Abstract
The nitrogen cycle plays a major role in aquatic nitrogen transformations, including in the terrestrial subsurface. However, the variety of transformations remains understudied. To determine how nitrogen cycling microorganisms respond to different aquifer chemistries, we sampled groundwater with varying nutrient and oxygen contents. Genes and transcripts involved in major nitrogen-cycling pathways were quantified from 55 and 26 sites, respectively, and metagenomes and metatranscriptomes were analyzed from a subset of oxic and dysoxic sites (0.3-1.1 mg/L bulk dissolved oxygen). Nitrogen-cycling mechanisms (e.g. ammonia oxidation, denitrification, dissimilatory nitrate reduction to ammonium) were prevalent and highly redundant, regardless of site-specific physicochemistry or nitrate availability, and present in 40% of reconstructed genomes, suggesting that nitrogen cycling is a core function of aquifer communities. Transcriptional activity for nitrification, denitrification, nitrite-dependent anaerobic methane oxidation and anaerobic ammonia oxidation (anammox) occurred simultaneously in oxic and dysoxic groundwater, indicating the availability of oxic-anoxic interfaces. Concurrent activity by these microorganisms indicates potential synergisms through metabolite exchange across these interfaces (e.g. nitrite and oxygen). Fragmented denitrification pathway encoding and transcription was widespread among groundwater bacteria, although a considerable proportion of associated transcriptional activity was driven by complete denitrifiers, especially under dysoxic conditions. Despite large differences in transcription, the capacity for the final steps of denitrification was largely invariant to aquifer conditions, and most genes and transcripts encoding N2O reductases were the atypical Sec-dependant type, suggesting energy-efficiency prioritization. Results provide insights into the capacity for cooperative relationships in groundwater communities, and the richness and complexity of metabolic mechanisms leading to the loss of fixed nitrogen.
Subject terms: Freshwater ecology, Environmental chemistry, Microbial communities, Water microbiology
Introduction
Groundwater represents the largest accessible freshwater source on Earth and is stored in permeable geological units known as aquifers that are generally characterized by long water residence times, low organic matter, and slow water exchange rates [1–3]. Natural stores of inorganic nitrogen (nitrate, nitrite, ammonium) are typically present in low concentrations [4]. However, long residence times and close links to surface water (e.g., lakes, rivers, and wetlands) make groundwater susceptible to pollution from nitrogen-based fertilisers [5]. Nitrogen (N) contamination in the terrestrial subsurface has become a global problem [6–8], presenting health risks associated with nitrate in drinking water, such as methaemoglobinaemia and cancer [9], along with eutrophication of surface waters [10].
Groundwater microbial communities contain extensive phylogenetic novelty [11]. While the metabolic capabilities of many groundwater microorganisms remain to be tested in laboratory conditions, genetic evidence suggests that a diverse collection of bacteria and archaea transform nitrogen in groundwater, including novel candidate phyla [12], diverse bacterial taxa [11, 13], archaeal ammonium oxidizers [13, 14], and novel aquifer-adapted clades of anammox bacteria [15]. Research also suggests that numerous aquifer organisms are equipped with genes encoding partial nitrogen cycle pathways, such as nitrite reduction [11, 16]. This has been shown to be a common feature of bacteria and archaea from other habitats [17], and suggests that cooperative interactions are commonly employed to complete individual nitrogen-cycling pathways.
The microbial nitrogen cycle comprises six distinct N-transformation processes, including ammonification, nitrogen fixation, nitrification, denitrification, anaerobic ammonium oxidation (anammox), and assimilation [18, 19]. Microorganisms that perform these processes can be sources and sinks of nitrate. Despite distinct requirements (e.g. for oxygen), many reactions in the nitrogen cycle tend to co-occur in the environment, leading to efficient nitrogen recycling [19], competition for the same resource (e.g. by respiratory ammonifiers and denitrifiers [20]), cooperative completion of the modular denitrification pathway [19], and coupled processes, such as nitrification—denitrification [21] or nitrification—anammox [22]. Accordingly, biological processes derived from networks of microorganisms in the terrestrial subsurface play a dominant role in N-transformations [23].
Denitrification is the most studied nitrogen cycling process in groundwater to-date, due to its importance for nitrogen pollution removal [5], although operation of the truncated pathway produces less desirable forms of inorganic nitrogen - NO2- due to its toxicity [24], and greenhouse gases NO and N2O [5]. Microbial denitrification is typically linked to dissolved organic carbon concentrations in aquifers, but is also fuelled by inorganic electron donors, such as reduced forms of iron or sulfur [5, 25]. Inorganic donors may be the primary source of electrons for nitrogen-cycling taxa given widespread organic carbon-limitation in aquifers [26, 27]. Accordingly, nitrification and anammox appear to be typical features of shallow oxic or partially oxic aquifers [15, 26], and carbon-limitation can create an opportunity for anammox to outcompete denitrification [28]. However, the occurrence of, or capacity for, these processes may not to be ubiquitous. A scarcity or lack of organisms capable of some processes, including nitrogen fixation, ammonia oxidation, and nitrous oxide reduction, has been reported from one low-oxygen aquifer [11]. Further work is needed to determine the distribution of nitrogen-cycling processes across different aquifers, including aquifers defined based on redox conditions and nutrient characteristics, such as pristine or N-contaminated.
This study investigates the microbial nitrogen cycle in aquifers traversing a wide range of nitrogen, organic carbon, and oxygen concentrations. We determined the metabolic capacity for each pathway in oxic and dysoxic groundwaters, and the transcriptional activity associated with these pathways (understudied in aquifers due to low cell densities) [29]. As aquifers comprise both suspended and attached communities, with distinct compositions and capacities for biogeochemical cycling [30], analyses included both groundwater (planktonic fraction) and groundwater enriched with the sediment-attached fraction. To further characterize reactions leading to nitrogen loss, we quantified ammonia monooxygenase (archaeal and bacterial ammonia oxidation), nitrous oxide reductase (final step in denitrification), and hydrazine synthase (anammox) genes in 64 groundwater samples (from 59 wells) and transcripts in 26, collected up to 860 km apart. Results give insights into environmental factors influencing the presence, co-occurrence, and transcriptional activity of nitrogen-cycling mechanisms, which determine the fate of nitrogen in aquifers.
Materials and methods
Study sites and sample collection
Eighty samples were collected from 59 wells, spanning 10 aquifers (mostly sandy-gravel) in the Auckland, Waikato, Wellington, and Canterbury regions, New Zealand (Fig. S1; Table S1). Wells were 4.5–114.6 m deep (18.9 m on average, Table S1). Wells were purged (~3–5 borehole volumes). Then, 3–90 L of groundwater (67 samples) or 0.5–15 L of attached-fraction enriched groundwater (13 samples, Canterbury sites A–D) were collected and immediately filtered on-site. The biofilm or “attached” fraction enriched groundwater (i.e combining planktonic and biofilm aquifer fractions) was collected directly following standard groundwater collection. Prior to collection of these samples, a low-frequency custom sonicator, as described by Close et al. [31] (2.43 kW) was applied for 2 min to detach biofilms and particles from the surrounding aquifer. Biomass was captured onto 142 mm diameter mixed cellulose ester membrane filters (1.2 µm pore size pre-filter over a 0.22 µm filter) using a 142 mm stainless steel filter holder (Merck Millipore Ltd, Cork, Ireland). Both filters were immediately submerged in RNAlater (ThermoFisher Scientific, Waltham, MA, USA), transported on dry ice, and stored at −80 °C. All samples were used to generate amplicon data (×80). A subset was used to quantify functional genes (×64) and transcripts (×26). A subset from Canterbury was used for metagenomics (×16) and metatranscriptomics (×6).
Dissolved oxygen (DO), water temperature, pH, oxidation-reduction potential (ORP), and specific conductance were collected on site using flow-through cell and field probes (YSI EXO sonde 2, YSI PRO+ and YSI ProDSS, Yellow Springs, OH, USA). Samples were grouped into categories based on DO concentrations: anoxic (0 mg/L), suboxic (<0.3 mg/L), dysoxic (0.3–3 mg/L or 9.4–93.8 μM), and oxic (>3 mg/L) as previously proposed for groundwater [32]. Unfiltered groundwater samples were analyzed for P, N, C, S, Fe, Cu, and alkalinity at Hill Laboratories (Hamilton, New Zealand; Supplementary Information).
Nucleic acid extraction, sequencing and genome assembly
Nucleic acid extraction for droplet digital PCR (ddPCR), 16S rRNA gene amplicons, metagenomes, metatranscriptomes, sequencing, metagenome-assembled genome generation, and transcript mapping, are as described previously [15], and detailed in the Supplementary Information. Only RNA samples with RIN ≥ 6 or DV200 > 30% (i.e. fragments >200 nucleotides) were used for downstream analyses. Metagenome-assembled genome (MAG) completeness and contamination were estimated using CheckM v1.0.12 [33], and MAGs were classified using the Genome Taxonomy Database taxonomic classification tool, GTDB-Tk v0.2.1 [34]. Metatranscriptomic reads were mapped to MAGs using Bowtie2 [35] (v2.3.5, --end-to-end --very_sensitive). Read counts were determined using featureCounts [36] (v1.6.3, -F SAF). Singleton mapped reads per gene were removed. Read counts were normalized to a modified version of transcripts per kilobase per million reads mapped (modified-TPM) [37] via (number of reads mapped to gene)*(1000/gene length)*(1000000/library size).
Quantitative PCR
Droplet digital PCR (ddPCR) of hydrazine synthase (hzsB), ammonia monoxygenase (amoA), nosZ clade I genes, and transcripts used the QX200 platform with 20 µl reactions, 10 µl 2× EvaGreen Supermix (Biorad, Hercules, CA, US), and 1 µl DNA or cDNA. RNA (24 pg–1.1 µg) was converted to cDNA using Superscript III Supermix (Invitrogen). DPEC-treated water was used for negative controls and gBlock dsDNA fragments were used as positive controls (Table S2; Integrated DNA Technologies, Coralville, IA, USA). Primers and PCR conditions are described in Table S2. Data were analyzed using the QuantaSoft software package v1.0 (Bio-Rad). Positive droplet thresholds were set based on negative and positive droplet fluorescence amplitudes using the positive control as reference.
