Abstract
Many ecosystems have been shown to retain a memory of past conditions, which in turn affects how they respond to future stimuli. In microbial ecosystems, community disturbance has been associated with lasting impacts on microbiome structure. However, whether microbial communities alter their response to repeated stimulus remains incompletely understood. Using the human gut microbiome as a model, we show that bacterial communities retain an “ecological memory” of past carbohydrate exposures. Memory of the prebiotic inulin was encoded within a day of supplementation among a cohort of human study participants. Using in vitro gut microbial models, we demonstrated that the strength of ecological memory scales with nutrient dose and persists for days. We found evidence that memory is seeded by transcriptional changes among primary degraders of inulin within hours of nutrient exposure, and that subsequent changes in the activity and abundance of these taxa are sufficient to enhance overall community nutrient metabolism. We also observed that ecological memory of one carbohydrate species impacts microbiome response to other carbohydrates, and that an individual’s habitual exposure to dietary fiber was associated with their gut microbiome’s efficiency at digesting inulin. Together, these findings suggest that the human gut microbiome’s metabolic potential reflects dietary exposures over preceding days and changes within hours of exposure to a novel nutrient. The dynamics of this ecological memory also highlight the potential for intra-individual microbiome variation to affect the design and interpretation of interventions involving the gut microbiome.
Subject terms: Microbial ecology, Microbiome
Ecological memory describes a broad range of phenomena in which past disturbances experienced by an ecosystem influence community responses in the present [1]. Such responses may reflect prior abiotic experiences (e.g. past rain or fires shaping reproduction strategies in plants) or biotic ones (e.g. species extinction altering competitive phenotypes among extant organisms) [2]. Knowing that ecosystems retain memory has helped shape frameworks for reintroducing locally extinct species and promoting species diversity in settings like hardwood forests [2, 3]. Ecological memory may also amplify the severity of ecosystem damage caused by climate change [4].
Despite the power of ecological memory to understand and manage ecological dynamics, our knowledge of how this concept applies to microbial ecosystems remains sparse. Within individual microbial species, evidence exists for memory-like processes. Among bacterial monocultures, past environmental conditions like nutrient availability affect metabolic potential [5–7], or the ability to utilize a substrate of interest. Yet, at the microbial community level, our understanding of how past environmental conditions affect future ecosystem function is limited. Lasting changes in the abundance of taxa result from disturbances like oil spills [8, 9], antibiotic administration [10], dietary oscillations [11], and infection [12]. In the human gut microbiome, it has been demonstrated that infection [13] and obesogenic diet exposure [14] induce a memory that affects the ecological outcome of subsequent perturbations. Still, it is not yet known what independent role microbes play in this memory (i.e. in the absence of host factors), nor the mechanism and properties of this memory. Furthermore, the fastest time scales on which microbial communities form ecological memory remain undefined. Due to the reproductive rates of bacteria, ecological memory may form in microbial ecosystems orders of magnitude faster than in communities of plants and animals as with prior observations of ecological succession in microbial ecosystems [12]. Understanding how and on what time scales memory forms should therefore provide fundamental insights into the ecological processes governing microbial community composition and metabolism.
Results
Nutrient metabolism in the human gut provides an opportunity to test for ecological memory in a microbial community setting. Nutrient availability plays a crucial role in determining the form of cooperative and competitive interactions in the gut [15], and host intake of nutrients varies on a daily basis [16]. We thus reasoned that if the metabolic potential of the human gut microbiome reflects recent nutritional availability, newly introduced nutrients would be incompletely metabolized and require multiple exposures to be fully utilized by intestinal bacteria. To test this hypothesis, we analyzed samples from 40 participants enrolled in a randomized placebo-controlled study. Individuals in the prebiotic group consumed 18 g/day of inulin (Fig. S1A–D), a nutrient that can be metabolized by the gut microbiome [17], but which is typically consumed by individuals at low amounts (1–4 g/day) [18]. These participants were not subject to any dietary restrictions for this study, and diet was monitored throughout the study by an initial Diet History Questionnaire III (DHQ3) and two Automated Self-Administered 24-Hour Dietary Assessments (ASA24).
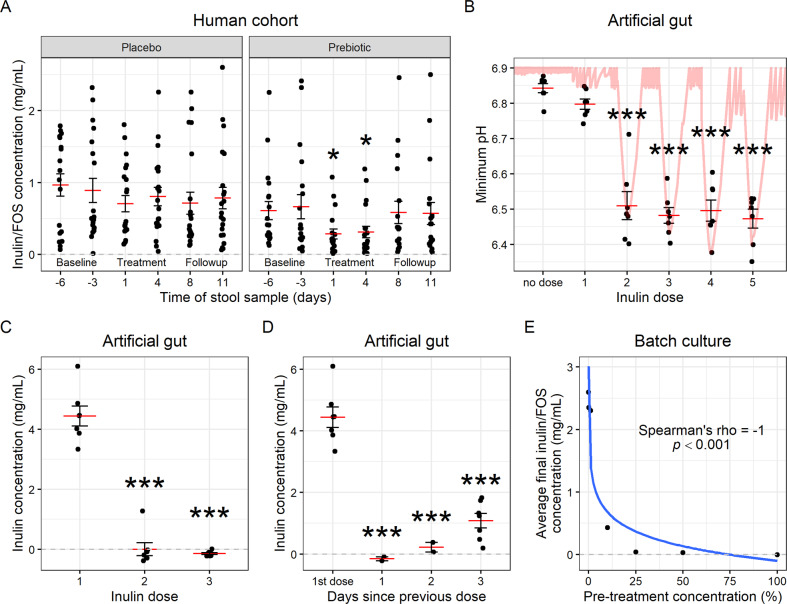
We measured metabolic potential before and after inulin exposure using an ex vivo assay of the capacity for the fecal microbiome to degrade carbohydrates over the course of 24 h [19]. We observed a significant increase in inulin degradation after participants in the prebiotic group consumed inulin for one day (p = 0.028, mixed-effects GLM; Figs. 1A, S2A). An improvement in average metabolic potential between Week 1 to Week 2 was observed for 17/19 (90%) of participants in the prebiotic group (Fig. S2A). We did not identify any specific taxa in this dietary intervention cohort significantly altered by treatment (Benjamini-Hochberg adjusted p > 0.05, ALDEx2 GLM; Fig. S3A–C), an outcome we consider likely due to different microbial species sharing the same functional role across individuals [20] as well as the short-term nature of our intervention. These findings suggest that within 24 h of a nutrient shift, the human gut microbiome alters its metabolic potential to favor metabolism of the added nutrient. Importantly, since this was a free-eating cohort, our findings demonstrate the practical capability of supplemental fiber to induce measurable metabolic changes in vivo within the context of typical day-to-day dietary variation.
Fig. 1. Ecological memory of prior nutrient exposure is encoded within 24 h.
A Inulin/FOS concentrations after incubation of slurried stool samples with inulin for 24 h ex vivo. (p = 0.028 and 0.019, mixed-effects GLM; n = 21 participants in placebo group, 19 in prebiotic group.) B Minimum pH reached over the 24 h following each inulin dose. Continuous pH trace for Vessel 1 shown. (Mixed-effects linear model with “no dose” as intercept; n = 7 vessels.) C Concentration of inulin remaining in each artificial gut vessel 6 h after dosing. (Mixed-effects linear model with dose 1 as intercept; n = 7 artificial gut vessels.) D Concentration of inulin remaining in each vessel after 6 h by time since previous dose. (Mixed-effects linear model with 1st dose as intercept; n = 7, 2, 2, and 7 vessels, respectively. Setting the model intercept to 1 day shows a significant difference, p = 0.048, between 1 and 3 days.) E Average final inulin concentration (of triplicate batch cultures) after 6 h incubation on inulin, preceded by pre-treatment with an inulin dose of varying concentration. Log2 regression line plotted. (n = 7 conditions.) A–D Mean and standard error plotted. *p < 0.05, **p < 0.01, ***p < 0.001.
