Abstract
In HNSCC, few studies have focused on the relationship between wild-type TP53 and mutant TP53-related immunity and prognosis. Our objective was to explore how TP53 mutation regulates the immunophenotype of HNSCC and thus affects the prognosis of HNSCC. Cox and Lasso regression were used to establish a prognostic model of TP53-related immune genes, on which basis a nomogram was used to establish a clinical prediction model, and ROC curves were further used to evaluate the effectiveness of the model. The risk of death in the TP53WT group was only 0.68 times that in the TP53Mut group (HR = 0.68, CI: 0.5–0.91, P < 0.05). T cells, CD8 T cells, cytotoxic lymphocytes, B lineage, NK cells, myeloid dendritic cells, and fibroblasts were significantly different between the TP53Mut and TP53WT groups (all P < 0.05). Time - dependent ROC curves of nomogram were plotted for 1-, 3-, and 5-year survival to further verify the predictive power of the nomogram for prognosis, and the AUCs were 0.78, 0.82, and 0.83, respectively. We showed there are significant differences in the immune microenvironment associated with wild-type TP53 and mutant TP53. The immune model associated with TP53 mutation has a good prediction ability for the prognosis of HNSCC and may be of reference value for other tumors with high mutation rate of TP53. Notably, the effect of TP53 mutation on the prognosis of HNSCC could be illustrated from an immunologic perspective.
Keywords: Head and neck squamous cell carcinoma, TP53 gene, Immune infiltration, Prognosis
Highlights
-
•
The differences in the immune microenvironment associated with wild-type TP53 and mutant TP53, affect the patient's sensitivity to immunotherapy.
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) is a common malignancy that accounts for 5.7% of cancer-related deaths worldwide [1]. HNSCC originates from the mucosal surface of the upper respiratory tract and mainly includes oral cancer, hypopharyngeal cancer and laryngeal cancer. The related risk factors include smoking, drinking alcohol, and HPV infection [2]. The therapeutic scheme of HNSCC is mainly a surgical combined therapy (radiotherapy, chemotherapy and biological treatment) based on TNM staging. However, the operation does cause irreversible loss of function, such as dysphagia, inability to pronounce, and even disfigurement, seriously affecting the quality of life of the patients [3]. Checkpoint inhibitors have been an important modality for the treatment of advanced or recurrent HNSCC [4]. Tumor cells can cause immune escape through the infiltration of T cells with high expression of PD-L1 or the recruitment of PD-1, and immune checkpoint inhibitors can block the process and reactivate the antitumor activity of T cells, thereby improving the local control rate and patient survival rate [5]. However, only a few patients respond to anti-PD-1/PD-L1 and other immunotherapies [6]. Although the exact mechanistic basis remains unclear, this may be attributed to factors in the tumor microenvironment, such as the lack of suitable rejection antigens, immune surveillance defects, or the presence of immunosuppressive mediators [7]. In addition, immune cell infiltration is considered a possible prognostic parameter for malignant tumors [8]. Finding a suited prognosis or predictive indicators is the key to the development of individualized immunotherapy for HNSCC.
TP53 is the most common somatically mutated gene in HNSCC [9], with a mutation rate of 65–85% [10]. Wild-type TP53 is a tumor suppressor gene that plays a key role in many cell pathways and regulates basic activities, such as proliferation, differentiation, cell death, DNA repair and angiogenesis [11,12]. Mutant TP53 is closely related to the occurrence, development and prognosis of tumor development, progression, and prognosis [13,14]. Studies have shown that TP53-mutant HNSCC patients have a worse overall survival prognosis than TP53 wild-type HNSCC patients [15]. TP53, as a transcription factor, mediates its tumor inhibition function to activate multiple target genes [16]. Mutation and deletion of the TP53 gene occurs frequently in cancer cells, leading to disorder of the intracellular signaling pathway, uncontrolled cell growth and escape from apoptosis [17]. In addition, some studies have proven that TP53 is also associated with tumor immune regulation. Yoon et al. determined that TP53 operates in a signaling pathway that protects against a systemic, life-threatening autoimmune disease [18]. Sonja et al. found that TP53 was involved in the regulation of specific NKG2D ligands and enhanced NK cell-mediated targeted recognition [19]. Both immunity and TP53 play vital roles in HNSCC. Few studies have explored the differences in related immune infiltration between wild-type TP53 and mutant TP53 in HNSCC. Therefore, our study aimed to explore how TP53 mutation regulates the immunophenotype of HNSCC and thus affects the prognosis of HNSCC.
2. Materials and methods
2.1. Data source
The unified and standardized TCGA-HNSCC gene expression data set was downloaded from the UCSC (https://xenabrowser.net/) database. The v33 version of gff3 has been downloaded from the gencode file at the website (http://ftp. ebi. ac. uk/pub/databases/gencode/gencode _ human/release_33/Gencode.v33.annotation.gff3.gz), and ENSG_ID is mapped onto GeneSymbol. Finally, the normalized gene expression profile matrix of 546 HNSCC samples was obtained. The corresponding clinical datasheets for 524 HCC samples were obtained from The Cancer Genome Atlas (TCGA) website (https://portal.gdc.cancer.gov/repository). Supplementary Fig. 1 showed the workflow diagram of this study.
2.2. Gene mutation analysis
The Simple Nucleotide Variation data set of TCGA samples processed by MuTect2 software from GDC (https://portal.gdc.cancer.gov/) was downloaded, and the mutation data of samples were integrated. The protein domain information was obtained from the R software package ‘maftools’ (version 2.2.10). According to the TP53 expression level, the samples were divided into HighExp group and LowExp group with the median as the cutoff value. We assessed the difference in gene mutation frequency within each group using the chi-square test, and the difference test results for all genes were visualized using the maftools package. Samples with TP53 somatic mutations, regardless of mutation type, were defined as the TP53 mutation group (TP53Mut group); Patients without any TP53 mutations were defined as the TP53 wild group (TP53WT group).
