Abstract
Simple Summary
The combination of androgen deprivation therapy (ADT) with docetaxel (DX) or/and with novel anti-androgen receptor therapies have become standards for the treatment of patients with metastatic hormone-sensitive prostate cancer (mHSPC). However, metastatic PC remains incurable, and biomarkers for individual treatment selection are needed. We propose here that molecular alterations associated with castration resistance may predict the clinical evolution of mHSPC patients. To test this hypothesis, we designed a custom expression panel of 184 genes and tested it in tumor biopsies from patients with mHSPC treated with ADT+DX. We found that AR and ESR signatures and ESR2 gene expression correlate with a good prognosis. The lower expression of TSG (PTEN, TP53 and RB1) signature, as well as high ARV7 and low RB1 gene expression, were associated with adverse clinical outcomes. The usefulness of transcriptomic analysis of such signatures as a strategy for personalized treatment selection should be further explored.
Abstract
(1) Background: Androgen deprivation therapy (ADT) and docetaxel (DX) combination is a standard therapy for metastatic hormone-sensitive prostate cancer (mHSPC) patients. (2) Methods: We investigate if tumor transcriptomic analysis predicts mHSPC evolution in a multicenter retrospective biomarker study. A customized panel of 184 genes was tested in mRNA from tumor samples by the nCounter platform in 125 mHSPC patients treated with ADT+DX. Gene expression was correlated with castration-resistant prostate cancer-free survival (CRPC-FS) and overall survival (OS). (3) Results: High expression of androgen receptor (AR) signature was independently associated with longer CRPC-FS (hazard ratio (HR) 0.6, 95% confidence interval (CI) 0.3–0.9; p = 0.015), high expression of estrogen receptor (ESR) signature with longer CRPC-FS (HR 0.6, 95% CI 0.4–0.9; p = 0.019) and OS (HR 0.5, 95% CI 0.2–0.9, p = 0.024), and lower expression of tumor suppressor genes (TSG) (RB1, PTEN and TP53) with shorter OS (HR 2, 95% CI 1–3.8; p = 0.044). ARV7 expression was independently associated with shorter CRPC-FS (HR 1.5, 95% CI 1.1–2.1, p = 0.008) and OS (HR 1.8, 95% CI 1.2–2.6, p = 0.004), high ESR2 was associated with longer OS (HR 0.5, 95% CI 0.2–1, p = 0.048) and low expression of RB1 was independently associated with shorter OS (HR 1.9, 95% CI 1.1–3.2, p = 0.014). (4) Conclusions: AR, ESR, and TSG expression signatures, as well as ARV7, RB1, and ESR2 expression, have a prognostic value in mHSPC patients treated with ADT+DX.
Keywords: metastatic prostate cancer, predictive biomarkers, hormonal therapy, chemotherapy, androgen receptor, estrogen receptor, tumor suppressor genes
1. Introduction
Prostate cancer (PC) is ranked second in cancer incidence and represents the fifth cause of cancer death in men worldwide [1]. Androgen deprivation therapy (ADT) in combination with docetaxel (DX) or anti-androgen receptor therapies (ART) are standard upfront treatments in metastatic hormone-sensitive prostate cancer (mHSPC) based on meaningful improvement in overall survival (OS) compared to ADT alone [2,3,4,5,6,7,8]. However, metastatic PC remains an incurable disease with heterogeneous clinical evolution, and treatment selection for individual patients remains a challenge. Recently, it has been shown that the transcriptional profile of primary tumors may determine a distinct clinical evolution of mHSPC patients treated with ADT alone or ADT+DX [9].
Molecular alterations in several genes such as those AR-related tumor suppressor genes (TSG) (RB1, PTEN and TP53), DNA-repair genes [10,11], and cell plasticity (neuroendocrine (NE), epithelial-mesenchymal transition (EMT))-related genes [12,13,14], have been associated with treatment resistance and aggressive clinical evolution of metastatic castration-resistant prostate cancer (CRPC). We hypothesized that if gene expression deregulation on those genes were present in non-castrated tumors, they could also predict clinical evolution and treatment benefit. To test this hypothesis, we designed a custom expression panel of 184 genes that may be relevant in PC biology, including genes commonly altered in CRPC, and tested it in tumor biopsies from patients with mHSPC.
We present here the results of a multicenter retrospective biomarker study in mHSPC patients treated with ADT+DX as standard clinical practice in different hospitals in Spain. The ultimate goal was the identification of gene expression signatures related to adverse outcomes that could identify patient candidates for the exploration of novel treatment strategies.
2. Materials and Methods
2.1. Design, Patients and Samples
This is a multicenter retrospective biomarker study in patients with mHSPC. Key inclusion criteria were prostate adenocarcinoma diagnosis with available formalin-fixed paraffin-embedded (FFPE) biopsy of the primary tumor or a metastatic site in the hormone-sensitive setting that was considered by the pathologist to have enough material for molecular analysis Treatment for mHSPC was ADT (i.e., luteinizing hormone-releasing hormone (LHRH) analogs) in combination with DX (75 mg/m2 in combination with prednisone 10 mg/day every 21 days for six cycles). Patients with primary NE tumors were excluded. Clinical variables were collected from patients’ electronic records. The volume of disease was defined according to the CHAARTED trial criteria, which considers the presence of visceral metastases or ≥4 bone lesions with ≥1 outside the spine or pelvis as high-volume disease [2].
The primary endpoint of the study was to correlate the gene expression profiles with CRPC-free survival (CRPC-FS). Secondary endpoints included overall survival (OS) and response to treatment.
2.2. Formalin-Fixed Paraffin-Embedded Tissue Preparation
Tissue samples were fixed in 10% neutral buffered formalin. For small biopsy samples, 6 h of fixation was required, and 12–48 h was required for surgical resection. Samples were then processed in a fluid-transfer advanced automatic tissue processor. To create paraffin blocks, a tissue embedding center (HistoStar, Thermo Scientific, Runcorn, Cheshire, UK) that contained a paraffin reservoir and dispenser, as well as warm and cold plates, was used. The first step was to pour melted paraffin until the stainless-steel mold was partially filled. The tissue samples were removed from the plastic cassettes and transferred into the bottom of the mold (the cutting surface faced down) on the warm plate. Then the tissue was oriented and pressed using a HistoPress. The labeled plastic cassette was placed on top of the mold. Finally, the blocks were cooled on the cold plate and detached from the mold. Once the paraffin-embedded blocks were made, histological sections could already be performed.
