Abstract
Plant viruses are known to infect most economically important crops and pose a major threat to global food security. Currently, few resistant host phenotypes have been delineated, and while chemicals are used for crop protection against insect pests and bacterial or fungal diseases, these are inefficient against viral diseases. Genetic engineering emerged as a way of modifying the plant genome by introducing functional genes in plants to improve crop productivity under adverse environmental conditions. Recently, new breeding technologies, and in particular the exciting CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR‐associated proteins) technology, was shown to be a powerful alternative to engineer resistance against plant viruses, thus has great potential for reducing crop losses and improving plant productivity to directly contribute to food security. Indeed, it could circumvent the “Genetic modification” issues because it allows for genome editing without the integration of foreign DNA or RNA into the genome of the host plant, and it is simpler and more versatile than other new breeding technologies. In this review, we describe the predominant features of the major CRISPR/Cas systems and outline strategies for the delivery of CRISPR/Cas reagents to plant cells. We also provide an overview of recent advances that have engineered CRISPR/Cas‐based resistance against DNA and RNA viruses in plants through the targeted manipulation of either the viral genome or susceptibility factors of the host plant genome. Finally, we provide insight into the limitations and challenges that CRISPR/Cas technology currently faces and discuss a few alternative applications of the technology in virus research.
Keywords: CRISPR/Cas, crop, genome editing, plant viruses
Plant viruses pose a major threat to global food security. In this review we provide an overview of the use and limitations of CRISPR/Cas‐based technology for resistance against DNA and RNA viruses in plants.
1. INTRODUCTION
Plant viruses are nucleoprotein complexes that rely mostly on host cells for their propagation. A large fraction of emerging plant diseases is caused by viruses, mostly because of their ability to adapt to changing environmental conditions and their effective dissemination facilitated by vector transmission (Anderson et al., 2004). Most economically important crops get infected with viruses, causing serious viral diseases that are responsible for significant decreases in both the yield and quality of harvests worldwide. It is estimated that 15% of global crop production is lost due to plant diseases, of which one‐third is accounted for by viruses (Boualem et al., 2016; Yadav & Chhibbar, 2018). Plant viruses therefore threaten global food security and agricultural productivity for the ever‐increasing world population.
Plant viruses have small genomes (4–20 kb) composed of DNA or RNA that encode conserved essential proteins such as the coat protein, movement protein, and replication‐associated enzymes, as well as a number of additional less‐conserved proteins (Awasthi et al., 2016). Viral replication and transcription are dependent on the host's cellular machinery, making plant viruses obligate parasites. Plant viruses are transmitted by exposure to wounds, seeds, and pollen or by a diverse range of vectors including insects, nematodes, soil fungi or mites (Bragard et al., 2013; Lefeuvre et al., 2019). Unlike other plant pathogens, viruses cannot be controlled directly by chemical applications on infected material, making preventative sanitary measures the only approach to manage infections. Currently, control measures include planting virus‐free material, the eradication of infected material that was detected early enough, crop rotation, and pesticides to control transmission vectors (Fereres & Raccah, 2015; Tavazza et al., 2017). While agricultural practices often depend on pesticides, the extensive use of these has been shown to have many adverse effects on the environment and has given rise to insecticide resistance in virus‐vector populations (Bragard et al., 2013).
The use of plant varieties that have natural genetic resistance factors constitutes the most efficient and sustainable approach to control viral infections. The first virus resistance gene was cloned and isolated from Nicotiana glutinosa. Named the N gene of tobacco, it confers a gene‐for‐gene resistance to the viral pathogen tobacco mosaic virus (TMV) in both tobacco and tomato transgenic plants (Whitham et al., 1994, 1996). Its cloning ultimately led to a better understanding of plant virus immune systems. By the introgression of resistance genes from wild to cultivated plants, a number of these plants were improved over the past decades and made commercially available. Unfortunately, for many plant–virus combinations the transfer of a resistance trait to a desired cultivar faces complex genetic constraints, such as linkage drag and high levels of heterozygosity (Kang et al., 2005). In addition, this approach requires a long generation time and is not cost‐effective for most breeding programmes. Viruses, like other pathogens, are able to evolve rapidly through recombination, mutations, and reassortment, making molecular advances in providing new tools for crop improvement and durable resistance vital.
In the 1980s, when alternative transgenic approaches were being explored, it was discovered that the inhibition of gene expression could be engineered by the expression of antisense RNA in plant cells, a phenomenon named pathogen‐derived resistance (PDR) (Sanford & Johnston, 1985). As further studies were conducted, resistance was obtained through the expression of partial or noncoding virus sequences, leading to successful developments of virus‐resistant crops (Lomonossoff, 1995; Wilson, 1993), even though the mechanisms behind PDR were not completely understood (Baulcombe, 1996). The RNA‐mediated mechanism behind PDR was later shown to be an antivirus response from plants, a strategy termed RNA silencing (Tenllado, 2004). By means of regulating gene expression, the RNA silencing strategy was a breakthrough for antiviral breeding and has for more than a decade been used to engineer resistance in several crops against more than 60 different viruses (Zhao et al., 2019). More recently, genome‐editing technology has emerged and can be used for virus resistance following two broad strategies: the first approach targets the virus directly, as with RNA silencing; while the second approach targets endogenous host plant susceptibility (S) factors (van Schie & Takken, 2014). S factors are host factors that viruses use to replicate and complete their lifecycle in the plant. By modifying these S factors with genome editing, we can limit their availability and therefore mitigate the pathogenicity of viruses in plants (Dong & Ronald, 2019).
2. PLANT GENOME‐EDITING TECHNOLOGIES
Since the emergence of genome‐editing technology, the achievements using this approach have revolutionized the fields of functional genomics and crop improvement. Essentially, genome‐editing technology is the use of sequence‐specific nucleases for recognizing specific DNA sequences and producing DNA double‐stranded breaks (DSBs) at targeted sites in chromosomal loci (Yin & Qiu, 2019). In almost all cell and organism types, a nuclease‐induced break is repaired via either nonhomologous end‐joining (NHEJ) or homology‐directed repair (HDR) (Sonoda et al., 2006). These two pathways differ in their efficiency and the mechanisms they require to repair the chromosome. If a repair template is absent, the error‐prone NHEJ operates, resulting in the introduction of a single or multiple insertion/deletion (indel) mutations after a DSB (Figure 1). These indels can cause a frameshift mutation as they disrupt either a translational reading frame or the binding sites of trans‐acting factors. Therefore, gene knockouts are created (Song et al., 2016). Alternatively, the high‐fidelity HDR method uses an intact homologous sequence as a donor template to enable sequence insertions or introduce point mutations by means of loci recombination (Belhaj et al., 2013).
FIGURE 1.
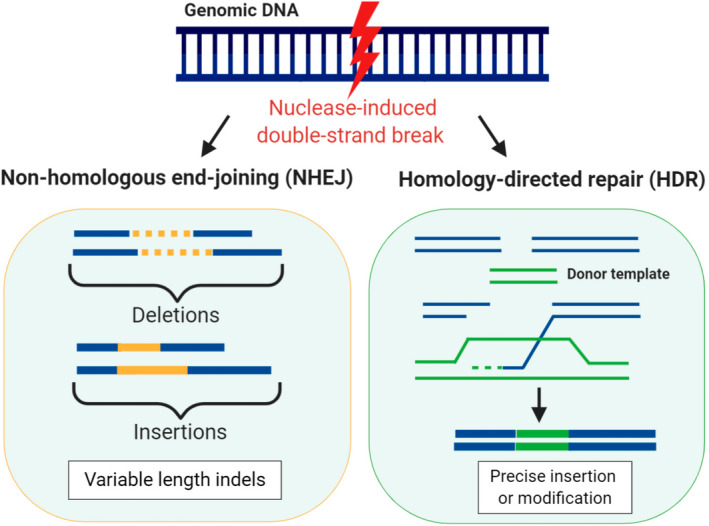
Repair pathways for nuclease‐induced double‐stranded breaks. Nonhomologous end‐joining leads to the introduction of random indel (insertion/deletion) mutations, whereas homology‐directed repair can introduce point mutations or sequence insertions through recombination using a donor template. This figure was created using Biorender.
Previously, the leading genome‐editing tools available were zinc finger nucleases (ZFNs) and transcription activator‐like effector nucleases (TALENs) (Boch et al., 2009; Kim et al., 1996). Both of these nucleases are chimeric proteins created by fusing their respective DNA‐binding domains (DBD) with the DNA cleavage domain of the FokI restriction enzyme. Sequence specificity of the target DNA is conferred by the DBD, while the FokI cleavage domain produces the DSBs in the targeted site (Christian et al., 2010; Kim et al., 1996). ZFN‐ and TALENS‐based genome editing has been used with variable success in several plant species (Cai et al., 2009; Curtin et al., 2011; Shan et al., 2013; Zhang et al., 2012). Although the use of ZFNs and TALENs led to important advances, the customization of these two genome‐editing platforms requires protein engineering for each new target, making it a time‐consuming and resource‐intensive process (Gaj et al., 2013). During the last decade, a new genome‐editing platform naturally surpassed ZFNs and TALENs and their applications in plants.
2.1. CRISPR/Cas systems
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR‐associated (Cas) proteins form the CRISPR/Cas system, which evolved in archaea and bacteria as an adaptive immune system against invading foreign nucleic acids originating from viral or plasmid pathogens (Barrangou et al., 2007; Barrangou & Marraffini, 2014; Brouns et al., 2008). While reported for the first time in 1987 by Ishino et al. (1987), it was only in 2012 that the potential of this system was realized by the fusion of the CRISPR‐RNA (crRNA) and trans‐acting CRISPR‐RNA (tracrRNA) to form guide‐RNA, an invention that would earn Jennifer Doudna and Emmanuelle Charpentier a Nobel prize in 2020 (Jinek et al., 2012). The mechanism of CRISPR/Cas‐mediated immunity (Figure S1) has been the topic of a multitude of research and review papers over the last decade (for recent reviews, see Nussenzweig & Marraffini (2020) and Nidhi et al. (2021)).
Since the discovery of the CRISPR/Cas9 system in Streptococcus pyogenes, related systems in many different bacterial and archaeal species have been discovered. These have been classified in different classes, types and subtypes, based mainly on the functionality of the effector molecules (Koonin et al., 2017; Shmakov et al., 2015, 2017).
The knowledge of all aspects of CRISPR/Cas systems is constantly expanding. In this review application of the better‐known members in the CRISPR toolbox are described, especially in the context of their use in virus interference.
2.2. CRISPR/Cas for DNA editing
2.2.1. CRISPR/Cas9
The class 2 type II endonuclease Cas9 from Streptococcus pyogenes (SpCas9) was the first Cas effector to be adapted as a genome engineering tool. In the natural system, the pre‐crRNA is processed into mature crRNAs by a tracrRNA and the bacterial RNase III. An RNA complex comprising a crRNA and tracrRNA then directs a cluster of Cas9 nuclease proteins to cleave invading double‐stranded DNA (dsDNA), which essentially gives rise to a target interference (Van Der Oost et al., 2014). For most applications in genome editing, the crRNA and tracrRNA are fused into a single guide RNA (sgRNA) with a specific 20 nucleotide (nt) spacer sequence complementary to a DNA target (Figure 2a). A prerequisite for Cas9 cleavage is the presence of a short G‐rich protospacer adjacent motif (PAM), found immediately downstream of a DNA target sequence. The Cas9 protein comprises two nuclease domains, RuvC and HNH, each responsible for the cleavage of one strand of the dsDNA target, generating either a blunt DSB (Ran et al., 2013; Sander & Joung, 2014) or a staggered DSB, as more recently discovered (Molla & Yang, 2020).
FIGURE 2.
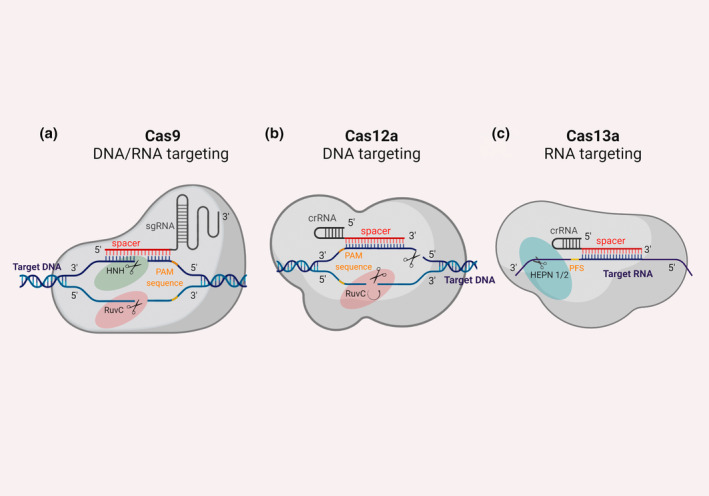
A schematic comparison of the class 2 CRISPR/Cas systems. (a) Cas9 represents a type II system and is guided by an sgRNA encoding a spacer bound to a dsDNA target adjacent to a PAM. The HNH and RuvC nuclease domains are activated when the correct base‐pairing occurs and cleave both DNA strands. (b) Cas12a represents a type V system and binds to the DNA sequence complementary to the single crRNA spacer and adjacent to a PAM. The RuvC nuclease domain is activated when the correct base‐pairing occurs and ssDNase activity cleaves both strands. (c) Cas13 represents a type VI system and binds to a ssRNA sequence complementary to the crRNA spacer. The HEPN domains are activated when the correct base‐pairing occurs for ssRNase activity. This figure was created using Biorender.