2.3. Gene set enrichment (GSEA)
GSEA software (version 3.0) is available from the GSEA (http://software.broadinstitute.org/GSEA/index.jsp) website. Samples were divided into two groups (TP53Mut group, TP53WT group) based on TP53 mutation or not and analyzed from the Molecular Signals Database (http://www.gsea-msigdb.org/gsea/downloads.jsp) download a “C7. immunesib. v7.4. SYMBOL. The GMT” subset was used to evaluate relevant pathways and molecular mechanisms, setting the minimum gene set to 5, the maximum gene set to 5000, one thousand subsamplings, a P value of <0.05 and an FDR of <0.25, which were considered statistically significant.
2.4. Analysis of differentially expressed genes (DEGs)
The “limma” package and “edgeR” package (version 3.14.3) in R were loaded. The gene expression data were converted into log2 values using the “calcNormFactors function”. The t.test function was used to evaluate the significant difference in the TP53 gene in the mutation group and the control group. The FDR value was calculated using the “p.adjust” function. Finally, the difference information of each gene was obtained, and the result was visualized. The significance threshold was set to P < 0.01, 1.5-fold difference.
2.5. Annotation of gene enrichment functions with GO and KEGG
We used genes from the R software package “org.Hs.e.g.db” (version 3.1.0) for GO annotation and the “KEGG rest API” (https://www.kegg.jp/kegg/rest/keggapi.html) for the latest pathway gene annotation. Taking the annotation as the background, the genes are mapped into the background set. Enrichment analysis was performed using the R software package “clusterProfiler” (version 3.14.3) to obtain results. The minimum gene was set to 5, the maximum gene was set to 5000, and a p value of <0.05 and an FDR-value of <0.25 were considered statistically significant.
2.6. Tumor immune microenvironment associated with TP53 in HNSCC
Immune infiltration score: The immune infiltration score was calculated by using “ESTIMATE”, an R software package. “ESTIMATE” (Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data) is a tool for predicting tumor purity, and the presence of infiltrating stromal/immune cells in tumor tissues using gene expression data. “ESTIMATE”is based on public data websites, such as Agilent, Affymetirx, Illumina RNASQ platform, and two signatures are screened out from transcripts, one is “Stromal signature”, and the other is “Immune signature”. The two signatures have 141 genes respectively. Then, “Stromal score”, “Immune score” and “ESTIMATE score” were calculated by gene set enrichment analysis of a single sample [20]. The Pearson correlation coefficient of genes in each tumor and the immune infiltration score were further calculated by using the corr.test function of the “psych” package (version 2.1.6).
Analysis of immune infiltrating cells: The “CIBERSORT” of the R software package was used to re-evaluate the infiltration scores of fibroblast cells, T cells, CD8 T cells, endothelial cells, neutrophils, myeloid dendritic cells, B lineage, cytotoxic lymphocytes and monocyte lineage of HNSCC patients [21]. “CIBERSORT” is a commonly used immuno infiltration analysis method. Based on the known data set, this method analyzes the types and distribution of various immune cells in the sample through the differential expression of marker genes in different immune cells. CIBERSORT converts RNAseq data of TCGA into MicroArray data set, and then uses deconvolution method to calculate immune infiltration [22]. We calculated the Pearson's correlation coefficient for the TP53 gene and immune cell infiltration scores using the R software package “psych” (version 2.1.6).
2.7. Nomogram
Combining the data of survival time, survival status, sex, age, TNM stage, tumor stage and Riskscore and taking survival status as the outcome index, the prognostic significance of these characteristics was evaluated by the Cox method. Loading the R package “rms” established a nomogram for clinical prognosis prediction.
2.8. Statistical methods
For the analysis of differences in clinical information, gene expression, and immune scores between the mutant group and the wild-type group, the chi-square test or unpaired Wilcoxon rank sum test was selected according to the data characteristics. The survival time, survival state and gene expression data were integrated, the prognostic significance of differentially expressed genes between the wild-type group and the mutation group was assessed using the Cox regression, and the riskscore of each sample was obtained. In previous work, Lasso regression has obvious advantages in dealing with genes with a large amount of data. Firstly, Ranstam et al. [23] uses Lasso regression to obtain a more refined model by constructing a penalty function, so that the sum of absolute values of mandatory coefficients is less than a certain fixed value; meanwhile, some regression coefficients are set to zero. Li et al. [24] Retained the advantages of subset shrinkage while better handling biased estimates with complex collinear data, Lasso regression has obvious advantages in dealing with genes with a large amount of data. For genes significantly affecting prognosis, regression analysis was performed using the lasso method, and 10-fold cross-validation was performed to obtain the optimal model. The riskscore was divided into a high group (H group) and a low group (L group) by the optimal cutoff value. The difference in survival state between the two groups was analyzed using “survfit” in the R software package. The ROC curve was used to evaluate the ability of variables to predict prognosis. The calibration curve and AUC were used to evaluate the accuracy of the nomogram predictions. All statistical tests were two-tailed with a statistical significance level set at 0.05 in this study.
