Abstract
Ethnopharmacological relevance
Acute lung injury (ALI) is a common manifestation of COVID-19. Xuanfei Baidu Formula(XFBD) is used in China to treat mild or common damp-toxin obstructive pulmonary syndrome in COVID-19 patients. However, the active ingredients of XFBD have not been extensively studied, and its mechanism of action in the treatment of ALI is not well understood.
Aim of the study
The purpose of this study was to investigate the mechanism of action of XFBD in treating ALI in rats, by evaluating its active components.
Materials and methods
Firstly, the chemical composition of XFBD was identified using ultra-high performance liquid chromatography with quadrupole time-of-flight mass spectrometry. The potential targets of XFBD for ALI treatment were predicted using network pharmacological analysis. Finally, the molecular mechanism of XFBD was validated using a RAW264.7 cell inflammation model and a mouse ALI model.
Results
A total of 113 compounds were identified in XFBD. Network pharmacology revealed 34 hub targets between the 113 compounds and ALI. The results of Kyoto Encyclopedia of Genes and Genomes and gene ontology analyses indicated that the NF-κB signaling pathway was the main pathway for XFBD in the treatment of ALI. We found that XFBD reduced proinflammatory factor levels in LPS-induced cellular models. By examining the lung wet/dry weight ratio and pathological sections in vivo, XFBD was found that XFBD could alleviate ALI. Immunohistochemistry results showed that XFBD inhibited ALI-induced increases in p-IKK, p–NF–κB p65, and iNOS proteins. In vitro experiments demonstrated that XFBD inhibited LPS-induced activation of the NF-κB pathway.
Conclusion
This study identified the potential practical components of XFBD, combined with network pharmacology and experimental validation to demonstrate that XFBD can alleviate lung injury caused by ALI by inhibiting the NF-κB signaling pathway.
Keywords: Xuanfei baidu formula, Acute lung injury, Component analysis, Network pharmacology, NF-κB signaling pathway
Graphical abstract
1. Introduction
Acute lung injury (ALI) is mainly characterized by acute, progressive respiratory failure and hypoxemia and is clinically typical in thoracic surgery diseases (Yin et al., 2021). Adult respiratory distress syndrome is characterized by a cascade of pro-inflammatory mediators due to immune system overactivation and is one of the most severe symptoms of ALI (Liu et al., 2021b). Because most researchers believe that inflammatory response plays a central role in ALI, treatment approaches are still focused on effective controlling the degree of the inflammatory response. Research on ALI pathogenesis has mainly focused on the nuclear factor-κB (NF-κB) inflammatory pathway (Ye et al., 2021; Liu et al., 2021). In clinical practice, commonly used anti-inflammatory drugs for treating ALI, such as glucocorticoids, cause symptoms such as skin atrophy, decreased bone density, and gastrointestinal discomfort. Although strategies for treating ALI are constantly improving, there remains a lack of specific drugs, and poor prognosis remains a significant challenge to supporting patients’ health (Reichardt et al., 2021). Therefore, there is an urgent need to develop specific drugs for ALI treatment. At present, research on traditional Chinese medicine (TCM) has emerged as a novel area of research for developing ALI treatments, and the anti-inflammatory mechanism and influence on the immune function of various TCM components are continuously being explored.
Xuanfei Baidu (XFBD) is a prescription recommended by the State Administration of Traditional Chinese Medicine for the clinical treatment of COVID-19 (Zhao et al., 2021). This prescription is a traditional Chinese medicine formula proposed by Academician Zhang Boli and Professor Liu Qingquan on the anti-epidemic front line. It is composed of 13 TCM compounds, listed as follows with their materials of origin: Semen Armeniacae Amarum from Prunus sibirica L., Verbenae Herba from Verbena officinalis L., Ephedrae Herba from Ephedra sinica Stapf, Atractylodis Rhizoma from Atractylodes chinensis (DC.) Koidz, Glycyrrhizae Radix et Rhizoma from Glycyrrhiza uralensis Fisch., Descurainiae Semen from Descurainia sophia (L.) Webb. ex Prantl., Polygoni Cuspidati Rhizoma et Radix from Polygonum cuspidatum Siebold & Zucc., Phragmitis Rhizoma from Phragmites communis Trin., Coicis Semen from Coix lacryma-jobi L. var mayuen (Roman.) Stapf, Citri Grandis Exocarpium from Citrus grandis (L.) Osbeck, Artemisia Annua Herba from Artemisia annua L., Pogostemonis Herba from Pogostemon cablin (Blanco) Benth., and Gypsum Fibrosum. Clinical trials have demonstrated that XFBD can restore white blood cell and lymphocytes levels to normal (Wu et al., 2020). However, there are few studies on the active components of XFBD, and the mechanism of action of XFBD in treating ALI is unclear.
The lung is a critical organ for gas exchange, and airborne pathogens, allergens, and other toxins increase susceptibility ALI during lung infection or inflammation. The ALI response to severe pulmonary microbial infection is the result of the immunological recognition of pathogens responsible for inducing a pro-inflammatory immune response (Rezoagli et al., 2017). ALI can cause signigicant tissue damage, and in severe cases, irreversible lung damage can lead to death. Macrophages play a crucial role in the pathogenesis of bacterial pneumonia and associated ALI(Wu et al., 2021). For example, in Gram-negative bacterial pneumonia, macrophages produce TNF-α, which induces granulocyte-macrophage colony-stimulating factor (GM-CSF) in AECs, causing proliferative signals in AECs through autocrine stimulation, thereby contributing to restoration of the alveolar epithelial barrier(Kumar, 2020). In this study, we used lipopolysaccharide (LPS; a unique component in the cell wall of gram-negative bacteria) to stimulate mouse mononuclear macrophage leukemia cells (RAW264.7) as a cell experimental model to verify that XFBD alleviates the acute mechanism of action in lung injury.
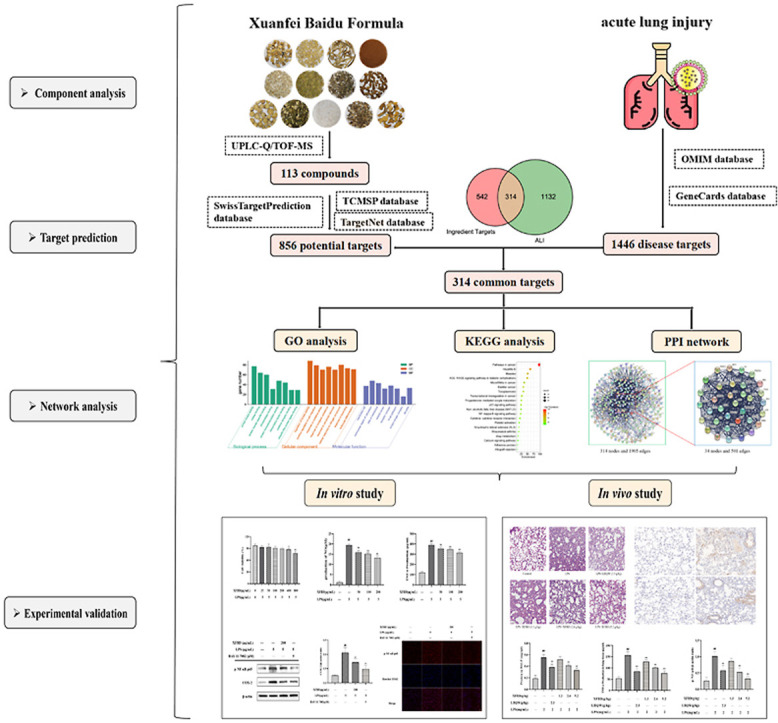
In this study, we analyzed the active components of XFBD, predicted and verified the targets of XFBD according to its active ingredients, and elucidated its mechanism of action in treating ALI. A graphical summary of this study is provided.
2. Materials and methods
2.1. Reagents and materials
XFBD powder was provided by Tianjin Modern TCM Innovation Center (Batch No: XF210204, HPLC detection: 1.21 mg/g for ephedrine and pseudoephedrine, 25.30 mg/g for naringin, and 1.83 mg/g for glycyrrhizic acid in XFBD powder). The production method is to soak the 13 TCM samples (Lepidium apetalum Willd bag decoction) in the prescription in water (1:4 ratio) for 30 min, heat the mixture to boiling, maintain the mixture overheat for 40 min, filter the decoction, and concentrate the filtrate to a relative density of 1.02–1.10 (60 °C), spray dried. Lianhua Qingwen Granules (LHQW) was purchased from Beijing Yiling Pharmaceutical Co., Ltd (Beijing, China). MS-grade methanol and acetonitrile were obtained from Thermo Fisher Scientific Co., Ltd (Shanghai, China). LC-MS/HPLC-grade formic acid was obtained from Anaqua Chemicals Supply (Wilmington, USA). Purified water was purchased from Watsons Co., Ltd (Guangdong, China). A CCK-8 Cell Proliferation Kit and BCA Protein Assay Kit were purchased from Beijing Solarbio Science & Technology Co., Ltd (Beijing, China). LPS was purchased from Sigma-Aldrich trading Co., Ltd (MO, USA). Dulbecco's modified Eagle medium (DMEM) was purchased from Meilunbio (Dalian, China). IL-6, TNF-α, IL-1β, IL-10, monocyte chemotactic protein 1 (MCP-1), and fetal bovine serum (FBS) were purchased from ExCell Biotech Co., Ltd (Shanghai, China). The NO assay kit was purchased from Beyotime Biote Co., Ltd (Shanghai, China). Myeloperoxidase (MPO) and cyclooxygenase-2 (COX-2) were purchased from Shanghaimeilian Biotech Co., Ltd (Shanghai, China). BAY 11–7082 was purchased from Selleck. CN (Shanghai, China). All antibodies used in this study were purchased from Affinity Biosciences LTD (Jiangsu, China).
