Significance
Unsaturated fatty acids are critical for maintaining membrane fluidity, cellular signaling, and lipid storage. Stearoyl-CoA desaturase (SCD) regulates the dynamics between saturated and unsaturated fatty acids. How SCD is involved in cancer cell fate decisions remains incompletely understood. Here, we leveraged transcriptomics, lipidomics, and single-cell stimulated Raman scattering microscopy to show that increased levels of lipid unsaturation promoted by SCD protected cancer cells against endoplasmic reticulum stress-induced apoptosis. Reduced levels of unsaturated fat caused either by SCD depletion or by SCD inhibition coupled with a diet rich in saturated fat blocked cancer cell growth in vitro and tumor progression in vivo. These findings support the concept of targeting the lipid balance as a cancer target.
Keywords: ovarian cancer, lipid metabolism, fatty acids, ER stress, SRS imaging
Abstract
Fatty acids are an important source of energy and a key component of phospholipids in membranes and organelles. Saturated fatty acids (SFAs) are converted into unsaturated fatty acids (UFAs) by stearoyl Co-A desaturase (SCD), an enzyme active in cancer. Here, we studied how the dynamics between SFAs and UFAs regulated by SCD impacts ovarian cancer cell survival and tumor progression. SCD depletion or inhibition caused lower levels of UFAs vs. SFAs and altered fatty acyl chain plasticity, as demonstrated by lipidomics and stimulated Raman scattering (SRS) microscopy. Further, increased levels of SFAs resulting from SCD knockdown triggered endoplasmic reticulum (ER) stress response with brisk activation of IRE1α/XBP1 and PERK/eIF2α/ATF4 axes. Disorganized ER membrane was visualized by electron microscopy and SRS imaging in ovarian cancer cells in which SCD was knocked down. The induction of long-term mild ER stress or short-time severe ER stress by the increased levels of SFAs and loss of UFAs led to cell death. However, ER stress and apoptosis could be readily rescued by supplementation with UFAs and reequilibration of SFA/UFA levels. The effects of SCD knockdown or inhibition observed in vitro translated into suppression of intraperitoneal tumor growth in ovarian cancer xenograft models. Furthermore, a combined intervention using an SCD inhibitor and an SFA-enriched diet initiated ER stress in tumors growing in vivo and potently blocked their dissemination. In all, our data support SCD as a key regulator of the cancer cell fate under metabolic stress and point to treatment strategies targeting the lipid balance.
To keep up with the demands of limitless proliferation and metastatic spread, cancer cells thwart physiological metabolic pathways to meet their augmented energetic needs. When glucose and oxygen are in short supply, fat becomes convenient fuel. Whether taken up from the tumor microenvironment or newly synthesized, lipids function as alternative energy source for rapidly growing tumors. In addition, lipids are key constituents of membranes and intracellular organelles, enabling the smooth functioning of signaling circuitries and the homeostasis of cells proliferating under stressful conditions (1). A unique property of ovarian cancer (OC) is its tropism to the omentum, a fat-laden organ, which has been thought to function as feeding soil for rapidly expanding tumors (2, 3). While lipid uptake by cancer cells has been studied to some extent (2–4), the roles of de novo lipogenesis and of the ensuing balance between unsaturated and saturated lipids remain not fully understood.
Within the de novo lipogenesis pathway, lipid desaturation is a key step required for the generation of unsaturated lipids to maintain the membrane fluidity, the integrity of cellular signaling, and the lipid pool for β-oxidation (5). Fatty acid (FA) desaturases catalyze the addition of double carbon bonds in acyl chains, regulating the formation of monounsaturated and polyunsaturated FAs (MUFAs and PUFAs, respectively). Stearoyl-CoA desaturase (SCD) converts saturated FA (SFA) to MUFAs and palmitic and stearic acid to palmitoleic and oleic acid (OA), respectively. SCD is up-regulated in cancer (6), and its inhibition was shown to suppress cancer cell proliferation, in conditions depleted of exogenous lipids (7). SCD was also shown to protect cancer cells from lipid peroxidation leading to ferroptosis, through a mechanism dependent on the antioxidant coenzyme CoQ10 (8). Using stimulated Raman scattering (SRS) microscopy (9, 10), which enables analysis of lipid species in rare cell populations, we recently demonstrated that ovarian cancer stem cells (CSCs) are enriched in UFAs (11). We showed that SCD small molecule inhibitors or short hairpin RNA (shRNA)-mediated knockdown (KD) eliminated ovarian CSCs, delaying tumor initiation (11). However, the mechanisms by which SCD regulates cancer cells’ and CSCs’ survival under stress conditions are not elucidated.
Through a combined lipidomic, transcriptomic and single cell imaging approach, we focused on how the balance between UFAs and SFAs regulates cell survival and tumor progression in OC models. We observed that an increase in SFAs caused by SFA supplementation, or by SCD depletion or inhibition, led to significant endoplasmic reticulum (ER) stress inducing apoptosis. The ER stress likely caused by direct effects of SFAs on the ER membrane fluidity, causing activation of the sensor proteins IRE1α and Protein Kinase R-like ER Kinase (PERK), could be rescued by addition of UFAs and restoration of the required SFA/UFA equilibrium. SCD KD inhibited tumor progression in vivo, and pharmacological inhibition of the enzyme coupled with a diet enriched in SFAs had potent antitumorigenic effects. These results point to the role of SCD as a gatekeeper ensuring the survival of OC cells under the metabolic stress imposed by nonstop proliferation and to the therapeutic potential of targeting the lipid balance.
Results
SCD Is Highly Expressed in OC Cell Lines and Tumors.
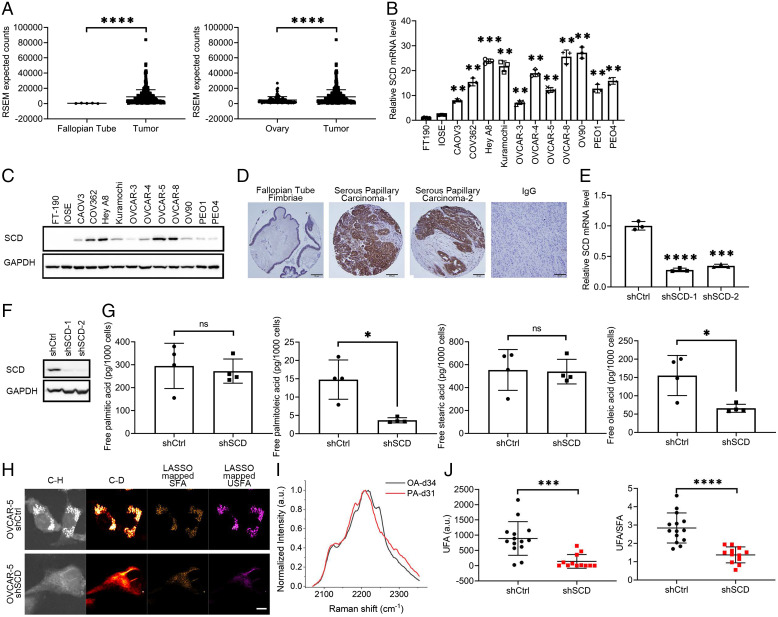
SCD expression was assessed by using representative cancer cell lines, human OC specimens included in two tissue microarrays (TMAs), and RNA sequencing (RNA-seq) data from human specimens profiled by The Cancer Genome Atlas (TCGA) (4) and the Genotype-Tissue Expression (GTEx) Project. SCD expression was significantly higher in high-grade serous ovarian tumors (HGSOC) profiled by the TCGA (n = 427) compared to normal fallopian tube epithelium (FTE, n = 5; Fig. 1 A, Left) and normal ovarian tissue (n = 93; Fig. 1 A, Right). Further, SCD was highly expressed in OC cell lines as compared to immortalized fallopian tube epithelial (FT-190) cells and immortalized ovarian surface epithelial (IOSE) cells at both messenger RNA (mRNA) and protein levels (Fig. 1 B and C). Immunohistochemistry (IHC) analysis demonstrated significant up-regulation of SCD in HGSOC tumors (n = 62) vs. FTE (n = 15), with 46 of 62 tumors displaying intense staining vs. FTE (Fig. 1D and SI Appendix, Table S1; P = 0.0016). Other histological subtypes of OC also displayed increased SCD expression compared to FTE (P = 0.0027), while nonmalignant ovarian tumors (borderline tumors and cystadenoma) did not overexpress SCD (SI Appendix, Table S1). Additionally, analysis of mRNA expression data from a publicly available dataset [Ovarian cancer database of Cancer Science Institute Singapore (CSIOVDB) (12)] showed a significant difference in SCD expression levels between all and each histological subtype of OC vs. low malignant ovarian tumors, (SI Appendix, Fig. S1A and Table S2), further supporting the significance of SCD in OC.
