Abstract
Attributes contributing to the current monkeypox virus (MPXV) outbreak remain unknown. It has been established that mutations in viral proteins may alter phenotype and pathogenicity. To assess if mutations in the MPXV DNA replication complex (RC) contribute to the outbreak, we conducted a temporal analysis of available MPXV sequences to identify mutations, generated a DNA replication complex (RC) using structures of related viral and eukaryotic proteins, and structure prediction method AlphaFold. Ten mutations within the RC were identified and mapped onto the RC to infer role of mutations. Two mutations in F8L (RC catalytic subunit), and two in G9R (a processivity factor) were ∼100% prevalent in the 2022 sequences. F8L mutation L108F emerged in 2022, whereas W411L emerged in 2018, and persisted in 2022. L108 is topologically located to enhance DNA binding affinity of F8L. Therefore, mutation L108F can change the fidelity, sensitivity to nucleoside inhibitors, and processivity of F8L. Surface exposed W411L likely affects the binding of regulatory factor(s). G9R mutations S30L and D88 N in G9R emerged in 2022, and may impact the interaction of G9R with E4R (uracil DNA glycosylase). The remaining six mutations that appeared in 2001, reverted to the first (1965 Rotterdam) isolate. Two nucleoside inhibitors brincidofovir and cidofovir have been approved for MPXV treatment. Cidofovir resistance in vaccinia virus is achieved by A314T and A684V mutations. Both A314 and A684 are conserved in MPXV. Therefore, resistance to these drugs in MPXV may arise through similar mechanisms.
Keywords: Monkeypox virus, Mutations, DNA replication complex, F8L, G9R, Family B polymerase
1. Introduction
According to the Centers for Disease Control and Prevention (CDC), the recent Monkeypox virus (MPXV) outbreak has spread to 106 countries (as of September 26, 2022), of which 99 had not previously reported monkeypox (MPX) cases (https://www.cdc.gov/poxvirus/monkeypox/response/2022/world-map.html). A total of 65,415 confirmed cases have been reported as of September 26, 2022 around the world. Due to the rapid spread of MPXV, the FDA has granted emergency use authorization of the Jynneos smallpox vaccine, and drugs tecovirimat, brincidofovir, and cidofovir for MPXV. The Jynneos vaccine, like all smallpox vaccines, is an attenuated vaccinia virus (VACV) [1,2].
MPXV (genus Orthopoxvirus; family Poxviridae) has a double-stranded DNA genome of ∼196,858 base pairs that encodes ∼200 proteins [3], including F8L, which is a family B DNA polymerase (DNA pol). DNA polymerases are critical enzymes for the replication and repair of genomic DNA among various organisms, spanning from archaea to mammals. These enzymes synthesize DNA in template-dependent and -independent manners. DNA polymerases have been classified into seven families: A, B, C, D, X, Y, and RT (reverse transcriptase), based on amino acid conservation and structural homology [[4], [5], [6]]. DNA polymerases that showed sequence homology with E.coli DNA pols I, II, and III were classified into families A, B, and C, respectively. Family D pols are unique to archaea, whereas eukaryotic DNA pols β, λ, μ, and TdT (terminal transferases) belong to family X. DNA pols sharing sequence homology with E.coli pols IV/V belong to family Y. The RT family encompasses DNA pols from retroviruses and eukaryotic telomerase. Regardless of the family, all DNA pols have conserved active site and template-primer (TP) binding modes [7,8]. Family B DNA pols are found in a wide range of organisms (eukaryotes, prokaryotes, bacteriophages, viruses, etc.) [5]. In eukaryotes, at least four DNA pols (α, δ, ε, and ζ) belong to family B. Additionally, viruses such as herpes simplex virus 1 (HSV1), human cytomegalovirus (HCMV), and poxviruses, encode family B DNA pols.
Mutations in DNA pols can alter the phenotype of the virus. For example, a point mutation, N752D, in equine herpes virus 1 (EHV-1) polymerase causes inflammation in the central nervous system and causes poor muscle control leading to gauche movements termed ataxia [9]. In VACV E9 (DNA polymerase catalytic subunit), mutations A498V, A684V, and S851Y in VACV confer an increased mutation frequency in forward-mutagenesis screens [10,11]. The three residues A498, A684 and S951 are conserved between MPXV F8L (DNA polymerase) and VACV E9. To assess if such mutations exist in the MPXV DNA RC, and whether these mutations are contributing to the current outbreak, we conducted a temporal sequence analysis of available MPXV isolates to identify mutations. We leveraged existing data including the crystal structure of VACV E9 protein (family B pol) [12], which shares ∼98% homology with MPXV F8L, to generate a molecular model of MPXV F8L. Additionally, a number of solved structures comprising pols α, δ, ε, ζ, HSV1 pol in complex with TP and inhibitor [[13], [14], [15], [16], [17]], and a state-of-the-art protein structure prediction method, AlphaFold [18], was used to build a DNA RC, and to infer the impact of mutations in 2022 MPXV viruses as well as predict the efficacy of existing vaccines and antivirals in mitigating the 2022 MPXV outbreak.