To examine how inulin exposure altered the metabolic potential of the human gut microbiome, we considered three primary facets of ecological memory: lag, duration, and strength [1]. Lag refers to the amount of time after a disturbance before the event is translated into a differential response to future stimuli. To measure memory lag with greater resolution and control than possible among humans in vivo, we employed an “artificial gut” model of the distal colon seeded with a fecal microbiome sample from a healthy human donor (Fig. S1E, F) [21]. To measure lag, we dosed each artificial gut vessel with a single 2 g dose of inulin each day (calculated initial concentration = 5 mg/mL). We measured differential responses to inulin supplementation by direct measurement of inulin concentration at −2 h and +6 h from each dose, as well as by continuous monitoring of pH, which decreases as inulin is metabolized [22] and impacts the overall ecology of the gut microbiome [23]. These measurements revealed that by the second day of inulin treatment, gut microbial metabolism was altered, as ecosystem pH reached a significantly lower minimum value on the second and subsequent days of dosing compared to the first (p < 0.001, mixed-effects linear model, Fig. 1B). Likewise, significantly less inulin remained in vessels at +6 h on the second and third days of dosing (p < 0.001, mixed-effects linear model, Fig. 1C). While nearly the entire initial concentration of inulin remained at +6 h from dose 1, the full amount had been either metabolized or washed out (by the continuous flow of spent media out and fresh media in) of the vessel at +22 h (Fig. S2B). Together, these findings suggest the gut microbiome can encode memory to a nutritional stimulus within a day of exposure.
We next investigated the duration and strength of microbiome memory. We added a second dosing week to our artificial gut model in which we varied the length between doses (Fig. S1E). Extending this period to two days between doses reduced, but did not entirely negate, the enhanced pH response (Fig. S1C). Microbiome potential to degrade inulin persisted even longer and remained enhanced when doses were separated by three days (p < 0.001, mixed-effects linear model; Fig. 1D), although the response at 3 days was less than that at 1 day (p = 0.048; mixed-effects linear model with 1 day as intercept). We then set up a high-throughput in vitro anaerobic batch culture system to measure how the strength of microbiome memory varies following a wide range of inulin exposures [24]. We observed a negative dose-dependence between concentration of inulin pre-treatment and subsequent metabolic memory (Spearman correlation p < 0.001; Fig. 1E, S2D). Notably, maximal inulin breakdown efficiency and acidification were reached below our original dose, which suggested that the gut microbiome’s ability to be primed for inulin metabolism could be saturated.
Having observed changes in community-level metabolism, we employed a multi-omics approach to determine the extent to which repeated nutrient exposure impacted community composition and function. While none of the 103 taxa analyzed by 16 S rRNA gene amplicon sequencing were found to have changed in abundance following the first inulin exposure (dose 1 h 6), forty-nine (48%) had altered abundance following the second exposure (95% credible interval excluding zero, Bayesian multinomial logistic-normal model; Figs. 2A, S4C). Similarly, RNA sequencing revealed that across the global transcriptome (i.e. the collective transcriptome of the whole community), only two gene groups were differentially expressed after the first inulin dose; following the second dose, 18 genes were differentially expressed (p < 0.05, ALDEx2 GLM with Benjamini-Hochberg correction; Fig. 2B). The two rapidly upregulated gene groups (K01212, levanase [EC:3.2.1.65]; and K01238, SUN family beta-glucosidase [EC:3.2.1.-]) both encode glycoside hydrolases. Analyzing transcriptional changes within taxa, we found that the upregulation of these genes came from three Bacteroides taxa: B. caccae, B. ovatus, and B. uniformis (Fig. 2C), including previously studied inulin utilization loci within these taxa [25, 26]. Furthermore, the initial set of differentially expressed transcripts was significantly enriched for carbohydrate metabolism and transport functions (COG category “G”; p = 0.020, chi-squared test; Fig. S5D). Following dose 2, there was a 2.6-fold change in the number of genes with expression changes (p < 0.05, ALDEx2 GLM with Benjamini-Hochberg correction), and an additional 19 taxa (a 50% increase) had at least one such transcript (Fig. S3B).
Fig. 2. Compositional, transcriptomic, and metabolic changes reflect nutritional memory in artificial guts.
A Examples of taxa found to be significantly altered in abundance with inulin treatment by 16S rRNA gene amplicon sequencing (of 59 total differentially abundant taxa shown in Fig. S4), selected to highlight the different outcomes observed. B. caccae (ASV 13) and Lachnospiraceae NK4A136 group sp. (ASV 37) increased by the dose 2 h −2 (positive β2, β3, and β4), Bacteroides sp. (ASV 22) increased by dose 2 h +6 (positive β3 and β4), Hungatella sp. (ASV 24) and Erysipelatoclostridium ramosum (ASV 36) increased transiently for the second and subsequent doses (positive β3 only), and Lachnoclostridium sp. (ASV 10) decreased after the second dose (negative β3 and β4). Dashed lines represent times of inulin dosing. (Multinomial logistic-normal model; n = 6 vessels.) B Heatmap of KEGG gene functions found to be differentially expressed by time point in the global transcriptome. Color scale denotes ALDEx2 effect size estimate relative to baseline (ALDEx2 GLM significance from baseline shown; n = 5 vessels.) C Gene transcripts significantly differentially expressed in the within-taxon analysis and mapping to KEGG categories increased at the earliest time point in panel (B). D Total SCFA concentration in artificial gut vessels over time. (Mixed-effects linear model with dose 1 h −2 (day 1.42 on x-axis) as intercept; n = 7 vessels.) E Change in concentration of select metabolites relative to baseline. Top row: fructose metabolism intermediates. Bottom row: amino acids. (Mixed-effects linear model significance from baseline shown; n = 6 vessels.) A, C–E Dot colors are used to distinguish individual artificial gut vessels. Mean and standard error plotted. *p < 0.05, **p < 0.01, ***p < 0.001.
Metabolomic analyses further confirmed widespread alterations to microbial biochemical activity and environment following repeated nutrient exposure. Total short-chain fatty-acid (SCFA) content was not significantly altered after the initial inulin dose, but was significantly increased after the second (p < 0.001, mixed-effects linear model; Fig. 2D), a trend which was driven by increases in acetate and butyrate (Fig. S6A). Since SCFA production is a major driver of acidification in the gut [27], these observations may explain the observed lag in pH decrease (Figs. 1B, S2C). Untargeted metabolomics revealed a 15.5-fold increase (2–31) in the number of metabolites whose levels changed after the second dose of inulin relative to the first (p < 0.05, mixed-effects linear model; Figs. 2E, S6B). Notably, the set of metabolites that increased after the second dose included fructose breakdown intermediates and five amino acids (p < 0.05, mixed-effect linear model; Figs. 2E, S6B), which suggests a shift from proteolytic metabolism towards a more saccharolytic state [28, 29]. More broadly, our multi-omic analyses point to significantly greater changes in microbiome composition and activity as being associated with repeated nutrient exposure.
We investigated the specific ecological shifts that could amplify microbiome responses to a second inulin dose. We did not observe evidence that extracellular secretions induced memory, as inulin metabolism among inulin-naïve cultures could not be enhanced by adding conditioned media from inulin-treated cultures (Fig. S7A). We also did not find that changes to overall cell density in culture were related to inulin metabolic rates (Fig. S7B-C). We therefore hypothesized that nutrient-induced changes in transcription and abundance, which have been well documented in individual taxa [5–7, 30], drove the observed ecological memory. Indeed, we were able to positively associate community taxonomic changes with microbiome memory to inulin exposure. Our 16S rRNA gene amplicon sequence analysis of artificial gut communities in the two hours prior to the second inulin exposure revealed that 19 out of 103 analyzed amplicon sequence variants (ASVs) were significantly altered in abundance (95% credible interval excluding zero, Bayesian multinomial logistic-normal model; Fig. S4) relative to two hours prior to the first dose. These ASVs included Bacteroides caccae, a known primary degrader of inulin (Virtual Metabolic Human [VMH]) [31], as well as Bacteroides sp. and Bifidobacterium sp., two genera that contain inulin-degrading species, and taxa such as Lachnospiraceae NK4A136 group previously shown to be associated with intestinal SCFA levels [32] (Figs. 2A, S4C). To test whether increased relative abundance of a single primary degrader could confer enhanced metabolic potential, we added an inulin-naïve B. caccae isolate derived from our artificial gut community (since we had observed both transcriptional and abundance changes in this taxon) to inulin-naïve stool-derived mixed community cultures. B. caccae spike-in was sufficient to enhance inulin metabolism (Fig. S7D), supporting the hypothesis that changes in the abundance of primary degraders may be sufficient to drive memory in microbial communities.