2.3. Gene Expression Panel Design
We configured a gene expression nCounter panel (Nanostring Technologies, Seattle, WA, USA) representing signatures described to be related to CRPC development and androgen suppression or taxane resistance [10,12,13,15,16,17,18,19,20,21,22]. The panel consisted of 184 genes, including 5 housekeeping genes (ACTB, GAPDH, GUSB, HPRT1, and RPL13A), and a total of 192 probe sets, with 2 site-specific probes for the isoforms III and VI of TMPRSS2-ERG and 8 probes for the detection of ERG gene expression imbalance between the 3′ and 5′ regions of mRNAs, allowing recognition of any fusion of TMPRSS2-ERG (Supplementary Table S1).
2.4. RNA Extraction
Formalin-fixed paraffin-embedded sections of PC tissues were examined with hematoxylin and eosin staining to determine the tumor area. Macrodissection was performed to avoid contamination with stroma or normal prostatic tissue. At least two 10 μm FFPE slides were used to extract total RNA by using the AllPrep DNA/RNA FFPE Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. RNA was quantified by a Nanodrop Spectrophotometer ND-1000 (Thermo Scientific, Wilmington, MA, USA).
2.5. Gene Expression Analysis
A minimum of ~100 ng of total RNA was used to measure gene expression using the nCounter platform according to the manufacturer’s protocol (Nanostring Technologies, Seattle, WA, USA). Briefly, RNA was hybridized into 192 probe sets for 18 h at 65 °C. Samples were then processed in an automated nCounter Prep Station and imaged on a nCounter Digital Analyzer (Nanostring Technologies, Seattle, WA, USA). Raw expression counts (Supplementary Table S2) were collected, normalized, and log2 transformed using the nSolver 4.0 software. Counts normalization steps consisted of background thresholding of the mean of negative control probe counts +2 standard deviations, normalization by a factor obtained from the geometric mean of the positive control probe counts, and finally a normalization by a factor obtained from the geometric mean of the housekeeping probe counts.
TMPRSS2-ERG expression was assessed by the imbalance of eight ERG probes, four at 3′ and four at 5′. We represented the ratio between the mean of ERG 3′ probe counts and the mean of ERG 5′ counts (ERG 3′/5′) for each patient. These ratios were compared with previous real-time quantitative reverse-transcription PCR data of the isoform III of the TMPRSS2-ERG gene obtained from 77 RNA samples analyzed in a previous study [23] and the counts from a site-specific probe for the isoform III in these patients. A score threshold for the ratio ERG 3′/5′ of 3.4 was established to consider the presence of TMPRSS2-ERG alteration (Supplementary Figure S1).
2.6. Bioinformatics and Statistical Analysis
Hierarchical cluster analysis of the expression values of the whole gene panel (excluding specific and imbalance TMPRSS2-ERG probes) or the signatures was performed using Cluster 3.0 [24], and results were visualized in Java TreeView [25].
Tertiles were applied to gene expression data to categorize the samples as high-, middle-, or low-expression groups. Clinical variables such as stage at diagnosis, Gleason at diagnosis, the presence of visceral metastasis, bone metastasis, the disease volume at ADT start time, and the time from ADT to docetaxel (<3 vs. ≥3 months) were evaluated as dichotomic. Lactate dehydrogenase (LDH) levels were evaluated as a continuous variable.
CRPC-FS, calculated from the date of start of ADT to the time of developing CRPC, and OS, calculated from the date of start of ADT to the time of death or last follow-up visit, were analyzed by the Kaplan–Meier method and compared by log-rank test. CRPC-FS definition, treatment-response criteria and progressive-disease definitions followed Prostate Cancer Working Group 2 criteria [26]. Univariate analysis of variables of interest was performed by Cox regression analysis; p < 0.1 was required for inclusion in the multivariate model. When considering all the individual genes of a signature, their expression levels were evaluated as continuous variables, and significant genes were selected if they accomplished a false discovery rate (FDR) < 0.2. Fisher’s exact test and the Wilcoxon Mann–Whitney test were used to compare the proportions of qualitative and continuous clinical variables between groups, respectively. Correlations between expression levels as continuous variables were measured by calculating Pearson’s coefficient. Significant differently expressed genes between groups were selected if they accomplished a fold change (|FC|) ≥ 1.5 and FDR < 0.05.
In order to compare the expression of a signature between groups, single-sample GSEA (ssGSEA) [27] from the GSVA R package [28] was used to calculate a gene-set-enrichment score per patient for each group, and a Wilcoxon Mann–Whitney test was then applied to test for statistical differences between groups.
Analyses were performed with R software (v.3.6.3) [29].
3. Results
3.1. Patients and Samples
A total of 133 patients were enrolled in this study: 125 of them were eligible, and 8 were excluded due to insufficient tumor sample (N = 4) or lack of RNA availability (N = 4). Table 1 summarizes the baseline clinical characteristics of the eligible patients. Of note, 92.8% (N = 116) of patients had de novo mHSPC disease, 20% (N = 25) had visceral metastasis, 78.4% (N = 98) were considered to have high-volume disease [2], and a Gleason score ≥8 was reported in 81.6% (N = 102) of patients. The number of patients who received ART (abiraterone or enzalutamide) as first-line treatment in CRPC was 77 (80.2%). We collected FFPE samples mostly from primary tumors (N = 117, 93.6%). The remaining biopsies were obtained from metastatic sites (N = 8, 6.4%).
Table 1.
