Abstract
Herein, we present an unconventional method for multimodal characterization of three-dimensional cardiac organoids. This method can monitor and control the mechanophysiological parameters of organoids within a single device. In this method, local pressure distributions of human-induced pluripotent stem-cell-derived cardiac organoids are visualized spatiotemporally by an active-matrix array of pressure-sensitive transistors. This array is integrated with three-dimensional electrodes formed by the high-resolution printing of liquid metal. These liquid-metal electrodes are inserted inside an organoid to form the intraorganoid interface for simultaneous electrophysiological recording and stimulation. The low mechanical modulus and low impedance of the liquid-metal electrodes are compatible with organoids’ soft biological tissue, which enables stable electric pacing at low thresholds. In contrast to conventional electrophysiological methods, this measurement of a cardiac organoid’s beating pressures enabled simultaneous treatment of electrical therapeutics using a single device without any interference between the pressure signals and electrical pulses from pacing electrodes, even in wet organoid conditions.
Keywords: bioelectronics, liquid metal, cardiac organoid, pressure sensor, multimodal characterization
Organoids are simplified and miniaturized in vitro model systems of organs and they recently have attracted a lot of attention because of their applications in tissue development, disease modeling, and drug screening.1 Despite rigorous advances in the culturing of physiologically optimized organoids in the relevant field, there is limited applicability due to inadequate tools for practical clinical translation. The integration of engineering innovation into complex organoid systems for more in-depth control and recording of mechanophysiological parameters has proved to be a problematic task.2−4 The high culturing variability of organoids can be managed by engineering a potential design for physical cues during the self-organization processes.5−8 Modern organs-on-a-chip devices have established systemic characterization and controlled synthesis of target tissue elements, which has improved the replicability of functional readouts significantly and extended their lifespans.9−11 However, these devices restrict the natural self-organizing process of growing cell aggregates. The expression of the physiological properties is hampered when using such devices with those constraining conditions. These cell lines fundamentally follow the mechanophysiological properties set out by the architectural design of the respective organs-on-a-chip devices. Hence, when paired with such devices, the physiological relevance of these organoids ultimately is reduced.12−14
For the translation of the mechanophysiological parameters of cardiac organoids, the physiological parameters associated with the beating motions of the organoid need to be captured. Current analytic models of cardiac organoids have been limited to the electrophysiological, genomic, and optical characterization of their physiologic functionality.15 It is difficult for most commonly used optical monitoring methods to provide full, viable information about the morphological property of the organoids.16,17 A design that controls the explicit locations of the organoids without restricting their natural structure during electrophysiological characterization is an optimal setup that resolves the limitations of translatability.
Herein, we present an unconventional method for the multimodal characterization and stimulation of human-induced pluripotent stem cell (hiPSC)-derived cardiac organoids. Our device establishes unrestricted development of cardiac organoids while comprehensively monitoring the readout of their functional activity. Our device can record and control essential cardiac functions that are indispensable for physiological analysis without the reductionist engineering approach presented in modern organs-on-a-chip devices. Unlike organs-on-a-chip devices, which comprise premeditated properties of the in vivo microenvironment, our device unrestrictedly monitors the complex cellular architecture of cardiac organoids. The variation of the pressure of the organoid’s beating motions that were recorded using the pressure-sensitive transistors can correspond to electrocardiogram (ECG) traces and can generate negligible artifacts in the sensing signal even when an electrical impulse was delivered for cardiac organoid stimulation. Table S1 compares and differentiates our device from previous works on pressure sensors for biomedical applications.
Integration of Pressure-Sensitive Transistors and 3D Liquid-Metal Electrodes
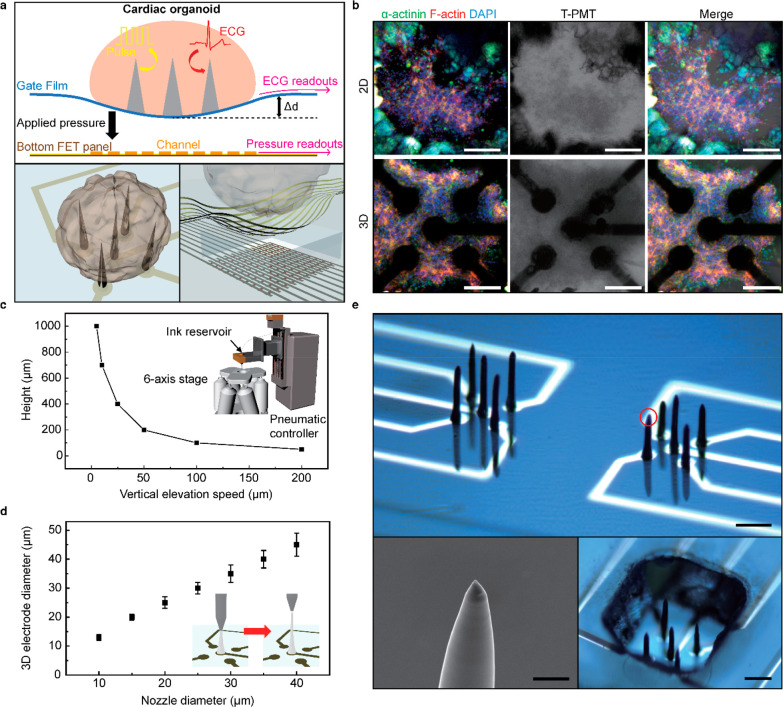
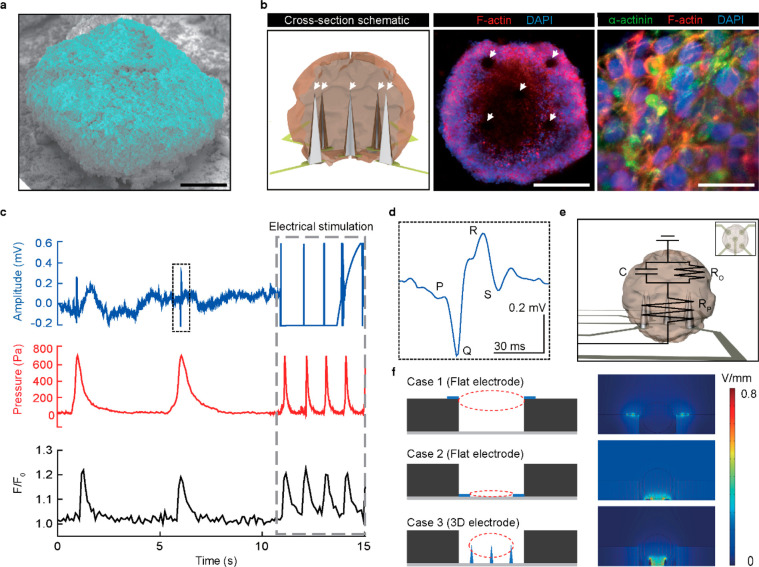
Figure 1a shows the schematic illustrations of the sensing mechanism of our multimodal device, which includes the pressure-sensitive transistor array, and the integrated 3D liquid-metal electrodes. When a cardiac organoid is located on this device, the 3D electrodes can be inserted naturally inside the cardiac organoid. Simultaneously, the active-matrix array of pressure-sensitive field-effect transistors (FET) can monitor the beating motions of this cardiac organoid mechanophysiologically by detecting the spatiotemporal change in compressive pressures. In contrast to previous surface-type electrodes, this 3D electrode can stimulate and record the interior area of an organoid directly by forming the intraorganoid interface. In addition, the 3D electrodes serve as physical cues that support the growth of the cardiac organoids. For example, Figure 1b shows immunofluorescence images of a hiPSC-derived cardiac organoid on 2D (flat) and 3D electrodes. We observed that cardiac cells cluster around these 3D electrodes during their early initiation and conformally surround the electrodes during their maturation process.
