Significance
Many viruses of importance to plant health rely on insect vectors for transmission, but how plant viruses interact with vectors to establish the well-adapted virus–vector combinations remains poorly understood. A detailed knowledge of the interactions will improve our understanding of virus–vector coevolution. Here, our study reveals that an insect vector protects itself against infection by a plant virus through the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway, whereas the virus maintains its transmission by inhibiting the insect’s JAK/STAT pathway, thus striking a balance between vector fitness and virus transmission. Our study sheds light on the virus–vector arms race and provides insights into how viruses and vectors can adapt to coexisting with each other.
Keywords: begomovirus, insect vector, JAK/STAT, virus transmission, whitefly
Abstract
Viruses pose a great threat to animal and plant health worldwide, with many being dependent on insect vectors for transmission between hosts. While the virus–host arms race has been well established, how viruses and insect vectors adapt to each other remains poorly understood. Begomoviruses comprise the largest genus of plant-infecting DNA viruses and are exclusively transmitted by the whitefly Bemisia tabaci. Here, we show that the vector Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway plays an important role in mediating the adaptation between the begomovirus tomato yellow leaf curl virus (TYLCV) and whiteflies. We found that the JAK/STAT pathway in B. tabaci functions as an antiviral mechanism against TYLCV infection in whiteflies as evidenced by the increase in viral DNA and coat protein (CP) levels after inhibiting JAK/STAT signaling. Two STAT-activated effector genes, BtCD109-2 and BtCD109-3, mediate this anti-TYLCV activity. To counteract this vector immunity, TYLCV has evolved strategies that impair the whitefly JAK/STAT pathway. Infection of TYLCV is associated with a reduction of JAK/STAT pathway activity in whiteflies. Moreover, TYLCV CP binds to STAT and blocks its nuclear translocation, thus, abrogating the STAT-dependent transactivation of target genes. We further show that inhibition of the whitefly JAK/STAT pathway facilitates TYLCV transmission but reduces whitefly survival and fecundity, indicating that this JAK/STAT-dependent TYLCV–whitefly interaction plays an important role in keeping a balance between whitefly fitness and TYLCV transmission. This study reveals a mechanism of plant virus–insect vector coadaptation in relation to vector survival and virus transmission.
Viruses are obligate intracellular pathogens that harness the host’s cellular resources during infection (1, 2). In the millions of years of what is often called a virus–host arms race, the hosts have developed highly sophisticated immune systems that protect themselves from virus attacks, and viruses have accordingly evolved diverse mechanisms that counteract the host immune responses (3–5). Although the critical role of arthropod vectors in mediating the host-to-host transmission of many viruses has been well recognized, the arms race between viruses and arthropod vectors remains poorly understood (6–8). Because the spread of arthropod-borne viruses is contingent upon the health of their vectors, it seems apparent that some adaptation between the virus and the vector must exist that ensures vector survival and facilitates virus transmission at the same time. However, the mechanisms underlying the adaptation between viruses and their arthropod vectors are largely unknown.
Begomovirus (family Geminiviridae), with over 400 species of plant-infecting DNA viruses, is the largest known genus that has caused serious crop losses worldwide (9, 10). These viruses are transmitted in a circulative manner by specific whitefly vectors of the Bemisia tabaci cryptic species complex (11, 12). During the past 30 y, with the global invasion of two species of the B. tabaci complex, Middle East Asia Minor 1 (previously biotype B) and Mediterranean (previously biotype Q), damages caused by begomoviruses have emerged as serious constraints to the cultivation of a variety of economically important crops (11, 13). Tomato yellow leaf curl virus (TYLCV) is one of the most devastating plant viruses affecting tomato crops worldwide (14). After acquisition from plant sap ingested by whiteflies, TYLCV first moves across the midgut wall into the hemolymph. From there, it translocates into the primary salivary glands (PSGs) and is finally secreted into the host plants during insect feeding (15, 16). TYLCV also invades the female ovaries and can be transmitted to whitefly offspring (17). In addition, TYLCV has been detected in the whitefly fat body and nervous system (18, 19). TYLCV can also replicate within whiteflies, which contributes to viral infectivity persistence (15, 20). Of note, infection by TYLCV results in a reduction of whitefly fecundity and longevity, suggesting that TYLCV has some features reminiscent of an insect pathogen (21). In response to TYLCV infection, the whitefly autophagy pathway is activated and participates in resistance to TYLCV (22). However, whether other immune pathways are also involved in protecting whiteflies against TYLCV infection is unclear. More importantly, whether and how TYLCV has evolved mechanisms that neutralize the vector’s antiviral immunity and promote virus transmission remain unknown.
Insects rely solely on innate immunity for protection against infection by pathogens, including viruses (23, 24). The Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway is an important innate immune signaling system that functions in insect antiviral defense by regulating the production of downstream effector molecules (24, 25). In Drosophila melanogaster, the JAK/STAT pathway is involved in resistance to many RNA viruses, including Drosophila C virus, Drosophila X virus, and cricket paralysis virus as well as a DNA virus invertebrate iridescent virus 6 (26–28). Activation of the canonical JAK/STAT pathway is initiated by the binding of secreted cytokines of the unpaired (UPD) family to the cell surface receptor domeless (DOME) that induces phosphorylation of the tyrosine kinase hopscotch (Hop/JAK). The activated Hop then phosphorylates DOME and forms the DOME/Hop complex to which the transcription factor STAT92E binds and undergoes phosphorylation with subsequent dimerization and translocation into the nucleus. There, it activates the expression of genes that regulate cell proliferation, stem cell renewal, development, and immunity (29, 30). In Culex quinquefasciatus cells, a secreted Vago restricts West Nile virus (WNV) infection by activating the JAK/STAT pathway (31). The JAK/STAT pathway in Aedes aegypti mosquitoes is also responsible for immune response against many flaviviruses, such as WNV, dengue, and yellow fever viruses (32, 33). However, little is known about the role of JAK/STAT signaling in the interactions between insect vectors and the plant viruses they transmit.
Here, we found that the JAK/STAT pathway plays an important role in protecting whiteflies against the deleterious effects of TYLCV by restricting its accumulation in whiteflies. Reciprocally, TYLCV can inhibit the whitefly JAK/STAT pathway as a means of achieving efficient transmission. Our studies also revealed that the TYLCV coat protein (CP) binds to B. tabaci STAT (BtSTAT) and blocks its nuclear translocation, resulting in the inhibition of BtSTAT-mediated transactivation. These results support an emergent mechanism of adaptation between TYLCV and its B. tabaci vector in mediating a balance between vector survival and virus transmission.
Results
Identification of BtSTAT as a TYLCV CP–Interacting Protein in Whiteflies.
To identify whitefly proteins that interact with TYLCV, we used TYLCV CP as a bait to screen the cDNA library of whiteflies in a yeast two-hybrid system. A 647-base pair (bp) fragment encoding partial STAT, which is the core transcription factor of the conserved JAK/STAT pathway in animals (34), was isolated. Yeast transformants with the plasmids pGBKT7-TYLCV CP and pGADT7-STAT were able to grow on quadruple dropout medium, whereas yeast transformants carrying two control constructs were unable to do so (SI Appendix, Fig. S1A).
After a BLAST search of the B. tabaci genome (35) and transcriptome (36) with the partial STAT sequence, the full-length open reading frame of BtSTAT was identified and cloned (GenBank accession number MN058988). Its 2,274 nucleotides encode a 757 amino acid protein with a predicted molecular mass of 85.8 kDa. Domain architecture analysis of BtSTAT showed the presence of four conserved STAT functional domains: the STAT_int, STAT_alpha, STAT_bind, and SH2 domains (SI Appendix, Fig. S2A) (30). The partial STAT sequence identified by yeast two-hybrid matched with deduced amino acids (position 127 to 343) encoding the entire STAT_alpha domain of full-length BtSTAT (SI Appendix, Fig. S2A). Phylogenetic analysis of protein sequences supported the identity of the cloned BtSTAT gene with those encoding other insects’ STAT (SI Appendix, Fig. S2B). We next investigated whether the full-length BtSTAT interacted with TYLCV CP using a glutathione S-transferase (GST) pull-down assay. His-tagged BtSTAT could bind to GST-fused TYLCV CP, but none bound to GST alone (SI Appendix, Fig. S1B), confirming the binding of full-length BtSTAT with TYLCV CP in vitro. The interaction of BtSTAT with TYLCV CP in viruliferous whiteflies was further confirmed by coimmunoprecipitation assays. A significantly higher amount of BtSTAT was detected in anti-TYLCV CP immunoprecipitates than in preimmune serum controls (SI Appendix, Fig. S1C).
BtSTAT Inhibits TYLCV Accumulation in Whiteflies.