Metabolic predictions
Protein-coding gene sequences from MAGs and metagenomic reads were predicted, and annotated (Supplementary Information Table S3). Phylogenetic trees for HzsABC (see figure 8 in Mosley et al. [15]), AmoA, PmoA, NosZ, and NxrA were generated by aligning predicted protein (amino acid) sequences using MUSCLE with default parameters [38], and constructing trees using FastTree (for AmoA) or IQ-TREE (for others) with 1000 bootstraps [39, 40]. IslandViewer4 [41] was used to identify genomic islands in MAG nzgw5.
Statistical analyses
Significant environmental factors were determined using R package vegan v2.5.6 functions (metaMDS, adonis, vegdist, and envfit) [42]. Bray-Curtis dissimilarities were constructed using vegan with relative abundances of MAGs or protein-coding sequences in metagenomic reads [42]. Heatmap Z-scores were calculated using Heatmap.2 from the gplots package [43]. Linear discriminant analysis and Kruskal–Wallis tests were determined using the Galaxy computational tool (http://huttenhower.sph.harvard.edu/galaxy/). Adonis permutation tests were undertaken using Bray-Curtis dissimilarities. Spearman’s rank correlations (r) were calculated using ‘rcorr’ from the Hmisc package, and p values were adjusted using the Benjamini-Hochberg method. Statistical analyses were considered significant with p < 0.05.
Results and discussion
Distribution of nitrogen-cycling pathways in groundwater
Differences in nitrogen-cycling processes based on oxygen and nitrate concentrations
Sixteen metagenomes (Table S4) were obtained from duplicate wells at four sites (A–D) from two unconfined alluvial aquifers (Canterbury, Fig. S1). These sites encompassed varied nitrate (0.45–12.6 g/m3), DO (0.37–7.5 mg/L), and dissolved organic carbon (DOC) (0–26 g/m3) concentrations (Fig. 1A; Table S1). Nitrate concentrations were pristine (site C) to N-contaminated (sites A, B, D) [4]. Sites A–C were oxic and had low DOC (typical of groundwaters), whereas site D was dysoxic with relatively high DOC. Metagenomes from groundwater wells comprised pairs, representing the planktonic and sediment-attached fractions. Over 70 Gbp of raw sequence was generated per site (390 Gbp overall, 322 Gbp trimmed). However, <10% of trimmed reads per sample assembled into contigs >2Kb long and only 0.64–8.14% of reads (3.8% on average) mapped to MAGs (Table S4), reflecting the complexity of microbial communities in the terrestrial subsurface [11]. To capture this diversity, metagenomic reads are first used here to determine the distribution of N metabolisms.
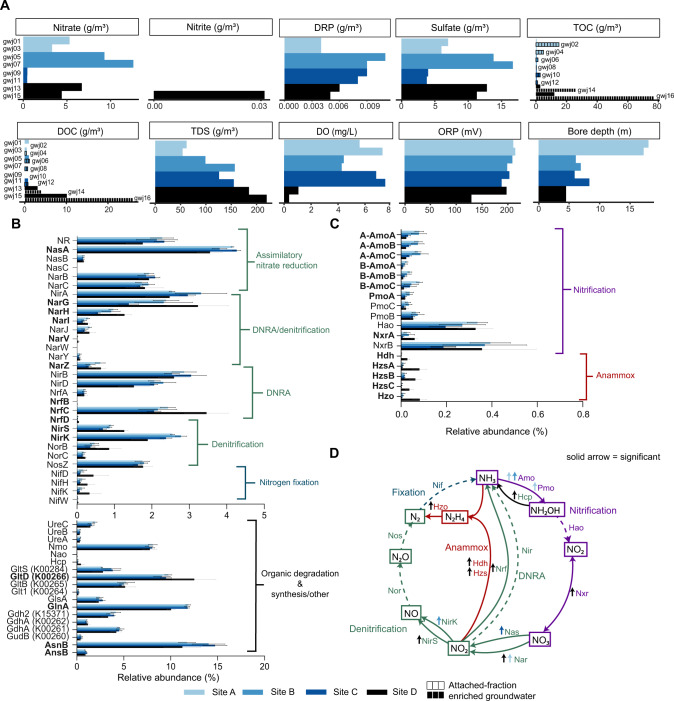
Fig. 1. Geochemistry and protein-coding sequences (based on reads) involved in nitrogen cycling that are significantly different among sites used for metagenomics.
A Bar plots showing geochemical data from groundwater samples, coloured according to site. Solid bar colour = groundwater samples. Grid lines = attached-fraction enriched groundwater. All samples from site D were characterized as dysoxic, although gwj15-16 contained 0.37 mg/L DO, which are near suboxic levels (i.e. <0.3 mg/L). For all samples shown, ammoniacal-N values were below detection. B, C Bar plots showing the abundance (average of four wells per site and standard deviation) of sequence reads encoding dissimilatory and assimilatory nitrogen-cycling proteins relative to all nitrogen-cycling processes. Predicted proteins that were statistically different between sites are bolded (y-axis). D Schematic of the nitrogen cycle displaying statistically significant differences between sites (LEfSe, Kruskal–Wallis test, p < 0.05). Solid lines depict pathways that were significantly more abundant across the sites, whereas dashed lines indicate no significant difference. Arrows indicate the site with significantly more genes. Abbreviations: A-Amo Archaeal Amo, B-Amo Bacterial Amo, Amo ammonia monooxygenase, Pmo Particulate methane monooxygenase, Hao hydroxylamine oxidoreductase, Nxr nitrite oxidoreductase, Nar nitrate reductase (dissimilatory), Nas nitrate reductase (assimilatory), NirK copper-containing nitrite reductase, NirS cytochrome cd1-containing nitrite reductase, Nor nitric oxide reductase, Nos nitrous oxide reductase, Nif nitrogenase (various), Hcp hydroxylamine reductase, Nir NADPH-nitrite reductase, Nrf nitrate reductase (associated with Nap), Hdh hydrazine hydrogenase, Hzs hydrazine synthase, Hzo hydrazine oxidoreductase.
A total of 4,462,950 sequences encoding nitrogen-cycling proteins were identified from metagenomic reads (1,013,871–1,211,591/site, Table S5). The relative abundance of 29/68 nitrogen-cycling gene (sub)families in NCycDB [44] differed significantly among sites (Fig. 1B, C; LEfSe, Table S6). Most of these encoded nitrification and anammox pathways (p < 0.05). Sequences encoding other pathways, such as nitrogen fixation and the final steps of denitrification remained consistent across sites (p > 0.05).
Results indicate the presence of ecologically robust mechanisms for nitrogen removal under the reduced DO conditions of dysoxic site D. In addition to the presence of metagenomic sequences encoding complete nitrification-denitrification, the relative abundances of sequences encoding anammox genes were all significantly higher at this site (LEfSe, Kruskal–Wallis test, p < 0.05). Sequences encoding genes involved in nitrate reduction to nitrite (narGHI), and those encoding mechanisms for nitrite reduction in the dissimilatory nitrate reduction to ammonia (DNRA) (nrfBCD), and denitrification (nirS) pathways were also higher. Anammox converts NH4+ and NO2- into N2 [45] and can be fuelled by ammonium replenished by DNRA, while anammox and denitrification (coupled to nitrification) represent alternative N2-generating pathways. Transcriptomic data showed a relatively high proportion of N-cycling gene expression derived from both anammox (hydrazine synthesis and oxidation) and denitrification (nitrous oxide reduction) in dysoxic groundwater (higher than that indicated by gene relative abundances), and showed that nitrifiers were transcriptionally active (Fig. 2A, B).
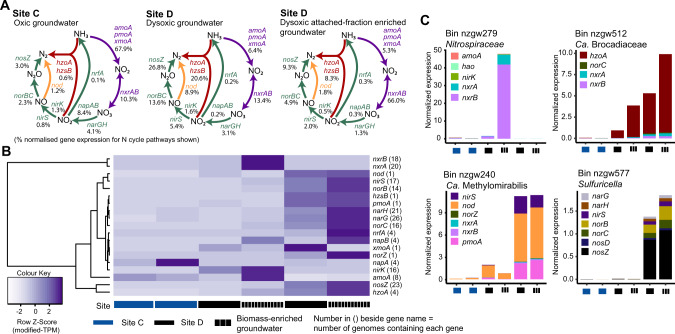
Fig. 2. Nitrogen-cycling gene transcription at site C groundwater and site D groundwater and attached-fraction enriched groundwater.
A Nitrogen cycle schematics display the average abundance of nitrogen-cycling transcripts (based on modified-TPM values) per site and sample type (relative to nitrogen-cycling pathways overall (as shown). The percentage of gene transcripts associated with each pathway component is shown in black font. Coloured arrows represent pathways (purple = nitrification, green = denitrification and red = anammox). Only NrfA and not NirBD are shown for the DNRA pathway. B Heatmap shows nitrogen-cycling modified transcripts per million (modified-TPM) at each site (ordered gwj9, gwj11, gwj13-gwj16), scaled by row (Z-Score). Solid coloured blocks represent groundwater, black grid blocks represent the attached-fraction (or biomass) enriched groundwater. C Stacked bar plots display four active nitrogen-cycling genomes and the relative abundance (modified-TPM normalized to genome coverage) of their nitrogen-cycling gene transcripts across each site. Abbreviations: amo ammonia monooxygenase, pmo particulate methane monooxygenase, xmo copper-containing membrane monooxygenase, nod nitric oxide dismutase, nxr nitrite oxidoreductase, nar nitrate reductase (dissimilatory), nap periplasmic nitrate reductase, nirK copper-containing nitrite reductase, nirS cytochrome cd1-containing nitrite reductase, nor nitric oxide reductase, nos nitrous oxide reductase, nrf nitrate reductase, hzo hydrazine oxidoreductase, hao hydroxylamine oxidoreductase.
Oxygen availability (oxic vs suboxic/anoxic) is linked to archaeal and bacterial ammonia-oxidizer abundance or absence in aquifers [11, 26]. As may be expected for an aerobic process, there were significantly more metagenomic sequences encoding the first step of nitrification (bacterial and archaeal amoABC) at oxic sites A and B (LEfSe, Kruskal–Wallis test), and some of the medium-high concentrations of nitrate at these sites is likely to have been re-generated by nitrifiers [19]. Regardless, our data indicate ammonia-oxidizers were present and transcriptionally active under all conditions, including dysoxic and pristine (low N and DOC, site C). AOA and AOB can be active at DO concentrations <1 mg/L [46] comparable to, or below, those at the dysoxic site in this study, and as may be found in oxygen-depleted niches within aquifer biofilms [47]. Although, under oxygen depleted conditions such as these, AOB could instead undertake nitrite-dependent ammonia oxidation (‘nitrifier denitrification’), generating N2O [48, 49].
