Correction: Genome Biol 23, 114 (2022)
https://doi.org/10.1186/s13059-022-02682-2
Following publication of the original paper [1], the authors have reported an error in reference genome version of the HiC data used for validation of the RE-TG interactions. After using the correct version of the HiC anchor locations, the result in Fig. 4D, supplementary Figure S11, and S12 are all improved.
Fig. 4.
D. Validation of RE-TG prediction by HiC data on Naïve CD4 T cell
Supplementary Information
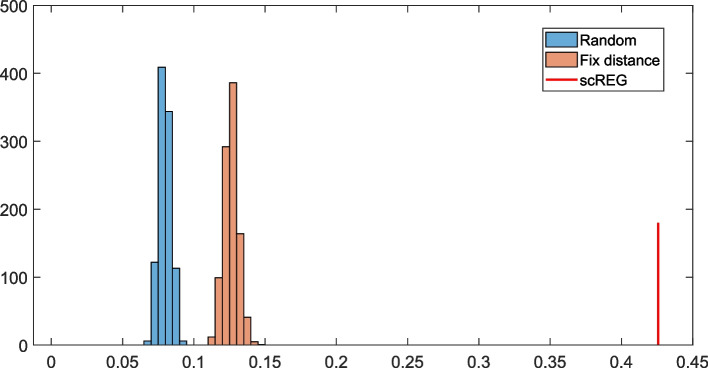
Additional file 1: Supplementary Figure S11. Validation of RE-TG prediction by HiC data. Consistency ratio of predicted RE and promoter capture HiC data on different cell types of. We can see in all cell type, scREG predict the greatest number of same RE-TG pairs as previously found promoter capture HiC data. set select distribution distance same with scREG, does improve the performance. Supplementary Figure S12. AUROC and AUPR of RE-TG predictio by taking HiC data as ground truth.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhana Duren and Fengge Chang contributed equally to this work.
Contributor Information
Zhana Duren, Email: zduren@clemson.edu.
Wing Hung Wong, Email: whwong@stanford.edu.
Reference
- 1.Duren Z, Chang F, Naqing F, Xin J, Liu Q, Wong WH. Regulatory analysis of single cell multiome gene expression and chromatin accessibility data with scREG. Genome Biol. 2022;23:114. doi: 10.1186/s13059-022-02682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure S11. Validation of RE-TG prediction by HiC data. Consistency ratio of predicted RE and promoter capture HiC data on different cell types of. We can see in all cell type, scREG predict the greatest number of same RE-TG pairs as previously found promoter capture HiC data. set select distribution distance same with scREG, does improve the performance. Supplementary Figure S12. AUROC and AUPR of RE-TG predictio by taking HiC data as ground truth.