Patients’ characteristics. N: number of cases; ADT: androgen deprivation therapy; ECOG: Eastern Cooperative Oncology Group; PSA: prostate-specific antigen; CRPC: castration-resistant prostate cancer; NA: not available.
| Patients Eligible (Enrolled), N | 125 (133) |
|---|---|
| Age (years) | |
| Median (range) | 66.6 (46.3–83.4) |
| Tumor origin, N (%) | |
| Primary | 117 (93.6) |
| Metastatic | 8 (6.4) |
| Stage at diagnosis, N (%) | |
| <IV | 9 (7.2) |
| IV | 116 (92.8) |
| Gleason sum at diagnosis, N (%) | |
| ≤7 | 22 (17.6) |
| ≥8 | 102 (81.6) |
| NA | 1 (0.8) |
| Presence of bone metastases, N (%) | |
| Yes | 112 (89.6) |
| No | 13 (10.4) |
| Presence of visceral metastases, N (%) | |
| Yes | 25 (20) |
| No | 100 (80) |
| Location of visceral metastases, N (%) | |
| Lung | 20 (80) |
| Liver | 7 (28) |
| Pleural | 1 (4) |
| NA | 1 (4) |
| Disease volume, N (%) | |
| High | 98 (78.4) |
| Low | 26 (20.8) |
| NA | 1 (0.8) |
| ECOG performance status score, N (%) | |
| 0 | 54 (43.2) |
| 1 or 2 | 69 (55.2) |
| NA | 2 (1.6) |
| Baseline PSA (ng/mL) at diagnosis | |
| Median (range) | 83.2 (1.8–7448) |
| Baseline lactate dehydrogenase (U/L) | |
| Median (range) | 316 (116–1023) |
| Time from ADT to docetaxel treatment, N (%) | |
| <3 months | 106 (84.8) |
| ≥3 months | 19 (15.2) |
| First line treatment in CRPC, N (%) | |
| Abiraterone or enzalutamide | 77 (80.2) |
| Taxanes | 5 (5.2) |
| Other treatments | 5 (5.2) |
| No treatment | 4 (4.2) |
| NA | 5 (5.2) |
3.2. Clinical Outcomes
The median follow-up time was 36.1 months (range 6.7–78.6), and 96 patients (76.8%) developed CRPC. Median CRPC-FS was 19.3 months (95% confidence interval (CI) 15.8–23.7), and median OS was 53 months (95% CI 40.4–72.6).
3.3. Gene Expression and Clinical Outcomes
3.3.1. Global Gene Expression Analysis
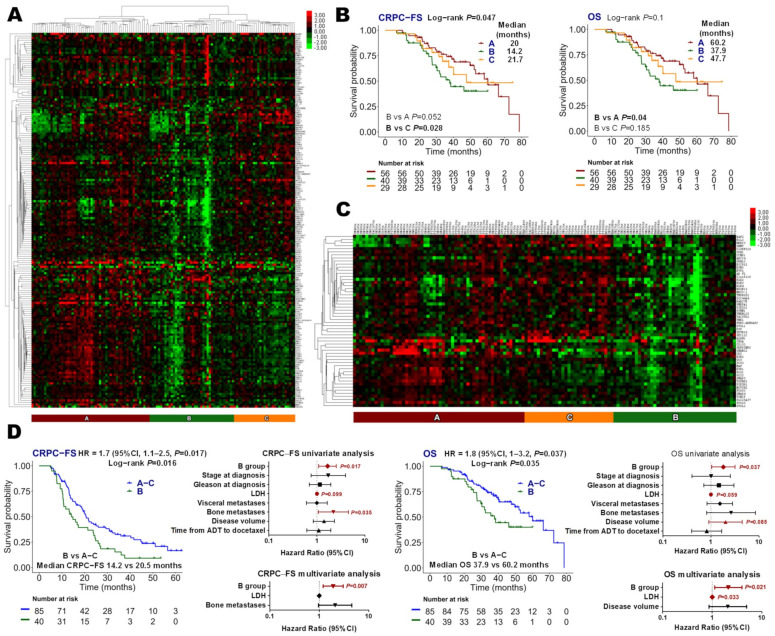
An unsupervised hierarchical cluster of the whole studied panel grouped patients into three main groups, designed as A (N = 59, 44.8%), B (N = 40, 32%), and C (N = 29, 23.2%) (Figure 1A). Cluster B presented a shorter CRPC-FS (hazard ratio (HR) 1.9, 95% CI 1.1.3, p = 0.028, with respect to cluster C) and OS (HR 1.9, 95% CI 1–3.5, p = 0.04, with respect to cluster A) (Figure 1B). Cluster B showed a down-expression of 56 genes (vs. the other two clusters) including AR-related genes such as KLK3, TMPRSS2 and ARFL, as well as ESR1 and ESR2 (Figure 1C). In this comparison, cluster B was independently associated with shorter CRPC-FS (HR 1.9, 95% CI 1.2–3, p = 0.007) and OS (HR 2.1, 95% CI 1.1–4, p = 0.021) (Figure 1D).
Figure 1.
Global gene expression analysis. (A) Hierarchical clustering expression heatmap for expression values of the global genes panel, excluding TMPRSS2-ERG-specific and imbalance probes; (B) Kaplan–Meier curves representing CRPC-free survival (CRPC-FS) and overall survival (OS) according to groups defined from the global genes panel; (C) hierarchical clustering expression heatmap for expression values of differentially expressed genes in group B vs. groups A–C (|FC| ≥ 1.5 and FDR < 0.05); (D) Kaplan–Meier curves representing CRPC-FS and OS according to group B and groups A–C and forest plots representing the univariate and multivariate analysis. LDH: lactate dehydrogenase; HR: hazard ratio; CI, confidence interval.
3.3.2. Androgen Receptor Signature
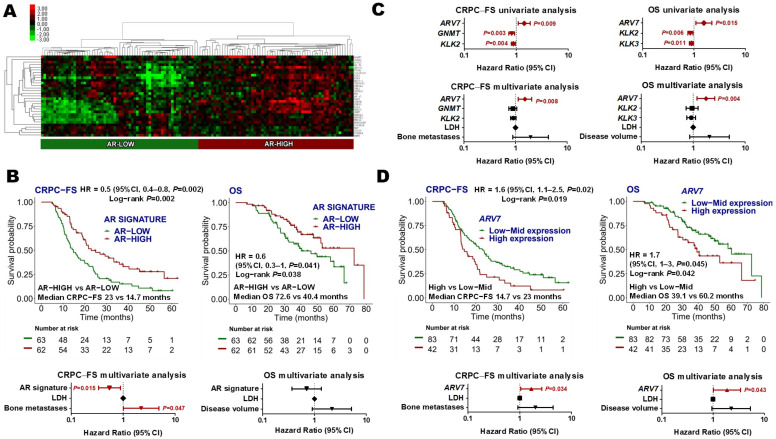
Thirty-one AR-related genes were analyzed (Supplementary Table S1). Hierarchical non-supervised clustering classified patients in AR-low (N = 63, 50.4%) and AR-high (N = 62, 49.6%) categories (Figure 2A). AR-high group was associated with lower LDH levels (p = 0.009) (Supplementary Figure S2A). No differences in AR signature expression were observed between patients with high- or low-volume disease or de novo mHSPC vs. recurrent disease (Supplementary Figure S3). Patients with high AR signature expression had longer CRPC-FS (HR 0.5, 95% CI 0.3–0.8, p = 0.002) and OS (HR 0.6, 95% CI 0.3–1; p = 0.041) than patients with low AR signature expression. AR signature was independently associated with CRPC-FS (HR 0.6, 95% CI 0.3–0.9; p = 0.015) (Figure 2B).