Among non-assimilatory pathways, ammonia-oxidation prevailed in pristine groundwater. Site C contained significantly more nasA genes, which comprises part of the assimilatory nitrate reduction pathway (p < 0.05), along with genes involved in amino acid biosynthesis such as asparagine synthetase (asnB) and L-asparaginase II (ansB) [50]. This suggests site C had enhanced potential for N uptake and storage, likely due to the large investment required to scavenge nitrogen for cellular maintenance with low nitrate and ammonia concentrations [51].
Nitrogen cycling is a core function of groundwater microbiomes, but community compositions are site-specific
To link pathways to organisms we analyzed 396 non-redundant medium-high to high-quality MAGs [52]. We reconstructed 7695 metagenome-assembled genomes (MAGs) of which 626 were non-redundant (ANI threshold of 99%), and 396 of these were estimated to be 70–100% complete, with 0–5% contamination. Nitrogen-cycling genes, specifically those involved in non-assimilatory redox reactions, such as nitrification, anammox, and complete or incomplete DNRA and denitrification (starting from nitrate reduction), were present in 40% of MAGs (Figs. 3, 4A, and Table S7). The capacity for diverse nitrogen-cycling processes was again observed to be pervasive across sites, and the overall richness of taxa capable of nitrogen cycling remained comparably diverse over most sites (Fig. 5A), regardless of large differences in measured inorganic N contents (>10-fold difference in nitrate-N, site averages 0–0.007 gm3 nitrite-N and 0–0.018 g/m3 ammoniacal-N). Analysis of 16S rRNA gene amplicons across 59 groundwater wells likewise shows that taxa linked through phylogenetic affiliation to nitrogen-cycling processes comprise a notable fraction (0.3–26.3%) of complex oxic to anoxic groundwater communities (Fig. S2). Functional redundancy was common among N-cycling microorganisms. Multiple MAGs recovered from each sample had the collective capacity for DNRA and actively expressed genes associated with each of the major steps of nitrification and denitrification (Fig. 4B, C).
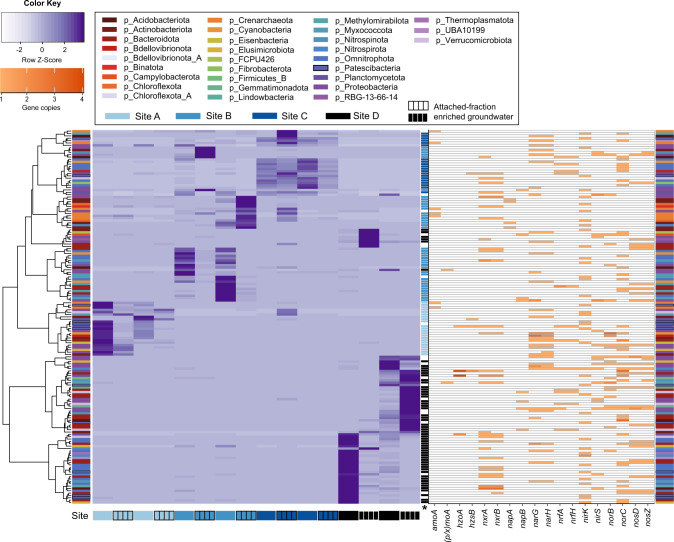
Fig. 3. Heatmap showing 159 MAGs, coloured according to phylum, that contain nitrogen-cycling genes involved in non-assimilatory reduction and oxidation of N species in groundwater.
Purple gradient (right) represents genome coverage scaled by row (Z-Score) across sites A–D (ordered gwj01-16). Rows = MAGs; columns = samples per site (groundwater and attached-fraction enriched groundwater). Orange gradient (right) represents number of nitrogen-cycling gene copies per genome. Microorganisms are ordered based on hierarchical clustering of abundance based Bray-Curtis dissimilarity matrix with ward.D2 clustering method. Final column (labelled with asterisk) indicates genomes that were significantly more abundant at a particular site (coloured rectangle) based on LEfSe analysis.
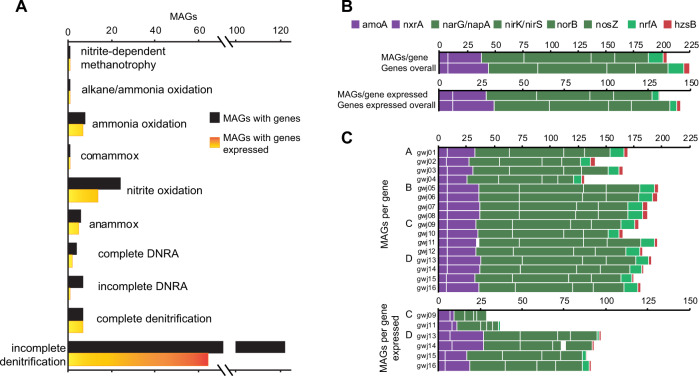
Fig. 4. Plots showing the distribution of N cycling mechanisms across MAGs, including DNRA and denitrification pathway fragmentation.
A Number of MAGs with genes or genes expressed per pathway, with indicative genes required for each step from nitrate to ammonia or N2 to denote complete DNRA or denitrification potential, respectively. B Number of MAGs with marker genes and marker genes overall for key N cycling processes: two steps for nitrification (purple), four steps for denitrification (dark green), DNRA (light green), and anammox (red). C Number of MAGs with at least one copy of each marker gene or marker gene expressed across groundwater samples and sites (A–D).
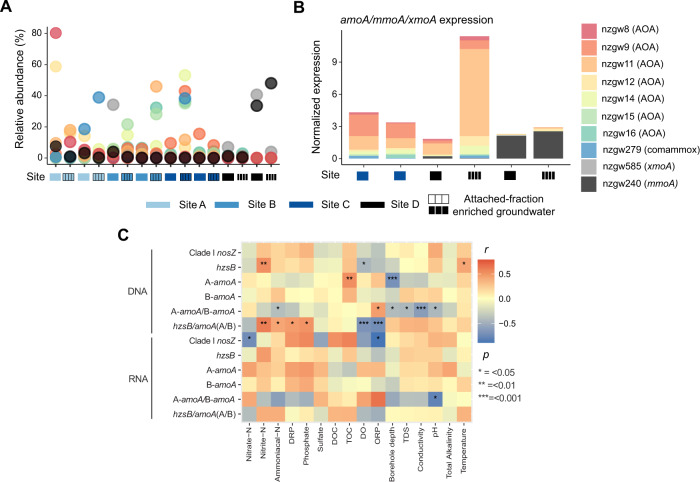
Fig. 5. Diversity of community fractions with N cycling capacity.
A Boxplots showing the median and interquartile range of richness of MAGs capable of nitrogen-cycling. Black solid circles show actual sample richness. Sites with significantly different richness are indicated by an overlying line and asterisk (Wilcoxon test, p < 0.05). B Non-metric Multi-dimensional Scaling plot showing groundwater community relatedness based on a Bray–Curtis dissimilarity matrix constructed using the relative abundance of MAGs with nitrogen-cycling genes. C Non-metric Multi-dimensional Scaling plot based on a Bray–Curtis dissimilarity matrix constructed using the relative abundance of protein-coding sequences in metagenomic reads. For ordinations environmental variables (Table S1) were fitted using envfit, and significant variables are indicated with an asterisk = p < 0.05. Ammoniacal-N and nitrite are not shown as >50% of values were below the detection limit and >50% of TKN values are missing.
Results suggest that nitrogen cycling is a core function of aquifer microbiomes, despite typically low levels of inorganic N in groundwater [4]. This conjecture is supported by prior evidence from ammonium- or nitrate-poor groundwater of microorganisms capable of, or actively engaged in, nitrification, anammox, or denitrification [25, 26, 28, 53, 54]. Microbial nitrogen cycling is likely to be a significant factor governing nitrogen availability in typically oligotrophic habitats, such as groundwater, the open ocean, and lakes. Indeed, in oligotrophic ocean waters with low primary production, the turnover of the dissolved inorganic nitrogen pool via microbial ammonium regeneration and nitrification is rapid [55]. Moreover, numerous microorganisms have evolved high affinities for nitrogen compounds, conferring them with competitive advantages under N-limited conditions [47, 56–59].
Analysis of the spatial distributions of MAGs showed distinct site-specific compositions of bacteria and archaea capable of nitrogen cycling (Fig. 3), a feature also observed in groundwater microbial communities as a whole [13]. Bray–Curtis dissimilarities, calculated based on relative abundance for MAGs and metagenomic reads encoding nitrogen-cycling proteins, revealed that spatial differences significantly influenced community composition (R2 = 0.49 for MAGs, R2 = 0.42 for metagenomic reads), more than DO, DOC, nitrate, and sample type (groundwater or attached-fraction enriched) (Fig. 5A, B; Table S8). Most MAGs inferred to undertake nitrogen cycling (87.4%) were significantly more abundant at a specific site (22–57 MAGs/site; LEfSe, Kruskal–Wallis test). Results therefore show site-specific environmental conditions drive species selection, and the capacity for certain reactions, such as ammonia oxidation (greater in oxic groundwater) or complete denitrification (greater in dysoxic groundwater) (Fig. S3).
Nitrifier diversity and activity in groundwater, and habitat specificity
Archaeal and bacterial ammonia-oxidizers exhibited distinct niche preferences, but higher similarity in transcriptional activity
Ammonia-oxidizing archaea (AOA) and bacteria (AOB) convert ammonia to nitrite using ammonia monooxygenase (Amo) and hydroxylamine oxidoreductase (Hao), and perform the rate-limiting step in nitrification [60]. Niche differentiation between the two domains is not clearly defined [57]. However, AOA usually have a higher affinity for ammonia than AOB [47, 49, 56], and typically outnumber AOB in oligotrophic environments with low ammonia concentrations and salinity, consistent with groundwater in this study (mean ammoniacal-N 0.36 g/m3 ± 1.4 SD; conductivity 220 µS/cm ± 142 SD). We found AOA and AOB exhibited distinct spatial trends in relative and absolute abundance, and activity related to various geochemical parameters in groundwater (e.g. ammonia, oxygen availability, and conductivity) (Fig. 6).
