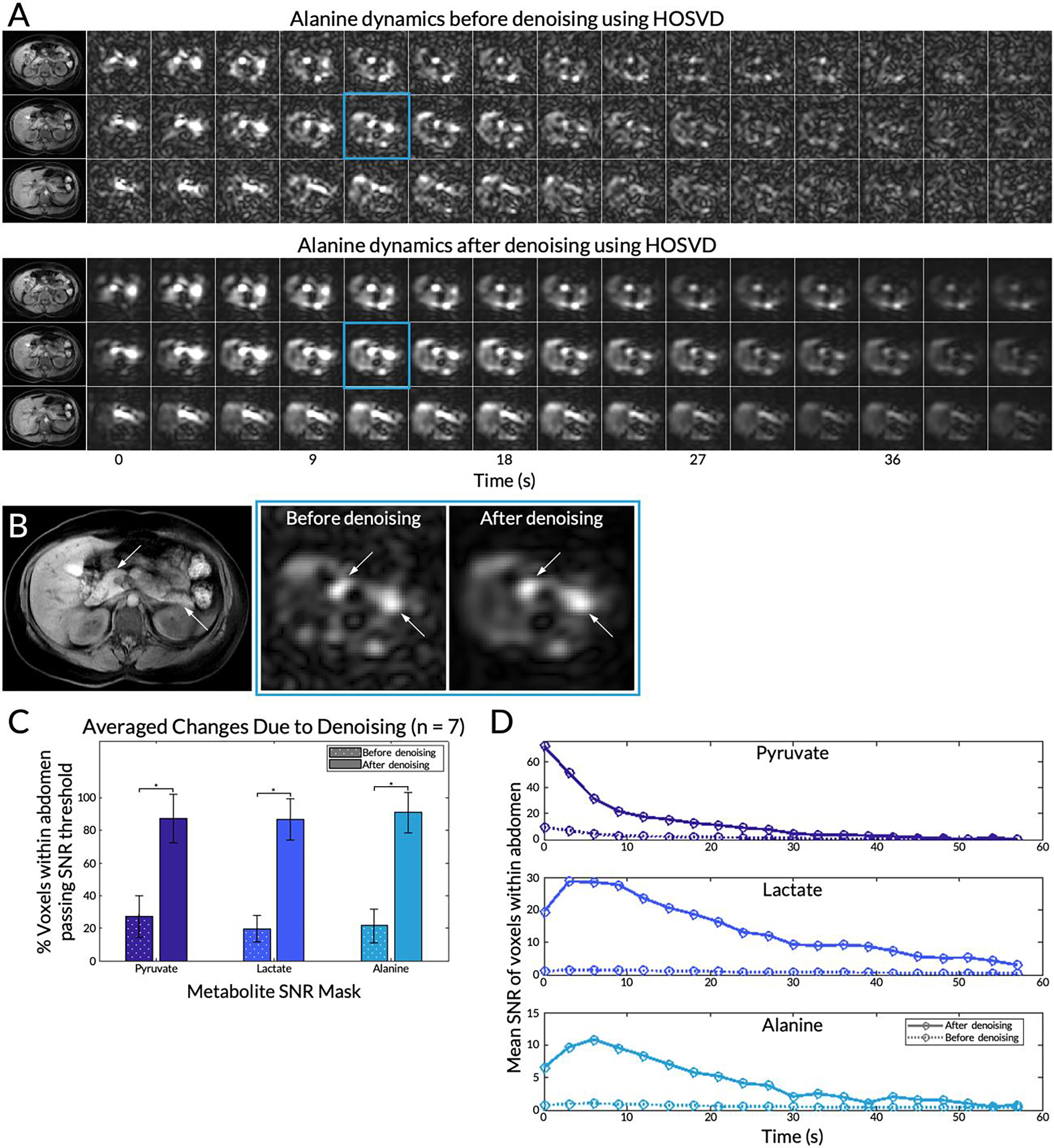
(A) The first 42 seconds of the alanine dynamics before and after patch-based higher-order singular value decomposition (HOSVD) denoising for three slices from a representative subject. Corresponding T1-weighted anatomical references are on the left. (B) Detailed view of the fifth timepoint images (time = 12 s) of the central 13C slice before and after denoising enlarged. Left and right arrows point to the head and tail of the pancreas, respectively. (C) Percent of voxels passing the SNR thresholds for each metabolite before and after denoising. Error bars indicate standard deviations. A single asterisk * denotes statistical significance. (D) For the same slice highlighted in (B), the mean SNR of voxels within the whole abdomen before and after denoising over the course of the 20 timepoints.
Metabolite Dynamics in Each Organ
Plots of the dynamic curves for each metabolite in the right kidney, liver, spleen, and pancreas from a representative EPI scan are shown in Figure 5A. The raw signals are plotted with circles and dashed line while the fitted curves, computed by the inputless two-site exchange model, are shown in solid lines. Of the seven subjects with EPI scans, data from subject #13 was not considered because acquisition began after the peak pyruvate signal in the kidneys had occurred. From the remaining six scans, the mean normalized pyruvate peak time in the liver was 2.9±−2.4 s (Figure 5B). Those of the kidneys were 0 s due to normalization. The time-to-peak in the pancreas and spleen were −0.7±0.7 s and 1.1±1.2 s, respectively. Decay time was highest in the liver and the spleen (P ≤ 0.052) at 4.6±1.8 s and 4.9±2.3 s, respectively. Tests for normality passed for both metrics in all organs (all P ≥ 0.52). One-way ANOVA tests for both metrics across organs were significant. However, comparisons of time-to-peak between organs were not statistically significant (all P ≥ 0.069). While comparisons of decay time in the liver and the spleen were not significantly different, comparisons between the liver and the other organs were.
