Abstract
By application of combinatorial library technology, we generated the first recombinant antibody fragments directed against the major capsid protein p24 of human immunodeficiency virus type 1 (HIV-1). A library of single-chain Fv fragments (scFvs) was constructed by using the antibody variable-region (V) genes of B cells derived from the spleen of a viral lysate-immunized mouse. Antibodies were selected by panning or by enrichment with biotinylated antigen, yielding four different families of antibody fragments. The different types of scFvs were characterized by affinity measurements, by antigen recognition on Western blots, and by pepscan analysis. The epitope of one of the scFvs is located near the residues involved in CypA binding, thereby making it an attractive candidate for therapeutic applications. Comparison of the V gene sequence of this scFV with that of a previously described monoclonal antibody reactive against this immunodominant epitope revealed the usage of the identical combination of VH and Vκ regions. Thus, this is one of the rare examples in which the original combination in a library-derived antibody fragment was retrieved. After appropriate affinity and format improvements, the best of our recombinant scFvs may form the basis for a sensitive p24 assay as a measure of viral load. In addition, anti-p24 scFvs could be expressed as intracellular antibodies (intrabodies) to aid in the treatment of HIV infections.
Human immunodeficiency virus (HIV) infection is diagnosed by detecting virus-specific antibodies (Abs), or the virus itself, by means of p24 antigen (Ag) detection or by quantitative amplification procedures such as PCR (38) or nucleic acid sequence-based amplification (62) or by coculturing and subsequent virus detection procedures. During a diagnostic window of 6 to 8 weeks after infection, Abs to HIV are undetectable, and alternative diagnostic methods would help to reduce the residual risk of transfusion transmission of HIV. Recently, the Food and Drug Administration recommended the implementation of p24 Ag tests in donor screening (20). The p24 capsid protein forms the viral core containing the single-stranded RNA genome and is abundantly present in the virus particle. Besides the structural role of the protein in forming the core of the mature virion, the molecule is essential during viral assembly; it plays a pivotal role in viral penetration or uncoating or both, a function which may be mediated by binding of p24 to the human cellular proline rotamase cyclophilin A (4, 41, 60). With current enzyme-linked immunosorbent assays (ELISAs), the presence of p24 Ag may be assessed 5 to 14 days earlier than could an Ab response measured by anti-HIV type 1 (anti-HIV-1) or anti-HIV-2 enzyme immunoassays (8, 9, 66). In addition, the capsid protein may be considered a marker for virus replication (3, 26, 65), and its detection in an extremely sensitive immunoassay would offer a cheap and generally applicable alternative to PCR-based assays for the diagnosis of reactivation during treatment of HIV-1-infected patients with (combinations of) nucleoside reverse transcriptase inhibitors or protease inhibitors (19, 59, 64). When reactivation, as the result of the evolution of drug-resistant HIV mutants, is detected, treatment may be changed to other drugs. Rapid and sensitive assays that can carefully detect the presence of p24 in serum are therefore crucial for early detection and monitoring of viral replication (66).
The sensitivity and specificity of the presently used anti-p24 immunoassays are limited by the affinity of the monoclonal Abs (MAbs) used for capturing and/or detection of the Ag, although by signal amplification in combination with heat denaturation, the sensitivity can be increased to the level obtained by PCR (6). The availability of the Ab genes in recombinant anti-p24 Abs allows the improvement of affinity by mutagenesis methods, as well as the engineering of avidity, thereby helping to improve the sensitivity of early virus detection. During in vivo maturation, the obtained affinities are limited by the off-rate, i.e., the rate at which the Ab-Ag complex dissociates. The off-rate of in vivo-matured Abs is on the order of 10−3 to 10−4 s−1, which permits endocytosis of membrane-bound Ab-Ag complexes on B cells (21). However, in vitro maturation with phage display allows the selection of Abs with lower off-rates, leading to affinities in the picomolar range (1, 57). Besides their obvious diagnostic application, it may also be possible to use anti-p24 single-chain Fv fragments (scFvs) for therapy. By expression with a retention signal for the endoplasmic reticulum, the scFvs may interfere with virus assembly in the infected cells, as was demonstrated with anti-gp120 (12) and anti-Tat (45) Abs. For intracellular expression, even murine Abs might be applied in humans: a human anti-mouse antibody (HAMA) response cannot be induced, since the fragments are shielded from the immune system.
In this context, we aimed to isolate anti-p24 scFvs from a phage Ab library made from a mouse immunized with HIV-1 viral lysate. Affinities were determined with surface plasmon resonance and compared with those of MAbs currently used in p24 assays. The specificity of the selected Ab fragments was determined by ELISA, Western blotting, and pepscan analysis. One of the recombinant Ab fragments recognized an epitope which was also detected by a previously described and sequenced murine MAb. Both the recombinant Ab and the hybridoma-derived Ab have nearly identical heavy and light chain variable regions, and, as such, the selected scFv D2 is one of the first examples in which the original combination of both VH and VL was maintained, in spite of the very low probability of finding this in a combinatorial library (23).
These newly described anti-p24 recombinant scFvs offer a starting point for Ab affinity maturation and engineering (for reviews, see 30, 67, and 68) and may be useful as intracellular Abs (intrabodies) for HIV therapy.
MATERIALS AND METHODS
Library construction.
A BALB/c mouse was immunized with 100 μg of protein from a viral lysate, which had been prepared from infected H9 cells (52) and purified by ultracentrifugation, plus Freund’s complete adjuvant. After 6 weeks, a booster with 100 μg of lysate proteins plus incomplete adjuvant was administered. Three days later, the animal was sacrificed and the spleen was removed. Upon extraction of the spleen, 20 μg of total RNA was isolated (13) and 5 μg of this was transcribed into random-primed cDNA.
The immunoglobulin G1 (IgG1)-derived VH- and Vκ-encoding DNA fragments as well as the linker fragment (the latter was cloned in pUC19, by using synthetic oligonucleotides which encoded the 15-residue linker described by Huston and colleagues [33]) were obtained by amplification (Amplitaq; Perkin-Elmer Cetus) with the primers (14) listed in Table 1. The fragments were purified from an agarose gel with the QIAEX kit (Qiagen, Hilden, Germany) and assembled in splicing by overlap extension-PCR (32).
TABLE 1.
Oligonucleotide primers used for construction of the library and sequencing
| Purpose | Primer name | Sequencea |
|---|---|---|
| VH | VH1back (a) | 5′aataggcccagccggccatggcc(C/G)AGGTTCA(A/G)(G/C)TGCAGCAG(C/T)CTGG3′ |
| VH2back (a) | 5′aataggcccagccggccatggccGAGGTG(C/A)AGCTT(G/C)T(G/C)GAGTCTGG3′ | |
| VH3back (a) | 5′aataggcccagccggccatggccCCGA(G/T)GTGCAGCTTCAGGAGTCAGG3′ | |
| VHfor | 5′CCTGAGGAGACGGTGACC(A/G/C)(G/T)GG(T/G)(T/C)CC3′ | |
| IgG1-CHfor | 5′TGGATAGACAGATGGGGG3′ | |
| Vκ | VK1back | 5′ggatctGACATC(C/A)AGATGACTCAG(T/A)CT3′ |
| VK2back | 5′ggatctGATATTGTGATGACGCAG(G/A)(C/A)T3′ | |
| VK3back | 5′ggatct(G/A)ACATTGTG(C/A)TGTCACAGTCTCC3′ | |
| VK4back | 5′ggatctGAAA(A/T)T(G/T)TTCTCACCCAGTCTCC3′ | |
| VK5back | 5′ggatctGATATT(G/T)TGATGACGCAG(C/T)CTGC3′ | |
| VKfor (a) | 5′aatggaattccgggcggccgcCCGTTTTA(T/G)(T/C)TCCAGCTT(G/T)GT(G/C)CC3′ | |
| CKfor | 5′AGTTGGTGCAGCATCAGCC3′ | |
| Linker | VHLINKback | 5′GGTCACCGTCTCCTCAgg3′ |
| VK1LINKfor | 5′AG(A/T)CTGAGTCATCT(G/T)GATGTCagatcc3′ | |
| VK2LINKfor | 5′CTGCGTCATCACAATATCagatcc3′ | |
| VK3LINKfor | 5′GGAGACTGTGACA(G/T)CACAATGT(C/T)agatcc3′ | |
| VK4LINKfor | 5′GGAGACTGGGTGAGAA(C/A)A(T/A)TTTCAGATagatcc3′ | |
| VK5LINKfor | 5′GCAG(G/A)CTGCGTCATCA(C/A)AATATCagatcc3′ | |
| Sequencing | geneIII | 5′TGAATTTTCTGTATGAGG3′ |
| linkfwd | 5′CTCGGGCGGTGGTGGGTC3′ | |
| linkrev | 5′GACCCACCACCGCCCGAG3′ |
The SfiI sites in the VHback primer sequences and the NotI site in the VKfor primer sequence are underlined. Nucleotides represented as uppercase characters are carried by the V gene; those in lowercase characters are carried by the vector.