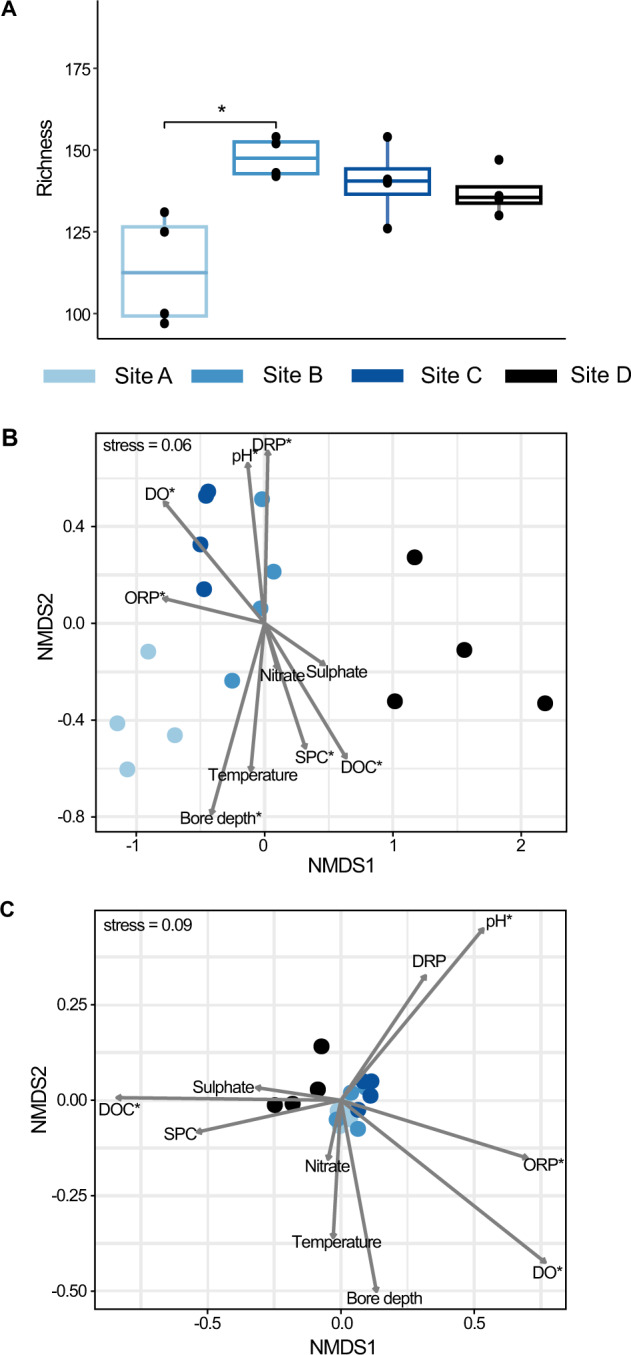
Fig. 6. Composition and transcriptional activity of ammonia-oxidizers, and relationship between ammonia-oxidizers, denitrifiers and anammox bacteria to geochemical and physical parameters.
A Relative abundance of each MAG capable of aerobic ammonia oxidation across the sites. B Stacked barplot showing modified transcripts per million, normalized to genome coverage, of amoA, pmoA and xmoA across sites (wells SR1, SR2, E1, N3). The 7 AOA are associated with 5 genera: Nitrosotenuis (nzgw11), UBA8516 (nzgw12–14), Nitrososphaera (nzgw16), Nitrosoarchaeum (nzgw8-9) and an unclassified genus in Nitrososphaeraceae (nzgw15). The xmoA is part of a gene cluster in MAG nzgw585, recovered from the dysoxic site and classified as Gammaproteobacteria genus Nevskia, which encodes a copper-containing membrane monooxygenase (CuMMO/xmoCAB). The alpha subunit had best hits to Polycyclovorans sp. SAT60 and gammaproteobacterial isolate MMS_B.mb.28 (85.71% amino acid identity, NCBI NR database). CuMMO catalyzes the oxidation of short-chain alkanes, ammonia or methane [103]. C Spearman’s rank correlations between the abundance (copies/L) of nitrogen-cycling genes (DNA, samples = 64) and transcripts (RNA, samples = 26 above detection limit) determined via ddPCR and geochemical parameters (Table S11). Significant correlations are indicated by * are based on Bonferroni adjusted p values (p).
Based on metagenomic reads and MAGs, protein-coding sequences for ammonia monooxygenase (Amo) subunits were most abundant at oxic site A (Figs. 1d, 6A; LEfSe, Kruskal–Wallis test). This trend was mostly driven by AOA, as there were significantly more sequences encoding AOA-AmoA than AOB-AmoA overall at oxic sites (Wilcoxon rank, p < 0.05), presumably due to lower ammonia regeneration (measured ammonia/ammonium concentrations were low across all sites regardless of oxygen content, >1 gm−3 in only 6% of wells, and most below detection, Table S1). While not all ammonia oxidizer diversity may have been captured by the ddPCR primer sets used (e.g. the AOA primer set has a known bias against some ammonia-oxidizing Thermoproteota/Thaumarchaeota) [61], quantification of amoA genes and transcripts demonstrated a similar relationship with oxygen across a wider set of groundwater sites (Fig. 6C). Archaeal/bacterial amoA gene ratios were significantly and positively correlated with ORP (Spearman’s r = 0.39), and negatively correlated with borehole depth (Spearman’s r = −0.35; Table S9), which is expected to become increasingly oxygen-depleted with depth [62]. These gene ratios were also significantly and negatively associated with ammonia concentrations, conductivity, TDS, and pH (Spearman’s r = −0.34 to 52). Transcript ratios exhibited similar trends, albeit significant only for pH.
Taken together, results indicate that AOA and AOB abundance and activity are governed by distinct environmental niches in groundwater, as found in soils [63]. However, while AOA amoA gene concentrations were on average 40x higher than AOB genes, this difference was ten-fold less for transcripts (Table S10). Moreover, the deficit in significant correlations between AOA/AOB amoA transcript ratios and geochemical/physical groundwater parameters (Fig. 6), suggests comparatively little difference in AOA and AOB activity overall.
Ammonia-oxidizers constituted several major lineages, including a single comammox bacterium
Commensurate with a greater abundance of AOA, we reconstructed seven AOA MAGs, along with one Nitrospiraceae MAG capable of complete ammonia oxidation (comammox). All contained at least one ammonia monooxygenase gene and six contained genes encoding all AmoABC subunits (Table S7). AOA genomes (and their amoA genes, Figs. S4-S6) were phylogenetically diverse, with the MAGs comprising five different genera (Fig. 6a). Of these, nzgw14 (UBA8516) was the most abundant nitrogen cycling MAG overall (Fig. S7), and was most abundant at oxic sites, along with other AOA MAGs. However, AOA MAG relative abundances did not reflect their transcriptional activity (Fig. 6B).
Recently characterized comammox bacteria (Nitrospira, phylum Nitrospirae) [64] oxidize ammonia to nitrate in three steps (ammonia → hydroxylamine → nitrite → nitrate). Nitrospiraceae MAG (nzgw279) possesses genes for ammonia oxidation (amoABC), hydroxylamine oxidation (haoAB), and nitrite oxidoreductase (nxrAB), consistent with comammox. It also possesses a dissimilatory nitrite reductase (nirK) present in comammox bacteria elsewhere [65, 66]. Based on 120 concatenated bacterial marker genes (GTDB-Tk) and the AmoA subunit, nzgw279 is closely related to clade B sublineage II comammox Nitrospira, and is most similar to Nitrospira sp. RCB obtained from an aquifer in Colorado (USA) [66] (Fig. S5), indicating strong habitat driven selection, independent of geographical distance. As expected for comammox, nzgw279 relative abundance was positively correlated with ORP and DO (Spearman’s r = 0.87 and 0.63; respectively). Although comammox bacteria can be the most abundant ammonia-oxidizers in some settings (e.g. groundwater-fed sand filters, forest soils and biofilters [67–69]), AOA genomes were more abundant overall here (Figs. 6, S7). Nevertheless, nzgw279 was highly active in terms of nxr (although not amoA) gene expression (Fig. 2C).
Diverse taxa with nitrite oxidoreductase homologues, including Nitrososphaerales
In addition to comammox bacterium nzgw279, we recovered the genomes of two Nitrospiraceae that we predict are canonical nitrite oxidizers, nzgw274 (no genus designation; nxrA gene present), and nzgw276 (genus=40CM-3-62-11; nxrAB present) (Table S7). Neither are affiliated with known comammox bacteria (Nitrospira spp. [66]). The nxrA from nzgw274 was expressed highest at dysoxic site D in attached-fraction enriched groundwater (nxrA, planktonic-fraction = 0.36, attached-fraction = 8.97 TPM). MAG nzgw276 transcripts were also present, but at a much lower level. Results suggest that the well (E1) had sufficient oxygen for NOB to exist. NOB can be active at nanomolar-to-micromolar concentrations of DO [70], and thereby compete for nitrite alongside anammox and denitrifying bacteria.
Known NXR are reported to be genetically diverse [71, 72]. The known diversity of nitrifiers continues to grow [73] and NXR has alternative uses, for example, as a nitrate reductase [71, 72]. Here, homologues were present in a diverse range of other MAGs, including several anammox bacteria (Planctomycetota, class Ca. Brocadiae, Table S7) [15], which typically use NXR to oxidize a small amount nitrite to nitrate during anammox [74], an archaeal Nitrososphaerales (nzgw5; nxrABC present), which belongs to a taxonomic group more typically associated with ammonia oxidation (this dataset, Table S7) [74], and various other bacterial phyla, which represent a pool of potentially novel nitrifiers (Fig. S8a).
To further explore NXR in the archaeal Nitrososphaerales MAG (nzgw5), we evaluated the 14,591 bp long contig (3135) on which the genes were found. The contig primarily comprises protein-coding genes with closest homology to archaea (based on the NCBI nr database) that are located on either side of a syntenous bacterial-like nxrABC gene cluster (Fig. S8b; Table S11), including chaperone-encoding torD directly adjacent to nxrC, as found in Nitrospina gracilis, which is thought to facilitate Mo cofactor maturation and insertion into NxrA [75]. To determine whether nxr genes were reproducibly present in other closely related Nitrososphaerales genomes, we searched for dereplicated MAGs sharing >99% ANI with nzgw5. We found one “replicate” Nitrososphaerales MAG (nzgw5-b) sharing 99.2% ANI, which was recovered from site B (nzgw5 derived from a co-assembly of site B samples), and that possessed a similar nxrABC gene cluster (Table S11).