Figure 1.
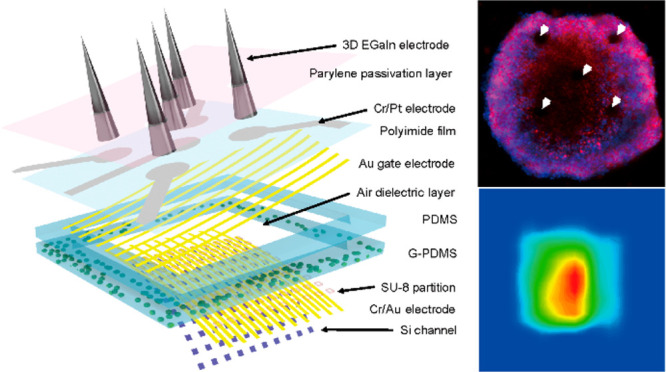
Multimodal sensing device using directly printed 3D electrodes and pressure-sensitive transistor arrays. (a) Schematic illustration of the sensing mechanism of the multimodal device (top). Illustration of the integrated 3D liquid-metal electrodes (bottom left) and pressure-sensitive transistor array (bottom right). (b) Immunofluorescence images of hiPSC-derived cardiac organoid that is grown separately on 2D (flat) and 3D electrodes. Scale bar, 100 μm. (c) Plot of the maximum height of the 3D electrode versus the speed of vertical elevation. Inset: Schematic illustration of the electrode printing system. (d) Plot of the diameter of the electrode versus the diameter of the nozzle. Inset: Schematic illustration of the 3D electrode printing. Error bars in (d) indicate the standard deviations. (e) Photograph of the 3D electrodes after high-resolution direct printing (top). Scale bar, 600 μm. The SEM image of the 3D electrode tip (indicated by the red circle, bottom left). Scale bar, 5 μm. The photograph of the directly printed 3D electrodes with PDMS microwell for cardiac organoid culturing (bottom right). Scale bar, 150 μm.
The use of liquid metals in deformable electronic devices has been prominent due to their good processability at ambient conditions and superb electrical conductivities.18−23 Their intrinsically low modulus and superb stretchability also can be advantageous for their use as biointerfacing electrodes, as conventional rigid solid metals can damage cellular interfaces because of the modulus mismatch between metallic solids and biological cells.24 We used eutectic gallium–indium (EGaIn) because of its low toxicity and negligible volatility. Figure S1 compares the Young’s modulus of EGaIn with conventional solid-phase conductive materials. The modulus of EGaIn is about 1000 times lower than the values of solid conductors and it is comparable to the modulus of human cardiac tissues.25 Upon exposure to air, EGaIn instantaneously forms a thin solid layer (∼1 nm) of gallium oxide on its surfaces under atmospheric oxygen levels. This oxide skin is thin enough to avoid damaging the cellular interfaces substantially and solid enough to maintain its 3D shape against gravity and surface tension. The printing setup consists of a printing nozzle connected to an ink reservoir that is connected to an external pneumatic pressure controller (see Supporting Information). By adjusting the stage from a fixed position to the direction of the z-axis, the 3D electrode can be printed (Video S1). Figure 1c shows the speed of the vertical elevation of a nozzle that presets the height of the printed 3D electrode. Also, the diameter of 3D electrode can be determined by the inner diameter of a nozzle (Figure 1d). For example, 3D electrodes with a diameter of 12 μm were printed using our nozzle of 10 μm in diameter (Figure S2), and the electrode diameter did not significantly degrade the electrode performance (Figure S3). Figure S4 shows the printing duration required to form a single 3D electrode, and 10 s typically were required to print a single 3D electrode with a height of 250 μm. Figure 1e shows optical micrographs of the 3D electrodes printed on a thin flexible polyimide (PI) film. The bottom-left inset shows a scanning electron microscope (SEM) image of the conical tip part of an EGaIn electrode. As shown in the bottom-right inset, a polydimethylsiloxane (PDMS) layer was perforated with a hole using a laser ablation machine as a microwell. Here, 3D electrodes were placed within the vacant hole of this PDMS microwell to promote the localization of the cell aggregate around the 3D electrodes during organogenesis.
Performance Characterization of the Multimodal Sensing Device
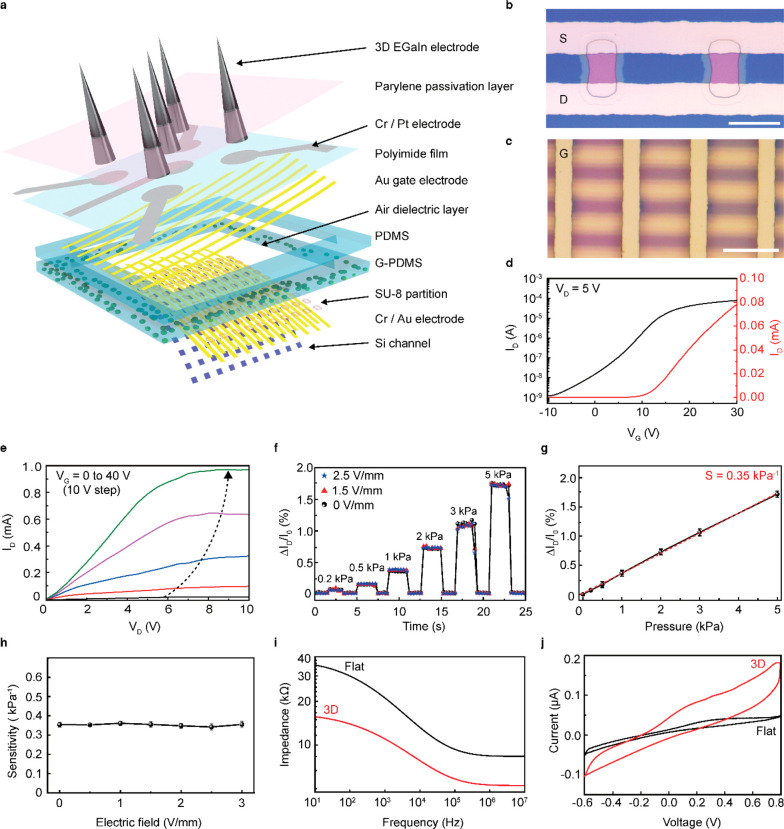
The schematic layouts of this multimodal characterization device for a cardiac organoid are shown in Figure 2a. This device is composed of two main components: (i) the bottom part comprises an array of pressure-sensitive FETs (as pressure sensors) for detecting the mechanophysiological compressive pressures, and (ii) the top part comprises 3D electrodes for the recording of electrophysiological signals and delivering of electrical stimulation. The image of the device and its relevant components are shown in Figure S5. Figure S6 illustrates the overall fabrication process of this device. We used an ultrathin, single-crystalline Si membrane as the FET channel by etching the top Si layer of a silicon-on-insulator wafer (see Supporting Information for detailed fabrication steps). For the elastomer layer, the embedding of glycerol microdroplets inside PDMS can drastically lower the value of the modulus of the elastomeric layer, which leads to large elastic deformation even at low compressive pressures.26 Then, hollow holes were produced inside the elastomer via laser ablation. Figure S7 shows the PI film with the gate electrodes and Pt interconnects. The 3D electrodes were printed directly onto the open area of the Pt interconnects.