Fig. 1.
SCD is highly expressed in OC and is associated with increased levels of free UFAs. (A) SCD expression from RNA-seq data in (Left) fallopian tube and (Right) normal ovary tissue compared to OC tumors in patients from the UCSC Xena Browser. Expression is shown as RSEM expected counts. (B) Real-time qRT-PCR analysis of SCD expression (mean ± SD, n = 3) and (C) Western blot measurements of SCD protein levels in FT-190, IOSE, and 11 OC cell lines. (D) Representative images of IHC staining for SCD under 20× magnification in fallopian tube fimbriae (first panel) and HGSOC specimens (second and third panels). Negative control (IgG) is shown in the fourth panel (tumor tissue). (Scale bar, 50 μm.) (E) SCD expression measured by qRT-PCR (mean ± SD, n = 3) in OVCAR-5 cells transduced with shRNAs (1 or 2) targeting SCD (shSCD) or control shRNA (shCtrl). (F) Western blot of SCD in shCtrl and shSCD OVCAR-5 cells. (G) Lipidomics analysis of PA (16:0), palmitoleic acid (16:1), stearic acid (18:0), and OA (18:1) in shCtrl and shSCD OVCAR-5 cells cultured in medium containing low serum (1% FBS) for 48 h (means ± SD, n = 4). (H) Representative SRS images in C-H and C-D regions and LASSO mappings of SFA and UFAs (USFA) in shSCD vs. shCtrl OVCAR-5 cells treated with 12.5μM PA-d31 and cultured in low serum conditions. (Scale bar, 10 µm.) (I) Normalized SRS spectra of OA-d34 and PA-d31 (PA-d31) as reference spectra of UFA and SFA, respectively, for LASSO analysis. (J) Quantitative analysis of LASSO mapped USFA and ratio between USFA and SFA in shSCD vs. shCtrl OVCAR-5 cells. Each data point represents quantitative result from a single cell (n = 12 to 14). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
SCD Regulates the Balance between SFAs and UFAs in OC Cells.
For functional studies, two shRNA sequences targeting SCD (shSCD-1 and shSCD-2) or control shRNA (shCtrl) were stably transduced in OVCAR-5, OVCAR-8, OVCAR-3, and PEO1 cells. SCD KD was verified by quantitative PCR and Western blotting (Fig. 1 E and F and SI Appendix, Fig. S1 B–E). Lipidomics and isotopic hyperspectral SRS (hSRS) assessed the abundance of SFAs and UFAs in OC cells transduced with shRNA targeting SCD vs. control shRNA and cultured under low-serum conditions to limit the impact of exogenous lipid uptake. A significant reduction in UFAs (palmitoleic and OAs) but no difference in abundance of SFAs (palmitic and stearic acids, Fig. 1G) was recorded in cells transduced with shRNA targeting SCD vs. control shRNA. Further, isotopic hSRS imaging compared the levels of UFAs converted from newly imported SFAs in shSCD vs. shCtrl transduced cells (Fig. 1 H–J). After being maintained in low-serum medium for 24 h, cells were cultured with deuterated SFA (PA-d31) for 24 h. The C-D bond signal, corresponding to UFAs, was measured by hSRS imaging (Fig. 1I), and intracellular deuterated SFA and UFA levels were distinguished through a least absolute shrinkage and selection operator (LASSO) unmixing analysis using the distinctive Raman spectra of PA-d31 and OA-d34, as reference for SFA and UFA, respectively (Fig. 1H). Images and quantitative analyses of UFA levels and UFA/SFA ratio (Fig. 1J) show that cells in which SCD was knocked down contain significantly less deuterated UFA synthesized from newly imported PA-d31, indicating diminished efficiency of converting SFA to UFA in cells depleted of SCD compared to controls.
To understand which lipid species were most affected by the decrease of UFAs caused by SCD depletion, lipidomics by multiple reaction monitoring (MRM) profiling (13) measured the abundance of phosphatidylcholines (PC), phosphatidylethanolamine (PE), sphingomyelin (SM), and triglycerides (triacylglycerols [TAGs]) in OVCAR-5 cells stably transduced with control or shRNA targeting SCD and cultured under low-serum conditions for 48 h. After data normalization, principal component analysis confirmed the separation of the two groups (i.e., shCtrl and shSCD) and heat maps for each lipid species generated by using unsupervised hierarchical clustering demonstrated separation and clustering of the two groups (SI Appendix, Figs. S2–S5). The most affected (fold change) lipid species by SCD depletion were PC and SM, followed by triacylglycerol (TAG) and PE (Fig. 2A and Dataset S1). A deeper examination of each lipid class revealed that both PC and SM phospholipids with 1° plasticity (one or two carbon–carbon double bonds within the FA tail) (14) were significantly less abundant in shSCD compared with shCtrl transduced cells (P < 0.0001; Fig. 2B). Further, a significant reduction of 16:1 or 18:1 fatty acyl chains integrated TAGs was observed in OC cells stably transduced with shRNA targeting SCD compared to cells transduced with control shRNA (P = 0.0382; Fig. 2C). Combined, the lipidomics analyses coupled with hSRS imaging demonstrate that cells depleted of this key desaturase contain less abundant UFAs, and less plastic lipid species, particularly PC and SM.
Fig. 2.
Restricting MUFAs activates the ER stress response pathway. (A) Volcano plot of changes in PC and SM (green), PE (blue), and TAG (red) relative peak intensities measured by lipidomics profiling analysis in shSCD vs. shCtrl OVCAR-5 cells cultured in low-serum conditions for 48 h. (B) Normalized peak intensities of PC lipids with differing fatty chain plasticity (0°, no carbon–carbon double bond; 1°, 1 or 2 carbon–carbon double bonds; 2°, more than 2 carbon–carbon double bonds) in shSCD vs. shCtrl OVCAR-5 cells. ShSCD cells have significantly fewer 1° PC lipids than shCtrl cells, whereas no difference was observed for 0° or 2° PC lipids. (C) Fold change of TAG lipids containing 16:0 or 18:0 fatty acyl chains integration and 16:1 or 18:1 fatty acyl chains integration determined by lipidomics profiling analysis in shSCD vs. shCtrl OVCAR-5 cells. (D) Volcano plot of changes in gene expression measured by RNA-seq in shSCD vs. shCtrl OVCAR-5 cells. Red dots represent up-regulated genes, and blue dots represent down-regulated genes. Genes of the ER stress response pathway are indicated by lines. (E) Dot plot of the top 15 enriched Gene Ontology terms identified by analysis of up-regulated genes (RNA-seq) in shSCD vs. shCtrl OVCAR-5 cells. Dot size corresponds to ratio of genes on the pathway vs. total number of up-regulated genes. Background color corresponds to FDR q value. (F) Enrichment plots generated by Gene Set Enrichment Analysis of gene expression (RNA-seq normalized counts) in OVCAR-5 shSCD vs. shCtrl using Hallmark and C2 gene sets from Molecular Signatures Database. (G) Top 10 significant upstream regulators identified by Ingenuity Pathway Analysis using DEGs from RNA-seq analysis of OVCAR-5 shSCD vs. shCtrl cells. Transcription factors involved in the ER stress response pathway are in bold characters. (H) Heat map of expression (RNA-seq normalized counts) and supervised hierarchical clustering of significant genes of the ER stress response pathway in shCtrl and shSCD OVCAR-5 cells (n = 3). *P < 0.05, ****P < 0.0001.
SCD KD Triggers ER Stress Response.