The MAG nzgw5 shares NxrA protein sequence similarity with bacteria as phylogenetically diverse as Nitrospira defluvii, Nitrospina gracilis, and Candidatus Brocadia (Fig. S8a), but the highest NCBI nr database matches were to other aquifer bacteria, Candidate division Zixibacteria bacterium RBG-1 (58.56% identity) and Planctomycetes bacterium RIFCSPHIGHO2_02_FULL_52_58 (56.7% identity)—both originally recovered genomically from an aquifer in Rifle, CO, USA [11, 76]. NxrB similarly had the highest match with other aquifer-derived genomes (also U.S.A.), including one Nitrospinae bacterium (65.97% identity), and most notably, two archaea affiliated with the Thermoproteota (previously Thaumarchaeota) phylum—another Nitrososphaerales, and Thermoproteota bacterium (68.71–71.87% identity) [13]. While the gene cluster appears to have been horizontally acquired, we found no identifiable genomic island associated with contig 3135. However, lateral gene transfer occurs more frequently between organisms, including unrelated taxa, that share a habitat [77]. Results suggest aquifer-adapted Nitrososphaerales acquired nxr genes, and potentially also the ability for nitrite oxidation/reduction, from a co-occurring bacterium.
Nitrite-dependent methanotroph (Ca. Methylomirabilis)
Nitrite-dependent methanotrophs were prevalent
Analysis of metagenomic reads revealed that copper-containing, membrane-associated particulate methane monooxygenase subunit A (PmoA) sequences (a closely related AmoA homologue [78]) were ubiquitous, but significantly more abundant at oxic site A (Fig. 1C). This protein is associated with aerobic and anaerobic nitrite-dependent methanotrophs that are able to oxidize methane to CO2, using methane as a sole carbon and energy source [79]. Methanotrophic bacteria can also oxidize ammonia to nitrite using particulate methane monooxygenase (pMMO) and a unique hydroxylamine oxidoreductase, HAO [80]. Interactions between methanotrophs and ammonia-oxidizers in aquifers are poorly understood. They are associated with opposite gradients of ammonia and methane, as ammonia inhibits the activity of methanotrophs and methane acts as a competitive inhibitor for ammonia oxidizers [81, 82]. Analysis of 16S rRNA gene amplicons across a wide distribution of aquifer samples (n = 80) showed that Ca. Methylomirabilis relative abundance was positively correlated with ammonia-oxidizers Nitrosotaleacae, Nitrosomonadaceae, and Nitrosopumilaceae (Spearman’s r = 0.42, r = 0.33 and r = 0.37, respectively). Aerobic ammonia-oxidizers consume O2 and may provide a habitable environment for Ca. Methylomirabilis at oxic/anoxic interfaces and produce nitrite, which could potentially be directly used by these nitrite-dependent methanotrophs [83].
We recovered one nitrogen cycling methanotrophic MAG (nzgw240), related to the anaerobic methane-oxidizing genus Ca. Methylomirabilis (Figs. 6, S6) [84]. Members of Ca. Methylomirabilis perform NO2− dependent anaerobic methane oxidation through an intra‐aerobic pathway involving the dismutation of NO into O2 and N2 [79, 84], and play an important role in controlling N2O and methane emissions from natural ecosystems [79]. All pmoABC subunits for methane oxidation [79] were present in nzgw240, and were closely related to those from Ca. Methylomirabilis lanthanidiphila (74.80–95.5% amino acid identity, Fig S6), a methanotroph that dominated an enrichment culture after addition of rare-earth metal cerium [85]. Several genes involved in nitrogen cycling, such as nitrate oxidoreductase (nxrAB), nitrite reductase (nirS), putative NO dismutase (nod) and nitric oxide reductase (norZ) were also identified in MAG nzgw240, and were expressed at the dysoxic site alongside pmoA (Figs. 2c, 6b), where dissolved methane was also detected [86]. The first of two nitric oxide-like reductases shares 95.6% amino acid identity with Nod (DAMO_2434) in Ca. Methylomirabilis oxyfera [87]. This enyzme is homologous to the quinol-dependent NO reductases (qNOR) [87], however experimental validation is still required to prove nitric oxide disproportionation. The second is a NO reductase sharing 88.34% amino acid identity with NorZ (DAMO_1889).
Final-step of denitrification
Atypical NosZ was more common than typical NosZ
Thirty MAGs, spanning 10 bacterial phyla, contained nosZ genes (Fig. 3). The NosZ protein catalyzes the conversion of green-house gas N2O to N2 in the last step of denitrification. Typical NosZ proteins (clade I) contain a twin-arginine translocation (Tat) signal peptide, and to date are affiliated exclusively with Proteobacteria, which usually perform complete denitrification [88]. A maximum-likelihood tree revealed that NosZ predicted protein sequences comprised both typical clade I twin-arginine (Tat) dependant N2O reductase with Proteobacteria (2 MAGs) and atypical clade II secretory (Sec) dependent N2O reductase proteins (23 MAGs) (Fig. S9) [88]. The tree also shows a novel clade of Nitrospirota and Nitrospinae NosZ Sec-dependant sequences, including four NosZ sequences from this study (Fig. S9) that were transcriptionally active (Fig. 7). Members of clade II are considered non-denitrifiers, typically performing just the final step of denitrification [89]. However, 4/7 MAGs capable of complete denitrification encode clade II NosZ, demonstrating complete denitrifiers are present across both clades. Sec-dependent protein translocation is considered more energetically favourable than Tat, requiring between 700 and 5,000 molecules (or equivalent) of ATP per protein translocation across the membrane, whereas Tat requires the equivalent of ~10,000 molecules of ATP [90]. A greater proportion of Sec signal pathways in low nutrient groundwater would be favourable for energy conservation.
Fig. 7. Nitrous-oxide reductase (nosZ) gene transcripts in sites C and D groundwater and attached-fraction enriched groundwater.
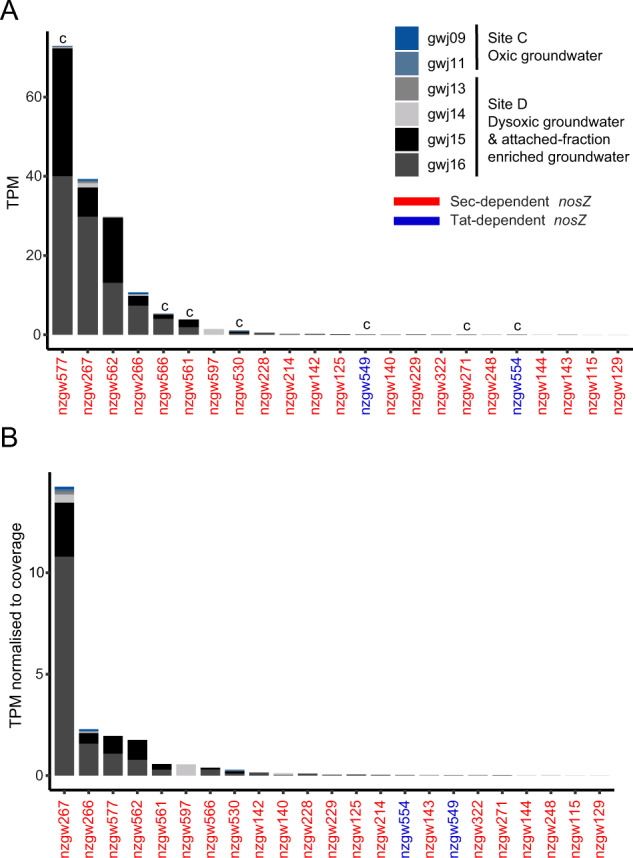
A Stacked barplot shows modified-TPM of Sec- and Tat- dependent nosZ genes at each site. B Stacked barplot shows modified-TPM normalized to genome coverage of Sec- and Tat-dependent nosZ genes at each site. While complete denitrifier, Sulfuricella MAG nzgw577, contributed the most transcripts, after normalizing to MAG relative abundance, nosZ genes from two novel Nitrospinota MAGs (nzgw266-267, class UBA7883 [104]) were transcriptionally most active. c = MAGs which contain genes for the complete denitrification pathway (nzgw numbers: 271, 530, 549, 554, 561, 566, and 577).
Nitrous oxide reductase gene expression was strongly associated with oxygen availability and not limited by pathway fragmentation
Based on ddPCR, nosZ clade I genes (3 × 101–6 × 105 copies/L) and transcripts (1 × 101–7 × 104 copies/L) were detected across most aquifer samples tested (Table S10). Expression was significantly and negatively correlated with ORP (Fig. 6), reflecting observations elsewhere that the rate of denitrification decreases linearly with increasing ORP [91]. Accordingly, at the dysoxic site there was also a proportionally higher abundance of MAGs with nosZ genes of any type (Fig. 3), and of complete denitrifier MAGs, which comprised up to 42% of the nitrogen cycling community (4–31× more on average than oxic sites) (Fig. S3).
N2O generation due to incomplete denitrification has been shown to be highest under oxic conditions in groundwater [92], comparable to sites A-C here. A fragmented denitrification pathway may explain higher N2O concentrations in some oxic groundwaters. Fragmented genetic potential for biogeochemical cycling processes, such as denitrification, appears to be a common trait in aquifer bacteria (Fig. 4) [11], necessitating metabolic handoffs among individuals to complete pathways. Our transcriptomic data points to active collaboration among incomplete denitrifiers for generation and removal of N2O (although in situ measurements would be required to determine the presence or absence of NO or N2O emissions). Transcriptional activity associated with N2O reduction was at least equivalent to that for generation, regardless of groundwater oxygen-content or the portion of pathway fragmentation (Fig. 2A).