Figure 2.
Androgen receptor (AR) signature. (A) Hierarchical clustering expression heatmap for expression values of the AR signature; (B) Kaplan–Meier curves representing CRPC-free survival (CRPC-FS) and overall survival (OS) according to AR signature and forest plots representing the multivariate analysis; (C) forest plots representing the univariate and multivariate analysis of the individual genes of the AR signature as continuous variables for CRPC-FS and OS. An FDR < 0.2 was applied (only genes that accomplished this condition are represented in the univariate forest plot); (D) Kaplan–Meier curves representing CRPC-FS and OS according to ARV7 expression segregated into tertiles and forest plots representing the multivariate analysis. LDH: lactate dehydrogenase; HR: hazard ratio; CI, confidence interval.
Considering AR signature individual genes as continuous variables in a multivariate analysis, high ARV7 expression was independently associated with shorter CRPC-FS (HR 1.5, 95% CI 1.1–2.1, p = 0.008) and OS (HR 1.8, 95% CI 1.2–2.6, p = 0.004) (Figure 2C). When segregating ARV7 expression levels according to tertiles, upper tertile ARV7 expression was independently associated with shorter CRPC-FS (HR 1.7, 95% CI 1–2.7, p = 0.034) and OS (HR 1.9, 95% CI 1–3.4, p = 0.043) (Figure 2D). No correlation between ARV7 levels and clinical variables was observed.
3.3.3. Estrogen Receptor Signature
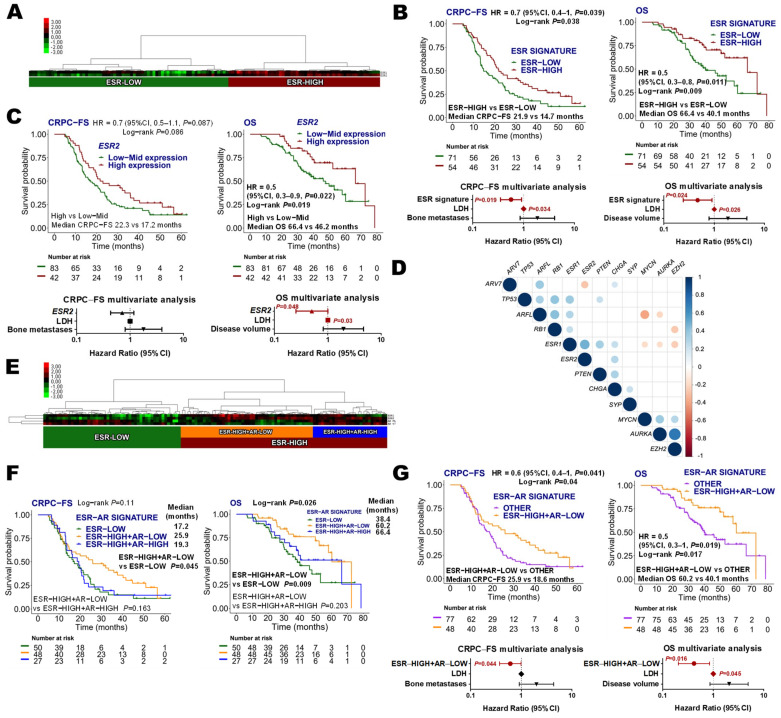
Estrogen receptor (ESR) signature was comprised of ESR1 and ESR2. The hierarchical unsupervised cluster analysis of these genes grouped patients as ESR-low (N = 71, 56.8%) and ESR-high (N = 54, 43.2%) (Figure 3A). No association between ESR signature groups and clinical factors was found, and no differences in ESR signature expression were observed between patients with high or low-volume disease, or de novo mHSPC vs. recurrent disease (Supplementary Figure S3). ESR-high was independently associated with longer CRPC-FS (HR 0.6, 95% CI 0.4–0.9; p = 0.019) and OS (HR 0.5, 95% CI 0.2–0.9, p = 0.024) (Figure 3B). Taking into account individual expression of ESR genes, upper tertile ESR2 expression was independently associated with longer OS (HR 0.5, 95% CI 0.2–1, p = 0.048) (Figure 3C). ESR1 expression was not related to clinical outcomes (Supplementary Figure S4A).
Figure 3.
Estrogen receptor (ESR) signature. (A) Hierarchical clustering expression heatmap for expression values of the ESR signature; (B) Kaplan–Meier curves representing CRPC-free survival (CRPC-FS) and overall survival (OS) according to ESR signature and forest plots representing the multivariate analysis; (C) Kaplan–Meier curves representing CRPC-FS and OS according to ESR2 expression segregated into tertiles and forest plots representing the multivariate analysis; (D) correlation matrix of ESR, AR, neuroendocrine (AURKA, CHGA, SYP, MYCN, and EZH2) and tumor suppressor (TP53, RB1 and PTEN) genes. Correlation coefficients (r) between expression values are represented when p < 0.05; (E) hierarchical clustering expression heatmap for expression values of the ESR and AR genes; (F) Kaplan–Meier curves representing CRPC-FS and OS according to ESR and AR genes; (G) Kaplan–Meier curves representing CRPC-FS and OS according to ESR-High+AR-low and the other groups and forest plots representing the multivariate analysis. LDH: lactate dehydrogenase; HR: hazard ratio; CI, confidence interval.