Several online bioinformatic software tools have been developed to predict the effectiveness of sgRNAs from whole‐genome information. A well‐designed sgRNA should be specific to the DNA target, meaning it should tolerate as few mismatches as possible, and none in the 8–12 nt seed region adjacent to the PAM, to reduce the possible off‐target activity of the attached Cas nuclease (Zhang et al., 2015). Both on‐target efficiency and off‐target activity are affected by the unique nucleotide sequence as well as possible secondary structure of a sgRNA (Uniyal et al., 2019). When applying the CRISPR/Cas9 system, it is still a concern that high frequencies of off‐target mutations may cause genomic instability (Wolt et al., 2016).
The cleavage action of the Cas9 protein is a crucial step in targeted genome editing because this introduces a DSB in the genomic sequence of interest. Owing to its higher mutation efficiency and design simplicity, the CRISPR/Cas9 system dominates applications in plants compared to ZFNs and TALENs. Initially, CRISPR/Cas9‐mediated genome editing was typically used to target one or two gene loci at the same time. To target multiple genomic loci simultaneously, multiplex CRISPR/Cas9 systems were designed to allow the co‐expression of several sgRNAs (Mushtaq et al., 2018). This multiplex genome‐editing approach is valuable for functional gene knockouts in plants.
In an alternative application called base editing, an inactivated CRISPR–Cas9 effector (Cas9 variants, dCas9 or Cas9 nickase) is fused to a cytosine or adenosine deaminase, and the CRISPR‐Cas9 system can be used to introduce point mutations in the target sequence without generating a DSB. The change of one base to another has the potential of generating new crop varieties, thereby enhancing crop improvement processes (Azameti & Dauda, 2021). A recent evolution of Cas9 applications is prime editing, which allows for the insertion of a desired sequence at a target site without making use of HDR. The technology relies on a novel CRISPR/Cas9 complex, which is composed of a protein consisting of a Cas9 nickase (H840A) fused to a reverse transcriptase and a prime editing guide RNA (pegRNA). The pegRNA consists of a reverse transcriptase template and a primer‐binding site at the 3′ end of the sgRNA. This primes the reverse transcription and incorporates the genetic information from the reverse transcriptase template into the genome (Azameti & Dauda, 2021). Prime editing, although very promising and versatile, still suffers from low efficiency in plants.
2.2.2. CRISPR/Cas12a
A second class 2 effector, Cas12a (formerly Cpf1), was later identified and categorized as a type V CRISPR/Cas. In contrast to Cas9, Cas12a contains an RuvC domain but not an HNH domain and generates a staggered DSB distal from a T‐rich PAM located upstream from the guide sequence (Makarova et al., 2015; Zetsche et al., 2015). The staggered DSB is situated close to the 3′‐end of the complementary target sequence, creating a 5′‐overhang. The production of DSBs with staggered ends by Cas12a may be advantageous for knock‐in applications, indeed being able to programme the exact sequence of a sticky end would allow researchers to design the DNA insert such that it integrates into the genome in the proper orientation and at precise positions by using complementary DNA ends through HDR (Zetsche et al., 2015). Furthermore, Cas12a requires a shorter crRNA than Cas9 and there is no evidence that a tracrRNA is required (Fonfara et al., 2016). Guided by a single crRNA and the presence of a T‐rich PAM sequence (5′‐TTN‐3′), the Cas12a effector can target either ssDNA or dsDNA. As shown in Figure 2b, the crRNA scaffold is located on the 5′‐end, as opposed to the 3′‐end in type II CRISPR/Cas systems. The Cas12a proteins also have RNase activity, used to process pre‐crRNAs into mature crRNAs (Jeon et al., 2018). This crRNA processing feature can be exploited to simplify multiplexed genome editing through the use of a single customized crRNA array. The potential of Cas12a as an alternative to the Cas9 endonuclease has been demonstrated in mammalian, plant, and microbial cells (Yan et al., 2017; Zaidi et al., 2017; Zetsche et al., 2015). Another interesting characteristic of Cas12a (and a few other type V Cas nucleases) is that specific cis‐cleavage of the target DNA induces collateral trans‐cleavage of nontarget ssDNA, a feature that has been exploited in the development of highly specific and sensitive nucleic acid detection systems. Recently, another Cas12‐variant was discovered in the genomes of some archaea (Harrington et al., 2018). This nuclease, named Cas14, is small (40–70 kDa) compared to Cas9 and Cas12, cleaves ssDNA, and also demonstrates indiscriminate trans‐cleaving of ssDNA.
2.3. CRISPR/Cas for RNA editing
2.3.1. RCas9 variant
The Cas9 effector derived from Streptococcus pyogenes (SpCas9) has been extensively utilized for dsDNA genome editing and has shown that it can be easily reprogrammed for efficient cleavage, making it a suitable candidate to be repurposed for ssRNA targeting and manipulation. This was shown by O'Connell et al., who demonstrated the SpCas9 binding and cleavage of ssRNA in vivo (O'Connell et al., 2014). In contrast to the native dependence of SpCas9 for a PAM sequence, when synthetic PAM sequences (PAMmers) were supplied exogenously, the SpCas9 was successfully redirected to target the ssRNA sequence complementary to the PAMmers (O'Connell et al., 2014). This indication of RNA targeting was further tested by including dsDNA with the ssRNA targets and PAMmers. Interestingly, the SpCas9 and its crRNA targeting counterpart exclusively targeted the ssRNA, avoiding the corresponding DNA in vitro. Thereafter denoted as an RNA targeting Cas9 (RCas9), this effector can be used as a programmable RNA binding platform. While RCas9 shows promise for further applications, a concern to consider is the costly synthesis of PAMmers, as well as the chemical modifications required to stabilize them in living cells (Nelles et al., 2016).
2.3.2. FnCas9 variant
Previously shown to mediate DNA cleavage, a Cas9 effector encoded from Francisella novicida (FnCas9) was identified and applied for targeted RNA cleavage in vivo (Price et al., 2015; Sampson et al., 2013; Zhang, Zheng et al., 2018b). Discovered in 2013 (Sampson et al., 2013), the enzyme was shown to target bacterial mRNA and target gene expression. This novel feature of FnCas9 led to its use for the targeting of several eukaryotic viruses such as the human hepatitis C virus (HCV), and cucumber mosaic virus (CMV) and TMV in plants, with different degrees of successful interference (Price et al., 2015; Zhang, Zheng, et al., 2018b). In addition to the crRNA and tracrRNA, the CRISPR/FnCas9 system also requires a small CRISPR/Cas‐associated RNA (scaRNA) that hybridizes with tracrRNA, forming a duplex that promotes RNA targeting. Unlike the RCas9 system, the RNA targeting action of FnCas9 does not depend on a PAM. While some studies highlight the potential that the CRISPR/FnCas9 system holds for specific RNA targeting, there are still underlying mechanisms of FnCas9 that remain unknown. Due to its dual DNA and RNA targeting ability, like the RCas9 system, FnCas9 is less likely to be selected for RNA manipulation in the nucleus (Price et al., 2015).
2.4. CRISPR/Cas13
Using data mining and bioinformatic approaches, three novel class 2 CRISPR systems besides the common Cas9 effector were discovered, namely C2c1, C2c2, and C2c3 (Shmakov et al., 2015). Similar to Cas12a, C2c1 and C2c3 contained RuvC‐like endonucleases and were therefore classified as type V‐B Cas12b and type V‐C Cas12c, respectively. Notably, C2c2 was shown to have unique properties compared to all other Cas proteins. Thereafter designated as Cas13, the putative effector was assigned to a novel type, class 2 type VI (Shmakov et al., 2017). Cas13a was the first class 2 effector found to solely function as a single RNA‐guided RNA‐targeting protein. Analysis of the Cas13a protein sequence resulted in the detection of two “higher eukaryotes and prokaryotes nucleotide‐binding” (HEPN) domains, which are exclusively associated with RNase activity (Anantharaman et al., 2013). The two structurally different HEPN domains, HEPN1 and HEPN2, are located on the outer surface and when activated lead to the cleavage of the target RNA outside of the binding region (Figure 2c). The exposed catalytic site of HEPN is available to all RNAs in a solution, thus explaining why unspecific cleavage of RNA was detected in bacterial cells (Liu et al., 2017). Further characterization of the RNA cleavage activity of Cas13a elucidated that Cas13a is guided by a crRNA containing a 28‐nt spacer sequence, an interaction maintained by the presence of a protospacer flanking sequence (PFS) of A, C, or U (Abudayyeh et al., 2016). As shown by the Cas12 system, Cas13a proteins can autonomously process their own pre‐crRNAs without the involvement of tracrRNA. This crRNA maturation activity is catalysed by a domain called Helical1 and can be harnessed for multiplexed processing (Abudayyeh et al., 2017; East‐Seletsky et al., 2016). The sensitivity of the Cas13a system to single and double mismatches was analysed and revealed that a central mismatch sensitive “seed” region is present in the crRNA, opposed to the 5′‐seed regions found in type I and II systems (Abudayyeh et al., 2016; Liu et al., 2017; O'Connell, 2019). Consistent with other type V Cas nucleases, Cas13 has nonspecific collateral trans‐cleaving activity, but in this case on ssRNA molecules. The Cas13 nuclease family was shown to contain three experimentally characterized subtypes, Cas13a, Cas13b, and Cas13d (Table 1).
TABLE 1.
Classification of the main CRISPR/Cas RNA‐targeting systems
| Effector | Class and type | Organism(S) harbouring respective types | Signature components | Protospacer requirements | References |
|---|---|---|---|---|---|
| RCas9 | Class 2, type II | Streptococcus pyogenes |
Cas9 PAMmer sgRNA |
5’‐NGG‐3′ | O'Connell et al. (2014) |
| FnCas9 | Class 2, type II | Francisella novicida |
Cas9 sgRNA |
5’‐NG‐3′ |
Price et al. (2015) Sampson et al. (2013) |
| Cas13a | Class 2, type VI‐A |
Leptotrichia shahii Leptotrichia wadei |
Cas13 crRNA |
3′‐A/U/C‐5′ (not required by all orthologues) |
Abudayyeh et al. (2016) Abudayyeh et al. (2017) |
| Cas13b | Class 2, type VI‐B |
Prevotella buccae Bergeyella zoohelcum |
Cas13 crRNA |
5′‐A/U/G and 3′‐NAN or NNA (not required by all orthologues) | Smargon et al. (2017) |
| Cas13d | Class 2, type VI‐D |
Ruminococcus flavefaciens Eubacterium siraeum |
CasRx crRNA |
– |
Konermann et al. (2018) Yan et al. (2018) |
2.4.1. Cas13 variants
In its native form, Cas13 can be used for targeted RNA cleavage such as down‐regulation of a specific transcript. The pioneer study that characterized the functionality of the first Cas13 family representative, Leptotrichia shahii Cas13a (LshCas13a), later confirmed that Cas13a solely cleaves ssRNA (Abudayyeh et al., 2016). It was shown that LshCas13a could provide interference against an MS2 lytic ssRNA phage in Escherichia coli. Interestingly, this study also identified that once activation by the target RNA was completed, nonspecific cleavage of RNAs other than the target RNA occurred. This suggests that LshCas13a elicits programmed cell death or dormancy in the natural system. Fortunately, this type of “collateral activity” was not detected in eukaryotic cells (Abudayyeh et al., 2017; Cox et al., 2017). Subsequently, a screening of various Cas13a proteins identified LwaCas13a from Leptotrichia wadei and the knockdown ability of LwaCas13a was demonstrated in mammalian cells with no evidence of collateral RNA cleavage (Abudayyeh et al., 2017). Abudayyeh et al. also verified the functionality of RNA knockdown by LwaCas13a in plants, with almost all guides exceeding 50% RNA knockdown in rice protoplasts, suggesting that a wide range of organisms can be edited using this system (Abudayyeh et al., 2017). Although the LshCas13a orthologue requires a biochemical PFS, analogous to the PAM for Cas9, LwaCas13a was shown to be exempt from this restriction in mammalian cells (Abudayyeh et al., 2017; Cox et al., 2017).