To further understand the global effects of UFA restriction in OC cells, RNA-seq compared the transcriptomic profiles of cells transduced with shRNA targeting SCD or control shRNA. Quality control of RNA-seq is included in SI Appendix, Fig. S6. Cells were cultured under low-serum conditions for 48 h. There were 1,513 differentially expressed genes (DEGs; false discovery rate (FDR) < 0.05), of which 707 were up-regulated and 806 were down-regulated between OVCAR-5 cells stably transduced with control or shRNA targeting SCD (Fig. 2D and Dataset S2). Gene Ontology analysis of significantly up-regulated genes revealed that the ER stress response was the top affected pathway by SCD KD (Fig. 2E). Gene Set Enrichment Analysis of all genes also indicated that ER stress response gene sets (Hallmark Unfolded Protein Response; Reactome_IRE1α Activities Chaperones, Reactome PERK Regulates Gene Expression, Reactome ATF4 Activities Genes in Response to Endoplasmic Reticulum Stress) were up-regulated in cancer cells in which SCD was knocked down (Fig. 2F). Several genes (DDIT3, ATF3, PPP1R15A, and CEBPB) involved in ER stress response are highlighted in the volcano plot depicting DEGs between SCD KD and control cells (Fig. 2D). Furthermore, upstream regulator analysis performed with Ingenuity Pathway Analysis (IPA) software identified several transcription factors (ATF4, DDIT3, and NUPR1) involved in the ER stress response pathway among the top ones predicted to be activated in OC cells transduced with shRNA targeting SCD vs. control cells (Fig. 2G). A heat map including significant genes in the ER stress response pathway (n = 64 genes) illustrates clear differences in expression levels for these transcripts between cells in which SCD was knocked down vs. control cells (Fig. 2H). Further, exploration of the transcriptomic dataset associated with the Cancer Dependence Map (15) and Cancer Cell Line Encyclopedia (16) shows that dependency score of key transcription regulators in the ER stress response pathway such as ATF4 and DDIT3 were positively correlated with that of SCD (r = 0.3117 and 0.3878, respectively; SI Appendix, Fig. S7 A and B), indicating that OC cells that are highly dependent on SCD also have higher dependency on ATF4/DDIT3. Additionally, expression levels of several important ER stress response genes (IGFBP1, ERN1, TXNIP, and NABP1) were negatively correlated with SCD (SI Appendix, Fig. S7 C–F), corroborating the association between SCD and ER stress response found through transcriptomic analysis.
Excess SFAs Caused by Depletion or Pharmacological Inhibition of SCD Induces ER Stress.
To further investigate how the imbalance between SFAs and UFAs caused by SCD depletion or inhibition in cancer cells activates the ER stress responses, we next evaluated the two key sensing mechanisms of this pathway: activation of IRE1α and PERK. ER-spanning transmembrane domain of IRE1α and PERK are critical sensors of the levels of lipid saturation in the cell and activate the ER stress response (17). IRE1α causes splicing of the X-box–binding protein 1 (XBP1) mRNA, which leads to a spliced form (XBP1s) with potent transcriptional activity (18). PERK autophosphorylates itself upon ER stress and then phosphorylates the eukaryotic translation initiation factor 2α (eIF2α) to halt translation of proteins (19). Translation of ATF4 mRNA, due to its unique 5′ UTR sequence, is up-regulated upon phosphorylation of eIF2α (20).
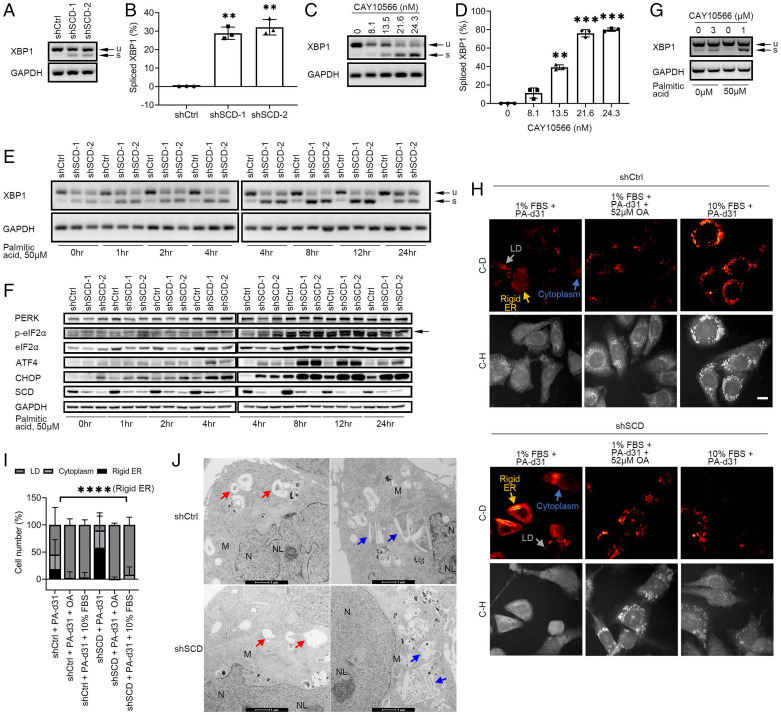
XBP1 splicing was assessed in OVCAR-5 cells stably transduced with shRNA targeting SCD or control shRNA or in cells treated with the SCD inhibitor, CAY10566. Increased baseline XBP1s levels were noted in OVCAR-5 and OVCAR-8 cells transduced with shRNA targeting SCD vs. control and cultured in low-serum medium or in 10% lipid depleted serum for 48 h (Fig. 3 A and B and SI Appendix, Fig. S8 A and B). XBP1 splicing was also increased in OVCAR-5 and OVCAR-8 cells transduced with shRNA targeting SCD vs. control even under full-serum conditions but to a lesser degree (SI Appendix, Fig. S8 C and D). Additionally, activation of PERK/CHOP was detected in shSCD vs. shCtrl stably transduced OVCAR-8 cells (SI Appendix, Fig. S8E) under low-serum conditions.
Fig. 3.
The IRE1α/XBP1 and PERK/eIF2α/ATF4 axes of the ER stress response pathway are activated in OC cells under restricted availability of UFA. (A) XBP1 splicing (u, unspliced transcript; s, spliced transcript) measured by RT-PCR and agarose-gel electrophoresis in OVCAR-5 cells transduced with control shRNA (shCtrl) or shRNAs (1 or 2) targeting SCD (shSCD) and cultured in low-serum conditions (1% FBS) for 48 h. (B) Densitometric analysis of XBP1 splicing products shown in A. Bars represent percentage of spliced XBP1 relative to total XBP1 (mean ± SD, n = 3). (C) XBP1 splicing in OVCAR-5 cells cultured in low-serum conditions and treated with SCD inhibitor CAY10566 for 48 h. (D) Percentage of spliced XBP1 isoform measured by densitometric analysis of PCR products shown in C (mean ± SD, n = 3). (E) XBP1 splicing and (F) Western blot of proteins of the PERK/eIF2α/ATF4 axis in shCtrl and shSCD OVCAR-5 cells cultured under low-serum conditions and treated with 50 μM PA for the time periods indicated. Arrows indicate the band of interest. (G) XBP1 splicing in primary cells from tumors of OC patients cultured in low-serum conditions and treated with 3 μM CAY10566 for 48 h or with 1 μM CAY10566 and 50 μM PA for 12 h. (H) Representative SRS images in the C-H and C-D regions of OVCAR-5 shCtrl and shSCD cells cultured in low-serum conditions (1% FBS) and treated with 12.5 μM PA-d31, with or without 52 μM OA, or cultured in medium with full serum (10% FBS) and treated with PA-d31 for 24 h. Yellow arrows indicate rigid ER, gray arrows indicate LDs, and blue arrows indicate cytoplasm. (Scale bar, 20 µm.) (I) Percentages of shCtrl and shSCD OVCAR-5 cells treated as described in H showing C-D SRS signal mainly in rigid ER, LD, and cytoplasm (n = 139 to 191). (J) TEM imaging of smooth ER (donut shaped, red arrows; ski board shaped, blue arrows) in OVCAR-5 shCtrl vs. shSCD cells cultured in low-serum conditions for 48 h. Other organelles are highlighted (M, mitochondria; N, nucleus; NL, nucleolus). **P < 0.01, ***P < 0.001, ****P < 0.0001.
