Summary
Here, we describe a protocol for modulating the dynamics of the extracellular signal-regulated kinases (ERK) pathway in a customized alternating current (AC) electric field stimulation chamber. We use an ERK translocation reporter that can accurately represent the intracellular ERK activity in real time without chemical agents or gene disruption. ERK activation is assessed by comparing the relative intensity of nuclear fluorescence to cytosolic fluorescence in live-cell conditions. The approach can be applied to other signaling pathways as well.
For complete details on the use and execution of this protocol, please refer to Guo et al. (2021).
Subject areas: Cell biology, Cell culture, Microscopy, Signal transduction, Biotechnology and bioengineering
Graphical abstract

Highlights
-
•
Protocol for robust electrical activation of intracellular signaling pathways
-
•
Step-by-step guide for delivering AC electric field in a customized chamber
-
•
Rapid quantification of single-cell ERK dynamics in time-lapse fluorescence images
-
•
Precise control of ERK dynamics by stimulation with different scheme
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we describe a protocol for modulating the dynamics of the extracellular signal-regulated kinases (ERK) pathway in a customized alternating current (AC) electric field stimulation chamber. We use an ERK translocation reporter that can accurately represent the intracellular ERK activity in real time without chemical agents or gene disruption. ERK activation is assessed by comparing the relative intensity of nuclear fluorescence to cytosolic fluorescence in live-cell conditions. The approach can be applied to other signaling pathways as well.
Before you begin
Protocol overview
Critical cell behaviors, including migration, proliferation, and differentiation are controlled by the activation of intracellular signaling pathways. Recent research using fluorescence reporters reveals that every signaling pathway varies in activities with diverse dynamic phenotypes, such as activation magnitude, frequency and duration. Such dynamics encode distinct information which can exert differential gene expression that controls cell proliferation and differentiation. Controlling the intracellular signaling dynamics practically with translational potentials therefore is highly desirable for in vivo applications. Here, we demonstrate our technique using ERK as a model on how to accomplish efficient and practical control of signaling dynamics through electrical stimulation.
The protocol below describes a step-by-step guide for the electrical modulation of the dynamics of one important intracellular signaling pathway — ERK in a customized AC (alternating current) EF (electric field) stimulation chamber. We here use an ERK translocation reporter that can accurately represent the intracellular ERK activity in real time. ERK activation is assessed by comparing the relative intensity of nuclear fluorescence to cytosolic fluorescence in live-cell conditions. Quantitative ERK dynamic data can be extracted from the raw images for further analysis. Using our controlled electrical delivery techniques, the amplitude, frequency, and duration of ERK activation can be precisely dominated in cell lines without the need of adding chemical agents or manipulating gene expression. This technique can be used to synchronize and modulate intracellular signaling dynamics in vivo.
Fabrication of AC EF stimulation chamber
Timing: 5 h
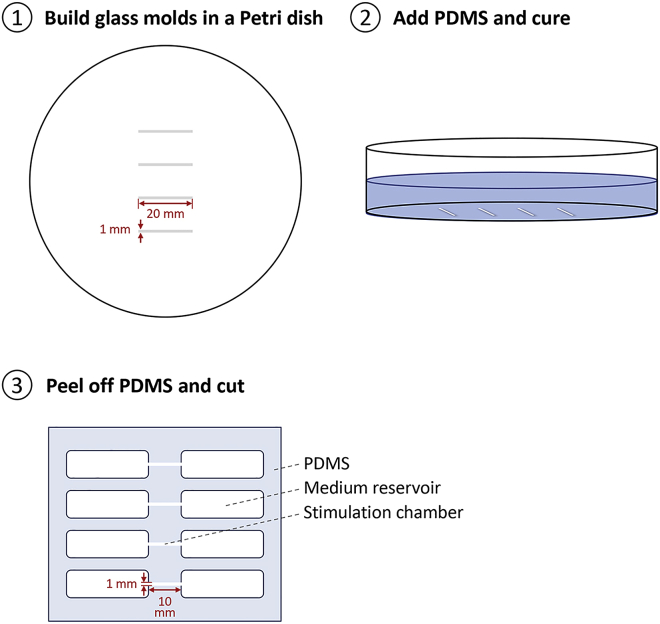
Preparation of multi-channel AC stimulation chamber by soft lithography (Figure 1).
-
1.Build a master for the multi-chamber AC EF stimulation device.
-
a.Precisely cut the glass coverslip (0.15–0.19 mm in thickness) as 1 mm wide and 20 mm length strips with a carbide glass cutter for thin (0.15 mm–0.5 mm) materials.
-
b.Molds of the chambers for AC stimulation were created by attaching glass strips to the base of a Petri dish (100 mm) with Dow Corning curing silicone rubber 3140 glue.