Figure 5.
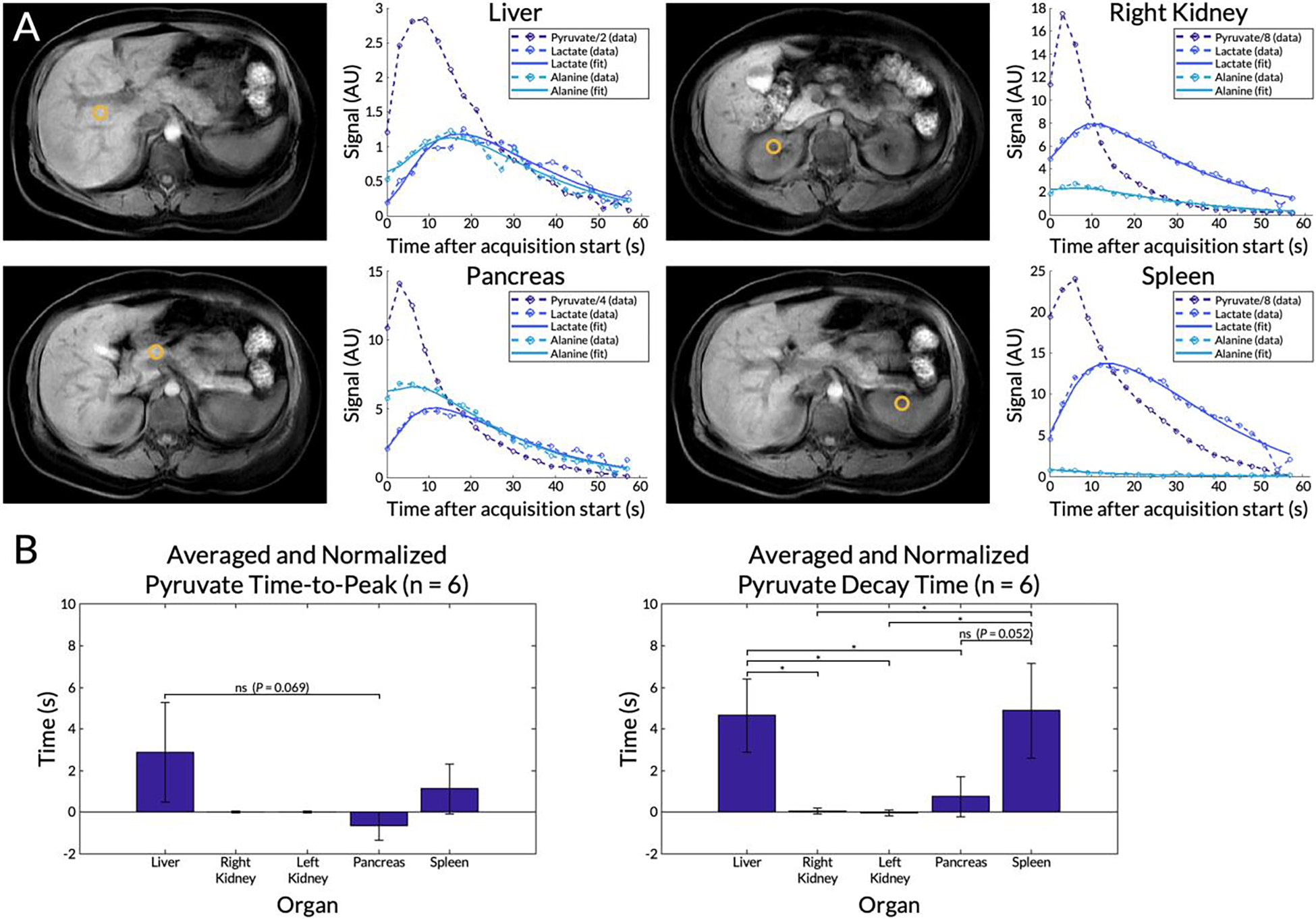
(A) Dynamic curves for each metabolite from a selected voxel in the liver, right kidney, pancreas, and spleen from a representative EPI scan. Locations of the voxels are shown in the corresponding T1-weighted anatomical images. Measured signals (circles and dashed lines) are shown together with curve fits computed using an inputless two-site exchange model (solid lines). The pyruvate signals were scaled for display. (B) Mean pyruvate time-to-peak and decay time for each organ averaged across six subjects with EPI scans (subject #9 was excluded). Error bars indicate standard deviations. A single asterisk * denotes statistical signiifcance, while “ns” denotes not statistically significant. Unlabeled comparisons were also not statistically significant.