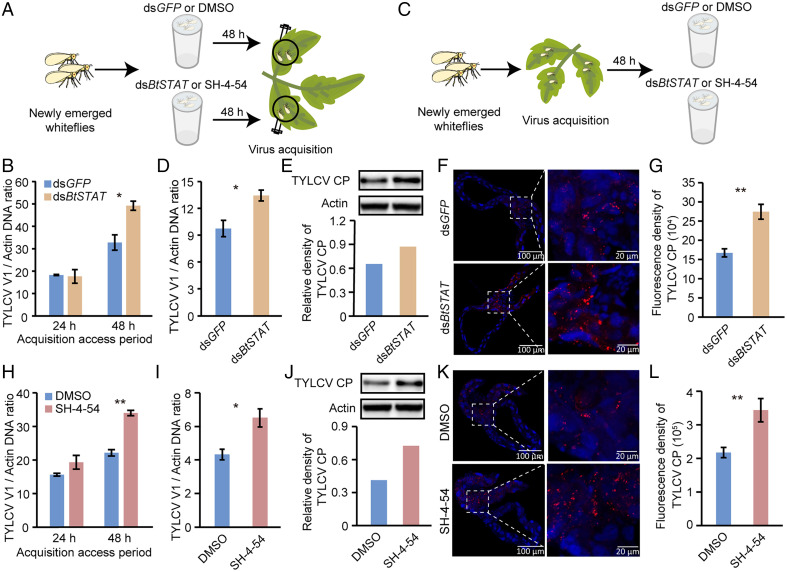
To investigate the role of BtSTAT in the interaction between TYLCV and whiteflies, we first examined the expression of BtSTAT in three different areas of the adult whitefly digestive and circulatory systems by qRT-PCR. The results showed that BtSTAT was expressed in all the tissues tested, with the highest levels in the midgut followed by the hemolymph and PSGs (SI Appendix, Fig. S2C). We then knocked down the expression of BtSTAT using RNA interference (RNAi). Whiteflies were first treated with double-stranded BtSTAT (dsBtSTAT) or green fluorescent protein (dsGFP) (control) for 48 h, causing a 36% reduction of BtSTAT expression compared with the dsGFP control (SI Appendix, Fig. S3 A and B), and were then transferred onto TYLCV-infected tomato plants for virus acquisition (Fig. 1A). qPCR analysis showed that the dsBtSTAT- or dsGFP-treated whiteflies acquired similar amounts of virus after a 24-h acquisition access period (AAP), suggesting that dsBtSTAT treatment had no negative effects on whitefly feeding behavior. However, the abundance of TYLCV DNA was significantly higher in dsBtSTAT-treated whiteflies than in the controls after a 48-h AAP (Fig. 1B), indicating that BtSTAT most likely had an inhibitory effect on TYLCV accumulation in whiteflies. To further test this hypothesis, whiteflies were first allowed to feed on TYLCV-infected tomato plants for 48 h. Next, the viruliferous whiteflies were divided randomly into two groups and treated with dsBtSTAT and dsGFP, respectively, for another 48 h (Fig. 1C). qPCR and Western blotting analyses revealed that the viral DNA and CP levels increased in the dsBtSTAT-treated whiteflies compared with the controls (Fig. 1 D and E and SI Appendix, Fig. S3 C and D). Immunofluorescence assays using the anti-TYLCV CP antibody further showed that the intensity of viral signal in the midguts of dsBtSTAT-treated whiteflies was 1.6-fold higher than that in the dsGFP-treated controls (Fig. 1 F and G).
Fig. 1.
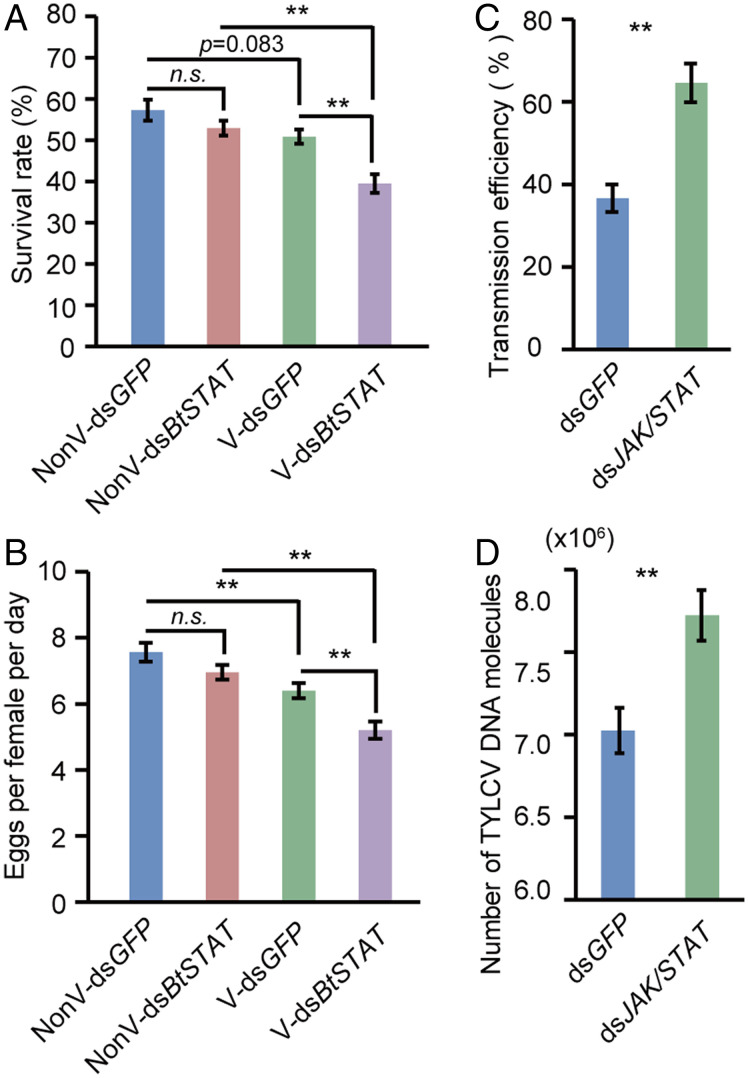
Effect of BtSTAT deficiency on virus accumulation in whiteflies. (A and B) Effect of BtSTAT silencing on virus acquisition by whiteflies. (B) Viral loads in whiteflies fed on TYLCV-infected plants for 24 h or 48 h after feeding with dsGFP or dsBtSTAT as determined by qPCR. (C–G) Effect of BtSTAT silencing on virus accumulation in whiteflies. (D and E) Quantitative analysis of TYLCV DNA (D) and immunoblotting analysis of TYLCV CP (E) in whiteflies that were fed with dsGFP or dsBtSTAT for 48 h following a 48-h AAP on TYLCV-infected plants. The relative densities of TYLCV CP were normalized with those of actin in E. (F) Immunofluorescence staining of TYLCV CP in midguts of dsGFP- or dsBtSTAT-treated whiteflies. (G) Fluorescence density of TYLCV CP signal in the whitefly midguts (F). (H) Quantitative analysis of viral DNA in whiteflies fed on TYLCV-infected plants for 24 h or 48 h after SH-4-54 or DMSO (control) treatment. (I and J) Quantitative analysis of TYLCV DNA (I) and immunoblotting analysis of TYLCV CP (J) in whiteflies that fed with SH-4-54 or DMSO for 48 h following a 48-h AAP on TYLCV-infected plants. The relative densities of TYLCV CP were normalized with those of actin in J. (K) Immunofluorescence staining of TYLCV CP in midguts of SH-4-54– or DMSO-treated whiteflies. (L) Fluorescence density of TYLCV CP signal in the whitefly midguts (K). Data in B, D, H, and I are shown as mean ± SEM from three independent experiments with 20 whiteflies in each replicate; *P < 0.05 and **P < 0.01 (independent-samples t test). In F and K, TYLCV CP was detected using a mouse anti-CP monoclonal antibody and goat anti-mouse IgG labeled with Dylight 549 (red) secondary antibody. Cell nuclei were stained with DAPI (blue). Images are representative of 16 whiteflies analyzed for each treatment. Data in G and L show mean ± SEM from 16 whiteflies; **P < 0.01 (nonparametric Mann–Whitney U test).
To ascertain the role of BtSTAT in TYLCV accumulation in whiteflies, we further inhibited BtSTAT by feeding whiteflies with the STAT phosphorylation inhibitor SH-4-54 (37). Whiteflies were first treated with SH-4-54 (prepared in dimethyl sulfoxide [DMSO]) or DMSO alone and then transferred onto TYLCV-infected tomato plants for virus acquisition. After a 24-h AAP, the amounts of virus were similar between the treatment and control groups, suggesting that the inhibitor did not affect whitefly feeding behavior. However, after a 48-h AAP, the amount of TYLCV was 1.5-fold higher in the inhibitor-treated whiteflies than in the controls (Fig. 1H), indicating that BtSTAT had an inhibitory effect on TYLCV accumulation in whiteflies. Feeding whiteflies with SH-4-54 following a 48-h AAP further confirmed the antiviral role of STAT in whiteflies. Compared with the controls, the virus DNA (Fig. 1I) and CP levels (Fig. 1J and SI Appendix, Fig. S3 E and F) greatly increased in SH-4-54–treated whiteflies, as revealed by qPCR and Western blotting. As expected, the intensity of viral signal was significantly higher in the midguts of SH-4-54–treated whiteflies than in the controls (Fig. 1 K and L). Together, these results demonstrated that BtSTAT inhibits TYLCV accumulation in whiteflies.