Likewise, a dose-dependent increase in XBP1s levels was observed in OVCAR-5 cells treated with CAY10566 under low-serum conditions (Fig. 3 C and D), in 10% lipid depleted serum (SI Appendix, Fig. S8F), and in full serum (SI Appendix, Fig. S8G). Having hypothesized that the major trigger of ER stress induced by SCD depletion is the excess of SFAs, XBP1 splicing and activation of PERK/eIF2α/ATF4 were measured in OC cells in which SCD was either knocked down or pharmacologically inhibited in the presence of exogenously added SFA (palmitic acid [PA]). We chose a concentration of PA (50 μM) equivalent to that found in media supplemented by 10% fetal bovine serum (FBS) (21). Further, as the consequences of ER stress depend on both the length of exposure to cellular stress and the severity of the stress (22), time dependency was assessed along both the IRE1α/XBP1 and the PERK/eIF2α/ATF4 axes in OC cells supplemented with PA. The XBP1 splicing signal was increased in OC cells in which SCD was knocked down compared with control cells after supplementation with 50 μM of PA, peaking at ∼8 to 12 h and diminishing at 24 h (Fig. 3E). Likewise, Western blotting analysis of the PERK/eIF2α/ATF4 axis showed increased phosphorylation levels of eIF2α in OC cells in which SCD was knocked down, starting 2 h after addition of PA and peaking at 8 to 12 h (Fig. 3F). The downstream transcription factors ATF4 and CHOP were up-regulated starting 4 h after addition of PA and remaining persistently high up to 24 h. To confirm these observations in human specimens, we used primary tumor cells dissociated from freshly obtained HGSOC tumors (SI Appendix, Table S3). Treatment with CAY10566 induced XBP1 splicing in primary HGSOC cells, and the XBP1s levels were further augmented by addition of PA (Fig. 3G and SI Appendix, Fig. S8H).
Next, to visualize the status of the ER under these conditions, hSRS imaging was performed after treating OVCAR-5 cells stably transduced with control or shRNA targeting SCD with PA-d31 under low-serum conditions (Fig. 3H). Most of OVCAR-5 shCtrl cells displayed C-D signal derived from PA-d31 in lipid droplets (LDs) (depicted as a dot, gray arrow), while a considerable portion of OVCAR-5 shSCD cells displayed the C-D signal on rigid ER (shown as a linear structure, yellow arrow), consistent with an ER stress-related morphology (23). Both cell lines contained a small portion of cells harboring detectable C-D signal all over the cytoplasm (Fig. 3H, blue arrow). After PA-d31 treatment alone under low-serum conditions, the cell population with rigid ER structures represented the major population of OVCAR-5 shSCD cells, while the cell population with detectable C-D signal in LD was the majority in OVCAR-5 shCtrl cells (Fig. 3 H and I). Rescue treatment with UFA (OA or full serum medium) led to the disappearance of rigid ER and increased PA-d31 accumulation in LDs for both shSCD and shCtrl OC cells (Fig. 3 H and I), suggesting that supplementation with UFAs can rescue SFA-induced ER stress and facilitate SFA storage in LD. These data support that the balance of intracellular SFA and UFA is essential to prevent ER stress.
Further, examination of OVCAR-5 cells stably transduced with control or shRNA targeting SCD cultured in low-serum medium for 48 h by using transmission electron microscopy (TEM) demonstrated smooth ER membrane disorganization and irregular and compromised smooth ER structure in OVCAR-5 shSCD cells compared to shCtrl cells (Fig. 3J), confirming the biochemical and hSRS imaging results.
UFAs Prevent ER Stress in OC Cells.
To exclude the contribution of potential desaturase-independent functions of SCD to the induction of ER stress in OC cells, we examined whether the effects of SCD depletion or inhibition could be rescued by OA, its main enzymatic product. Supplementation with OA reversed XBP1 splicing in OVCAR-5 cells in which SCD was knocked down (Fig. 4A and SI Appendix, Fig. S8B), as well as in OVCAR-5 cells depleted of SCD and treated with PA (Fig. 4B). Likewise, supplementation with OA reduced the up-regulation of ATF4 and CHOP along the PERK/eIF2α/ATF4 axis observed in OVCAR-5 cells stably transduced with shRNA targeting SCD and maintained under low-serum conditions (Fig. 4 C and D). Similar rescue effects of OA on XBP1 splicing were observed in OVCAR-8 and PEO1 cells stably transduced with shRNA targeting SCD (SI Appendix, Fig. S9 A and B). Likewise, supplementation with OA reduced XBP1s levels in OVCAR-5 cells treated with CAY10566 (Fig. 4E and SI Appendix, Fig. S8F), as well as XBP1s levels in cells treated with CAY10566 and PA (Fig. 4F). OA also reduced the up-regulation of ATF4 and CHOP induced by the SCD inhibitor in OVCAR-5 cells maintained under low-serum conditions (Fig. 4G) or in cells treated with 50 μM PA (Fig. 4H). Last, to test the sufficiency of SCD in regulating ER stress triggered by an imbalance of SFA and UFA in OC cells, the enzyme was overexpressed in OVCAR-5 cells. Increased SCD mRNA and protein levels were observed in cells stably transfected with pLenti-SCD compared to pLenti-Ctrl (Fig. 4 I and J). XBP1 splicing induced by addition of PA in control cells was significantly reduced by SCD overexpression (Fig. 4 K and L; P = 0.0277). Collectively, our data support the significance of the balance between SFAs and UFAs regulated by SCD in fine tuning the functions of the ER, likely mediated through direct effects on the ER membrane.
Fig. 4.
UFA-induced ER stress response is reversed by exogenous OA. XBP1 splicing measured by RT-PCR and agarose gel electrophoresis in OVCAR-5 cells transduced with control shRNA (shCtrl) or shRNAs (1 or 2) targeting SCD (shSCD), cultured in medium containing low serum, and treated (A) with indicated doses of OA for 48 h or (B) with 50 μM PA and indicated doses of OA for 12 h. Western blot of SCD and proteins of the PERK/eIF2α/ATF4 axis in shCtrl and shSCD cells cultured in low serum medium and treated (C) with different doses of OA for 48 h or (D) with 50μM PA and indicated doses of OA for 12 h. XBP1 splicing in OVCAR-5 cells cultured in medium containing low serum and treated (E) with 21.6 nM CAY10566 and indicated doses of OA for 48 h or (F) with 8.1 nM CAY10566, 50 μM PA, and indicated doses of OA for 12 h. (G) Proteins of the PERK/eIF2α/ATF4 pathway measured by Western blot in OVCAR-5 cells treated as described in C. (H) Western blot of proteins of the PERK/eIF2α/ATF4 pathway in OVCAR-5 cells treated as described in D. Arrows indicate the band of interest. Verification of SCD overexpression by (I) qRT-PCR and (J) Western blotting in OVCAR-5 cells transduced with a SCD expression vector (pLenti-SCD). Cells transduced with empty vector (pLenti-Ctrl) served as control. Bars represent means ± SD, n = 3. (K) XBP1 splicing in pLenti-SCD vs. pLenti-Ctrl cells cultured in low-serum conditions and treated with 50 μM PA for 12 h. (L) Percentage of spliced isoform (mean ± SD, n = 3) calculated by densitometric analysis of PCR products shown in K. *P < 0.05, ***P < 0.001.
SCD Depletion or Inhibition Induces Apoptosis of OC Cells.