3. Results
3.1. Gene mutation landscape in HNSCC and GSEA
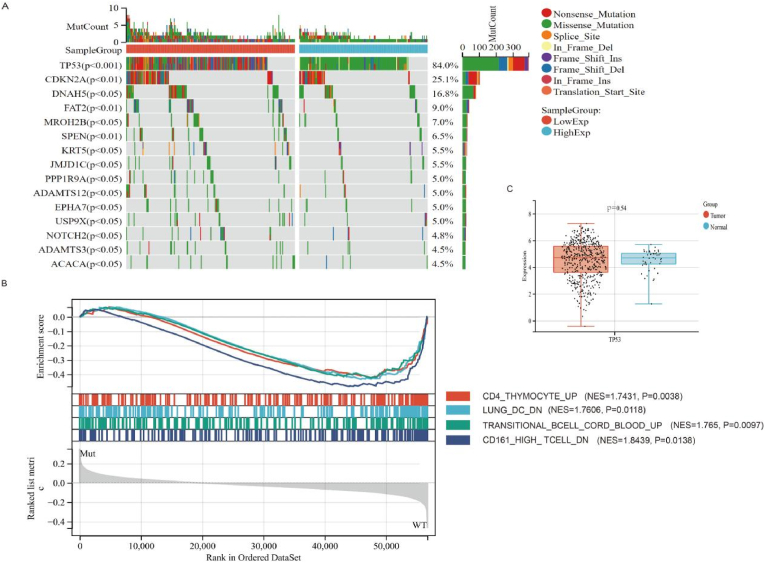
A total of 508 samples with mutations were detected, of which 481 (94.7%) were drawn samples. The first 20 mutant genes were TP53, TTN, FAT1, CDKN2A, MUC16, CSMD3, NOTCH1, PIK3CA, SYNE1, LRP1B, KMT2D, PCLO, DNAH5, FLG, USH2A, NSD1, RYR2, CASP8, PKHD1L1, and SI. The highest mutated gene in HNSCC was TP53, with a mutation rate of 84%. Mutation types included missense mutation, frameshift del, nonsense mutation, frameshift ins, splice site, in frame ins, in frame del, translation start site and nonstop mutation (Fig. 1A). The Gene set enrichment analysis (GSEA) was employed to explore the difference of biologic progress between TP53WT and TP53Mut. The result showed that the TP53WT group was significantly enriched in 1000 biological processes (Supplementary Table 1),4 immune-related biological processes were selected: CD4_THYMOCYTE_UP (NES = 1.7431, P = 0.0038), LUNG_DC_DN (NES = 1.7606, P = 0.0118), TRANSITIONAL_BCELL_CORD_BLOOD_UP (NES = 1.765, P = 0.0097), and CD161_HIGH_ TCELL_DN (NES = 1.8439, P = 0.0138) (Fig. 1B). In contrast, these immune-related biological processes were not enriched in TP53 WT group HNSCC patients, suggesting TP53Mut is closely associate with immune process. The expression levels of TP53 in HNSCC and normal tissues were compared, and there was no significant difference between the two groups (P = 0.54) (Fig. 1C). Moreover, the survival status analysis showed that the risk of death in the TP53WT group was only 0.68 times that in the TP53Mut group (HR = 0.68, CI: 0.5–0.91, P < 0.05) (Supplementary Fig. 1A). These results suggested that TP53 mutation may be a prognostic factor in HNSCC, not expression level (high or low) of TP53.
Fig. 1.
Gene mutation in HNSC.
3.2. Clinical information of the TP53Mut group and TP53WT group
To investigate whether there is a difference of clinical characterization between TP53Mut and TP53WT. We matched the clinical characteristics of patients with biological sample information, and finally included 239 patients. Comparing the clinical information of the TP53Mut group and TP53WT group, the results showed that there was no significant difference in age, sex, T stage, lymph node metastasis, distant metastasis, cancer stage or cancer grade (P > 0.05) (Table 1). TP53 acts as a transcription factor to mediates its tumor inhibition function and biological progresses [16]. Thus, we make a hypothesis that TP53 mutation resulting in tumor inhibition function loss is an important reason of prognosis in HNSCC patients.
Table 1.
Clinical characteristics of the TP53Mut group and TP53WT group.
| Characteristics | Mut(N = 76) | WT(N = 163) | Total(N = 239) | P value |
|---|---|---|---|---|
| Age(Mean±SD) | 61.92 ± 12.00 | 61.21 ± 12.08 | 61.44 ± 12.03 | 0.83 |
| Sex | 0.16 | |||
| FEMALE | 25(10.46%) | 38(15.90%) | 63(26.36%) | |
| MALE | 51(21.34%) | 125(52.30%) | 176(73.64%) | |
| T | 0.18 | |||
| T1 | 7(2.93%) | 15(6.28%) | 22(9.21%) | |
| T2 | 18(7.53%) | 54(22.59%) | 72(30.13%) | |
| T3 | 24(10.04%) | 30(12.55%) | 54(22.59%) | |
| T4 | 24(10.04%) | 60(25.10%) | 84(35.15%) | |