BtSTAT-Regulated Genes BtCD109-2 and BtCD109-3 Participate in Resistance to TYLCV in Whiteflies.
In many systems, the antiviral effect of the JAK/STAT pathway is mediated by STAT-regulated effector genes (38). To better understand the role of STAT in TYLCV–whitefly interactions, we sought to identify BtSTAT-regulated effector genes in whiteflies. Several STAT downstream genes that are involved in the response to immune challenge have previously been identified in D. melanogaster (Turandot A [TotA], TotC, TotM, virus-induced RNA 1 [Vir-1], and thiolester-containing protein 1 [Tep1]) and Ae. aegypti (dengue virus restriction factors DVRF1 and DVRF2) (28, 29, 32, 39–42) (SI Appendix, Table S1). After a search of the whitefly genome database (www.whiteflygenomics.org/), three CD109 antigen proteins (BtCD109-1 [Bta08339], BtCD109-2 [Bta03341], and BtCD109-3 [Bta09750]) were identified as B. tabaci orthologs of D. melanogaster Tep1, while no ortholog of the D. melanogaster Tot proteins or Vir-1 was found (SI Appendix, Table S1). Membrane magnesium transporter 1 (BtMgT1, Bta12654) was identified as an ortholog of Ae. aegypti DVRF1, and two cuticle proteins (BtCP67 [Bta12955] and BtLPCP23 [Bta13231]) were identified as the orthologs of Ae. aegypti DVRF2 in the B. tabaci genome (SI Appendix, Table S1). Domain architecture analysis of the deduced amino acid sequences of these B. tabaci genes showed the presence of conserved functional domains as their D. melanogaster and Ae. aegypti orthologs (SI Appendix, Fig. S4). In addition, at least two putative STAT-binding sites were predicted in the 2-kilobase (kb) promoter regions of these B. tabaci genes (SI Appendix, Table S1), indicating that they may be regulated by STAT.
To determine whether the six whitefly genes isolated above are regulated by BtSTAT, we examined their transcript levels after knocking down the expression of BtSTAT in nonviruliferous whiteflies. Compared with the dsGFP-treated group, the transcript levels of four genes (BtCD109-2, BtCD109-3, BtMgT1, and BtCP67) were significantly lower in dsBtSTAT-treated whiteflies (SI Appendix, Fig. S5A). We further tested the effect of STAT phosphorylation inhibition on the expression of these genes. After feeding whiteflies with the STAT phosphorylation inhibitor SH-4-54, the expression levels of the same four genes were reduced compared with the DMSO-treated control (SI Appendix, Fig. S5B). These results thus demonstrated that four of the six genes we identified are activated by BtSTAT in whiteflies.
To investigate whether BtSTAT directly activates the expression of these four genes, we performed dual luciferase assays. HEK293 cells were cotransfected with BtSTAT expression vector and the luciferase reporter construct that harbors the 2-kb promoter region of BtCD109-2 (BtCD109-22kb-Luc), BtCD109-3 (BtCD109-32kb-Luc), BtMgT1 (BtMgT12kb-Luc), or BtCP67 (BtCP672kb-Luc). Compared with cells transfected with empty expression vector, the BtCD109-22kb-Luc reporter activity increased by 3.5-fold, and the BtMgT12kb-Luc reporter activity increased by 1.5-fold in cells transfected with the expression vector for BtSTAT. By contrast, the reporter activities of BtCD109-32kb-Luc and BtCP672kb-Luc were not markedly changed by the overexpression of BtSTAT compared with the control (SI Appendix, Fig. S5 C and D). Therefore, of the four BtSTAT-regulated genes, BtCD109-2 and BtMgT1 are likely to be directly activated by BtSTAT.
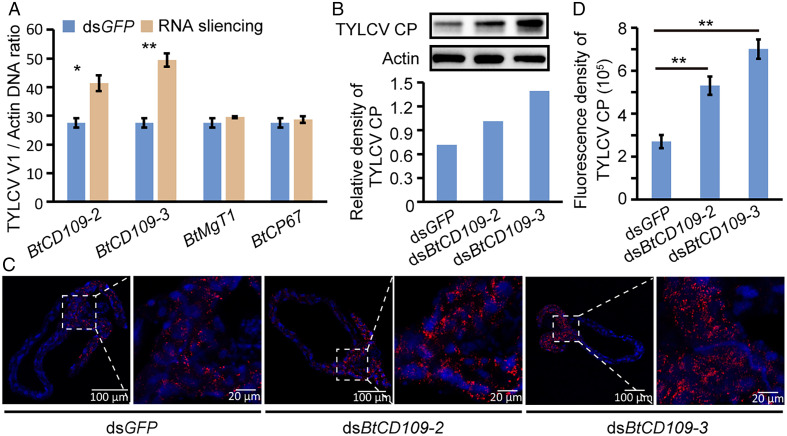
To determine whether the four BtSTAT-regulated genes participate in resistance to TYLCV in whiteflies, we knocked down the expression of these genes using RNAi. Whiteflies were first given a 48-h AAP on TYLCV-infected tomato plants and then treated with dsRNA corresponding to the four genes, respectively, for another 48 h. Compared with the control, the transcript levels of the four genes were significantly lower in the treatment groups (SI Appendix, Fig. S6A). Whereas the TYLCV DNA level was not markedly changed in BtMgT1- or BtCP67-depleted whiteflies, it was significantly higher in BtCD109-2- or BtCD109-3-depleted whiteflies than in the controls (Fig. 2A). To validate the antiviral role of BtCD109-2 and BtCD109-3, we further synthesized dsRNAs targeting other regions of these two genes and tested their effect on virus accumulation in whiteflies. Both dsRNAs led to a significant decrease of the transcript levels of the corresponding gene in whiteflies compared with the control group (SI Appendix, Fig. S6B). Consistent with the previous results, the amount of TYLCV DNA was significantly increased in the dsBtCD109-2- or dsBtCD109-3-treated whiteflies (SI Appendix, Fig. S6C), conforming the antiviral function of these two genes. The virus CP level in dsBtCD109-2- or dsBtCD109-3-treated whiteflies also increased, as determined by Western blotting using the anti-CP antibody (Fig. 2B and SI Appendix, Fig. S6 D and E). Immunofluorescence assays further showed that the intensity of viral signal in the midguts of BtCD109-2– or BtCD109-3–depleted whiteflies was higher than in the controls (Fig. 2 C and D). Taken together, these results suggest that the BtSTAT-regulated genes BtCD109-2 and BtCD109-3 mediate the antiviral effect of BtSTAT in whiteflies.
Fig. 2.
Effect of BtSTAT-regulated gene silencing on virus accumulation in whiteflies. (A and B) Quantitative analysis of TYLCV DNA (A) and immunoblotting analysis of TYLCV CP (B) in whiteflies that were fed with dsRNAs for 48 h following a 48-h AAP on TYLCV-infected plants. Data in A are represented as mean ± SEM from three independent experiments with 20 whiteflies in each replicate; *P < 0.05 and **P < 0.01 (independent-samples t test). The relative densities of TYLCV CP were normalized with those of actin in B. (C) Immunofluorescence staining of TYLCV CP in midguts of dsRNA-treated whiteflies. TYLCV CP was detected using a mouse anti-CP monoclonal antibody and goat anti-mouse IgG labeled with Dylight 549 (red) secondary antibody. Cell nuclei were stained with DAPI (blue). Images are representative of 16 whiteflies analyzed for each treatment. (D) Fluorescence density of TYLCV CP signal in the whitefly midguts (C). Data are shown as mean ± SEM from 16 whiteflies; **P < 0.01 (nonparametric Mann–Whitney U test).
The JAK/STAT Pathway Functions as an Antiviral Mechanism against TYLCV Accumulation in Whiteflies.