Transcriptional activity of co-occurring aerobic and anaerobic nitrogen-cycling pathways in oxic versus dysoxic groundwater
Based on transcripts mapped to MAGs, there was less nitrogen-cycling transcriptional activity at the pristine oxic site compared to the dysoxic site (average modified-TPM 69 ± 13 site C versus 359 ± 362 site D) (Fig. 2B). This is consistent with quantification of 8x more nitrogen-cycling transcripts by ddPCR in dysoxic versus oxic groundwater, across a wider set of groundwaters (on average 7 × 104 transcripts/L in dysoxic and 8 × 103 transcripts/L in oxic groundwater; Table S11). The greatest proportion of mapped transcripts at the oxic site was associated with ammonia oxidation to nitrate and re-reduction to nitrite, based on ammonia monooxygenase (amo), nitrite oxidoreductase (nxr), and periplasmic nitrate reductase (nap) gene transcripts (Fig. 2A). Expression of nap genes is suggestive of aerobic denitrification at this site, as nitrate reduction in the periplasm is not inhibited by oxygen [93].
At the dysoxic site, the greatest portion of gene expression in groundwater and attached-fraction enriched groundwater, based on mapped transcripts was associated with, again, nitrite oxidation (nxr), but also anammox (hzo, hzs), and denitrification (nor and nos) (Fig. 2A). These genes each contributed up to 8−66% of nitrogen-cycling transcripts at site D. Hydrazine synthase hzsB transcripts (quantified by ddPCR) were also highest at the dysoxic site (April 2018), one of only seven sites in the study with detectable nitrite concentrations. Contemporaneous measurements of excess N2 indicated active N2 generation in dysoxic groundwater from this site (wells E1 and N3; Table S12) due denitrification and/or anammox [15]—the technique cannot distinguish between N2 produced by these processes. Nitrate-based δ18O against δ15N measurements from groundwater in well N3 also indicated the occurrence of denitrification [86]. The majority of hzoA and hzsB genes were expressed by just two Planctomycetes MAGs (nzgw511–512) at this site (37% and 58%, respectively). However, when considering hzsB transcript concentrations in the wider aquifer dataset, we observed no relationship with DOC or oxygen availability (DO, ORP or borehole depth) (Fig. 6), indicating these bacteria are active under a wide variety of groundwater conditions, including those considered unfavourable for anammox (i.e. high DOC and DO). Ca. Brocadiae genomes recovered from Sites A–D were previously found to have a broad range of ABC transport systems and variations in substrate importation such as phosphate, cobalt, nickel, iron(III), zinc, sulfate, molybdate, lipoproteins, ribose, rhamnose, polysaccharides, and oligopeptides, suggesting that they may not just be autotrophic specialists [15]. We found evidence for greater competition among N2O reducers (Fig. 2B), although most nosZ transcripts overall (81%) were expressed by a single complete denitrifier, MAG nzgw577 (genus Sulfuricella; Fig. 7) at the dysoxic site (Fig. 2C), despite it being only the third most active in terms of nosZ expression after normalizing to MAG coverage (Fig. 7). Members of this genus are reported to perform autotrophic denitrification coupled with the oxidation of reduced sulfur compounds [94].
Several aerobic ammonia-oxidizers, for which we recovered MAGs, were also active at the dysoxic site, contributing 5-6% of nitrogen-cycling transcripts mapped (Fig. 2B). This suggests the potential for simultaneous nitrification and denitrification, and partial coupling of these pathways (overall modified-TPM 1:2.7 amoA:nosZ, 1:2.3 nosZ:nxrA). Most (88%) amoA transcriptional activity at this dysoxic site was attributed to four AOA MAGs affiliated with Nitrosopumilaceae. The greatest proportion of ammonia monooxygenase transcription was associated with the attached-fraction enriched groundwater from this site (Fig. 6B), consistent with preferences previously reported for the ammonia-oxidizing genera, Nitrososphaera and Nitrosopumilus, in groundwater [30]. Prior findings from karstic aquifers also suggest that soil-derived ammonia oxidizers may be imported into groundwater [26], which may be important for shallow aquifers that directly receive leachate from the soil zone, along with any microorganisms it carries.
The most transcribed nitrogen-cycling gene among all MAGs was nxrB from comammox Nitrospiraceae nzgw279, which showed the highest expression in attached-fraction enriched groundwater at site D. This groundwater contained more total suspended solids (Table S1), and therefore more sediment particles coated in biofilms [95]. Comammox Nitrospira populations have previously been found to dominate biofilms in wastewater, outnumbering all other nitrifiers [96]. As nzgw279 was associated with fewer amoA transcripts than other ammonia-oxidizers (average modified-TPM 0.77 ± 1.02 SD vs 2.01 ± 3.22 SD), and appeared to act largely as a canonical nitrifier [97], it potentially received nitrite as a by-product from the several active AOAs.
Results show methanotroph methane monooxygenase gene transcription occurred alongside gene expression associated with nitrification, anammox, and denitrification at the dysoxic site. Methanotrophs and ammonia-oxidizers share many metabolic similarities based on a common evolutionary history, and supported by the structural similarities of ammonia and methane monooxygenases [81], methanotrophs have been implicated in both methane and ammonia oxidation in groundwater [98]. In this study, Ca. Methylomirabilis (nzgw240) expressed genes, associated with concurrent methane oxidation (pmoA) and nitric oxide dismutation (NO2- reductase nirS and NO dismutase nod) to N2 and O2 (Fig. 2C) [84]. Co-occurring gene expression reveals a potential interaction among AOA, Ca. Methylomirabilis, and anammox bacteria, whereby nitrite produced from aerobic ammonia oxidation by AOA, drives anammox and nitrite-dependent anaerobic methane oxidation by Ca. Methylomirabilis. Ca. Methylomirabilis produces oxygen which could create an interface whereby AOA and comammox can co-exist. Oxygen consumption and nitrite provisioning by AOA could represent synergism with anammox in the terrestrial subsurface, as previously predicted to occur in unconfined aquifer soils [99]. Indeed, ammonia oxidizer and anammox activities, based on transcript copy numbers, were found to be tightly linked across distinct groundwater chemistries in the wider set of samples [15]. These heterogeneous reactions at the dysoxic site indicate that it likely contained mixed redox conditions in situ. This could be due to oxygen penetration from above [100], and vertical stratification of electron donors [101], geochemical gradients created by biofilm formation [102], or oxygen produced by the intra-aerobic pathway of Ca. Methylomirabilis species [84].
Conclusion
Results show that the capacity for non-assimilatory nitrogen-cycling reactions, such as ammonia oxidation and denitrification, was prevalent in groundwater regardless of site-specific physicochemistry, although the relative abundance of each pathway differed. Phylogenetically diverse AOA and AOB were associated with distinct environmental niches in groundwater, and AOA-amoA genes and transcripts were more abundant overall. While incomplete denitrifiers were numerous, complete denitrifiers contributed to a substantial fraction of transcriptional activity under dysoxic conditions, where activity associated with denitrification, and N-cycling transcripts was greatest. Gene expression associated with nitrification, denitrification, nitrite-dependent methane oxidation, and anammox occurred simultaneously in dysoxic groundwater, such that nitrite (or nitrate) produced by AOA or comammox could fuel anammox, denitrification, and methanotrophy by Ca. Methylomirabilis. Results provide insights into microbial N-transformations in groundwater with distinct chemical characteristics (such as oxygen availability and DOC), and potential metabolic “handoffs” among nitrogen-cycling organisms.
Supplementary information
Acknowledgements
We thank P. Abraham (ESR), H.S. Tee and J.S. Boey for help sampling wells in Canterbury and council staff in Auckland (C. Foster), Wellington (D. McQueen), Waikato (S. Herath) and Canterbury (D. Evans and R. Cressy). We thank D. Waite and J.S. Boey for bioinformatics support, and R. van der Raaij and H. Martindale for excess N2 measurements. Computational resources were provided by New Zealand eScience Infrastructure. Research was supported by a Smart Ideas grant, from the Ministry of Business, Innovation and Employment, awarded to KMH (project UOAX1720).
Author contributions
KMH and CD conceived the study, and KMH, CD, LW, and MC obtained funding. OEM, EG and KMH collected samples. MC contributed technical expertise for in situ sonication. OEM and EG performed laboratory procedures and bioinformatic analyses. OEM led the analysis of data and writing of the manuscript with KMH. All authors reviewed the manuscript.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Data availability
Sequences are deposited with NCBI under BioProject PRJNA699054.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01300-0.
References
- 1.Taylor RG, Scanlon B, Döll P, Rodell M, Van Beek R, Wada Y, et al. Groundwater and climate change. Nat Clim Chang. 2013;3:322–9. doi: 10.1038/nclimate1744. [DOI] [Google Scholar]
- 2.Aeschbach-Hertig W, Gleeson T. Regional strategies for the accelerating global problem of groundwater depletion. Nat Geosci. 2012;5:853–61. doi: 10.1038/ngeo1617. [DOI] [Google Scholar]
- 3.Alley WM, Healy RW, LaBaugh JW, Reilly TE. Flow and storage in groundwater systems. Science. 2002;296:1985–90. doi: 10.1126/science.1067123. [DOI] [PubMed] [Google Scholar]
- 4.Morgenstern U, Daughney CJ. Groundwater age for identification of baseline groundwater quality and impacts of land-use intensification - The National Groundwater Monitoring Programme of New Zealand. J Hydrol. 2012;456–457:79–93. doi: 10.1016/j.jhydrol.2012.06.010. [DOI] [Google Scholar]
- 5.Rivett MO, Buss SR, Morgan P, Smith JWN, Bemment CD. Nitrate attenuation in groundwater: a review of biogeochemical controlling processes. Water Res. 2008;42:4215–32. doi: 10.1016/j.watres.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 6.Rivett MO, Smith JWN, Buss SR, Morgan P. Nitrate occurrence and attenuation in the major aquifers of England and Wales. Q J Eng Geol Hydrogeol. 2007;40:335–52. doi: 10.1144/1470-9236/07-032. [DOI] [Google Scholar]
- 7.Yu G, Wang J, Liu L, Li Y, Zhang Y, Wang S. The analysis of groundwater nitrate pollution and health risk assessment in rural areas of Yantai, China. BMC Public Health. 2020;20:437. doi: 10.1186/s12889-020-08583-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundermann G, Wägner N, Cullmann A, von Hirschhausen CR, Kemfert C. Nitrate pollution of groundwater long exceeding trigger value: Fertilization practices require more transparency and oversight. DIW Wkly Rep. 2020;10:61–72. [Google Scholar]
- 9.Nitrate and nitrite in drinking-water Background document for development of WHO Guidelines for Drinking-water Quality. World Health Organization; 2003. https://apps.who.int/iris/handle/10665/75380.