The assembled scFv-encoding DNA fragments were also purified from the gel, digested with SfiI and NotI (Pharmacia, Lund, Sweden), and cloned into the phagemid vector pV1, which is identical to pHEN1 (31) but with a FLAG-tag (53) instead of a Myc-tag. Transformations were performed by electroporation in JM101 (17). The library consisted of 107 clones, of which 60 to 70% contained a complete insert as was deduced from PCR analysis.
Selection of libraries.
The rescue of phagemid particles was performed as described before (43). The capsid protein p24, expressed as full-length product in Escherichia coli under control of the inducible lac promoter (constructed at Organon Teknika [3a]) and encoding the HIV-1 strain RF sequence, was affinity purified with MAb 39B (generated at Organon Teknika [unpublished data]). A six-well tissue culture plate (Costar, Cambridge, Mass.) was coated with Ag at a concentration of 3 μg/ml in 50 mM NaHCO3 (pH 9.6). Selection, washing, and elution were performed as described elsewhere (43).
For the construction of a light chain shuffling library of clone D2, the VH segment was amplified on plasmid DNA. The VH fragment was recombined with the linkers and the corresponding kappa light chain fragments, initially used for the generation of the library, yielding the shuffling library containing 106 clones. Selection was performed with p24 biotinylated with N-hydroxy-succinimide–biotin (Pierce, Rockford, Ill.); phage Ag complexes were captured with streptavidin-coated paramagnetic beads (Promega, Madison, Wis.).
Screening and sequencing of clones.
Soluble scFv was produced from individual clones by growth in V-shaped microtiter plates in 2*TYE medium as described before (43). The described method was used for constructs transformed in the nonsuppressor strain TOP F′ as well as in the suppressor strain JM101. Suppression is incomplete and results in the leakage of free scFv in the culture medium, while the expression of gene III makes the cellular membranes more permeable (5), leading to higher concentrations of Ab fragments in the medium.
Microtiter plates were coated with 3 μg of recombinant p24 per ml (see above) or 5 μg of viral lysate per ml in 50 mM NaHCO3 (pH 9.6) for 16 h at room temperature. The plates were blocked for 2 h at room temperature with bovine serum albumin (BSA; 0.2% [wt/vol] in 0.1 M Tris [pH 7.4]–30 mM KI). The culture supernatant was diluted fivefold in sample diluent (phosphate-buffered saline [PBS], 20% normal goat serum, 1.1% Triton X-100). After incubation at room temperature for 2 h, the plates were washed four times with PBS-Tween 20 (0.05%), and bound scFv was detected with a mixture of a 1/4,000 dilution of anti-FLAG Ab M2 (Kodak IBI, New Haven, Conn.) and a 1/2,000 dilution of anti-mouse Ab-horseradish peroxidase conjugate (DAKO, Glostrup, Denmark). Following a 1-h incubation and washing as before, staining was performed with tetramethylbenzidine and ureaperoxide as the substrate and stopped by adding an equal volume of 1 M H2SO4; the optical density was measured at 450 nm.
Clones giving positive signals and with a unique BstNI fingerprint (New England Biolabs, Beverly, Mass.) were analyzed by sequencing with the T7 sequencing kit (Pharmacia) with the M13rev primer, the geneIII primer, and two primers located in the linker (linkfwd and linkrev) (Table 1). Plasmid DNA was purified with the QIAGEN kit (Qiagen).
Characterization of scFvs.
For the preparation of periplasmic fractions by the borate-buffered saline shock procedure (58), clones obtained by transfection to the nonsuppressor strain TOP F′ were cultured on a 50-ml scale and induced as described previously (43). The fractions were analyzed on 15% polyacrylamide gels and then blotted onto nitrocellulose. After blocking with a skim milk solution (5% [wt/vol] in Tris-buffered saline [TBS]), the scFv was detected with anti-FLAG M2 diluted 1/4,000 in TBS–0.05% Tween (TBST). After a 2-h incubation period, the anti-FLAG was detected with anti-mouse Ab-horseradish peroxidase and the ECL detection kit (Amersham, Buckinghamshire, United Kingdom).
For the purification of scFv, refolding was used in combination with affinity chromatography on p24 columns. The pelleted cells from a 50-ml culture were resuspended in 8 ml of an 8 M urea solution (in PBS) and sonicated. The mixture was rotated head-over-head for 30 min, and insoluble material was removed (centrifugation for 30 min at 13,000 × g). The supernatant was dialyzed against PBS with four buffer changes. Insoluble proteins were removed by centrifugation, and after passage through a 0.2-μm-pore-size filter, the flowthrough was immediately loaded on a p24 column (bed volume, 0.3 ml). The column material was prepared by coupling 8.4 mg of protein to 1 g of Tresyl Sepharose in accordance with the supplier’s instructions (Pierce). Washing and elution were performed as in the panning procedure. The yield was determined by measuring the optical density at 280 nm, assuming that an scFv has a molar extinction coefficient (E2801%) of 14.3 (27).
To establish the valencies of the scFvs, the molecular weights of the Ag binding molecules were determined by gel filtration (27) on a Superdex 75HR column (Pharmacia), which was calibrated with a mixture of BSA, chymotrypsin, ovalbumin, and RNase A (Pharmacia). As a control, an anti-human chorionic gonadotropin (anti-hCG) scFv selected from a murine immune library was used (unpublished data). Either 20% of a periplasmic fraction from a 50-ml culture or affinity-purified scFv was injected onto a calibrated column. Fractions (0.5 ml) were collected and analyzed on Western blots.
Epitope mapping was performed with the pepscan method (22). Overlapping decapeptides of p24 from HIV strain ANT70, coupled to a solid support and obtained from the Department of Molecular Recognition of the Institute for Animal Science and Health (IDO-DLO), were tested with the scFv. The periplasmic fraction of clone D2 was diluted 100-fold in Super Q buffer (PBS containing 5% [vol/vol] horse serum, 5% [wt/vol] BSA, and 1% [vol/vol] Tween 20) to a concentration of 80 ng/ml. After 16 h of incubation at 4°C, scFv bound to peptides on the solid support was detected as described above.
Affinity measurements in solution.