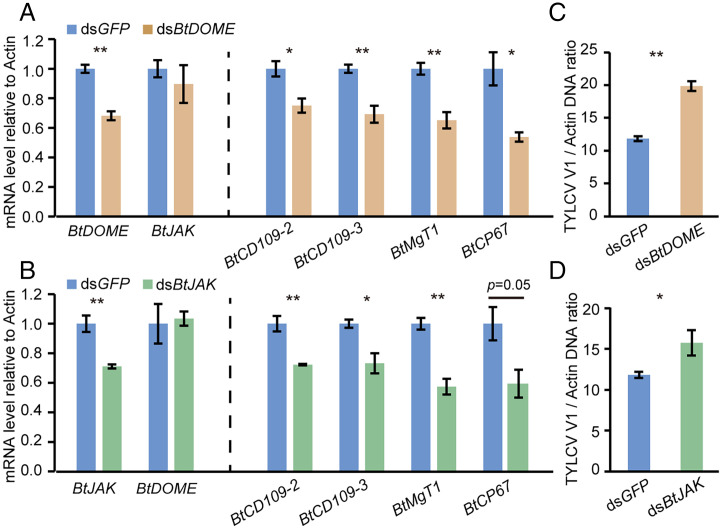
In insects, the most common mode of STAT activation is via tyrosine phosphorylation by phosphorylated DOME/JAK complex (25). To examine if the whitefly JAK/STAT pathway has a role in TYLCV accumulation in whiteflies, we identified whitefly DOME and JAK and silenced their expression using RNAi. The full-length genes DOME (Bta07497) and JAK (Bta12850) in the B. tabaci genome encode proteins of 1,131 and 1,138 amino acids, respectively, with high pairwise amino acid similarity to members of the DOME or JAK family from other organisms (SI Appendix, Figs. S7 and S8). Compared with dsGFP treatment, the transcript levels of BtDOME and the four BtSTAT-activated genes all decreased in dsBtDOME-treated nonviruliferous whiteflies (Fig. 3A). Similarly, the transcript levels of BtJAK and the four BtSTAT-activated genes were significantly lower in dsBtJAK-treated insects than in the controls (Fig. 3B). These results indicate that both BtDOME and BtJAK are required for the activation of STAT pathway in whiteflies. To assess the role of BtDOME and BtJAK in TYLCV accumulation, whiteflies were fed with the corresponding dsRNA following a 48-h AAP on TYLCV-infected tomato plants. The amount of TYLCV DNA was significantly higher in the dsBtDOME- or dsBtJAK-treated whiteflies than in the dsGFP treatments (Fig. 3 C and D), confirming that the JAK/STAT pathway functions as an antiviral mechanism against TYLCV accumulation in whiteflies.
Fig. 3.
Effect of JAK/STAT pathway inhibition on virus accumulation in whiteflies. (A and B) Relative mRNA levels of BtDOME, BtJAK, and JAK/STAT pathway downstream genes BtCD109-2, BtCD109-3, BtMgT1, and BtCP67 in whiteflies after dsRNA treatment. Data are represented as mean ± SEM from three independent experiments with 40 whiteflies in each replicate. (C and D) Quantitative analysis of viral DNA in whiteflies that were fed with dsRNA for 48 h following a 48-h AAP on TYLCV-infected plants. Data are represented as mean ± SEM from three independent experiments with 20 whiteflies in each replicate; *P < 0.05 and **P < 0.01 (independent-samples t test).
TYLCV Infection Suppresses the Whitefly JAK/STAT Pathway.
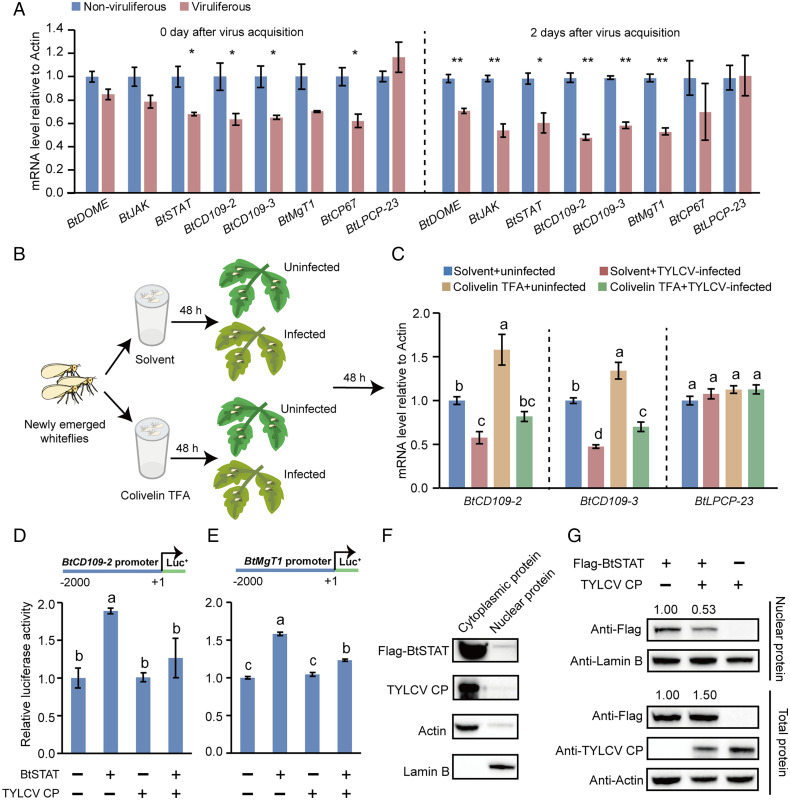
To investigate the effect of TYLCV infection on JAK/STAT signaling in whiteflies, we examined the transcript levels of BtDOME, BtJAK, BtSTAT, and the four BtSTAT-activated genes after TYLCV infection. Whiteflies were first fed on TYLCV-infected or uninfected tomato plants for 48 h and then transferred onto cotton, a TYLCV nonhost plant (20), to exclude the plant-mediated interactions between TYLCV and whiteflies (43). At the time of transfer (0 d after virus acquisition), the transcript levels of BtSTAT and three of the BtSTAT-activated genes (BtCD109-2, BtCD109-3, and BtCP67) were significantly lower in viruliferous whiteflies than in nonviruliferous whiteflies. At 2 d after virus acquisition, the expression of BtDOME and BtJAK was also down-regulated in viruliferous insects. Moreover, the majority of BtSTAT-activated genes still showed repressed expression in viruliferous insects after 2 d of feeding on cotton (Fig. 4A). By contrast, the transcript levels of BtLPCP-23, which is unregulated by BtSTAT (SI Appendix, Fig. S5 A and B), were always similar between viruliferous and nonviruliferous whiteflies (Fig. 4A).
Fig. 4.
TYLCV infection inhibits the JAK/STAT pathway. (A) Effect of TYLCV infection on the expression of BtDOME, BtJAK, BtSTAT, and BtSTAT-regulated genes BtCD109-2, BtCD109-3, BtMgT1, and BtCP67 in whiteflies as detected by qRT-PCR. The mRNA levels of BtLPCP-23, which is not regulated by BtSTAT, were detected in parallel as a control. Whiteflies were sampled at the time points as indicated after transfer onto cotton plants following a 48-h AAP on TYLCV-infected or uninfected tomato plants. Data are represented as mean ± SEM from three independent experiments with 40 whiteflies in each replicate; *P < 0.05 and **P < 0.01 (independent-samples t test). (B and C) Effect of TYLCV infection on the expression of BtSTAT-regulated genes BtCD109-2 and BtCD109-3 in colivelin TFA– or solvent (H2O)-treated whiteflies. The mRNA levels of BtLPCP-23 were detected in parallel as a control. Data are represented as mean ± SEM from three independent experiments with 40 whiteflies in each replicate; P < 0.05 (one-way ANOVA, least significant difference [LSD] test). (D and E) TYLCV CP suppresses BtSTAT-mediated transactivation. HEK293 cells were transfected with the expression vectors for TYLCV CP and/or BtSTAT together with the reporter construct BtCD109-22kb-Luc (D) or BtMgT12kb-Luc (E). Treatments with empty expression vector served as controls. A Renilla luciferase reporter construct was cotransfected in each well as an internal reference. Data represent normalized luciferase activity (firefly/Renilla). Data represent mean ± SEM from three independent experiments; P < 0.05 (one-way ANOVA, LSD test). (F and G) TYLCV CP blocks BtSTAT nuclear translocation. (F) Immunoblotting analysis of Flag-BtSTAT, TYLCV CP, actin, and lamin B in the cytoplasm and nucleus of HEK293 cells coexpressing Flag-BtSTAT and TYLCV CP. (G) HEK293 cells were transfected with the expression vectors for TYLCV CP and/or BtSTAT. Total proteins and nuclear proteins were extracted and analyzed by immunoblotting using Flag, TYLCV CP, actin, and lamin B antibodies. Lamin B served as a loading control for nuclear protein, while actin served as a loading control for total protein.
Based on the above results, we hypothesized that TYLCV infection specifically suppresses the whitefly JAK/STAT pathway. To verify this hypothesis, we activated JAK/STAT signaling by feeding nonviruliferous whiteflies with colivelin TFA, a STAT activator (44). The mRNA levels of four BtSTAT-activated genes were significantly higher in colivelin TFA–treated insects than in the solvent (H2O)-treated controls (SI Appendix, Fig. S9), confirming activation of the JAK/STAT pathway. The colivelin TFA–treated whiteflies were then given a 48-h AAP on TYLCV-infected or uninfected tomato plants (Fig. 4B), and the expression levels of two BtSTAT-activated genes (BtCD109-2 and BtCD109-3) and an unregulated gene (BtLPCP-23) were determined by qRT-PCR. Both the basal and the colivelin TFA–induced expression of BtCD109-2 and BtCD109-3 were repressed by the acquisition of TYLCV; however, the transcript levels of BtLPCP-23 were comparable among all treatments (Fig. 4C), clearly demonstrating that TYLCV infection specifically suppresses the whitefly JAK/STAT pathway.
TYLCV CP Attenuates BtSTAT-Mediated Transactivation by Blocking BtSTAT Nuclear Translocation.
