Abstract
RAK antigens p120, p42, and p25 exhibit molecular and immunological similarity to the proteins encoded by human immunodeficiency virus type 1 (HIV-1) and are expressed by 95% of breast and gynecological cancer cases in women and prostate cancer cases in men. The binding of an epitope-specific anti-HIV-1 gp120 monoclonal antibody (MAb) (amino acids 308 to 322) to cancer RAK antigens has been found to be inhibited by a peptide derived from variable loop V3 of HIV-1. Breast cancer DNAs of 40 patients were PCR amplified with HIV-1 gp41-derived primers, and all of the samples were found to be positive. The DNA fragments amplified in seven blindly selected breast cancer samples were sequenced. The breast cancer DNA sequences showed at least 90% homology to the HIV-1 gene for gp41. Antisense oligonucleotides complementary to the HIV-1-like sequences inhibited reverse transcriptase activity and inhibited the growth of breast cancer cells in vitro. Viral particles detected in breast cancer cell lines were strongly immunogold labeled with the anti-HIV-1 gp120 MAb. The results obtained strongly suggest that the long-postulated breast cancer virus may, in fact, be related to HIV-1.
Breast cancer affects 1 in every 8 to 10 women in the United States (10, 19, 32, 35, 36). Approximately 10% of breast cancer patients exhibit a genetic inheritance pattern, while the overwhelming majority of women develop breast cancer for an unpredictable reason (2, 6, 7, 14, 20, 23, 37, 43). Recent enthusiasm following the characterization of the BRCA1 and BRCA2 genes, which are associated with some inherited forms of breast and ovarian cancer, has been diminished by the fact that only 1 of 800 women carries the mutated BRCA gene, while at least 80 to 100 of 800 women will develop breast cancer (17, 21, 41, 42).
Involvement of a viral factor in the etiology of human breast cancer has been considered by several laboratories (1, 5, 9, 11–13, 18, 39). Special attention was focused on human DNA sequences with homology to mouse mammary tumor virus (MMTV) (1, 38, 40). Retrovirus-like particles with immunological similarity to MMTV proteins were found in human breast carcinoma cell lines (13), peripheral blood monocytes of breast cancer patients (11), pleural effusion fluids from breast adenocarcinoma patients (39), human breast cancer tissue (5, 9, 12, 39), and breast milk (18). No viral agent has been identified as a causative agent of breast cancer in humans.
Except for cervical cancer, which is associated predominantly with human papillomavirus and/or herpes simplex virus infection (3), the viral etiology of other types of female reproductive tract cancer remains unverified. Recent studies on herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma (4, 22, 25, 33) strongly suggest that the role of retroviruses in human cancer was underestimated for a long time.
We have recently identified a new class of breast and gynecological cancer markers, which have been named Rakowicz markers or, briefly, RAK markers (25–30). RAK antigens p120, p42, and p25 express epitopes in common with envelope protein gp120 of human immunodeficiency virus type 1 (HIV-1) and can be detected either by an epitope-specific anti-HIV-1 gp120 monoclonal antibody (MAb) (25–27), or by MAb RAK-BrI, which is directed against breast cancer (28, 29). RAK antigens are absent in normal breast tissue and other normal tissues (28), which suggests the strong diagnostic potential of these unique markers. Very recently, RAK markers were also found in prostate cancer and in a number of benign hyperplasia cases (31).
One of the RAK antigens, p160, corresponding in size to HIV-1 gp160 (precursor of gp120 and gp41), was detected in the blood of 70% of gynecological cancer patients before surgery, in 40 to 50% of breast cancer patients after surgery, in 20% of healthy women with a family history of breast cancer, and in 13% of healthy women without a family history of breast or gynecological cancer (29, 30).
The similarity of RAK breast cancer antigens to HIV-1 major proteins led to speculation that HIV-1-like DNA sequences encoding RAK antigens might also be localized in cancer DNA. That hypothesis was confirmed by the finding that HIV-1 gp41-derived primers SK68 and SK69 initiated a PCR with breast cancer DNA but not with normal breast DNA (28). It is noteworthy that the same HIV-1-derived primers amplified prostate cancer DNA but not normal prostate DNA (31). The prostate cancer DNA fragments amplified exhibited strong homology to HIV-1 (31). The study described in this report revealed strong homology between HIV-1 and breast cancer DNA sequences. Viral particles cross-reactive with the anti-HIV-1 gp120 MAb were also detected in breast cancer cells, supporting the hypothesis of a potential viral origin of breast cancer.
MATERIALS AND METHODS
Breast cancer tissue.
Breast cancer tissue, breast cancer adjacent tissue (NAT), and histologically normal breast tissue samples were provided by the Cooperative Human Tissue Network.
Cell lines.
Breast carcinoma cell lines SKBr3 and MCF7 and cervical cancer cell line SiHa, obtained from the American Type Culture Collection (ATCC), were grown in Eagle’s minimal essential medium-Leibovitz’s L15 medium (3:4) or other recommended medium supplemented with 10% fetal bovine serum.
MAbs.
MAb RAK-BrI, which is directed against breast cancer antigens but also detects gynecological cancer antigens, was described before (28, 29). The anti-HIV-1 gp120 MAb (amino acids 308 to 322) was purchased from Du Pont. The ability of that MAb to detect a cancer antigen was reported before (25, 26). Control antibodies, which do not recognize cancer antigens, including another epitope-specific anti-HIV-1 gp120 MAb (25, 27) and an anti-HIV-1 p25 MAb, were from Du Pont.
Tissue fractionation.
Fractionation of tissue was described before (25–29). Briefly, cancer and normal tissue samples were homogenized in 0.35 M sucrose–10 mM KCl–1.5 mM MgCl2–10 mM Tris-HCl (pH 7.6)–0.12% Triton X-100–12 mM 2-mercaptoethanol and centrifuged at 600 × g for 10 min. The supernatant was defined as the cytoplasmic fraction.
Electrophoresis of proteins.
Cytoplasmic and chromatin proteins were analyzed by electrophoresis in a 10% polyacrylamide gel with 0.1% sodium dodecyl sulfate (SDS) in a buffer containing 250 mM Tris-HCl (pH 8.3), 195 mM glycine, and 0.1% SDS as described by Laemmli (15).
Western blotting.