2. Materials and methods
2.1. Sequence acquisition
The sequences used in the analysis were acquired from the NCBI nucleotide sequence database through the NCBI Virus portal. As of June 28, 2022, all available and complete MPXV nucleotide sequences were downloaded from the portal (n = 205). The sequence dataset included sequences from over 20 countries, during several outbreaks. This included the earliest MPXV sequence in 1965 from Rotterdam, Netherlands, and several sequences from the 2022 outbreak (n = 122).
2.2. Sequence processing and analysis
Using Python, an in-house bioinformatics pipeline was developed to rapidly process, and align MPXV sequences [19]. All sequences were compared to a monkeypox virus sequence that was isolated at a Rotterdam Zoo in 1965 (referred to as 1965 isolate hereafter) (NCBI: KJ642614.1). The sequences were cropped to the gene in focus using coordinates provided by NCBI and translated to protein sequences automatically through Expasy [20]. After translation, each sequence was compared with that of the reference protein sequence using the Biopython library [21]. Comparisons were deposited into a data file for further interpretation and the process was applied for the various genes studied (F8L, A22R, E5R, E4R, G9R). The pipeline code is available at GitHub (https://github.com/bluesk1/RapidSequenceAnalysis-MPXV). Using the resultant data files from the pipeline, mutations in 2001 (NC_003310.1), and 2018 (NC_063383.1) were mapped. For the 2022 outbreak, the most prevalent mutations (i.e., present in ≥50% of sequences) in 2022 sequences were included.
Sequences with collection in our dataset were divided into four time periods: 1965–1999 (n = 11), 2000–2009 (n = 26), 2010–2020 (n = 17), and 2021–2022 (n = 126). The sequences from each time period were independently processed through the above-mentioned pipeline to identify mutation prevalence. An in-house circos configuration script (available upon request) was used for the generation of the circos representation of mutations within each time period.
2.3. Construction of MPXV replication complex
A low-resolution structure of VACV DNA replication machinery consisting of E9, A20, D4, and D5 has been reported [22]. A PCNA ortholog in VACV has also been identified [23]. Since PCNA is an integral part of the eukaryotic DNA replication machinery, the PCNA ortholog in MPXV (G9R) is likely a part of the MPXV replication fork. Therefore, G9R was added as a fifth component of the MPXV replication machinery, and a minimal MPXV replication fork was constructed in multiple steps as detailed below.
2.3.1. Modeling of the structures of replication complex components
Due to high homology between VACV E9 and MPXV F8L, a homology-derived structure of F8L was constructed by Modeller software [24] using the crystal structure of VACV E9 DNA pol (PDB entry 5N2E) [12] as a template. Similarly, the structure of MPXV E4R in complex with A22R (1-50) was modeled using the crystal structure of the VACV D4/A20 N-terminal (1-50) complex [25]. The structure of the A22R homolog in VACV (A20) is only partially solved. Therefore, we used AlphaFold [18], a state-of-the-art deep-learning molecular modeling program, to generate a molecular model of A22R. To generate a trimeric structure of G9R, we used ColabFold [26], which is based on AlphaFold. Since a high-resolution structure of VACV helicase is not known, and AlphaFold is computationally expensive for generation of a hexamer of VACV/MPXV helicase, we omitted MPXV E5R in our analyses.
2.3.2. Assembly of the RC
To assemble a reliable RC, we first superposed the VACV E9-insert 3 peptide fused to the C-terminal domain of VACV A20 (PDB entry 6ZXP) [27] onto the helix of insert 3 in the modeled structure of MPXV F8L. Next, AlphaFold-generated A22R was superposed onto the A20/E9-insert 3 peptide (PDB entry 6ZXP). Following this step, the crystal structure of the VACV D4 in complex with the A20 N-terminus (PDB entry 4OD8) [25] was superposed on A22R. To this structure, the molecular model of MPXV E4R in complex with the N-terminus of A22R was superposed. Thus, we obtained a complex consisting of F8L, A22R and E4R. To obtain the position of TP and G9R, we superposed the palm subdomain of the S.cerevisiae pol δ holoenzyme consisting of TP, accessory subunits (Pol31 and Pol32), and the PCNA clamp (PDB entry 7KC0) [13] onto the palm subdomain of modeled MPXV F8L. We then superposed the modeled structure of G9R on the PCNA structure. This superposition provided an approximate position of G9R in the MPXV RC. Minor adjustments were made to accommodate for bad contacts of G9R with A22R. A similar guide for the helicase position in VACV, and therefore in MPXV was not available. Hence, MPXV E5R was not included in the MPXV RC.