Given that TYLCV CP interacts with BtSTAT (SI Appendix, Fig. S1), we thus investigated whether TYLCV CP could affect BtSTAT-mediated transactivation by performing luciferase assays with the two BtSTAT-responsive reporter constructs BtCD109-22kb-Luc and BtMgT12kb-Luc. As controls, the two BtSTAT-nonresponsive constructs, BtCD109-32kb-Luc and BtCP672kb-Luc, were tested in parallel. HEK293 cells were transfected with the desired luciferase reporter construct and the expression vectors for BtSTAT and TYLCV CP. The successful expression of BtSTAT and TYLCV CP in HEK293 cells was confirmed by Western blotting using the anti-BtSTAT and anti-TYLCV CP antibodies (SI Appendix, Fig. S10 A and B). As expected, the activity of the BtCD109-22kb-Luc reporter was significantly induced by the overexpression of BtSTAT. Overexpression of TYLCV CP alone did not markedly affect the basal activity of the reporter gene. However, the induction of reporter activity by BtSTAT was drastically reduced in cells coexpressed with BtSTAT and TYLCV CP. Similar results were obtained with the BtMgT12kb-Luc reporter construct (Fig. 4 D and E). Conversely, the activities of the two BtSTAT-nonresponsive reporter constructs were always similar under all these treatments (SI Appendix, Fig. S10 C and D). These data indicate that TYLCV CP specifically inhibits the BtSTAT-mediated transactivation.
STAT nuclear translocation following activation is critical for the transactivation of its target genes (30). Cellular distribution analysis of TYLCV CP and BtSTAT in HEK293 cells showed that nearly all TYLCV CP and most of BtSTAT were localized in the cytoplasm (Fig. 4F). We thus tested whether the interaction between TYLCV CP and BtSTAT affects the nuclear translocation of BtSTAT. Western blotting assays revealed that while the total amount of BtSTAT was even higher, the amount of BtSTAT in the nucleus was much lower in cells coexpressing TYLCV CP and BtSTAT than in cells expressing BtSTAT alone (Fig. 4G). These results suggest that BtSTAT’s interaction with TYLCV CP likely blocks its nuclear translocation, thus inhibiting the BtSTAT-mediated transactivation.
The JAK/STAT Pathway Promotes Whitefly Fitness under TYLCV Infection.
It has been shown that infection by TYLCV is associated with a reduction in whitefly fecundity and longevity, indicating that TYLCV has some features reminiscent of an insect pathogen (21). Considering that the JAK/STAT pathway imposes a negative effect on TYLCV accumulation, we then asked whether it promotes whitefly fitness in the presence of TYLCV. To answer this question, we examined the effect of BtSTAT knockdown on whitefly survival and fecundity. Whiteflies were treated with dsBtSTAT following a 48-h AAP on TYLCV-infected or uninfected tomato plants. Consistent with the previous study (21), TYLCV infection caused a decrease of whitefly fitness. Notably, whereas the survival and fecundity of nonviruliferous whiteflies were not markedly affected by dsBtSTAT treatment, those of viruliferous insects were significantly decreased after dsBtSTAT treatment compared with those of the dsGFP control (Fig. 5 A and B and SI Appendix, Fig. S11A). These data indicate that the JAK/STAT pathway promotes the fitness of the viruliferous whiteflies by suppressing virus accumulation.
Fig. 5.
Effect of inhibiting the JAK/STAT pathway on whitefly fitness and virus transmission. (A) Survival rate of viruliferous (V) and nonviruliferous (NonV) whiteflies feeding on an artificial diet containing dsGFP or dsBtSTAT for 6 d. Data represent four biological replicates with 100 adults in each replication. (B) Egg numbers laid per female per day on cotton by dsGFP- or dsBtSTAT-treated whiteflies. Data represent 24 biological replicates with 4 females in each replication. (C and D) Effect of inhibiting JAK/STAT pathway on virus transmission. The percentage of test plants with virus genomic DNA (C) and absolute quantification of TYLCV DNA molecules in plants (D) at 30 d after inoculation by dsGFP- or dsJAK/STAT (a mixture of dsBtJAK and dsBtSTAT)-treated whiteflies. Eight to 10 plants were used per replicate, and three replicates were used to calculate the disease incidence rate and viral DNA copies. Data in A to D represent mean ± SEM; *P < 0.05 and **P < 0.01 (independent-samples t test).
Inhibiting the Whitefly JAK/STAT Pathway Facilitates TYLCV Transmission.
A biological relevance of the inhibition of JAK/STAT signaling by TYLCV could be the impact that it has on virus transmission. We therefore investigated whether TYLCV transmission was affected by inhibiting the whitefly JAK/STAT pathway. Whiteflies fed with dsBtSTAT following a 48-h AAP were used for virus inoculation to target plants. At 30 d after inoculation, we first examined the virus infection rate by PCR, a classical and economical method for virus qualitative analysis in plants (45). The TYLCV infection rate in tomato plants was slightly increased (by about 15%) in the dsBtSTAT treatment (SI Appendix, Fig. S11B). We then analyzed the amount of viral DNA molecules in plants using the more sensitive qPCR method. The amount of TYLCV DNA in plants was significantly elevated compared with that of the dsGFP control (SI Appendix, Fig. S11 C and D). To achieve a thorough inhibition of the JAK/STAT pathway, viruliferous whiteflies used to transmit TYLCV were fed with a mixture of dsBtJAK and dsBtSTAT (1:1) to knockdown the expression of BtJAK and BtSTAT simultaneously. The TYLCV infection rate in the dsJAK/STAT treatment was 28% higher than that of the control (Fig. 5C), and the amount of TYLCV DNA in plants was increased in the dsJAK/STAT treatment (Fig. 5D). These results indicate that inhibition of whitefly JAK/STAT signaling by TYLCV facilitates its transmission.
Discussion
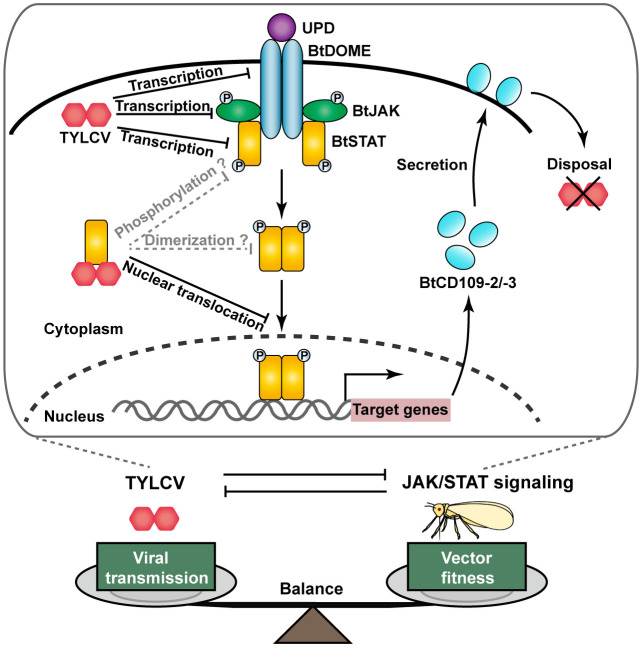
During the long-term evolution, mammals and plants have developed highly sophisticated immune systems that protect themselves from virus attack, whereas viruses have evolved diverse mechanisms capable of counteracting the host immune responses, showing a fierce arms race between viruses and their hosts (3–5). However, whether such an arms race has occurred between viruses and their insect vectors and, if so, the underlying mechanisms and the significance to virus spread remain largely unknown. Our results showed that the JAK/STAT signaling pathway protects whiteflies from TYLCV infection through two BtSTAT-activated antiviral effector genes, BtCD109-2 and BtCD109-3, whereas TYLCV has acquired strategies that inhibit the whitefly JAK/STAT pathway, thus ensuring its own transmission, finally leading to a balance between vector fitness and virus transmission (Fig. 6). Therefore, this coevolutionary arms race between TYLCV and whiteflies led to an adaptation between the plant DNA virus and its insect vector, which may help to explain the global spread of this devastating virus.
Fig. 6.
Model depicting the JAK/STAT-dependent TYLCV–whitefly arms race. Activation of the JAK/STAT pathway is initiated by the binding of secreted cytokines of the UPD family to the cell surface receptor BtDOME that induces phosphorylation of the tyrosine kinase BtJAK. The activated BtJAK then phosphorylates BtDOME and forms the BtDOME/BtJAK complex that serves as docking sites for the transcription factor BtSTAT, which then is phosphorylated and dimerizes and translocates into the nucleus where it activates the expression of antiviral effector genes (BtCD109-2/BtCD109-3). Infection with TYLCV inhibits the JAK/STAT pathway by reducing the transcription levels of BtDOME, BtJAK, and BtSTAT. Moreover, TYLCV CP binds to BtSTAT and abrogates the BtSTAT-mediated transactivation of target genes by blocking BtSTAT nuclear translocation. Whether the interaction between TYLCV CP and BtSTAT blocks the phosphorylation and/or dimerization of BtSTAT in whiteflies remains to be determined. This JAK/STAT-dependent TYLCV–whitefly interaction plays an important role in keeping a balance between vector fitness and virus transmission.