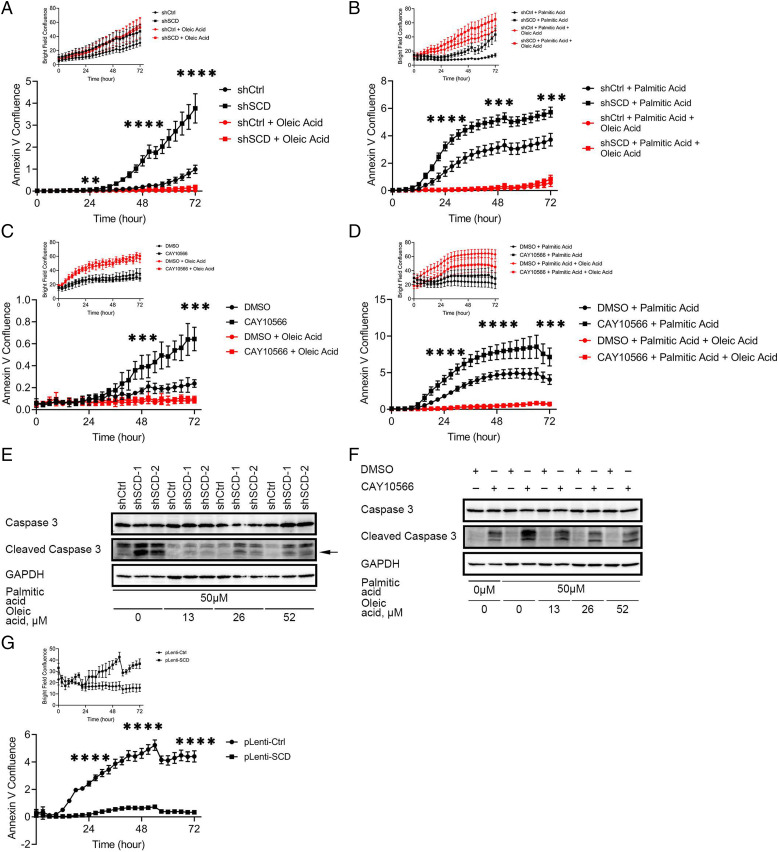
It is accepted that long-term exposure to mild ER stress or short-term exposure to severe ER stress leads to CHOP-mediated apoptosis (22). We therefore hypothesized that OC cells depleted of SCD and cultured under low-serum conditions over a long period of time would undergo apoptosis. IncuCyte imaging examining Annexin V staining showed increased percentage of apoptotic cells among OVCAR-5 SCD KD cells cultured in low-serum medium compared with control cells. Supplementation with OA rescued the phenotype (Fig. 5A). Further, addition of PA (50 μM) induced early onset of apoptosis in shSCD cells as compared to shCtrl cells, and supplementation with OA reversed the phenotype (Fig. 5B). Likewise, CAY10566 induced apoptosis in OVCAR-5 cells, while OA supplementation blocked this effect (Fig. 5C). Addition of PA to CAY10566 caused earlier onset of apoptosis (less than 24 h) and augmented cell death (Fig. 5D), while OA rescued the phenotype (Fig. 5D). To confirm the apoptosis phenotype, we examined cleaved caspase-3 levels. Increased caspase-3 cleavage was observed in OVCAR-5 shSCD compared to shCtrl cells (Fig. 5E), and this was reversed by OA (Fig. 5E). Caspase-3 cleavage was also induced by CAY10566, augmented by addition of PA and rescued by repletion of OA (Fig. 5F).
Fig. 5.
Apoptosis induced by SCD inhibition, or treatment with PA, is attenuated by exogenous OA or SCD overexpression. Time-lapse imaging of Annexin V staining (bright field signal as shown in Top) to measure apoptosis in OVCAR-5 cells transduced with control shRNA (shCtrl) or shRNA targeting SCD (shSCD), cultured in low-serum conditions, and treated (A) with 52μM OA or (B) with 50 μM PA alone or in combination with 52 μM OA. Time-lapse of Annexin V imaging (bright field signal as shown in Top) to measure apoptosis in OVCAR-5 cells cultured in low-serum conditions and treated (C) with 21.6 nM CAY10566, 52 μM OA or combination, or (D) with 8.1 nM CAY10566, 50 μM PA, 52 μM OA, or combinations. (E) Western blot of full-length and cleaved caspase-3 in shSCD vs. shCtrl cells cultured under low-serum medium and treated with 50 μM PA and indicated doses of OA for 12 h. Arrow indicates the band of interest. (F) Western blot of full-length and cleaved caspase-3 in OVCAR-5 cells cultured in low-serum medium and treated with 8.1 nM CAY10566, 50 μM PA, and indicated doses of OA for 12 h. (G) Time-lapse of Annexin V imaging (bright field signal as shown in Top) to determine apoptosis in OVCAR-5 cells overexpressing SCD (pLenti-SCD) and control cells (pLenti-Ctrl), cultured in medium containing low serum and treated with 50 μM PA. Values in A–D and G are means ± SD, n = 6. **P < 0.01, ***P < 0.001, ****P < 0.0001.
As the numbers of apoptotic cells appeared insufficient to explain the decreased numbers of surviving cells (Fig. 5 A and C, Top) and SCD inhibition had been shown to induce nonapoptotic cell death through ferroptosis (8), we examined whether SCD depletion or blockade could promote cell death through a combination of apoptosis and ferroptosis. C11 BODIPY staining measured lipid peroxidation in cells in which SCD was knocked down or inhibited by CAY10566. A clear peak shift in the oxidized lipid channel was observed after SCD KD or inhibition (SI Appendix, Fig. S9C), supporting induction of ferroptosis. Cell viability inhibited by CAY10566 was not rescued either by a caspase 3 inhibitor (Z-VAD-FMK) or by ferrostatin-1 (ferroptosis inhibitor) alone but was blocked by the combination of Z-VAD-FMK and ferrostatin-1 (SI Appendix, Fig. S9D). Together these data support that decreased UFAs induced by SCD blockade or depletion block cell survival through both apoptosis and nonapoptotic cell death.
Last, to test the sufficiency of SCD to this phenotype, OVCAR-5 cells in which SCD was overexpressed (pLenti-SCD) vs. control cells (pLenti-Ctrl) were maintained under low serum and treated with PA. The percentage of apoptotic cells was significantly reduced in cells in which SCD was overexpressed compared to controls (Fig. 5G), supporting that OC cells with higher SCD expression could survive the stress induced by SFAs. Together, the data demonstrate that the UFAs, the direct product of SCD, act as a buffer against SFAs, promoting the survival of OC cells.
SCD Depletion or Inhibition Suppresses Tumor Growth In Vivo.
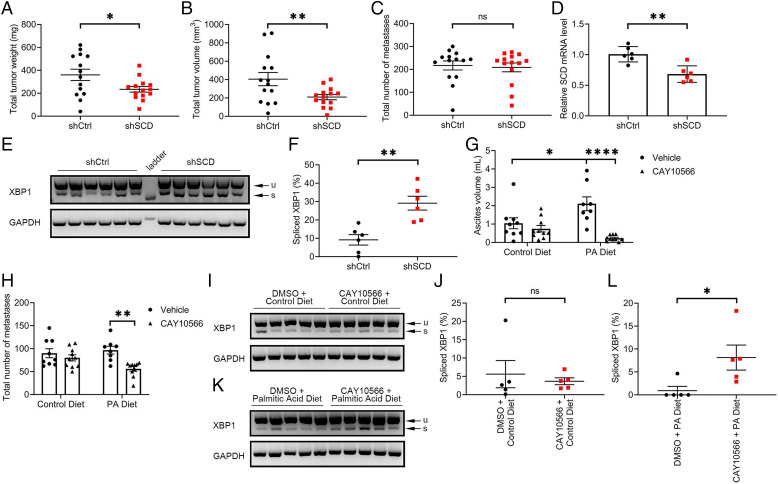
Whether the observed effects of SCD on cancer cell survival impact tumor progression in vivo remains unknown. Tumorigenicity of OC cells depleted of SCD was tested by using an intraperitoneal (ip) xenograft model in athymic nude mice. SCD KD compared to control OVCAR-5 cells led to significantly reduced tumor weight (234.3 mg vs. 360.1 mg, P = 0.0343) and tumor volume (209.2 mm3 vs. 404.8 mm3, P = 0.0212; Fig. 6 A and B), although the numbers of peritoneal metastases (Fig. 6C; 208.8 vs. 217.3, P = 0.7602) and ascites volume (SI Appendix, Fig. S10A) were similar between groups. SCD KD was maintained in vivo, as demonstrated by qRT-PCR (Fig. 6D; P = 0.0015), and increased XBP1s levels were detected in xenografts derived from OVCAR-5-shSCD compared to controls (Fig. 6 E and F; P = 0.0018), supporting that an increased susceptibility to ER stress is maintained in vivo in tumors depleted of the enzyme. Similar inhibitory effects on tumor growth were observed in a subcutaneous mouse xenograft model derived from OVCAR-3 cells stably transduced with shRNA targeting SCD vs. shRNA control (SI Appendix, Fig. S10 B and C). Interestingly, HGSOC patients with high (n = 590) vs. low (n = 411) SCD expressing tumors in the TCGA database (24) had shorter progression free survival (SI Appendix, Fig. S10D; P = 0.015), supporting the potential functional relevance of SCD to OC progression.