Blotting of proteins from the polyacrylamide gel to a polyvinylidene difluoride membrane was performed in 25 mM Tris-HCl (pH 8.6)–192 mM glycine buffer containing 10% methanol. Filters were incubated with 1% bovine serum albumin for 16 h at 0°C and then with MAb 5023 (2 μg/ml) or MAb RAK-BrI (0.1 μg/ml), washed with Tris-glycine buffer, and incubated with alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G for 1 h. After washing with TBST, membranes were incubated with 0.1% 1-naphthyl-phosphate and Fast Red. In some experiments, the anti-HIV-1 gp120 or RAK-BrI MAb (5 μg/ml) was preincubated with peptide RIQRGPGRAFVTIGK, RIQRGPGRKFVTIGK, or RIQRGPGRVVVTGK (18 μg/ml) for 1 h on ice and then incubated with blots.
DNA isolation.
Sections of breast cancer tissue, NAT, or normal tissues (obtained during standard surgical procedures and stored at −80°C) were lysed in 100 mM NaCl–10 mM Tris-HCl (pH 8.0)–25 mM EDTA (pH 8.0)–0.5% SDS. The lysate was digested with proteinase K (0.1 mg/ml), extracted with phenol-chloroform, and immunoprecipitated with ethanol. DNA from HIV-1-infected cells (positive control) was obtained from Advanced Biotechnology Services, Inc.
PCR.
PCR occurred in a solution containing 10 mM KCl; 10 mM (NH4)SO4, 20 mM Tris-HCl; 5 mM MgCl2; 0.1% Triton X-100; dATP, dTTP, dCTP, and dGTP at 0.2 mM each; each primer at 0.5 μM; and 2.5 U of Taq polymerase. The amount of template DNA used was 1.0 μg/50 μl of the reaction mixture. The reactions ran for 30 cycles in a Perkin Elmer 9600 thermal cycler. The PCR was done by using HIV-1-derived primers SK68 (7801-to-7820 region of gp41 Env) [5′-AGCAGCAGGAAGCACTATGG-3′]) and SK69 (7922-to-7942 region of gp41 Env) [5′-CCAGACTGTGAGTTGCAAGAG-3′]). All DNA samples which tested negative with primers SK68 and SK69, as well as approximately 50% of the PCR-positive samples, were tested with a control set of primers derived from the globin gene. Each set of PCRs included DNA isolated from HIV-1-infected lymphocytes (positive control). The PCR mixture was electrophoretically analyzed in a 1.5% agarose gel. DNA was visualized by UV fluorescence after staining with ethidium bromide. Each cancer or normal DNA sample was tested by PCR three to seven times in different sets. Samples selected for DNA sequencing were separated in a 4% agarose gel. PCR bands were encoded by number and submitted for blind sequencing with primers SK68 and SK69. As a positive control, the amplified HIV-1 DNA fragment was used for sequencing.
Test for RNA-dependent DNA polymerase activity.
Cell culture media from the cells not exposed (control) or exposed for 4 days to RAK I (100 μg/ml) were ultracentrifuged (160,000 × g, 60 min). The pellet was resuspended in 137 mM NaCl–3 mM KCl–10 mM phosphate buffer (pH 7.4)–0.6 mM EDTA. The assay mixture contained 5 mM Tris-HCl (pH 8.3); 0.6 mM magnesium acetate; 6 mM NaCl; 2 mM dithiothreitol; 0.08 mM each dATP, dCTP, and dGTP; and 0.001 mM [methyl-3H]TTP. A viral pellet sample corresponding to 4 × 106 viral particles was added to 500 μl of the assay mixture, which was then incubated for 45 min at 37°C. The reaction was stopped by adding 5 ml of 20% trichloroacetic acid (34).
Nucleotide sequence accession numbers.
The DNA sequences identified in this study have been assigned accession no. AF073463 to AF073469.
RESULTS
Detection of RAK antigens in breast cancer tissue.
Breast cancer tissue samples, NAT, and normal breast tissue were electrophoretically separated in a polyacrylamide gel with SDS, transferred onto a polyvinylidene difluoride membrane, and Western blot hybridized with MAb RAK-BrI (Fig. 1) or an anti-HIV-1 MAb (Fig. 2). Of the 125 breast cancer cases tested, 95.2% were strongly RAK p120, p42, and p25 positive and the rest were weakly positive (Table 1). Of 70 NAT samples, only 8 (11.4%) tested moderately RAK positive and an additional 9 (12.9%) were weakly positive. Of 40 samples obtained from histologically “normal” parts of cancer-affected breasts, only 3 tested positive and 1 was weakly positive (7.5 and 2.5%, respectively). In women with advanced fibrocystic disease, which qualified them for either partial breast removal or complete mastectomy, only 5.7% expressed RAK antigens at a moderate level and 2.8% expressed RAK antigens at a low level. Of the 10 cases of breast reduction, only 1 tested weakly RAK positive. Breast milk was used as the source of normal breast epithelial cells, and all 10 samples tested RAK negative. Control samples of skin and placenta tested negative. Other RAK-negative normal samples were described before (30, 31). The results confirmed a clear association of RAK antigens p120, p42, and p25 with breast cancer.
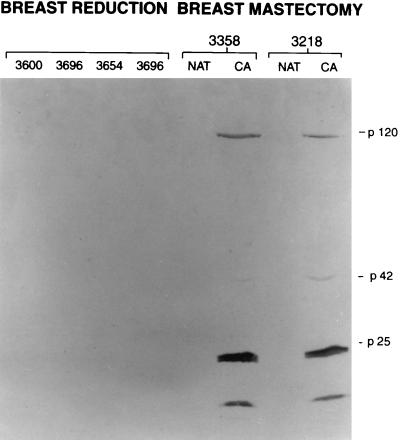
FIG. 1.
Electrophoretic analysis (10% polyacrylamide gel) and Western blotting of cytoplasmic proteins with MAb RAK-BrI. In a blind experiment, both breast cancers (CA) (3358 and 3218) tested RAK antigen positive and NAT samples tested RAK antigen negative. No RAK antigens were detected in the four breast tissue samples obtained during breast reduction (3600, 3696, 3654, and 3696).
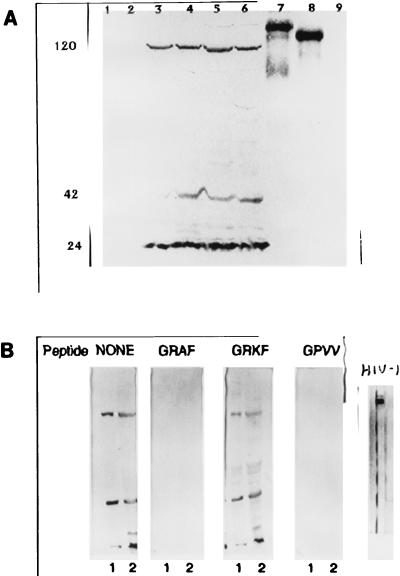
FIG. 2.