The following 37 standard compounds were used as reference standards (purity≥98%). Isoquercitrin, ephedrine, catechin, caffeic acid, vanillic acid, pseudoephedrine, vitexin, liquiritin, hesperidin, atractylenolide II, scopoletin, wogonin, amygdalin, adenosine, isochlorogenic acid C, polydatin, naringin, rhoifolin, atractylenolide III, hastatoside, physcion, verbenalin and glycyrrhetinic acid were purchased from China National Institute for Food and Drug Control (Beijing, China). Glycyrrhizic acid, scopolin, verbascoside, ferulic acid, ononin, apigenin-7-O-diglucuronide, vicenin-2, cryptochlorogenic acid, torachrysone 8-O-glucoside, liquiritin apioside, emodin and apigenin-7-O-glucoside were purchased from Shanghai Shidande Standard Technical Service Co., Ltd (Shanghai, China). Kaempferol 3-O-β-D-glucoside and naringenin were purchased from Chengdu Aifa Biological Technology Co., Ltd (Chengdu, China).
2.2. Ultra-high performance liquid chromatography with quadrupole time-of-flight mass spectrometry (UPLC-Q/TOF-MS)
The UPLC-Q/TOF-MS method employed for analyzing XFBD was the same as that used in our previous study (Wang et al., 2022).
2.3. Gene networks analysis
The identified compounds were imported into the traditional Chinese medicine ayatems pharmacology database and analysis platform (TCMSP; https://old.tcmsp-e.com/tcmsp.php), TargetNet (http://targetnet.scbdd.com), and SwissTargetPrediction database (http://www. swisstargetprediction.ch/) to predict the active ingredient targets. The keyword “acute lung injury” was searched in the Gene Cards (https://www.genecards.org/, date of visit: December 24, 2021) and OMIM (http://omim.org/) databases to summarize disease targets. Overlap targets were screened by comparing ALI-associated targets with active compound targets to indentify potential targets for ALI treatmet. Kyoto Encyclopedia of Genes and Genomics (KEGG) pathway enrichment analysis and gene ontology (GO) analysis in the Metascape database (https://metascape.org) were performed to investigate gene function. GO functionally annotated key genes as molecular functions (MFs), cellular components (CCs) and biological processes (BPs). Cytoscape 3.7.0 software (http://www.cytoscape.org/) and the String website (http://www.string-db.org/) were used to further analyze the overlapping targets and obtain hub targets (Wang et al., 2021). The lowest interaction score in the protein-protein interaction (PPI) network is 0.900. According to previous research, nodes with a degree more than twice the median degree of all nodes were selected as hub nodes (Chen et al., 2021, Chen et al., 2021).
2.4. Cell experiments
2.4.1. Cell culture and treatment
RAW264.7 cells were obtained from the American Type Culture Collection (ATCC, Manassas, USA) and grown in DMEM containing 10% FBS in a humidified incubator at 37 °C and 5% CO2.
2.4.2. CCK-8 assay
Cells were seeded at a density of 1 × 104 cells/well in 96-well plates for 24 h. XFBD was dissolved in the medium, and 100 μL of medium was added at different concentrations (0, 25, 50, 100, 200, 400, 800 μg/mL) to culture cells for 24 h. After administration, 10 μL of the CCK-8 solution was added to each well and incubated for 2 h. The 96-well plate was removed, and the absorbance was measure at 450 nm.
2.4.3. Detection of inflammatory cytokines
Cells were seeded in 96-well plates at a density of 1 × 104 cells/well for 24 h. The cells were divided into control, LPS, and administration groups (50, 100, and 200 μg/mL). The model group was cultured for 24 h in medium containing 5 μg/mL LPS, and the administration group was cultured for 24 h in the medium containing 5 μg/mL LPS and the corresponding concentration of XFBD, and the cytokines were measured after the culture. NO, IL-10, MPO, TNF-α, COX-2, IL-6, MCP-1, and IL-1β levels were observed according to the recommended instruction. Absorbance was measured using by an automatic microplate reader.
2.4.4. Immunofluorescence assay
For immunofluorescence analysis, cells were fixed, permeabilized, and blocked with 5% bovine serum albumin for 1 h. The cells were soaked in a 4 °C refrigerator with a p–NF–κB p65 (1:400) antibody for 12 h. Sections were washed 3 times with PBS to remove residual antibody, and then labelled with anti-rabbit IgG (H + L) (Alexa Fluo 555) for 2 h at room temperature. The cleaning procedure was repeated, and the cells were stained with Hoechst 33342 for 10 min. After repeating the cleaning, images were acquired by Operetta CLS (PerkinElmer, USA) (Li et al., 2020a, Li et al., 2020b).
2.4.5. Western blot analysis
Cells were seeded in 96-well plates at a density of 1 × 104 cells/well for 24 h. The cells were divided into control, LPS, and treatment groups (50, 100, and 200 μg/mL). The model group was cultured for 24 h with the medium containing 5 μg/mL LPS, and the administration group was cultured with the medium containing 5 μg/mL LPS and the corresponding concentration of XFBD for 24 h. The cell proteins were then extracted for western blotting experiments. Western blotting was performed as previously described (Li et al., 2020a, Li et al., 2020b). The following primary antibodies were used in the RAW264.7 cell experiments: Phospho–NF–κB-p65 (1:1000 dilution), NF-κB-p65 (1:1000 dilution), COX-2 (1:1000 dilution), and β-actin protein (1:10000 dilution).
2.5. Animal experiment
2.5.1. Model design
Male C57BL/6 mice (n = 72, body weight: 18–22 g, six weeks old) were obtained from SPF Biotechnology Co., Ltd. (license number: SCXK (Tianjin) 2019-0002). All experimental procedures were approved by the Animal Care and Use Committee of the Tianjin University of Traditional Chinese Medicine (authorization number: TCM-LAEC2021186). All mice were kept at 22–26 °C and 55 ± 5% humidity with a light/dark cycle for 12 h, acclimated for 7 days before the experiment, and given free access to food and water.
The 72 mice were randomly divided into control, LPS (2 mg/mL), LPS + LHQW (2.3 g/kg), and LPS + XFBD (1.3, 2.6, and 5.2 g/kg) groups. Different groups of mice were administered XFBD by gavage according to the experimental design. On the 8th day, 1 h after gavage, LPS solution (0.5 μL/g) was injected into the nasal cavity. Mice were anesthetized 6 h after instillation, and lung tissue, serum and bronchoalveolar lavage fluid (BALF) were collected. LPS, LHQW and XFBD administrations were formulated using deionized water.
2.5.2. Lung wet/dry weight (W/D) ratio
The surface layer of the upper lobe of the left lung was wiped dry and then weighed. Subsequently, the lung tissue was dried in an oven at 80 °C for 3 days and weighed again. The W/D ratios were recorded for all groups.
2.5.3. Protein concentration determination in BALF
After death, the chest of each mouse was opened, and the trachea was bluntly separated and exposed. A small “inverted V″ cut was made in the trachea with surgical scissors, the catheter was carefully inserted through the incision, and lavage was performed with a 1 mL syringe. The lungs were lavaged 3 times with 500 μL of sterile PBS (An et al., 2021). The three lavages were pooled and centrifuged at 3000 rpm for 10 min at 4 °C, and the supernatant was used for BCA determination of total protein concentration.
2.5.4. Inflammatory cytokines detections
Tissues were rinsed with pre-chilled PBS, the blood removed, and the tissues were dried with filter paper. Then, 1 g of tissue were weighed and homogenized by adding 1 ml of PBS. The prepared tissue homogenate was centrifuged at 4 °C and 10,000 rpm for 10 min. The sediment was discarded, and the supernatant was retained. NO, IL-10, MPO, TNF-α, COX-2, IL-6, MCP-1, and IL-1β levels were observed according to the recommended instructions. Absorbance value was measured using by an automatic microplate reader.
2.5.5. Histopathology evaluation
Lung tissue was soaked in 4% paraformaldehyde for 24 h. The tissue was dehydrated, embedded and sectioned (5 μm). The tissue sections were stained using the Hematoxylin and eosin staining (H&E) method (Ying et al., 2013). The structure of the lung tissues was photographed using a microscope (Leica, GER).
2.5.6. Immunohistochemistry assay
Immunohistochemical studies were performed to detect p-IKK, p–NF–κB p65, and iNOS expression. The lung tissue sections were deparaffinized, rehydrated, and incubated in 10 mM citric acid buffer at 120 °C for 5 min. Then, 10% goat serum was sectioned at room temperature for 1 h and incubated with antibodies p-IKK, p–NF–κB p65, and iNOS (1:200 dilution) at 4 °C overnight. Finally, the sections were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (1:1000 dilution) was sectioned at room temperature for 1 h and photographed.
2.6. Statistical analysis
Statistical analysis was performed using the GraphPad Prism software (version 6.07, San Diego, CA, USA). Statistical analysis was performed using one-way analysis of variance (ANOVA) with post hoc Tukey's test. Data are shown as the mean ± standard deviation. Statistical significance was set at P < 0.05.
3. Results
3.1. Compound identification
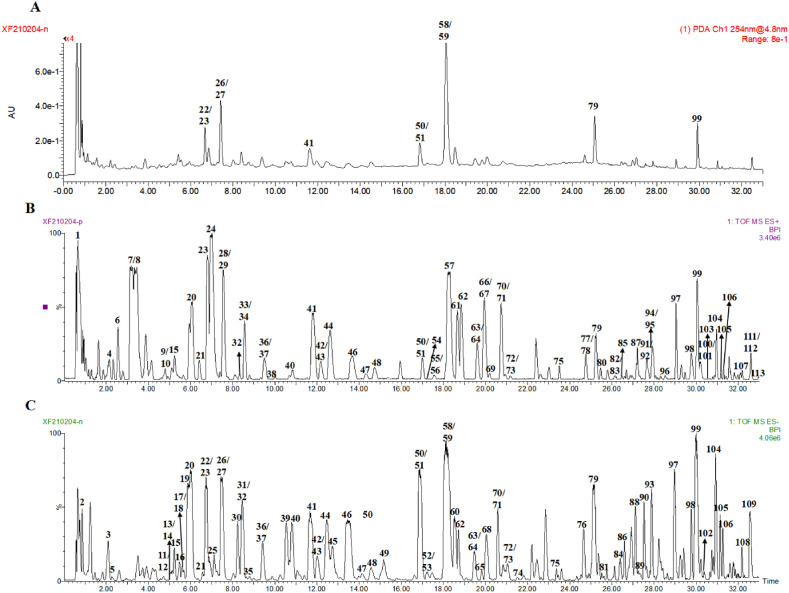
In total, 113 compounds were analyzed and identified from the XFBD by UPLC-Q/TOF-MS, including 48 flavonoids, 6 alkaloids, 4 triterpenes, 5 anthraquinones, 5 iridoid glycosides, 5 organic acids, 7 sesquiterpene glycosides, 6 coumarins, 11 phenylpropanoids class, 4 phenols, 9 glycosides, and 3 other compounds (Sun et al., 2021; Xu et al., 2021; Zhang et al., 2021; Gao et al., 2021; Ming et al., 2021; Dong et al., 2021). The results are presented in Fig. 1 and Table 1 . Of these, 37 were verified using reference materials.