The affinities measured by the “in solution” method were determined with surface plasmon resonance on BIAcore (Biacore AB, Uppsala, Sweden) in accordance with the suggested procedures described in the BIAevaluation software. A high-density p24-coated (3,777 Response Units [RUs] of immobilized Ag) CM chip (Biacore AB, Uppsala, Sweden) was prepared by covalent coupling as described in the supplier’s recommendations. A fixed amount of scFv or MAb, yielding a signal of between 1,000 and 1,500 RUs when injected onto the p24 chip, was mixed with a variable amount of Ag in a volume of 60 μl by using HEPES-buffered saline (10 mM HEPES, 3.4 mM EDTA, 150 mM NaCl, 0.05% surfactant P20 [pH 7.4]) as the dilution buffer. After a 1-h period, needed to achieve equilibrium, the samples were injected. For the quantification of free Ab, a standard curve was prepared by injection of a dilution series of the tested Ab on the same flow cell (37).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences of the antibodies reported in this study are as follows: D2 VH, AF083186; A2 VH, AF081541; A3 VH, AF081545; A5 VH, AF081543; D1 VH, AF083189; D2 VL, AF083188; A2 VL, AF081542; A3 VL, AF083185; A5 VL, AF081544; D1 VL, AF083187.
RESULTS
Library construction and selections.
Our major aim was to evaluate the library technology as a substitute for hybridoma technology for the generation of anti-HIV-1 Abs, which might be useful for in vitro diagnostics. Therefore, we immunized a mouse with a lysate prepared from HIV-1-infected H9 cells (52). After amplification of the V genes of the murine spleen B cells and cloning in the phagemid vector pVI, a library containing 107 clones was obtained. Seventy-five percent of the clones expressed scFvs, as was determined by PCR screening and by inspecting the morphology of the colonies: clones which express gene III (fusions) are usually visible as flat colonies, while those having an interrupted frame exhibit a more solid appearance (14a).
The library was subjected to four rounds of panning on recombinant p24. Four rounds of panning produced a 100-fold enrichment in the number of eluted phages, indicating the presence of Ag-specific phage Abs. The scFvs produced by 48 individual clones after round 4 were analyzed by ELISA (43). Seventy percent of the tested scFvs produced high signals on viral lysate and p24 Ag, whereas no responses were found on BSA. After 24 clones were DNA fingerprinted by BstNI digestion (43), two different patterns were identified; five clones representing the two patterns were selected for sequence analysis and further binding analysis. All five clones analyzed had minor differences in sequences and were classified as scFv D2 (Fig. 1).
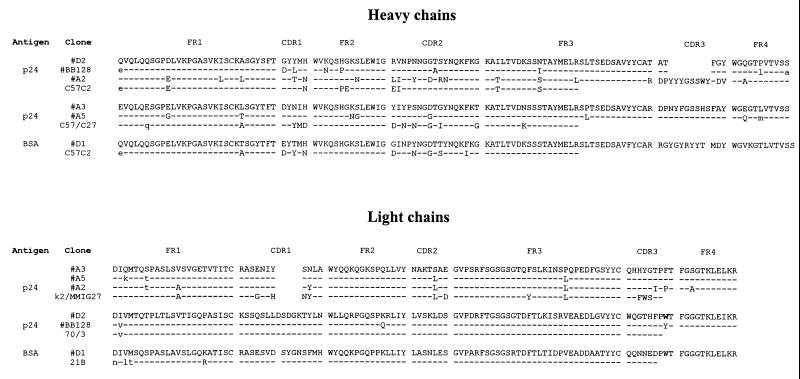
FIG. 1.
Deduced amino acid sequences of the variable heavy and light chain regions and germ line origin of the p24 Abs derived from the murine library. Identical residues are indicated by dashes, while different residues are indicated by uppercase characters and primer-encoded differences are indicated by lowercase characters.
Immune libraries usually contain a very diverse set of Ag-specific Abs (14, 44); thus, we modified our method of selection to retrieve phage Abs other than the ones found before. Selections were repeated with soluble (biotinylated) recombinant p24 (Sol-p24) and with p24-coated magnetic particles (Bead-p24). After four rounds, enrichment factors of 100 and 10,000 were found for Sol-p24 and Bead-p24, respectively. Concomitant with this enrichment in the absolute number of eluted phages, we found an enrichment in the fraction of clones producing scFv, as determined by colony morphology. The percentage of scFv-producing clones increased from 50 to 75% of the clones before selection and after round 1 to more than 99% after round 4. At this stage, 8 of 11 clones of Sol-p24 and 12 of 12 clones of Bead-p24 contained a complete insert, whereas BstNI fingerprinting revealed four different but closely related clones for Sol-p24 and only one type for Bead-p24. The four Sol-p24 scFv clones reacted positively on p24 and negatively on BSA, while the Bead-p24 clones gave high-level responses on p24 and BSA. Obviously, panning with the beads resulted in the isolation of an scFv specific for the blocking reagent BSA, which was also used for blocking the microtiter plates, which explains the positive signals in the p24 ELISA. The selection of BSA-specific clones can be explained by the presence of this Ag in the viral lysate used for immunization and its use for blocking the beads and the microtiter wells. We chose three different p24 binders of Sol-p24 and the Bead-p24 BSA binder for sequencing and further characterization. For Sol-p24, the clones designated A2, A3, and A5 were used; for Bead-p24, the clone used was named D1.
Characterization of isolated scFvs.
Figure 1 shows the deduced amino acid sequences of the VH and VL gene products of the clones derived from panning the murine library with Ag-coated polystyrene plates (D2) and with biotinylated Ag (A2, A3, and A5) together with the most related mouse germ line analogues (V regions). The sequence of the BSA binding antibody (D1) selected with the beads is also included.
The four anti-p24 scFvs use highly homologous VH gene segments. They belong to class IIA according to Kabat’s classification (36) and use two germ line gene segments only. Clones A2 and D2 and clones A3 and A5 can be grouped in two classes depending on the germ line used. D2 has a divergent and shorter CDR3 than clone A2 and thus is clonally not related. The light chain of clone D2 could be matched adequately with a germ line segment and is very different from the A2 light chain. The kappa light chains of the three antibodies A2, A3, and A5 use a nearly identical Vκ gene segment, derived from the germ line segment coded k2/MMIG27 (for classification and the codes used in Fig. 1, see “Germline gene directories of the mouse” homepage (http://www.ibt.unam.mx/∼almagro/V_mice.html)).
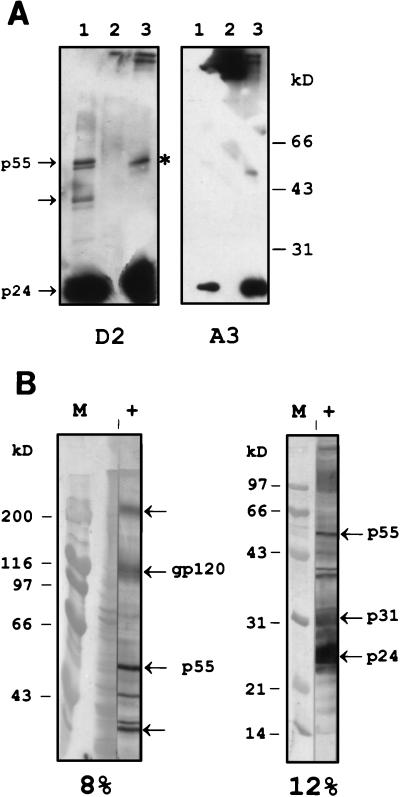
The specificities of the scFvs were determined by Western blot analysis (Fig. 2). Periplasmic fractions containing scFv were used for the detection of viral and recombinant p24. Two different recognition patterns were obtained. Abs A2, A3, and A5 reacted with (the reduced form of) p24, in a viral lysate, and as a recombinant product (for example, the blot incubated with scFv A3 is shown in Fig. 2A). ScFv D2 recognizes p24, but it also detects the viral Gag polyprotein p55 (Fig. 2A, left panel, lane 1) from which the matrix protein p17, the nucleocapsid p7, and the capsid protein p24 are proteolytically processed. Another product with a molecular mass of approximately 40 kDa is visible. This protein seems also to be recognized by polyclonal serum (Fig. 2B) obtained from the mouse that was used for the construction of the library. The p24 dimer, which is the predominant form of the native protein as was observed by gel filtration (results not shown), might also be detected with mouse serum (Fig. 2B, left panel, band between p55 and the 40-kDa derivative). Indeed, the native p24 protein can form a complex of oligomers in solution, including dimers, tetramers, dodecamers, spheres, fibers, and tubes (18). An intense reactivity of the original murine polyclonal serum against p24 and its precursors can be observed, while the mouse seems to have a lower Ab titer against the envelope protein gp120. Thus, the original library was biased in its immune response towards p24, which is probably caused by the abundance of the antigen in the viral lysate used for immunization (10, 63).