Reactivity of an anti-HIV-1 gp120 MAb with breast and gynecological cancer cytoplasmic proteins and HIV-1 proteins. (A) Lanes: 1 and 2, normal breast and normal uterine proteins, respectively; 3 to 5, breast cancer tissues from three different patients; 6, uterine cancer; 7 and 8, HIV-1 gp160 and gp120, respectively; 9, HIV-1 p24 (negative control). The anti-HIV-1 MAb reacted with three cancer proteins (RAK p120, p42, and p25) but also with HIV-1 gp120 and its precursor gp160. The positions of p120, p42, and p24 are shown on the left. (B) Reactivity of the anti-HIV-1 gp120 MAb with breast cancer proteins obtained from two different patients (lanes 1 and 2) before and after preincubation with the indicated peptides. The peptides containing the consensus sequence GRAF or GRVV inhibited the binding of the anti-HIV-1 MAb to all three cancer antigens. Positively charged lysine in the peptide GRKF did not allow MAb binding, and the peptide did not affect interaction with cancer antigens.
TABLE 1.
Expression of RAK antigens in women with breast cancer
| Tissue or cells | No. of samples tested/no. (%) positive for RAK antigen |
|---|---|
| Breast cancer | 125/119a 6b (95.2, 4.8) |
| NAT | 70/8a 9b (11.4, 12.9) |
| Normal breastc | 40/3a 1b (7.5, 2.5) |
| Breast with fibrocystic disease | 35/2a 1b (5.7, 2.8) |
| Breast reductiond | 10/1b (10) |
| Breast milk | 10/0 (0) |
| Skin | 10/0 (0) |
| Placenta | 2/0 (0) |
| MCF 7, SiHae | A,a B,a Ca |
Moderate to very high expression of RAK p120, p42, and p25.
Low expression of all three antigens or expression of only two antigens.
The majority of “normal” tissues were obtained from the cancer-affected breast.
Breast reduction cases are theoretically normal cases. In the United States, one of every eight women will develop breast cancer during her lifetime; therefore, some premalignant changes may occur.
MCF 7 and SiHa are breast and cervical cell lines, respectively, grown under laboratory conditions for 1 year (A) or 2 to 3 weeks (B) or freshly supplied by ATCC.
The expression of all three RAK antigens in several breast and cervical cancer cell lines was described before (25, 27). In the current experiments, two cell lines, breast cancer MCF 7 and cervical cancer SiHa, were used as models. Expression of RAK antigens was compared in (i) cells remaining in cell culture for 1 year, (ii) cells which were obtained from ATCC as a frozen stock and grown in the laboratory for 3 to 4 weeks, and (iii) cells which were grown to a monolayer at ATCC facilities and immediately processed after receipt at our laboratory. No differences in the expression of RAK antigens was observed between groups ii and iii. Expression of RAK antigens in group i was slightly lower than in groups ii and iii, which is consistent with the observation that expression of several proteins is lower in cells remaining in culture for a long time. The last experiments also clearly indicated that expression of RAK markers is constitutively associated with MCF 7 and SiHa cells and does not represent laboratory contamination.
Mapping of cancer epitopes in common with HIV-1.
The anti-HIV-1 MAb, which recognizes breast and gynecological cancer antigens but does not recognize normal cells (Fig. 2A, lanes 1 to 6), was developed against amino acids 308 to 322 (RIQRGPGRAFVTIGK) of the variable loop of HIV-1 gp120, and this MAb binds to the GRAF epitope of HIV-1 gp160 and gp120 (8, 16, 24) (Fig. 2A, lanes 7 and 8). To assess the specificity of anti-HIV-1 gp120 MAb binding to the cancer cell epitopes, Western blots were incubated with a MAb which was either not preincubated or preincubated with the peptide RIQRGPGRAFVTIGK (Fig. 2B). A peptide derived from HIV-1 gp120 which contained the consensus sequence GRAF competitively blocked the binding of the anti-HIV-1 MAb to all three cancer antigens. A peptide with the GRAF sequence replaced with GRVV also competitively blocked the binding of MAb RAK-BrI to cancer antigens, while a peptide containing the sequence GRKF did not inhibit MAb RAK-BrI binding to cancer antigens. The results suggest that either the GRAF epitope or a very similar epitope is present in cancer antigens. Positively charged lysine (K) destroys the binding site for MAb RAK-BrI. Since the sequence alanine-phenylalanine (AF) might be replaced with valine-valine (VV), it is likely that the cancer epitope is conformational and not identical to the HIV epitope. The G preceding RAF is critical for cancer antigen binding, since another anti-HIV-1 MAb which recognizes RAF but forms weak interactions with G does not recognize cancer antigens (25) and has a low affinity for HIV-1 gp120 (8, 16, 24). Another control MAb directed against HIV-1 gp25 did not recognize any of the cancer antigens, which confirmed that there is limited homology between cancer and HIV-1 proteins, but cancer antigens are very distinct from HIV-1 proteins and cannot originate from HIV-1 contamination.
PCR detection of HIV-1-like DNA sequences in breast cancer DNA.
Primers SK68 and SK69, derived from the HIV-1 genome (region gp41), specifically initiated amplification of breast cancer DNA (Fig. 3A to C). All 40 of the breast cancer cases tested were strongly PCR positive (Table 2). Of 39 samples of DNA isolated from NAT, only 41% tested moderately to highly positive and an additional 12.8% tested weakly positive. Of 30 DNA samples obtained from histologically normal tissue of a cancer-affected breast, 26.7% tested moderately positive and 16.7% were weakly positive. Of the 10 DNA samples obtained from breast reduction, only 1 tested very weakly positive. DNA extracted from breast milk of healthy women tested PCR negative. Other control normal tissues also tested PCR negative (Table 2).
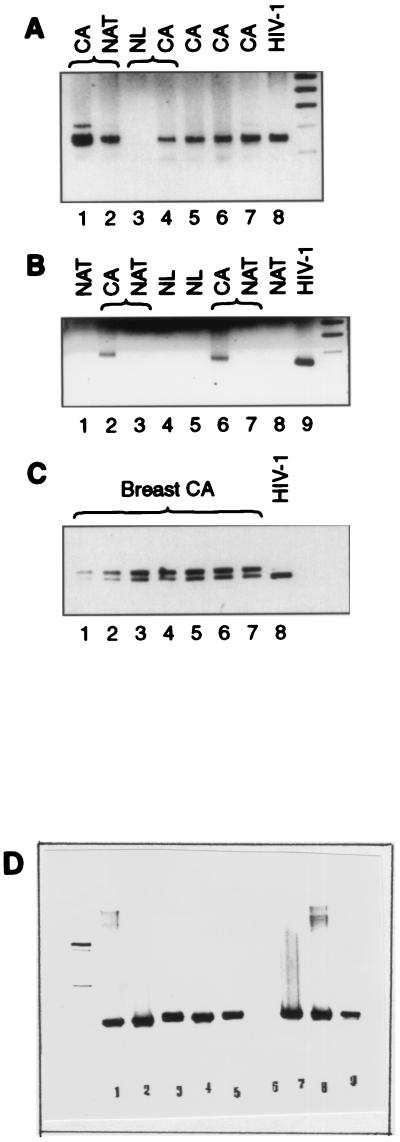
FIG. 3.