3. Results
3.1. Mutations in the MPXV RC
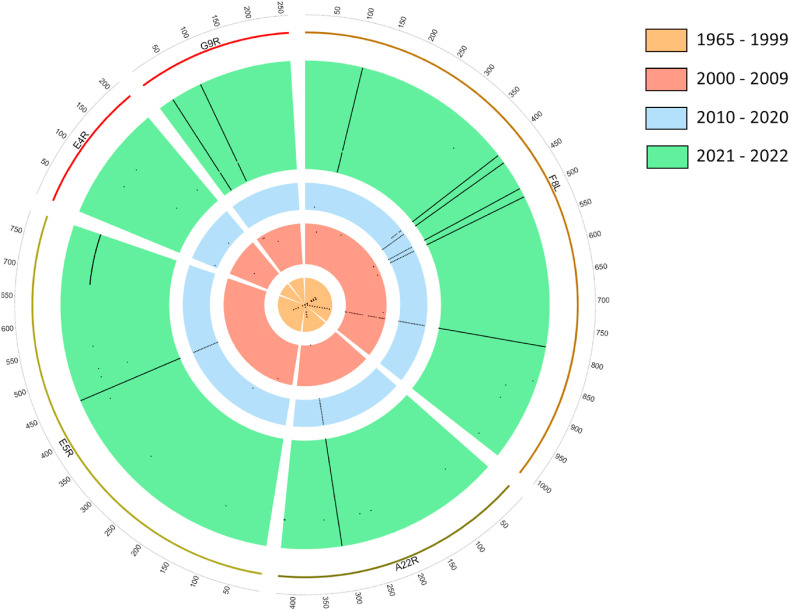
We conducted a temporal analysis of available MPXV sequences (n = 204) and identified mutations within MPXV RC components F8L, E5R, A22R, E4R, and G9R (see below) with respect to the 1965 isolate (Table 1 ), which was considered here as a reference isolate. A total of 10 residue positions, where mutations emerged at different time periods were identified (Table 1). Four mutations (2 in F8L, and 2 in G9R) were most prevalent in the 2022 isolates. F8L mutation L108F emerged in 2022, whereas mutation W411L emerged in 2018 and persisted in 2022 isolates. G9R mutations S30L and D88 N emerged in 2022 isolates. Six mutations that appeared in the 2001 outbreak reverted to the 1965 sequence, and revertant mutations persisted in 2022 isolates (Table 1, Fig. 1 ).
Table 1.
Mutations in MPXV over various periods.
| Gene | Residue | 1965 | 2001 | 2018 | 2022 | Epitopea |
|---|---|---|---|---|---|---|
| F8L MPXV DNA Polymerase |
108 | L | L | L | F | ISPDGCYSL (epitope 1) |
| 411 | W | W | L | L | LTFDYVVTF | |
| 428 | I | T | I | I | (epitope 2) | |
| 484 | A | S | A | A | ||
| 501 | I | V | I | I | ||
| 785 | N | D | N | N | ||
| A22R Processivity factor |
313 | I | S | I | I | – |
| E5R Helicase (NTPase) |
454 | D | N | D | D | – |
| E4R Uracil Glycosylase |
– | – | – | – | – | – |
| G9R PCNA ortholog |
30 | S | S | S | L | - |
| 88 | D | D | D | N | - |
Based on Song et al., 2013.
Fig. 1.
Circos diagram showing temporal analyses of mutations in the MPXV replication complex: This figure was generated using an in-house Circos configuration script (available upon request) in conjunction with our bioinformatics pipeline in Python (code available at https://github.com/bluesk1/RapidSequenceAnalysis-MPXV). The thickness of the band represents the number of sequences. Each black dot represents a mutation in the MPXV gene.