Figure 2.
Characterization of the multimodal sensing device for cardiac organoid detection. (a) Schematic layouts of the multimodal sensor that is composed of an active-matrix pressure-sensitive transistor array with integrated soft liquid-metal electrodes. (b) Optical micrograph of the two adjacent transistors. S and D denote the source electrode and drain electrode, respectively. Scale bar, 20 μm. (c) Optical micrograph after the gate electrode (denoted as G) assembly. Scale bar, 50 μm. (d) Representative transfer characteristics of Si FET (VD = 5 V) of the device at ambient conditions. (e) Output characteristic of an air-dielectric FET (VG = 0 to 40 V, 10 V step) at ambient conditions. (f) Real-time measurements of normalized ID changes for the applied compressive pressures during electrical stimulation at different electrical fields (VG = 20 V, VD = 5 V). (g) Plot of the relative changes in ID versus applied compressive pressure to define pressure sensitivity (denoted as S). Error bars indicate the standard deviations. (h) Plot of the pressure sensitivity versus the different magnitudes of the electric field. (i) Impedance of the 3D EGaIn electrode (height, 250 μm; diameter, 60 μm; red line) and flat electrode (black line) over the frequency range from 10 Hz to 10 MHz. (j) Cyclic voltammetry plot of 3D EGaIn electrode (height, 250 μm; diameter, 60 μm; red line) and flat electrode (black line) over a range from −0.6 to 0.8 V at a sweep rate of 50 mV s–1.
For the spatiotemporal pressure mapping, a 10 × 10 active-matrix array of pressure-sensitive air-dielectric FETs was fabricated. On the basis of the average size of the cardiac organoid, the total size of the pressure-sensing region was optimized to 500 × 500 μm. Also, the channel size of the individual FET was designed as 10 × 10 μm, and the total distance between adjacent FETs (i.e., pixel resolution) was 50 μm. This active-matrix circuitry consists of scan and data lines in which a targeted pressure sensor (or pixel) was designed to operate separately to give direct electrical responses, according to the selection of the combination of rows and columns.27−30Figure 2b shows an optical micrograph of the Si channel with the patterned source/drain electrodes. Accordingly, the assembly of the gate electrode results in the formation of the air-dielectric FETs (Figure 2c). Figure 2d,e plots the representative transfer and output curves of this transistor at ambient conditions, respectively. The on/off ratio (Ion/Ioff) and threshold voltage (Vth) were 105 and 10.5 V, respectively. As shown in Figure S8, the statistical measurement data fit the Gaussian distribution profile by presenting outstanding homogeneity of sensitivity (average sensitivity: 0.35 ± 0.01 kPa–1) in all pixels. The clean interface between the Si channel and the air dielectric can exhibit this outstanding homogeneity at ambient conditions.31−33 In particular, the air-dielectric structure can result in negligible hysteresis during the operation of the FET.34 Our air-dielectric FET exhibited a response time of 48 ms and a recovery time of 42 ms (Figure S9).
Figure 2f shows the real-time detection of the relative change in ID [ΔID/I0 (%)], where ID is the drain current, I0 is the current at zero Pascal, and ΔID= I – I0, which denotes the variation of the ID during stepwise pressure loading. The sensitivity (S) of the transistor was obtained by plotting the relative change in ID concerning the applied pressure (Figure 2g) and was expressed as [ΔID/I0 (%)]/ΔP, where ΔP denotes the applied pressure. ΔID/I0 linearly increased within the pressure range from 1 to 5 kPa, where the sensitivity was approximately calculated as ∼0.35 kPa–1. In addition, the ΔID/I0 values measured under desired pressure loading showed negligible variation when an electrical field was applied simultaneously to the 3D electrodes during pressure sensing (Figure 2f). As shown in Figure 2h, the application of an electric field did not change the S value.
The significance of the 3D electrodes was validated by using electrochemical impedance spectroscopy (EIS) and cyclic voltammetry (CV) analyses to compare the impedance of the 3D EGaIn electrode (height: 250 μm) with a flat surface Pt electrode using a potentiostat. The areal dimensions of both electrodes, i.e., 3D EGaIn and flat Pt, were identical, but the 3D electrode exhibited 250 μm in height. Both electrodes were submerged in a phosphate-buffered saline (PBS) solution with a reference electrode (Ag/AgCl) and a counter electrode (Pt sheet) at ambient conditions (pH 7.4, 25 °C). As plotted in Figure 2i, the 3D electrode and flat electrode exhibited impedance values (at 103 Hz) of 10.5 and 20.7 kΩ, respectively. The charge storage characteristic was tested via CV analysis conducted at a scan rate of 50 mV/s with a potential limit range from −0.6 to 0.8 V (Figure 2j). The 3D electrode showed an outstanding charge storage characteristic, which was notable in the large area of the enclosed CV curve. Overall, the 3D electrodes had lower impedance and a better charge storage characteristic, which indicates efficient performance for electrode-to-cell signal transfer.