-
a.
Optional: Clean the glass coverslip/strips by sonication in ethanol and washing with deionized water followed by drying.
-
2.Prepare PDMS (Polydimethylsiloxane) for fabricating AC EF stimulation chambers.
-
a.Mix PDMS prepolymer with curing reagent at a ratio of 10:1.
-
b.Pour the above-mixed PDMS onto the Petri dish with glass molds on it to form a layer of 3–5 mm thick and degas in a vacuum chamber for 1 h.
-
c.Cure the PDMS at 70°C–90°C in an incubator for 1–3 h.
-
a.
-
3.Cut the PDMS precisely as designed (Figure 1, panel 3):
-
a.Gently peel off the cured PDMS from the dish and cut into a 6 × 6 cm2 square with a blade.
-
b.Punch 2 rectangular reservoirs with a biopsy punch on both ends of each chamber. These act to hold the cell culture medium for applying electric stimulation.
-
c.Autoclave the PDMS at 121°C for 60 min using a dry cycle run.
-
d.After sterilization, assemble the chamber in a Petri dish.
-
a.
Note: The chambers and reservoirs of the devices are designed to be sealed and leak-proof. Keep the PDMS free of dust particles for better sealing performance.
Optional: The AC EF stimulation device can be reused after careful cleaning. Gently peel off the PDMS chamber from a petri dish after each experiment and keep it in 70% ethanol. Clean the device in an ultrasonic cleaner for 30 min at 50°C before autoclaved to remove any dust particles on the surface.
Pause point: Autoclaved PDMS devices can be stored at room temperature (22°C–25°C) until further use.
Figure 1.
Steps for fabricating the multi-channel EF stimulation device
Build mold for the multi-channel device in a petri dish with pre-cut glass strips. Pour PDMS onto the mold cure for 1–3 h in a heated incubator. Peel off and cut the PDMS device bearing the multi-channel design.
Preparation of cell lines stably expressing kinase translocation reporter (KTR) for monitoring ERK activation dynamics
Timing: 2 weeks
We here provide a technical procedure for constructing a stable cell line expressing ERK Kinase Translocation Reporters (ERKTR-mCherry) (Sparta et al., 2015) by using viral transfection.
-
4.Prepare purified viral particles carrying desired reporter(s).
-
a.Culture 293T cells in a 6-well plate at ∼70% confluence.
-
b.Co-transfecting 293T cells with pMSCV-puro-ERKTR-mCherry and pCL-Ampho. Combine reagents in a 1.5 mL reaction tube in the following order to prepare the transfection mix for each well (Pargett et al., 2017): 100 μL Opti-MEM, 3 μL FuGENE HD, 1 μg packaging plasmid, and 1 μg retroviral plasmid DNA.
-
c.Add transfection mix drop-wise to each well of the 6-well plate and incubate cells at 37°C with 5% CO2 for 12–18 h.
-
d.Remove the transfection mix and replace with 1 mL DMEM medium supplemented with 10% fetal bovine serum (FBS).
-
e.Collect the growth medium with viral particles and pass it through a 0.45 μm syringe filter, 24 and 48 h after removing the transfection mix.
-
f.Collect and store the filtered viral containing medium at 4°C for later use up to two weeks or at −80°C for up to 1 year.
-
a.
Note: Virus titer may vary due to the differences in DNA purity, virus system, and other experimental factors. The volume of the viral containing medium used in the following viral transduction need to be adjusted to achieve a >70% success rate of stable reporter integration.
-
5.Prepare cells for transduction.
-
a.Seed cells of interest in a 6-well plate at an optimized seeding density.Note: MCF-10A cells were used in this protocol as an example but can be replaced with your cell line of interest. We use a seeding density of 100,000 cells per well. At this seeding density, cells within the linear phase of cell growth will become confluent four days post-seeding.
-
b.Incubate cells for 12–18 h at 37°C with 5% CO2.
-
a.
-
6.Viral Transduction.
-
a.Prepare the transduction mixture by combining 500 μL of cell growth medium, 500 μL of filtered viral containing medium and 1 μL of 4 mg/mL polybrene and mix gently by pipetting.Note: The volume of virus-containing medium and concentration of polybrene can be adjusted as needed to promote the efficiency of transduction.
-
b.Aspirate medium from target cells.
-
c.Add transduction mixture dropwise to MCF10A cells and incubate for 12–18 h at 37°C with 5% CO2.
-
d.Remove the medium with virus and add 2 mL fresh growth medium.
-
e.Incubate at 37°C with 5% CO2.
-
a.
-
7.Reporter Cell selection.
-
a.Check cells for reporter expression using a fluorescence microscope after incubating cells for about 24-h post transduction.