Another recently categorized CRISPR/Cas13 system identified from computational sequence data mining is Cas13b (previously C2c6), assigned to class 2 and type VI‐B (Smargon et al., 2017). Although Cas13b also contains two HEPN domains and actively targets ssRNA, it has a novel protein sequence that differs significantly from Cas13a. In an E. coli essential gene screen, RNA cleavage by Cas13b was shown to be dependent on a double‐sided PFS, one on each of the 5′‐ and 3′‐ends of the protospacer target sequence (Smargon et al., 2017). Another interesting finding indicated that Cas13b interacts with two novel proteins, Csx27 and Csx28, of which Csx27 can repress RNA targeting and Csx28 can enhance RNA cleavage (Smargon et al., 2017). A study by Cox et al. evaluated a subset of Cas13 enzymes and found that the Cas13b orthologue from a Prevotella sp. P5‐125 (PspCas13b) exhibited a higher level of knockdown efficiency and specificity than the previously characterized LwaCas13a (Cox et al., 2017). Similar to LwaCas13a, however, PspCas13b showed no collateral RNase activity or PFS preference in mammalian cells (Cox et al., 2017).
The most recent addition to the Cas13 subtypes is type VI‐D, which appears to be more distantly related on a primary sequence level to previous Cas13 effectors (Yan et al., 2018). Cas13d enzymes are about 20–30% smaller than all the previously reported subtypes Cas13a to Cas13c, and the orthologues from Eubacterium siraeum (Es) and Ruminococcus spp. (Rsp) possess associated WYL‐domain‐containing accessory proteins for enhanced binding and cleavage activity (Konermann et al., 2018; Yan et al., 2018; Zhang, Konermann, et al., 2018a). Similar to the Cas13 subtypes LwaCas13a and PspCas13b, Cas13d shares many commonalities, such as the lack of PFS requirements for target sequence selection and the innate ability to process pre‐crRNAs (Zhang, Ye, et al., 2019a). Notably, the high RNase activity of the Cas13d orthologue from Ruminococcus flavefaciens (CasRx) was shown to provide specificity and robust activity in both mammalian and plant cells when compared to other Cas13 proteins, such as LwaCas13a and PspCas13b (Konermann et al., 2018; Mahas et al., 2019). In addition, while a seed region was previously not reported for Cas13d, researchers found a critical seed region for optimal Cas13d knockdown efficiency between nucleotides 15 and 21 of the gRNA. By performing a set of pooled screens for CRISPR/Cas13d, they identified optimal gRNA design rules for Cas13d and developed a predictive model to select gRNAs with optimal efficiency (Guo et al., 2021; Wessels et al., 2020). With reports of favourable RNA targeting efficiency, this enzyme will enable a wide scope of RNA manipulations in plants. Already, a CRISPR/CasRx activity prediction tool for gRNA target design has been developed for mammalian cell culture applications (Wessels et al., 2020).
3. CRISPR/CAS‐BASED PLANT VIRUS RESISTANCE
Plant viruses infect a wide range of plant species and are responsible for substantial losses in the yield and quality of staple crops (Nicaise, 2014; Oerke & Dehne, 2004). The first studies that looked at using the genome‐editing tool CRISPR/Cas for plant viruses were designed to target DNA viruses. However, the majority of plant viruses have RNA genomes and often plant DNA viruses have an intermediate RNA stage in their life cycle (Roossinck, 2003), making effectors with RNA specificity the systems of choice for this application (Figure 3).
FIGURE 3.
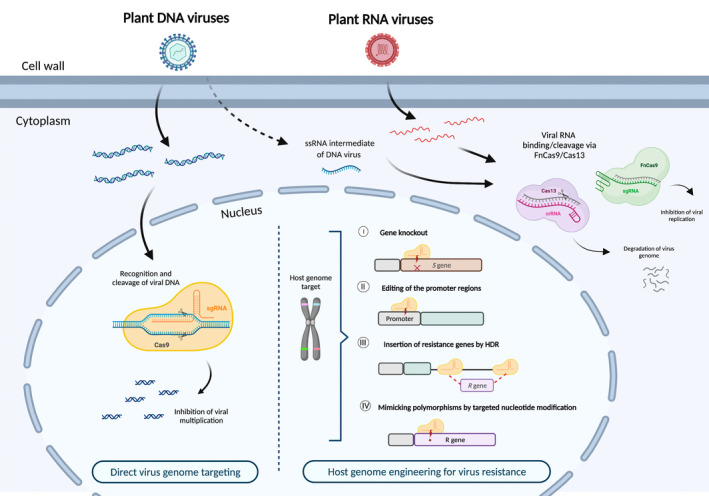
Schematic diagram of class 2 CRISPR/Cas strategies against viruses and targeting the host genomic DNA. On DNA virus entry into the plant cell, the Cas9/sgRNA complex binds to and cleaves DNA target sites. For RNA viruses or the RNA transcripts of pathogens with DNA genomes, both FnCas9 and Cas13a proteins guided by their cognate sgRNA or crRNA, respectively, have been proven to target and cleave the virus genome or transcripts. Alternatively, host susceptibility factors can be disrupted by CRISPR/Cas9 to perturb viral infection. The plant susceptibility (S) genes can be altered by directly targeting their coding regions or by modifying the promoter region sequences to prevent pathogen‐effector binding. In instances where the outcome of disturbing S genes is not extensively studied, the CRISPR toolkit can be used to introduce resistance (R) genes. Using the cellular homology‐directed repair (HDR) machinery, Cas9 can mediate the insertion of an R gene. To avoid whole‐gene disruption, Cas9 base‐editing technology can be used to make specific mutations that are associated with a disease‐resistant trait. This figure was created using Biorender.
3.1. Inactivation of plant DNA viruses
It has been demonstrated that CRISPR/Cas9‐mediated DNA editing can be used as a successful defence mechanism against plant DNA viruses (Ali, Abulfaraj, Idris, et al., 2015a). The members of the plant virus family Geminiviridae are composed of ssDNA genomes but also contain replicative intermediates of dsDNA, making them suitable candidates for CRISPR/Cas9 targeting. Three previous studies reported the successful use of CRISPR/Cas9 to generate geminivirus resistance in the model plants Nicotiana benthamiana and Arabidopsis thaliana (Table 2; Ali, Abul‐faraj, Li, et al., 2015b; Baltes et al., 2015; Ji et al., 2015). In these, target regions within the virus genomes, such as the replicase, coat protein, or intergenic region, were selected to design sgRNAs. As expected, all of these studies showed that the transgenic plants that expressed the CRISPR/Cas9 components and were challenged with the respective virus had reduced virus loads and symptoms (Ali, Abul‐faraj, Li, et al., 2015b; Baltes et al., 2015; Ji et al., 2015). In another approach targeting a monopartite geminivirus, Yin et al. used transgenic N. benthamiana plants expressing Cas9 and sgRNAs that simultaneously targeted two different sequences in the genome of cotton leaf curl Multan virus (CLCuMuV). This led to the plants being completely resistant to CLCuMuV (Yin et al., 2019). The effectiveness of using a multiplexed gRNA approach to minimize mutant escape formation as much as possible was also confirmed by successful attenuation of chilli leaf curl virus (ChiLCV) (Roy et al., 2019).
TABLE 2.
Major applications of CRISPR/Cas technology for DNA and RNA virus resistance in plants
| Virus genome | CRISPR system | Virus family | Virus genus | Virus name | Plant species | References |
|---|---|---|---|---|---|---|
| ssDNA | SpCas9 | Geminiviridae | Mastrevirus | Bean yellow dwarf virus (BeYDV) | Nicotiana benthamiana | Baltes et al. (2015) |
| Curtovirus | Beet severe curly top virus (BSCTV) |
N. benthamiana Arabidopsis thaliana |
Ji et al. (2015); Ali, Abulfaraj, Idris, et al. (2015a) | |||
| Begomovirus |
Tomato yellow leaf curl virus (TYLCV) Cotton leaf curl Kokhran virus (CLCuKoV) Merremia mosaic virus (MeMV) |
N. benthamiana | Ali, Abulfaraj, Idris, et al. (2015a); Ali et al. (2016) | |||
| Begomovirus | Cotton leaf curl Multan virus (CLCuMuV) | N. benthamiana | Yin et al. (2019) | |||
| Mastrevirus | Wheat dwarf virus (WDV) | Hordeum vulgare (barley) | Kis et al. (2019) | |||
| Begomovirus | Tomato yellow leaf curl virus (TYLCV) |
Solanum lycopersicum (tomato) N. benthamiana |
Tashkandi et al. (2018) | |||
| Begomovirus | African cassava mosaic virus (ACMV) | Manihot esculenta (cassava) | Mehta et al. (2019) | |||
| Begomovirus | Chilli leaf curl virus (ChiLCV) | N. benthamiana | Roy et al. (2019) | |||
| dsDNA | Caulimoviridae | Caulimovirus | Cauliflower mosaic virus (CaMV) | A. thaliana | Liu et al. (2018) | |
| Badnavirus | Banana streak virus (BSV) | Musa balbisiana (banana) | Tripathi et al. (2019) | |||
| +ssRNA | Alphaflexiviridae | Potexvirus | Potato virus X (PVX) | S. lycopersicum (tomato) | Wang et al. (2018) | |
| Virgaviridae | Tobamovirus | Tobacco mosaic virus (TMV) | ||||
| Potyviridae | Potyvirus | Soybean mosaic virus (SMV) | Glycine max (soy‐bean) | Zhang et al. (2020) | ||
| FnCas9 | Virgaviridae | Tobamovirus | Tobacco mosaic virus (TMV) |
N. benthamiana A. thaliana |
Zhang, Zheng, et al. (2018b) | |
| Bromoviridae | Cucumovirus | Cucumber mosaic virus (CMV) | ||||
| LshCas13a | Potyviridae | Potyvirus | Turnip mosaic virus (TuMV) |
N. benthamiana A. thaliana |
Aman, Ali, et al. (2018a); Aman, Mahas, et al. (2018b) | |
| dsRNA | LshCas13a | Reoviridae | Fijivirus | Southern rice black‐streaked dwarf virus (SRBSDV) | Oryza sativa (rice) | Zhang, Zhao, et al. (2019b) |
| −ssRNA | LshCas13a | Rhabdoviridae | Cytorhabdovirus | Rice stripe mosaic virus (RSMV) | ||
| +ssRNA | LshCas13a | Virgaviridae | Tobamovirus | Tobacco mosaic virus (TMV) | N. benthamiana | |
| Potyviridae | Potyvirus | Potato virus Y (PVY) | Solanum tuberosum (potato) | Zhan et al. (2019) | ||
|
LshCas13a LwaCas13a BzCas13b PspCas13b CasRx |
Virgaviridae | Tobamovirus | Tobacco mosaic virus (TMV) | N. benthamiana | Mahas et al. (2019) | |
| Potyviridae | Potyvirus | Turnip mosaic virus (TuMV) | ||||
| CasRx | Potyviridae | Potyvirus | Turnip mosaic virus (TuMV) | N. benthamiana | Cao et al. (2021) | |
| Virgaviridae | Tobamovirus | Tobacco mosaic virus (TMV) | ||||
| Bromoviridae | Cucumovirus | Cucumber mosaic virus (CMV) | ||||
|
LshCas13a FnCas9 |
Closteroviridae | Ampelovirus | Grapevine leafroll‐associated virus 3 (GLRaV‐3) | Vitis vinifera (grapevine) | Jiao et al. (2022) |
In addition to the Geminiviridae family, strong virus resistance was achieved against cauliflower mosaic virus (CaMV), a plant pararetrovirus with a dsDNA genome. Here, the expression of multiple sgRNAs, targeting the coat protein region, conferred successful resistance in transgenic Arabidopsis plants (Liu et al., 2018). There are some pararetroviruses, such as banana streak virus (BSV), that may integrate their DNA into the nuclear genome of the plant host, forming an endogenous virus (eBSV) that can also induce infections under stress conditions. By generating transgenic banana plants expressing Cas9 and sgRNAs targeting integrated regions of the eBSV genome, Tripathi et al. demonstrated the inactivation of the virus. When the transgenic plants were challenged under water stress conditions, they were resistant to reactivation of the virus when compared to the nontransgenic control plants (Tripathi et al., 2019).
Recently, some studies have translated CRISPR/Cas‐mediated resistance against geminiviruses from model plants to crop plants. For example, Tashkandi et al. engineered the CRISPR/Cas9 machinery in tomato plants to target tomato yellow leaf curl virus (TYLCV) genomic sequences, resulting in robust interference of TYLCV in all tomato plants from the T2 to the homozygous T3 generation (Tashkandi et al., 2018). Later, another study showed effective resistance against wheat dwarf virus (WDV) in the monocot plant barley. The sgRNA‐Cas9 construct was developed to introduce mutations at multiple sites within conserved regions of two WDV strains (Kis et al., 2019). In contrast, Mehta et al. attempted to engineer resistance to an important geminivirus using CRISPR/Cas9 in cassava, but failed to induce effective resistance against African cassava mosaic virus (ACMV) in transgenic cassava plants expressing Cas9 and sgRNAs that targeted regions of the virus genome (Mehta et al., 2019). The probable reason for this is the fact that ACMV replication was more efficient than CRISPR‐cleavage, and that this potentially leads to the emergence of novel virus mutants that cannot be cleaved by the original CRISPR/Cas9 system again. In an interesting commentary, Rybicki suggested that the conclusion by these authors may be premature, because the study lacked a number of important controls, such as lower concentrations of the challenging virus and the use of multiple sgRNAs (Rybicki, 2019). However, the Mehta et al. study highlights the risks surrounding transgenic CRISPR/Cas9 plants, given that they may accelerate the evolution of novel virus genomes that can escape engineered resistance if they are not monitored correctly (Mehta et al., 2019).