Fig. 1.
Ultra-high performance liquid chromatography with quadrupole time-of-flight mass spectrometry (UPLC-Q/TOF-MS) analysis of Xuanfei Baidu Formula (XFBD). (A) Chromatogram of XFBD in 254 nm mode. (B) Base peak intensity chromatograms of XFBD in positive and (C) negative ESI mode.
Table 1.
Chemical constituents identified in XFBD based on UPLC-Q/TOF-MS.
| NO. | RT (min) | Identity | Molecular Formula |
Observed MS1 (m/z) | MS2 (m/z) | type | Source |
|---|---|---|---|---|---|---|---|
| 1 | 0.65 | Arginine | C6H14N4O2 | 175.1190 [M+H]+ | 158.0923, 116.0715 | Miscellaneous | — |
| 2 | 0.82 | Maltose/Sucrose | C12H22O11 | 341.1009 [M-H]- | 221.0647, 179.0517, 161.0440 | Miscellaneous | — |
| 3 | 2.09 | Amygdalin amide | C20H29NO12 | 520.1565 [M + HCOO]- | 474.1499, 312.1021 | Miscellaneous | C |
| 4 | 2.12 | Adenosinea | C10H13N5O4 | 268.1049 [M+H]+ | 136.0620, 119.0362 | Glycosides | L |
| 5 | 2.25 | Protocatechuic acid | C7H6O4 | 153.0158 [M-H]- | 109.0270 | Organic acid | A/E/H/J |
| 6 | 2.77 | Norephedrine/Norpseudoephedrine | C9H13NO | 152.1072 [M+H]+ | 134.0974, 117.0691, 115.0539 | Alkaloids | A |
| 7 | 3.47 | Ephedrinea | C10H15NO | 166.1243 [M+H]+ | 149.1159, 133.0873, 117.0669 | Alkaloids | A |
| 8 | 3.61 | Pseudoephedrinea | C10H15NO | 166.1243 [M+H]+ | 149.1159, 133.0874, 117.0669 | Alkaloids | A |
| 9 | 4.67 | Atractyloside A | C21H36O10 | 471.2218 [M+Na]+ | 448.2250, 285.1708 | Sesquiterpene glycosides | G |
| 10 | 4.71 | Methylephedrine | C11H17NO | 180.1368 [M+H]+ | 162.1280, 117.0711, 115.0551 | Alkaloids | A |
| 11 | 4.73 | Vanillic acida | C8H8O4 | 167.0343 [M-H]- | 152.0106, 123.0440 | Organic acid | A/D |
| 12 | 4.75 | Scopolina | C16H18O9 | 353.0875 [M-H]- | 191.0548 | Coumarins | C |
| 13 | 4.97 | 3,4-Dihydroharpagide | C15H26O10 | 365.1447 [M-H]- | 221.1020, 161.0441 | Glycosides | — |
| 14 | 5.05 | Catechina | C15H14O6 | 289.0716 [M-H]- | 254.0820, 123.0450, 109.0292 | Phenols | A/D/E |
| 15 | 5.07 | Chlorogenic acid | C16H18O9 | 353.0867 [M-H]- | 191.0550, 173.0460 | Phenylpropanoids | A/D/E/H |
| 16 | 5.54 |
Quercetin 3-O-β-D-glucose -7-O-β-D-gentiobioside |
C33H40O22 | 787.1265 [M-H]- | 625.1518, 463.0920, 301.0360 | Flavonoids | K |
| 17 | 5.60 | Caffeic acida | C9H8O4 | 179.0363 [M-H]- | 135.0474, 117.0360, 107.0528 | Organic acid | L |
| 18 | 5.74 | Cryptochlorogenic acida | C16H18O9 | 353.0859 [M-H]- | 191.0556, 173.0450 | Phenylpropanoids | D |
| 19 | 5.89 | Amygdalina | C20H27NO11 | 456.1451 [M-H]- | 323.0934, 161.0417 | Glycosides | C |
| 20 | 6.05 | Prunasin | C14H17NO6 | 296.1134 [M+H]+ | 340.1045 | Glycosides | C |
| 21 | 6.73 | Hastatosidea | C17H24O11 | 449.1310 [M + HCOO]- | 241.0668, 223.0586, 139.0388 | Iridoid glycosides | I |
| 22 | 6.81 | 3,4-Dihydroverbenalin | C17H26O11 | 435.1512 [M-H + FA]- | 227.0918, 179.0558, 119.0352 | Iridoid glycosides | I |
| 23 | 6.81 | Ferulic acida | C10H10O4 | 193.0483 [M-H]- | 178.0246, 134.0355 | Phenylpropanoids | A/D/E/L |
| 24 | 6.97 | Sinapine | C16H24NO5+ | 310.1669 [M+H]+ | 251.0922, 207.0653, 175.0405, 147.0439 | Alkaloids | K |
| 25 | 7.36 | 1-β-D-glucopyranosyloxy-3,5-dihydroxybenzene | C12H16O8 | 287.0767 [M-H]- | 149.0238, 125.0248, 83.0151 | Glycosides | — |
| 26 | 7.46 | Verbenalina | C17H24O10 | 433.1366 [M + HCOO]- | 387.1633, 225.0755, 193.0483 | Iridoid glycosides | I |
| 27 | 7.47 | Swertiamarin | C16H22O10 | 433.1350 [M + HCOO]- | 101.0299 | Iridoid glycosides | I |
| 28 | 7.49 |
Isorhamnetin 3-O-β-D-glucose-7-O-β-D -gentiobioside |
C34H42O22 | 803.2251 [M+H]+ | 641.1687, 479.1253, | Flavonoids | K |
| 29 | 7.55 | Gentiopicroside | C16H20O9 | 357.1198 [M+H]+ | 195.0670, 177.0553, 149.0610 | Iridoid glycosides | I |
| 30 | 8.23 | Schaftoside | C26H28O14 | 563.1545 [M-H]- | 473.1130, 443.1016, 383.0792 | Flavonoids | B |
| 31 | 8.34 | 5-O-Feruloylquinic acid | C17H20O9 | 367.0988 [M-H]- | 191.0534, 173.0420, 134.0333 | Phenylpropanoids | A/D |
| 32 | 8.44 | Vicenin-2a | C27H30O15 | 593.1474 [M-H]- | 503.1119, 473.1027, 383.0724, 353.0642 | Flavonoids | B/J |
| 33 | 8.52 | Isoferulic acid | C10H10O9 | 195.0654 [M+H]+ | 177.0527, 145.0257, 117.0317 | Phenylpropanoids | A/D/E/L |
| 34 | 8.57 | Nicotiflorin | C27H30O15 | 595.1654 [M+H]+ | 449.1094, 287.0565 | Flavonoids | B |
| 35 | 8.63 | 3-O-Feruloylquinic acid | C17H20O9 | 367.0949 [M-H]- | 193.0455, 173.0420, 134.0333 | Phenylpropanoids | A/D |
| 36 | 9.53 | Luteolin-7-O-diglucuronide | C27H26O18 | 639.1160 [M+H]+ | 463.0872, 287.0565 | Flavonoids | I |
| 37 | 9.53 | Scopoletina | C10H8O4 | 193.0485 [M+H]+ | 178.0261, 137.0594, 133.0283, 122.0366 | Coumarins | F |
| 38 | 9.87 | Sinapinic acid | C11H12O5 | 225.0769 [M+H]+ | 207.0662, 179.0713, 147.0452 | Organic acid | K |
| 39 | 10.58 | Neochlorogenic acid | C16H18O9 | 353.0896 [M-H]- | 191.0559, 179.0349, 135.0453 | Phenylpropanoids | H/C |
| 40 | 10.67 | Quercetin 3-O-β-D-glucuronopyranoside | C21H18O13 | 479.0777 [M+H]+ | 303.0501 | Flavonoids | E |
| 41 | 11.69 | Polydatina | C20H22O8 | 389.1200 [M-H]- | 227.0706, 143.0489 | Phenols | E |
| 42 | 12.00 | kaempferol 3-O-β-D-glucosidea | C21H20O11 | 447.0924 [M-H]- | 207.0768 | Flavonoids | A |
| 43 | 12.03 | Apigenin-7-O-diglucuronide | C27H26O17 | 621.1031 [M-H]- | 351.0508, 269.0403, 193.0313 | Flavonoids | I |
| 44 | 12.50 | Liquiritina | C21H22O9 | 417.1143 [M-H]- | 255.0627, 119.0487, 135.0071 | Flavonoids | B |
| 45 | 12.67 | Isoliquiritigenin | C15H12O4 | 257.0828 [M+H]+ | 239.0719, 137.0248 | Flavonoids | B |
| 46 | 13.66 | Liquiritin apiosidea | C26H30O13 | 573.1596 [M+Na]+ | 257.0815, 137.0234, 119.0481 | Flavonoids | B |
| 47 | 14.36 | Luteolin 7-O-glucuronide | C21H18O12 | 463.0872 [M+H]+ | 287.0565 | Flavonoids | I |
| 48 | 14.74 | Isoquercitrina | C21H20O12 | 463.0887 [M-H]- | 301.0354, 245.0450 | Flavonoids | I/K |
| 49 | 15.38 | Patuletin 3-O-glucoside | C22H22O13 | 495.1142 [M+H]+ | 435.0918, 303.0501 | Flavonoids | F |
| 50 | 16.88 | Verbascosidea | C29H36O15 | 623.2024 [M-H]- | 461.1620, 161.0235 | Glycosides | I/H |
| 51 | 16.99 | Galloyl piceid | C27H26O12 | 541.1277 [M-H]- | 313.0542, 227.0676 | Phenols | E |
| 52 | 17.20 | Isochlorogenic acid B | C25H24O12 | 515.1218 [M-H]- | 353.0904, 335.0793, 191.0564, 179.0353 | Phenylpropanoids | D |
| 53 | 17.22 | 3,5-O-dicaffeoylquinic acid | C25H24O12 | 515.1208 [M-H]- | 353.0898, 191.0560 | Phenylpropanoids | D |
| 54 | 17.27 | Neoliquiritin | C21H22O9 | 419.1335 [M+H]+ | 257.0801 | Flavonoids | B |
