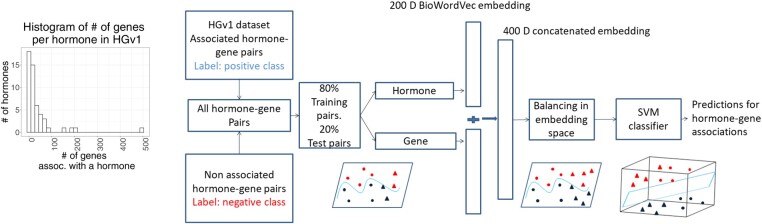
Fig. 1.
BioEmbedS model overview: our BioEmbedS model predicts if a hormone–gene pair is associated or not from D-dimensional word embedding vectors of the hormone name and the gene symbol. Our HGv1 dataset is crucial for systematic training/evaluation of our model, after its proper balancing to handle variability in available information for different hormones (see inset histogram; ‘assoc.’ stands for associated). In the toy-example shown, circles and triangles indicate hormone–gene pairs for two illustrative hormones; and below-the-boundary blue and above-the-boundary red symbols, respectively, denote the positive (associated) and negative (non-associated) genes for each hormone. The positive/negative classes are balanced across the two hormones, before separating them in a higher dimensional space using a SVM classifier. BioEmbedS-TS model has source and target genes for a hormone in place of positive and negative genes (A color version of this figure appears in the online version of this article.)