Formation of Human Cardiac Organoids Using Cardiomyocytes Differentiated from hiPSCs
The shape and duration of the action potentials are incoherent between animal and human heart. Hence, animal modeling is often inaccurate in translation to human clinical setting in cardiology research.35,36 Cardiomyocyte differentiation of hiPSCs has been employed to overcome these limitations and to recapitulate action potentials and functional structures of human cardiomyocytes. It is more efficient to obtain cardiomyocytes from hiPSCs than other sources since human fetal cardiac tissues are difficult to obtain, and the ability of adult stem cells to proliferate and differentiate into functional cardiomyocytes is known to be limited.37 More importantly, when hiPSC-derived cardiomyocytes are cultivated in a 3D condition, their electrophysiological properties are further enhanced. Their phenotypic characteristics become closer to that of actual human heart tissue as the 3D structural environment increases the cellular interactions and enhances the differentiation and maturation of the cardiomyocytes.38
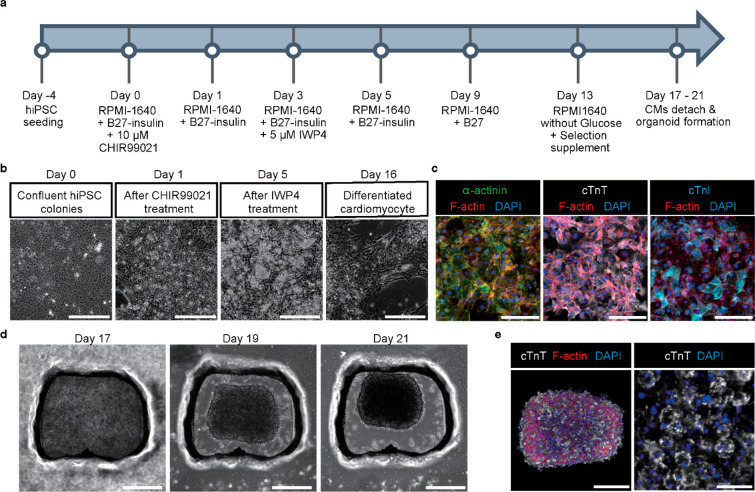
hiPSCs were seeded on a 6-well plate to differentiate hiPSCs into cardiomyocytes, and cardiomyocyte differentiation was induced according to the protocol established by Hoang et al. (Figure 3a).39 Four days after hiPSC seeding, the differentiation process was initiated by replacing the culture medium with a differentiation medium (day 0). The differentiated cells with cardiomyocyte-like morphology appeared over culture time up to 16 days (Figure 3b). The immunofluorescence staining to check cardiomyocyte differentiation revealed that three cardiac markers, α-actinin, cardiac troponin T (cTnT), and cardiac troponin I (cTnI), were highly expressed in the differentiated cardiomyocytes (Figure 3c). Then, cardiac organoids were generated using hiPSC-derived cardiomyocytes that had started beating on day 17 to day 21. The cardiac organoid development in the specially designed PDMS microwell was monitored over culture time (Figure 3d). We confirmed the high level of cardiac marker (cTnT) expression in human cardiac organoids at day 21 (Figure 3e). Moreover, human cardiac organoids showed autonomous beating motion in the microwell (Video S2). Additionally, a live/dead staining assay conducted 3 days after the generation of cardiac organoids with the 3D electrodes showed good biocompatibility (Figure S10).
Figure 3.
Formation of human cardiac organoids using human-induced pluripotent stem cell (hiPSC). (a) Timeline and culture conditions for cardiomyocyte differentiation from hiPSC. (b) Light microscopic observation of differentiated cardiomyocytes over culture time. Scale bar, 300 μm. (c) Immunofluorescent images of α-actinin, cardiac troponin T (cTnT), cardiac troponin I (cTnl), and F-actin in hiPSC-derived cardiomyocytes. DAPI was used for nucleus staining. Scale bars, 150 μm. (d) Light microscopic observation of 3D human cardiac organoids generated with differentiated cardiomyocytes over culture time. Scale bar, 300 μm. (e) Immunofluorescent image of cTnT and F-actin in human cardiac organoid (left; scale bar, 200 μm) and high-magnified image of cTnT in human cardiac organoid (right; scale bar, 30 μm). DAPI was used for nucleus staining.
Multimodal Characterization of the Cardiac Organoids
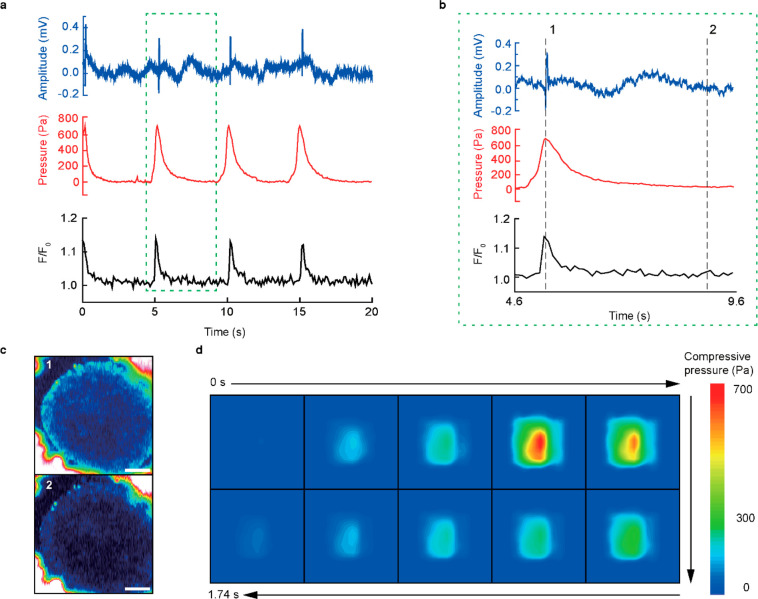
Since the average size of the cardiac organoids was no larger than 400 μm, the organoid spanned over multiple FET, which allowed detection of spatiotemporal changes in local pressure distribution during its beating. As shown in Figure 4a, our device simultaneously monitored the beating activity of a cardiac organoid by ECG (blue), pressure (red), and calcium imaging (black). The ECG signal (unit: mV) was recorded using the 3D electrodes and an electrophysiological measurement system that was comprised of a multielectrode array and a data processor with a real-time controller. The measurement of beating pressure was obtained by using automated instrumentation with a custom-made jig (Figure S11) connected to separate modules using two sourcemeters, a system switch, and a relay card. The readouts of each FET were converted to corresponding compressive pressure values. The organoid was stained using Fluo-4 AM for calcium imaging. The normalized fluorescence intensity at a specific region of interest (F/F0) of the calcium flux was plotted against time. All modes of characterization showed a consistent beating frequency of 12.1 beats per minute (bpm) with a consecutive beating interval of 4.94 s. Figure 4b presents the relative changes in the three characterization data during a single beat (indicated as the green dashed box in Figure 4a). Figure 4c shows pseudocolor fluorescence images of this organoid, which represents the changes of the calcium indicator at the time phases indicated in Figure 4b. These images indicated the depolarization (top) and resting state (bottom) of the organoid. The ECG signal recorded using the 3D electrodes did not significantly deteriorate for 7 days (Figure S12). Figure 4d visualizes the spatiotemporal changes in the beating pressure distribution as 2D color gradation contour plots. Video S3 presents the synchronization of this spatiotemporal pressure mapping with the calcium imaging during beating motions.