- 10.Smolders AJP, Lucassen ECHET, Bobbink R, Roelofs JGM, Lamers LPM. How nitrate leaching from agricultural lands provokes phosphate eutrophication in groundwater fed wetlands: the sulphur bridge. Biogeochemistry. 2010;98:1–7. doi: 10.1007/s10533-009-9387-8. [DOI] [Google Scholar]
- 11.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danczak RE, Johnston MD, Kenah C, Slattery M, Wrighton KC, Wilkins MJ. Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome. 2017;5:112. doi: 10.1186/s40168-017-0331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He C, Keren R, Whittaker ML, Farag IF, Doudna JA, Cate JHD, et al. Genome-resolved metagenomics reveals site-specific diversity of episymbiotic CPR bacteria and DPANN archaea in groundwater ecosystems. Nat Microbiol. 2021;3:354–65. doi: 10.1038/s41564-020-00840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardarelli EL, Bargar JR, Francis CA. Diverse Thaumarchaeota dominate subsurface ammonia-oxidizing communities in semi-arid floodplains in the western united states. Microb Ecol. 2020;80:778–92. doi: 10.1007/s00248-020-01534-5. [DOI] [PubMed] [Google Scholar]
- 15.Mosley OE, Gios E, Weaver L, Close M, Daughney C, Van Der Raaij R, et al. Metabolic diversity and aero-tolerance in anammox bacteria from geochemically distinct aquifers. mSystems. 2022;7:e0125521. doi: 10.1128/msystems.01255-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haruta S, Kato S, Yamamoto K, Igarashi Y. Intertwined interspecies relationships: approaches to untangle the microbial network. Environ Microbiol. 2009;11:2963–9. doi: 10.1111/j.1462-2920.2009.01956.x. [DOI] [PubMed] [Google Scholar]
- 17.Albright MBN, Timalsina B, Martiny JBH, Dunbar J. Comparative genomics of nitrogen cycling pathways in bacteria and archaea. Microb Ecol. 2019;77:597–606. doi: 10.1007/s00248-018-1239-4. [DOI] [PubMed] [Google Scholar]
- 18.Stein LY, Klotz MG. The nitrogen cycle. Curr Biol. 2016;26:R94–R98. doi: 10.1016/j.cub.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nat Rev Microbiol. 2018;16:263–76. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 20.Yoon S, Cruz-García C, Sanford R, Ritalahti KM, Löffler FE. Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3-/NO2- reduction pathways in Shewanella loihica strain PV-4. ISME J. 2015;9:1093–104. doi: 10.1038/ismej.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu W, Wang C, Hill J, He Y, Tao B, Mao Z, et al. A missing link in the estuarine nitrogen cycle? Coupled nitrification-denitrification mediated by suspended particulate matter. Sci Rep. 2018;8:2282. doi: 10.1038/s41598-018-20688-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam P, Jensen MM, Lavik G, McGinnis DF, Müller B, Schubert CJ, et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA. 2007;104:7104–9. doi: 10.1073/pnas.0611081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huno SKM, Rene ER, Van Hullebusch ED, Annachhatre AP. Nitrate removal from groundwater: a review of natural and engineered processes. J Water Supply Res Technol - AQUA. 2018;67:885–902. doi: 10.2166/aqua.2018.194. [DOI] [Google Scholar]
- 24.Killingstad MW, Widdowson MA, Smith RL. Modeling enhanced in situ denitrification in groundwater. J Environ Eng. 2002;128:491–504. doi: 10.1061/(ASCE)0733-9372(2002)128:6(491). [DOI] [Google Scholar]
- 25.Handley KM, Verberkmoes NC, Steefel CI, Williams KH, Sharon I, Miller CS, et al. Biostimulation induces syntrophic interactions that impact C, S and N cycling in a sediment microbial community. ISME J. 2013;7:800–16. doi: 10.1038/ismej.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opitz S, Küsel K, Spott O, Totsche KU, Herrmann M. Oxygen availability and distance to surface environments determine community composition and abundance of ammonia-oxidizing prokaroytes in two superimposed pristine limestone aquifers in the Hainich region, Germany. FEMS Microbiol Ecol. 2014;90:39–53. doi: 10.1111/1574-6941.12370. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann M, Rusznyák A, Akob DM, Schulze I, Opitz S, Totsche KU, et al. Large fractions of CO2-fixing microorganisms in pristine limestone aquifers appear to be involved in the oxidation of reduced sulfur and nitrogen compounds. Appl Environ Microbiol. 2015;81:2384–94. doi: 10.1128/AEM.03269-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Herrmann M, Blohm A, Hilke I, Frosch T, Trumbore SE, et al. Thiosulfate- and hydrogen-driven autotrophic denitrification by a microbial consortium enriched from groundwater of an oligotrophic limestone aquifer. FEMS Microbiol Ecol. 2018;94:10. doi: 10.1093/femsec/fiy141. [DOI] [PubMed] [Google Scholar]
- 29.Pan D, Nolan J, Williams KH, Robbins MJ, Weber KA. Abundance and distribution of microbial cells and viruses in an alluvial aquifer. Front Microbiol. 2017;8:1199. doi: 10.3389/fmicb.2017.01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flynn TM, Sanford RA, Ryu H, Bethke CM, Levine AD, Ashbolt NJ, et al. Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol. 2013;13:146. doi: 10.1186/1471-2180-13-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Close M, Abraham P, Webber J, Cowey E, Humphries B, Fenwick G, et al. Use of sonication for enhanced sampling of attached microbes from groundwater systems. Groundwater. 2020;58:901–12. doi: 10.1111/gwat.12984. [DOI] [PubMed] [Google Scholar]
- 32.Malard F, Hervant F. Oxygen supply and the adaptations of animals in groundwater. Freshw Biol. 1999;41:1–30. doi: 10.1046/j.1365-2427.1999.00379.x. [DOI] [Google Scholar]
- 33.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36:1925–7. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 37.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–5. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 38.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinforma. 2004;5:115. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, Mcglinn D, et al. vegan: Community ecology. R Package version 25-6. 2019;2:1–297.
- 43.Warnes, GR Gplots: Various R Programming Tools for Plotting Data. 2011. http://cran.r-project.org/web/packages/gplots/index.html.
- 44.Tu Q, Lin L, Cheng L, Deng Y, He Z. NCycDB: a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. Bioinformatics. 2019;35:1040–8. doi: 10.1093/bioinformatics/bty741. [DOI] [PubMed] [Google Scholar]
- 45.Kartal B, Keltjens JT. Anammox biochemistry: a tale of heme c proteins. Trends Biochem Sci. 2016;41:998–1011. doi: 10.1016/j.tibs.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 46.Yu R, Chandran K. Strategies of nitrosomonas europaea 19718 to counter low dissolved oxygen and high nitrite concentrations. BMC Microbiol. 2010;10:1–11. doi: 10.1186/1471-2180-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Straka LL, Meinhardt KA, Bollmann A, Stahl DA, Winkler MKH. Affinity informs environmental cooperation between ammonia-oxidizing archaea (AOA) and anaerobic ammonia-oxidizing (Anammox) bacteria. ISME J. 2019;13:1997–2004. doi: 10.1038/s41396-019-0408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arp DJ, Stein LY. Metabolism of inorganic n compounds by ammonia-oxidizing bacteria. Crit Rev Biochem Mol Biol. 2003;38:471–95. doi: 10.1080/10409230390267446. [DOI] [PubMed] [Google Scholar]
- 49.Hink L, Lycus P, Gubry-Rangin C, Frostegård Å, Nicol GW, Prosser JI, et al. Kinetics of NH3-oxidation, NO-turnover, N2O-production and electron flow during oxygen depletion in model bacterial and archaeal ammonia oxidisers. Environ Microbiol. 2017;19:4882–96. doi: 10.1111/1462-2920.13914. [DOI] [PubMed] [Google Scholar]
- 50.Singh SN, Tripathi RD, editors. Environmental bioremediation technologies. 1st edn. Springer-Verlag Berlin Heidelberg; 2007.