Summed Metabolite Signals and Metabolite Kinetics in Each Organ
From a representative subject, the metabolite maps summed through time are displayed in Figure 6A, overlaid on top of a T1-weighted anatomical reference. The mean pyruvate, lactate, and alanine signals within each organ for this subject are shown in Figure 6B. In this subject, mean pyruvate signals were highest in the spleen (1.8±0.69 AU) and the kidneys (right: 1.2±0.29 AU, left: 1.4±0.32 AU) (Figure 6B). The same organs also exhibited the highest lactate signals (spleen: 0.28±0.083 AU, right kidney: 0.18±0.043 AU, and left kidney: 0.23±0.050 AU). The liver. had the lowest overall pyruvate and lactate signals and it also had the lowest standard deviation for the two metabolites (pyruvate = 0.13 AU, lactate = 0.028 AU) compared to the other organs. The highest alanine signal was observed in the pancreas (0.14±0.031 AU) while the lowest were in the liver (0.045±0.019 AU) and spleen (0.039±0.021 AU). Though not measured using an ROI, elevated alanine signals were observed in the paraspinal muscles compared to the background (Figure 6A). Although the right kidney is not shown in the anatomical reference, pyruvate signal from that kidney is present in this axial slice due to partial volume effects and the 20 mm slice thickness.
Figure 6.
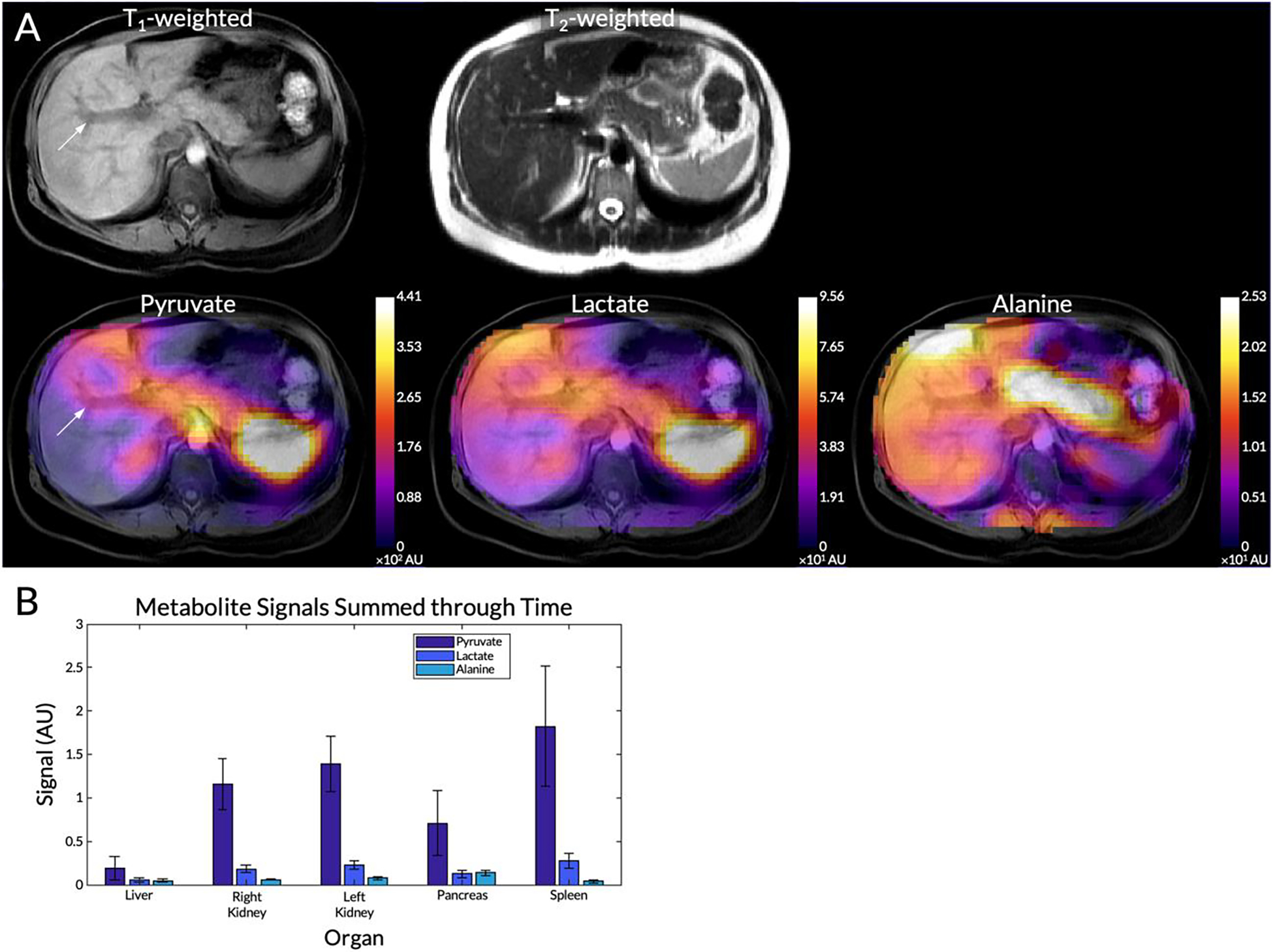
(A) Pyruvate, lactate, and alanine maps summed through time from a representative subject overlaid on top of a T1-weighted anatomical reference. The T2-weighted anatomical reference is also shown in the top row. An arrow highlights the portal vein. (B) Mean pyruvate, lactate, and alanine signals within each organ for this subject. Error bars indicate intra-organ standard deviation.