Figure 4.
Multimodal characterization of cardiac organoids. (a) Real-time characterization plot of the ECG amplitude (blue), compressive pressure (red), and normalized fluorescence intensity (black) of the hiPSC-derived cardiac organoid by the multimodal characterization device. (b) Magnified plot of multimodal characterization data versus time which defines one beating cycle of the cardiac organoid, denoted as the green dotted box in (a). (c) Pseudocolor fluorescence images of the cardiac organoid at the instance of depolarization (top), labeled as time frame 1 in (b) and 3.8 s afterward (bottom) labeled as time frame 2 in (b). Scale bar, 100 μm. (d) Time-lapsed spatiotemporal color maps of compressive pressure distribution of the hiPSC-derived cardiac organoid to capture the instances of movement during a single beating. Each motion frame of the cardiac organoid was taken every 174 ms.
Electrical Stimulation of the Cardiac Organoids
Unlike previous studies that measured electrophysiological signals of an organoid by attaching thin, flexible electrodes to the outer surfaces of the organoid (extraorganoid sensing),5,6,40,41 the 3D electrodes of our device were embedded inside a cardiac organoid for the intraorganoid electrophysiological recording. Modern methods of confinement are microfluidic devices or arrays of microwells, which can diminish the quality of physiological signals or are ill-equipped to make high-quality characterizations.42−44 In contrast, our device was advantageous for localizing the organoid development toward the sensing site without any elements of constraint that potentially diminish the significance of the readouts. These 3D electrodes do not force organoids into a final specific shape; rather, they naturally guide the formation of the shape of the hiPSC-derived cardiac organoids during their growth. The SEM image of the cardiac organoid developed on these 3D electrodes is shown in Figure 5a. The cross section schematic inset in Figure 5b (left) illustrates the intraorganoid interface between the cardiac organoid and the 3D electrodes. In Figure S13, the immunofluorescence images of the cryosectioned cardiac organoid showed the positional traces of the 3D electrodes in the cardiac organoid, which represent the intraorganoid interfaces with no signs of bending or displacement. Figure 5b shows the immunofluorescent images of the organoid with the 3D electrodes (middle inset) and close-up of the surface of the organoid (right).
Figure 5.
Simultaneous electrical stimulation of cardiac organoids with multimodal sensing. (a) Colorized SEM image of the hiPSC-derived cardiac organoid developed on the 3D liquid-metal electrodes. Scale bar, 100 μm. (b) Cross section schematics of the intraorganoid interface between the cardiac organoid and the 3D electrodes (left). The immunofluorescence images of the matured cardiac organoid with the 3D electrodes (middle inset) and close-up of the surface of the hiPSC-derived cardiac organoid (right). Scale bar, 200 and 20 μm, respectively. (c) Real-time characterization plot of the ECG amplitude (blue), compressive pressure (red), and normalized fluorescence intensity (black) of the hiPSC-derived cardiac organoid before and during electrical stimulation (gray dotted line, 0.8 V/mm). (d) Magnified single ECG signal denoted as the black dotted box indicated in (c). (e) Electrical circuit of cardiac organoid with 3D electrodes for electrical stimulation and characterization. The C, RP, and RO represent the capacitance of the organoid, the resistance of the probe, and the resistance of the organoid, respectively. Inset: Aerial schematics of the cardiac organoid. (f) Finite element analysis (FEA) simulation of the electric field for different cases of electrodes for the optimization of the organoid stimulation. Case 1 and 2 define the electric field simulation of the flat electrode at the top and bottom position, respectively. Case 3 defines the electric field simulation of the 3D electrodes located at the bottom base.
For the modulation of the organoid’s beating, it was subjected to an electrical stimulation pulse of 0.8 V/mm (pulse width, 1 ms; frequency, 1 Hz) using an externally connected pulse generator. Figure 5c shows the simultaneous multimodal characterizations (ECG, pressure, and calcium flux) of the organoid before and after electrical stimulation (gray-dashed box). A single ECG peak before applying the electrical impulses (black-dashed box in Figure 5c) is shown in Figure 5d. As seen, the depolarization of a cardiac organoid was indicated by a distinct P wave in the PQRS complex of the ECG signal. However, during the electric pacing, the electric pulses significantly disrupted the ECG trace and essentially rendered the interpretation of the ECG impossible. There was an undesirable interference with the electric signals due to the leakage through the organoid’s electrical conduction system, thereby limiting the simultaneous functions of ECG recording and electrical pacing. After the end of electrical stimulation, the ECG signal was returned to the normal state (Figure S14). Figure 5e shows a diagram of the electrical circuit of this cardiac organoid with 3D electrodes. In contrast, our mechanophysiological sensing enabled the recording of the organoid’s beating motions with no signal disruption during the application of electrical impulses in electrical therapeutics. The intraorganoid interface between the 3D electrodes and the organoid enlarged the contact area relatively to ensure efficient charge transfer. Also, as shown in the computational simulation of the electric potential for the shape and placement of electrodes inside a microwell (Figure 5f), the 3D microstructure of the electrodes for our device can be effective to focus and locally distribute the electric field near the pacing region (case 3), compared with the flat geometries of the other electrodes (case 1 and case 2).
Recent research related to organoids has performed exceedingly well in the recapitulation of cellular diversity and microscale tissue architecture to bring out critical functions of their in vivo counterparts. However, the issues of translational relevance and high-throughput functional readout remain crucial challenges in practical utilization for clinical applications.43,45 In traditional culture systems, there is little or no integration of biophysical and topological parameters that can support the self-organization of organoids.46 Hence, the growth environment of these organoids usually is versatile and ill-defined, which leads to high variability in organoid phenotypes. Although optical analysis methods have been used mainly for cellular characterization, organoids have 3D shapes and versatile motion displacements, which makes automated live imaging very challenging. The optical imaging method also holds little functional insight and relies on reporter lines for the assessment of the expression of specific markers. The multimodal sensing mechanism can provide a capacity for deeper contextual understanding of physiological parameters for more profound analytical studies. Furthermore, the implementation of each of the integrated methods was conducted independently and does not effectively affect the performance of the other methods. The straightforward microfabrication steps and the simplicity of the direct printing of 3D liquid-metal electrodes allow a smooth transition into large-scale microcompartmentalization for automated screenings. This multimodal sensory device suggests the future promise of an all-in-one analysis platform for a higher level of biomedical modeling of cardiac organoids in various areas, such as personalized medicine, disease modeling, and drug cardiotoxicity.