3.2. Inhibition of plant RNA viruses
The majority (more than 60%) of plant‐infecting viruses have RNA genomes and pose a serious threat to agricultural production (Lefkowitz et al., 2017). The discovery of CRISPR/Cas variants from various bacterial strains such as RCas9, FnCas9, and Cas13a/b/d have led to these being used to target RNA in vivo (Abudayyeh et al., 2017; O'Connell et al., 2014; Sampson et al., 2013). The first report of CRISPR/Cas9‐engineered plant immunity for an RNA virus was performed by a group that targeted CMV and TMV using FnCas9 and observed a reduction in virus accumulation in both transgenic tobacco and Arabidopsis plants (Table 2) (Zhang, Zheng, et al., 2018b). Applications of RNA virus interference by CRISPR/Cas13 in plants have been described in recent literature. Aman et al. first demonstrated the RNA targeting ability of CRISPR/Cas13 as a tool to combat viruses in plants (Aman, Ali et al., 2018a). The study used LshCas13a for engineered interference against a green fluorescent protein (GFP)‐expressing turnip mosaic virus (TuMV), a member of the Potyvirus genus, in N. benthamiana. Leaves of plants stably transformed with a codon‐optimized LshCas13a were infiltrated with mixed Agrobacterium cultures carrying TuMV‐GFP and crRNAs that target different regions of the virus genome. After infiltration, a c.50% reduction in GFP signal was detected in the leaves for two of the tested crRNAs targets. These initial results indicated the functional capacity for CRISPR/Cas13 in plants. The same group conducted a study with the same objectives in A. thaliana (Aman, Mahas et al., 2018b). RNA interference against TuMV‐GFP virus replication was successful in A. thaliana too.
A preliminary study demonstrated that the LshCas13a system can target and degrade viral RNA genomes and confer resistance to an RNA virus in a monocot grain plant (Zhang, Zhao, et al., 2019b). Transgenic rice plants harbouring the CRISPR/Cas13a system were generated, with three crRNAs each targeting the RNA genomes of southern rice black‐streaked dwarf virus (SRBSDV) and rice stripe mosaic virus (RSMV). Inhibition of viral infection was confirmed in the transgenic rice plants, indicating that CRISPR/Cas13a can effectively target viral RNA in monocot plants too. Zhan et al. verified that the CRISPR/Cas13a system can be engineered to deliver broad‐spectrum resistance to transgenic potato plants against multiple potato virus Y (PVY) strains (Zhan et al., 2019). Confirmed by enzyme‐linked immunosorbent assays (ELISA) and reverse transcription quantitative polymerase chain reaction (RT‐qPCR), the transgenic potato plants expressing Cas13/sgRNA showed a significant reduction in PVY accumulation. In a recent study in grapevine, Jiao et al. compared FnCas9 and LshCas13a for efficacy against grapevine leafroll‐associated virus 3 (GLRaV‐3) and demonstrated that while both systems could confer resistance, the latter provided better interference efficiency against this virus (Jiao et al., 2022).
In another study, Mahas et al. characterized multiple Cas13 proteins from three different Cas13 subtypes (a, b, and d) for their efficiency to target viral RNA in N. benthamiana (Mahas et al., 2019). To improve cellular localization, each Cas13 orthologue was fused to either a nuclear localization signal or a nuclear export signal. Transient and stable overexpression assays were conducted using a TMV‐RNA‐based overexpression (TRBO‐G) system expressing GFP, as well as a GFP‐expressing TuMV, as interference targets. The TRBO‐GFP construct served as a reporter system that is not capable of systemic movement, while the TuMV‐GFP virus was used to test whether the variants could limit systemic spread efficiently (Lindbo, 2007). Overall, while the variants LwaCas13a, PspCas13b, and CasRx all showed high interference activities (over c.50% virus reduction), CasRx mediated the most robust interference in both stable and transient assays (Mahas et al., 2019). In addition, it was shown that CasRx can target either one or two RNA viruses simultaneously, making CasRx a variant that is potentially amenable to multiplex targeting of RNA plant viruses. Likewise, Cao et al. recently expanded on the applicability of CasRx and was able to show a CRISPR/CasRx‐mediated RNA interference against an array of RNA viruses (Cao et al., 2021).
For viral interference applications, these reports are encouraging for future multiplex strategies that can simultaneously either target multiple species of viruses within the same family to provide broad virus protection or target multiple regions of a single virus genome to evade the possibility of evolutionary resistance to the CRISPR/Cas system from occurring. A recent study showed that single polyvalent gRNAs (pgRNAs), designed for one spacer to be able to target multiple viral target sequences, in complex with the CasRx effector can effectively suppress virus spread and gene expression in planta, better than those with a monovalent gRNA counterpart (Bagchi et al., 2022). This observation of enhanced antiviral suppression is related to improvements reported by CRISPR antiviral treatments with multiple gRNAs, and future studies could now also use multiple pgRNAs to further increase the number of target sites. The impressive catalytic activity and high specificity of CasRx therefore enables diverse RNA manipulations in plants and it appears that it will continue to be favoured for viral RNA genome degradation. In a recent surprising finding, Sharma et al. reported a system that they termed “Cas13‐independent guide‐induced gene silencing (GIGS)”. Using a CRISPR/Cas13 system, they demonstrated effective gene silencing in three plant species when multiple gRNAs were expressed in the absence of Cas13 (Sharma et al., 2022).
3.3. Editing of host plant factors for virus resistance
As mentioned earlier, another approach to obtain virus resistance in plants is the targeting of the so‐called susceptibility (S) genes. Indeed, the success of viral infection depends on the deployment of host cell machinery, including host‐encoded virus‐compatible proteins or S factors, and whose modification may cause loss of susceptibility, passive resistance, or recessive resistance (Kan et al., 2022).
A well‐known S factor is the eukaryotic translation initiation factor 4E (eIF4E). As shown in Table 3, there are a number of examples in literature where the gene(s) encoding eIF4E have been edited and subsequent resistance to viruses observed. For example, Chandrasekaran et al. (2016) used the CRISPR/Cas9 system to induce mutations in the elF4E gene of cucumber and reported the effective resistance of these cucumber plants against three different potyviruses, which have RNA genomes, namely zucchini yellow mosaic virus (ZYMV), cucumber vein yellowing virus (CVYV), and papaya ringspot mosaic virus‐W (PRSV‐W) (Chandrasekaran et al., 2016). After three generations of backcrossing, homozygous nontransgenic cucumber plants showed broad virus resistance. Similarly, by introducing site‐specific mutations in the A. thaliana eIF(iso)4E locus using CRISPR/Cas9, resistance to the potyvirus TuMV was conferred (Pyott et al., 2016). An investigation by Macovei et al. (2018) demonstrated novel sources of resistance against rice tungro spherical virus (RTSV) in rice (Oryza sativa) through biomimicking of eIF4G alleles (Macovei et al., 2018). T2 plants selected from this study were resistant to RSTV and tested negative for the presence of Cas9. More recently, the CRISPR/Cas9‐based targeting of two of the five cassava eIF4E isoforms (nCBP‐1 and nCBP‐2), found to interact with the viral genome‐linked protein of cassava brown streak virus (CBSV), significantly suppressed the symptoms of the disease in cassava plants (Gomez et al., 2019). In addition to this, these plants did not exhibit any observed mutations in potential off‐target sites. In barley, thanks to the rym4 and rym5 allelic variants of the HveIF4E gene, more than two‐thirds of current European winter barley cultivars are resistant to the bymoviruses barley yellow mosaic virus (BaYMV) and barley mild mosaic virus (BaMMV). However, several strains of BaYMV and BaMMV have already overcome rym4‐ and rym5‐mediated resistance. For this reason, Hoffie and colleagues saw the need to generate new resistance alleles using CRISPR/Cas9 (Hoffie et al., 2021). In this work, a homozygous mutation in the first exon generated a premature stop codon and presumably a nonfunctional protein, and the plants showed resistance to mechanical inoculation with BaMMV. Surprisingly, the plant yield was not affected in greenhouse conditions.
TABLE 3.
Summary of studies that have employed CRISPR/Cas9 strategies for the targeting of host susceptibility genes
| Plant species | Name of the susceptibility (S) gene targeted | Virus name | Reference |
|---|---|---|---|
| Arabidopsis thaliana | AteIF(iso)4E | Turnip mosaic virus (TMV) | Pyott et al. (2016) |
| eIF4E1 | Clover yellow vein virus (CYVV) | Bastet et al. (2019) | |
| Hordeum vulgare (barley) | eIF4E1 | Barley mild mosaic virus (BaMMV) | Hoffie et al. (2021) |
| Manihot esculenta (cassava) | nCBP‐1/2 | Cassava brown streak virus (CBSV) | Gomez et al. (2019) |
| Cucumis sativus (cucumber) | CseIF4E |
Zucchini yellow mosaic virus (ZYMV) Cucumber vein yellowing virus (CVYV) Papaya ring spot mosaic virus‐W (PRSV‐W) |
Chandrasekaran et al. (2016) |
| Nicotiana benthamiana | CLC‐Nb1a/b | Potato virus Y (PVY) | Sun et al. (2018) |
| Oryza sativa (rice) | OseIF4G | Rice tungro spherical virus (RTSV) | Macovei et al. (2018) |
| Solanum tuberosum (potato) | Coilin | Potato virus Y (PVY) | Makhotenko et al. (2019) |
| Glycine max (soybean) | GmF3H1/2, FNSII‐1 | Soybean mosaic virus (SMV) | Zhang et al. (2020) |
| Solanum lycopersicum (tomato) | TOM1 | Tomato brown rugose fruit virus (ToBRFV) | Ishikawa et al. (2022) |
| eIF4E1 | Pepper mottle virus (PepMoV) | Yoon et al. (2020) | |
| eIF4E1 |
Cucumber mosaic virus (CMV) Potato virus Y (PVY) |
Atarashi et al. (2020) | |
| eIF4E1 | Pepper veinal mottle virus (PVMV) | Kuroiwa et al. (2022) | |
| SleIF4E1, SleIF4E2 | Potato virus Y (PVY) | Kumar et al. (2022) | |
| Triticum aestivum (wheat) | TaPDIL5‐1 | Wheat yellow mosaic virus (WYMV) | Kan et al. (2022) |
The editing of eIF4E in tomato has been reported in four different publications in the last two years (Table 3), with all results demonstrating resistance to viruses. It is interesting to note that tomato contains three eIF4E genes, eIF4E1, eIF4E2, and eIF(iso)4E, and from these studies it is evident that the level of susceptibility to a virus is related to the specific eIF4E gene editing. For example, the editing of eIF4E1 reduced susceptibility to the pepper mottle virus (PepMoV) (Yoon et al., 2020), potato virus Y N strain (PVYN), and cucumber mosaic virus (CMV) (Atarashi et al., 2020), while the knockout of eIF4E2 produced full resistance to one isolate of pepper veinal mottle virus (PVMV) but only partial resistance to another isolate (Kuroiwa et al., 2022). A double knockout of eIF4E1 and eIF4E2, as well as the single mutants for these genes, was further analysed in a study by Kumar et al. (2022) and it was confirmed that eIF4E1 is responsible for resistance to PVY and that the editing of both genes affected plant growth (Kumar et al., 2022). The mutants were also challenged with CMV, eggplant mild leaf mottle virus (EMLMV), pepino mosaic virus (PepMV), and tomato brown rugose fruit virus (ToBRFV) to verify if a broad‐spectrum resistance was obtained, but the edited plants did not show reduced susceptibility to these viruses. Surprisingly, the authors noticed a higher accumulation of the coat proteins of CMV and PepMV in the mutants compared to wild‐type plants. These findings link to a previous study by Zafirov et al. (2021) in which it was found that the loss of function of eIF4E1 in A. thaliana led to higher susceptibility to turnip mosaic virus (TuMV). The authors suggest that knockout of eIF4E genes could expose the plant to the severe threat of potyviruses able to recruit alternative eIF4E copies and that a better strategy could be the use of CRISPR base‐editing technology to create functional alleles. A nice example of this type of approach was given by Bastet et al. (2019) in A. thaliana where CRISPR‐Cas9 cytidine deaminase was successfully used to introduce the N176K mutation in the eIF4E gene to confer transgene‐free resistance to clover yellow vein virus (ClYVV). Using base editing to create additional mutations in a resistance allele can lead to resistance pyramiding against several viruses, thus expanding the resistance spectrum and/or increasing resistance durability (Bastet et al., 2019).