Given the known role of polysaccharide utilization loci (PULs) [33] in carbohydrate sensing among individual gut microbes [34] and in the context of cooperative and competitive interactions in the gut microbiome [25, 26], we also expected to observe changes among select microbes after initial exposure to inulin in our artificial gut. Indeed, in our metatranscriptomics results, we found activation/repression of PULs in twelve Bacteroides taxa in the hours preceding the second inulin dose (Figs. 3A, S8). In order to circumvent the influence of compositional changes on our interpretation of the metatranscriptome, we ran an individual ALDEx2 GLM for each taxon; thus, these differences reflected changes in the expression of genes within a given genome and not simply changes in bacterial taxonomic abundance. The PULs identified included genes encoding inulin-degradative glycoside hydrolase family 32 (GH32) enzymes, which were also upregulated in the global transcriptome after a second inulin dose (Fig. 2B, C), as well as transcriptionally linked SusC/SusD homologs, which work together to bind and import oligosaccharides [34] (Figs. 3A, S8). Given, however, that these PULs were activated even earlier, at dose 1 h 6 (Figs. 2C, 3A, S8), we suspected that transcriptional changes played a role in enabling growth on the new substrate. In support of this hypothesis, we found that even after controlling for starting cell density, inulin pre-treatment enhanced the ability of B. caccae to grow on inulin (Fig. S7E, F). We therefore suspect that patterns of transcriptional change observed when monocultures of bacteria leave lag phase during diauxic shifts [30] are likely to also occur among Bacteroides species in a mixed species setting.
Fig. 3. Bacteroides PUL activation precedes broader transcriptional and compositional changes in artificial guts.
A Activation and repression of select PULs in Bacteroides by time point. (ALDEx2 GLM; n = 5 artificial gut vessels). Gene text color depicts functional categories from PULDB, notably SusC/SusD systems and glycoside hydrolase family 32 (GH32) enzymes (GH glycoside hydrolase family, HTCS hybrid two-component system, MFS major facilitator superfamily transporter, unk unknown). Not all distances between genes to scale. B Corrected p values by time point for all genes analyzed within taxa by ALDEx2 GLM. Taxa clustered along x-axis by output of phylogenetic tree clustering. At Dose 1 h +6, we detected 420 differentially expressed genes among 38 taxa (17 of which were Bacteroides), from a post-filtering set of 62,181 genes in 178 taxa (37 Bacteroides). At Dose 2 h −2, there were 355 genes from 44 taxa (23 Bacteroides). At Dose 2 h +6, there were 1082 genes from 57 taxa (25 Bacteroides). *p < 0.05, **p < 0.01, ***p < 0.001.
We also identified upregulated sets of syntenic genes outside of Bacteroides, identifying several putative carbohydrate-associated loci (p < 0.05, ALDEx2 GLM with Benjamini–Hochberg correction; Fig. S9) in taxa that degrade products of inulin hydrolysis (glucose, fructose, sucrose, or short-chain fructo-oligosaccharides [scFOS]) [31]. Seven Bacteroides and two Clostridium species had loci with at least one gene still significantly upregulated two hours prior to the second prebiotic dose (Figs. S8 and 9). Three of these Bacteroides taxa maintained activation of genes encoding inulin-degrading enzymes, and B. caccae also continued to upregulate a fructokinase gene (p < 0.05, ALDEx2 GLM with Benjamini-Hochberg correction; Figs. S8, S5E). This transcriptional activity, coupled with enhanced degradation of inulin (Fig. 1C), may have enabled additional taxa to upregulate genes involved in pathways downstream of inulin processing [35] (Figs. 2B, S5E) and increase SCFA production (Figs. 2D, S6A). Our metagenomic and metabolomic data suggest that sustained expression of GH32 enzymes, along with increased relative abundance of the taxa that produce them (Fig. 2A), prime microbial communities for a more widespread metabolic response to inulin upon re-exposure (Fig. S5E).
Given that expression of a particular PUL or growth of a certain taxon may be activated by multiple substrates [25], we reasoned that ecological memory of inulin may trigger, or be triggered by, ecological memory of related carbohydrate compounds. We found evidence for this model in our human dietary intervention study. Participants’ total baseline dietary fiber intake, as estimated by Diet History Questionnaire III (DHQ3), was negatively correlated with fecal inulin/FOS content in donor stool one day after the start of treatment in the prebiotic group (p = 0.023, Spearman correlation) but not the placebo group (p = 0.40, Spearman correlation; Fig. 4A). Thus, participants who habitually consumed a high rate of dietary fiber initially tended to excrete less undigested inulin than individuals with low habitual fiber consumption. Furthermore, a positive correlation was found between the change in fecal inulin/FOS content from treatment day 1 to treatment day 4 in the prebiotic group only (p = 0.024, Spearman correlation; Fig. 4B). Baseline fecal inulin/FOS concentrations did not differ between groups (Fig. S2E), and therefore does not explain these differences. This correlation suggests that low habitual fiber consumers stand to receive the greatest benefit in metabolic potential from supplementation, consistent with prior findings from our lab [36].
Fig. 4. Ecological memory is cross-reactive.
A, B Correlation between participant baseline dietary fiber (DHQ3) intake and fecal inulin/FOS content on treatment day 1 (A) or the change in fecal inulin/FOS from treatment day 1 to treatment day 4 (B). (n = 21 participants in placebo group, 16 in prebiotic group.) C Correlation between average degree of polymerization (DP) of pre-treatment inulin or constituent sugar and average final inulin concentration (of triplicate cultures; n = 6 conditions). D Final pH after 6 h fermentation on dextrin, GOS, or inulin, with and without pre-treatment of the same prebiotic. (t-test; n = 3 batch cultures.) Final inulin/FOS concentration (E) and pH (F) after 6 h fermentation on inulin, after pre-treatment with different prebiotics. (Linear models with control as intercept; n = 3 batch cultures.) Mean and standard error plotted. *p < 0.05, **p < 0.01, ***p < 0.001.
We tested the cross-reactivity of ecological memory to inulin degradation using in vitro batch cultures with the same inoculum used for the artificial gut experiments. We treated human gut microbiome cultures with the constituent components of inulin (fructose, glucose, sucrose, and FOS/inulin of various chain lengths). This treatment indeed enhanced subsequent inulin metabolism to varying degrees (p < 0.05, linear model; Fig. S10A, B). The magnitude of this effect was correlated with component chain length (p = 0.0499, Spearman correlation; Fig. 4C). To test ecological memory responses to non-fructan polysaccharides, we fermented batch cultures with dextrin, galacto-oligosaccharides (GOS), inulin, or control, performing every combination of pre-treatment and second treatment (16 combinations total). We found evidence of prebiotic memory within both dextrin and GOS based on final culture pH (p < 0.05, t-test; Fig. 4D). We additionally found that exposure to GOS or dextrin resulted in partial, significant increases in inulin degradation capacity (p < 0.01, linear model; Fig. 4E) and significant decreases in culture pH relative to control following inulin treatment (p < 0.001, linear model; Fig. 4F). Community analysis of microbiome cultures revealed that while no taxa were significantly correlated with the amount of inulin remaining in culture (Benjamini–Hochberg corrected ALDEx2 Spearman correlation p > 0.05, Fig. S10C), we did identify taxa that significantly correlated with pH (Fig. S10D). In both analyses, the top correlated taxon was Erysipelatoclostridium ramosum, which was correlated with subsequent inulin response in terms of pH (p = 0. 0.0036, Benjamini-Hochberg corrected ALDEx2 Spearman correlation; Fig. S10E, F), and significantly increased in abundance by pre-treatment with each of the three prebiotics tested (p < 0.001, Benjamini-Hochberg corrected ALDEx2 GLM; Fig. S10G). E. ramosum is a known degrader of scFOS, as well as the component mono- and di-saccharides of GOS and dextrin, respectively [31], which suggests that cross-reactivity may be driven by increased abundance of generalist taxa. These findings support a model where ecological memory within a microbial community can exhibit cross-reactive properties in which altering the abundance of a nutrient-degrading microbial species ultimately impacts the rates at which related nutrients will subsequently be metabolized. Furthermore, which taxa encode ecological memory is likely dependent both on the specific nutrient and which primary degraders are present in the starting community.
Discussion
In concert, our findings demonstrate the existence of ecological memory of past nutrient exposure in the human gut microbiome. Previous microbiome studies have also shown that past disturbances to the gut microbiome induce memory involving host processes (e.g. host metabolite production in response to infection [13], host weight gain [14]). However, it had not been previously shown that nutrient memory occurs as a community-level emergent property. Here, we elucidate fundamental processes underlying ecological memory in microbial communities. In particular, while microbiome metabolism is often measured by SCFA production or transcriptional analysis, we directly measured inulin breakdown over time (Fig. 1A, C, D), enabling us to assess both the immediate action of inulin degraders as well as the effects of broader cross-feeding interactions on the metabolome (Figs. 2D, E, S6). Our results support a working model in which transcriptional changes among primary prebiotic degraders after nutrient exposure allow levels of these degraders to expand; following expansion, the elevated abundance of primary degraders is sufficient to encode nutrient memory (Figs. 2A–C, S7D–F). We hypothesize these shifts among primary degraders initiate a cascade of nutrient sensing events driven by cross-feeding interactions that leads to further changes in community composition (Figs. 2A, S4C) nutrient breakdown (i.e. increased saccharolytic and reduced proteolytic fermentation; Figs. 2E, S5E, S6B), and metabolite production (e.g. SCFA production; Figs. 2D, S6A).