3.3.4. Estrogen and Androgen Receptor Correlations
Since a negative regulation of AR signaling mediated by ESR2 has been documented [30,31], we decided to analyze the correlations between the expression of ESR and AR genes. A gene expression correlation matrix showed a significant negative correlation between ESR2 and ARV7 and a positive correlation between ESR1-ESR2, ARFL-ARV7, and ESR1-ARFL (Figure 3D).
Moreover, a hierarchical non-supervised cluster analysis distributed patients according to three main categories: ESR-low (N = 50, 40%), ESR-high+AR-low (N = 48, 38.4%); and ESR-high+AR-high (N = 27, 21.6%) (Figure 3E). ESR-high+AR-low was the group with longer CRPC-FS (HR 0.6, 95% CI 0.4–1, p = 0.045, with respect to the ESR-low group) and OS (HR 0.4, 95% CI 0.2–0.8, p = 0.009, with respect to the ESR-low group) (Figure 3F). When comparing the ESR-high+AR-low group vs. the other groups together, it was independently correlated with longer CRPC-FS (HR 0.6, 95% CI 0.4–1, p = 0.044) and OS (HR 0.4, 95% CI 0.2–0.9, p = 0.016) (Figure 3G).
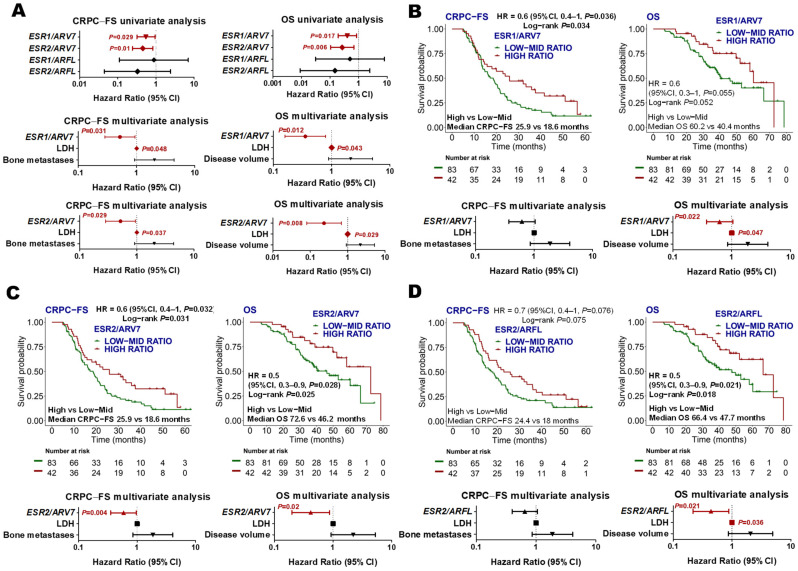
Based on these observations, we decided to further explore if the relationship between AR and ESR could be associated with clinical outcomes by establishing ESR/AR expression ratios. Taking expression ratios as continuous variables, we found that high ESR1/ARV7 and ESR2/ARV7 were independently associated with longer CRPC-FS (HR 0.5, 95% CI 0.3–0.9, p = 0.031; HR 0.5, 95% CI 0.2–0.9, p = 0.029; respectively) and OS (HR 0.4, 95% CI 0.2–0.8, p = 0.012; HR 0.2, 95% CI 0.1–0.7, p = 0.008, respectively) (Figure 4A). Upper tertile ESR1/ARV7 expression ratio independently correlated with longer OS (HR 0.4, 95% CI 0.2–0.9, p = 0.022), and upper tertile ESR2/ARV7 correlated with longer CRPC-FS (HR 0.6, 95% CI 0.4–1, p = 0.04) and OS (HR 0.4, 95% CI 0.2–0.9, p = 0.02) (Figure 4B,C). Moreover, upper tertile ESR2/ARFL expression was independently associated with longer OS (HR 0.5, 95% CI 0.3–0.9, p = 0.021) (Figure 4D), while ESR1/ARFL did not correlate with clinical outcomes (Supplementary Figure S4B).
Figure 4.
ESR/AR expression ratios. (A) Forest plots representing the univariate and multivariate analysis of the ratios between ESR and AR genes as continuous variables for CRPC-FS and OS. Those ratios with p < 0.1 in the univariate were included in the multivariate analysis; (B-D) Kaplan–Meier curves representing CRPC-FS and OS according to ESR1/ARV7 (B), ESR2/ARV7 (C), and ESR2/ARFL (D) ratios segregated into tertiles and forest plots representing the multivariate analysis. LDH: lactate dehydrogenase; HR: hazard ratio; CI, confidence interval.
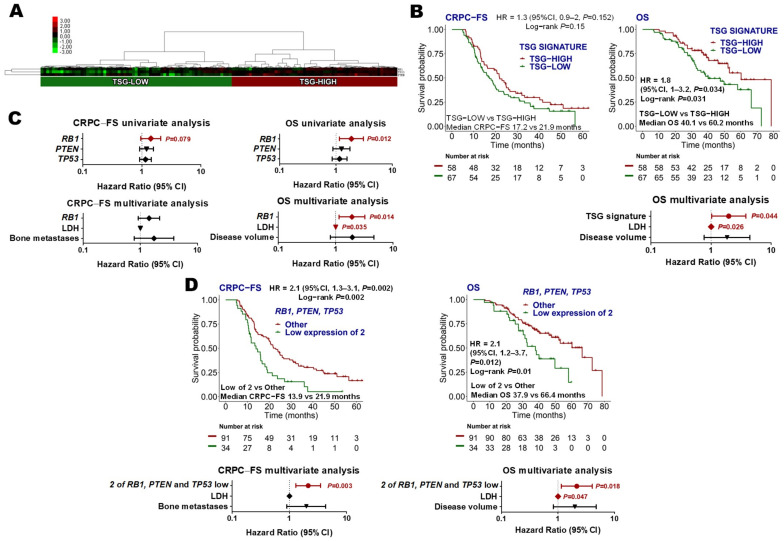
3.3.5. Tumor Suppressor Gene (TP53, RB1 and PTEN) Signature
The expression of TSG, which associates with aggressive CRPC clinical evolution [11,32,33], was tested in this cohort of mHSPC patients. Hierarchical non-supervised cluster analysis classified patients as “TSG-low” (N = 67, 53.6%) or “TSG-high” (N = 58, 46.4%) categories (Figure 5A). No association between TSG signature groups and clinical factors was found, and no differences in TSG signature expression were observed between patients with high- or low-volume disease or de novo mHSPC vs. recurrent disease (Supplementary Figure S3). Low PTEN levels correlated with the presence of visceral metastasis (p = 0.044) (Supplementary Figure S2B).