3.2. MPXV RC
The crystal structure of VACV E9 has revealed the topological positions of poxvirus-specific inserts, inserts 0 to 4 [12]. These inserts are also present in MPXV F8L (Fig. 2 a). Compared to the VACV E9 sequence, inserts 0, 1, and 3 are 100% identical, insert 2 has four substitutions, and insert 4 has one substitution. In VACV, only insert 3 has been demonstrated to interact with processivity factor A20 [12]. Therefore, insert 3 in MPXV is also expected to interact with A22R (VACV A20 homolog). Insert 2 is proximal to the ‘fingers’ subdomain (Fig. 2b), suggesting this insert may interact with the fingers as does ‘polymerase associated domain’ (PAD) in Y-family polymerases upon substrate binding [28]. The interactions of inserts 0, 1 and 4 could not be inferred from the modeled RC. The modeled MPXV RC shows that G9R interacts with A22R and the thumb subdomain of F8L. Similar interactions of PCNA (MPXV G9R ortholog) with the thumb subdomain of pol δ have been reported [13].
Fig. 2.
Structural details of MPXV DNA pol (F8L) and proposed components of the MPXV replication complex:Panel a shows the ribbon representation of the homology-derived molecular model of F8L. This model was constructed by Modeller [24] using the crystal structure of VACV DNA pol (E9) (PDB entry 5N2E) [12] as a template. Classical polymerase subdomains, namely the palm, fingers, and thumb are colored in red, blue, and green, respectively. The N-terminal domain is colored gray, whereas the 3′-5′ exonuclease (marked as 3′-5′ Exo) is colored olive-green. Poxvirus-specific structures designated as inserts 0, 1, 2, and 3 are shown in (pink, purple, magenta, and deep taupe, respectively). Insert 4 (red) as it is part of the palm subdomain. The template (marked as Temp) and primer (marked as Pri) were derived from the crystal structure of HSV1 (PDB entry 7LUF) [17] by superposing the palm subdomains of HSV1 pol and F8L are colored as cyan (template) and orange (primer). The epitopes of the antibodies induced by VACV-derived vaccines within F8L are marked as Epitope 1 and Epitope 2 (for sequences of these epitopes see Table 1). Epitope 1 (pastel purple) is within the N-terminal domain, whereas Epitope 2 (plum purple) is within the 3′-5′ exonuclease domain. Panel b shows the positions of G9R as derived from the superposition of pol δ in complex with subunits Pol31, Pol32, and PCNA with respect to the structure of F8L. This model was generated by superposing the palm subdomain of pol δ (PDB entry 7KC0) [13] onto the modeled structure of F8L. The PCNA ortholog in MPXV, G9R was constructed using ColabFold [26], a variant of AlphaFold. A22R was constructed using AlphaFold [18] protein structure prediction. The superposition of the G9R molecular model onto PCNA in the pol δ holoenzyme (PDB entry 7KC0), positions G9R close to the thumb subdomain of F8L.
Insert 2 is one of the least conserved sequences between VACV E9 and MPXV F8L with four substitutions within this region. However, insert 2 contains mutations C356Y, G372D and G380S, which confer resistance to phosphonoacetic acid (PAA), a broad-spectrum family B DNA pol inhibitor. C356, G372 and G380 are conserved between VACV E9 and MPXV F8L. These residues are expected to become proximal to the ‘fingers subdomain’ upon ternary complex formation, which induces conformational changes when a polymerase forms a ternary complex consisting of TP, dNTP and divalent cations [[29], [30], [31]]. Thus, PAA resistance mutations may impact the interaction of the ‘fingers’ subdomain with insert 2, and thereby reduce the binding of PAA.
3.3. Mutations in the MPXV RC
We sought to identify mutations in plausible MPXV RC constituents and conducted structural analyses to understand whether mutations are located at critical positions to influence the function of the replication complex.
3.3.1. Mutation L108F
Mutation L108F was consistently identified in 2022 MPXV isolates. Residue L108 is located in the N-terminal domain of MPXV F8L (or VACV E9) on a loop that is close to the ‘fingers’ subdomain (Fig. 3 a). A rigid-body docking of TP from HSV1 DNA polymerase in complex with DNA (PDB entry 7LUF) and inhibitor PNU-183792 showed that L108 is close to the base moiety of the ‘flipped’ template overhang nucleotide (Fig. 3a). The phenyl group in L108F is closer to the ‘flipped’ base compared to leucine, and would be predicted to increase the binding affinity of polymerase and TP due to increased hydrophobicity. To generate more direct evidence of enhanced TP binding affinity with the L108F mutation, we determined the buried surface area of L108 and F108, and converted this buried surface area into the Gibbs free energy change (ΔG) of binding using the correlation of Chothia [32]. We then used ΔG to calculate equilibrium binding constant in the following equation:
Fig. 3.