FIG. 2.
Western blot detection of p24 by scFvs and mouse polyclonal Abs (A) Detection with scFv D2 (left panel) and A3 (right panel). Viral lysate (lanes 1), BSA as a negative control (lanes 2), and purified recombinant p24 (lanes 3) were analyzed on a 12% gel. The band indicated with an asterisk represents the dimer of p24, present in the recombinant and in the viral product. (B) Blot analysis of viral lysate with polyclonal serum (lanes labeled +) obtained from the mouse used for library construction (left blot made with an 8% gel and right one made with a 12% gel). Amido black-stained markers (lanes labeled M) are shown. The Gag precursor p55 and its derivative p24, the envelope protein gp120, and the polymerase p31 are indicated with arrows. The slow-migrating protein indicated by the unlabeled arrow at the top of the left blot is probably the envelope precursor gp160. The other marked protein (bottom unlabeled arrow) is a cleavage product of Gag, which is also recognized by scFv D2 (Fig. 2A).
D2 is a diabody.
After performing the sequence analysis of clone D2, selected on immobilized p24, we noticed that it contained a linker consisting of 2 amino acids (GlySer) rather than 15 amino acids [(Gly4Ser)3]. As suggested in the literature (29), shortened linkers may lead to the formation of dimeric molecules with two functional binding sites, the so-called “diabodies.” The shorter linker in clone D2 might therefore be responsible for the formation of a bivalent scFv molecule, and this type of molecule might be strongly enriched over the other anti-p24 scFvs by its greater avidity in the panning selection. We generated a number of D2 derivatives and characterized their behavior.
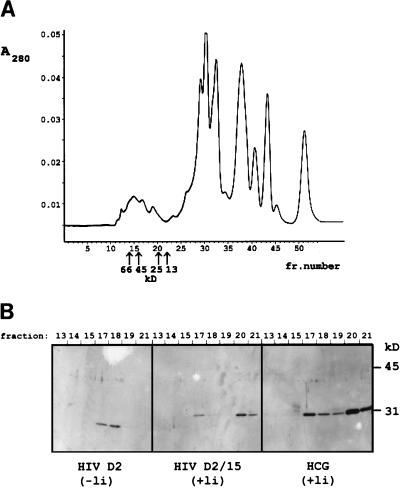
From a light chain shuffling library, we selected a derivative of clone D2 that had an identical light chain and a complete 15-amino-acid linker; the clone was designated D2/15. We examined the molecular weight of the antibody fragments present in the periplasmic space of the producing bacteria by gel filtration on a calibrated Superdex 75HR column (27); the obtained chromatogram of clone D2 is shown in Fig. 3A. The scFv with a two-amino-acid linker emerged from the column as a single peak with a molecular mass of 45 kDa, as could be deduced from Western blot analysis with the fractions collected during gel filtration (Fig. 3B). Both of the scFvs, D2/15 and a control anti-hCG Ab fragment with a 15-residue linker, gave two peaks: the dimeric fraction at 45 kDa and a monomeric fraction at 25 kDa.
FIG. 3.
Gel filtration of periplasmic fractions on Superdex 75HR and analysis of the collected fractions on a Western blot with anti-FLAG M2. (A) Chromatogram, obtained by plotting A280 against the fraction number, illustrating the separation of the periplasmic proteins from clone D2. The retention times of the marker proteins are indicated with arrows. (B) Western blot analysis (15% gel) of the collected fractions (fraction number is shown above the blots) obtained from gel filtration of anti-p24 clones and of an anti-hCG scFv-producing clone. Two peak fractions can be discriminated in the scFvs having a complete linker (the derivative D2/15 and the anti-hCG): a main peak of the monomer (fraction 20; molecular mass, 25 kDa) and a dimer peak (fraction 17; molecular mass, 45 kDa). scFv of clone D2 emerged from the column in a single peak corresponding to the dimeric product.
By chain shuffling the light chain of the D2 diabody, clone D2/15 was isolated by using an excess of Ag during selection. By increasing the stringency of selection, i.e., decreasing the Ag concentration from 100 nM to 100 pM, the monomeric clone D2/15 was completely lost and the selected population was overtaken by (avid) diabodies. We retrieved no D2 derivatives with other light chains, not even the one of clone A2, in spite of its highly related VH gene, as was also reported by others (56). However, it should be emphasized that the diabody’s CDR3 sequence is completely different from the CDR3 sequence of clone A2 in length as well as in sequence. Due to the close proximity of H3 to the light chain variable-region loops, this may determine the pairing light chain. In the course of the shuffling experiment, we isolated one clone, D2/L3mut, which appeared to have a 15-residue linker but still produced predominantly dimeric scFvs. Sequencing revealed a single mutation from the D2 sequence, of residue W (shown in boldface type in Fig. 1) to Y, in the third hypervariable region of the light chain. Since this region of the Ab is in close contact with the VH region, the mutation may destabilize the VH-VL association, thereby promoting dissociation and intermolecular pairing (56).
Affinity measurements.
For affinity measurement, the scFvs were purified by affinity chromatography on Ag columns. The scFv fragments were refolded after denaturation of all cell proteins with urea by subsequent dialysis against PBS. Although the efficiency of refolding is limited, the yield of functional scFv is much higher than that in periplasmic fractions obtained by the osmotic shock procedure (58). The purity was analyzed on a Coomassie blue-stained gel (data not shown).
All purified scFvs were analyzed by gel filtration chromatography, as was already performed with the periplasmic fraction of clone D2 (Fig. 3); the purified product of the D2 diabody contains dimers only, while the analogue with the complete linker, D2/15, is predominantly monomeric. Clones A3 and A5 produce monomeric scFv fragments only (Table 2).
TABLE 2.
Affinities of the anti-p24 MAbs and scFvs
| Ab type | Ab | Forma | linker length (aa)b | Kd (M) |
|---|---|---|---|---|
| mAb | 5B4 | 3.6 × 10−8 | ||
| 34A | 8.0 × 10−9 | |||
| 39B | 8.0 × 10−9 | |||
| scFv | A2 | NDc | 15 | 2.0 × 10−6 |
| A3 | ND | 15 | 2.5 × 10−8 | |
| A5 | Monomer | 15 | 3.0 × 10−8 | |
| D2 | Dimer | 2 | 1.0 × 10−6 | |
| D2/15 | Monomer | 15 | 3.0 × 10−6 | |
| D2/L3mutd | Dimer | 15 | 1.3 × 10−6 |
As determined by gel filtration.
aa, amino acids.
ND, not determined.
This clone is identical to clone D2 but with a complete linker and a single mutation within CDR3 of Vκ (see text).