Electrophoretic pattern of PCR products amplified by HIV-1 (gp41 Env)-derived primers SK68 and SK69 (A to C) or globin primers (D), separated in a 1.5% (A and B) or a 4% (C) agarose gel and stained with ethidium bromide. CA, cancer; NL, normal breast tissue. (A) Lanes: 1 and 2, CA and NAT samples from one patient that both tested positive (but the PCR with NAT was weaker); 3 and 4, NL sample that tested negative and CA sample from the same patient that tested positive; 5, 6, and 7, CA samples from different patients that tested positive; 8, HIV-1-positive control. (B) Lanes 1 and 8, NAT samples from two different patients that tested negative; 2, 3, 6, and 7, CA samples that tested positive and NAT samples that tested negative; 4 and 5, NL samples from different patients that tested negative; 9, HIV-1-positive control. (C) PCR amplification patterns of breast cancer DNAs selected for sequencing. The lower band (142 bp) corresponded in size to the HIV-1 band. (D) PCR amplification patterns obtained with globin primers. Lanes: 1 to 3, normal breast DNA; 4, breast milk DNA; 5, NAT sample DNA; 7 to 9, breast cancer DNA. Sample 6 tested globin negative and was discarded. Globin-positive samples 1 to 5 were all negative with primers SK68 and SK69.
TABLE 2.
PCR amplification of breast cancer DNA with HIV-1-derived primers
| Tissue or cells | No. of samples tested by PCR/no. (%) positive for RAK antigen |
|---|---|
| Breast cancer | 40/40a (100) |
| NAT | 39/16,a 5b (41, 12.8) |
| Normal breastc | 30/8a 5b (26.7, 16.7) |
| Breast reductiond | 10/1b (10) |
| Breast milk | 10/0a (0) |
| MCF 7, SiHae | A,a B,a Ca |
| Cervix | 2/0a (0) |
| Ovary | 2/0a (0) |
| Uterus | 2/0a (0) |
| Vagina | 2/0a (0) |
| Colon | 3/0a (0) |
| Skin | 6/0a (0) |
| Placenta | 2/0a (0) |
To eliminate the possibility that a negative PCR of normal tissue DNA had been caused by nonspecific inhibition of the amplification reaction, each DNA sample was also amplified with globin primers (Fig. 3D). One of 11 DNA samples obtained from normal breast tissue, one DNA sample obtained from breast milk, and one skin DNA sample tested negative with both globin primers and with the SK68 and SK69 primers. These globin-negative samples were discarded. All of the SK68-SK69-negative samples included in Table 2 were PCR positive with the globin primers (Fig. 3D, lanes 1 to 3), similar to SK68-SK69-positive cancer DNA (Fig. 3D, lanes 7 to 9).
In a 1.5% agarose gel, a single amplification band was observed in HIV-1 and breast cancer DNAs (Fig. 3A and B). The size of the amplified DNA region (142 bp) was similar to the size of the DNA fragment amplified in DNA isolated from HIV-1-infected cells. For DNA sequencing, seven breast cancer DNA samples were blindly selected, PCR amplified, and separated in a 4% agarose gel (Fig. 3C). The PCR band, which seemed to be homogeneous in a 1.5% agarose, separated into a dominant band with mobility equal to that of the PCR fragment amplified in HIV-1 and an additional band migrating slightly slower (∼160 bp).
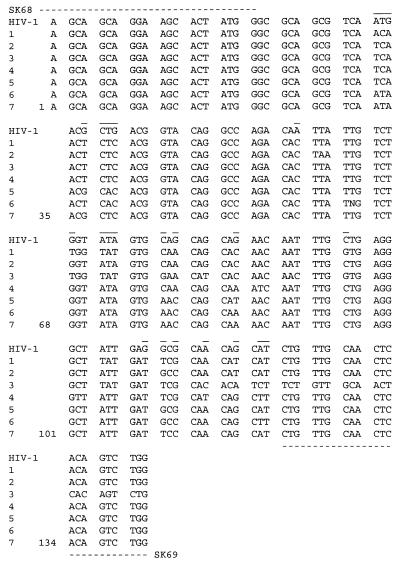
Both bands were isolated from the gel and sequenced. Each fragment was sequenced at least four times, twice with primer SK68 (forward) and twice with the SK69 (reverse) primer. The overlapping sequences of the lower band showed a perfect match with HIV-1 sequences. Single point mutations were observed in DNAs of breast cancer patients, compared to HIV-1 sequences. The average similarity to HIV-1 in all of the breast cancer DNAs tested was at least 90% (Fig. 4), which indicated a strong homology of the identified cancer sequences to HIV-1.
FIG. 4.
DNA sequences amplified in breast cancer samples from seven different patients with HIV-1 gp41-derived primers SK68 and SK69. Broken lines indicate primer locations. Lines over the sequences indicate variable sequences that are different in at least two patients from that of HIV-1.
The upper PCR band seemed to represent an artifact occurring by partial homology of HIV-1 primers to some repetitive sequences of the human genome. No correlation of that fragment with HIV-1 or any other sequences has been found.
Electron microscopic detection of viral particles.