Acknowledgments
This work was supported by the Ministry of Science & ICT (MSIT), the Ministry of Trade, Industry and Energy (MOTIE), the Ministry of Health & Welfare, and the Ministry of Food and Drug Safety of Korea through the National Research Foundation for Nano Material Technology Development Program (2021M3D1A204991411), the Bio & Medical Technology Development Program (2018M3A9F1021649), the Wearable Platform Materials Technology Center ERC Program (2022R1A5A6000846), the Korea Initiative for fostering University of Research and Innovation (KIURI) Program (2020M3H1A1077207), the Technology Innovation Program (20013621, Center for Super Critical Material Industrial Technology), and the Korea Medical Device Development Fund grant (RMS 2022-11-1209/KMDF RS-2022-00141392). Also, the authors thank the financial support by the Samsung Research Funding & Incubation Center of Samsung Electronics (SRFC-TC2003-03) and the Institute for Basic Science (IBS-R026-D1).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.2c02790.
Video S1: fabrication of soft 3D EGaIn electrode using direct printing method (MP4)
Video S2: beating of hiPSC cardiac organoid after cultivation in PDMS stencil (MP4)
Video S3: time-lapsed 2D pressure distribution of the cardiac organoid with calcium flux imaging (MP4)
Discussion of materials used in this work, fabrication of cardiac organoid, preparation of multimodal sensor, direct printing of 3D electrodes, calculation of pressure of air-dielectric Si pressure sensor, conditioning of organoid for fluorescent immunostaining, organoid dehydration process for scanning electron microscopy, organoid preparation for cryosectioning, assessment of biocompatibility, modulus values of conventional materials used for bioelectronic electrodes, SEM images of 3D electrodes, plot of the ECG SNR versus the diameter of 3D electrodes, plot of the electrode height versus printing duration, digital photos of the device, schematic illustrations for the fabrication method of multimodal organoid sensor, photograph of the gate electrode and interconnect contact pad, electrical property analysis of FET arrays, response time and recovery time of pressure sensor, biocompatibility data of our device, data acquisition system and circuit diagram, ECG SNR variation for 7 days, cross-sectional immunofluorescent image of cardiac organoid, and tables of previous pressure sensors for biomedical applications and chemical and fluorescence agents used in this work (PDF)
Author Contributions
⊥ M.K., J.C.H., and S.M. contributed equally to this work. M.K. and J.C.H. carried out the experiments, analyzed the data, wrote the manuscript. S.M. and S.K. oversaw the culturing and maturation of hiPSC-derived cardiac organoid and conducted its immunofluorescence characterization. Y.-G.P., E.K., H.S., J.L., and W.G.C. contributed the device fabrications. S.-W.C. and J.-U.P. wrote the paper. J.-U.P. oversaw all phases of this research. All authors discussed and commented on the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- McCauley H. A.; Wells J. M. Pluripotent Stem Cell-Derived Organoids: Using Principles of Developmental Biology to Grow Human Tissues in a Dish. Development 2017, 144 (6), 958–962. 10.1242/dev.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. S.; Aleman J.; Shin S. R.; Kilic T.; Kim D.; Shaegh S. A. M.; Massa S.; Riahi R.; Chae S.; Hu N.; Avci H.; Zhang W.; Silvestri A.; Nezhad A. S.; Manbohi A.; Ferrari F. D.; Polini A.; Calzone G.; Shaikh N.; Alerasool P.; Budina E.; Kang J.; Bhise N.; Ribas J.; Pourmand A.; Skardal A.; Shupe T.; Bishop C. E.; Dokmeci M. R.; Atala A.; Khademhosseini A. Multisensor-Integrated Organs-on-Chips Platform for Automated and Continual in Situ Monitoring of Organoid Behaviors. Proc. Natl. Acad. Sci. U.S.A. 2017, 114 (12), E2293–E2302. 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumann E. N.; Tian B. Biological Interfaces, Modulation, and Sensing with Inorganic Nano-Bioelectronic Materials. Small Methods 2020, 4 (5), 1900868. 10.1002/smtd.201900868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. S.; Yu C. Towards Engineering Integrated Cardiac Organoids: Beating Recorded. J. Thorac. Dis. 2016, 8 (12), E1683–E1687. 10.21037/jtd.2016.12.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H.; Park Y.; Luan H.; Dalgin G.; Jeffris K.; Yoon H.-J.; Chung T. S.; Kim J. U.; Kwak S. S.; Lee G.; Jeong H.; Kim J.; Bai W.; Kim J.; Jung Y. H.; Tryba A. K.; Song J. W.; Huang Y.; Philipson L. H.; Finan J. D.; Rogers J. A. Transparent, Compliant 3D Mesostructures for Precise Evaluation of Mechanical Characteristics of Organoids. Adv. Mater. 2021, 33 (25), 2100026. 10.1002/adma.202100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.; Franz C. K.; Ryu H.; Luan H.; Cotton K. Y.; Kim J. U.; Chung T. S.; Zhao S.; Vazquez-Guardado A.; Yang D. S.; Li K.; Avila R.; Phillips J. K.; Quezada M. J.; Jang H.; Kwak S. S.; Won S. M.; Kwon K.; Jeong H.; Bandodkar A. J.; Han M.; Zhao H.; Osher G. R.; Wang H.; Lee K.; Zhang Y.; Huang Y.; Finan J. D.; Rogers J. A. Three-Dimensional, Multifunctional Neural Interfaces for Cortical Spheroids and Engineered Assembloids. Sci. Adv. 2021, 7 (12), eabf9153. 10.1126/sciadv.abf9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H.; Zhang Q.; Liu T.-L.; Wang X.; Zhao S.; Wang H.; Yao S.; Xue Y.; Kwak J. W.; Bai W.; Xu Y.; Han M.; Li K.; Li Z.; Ni X.; Ye J.; Choi D.; Yang Q.; Kim J.-H.; Li S.; Chen S.; Wu C.; Lu D.; Chang J.-K.; Xie Z.; Huang Y.; Rogers J. A. Complex 3D Microfluidic Architectures Formed by Mechanically Guided Compressive Buckling. Sci. Adv. 2021, 7 (43), eabj3686. 10.1126/sciadv.abj3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Nan K.; Le Floch P.; Lin Z.; Sheng H.; Blum T. S.; Liu J. Cyborg Organoids: Implantation of Nanoelectronics via Organogenesis for Tissue-Wide Electrophysiology. Nano Lett. 2019, 19 (8), 5781–5789. 10.1021/acs.nanolett.9b02512. [DOI] [PubMed] [Google Scholar]