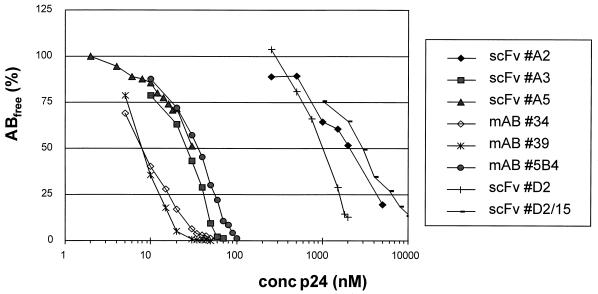
We compared the affinities of the library-derived scFvs with the binding strength of hybridoma-produced MAbs. We chose to measure the kinetics of binding by surface plasmon resonance on a BIAcore machine. When using Ag immobilized on Sensorchips, we always observed biphasic dissociations, probably caused by the oligomerization of p24 (18), resulting in a heterogeneous coat. The abnormal sensorgrams were also noted before by others (25). Therefore, the affinity was determined in solution (47), by injecting a preestablished equilibrium mixture of a fixed amount of Ab and a variable amount of Ag on a sensorchip with a high-density coat of Ag. Under mass transfer limiting conditions, the amount of free Ab (Abfree) is proportional to the binding rate. Subsequently, the Kd is obtained by plotting the concentration of Abfree (or the fraction Abfree/Ab0; Ab0 is the concentration of Ab when no Ag has been added) against the total concentration of Ag. In Fig. 4, the binding curves derived from the MAbs and scFvs are shown; Table 2 gives the corresponding Kds. The affinities of MAbs 34A and 39B are threefold higher than those of scFvs A3 and A5, both selected on biotinylated Ag, which in turn have slightly higher affinities than that of MAb 5B4. The D2 diabody, selected on immobilized p24 via panning, has a much lower affinity (micromolar range). The monomeric version of D2, D2/15, has a threefold-lower affinity than that of its dimeric counterpart, demonstrating that the effect of avidity on the apparent affinity determined in this assay is limited. Finally, clone D2/L3mut, which expressed mainly dimeric scFv (as gel filtration of affinity-purified scFv revealed [results not shown]), had an affinity approaching that of clone D2.
FIG. 4.
Percentage of Abfree as a function of Ag concentration (logarithmic scale). The amount of Abfree (i.e., not bound to soluble Ag) was determined by surface plasmon resonance and related to the input amount of Ab (no Ag added). The Kd was derived from the plots at 50% binding.
Epitope recognition.
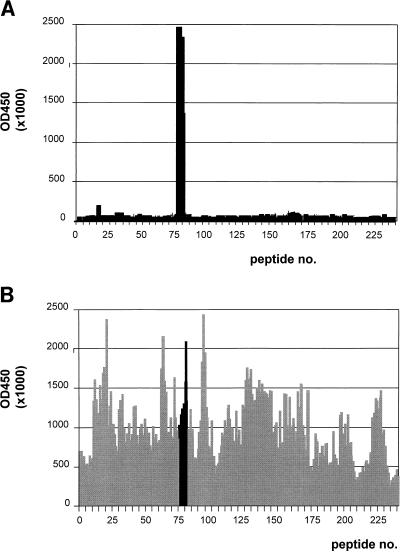
Pepscan analysis (22) was used in the identification of the epitopes of the scFvs. Although all recombinant fragments recognized denatured Ag on Western blots, only the diabody type D2 gave a good response. The results obtained with the overlapping decapeptides based on the sequence of HIV-1 subtype O strain ANT70 and the scFv-containing periplasmic fraction are shown in Fig. 5A. The Ab reacted with the peptides starting with the sequence INEEAVEEWDRTH and ending with EWDRTHPPPVGP, thereby having in common the sequence EWDRTH. This sequence is also recognized by serum Abs from an HIV-1 patient (Fig. 5B), indicating that the immune system of the mouse recognizes the same epitope on the immunized Ag as that of humans after virus infection. It should be noted that the p24 used for immunization and selection is derived from strain IIIB, while the pepscan is based on strain ANT70. For the A2, A3, and A5 Abs, the pepscan based on the ANT70 and the IIIB sequences did not reveal any unique epitopes (results not shown), indicating that these Abs may recognize a conformation-sensitive epitope.
FIG. 5.
Pepscan results obtained with decapeptides of p24 of HIV-1 strain ANT70 with scFv D2 (A) and serum Abs of an HIV-1 patient (B). The numbering on the horizontal axis corresponds with the amino-terminal amino acid of the peptide: the first peptide starts with a Q at −3 relative to the cleavage site of p17 and p24, and the last peptide (no. 240) ends with an A located at −3 relative to the cleavage site of p2 with p7. On the vertical axis, the optical density at 450 nm obtained for each peptide in ELISA has been plotted. The common sequence recognized by the human serum and scFv D2, starting with peptide 76 and ending with peptide 82, is indicated in panel B by solid bars.
Inspection of the HIV-1 B-cell epitope database (Los Alamos) showed us that several MAbs that share the epitope with scFv D2 have been found. In one of these, hybridoma BB128γ (61), the V gene sequence contains a nearly identical heavy chain variable region, with a remarkably short H3 loop, and, even more surprisingly, a light chain variable region with only one residue difference in the germ line-encoded region from the sequence of D2 (Fig. 1).
DISCUSSION
Isolation of different sets of anti-p24 Abs dependent on the selection procedure.
When we used the combinatorial library technology for the generation of MAbs, we found that the selection method determines to a great extent what type of Ab will be found. By using Ag-coated polystyrene, the high Ag density probably favors the selection of scFvs which are avid, exemplified by the selection of the D2 clone, while selection with soluble Ag leads to the isolation of monomeric Ab fragments produced by clones A2, A3, and A5. In the course of the assembly reaction between VH, linker, and VL to generate scFvs with 15-residue linkers (14), mispriming may infrequently lead to short-linker versions of VH and VL as reported previously (42). In the resulting Abs with a 5- or 10-residue linker, structural constraints prevent the intramolecular combination of VH and VL. Instead, the product forms an intermolecular pair with another VH-VL fusion product, yielding a bivalent scFv molecule, the so-called diabody (29, 44). The D2 diabody is heavily dependent on avidity for its selection; when a selection procedure with soluble p24 is used, the avidity effect is less pronounced and other clones (A3 and A5) are selected on the basis of affinity. The avidity effect may be more pronounced also by the intrinsic aggregation behavior of p24, which upon coating may lead to epitopes that are spatially ordered such that they are more prone to cross-linking by scFv Abs. Alternatively, the epitope recognized by clones A3 and A5 might be shielded or modified on p24 adsorbed onto polystyrene, although this hypothesis was not supported by the high ELISA signals. Our data indicate that selections should be carefully fine-tuned to retrieve the desired Abs. To obtain a wider panel of anti-p24 Abs, the various selection procedures used here may be altered or extended, for example, by using epitope-shielding Abs as described elsewhere (16).
The affinities of the Abs were determined with an in-solution method. The multimerization behavior of p24 complicated the affinity determination by direct kinetic measurement, while the alternative assay gave reliable data with only a minor effect of avidity. The affinities of the monomeric scFvs A3 and A5 found by selection with soluble Ag are in the nanomolar range and are slightly better than the affinity of one of the studied MAbs, which was taken from a panel of hybridoma-produced MAbs that are currently used in diagnostic assays. Their affinity is still threefold lower than of the best MAbs tested, but this might be due to the avidity of the MAbs. Indeed, a threefold difference in affinity was also found between the dimeric and monomeric versions of clone D2.
The present study showed that Ab fragments obtained from the murine library have affinities comparable to those of their hybridoma-derived analogues. For obtaining high-affinity Abs useful as diagnostic reagents, avidity may be recruited, for example, by making diabodies (29), scFv trimers (39), or tetramers (for a review, see reference 51). The affinities of the best Abs selected, A3 and A5, may be improved, for example, by targeted mutagenesis (1, 30, 57). Finally, additional alternative selections should be employed to further explore the diversity of the murine phage library that was constructed.