-
b.Add the appropriate concentration of the selection antibiotic to the target cells. Treat noninfected MCF10A cells with a range of puromycin concentrations to find the lowest dose for antibiotic selection. We typically use 3 μg/mL puromycin as an optimal selection concentration.
-
c.Change medium containing the selection antibiotic every 2–3 days to removed dead cells during antibiotic treatment.
-
d.Monitor cells daily after the 1st week of antibiotic selection. Only cells with stable expression of the reporter construct will survive past the puromycin selection process.
-
a.
Optional: The reporter expression may remain heterogeneous within the cell population following antibiotic selection, further selection can be performed to generate a monoclonal cell population by limited dilution or flow cytometry.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| EGF | Thermo Fisher Scientific | Cat#PHG0311 |
| Horse Serum | Thermo Fisher Scientific | Cat#26050088 |
| Insulin | Sigma-Aldrich | Cat#I9278 |
| Cholera Toxin | Sigma-Aldrich | Cat#C8052 |
| Hydrocortisone | Sigma-Aldrich | Cat#H0888 |
| PBS | Thermo Fisher Scientific | Cat#10010023 |
| Opti-MEM | Thermo Fisher Scientific | Cat#31985070 |
| DMEM/F12 | Thermo Fisher Scientific | Cat#11320033 |
| DMEM/F12, no phenol red | Thermo Fisher Scientific | Cat#21041025 |
| Bovine Serum Albumin solution | Sigma | Cat#9048-46-8 |
| FuGENE HD | Promega | Cat#E231A |
| Puromycin | Thermo Fisher Scientific | Cat#A1113803 |
| Collagen I | Sigma | Cat#9007-34-5 |
| FNC Coating Mix | Athena Enzyme Systems | Cat#0407 |
| Experimental models: Cell lines | ||
| HEK 293T | ATCC | CRL-11268 |
| MCF10A | ATCC | CRL-10317 |
| Recombinant DNA | ||
| Plasmid: ERKTR-mCherry | Sparta et al. (2015) | N/A |
| Plasmid: EKAR3-FRET | Sparta et al. (2015) | N/A |
| Software and algorithms | ||
| MetaMorph NX 7.10 | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy |
| ImageJ | ImageJ | https://imagej.nih.gov/ij/ |
| Other | ||
| PDMS | Dow Corning | Cat#0008691565 |
| Cover glass | Fisher Scientific | Cat#12-548-BP |
| Precision glass cutters | Structure Probe Inc. | Cat#07635-AB |
| 3140 Silicone rubber | Dow Corning | Cat#121-353-ML |
| Biopsy Punch | Integra | Cat#12-460-413 |
| Silver electrode | Advent Research Materials Ltd | Cat#AG549112 |
| True-rms digital multimeter | RadioShack | Cat#2200087 |
Note: Alternative imaging software might be needed depending on the hardware available to users.
Materials and equipment
Cell growth medium formulation
In a 50 mL conical tube, combine the following supplements, mix well and filter through a 0.22 μm flip-top filter.
| Reagent | Concentration of stock solution | Amount |
|---|---|---|
| Horse serum ∗ | N/A | 25 mL |
| Penicillin-Streptomycin | 100×, 10,000 U/mL | 5 mL |
| Cholera Toxin ∗ | 1 mg/mL in ddH2O | 50 μL |
| EGF ∗ | 100 μg/mL in ddH2O | 100 μL |
| Hydrocortisone ∗ | 1 mg/mL in alcohol | 250 μL |
| Insulin ∗ | 10 mg/mL | 500 μL |
Add the filtered mixture to a new bottle of DMEM/F12 (500 mL). Keep at 4°C for up to 3 months.
Low fluorescence imaging medium formulation
Combine the following supplements in a 250 mL filter-top bottle.
| Reagent | Concentration of stock solution | Amount |
|---|---|---|
| Penicillin-Streptomycin | 100×, 10,000 U/mL | 2 mL |
| Cholera Toxin ∗ | 1 mg/mL in ddH2O | 20 μL |
| Hydrocortisone ∗ | 1 mg/mL in alcohol | 100 μL |
| BSA | 30% w/v | 2 mL |
| DMEM/F12 without phenol red | N/A | 200 mL |
Filter into a bottle and discard filter.
Keep at 4°C for up to 3 months.
Note: Media components with a ∗ are specific for the MCF10A cell line.
Steinberg’s solution (10×)
| Reagent | Final concentration of stock solution | Amount |
|---|---|---|
| NaCl | 580 mM | 34 g |
| KCl | 6.7 mM | 0.5 g |
| Ca(NO3)2 · 4H2O | 4.4 mM | 1.0 g |
| MgSO4 · 7H2O | 13 mM | 3.13 g |
| Tris | 46 mM | 5.52 g |
| ddH2O | N/A | ∼1 L |
| Total | N/A | 1 L |
Note: After autoclave, the buffer can be stored up to 3 months at room temperature (22°C–25°C).