The location of mutations in MPXV F8L and functional implications:Panel a indicates the position of the L108F mutation in F8L and the interaction of the residue at position 108 with the ‘flipped’ template overhang (rendered as ball-and-sticks). The template nucleotide atoms, carbon, oxygen, nitrogen, and phosphorus, are colored in cyan, red, blue, and orange, respectively. The wild-type residue L108 is shown as brown carbons whereas F108 is shown as gray carbons. This figure also shows the proximity of residue I501, which was mutated to V501 in the 2001 MPXV isolate but reverted to I501 in the 2018 and 2022 isolates. Panel B shows the position of mutations in insert 2 of MPXV F8L. The wild-type residues are colored in magenta carbons, whereas mutations are colored as gray carbons. It is clear from this figure that L411 is exposed to the surface of the protein to have hydrophobic interactions with another protein.
Here, R is the Gas constant, T is temperature in Kelvin, and K is equilibrium dissociation constant. This analysis showed that F108 has ∼2.8-fold greater binding affinity to TP compared to L108. While this difference in binding affinity does not account for the change in entropy compensation, it does suggest that the binding affinity of F108 containing F8L is higher than L108-containing F8L (wild-type). A crucial functional implication of the change in TP binding affinity is that pol processivity is expected to increase [33]. The interactions involving flipped nucleotides with phenylalanine and their biochemical impact have been previously reported in various polymerases [[34], [35], [36], [37], [38], [39]]. It is anticipated that the interaction of F108 with the ‘flipped’ nucleotide will change the biochemical properties of F8L including fidelity, sensitivity to nucleoside inhibitors, and processivity. In different polymerases, these properties were modulated based on the particular mutation and shorter sidechains [37,[39], [40], [41]]. Therefore, mutation L108F in MPXV is a ‘gain-of-function’ mutation, which most likely enhances processivity, changes sensitivity to nucleoside inhibitors, and fidelity of DNA synthesis.
3.3.2. Mutation W411L
W411 resides within F8L insert 2 and is surface exposed, making it ideally positioned to interact with the aforementioned ‘unidentified’ regulatory factor of poxviruses. Hydrophobic residues are typically buried within the protein core; however, F411L would expose a hydrophobic residue to the surface (Fig. 3b). W411L may have evolved to enhance the interaction between insert 2 and a regulatory/processivity factor(s), and the absence of strong electrostatic features of this insert [12] further support our prediction.
3.3.3. Other mutations in F8L
Four mutations in F8L (I428T, A484S, I501V and N785D) emerged in the 2001 outbreak, but reverted to the 1965 isolate sequences in 2018 and 2022. I428 is also in insert 2 and proximal to the PAA resistance mutation (G380S) in VACV E9 [42]. Due to the proximity of insert 2 with the fingers subdomain and the role of the fingers subdomain in nucleotide selection, I428 may be participating in nucleotide selection. Residue S484 is within the 3′-5′ exonuclease domain of F8L and proximal to epitope 1 [43] (Table 1 and Fig. 2). Residue I501 is within the ‘fingers’ domain close to the template nucleotide (Fig. 3a). I501 might function in conjunction with F108 and contribute to the binding of TP to enhance the processivity. Residue N785 is at the bottom of the ‘palm’ subdomain and its functional significance remains unclear.
3.4. Mutations in other components of the replication complex
No mutations identified in the E4R (uracil DNA glycosylase) protein. Processivity factor A22R has one mutation that emerged in the 2001 isolate (I313S) but reverted to the 1965 Rotterdam isolate (I313). Similarly, the D454 N mutation in the E5R protein appeared in 2001 and reverted to D454. While a low-resolution structure of VACV (D5), the E5R homolog, has been reported [44], the atomic details and topological position of the D454 equivalent residue in VACV D5 remains unclear. The MPXV G9R homolog in VACV is G8R, which has been reported to be a structural ortholog of human PCNA [23]. There are two mutations in G9R: S30L and D88N that appeared in the 2022 outbreak (Table 1). To understand the function of these residues in MPXV replication, we generated a model of G9R using ColabFold [26]. We superposed the S.cerevisiae pol δ replication complex containing TP [13] and PCNA, which was pre-superposed on the MPXV F8L model (using the palm subdomain). This structure modeling showed that S30 and D88 are in the close vicinity of E4R. Therefore, these mutations may impact the interaction of G9R with E4R. Similar to the F8L mutations L108F and W411L, G9R mutations S30L and D88 N are ∼100% prevalent in 2022 isolates, suggesting a strong correlation among these mutations.