D2 recognizes an immunodominant epitope closely located at the CypA binding site.
All selected scFvs recognized linear epitopes of p24, as was concluded from Western blot studies. p24 expressed in E. coli as well as Ag, derived from cultured virus and presented in a reduced form was reactive on Western blots. Pepscan analysis with overlapping decapeptides yielded the epitope of the scFv produced by clone D2. The three-dimensional structure of the amino-terminal core domain of the HIV-1 capsid protein is known (24); it reveals the presence of an arrowhead-shaped domain, with seven helices and two β hairpins, and an exposed partially ordered loop involved in binding cyclophilin A. Recently, the binding domain was analyzed in more detail by crystallization of a peptide fragment of p24 with cyclophilin A (69), indicating the direct interaction of residues Ala 88, Gly 89, Pro 90, and Ile 91 with residues from the cellular enzyme. The epitope of D2 is located within the carboxy-terminal part of helix IV, ending at His 84 (or Leu in strain IIIB), and it is separated by three residues from the residues interacting with cyclophilin A.
The interaction of p24 with cyclophilin A appears to result in the packaging of approximately 200 copies of CypA into each HIV-1 virion. Although the function of CypA remains unclear, virions lacking the enzyme are known to be poorly infectious (7). Cyclosporins capable of binding to cyclophilin A are effective in inhibiting virus replication, as was reported before (4, 41, 60). Agents that interfere with the CypA binding function of p24 may be useful in reducing viral load; application of our scFv D2 as intracellularly expressed Ab may therefore prevent binding of the enzyme in infected cells, rendering the progeny virus less infectious or noninfectious, as was done successfully with anti-Tat (45) and anti-gp120 (12) Abs.
Retained VH-VL pairing.
Several epitopes have been mapped within the CypA loop region, which by its exposed orientation is accessible for reaction with Abs. Other MAbs that recognize the same epitope have been reported (28, 34, 48–50, 54, 55). In addition, the structure of a complex between an anti-p24 Ab and its Ag was determined with an Ab with nearly identical epitope recognition as that of Ab D2 (46). Moreover, the epitope is also reactive with human Abs, as indicated by our pepscan with serum from an HIV-1-infected patient and as was also concluded in a study in which the peptide was incorporated in a diagnostic assay (35).
Despite the recognition of the same epitope by the Abs mentioned above, their heavy chain V genes have little homology to each other, except that they both use a CDR3 of the same size and a nearly identical CDR1. The most striking resemblance exists between scFv D2 and anti-p24 MAb BB128γ (61), which have almost identical VH and Vκ sequences (differences of six and two amino acid residues, respectively). Clones isolated from a Vκ light chain shuffling library were found to have the original MAb light chain sequence, including the tyrosine (replacing the tryptophan present in clone D2), indicating a favorable selection for original pairings. It has been argued that the changes of retrieving the original pairing from a combinatorial library are very low, as the result of RNA isolation (67), and that it is particularly difficult to retrieve the original light chain (2). During the early days of combinatorial library construction with the bacteriophage lambda system, an original VH-VL combination identical to the one found in a hybridoma antibody was reported (11). However, the identity was less pronounced than that with our p24 scFv: in the partially sequenced VH, containing CDR2, FR3, and a part of FR2, there were eight mismatches present and the light chain comprised four differences. Moreover, the VH could pair with a different VL, as was also found by Barbas and colleagues (2). Considering the results from the light chain shuffling library, the D2 VH is strictly monogamous, i.e., it can pair with its original light chain only. When a mouse has 106 different B cells, a library with at least 1012 clones must be made to have a chance to identify an original VH-VL combination. After immunization, the number of anti-p24-expressing B lymphocytes will be larger, but still a rather huge library would be needed to find original combinations. One of the reasons that we isolated the original clone may be because of a high titer of these Abs, since pepscan analysis with the polyclonal mouse serum gave a major response on the peptides containing the EWDRTH sequence (data not shown).
Diagnostic applications.
The selected anti-p24 Abs could be used in a sensitive p24 Ag assay in which HIV-1 infection could be detected in an early phase (20). The correlation between affinity and sensitivity in ELISA has been clearly demonstrated (15, 40). The affinities of the selected scFvs approach the values of MAbs, which can detect 5 to 10 pg of p24 per ml of serum, when combined in a capture assay. An exception is scFv D2, with its micromolar level of affinity. It has been reported that the closely related MAb BB128γ is a synergistic Ab, meaning that its affinity is enhanced dramatically (up to 1,000-fold) when combined with MAbs, which recognize epitopes on the opposite side of the Ag. In vitro affinity maturation (1, 30, 57) of the Abs might improve the sensitivity to a level comparable with that of PCR detection (6). The format of the Abs could be adapted to their application; for instance, a high-affinity monovalent fragment might be useful for capturing Ag, while a multivalent scFv might be more suitable for the sensitive detection of high-density Ag arrested on a solid surface. Therefore, the application of engineered Ab fragments should be very useful for the development of new generations of diagnostics.
REFERENCES
- 1.Barbas C F, Burton D R. Selection and evolution of high-affinity human anti-viral antibodies. Trends Biotechnol. 1996;14:230–234. doi: 10.1016/0167-7799(96)10029-9. [DOI] [PubMed] [Google Scholar]
- 2.Barbas C F, Collet T A, Amberg W, Roben P, Binley J M, Hoelstra D, Cababa D, Jones T M, Williamson A, Pilkington G R, Haigwood N L, Cabezas E, Satterhwait A C, Sanz I, Burton D R. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230:812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- 3.Baumberger, C., S. Kinloch-de-Loes, S. Yerly, B. Hirschel, and L. Perrin. 1993. High levels of circulating RNA in patients with symptomatic HIV-1 infection. AIDS 7(Suppl. 2):S59–S64. [DOI] [PubMed]
- 3a.Berman, M. Unpublished data.
- 4.Billich A, Hammerschmid F, Peichl P, Wenger R, Zenke G, Quesniaux V, Rosenwirth B. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: interference with HIV protein-cyclophilin A interactions. J Virol. 1995;69:2451–2461. doi: 10.1128/jvi.69.4.2451-2461.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke J D, Model P, Zinder N D. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol Gen Genet. 1982;186:185–192. doi: 10.1007/BF00331849. [DOI] [PubMed] [Google Scholar]
- 6.Boni J, Opravil M, Tomasik Z, Rothen M, Bisset L, Grob P J, Luthy R, Schupbach J. Simple monitoring of antiretroviral therapy with a signal-amplification-boosted HIV-1 p24 antigen assay with heat-denatured plasma. AIDS. 1997;11:F47–F52. doi: 10.1097/00002030-199706000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brookmeyer R, Quinn T C. Estimation of current human immunodeficiency virus incidence rates from a cross-sectional survey using early diagnostic tests. Am J Epidemiol. 1995;141:166–172. doi: 10.1093/oxfordjournals.aje.a117404. [DOI] [PubMed] [Google Scholar]
- 9.Busch M P, Lee L L, Satten G A, Henrard D R, Farzadegan H, Nelson K E, Read S, Dodd R Y, Petersen L R. Time course of detection of viral and serologic markers preceding human immunodeficiency virus type 1 seroconversion: implications for screening of blood and tissue donors. Transfusion. 1995;35:91–97. doi: 10.1046/j.1537-2995.1995.35295125745.x. [DOI] [PubMed] [Google Scholar]
- 10.Casey J M, Kim Y, Andersen P R, Watson K F, Fox J L, Devare S G. Human T-cell lymphotropic virus type III: immunologic characterization and primary structure analysis of the major internal protein, p24. J Virol. 1985;55:417–423. doi: 10.1128/jvi.55.2.417-423.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caton A J, Koprowski H. Influenza virus hemagglutinin-specific antibodies isolated from a combinatorial expression library are closely related to the immune response of the donor. Proc Natl Acad Sci USA. 1990;87:6450–6454. doi: 10.1073/pnas.87.16.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S Y, Khouri Y, Bagley J, Marasco W A. Combined intra- and extracellular immunization against human immunodeficiency virus type 1 infection with a human anti-gp120 antibody. Proc Natl Acad Sci USA. 1994;91:5932–5936. doi: 10.1073/pnas.91.13.5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 14.Clackson T, Hoogenboom H R, Griffiths A D, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352:624–628. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 14a.de Haard, H. J. W. Unpublished data.