- 51.Ruan Y, Ma B, Cai C, Cai L, Gu J, Lu HF, et al. Kinetic affinity index informs the divisions of nitrate flux in aerobic denitrification. Bioresour Technol. 2020;309:123345. doi: 10.1016/j.biortech.2020.123345. [DOI] [PubMed] [Google Scholar]
- 52.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 2017;35:725–31. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Handley KM, Bartels D, O’Loughlin EJ, Williams KH, Trimble WL, Skinner K, et al. The complete genome sequence for putative H2- and S- oxidizer Candidatus Sulfuricurvum sp., assembled de novo from an aquifer-derived metagenome. Environ Microbiol. 2014;16:3443–62. doi: 10.1111/1462-2920.12453. [DOI] [PubMed] [Google Scholar]
- 54.Bell E, Lamminmäki T, Alneberg J, Andersson AF, Qian C, Xiong W, et al. Biogeochemical cycling by a low-diversity microbial community in deep groundwater. Front Microbiol. 2018;9:2129. doi: 10.3389/fmicb.2018.02129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark DR, Rees AP, Joint I. Ammonium regeneration and nitrification rates in the oligotrophic Atlantic Ocean: Implications for new production estimates. Limnol Oceanogr. 2008;53:52–62. doi: 10.4319/lo.2008.53.1.0052. [DOI] [Google Scholar]
- 56.Martens-Habbena W, Berube PM, Urakawa H, De La Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–9. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 57.Prosser JI, Nicol GW. Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 2012;20:523–31. doi: 10.1016/j.tim.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 58.Horak REA, Qin W, Schauer AJ, Armbrust EV, Ingalls AE, Moffett JW, et al. Ammonia oxidation kinetics and temperature sensitivity of a natural marine community dominated by Archaea. ISME J. 2013;7:2023–33. doi: 10.1038/ismej.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dimitri Kits K, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature. 2017;549:269–72. doi: 10.1038/nature23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alves RJE, Wanek W, Zappe A, Richter A, Svenning MM, Schleper C, et al. Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J. 2013;7:1620–31. doi: 10.1038/ismej.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sintes E, Bergauer K, De Corte D, Yokokawa T, Herndl GJ. Archaeal amoA gene diversity points to distinct biogeography of ammonia-oxidizing Crenarchaeota in the ocean. Environ Microbiol. 2013;15:1647–58. doi: 10.1111/j.1462-2920.2012.02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McDonough LK, Santos IR, Andersen MS, O’Carroll DM, Rutlidge H, Meredith K, et al. Changes in global groundwater organic carbon driven by climate change and urbanization. Nat Commun. 2020;11:1–10. doi: 10.1038/s41467-020-14946-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–17. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–9. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sakoula D, Koch H, Frank J, Jetten MSM, Van Kessel MAHJ, Lücker S. Enrichment and physiological characterization of a novel comammox Nitrospira indicates ammonium inhibition of complete nitrification. ISME J. 2021;15:1010–24. doi: 10.1038/s41396-020-00827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poghosyan L, Koch H, Lavy A, Frank J, Kessel MAHJ, Jetten MSM, et al. Metagenomic recovery of two distinct comammox Nitrospira from the terrestrial subsurface. Environ Microbiol. 2019;21:3627–37. doi: 10.1111/1462-2920.14691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fowler SJ, Palomo A, Dechesne A, Mines PD, Smets BF. Comammox Nitrospira are abundant ammonia oxidizers in diverse groundwater-fed rapid sand filter communities. Environ Microbiol. 2018;20:1002–15. doi: 10.1111/1462-2920.14033. [DOI] [PubMed] [Google Scholar]
- 68.Xia F, Wang JG, Zhu T, Zou B, Rhee SK, Quan ZX. Ubiquity and diversity of complete ammonia oxidizers (comammox) Appl Environ Microbiol. 2018;84:e01390–18. doi: 10.1128/AEM.01390-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu HW, He JZ. Comammox—a newly discovered nitrification process in the terrestrial nitrogen cycle. J Soils Sediment. 2017;17:2709–17. doi: 10.1007/s11368-017-1851-9. [DOI] [Google Scholar]
- 70.Bristow LA, Dalsgaard T, Tiano L, Mills DB, Bertagnolli AD, Wright JJ, et al. Ammonium and nitrite oxidation at nanomolar oxygen concentrations in oxygen minimum zone waters. Proc Natl Acad Sci USA. 2016;113:10601–6. doi: 10.1073/pnas.1600359113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chicano TM, Dietrich L, de Almeida NM, Akram M, Hartmann E, Leidreiter F, et al. Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex. Nat Microbiol. 2021;6:1129–39. doi: 10.1038/s41564-021-00934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci USA. 2015;112:11371–6. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park SJ, Andrei AŞ, Bulzu PA, Kavagutti VS, Ghai R, Mosier AC. Expanded diversity and metabolic versatility of marine nitrite-oxidizing bacteria revealed by cultivation- and genomics-based approaches. Appl Environ Microbiol. 2020;86:1–17. doi: 10.1128/AEM.01667-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Almeida NM, Wessels HJCT, de Graaf RM, Ferousi C, Jetten MSM, Keltjens JT, et al. Membrane-bound electron transport systems of an anammox bacterium: a complexome analysis. Biochim Biophys Acta Bioenergy. 2016;1857:1694–704. doi: 10.1016/j.bbabio.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 75.Lücker S, Nowka B, Rattei T, Spieck E, Daims H. The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol. 2013;4:27. doi: 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Castelle CJ, Hug LA, Wrighton KC, Thomas BC, Williams KH, Wu D, et al. Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat Commun. 2013;4:1–10. doi: 10.1038/ncomms3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hooper SD, Mavromatis K, Kyrpides NC. Microbial co-habitation and lateral gene transfer: What transposases can tell us. Genome Biol. 2009;10:1–12. doi: 10.1186/gb-2009-10-4-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ricke P, Erkel C, Kube M, Reinhardt R, Liesack W. Comparative analysis of the conventional and novel pmo (particulate methane monooxygenase) operons from Methylocystis strain SC2. Appl Environ Microbiol. 2004;70:3055–63. doi: 10.1128/AEM.70.5.3055-3063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–8. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 80.Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–8. doi: 10.1111/j.1574-6968.1995.tb07834.x. [DOI] [PubMed] [Google Scholar]
- 81.Stein LY, Roy R, Dunfield PF. Aerobic methanotrophy and nitrification: processes and connections. eLS. Chichester, UK: John Wiley & Sons, Ltd; 2012.
- 82.Nyerges G, Stein LY. Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol Lett. 2009;297:131–6. doi: 10.1111/j.1574-6968.2009.01674.x. [DOI] [PubMed] [Google Scholar]
- 83.Welte CU, Rasigraf O, Vaksmaa A, Versantvoort W, Arshad A, Op den Camp HJM, et al. Nitrate- and nitrite-dependent anaerobic oxidation of methane. Environ Microbiol Rep. 2016;8:941–55. doi: 10.1111/1758-2229.12487. [DOI] [PubMed] [Google Scholar]
- 84.Wu ML, Ettwig KF, Jetten MSM, Strous M, Keltjens JT, Van, et al. A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus ‘Methylomirabilis oxyfera’. Biochem Soc Trans. 2011;39:243–8. doi: 10.1042/BST0390243. [DOI] [PubMed] [Google Scholar]
- 85.Versantvoort W, Guerrero-Cruz S, Speth DR, Frank J, Gambelli L, Cremers G, et al. Comparative genomics of Candidatus Methylomirabilis species and description of Ca. Methylomirabilis lanthanidiphila. Front Microbiol. 2018;9:1672. doi: 10.3389/fmicb.2018.01672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martindale H, van der Raaij RW, Daughney CJ, Morgenstern U, Hadfield J. Measurement of neon in groundwaters for quantification of denitrification in aquifers. Lower Hutt (NZ): GNS Science. 44p. (GNS Science report; 2018/34). 2018. 10.21420/FR4X-J821
- 87.Ettwig KF, Speth DR, Reimann J, Wu ML, Jetten MSM, Keltjens JT. Bacterial oxygen production in the dark. Front Microbiol. 2012;3:273. doi: 10.3389/fmicb.2012.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hallin S, Philippot L, Löffler FE, Sanford RA, Jones CM. Genomics and ecology of novel N2O-reducing microorganisms. Trends Microbiol. 2018;26:43–55. doi: 10.1016/j.tim.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 89.Conthe M, Wittorf L, Kuenen JG, Kleerebezem R, Van Loosdrecht MCM, Hallin S. Life on N2O: Deciphering the ecophysiology of N2O respiring bacterial communities in a continuous culture. ISME J. 2018;12:1142–53. doi: 10.1038/s41396-018-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi LX, Theg SM. Energetic cost of protein import across the envelope membranes of chloroplasts. Proc Natl Acad Sci USA. 2013;110:930–5. doi: 10.1073/pnas.1115886110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lie E, Welander T. Influence of dissolved oxygen and oxidation-reduction potential on the denitrification rate of activated sludge. Water Sci Technol. 1994;30:91–100. doi: 10.2166/wst.1994.0256. [DOI] [Google Scholar]
- 92.McAleer EB, Coxon CE, Richards KG, Jahangir MMR, Grant J, Mellander PE. Groundwater nitrate reduction versus dissolved gas production: a tale of two catchments. Sci Total Environ. 2017;586:372–89. doi: 10.1016/j.scitotenv.2016.11.083. [DOI] [PubMed] [Google Scholar]
- 93.Rajta A, Bhatia R, Setia H, Pathania P. Role of heterotrophic aerobic denitrifying bacteria in nitrate removal from wastewater. J Appl Microbiol. 2020;128:1261–78. doi: 10.1111/jam.14476. [DOI] [PubMed] [Google Scholar]
- 94.Watanabe T, Kojima H, Fukui M. Complete genomes of freshwater sulfur oxidizers Sulfuricella denitrificans skB26 and Sulfuritalea hydrogenivorans sk43H: Genetic insights into the sulfur oxidation pathway of betaproteobacteria. Syst Appl Microbiol. 2014;37:387–95. doi: 10.1016/j.syapm.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 95.Williamson WM, Close ME, Leonard MM, Webber JB, Lin S. Groundwater biofilm dynamics grown in situ along a nutrient gradient. Ground Water. 2012;50:690–703. doi: 10.1111/j.1745-6584.2011.00904.x. [DOI] [PubMed] [Google Scholar]
- 96.Spasov E, Tsuji JM, Hug LA, Doxey AC, Sauder LA, Parker WJ, et al. High functional diversity among Nitrospira populations that dominate rotating biological contactor microbial communities in a municipal wastewater treatment plant. ISME J. 2020;14:1857–72. doi: 10.1038/s41396-020-0650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koch H, van Kessel MAHJ, Lücker S. Complete nitrification: insights into the ecophysiology of comammox Nitrospira. Appl Microbiol Biotechnol. 2019;103:177–89. doi: 10.1007/s00253-018-9486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kutvonen H, Rajala P, Carpén L, Bomberg M. Nitrate and ammonia as nitrogen sources for deep subsurface microorganisms. Front Microbiol. 2015;6:1079. doi: 10.3389/fmicb.2015.01079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang S, Radny D, Huang S, Zhuang L, Zhao S, Berg M, et al. Nitrogen loss by anaerobic ammonium oxidation in unconfined aquifer soils. Sci Rep. 2017;7:1–10. doi: 10.1038/srep40173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schmidt CM, Fisher AT, Racz AJ, Lockwood BS, Huertos ML. Linking denitrification and infiltration rates during managed groundwater recharge. Environ Sci Technol. 2011;45:9634–40. doi: 10.1021/es2023626. [DOI] [PubMed] [Google Scholar]
- 101.Kolbe T, De Dreuzy JR, Abbott BW, Aquilina L, Babey T, Green CT, et al. Stratification of reactivity determines nitrate removal in groundwater. Proc Natl Acad Sci USA. 2019;116:2494–9. doi: 10.1073/pnas.1816892116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bishop PL, Yu T. A microelectrode study of redox potential change in biofilms. Water Sci Technol. 1999;39:179–85. doi: 10.2166/wst.1999.0357. [DOI] [Google Scholar]
- 103.Rochman FF, Kwon M, Khadka R, Tamas I, Lopez-Jauregui AA, Sheremet A, et al. Novel copper-containing membrane monooxygenases (CuMMOs) encoded by alkane-utilizing Betaproteobacteria. ISME J. 2020;14:714–26. doi: 10.1038/s41396-019-0561-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences are deposited with NCBI under BioProject PRJNA699054.