Figure 5.
Tumor suppressor genes (TSG) signature. (A) Hierarchical clustering expression heatmap for expression values of the TSG signature; (B) Kaplan–Meier curves representing CRPC-free survival (CRPC-FS) and overall survival (OS) according to TSG signature and forest plot representing the multivariate analysis for OS; (C) forest plots representing the univariate and multivariate analysis of the individual genes of the TSG signature as continuous variables for CRPC-FS and OS. The reciprocal of the hazard ratio (HR) and the associated confidence interval (CI) were calculated; (D) Kaplan–Meier curves representing CRPC-FS and OS according to low expression of two out of the three TSG, and forest plots representing the multivariate analysis. Expression levels were segregated into tertiles to establish the cut-offs. LDH: lactate dehydrogenase.
TSG-low expression was independently associated with shorter OS (HR 2, 95% CI 1–3.8; p = 0.044) (Figure 5B). Considering TSG individual genes as continuous variables in a multivariate analysis, low expression of RB1 was independently associated with shorter OS (HR 1.9, 95% CI 1.1–3.2, p = 0.014) (Figure 5C). Moreover, the lower tertile expression of 2 out of the 3 TSG independently correlated with shorter CRPC-FS (HR 2.1, 95% CI 1.3–3.5, p = 0.003) and OS (HR 2.2, 95% CI 1.1–4.1, p = 0.018) (Figure 5D).
As alterations in TSG have been associated with low AR activity and NE dedifferentiation, we explored how these signatures were correlated in our series. ARFL was positively correlated with RB1, TP53 and ARV7 expression and negatively with MYCN and AURKA. Moreover, a negative correlation between RB1 and EZH2 expression was found. Additionally, a significant positive correlation between TSG and ESR1 was observed (Figure 3D).
3.3.6. Neuroendocrine and Other Signatures
The expression of forty-five NE-related genes was analyzed (Supplementary Table S1), and no correlation with clinical outcomes was observed (Supplementary Figure S5A). No correlation between other signatures or TMPRSS2-ERG expression and clinical outcomes was found (Supplementary Figures S5 and S6).
3.3.7. Joint Analysis of Gene Expression Signatures
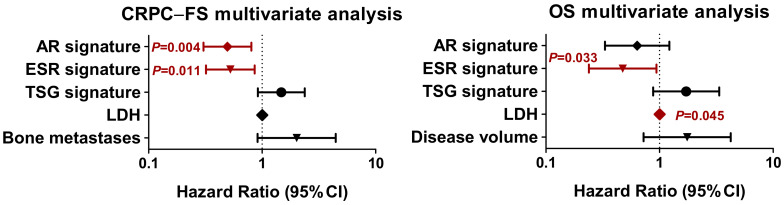
Next, we assessed the significance of AR, ESR, and GST signatures together. The multivariate analysis including significant molecular signatures and clinical factors showed that high expression of AR (HR 0.5, 95% CI 0.3–0.8, p = 0.004) and ESR (HR 0.5, 95% CI 0.3–0.9, p = 0.011) signatures correlated with longer CRPC-FS, and high expression of ESR signature (HR 0.5, 95% CI 0.2–0.9, p = 0.033) correlated with longer OS (Figure 6).
Figure 6.
Multivariate analysis for androgen receptor (AR), estrogen receptor (ESR) and tumor suppressor gene (TSG) signatures. Forest plots represent the multivariate analysis of AR, ESR and TSG signatures for CRPC-free survival (CRPC-FS) and overall survival (OS). LDH: lactate dehydrogenase; HR: hazard ratio; CI, confidence interval.
4. Discussion
In this study, we show that the expression of AR, ESR and the TSG (PTEN, RB1 and TP53) signatures are associated with the clinical evolution of patients with mHSPC treated with the combination of ADT+DX.
While AR overexpression and pathway activation has been demonstrated as one of the fundamental mechanisms of progression and resistance to therapy in CRPC patients [10,34,35,36], its role in HSPC has to be defined. In our series, we found that a high AR signature was associated with longer OS and, independently, predicted longer CRPC-FS. The molecular analysis of 160 mHSPC patients included in the phase III CHAARTED trial [9] that compared ADT vs. ADT+DX therapy showed that different PAM50 molecular subtypes have distinct treatment benefits: luminal B subtype was associated with a poorer prognosis on ADT alone but benefited significantly from ADT+DX (OS: HR 0.45, p = 0.007), in contrast to the basal subtype, which showed no OS benefit (HR 0.85, p = 0.58). These results were in contrast with a previous study where, in a subset of non-metastatic 315 patients, the luminal B subtype was the only group that benefited from postoperative response to ADT [37]. These discrepant results may be explained by the different patient populations included in both studies. As luminal expression profile is associated with high AR signaling and steroid hormone receptor processing [9], results from the CHAARTED trial may be in concordance with our data, showing better outcomes for patients with high AR-related expression when treated with the combination therapy. However, only 7 genes from the PAM50 gene set were represented in our signature, and for that reason, no definitive conclusions can be drawn about the PAM50 molecular subtypes in our cohort.
The AR splicing variant ARV7, which lacks the ligand-binding domain, may be constitutively activated in the absence of androgens and acts as a transcription factor repressing crucial tumor suppressor genes and promoting PC progression [38]. It has been recently shown that its detection by IHC correlates with poor prognosis and short response to ADT in mHSPC patients [39]. In our study, high ARV7 expression was independently associated with shorter CRPC-FS and OS, supporting that ARV7 also confers adverse prognosis in patients treated with combined therapy.