| TX | 3(1.26%) | 4(1.67%) | 7(2.93%) | |
| N | 0.33 | |||
| N0 | 39(16.32%) | 69(28.87%) | 108(45.19%) | |
| N1 | 14(5.86%) | 29(12.13%) | 43(17.99%) | |
| N2 | 18(7.53%) | 57(23.85%) | 75(31.38%) | |
| N3 | 1(0.42%) | 4(1.67%) | 5(2.09%) | |
| NX | 4(1.67%) | 4(1.67%) | 8(3.35%) | |
| M | 0.59 | |||
| M0 | 73(30.54%) | 156(65.27%) | 229(95.82%) | |
| M1 | 0(0.0e+0%) | 2(0.84%) | 2(0.84%) | |
| MX | 3(1.26%) | 5(2.09%) | 8(3.35%) | |
| Stage | 0.86 | |||
| Stage I | 4(1.67%) | 6(2.51%) | 10(4.18%) | |
| Stage II | 15(6.28%) | 29(12.13%) | 44(18.41%) | |
| Stage III | 14(5.86%) | 27(11.30%) | 41(17.15%) | |
| Stage IV | 43(17.99%) | 101(42.26%) | 144(60.25%) | |
| Grade | 0.17 | |||
| G1 | 9(3.77%) | 19(7.95%) | 28(11.72%) | |
| G2 | 51(21.34%) | 85(35.56%) | 136(56.90%) | |
| G3 | 13(5.44%) | 49(20.50%) | 62(25.94%) | |
| G4 | 0(0.0e+0%) | 2(0.84%) | 2(0.84%) | |
| GX | 3(1.26%) | 8(3.35%) | 11(4.60%) |
3.3. Immune landscape in the TP53Mut and TP53 WT groups
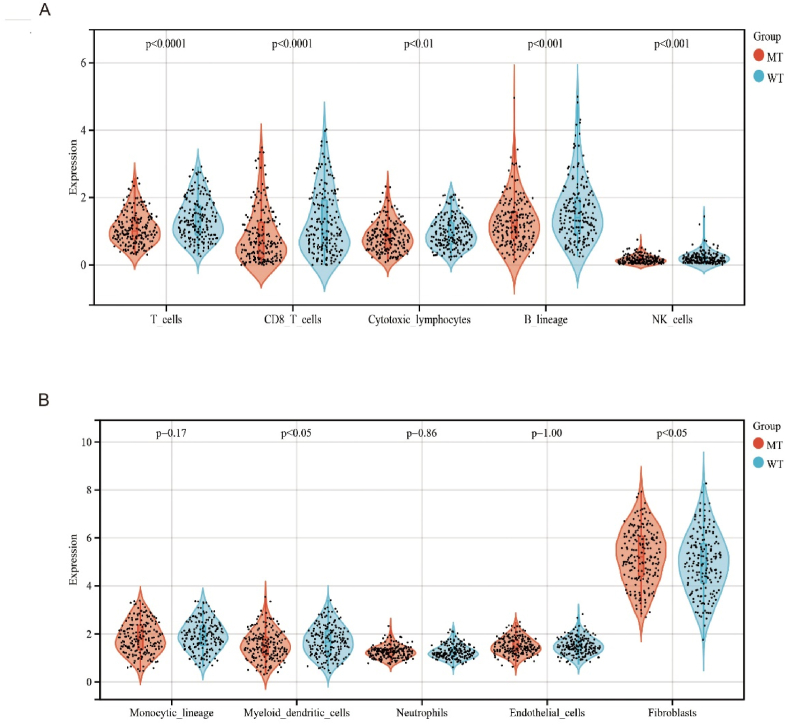
A previous study also demonstrate TP53Mut was closely related to immune-related biological processes [25], Thus,the differences in survival status and the immune microenvironment were further analyzed in the TP53Mut and TP53WT groups (Supplementary Fig. 2A). In HNSCC, TP53 gene expression, including mutant and wild type, was significantly correlated with immune cells and stromal cells (ImmuneScore, StromalScore, and ESTIMATEScore) (all P < 0.05) (Supplementary Fig. 2B)."CIBERSORT” is a tool for deconvolution of the expression matrix of human immune cell subtypes based on the linear support vector regression principle. For chip expression matrix and sequencing expression matrix, deconvolution analysis of expression matrix can be used to estimate the proportion of immune cells. We found that TP53 was significantly associated with fibroblast (r = 0.10, P = 0.02), CD8 T cell(r = 0.32,P < 0.01), T cell (r = 0.16,P < 0.01), neutrophils (r = 0.12,P < 0.01), NK cell(r = 0.25,<0.01), B cell(r = 0.19,P < 0.01), endothelial cell(r = 0.14,P < 0.01) (Supplementary Fig. 2C). Using the “CIBERSORT” method in combination with an immunocyte characteristic matrix, we also estimated 10 kinds of immunocyte infiltration scores between patients in the TP53Mut and TP53WT groups in HNSCC. Multi-immune cell types were significantly different between the TP53Mut and TP53WT groups (P < 0.05) (Fig. 2). It was worth noting that TP53Mut had a significantly lower abundance of T cells, CD8 T cells, cytotoxic lymphocytes, B lineage, NK cells, and myeloid dendritic cells as well as higher abundance of fibroblasts (Fig. 2A). The heterogeneity of immunocyte in HNSCC immune infiltration indicated adaptive immune may be served potential prognostic biomarkers, immunotherapy targets, and might exhibit important clinical significance. There were no significant differences among Monocytic lineage, Neutrophils, and Endothelial cells (Fig. 2B) (Supplementary Table2). Above data also demonstrated that TP53Mut might result in downregulation of adaptive immunity to influence HSNCC patient prognosis.
Fig. 2.
Immune landscape in the TP53Mut and TP53WT groups.