Previously, metagenomic studies have inferred functional changes on the order of days or weeks based on changes in genes detected, often limited by sampling frequency [37, 38]. Changes in expression of metabolism-related genes have been shown by metatranscriptomic analysis in participants during a five day dietary intervention [39]. Our findings suggest ecological memory can be encoded even more rapidly, on sub-daily time scales for the human gut microbiome (Fig. 2B, C), consistent with past findings showing that fecal SCFA content varies by the time of day samples are taken, which suggested the possibility of sub-daily functional shifts [40]. This response time is consistent with how quickly individual microbes are known to undergo diauxic shifts, which also occur on the order of hours [41]. Importantly, these time scales are relevant to human nutrition, as we showed that low-fiber consuming individuals excreted more inulin on the day after their first dose, but that just one day was sufficient time to improve the metabolic potential of the microbiome of participants (Figs. 1A, 4A, B). These findings demonstrate that intrinsic ecological processes of microbial communities may exhibit memory, and suggest a potential for such fundamental processes to underlie downstream host outcomes.
Selective pressures for rapid microbial metabolic change may exist in environments like the mammalian gut, where community members replicate on short time scales and nutrient availability varies in both stochastic and rhythmic manners within a single day [42]. Such adaptation may also benefit hosts by providing them with an adaptive microbial response to dietary shifts [40]. Yet, given the benefits to rapid metabolic plasticity, it is perhaps surprising that we also observe evidence that after a nutrient is withdrawn, ecological memory persists for days and therefore likely across multiple generations of bacteria. Bacterial communities or their members may benefit from “bet hedging” strategies that balance the odds that a withdrawn nutrient is reintroduced [30]. It is also possible that once the transcriptional, compositional, and metabolomic landscape of a microbial community has been altered, restoring to an original starting state will encounter delay [43]. Understanding these temporal dynamics can aid in the rational design of prebiotic treatments by highlighting the importance of recent nutritional exposures and the frequency of treatment dosage.
Our finding of ecological memory in human gut microbial systems also suggests avenues for the rational design of treatments that alter how the gut microbiome harvests energy [44] or metabolizes drugs [45, 46]. To date, individualized therapies have accounted for inter-individual variation in microbiome composition and function, which have been linked to fixed population differences in both overall diversity and specific taxonomic differences [47, 48]. Yet, if the metabolic potential of the gut microbiome is plastic, observed microbiome heterogeneity may also reflect recent intra-individual variation in behavior or lifestyle. Indeed, it was recently shown that antibiotic administration drives changes in metabolic potential to degrade dietary fiber [49]. Conversely, given our observations on prebiotic cross-reactivity (Figs. 4, S10), it is possible that prebiotic administration may be used to broadly alter a gut microbiome’s metabolic state prior to an unrelated intervention in order to enhance positive effects or reduce undesired ones. Rationally designed therapies may therefore benefit from monitoring changes to, and even manipulating, microbiome metabolic potential over time [50]. For example, our work suggests that microbiome-targeting nutritional interventions have the most potential to impact the microbial metabolism of individuals who are normally deficient in that nutrient’s intake (Fig. 4A, B). Moreover, our finding that microbiome memory does not persist indefinitely suggests that such interventions will require repeated administration to sustain their microbial ecological impact.
Materials and methods
Carbohydrate sources
The following sources of fructan prebiotics were used in this study: Synergy1 inulin (Orafti), inulin from dahlia tubers (Sigma), fructo-oligosaccharides from chicory (Sigma), inulin (NOW), and inulin FOS (Jarrow) (Fig. S11). Orafti Synergy1 inulin was used for the human study as a food-safe ingredient that could be added to the snack bars, Sigma inulin from dahlia tubers was used for in vitro work including the artificial gut run as a relatively pure long-chain inulin. Jarrow inulin FOS was used for its greater solubility in select in vitro experiments (Figs. 4D, E, S7B, C, E, S10C–G), where it was necessary to measure absorbance and/or ensure that pre-treatment inulin could be removed (via supernatant) prior to the treatment phase.
The following sources of non-fructan prebiotics were used: dextrin (Benefiber) and galacto-oligosaccharides (Bimuno). The following simple sugars were also used: dextrose (Amresco), fructose (Amresco), and sucrose (Sigma).
Participant recruitment and sample collection
Human participant data presented in this paper is a secondary, post hoc analysis of a cohort recruited primarily to test a link between the gut microbiome and aspects of cognition and behavior. The original study protocol was approved by the Duke Health Institutional Review Board (IRB) at Duke University under protocol number Pro00093322, and registered on ClinicalTrials.gov, with the identified number NCT04055246. We recruited 40 healthy subjects by use of flyers on Duke University campus as well as electronic postings on DukeList (a university-internal classifieds website), a lab website, and ClinicalTrials.gov. All patients provided written informed consent via an electronic consent form (eConsent) prior to participation in the study protocol.
All participants were between the ages of 18 and 35, weighed at least 110 pounds (50 kg), and had a body mass index (BMI) between 17.0 and 27.5. Furthermore, individuals were excluded from participation if they: had a diagnosis of psychiatric or neurological disorder, were currently on steroid medications, had used recreational drugs within the past month, consumed at least 2 alcoholic beverages per day on average, had dietary restrictions of milk or dairy products, had food allergies to wheat/gluten/nuts/soy, or were currently pregnant or breastfeeding. A secondary screen further excluded individuals who scored greater than 9 on a modified Patient Health Questionnaire-9 (PHQ-9, a depression severity questionnaire), had a colonoscopy or oral antibiotics within the past month, or had a history or current diagnosis of any of the following: irritable bowel syndrome, inflammatory bowel disease, type 2 diabetes, chronic kidney disease with decreased kidney function, intestinal obstruction, or untreated colorectal cancer.
Participants were 60% female (24/40) and 40% male (16/40). Participants were 57.5% white (23/40), 40% Asian (16/40), 2.5% Black (1/40), 2.5% Native American (1/40), 2.5% Native Hawaiian or Other Pacific Islander (1/40), and 12.5% Hispanic or Latino (5/40). Average age at time of enrollment was 25.6 ± 4.9 years (mean ± stdev). Most participants (67.5%; 27/40) were omnivores, 17.5% were vegetarian (7/40), 12.5% ate everything except red meat (5/40), 2.5% did not respond to this question (1/40), and none were vegan. Average weight was 147.9 ± 21.6 lb and average BMI was 23.2 ± 2.2.
Participants provided stool samples twice weekly on Tuesday and Friday over a 3-week study period (Fig. S1A). Samples were self-collected using polypropylene scoop-cap tubes (Globe Scientific), and participants were instructed to keep samples in personal freezers until ready to transport to lab using a provided insulated container and ice pack. Once arrived, samples were kept at −20 °C for up to a week and then moved to −80 °C until further processing. If a participant was unable to produce a sample on the requested day, they were instructed to provide the next available sample (on Wednesday or Thursday in place of a Tuesday sample, or on Saturday or Sunday in place of a Friday sample). For statistical analysis purposes, we treated any such samples as Tuesday or Friday samples, since they still represented the first possible measurement after 1 or 4 days, respectively, of consuming the snack bars.
On the second study week, participants consumed treatment bars (Fig. S1B) twice daily for five days (Monday through Friday) containing either 9 g/bar inulin (prebiotic group, n = 19) or 9 g/bar maltodextrin (placebo group, n = 21) (Fig. S1D). Participants were blinded to which group they were in (Fig. S1C). The bars were manufactured by the Natick Soldier Research Development and Engineering Center (NSRDEC) in Natick, MA, USA, based on a modified formula of the First Strike bar manufactured at NSRDEC. This formulation does include fructose, the constituent monosaccharide of inulin (Fig. S1D). Due to the absorption of fructose by the upper GI tract, gut microbes, particularly colonic microbes, are not exposed to significant levels of free fructose from diet; inulin, however, is recalcitrant to breakdown and absorption by human enzymes and does serve as a nutrient source for colonic gut microbes in vivo [51].