| 55 | 17.57 | Campneoside i | C30H38O16 | 655.2574 [M+H]+ | 653.2673 | Flavonoids | J |
| 56 | 17.58 | Prunin | C21H22O10 | 435.1295 [M+H]+ | 273.0745, 153.0184 | Flavonoids | B/J |
| 57 | 17.98 | Apigenin-7-O-glucuronidea | C21H18O11 | 447.0933 [M+H]+ | 271.0610, 153.0170 | Flavonoids | I |
| 58 | 18.14 | Isoacteoside | C29H36O15 | 623.1945 [M-H]- | 461.1694, 161.0241 | Glycosides | I |
| 59 | 18.24 | Naringina | C27H32O14 | 579.1768 [M-H]- | 459.1151, 271.0593, 151.0011 | Flavonoids | J |
| 60 | 18.55 | Rhoifolina | C27H30O14 | 577.1580 [M-H]- | 269.0437, 227.0676 | Flavonoids | J |
| 61 | 18.58 | Apigenin-5-rhamnoside | C21H20O9 | 417.1190 [M+H]+ | 271.0610, 177.0547 | Flavonoids | B |
| 62 | 18.65 | Vitexin-2″-O-α-L-rhamno side | C27H30O14 | 579.1700 [M+H]+ | 433.1110 | Flavonoids | A |
| 63 | 19.34 | Hesperidina | C28H34O15 | 609.1854 [M-H]- | 301.0728 | Flavonoids | A |
| 64 | 19.54 | Isochlorogenic acid Ca | C25H24O12 | 515.1212 [M-H]- | 353.0896, 191.0556, 179.0556, 173.0451 | Phenylpropanoids | D |
| 65 | 19.60 | Pedaliin | C22H22O12 | 477.1042 [M-H]- | 315.0479, 299.0551 | Flavonoids | I |
| 66 | 19.94 | Meranzin hydrate | C15H18O5 | 301.1050 [M+H]+ | 296.1484, 261.1108 | Coumarins | J |
| 67 | 20.04 | Vitexina | C21H20O10 | 433.1120 [M+H]+ | 313.0684, 283.0589 | Flavonoids | A |
| 68 | 20.05 | Apigenin-7-O-glucosidea | C21H20O10 | 431.0977 [M-H]- | 269.0437, 240.0410 | Flavonoids | I/H |
| 69 | 20.05 | Kaempferol-3-O-rhamnoside | C21H20O10 | 455.0947 [M+H]+ | 486.0246, 271.0600 | Flavonoids | A |
| 70 | 20.77 | Osthol | C15H16O3 | 245.1174 [M+H]+ | 243.77830 | Coumarins | B |
| 71 | 20.95 | Isoliquiritin apioside | C26H30O13 | 551.1791 [M+H]+ | 419.1334, 257.0815, | Flavonoids | B |
| 72 | 21.03 | Isoliquiritin | C21H22O9 | 417.1185 [M-H]- | 225.0627, 135.0071, 148.0141 | Flavonoids | B |
| 73 | 21.04 | Isoglycyrrhizin | C26H30O13 | 549.1599 [M-H]- | 417.1185, 255.0627, 148.0141, 135.0071 | Flavonoids | B |
| 74 | 21.62 | Physion 8-O-β-D-glucoside | C22H22O10 | 447.1277 [M+H]+ | 285.0744 | Flavonoids | E |
| 75 | 23.81 | Ononina | C22H22O9 | 431.1359 [M+H]+ | 269.0813, 254.0578 | Flavonoids | B |
| 76 | 24.67 | Torachrysone-8-O-glucosidea | C20H24O9 | 407.1334 [M-H]- | 245.0830, 215.0337 | Flavonoids | E |
| 77 | 24.67 | Helveticoside | C29H42O9 | 535.646 [M+H]+ | 557.2698, 475.1924 | Flavonoids | E |
| 78 | 24.92 | Evomonoside | C29H44O8 | 521.3120 [M+H]+ | 568.4443, 354.2845 | Glycosides | K |
| 79 | 25.13 | Emodin 8-O-β-D-glucoside | C21H20O10 | 433.8764 [M+H]+ | 283.0618, 240.0411 | Anthraquinones | E |
| 80 | 25.25 | Apigenin | C15H10O5 | 271.0616 [M+H]+ | 153.0173, 173.0587, 119.0504 | Flavonoids | A |
| 81 | 25.54 | Macranthoin G | C26H26O12 | 529.1360 [M-H]- | 367.1052, 353.0895, 191.0552, 179.0343 | Phenylpropanoids | D |
| 82 | 26.05 | 4′-Hydroxywogonin | C16H12O6 | 301.0708 [M+H]+ | 299.0565 | Flavonoids | B |
| 83 | 26.05 | Diosmetin | C16H12O6 | 299.0535 [M-H]- | 285.0367 | Flavonoids | I |
| 84 | 26.41 | 6″-O-malonylgenistin | C24H22O13 | 517.1008 [M-H]- | 473.1139, 311.0571, 269. 0456 | Glycosides | — |
| 85 | 26.45 | Naringenina | C15H12O5 | 273.0770 [M+H]+ | 153.0184, 119.0505 | Flavonoids | A/J |
| 86 | 26.51 | Physcion 1-glucoside | C22H22O10 | 445.1149 [M-H]- | 283.0614, 240.0419 | Anthraquinones | E |
| 87 | 27.18 | Physciona | C16H12O5 | 285.0765 [M+H]+ | 242.0590, 211.0749 | Anthraquinones | E |
| 88 | 27.25 | Isoarctigenin | C21H24O6 | 371.1509 [M-H]- | 254.0583, 225.0549, 210.0679 | Phenols | C |
| 89 | 27.33 | Magnograndiolide | C15H22O4 | 265.1456 [M-H]- | 221.1541, 203.1433, 151.1119 | Sesquiterpene glycosides | F |
| 90 | 27.63 | Eupatin | C18H16O8 | 359.0775 [M-H]- | 344.0547, 329.0309, 286.0123, 175.0021 | Flavonoids | F |
| 91 | 27.66 | Notopterol | C21H22O5 | 355.1546 [M+H]+ | 337.1470, 229.0501, 215.0343 | Coumarins | B/I |
| 92 | 27.73 | Isomeranzin | C15H16O4 | 261.1137 [M+H]+ | 189.0543, 125.9860 | Coumarins | J |
| 93 | 27.83 | Rosmarinic acid | C18H16O8 | 359.0774 [M-H]- | 197.0458, 179.0355, 161.0254 | Organic acid | A |
| 94 | 27.97 | Glabrolide | C30H44O4 | 469.3323 [M+H]+ | 451. 3203, 433.3126, 405. 3137 | Flavonoids | B |
| 95 | 28.01 | Atractylenolide IIa | C15H20O2 | 233.1530 [M+H]+ | 189.0905, 215.1438 | Sesquiterpene glycosides | G |
| 96 | 28.53 | Wogonina | C16H12O5 | 285.0762 [M+H]+ | 291.2162, 226.1796 | Flavonoids | G |
| 97 | 29.25 | Limonin | C26H30O8 | 471.2021 [M+H]+ | 453.3379, 235.1696 | Triterpenoids | G |
| 98 | 29.93 | Semilicoisoflavone B | C20H16O6 | 351.0884 [M-H]- | 283.0978, 265.0866, 241.0867, 199.0761 | Flavonoids | B |
| 99 | 30.05 | Glycyrrhizic acida | C42H62O16 | 823.4174 [M+H]+ | 647.3793, 471.3462, 453.3400 | Triterpenoids | B |
| 100 | 30.21 | Casticin | C19H18O8 | 375.1081 [M+H]+ | 373.0675, 161.0593 | Flavonoids | H |
| 101 | 30.22 | Nomilinoate A-ring lactone | C28H36O10 | 531.2225 [M+H]+ | 427.2035, 329.2324 | Triterpenoids | J |
| 102 | 30.41 | Arteannuin B | C15H20O3 | 249.1471 [M+H]+ | 128.0608 | Sesquiterpene glycosides | F |
| 103 | 30.58 | Vulgarin | C15H20O4 | 265.1441 [M+H]+ | 219.1381, 201.1276 | Sesquiterpene glycosides | C |
| 104 | 31.09 | Atractylenolide IIIa | C15H20O3 | 249.1488 [M+H]+ | 231.1387, 213.1282, 203.1434 | Sesquiterpene glycosides | G |
| 105 | 31.27 | Atractylone | C15H20O | 217.1575 [M+H]+ | — | Sesquiterpene glycosides | G |
| 106 | 31.57 | Artemitin | C20H20O8 | 389.1244 [M+H]+ | 318.3002, 169.9772 | Flavonoids | H |
| 107 | 32.10 | Glabrone | C20H16O5 | 335.0916 [M-H]- | 293.1776, 134.8929 | Flavonoids | B |
| 108 | 32.29 | Licochalcone C | C21H22O4 | 337.1414 [M-H]- | 305.1181, 229.0857 | Flavonoids | B |
| 109 | 32.56 | Emodina | C15H10O5 | 269.0470 [M-H]- | 241.0540, 225.0540, 197.0578 | Anthraquinones | E |
| 110 | 32.59 | Glycyrrhetinic acida | C30H46O4 | 471.3485 [M+H]+ | 425.3422, 317.2115, 235.1693, 189.1636 | Triterpenoids | B |
| 111 | 32.60 | Retusine | C19H18O7 | 359.2385 [M+H]+ | — | Alkaloids | H |
| 112 | 32.70 | Rhein | C15H8O6 | 283.0254 [M+H]+ | — | Anthraquinones | E |
| 113 | 32.91 | Glabridin | C20H20O4 | 323.1265 [M-H]- | — | Flavonoids | B |
A: Ephedrae Herba, B: Glycyrrhizae Radix et Rhizoma, C: Armeniacae Semen Amarum, D: Coicis Semen, E: Polygoni Cuspidati Rhizoma et Radix, F: Artemisia Annua Herba, G: Atractylodis Rhizoma, H: Pogostemonis Herba, I: Verbenae Herba, J: Citri Grandis Exocarpium, K: Descurainiae Semen Lepidii Semen, L: Phragmitis Rhizoma.