In mammals, induction of the JAK/STAT signaling pathway by interferons (IFNs) gives rise to the expression of hundreds of genes, many of which have the ability to inhibit and kill the invading viruses (46, 47). In insects, the JAK/STAT pathway was best characterized in fly and mosquito models, and several downstream effector genes involved in the response to immune challenge have been identified. In D. melanogaster, the JAK/STAT pathway induces the expression of several members of the Tot gene family (TotA, TotC, and TotM) upon septic injury (29, 39). During Drosophila C virus infection, the JAK/STAT pathway is activated and results in the up-regulation of Vir-1 (28). Tep1 was also shown to be up-regulated by the JAK/STAT pathway in D. melanogaster and is involved in antimicrobial defenses in Anopheles gambiae (40–42). In addition, dengue virus infection in Ae. aegypti activates the JAK/STAT pathway. In turn, the JAK/STAT pathway controls dengue virus infection by inducing the expression of DVRF1 and DVRF2 (32). To identify BtSTAT-regulated effector genes involved in resistance to TYLCV, we searched the whitefly genome for orthologs of those STAT downstream genes. No ortholog of D. melanogaster TotA, TotC, TotM, or Vir-1 was found in the whitefly genome. A previous study found that the Drosophila Tot genes are not similar to known genes from other organisms, indicating that this gene family may be limited to Drosophila species or may undergo rapid evolution, and homologs are therefore difficult to find (48). By contrast, three whitefly orthologs of D. melanogaster Tep1 were identified, and two of them (BtCD109-2 and BtCD109-3) are activated by BtSTAT and responsive to TYLCV infection. More importantly, both BtCD109-2 and BtCD109-3 participate in resistance to TYLCV in whiteflies. In D. melanogaster, the Tep family is composed of 6 genes named Tep1 to Tep6 (49). A comparison of the amino acid sequences of BtCD109s and Drosophila Teps revealed that BtCD109-1 is more similar to Tep2, whereas BtCD109-2 and BtCD109-3 are more similar to Tep6 (SI Appendix, Table S4). It has been shown that the expression of Tep6 is induced by Photorhabdus infection and is involved in the Drosophila antibacterial immune response (50). The mammalian CD109s are glycosylphosphatidylinositol (GPI)-anchored proteins (51). Analysis with several online prediction programs revealed that both BtCD109-2 and BtCD109-3 are likely to be GPI-anchored proteins (SI Appendix, Table S5). The vertebrate orthologs of insect Teps comprise the universal protease inhibitors α2-macroglobulins and the complement factors C3/C4/C5, which are involved in labeling pathogens, including viruses, and promoting their destruction through phagocytosis or cell lysis (42, 52). Therefore, BtCD109-2 and BtCD109-3 may recognize TYLCV and induce virus disposal in whiteflies, although future investigations are needed to confirm this hypothesis. As to the whitefly orthologs of Ae. aegypti DVRF1 (BtMgT1) and DVRF2 (BtCP67 and BtLPCP23), BtMgT1 and BtCP67 are regulated by BtSTAT; however, none of them were involved in resistance to TYLCV in whiteflies. Thus, the antiviral activity of these two genes seems to be virus specific.
Our study showed that inhibition of the JAK/STAT pathway could facilitate TYLCV transmission by whiteflies. However, the TYLCV infection rate in tomato plants was only slightly increased (by about 15%) in the dsBtSTAT treatment when analyzed by classical PCR. Further analysis by the more sensitive qPCR approach revealed a significant increase (by 34%) in virus amount of test plants in the dsBtSTAT treatment compared with that of the dsGFP control. According to our results, the JAK/STAT pathway is already partially inhibited by TYLCV in viruliferous whiteflies, which may be responsible, at least in part, for the modest effects of further interfering this pathway on virus transmission by whiteflies. In addition, the limited effect of silencing BtSTAT on the virus transmission capacity of whiteflies implies that other mechanisms may be involved in inhibiting viral infection in whiteflies. Indeed, a previous study has proven that infection with TYLCV activates the autophagy pathway in whiteflies, which leads to the subsequent degradation of TYLCV (22). Thus, at least two mechanisms are involved in inhibiting TYLCV infection in whiteflies. The RNAi pathway is considered an important antiviral defense mechanism in Drosophila and mosquitoes (53, 54). In virus-infected plants, RNA silencing also functions as a powerful mechanism against TYLCV infection (55, 56). Therefore, the RNAi pathway may also inhibit TYLCV infection in whiteflies. In addition, TYLCV infection in whiteflies could activate the apoptosis pathway resulting in enhanced virus transmission (57). Interestingly, the JAK/STAT pathway could positively regulate an apoptosis inhibitor, leading to the suppression of stress-induced apoptosis in Drosophila (58, 59). Similar cross-talk among innate immunity pathways for countering pathogen infection or maintaining an immune homeostasis has been found (60–62). Further studies are needed to decipher whether and how the whitefly JAK/STAT pathway interacts with other immune signaling pathways.
In addition to its critical role in immune response, JAK/STAT signaling is essential for numerous developmental and homeostatic processes, including hematopoiesis, stem cell maintenance, organismal growth, and mammary gland development in vertebrates (34). Similarly, this pathway functions in the regulation of cellular proliferation and oogenesis in D. melanogaster, including generation of the somatic niche and patterning of follicle cells (30). Inhibition of the whitefly JAK/STAT pathway by TYLCV infection suggests that a basal activity of JAK/STAT signaling is present in nonviruliferous whiteflies and may be required for whitefly development and reproduction. The reduced activity of this pathway may be responsible, at least in part, for the decreased fecundity and longevity in TYLCV-infected whiteflies (21).
Components of the JAK/STAT pathway involved in antiviral responses are targets of antagonism by mammalian viruses (63). The majority of viruses that impair JAK/STAT signaling have acquired mechanisms of STAT1 and STAT2 antagonism, including degradation of STATs through the proteasome, blocking phosphorylation of STATs, sequestration of STATs in high-molecular–weight complexes and prevention of nuclear translocation of STATs (63). For instance, the Nipah virus–encoded V protein binds tightly with both STAT1 and STAT2 in the cytoplasm. As a result, the proteins are retained in the cytoplasm, preventing both IFN-induced STAT activation and nuclear translocation (64). The C protein of Sendai virus binds to the N-terminal domain of STAT1 and interferes with the domain arrangement of the STAT1 dimer, leading to the formation of high-molecular–weight complexes and the accumulation of phosphorylated STAT1 in the cytoplasm (65). In insects, although the important role of JAK/STAT pathways in antiviral responses has been well established, whether and how viruses have evolved mechanisms suppressing this pathway remain largely unexplored. Here, we found that a plant DNA virus, TYLCV, has acquired the ability to inhibit whitefly JAK/STAT signaling by targeting STAT. TYLCV CP binds to BtSTAT and inhibits its nuclear translocation, resulting in down-regulation of BtSTAT-activated effector genes. These results not only indicate that virus has also evolved tactics for manipulating the JAK/STAT signaling of its insect vector but also suggest that plant viruses may utilize similar mechanisms as mammalian viruses to impair the host/vector’s JAK/STAT pathway. Nevertheless, whether the interaction between TYLCV CP and BtSTAT blocks the phosphorylation and/or dimerization of BtSTAT in whiteflies remains to be determined.
Notably, in addition to the down-regulation of BtSTAT-activated downstream genes, a significant decrease in the transcript levels of BtDOME, BtJAK, and BtSTAT was observed in TYLCV-infected whiteflies, indicating that TYLCV also inhibits JAK/STAT signaling by reducing the expression levels of these key components. Similarly, influenza A viruses impair JAK/STAT signaling in HeLa cells in part by reducing IFN receptor expression at the transcriptional level by the nonstructural protein 1 (66). Adenovirus type 5 was shown to inhibit JAK/STAT signaling by decreasing JAK1 mRNA levels in human tracheobronchial epithelial cells (67). It would be very interesting to reveal how TYLCV disrupts BtDOME, BtJAK, and BtSTAT at the mRNA level in the whitefly vectors. Given the limited knowledge on viral inhibition of insect vector immune responses, the results from future studies in this regard will not only shed new light on the understanding of TYLCV–whitefly interactions but also provide novel insights into the interactions between other insect vectors and the animal/plant viruses that they transmit.
Materials and Methods
GST Pull-down, Coimmunoprecipitation, and Western Blotting Assays.
For the GST pull-down assay, the fragment of TYLCV CP was amplified and cloned into pGEX-6p-1 for fusion with GST. The recombinant protein was expressed in Escherichia coli strain BL21 (DE3). The GST–TYLCV CP was incubated with glutathione Sepharose beads (GE Healthcare) for 2 h at 4 °C, the mixture was centrifuged for 5 min at 100 × g, and the supernatants were discarded. After being centrifuged and washed five times with phosphate-buffered saline (PBS) buffer, the beads were used for the pull-down assay. The protein extracts of S2 cells expressing His-BtSTAT were added to the beads and gently mixed for 4 h at 4 °C. After being centrifuged and washed five times with PBS, the bead-bound proteins were eluted by boiling in protein loading buffer for 5 min and then separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and detected by anti-His antibody (Abcam, ab213204).