Agar gel
Dissolve 1.5% (wt/vol) agar into Steinberg’s solution and autoclave the mixture. The agar gel can be kept in a sterilized condition at room temperature (22°C–25°C) for up to a month.
AC power supply
An Instek APS-7100 AC power supply device was used in this study. This fully programmable device is preferable for its ability to maintain high precision when outputting different voltages in a custom sequence. Options include selecting arbitrary waveforms, voltage, or frequency sweep. Any alternative AC power source that can generate an AC EF with a frequency equal to or greater than 50 Hz can be used for this study.
Step-by-step method details
Plating cells for live-cell microscopy
Timing: 1 day
Seeding of cells inside the AC stimulation chamber to investigate the effects of AC EFs on intracellular signaling pathways.
-
1.Pre-coat the chambers to accelerate cell adhesion.
-
a.Use a 20 μL pipette to load 5 μL of coating solution (Collagen or FNC Coating Mix®) from one side of each chamber (Figure 2).
-
b.Use a Pasteur pipette attached to a vacuum line to aspirate any air bubble remaining in the chamber (troubleshooting 1).
-
c.Cover the plate and incubate for 5 (for coating with FNC) to 30 (for coating with 20 μg/mL collagen I) min in a sterile hood.
-
d.Remove the coating solution by aspirating.
-
e.Gently rinse the chamber once with sterile PBS.
-
a.
Note: Multiple rinses may be needed for different coating materials that are dissolved in an acidic or alkaline solution.
-
2.Prepare target cell suspension.
-
a.Remove medium and rinse the MCF10A cells expressing ERKTR-mCherry with 3 mL PBS twice in a T25 flask.
-
b.Trypsinize MCF10A cells expressing ERKTR-mCherry with 2 mL 0.05% trypsin, 0.53 mM EDTA for 15–20 min following the ATCC’s protocol (ATCC # CRL-10317).
-
c.Neutralize the trypsin by adding 4 mL culture medium or 2 mL 0.1% soybean trypsin inhibitor.
-
d.Collect the single cell suspension after gentle pipetting.
-
e.Adjust cell concentration after counting to approximately 5 × 105/mL.
-
a.
-
3.Seed cells in the stimulation chamber.
-
a.Remix the cell suspension gently.Note: Mixing the cell suspension might introduce stress and reduce cell viability. We suggest to only mix the cell suspension every 2 min by gently shaking or by using a large gauge pipette tip and pipetting gently. This will prevent natural sedimentation caused by gravity while minimizing unnecessary stress caused by mixing.
-
b.Load 10 μL cell suspension through one side of the chamber (Figure 2).
 CRITICAL: To maintain a consistent environment for the cells, it is necessary to limit total cell numbers that does not result in nutrient depletion during the experiment. We typically load 5,000 cells in 10 μL medium to each chamber in the dish which only covers the 10 mm-channel area for imaging. The cells should be about 50% confluent the next day. It would be difficult to distinguish each cell in the images of high confluency during ERK quantification.
CRITICAL: To maintain a consistent environment for the cells, it is necessary to limit total cell numbers that does not result in nutrient depletion during the experiment. We typically load 5,000 cells in 10 μL medium to each chamber in the dish which only covers the 10 mm-channel area for imaging. The cells should be about 50% confluent the next day. It would be difficult to distinguish each cell in the images of high confluency during ERK quantification. -
c.Aspirate using a 2 μL pipette from the other side if needed to remove air bubbles.
-
d.Gently add 200 μL cell culture medium to the far end of each reservoir without connecting to the cell suspension in the chamber in order to raise the humidity which helps prevent drying of the medium (troubleshooting 2).
-
e.Add PBS to all spaces in the plate between the PDMS device and the dish to further increase the humidity in the plate.
-
f.Cover the plate and incubate at 37°C with 5% CO2 for about 2 h.
-
g.When cells are attached to the substrate, gently add additional 0.5 mL growth medium to each medium reservoir.
 CRITICAL: Check cells using an inverted microscope. Add enough medium to fulfill the two reservoirs when cells are fully attached, i.e., flattened and spread out. But if the cells are still spherical, they would need more time to attach. View with a microscope to make sure cells remain in the chamber with an expected density (∼150 cells/mm2 in our cases) after adding the medium.
CRITICAL: Check cells using an inverted microscope. Add enough medium to fulfill the two reservoirs when cells are fully attached, i.e., flattened and spread out. But if the cells are still spherical, they would need more time to attach. View with a microscope to make sure cells remain in the chamber with an expected density (∼150 cells/mm2 in our cases) after adding the medium. -
h.Incubate cells 12–18 h at 37°C with 5% CO2 to allow cells to fully attach.