Participants were not asked to restrict their diet for the study, and instead were instructed to continue their typical diet. All participants completed three dietary surveys. The Diet History Questionnaire III (DHQ3) was administered prior to the Baseline and assessed participants’ eating habits over the past month. The Automated Self-Administered 24-h Dietary Assessment Tool (ASA24-2018) was administered twice, once during the baseline week and once during the treatment week.
In some instances, a participant did not provide a stool sample on the day requested or not enough sample was present for all analyses, and one participant did not complete the DHQ3. Thus, n is not always 19 for the placebo group and 21 for the prebiotic group; otherwise, no samples were intentionally excluded from analysis.
Measurement of metabolic potential in stool samples
To measure the capacity of microbial communities to degrade prebiotic substrate, we used a previously described stool fermentation assay [19]. Briefly, stool samples were weighed out in an anaerobic chamber and combined with pre-reduced phosphate-buffered saline (PBS) at 10% weight/volume in polyethylene filter bags with 0.33-mm pore size (Whirl-Pak B01385). Samples were then homogenized (aerobically) using a stomacher (Seward Stomacher 80 MicroBiomaster) set to medium for 60 s. Total time exposed to oxygen did not exceed 10 min.
Either 1 mL of either PBS (control) or 1% (10 mg/mL) solution of Orafti Synergy1 inulin was added to 24-well plate wells. Then, 1 mL of stool slurry was aliquoted into each well (final concentration of inulin 5 mg/mL). Plates were sealed with adhesive foil seals and incubated anaerobically for 24 h at 37 °C. Aliquots were taken from each well and saved at −80 °C.
Artificial gut culturing and sampling
Artificial guts were run according to a previously established protocol [21]. An eight-vessel continuous flow artificial gut system (Multifors 2, Infors) was used to culture gut microbes seeded from human stool samples. One of the vessels (vessel 5) was not inoculated for this run due to a component that was found to be non-functioning prior to inoculation. Replicate artificial gut vessels were uniformly inoculated with a starting community derived from a single healthy stool donor so that our analyses could assume measurement variation was due to technical sources of noise [21]. Since these vessels maintain stool-derived cultures, we consider them as models of the distal colon [52]. Vessels were sterilized and prepared with 300 mL of fresh modified Gifu Anaerobic Medium (mGAM) [53]. Nitrogen was sparged into the vessels at 1 L/min to maintain positive pressure and create an anaerobic environment [54]. Vessels were inoculated using culture aliquots saved at −80 °C from a previous artificial gut run (original fecal samples obtained from a healthy volunteer who provided written informed consent per Duke Health IRB Pro00049498). From each of 12 frozen aliquots, 1.5 mL was added to 5 mL mGAM media and incubated for 6 h at 37 °C in an anaerobic chamber (Coy). Five milliliters of these cultures were then transferred to media bottles containing 100 mL mGAM and incubated overnight (17 h) at 37 °C. After this final incubation, cultures were combined aseptically in a biosafety cabinet and loaded into syringes, with two 50 mL aliquots inoculated into each vessel via a septum in the vessel lid, for a final working volume of 400 mL. The media feed was started 24 h after inoculation at a constant rate of 400 mL per day (a rough approximation of 24 h average passage time in the human GI tract). Dosing began on day 14 after inoculation; the length of this “burn-in” period is based on prior studies suggesting that it takes approximately 2 weeks for the community composition to stabilize [55].
The pH of each vessel was monitored and controlled by the IRIS software (v6, Infors), and pH was measured continuously with Hamilton EasyFerm Plus PHI ARC 225 probes. The pH probes were calibrated with a two-point calibration with standardized pH buffers at 4.00 ± 0.1 and 10.00 ± 0.1 (BDH). Vessel pH was maintained within the ranges of 6.8 ± 0.1 during the burn-in phase and 6.2 ± 0.7 thereafter (to enable us to observe fermentation-induced pH shifts) using a 1 N HCl solution and a 1 N H3PO4 solution. Prior to dosing, pH was stable at around 6.9. Vessels were maintained at 37 °C via the Infors’s onboard temperature control system. Vessels were continuously stirred at 100 rpm using magnetic impeller stir shafts.
Prior to sampling, sampling lines were cleared with a 0.2 μm filter-tipped syringe and wiped clean with 70% ethanol. Sampling consisted of the collection of 7 mL of active artificial gut culture via sterile syringe and then immediate storage in labeled cryovials at −80 °C. Dosing was done by combining 16 g inulin with 80 mL PBS in a biosafety cabinet, and administering 10 mL of the mixture to each vessel by needle-equipped syringe through a septum at the top of the vessel.
Samples from all seven inoculated vessels were analyzed for inulin content, pH, and SCFA content, as described below. Due to an issue with media flow on the final day of treatment (day 11; Fig. S1E), samples from this day were not analyzed for inulin content, and instead, samples from all 7 vessels on day 8 were used to test the effect of 3 days between doses (Fig. 1D). Due to analytical pipeline throughput limitations, we prioritized samples from six of the vessels for metatranscriptomic sequencing and untargeted metabolomics. Additionally, DNA/RNA sequencing data from one of the artificial guts (vessel 3) was excluded from statistical analyses involving the origins of ecological memory, due to this replicate exhibiting evidence for a further delayed memory effect (outlier point at dose 2 in Fig. 1B, C).
Small-batch bacterial culturing
To perform controlled in vitro experiments in a more high-throughput way than the artificial gut allowed, we carried out small-batch anaerobic culture experiments based on previously established protocols [24]. Cultures were started from a 1 mL frozen aliquot from a previous artificial gut run added to 4 mL mGAM, which was grown anaerobically for 6–8 h at 37 °C in a 14 mL polystyrene round-bottom tube. This 5 mL culture was then added to 45 mL mGAM and grown overnight (16–18 h) at 37 °C. Following this overnight culture, culture tubes each containing 3 mL mGAM were inoculated with 1 mL overnight culture for a total volume of 4 mL. Cultures were pre-treated by adding 200 μL of carbohydrate at 100 mg/mL (except for the dose-response experiment), for a final concentration of 4.8 mg/mL. After 24 h incubation, cells were spun down 10 min at 3000 × g and resuspended in fresh mGAM along with 200 μL of inulin at 100 mg/mL. Samples were collected after a final 6 h incubation. Inulin concentration was analyzed by HPAEC-PAD as described below and endpoint pH was measured by Accument AR-15 pH meter (Fisher Scientific).
Conditioned media was generated by growing stool-derived microbiota cultures on mGAM overnight and filter-sterilizing the resultant culture. The B. caccae isolate used was previously isolated from the community used for the artificial gut experiment. For the spike-in experiment (Fig. S7D), cultures were grown overnight, then diluted 1:10 and grown for 6 h with or without inulin. OD600 was measured, and diluted with fresh mGAM to a combined calculated OD600 of 1.0 in each 4 mL culture, with B. caccae and whole community cultures mixed at 10/90 or 50/50% by OD600.
For the growth curve experiment (Fig. S7E, F), cultures were pre-treated for 9 h on (inulin-naïve) conditioned mGAM supplemented with 4.8 mg/mL glucose or inulin/FOS (Jarrow) at which point OD600 was measured and cultures were diluted to a calculated OD600 of 0.5 into 200 μL conditioned mGAM plus 10 μL of 100 mg/mL glucose of inulin (final concentration of 4.8 mg/mL) in a 96-well plate. Absorbance was measured by SPECTROstar Nano Microplate Reader (BMG Labtech) in the anaerobic chamber, with 48 readings taken at 30 min intervals, with the plate shaken before each reading (OD600). A growth curve was fit using the growthcurver package in R (Fig. S7F).
Quantification of inulin
Analysis of inulin content was performed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD). Briefly, 200 µL of each sample (in randomized order) was added to 800 µL of deionized (DI) water, ACS reagent grade, 18 MΩ-cm resistivity. Samples were then centrifuged at 14,000 × g for 5 min at 4 °C. The resulting supernatant was transferred to a 0.22 µm spin filter column and centrifuged at 14,000 × g for 5 min at 4 °C. The filtrate was analyzed on a Dionex ICS-6000 HPIC system equipped with a Dionex AS-AP autosampler and a pulsed electrochemical detector consisting of an amperometric flow-through cell and a silver/silver chloride reference electrode. The electrochemical detector provided the following waveform: E1 = + 0.1 V (t1 = 0.00–0.40 s), integration from 0.20 to 0.40 s, E2 = −2.0 V (t2 = 0.41–0.42 s), E3 = + 0.6 V (t3 = 0.43 s), E4 = −0.1 V (t4 = 0.44–0.50 s).