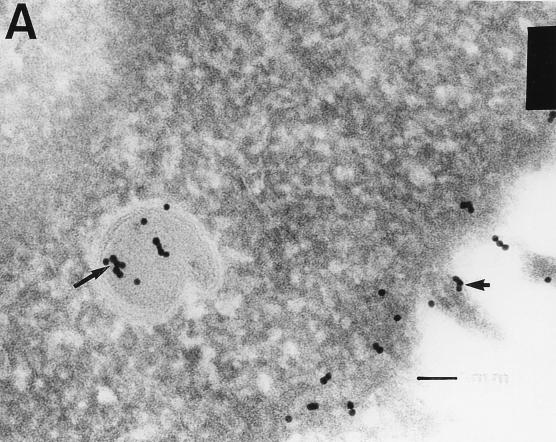
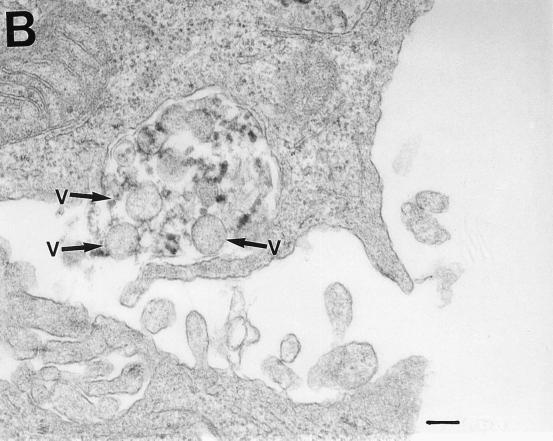
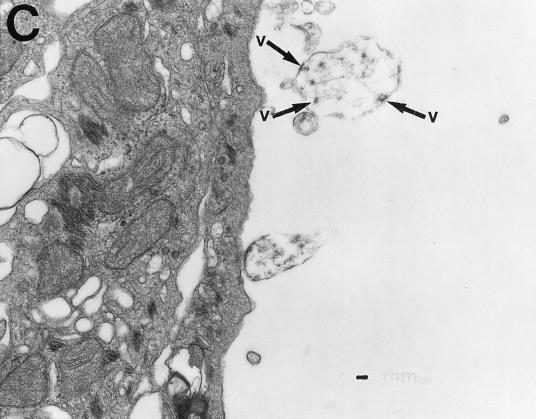
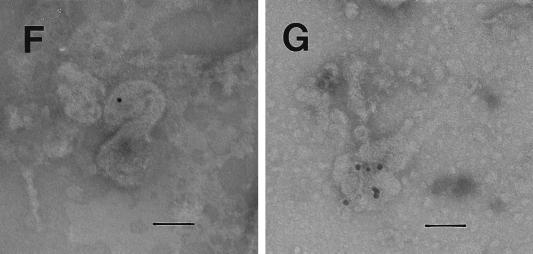
Electron microscopic analysis of thin sections of SiHa cervical cancer and MCF 7 breast cancer cells revealed large, membrane-coated vesicles (vacuoles) that were immunogold labeled with the anti-HIV-1 gp120 MAb and localized in the cytoplasm (Fig. 5A), at the edges of cells (Fig. 5B), and outside of cells (Fig. 5C). Strong immunogold labeling of the villus-like structures on the cell surface, as well as of the intracellular and extracellular particles, was also observed (Fig. 5A).
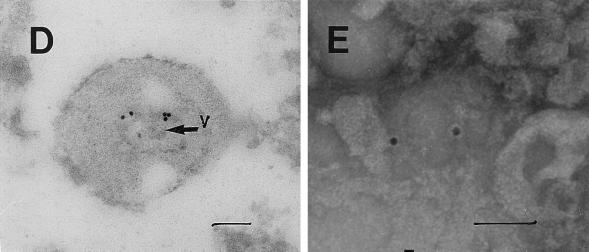
FIG. 5.
Transmission electron micrographs of cellular and extracellular vacuoles carrying viral particles in SiHa (A, C, and D) and MCF 7 (B) cells. Viral particles were obtained by ultracentrifugation (100,000 × g, 1 h) of cell culture media and negatively stained with uranyl acetate (E to G). In A and D to G, samples were also immunogold labeled with an anti-HIV-1 gp120 MAb. The sizes of the immunogold particles (arrows in A) were 15 (A and D) and 10 (E) nm. V, virus-like particles. Bars, 100 nm. The original magnification was 30,000 (C) or 75,000 (A, B, and D to G).
A cross section (Fig. 5B) of a large cytoplasmic vesicle located close to the cell surface revealed several oval-shaped structures localized within the vesicle. The virus-like particles were also “budding” from the extracellular vacuoles (Fig. 5C). Immunogold staining of the membrane-coated vesicles obtained by ultracentrifugation (100,000 × g for 1 h) of SiHa or MCF 7 cell culture medium revealed virus-like particles localized inside the membrane-covered vesicles (Fig. 5D). Negative staining of purified viral particles is shown in Fig. 5E to G. The size of the virus-like particles seems to be 120 nm. The tail-and-head structure of the anti-HIV-1 gp120 MAb-labeled viral particles was frequently visible (Fig. 5E to G). It is noteworthy that labeling of MCF 7 and SiHa cells with the anti-HIV-1 gp120 MAb was stronger in cells supplied by ATCC as a monolayer than in those remaining under laboratory conditions for 1 year. This observation was consistent with lower expression of RAK antigens in the aging cell cultures. Both MCF 7 and SiHa cells were tested by ATCC and found to be mycoplasma negative and tested by Advanced Biotechnology Inc. and found to be HIV-1 negative and mycoplasma and bacterium free. Thus, the identified particles are likely to belong to a virus which expresses epitopes similar to HIV-1 gp120 but completely different in structure and function.
Effect of antisense oligonucleotides on cancer cell growth in vitro.
To evaluate whether the identified cancer DNA sequences, which are absent in normal tissues and may belong to a retrovirus, play a role in malignant cell growth, antisense oligonucleotides have been synthesized and used to treat SiHa and MCF 7 cells in vitro. Antisense oligonucleotide RAK-I (21-mer 5′-CCAGACTGTGAGTTGCAACAG-3′), complementary to the 3′ end of the cancer DNA sequences amplified with HIV-1 Env-derived primers, inhibited the growth of breast cancer cells by 70% within 4 days (Fig. 6) and inhibited reverse transcriptase activity in the cell culture medium by 75% (Table 3). The control antisense oligonucleotide (5′-TGTGACATCAGGCTCAAATC-3′) neither inhibited cell growth nor affected reverse transcriptase activity. These results suggest that a cancer antigen(s) encoded by the gene related to HIV-1 gp41 is critical for the growth of breast and cervical cancer cells. Reverse transcriptase inhibition suggests that the identified DNA sequences may, in fact, belong to a retrovirus.
FIG. 6.
Effect of antisense oligonucleotide RAK-I on growth of breast cancer cell line MCF 7. Breast cancer MCF 7 cells were grown for 4 days in the absence (A) or presence (B) of antisense oligonucleotide RAK-I (5′-CCAGACTGTGAGTTGCAACAG-3′), which was added daily to concentrations of 100 μg/ml (day 1) and 50 μg/ml (days 2 and 3). The oligonucleotide (5′-TGTGACATCAGGCTCAAATC-3′) used in control experiments did not affect cell growth (data not shown).
TABLE 3.