 Pause point: Cells seeded in the chambers can be used for experiments in the next 2–3 days before cells get too confluent. Cell density might be a confounding factor that affects the basal ERK activity as cells actively deplete nutrients from the medium. Performing the experiments with a certain density range is important for data comparison. We typically use a monolayer with ∼50%–70% confluence in the experiments.
Pause point: Cells seeded in the chambers can be used for experiments in the next 2–3 days before cells get too confluent. Cell density might be a confounding factor that affects the basal ERK activity as cells actively deplete nutrients from the medium. Performing the experiments with a certain density range is important for data comparison. We typically use a monolayer with ∼50%–70% confluence in the experiments.
-
a.
-
4.Cell starvation to reduce the basal ERK level.
-
a.After cells are fully attached and adapted to the chamber, aspirate culture medium from both reservoirs of each chamber.Note: Cells need some time to recover from trypsinization, we typically incubate cells overnight (12–18 h) before starvation.
-
b.Rinse the chamber 3 times with PBS to remove unattached cells and growth factors in the old medium.Note: Rinsing the chamber with the flow of solution may cause significant shearing stress on the cell surface. We suggest reducing fluid pressure difference between two reservoirs to minimize the shear stress when rinsing the chamber. For cells with relatively weak attachment to the dish, such as PC12 cells, extra coating with appropriate proteins or peptides (e.g., Collagen IV for PC12 cells) can help reduce the risk of cell detachment.
-
c.Fill the chambers with serum-free, growth factor-free medium to starve cells.
-
d.Incubate cells at 37°C with 5% CO2 for at least 3 h.
-
a.
Figure 2.
Cell seeding in the AC stimulation chamber
Seed cells from one side of the chamber after coating. Aspirate on the other side to remove air bubbles. Add 200 μL culture medium to the far end of each reservoir. Allow cells to adhere for around 2–3 h and fill the reservoirs with more medium after cells are attached.
Monitoring ERK dynamics using time-lapse microscopy during AC stimulation
Timing: 1–3 days
This section describes a step-by-step walkthrough of live-cell fluorescence imaging of ERKTR during AC stimulation with enhanced sensitivity and temporal resolution.
Note: The protocol below describes the specific steps for using ERKTR-mCherry to monitor ERK activation dynamics. However, we have also used a pair of fluorescence resonance energy transfer (FRET)-based kinase biosensors, EKAR3, in MCF10A cells to monitor ERK dynamics following this protocol.
-
5.Experimental setup for AC stimulation.
-
a.Carefully aspirate the medium from the imaging chamber and wash the reservoirs twice with 1 mL of PBS (troubleshooting 3).
-
b.Add 1 mL of low fluorescence imaging medium to each reservoir.
-
c.Transfer the dish containing starved cells to be studied to the temperature-controlled chamber of the imaging system and mount the dish securely on the microscope stage.Note: Our Zeiss Observer Z1 microscope has separated temperature and CO2 control module. Turn on the microscope and the control module. Set the temperature to 37°C and allow the temperature to stabilize for 20–30 min before mounting the dish. Switch on the CO2 control module when needed.
-
d.Cover the dish using a sterilized lid with holes on it. Align the holes to the reservoirs.
-
e.Mount the Ag/AgCl electrodes with one on each reservoir and dip the tip of the electrodes in the medium (Figure 3A).
-
f.Connect the electrodes with the AC power supply via electric cables (Figure 3B).Optional: Electrical stimulation may cause electrochemical reactions in the medium near electrodes. For long-term stimulation, we recommend using agar-salt bridges to minimize the electrochemical byproducts in the medium (Song et al., 2007). Fill two beakers with Steinberg’s solution and immerse electrodes in the solution. Connect the beakers of Steinberg’s solution to the medium reservoirs of the stimulation chambers with the pre-made agar glass bridges through the holes on the lid.
-
g.Switch on and adjust the voltage across the AC chamber using a standard voltage meter.Optional: For monitoring voltages during experiments, mount platinum wires through an extra pair of holes between the electrodes and seal the holes with 3140 silicone rubber if the platinum wires are too thin to prevent evaporation.
-
a.
-
6.Perform real-time imaging with precisely controlled AC stimulation.
-
a.Start the image acquisition software and set up parameters required for the time-lapse recording.
-
b.Select the objective according to the experimental demand. We typically use a low magnification (10×) lens for quick evaluations of the ERK responses with different stimulation and a 40× oil immersion lens for collecting high resolution images of single cells.
-
c.Bring the imaging window of the stimulation chamber into focus.