- Park S. E.; Georgescu A.; Huh D. Organoids-on-a-Chip. Science 2019, 364 (6444), 960–965. 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair V.; Yi J.; Isheim D.; Rotenberg M.; Meng L.; Shi F.; Chen X.; Gao X.; Prominski A.; Jiang Y.; Yue J.; Gallagher C. T.; Seidman D. N.; Tian B. Laser Writing of Nitrogen-Doped Silicon Carbide for Biological Modulation. Sci. Adv. 2020, 6 (34), eaaz2743. 10.1126/sciadv.aaz2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykov A.; Huang C.; Bliley J.; Shiwarski D.; Tashman J.; Abdullah A.; Rastogi S. K.; Shukla S.; Mataev E.; Feinberg A. W.; Hsia K. J.; Cohen-Karni T. Organ-on-e-Chip: Three-Dimensional Self-Rolled Biosensor Array for Electrical Interrogations of Human Electrogenic Spheroids. Sci. Adv. 2019, 5 (8), eaax0729. 10.1126/sciadv.aax0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musah S.; Mammoto A.; Ferrante T. C.; Jeanty S. S. F.; Hirano-Kobayashi M.; Mammoto T.; Roberts K.; Chung S.; Novak R.; Ingram M.; Fatanat-Didar T.; Koshy S.; Weaver J. C.; Church G. M.; Ingber D. E. Mature Induced-Pluripotent-Stem-Cell-Derived Human Podocytes Reconstitute Kidney Glomerular-Capillary-Wall Function on a Chip. Nat. Biomed. Eng. 2017, 1 (5), 0069. 10.1038/s41551-017-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J.; Huh D.; Hamilton G.; Ingber D. E. Human Gut-on-a-Chip Inhabited by Microbial Flora That Experiences Intestinal Peristalsis-like Motions and Flow. Lab Chip 2012, 12 (12), 2165–2174. 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- Wang X.; Feiner R.; Luan H.; Zhang Q.; Zhao S.; Zhang Y.; Han M.; Li Y.; Sun R.; Wang H.; Liu T.-L.; Guo X.; Oved H.; Noor N.; Shapira A.; Zhang Y.; Huang Y.; Dvir T.; Rogers J. A. Three-Dimensional Electronic Scaffolds for Monitoring and Regulation of Multifunctional Hybrid Tissues. Extreme Mech. Lett. 2020, 35, 100634. 10.1016/j.eml.2020.100634. [DOI] [Google Scholar]
- Kim G.-A.; Ginga N. J.; Takayama S. Integration of Sensors in Gastrointestinal Organoid Culture for Biological Analysis. Cell. Mol. Gastroenterol. Hepatol. 2018, 6 (1), 123–131. 10.1016/j.jcmgh.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers J. F.; Wiegerinck C. L.; de Jonge H. R.; Bronsveld I.; Janssens H. M.; de Winter-de Groot K. M.; Brandsma A. M.; de Jong N. W. M.; Bijvelds M. J. C.; Scholte B. J.; Nieuwenhuis E. E. S.; van den Brink S.; Clevers H.; van der Ent C. K.; Middendorp S.; Beekman J. M. A Functional CFTR Assay Using Primary Cystic Fibrosis Intestinal Organoids. Nat. Med. 2013, 19 (7), 939–945. 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- Okkelman I. A.; Foley T.; Papkovsky D. B.; Dmitriev R. I. Live Cell Imaging of Mouse Intestinal Organoids Reveals Heterogeneity in Their Oxygenation. Biomaterials 2017, 146, 86–96. 10.1016/j.biomaterials.2017.08.043. [DOI] [PubMed] [Google Scholar]
- Park Y.-G.; An H. S.; Kim J.-Y.; Park J.-U. High-Resolution, Reconfigurable Printing of Liquid Metals with Three-Dimensional Structures. Sci. Adv. 2019, 5 (6), eaaw2844. 10.1126/sciadv.aaw2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.-G.; Min H.; Kim H.; Zhexembekova A.; Lee C. Y.; Park J.-U. Three-Dimensional, High-Resolution Printing of Carbon Nanotube/Liquid Metal Composites with Mechanical and Electrical Reinforcement. Nano Lett. 2019, 19 (8), 4866–4872. 10.1021/acs.nanolett.9b00150. [DOI] [PubMed] [Google Scholar]
- Park Y.-G.; Yun I.; Chung W. G.; Park W.; Lee D. H.; Park J.-U. High-Resolution 3D Printing for Electronics. Adv. Sci. 2022, 9 (8), 2104623. 10.1002/advs.202104623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.-G.; Lee S.; Park J.-U. Recent Progress in Wireless Sensors for Wearable Electronics. Sensors 2019, 19 (20), 4353. 10.3390/s19204353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.; Hwang J. C.; Kim G. G.; Park J.-U. Flexible Electronics Based on One-Dimensional and Two-Dimensional Hybrid Nanomaterials. InfoMat 2020, 2 (1), 33–56. 10.1002/inf2.12047. [DOI] [Google Scholar]
- Park J.; Hyun B. G.; An B. W.; Im H.-G.; Park Y.-G.; Jang J.; Park J.-U.; Bae B.-S. Flexible Transparent Conductive Films with High Performance and Reliability Using Hybrid Structures of Continuous Metal Nanofiber Networks for Flexible Optoelectronics. ACS Appl. Mater. Interfaces 2017, 9 (24), 20299–20305. 10.1021/acsami.7b04314. [DOI] [PubMed] [Google Scholar]
- Park Y.-G.; Lee G.-Y.; Jang J.; Yun S. M.; Kim E.; Park J.-U. Liquid Metal-Based Soft Electronics for Wearable Healthcare. Adv. Healthc. Mater. 2021, 10 (17), 2002280. 10.1002/adhm.202002280. [DOI] [PubMed] [Google Scholar]
- Mathur A. B.; Collinsworth A. M.; Reichert W. M.; Kraus W. E.; Truskey G. A. Endothelial, Cardiac Muscle and Skeletal Muscle Exhibit Different Viscous and Elastic Properties as Determined by Atomic Force Microscopy. J. Biomech. 2001, 34 (12), 1545–1553. 10.1016/S0021-9290(01)00149-X. [DOI] [PubMed] [Google Scholar]
- Jang J.; Kim H.; Ji S.; Kim H. J.; Kang M. S.; Kim T. S.; Won J.; Lee J.-H.; Cheon J.; Kang K.; Im W. B.; Park J.-U. Mechanoluminescent, Air-Dielectric MoS2 Transistors as Active-Matrix Pressure Sensors for Wide Detection Ranges from Footsteps to Cellular Motions. Nano Lett. 2020, 20 (1), 66–74. 10.1021/acs.nanolett.9b02978. [DOI] [PubMed] [Google Scholar]
- Jang J.; Park Y.-G.; Cha E.; Ji S.; Hwang H.; Kim G. G.; Jin J.; Park J.-U. 3D Heterogeneous Device Arrays for Multiplexed Sensing Platforms Using Transfer of Perovskites. Adv. Mater. 2021, 33 (30), e2101093. 10.1002/adma.202101093. [DOI] [PubMed] [Google Scholar]