A novel and relevant result of our study is that the expression of the ESR signature is independently associated with a better outcome. When analyzing the significant signatures (AR, ESR and TSG) together, only the ESR signature was independently associated with CRPC-FS and OS. The ESR subfamily proteins are composed of two main subtypes of receptors, ESR1 and ESR2. ESR1 may be expressed in prostate stem cells and is up-regulated during malignant transformation of the prostatic epithelium, in high-grade PIN, in metastatic lesions, and in CRPC. In contrast, ESR2 is expressed at high levels in the luminal cells of the prostatic epithelium and may be partly lost in the high-grade PIN. ESR2 may function in PC as a tumor suppression gene; it preferentially binds phytoestrogens and is likely to protect the prostate epithelium from malignant transformation [40]. Notably, when analyzing individual genes, ESR2 was independently associated with a longer CRPC-FS and OS. In pre-clinical models, ESR2 down-regulates AR signaling [30,31] and up-regulates PTEN [30]. Moreover, it has been shown that androgen deprivation and/or long-term abiraterone therapy induces the loss of ESR2 and PTEN, and the addition of ESR2 agonists together with abiraterone has been proposed as a strategy to sustain the expression of ESR2 and offer some benefit to patients [41]. In our series, we also found an inverse correlation between ESR2 and ARV7 gene expression. Notably, a high ESR2/ARV7 ratio was independently associated with a better clinical evolution. In pre-clinical models, ESR2 stimulation reduced ARV7 expression [42]. Overall, this may suggest that ESR2 stimulation may be a potential strategy to revert or prevent ARV7-related resistance. Of note, ESR1 expression positively correlated with PTEN, TP53 and RB1, which may also explain the good prognosis of patients with a high-ESR signature. Globally, these results suggest that the transcriptional program associated with ESR regulates PC essential genes and support further investigation of its role as a biomarker and as a therapeutic target.
Alterations in the tumor suppression genes PTEN, TP53 and RB1 have been associated with aggressive clinical cancer evolution and resistance to conventional therapy in CRPC patients [11,32,33]. Few studies have investigated the role of TSG genomic alterations in HSPC. Gilson et al. explored the genomic landscape of mHSPC and found that the most prevalent mutations were located in PTEN or TP53 [43]. Mateo et al. studied genomic aberrations in primary PC biopsies from patients who developed mCRPC [44]. They found that patients with lower expression of RB1 had a worse prognosis, in concordance with our work where the low expression of RB1 was independently associated with shorter OS. Another study of targeted sequencing TSG in localized and metastatic tumors reported that altered TSG increased with advanced disease, which was associated with an increase in the risk of relapse and death in mHSPC [32,45]. To our knowledge, this is the first study that investigates the prognostic value of mRNA expression of TSG in mHSPC. We found that the low expression of TSG signature was independently associated with shorter OS. Considering individual TSG genes as continuous variables, low expression of RB1 was independently associated with shorter OSand the lower tertile expression of 2 out of the 3 TSG independently correlated with shorter CRPC-FS and OS. Our results support that the transcriptomic analysis of TSG may define the mHSPC group of patients with aggressive clinical evolution. In that sense, there is evidence that the administration of platinum-based chemotherapy may be more active than taxanes alone in aggressive mCRPC [46]. Moreover, the AKT inhibitors have shown promising results in mCRPC patients with PTEN alterations in combination with abiraterone [47] or DX [48]. Exploring these strategies in mHSPC with TSG alterations may be warranted.
In our study, only 6% of patients were excluded from molecular analysis due to insufficient tumor samples or lack of RNA availability. A possible explanation is that an inclusion criterion for participation in the study was to have available FFPE samples that were considered by the pathologist to have enough material for molecular analysis. Moreover, the use of the nCounter technology may also represent an advantage over other methodologies for FFPE sample molecular analysis. Our laboratory has much experience in the use of the nCounter technology in the study of transcriptional signatures in breast cancer [49] and other tumor types [50]. This technology has demonstrated high profitability for the analysis of mRNA from FFPE-tumor samples with low RNA quantity and high reproducibility [49,51,52], which has led to its clinical application in breast cancer [49]. To expand its investigation into prostate cancer is warranted.
The main limitation of this work relied on the lack of independent validation of the results. In addition, the combination of ADT+ART is another current standard treatment for patients with mHSPC and has not been explored in the present study. However, due to the potential interest that they could arise in other groups, we presented these results while we were working on independent series of mHSPC patients receiving different treatment strategies in order to validate its prognostic value and to explore its potential usefulness for treatment selection.
5. Conclusions
Our study suggests that AR and ESR signatures and ESR2 gene expression correlate with good prognosis in patients receiving ADT+DX. Moreover, the lower expression of TSG (PTEN, TP53 and RB1) signature, as well as high ARV7 and low RB1 gene expression, are associated with adverse clinical outcomes. The usefulness of transcriptomic analysis of such signatures as a strategy for personalized treatment selection should be further explored.
Acknowledgments
The authors thank Esther Barnadas for her kind organization of sample collection from Hospital Clínic and her excellent technical assistance with FFPE tumor sections. We want to acknowledge Parc de Salut MAR Biobank (MARBiobanc) (RD09/0076/00036, PT17/0015/0011) and IGTP-HUGTP Biobank [PT13/0010/0009, PT17/0015/0045] integrated into the Spanish National Biobanks Network and Tumor Bank Network of Catalonia; and BioBank FIVO (PT17/0015/0051) integrated into the Spanish National Biobanks Network and in the Valencian Biobanking Network for their collaboration in providing samples. The authors are also deeply indebted to all patients that agreed to be involved in the study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14194757/s1, Figure S1: TMPRSS2-ERG expression threshold determination; Figure S2: Correlations with clinical factors; Figure S3: Androgen receptor (AR), estrogen receptor (ESR), and tumor suppressor gene (TSG) signature expression between patients; Figure S4: Clinical outcomes according to ESR1 expression and ESR1/ARFL expression ratio; Figure S5: Neuroendocrine (NE), epithelial-mesenchymal transition (EMT), stemness, and immune signatures; Figure S6: Cell cycle (CC), PI3K, DNA damage response (DDR), Notch, and Hedgehog signatures, and TMPRSS2-ERG; Table S1: Customized gene expression nCounter panel, Table S2: nCounter raw count data.