For the same subject, values of kPL and kPA were greatest in the parenchymal tissue of the liver, with voids around blood vessels as expected (highlighted with arrows in Figure 7). Low kPA was observed in the spleen. Mean kPL and kPA values for the seven successful EPI scans are shown in Figure 8. Tests for normality passed for both metrics in all organs (all P ≥ 0.23). Mean and standard deviation values and comparisons between organs with statistical significance values are displayed in Table 2. One-way ANOVA tests for both kPL and kPA across organs were significant. All comparisons of kPL and kPA between the liver and those of other organs were statistically significant. Hence, although the absolute lactate and alanine signals were lower in the liver compared to other organs (Figure 6B), the liver exhibited the highest rate of conversion from pyruvate to lactate and to alanine (kPL = 0.019±0.0076 s−1, kPA = 0.012±0.0037 s−1). Although not statistically significant, the pancreas did exhibit a trend of higher alanine production compared to the kidneys (right: P = 0.12, left: P = 0.078) and the spleen (P = 0.057).
Figure 7.
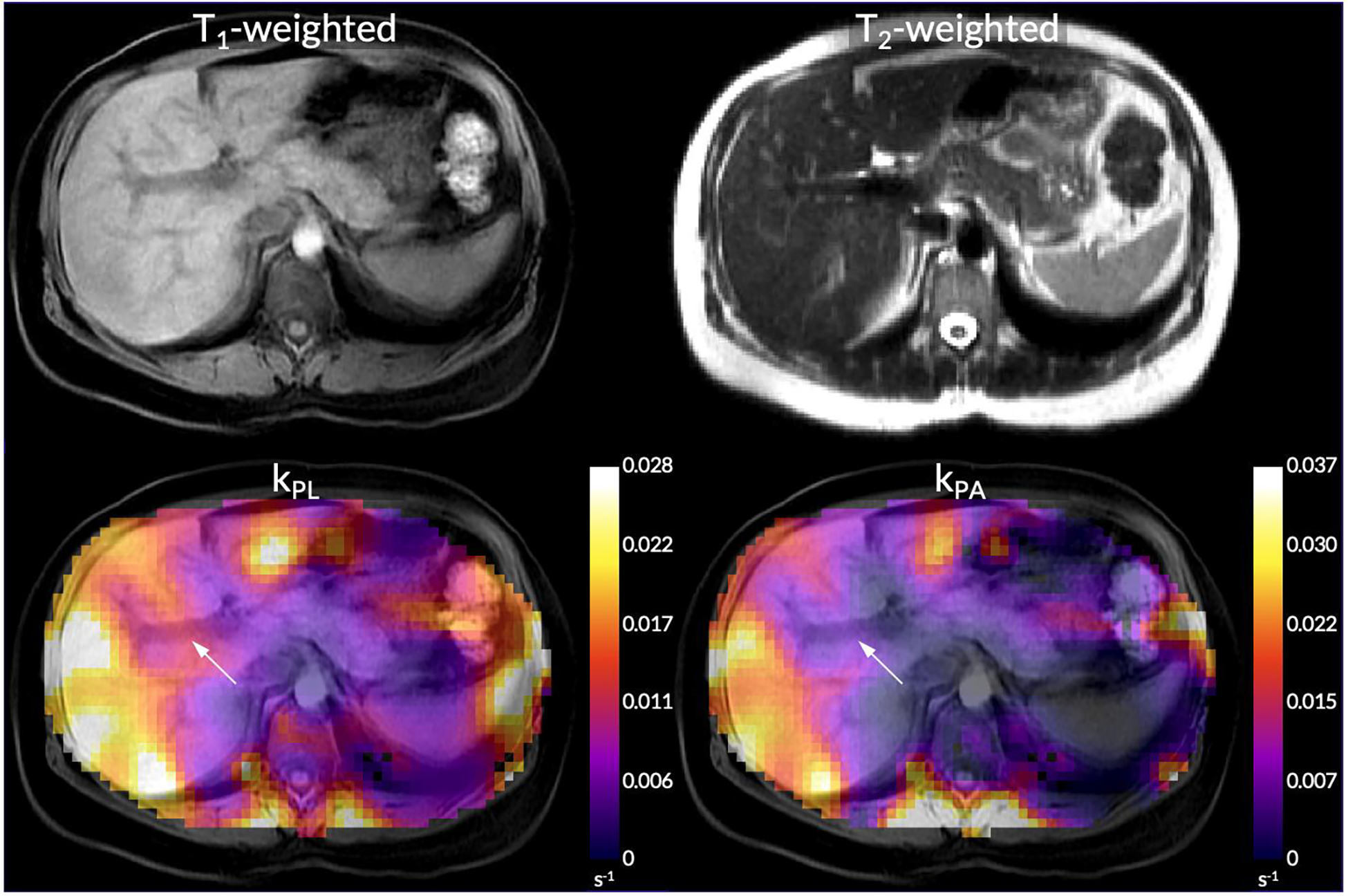
kPL and kPA maps overlaid on top of a T1-weighted anatomical reference from the same subject shown in Figure 6. The T2-weighted anatomical reference is also shown in the top row. An arrow highlight the portal vein.
Figure 8.