3.5. Approved MPXV treatments targeting replication complex
Currently, three antiviral drugs (tecovirimat, brincidofovir, and cidofovir) have been approved for use in MPXV infection. Brincidofovir and cidofovir target the DNA pol (F8L). Brincidofovir is the oral lipid prodrug of cidofovir which must be metabolized into the triphosphate form to be used as a substrate for F8L. Since these drugs are nucleoside analogs, they bind at the dNTP binding site. The triphosphate form of cidofovir [45] has a high affinity with DNA pol and competes with its natural substrate, dCTP. Due to the presence of a 3′OH group, cidofovir does not act as a chain terminator, in contrast to all approved anti-HIV nucleoside inhibitors.
The mechanism of resistance for Brincidofovir and cidofovir has been established. VACV E9 mutations A314T and A684V are cidofovir resistance mutations [45]. Structural modeling and proximity of A314 near the primer of TP suggests that A314T interferes with the shuttling of primer to the exonuclease domain, whereas A684V is in the ‘palm’ subdomain and likely interferes with template positioning, which, in turn, alters the cidofovir-triphosphate binding at the dNTP binding pocket. Therefore, these mutations may have emerged due to cidofovir usage to treat cytomegalovirus-induced retinitis in HIV-infected individuals, who are immunocompromised and resistant to cidofovir.
4. Conclusions
Of the total 10 mutations in the MPXV RC, 6 reverted to the 1965 isolate. I428 is within insert 2, which also carries PAA resistance mutations but the resistance mutation is not at position 428. It is possible that I428T evolved as a secondary mutation to functionally compensate for the fitness of the viruses that were resistant to PAA (C356T, G372D and G380S). Revertant mutations at positions 484 and 501 are in close proximity to L108. Therefore, it appears that mutations at 484 and 501 reverted to compensate for mutation L108F.
In addition to the mutations in the DNA RC discussed here, other viral proteins from 2022 isolates also contain a variety of mutations. For example, there are four mutations (V6D, D282 N, T601 M, D633E) in the DNA dependent RNA polymerase gene (OPG151) in 2022 isolates. However, due to the mutation in the DNA RC, we have not discussed the mutations in other genes of MPXV. A low-resolution structure of VACV consisting of E9, A29, D4 and D5 has been reported [22]. A PCNA ortholog in VACV (G8R) has also been reported [23]. Reported low-resolution structure does not contain G8R. It is well-known that PCNA and its homologs or orthologs are integral parts of the replication machinery of many species. It is highly likely that VACV G8R and MPXV G9R are also part of the RC in two poxviruses. Our structural modeling shows that G9R can be easily accommodated between F8L and E4R. Additionally, AlphaFold accurately predicted the N-terminal helices of A22R, as seen in the crystal structure of VACV D4 in complex with the A20 N-terminus [25]. These helices were used for the superposition to obtain the position of E4R without any modification in the orientation of the two helices. It is possible that these two helices adopt a different orientation, which may reposition E4R. However, regardless of the position of E4R, there remains enough space in the RC to accommodate G9R within the MPXV G9R.
In summary, we presented the minimum components of the MPXV replication complex and identified mutations in this complex that likely contribute to the 2022 MPX outbreak. Mutation L108F in F8L, which appeared in the 2022 outbreak, is close to the ‘flipped’ template nucleotide and will enhance F8L processivity, whereas W411L likely enhances the binding of a component of the replication complex by altering the surface exposed region. Due to the high sequence and structural similarity among viral B-type DNA pols, the currently-approved antiviral treatments are expected to retain efficacy against the current iteration of MPXV. However, the evolution of resistance mutations remains possible since critical functional pathways have already been susceptible to functional mutations within the viral proteins.
Author contributions
KS conceptualized the study; KS, SRK, and SS wrote the first draft, and KS, SS, SRK, and CLL wrote the final manuscript. SRK, SLK, AR, and KS conducted genetic analyses, and wrote required programs either in R or in Python; KS and SRK conducted structural analysis; KS, SS, SNB, and CLL edited the manuscript and contributed to understanding the pathogenicity of MPXV. All authors approved the final manuscript.
Funding
K. Singh was partially funded by the Bond Life Sciences Center (Early Concept grant) and the University of Missouri startup support.
Data sharing
Genomic sequences used in this study were obtained from NCBI GenBank.
Institutional review board statement
Not Applicable.
Informed consent statement
Not Applicable.
Declaration of competing interest
CLL is co-founder and Chief Scientific Officer of Shift Pharmaceuticals, Overland Park, KS, USA. KS is a consultant for Sanctum Therapeutics Corporation, Sunnyvale, CA, USA.