For coimmunoprecipitation assays, whiteflies were first given a 7-d virus AAP, and then soluble proteins were extracted by cell lysis buffer, as described in the manufacturer’s instructions (Invent Biotechnologies). Two micrograms of anti-TYLCV CP monoclonal antibody (68) or preimmune sera (control sera) were incubated overnight at 4 °C with 500 μL of whitefly soluble protein extracts. The protein extracts were added to 50 μL of Protein G Sepharose 4 Fast Flow suspension (GE Healthcare) and gently mixed for 4 h at 4 °C. After being centrifuged and washed five times with PBS, the bead-bound proteins were eluted by boiling in protein loading buffer for 5 min, separated by SDS–PAGE, and detected by anti-BtSTAT antibody.
For detection of proteins extracted from whiteflies, S2 cell or HEK293 cell protein samples were separated using 4 to 20% SDS–PAGE and then transferred onto polyvinylidene fluoride membranes. The membrane was blocked with 5% nonfat milk in Tris-buffered saline (10 mM Tris-HCl and 150 mM sodium chloride, pH 7.5) with 0.1% Tween-20 and then incubated with anti-TYLCV CP, anti-β-actin (EarthOx E021020-03), anti-BtSTAT, anti-GST (CST, 2624S), anti-His, or anti-Flag antibody (Sigma-Aldrich, F1804). After incubation with horseradish peroxidase–conjugated secondary antibody, blots were visualized with the ECL Plus Detection system (Bio-Rad).
Gene Silencing by Oral Ingestion of DsRNA.
DsRNA was synthesized using the T7 high-yield RNA transcription kit (Vazyme) following the manufacturer’s instructions. The synthesized dsRNA was purified via phenol–chloroform precipitation and resuspended in nuclease-free water, and the concentration of dsRNA was determined by spectrophotometry using the NanoDrop 2000 system (Thermo Fisher Scientific). The quality of dsRNA was verified by electrophoresis on a 2% agarose gel. Gene silencing was performed as previously described (20). Briefly, dsRNA was diluted into 15% (wt/vol) sucrose solution at the concentration of 250 ng/µL. Approximately 100 adult whiteflies were released into glass tubes with a diameter of 1.5 cm and a length of 10 cm. One opening of the tube was covered with double layers of parafilm filled with a diet solution containing dsRNA and the other was covered with gauze. After a 48-h feeding, groups of 20 whiteflies were collected for DNA extraction, groups of 40 whiteflies for RNA extraction, and groups of 100 whiteflies for protein extraction.
Protein Expression in Cells and Luciferase Assay.
Drosophila Schneider S2 cells were maintained at 27 °C in Drosophila serum-free medium supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 1% penicillin–streptomycin (Gibco). The HEK293 cells were cultured in Dulbecco's modified Eagle medium/high-glucose medium (Gibco) containing 10% fetal bovine serum (Gibco) and 1% penicillin–streptomycin (Gibco) at 37 °C in a 5% CO2 incubator. pAc5.1/V5-His B vector (Invitrogen) was used for the expression of full-length BtSTAT in S2 cells. pcDNA3.1 vector (Invitrogen) was used for the expression of full-length BtSTAT and that of TYLCV CP in HEK293 cells. The promoter regions of BtCD109-2, BtCD109-3, BtMgT1, and BtCP67 were subcloned into pGL3-Basic vector (Promega) to construct the reporter plasmids. For pull-down assays, S2 cells were seeded in a six-well plate at 1 × 106 cells per well and transfected 12 h later. At 72 h posttransfection, the cells were harvested, and total proteins were extracted for the pull-down assay (Invent Biotechnologies). For the luciferase assay, HEK293 cells were seeded in a 24-well plate at 2 × 105 cells per well and transfected 12 h later. At 48 h posttransfection, the cells were harvested and processed with the dual-luciferase reporter assay kit (Vazyme) according to the manufacturer’s protocol. All transfections were conducted using Lipofectamine 3000 (Invitrogen). The luciferase activities were measured by the FlexStation-3 microplate reader (Molecular Devices).
To examine the impact of TYLCV CP on BtSTAT’s nuclear translocation, HEK293 cells were seeded in a six-well plate at 106 cells per well and transfected with the expression vectors for Flag-BtSTAT and TYLCV CP 12 h later. At 48 h posttransfection, the cells were harvested, and the total protein (Invent Biotechnologies), nuclear protein, and cytoplasmic protein (Thermo Scientific) were extracted, respectively, for Western blotting assays.
STAT Inhibitor and Activator Treatment.
The STAT inhibitor SH-4-54 (37) was used to inhibit BtSTAT. A 40-mM SH-4-54 stock solution was prepared by dissolving SH-4-54 powder (MCE) in DMSO. A 40-µM SH-4-54 working solution was prepared by dissolving 2 µL of SH-4-54 stock solution in 2 mL of 15% sucrose solution immediately before use. A control (DMSO) working solution was prepared by dissolving 2 µL DMSO in 2 mL of 15% sucrose solution. For virus acquisition analysis, ∼100 newly emerged whiteflies were released into each feeding chamber to feed on the SH-4-54 or DMSO solution for 48 h. Afterward, the whiteflies were confined to clip cages on two opposite leaves of a TYLCV-infected tomato plant for 24 and 48 h (Fig. 1A), and groups of 20 whiteflies were collected for virus quantitative analysis. For virus accumulation analysis, newly emerged whiteflies were first fed on a TYLCV-infected tomato plant for 48 h and then randomly divided into groups of ∼100 insects for another 48-h feeding of the SH-4-54 or DMSO solution (Fig. 1C). Subsequently, groups of 20 whiteflies were collected for virus quantitative analysis, and groups of 100 whiteflies were collected for Western blotting.
The STAT activator colivelin TFA (44) was used to activate BtSTAT. A 5-mM colivelin TFA stock solution was prepared by dissolving colivelin TFA powder (MCE) in distilled water. A 10-µM colivelin TFA working solution was prepared by dissolving 2 µL of colivelin TFA stock solution in 1 mL of 15% sucrose solution immediately before use. A control (solvent) working solution was prepared by adding 2 µL of water into 1 mL of 15% of sucrose solution. Approximately 100 newly emerged whiteflies were released into each feeding chamber to feed on the colivelin TFA solution or solvent solution for 48 h. The treated whiteflies were allowed to feed on TYLCV-infected or uninfected tomato plants for another 48 h, and then groups of 40 whiteflies were collected for detection of gene expression levels.
Analysis of Whitefly Survival and Fecundity.
Newly emerged whiteflies were first given a 48-h AAP on TYLCV-infected or uninfected tomato plants. For whitefly survival analysis, groups of 100 whiteflies were randomly collected and fed with 15% sucrose solution containing 250 ng/μL dsBtSTAT or dsGFP for 6 d in feeding chambers. Whitefly survival was recorded every 2 d. Four replicates were conducted for each treatment, and the sucrose solution was changed every 2 d. For the fecundity analysis, the viruliferous or nonviruliferous whiteflies were fed with the above dsRNA preparations for 48 h, and four female adults were released into a clip cage secured to the abaxial surface of a cotton plant leaf. Two days later, the numbers of eggs laid on the leaf were counted under a dissection microscope.
Transmission of TYLCV to Plants by Whiteflies.
Whiteflies fed with dsRNA for 48 h (following a 48-h AAP on TYLCV-infected tomato plants) were collected in groups of four (female:male = 1:1). Each group of whiteflies was confined to a clip cage secured to the top second leaf of an uninfected tomato plant at the 3- to 4-true–leaf stage (ca. 3 wk after sowing) for a 72-h inoculation access period. The plants were sprayed with imidacloprid at a concentration of 20 mg/L and maintained until symptoms had developed. The top fully expanded leaf of tomato plants was harvested at 30 d after whitefly inoculation, and the genomic DNA was then extracted. The virus infection status of the test plants was determined by PCR. The amount of virus DNA in the test plants was determined by qPCR.