In addition to elF4E, other host S genes have been edited. For example, coilin encodes a major structural scaffolding protein necessary for Cajal body formation and mediates plant–virus interactions. By deploying CRISPR/Cas9‐based ribonucleoprotein (RNP) complexes to the apical meristematic tissue of potato plants using biolistic bombardment, the editing of at least a single allele of coilin in the tetraploid potato genome resulted in successful resistance against PVY (Makhotenko et al., 2019). Isoflavonoids are an essential group of secondary metabolites in leguminous plants and also play an important role in the regulation of plant–environment interactions. The multiplexed CRISPR/Cas9 targeting of GmF3H1, GmF3H2, and GmFNSII‐1 from the phenylpropanoid pathway resulted in both increased isoflavone content and enhanced resistance to soybean mosaic virus (SMV) in soybean plants (Zhang et al., 2020). A recent addition to the list of S genes targeted with genome editing is TOBAMOVIRUS MULTIPLICATION1 (TOM1). TOM1 is necessary for efficient multiplication of tobamoviruses in Arabidopsis (Ishikawa et al., 2022). The authors described how the simultaneous knockout of the four TOM1 homologues in tomato confers strong resistance to ToBRFV and that obvious defects in growth or fruit production were not observed. Importantly, it was noticed that when only three of the four TOM1 homologues were disrupted, ToBRFV coat protein accumulation was detectable but greatly reduced, but this led to the emergence of mutant viruses capable of more efficient multiplication. Ishikawa and colleagues hypothesized that the emergence and spread of resistance‐breaking mutants as observed in Sltom1acd triple mutant plants is caused by the low rate of viral accumulation in these plants, and that can be avoided with the complete knockout of all functional TOM1 homologues.
In barley, the protein disulphide isomerase‐like 5–1 (HvPDIL5‐1) gene encodes a chaperone protein involved in the quality check system of correct protein folding. Indeed, the loss of HvPDIL5‐1 confers broad‐spectrum resistance to multiple strains of bymoviruses, including BaMMV and BaYMV. Kan et al. (2022) edited the wheat orthologues TaPDIL5‐1‐4A, TaPDIL5‐1‐4B, and TaPDIL5‐1‐4D, and demonstrated that triple editing of the three TaPDIL5‐1 homoeoalleles was sufficient to achieve reliable resistance against wheat yellow mosaic virus (WYMV) in hexaploid wheat and that it did not affect agronomic performance, thus providing another option for genome editing with CRISPR/Cas9 to achieve virus resistance in crops (Kan et al., 2022).
To advance the practical applications of this CRISPR technology approach for plant virus resistance, there is an urgent demand for the identification of novel virus S genes from our understanding of plant–virus interactions, as is well illustrated in the reviews by Mäkinen (2020) and Hashimoto et al. (2016).
4. CRISPR/CAS COMPONENT DELIVERY AND EXPRESSION IN PLANTS
The effective delivery and subsequent expression of CRISPR/Cas components in plant cells are crucially important for successful editing in plants. The three primary methods for delivery are Agrobacterium‐mediated transformation, a physical means such as biolistic bombardment, or protoplast transfection. These all depend on plasmids, viruses, or RNPs to carry the required coding sequences or the functional proteins into cells (Kuluev et al., 2019). Agrobacterium‐mediated delivery of CRISPR/Cas components is the most common approach, but generates transgenic plants because desired sequences are integrated into the host genome. Likewise, biolistic bombardment of microprojectiles coated with a plasmid vector encoding the CRISPR components allows for the random integration of these sequences in the plant genome, potentially leading to multiple copies of the introduced genes (Sandhya et al., 2020).
The negative perceptions and onerous regulatory processes associated with transgenic plants has led to the development of a number of effective transgene‐free (or DNA‐free) delivery methods. The most widely used is the transient expression of the CRISPR‐related transgenes, which can be achieved by agroinfiltration of plasmids containing these sequences (Nester, 2015). In one such application, Agrobacterium‐mediated transient expression of a base editor targeting specific genes allowed for the regeneration of T‐DNA‐free edited tomato and potato plants (Veillet et al., 2019). Moreover, DNA‐free genome‐editing approaches based on the biolistic delivery of an RNP complex have been developed. These RNPs comprise a Cas nuclease complexed with one or more sgRNA(s), and have shown significantly improved editing efficiency (Kuluev et al., 2019). Yet another approach is the direct transformation of protoplasts, using a transfecting agent like polyethylene glycol (PEG) or by electroporation. The transfection of the CRISPR/Cas components, either a plasmid or a pre‐assembled Cas/sgRNA RNP, into protoplasts and the subsequent regeneration of transgenic or nontransgenic plants, respectively, has allowed for the successful introduction of desired mutations in several plant species such as rice, tobacco, and lettuce (Woo et al., 2015; Xie & Yang, 2013). Protoplast transfection with RNPs is a relatively quick process, making it useful to validate the mutagenesis efficiency of a CRISPR/Cas system (Yue et al., 2020). However, protoplast isolation and whole‐plant regeneration from protoplasts remains a challenge for many plant species, especially crop plants (Sandhya et al., 2020).
Harnessing plant viruses to act as delivery vectors is a promising approach to obtain CRISPR/Cas‐edited plants without the challenges that accompany transgene delivery. When viruses infect plants, they replicate and move systemically in their hosts, making them excellent vehicles for the delivery, high‐level expression, and distribution of CRISPR components in plants, leading to overall improved genome‐editing efficiency (Varanda et al., 2021). Recently, a number of viral vectors for the delivery of genome‐editing components to plant cells have been developed (Zaidi & Mansoor, 2017). Among these are the RNA viruses tobacco rattle virus (TRV) (Ali, Abul‐faraj, Li, et al., 2015b; Ghoshal et al., 2020), TMV (Cody et al., 2017), pea early‐browning virus (PEBV) (Ali et al., 2018), and beet necrotic yellow vein virus (BNYVV) (Jiang et al., 2019); all these have been shown to be efficient vectors in delivering CRISPR/Cas components to N. benthamiana, A. thaliana, and Beta macrocarpa plants. Virus‐mediated gRNA delivery provides a number of advantages compared to the conventional promoter‐driven expression of gRNAs, such as the rapid replication and systemic spread of the virus, which ensures effective amplification of the gRNA, while the small genome size allows for multiplexing and simple cloning strategies (Ali, Abul‐Faraj, Piatek, et al., 2015c). Single‐stranded DNA viruses, typically geminiviruses, have also been modified to carry heterologous coding sequences for increased protein expression in plants (Gil‐Humanes et al., 2017; Yin et al., 2015). Interestingly, a recent study found that a geminiviral replicon‐based expression vector was more efficient at LwaCas13a‐mediated RNA targeting than a regular binary vector in N. benthamiana (Yu et al., 2020).
It is therefore clear that the method employed to deliver and express CRISPR/Cas components can significantly affect the efficiency of CRISPR/Cas‐based genome editing. Notably, the success of a delivery method in plants is dependent on the species, the tissue type, and its totipotency.
5. CHALLENGES AND FUTURE PROSPECTS
5.1. Virus evolution and development of resistance
Despite the many technological advantages and an impressive track record in terms of applications in modern commercial agriculture, CRISPR/Cas technology still has a number of limitations and concerns, specifically for the development of disease‐resistant crop plants and especially for virus resistance. Mutation and recombination are the main driving forces in the evolution of plant DNA and RNA viruses, therefore the deliberate direct targeting and mutating of virus genomes may contribute to accelerated virus evolution, as demonstrated in a study by Mehta et al. that reported the failure of African cassava mosaic virus (ACMV) resistance in transgenic cassava plants expressing Cas9 and an sgRNA targeting a genome region of the virus (Mehta et al., 2019). The study suggested that between 33% and 48% of the edited ACMV genomes evolved a conserved single‐nucleotide mutation that protected the virus against CRISPR/Cas9 cleavage. In a subsequent analysis of this work, Rybicki commented on the novel aspects of this study, but also highlighted several shortcomings and limitations, among others the fact that only Cas9 was tested and also a single gRNA. He suggested that the conclusions of the Mehta study probably only hold true for single‐stranded DNA viruses and specifically for geminiviruses (Rybicki, 2019). Strategies to avoid the occurrence of new viruses, like the targeting of multiple viral genome regions, the deletion of larger genome stretches using CRISPR‐based nickases, or the use of alternative RNA‐targeting nucleases, should be investigated in both DNA and RNA viruses. Multiplexing of gRNAs has the further potential to establish resistance to multiple viruses in crops. Moreover, current strategies to introduce CRISPR/Cas‐based virus resistance by directly targeting the virus DNA or RNA genomes requires constitutive maintenance and expression of CRISPR components in plants. Such approaches not only pose the risk of subsequent unwanted mutations, but also potentially add a “GMO” label to the plant. Finally, the effectivity and durability of CRISPR/Cas‐based engineered virus resistance remains to be evaluated, especially under natural conditions in field trials.
5.2. Fitness cost to host plants
The targeting of host factors like S genes may have a fitness cost because these are often involved in essential endogenous processes like plant growth and development. In the case of elF4E, the gene product is an essential eukaryotic translation initiation factor (also known as a cap‐binding protein), responsible for directing ribosomes to the 7‐methyl‐guanosine cap of mRNAs for subsequent translation. In this case, the potentially lethal scenario is mitigated by the presence of an eIF(iso)4E paralogue in the genomes of many crop plants. Exclusively mutating only one of the elF4E isoforms has been shown to be an efficient way to introduce virus resistance without incurring a fitness penalty to the host plant (Pyott et al., 2016). It is generally accepted that the recessive resistance acquired by knocking down a host susceptibility factor is more durable than dominant resistance genes because of lower selective pressures on the pathogen to evolve counterdefence strategies (de Ronde et al., 2014). The elF4E gene is by far the best characterized virus‐related S gene, and while several other host factors have been identified (Garcia‐Ruiz, 2018), an urgent need for the identification of novel virus‐related S genes remains. An alternative approach may be the identification of host susceptibility genes for insect vectors of plant viruses. CRISPR/Cas‐based inhibition of such S genes may be an effective strategy to prevent the spread and dissemination of economically important viruses.
5.3. Off‐target effects
One of the biggest concerns regarding genome editing by CRISPR/Cas is the occurrence of off‐target mutations (Hahn & Nekrasov, 2019). These are DNA edits at unintended and nonspecific genomic sites as a result of the tolerance of gRNA sequence mismatches (Tsai & Joung, 2016) or even modifications in a gRNA‐independent manner (Jin et al., 2019). Off‐target editing is also a critical factor for the CRISPR/Cas13 system, although it has been shown to produce significantly lower off‐target effects compared to the existing RNA‐targeting method, RNAi (Abudayyeh et al., 2017; Cox et al., 2017). It is suspected that minimal off‐target modifications to a host plant's transcriptome occur when the RNA cleavage activity of the CRISPR/Cas13 system is engineered for the specific targeting of RNA viruses or RNA intermediates of DNA viruses. At present, the extent to which CRISPR/Cas13‐based RNA editing can give rise to transcriptomic irregularities is not completely understood and further research into this is required. Efforts to improve the design rules for the generation of gRNAs with higher fidelity comprise both experimental and computational approaches. Experimental techniques include methods to detect Cas binding to its target, the detection of Cas‐induced DSBs, and the detection of repair products that result from Cas‐induced DSBs (Bao et al., 2021). Numerous bioinformatic tools have been developed to identify potential CRISPR/Cas off‐target sites (for a comprehensive review of these, see Bao et al., 2021). These authors concluded that in their experience, none of the bioinformatic tools was able to accurately predict low‐frequency off‐target editing and recommended the use of at least one bioinformatic tool in tandem with an experimental approach for the prediction of potential off‐target sites (Bao et al., 2021).
5.4. GMO status and public acceptance
As stated, if the viral targeting activity of Cas nucleases is intended for heritable purposes, the permanent expression of the CRISPR/Cas components would be required, which can only be achieved through the generation of transgenic plants (Taliansky et al., 2021). Due to current regulation of genetically modified organisms (GMOs), the practical application of this technology may therefore be challenged by these regulatory constraints and thus be a limitation for the development of commercial crop varieties (Kalinina et al., 2020; Khatodia et al., 2017). However, by opting for RNA targeting over DNA targeting in plants, it is possible to confer a temporary or reversible modulation of gene expression, rather than knockout mutagenesis, which can often be lethal or have pleiotropic effects (Zhu et al., 2020). Without permanently editing the genome, CRISPR/Cas13 allows researchers to investigate gene function more systematically and could rather be harnessed as a “treatment” application for the transient inhibition of viruses in important crops. Therefore, the temporary nature of RNA editing overcomes major limitations relating to DNA targeting and its broad application in plant virology could potentially help overcome GMO regulatory hurdles. Despite this, the failure of genome‐edited food crops to gain acceptance by the general public, as well as to appease the regulators globally, is preventing this technology from reaping the fruits of success that it deserves. While the lay public in general are uninformed about the technology, a direct connotation with first‐generation genetic engineering persists, a situation that sadly has been “vindicated” by instances like the EU's decision to declare genome‐edited crops as GMOs. Fortunately, a few progressive‐thinking governments have relaxed regulation of genome‐edited crops significantly, with the result that the first genome‐edited crops, including mushroom, rice, maize, soybean, bristle grass, flax, wheat, and tomato, have been or are on their way to be commercialized (Menz et al., 2020). The early detection of plant viruses in combination with virus prevention strategies is of major economic importance in any crop production system.