3.4. Limma, Cox and Lasso regression analyses of DEGs in HNSCC patients
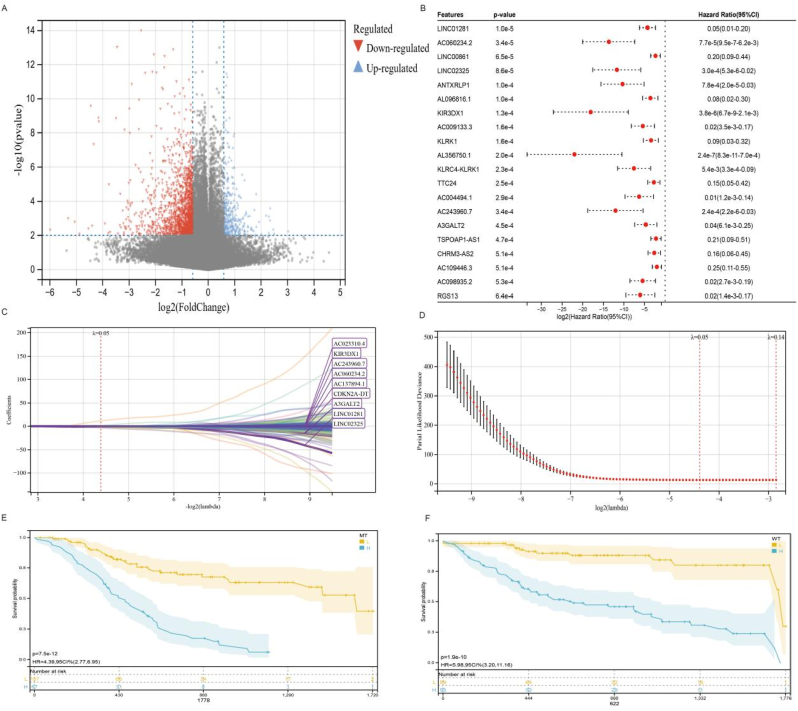
The TP53Mut group and TP53WT group were subjected to limma analysis, and 325 upregulated genes and 1560 downregulated genes were obtained (threshold: P < 0.01, 1.5-fold difference) (Fig. 3A). The differential genes obtained with different threshold settings are shown inSupplementary Table3 and 413 genes were screened out (P < 0.05) (Fig. 3B). We screened out significant differential genes, and then carried out COX regression analysis on each differential gene to judge the influence of each gene on the prognosis. HR < 1 mean that the high expression of these genes reduces the hazard ratio. The differentially expressed genes (n = 178) with significance less than 0.001 were further regressed by lasso, and the λ value was set to 0.048. Finally, 34genes(LINC01281, AC060234.2, LINC02325, KIR3DX1, CDKN2A-DT,AC137894.1, AC243960.7, AC023310.4, A3GALT2, AC013726.2, AL590822.1, NBEAP1, RAP2CP1, PROSER2-AS1, AC092580.1, TRAJ4, MYCNOS, RPL7P51, SHANK2, AP003041.1, RNU6-173P, AC024884.1, LRRC9, AC020978.6, AC005291.1, AC133485.4, AC243829.2, AL139413.1, RN7SKP64, LINC01597, AC136618.2, KLHL6-AS1, SCGB3A1, AL079307.2) and riskscores of 227 patients were obtained(Fig. 3C,D). The model formula of Lasso regression as below:
Fig. 3.
Limma, Cox regression and Lasso regression analyses.
RiskScore = - 0.49 * LINC01281–1.27 * AC060234.2–0.82 * LINC02325–0.86 * KIR3DX1 -0.32 * CDKN2A - DT - 0.12 * AC137894.1–0.39 * AC243960.7 + 0.13 * AC023310.4–0.31 * A3GALT2 - 3.34 * AC013726.2–0.12 * AL590822.1 + 0.02 * NBEAP1 - 0.03 * RAP2CP1 - 0.1 * PROSER2 - AS1 - 0.38 * AC092580.1–0.08 * TRAJ4 - 0.13 * MYCNOS - 0.88 * RPL7P51 + 0.04 * SHANK2 - 0.24 * AP003041.1 + 0.19 * RNU6 - 173P - 0.001 * AC024884.1 + 0.54 * LRRC9 + 11.43 * AC020978.6 + 0.02 * AC005291.1–2 * AC133485.4–0.17 * AC243829.2 + 0.22 * AL139413.1–1.42 * RN7SKP64 + 0.34 * LINC01597 + 1.26 * AC136618.2 + 0.11 * KLHL6 - AS1 - 0.01 * SCGB3A1 - 0.43 * AL079307.2.
In the TP53Mut group, the risk of death was 4.39 times higher in the H group than in the L group (HR = 4.39, 95% CI: 2.77–6.95, P < 0.01). In the TP53Mut group, there was also a significant difference in prognosis between the H and L groups (HR = 5.98, 95% CI 3.2–11.6, P < 0.01) (Fig. 3E,F).
3.5. Gene expression difference and functional enrichment analysis in the H and L groups
We divided the riskscore into two groups by the optimal cutoff value: High group (H group) and Low group (L group). The H group and L group performed limma analysis and obtained 30 upregulated genes and 1587 downregulated genes (threshold: FDR<0.05, 1.5-fold difference) (Supplementary Fig. 3A). The differential genes obtained with different threshold settings are shown in Supplementary Table4. The functions in GO analysis included the following: biological process (BP): adaptive immune response, regulation of immune system response, positive regulation of immune response, immune response-regulating cell surface receptor signaling pathway, lymphocyte mediated immunity; cellular component (CC): intrinsic component of membrane; intrinsic component of plasma membrane; integral component of plasma membrane; cell surface; side of membrane; external side of plasma membrane; blood microparticle; plasma membrane receptor complex; T-cell receptor complex; molecular function (MP): signaling receptor activity, molecular transducer activity, antigen binding, signaling receptor binding, transmembrane signaling receptor activity, cytokine receptor activity, G protein-coupled peptide receptor activity; MHC protein binding, G protein-coupled chemoattractant receptor activity (Supplementary Fig. 3B). Pathways in KEGG included primary immunodeficiency, cytokine–cytokine receptor signaling interaction, T-cell receptor signaling pathway, cell adhesion molecules, natural killer cell mediated cytotoxicity, chemokine signaling pathway, Th17-cell differentiation, hematopoietic cell lineage, viral protein interaction with cytokine and cytokine receptor, and Th1 and Th2 cell differentiation (Supplementary Fig. 3C).