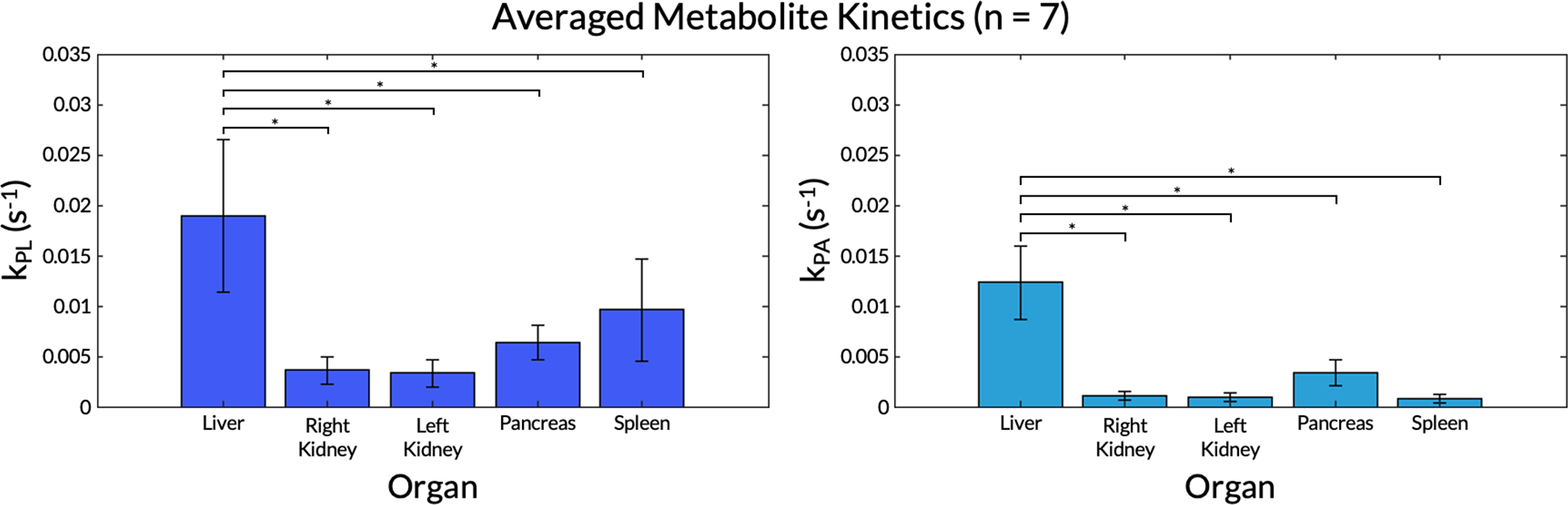
kPL and kPA values for each organ averaged across all seven subjects with EPI scans. Error bars indicate standard deviations. A single asterisk * denotes statistical signiifcance. Unlabeled comparisons were not statistically significant.
Table 2.
Summary of averaged kPL and kPA values across successful echo-planar imaging (EPI) scans, including mean and standard deviation and comparisons between organs with corresponding P-values.
| Kinetic Parameter | Organ | Mean±SD (n = 7) | Comparison | P ≤ 0.05? |
|---|---|---|---|---|
| kPL | Liver | 0.019±0.0076 | Liver vs. right kidney | Yes |
| Liver vs. left kidney | Yes | |||
| Liver vs. pancreas | Yes | |||
| Liver vs. spleen | Yes | |||
| Right kidney | 0.0036±0.0013 | Right kidney vs. left kidney | No, P > 0.99 | |
| Right kidney vs. pancreas | No, P = 0.75 | |||
| Right kidney vs. spleen | No, P = 0.085 | |||
| Left kidney | 0.0033±0.0014 | Left kidney vs. pancreas | No, P = 0.67 | |
| Left kidney vs. spleen | No, P = 0.064 | |||
| Pancreas | 0.0063±0.0017 | Pancreas vs. spleen | No, P = 0.56 | |
| Spleen | 0.0096±0.0050 | |||
| Ordinary one-way ANOVA | Yes | |||
| kPA | Liver | 0.012±0.0037 | Liver vs. right kidney | Yes |
| Liver vs. left kidney | Yes | |||
| Liver vs. pancreas | Yes | |||
| Liver vs. spleen | Yes | |||
| Right kidney | 0.0011±0.00039 | Right kidney vs. left kidney | No, P > 0.99 | |
| Right kidney vs. pancreas | No, P = 0.12 | |||
| Right kidney vs. spleen | No, P > 0.99 | |||
| Left kidney | 0.00087±0.00042 | Left kidney vs. pancreas | No, P = 0.078 | |
| Left kidney vs. spleen | No, P > 0.99 | |||
| Pancreas | 0.0034±0.0013 | Pancreas vs. spleen | No, P = 0.057 | |
| Spleen | 0.00073±0.00044 | |||
| Ordinary one-way ANOVA | Yes | |||
DISCUSSION
To effectively translate HP [1-13C]pyruvate MRI into the clinic for assessing neoplastic and metabolic disease in the abdomen, a specialized whole-abdomen imaging approach is required. This method must have sufficient coverage, be insensitive to respiratory motion, and address B0 inhomogeneities present due to air-tissue interfaces. Post-processing of the data must address low SNR through denoising methods, and compute key metabolic metrics, such as kPL and kPA, to metabolically characterize organs of interest. In this study, a whole-abdomen HP [1-13C]pyruvate metabolic imaging protocol was developed and used to characterize normal metabolic energetics in the human liver, kidneys, pancreas, and spleen.
This imaging protocol did not seek to address all challenges that can occur when conducting HP 13C MRI studies, such as dissolution hardware failures. Additionally, to avoid confounding factors, data from spiral gradient echo acquisition schemes were not used, although data acquisition was itself successful. Only for one scan was there a QTAR coil hardware failure. Therefore, given the scope of this pilot study and its aims, with only one acquisition failing against twelve successful scans, this imaging protocol was considered reliable for future studies.