5.5. Alternative applications
Some features of the CRISPR/Cas systems have exciting potential for pathogen detection and the development of reliable diagnostic systems, none more than the so‐called “collateral activity” of the Cas12, ‐13 and ‐14 nucleases. As mentioned previously, after the recognition of its cognate target (dsDNA for Cas12, RNA for Cas13, ssDNA for Cas14a), the nuclease is activated, resulting in target cleavage and subsequent indiscriminate cleavage of nearby nucleic acids (ssRNA for Cas13, ssDNA for Cas12 and Cas14). Interestingly, this collateral cleavage is only seen in bacterial cells (Abudayyeh et al., 2016). This feature has since been combined with different preamplification technologies, like different versions of PCR, recombinase polymerase amplification (RPA), or loop‐mediated isothermal amplification (LAMP) to develop detection systems such as specific high‐sensitivity enzymatic reporter unlocking (SHERLOCK) (Gootenberg et al., 2017), DNA endonuclease‐targeted CRISPR trans reporter (DETECTR) (Chen et al., 2018), and the 1‐h low‐cost multipurpose highly efficient system (HOLMES) (Li et al., 2018). These detection systems can employ different read‐out formats, like colourimetric visual display, fluorescent detection, or lateral flow dipstick display, with limit‐of‐detection sensitivities reaching attomolar (DETECTR and HOLMES) to zeptomolar (SHERLOCK) levels (Kaminski et al., 2021), demonstrating that CRISPR‐based detection systems have evolved in a few years from experimental nucleic acid sensing tools to a dominant diagnostic technology for the fast, affordable, and ultrasensitive detection of pathogens, including RNA and DNA viruses in the clinical and agricultural sectors.
Another exciting example of a very recent alternative application of CRISPR/Cas technology is that of “live” molecular imaging of macromolecules in cells. By fusing a catalytically inactive Cas9 (dCas9), complexed with gRNAs, to three different fluorescent proteins, Ma et al. were able to image multiple genomic loci in live human cells (Ma et al., 2016). Likewise, Abudayyeh et al. deactivated the two catalytic residues in the HEPN domains of Cas13 to create a dCas13, which, complexed to gRNAs, was fused to fluorescent proteins to image RNA transcripts in live cells (Abudayyeh et al., 2017). For applications in plant virology, this approach can allow the direct visualization of viral replication with high precision.
6. CONCLUDING REMARKS
In less than a decade, CRISPR/Cas systems have demonstrated their immense potential as a genome‐editing technology to overcome the limitations of conventional breeding for the development of resistance to both biotic and abiotic stresses in crop plants. Moreover, the fact that Cas nucleases are guided by RNA rather than protein circumvents the major limitations of TALENs and ZFNs. Collectively, the CRISPR/Cas systems present much promise as simple, robust, precise, and scalable DNA and RNA targeting platforms and can be efficiently exploited to achieve virus resistance in plants. The ability for multiplex targeting at both the DNA and RNA level to create one‐off mutations in a transgene‐free manner is a huge added bonus. CRISPR/Cas13 systems can target specific endogenous RNAs, viral RNAs, and RNA intermediates of DNA viruses in plants and thus increase possibilities for their application in agriculture. It is our belief that CRISPR/Cas technology will play a major role in the creation of disease‐resistant food crops in the near future, thereby contributing significantly to securing the sustainable food supplies urgently needed to support the world population expansion towards the middle of this century.
Supporting information
FIGURE S1 The three stages of CRISPR/Cas adaptive immunity in bacterial cells. (i) The first process, termed adaptation, is where the foreign genetic material is acquired by Cas1‐Cas2 and integrated into the CRISPR array. (ii) The associated Cas proteins are expressed and the CRISPR array is processed into mature crRNA to provide targeting specificity. (iii) The crRNA guides the effector nucleases to foreign genetic elements, leading to target interference and immunity. This figure was created using Biorender
ACKNOWLEDGEMENTS
This work was supported by a grant from Winetech, South Africa (GenUS17/1).
Robertson, G. , Burger, J. & Campa, M. (2022) CRISPR/Cas‐based tools for the targeted control of plant viruses. Molecular Plant Pathology, 23, 1701–1718. Available from: 10.1111/mpp.13252
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed.
REFERENCES
- Abudayyeh, O.O. , Gootenberg, J.S. , Konermann, S. , Joung, J. , Slaymaker, I.M. , Cox, D.B. et al. (2016) C2c2 is a single‐component programmable RNA‐guided RNA‐targeting CRISPR effector. Science, 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh, O.O. , Gootenberg, J.S. , Essletzbichler, P. , Han, S. , Joung, J. , Belanto, J.J. et al. (2017) RNA targeting with CRISPR‐Cas13. Nature, 550, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, Z. , Abulfaraj, A. , Idris, A. , Ali, S. , Tashkandi, M. & Mahfouz, M.M. (2015) CRISPR/Cas9‐mediated viral interference in plants. Genome Biology, 16, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, Z. , Abul‐faraj, A. , Li, L. , Ghosh, N. , Piatek, M. , Mahjoub, A. et al. (2015) Efficient virus‐mediated genome editing in plants using the CRISPR/Cas9 system. Molecular Plant, 8, 1288–1291. [DOI] [PubMed] [Google Scholar]
- Ali, Z. , Abul‐Faraj, A. , Piatek, M. & Mahfouz, M.M. (2015) Activity and specificity of TRV‐mediated gene editing in plants. Plant Signaling & Behavior, 10, e1044191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, Z. , Ali, S. , Tashkandi, M. , Zaidi, S.S.‐A. & Mahfouz, M.M. (2016) CRISPR/Cas9‐mediated immunity to geminiviruses: differential interference and evasion. Scientific Reports, 6, 26912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, Z. , Eid, A. , Ali, S. & Mahfouz, M.M. (2018) Pea early‐browning virus‐mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis . Virus Research, 244, 333–337. [DOI] [PubMed] [Google Scholar]
- Aman, R. , Ali, Z. , Butt, H. , Mahas, A. , Aljedaani, F. , Khan, M.Z. et al. (201a) RNA virus interference via CRISPR/Cas13a system in plants. Genome Biology, 19, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman, R. , Mahas, A. , Butt, H. , Aljedaani, F. & Mahfouz, M. (2018) Engineering RNA virus interference via the CRISPR/Cas13 machinery in Arabidopsis . Viruses, 10, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman, V. , Makarova, K.S. , Burroughs, A.M. , Koonin, E.V. & Aravind, L. (2013) Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra‐genomic conflicts, defense, pathogenesis and RNA processing. Biology Direct, 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P.K. , Cunningham, A.A. , Patel, N.G. , Morales, F.J. , Epstein, P.R. & Daszak, P. (2004) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends in Ecology & Evolution, 19, 535–544. [DOI] [PubMed] [Google Scholar]
- Atarashi, H. , Jayasinghe, W.H. , Kwon, J. , Kim, H. , Taninaka, Y. , Igarashi, M. et al. (2020) Artificially edited alleles of the eukaryotic translation initiation factor 4E1 gene differentially reduce susceptibility to cucumber mosaic virus and potato virus Y in tomato. Frontiers in Microbiology, 11, 564310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi, S. , Chauhan, R. & Narayan, R.P. (2016) Plant viruses: history and taxonomy. In: Gaur, R. (Ed.) Plant viruses: evolution and management. Singapore: Springer, pp. 1–17. [Google Scholar]
- Azameti, M.K. & Dauda, W.P. (2021) Base editing in plants: applications, challenges, and future prospects. Frontiers in Plant Science, 12, 664997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi, R. , Tinker‐Kulberg, R. , Salehin, M. , Supakar, T. , Chamberlain, S. , Ligaba‐Osena, A. , et al. (2022) Polyvalent guide RNAs for CRISPR antivirals. Biophysical Journal, 121, 422a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes, N.J. , Hummel, A.W. , Konecna, E. , Cegan, R. , Bruns, A.N. , Bisaro, D.M. et al. (2015) Conferring resistance to geminiviruses with the CRISPR‐Cas prokaryotic immune system. Nature Plants, 1, 4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, X.R. , Pan, Y. , Lee, C.M. , Davis, T.H. & Bao, G. (2021) Tools for experimental and computational analyses of off‐target editing by programmable nucleases. Nature Protocols, 16, 10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou, R. & Marraffini, L.A. (2014) CRISPR‐cas systems: prokaryotes upgrade to adaptive immunity. Molecular Cell, 54, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou, R. , Fremaux, C. , Deveau, H. , Richards, M. , Boyaval, P. , Moineau, S. et al. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science, 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Bastet, A. , Zafirov, D. , Giovinazzo, N. , Guyon‐Debast, A. , Nogué, F. , Robaglia, C. et al. (2019) Mimicking natural polymorphism in eIF4E by CRISPR‐Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnology Journal, 17, 1736–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D.C. (1996) Mechanisms of pathogen‐derived resistance to viruses in transgenic plants. The Plant Cell, 8, 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhaj, K. , Chaparro‐Garcia, A. , Kamoun, S. & Nekrasov, V. (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods, 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch, J. , Scholze, H. , Schornack, S. , Landgraf, A. , Hahn, S. , Kay, S. et al. (2009) Breaking the code of DNA binding specificity of TAL‐type III effectors. Science, 326, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Boualem, A. , Dogimont, C. & Bendahmane, A. (2016) The battle for survival between viruses and their host plants. Current Opinion in Virology, 17, 32–38. [DOI] [PubMed] [Google Scholar]
- Bragard, C. , Caciagli, P. , Lemaire, O. , Lopez‐Moya, J.J. , MacFarlane, S. , Peters, D. et al. (2013) Status and prospects of plant virus control through interference with vector transmission. Annual Review of Phytopathology, 51, 177–201. [DOI] [PubMed] [Google Scholar]
- Brouns, S.J.J. , Jore, M.M. , Lundgren, M. , Westra, E.R. , Slijkhuis, R.J. , Snijders, A.P. et al. (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science, 321, 960–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, C.Q. , Doyon, Y. , Ainley, W.M. , Miller, J.C. , DeKelver, R.C. , Moehle, E.A. et al. (2009) Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Molecular Biology, 69, 699–709. [DOI] [PubMed] [Google Scholar]
- Cao, Y. , Zhou, H. , Zhou, X. & Li, F. (2021) Conferring resistance to plant RNA viruses with the CRISPR/CasRx system. Virologica Sinica, 34, 814–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran, J. , Brumin, M. , Wolf, D. , Leibman, D. , Klap, C. , Pearlsman, M. et al. (2016) Development of broad virus resistance in non‐transgenic cucumber using CRISPR/Cas9 technology. Molecular Plant Pathology, 17, 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.S. , Ma, E. , Harrington, L.B. , Da Costa, M. , Tian, X. , Palefsky, J.M. et al. (2018) CRISPR‐Cas12a target binding unleashes indiscriminate single‐stranded DNase activity. Science, 360, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian, M. , Cermak, T. , Doyle, E.L. , Schmidt, C. , Zhang, F. , Hummel, A. et al. (2010) Targeting DNA double‐strand breaks with TAL effector nucleases. Genetics, 186, 756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody, W.B. , Scholthof, H.B. & Mirkov, T.E. (2017) Multiplexed gene editing and protein overexpression using a tobacco mosaic virus viral vector. Plant Physiology, 175, 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D.B.T. , Gootenberg, J.S. , Abudayyeh, O.O. , Franklin, B. , Kellner, M.J. , Joung, J. et al. (2017) RNA editing with CRISPR‐Cas13. Science, 358, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin, S.J. , Zhang, F. , Sander, J.D. , Haun, W.J. , Starker, C. , Baltes, N.J. et al. (2011) Targeted mutagenesis of duplicated genes in soybean with zinc‐finger nucleases. Plant Physiology, 156, 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, O.X. & Ronald, P.C. (2019) Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiology, 180, 26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East‐Seletsky, A. , O'Connell, M.R. , Knight, S.C. , Burstein, D. , Cate, J.H.D. , Tjian, R. et al. (2016) Two distinct RNase activities of CRISPR‐C2c2 enable guide‐RNA processing and RNA detection. Nature, 538, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fereres, A. & Raccah, B. (2015) Plant virus transmission by insects. In: Encyclopedia of life sciences (eLS). Chichester, UK: John Wiley & Sons, Ltd. 10.1002/9780470015902.a0000760.pub3 [DOI] [Google Scholar]
- Fonfara, I. , Richter, H. , BratoviÄ, M. , Le Rhun, A. & Charpentier, E. (2016) The CRISPR‐associated DNA‐cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature, 532, 517–521. [DOI] [PubMed] [Google Scholar]