Fig. 6.
SCD KD inhibited growth of OC xenografts in mice. (A) Total tumor weight, (B) total tumor volume, and (C) total number of metastases in athymic nude mice intraperitoneally injected with OVCAR-5 cells transduced with control shRNA (shCtrl) or shRNA targeting SCD (shSCD) and evaluated after 28 d (values are means ± SE, n = 14 per group). (D) qRT-PCR measurements of SCD expression (mean ± SD, n = 6) in a random sample of tumor xenografts described in A. (E) Agarose gel electrophoresis of XBP1 splicing products and (F) percentage spliced XBP1 isoform estimated by image analysis of transcript bands (mean ± SE, n = 6) in a random sample of tumor xenografts described in A. (G) Ascites volume and (H) total number of metastases in athymic nude mice intraperitoneally injected with OVCAR-5 cells, fed with a PA-rich diet or control diet, and treated with SCD inhibitor CAY10566 or vehicle for 28 d. Values are means ± SE, n = 10. (I–L) XBP1 splicing products (I and K) and percentage of intensity of the spliced transcript estimated by image analysis (J and L) in a random sample (n = 5) of tumor xenografts described in F. Values are means ± SE. *P < 0.05, **P < 0.01, ****P < 0.0001.
Our observations that ER stress and apoptosis are augmented in vitro by inhibition of the desaturase coupled with an excess of SFAs led us to hypothesize that the effects of a pharmacological inhibitor targeting this pathway would be enhanced through dietary modifications tilting the FA balance toward increased saturation levels. To test this hypothesis, we used an ip model fed a PA-rich or control diet and treatment with CAY10566 or diluent. While CAY10566 did not significantly alter the numbers of peritoneal metastases and the ascites volume in mice fed a control diet (Fig. 6 G and H), the inhibitor significantly decreased both the ascites volume (P < 0.0001) and the numbers of metastases (P = 0.0003) in mice fed PA-rich diet (Fig. 6 G and H). Interestingly, an unanticipated but significant increase in ascites volume was observed in the group receiving the SFA-rich diet compared with the control diet (Fig. 6G; P = 0.0252). No significant differences were observed in total tumor weights and total tumor volumes (SI Appendix, Fig. S10 E and F). XBP1s levels were not significantly different in tumors derived from mice fed the control diet and treated with vehicle vs. CAY10566 (Fig. 6 I and J; P = 0.6370) but were significantly increased in tumors derived from mice fed PA-rich diet and treated with CAY10566 vs. control (Fig. 6 K and L; P = 0.0369). The data support that SCD inhibition or depletion exerts antitumorigenic effects through an ER stress-dependent mechanism caused by decreased levels of lipid desaturation.
Discussion
Our data based on transcriptomic, lipidomic analyses, SRS, and EM imaging demonstrate that the imbalance between SFAs and UFAs caused by depletion or inhibition of the enzyme SCD has a key function in determining cancer cell survival or death and impacts tumor progression in vivo. These findings hone on a largely understudied metabolic process in cancer cells and suggest the possibility of using an intervention combining an SCD enzymatic inhibitor together with a dietary intervention to increase levels of lipid saturation in cancer cells and trigger a potent antitumor effect. Our findings have several important consequences.
First, our results point to the significance of the process of lipid unsaturation in cancer. Lipid synthesis includes the synthesis of FA from acetyl-coA mediated by FA synthase (FASN) and acyl-CoA synthase (ACSL), followed by addition of double carbon bonds catalyzed by FA desaturases. Among the three desaturases, Δ9 (stearoyl-CoA desaturase [SCD]) catalyzes the synthesis of MUFAs by adding one double bond to SFAs (mostly stearic acid), while the Δ6 and Δ5 desaturases are involved predominantly in the synthesis of PUFAs (25). The current study builds on our previous observations that highly tumorigenic ovarian CSCs are enriched in UFAs, caused by SCD up-regulation, which in turn induce prosurvival NF-κB signaling allowing CSCs to proliferate as spheres and effectively initiate tumors in immunodeficient mice (11). Here, we show that beyond the previously observed effects on CSCs, SCD KD, and to a lesser extent its inhibition, induces antitumor effects in ip xenografts. Overexpression of ACSL and SCD leading to increased UFA levels was described in association with epithelial to mesenchymal transition and aggressive clinical prognosis in colon cancer, and SCD up-regulation was observed in basal-type breast cancer and aggressive prostate cancer (26, 27). In renal cell carcinoma, triacylglycerides stored in LDs could release OA and counter the stress induced by SFAs (28). Depletion of OA (but not of palmitate) inhibited the proliferation of AML and lymphoma cells (29), and lipid saturation levels in plasma or tumor tissue were indirectly correlated with cancer risk in epidemiological studies (30, 31). However, it remains not clear how the balance between UFAs and SFAs alter cancer predisposition or cancer progression, a concept we addressed here.
A powerful tool that allowed us to quantify UFAs and SFAs at the single-cell level in this study was SRS microscopy, an innovative label-free chemical imaging technique which we have previously employed to characterize metabolic reprograming from glycolysis to FA uptake and β-oxidation in platinum-resistant cancer cells (32), increased lipid desaturation in ovarian CSCs (11), and cholesteryl ester accumulation as a metabolic marker for multiple aggressive cancers (33, 34). Combined with postprocessing methods such as least-square fitting, multivariate curve resolution, and phasor segmentation, hSRS can distinguish different intracellular biomolecules such as protein and FA (10, 35). Here, SRS imaging visualized SFA-induced ER stress in OC cells depleted of SCD and the rescue observed by manipulating the ratio between SFAs and UFAs. We recognize that highly spatially overlapping biomolecules with similar spectrum profiles such as SFAs and UFAs are difficult to decompose by using conventional analysis methods. We propose that pixel-wise LASSO unmixing can resolve such shortcomings, enabling the simultaneous evaluation of multiple metabolites, facilitating better assessment of cancer cell metabolism and metabolic changes in response to gene modification and drug treatment. The innovative hSRS-LASSO imaging method utilized here provides a way to explore cancer lipid metabolism in deeper detail.
Our findings demonstrate that the imbalance between SFAs and UFAs impedes cancer cell survival and in vivo tumor growth through induction of ER stress. We observed that two branches of ER stress response, regulated by the PERK and IRE1α, were activated in response to SCD depletion or inhibition and that this activation was augmented in the presence of SFA supplementation and could be rescued by addition of UFAs. Interestingly, noncancer cells, such as neural stem/progenitor cells, were found to be susceptible to lipid accumulation and consequent ER stress (36), and PA was shown to induce ER stress and apoptosis in human bone marrow–derived mesenchymal stem cells (37). Another study found that N-Myc driven hepatocellular cancer cells were dependent on lipid unsaturation through modulation of ER stress responses (38). While a definitive mechanistic explanation is still lacking, our hSRS and TEM imaging suggest that increased levels of lipid unsaturation in cancer cells alter the morphology of the ER, causing increased membrane rigidity and potentially direct activation of the sensor proteins.
The link between ER stress and tumor progression is emerging. In orthotopic cervical cancer xenografts, hypoxia-induced metastasis to lymph nodes was blocked by PERK inhibition (39). The PERK/eIF2α/ATF4 axis was also shown to be important for inducing epithelial-to-mesenchymal transition of breast cancer cells (40), while the IRE1α/XBP1 pathway was involved in tumor initiation and breast cancer progression by activating the transcription factor HIF1α (41). Our study, together with previous reports, reveals the biphasic role of PERK/eIF2α/ATF4 axis of the ER stress response, with the initial response being implicated in salvaging the stressed cells, whereas prolonged activation of the pathway leads to apoptosis. Future investigation of how lipid unsaturation triggers and modulates the ER stress response could provide new therapeutic opportunities to target cancer progression.