3.6. Survival analysis and gene expression in the high riskscore and low riskscore groups
The optimal cutoff value of the riskscore was −0.071, based on which the patients were divided into a high riskscore group (H group) and a low riskscore group (L group). The log-rank test was further used to assess the prognosis between the samples in different groups, and finally, a significant difference in prognosis was observed (P-value = 7.7e−30<0.001). The risk of death in the H group was 4.47 times higher than that in the L group (Supplementary Fig. 4A). Time-dependent ROC curves of riskscore for the first, third and fifth year were drawn, and AUC was 0.76(95%CI:0.7–0.81), 0.8(95%CI:0.75–0.85) and 0.81 (95%CI:0.75–0.87) respectively (Supplementary Fig. 4B). We analyzed the relationship between different riskscores and patients' follow-up time, outcome events and gene expression changes. It can be observed that with the increase in riskscores (Supplementary Fig. 4C upper picture), the survival rate of patients decreased significantly (Supplementary Fig. 4C middle picture), and the expression of most genes showed a downward trend with the increase in riskscores (Supplementary Fig. 4C lower picture).
3.7. Nomogram of riskscore combined with simple clinical information
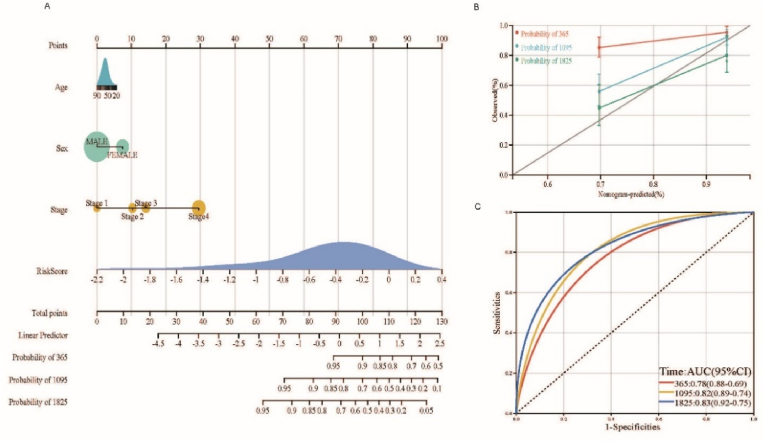
We established the formula for the prognostic riskscore model according to Lasso regression. Each patient was assigned a riskscore that was used as one of the univariates in the construction of the nomogram with other clinical information (sex, age, tumor stage) to assess the prognosis of patients with HNSCC. The overall C-index of the model was 0.77 (95% CI: 0.72–0.83), and P-value = 2.08e−23 < 0.01, indicating that the prediction ability of the model was excellent (Fig. 4A). The bias correction lines in the 1-,3- and 5-year calibration plots were close to the ideal curve (micro: 45 points), indicating good agreement between the prediction and observation (Fig. 4B). Time-dependent ROC curves were plotted for 1-, 3-, and 5-years to further verify the predictive power of the nomogram for prognosis, and the AUCs were 0.78 (95% CI: 0.69–0.88), 0.82 (95% CI: 0.74–0.89), and 0.83 (95% CI: 0.75–0.92), respectively (Fig. 4C).
Fig. 4.
Nomogram, calibration curves, and ROC curves.
4. Discussion
This study maps the mutational landscape of TP53 in HNSCC, and the results showed that the mutation rate of TP53 reached 84%. The mutation types included missense mutation, frameshift shift del, nonsense mutation, frame shift ins, splice site, in frame ins, in frame del, translation start site and nonstop mutation. However, compared with normal tissues, the expression of the TP53 gene was not significantly different from that in normal tissues. The TP53 gene is located on the broken arm of chromosome 17 (17p13.17) and is expressed in a variety of vertebrates [26]. The wild TP53 gene acts as a “monitor” during cell growth. When cellular DNA is damaged, TP53 can cause cell division to terminate in G1/S phase, inhibiting cell division so that there is enough time to repair the damage; for DNA that cannot be repaired, TP53 can initiate programmed death of cells to prevent abnormal cell proliferation [27]. Mutant TP53 not only loses its normal tumor suppressor function but also functions as an oncogene [28]. This is because the protein product of the mutant TP53 gene can combine with the wild TP53 protein to form a hetero-oligomeric complex, which interferes with the function of the TP53 protein [29]. GSEA showed that TP53-related immune pathways were mainly involved in the regulation of T cells, B cells and DCs, indicating that TP53 was closely related to immune infiltration. Many studies have shown that TP53 plays an important role in the recognition of tumors by the immune system, and TPP53 plays an active role in antitumor immune surveillance [18,30].
We analyzed the differentially expressed genes between the TP53WT and TP53Mut groups and then filtered the genes that significantly affected the prognosis by Cox regression. Then, we established the final prognostic model further by lasso regression to obtain the riskscore of each patient. We divided the patients into a high riskscore group and a low riskscore group according to the riskscore. Stratification analyses were performed to test whether the prognostic value of the model was independent of TP53 status. It was found that the prognostic model could classify high-risk and low-risk patients, regardless of whether TP53 was mutated. Based on this model, we added simple clinical information to construct the nomogram. The results showed that the predicted AUC values of the model for the first, third and fifth years were 0.78, 0.82 and 0.83, respectively. An AUC value greater than 7 indicated that the model had good prediction ability. The results showed that the model has a good effect in predicting the prognosis of patients. On this basis, we analyzed the functions of the differentially expressed genes in the high riskscore and low riskscore groups and found that the enrichment of these genes was mainly concentrated in immune-related pathways.