-
d.Navigate to an XY position to select 2 or more regions of interest (ROI) in each chamber for multi-field time-lapse recording. Try to avoid regions where cells are overcrowded.
 CRITICAL: Ensure that the ROI positions are located sufficiently apart to minimize photobleaching and phototoxicity.
CRITICAL: Ensure that the ROI positions are located sufficiently apart to minimize photobleaching and phototoxicity. -
e.Select appropriate filters and dichromatic mirrors to acquire differential interference contrast (DIC) images and images of Cyan-fluorescent and red-fluorescent protein fusions.
-
f.Determine optimal exposure time for each channel. We use the following exposure times in our settings: DIC 100 msec, RFP 300 msec, CFP 500 msec.
-
g.Adjust the focus manually at each ROI as necessary.
-
h.Set auto-focus for DIC channel with a step accuracy of 0.5–1 μm in a range of 3 μm. Run autofocus routinely for each time point to correct for focal drift. Set the autofocus strategy to “update z-positions after each acquisition” if available (troubleshooting 4).
-
i.Set up a time series to acquire images at the desired time points in sequence. For investigating the effects of AC stimulation of different voltages, durations, frequencies, waveforms, or duty ratios on ERK activation, select a 30–60 s time interval over a 2 h period. For long-term monitoring ERK dynamics controlled by the AC with optimized parameters, select a 5 min time interval over a 5–24 h period.
-
j.Give it a dry run by clicking the acquisition button without any AC stimulation.
-
k.Stop image recording after finishing the second frame of all ROIs. Check the images to confirm that the image focus and exposure are set correctly and have not shifted during multi-position acquisition.
-
l.Start the time-lapse experiment by clicking the acquisition button again.
-
m.Image the cells for 5–30 min before any stimulation to acquire a baseline signal.
 CRITICAL: A minimum baseline recording of 5 min is required to determine the effect of AC stimulation. Longer times may be required if a drift in the baseline is observed (troubleshooting 5).
CRITICAL: A minimum baseline recording of 5 min is required to determine the effect of AC stimulation. Longer times may be required if a drift in the baseline is observed (troubleshooting 5). -
n.Switch on the AC with desired voltage, frequency, and waveform to stimulate cells in each chamber. Use a time relay to automatically control the on and off duration of AC stimulation.
-
o.Monitor the voltage during stimulation and adjust it if needed.
-
p.Check that all stage positions are still in focus at later points.Optional: Stage drift is a common complication when performing time-lapse recordings that span multiple hours. Even with addition of autofocus by the microscope software, stage drift can still occur and thus the following is recommended if it becomes problematic. If stage positions need to be refocused, pause imaging acquisition at an appropriate time point, adjust the focus and resume imaging within the defined imaging time interval.
-
q.Stop image acquisition at a defined time point and shut down the computer and microscope.Optional: Remove the PDMS chamber and fix the cells immediately after stimulation for immunostaining if needed.
-
a.
-
7.Generation of time-lapse movies using ImageJ software.
-
a.Launch ImageJ program and import the imaging file using “File>Import>Image sequence”. In the “Image sequence”, adjust the numbers of “Starting image” and “Increment” to obtain a separate image stack for each illumination channel (DIC, RFP, CFP) of each ROI position.Optional: Adjust the brightness and/or contrast to attain a better view of the images by clicking “Image>Adjust>Brightness/Contrast” or “Ctrl”+“Shift”+“C”.
-
b.Crop the images to exclude non-cell areas and to minimize the file size to accelerate subsequent quantification processes. Use the “Rectangular Selection” tool to select a smaller ROI in the DIC image stack and duplicate the stack of ROI with “Ctrl”+“Shift”+“D”.
-
c.Crop the same ROI for RFP and CFP stacks using "Edit>Selection>Restore Selection" and “Ctrl”+“Shift”+“D”.
-
d.Save cropped image stacks as single “.tiff” files.
-
a.
-
8.Data analysis to extract single-cell kinetics.
-
a.Import .tiff stacks to MATLAB for image segmentation, tracking, and intensity quantitation as described previously (Pargett et al., 2017).
-
b.Identify the nuclear region based on the nuclear localization of EKAR3 which contains a nuclear import sequence.
-
c.Define the cytosolic region as the neighboring 7-pixel outside the nuclear region.
-
d.Calculate cell ERKTR level as Fc/Fn, where Fc and Fn stand for the RFP fluorescence intensity of cytosol and nucleus, respectively.
-
a.
Figure 3.
Schematic depicting the experimental set-up
(A) Connecting the AC EF stimulation chamber with the AC power supply through a pair of agar salt bridges.