Indicates that the compound has been compared with the reference substance.
3.2. Compound and disease targets predictions
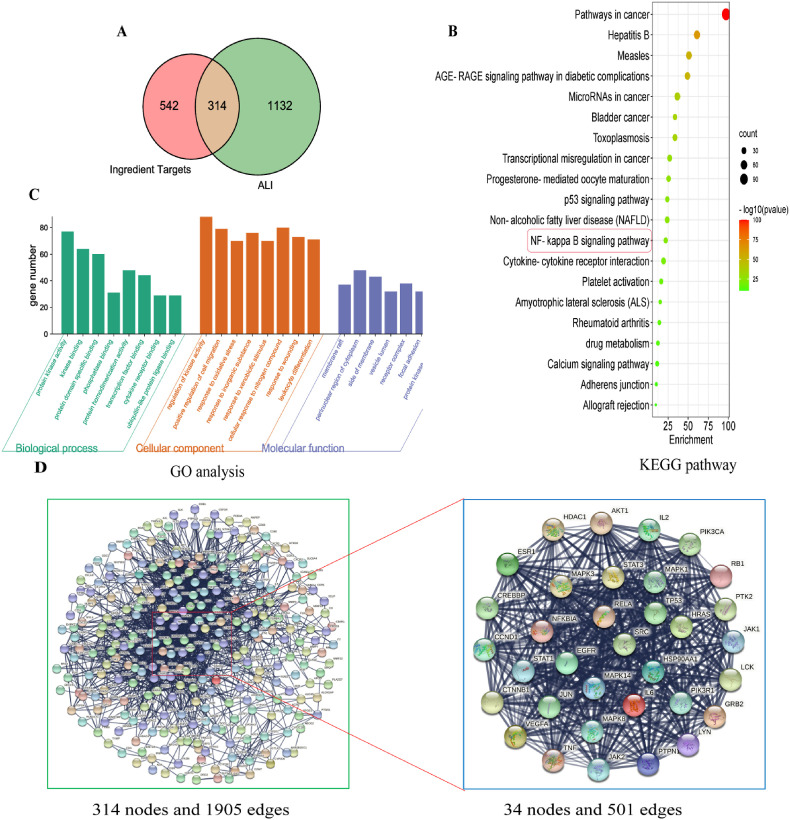
We explored the mechanisms by which XFBD treats ALI by querying several databases. Searching 113 chemical constituents through SwissTargetPrediction and TCMSP databases resulted in 856 candidate compound targets. By searching the OMIM and Gene Card databases, 1446 ALI-associated targets were retrieved. The 314 potential therapeutic targets of XFBD for ALI were identified using Venn diagrams (Fig. 2 A).
Fig. 2.
Network pharmacology analysis of potential XFBD mechanisms in treating Acute lung injury (ALI). (A) Venn diagram showing overlap of ALI genes and predicted compound targets. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. (C) Gene Ontology (GO) analysis of biological processes. (D) Protein-protein interaction (PPI) results and hub targets of compound targets were analyzed by Cytoscape software.
3.3. Pathway enrichment analysis
KEGG analysis showed that 314 genes were enriched in 20 pathways, including disease pathways and biological communication (Fig. 2B). GO analysis revealed that these molecular functions were primarily concentrated on multiple processes, such as protein kinase activity, transcription factor binding, leukocyte differentiation, and cytokine receptor binding (Fig. 2C). The PPI network is interrelated for visualizing the role of diverse essential proteins in diseases, and targets with a higher degree have a more significant impact on central associations (Zou et al., 2020). The 314 targets were screened by twice the median degree, and 34 hub targets were identified: SRC, TP53, PIK3R1, STAT3, MAPK3, HSP90AA1, GRB2, MAPK1, PIK3CA, AKT1, HRAS, RELA, PTPN11, CTNNB1, EGFR, LYN, JAK2, JUN, LCK, CREBBP, VEGFA, ESR1, MAPK8, HDAC1, STAT1, MAPK14, IL6, PTK2, JAK1, IL2, CCND1, RB1, NFKBIA, and TNF (Fig. 2D). KEGG pathway enrichment analysis showed that the NF-κB pathway plays an important role in ALI and the PPI network revealed that the RELA (P65) gene occupies a central position in the composite target. RelA is a subunit of NF-κB, and upon release of NF-κB from an inhibitory interaction with IκB, the dimer containing RelA and cRel translocates to the nucleus, where it activates the transcription of specific NF-κB target genes. The combination of the two suggests that the NF-κB signaling pathway might be an effective way for XFBD action for treat XFBD-induced ALI. Therefore, the NF-κB pathway was selected for subsequent verification.
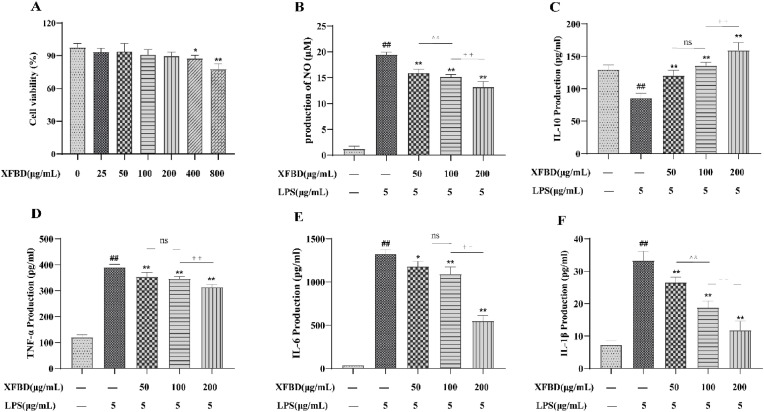
3.4. Effect of XFBD on LPS-stimulated inflammatory cytokines in RAW264.7 cells
The XFBD toxicity test showed that 0–200 μg/mL of XFBD had no significant effect (Fig. 3 A). We examined the effect of XFBD on LPS-stimulated inflammatory cytokines. After LPS stimulation, cellular NO, TNF-α, IL-6, and IL-1β concentrations were significantly increased and IL-10 concentrations were decreased, indicating that cells in the LPS group exhibited severe inflammatory damage. XFBD can modulate these factors to reduce inflammatory damage. Moreover, the therapeutic effect of XFBD was found to be dose-dependent (Fig. 3B–F).
Fig. 3.
Effect of XFBD on lipopolysaccharide (LPS)-stimulated inflammatory cytokines in RAW264.7 cells. (A) XFBD cytotoxicity in RAW264.7 cells was determined by the CCK-8 assay. (B–F) Nitric oxide (NO), interleukin-10 (IL-10), tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), and interleukin-1β (IL-1β) levels in RAW264.7 cells. Data are shown as mean ± standard deviation (SD) by one-way analysis of variance (ANOVA). n = 4. #p < 0.05, ##p < 0.01 vs control group, *p < 0.05, **p < 0.01 vs LPS group, ∧∧p < 0.01 vs administration group (50 μg/mL), ††p < 0.01 vs administration group (100 μg/mL).
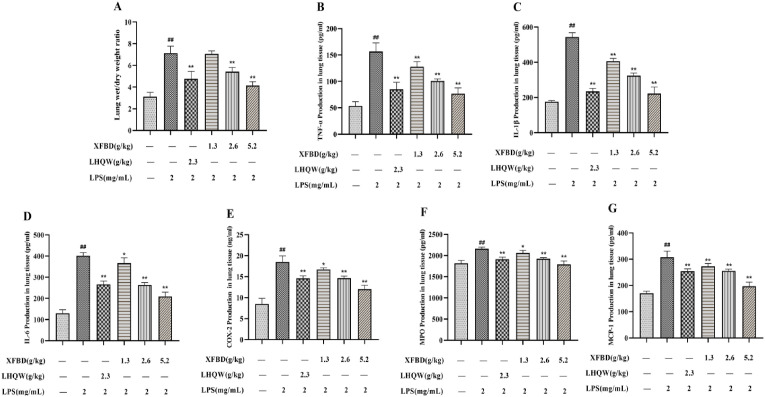
3.5. Effects of XFBD on W/D ratio of lung tissue in ALI model
The lung W/D ratio directly responds to the degree of pulmonary edema. As shown in Fig. 4, Fig. 6 h after modelling, the lung W/D ratio in the model group increased. Treatment with XFBD (2.6 and 5.2 g/kg) and LHQW (2.3 g/kg) effectively reduced tissue edema.
Fig. 4.
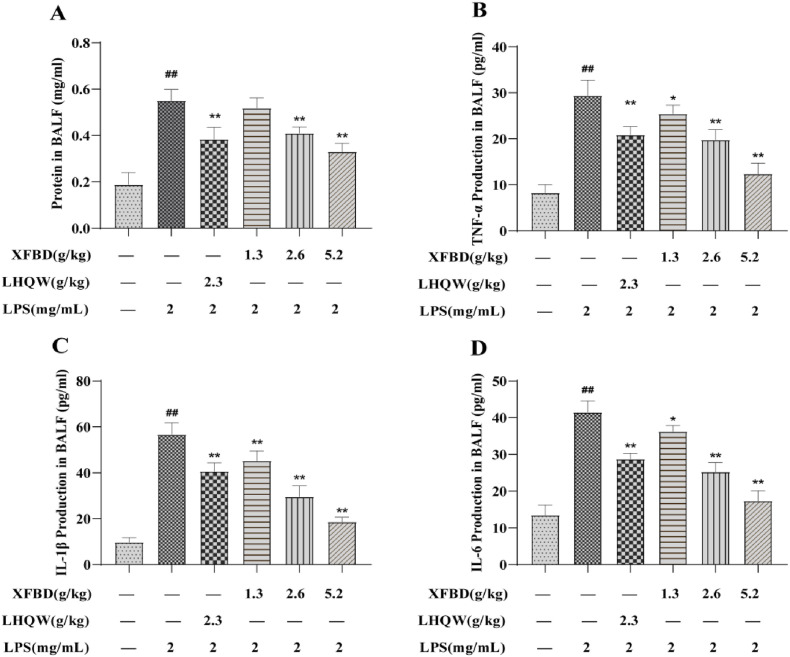
Effects of XFBD on inflammatory response in lung tissue of LPS-induced ALI model mice. (A) Wet/dry ratio. (B–G) TNF-α, IL-1β, IL-6, COX-2, MPO andMCP-1 levels in lung tissue. Data are shown as mean ± SD by ANOVA. n = 5. #p < 0.05, ##p < 0.01 vs control group, *p < 0.05, **p < 0.01 vs LPS group.