TP53 expression is correlated with immune cell infiltration [31,32]. In this study, the main immune cells of HNSCC include fibroblasts, CD8 T cells, T cells, neutrophils, NK cells, B cells, and endothelial cells. For the TP53 mutant group, the infiltration of T cells, CD8 T cells, cytotoxic lymphocytes, B lineage, NK cells, myeloid dendritic cells, and fibroblasts was significantly lower than that in the TP53 WT group. This difference suggested that the immune microenvironment after TP53 mutation was different from that of the wild type. The worse prognosis in the mutant group was probably due to the relatively low immune function. Lu et al. demonstrated that wild-type TP53 colorectal cancer exhibited enhanced T-cell recognition of cancer cells and a higher proportion of effector memory CD8+ T cells, and TP53 mutation might inhibit tumor immune cell infiltration [33]. Therefore, these results showed that the heterogeneity of immune infiltration in HNSCC may serve as a prognostic indicator and target for immunotherapy and may have significant clinical implications. Immune checkpoints are a class of immunosuppressive molecules [34], and tumor cells can evade immune surveillance and progression by activating immune checkpoints that suppress antitumor immune responses. Immune checkpoint inhibitors enhance antitumor immune responses by disrupting coinhibitory signaling and promote the elimination of immune-mediated tumor cells [35]. Some research has shown that TP53 interacts with immune responses by adjusting inflammatory cytokines, toll-like receptors, and IFN signaling [36,37]. Sonja et al. proposed that NK-cell-based immunotherapy might be an effective treatment strategy for wild-type TP53 cancer with preserved function [19]. The effect of immunotherapy in HNSCC varies greatly among individuals, and only a few patients benefit from it. Differences in the immune microenvironment of HNSCC patients with mutant TP53 and wild-type TP53 may cause differences in immune checkpoints and may also affect patient sensitivity to immunotherapy. Studies have reported that wild-type TP53 can enhance the role of cellular immunogenicity and indirectly enhance the body's antitumor ability [38]. Therefore, the combination of TP53 gene treatment and immunotherapy can be considered a method for the treatment of HNSCC. Our study has some limitations, firstly based on that analysis done by the TCGA public database without our own collect cohort as validation. Second, the best choice for studying cell heterogeneity is to use single-cell sequencing data. Thirdly, TP53 mutations are of various types, and different mutations may have different phenotypes. Therefore, TP53 mutations should be further grouped to better clarify the effects of TP53 mutations on the prognosis and immunity of HNSCC. In the future, there will be more information about HNSCC single-cell databases, and we will further study the heterogeneity of TP53-related immune cell types and differentiation.
5. Conclusions
The immune model associated with TP53 mutation has a good prediction ability for the prognosis of HNSCC and may be of reference value for other tumors with high mutation rate of TP53. Notably, the effect of TP53 mutation on the prognosis of HNSCC could be illustrated from an immunologic perspective.
Author contributions
WK: design of the research, collection of data, writing up of article; HY and HG: collection of data; YH: analysis of experiment data; YZ: design of the research, revising the article. All authors read and approved the final manuscript.
Funding information
None.
Ethical permissions
Not applicable.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank Daohong Meng, Professor of Public Health and Epidemiology in University of South Florida, for his suggestion on the study and discussion of the results.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101359.
Contributor Information
Weili Kong, Email: kong3320@wchscu.cn.
Yinze Han, Email: aiker_swu@163.com.
Hailing Gu, Email: hailing_gu@163.com.
Hui Yang, Email: yh8806@163.com.
Yi Zang, Email: zangyi_2022@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The data that has been used is confidential.
References
- 1.Patterson R.H., Fischman V.G., Wasserman I., et al. Global burden of head and neck cancer: economic consequences, health, and the role of surgery. Otolaryngology-Head Neck Surg. (Tokyo) 2020;162(3):296–303. doi: 10.1177/0194599819897265. [DOI] [PubMed] [Google Scholar]
- 2.Dhull A.K., Atri R., Dhankhar R., et al. Major risk factors in head and neck cancer: a retrospective analysis of 12-year experiences. World J. Oncol. 2018;9(3):80. doi: 10.14740/wjon1104w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martina, Pezdirec, Primoz, et al. Swallowing disorders after treatment for head and neck cancer. Radiol. Oncol. 2019;53(2):225–230. doi: 10.2478/raon-2019-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seiwert T.Y., Burtness B., Mehra R., et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicenter, phase 1b trial. Lancet Oncol. 2016:956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 5.Solomon B., Young R.J., Rischin D. Seminars in Cancer Biology. Elsevier; 2018. Head and neck squamous cell carcinoma: genomics and emerging biomarkers for immunomodulatory cancer treatments; pp. 228–240. 2018. [DOI] [PubMed] [Google Scholar]
- 6.Sim F., Leidner R., Bell R.B. Immunotherapy for head and neck cancer. Oral and Maxillofacial Surgery Clinics. 2019;31(1):85–100. doi: 10.1016/j.coms.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Poggio M., Hu T., Pai C.-C., et al. Suppression of exosomal PD-L1 induces systemic antitumor immunity and memory. Cell. 2019;177(2):414–427. doi: 10.1016/j.cell.2019.02.016. e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassetta L., Fragkogianni S., Sims A.H., et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. 2019;35(4):588–602. doi: 10.1016/j.ccell.2019.02.009. e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caponio V.C.A., Troiano G., Adipietro I., et al. Computational analysis of TP53 mutational landscape unveils key prognostic signatures and distinct pathobiological pathways in head and neck squamous cell cancer. Br. J. Cancer. 