(B) Cells seeded in the AC stimulation chambers are imaged using a fully automated fluorescence microscope with an incubation chamber to maintain an optimal culture condition. AC EFs are applied through a pair of Ag electrodes with the desired stimulation scheme precisely controlled by a digital time relay. A voltmeter is connected in parallel with the stimulation chamber to monitor the output voltage. The time-lapse images are recorded using Metamorph.
Expected outcomes
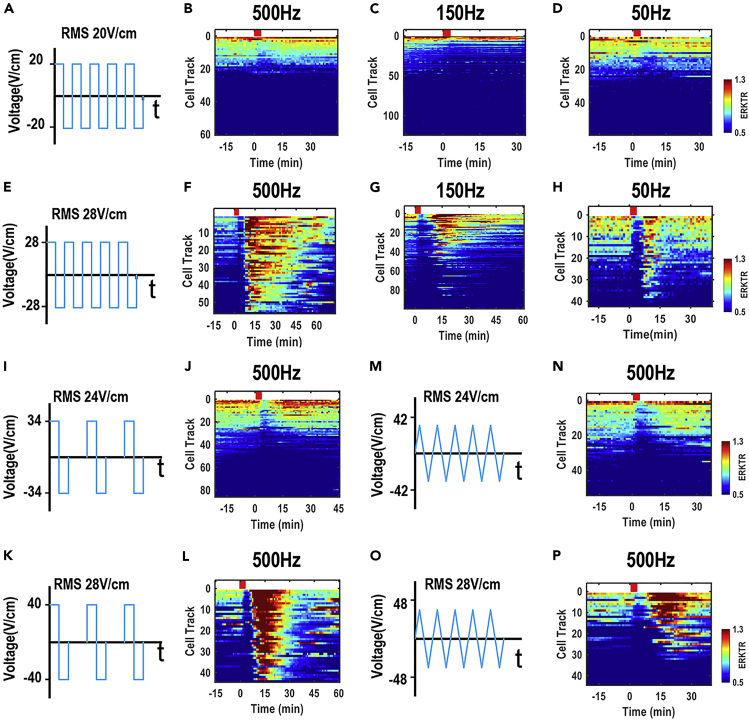
At the end of the protocol, the time-lapse data can be analyzed to evaluate the effect of AC stimulation on the activation dynamics of intracellular signaling, e.g., ERK (Figure 4). Single cell dynamics can be plotted in a heatmap showing the time courses of ERK activities (Figure 4A). The population responses of ERK activity (Figure 4B) can be calculated and compared with cells treated with different voltages, frequencies, duty cycles, and waveforms (Figure 5).
Figure 4.
AC modulates ERK dynamics by inducing robust ERK activation
(A) Representative time-lapse fluorescence images of MCF10A cells expressing ERKTR. Starved cells were exposed to a periodical 3-min AC stimulations with a 1 h interval for 6 h. ERKTR translocated from the nucleus to the cytosol after each stimulation, reporting the activation of ERK.
(B) Composite heatmap of the time courses of ERK activities as quantified by as the ratio of cytosolic fluorescence (Fc) to nuclear fluorescence (Fn). Each horizontal line represents the ERK activity time course of one single cell.
(C) Population responses of ERK activity. The thick blue line indicates the mean ERKTR ratio from a population of ∼250 cells with a blue shade showing the standard deviation range (SD). Note: Horizontal red bars indicate the duration of AC stimulation. EF= 28 V/cm (RMS), 500 Hz, bipolar square wave. Scale bars, 30 μm.
Figure 5.
Voltage, frequency, duty cycle, and waveform dependence of ERK activation
(A–H) AC-induced ERK activation is voltage-dependent and frequency-independent. 20 VRMS/cm AC of a square wave with 100% duty cycle (A) did not induce ERK activation at a frequency of 500 Hz (B), 150 Hz (C), or 50 Hz (D), respectively, while 28 VRMS/cm of the same waveform (E) activated ERK after 3 min stimulation at all frequencies (F–H).
(I–L) Duty cycle-independence of AC-induced ERK activation. 24 VRMS/cm square wave AC of 50% duty cycle (I, peak voltage = 34 V/cm) did not induce ERK activation at a frequency of 500 Hz (J), while 28 VRMS/cm (K, peak voltage = 40 V/cm) activated ERK (L).
(M–P) Waveform-independence of AC-induced ERK activation. 24 VRMS/cm AC of triangle wave (M, peak voltage = 42 V/cm) did not induce ERK activation at a frequency of 500 Hz (N), while 28 VRMS/cm (O, peak voltage = 48 V/cm) activated ERK (P). Note: Red bars indicate the duration of AC stimulation. n>40.
Limitations
At this stage, a very well-configurated cell culture chamber is required for delivering AC stimulation.