Separation was carried out on a CarboPac PA200 analytical column equipped with a CarboPac PA200 guard column at 30 °C. The autosampler was kept at 4 °C to prevent degradation of carbohydrates. A gradient elution was performed using the following eluents: 100 mM NaOH (eluent A) and 1 M NaOAc, 100 mM NaOH (eluent B). The eluents were kept blanketed under nitrogen at 6 psi to prevent the formation of carbonate. A flow rate of 0.5 mL/min was used and the linear gradient was set up as follows: 98% eluent A and 2% eluent B from −5 to 15 min, ramping to 50% eluent A and 50% eluent B at 70 min, then ramping back to the original parameters at 70.1 min and remaining that way until the run was ended at 75 min.
The data were acquired and processed using Thermo Scientific Dionex Chromeleon Chormatography Data System software, using inulin (inulin from dahlia tubers, Sigma), glucose (Amresco), fructose (Amresco), sucrose (Sigma), 1-kestose (≥98%, Sigma), and nystose (≥98%, Sigma) standards to identify peaks. Resulting data was processed in R by transforming peak areas to concentrations based on the inulin standard for peaks with degree of polymerization (DP) > 10, and based on an average coefficient derived from kestose and nystose standards for DP3–10. Peak values for negative control samples containing no inulin were subtracted from bacterial culture samples. Negative controls were unavailable for stool samples due to the unique nature of each sample. Since Sigma dahlia inulin contained almost exclusively long-chain inulin (DP 11+; Fig. S11B), we analyzed only this fraction for experiments using this inulin source. Otherwise, (i.e. for Orafti Synergy1 inulin and Jarrow inulin FOS), we included all inulin/FOS of DP3+ in our analysis.
16S rRNA gene amplicon sequencing
For all samples, 16S rRNA gene amplicon sequencing was performed using custom barcoded primers targeting the V4 region of the gene according to previously published protocols [19, 21, 56, 57]. Samples were randomized and DNA extractions were performed using the Qiagen DNeasy PowerSoil DNA extraction kit (ID 12888-100). Sequencing runs were standardized to 10 nM and sequenced using a MiniSeq (Illumina) with paired end 150 bp reads. DADA2 was used to identify and quantify amplicon sequence variants (ASVs) in our dataset, using version 138 of the Silva database [58]. We retained only samples with more than 5000 read counts to remove outlying samples that may have been subject to library preparation or sequencing artifacts [21], and to only retain taxa that appeared more than three times in at least ten percent of samples.
Metatranscriptomics
RNA was extracted from samples using Quick-RNA Fungal/Bacterial Miniprep Kit (Zymo Research R2014) according to the manufacturer’s instructions. Samples were then shipped to Novogene (Sacramento, CA) for further processing. mRNA was enriched using the Ribo-Zero Gold rRNA Removal Kit, Epidemiology (Illumina) and 300 bp reads were sequenced on a HiSeq (Illumina).
Analysis of sequencing reads was carried out using a pipeline adapted from previous metagenomic work [12, 39]. Briefly, reads were matched using the Bowtie 2 sequence alignment program to a reference survey from version 3.5 of the Integrated Microbial Genomes system [59]. We used the system’s annotations to the COG, [60] KEGG [61], and EC [62] databases for functional analyses. For single-species level analyses, reads that mapped to more than 1 reference genome were discarded. For global analyses, however, reads that mapped to multiple reference genomes were still counted.
We detected RNA from 1394 taxa initially; this was reduced to 343 after retaining only taxa that appeared more than three times in at least 10% of samples. The same filter was applied to genes within taxa; only genes that appeared more than three times in at least 10% of samples were retained. Finally, we could only perform our analysis (ALDEx2 GLM and PERMANOVA) on taxa that had at least two genes present (non-zero) in every sample. This filter brought the final number of taxa analyzed down to 178.
Metabolomic profiling
SCFAs were quantified by GC as previously described [19]. Briefly, samples were acidified by adding 50 μL 6 N HCl to lower the pH below 3. The mixture was vortexed and then centrifuged at 14,000 × g for 5 min at 4 °C. 750 μL of this supernatant was passed through a 0.22 μm spin column filter. The resulting filtrate was then transferred to a glass autosampler vial (VWR, part 66009-882). Filtrates were analyzed on an Agilent 7890b gas chromatograph equipped with a flame ionization detector and an Agilent HP-FFAP free fatty-acid column (25 m x 0.2 mm [inner diameter] × 0.3 μm film). A volume of 0.5 μL of the filtrate was injected into a sampling port heated to 220 °C and equipped with a split injection liner. The column temperature was maintained at 120 °C for 1 min, ramped to 170 °C at a rate of 10 °C/min, and then maintained at 170 °C for 1 min. The helium carrier gas was run at a constant flow rate of 1 mL/min, giving an average velocity of 35 cm/s. After each sample, we ran a 1-min postrun at 220 °C and a carrier gas flow rate of 1 mL/min to clear any residual sample. All C2:C5short-chain fatty acids were identified and quantified in each sample by comparing to an eight-point standard curve.
Additional metabolites were quantified by untargeted GC/MS similar to previously described work [63] with some differences noted below. As above, samples were pre-processed by vortexing, centrifuging at 14,000 × g for 5 min at 4 °C, and passing 750 μL of this supernatant through a 0.22 μm spin column filter. Proteins were then precipitated by adding 750 μL methanol (containing 6.25 mg/L C14:0-D27 myristic acid as retention time standard) to 100 μL of each sample in a glass vial. After additional vortexing and centrifugation, 700 μL of each supernatant was transferred into a fresh glass vial for 4–6 h of drying on a SpeedVac. Next, 100 μL toluene was added for a final step of azeotropic drying for 30 min, and the dried samples were stored at −80 °C for later derivatization. Subsequent steps included addition of 25 μL methoxyamine HCl with incubation at 50 °C for 30 min (for methoximation of reactive carbonyl groups) followed by addition of 75 μL N-methyl-N- (trimethylsilyl) trifluroacetamide (MSTFA, Cerilliant M-132) and similar incubation for trimethylsilyl replacement of exchangeable protons. Supernatants were transferred to GC vials and analyzed on a 7890B GC-5977B ei-MS (Agilent Corp., Santa Clara, CA). We detected 547 metabolite features that were present in at least 10% of samples, from which 206 were annotated using our GC/MS spectral library [63, 64] for use in subsequent statistical analysis (the remaining 341 included 280 unannotated, and 61 that either had uncertain annotation or were suspected contaminants). Levels for each metabolite were given as log-2 integrated peak area. For the purposes of statistical testing, we set the values of sample-metabolites that were missing, assumed below the limit of detection, to a pseudocount based on the minimum value observed for that particular feature.
Linear model design
Except where otherwise noted, statistics were performed using linear models implemented with the lme4 package in R and the lmerTest package to generate p values. For data with repeated measures from the same individual/vessel (human participants, artificial gut), a mixed-effects model was used by including a random intercept term (1 | ID) in the model formula within the lmer() function; otherwise (for in vitro cross-reactivity experiments, for which measurements were taken from distinct cultures) the lm() function was used. For the fecal metabolic potential experiment, the data were found to be non-normal by using the descdist() function in the fitdistrplus package; in this case, we therefore used a generalized mixed-effects linear model with a Gamma distribution, implemented with the following call:
For metatranscriptomic data, analysis was done using an ALDEx2 generalized linear model using the aldex.glm() function [65] with vessel and time as factors. For the within-taxa analysis, an individual GLM was constructed for each taxon in order to account for changes in the total amount of RNA contributed by a given taxon (i.e. to avoid seeing apparent changes in a transcript just because the relative compositional abundance of that taxon changed). In effect, transcript reads were treated as relative abundances within each taxon. Multiple comparisons correction was performed by the Benjamini–Hochberg method. To validate this approach, we re-analyzed four key transcripts of interest from B. caccae found to be upregulated in response to inulin (Figs 2C, 3A), by calculating the ratio of transcript reads to reads of four housekeeping genes: dnaJ (BC01081), gyrB (BC03484), recA (BC02059), and rpoB (BC00195) [66]. Mixed-effects GLMs were generally consistent with our original findings. Particularly, in the case of BC02731, analyses with all four housekeeping genes indicated sustained upregulation at each of the three post-intervention time points, consistent with our original method (Fig. S12).