- Gaj, T. , Gersbach, C.A. & Barbas, C.F. (2013) ZFN, TALEN, and CRISPR/Cas‐based methods for genome engineering. Trends in Biotechnology, 31, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. (2018) Susceptibility genes to plant viruses. Viruses, 10, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal, B. , Vong, B. , Picard, C. & Feng, S. (2020) A viral guide RNA delivery system for CRISPR‐based transcriptional activation and heritable targeted DNA demethylation in Arabidopsis thaliana . PLoS Genetics, 16, e1008983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil‐Humanes, J. , Wang, Y. , Liang, Z. , Shan, Q. , Ozuna, C.V. , Sánchez‐León, S. et al. (2017) High‐efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. The Plant Journal, 89, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, M.A. , Lin, Z.D. , Moll, T. , Chauhan, R.D. , Hayden, L. , Renninger, K. et al. (2019) Simultaneous CRISPR/Cas9‐mediated editing of cassava eIF4E isoforms nCBP‐1 and nCBP‐2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnology Journal, 17, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg, J.S. , Abudayyeh, O.O. , Lee, J.W. , Essletzbichler, P. , Dy, A.J. , Joung, J. et al. (2017) Nucleic acid detection with CRISPR‐Cas13a/C2c2. Science, 356, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X. , Rahman, J.A. , Wessels, H.‐H. , Méndez‐Mancilla, A. , Haro, D. , Chen, X. , et al. (2021) Transcriptome‐wide Cas13 guide RNA design for model organisms and viral RNA pathogens. Cell Genomics, 1, 100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, F. & Nekrasov, V. (2019) CRISPR/Cas precision: do we need to worry about off‐targeting in plants? Plant Cell Reports, 38, 437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L.B. , Burstein, D. , Chen, J.S. , Paez‐Espino, D. , Ma, E. , Witte, I.P. et al. (2018) Programmed DNA destruction by miniature CRISPR‐Cas14 enzymes. Science, 362, 839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M. , Neriya, Y. , Yamaji, Y. & Namba, S. (2016) Recessive resistance to plant viruses: potential resistance genes beyond translation initiation factors. Frontiers in Microbiology, 7, 1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffie, R.E. , Otto, I. , Perovic, D. , Budhagatapalli, N. , Habekuß, A. , Ordon, F. et al. (2021) Targeted knockout of eukaryotic translation initiation factor 4E confers bymovirus resistance in winter barley. Frontiers in Genome Editing, 3, 784233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, M. , Yoshida, T. , Matsuyama, M. , Kouzai, Y. , Kano, A. & Ishibashi, K. (2022) Tomato brown rugose fruit virus resistance generated by quadruple knockout of homologs of TOBAMOVIRUS MULTIPLICATION1 in tomato. Plant Physiology, 189, 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino, Y. , Shinagawa, H. , Makino, K. , Amemura, M. & Nakata, A. (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of Bacteriology, 169, 5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, Y. , Choi, Y.H. , Jang, Y. , Yu, J. , Goo, J. , Lee, G. et al. (2018) Direct observation of DNA target searching and cleavage by CRISPR‐Cas12a. Nature Communications, 9, 2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, X. , Zhang, H. , Zhang, Y. , Wang, Y. & Gao, C. (2015) Establishing a CRISPR–Cas‐like immune system conferring DNA virus resistance in plants. Nature Plants, 1, 15144. [DOI] [PubMed] [Google Scholar]
- Jiang, N. , Zhang, C. , Liu, J. , Guo, Z. , Zhang, Z. , Han, C. et al. (2019) Development of Beet necrotic yellow vein virus‐based vectors for multiple‐gene expression and guide RNA delivery in plant genome editing. Plant Biotechnology Journal, 17, 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, B. , Hao, X. , Liu, Z. , Liu, M. , Wang, J. , Liu, L. et al. (2022) Engineering CRISPR immune systems conferring GLRaV‐3 resistance in grapevine. Horticulture Research, 9, uhab023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, S. , Zong, Y. , Gao, Q. , Zhu, Z. , Wang, Y. , Qin, P. et al. (2019) Cytosine, but not adenine, base editors induce genome‐wide off‐target mutations in rice. Science, 364, 292–295. [DOI] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. & Charpentier, E. (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina, N.O. , Khromov, A. , Love, A.J. & Taliansky, M.E. (2020) CRISPR applications in plant virology: virus resistance and beyond. Phytopathology, 110, 18–28. [DOI] [PubMed] [Google Scholar]
- Kaminski, M.M. , Abudayyeh, O.O. , Gootenberg, J.S. , Zhang, F. & Collins, J.J. (2021) CRISPR‐based diagnostics. Nature Biomedical Engineering, 5, 643–656. [DOI] [PubMed] [Google Scholar]
- Kan, J. , Cai, Y. , Cheng, C. , Jiang, C. , Jin, Y. & Yang, P. (2022) Simultaneous editing of host factor gene TaPDIL5‐1 homoeoalleles confers wheat yellow mosaic virus resistance in hexaploid wheat. New Phytologist, 234, 340–344. [DOI] [PubMed] [Google Scholar]
- Kang, B.‐C. , Yeam, I. & Jahn, M.M. (2005) Genetics of plant virus resistance. Annual Review of Phytopathology, 43, 581–621. [DOI] [PubMed] [Google Scholar]
- Khatodia, S. , Bhatotia, K. & Tuteja, N. (2017) Development of CRISPR/Cas9 mediated virus resistance in agriculturally important crops. Bioengineered, 8, 274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.G. , Cha, J. & Chandrasegaran, S. (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proceedings of the National Academy of Sciences of the United States of America, 93, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis, A. , Hamar, É. , Tholt, G. , Bán, R. & Havelda, Z. (2019) Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnology Journal, 17, 1004–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann, S. , Lotfy, P. , Brideau, N.J. , Oki, J. , Shokhirev, M.N. & Hsu, P.D. (2018) Transcriptome engineering with RNA‐targeting type VI‐D CRISPR effectors. Cell, 173, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V. , Makarova, K.S. & Zhang, F. (2017) Diversity, classification and evolution of CRISPR‐Cas systems. Current Opinion in Microbiology, 37, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuluev, B.R. , Gumerova, G.R. , Mikhaylova, E.V. , Gerashchenkov, G.A. , Rozhnova, N.A. , Vershinina, Z.R. et al. (2019) Delivery of CRISPR/Cas components into higher plant cells for genome editing. Russian Journal of Plant Physiology, 66, 694–706. [Google Scholar]
- Kumar, S. , Abebie, B. , Kumari, R. , Kravchik, M. , Shnaider, Y. , Leibman, D. et al. (2022) Development of PVY resistance in tomato by knockout of host eukaryotic initiation factors by CRISPR‐Cas9. Phytoparasitica. 10.1007/s12600-022-00991-7 [DOI] [Google Scholar]
- Kuroiwa, K. , Thenault, C. , Nogué, F. , Perrot, L. , Mazier, M. & Gallois, J.L. (2022) CRISPR‐based knock‐out of eIF4E2 in a cherry tomato background successfully recapitulates resistance to pepper veinal mottle virus. Plant Science, 316, 111160. [DOI] [PubMed] [Google Scholar]
- Lefeuvre, P. , Martin, D.P. , Elena, S.F. , Shepherd, D.N. , Roumagnac, P. & Varsani, A. (2019) Evolution and ecology of plant viruses. Nature Reviews Microbiology, 17, 632–644. [DOI] [PubMed] [Google Scholar]
- Lefkowitz, E.J. , Dempsey, D.M. , Hendrickson, R.C. , Orton, R.J. , Siddell, S.G. & Smith, D.B. (2017) Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Research, 46(D1), D708–D717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.Y. , Cheng, Q.X. , Li, X.Y. , Zhang, Z.L. , Gao, S. , Cao, R.B. et al. (2018) CRISPR‐Cas12a‐assisted nucleic acid detection. Cell Discovery, 4, 18–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo, J.A. (2007) TRBO: a high‐efficiency tobacco mosaic virus RNA‐based overexpression vector. Plant Physiology, 145, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Li, X. , Ma, J. , Li, Z. , You, L. , Wang, J. et al. (2017) The molecular architecture for RNA‐guided RNA cleavage by Cas13a. Cell, 170, 714–726.e10. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Soyars, C.L. , Li, J. , Fei, Q. , He, G. , Peterson, B.A. et al. (2018) CRISPR/Cas9‐mediated resistance to cauliflower mosaic virus. Plant Direct, 2, e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff, G.P. (1995) Pathogen‐derived resistance to plant viruses. Annual Review of Phytopathology, 33, 323–343. [DOI] [PubMed] [Google Scholar]
- Ma, H. , Tu, L.‐C. , Naseri, A. , Huisman, M. , Zhang, S. , Grunwald, D. et al. (2016) Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nature Biotechnology, 34, 528–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei, A. , Sevilla, N.R. , Cantos, C. , Jonson, G.B. , Slamet‐Loedin, I. , Čermák, T. et al. (2018) Novel alleles of rice eIF4G generated by CRISPR/Cas9‐targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnology Journal, 16, 1918–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahas, A. , Aman, R. & Mahfouz, M. (2019) CRISPR‐Cas13d mediates robust RNA virus interference in plants. Genome Biology, 20, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova, K.S. , Wolf, Y.I. , Alkhnbashi, O.S. , Costa, F. , Shah, S.A. , Saunders, S.J. et al. (2015) An updated evolutionary classification of CRISPR‐Cas systems. Nature Reviews Microbiology, 13, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhotenko, A.V. , Khromov, A.V. , Snigir, E.A. , Makarova, S.S. , Makarov, V.V. , Suprunova, T.P. et al. (2019) Functional analysis of coilin in virus resistance and stress tolerance of potato Solanum tuberosum using CRISPR‐Cas9 editing. Doklady. Biochemistry and Biophysics, 484, 88–91. [DOI] [PubMed] [Google Scholar]
- Mäkinen, K. (2020) Plant susceptibility genes as a source for potyvirus resistance. Annals of Applied Biology, 176, 122–129. [Google Scholar]
- Mehta, D. , Stürchler, A. , Anjanappa, R.B. , Zaidi, S.S.E.A. , Hirsch‐Hoffmann, M. , Gruissem, W. et al. (2019) Linking CRISPR‐Cas9 interference in cassava to the evolution of editing‐resistant geminiviruses. Genome Biology, 20, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz, J. , Modrzejewski, D. , Hartung, F. , Wilhelm, R. & Sprink, T. (2020) Genome edited crops touch the market: a view on the global development and regulatory environment. Frontiers in Plant Science, 11, 586027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla, K.A. & Yang, Y. (2020) Predicting CRISPR/Cas9‐induced mutations for precise genome editing. Trends in Biotechnology, 38, 136–141. [DOI] [PubMed] [Google Scholar]
- Mushtaq, M. , Bhat, J.A. , Mir, Z.A. , Sakina, A. , Ali, S. , Singh, A.K. et al. (2018) CRISPR/Cas approach: a new way of looking at plant‐abiotic interactions. Journal of Plant Physiology, 224–225, 156–162. [DOI] [PubMed] [Google Scholar]
- Nelles, D.A. , Fang, M.Y. , O'Connell, M.R. , Xu, J.L. , Markmiller, S.J. , Doudna, J.A. et al. (2016) Programmable RNA tracking in live cells with CRISPR/Cas9. Cell, 165, 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nester, E.W. (2015) Agrobacterium: nature's genetic engineer. Frontiers in Plant Science, 5, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise, V. (2014) Crop immunity against viruses: outcomes and future challenges. Frontiers in Plant Science, 5, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nidhi, S. , Anand, U. , Oleksak, P. , Tripathi, P. , Lal, J.A. , Thomas, G. et al. (2021) Novel CRISPR–Cas systems: an updated review of the current achievements, applications, and future research perspectives. International Journal of Molecular Sciences, 22, 3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig, P.M. & Marraffini, L.A. (2020) Molecular mechanisms of CRISPR‐Cas immunity in bacteria. Annual Review of Genetics, 54, 93–120. [DOI] [PubMed] [Google Scholar]
- O'Connell, M.R. (2019) Molecular mechanisms of RNA targeting by Cas13‐containing type VI CRISPR–Cas systems. Journal of Molecular Biology, 431, 66–87. [DOI] [PubMed] [Google Scholar]
- O'Connell, M.R. , Oakes, B.L. , Sternberg, S.H. , East‐Seletsky, A. , Kaplan, M. & Doudna, J.A. (2014) Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature, 516, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerke, E.C. & Dehne, H.W. (2004) Safeguarding production – losses in major crops and the role of crop protection. Crop Protection, 23, 275–285. [Google Scholar]
- Price, A.A. , Sampson, T.R. , Ratner, H.K. , Grakoui, A. & Weiss, D.S. (2015) Cas9‐mediated targeting of viral RNA in eukaryotic cells. Proceedings of the National Academy of Sciences of the United States of America, 112, 6164–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyott, D.E. , Sheehan, E. & Molnar, A. (2016) Engineering of CRISPR/Cas9‐mediated potyvirus resistance in transgene‐free Arabidopsis plants. Molecular Plant Pathology, 17, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A. , Hsu, P.D. , Wright, J. , Agarwala, V. , Scott, D.A. & Zhang, F. (2013) Genome engineering using the CRISPR‐Cas9 system. Nature Protocols, 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronde, D.d. , Butterbach, P. & Kormelink, R. (2014) Dominant resistance against plant viruses. Frontiers in Plant Science, 5, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck, M.J. (2003) Plant RNA virus evolution. Current Opinion in Microbiology, 6, 406–409. [DOI] [PubMed] [Google Scholar]