2020;123(8):1302–1314. doi: 10.1038/s41416-020-0984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindemann A., Patel A.A., Silver N.L., et al. COTI-2, a novel thiosemicarbazone derivative, exhibits antitumor activity in HNSCC through p53-dependent and-independent mechanisms. Clin. Cancer Res. 2019;25(18):5650–5662. doi: 10.1158/1078-0432.CCR-19-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vennin C., Mélénec P., Rouet R., et al. CAF hierarchy driven by pancreatic cancer cell p53-status creates a pro-metastatic and chemoresistant environment via perlecan. Nat. Commun. 2019;10(1):1–22. doi: 10.1038/s41467-019-10968-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aubrey B.J., Strasser A., Kelly G.L. Tumor-suppressor functions of the TP53 pathway. Cold Spring Harbor perspectives in medicine. 2016;6(5):a026062. doi: 10.1101/cshperspect.a026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soussi T., Wiman K. TP53: an oncogene in disguise. Cell Death Differ. 2015;22(8):1239–1249. doi: 10.1038/cdd.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher O.M., Lord S.J., Falkenback D., et al. The prognostic value of TP53 mutations in esophageal adenocarcinoma: a systematic review and meta-analysis. Gut. 2017;66(3):399–410. doi: 10.1136/gutjnl-2015-310888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poeta M.L., Manola J., Goldwasser M.A., et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2007;357(25):2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer M., Grossmann P., Padi M., et al. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016;44(13):6070–6086. doi: 10.1093/nar/gkw523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreeff M., Kelly K.R., Yee K., et al. Results of the phase I trial of RG7112, a small-molecule MDM2 antagonist in leukemia. Clin. Cancer Res. 2016;22(4):868–876. doi: 10.1158/1078-0432.CCR-15-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zitvogel L., Kroemer G. A p53-regulated immune checkpoint relevant to cancer. Science. 2015;349(6247):476–477. doi: 10.1126/science.aac8475. [DOI] [PubMed] [Google Scholar]
- 19.Textor S., Fiegler N., Arnold A., et al. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res. 2011;71(18):5998–6009. doi: 10.1158/0008-5472.CAN-10-3211. [DOI] [PubMed] [Google Scholar]
- 20.Chen B., Khodadoust M.S., Liu C.L., et al. Humana Press; New York, NY: 2018. Profiling Tumor Infiltrating Immune Cells with CIBERSORT[M]//Cancer Systems Biology; pp. 243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sui S., An X., Xu C., et al. An immune cell infiltration-based immune score model predicts prognosis and chemotherapy effects in breast cancer[J] Theranostics. 2020;10(26) doi: 10.7150/thno.49451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshihara K., Shahmoradgoli M., Martínez E., et al. Inferring tumour purity and stromal and immune cell admixture from expression data[J] Nat. Commun. 2013;4(1):1–11. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranstam J., Cook J.A. LASSO regression[J] Journal of British Surgery. 2018;105(10) 1348-1348. [Google Scholar]
- 24.Li W., Feng J., Jiang T. IsoLasso: a LASSO regression approach to RNA-Seq based transcriptome assembly[J] J. Comput. Biol. 2011;18(11):1693–1707. doi: 10.1089/cmb.2011.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X , Lv D , Cai C , et al. A TP53-associated immune prognostic signature for the prediction of overall survival and therapeutic responses in muscle-invasive bladder cancer[J]. Front. Immunol., 11:590618. [DOI] [PMC free article] [PubMed]
- 26.Yang S.X., Steinberg S.M., Nguyen D., et al. p53, HER2 and tumor cell apoptosis correlate with clinical outcome after neoadjuvant bevacizumab plus chemotherapy in breast cancer. Int. J. Oncol. 2011;38(5):1445–1452. doi: 10.3892/ijo.2011.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krell J., Stebbing J., Carissimi C., et al. TP53 regulates miRNA association with AGO2 to remodel the miRNA–mRNA interaction network. Genome Res. 2016;26(3):331–341. doi: 10.1101/gr.191759.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alam S.K., Yadav V.K., Bajaj S., et al. DNA damage-induced ephrin-B2 reverse signaling promotes chemoresistance and drives EMT in colorectal carcinoma harboring mutant p53. Cell Death Differ. 2016;23(4):707–722. doi: 10.1038/cdd.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nan K., Wang Y., Guo S., et al. Mutant TP53 G245C and R273H promote cellular malignancy in esophageal squamous cell carcinoma. BMC Cell Biol. 2018;19(1):16. doi: 10.1186/s12860-018-0167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao W., Nan D., Huang T., et al. TP53 mutation as potential negative predictor for response of anti-CTLA-4 therapy in metastatic melanoma. EBioMedicine. 2018;32 doi: 10.1016/j.ebiom.2018.05.019. S235239641830183X- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Junyu, Long Anqiang, et al. Ebiomedicine; 2019. Development and Validation of a TP53-Associated Immune Prognostic Model for Hepatocellular Carcinoma. [J] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J., Ma Q., Zhang M., et al. Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: evidence from a systematic review and meta-analysis[J] Eur. J. Cancer. 2012;48(15):2328–2338. doi: 10.1016/j.ejca.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Z., Shen L., Zhang H., et al. Effect of TP53 mutation on antitumor immunity and responsiveness to immunotherapy in colorectal cancer. ASCO. 2020:2020. [Google Scholar]
- 34.Abril-Rodriguez G., Ribas A. SnapShot: immune checkpoint inhibitors. Cancer Cell. 2017;31(6):848. doi: 10.1016/j.ccell.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 35.Darvin P., Toor S.M., Sasidharan Nair V., et al. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp. Mol. Med. 2018;50(12) doi: 10.1038/s12276-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daniel M., Maria S., Kathleen A., et al. The Toll-like receptor gene family is integrated into human DNA damage and p53 networks. PLoS Genet. 2011;7(3) doi: 10.1371/journal.pgen.1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takaoka A., Hayakawa S., Yanai H., et al. Integration of interferon-|[alpha]|/|[beta]| signaling to p53 responses in tumor suppression and antiviral defense. Nature. 2003;424(6948):516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 38.Moore E C, Sun L, Clavijo P E, et al. Nanocomplex-based TP53 gene therapy promotes anti-tumor immunity through TP53- and STING-dependent mechanisms. Vol. 7. Taylor & Francis; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.