The 16 S rRNA gene amplicon sequence data from the artificial gut required a more complex model to account for the multivariate and compositional nature of the data as well as the high density of repeated measures samples. We designed an autoregressive Bayesian multinomial logistic-normal model using a “pibble” model of the following form in the fido R package [67]:
where CLR is the centered log-ratio transform of the data, AR(v,t) is the first order autoregressive AR(1) portion of this model in which each time point is related to all other time points within the same vessel according to a decaying factor, β variables are effect parameters to be calculated by the model, Δt is the change in time from the previous time point in that vessel (to account for consistent drift), A1 represents samples taken on afternoon 1 (dose 1 h +6), M1 samples taken on morning 1 (dose 2 h −2), A2:5 represents samples taken on afternoons 2–5, M2:4 represents samples taken on mornings 2–4 (no morning sample was collected after dose 5), and ∈ is the model intercept.
The autoregressive portion of the model was designed by creating a variable for each sample and setting the prior covariance of samples taken from the same vessel to 0.8∆t/8, where ∆t is the number of hours elapsed between the samples. Samples taken from different vessels had a covariance of 0, and all samples had a covariance of 1 with themselves. The values of the four experimental variables (A and M) supplied in the covariate matrix were all binary, set to 1 only for samples taken at the relevant time point(s) and 0 for all other samples. The prior covariance of these variables was set to 0 with each other and 1 with themselves. Essentially, the variables were designed in order distinguish the effects at +6 h and +22 h, and to uniquely distinguish the effects on day 1, since we had previously identified unique metabolic activity (or a lack thereof) on this day (Fig. 1B). The model was robust to changes in the priors.
Supplementary information
Acknowledgements
We would like to thank Justin Silverman for assistance with sequencing and model design; Andrew Grover for help sampling from the artificial guts; Danielle Anderson and Ken Racicot for providing the snack bars used in the study; Neil Surana for helpful guidance related to our literature review; Michelle Kirtley for comments on manuscript revision; Tonya Snipes, Lisa Alston-Latta, and Margaret Huggins for keeping our lab spaces and glassware clean; and our study volunteers for their participation. This work was supported by National Institutes of Health grant 1R01DK116187, Office of Naval Research grant N00014-18-1-2616, Translational Research Institute through Cooperative Agreement NNX16AO69A, the Damon Runyon Cancer Research Foundation, and the UNC CGIBD (NIDDK P30DK034987). This study used a high-performance computing facility partially supported by grant 2016-IDG-1013 (HARDAC+: Reproducible HPC for Next-Generation Genomics) from the North Carolina Biotechnology Center. JRB received salary support from NIH 5P30DK124723, 5R01DK117491, 1U24DK129557, and 2P30AG027816.
Author contributions
Conceptualization: JL, ZCH, JRB, LAD; Data curation: JL, MJM; Formal analysis: JL; Funding acquisition: LAD; Investigation: JL, ZCH, EPD, HKD, SJ, SKG, ACM, MJM, VMC; Software: JL; Visualization: JL; Writing – original draft: JL; Writing – review & editing: JL, LAD.
Data availability
Data and code used to generate the figures presented in this paper are publicly available at https://github.com/jrletourneau/ecological_memory_microbiome. Sequence data are publicly available via the European Nucleotide Archive as demultiplexed reads with the accession numbers PRJEB45244 (metatranscriptomics), PRJEB45247 (artificial gut 16S rRNA gene amplicon sequence data), and PRJEB48837 (cross-reactivity experiment 16S rRNA gene amplicon sequence data), and PRJEB47805 (human cohort 16S rRNA gene amplicon sequence data). Our lab’s 16S rRNA gene amplicon sequence data processing pipeline is also available at https://github.com/jianhong/16S_pipeline/.
Competing interests
LAD previously served on the Strategic Advisory Board and held equity in the company Kaleido Biosciences.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01292-x.
References
- 1.Ogle K, Barber JJ, Barron-Gafford GA, Bentley LP, Young JM, Huxman TE, et al. Quantifying ecological memory in plant and ecosystem processes. Ecol Lett. 2015;18:221–35. doi: 10.1111/ele.12399. [DOI] [PubMed] [Google Scholar]
- 2.Schweiger AH, Boulangeat I, Conradi T, Davis M, Svenning JC. The importance of ecological memory for trophic rewilding as an ecosystem restoration approach. Biol Rev. 2019;94:1–15. doi: 10.1111/brv.12432. [DOI] [PubMed] [Google Scholar]
- 3.Webster CR, Dickinson YL, Burton JI, Frelich LE, Jenkins MA, Kern CC, et al. Promoting and maintaining diversity in contemporary hardwood forests: confronting contemporary drivers of change and the loss of ecological memory. Ecol Manag. 2018;421:98–108. doi: 10.1016/j.foreco.2018.01.010. [DOI] [Google Scholar]
- 4.Hughes TP, Kerry JT, Connolly SR, Baird AH, Eakin CM, Heron SF, et al. Ecological memory modifies the cumulative impact of recurrent climate extremes. Nat Clim Change. 2019;9:40–43. doi: 10.1038/s41558-018-0351-2. [DOI] [Google Scholar]
- 5.Stockwell SR, Landry CR, Rifkin SA. The yeast galactose network as a quantitative model for cellular memory. Mol Biosyst. 2015;11:28–37. doi: 10.1039/C4MB00448E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf DM, Fontaine-Bodin L, Bischofs I, Price G, Keasling J, Arkin AP. Memory in microbes: quantifying history-dependent behavior in a bacterium. PLoS ONE. 2008;3:e1700. doi: 10.1371/journal.pone.0001700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyon P. The cognitive cell: bacterial behavior reconsidered. Front Microbiol. 2015;6:264. doi: 10.3389/fmicb.2015.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith MB, Rocha AM, Smillie CS, Olesen SW, Paradis C, Wu L, et al. Natural bacterial communities serve as quantitative geochemical biosensors. mBio. 2015;6:e00326–15. doi: 10.1128/mBio.00326-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordeiro MC, Garcia GD, Rocha AM, Tschoeke DA, Campeão ME, Appolinario LR, et al. Insights on the freshwater microbiomes metabolic changes associated with the world’s largest mining disaster. Sci Total Environ. 2019;654:1209–17. doi: 10.1016/j.scitotenv.2018.11.112. [DOI] [PubMed] [Google Scholar]
- 10.Kuster SP, Rudnick W, Shigayeva A, Green K, Baqi M, Gold WL, et al. Previous antibiotic exposure and antimicrobial resistance in invasive pneumococcal disease: results from prospective surveillance. Clin Infect Dis. 2014;59:944–52. doi: 10.1093/cid/ciu497. [DOI] [PubMed] [Google Scholar]
- 11.Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, Svenson KL, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David LA, Weil A, Ryan ET, Calderwood SB, Harris JB, Chowdhury F, et al. Gut microbial succession follows acute secretory diarrhea in humans. mBio. 2015;6:e00381–15. doi: 10.1128/mBio.00381-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stacy A, Andrade-Oliveira V, McCulloch JA, Hild B, Oh JH, Perez-Chaparro PJ, et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell. 2021;184:615–27. doi: 10.1016/j.cell.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thaiss CA, Itav S, Rothschild D, Meijer MT, Levy M, Moresi C, et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature. 2016;540:544–51. doi: 10.1038/nature20796. [DOI] [PubMed] [Google Scholar]
- 15.Coyte KZ, Rakoff-Nahoum S. Understanding competition and cooperation within the mammalian gut microbiome. Curr Biol. 2019;29:R538–R544. [DOI] [PMC free article] [PubMed]
- 16.Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, et al. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe. 2019;25:789–802. doi: 10.1016/j.chom.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Tarini J, Wolever TMS. The fermentable fibre inulin increases postprandial serum short-chain fatty acids and reduces free-fatty acids and ghrelin in healthy subjects. Appl Physiol Nutr Metab. 2010;35:9–16. doi: 10.1139/H09-119. [DOI] [PubMed] [Google Scholar]
- 18.van Loo J, Coussement P, de Leenheer L, Hoebreg H, Smits G. On the presence of inulin and oligofructose as natural ingredients in the western diet. Crit Rev Food Sci Nutr. 1995;35:525–52. doi: 10.1080/10408399509527714. [DOI] [PubMed] [Google Scholar]