A starting point toward this goal is our attempt to combine SCD inhibition with a dietary intervention. We demonstrate that this intervention resulting in accumulation of toxic SFAs has potent antitumor effects (numbers of metastases and ascites) and induces ER stress responses detectable in tumor tissue. Whether the effects of the combined intervention target peritoneal dissemination more than tumor growth, as observed in experiments using SCD KD, remains to be elucidated. The putative role of diet in cancer treatment has remained a “holy grail” and is based on the concept that altered metabolite levels in the tumor microenvironment could create metabolic challenges impeding tumor cells’ survival. The role of saturated lipids in cancer remains controversial, with the overwhelming belief that saturated fat is protumorigenic. Indeed, a recent study showed that dietary PA promoted metastasis in oral carcinoma and melanoma models (42). Consistent with these observations, we found that the PA-rich diet induced increased accumulation of malignant ascites, the main vehicle of OC metastasis. However, the combination of a PA-rich diet and CAY10566 caused a significant reduction in ascites and peritoneal implants. Given the plasticity and redundancy of metabolic pathways, which have by and large thwarted prior attempts to use metabolic interventions to target cancer, our results highlight the importance of combination strategies. Taken together, our findings support SCD’s key role in regulating cancer cell fate and tumorigenicity and point to modalities to block OC progression.
Materials and Methods
Reagents and Cell Culture.
Palmitic acid (cat. no. P0500), OA (cat. no. O3008), dimethyl sulphoxide (DMSO) (cat. no. D2650), and Tween 80 (cat. no. P4780) were purchased from MilliporeSigma. CAY10566 (cat. no. HY-15823) and PEG300 (cat. no. HY-Y0873) were purchased from MedChemExpress. OC cell lines culture conditions are included in SI Appendix.
Human Specimens.
Two OC TMAs containing OC specimens of different histological subtypes, benign ovarian tumors, and normal fallopian tube were obtained from the Cooperative Human Tissue Network (OvCa2) and the Lurie Cancer Center Pathology Core (SI Appendix). Primary tumors or ascites from high-grade serous OC patients were collected at Northwestern Memorial Hospital from consenting donors. Tumor tissues were minced into small pieces and digested as described in SI Appendix.
Cell transfection, construction of SCD expression vector, RNA extraction and real-time qPCR, Western blot, IHC, and TEM were performed using protocols described previously (43–45) and detailed in SI Appendix. Primers are included in SI Appendix, Table S4.
XBP1 Splicing Assay.
Synthesized cDNA as described above was used to measure levels of unspliced and spliced XBP1. mRNA was measured by regular PCR performed with GoTaq Green Master Mix (Promega cat. no. M7123) on a T100 Thermo Cycler (Bio-Rad, cat. no. 186-1096). The primers targeting the spliced XBP1 region (116 bp) (SI Appendix, Table S4) were designed according to a previous study (46). PCR products were resolved by agarose gel electrophoresis and visualized using GelGreen stain (Biotium, cat. no. 41005) on an ImageQuant LAS 4000 machine (GE Healthcare). Densitometric analysis of product bands was performed using the Gel Analyzer function in Fiji from NIH (47).
Lipidomics.
Cell pellets were obtained from OVCAR-5 cells transduced with shRNA targeting SCD vs. control shRNA cells used for analysis at the Bindley Bioscience Center, Purdue University. Lipidomic analysis was performed using MRM profiling, as detailed in SI Appendix. Statistical analysis was performed with MetaboAnalystR 3.0 (48).
SRS Imaging.
Stimulated Raman scattering (SRS) imaging was performed to measure isotope labeled cellular SFAs/UFAs on a previously described laboratory-built system with a femtosecond laser source operating at 80 MHz (InSight DeepSee, Spectra-Physics) (11, 32), as previously described (SI Appendix). Hyperspectral SRS images were analyzed using ImageJ and previously described pixel-wise LASSO regression algorithm (49). In brief, LASSO can effectively decompose hSRS imaging into maps of different biomolecules by introducing a sparsity constraint to suppress the cross-talk between different chemical maps. PA-d31 and OA-d34 were used to provide reference spectra of SFA and USFA, respectively, for LASSO unmixing analysis.
Xenograft Experiments.
Athymic female nude mice (strain Foxn1nu), 6 to 8 wk old, were obtained from Envigo. For the subcutaneous model, mice were injected with 2 × 106 OVCAR-3 cells stably transduced with shRNAs targeting SCD or control shRNA resuspended in 1/1 mix of RPMI 1640 basal medium and Matrigel (Corning, cat. no. 356234). Tumor length (L), width (W), and height (H) were measured every 3 d with digital calipers. Tumor volume was calculated using the formula V = L × H × W/2. For the ip model, 5 × 106 OVCAR-5 cells were injected ip. Mice were euthanized 28 d after injection of cells, and numbers of disseminated peritoneal implants were counted, volume of ascites was measured, and tumors >5 mm3 were weighed and measured by calipers to calculate volumes. Total tumor weight and volume per mouse was calculated by adding individual tumor sizes. To determine the effects of a PA-enriched diet and SCD inhibitor, nude mice were randomized to be fed with a PA-enriched or a control diet beginning 1 wk before ip injection of OVCAR-5 cells and continued for the duration of the experiment (28 d). Mice were treated ip with the SCD inhibitor CAY10566 (8 mg/kg body weight) or diluent (10% DMSO, 40% PEG300, 5% Tween-80, 45% saline), every other day during weekdays. The contents of PA in the PA-enriched diet and control were 30 and 11.7% of the total FAs, respectively. The levels of total fat (58 g/kg) were the same, and levels of saturated fat, monounsaturated fat, polyunsaturated fat, stearic acid, and OA were similar (<2% difference) between the PA-enriched and control diets. The diets were custom-made by Teklad (Envigo).
RNA-seq was performed as previously described (43) and detailed in SI Appendix. DEGs between experimental groups were determined and FDR corrected for multiple hypothesis testing with the edgeR package (50) in R. Pathway analysis based on the DEGs was performed using IPA software (QIAGEN). Data are deposited in GEO (GSE192442).
The GTEx and TCGA OC RNA-Seq Data Analysis.
Recomputed RNA-Seq by Expectation Maximization (RSEM) expected counts of fallopian tube samples (GTEx) and primary tumor samples from TCGA OC patients were downloaded from the University of California, Santa Cruz (UCSC) Xena Browser. Expression of SCD (Ensembl ID ENSG00000099194) was extracted from the matrix of all genes, and a Mann–Whitney u test was performed on log2 transformed RSEM expected counts between fallopian tube samples or normal ovary tissue and primary tumor samples.
Cancer Dependency Map Data Analysis.
Dependency score data from CRISPR screening in cancer cell lines was downloaded from Depmap Portal (15). OC cell lines were selected, and Pearson correlation score was calculated between SCD and transcriptional regulators identified from IPA.
Apoptosis Assay by IncuCyte Imaging.
Cells were seeded on 96-well plates at 500 per well and cultured in an IncuCyte S3 live-cell analysis system. Serum was reduced in the medium to 1% FBS, and Annexin V Green Dye (Sartorius, cat. no. 4642) was added for detection of apoptotic cells under different experimental conditions. Four images of bright field and green fluorescence were captured at 3-h intervals during a 72-h evaluation period. Cells were counted on the images and expressed as percentage of apoptotic cells (green cells per total cells under bright field).
Statistical Analyses.
Data were analyzed by two-tailed Student t test or nonparametric u test as necessary. Two-way ANOVA was used for the analyses of the in vivo experiment with CAY10566 and PA-enriched diet. IHC staining analysis employed the Fisher exact test to compare the groups. Tumor weights and volumes from the subcutaneous mouse xenograft experiment were log transformed as paired samples before statistical analysis. P < 0.05 was considered statistically significant. All statistical analyses except for RNA-seq and lipidomics were performed using GraphPad Prism v8.0.
Ethics Declarations.