Acknowledgments
KS acknowledges the computation facilities of the Molecular Interactions Core at the University of Missouri, Columbia, MO 65212. We thank the laboratories that have generously deposited sequences into the NCBI database.
Data availability
Data will be made available on request.
References
- 1.Weaver J.R., Isaacs S.N. Monkeypox virus and insights into its immunomodulatory proteins. Immunol. Rev. 2008;225:96–113. doi: 10.1111/j.1600-065X.2008.00691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar N., Acharya A., Gendelman H.E., Byrareddy S.N. The 2022 outbreak and the pathobiology of the monkeypox virus. J. Autoimmun. 2022;131 doi: 10.1016/j.jaut.2022.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shchelkunov S.N., Totmenin A.V., Safronov P.F., Mikheev M.V., Gutorov V.V., Ryazankina O.I., Petrov N.A., Babkin I.V., Uvarova E.A., Sandakhchiev L.S., Sisler J.R., Esposito J.J., Damon I.K., Jahrling P.B., Moss B. Analysis of the monkeypox virus genome. Virology. 2002;297:172–194. doi: 10.1006/viro.2002.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito J., Braithwaite D.K. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991;19:4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braithwaite D.K., Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 7.Steitz T.A. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 8.Singh K., Modak M.J. A unified DNA- and dNTP-binding mode for DNA polymerases. Trends Biochem. Sci. 1998;23:277–281. doi: 10.1016/s0968-0004(98)01250-x. [DOI] [PubMed] [Google Scholar]
- 9.Goodman L.B., Loregian A., Perkins G.A., Nugent J., Buckles E.L., Mercorelli B., Kydd J.H., Palu G., Smith K.C., Osterrieder N., Davis-Poynter N. A point mutation in a herpesvirus polymerase determines neuropathogenicity. PLoS Pathog. 2007;3:e160. doi: 10.1371/journal.ppat.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrei G., Gammon D.B., Fiten P., De Clercq E., Opdenakker G., Snoeck R., Evans D.H. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J. Virol. 2006;80:9391–9401. doi: 10.1128/JVI.00605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taddie J.A., Traktman P. Genetic characterization of the vaccinia virus DNA polymerase: identification of point mutations conferring altered drug sensitivities and reduced fidelity. J. Virol. 1991;65:869–879. doi: 10.1128/jvi.65.2.869-879.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarbouriech N., Ducournau C., Hutin S., Mas P.J., Man P., Forest E., Hart D.J., Peyrefitte C.N., Burmeister W.P., Iseni F. The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding. Nat. Commun. 2017;8:1455. doi: 10.1038/s41467-017-01542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng F., Georgescu R.E., Li H., O'Donnell M.E. Structure of eukaryotic DNA polymerase delta bound to the PCNA clamp while encircling DNA. Proc. Natl. Acad. Sci. U. S. A. 2020;117:30344–30353. doi: 10.1073/pnas.2017637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik R., Johnson R.E., Prakash L., Prakash S., Ubarretxena-Belandia I., Aggarwal A.K. Cryo-EM structure of translesion DNA synthesis polymerase zeta with a base pair mismatch. Nat. Commun. 2022;13:1050. doi: 10.1038/s41467-022-28644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan Z., Georgescu R., Schauer G.D., O'Donnell M.E., Li H. Structure of the polymerase epsilon holoenzyme and atomic model of the leading strand replisome. Nat. Commun. 2020;11:3156. doi: 10.1038/s41467-020-16910-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coloma J., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K. Human DNA polymerase alpha in binary complex with a DNA:DNA template-primer. Sci. Rep. 2016;6 doi: 10.1038/srep23784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes R.P., Heo M.R., Mason M., Reid J., Burlein C., Armacost K.A., Tellers D.M., Raheem I., Shaw A.W., Murray E., McKenna P.M., Abeywickrema P., Sharma S., Soisson S.M., Klein D. Structural understanding of non-nucleoside inhibition in an elongating herpesvirus polymerase. Nat. Commun. 2021;12:3040. doi: 10.1038/s41467-021-23312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cock P.J., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M.J. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sele C., Gabel F., Gutsche I., Ivanov I., Burmeister W.P., Iseni F., Tarbouriech N. Low-resolution structure of vaccinia virus DNA replication machinery. J. Virol. 2013;87:1679–1689. doi: 10.1128/JVI.01533-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Da Silva M., Upton C. Vaccinia virus G8R protein: a structural ortholog of proliferating cell nuclear antigen (PCNA) PLoS One. 2009;4:e5479. doi: 10.1371/journal.pone.0005479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 25.Contesto-Richefeu C., Tarbouriech N., Brazzolotto X., Betzi S., Morelli X., Burmeister W.P., Iseni F. Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminal domain. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirdita M., Schutze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat. Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bersch B., Tarbouriech N., Burmeister W.P., Iseni F. Solution structure of the C-terminal domain of A20, the missing brick for the characterization of the interface between vaccinia virus DNA polymerase and its processivity factor. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167009. [DOI] [PubMed] [Google Scholar]