- 19.Holmes ZC, Silverman JD, Dressman HK, Wei Z, Dallow EP, Armstrong SC, et al. Short-chain fatty acid production by gut microbiota from children with obesity differs according to prebiotic choice and bacterial community composition. mBio. 2020;11:e00914–20. doi: 10.1128/mBio.00914-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shafquat A, Joice R, Simmons SL, Huttenhower C. Functional and phylogenetic assembly of microbial communities in the human microbiome. Trends Microbiol. 2014;22:261–6. doi: 10.1016/j.tim.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silverman JD, Durand HK, Bloom RJ, Mukherjee S, David LA. Dynamic linear models guide design and analysis of microbiota studies within artificial human guts. Microbiome. 2018;6:1–20. doi: 10.1186/s40168-018-0584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pompei A, Cordisco L, Raimondi S, Amaretti A, Pagnoni UM, Matteuzzi D, et al. In vitro comparison of the prebiotic effects of two inulin-type fructans. Anaerobe. 2008;14:280–86. [DOI] [PubMed]
- 23.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–40. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reichardt N, Vollmer M, Holtrop G, Farquharson FM, Wefers D, Bunzel M, et al. Specific substrate-driven changes in human faecal microbiota composition contrast with functional redundancy in short-chain fatty acid production. ISME J. 2018;12:610–22. doi: 10.1038/ismej.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–52. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–9. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–43. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 28.van de Wiele T, Boon N, Possemiers S, Jacobs H, Verstraete W. Inulin-type fructans of longer degree of polymerization exert more pronounced in vitro prebiotic effects. J Appl Microbiol. 2007;102:452–60. doi: 10.1111/j.1365-2672.2006.03084.x. [DOI] [PubMed] [Google Scholar]
- 29.Aguirre M, Eck A, Koenen ME, Savelkoul PHM, Budding AE, Venema K. Diet drives quick changes in the metabolic activity and composition of human gut microbiota in a validated in vitro gut model. Res Microbiol. 2016;167:114–25. doi: 10.1016/j.resmic.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Solopova A, van Gestel J, Weissing FJ, Bachmann H, Teusink B, Kok J, et al. Bet-hedging during bacterial diauxic shift. Proc Natl Acad Sci USA. 2014;111:7427–32. doi: 10.1073/pnas.1320063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noronha A, Modamio J, Jarosz Y, Guerard E, Sompairac N, Preciat G, et al. The Virtual Metabolic Human database: Integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 2019;47:D614–D624. doi: 10.1093/nar/gky992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Liu F, Lu J, Shi J, Guan J, Yan F, et al. Probiotic mixture of Lactobacillus plantarum strains improves lipid metabolism and gut microbiota structure in high fat diet-fed mice. Front Microbiol. 2020;11:512. doi: 10.3389/fmicb.2020.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terrapon N, Lombard V, Drula É, Lapébie P, Al-Masaudi S, Gilbert HJ, et al. PULDB: the expanded database of polysaccharide utilization loci. Nucleic Acids Res. 2018;46:D677–D683. doi: 10.1093/nar/gkx1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolam DN, van den Berg B. TonB-dependent transport by the gut microbiota: novel aspects of an old problem. Curr Opin Struct Biol. 2018;51:35–43. doi: 10.1016/j.sbi.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, Flint HJ. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. 2004;91:915–23. doi: 10.1079/BJN20041150. [DOI] [PubMed] [Google Scholar]
- 36.Holmes ZC, Villa MM, Durand HK, Jiang S, Dallow EP, Petrone BL, et al. Microbiota responses to different prebiotics are conserved within individuals and associated with habitual fiber intake. bioRxiv. 2021. 10.1101/2021.06.26.450023. [DOI] [PMC free article] [PubMed]
- 37.Holscher HD, Gregory Caporaso J, Hooda S, Brulc JM, Fahey GC, Swanson KS. Fiber supplementation influences phylogenetic structure and functional capacity of the human intestinal microbiome: follow-up of a randomized controlled trial. Am J Clin Nutr. 2015;101:55–64. [DOI] [PubMed]
- 38.Liu H, Liao C, Wu L, Tang J, Chen J, Lei C, et al. Ecological dynamics of the gut microbiome in response to dietary fiber. ISME J. 2022;16:2040–55. [DOI] [PMC free article] [PubMed]
- 39.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaczmarek JL, Musaad SMA, Holscher HD. Time of day and eating behaviors are associated with the composition and function of the human gastrointestinal microbiota. Am J Clin Nutr. 2017;106:1220–31. [DOI] [PubMed]
- 41.Basan M, Honda T, Christodoulou D, Hörl M, Chang YF, Leoncini E, et al. A universal trade-off between growth and lag in fluctuating environments. Nature. 2020;584:470–4. doi: 10.1038/s41586-020-2505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matenchuk BA, Mandhane PJ, Kozyrskyj AL. Sleep, circadian rhythm, and gut microbiota. Sleep Med Rev. 2020;53:101340. doi: 10.1016/j.smrv.2020.101340. [DOI] [PubMed] [Google Scholar]
- 43.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–62. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 45.Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-019-14177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res. 2017;179:204–22. doi: 10.1016/j.trsl.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218–30. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wissel EF, Smith LK. Inter-individual variation shapes the human microbiome. Behav Brain Sci. 2019;42:E79.
- 49.Wurster JI, Peterson RL, Brown CE, Penumutchu S, Guzior DV, Neugebauer K, et al. Streptozotocin-induced hyperglycemia alters the cecal metabolome and exacerbates antibiotic-induced dysbiosis. Cell Rep. 2021;37:110113. doi: 10.1016/j.celrep.2021.110113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerimi A, Kraut NU, da Encarnacao JA, Williamson G. The gut microbiome drives inter- and intra-individual differences in metabolism of bioactive small molecules. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-020-76558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.di Bartolomeo F, van den Ende W. Fructose and fructans: opposite effects on health? Plant Foods Hum Nutr. 2015;70:227–37. [DOI] [PubMed]
- 52.Pereira FC, Berry D. Microbial nutrient niches in the gut. Environ Microbiol. 2017;19:1366–78. doi: 10.1111/1462-2920.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rettedal EA, Gumpert H, Sommer MOA. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat Commun. 2014;5:1–9. doi: 10.1038/ncomms5714. [DOI] [PubMed] [Google Scholar]
- 54.Oliphant K, Parreira VR, Cochrane K, Allen-Vercoe E. Drivers of human gut microbial community assembly: coadaptation, determinism and stochasticity. ISME J. 2019;13:3080–92. doi: 10.1038/s41396-019-0498-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Possemiers S, Verthé K, Uyttendaele S, Verstraete W. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol. 2004;49:495–507. doi: 10.1016/j.femsec.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(supplement_1):4516–22. [DOI] [PMC free article] [PubMed]
- 57.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–24. [DOI] [PMC free article] [PubMed]
- 58.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. [DOI] [PMC free article] [PubMed]
- 60.Bioinformatics B, Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. [DOI] [PMC free article] [PubMed]
- 61.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. [DOI] [PMC free article] [PubMed]
- 62.Webb EC. Enzyme nomenclature 1992: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the nomenclature and classification of Enzymes. Cambridge, MA, USA: Academic Press; 1992.
- 63.Enriquez-Hesles E, Smith DL, Maqani N, Wierman MB, Sutcliffe MD, Fine RD, et al. A cell-nonautonomous mechanism of yeast chronological aging regulated by caloric restriction and one-carbon metabolism. J Biol Chem. 2021;296:100125. [DOI] [PMC free article] [PubMed]
- 64.Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S, et al. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81:10038–48. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandes AD, Macklaim JM, Linn TG, Reid G, Gloor GB. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS ONE. 2013;8:e67019. doi: 10.1371/journal.pone.0067019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakamoto M, Ohkuma M. Identification and classification of the genus Bacteroides by multilocus sequence analysis. Microbiology. 2011;157:3388–97. doi: 10.1099/mic.0.052332-0. [DOI] [PubMed] [Google Scholar]
- 67.Silverman JD, Roche K, Holmes ZC, David LA, Mukherjee S. Bayesian multinomial logistic normal models through marginally latent matrix-T processes. J Mach Learn Res. 2022;23:1–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code used to generate the figures presented in this paper are publicly available at https://github.com/jrletourneau/ecological_memory_microbiome. Sequence data are publicly available via the European Nucleotide Archive as demultiplexed reads with the accession numbers PRJEB45244 (metatranscriptomics), PRJEB45247 (artificial gut 16S rRNA gene amplicon sequence data), and PRJEB48837 (cross-reactivity experiment 16S rRNA gene amplicon sequence data), and PRJEB47805 (human cohort 16S rRNA gene amplicon sequence data). Our lab’s 16S rRNA gene amplicon sequence data processing pipeline is also available at https://github.com/jianhong/16S_pipeline/.