- 15.Devey M E, Steward M W. The role of antibody affinity in the performance of solid phase assays. In: Kemeny D M, Challacombe S J, editors. ELISA and other solid phase immunoassays. London, United Kingdom: John Wiley & Sons Ltd.; 1988. pp. 135–153. [Google Scholar]
- 16.Ditzel H J, Binley J M, Moore J P, Sodroski J, Sullivan N, Sawyer L S, Hendry R M, Yang W P, Barbas C F, Burton D R. Neutralizing recombinant human antibodies to a conformational V2- and CD4-binding site-sensitive epitope of HIV-1 gp120 isolated by using an epitope-masking procedure. J Immunol. 1995;154:893–906. [PubMed] [Google Scholar]
- 17.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrlich L S, Agresta B E, Carter C A. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischl M A, Richman D D, Hansen N, Collier A C, Carey J T, Para M F, Hardy W D, Dolin R, Powderly W G, Allan J D, et al. The safety and efficacy of zidovudine (AZT) in the treatment of subjects with mildly symptomatic human immunodeficiency virus type 1 (HIV) infection. A double-blind, placebo-controlled trial. The AIDS Clinical Trials Group. Ann Intern Med. 1990;112:727–737. doi: 10.7326/0003-4819-112-10-727. [DOI] [PubMed] [Google Scholar]
- 20.Food and Drug Administration Center for Biologics Evaluation and Research. Recommendations for donor screening with a licensed test for HIV-1 antigen. Rockville, Md: Food and Drug Administration; 1995. [Google Scholar]
- 21.Foote J, Eisen H N. Kinetic and affinity limits on antibodies produced during immune responses. Proc Natl Acad Sci USA. 1995;92:1254–1256. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geysen H M, Meloen R H, Barteling S J. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc Natl Acad Sci USA. 1984;81:3998–4002. doi: 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gherardi E, Milstein C. Original and artificial antibodies. Nature. 1992;357:201–202. doi: 10.1038/357201a0. [DOI] [PubMed] [Google Scholar]
- 24.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 25.Glaser R W, Hausdorf G. Binding kinetics of an antibody against HIV p24 core protein measured with real-time biomolecular interaction analysis suggest a slow conformational change in antigen p24. J Immunol Methods. 1996;189:1–14. doi: 10.1016/0022-1759(95)00221-9. [DOI] [PubMed] [Google Scholar]
- 26.Graziosi C, Pantaleo G, Butini L, Demarest J F, Saag M S, Shaw G M, Fauci A S. Kinetics of human immunodeficiency virus type 1 (HIV-1) DNA and RNA synthesis during primary HIV-1 infection. Proc Natl Acad Sci USA. 1993;90:6405–6409. doi: 10.1073/pnas.90.14.6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths A D, Malmqvist M, Marks J D, Bye J M, Embleton M J, McCafferty J, Baier M, Holliger K P, Gorick B D, Hughes J N, Hoogenboom H R, Winter G. Human anti-self antibodies with high specificity from phage display libraries. EMBO J. 1993;12:725–734. doi: 10.1002/j.1460-2075.1993.tb05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinkula J, Rosen J, Sundqvist V A, Stigbrand T, Wahren B. Epitope mapping of the HIV-1 gag region with monoclonal antibodies. Mol Immunol. 1990;27:395–403. doi: 10.1016/0161-5890(90)90163-t. [DOI] [PubMed] [Google Scholar]
- 29.Holliger P, Prospero T, Winter G. “Diabodies”: small bivalent and bispecific antibody fragments. Proc Natl Acad Sci USA. 1993;90:6444–6448. doi: 10.1073/pnas.90.14.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoogenboom H R. Designing and optimizing library selection strategies for generating high-affinity antibodies. Trends Biotechnol. 1997;15:62–70. doi: 10.1016/S0167-7799(97)84205-9. [DOI] [PubMed] [Google Scholar]
- 31.Hoogenboom H R, Griffiths A D, Johnson K S, Chiswell D J, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19:4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 33.Huston J S, Levinson D, Mudgett H M, Tai M S, Novotny J, Margolies M N, Ridge R J, Bruccoleri R E, Haber E, Crea R, Opperman H. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janvier B, Archinard P, Mandrand B, Goudeau A, Barin F. Linear B-cell epitopes of the major core protein of human immunodeficiency virus types 1 and 2. J Virol. 1990;64:4258–4263. doi: 10.1128/jvi.64.9.4258-4263.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janvier B, Baillou A, Archinard P, Mounier M, Mandrand B, Goudeau A, Barin F. Immune response to a major epitope of p24 during infection with human immunodeficiency virus type 1 and implications for diagnosis and prognosis. J Clin Microbiol. 1991;29:488–492. doi: 10.1128/jcm.29.3.488-492.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Sequences of proteins of immunological interest. 5th ed. Bethesda, Md: National Institutes of Health; 1991. [Google Scholar]
- 37.Kazemier B, de Haard H, Boender P, van Gemen B, Hoogenboom H R. Determination of active single chain antibody concentrations in crude periplasmic fractions. J Immunol Methods. 1996;194:201–209. doi: 10.1016/0022-1759(96)00086-5. [DOI] [PubMed] [Google Scholar]
- 38.Kellogg D E, Sninsky J J, Kowk S. Quantitation of HIV-1 proviral DNA relative to cellular DNA by the polymerase chain reaction. Anal Biochem. 1990;189:202–208. doi: 10.1016/0003-2697(90)90108-l. [DOI] [PubMed] [Google Scholar]
- 39.Kortt A A, Lah M, Oddie G W, Gruen C L, Burns J E, Pearce L A, Atwell J L, McCoy A J, Howlett G J, Metzger D W, Webster R G, Hudson P J. Single-chain Fv fragments of anti-neuraminidase antibody NC10 containing five- and ten-residue linkers form dimers and with zero-residue linker a trimer. Protein Eng. 1997;10:423–433. doi: 10.1093/protein/10.4.423. [DOI] [PubMed] [Google Scholar]
- 40.Lew A M. The effect of epitope density and antibody affinity on the ELISA as analysed by monoclonal antibodies. J Immunol Methods. 1984;72:171–176. doi: 10.1016/0022-1759(84)90445-9. [DOI] [PubMed] [Google Scholar]
- 41.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 42.Marks J D, Griffiths A D, Malmqvist M, Clackson T P, Bye J M, Winter G. By-passing immunization: building high affinity human antibodies by chain shuffling. Bio/Technology. 1992;10:779–783. doi: 10.1038/nbt0792-779. [DOI] [PubMed] [Google Scholar]
- 43.Marks J D, Hoogenboom H R, Bonnert T P, McCafferty J, Griffiths A D, Winter G. By-passing immunization: human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 44.McGuinness B T, Walter G, FitzGerald K, Schuler P, Mahoney W, Duncan A R, Hoogenboom H R. Phage diabody repertoires for selection of large numbers of bispecific antibody fragments. Nat Biotechnol. 1996;14:1149–1154. doi: 10.1038/nbt0996-1149. [DOI] [PubMed] [Google Scholar]