This approach combined several innovations to address the challenges of performing HP 13C MR imaging in the abdomen: 1) a flexible multi-channel coil array and volumetric transmitter to broaden the imaging field of view, 2) a fast, multi-slice metabolite-specific EPI technique to mitigate the effects of motion, 3) careful shimming and frequency measurement to ensure B0 field uniformity over the liver, and 4) denoising using patch-based HOSVD to improve the overall apparent SNR.
The flexible large-volume transmit and receive coil used in this study was first characterized. While the homogeneity of the transmitter was sufficient for 13C acquisitions, higher transmit power was observed in the right-anterior and left-posterior regions in a phantom setup. These variations were likely in part due to the flexible transmitter design that resulted in a somewhat irregular transmit coil geometry. The transmitter had cutouts for the coil to fit under the subject’s arms at the cost of B1+ homogeneity. In addition, as the B1+ field is determined by the spatial positioning of the conductors in the transmitter, it was expected that the B1+ field of the QTAR coil would vary depending on how it was wrapped around people of different sizes (37). However, as the standard deviations of B1+ scale factors were less than 0.2 (or 20% of 100% transmit power) in each quadrant and in the central, circular 10 cm region of the central axial slice, the B1+ variation was acceptable for this pilot study and sufficient signal was still obtained in every organ of interest. Metabolite signals in the right kidney, which were in the region of lower transmit power, had high enough signal for metabolic characterization.
To obtain volumetric and dynamic HP [1-13C]pyruvate, [1-13C]lactate, and [1-13C]alanine signals, a multi-slice metabolite-specific EPI acquisition scheme was used to acquire twenty timepoints over one minute. This approach eliminated motion artifacts due to its short (64–72 ms) acquisition time per slice. One challenge of using metabolite-specific EPI acquisitions was the need to ensure that the relevant metabolites were centered in the 100 Hz passband of the spectral-spatial pulse used in this approach. For several subjects, the 13C frequency range was confirmed using separate HP 13C EPSI acquisitions that included the full chemical shift range for each voxel. As an alternative to these separate HP 13C acquisitions, a simplified 1H imaging approach with an IDEAL-IQ acquisition was used to measure B0 frequency shifts. This latter method had the advantage of not requiring a second injection. As expected, a strong linear correlation between the acquired 1H ΔB0 map and the EPSI-derived 13C ΔB0 map was observed. This allowed forgoing the EPSI acquisition for later subjects with each patient scan, instead using the IDEAL IQ-derived field map to ensure the correct 13C acquisition frequencies.
Measurement of the B0 field revealed large variations throughout the abdomen, both in-plane and through-plane. These large field variations were a result of the scanner’s automatic shimming process and risked placing several regions of the abdomen outside of the 100 Hz FWHM passband employed by the spectral-spatial pulse. After a localized shim volume was placed over the liver, followed by manual adjustments of through-plane frequency variations, frequency variations were greatly reduced throughout the abdomen. Before local shimming, four out of five organs of interest had a median frequency offset outside of the FWHM range of the spectral spatial pulse. After local shimming, all organs had a median frequency offset within the passband; also, the liver, right kidney, and pancreas had minimum and maximum values within the passband. The improvement was more noticeable in the liver and the right kidney than in other organs, likely because the localized shim box was placed over the liver. Therefore, frequency variations in more distant organs, such as in the left kidney and the spleen, were not as affected. Nevertheless, all organs were shifted closer to a 0 Hz median. For instance, the spleen had a median of −83.5 Hz which was corrected to 9.1 Hz. This was a critical improvement as otherwise voxels in these organs would have less than 50% of the expected RF power resulting in lower flip angles and would have experienced EPI chemical shift artifacts.
In addition to using a fast acquisition scheme and careful shimming to mitigate motion effects, patch-based denoising was used to greatly increase the quality of acquired metabolite images (34). More voxels of metabolite signals within the whole abdomen passed the empirical SNR thresholds after denoising. As observed in one subject, the mean SNR of voxels within the whole abdomen were above the SNR thresholds for a longer duration after denoising, demonstrating an improvement of HP 13C dynamics against the background noise.
The liver trended towards a longer time-to-peak, which was compounded upon a longer decay time, resulting in an overall broadened dynamic curve. The kidneys, pancreas, and spleen exhibited steeper drop-offs after reaching peak pyruvate signal. Thus, even though the spleen had a comparable decay time to the liver, the liver’s pyruvate uptake and dynamics remained distinct. This is perhaps reflective of the organ’s unique dual blood-supply, where the portal venous phase (75–80% of the blood entering the liver) occurs approximately 8 seconds after the hepatic arterial phase (38, 39).
Summed metabolite maps showed absolute metabolite signals from the kidneys and spleen that were 3–7 times higher than those from liver and pancreas, likely reflecting differences in perfusion and/or blood volume. However, smaller variance in pyruvate and lactate signals indicated more uniform distributions in the liver.
Although absolute metabolite signals were lower in the liver, kPL and kPA were both higher in the liver than in other abdominal organs, indicating higher relative production of these metabolites. Heterogeneity in the kPL and kPA maps of the liver was observed. In one subject, areas of low signal conformed to non-metabolically active regions such as the falciform ligament and the portal vein. In addition, heterogeneity may be caused by residual effects of transmit field non-uniformities as well as partial volume effects, for example due to the kidneys in nearby slices. These can be addressed in future work. Right and left kidney metabolism were symmetric as expected. The pancreas showed higher levels of alanine production relative to other organs, possibly due to high levels of protein synthesis related to its digestive enzyme and endocrine hormone functions.