Effects of antisense oligonucleotide RAK-I and a control oligonucleotide on reverse transcriptase activity
| Cell line | [3H]TTP incorporation (cpm)a (% inhibition)
|
||
|---|---|---|---|
| Controlb | RAK I | Control oligonucleotide | |
| MCF 7 | 10,000 | 2,550 (74.5) | 10,200 (0) |
| SKBr-3 | 8,300 | 2,100 (74.7) | 8,220 (0) |
Data are means from four experiments. The standard deviations were 5 to 15%.
No oligonucleotide.
DISCUSSION
RAK antigens p120, p42, and p25 are expressed in breast and gynecological cancer tissues in women (28, 29) and prostate cancer tissue in men (31) but are absent in normal tissues and in the majority of NATs. Although the nature of these unique proteins is not fully understood, the unusual association with cancer strongly suggests a potential role in the etiology of cancers of the reproductive system. Why cancers that differ in histological structure and originated from completely different tissues all express these unique proteins remains a medical and biological puzzle. If we assume that RAK antigens are encoded by a virus, then one of the possible mechanisms is hormonal regulation. One of the cancer antigens (RAK antigen p160) is expressed in the blood of the majority of breast, cervical, and ovarian cancer patients (29, 30). The molecular weight correlation of blood RAK antigen p160 with the precursor of HIV-1 envelope proteins gp120 and gp41 strongly supports a viral origin of this marker. Cancer-limited expression of RAK antigens suggests either that normal human genes are selectively transcribed in cancer or that a unique virus affects reproductive organs, leading to malignancy. Transcriptional regulation of human genes is very unlikely in light of the fact that HIV-1 gp41-derived primers PCR amplified breast (28) and prostate (31) cancer DNAs but not normal tissue DNA, including DNA extracted from NAT.
In a low-density agarose gel the electrophoretic migration of the breast cancer DNA fragment amplified was indistinguishable from that of the HIV-1 fragment amplified. High-resolution gel electrophoresis revealed one band (142 bp) common to HIV-1 and cancer DNAs and another PCR band greater in size (160 bp) that was present only in cancer DNA. In contrast to the smaller band, which exhibited over 90% homology to HIV-1 sequences encoding transmembrane protein gp41, the sequences of the larger band contained long A and T repeats and did not show homology to any known human gene. It is noteworthy that the larger band was never amplified in the absence of the smaller band (in normal tissue DNA). It is likely that further understanding of the structure of the putative virus and its sites of integration with the human genome will clarify the role of that 160-bp amplification product.
In 48% of breast cancer patients, HIV-1-like sequences were absent in histologically normal tissue of the cancer-affected breast, which eliminates the possibility of a random distribution of these sequences in the human genome and excludes the possibility that the identified sequences were of human origin. Whether the identified HIV-1 gp41-like sequences encode RAK antigen p42 cannot be established, but it seems very unlikely that HIV-1-like RAK antigens and HIV-1-like DNA sequences represent two unrelated phenomena, both exclusively associated with cancer.
Breast cancer RAK antigens p120, p42, and p25 exhibit molecular and immunological similarity to the proteins encoded by HIV-1. Moreover, RAK antigens express an amino acid region homologous to variable loop V3 of HIV-1 and cross-react with an epitope-specific anti-HIV-1 envelope protein gp120 MAb. MAb RAK-BrI, which was developed against RAK antigens, also cross-reacts with HIV-1 antigens gp160 and gp120, which confirms the nonaccidental similarity of cancer and HIV-1 proteins. Recent studies indicated that several antibodies developed against a nonglycosylated form of HIV-1 gp120 recognized RAK p120 in cancer cells, which implies that the homology of RAK antigen p120 to HIV-1 gp120 is expanded to various parts of the molecule but is probably limited to the primary structure of the protein (31a). Previous studies indicated that a MAb raised against HIV-1 gp41 is also able to recognize RAK p42 in breast and cervical cancer cell lines; however, limited studies were done with that MAb (27).
Although RAK antigens exhibit homology to HIV-1 antigens, these cancer markers can be easily distinguished from HIV-1 infection by using (i) any antibody directed against the glycosylated form of HIV-1 gp120, (ii) MAb 5025, or (iii) any other anti-HIV-1 MAb which does not cross-react with cancer antigens. Moreover, MAb RAK-BrI binds to RAK antigens p120, p42, and p25 in cancer tissue but only to gp160 and gp120 of HIV-1, which automatically eliminates the possibility of infection.
The similarity of breast cancer RAK antigens p120, p42, and p25 to HIV-1 major proteins, the fact that all three antigens are usually found together, and the exclusive cancer affiliation of these markers, further supported by the presence of HIV-1-like sequences in cancer DNA, strongly suggest that these antigens belong to a slow retrovirus, a fragment of which has been sequenced. Inhibition of cancer cell growth, in parallel with inhibition of reverse transcriptase activity in the presence of antisense oligonucleotides complementary to the HIV-1-like sequences, supports the viral nature of RAK markers. The mechanism of cancer growth promotion by the putative virus remains to be further investigated. The growth factor-like character of the RAK antigens is suggested by the previously described cancer growth activation of the anti-HIV-1 gp120 MAb (27).
A viral etiology of human breast cancer has been considered by several investigators; however, it has never been confirmed (1, 5, 9, 11–13, 18, 38–40). The assumption that a breast cancer virus would exhibit homology to MMTV was probably misleading, since the human genome contains numerous copies of genes with sequence homology to MMTV (1, 38, 40). MMTV sequences were found in both healthy persons and cancer patients, and proteins homologous to those of MMTV were found in both groups of women as well (1, 12, 38, 40). Our studies suggest that the breast cancer virus exhibits homology to HIV-1 rather than to MMTV.
Electron microscopic studies revealed some virus-like particles that were immunogold labeled with an anti-HIV-1 MAb. Labeling of breast cancer cells with an anti-HIV-1 gp120 MAb was also observed when the immunofluorescence technique was used (27). Electron microscopic analysis suggests that the virus buds into the intracytoplasmic vacuoles, which are then secreted to the cell surface, fuse with the membrane, and release viral particles through exocytosis. Exocytosis of the virus-containing vesicles might be responsible for the formation of the characteristic “peninsula-like” surface of the tested cancer cells, with very long villus-like structures. Alternatively, intact vesicles might be released by cells and the viral particles would thus be budding from the surface. Viral particles were observed in breast cancer cells by several researchers. It is noteworthy that the tail-and-head structure of the viral particles, with a strong morphological resemblance to MMTV, which was reported before by Moore et al. (18) in breast milk studies, was also frequently observed in our study (Fig. 5E to G). However, the particles detected by us in breast cancer cells were labeled with an anti-HIV-1 gp120 MAb, suggesting homology to HIV-1 rather than to MMTV. The possibility of cross-reactivity between the anti-HIV-1 gp120 MAb and MMTV was excluded, since MMTV antigens were not recognized by this MAb either in a Western blot or in microscopic studies. The possibility that HIV-1-derived primers SK68 and SK69 amplified MMTV sequences in the human genome may be also excluded, since (i) these primers did not amplify MMTV sequences in mouse cells and (ii) cancer DNA sequences amplified with HIV-1-derived primers exhibited no homology to MMTV. Further studies are needed to verify whether the identified RAK markers are expressed by the detected viral particles.