- 45.Mhashilkar A M, Bagley J, Chen S Y, Szilvay A M, Helland D G, Marasco W A. Inhibition of HIV-1 Tat-mediated LTR transactivation and HIV-1 infection by anti-Tat single chain intrabodies. EMBO J. 1995;14:1542–1551. doi: 10.1002/j.1460-2075.1995.tb07140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 47.Nieba L, Krebber A, Pluckthun A. Competition BIAcore for measuring true affinities: large differences from values determined from binding kinetics. Anal Biochem. 1996;234:155–165. doi: 10.1006/abio.1996.0067. [DOI] [PubMed] [Google Scholar]
- 48.Niedrig M, Hinkula J, Harthus H P, Broker M, Hopp L, Pauli G, Wahren B. Characterization of murine monoclonal antibodies directed against the core proteins of human immunodeficiency virus types 1 and 2. J Virol. 1991;65:4529–4533. doi: 10.1128/jvi.65.8.4529-4533.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niedrig M, Hinkula J, Weigelt W, L’Age-Stehr J, Pauli G, Rosen J, Wahren B. Epitope mapping of monoclonal antibodies against human immunodeficiency virus type 1 structural proteins by using peptides. J Virol. 1989;63:3525–3528. doi: 10.1128/jvi.63.8.3525-3528.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niedrig M, Rabanus J P, L’Age Stehr J, Gelderblom H R, Pauli G. Monoclonal antibodies directed against human immunodeficiency virus (HIV) gag proteins with specificity for conserved epitopes in HIV-1, HIV-2 and simian immunodeficiency virus. J Gen Virol. 1988;69:2109–2114. doi: 10.1099/0022-1317-69-8-2109. [DOI] [PubMed] [Google Scholar]
- 51.Pluckthun A, Pack P. New protein engineering approaches to multivalent and bispecific antibody fragments. Immunotechnology. 1997;3:83–105. doi: 10.1016/s1380-2933(97)00067-5. [DOI] [PubMed] [Google Scholar]
- 52.Popovic M, Sarin P S, Robert-Gurroff M K, Kalyanaraman V S, Mann D, Minowada J, Gallo R C. Isolation and transmission of human retrovirus (human t-cell leukemia virus) Science. 1983;219:856–859. doi: 10.1126/science.6600519. [DOI] [PubMed] [Google Scholar]
- 53.Prickett K S, Amberg D C, Hopp T P. A calcium-dependent antibody for identification and purification of recombinant proteins. BioTechniques. 1989;7:580–589. [PubMed] [Google Scholar]
- 54.Robert-Hebmann V, Emiliani S, Jean F, Resnicoff M, Traincard F, Devaux C. Clonal analysis of murine B cell response to the human immunodeficiency virus type 1 (HIV1)-gag p17 and p25 antigens. Mol Immunol. 1992;29:729–738. doi: 10.1016/0161-5890(92)90183-x. [DOI] [PubMed] [Google Scholar]
- 55.Robert-Hebmann V, Emiliani S, Resnicoff M, Jean F, Devaux C. Subtyping of human immunodeficiency virus isolates with a panel of monoclonal antibodies: identification of conserved and divergent epitopes on p17 and p25 core proteins. Mol Immunol. 1992;29:1175–1183. doi: 10.1016/0161-5890(92)90053-z. [DOI] [PubMed] [Google Scholar]
- 56.Schier R, Bye J, Apell G, McCall A, Adams G P, Malmqvist M, Weiner L M, Marks J D. Isolation of high-affinity monomeric human anti-c-erbB-2 single chain Fv using affinity-driven selection. J Mol Biol. 1996;255:28–43. doi: 10.1006/jmbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 57.Schier R, McCall A, Adams G P, Marshall K W, Merritt H, Yim M, Crawford R S, Weiner L M, Marks C, Marks J D. Isolation of picomolar affinity anti-c-erbB-2 single-chain Fv by molecular evolution of the complementarity determining regions in the center of the antibody binding site. J Mol Biol. 1996;263:551–567. doi: 10.1006/jmbi.1996.0598. [DOI] [PubMed] [Google Scholar]
- 58.Skerra A, Pluckthun A. Secretion and in vivo folding of the Fab fragment of the antibody McPC603 in Escherichia coli: influence of disulphides and cis-prolines. Protein Eng. 1991;4:971–979. doi: 10.1093/protein/4.8.971. [DOI] [PubMed] [Google Scholar]
- 59.Spector S A, Kennedy C, McCutchan J A, Bozzette S A, Straube R G, Connor J D, Richman D D. The antiviral effect of zidovudine and ribavirin in clinical trials and the use of p24 antigen levels as a virologic marker. J Infect Dis. 1989;159:822–828. doi: 10.1093/infdis/159.5.822. [DOI] [PubMed] [Google Scholar]
- 60.Steinkasserer A, Harrison R, Billich A, Hammerschmid F, Werner G, Wolff B, Peichl P, Palfi G, Schnitzel W, Mlynar E, et al. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus type 1 (HIV-1): interference with early and late events in HIV-1 replication. J Virol. 1995;69:814–824. doi: 10.1128/jvi.69.2.814-824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stevens P W, Francis C H, Jolley M E, Bethell D R. Monoclonal antibodies to HIV-1 p24 core protein include pairs which exhibit synergistic binding. Mol Immunol. 1993;30:243–254. doi: 10.1016/0161-5890(93)90053-e. [DOI] [PubMed] [Google Scholar]
- 62.van Gemen, B., T. Kievits, P. Nara, H. G. Huisman, S. Jurriaans, J. Goudsmit, and P. Lens. 1993. Qualitative and quantitative detection of HIV-1 RNA by nucleic acid sequence-based amplification. AIDS. 7(Suppl. 2):S107–S110. [DOI] [PubMed]
- 63.Veronese F D, Copeland T D, Oroszlan S, Gallo R C, Sarngadharan M G. Biochemical and immunological analysis of human immunodeficiency virus gag gene products p17 and p24. J Virol. 1988;62:795–801. doi: 10.1128/jvi.62.3.795-801.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Volberding P A, Lagakos S W, Koch M A, Pettinelli C, Myers M W, Booth D K, Balfour H H J, Reichman R C, Bartlett J A, Hirsch M S, et al. Zidovudine in asymptomatic human immunodeficiency virus infection. A controlled trial in persons with fewer than 500 CD4-positive cells per cubic millimeter. The AIDS Clinical Trials Group of the National Institute of Allergy and Infectious Diseases. N Engl J Med. 1990;322:941–949. doi: 10.1056/NEJM199004053221401. [DOI] [PubMed] [Google Scholar]
- 65.Von Sydow M, Gaines H, Sonnerborg A, Forsgren M, Pehrson P O, Strannegard O. Antigen detection in primary HIV infection. Br Med J Clin Res. 1988;296:238–240. doi: 10.1136/bmj.296.6617.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wehrly K, Chesebro B. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods. 1997;12:288–293. doi: 10.1006/meth.1997.0481. [DOI] [PubMed] [Google Scholar]
- 67.Winter G, Griffiths A D, Hawkins R E, Hoogenboom H R. Making antibody by phage display technology. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 68.Winter G, Milstein C. Man-made antibodies. Nature. 1991;349:293–299. doi: 10.1038/349293a0. [DOI] [PubMed] [Google Scholar]
- 69.Zhao Y, Chen Y, Schutkowski M, Fischer G, Ke H. Cyclophilin A complexed with a fragment of HIV-1 gag protein: insights into HIV-1 infectious activity. Structure. 1997;5:139–146. doi: 10.1016/s0969-2126(97)00172-x. [DOI] [PubMed] [Google Scholar]