Supplementary Material
Acknowledgments
Financial support for this paper was provided by the National Key Research and Development Program (2021YFC2600100), the National Natural Science Foundation of China (31925033), and the Hainan Major Science and Technology Project (ZDKJ2021007). We thank Ms. Rong Jin from Zhejiang University for greenhouse management.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122099119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Weaver S. C., Charlier C., Vasilakis N., Lecuit M., Zika, chikungunya, and other emerging vector-borne viral diseases. Annu. Rev. Med. 69, 395–408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones R. A. C., Naidu R. A., Global dimensions of plant virus diseases: Current status and future perspectives. Annu. Rev. Virol. 6, 387–409 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann H.-H., Schneider W. M., Rice C. M., Interferons and viruses: An evolutionary arms race of molecular interactions. Trends Immunol. 36, 124–138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao S., Wu Y., Wu J., Arms race between rice and viruses: A review of viral and host factors. Curr. Opin. Virol. 47, 38–44 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Ramesh S. V., Sahu P. P., Prasad M., Praveen S., Pappu H. R., Geminiviruses and plant hosts: A closer examination of the molecular arms race. Viruses 9, 256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray S. M., Banerjee N., Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63, 128–148 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia D., et al. , Vector mediated transmission of persistently transmitted plant viruses. Curr. Opin. Virol. 28, 127–132 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Huang Y.-J. S., Higgs S., Horne K. M., Vanlandingham D. L., Flavivirus-mosquito interactions. Viruses 6, 4703–4730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeske H., Geminiviruses. Curr. Top. Microbiol. Immunol. 331, 185–226 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Walker P. J., et al. , Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch. Virol. 164, 2417–2429 (2019). [DOI] [PubMed] [Google Scholar]
- 11.De Barro P. J., Liu S. S., Boykin L. M., Dinsdale A. B., Bemisia tabaci: A statement of species status. Annu. Rev. Entomol. 56, 1–19 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Rosen R., et al. , Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol. 15, 1–8 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Gilbertson R. L., Batuman O., Webster C. G., Adkins S., Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2, 67–93 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Scholthof K.-B. G., et al. , Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X. W., Blanc S., Insect transmission of plant single-stranded DNA viruses. Annu. Rev. Entomol. 66, 389–405 (2021). [DOI] [PubMed] [Google Scholar]
- 16.He Y. Z., et al. , Gut-expressed vitellogenin facilitates the movement of a plant virus across the midgut wall in its insect vector. mSystems 6, e0058121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei J., et al. , Vector development and vitellogenin determine the transovarial transmission of begomoviruses. Proc. Natl. Acad. Sci. U.S.A. 114, 6746–6751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luan J. B., et al. , Global analysis of the transcriptional response of whitefly to tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J. Virol. 85, 3330–3340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S., Guo H., Ge F., Sun Y., Apoptotic neurodegeneration in whitefly promotes the spread of TYLCV. eLife 9, e56168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Y. Z., et al. , A plant DNA virus replicates in the salivary glands of its insect vector via recruitment of host DNA synthesis machinery. Proc. Natl. Acad. Sci. U.S.A. 117, 16928–16937 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubinstein G., Czosnek H., Long-term association of tomato yellow leaf curl virus with its whitefly vector Bemisia tabaci: Effect on the insect transmission capacity, longevity and fecundity. J. Gen. Virol. 78, 2683–2689 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Wang L. L., et al. , The autophagy pathway participates in resistance to tomato yellow leaf curl virus infection in whiteflies. Autophagy 12, 1560–1574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baxter R. H. G., Contet A., Krueger K., Arthropod innate immune systems and vector-borne diseases. Biochemistry 56, 907–918 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingsolver M. B., Huang Z., Hardy R. W., Insect antiviral innate immunity: Pathways, effectors, and connections. J. Mol. Biol. 425, 4921–4936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bang I. S., JAK/STAT signaling in insect innate immunity. Entomol. Res. 49, 339–353 (2019). [Google Scholar]
- 26.Kemp C., et al. , Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 190, 650–658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West C., Silverman N., p38b and JAK-STAT signaling protect against invertebrate iridescent virus 6 infection in Drosophila. PLoS Pathog. 14, e1007020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dostert C., et al. , The JAK-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat. Immunol. 6, 946–953 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Agaisse H., Perrimon N., The roles of JAK/STAT signaling in Drosophila immune responses. Immunol. Rev. 198, 72–82 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Arbouzova N. I., Zeidler M. P., JAK/STAT signalling in Drosophila: Insights into conserved regulatory and cellular functions. Development 133, 2605–2616 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Paradkar P. N., Trinidad L., Voysey R., Duchemin J.-B., Walker P. J., Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the JAK-STAT pathway. Proc. Natl. Acad. Sci. U.S.A. 109, 18915–18920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Souza-Neto J. A., Sim S., Dimopoulos G., An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. U.S.A. 106, 17841–17846 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colpitts T. M., et al. , Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog. 7, e1002189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrison D. A., The JAK/STAT pathway. Cold Spring Harb. Perspect. Biol. 4, a011205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen W., et al. , The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 14, 110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geng L., et al. , Transcriptome profiling of whitefly guts in response to tomato yellow leaf curl virus infection. Virol. J. 15, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hindupur S. V., et al. , STAT3/5 inhibitors suppress proliferation in bladder cancer and enhance oncolytic adenovirus therapy. Int. J. Mol. Sci. 21, 1106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raftery N., Stevenson N. J., Advances in anti-viral immune defence: Revealing the importance of the IFN JAK/STAT pathway. Cell. Mol. Life Sci. 74, 2525–2535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agaisse H., Petersen U. M., Boutros M., Mathey-Prevot B., Perrimon N., Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 5, 441–450 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Lagueux M., Perrodou E., Levashina E. A., Capovilla M., Hoffmann J. A., Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 97, 11427–11432 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levashina E. A., et al. , Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell 104, 709–718 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Blandin S., et al. , Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116, 661–670 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Li P., et al. , Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog. 15, e1007607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan Z., et al. , Upregulation of HSP72 attenuates tendon adhesion by regulating fibroblast proliferation and collagen production via blockade of the STAT3 signaling pathway. Cell. Signal. 71, 109606 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Atzmon G., Oss H. V., Czosnek H., PCR-amplification of tomato yellow leaf curl virus (TYLCV) DNA from squashes of plants and whitefly vectors: Application to the study of TYLCV acquisition and transmission. Eur. J. Plant Pathol. 104, 189–194 (1998). [Google Scholar]
- 46.Darnell J. E. Jr., Kerr I. M., Stark G. R., JAK-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–1421 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Schindler C., Levy D. E., Decker T., JAK-STAT signaling: From interferons to cytokines. J. Biol. Chem. 282, 20059–20063 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Ekengren S., Hultmark D., A family of Turandot-related genes in the humoral stress response of Drosophila. Biochem. Biophys. Res. Commun. 284, 998–1003 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Bou Aoun R., et al. , Analysis of thioester-containing proteins during the innate immune response of Drosophila melanogaster. J. Innate Immun. 3, 52–64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shokal U., Kopydlowski H., Eleftherianos I., The distinct function of Tep2 and Tep6 in the immune defense of Drosophila melanogaster against the pathogen Photorhabdus. Virulence 8, 1668–1682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin M., et al. , Cell surface antigen CD109 is a novel member of the α(2) macroglobulin/C3, C4, C5 family of thioester-containing proteins. Blood 99, 1683–1691 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Cummings K. L., Waggoner S. N., Tacke R., Hahn Y. S., Role of complement in immune regulation and its exploitation by virus. Viral Immunol. 20, 505–524 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Olson K. E., Blair C. D., Arbovirus-mosquito interactions: RNAi pathway. Curr. Opin. Virol. 15, 119–126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swevers L., Liu J., Smagghe G., Defense mechanisms against viral infection in Drosophila: RNAi and non-RNAi. Viruses 10, 230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szittya G., Burgyán J., RNA interference-mediated intrinsic antiviral immunity in plants. Curr. Top. Microbiol. Immunol. 371, 153–181 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Rosas-Diaz T., et al. , A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc. Natl. Acad. Sci. U.S.A. 115, 1388–1393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X. R., et al. , Apoptosis in a whitefly vector activated by a begomovirus enhances viral transmission. mSystems 5, e00433-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Betz A., Ryoo H. D., Steller H., Darnell J. E. Jr., STAT92E is a positive regulator of Drosophila inhibitor of apoptosis 1 (DIAP/1) and protects against radiation-induced apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13805–13810 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borensztejn A., Mascaro A., Wharton K. A., JAK/STAT signaling prevents excessive apoptosis to ensure maintenance of the interfollicular stalk critical for Drosophila oogenesis. Dev. Biol. 438, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oeckinghaus A., Hayden M. S., Ghosh S., Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Czerkies M., et al. , Cell fate in antiviral response arises in the crosstalk of IRF, NF-κB and JAK/STAT pathways. Nat. Commun. 9, 493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu X., Chen J., Wang L., Ivashkiv L. B., Crosstalk among JAK-STAT, Toll-like receptor, and ITAM-dependent pathways in macrophage activation. J. Leukoc. Biol. 82, 237–243 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Fleming S. B., Viral inhibition of the IFN-induced JAK/STAT signalling pathway: Development of live attenuated vaccines by mutation of viral-encoded IFN-antagonists. Vaccines (Basel) 4, 23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez J. J., Parisien J.-P., Horvath C. M., Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76, 11476–11483 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oda K., et al. , Structural basis of the inhibition of STAT1 activity by Sendai virus C protein. J. Virol. 89, 11487–11499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jia D., et al. , Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS One 5, e13927 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi L., Ramaswamy M., Manzel L. J., Look D. C., Inhibition of JAK1-dependent signal transduction in airway epithelial cells infected with adenovirus. Am. J. Respir. Cell Mol. Biol. 37, 720–728 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu J. X., Shang H. L., Xie Y., Shen Q. T., Zhou X. P., Monoclonal antibodies against the whitefly-transmitted tomato yellow leaf curl virus and their application in virus detection. J. Integr. Agric. 11, 263–268 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.