The presence of HIV in cell cultures or in fresh cancer tissue may be eliminated since they tested negative for HIV-1 p24. The fact that of the over 1,000 cancer patients tested in this and other studies (28, 29) for the presence of RAK antigens in breast and gynecological tissue, 95% were RAK positive automatically excludes the HIV-1 origin of RAK antigens. Further studies are needed to fully characterize and classify the virus.
Independently of the viral or human origin of the novel cancer antigens, the specific association of RAK antigens with gynecological and breast cancer in women and prostate cancer in men and the lack of these markers in normal tissues strongly suggest that RAK markers have a critical value in the early diagnosis of cancers affecting reproductive organs. The diagnostic value of protein and PCR RAK markers was evaluated before (28, 31). It is suggested that protein RAK markers might improve the early detection of malignant or premalignant changes and would help in more effective evaluation of cancer margins, leading to a reduction in the number of unneeded mastectomies. PCR markers could be used to determine the predisposition of breast tissue to become malignant. Current studies on the identification of HIV-1-like sequences will definitely help to clone the virus and understand the etiology of reproductive tract cancers. If the structure of the new cancer virus were understood, then completely new approaches to cancer diagnosis, prevention, prognosis, and therapy could be developed. Cancer RAK antigen p120 and/or other RAK antigens would definitely play a critical role in the production of a breast cancer vaccine.
ACKNOWLEDGMENTS
This study was sponsored by the Leland J. and Dorothy H. Olson Foundation for Women’s Health.
We thank Martin Cane for immunogold labeling of cancer cells, Rick Vaughn for taking photographs of viral particles, and William Snyder for general technical support. We also thank Advanced Biotechnology Inc. for testing cells for contamination with known viruses, bacteria, or mycoplasmas.
REFERENCES
- 1.Andersson M L, Medstrand P, Yin H, Blomberg J. Differential expression of human endogenous retroviral sequences similar to mouse mammary tumor virus in normal peripheral blood mononuclear cells. AIDS Res Hum Retroviruses. 1996;12:833–840. doi: 10.1089/aid.1996.12.833. [DOI] [PubMed] [Google Scholar]
- 2.Biesecker B B, Boehnke M, Calzone K, Markel D S, Garber J E, Collins F S, Weber B L. Genetic counseling for families with inherited susceptibility to breast and ovarian cancer. JAMA. 1993;269:1970–1974. [PubMed] [Google Scholar]
- 3.Bouton A H, Parsons J T. Retroviruses and cancer: models for cancer in animals and humans. Cancer Invest. 1993;11:70–79. doi: 10.3109/07357909309020264. [DOI] [PubMed] [Google Scholar]
- 4.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpes virus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 5.Chopra H C, Feller W F. Virus-like particles in human breast cancer. Tex Rep Biol Med. 1969;27:945–954. [PubMed] [Google Scholar]
- 6.Claus E B, Risch N, Thompson W D. Genetic analysis of breast cancer in the cancer and steroid hormone study. Am J Hum Genet. 1991;48:232–242. [PMC free article] [PubMed] [Google Scholar]
- 7.Coles C, Condie A, Chetty U, Steel C M, Evans H J, Prosser J. p53 mutations in breast cancer. Cancer Res. 1992;52:5291–5298. [PubMed] [Google Scholar]
- 8.Durda P J, Bacheler L, Clapham P, Jenoski A M, Leece B, Matthews T J, McKnight A, Pomerantz R, Rayner M, Weinhold K J. HIV-1 neutralizing monoclonal antibodies induced by a synthetic peptide. AIDS Res Hum Retroviruses. 1988;6:1115–1118. doi: 10.1089/aid.1990.6.1115. [DOI] [PubMed] [Google Scholar]
- 9.Feller W F, Chopra H C. A small virus-like particle observed in human breast cancer by means of electron microscopy. J Natl Cancer Inst. 1968;40:1359–1373. [PubMed] [Google Scholar]
- 10.Garfinkel L, Boring C C, Heath C W., Jr Changing trends. An overview of breast cancer incidence and mortality. Cancer. 1994;74:222–227. doi: 10.1002/cncr.2820741304. [DOI] [PubMed] [Google Scholar]
- 11.Kahl L P, Carrol A R, Rhodes P, Wood J, Read N G. An evaluation of the putative human mammary tumor retrovirus associated with peripheral blood monocytes. Br J Cancer. 1991;63:534–540. doi: 10.1038/bjc.1991.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keydar I, Chou C S, Hareuveni M, Tsarfaty I, Sahar E, Selzer G, Chaichik S, Hizi A. Production and characterization of monoclonal antibodies identifying breast tumor-associated antigens. Proc Natl Acad Sci USA. 1989;86:1362–1366. doi: 10.1073/pnas.86.4.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keydar I, Ohno T, Nayak R, Sweet R, Simoni F, Weiss F, Karby S, Mesa-Tejada R, Spiegelman S. Properties of retrovirus-like particles produced by a human breast carcinoma cell line: immunological relationship with mouse mammary tumor virus proteins. Proc Natl Acad Sci USA. 1984;81:4188–4192. doi: 10.1073/pnas.81.13.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King M C, Rowell S, Love S M. Inherited breast and ovarian cancer. JAMA. 1993;269:1975–1980. [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1971;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Langedijk J P M, Back N K T, Durda P J, Goudsmit J, Meloen R H. Neutralizing activity of anti-peptide antibodies against the principal neutralization domain of human immunodeficiency virus type 1. J Gen Virol. 1991;72:2519–2526. doi: 10.1099/0022-1317-72-10-2519. [DOI] [PubMed] [Google Scholar]
- 17.Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, Tavtigian S, Liu Q, Cochrane C, Bennett L M, Ding W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 18.Moore D H, Charney J, Kramarsky B, Lasfargues E Y, Sarkar N H. Search for a human breast cancer virus. Nature. 1971;229:611–615. doi: 10.1038/229611a0. [DOI] [PubMed] [Google Scholar]