- Roy, A. , Zhai, Y. , Ortiz, J. , Neff, M. , Mandal, B. , Mukherjee, S.K. et al. (2019) Multiplexed editing of a begomovirus genome restricts escape mutant formation and disease development. PLoS One, 14, e0223765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybicki, E.P. (2019) CRISPR‐Cas9 strikes out in cassava. Nature Biotechnology, 37, 727–728. [DOI] [PubMed] [Google Scholar]
- Sampson, T.R. , Saroj, S.D. , Llewellyn, A.C. , Tzeng, Y.L. & Weiss, D.S. (2013) A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature, 497, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, J.D. & Joung, J.K. (2014) CRISPR‐Cas systems for editing, regulating and targeting genomes. Nature Biotechnology, 32, 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhya, D. , Jogam, P. , Allini, V.R. , Abbagani, S. & Alok, A. (2020) The present and potential future methods for delivering CRISPR/Cas9 components in plants. Journal of Genetic Engineering & Biotechnology, 18, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford, J.C. & Johnston, S.A. (1985) The concept of parasite‐derived resistance‐deriving resistance genes from the parasite's own genome. Journal of Theoretical Biology, 113, 395–405. [Google Scholar]
- van Schie, C.C.N. & Takken, F.L.W. (2014) Susceptibility genes 101: how to be a good host. Annual Review of Phytopathology, 52, 551–581. [DOI] [PubMed] [Google Scholar]
- Shan, Q. , Wang, Y. , Chen, K. , Liang, Z. , Li, J. , Zhang, Y. et al. (2013) Rapid and efficient gene modification in rice and Brachypodium using TALENs. Molecular Plant, 6, 1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, V.K. , Marla, S. , Zheng, W. , Mishra, D. , Huang, J. , Zhang, W. et al. (2022) CRISPR guides induce gene silencing in plants in the absence of Cas. Genome Biology, 23, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov, S. , Abudayyeh, O.O. , Makarova, K.S. , Wolf, Y.I. , Gootenberg, J.S. , Semenova, E. et al. (2015) Discovery and functional characterization of diverse class 2 CRISPR‐Cas systems. Molecular Cell, 60, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov, S. , Smargon, A. , Scott, D. , Cox, D. , Pyzocha, N. , Yan, W. et al. (2017) Diversity and evolution of class 2 CRISPR‐Cas systems. Nature Reviews Microbiology, 15, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon, A.A. , Cox, D.B.T. , Pyzocha, N.K. , Zheng, K. , Slaymaker, I.M. , Gootenberg, J.S. et al. (2017) Cas13b is a type VI‐B CRISPR‐associated RNA‐guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Molecular Cell, 65, 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, G. , Jia, M. , Chen, K. , Kong, X. , Khattak, B. , Xie, C. et al. (2016) CRISPR/Cas9: a powerful tool for crop genome editing. The Crop Journal, 4, 75–82. [Google Scholar]
- Sonoda, E. , Hochegger, H. , Saberi, A. , Taniguchi, Y. & Takeda, S. (2006) Differential usage of non‐homologous end‐joining and homologous recombination in double strand break repair. DNA Repair, 5, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Sun, H. , Shen, L. , Qin, Y. , Liu, X. , Hao, K. , Li, Y. et al. (2018) CLC‐Nt1 affects potato virus Y infection via regulation of endoplasmic reticulum luminal Ph. New Phytologist, 220, 539–552. [DOI] [PubMed] [Google Scholar]
- Taliansky, M. , Samarskaya, V. , Zavriev, S.K. , Fesenko, I. , Kalinina, N.O. & Love, A.J. (2021) RNA‐based technologies for engineering plant virus resistance. Plants, 10, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashkandi, M. , Ali, Z. , Aljedaani, F. , Shami, A. & Mahfouz, M.M. (2018) Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signaling & Behavior, 13, e1525996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazza, M. , Lucioli, A. & Ilardi, V. (2017) Gene silencing provides efficient protection against plant viruses. In: Dalmay, T. (Ed.) Plant gene silencing: mechanisms and applications. Wallingford: CABI, pp. 193–205. [Google Scholar]
- Tenllado, F. (2004) RNA interference as a new biotechnological tool for the control of virus diseases in plants. Virus Research, 102, 85–96. [DOI] [PubMed] [Google Scholar]
- Tripathi, J.N. , Ntui, V.O. , Ron, M. , Muiruri, S.K. , Britt, A. & Tripathi, L. (2019) CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Communications Biology, 2, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, S.Q. & Joung, J.K. (2016) Defining and improving the genome‐wide specificities of CRISPR–Cas9 nucleases. Nature Reviews Genetics, 17, 300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniyal, A.P. , Mansotra, K. , Yadav, S.K. & Kumar, V. (2019) An overview of designing and selection of sgRNAs for precise genome editing by the CRISPR‐Cas9 system in plants. 3 Biotech, 9, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Oost, J. , Westra, E.R. , Jackson, R.N. & Wiedenheft, B. (2014) Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nature Reviews Microbiology, 12, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varanda, C.M. , Félix, M.D.R. , Campos, M.D. , Patanita, M. & Materatski, P. (2021) Plant viruses: from targets to tools for CRISPR. Viruses, 13, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillet, F. , Perrot, L. , Chauvin, L. , Kermarrec, M.‐P. , Guyon‐Debast, A. , Chauvin, J.‐E. et al. (2019) Transgene‐free genome editing in tomato and potato plants using Agrobacterium‐mediated delivery of a CRISPR/Cas9 cytidine base editor. International Journal of Molecular Sciences, 20, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Hardcastle, T.J. , Pastor, A.C. , Yip, W.H. , Tang, S. & Baulcombe, D.C. (2018) A novel DCL2‐dependent miRNA pathway in tomato affects susceptibility to RNA viruses. Genes & Development, 32, 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, H.‐H. , Méndez‐Mancilla, A. , Guo, X. , Legut, M. , Daniloski, Z. & Sanjana, N.E. (2020) Massively parallel Cas13 screens reveal principles for guide RNA design. Nature Biotechnology, 38, 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S. , Dinesh‐Kumar, S.P. , Choi, D. , Hehl, R. , Corr, C. & Baker, B. (1994) The product of the tobacco mosaic virus resistance gene N: similarity to Toll and the interleukin‐1 receptor. Cell, 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Whitham, S. , McCormick, S. & Baker, B. (1996) The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proceedings of the National Academy of Sciences of the United States of America, 93, 8776–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T.M.A. (1993) Strategies to protect crop plants against viruses: pathogen‐derived resistance blossoms. Proceedings of the National Academy of Sciences of the United States of America, 90, 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- :Wolt, J.D. , Wang, K. , Sashital, D. & Lawrence‐Dill, C.J. (2016) Achieving plant CRISPR targeting that limits off‐target effects. The Plant Genome, 9. 10.3835/plantgenome2016.05.0047 [DOI] [PubMed] [Google Scholar]
- Woo, J.W. , Kim, J. , Kwon, S.I. , Corvalán, C. , Cho, S.W. , Kim, H. et al. (2015) DNA‐free genome editing in plants with preassembled CRISPR‐Cas9 ribonucleoproteins. Nature Biotechnology, 33, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Xie, K. & Yang, Y. (2013) RNA‐guided genome editing in plants using a CRISPR‐Cas system. Molecular Plant, 6, 1975–1983. [DOI] [PubMed] [Google Scholar]
- Yadav, S. & Chhibbar, A.K. (2018) Plant–virus interactions. In: Singh, A. & Singh, I. (Eds.) Molecular aspects of plant‐pathogen interaction. Singapore: Springer Singapore, pp. 43–77. [Google Scholar]
- Yan, M.‐Y. , Yan, H. , Ren, G. , Zhao, J. , Guo, X. & Sun, Y.‐C. (2017) CRISPR‐Cas12a‐assisted recombineering in bacteria. Applied and Environmental Microbiology, 83, e00947‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, W.X. , Chong, S. , Zhang, H. , Makarova, K.S. , Koonin, E.V. , Cheng, D.R. et al. (2018) Cas13d is a compact RNA‐targeting type VI CRISPR effector positively modulated by a WYL‐domain‐containing accessory protein. Molecular Cell, 70, 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, K. , Han, T. , Liu, G. , Chen, T. , Wang, Y. , Yu, A.Y.L. et al. (2015) A geminivirus‐based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Scientific Reports, 5, 14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, K. , Han, T. , Xie, K. , Zhao, J. , Song, J. & Liu, Y. (2019) Engineer complete resistance to Cotton Leaf Curl Multan virus by the CRISPR/Cas9 system in Nicotiana benthamiana . Phytopathology Research, 1, 9. [Google Scholar]
- Yin, K. & Qiu, J.‐L. (2019) Genome editing for plant disease resistance: applications and perspectives. Philosophical Transactions of the Royal Society B: Biological Sciences, 374, 20180322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, Y.‐J. , Venkatesh, J. , Lee, J.‐H. , Kim, J. , Lee, H.‐E. , Kim, D.‐S. et al. (2020) Genome editing of eIF4E1 in tomato confers resistance to pepper mottle virus. Frontiers in Plant Science, 11, 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Wang, X. , Sun, H. , Liang, Q. , Wang, W. , Zhang, C. et al. (2020) Improving CRISPR‐Cas‐mediated RNA targeting and gene editing using SPLCV replicon‐based expression vectors in Nicotiana benthamiana . Plant Biotechnology Journal, 18, 1993–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, J.‐J. , Hong, C.‐Y. , Wei, P. , Tsai, Y.‐C. & Lin, C.‐S. (2020) How to start your monocot CRISPR/Cas project: plasmid design, efficiency detection, and offspring analysis. Rice, 13, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafirov, D. , Giovinazzo, N. , Bastet, A. & Gallois, J. (2021) When a knockout is an Achilles' heel: resistance to one potyvirus species triggers hypersusceptibility to another one in Arabidopsis thaliana . Molecular Plant Pathology, 22, 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi, S.S. & Mansoor, S. (2017) Viral vectors for plant genome engineering. Frontiers in Plant Science, 8, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi, S.S.E.A. , Mahfouz, M.M. & Mansoor, S. (2017) CRISPR‐Cpf1: a new tool for plant genome editing. Trends in Plant Science, 22, 550–553. [DOI] [PubMed] [Google Scholar]
- Zetsche, B. , Gootenberg, J.S. , Abudayyeh, O.O. , Slaymaker, I.M. , Makarova, K.S. , Essletzbichler, P. et al. (2015) Cpf1 is a single RNA‐guided endonuclease of a class 2 CRISPR‐Cas system. Cell, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, X. , Zhang, F. , Zhong, Z. , Chen, R. , Wang, Y. , Chang, L. et al. (2019) Generation of virus‐resistant potato plants by RNA genome targeting. Plant Biotechnology Journal, 17, 1814–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, F. , Li, X. , Baller, J.A. , Qi, Y. , Starker, C.G. et al. (2012) Transcription activator‐like effector nucleases enable efficient plant genome engineering. Plant Physiology, 161, 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.‐H. , Tee, L.Y. , Wang, X.‐G. , Huang, Q.‐S. & Yang, S.‐H. (2015) Off‐target effects in CRISPR/Cas9‐mediated genome engineering. Molecular Therapy‐Nucleic Acids, 4, e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Konermann, S. , Brideau, N.J. , Lotfy, P. , Wu, X. , Novick, S.J. et al. (2018) Structural basis for the RNA‐guided ribonuclease activity of CRISPR‐Cas13d. Cell, 175, 212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Zheng, Q. , Yi, X. , An, H. , Zhao, Y. , Ma, S. et al. (2018) Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnology Journal, 16, 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Ye, Y. , Ye, W. , Perčulija, V. , Jiang, H. , Chen, Y. et al. (2019) Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d. Nature Communications, 10, 2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Zhao, Y. , Ye, J. , Cao, X. , Xu, C. , Chen, B. et al. (2019) Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnology Journal, 17, 1185–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Du, H. , Wang, J. , Pu, Y. , Yang, C. , Yan, R. et al. (2020) Multiplex CRISPR/Cas9‐mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnology Journal, 18, 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Yang, X. , Zhou, G. & Zhang, T. (2019) Engineering plant virus resistance: from RNA silencing to genome editing strategies. Plant Biotechnology Journal, 18, 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. , Li, C. & Gao, C. (2020) Applications of CRISPR–Cas in agriculture and plant biotechnology. Nature Reviews Molecular Cell Biology, 21, 661–677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 The three stages of CRISPR/Cas adaptive immunity in bacterial cells. (i) The first process, termed adaptation, is where the foreign genetic material is acquired by Cas1‐Cas2 and integrated into the CRISPR array. (ii) The associated Cas proteins are expressed and the CRISPR array is processed into mature crRNA to provide targeting specificity. (iii) The crRNA guides the effector nucleases to foreign genetic elements, leading to target interference and immunity. This figure was created using Biorender
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed.