Limitations
This study took place at a single center, using a single scanner of a single field strength from a single vendor. Additionally, there was a small number of subjects in this study. There was no available external reference for the measured pyruvate-to-lactate and pyruvate-to-alanine conversion rates. All of the subjects examined in this study were healthy volunteers. Although the RF coil used in this work was designed to have relatively uniform transmit and receive sensitivity over the entire imaging volume, residual variations due to non-uniformities in the transmit field and surface coil sensitivity profiles were present. In future studies, the effects of transmit field non-uniformities can be corrected through post-injection B1+ measurements or the future development and use of a built-in 13C full-volume body coil (23). Another limitation of this study was that it relied on a kinetic model that did not explicitly incorporate the effects of non-uniform excitation profiles. This has been shown to potentially cause overestimation of lactate exchange, particularly when large flip angles are used (40). This can also be explored in future work. As this was a technical development study, some minor variations in scan protocol were tested between different subjects. These variations mainly related to the degree of localized shimming that was performed as well as whether subjects received the EPSI acquisition, EPI acquisition, or both. None of the variations that were tested were expected to affect the overall conclusions drawn from the results.
Conclusions
In this study, a novel approach was developed and applied for acquiring HP 13C metabolic images of multiple organs, that utilized a multi-echo gradient echo 1H acquisition to inform 13C center frequencies and to overcome challenges of broad spatial coverage as well as B0 inhomogeneities. This enabled metabolically characterizing the healthy liver, kidneys, pancreas, and spleen. This normative data will be valuable in further investigations of cancer as well as metabolic liver diseases.
Supplementary Material
ACKNOWLEDGEMENTS:
We would like to acknowledge the assistance of Jim Slater, Andrew Riselli, Evelyn Escobar, Mary Frost, Kimberly Okamoto, and Jonathan Lin.
Grant Support:
This work was supported by NIH grants NIDDK 5R01DK115987 and P41 EB013598 and American Cancer Society Research Scholar Grant 131715-RSG-18-005-01-CCE.
REFERENCES
- 1.Kurhanewicz J, Vigneron DB, Ardenkjaer-Larsen JH, et al. : Hyperpolarized 13C MRI: Path to Clinical Translation in Oncology. Neoplasia (United States) 2019; 21:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggarwal R, Vigneron DB, Kurhanewicz J: Hyperpolarized 1-[13C]-Pyruvate Magnetic Resonance Imaging Detects an Early Metabolic Response to Androgen Ablation Therapy in Prostate Cancer. Eur Urol 2017; 72:1028–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen HY, Aggarwal R, Bok RA, et al. : Hyperpolarized 13C-pyruvate MRI detects real-time metabolic flux in prostate cancer metastases to bone and liver: a clinical feasibility study. Prostate Cancer Prostatic Dis 2020; 23:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park I, Larson PEZ, Gordon JW, et al. : Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies. Magn Reson Med 2018; 80:864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham CH, Lau JYC, Chen AP, et al. : Hyperpolarized 13C Metabolic MRI of the Human Heart: Initial Experience. Circ Res 2016; 119:1177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abeyakoon O, Latifoltojar A, Gong F, et al. : Hyperpolarised 13 C MRI: a new horizon for non-invasive diagnosis of aggressive breast cancer. BJR Case Rep 2019; 5:20190026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ardenkjær-Larsen JH, Golman K, Gram A, et al. : Increase of signal-to-noise of more than 10,000 times in liquid state NMR. Discov Med 2003; 100:10158–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warburg O: On the origin of cancer cells. Science (80- ) 1956; 123:309–314. [DOI] [PubMed] [Google Scholar]
- 9.Hu S, Balakrishnan A, Bok RA, et al. : 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab 2011; 14:131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keshari KR, Sriram R, Van Criekinge M, et al. : Metabolic reprogramming and validation of hyperpolarized 13C lactate as a prostate cancer biomarker using a human prostate tissue slice culture bioreactor. Prostate 2013; 73:1171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larson PEZ, Chen HY, Gordon JW, et al. : Investigation of analysis methods for hyperpolarized 13C-pyruvate metabolic MRI in prostate cancer patients. NMR Biomed 2018; 31:e3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crane JC, Gordon JW, Chen HY, et al. : Hyperpolarized 13C MRI data acquisition and analysis in prostate and brain at University of California, San Francisco. NMR Biomed 2021; 34:e4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang ZJ, Ohliger MA, Larson PEZ, et al. : Hyperpolarized 13C MRI: State of the art and future directions. Radiology 2019; 291:273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day SE, Kettunen MI, Gallagher FA, et al. : Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med 2007; 13:1382–7. [DOI] [PubMed] [Google Scholar]