- 19.Newcomb P A, Lantz P M. Recent trends in breast cancer incidence, mortality, and mammography. Breast Cancer Res Treat. 1993;28:97–106. doi: 10.1007/BF00666422. [DOI] [PubMed] [Google Scholar]
- 20.Newman B, Austin M A, Lee M, King M C. Inheritance of human breast cancer: for autosomal dominant transmission in high-risk families. Proc Natl Acad Sci USA. 1988;85:3044–3048. doi: 10.1073/pnas.85.9.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowak R. Breast cancer gene offers surprises. Science. 1994;265:1796–1799. doi: 10.1126/science.8091205. [DOI] [PubMed] [Google Scholar]
- 22.O’Leary J J, Kennedy M M, McGee J. Mol. Pathol. 1997;50:4–8. doi: 10.1136/mp.50.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peles E, Bacus S S, Koski R A, Lu H S, Wen D, Ogden S G, Levy R B, Yarden Y. Isolation of the Neu/HER-2 stimulatory ligand: a 44 kd glycoprotein that induces differentiation of mammary tumor cells. Cell. 1992;69:205–216. doi: 10.1016/0092-8674(92)90131-u. [DOI] [PubMed] [Google Scholar]
- 24.Rakowicz-Szulczynska E, Raso V, Kaczmarski W, Steimer K S, Durda P J. Internalization of anti-gp120 monoclonal antibody and human antibodies by HIV-1-infected T lymphocytes. Antib Immunoconjug Radiopharm. 1993;6:209–219. [Google Scholar]
- 25.Rakowicz-Szulczynska E M, Kaczmarski W, Steimer K S, Durda P J. Internalized antibodies as a potential tool against retroviral disease. In: Rakowicz-Szulczynska E M, editor. Nuclear localization of growth factors and of monoclonal antibodies. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 180–197. [Google Scholar]
- 26.Rakowicz-Szulczynska E M, McIntosh D G, Smith M L. Novel family of gynecological cancer antigens detected by anti-HIV antibody. Infect Dis Obstet Gynecol. 1994;2:171–178. doi: 10.1155/S1064744994000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakowicz-Szulczynska E M, McIntosh D G, Smith M L. Mechanisms of cancer growth promotion by HIV-I neutralizing antibodies. Cancer J. 1995;8:143–149. [Google Scholar]
- 28.Rakowicz-Szulczynska E M, Roszak A, Mackiewicz A, Markowska J, Karczewska A, Snyder W, Smith M L. New protein and PCR markers RAK for diagnosis, prognosis and surgery guidance for breast cancer. Cancer Lett. 1997;112:93–101. doi: 10.1016/S0304-3835(96)04550-8. [DOI] [PubMed] [Google Scholar]
- 29.Rakowicz-Szulczynska E M, Roszak A, Mackiewicz A, Markowska J, McIntosh D G, Karczewska A, Snyder W, Leary D, Smith M L. Diagnostic evaluation of cancer antigens RAK. I. Cervical and ovarian cancer. Int J Oncol. 1996;9:693–699. doi: 10.3892/ijo.9.4.693. [DOI] [PubMed] [Google Scholar]
- 30.Rakowicz-Szulczynska E M, McIntosh D G, Perry M J, Smith M L. Antigen RAK: a new breast cancer diagnostic marker. J Tumor Marker Oncol. 1995;10:25–37. [Google Scholar]
- 31.Rakowicz-Szulczynska E M, Jackson B, Snyder W. Prostate, breast, and gynecological cancer markers RAK with homology to HIV-1. Cancer Lett. 1998;124:213–223. doi: 10.1016/s0304-3835(97)00483-7. [DOI] [PubMed] [Google Scholar]
- 31a.Rakowicz-Szulczynska, E. M. Unpublished data.
- 32.Remington P L, Lantz P M. Using a population-based cancer reporting system to evaluate a breast cancer detection and awareness program. Cancer. 1992;42:367–371. doi: 10.3322/canjclin.42.6.367. [DOI] [PubMed] [Google Scholar]
- 33.Schalling M, Elkman M, Kaaya E E, Linde A, Biberfeld P. A role for a new herpes virus (KSHV) in different forms of Kaposis’s sarcoma. Nat Med. 1995;1:705–706. doi: 10.1038/nm0795-707. [DOI] [PubMed] [Google Scholar]
- 34.Scolnick E M, Aaronson S T, Todaro G J, Parks W P. RNA dependent polymerase activity in mammalian cells. Nature. 1971;229:318–321. doi: 10.1038/229318a0. [DOI] [PubMed] [Google Scholar]
- 35.Senie R T, Lesser M, Kinne D W, Rosen P P. Method of tumor detection influences disease-free survival of women with breast carcinoma. Cancer. 1994;73:1666–1672. doi: 10.1002/1097-0142(19940315)73:6<1666::aid-cncr2820730619>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 36.Silverberg E, Boring C C, Squires T S. Cancer statistics. Cancer. 1990;40:9–15. [PubMed] [Google Scholar]
- 37.Slamon D J, Godolphin W, Jones L A, Holt J A, Wong S G, Keith D E, Levin W J, Stuart S G, Udove J, Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;248:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 38.Sorhaug H, Grinde B. Evolution of mouse mammary tumor virus-related sequences in the human genome. Virus Res. 1993;30:53–61. doi: 10.1016/0168-1702(93)90015-f. [DOI] [PubMed] [Google Scholar]
- 39.Soule H R, Linder E, Edgington T S. Membrane 126-kilodalton phosphoglycoprotein associated with human carcinoma identified by a hybridoma antibody to mammary carcinoma cells. Proc Natl Acad Sci USA. 1983;80:1332–1336. doi: 10.1073/pnas.80.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szakacs J G, Moscinski L C. Sequence homology to deoxyribonucleic acid to mouse mammary tumor virus genome in human breast tumors. Ann Clin Lab Sci. 1991;21:402–412. [PubMed] [Google Scholar]
- 41.Tonin P, Serova O, Simard J, Lenoir G, Faunterin J, Morgan K, Lynch H, Narod S. The gene for hereditary breast-ovarian cancer, BRCA1, maps distal to EDH17B2 in chromosome region. Hum Mol Genet. 1994;3:1679–1682. doi: 10.1093/hmg/3.9.1679. [DOI] [PubMed] [Google Scholar]
- 42.Wooster R, Neuhausen S, Mangion J, Quirk Y, Ford D, Collins D, Nguyen K, Seal S, Tran T, Averill D. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 43.Zuppan P J, Hall J M, Lee M K, Ponglikitmongkol M, King M C. Possible linkage of the estrogen receptor genes to breast cancer in a family with late-onset disease. Am J Hum Genet. 1991;48:1065–1068. [PMC free article] [PubMed] [Google Scholar]