Fig. 6.
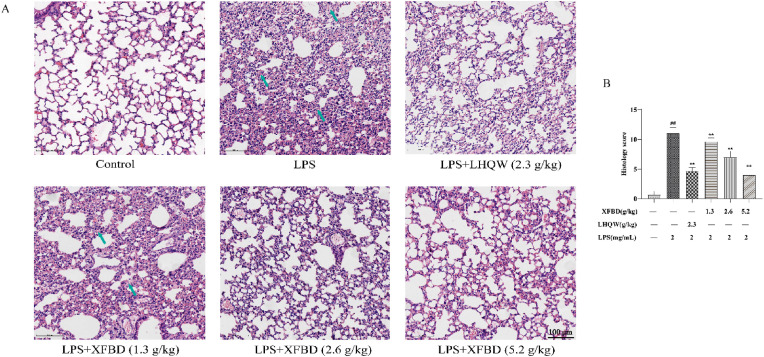
Effects of XFBD on pathological changes to lung tissue in LPS-induced ALI model mice revealed by Hematoxylin and eosin Staining. (A) Serial sections of lung tissues were stained with H&E Scale bar = 100 μm (magnification: x200). (B) Statistical results of lung pathological damage. Data are shown as mean ± SD by ANOVA. n = 3. ##p < 0.01 vs control group, **p < 0.01 vs LPS group.
3.6. Effects of XFBD on pulmonary inflammatory factors in ALI model mice
The levels of TNF-α, MPO, IL-1β, MCP-1, IL-6, and COX-2 were examined to detect the inflammatory response to XFBD in LPS-stimulated mice. The results indicated that TNF-α, MPO, IL-1β, MCP-1, IL-6, and COX-2 levels were notably increased in the model group. However, the increments were suppressed afte XFBD (1.3, 2.6, and 5.2 g/kg) and LHQW (2.3 g/kg) administration (Fig. 4B–G).
We also detected the protein concentration and inflammatory factors in mouse BALF. The results showed that the protein concentration, TNF-α, IL-1β, and IL-6 content (Fig. 5 A–D) in BALF of model group mice increased, while XFBD (1.3, 2.6 and 5.2 g/kg) and LHQW alleviated these increased. These results indicate that XFBD significantly attenuated inflammatory damage in the lungs.
Fig. 5.
Effects of XFBD on inflammatory response in bronchoalveolar lavage fluid (BALF) of LPS-induced ALI model mice. (A) Protein concentrations in BALF. (B–D) TNF-α, IL-1β, IL-6 levels in BALF. Data are shown as mean ± SD by ANOVA. n = 5. #p < 0.05, ##p < 0.01 vs control group, *p < 0.05, **p < 0.01 vs LPS group.
3.7. Effects of XFBD on pathological lung tissue in ALI model mice
We then examined the lung sections. Pathological damage, such as inflammatory cell infiltration, alveolar wall thickening, and alveolar destruction, was observed in the stained areas of the model group. However, treatment with XFBD (2.6 and 5.2 g/kg) and LHQW (2.3 g/kg) alleviated LPS-induced pathology damages (Fig. 6).
3.8. Effects of XFBD on the protein content of p-IKK, p–NF–κB p65, and iNOS protein in ALI model mice
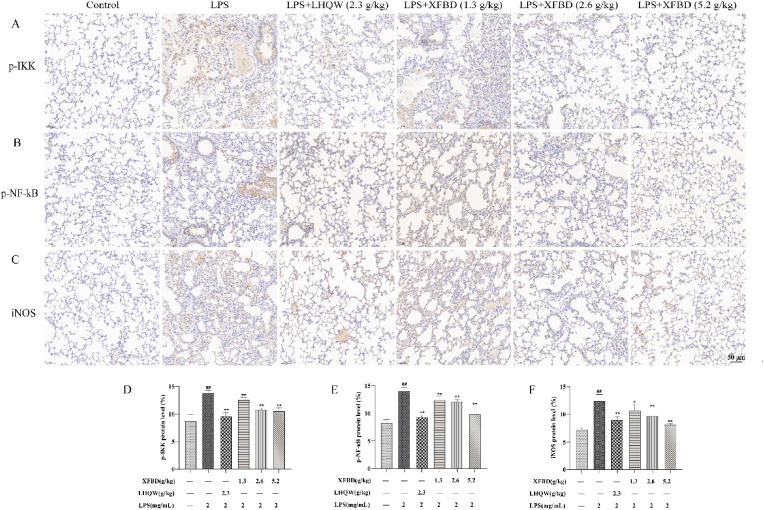
Previous results have indicated that the NF-κB pathway is a crucial pathway for XFBD in ALI treatment. Therefore, we performed immunohistochemistry to investigate the levels of p-IKK, p–NF–κB p65 and iNOS. The results showed that more positive nuclear p-IKK, p–NF–κB p65 and iNOS staining was detected in LPS-induced mouse lung tissue (Fig. 7 ). However, the number of the above-mentioned positive cells decreased by XFBD (2.6 and 5.2 g/kg) and LHQW (2.3 g/kg) administration.
Fig. 7.
Effects of XFBD on the expression of phosphorylated κB inhibits protein kinases (p-IKK), phosphorylated nucleus factor-κB (p–NF–κB) p65, and inducible nitric oxide synthase (iNOS) protein in LPS-induced ALI model mice. (A) Protein expression levels of p-IKK was determined by immunohistochemistry. (B) Protein expression levels of p–NF–κB p65 was determined by immunohistochemistry. (C) Protein expression levels of iNOS was determined by immunohistochemistry. Scale bar = 50 μm (magnification: x200). (D) Results were expressed of p–NF–κB p65. (E) Results were expressed of p–NF–κB p65. (F) Results were expressed of p–NF–κB p65. Data are shown as mean ± SD by ANOVA. n = 3. ##p < 0.01 vs control group, *p < 0.05, **p < 0.01 vs LPS group.
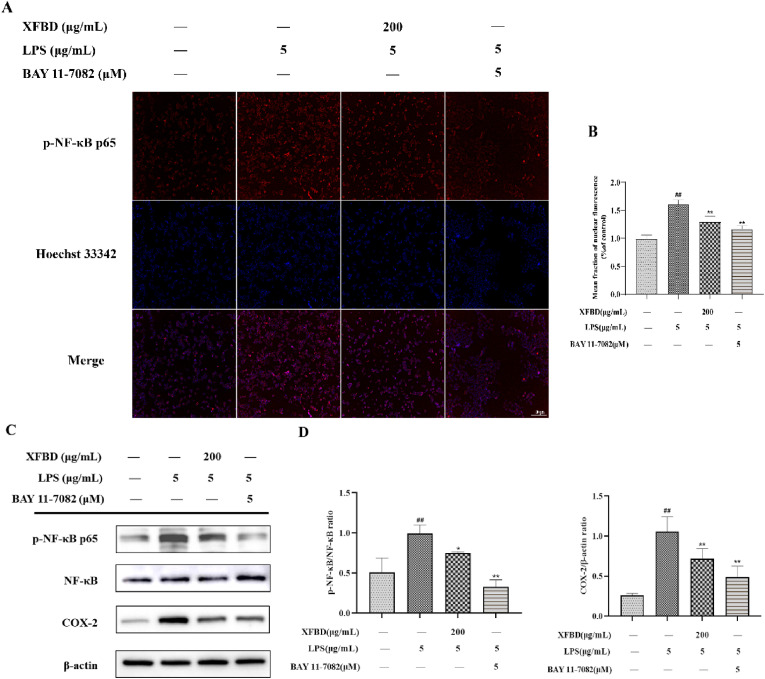
We revalidated the NF-κB pathway at the cellular level. As shown in Fig. 8 , both immunofluorescence and western blotting results indicated that the inhibitory effect of XFBD on the NF-κB pathway was similar to that of BAY 11–7082, and they were both able to inhibit the protein levels of p–NF–κB and COX-2 in the model group. These results confirm that XFBD can alleviate ALI in mice by inhibiting the NF-κB pathway.
Fig. 8.
Effect of XFBD on the expression of p–NF–κB p65 and COX-2 protein in LPS-induced RAW264.7 cells. (A) p–NF–κB p65 protein was determined by immunofluorescence staining. Scale bar = 100 μm. (B) Results were expressed of p–NF–κB p65. (C) Expression levels of p–NF–κB p65, NF-κB p65 and COX-2 were determined by western blotting. (D) Results were expressed as the ratios of p–NF–κB p65/NF-κB p65 and COX-2/β-actin, respectively. Data are shown as mean ± SD by ANOVA. n = 3. #p < 0.05, ##p < 0.01 vs control group, *p < 0.05, **p < 0.01 vs LPS group.