Human subject studies were conducted in accordance with the Declaration of Helsinki and approved by the Northwestern University Institutional Review Board (no. STU00202468). Animal studies were conducted according to a protocol approved by the Institutional Animal Care and Use Committee of Northwestern University (no. IS00008973).
Supplementary Material
Acknowledgments
This work was supported by grant R01 CA224275 to D.M. and J.X.C. and grant R33 CA223581 and NSF CHE1807106 grant to J.X.C. We thank Drs. Debabrata Chakravarti and Navdeep Chandel for valuable comments. Tumor specimens were procured through the Pathology Core, and sequencing was performed in the NUSeq Core supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. Lipidomics analysis was performed in Metabolite Profiling Facility at Bindley Bioscience Center, Purdue University. TEM was performed in the Center for Advanced Microscopy/Nikon Imaging Center (CAM) at Northwestern University supported by NCI grant CA060553. IncuCyte imaging was performed in the Analytical bioNanoTechnology Core Facility of the Simpson Querrey Institute for BioNanotechnology at Northwestern University (NSF grant ECCS-2025633). Flow cytometry analysis of ferroptosis was supported by the Northwestern University – Flow Cytometry Core Facility supported by Cancer Center Support Grant (NCI grant CA060553). This research was supported in part through the computational resources and staff contributions provided for the Quest high-performance computing facility at Northwestern University which is jointly supported by the Office of the Provost, the Office for Research, and Northwestern University Information Technology.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2203480119/-/DCSupplemental.
Data, Materials, and Software Availability
The RNA-seq data generated during the current study are available in the Gene Expression Omnibus repository (accession no. GSE192442) (51). OVCAR-5 SCD KD lipidomics profiling raw and significant lipids species list and OVCAR-5 SCD KD RNA-seq significant genes list are included in Dataset S2. All other study data are included in the article and/or supporting information.
References
- 1.Zhao G., Cardenas H., Matei D., Ovarian cancer—Why lipids matter. Cancers (Basel) 11, E1870 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ladanyi A., et al. , Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 37, 2285–2301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nieman K. M., et al. , Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 17, 1498–1503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network, Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peck B., Schulze A., Lipid desaturation—The next step in targeting lipogenesis in cancer? FEBS J. 283, 2767–2778 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Du X., et al. , FGFR3 stimulates stearoyl CoA desaturase 1 activity to promote bladder tumor growth. Cancer Res. 72, 5843–5855 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Roongta U. V., et al. , Cancer cell dependence on unsaturated fatty acids implicates stearoyl-CoA desaturase as a target for cancer therapy. Mol. Cancer Res. 9, 1551–1561 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Tesfay L., et al. , Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Res. 79, 5355–5366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng J. X., Xie X. S., Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 350, aaa8870 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Zhang D., Wang P., Slipchenko M. N., Cheng J. X., Fast vibrational imaging of single cells and tissues by stimulated Raman scattering microscopy. Acc. Chem. Res. 47, 2282–2290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J., et al. , Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell 20, 303–314.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan T. Z., et al. , CSIOVDB: A microarray gene expression database of epithelial ovarian cancer subtype. Oncotarget 6, 43843–43852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Z., Ferreira C. R., Virequ A. A., Cooks R. G., Multiple reaction monitoring profiling (MRM profiling): Small molecule exploratory analysis guided by chemical functionality. Chem. Phys. Lipids 235, 105048 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Surma M. A., et al. , Mouse lipidomics reveals inherent flexibility of a mammalian lipidome. Sci. Rep. 11, 19364 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsherniak A., et al. , Defining a cancer dependency map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghandi M., et al. , Next-generation characterization of the cancer cell line encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volmer R., van der Ploeg K., Ron D., Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. U.S.A. 110, 4628–4633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calfon M., et al. , IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Harding H. P., Zhang Y., Ron D., Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Harding H. P., et al. , Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Vriens K., et al. , Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature 566, 403–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hetz C., The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Wikstrom J. D., et al. , AMPK regulates ER morphology and function in stressed pancreatic β-cells via phosphorylation of DRP1. Mol. Endocrinol. 27, 1706–1723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gyorffy B., Lánczky A., Szállási Z., Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 19, 197–208 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Nakamura M. T., Nara T. Y., Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 24, 345–376 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Dória M. L., et al. , Fatty acid and phospholipid biosynthetic pathways are regulated throughout mammary epithelial cell differentiation and correlate to breast cancer survival. FASEB J. 28, 4247–4264 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Fritz V., et al. , Abrogation of de novo lipogenesis by stearoyl-CoA desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol. Cancer Ther. 9, 1740–1754 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ackerman D., et al. , Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep. 24, 2596–2605.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Southam A. D., et al. , Drug redeployment to kill leukemia and lymphoma cells by disrupting SCD1-mediated synthesis of monounsaturated fatty acids. Cancer Res. 75, 2530–2540 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Petrek J. A., Hudgins L. C., Ho M., Bajorunas D. R., Hirsch J., Fatty acid composition of adipose tissue, an indication of dietary fatty acids, and breast cancer prognosis. J. Clin. Oncol. 15, 1377–1384 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Chajès V., et al. , Fatty-acid composition in serum phospholipids and risk of breast cancer: An incident case-control study in Sweden. Int. J. Cancer 83, 585–590 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Tan Y., et al. , Metabolic reprogramming from glycolysis to fatty acid uptake and beta-oxidation in platinum-resistant cancer cells. Nature communications 13, 4554 (2022). [DOI] [PMC free article] [PubMed]
- 33.Yue S., et al. , Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 19, 393–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J., et al. , Abrogating cholesterol esterification suppresses growth and metastasis of pancreatic cancer. Oncogene 35, 6378–6388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang K. C., Li J., Zhang C., Tan Y., Cheng J. X., Multiplex stimulated Raman scattering imaging cytometry reveals lipid-rich protrusions in cancer cells under stress condition. iScience 23, 100953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowers M., et al. , FASN-dependent lipid metabolism links neurogenic stem/progenitor cell activity to learning and memory deficits. Cell Stem Cell 27, 98–109.e11 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Lu J., et al. , Palmitate causes endoplasmic reticulum stress and apoptosis in human mesenchymal stem cells: Prevention by AMPK activator. Endocrinology 153, 5275–5284 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Pinkham K., et al. , Stearoyl CoA desaturase is essential for regulation of endoplasmic reticulum homeostasis and tumor growth in glioblastoma cancer stem cells. Stem Cell Reports 12, 712–727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mujcic H., et al. , Hypoxic activation of the PERK/eIF2alpha arm of the unfolded protein response promotes metastasis through induction of LAMP3. Clin. Cancer Res. 19, 6126–6137 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Feng Y. X., et al. , Epithelial-to-mesenchymal transition activates PERK-eIF2α and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 4, 702–715 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Chen X., et al. , XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature 508, 103–107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pascual G., et al. , Dietary palmitic acid promotes a prometastatic memory via Schwann cells. Nature 599, 485–490 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Wang Y., et al. , Frizzled-7 identifies platinum-tolerant ovarian cancer cells susceptible to ferroptosis. Cancer Res. 81, 384–399 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Condello S., et al. , Tissue tranglutaminase regulates interactions between ovarian cancer stem cells and the tumor niche. Cancer Res. 78, 2990–3001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cardenas H., et al. , Interferon-γ signaling is associated with BRCA1 loss-of-function mutations in high grade serous ovarian cancer. NPJ Precis. Oncol. 3, 32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song M., et al. , IRE1α-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature 562, 423–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pang Z., Chong J., Li S., Xia J., MetaboAnalystR 3.0: Toward an optimized workflow for global metabolomics. Metabolites 10, E186 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin H., et al. , Microsecond fingerprint stimulated Raman spectroscopic imaging by ultrafast tuning and spatial-spectral learning. Nat. Commun. 12, 3052 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao G., et al. , Ovarian cancer cell fate is regulated by the balance between saturated and unsaturated fatty acids. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192442. Deposited 21 December 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated during the current study are available in the Gene Expression Omnibus repository (accession no. GSE192442) (51). OVCAR-5 SCD KD lipidomics profiling raw and significant lipids species list and OVCAR-5 SCD KD RNA-seq significant genes list are included in Dataset S2. All other study data are included in the article and/or supporting information.