Lack of spatial resolution. The stimulation is uniform and globally of the whole cell population. We are developing microelectrode arrays for electrical stimulation with precise spatial control. We expect that increasing the precision of the electrode position and size can accomplish 50–100 μm spatial resolution of intracellular signaling activation.
Troubleshooting
Problem 1
Difficulty removing bubbles during the coating or cell seeding stages.
Potential solution
Coating with collagen or FNC could enhance the surface hydrophilicity of PDMS. If some bubbles remain in the chamber, completely remove the solution after 5 min of coating by aspiration. Load the same volume of coating solution from the same side as the first time, and gently aspirate from the other side. Less or no bubble would be expected. Repeat the same procedure if bubbles are present during cell seeding.
Another option for enhancing surface hydrophilicity is to treat the PDMS chambers with ultraviolet (UV) light for 30 min before assembling to a petri dish.
Problem 2
Difficulty in maintaining cells/cell suspension in the chamber after seeding.
Potential solution
Level the dish and transfer it carefully after cell seeding. Any shaking during the transfer of the AC stimulation device from the clean hood to the incubator would cause the medium in the reservoirs to flow and subsequently to connect with the cell suspension in the chamber. Cells will be washed out before adherence.
Replacing growth medium with cell suspension when filling the reservoirs can also help.
Problem 3
Difficulty in removing media properly from the channels. If growth media remains in the channels without sufficient exchanged with the low fluorescence medium added in the reservoir, high RFP background may be observed during the live imaging.
Potential solution
Create a small fluid pressure difference between two reservoirs to enhance medium exchange by adding different volumes of solution. Incubating cells for an extra 30 min after the medium change can also help.
Problem 4
Focus instability of the multi-wavelength timelapse recording.
Potential solution
Automatic focusing of one single channel may result in out-of-focus images on other channels. Set a compensatory distance for each channel to adjust the focus after auto-focusing the first one. Another option is to set auto-focus for all wavelengths that will be examined during the experiment. But it will result in the microscope spending a longer time capturing images at each position.
Problem 5
Drifting baseline of ERK before AC experiments.
Potential solution
ERK is sensitive to extracellular environmental changes, e.g., temperature drop when transferring cells from the cell culture incubator to the microscope. Place the dish in the incubation chamber of the microscope 30 min in advance with well-controlled temperature and CO2 concentration would help stabilize ERK before experiments.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Min Zhao (minzhao@ucdavis.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work is supported by AFOSR MURI grant (FA9550-16-1-0052, Program Leader: Wolfgang Losert, University of Maryland), and also partially supported by DURIP award FA9550-22-1-0149 from the AFOSR, NIH 1R01EY019101, DARPA grant (D20AC00003, Program Leader: Marco Rolando, University of California Santa Cruz), and Adolph Coors Foundation grant.
Author contributions
K.Z., L.G., and M.Z. conceived the study. J.A. provided the MCF10A cell line and ERK reporter used in the research. K.Z. and L.G. designed the AC stimulation devices and experimental details. K.Z. and P.P. performed the experiments. K.Z. led the preparation of the manuscript with help from all authors. Q.Q. and W.L. contributed to the improvement of AC devices and data interpretation. M.Z. supervised the study.
Declaration of interests
M.Z., J.A., Q.Q., and L.G. are named inventors with Houpu Li of a U.S. Patent application no. 16/675,127, filed November 5, 2019. Cell signaling pathway activation by local AC electric field. The patent is under consideration. The manuscript used some techniques described in the manuscript to regulate signaling dynamics. Arizona State University and University of California are patent applicants.
Data and code availability
The published article includes all details required for this study.
References
- Guo L., Zhu K., Pargett M., Contreras A., Tsai P., Qing Q., Losert W., Albeck J., Zhao M. Electrically synchronizing and modulating the dynamics of ERK activation to regulate cell fate. iScience. 2021;24:103240. doi: 10.1016/j.isci.2021.103240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pargett M., Gillies T.E., Teragawa C.K., Sparta B., Albeck J.G. Single-cell imaging of ERK signaling using fluorescent biosensors. Methods Mol. Biol. 2017;1636:35–59. doi: 10.1007/978-1-4939-7154-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Gu Y., Pu J., Reid B., Zhao Z., Zhao M. Application of direct current electric fields to cells and tissues in vitro and modulation of wound electric field in vivo. Nat. Protoc. 2007;2:1479–1489. doi: 10.1038/nprot.2007.205. [DOI] [PubMed] [Google Scholar]
- Sparta B., Pargett M., Minguet M., Distor K., Bell G., Albeck J.G. Receptor level mechanisms are required for epidermal growth factor (EGF)-stimulated extracellular signal-regulated kinase (ERK) activity pulses. J. Biol. Chem. 2015;290:24784–24792. doi: 10.1074/jbc.M115.662247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article includes all details required for this study.