4. Discussion
The complexity of the herbal components that comprise XFBD complicate investigations into its active chemical components and their associated mechanisms of action. UPLC-Q/TOF-MS has the characteristics of high separation efficiency, fast scanning speed, high resolution, and high sensitivity and can rapidly characterize compounds in herbal medicines with accurate molecular mass and MSn multi-level mass spectrometry information (Chang et al., 2021; He et al., 2021). In this study, 113 chemical components of XFBD were analyzed using UPLC-Q/TOF-MS. Among them, there were 23 compounds from Glycyrrhizae Radix et Rhizoma, 20 from Ephedrae Herba, 16 from Verbenae Herba, 16 from Polygoni Cuspidati Rhizoma et Radix, 12 from Coicis Semen, 10 from Citri Grandis Exocarpium, 8 from Pogostemonis Herba, 7 from Armeniacae Semen Amarum, 6 from Descurainiae Semen Lepidii Semen, 5 from Atractylodis Rhizoma, 5 from Artemisia Annua Herba, and 4 from Phragmitis Rhizoma. Some active components of Glycyrrhizae Radix et Rhizoma, Polygoni Cuspidati Rhizoma et Radix, Verbenae Herba, and Ephedrae Herba have been found to exhibit anti-inflammatory properties. For example, ephedrine (7) from Ephedrae Herba contributes to immune homeostasis by balancing the production of inflammatory cytokines in TLR4 signaling (Zheng et al., 2012). Naringin (59) is an active component of Citri Grandis Exocarpium that alleviates pathological lung damage and tissue edema in ALI mice (Liu et al., 2011). Glycyrrhizic acid (99) is the main active ingredient of Glycyrrhizae Radix et Rhizoma, which can reduce lung inflammation by inhibiting the NF-κB protein (Liu et al., 2021). Amygdalin (19), an active component of Armeniacae Semen Amarum can reduce ALI damage in mice by inhibiting NF-κB and NLRP3 inflammatory bodies (Zhang et al., 2017). Treatment with polydatin (41), a primary active component of Polygoni Cuspidati Rhizoma et Radix, can reduce total cell number in BALF and improves cell permeability (Q. Jiang et al., 2015). In particular, polydatin exerts an anti-inflammatory effect on ALI by inhibiting the TLR4/MyD88/NF-κB p65 pathway (Zou et al., 2020). Verbenalin (26), bioactive compound identified in Verbenae Herba, also show notable anti-inflammatory effects in ALI (Yuan et al., 2018). Among the 113 compounds identified, 46 have been reported to possess anti-inflammatory activity.
Network pharmacology predictions were made based on the analysis of 113 components obtained from XFBD. The results revealed that 314 genes were mainly enriched in disease and biological pathway, such as those in cancers, hepatitis, and toxoplasmosis and the NF-κB, AGE-RAGE, and p53 signaling pathways, respectively. GO analysis revealed that the effects of XFBD may involve many biological processes, including protein kinase activity, leukocyte differentiation, cytokine receptor combination, and transcription factor binding, with many of these affecting inflammations. PPI network analysis revealed that RELA, the nuclear transcription factor p65 regulatory gene, has a high degree of value, and controls the expression of NF-κB p65 protein (Wang et al., 2021; X. Jiang et al., 2015). After integrating these results, we identified RELA, NFKBIA, TNF and IL6 as highly relevant targets in inflammation pathways.
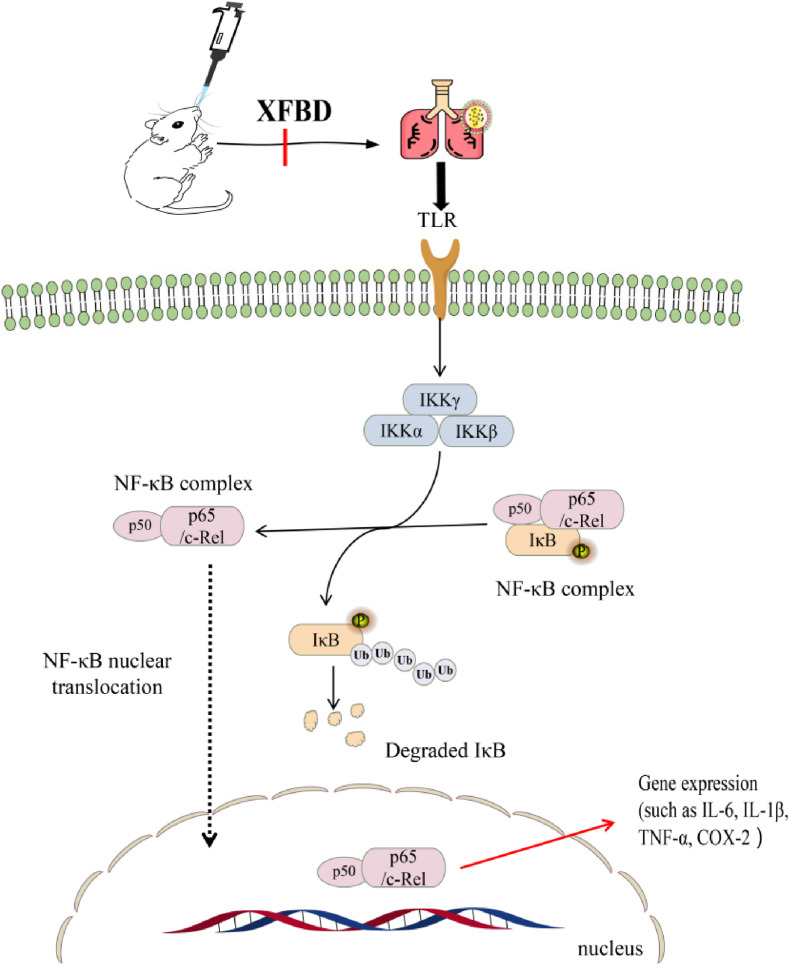
Network pharmacology predicts methods were employed to predict and validate the mechanism of action and relevant pathways associated with XFBD for treating ALI both in vivo and in vitro. XFBD markedly improved lung morphology, inhibited pulmonary edema formation, and prevented inflammatory cells filtration into the lung tissue, which also significantly inhibited MPO, COX-2, and MCP-1 production in the lung tissue induced by LPS and reduced the inflammatory factor content in BALF. In addition, we found that XFBD treatment effectively inhibited the LPS-stimulated activation of p-IKK, p–NF–κB p65, and COX-2. NF-κB is a well-known transcription factor related to inflammation, oxidative stress, and apoptosis. As a family of transcription factor proteins, NF-κB comprises 5 subunits: Rel, p65, RelB, p50, and p52 (Luan et al., 2016). It usually combines with its inhibitor IκB in the form of dimeric subunits (p65 and p50) to form a trimer. When cells are stimulated by inflammation, IκB kinase changes from an inactive state to an active state, catalyzing the phosphorylation of IκB protein and releasing the NF-κB protein (Jing et al., 2015). NF-κB enters the nucleus, thereby initiating downstream IL-6, TNF-α, COX-2, and other inflammatory factors (Zhang et al., 2017) (Fig. 9 ). In our study, we predicted that XFBD could inhibit the NF-κB pathway to alleviate lung injury. We also demonstrated that XFBD can inhibit the activity of κB kinase and reduce the release of NF-κB protein from IκB protein, thereby inhibiting the function of NF-κB protein and reducing inflammatory damage.
Fig. 9.
XFBD alleviates ALI by inhibiting the NF-κB pathway.
Although some of the 113 components analyzed have been confirmed to exhibit pharmacological properties such as anti-inflammatory and anti-fibrosis effects, this study did not independently verify the efficacy of the chemical components of XFBD for treating ALI. In the future, we will verify the efficacy of specific chemical components and explore their mechanisms of action. We will also evaluate the efficacy of pairing of different components to verify efficiency and efficacy of the formulation.
5. Conclusion
In this study, 113 chemical components were identified from XuanfeiBaidu Prescription, and the NF-κB signaling pathway was predicted by network pharmacology analysis of chemical components to be an effective way for XFBD to treat acute lung injury. Animal experiments and cell experiments confirmed that XFBD can reduce the lung injury caused by ALI by inhibiting the NF-κB pathway.
Ethics approval and consent to participate
All experimental procedures were approved by the Animal Care and Use Committee of Tianjin University of Traditional Chinese Medicine (authorization number: TCM-LAEC2021186).
Consent for publication
All authors agree to publish this article.
Availability of data and materials
The data analyzed in this study can be obtained from the corresponding author upon reasonable request.
Funding
The work was supported by the National Key Research and Development Project of China (2020YFA0708000) and (2020YFA0708004), and the National Natural Science Foundation of China (81673693) for financial support.
CRediT authorship contribution statement
Yanru Zhu: Formal analysis, conducted the experiments and analyses, drafted the manuscript, All authors approved the final manuscript. Lifei Luo: Formal analysis, and, Data curation, contributed to formal analysis and data curation, All authors approved the final manuscript. Meng Zhang: Formal analysis, and, Data curation, contributed to formal analysis and data curation, All authors approved the final manuscript. Xinbo Song: provided the experimental platform, All authors approved the final manuscript. Ping Wang: provided the experimental platform, All authors approved the final manuscript. Han Zhang: provided the experimental platform, All authors approved the final manuscript. Jingze Zhang: designed this study, revised the manuscript. All authors approved the final manuscript. Dailin Liu: designed this study, revised the manuscript, All authors approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Editage (www.editage.cn) for English language editing.
Abbreviations
- ALI
Acute lung injury
- BALF
Bronchoalveolar lavage fluid
- COVID-19
Coronavirus disease 2019
- COX-2
cyclooxygenase 2
- FBS
Fetal bovine serum
- GO
Gene Ontology
- H&E
Hematoxylin and eosin Staining
- IL-6
interleukin-6
- IL-1β
interleukin-1β
- IL-10
interleukin-10
- iNOS
inducible nitric oxide synthase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LPS
Lipopolysaccharide
- MCP-1
monocyte chemotactic protein 1
- MPO
myeloperoxidase
- NF-κB
nuclear factor kappaB
- NO
nitric oxide
- PPI
protein-protein interaction
- TCM
traditional Chinese medicine
- TNF-α
tumor necrosis factor-α
- UPLC-Q/TOF-MS
ultra-performance liquid chromatography-tandem quadrupole time-of-flight mass spectrometry
- XFBD
Xuanfei Baidu Formula
Data availability
Data will be made available on request.