- 15.Chaumeil MM, Ozawa T, Park IW, et al. : Hyperpolarized 13C MR spectroscopic imaging can be used to monitor Everolimus treatment in vivo in an orthotopic rodent model of glioblastoma. Neuroimage 2012; 59:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran M, Latifoltojar A, Neves JB, et al. : First-in-human in vivo non-invasive assessment of intra-tumoral metabolic heterogeneity in renal cell carcinoma. BJR|case reports 2019; 5:20190003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang S, Meng MV, Slater JB, et al. : Metabolic imaging with hyperpolarized 13C pyruvate magnetic resonance imaging in patients with renal tumors—Initial experience. Cancer 2021; 127:2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwanten W, Martinet W, Francque S: Autophagy in Non-Alcoholic Fatty Liver Disease (NAFLD). In Autophagy Curr Trends Cell Physiol Pathol; 2016:455–83. [Google Scholar]
- 19.Moon CM, Oh CH, Ahn KY, et al. : Metabolic biomarkers for non-alcoholic fatty liver disease induced by high-fat diet: In vivo magnetic resonance spectroscopy of hyperpolarized [1–13C] pyruvate. Biochem Biophys Res Commun 2017; 482:112–119. [DOI] [PubMed] [Google Scholar]
- 20.Wilson DM, Di Gialleonardo V, Wang ZJ, et al. : Hyperpolarized 13C spectroscopic evaluation of oxidative stress in a rodent model of steatohepatitis. Sci Rep 2017; 7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjærgaard U, Laustsen C, Nørlinger T, et al. : Hyperpolarized [1– 13 C] pyruvate as a possible diagnostic tool in liver disease. Physiol Rep 2018; 6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morze C Von, Allu PKR, Chang GY, et al. : Non-invasive detection of divergent metabolic signals in insulin deficiency vs. insulin resistance in vivo. Sci Rep 2018; 8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang S, Milshteyn E, Reed G, et al. : A regional bolus tracking and real-time B1 calibration method for hyperpolarized 13C MRI. Magn Reson Med 2019; 81:839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau AZ, Chen AP, Hurd RE, Cunningham CH: Spectral-spatial excitation for rapid imaging of DNP compounds. NMR Biomed 2011; 24:988–996. [DOI] [PubMed] [Google Scholar]
- 25.Schulte RF, Sacolick L, Deppe MH, et al. : Transmit gain calibration for nonproton MR using the Bloch-Siegert shift. NMR Biomed 2011; 24:1068–1072. [DOI] [PubMed] [Google Scholar]
- 26.Lau AZ, Chen AP, Cunningham CH: Integrated Bloch-Siegert B1 mapping and multislice imaging of hyperpolarized 13C pyruvate and bicarbonate in the heart. Magn Reson Med 2012; 67:62–71. [DOI] [PubMed] [Google Scholar]
- 27.Sacolick LI, Wiesinger F, Hancu I, Vogel MW: B1 mapping by Bloch-Siegert shift. Magn Reson Med 2010; 63:1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grist JT, Hansen ESS, Sánchez-Heredia JD, et al. : Creating a clinical platform for carbon-13 studies using the sodium-23 and proton resonances. Magn Reson Med 2020; 84:1817–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson SJ, Kurhanewicz J, Vigneron DB, et al. : Metabolic Imaging of Patients with Prostate Cancer Using Hyperpolarized [1–13C]Pyruvate. Sci Transl Med 2013; 5:198ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen HY, Larson PEZ, Gordon JW, et al. : Technique development of 3D dynamic CS-EPSI for hyperpolarized 13 C pyruvate MR molecular imaging of human prostate cancer. Magn Reson Med 2018; 80:2062–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee PM, Chen HY, Gordon JW, et al. : Specialized computational methods for denoising, B1 correction, and kinetic modeling in hyperpolarized 13C MR EPSI studies of liver tumors. Magn Reson Med 2021; 86:2402–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon JW, Chen HY, Autry A, et al. : Translation of Carbon-13 EPI for hyperpolarized MR molecular imaging of prostate and brain cancer patients. Magn Reson Med 2019; 81:2702–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z, Zhu X, Ohliger MA, et al. : Coil combination methods for multi-channel hyperpolarized 13 C imaging data from human studies. J Magn Reson 2019; 301:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y, Chen H, Autry AW, et al. : Denoising of hyperpolarized 13 C MR images of the human brain using patch-based higher-order singular value decomposition. Magn Reson Med 2021; 86:2497–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yushkevich PA, Piven J, Hazlett HC, et al. : User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006; 31:1116–1128. [DOI] [PubMed] [Google Scholar]
- 36.Gordon JW, Larson PEZ: Pulse sequences for hyperpolarized MRS. eMagRes 2016; 5:1229–1246. [Google Scholar]
- 37.Singerman R, Denison T, Wen H, Balaban R.: Simulation ofB1Field Distribution and Intrinsic Signal-to-Noise in Cardiac MRI as a Function of Static Magnetic Field. J Magn Reson 1997; 125:72–83. [DOI] [PubMed] [Google Scholar]
- 38.Rocha FG: Liver blood flow: Physiology, measurement, and clinical relevance. In Blumgart’s Surg Liver, Pancreas Biliary Tract. Fifth Edit. Edited by Jarnagin W, Blumgart L. Elsevier; 2012:74–86.e5. [Google Scholar]
- 39.Bader T, Prokesch R, Grabenwöger F: Timing of the Hepatic Arterial Phase During Contrast-Enhanced Computed Tomography of the Liver. Invest Radiol 2000; 35:486–492. [DOI] [PubMed] [Google Scholar]
- 40.Walker CM, Gordon JW, Xu Z, et al. : Slice profile effects on quantitative analysis of hyperpolarized pyruvate. NMR Biomed 2020; 33:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