- Jang J.; Ji S.; Grandhi G. K.; Cho H. B.; Im W. B.; Park J.-U. Multimodal Digital X-Ray Scanners with Synchronous Mapping of Tactile Pressure Distributions Using Perovskites. Adv. Mater. 2021, 33 (30), 2008539. 10.1002/adma.202008539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S.; Jang J.; Hwang J. C.; Lee Y.; Lee J.-H.; Park J.-U. Amorphous Oxide Semiconductor Transistors with Air Dielectrics for Transparent and Wearable Pressure Sensor Arrays. Adv. Mater. Technol. 2020, 5 (2), 1900928. 10.1002/admt.201900928. [DOI] [Google Scholar]
- Cheong W. H.; Oh B.; Kim S.-H.; Jang J.; Ji S.; Lee S.; Cheon J.; Yoo S.; Lee S.-Y.; Park J.-U. Platform for Wireless Pressure Sensing with Built-in Battery and Instant Visualization. Nano Energy 2019, 62, 230–238. 10.1016/j.nanoen.2019.05.047. [DOI] [Google Scholar]
- Jang J.; Jun Y. S.; Seo H.; Kim M.; Park J.-U. Motion Detection Using Tactile Sensors Based on Pressure-Sensitive Transistor Arrays. Sensors 2020, 20 (13), 3624. 10.3390/s20133624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S.-H.; Ji S.; Choi S.; Pyo K.-H.; Wan An B.; Park J.; Kim J.; Kim J.-Y.; Lee K.-S.; Kwon S.-Y.; Heo J.; Park B.-G.; Park J.-U. Integrated Arrays of Air-Dielectric Graphene Transistors as Transparent Active-Matrix Pressure Sensors for Wide Pressure Ranges. Nat. Commun. 2017, 8 (1), 14950. 10.1038/ncomms14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J.; Oh B.; Jo S.; Park S.; An H. S.; Lee S.; Cheong W. H.; Yoo S.; Park J.-U. Human-Interactive, Active-Matrix Displays for Visualization of Tactile Pressures. Adv. Mater. Technol. 2019, 4 (7), 1900082. 10.1002/admt.201900082. [DOI] [Google Scholar]
- Guo Y.; Wei X.; Shu J.; Liu B.; Yin J.; Guan C.; Han Y.; Gao S.; Chen Q. Charge Trapping at the MoS2-SiO2 Interface and Its Effects on the Characteristics of MoS2Metal-Oxide-Semiconductor Field Effect Transistors. Appl. Phys. Lett. 2015, 106 (10), 103109. 10.1063/1.4914968. [DOI] [Google Scholar]
- Zuppinger C. 3D Cardiac Cell Culture: A Critical Review of Current Technologies and Applications. Front. Cardiovasc. Med. 2019, 6, 87. 10.3389/fcvm.2019.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine M. D.; Mannhardt I.; Breckwoldt K.; Prondzynski M.; Flenner F.; Ulmer B.; Hirt M. N.; Neuber C.; Horváth A.; Kloth B.; Reichenspurner H.; Willems S.; Hansen A.; Eschenhagen T.; Christ T. Human IPSC-Derived Cardiomyocytes Cultured in 3D Engineered Heart Tissue Show Physiological Upstroke Velocity and Sodium Current Density. Sci. Rep. 2017, 7 (1), 5464. 10.1038/s41598-017-05600-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloch N. L.; Muskheli V.; Razumova M. V.; Korte F. S.; Regnier M.; Hauch K. D.; Pabon L.; Reinecke H.; Murry C. E. Growth of Engineered Human Myocardium With Mechanical Loading and Vascular Coculture. Circ. Res. 2011, 109 (1), 47–59. 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. C.; Hookway T. A.; Wu Q.; Jha R.; Preininger M. K.; Chen X.; Easley C. A.; Spearman P.; Deshpande S. R.; Maher K.; Wagner M. B.; McDevitt T. C.; Xu C. Microscale Generation of Cardiospheres Promotes Robust Enrichment of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cell Rep. 2014, 3 (2), 260–268. 10.1016/j.stemcr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang P.; Wang J.; Conklin B. R.; Healy K. E.; Ma Z. Generation of Spatial-Patterned Early-Developing Cardiac Organoids Using Human Pluripotent Stem Cells. Nat. Protoc. 2018, 13 (4), 723–737. 10.1038/nprot.2018.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair S. R.; Julian D.; Hartlaub A. M.; Pusuluri S. T.; Malik G.; Summerfied T. L.; Zhao G.; Hester A. B.; Ackerman W. E.; Hollingsworth E. W.; Ali M.; McElroy C. A.; Buhimschi I. A.; Imitola J.; Maitre N. L.; Bedrosian T. A.; Hester M. E. Electrophysiological Maturation of Cerebral Organoids Correlates with Dynamic Morphological and Cellular Development. Stem Cell Rep. 2020, 15 (4), 855–868. 10.1016/j.stemcr.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo C. A.; Gao R.; Negraes P. D.; Gu J.; Buchanan J.; Preissl S.; Wang A.; Wu W.; Haddad G. G.; Chaim I. A.; Domissy A.; Vandenberghe M.; Devor A.; Yeo G. W.; Voytek B.; Muotri A. R. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell 2019, 25 (4), 558–569. 10.1016/j.stem.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniecki S. M.; Cruz N. M.; Harder J. L.; Menon R.; Annis J.; Otto E. A.; Gulieva R. E.; Islas L. V.; Kim Y. K.; Tran L. M.; Martins T. J.; Pippin J. W.; Fu H.; Kretzler M.; Shankland S. J.; Himmelfarb J.; Moon R. T.; Paragas N.; Freedman B. S. High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell 2018, 22 (6), 929–940. 10.1016/j.stem.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberg N.; Hoehnel S.; Kuttler F.; Homicsko K.; Ceroni C.; Ringel T.; Gjorevski N.; Schwank G.; Coukos G.; Turcatti G.; Lutolf M. P. High-Throughput Automated Organoid Culture via Stem-Cell Aggregation in Microcavity Arrays. Nat. Biomed. Eng. 2020, 4 (9), 863–874. 10.1038/s41551-020-0565-2. [DOI] [PubMed] [Google Scholar]
- Jin B.-J.; Battula S.; Zachos N.; Kovbasnjuk O.; Fawlke-Abel J.; In J.; Donowitz M.; Verkman A. S. Microfluidics Platform for Measurement of Volume Changes in Immobilized Intestinal Enteroids. Biomicrofluidics 2014, 8 (2), 024106. 10.1063/1.4870400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukonin I.; Serra D.; Challet Meylan L.; Volkmann K.; Baaten J.; Zhao R.; Meeusen S.; Colman K.; Maurer F.; Stadler M. B.; Jenkins J.; Liberali P. Phenotypic Landscape of Intestinal Organoid Regeneration. Nature 2020, 586 (7828), 275–280. 10.1038/s41586-020-2776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz S. R. A.; Höll G.; Schuller P.; Ertl P.; Rothbauer M. Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems. Biosensors 2019, 9 (3), 110. 10.3390/bios9030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.