References
- An N., Yang T., Zhang X., Xu M. Bergamottin alleviates LPS-induced acute lung injury by inducing SIRT1 and suppressing NF-κB. Innate Immun. 2021;27:543–552. doi: 10.1177/17534259211062553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang G., Bo Y., Cui J., Xu L., Zhao Z., Wang W., Hou J. Main chemical constituents in aerial parts of Glycyrrhiza uralensis by UPLC-Q-Exactive Orbitrap-MS. Zhongguo Zhongyao Zazhi. 2021;46:1449–1459. doi: 10.19540/j.cnki.cjcmm.20201225.301. [DOI] [PubMed] [Google Scholar]
- Chen W., He L., Zhong L., Sun J., Zhang L., Wei D., Wu C. Identification of active compounds and mechanism of huangtu decoction for the treatment of ulcerative colitis by network pharmacology combined with experimental verification. Drug Des. Dev. Ther. 2021;15:4125–4140. doi: 10.2147/DDDT.S328333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Wang X., Ma L., Fang S., Li J., Boadi E., He J., Gao X., Wang Y., Chang Y. The network pharmacology integrated with pharmacokinetics to clarify the pharmacological mechanism of absorbed components from Viticis fructus extract. J. Ethnopharmacol. 2021;278 doi: 10.1016/j.jep.2021.114336. [DOI] [PubMed] [Google Scholar]
- Dong Y., Jia G., Hu J., Liu H., Wu T., Yang S., Li Y., Cai T. Determination of alkaloids and flavonoids in Sophora flavescens by UHPLC-Q/TOF/MS. J Anal Methods Chem. 2021;2021 doi: 10.1155/2021/9915027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Si N., Li M., Gu X., Zhang Y., Zhou Y., Wang H., Wei X., Bian B., Zhao H. The integrated study on the chemical profiling and in vivo course to explore the bioactive constituents and potential targets of Chinese classical formula Qingxin Lianzi Yin Decoction by UHPLC-MS and network pharmacology approaches. J. Ethnopharmacol. 2021;272 doi: 10.1016/j.jep.2021.113917. [DOI] [PubMed] [Google Scholar]
- He L., Jiang H., Lan T., Qiu Y., Yang K., Chen K., Yao X., Yao Z., Lu W. Chemical profile and potential mechanisms of Huo-Tan-Chu-Shi decoction in the treatment of coronary heart disease by UHPLC-Q/TOF-MS in combination with network pharmacology analysis and experimental verification. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2021;1175 doi: 10.1016/j.jchromb.2021.122729. [DOI] [PubMed] [Google Scholar]
- Jiang Q., Yi M., Guo Q., Wang C., Wang H., Meng S., Liu C., Fu Y., Ji H., Chen T. Protective effects of polydatin on lipopolysaccharide-induced acute lung injury through TLR4-MyD88-NF-κB pathway. Int. Immunopharm. 2015;29:370–376. doi: 10.1016/j.intimp.2015.10.027. [DOI] [PubMed] [Google Scholar]
- Jiang X., Lv B., Li P., Ma X., Wang T., Zhou Q., Wang X., Gao X. Bioactivity-integrated UPLC/Q-TOF-MS of Danhong injection to identify NF-κB inhibitors and anti-inflammatory targets based on endothelial cell culture and network pharmacology. J. Ethnopharmacol. 2015;174:270–276. doi: 10.1016/j.jep.2015.08.026. [DOI] [PubMed] [Google Scholar]
- Jing W., Chunhua M., Shumin W. Effects of acteoside on lipopolysaccharide-induced inflammation in acute lung injury via regulation of NF-κB pathway in vivo and in vitro. Toxicol. Appl. Pharmacol. 2015;285:128–135. doi: 10.1016/j.taap.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Kumar V. Pulmonary innate immune response determines the outcome of inflammation during pneumonia and sepsis-associated acute lung injury. Front. Immunol. 2020;11:1722. doi: 10.3389/fimmu.2020.01722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Huang R., Liu K., Li M., Luo H., Cui L., Huang L., Luo L. Fucoxanthin attenuates LPS-induced acute lung injury via inhibition of the TLR4/MyD88 signaling axis. Aging (Albany NY) 2020;13:2655–2667. doi: 10.18632/aging.202309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu R., Zhao Y., Gao N., Jin X., Gao X., Li T., Liu D. The extract from the roots of Rose odorata sweet var. gigantean (Coll. et Hemsl.) Rehd. et Wils attenuates DSS-induced ulcerative colitis by regulating the Nrf2/NF-κB signaling pathways. RSC Adv. 2020;10(16):9450–9461. doi: 10.1039/C9RA10747A. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu Y., Wu H., Nie Y., Chen J., Su W., Li P. Naringin attenuates acute lung injury in LPS-treated mice by inhibiting NF-κB pathway. Int. Immunopharm. 2011;11:1606–1612. doi: 10.1016/j.intimp.2011.05.022. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhou J., Luo Y., Li J., Shang L., Zhou F., Yang S. Honokiol alleviates LPS-induced acute lung injury by inhibiting NLRP3 inflammasome-mediated pyroptosis via Nrf2 activation in vitro and in vivo. Chin. Med. 2021;16:127. doi: 10.1186/s13020-021-00541-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan R., Meng X., Jiang W. Protective effects of apigenin against paraquat-induced acute lung injury in mice. Inflammation. 2016;39:752–758. doi: 10.1007/s10753-015-0302-2. [DOI] [PubMed] [Google Scholar]
- Ming J., Liu W., Wu H., Li Y., Yang E., Wang Z., Xiao H., Quan R., Hu X. The active ingredients and mechanisms of Longchai Jiangxue Formula in treating PV, based on UPLC/Q-TOF-MS/MS, systematic pharmacology, and molecular biology validation. Biomed. Pharmacother. 2021;140 doi: 10.1016/j.biopha.2021.111767. [DOI] [PubMed] [Google Scholar]
- Reichardt S., Amouret A., Muzzi C., Vettorazzi S., Tuckermann J., Lühder F., Reichardt H. The role of glucocorticoids in inflammatory diseases. Cells. 2021;10:2921. doi: 10.3390/cells10112921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezoagli E., Fumagalli R., Bellani G. Definition and epidemiology of acute respiratory distress syndrome. Ann. Transl. Med. 2017;(14):282. doi: 10.21037/atm.2017.06.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Yang H., Wang S., Wang C., Yang M., Gao Z., Xu X., Nie B. Analysis of the chemical components of simiao yongan decoction based on UPLC-LTQ-orbitrap-MS. Zhongguo Zhongyao Zazhi. 2021 doi: 10.19540/j.cnki.cjcmm.20210823.303. [DOI] [PubMed] [Google Scholar]
- Wang X., Quan S., Zhang H., Song X., Zhang J., Liu D. Development and validation of a sensitive UPLC-Q-TOF-MS/MS for the measurement of nine components in rat plasma and tissues and its application to pharmacokinetics and tissue distribution studies with Xuanfei Baidu granules. Curr. Drug Metabol. 2022;23(2):150–163. doi: 10.2174/1389200223666220215151245. [DOI] [PubMed] [Google Scholar]
- Wang Y., Chu F., Lin J., Li Y., Johnson N., Zhang J., Gai C., Su Z., Cheng H., Wang L., Ding X. Erianin, the main active ingredient of Dendrobium chrysotoxum Lindl, inhibits precancerous lesions of gastric cancer (PLGC) through suppression of the HRAS-PI3K-AKT signaling pathway as revealed by network pharmacology and in vitro experimental verification. J. Ethnopharmacol. 2021;279 doi: 10.1016/j.jep.2021.114399. [DOI] [PubMed] [Google Scholar]
- Wu D., Zhang H., Wu Q., Li F., Wang Y., Liu S., Wang J. Sestrin 2 protects against LPS-induced acute lung injury by inducing mitophagy in alveolar macrophages. Life Sci. 2021;267 doi: 10.1016/j.lfs.2020.118941. [DOI] [PubMed] [Google Scholar]
- Wu X., Wang G., Du J., Ai W. Efficacy of herbal medicine (Xuanfei Baidu decoction) combined with conventional drug in treating COVID-19: a pilot randomized clinical trial. Integr Med Res. 2020;9 doi: 10.1016/j.imr.2020.100489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Xu B., Yang J., Liu Y., Zeng Z., Xia L., Li Q., Zou G. Mucin1 relieves acute lung injury by inhibiting inflammation and oxidative stress. Eur. J. Histochem. 2021 doi: 10.4081/ejh.2021.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Zhu G., Wang Q., Fu Y., Wang J., Xu B. Ferroptosis, a new insight into acute lung injury. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.709538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying K., Zhen W., Jing L., Xiu L., Huimin Y. Stevioside protects LPS-induced acute lung injury in mice. Inflammation. 2013;36:242–250. doi: 10.1007/s10753-012-9540-8. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Liao Q., Xue M., Shi Y., Rong L., Song Z., Tong Z., Zheng W., Zhu Q., Cui X., Tao Z. Shufeng jiedu capsules alleviate lipopolysaccharide-induced acute lung inflammatory injury via activation of GPR18 by verbenalin. Cell. Physiol. Biochem. 2018;50:629–639. doi: 10.1159/000494184. [DOI] [PubMed] [Google Scholar]
- Zhang A., Pan W., Lv J., Wu H. Protective effect of amygdalin on LPS-induced acute lung injury by inhibiting NF-κB and NLRP3 signaling pathways. Inflammation. 2017;40:745–751. doi: 10.1007/s10753-017-0518-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhou Y., Gao W., Cui M., Bian B., Ni L., Wang K., Wang H., Si N., Zhao H. 2021. Based on UHPLC-LTQ-Orbitrap-MS/MS Technology Identification of Chemical Composition of Granules and Research on Internet Pharmacology. [DOI] [PubMed] [Google Scholar]
- Zhao J., Guo D., Fan M., Liu Y. Efficacy and safety of Xuanfei Baidu granules for treating COVID-19: a protocol for systematic review and meta-analysis. Medicine (Baltim.) 2021;100 doi: 10.1097/MD.0000000000025653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Guo Z., He W., Yang Y., Li Y., Zheng A., Li P., Zhang Y., Ma J., Wen M., Yang M., An H., Ji G., Yu Y. Ephedrine hydrochloride protects mice from LPS challenge by promoting IL-10 secretion and inhibiting proinflammatory cytokines. Int. Immunopharm. 2012;13:46–53. doi: 10.1016/j.intimp.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Zou M., Yang W., Niu L., Sun Y., Luo R., Wang Y., Peng X. Polydatin attenuates Mycoplasma gallisepticum (HS strain)-induced inflammation injury via inhibiting the TLR6/MyD88/NF-κB pathway. Microb. Pathog. 2020;149 doi: 10.1016/j.micpath.2020.104552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data analyzed in this study can be obtained from the corresponding author upon reasonable request.
Data will be made available on request.