- 28.Trincao J., Johnson R.E., Escalante C.R., Prakash S., Prakash L., Aggarwal A.K. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol. Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Kong Y., Korolev S., Waksman G. Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates. Protein Sci. 1998;7:1116–1123. doi: 10.1002/pro.5560070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H., Chopra R., Verdine G.L., Harrison S.C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 31.Doublie S., Tabor S., Long A.M., Richardson C.C., Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 32.Chothia C. Hydrophobic bonding and accessible surface area in proteins. Nature. 1974;248:338–339. doi: 10.1038/248338a0. [DOI] [PubMed] [Google Scholar]
- 33.Loh E., Loeb L.A. Mutability of DNA polymerase I: implications for the creation of mutant DNA polymerases. DNA Repair. 2005;4:1390–1398. doi: 10.1016/j.dnarep.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Patel P.H., Suzuki M., Adman E., Shinkai A., Loeb L.A. Prokaryotic DNA polymerase I: evolution, structure, and "base flipping" mechanism for nucleotide selection. J. Mol. Biol. 2001;308:823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 35.Srivastava A., Singh K., Modak M.J. Phe 771 of Escherichia coli DNA polymerase I (Klenow fragment) is the major site for the interaction with the template overhang and the stabilization of the pre-polymerase ternary complex. Biochemistry. 2003;42:3645–3654. doi: 10.1021/bi026699y. [DOI] [PubMed] [Google Scholar]
- 36.Purohit V., Grindley N.D., Joyce C.M. Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment) Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 37.Singh K., Srivastava A., Patel S.S., Modak M.J. Participation of the fingers subdomain of Escherichia coli DNA polymerase I in the strand displacement synthesis of DNA. J. Biol. Chem. 2007;282:10594–10604. doi: 10.1074/jbc.M611242200. [DOI] [PubMed] [Google Scholar]
- 38.Rutvisuttinunt W., Meyer P.R., Scott W.A. Interactions between HIV-1 reverse transcriptase and the downstream template strand in stable complexes with primer-template. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher T.S., Prasad V.R. Substitutions of Phe61 located in the vicinity of template 5'-overhang influence polymerase fidelity and nucleoside analog sensitivity of HIV-1 reverse transcriptase. J. Biol. Chem. 2002;277:22345–22352. doi: 10.1074/jbc.M200282200. [DOI] [PubMed] [Google Scholar]
- 40.Fisher T.S., Darden T., Prasad V.R. Substitutions at Phe61 in the beta3-beta4 hairpin of HIV-1 reverse transcriptase reveal a role for the Fingers subdomain in strand displacement DNA synthesis. J. Mol. Biol. 2003;325:443–459. doi: 10.1016/s0022-2836(02)01225-1. [DOI] [PubMed] [Google Scholar]
- 41.Takata K., Arana M.E., Seki M., Kunkel T.A., Wood R.D. Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic Acids Res. 2010;38:3233–3244. doi: 10.1093/nar/gkq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taddie J.A., Traktman P. Genetic characterization of the vaccinia virus DNA polymerase: cytosine arabinoside resistance requires a variable lesion conferring phosphonoacetate resistance in conjunction with an invariant mutation localized to the 3'-5' exonuclease domain. J. Virol. 1993;67:4323–4336. doi: 10.1128/jvi.67.7.4323-4336.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song H., Sidney J., Wiseman R.W., Josleyn N., Cohen M., Blaney J.E., Jahrling P.B., Sette A. Characterizing monkeypox virus specific CD8+ T cell epitopes in rhesus macaques. Virology. 2013;447:181–186. doi: 10.1016/j.virol.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hutin S., Ling W.L., Round A., Effantin G., Reich S., Iseni F., Tarbouriech N., Schoehn G., Burmeister W.P. Domain organization of vaccinia virus helicase-primase D5. J. Virol. 2016;90:4604–4613. doi: 10.1128/JVI.00044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrei G., Snoeck R. Cidofovir activity against poxvirus infections. Viruses. 2010;2:2803–2830. doi: 10.3390/v2122803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.