Author Contributions
Study conception and design: N.J., Ò.R., M.M.-A. and B.M. Development of methodology: N.J., Ò.R., M.M.-A., Ò.R. and B.M. Acquisition of data: N.J., Ò.R., M.M.-A., C.A., L.F.-M., A.F., A.R.-V., M.Á.C., S.C., I.C., M.D., M.F., E.G.-B., D.J.P., L.R.-C., M.G.d.H. and B.M. Analysis and interpretation of data: N.J., Ò.R., M.M.-A., C.A., L.F.-M., S.G.-E., M.J.R., A.P. and B.M. Funding acquisition: B.M. Supervision: B.M. Manuscript preparation and editing: N.J., Ò.R., M.M.-A. and B.M. All authors contributed to the manuscript revision and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the principles of the Declaration of Helsinki, and it was approved by the Institutional Ethics Committees of all participating centers.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The raw counts from NanoString nCounter gene expression data generated in this study are available in Supplementary Table S2.
Conflicts of Interest
Authors have provided the following conflicts to disclose (which may not be related to the subject matter of this manuscript): B.M.: research funding from Janssen, Roche, Bayer and Pfizer; speaker’s bureau for Roche, Sanofi, Janssen, Astellas, Pfizer, Novartis, and Bristol-Myers Squibb; and travel and accommodation expenses from Janssen and Pfizer. O.R.: consulting or advisory role by BMS; and travel and accommodation expenses from Ipsen and Pfizer. L.F.M.: speaker honoraria and travel accommodation expenses from Pfizer and Kyowa kirin. A.F.: research funding from Astra-Zeneca, Astellas, and Pierre Fabre; consulting or advisory role by Janssen, Sanofi, Astellas, Ipsen, Roche, and Astra-Zeneca; and travel and accommodation expenses from Roche, Sanofi, and Janssen. A.R.V.: research funding from Takeda, Pfizer, and Merck; and consulting or advisory role by Roche, MSD, Pfizer, BMS, Astellas, Janssen, Bayer, Clovis, Astra-Zeneca, and Sanofi. M.A.C.: honoraria from BMS, Astellas, Janssen, MSD, Sanofi, Bayer, Roche, Pfizer, Novartis, and Ipsen; consulting or advisory role by BMS, MSD, Bayer, EUNSA, Pfizer, Roche, Janssen, Pierre Fabre, and Ipsen; and travel and accommodation expenses from Janssen, Astellas, Roche, Ipsen, and MSD. S.C.: research funding from Pfizer and Janssen; and consulting or advisory role by GSK, Tesaro, Roche, Astra-Zeneca, Janssen, Pfizer, BMS, and Merck. I.C.: consultant or advisory board and speaker’s honoraria from Sanofi, Bayer, Astellas, Novartis, Astra-Zeneca, and Janssen; travel and accommodation expenses from Sanofi, Novartis, and Janssen. M.D.: advisory board by Sanofi. M.F.: research funding from Pfizer; honoraria from BMS, Pfizer, Merck, and Sanofi; and travel and accommodation expenses from Sanofi, Merck, BMS, and Novartis. E.G.B.: advisory board by Astra-Zeneca; and speaker’s bureau for Astellas, Bayer, and Janssen. M.J.R.: speaker honoraria from Astellas, Janssen, Ipsen, and Olympus. A.P.: advisory and consulting fees from Roche, Pfizer, Novartis, Amgen, BMS, Puma, Oncolytics Biotech, MSD, Guardant Health, Peptomyc, and Lilly; lecture fees from Roche, Pfizer, Novartis, Amgen, BMS, Daiichi Sankyo, and Nanostring technologies; institutional financial interests from Boehringer, Novartis, Roche, Nanostring technologies, Sysmex Europe GmbH, Medica Scientia Innovation Research, SL, Celgene, Astellas, and Pfizer; leadership role in Reveal Genomics, SL; patent PCT/EP2016/080056. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding Statement
This work was supported by Instituto de Salud Carlos III-Subdirección General de Evaluación y Fomento de la Investigación (PI18/714) and co-funded by the European Union. Institutional funding from CERCA Programme/Generalitat de Catalunya is gratefully acknowledged. This work was funded by a grant from Janssen-Pharmaceuticals (212082PCR4056). This work was developed at the Centro Esther Koplowitz, and CELLEX, Barcelona, Spain.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sweeney C.J., Chen Y.-H., Carducci M., Liu G., Jarrard D.F., Eisenberger M., Wong Y.-N., Hahn N., Kohli M., Cooney M.M., et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer. N. Engl. J. Med. 2015;373:737–746. doi: 10.1056/NEJMoa1503747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gravis G., Boher J.-M., Chen Y.-H., Liu G., Fizazi K., Carducci M.A., Oudard S., Joly F., Jarrard D.M., Soulie M., et al. Burden of Metastatic Castrate Naive Prostate Cancer Patients, to Identify Men More Likely to Benefit from Early Docetaxel: Further Analyses of CHAARTED and GETUG-AFU15 Studies. Eur. Urol. 2018;73:847–855. doi: 10.1016/j.eururo.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.James N.D., Sydes M.R., Clarke N.W., Mason M.D., Dearnaley D.P., Spears M.R., Ritchie A.W.S., Parker C.C., Russell J.M., Attard G., et al. Addition of Docetaxel, Zoledronic Acid, or Both to First-Line Long-Term Hormone Therapy in Prostate Cancer (STAMPEDE): Survival Results from an Adaptive, Multiarm, Multistage, Platform Randomised Controlled Trial. Lancet. 2016;387:1163–1177. doi: 10.1016/S0140-6736(15)01037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fizazi K., Tran N., Fein L., Matsubara N., Rodriguez-Antolin A., Alekseev B.Y., Özgüroğlu M., Ye D., Feyerabend S., Protheroe A., et al. Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. N. Engl. J. Med. 2017;377:352–360. doi: 10.1056/NEJMoa1704174. [DOI] [PubMed] [Google Scholar]
- 6.James N.D., de Bono J.S., Spears M.R., Clarke N.W., Mason M.D., Dearnaley D.P., Ritchie A.W.S., Amos C.L., Gilson C., Jones R.J., et al. Abiraterone for Prostate Cancer Not Previously Treated with Hormone Therapy. N. Engl. J. Med. 2017;377:338–351. doi: 10.1056/NEJMoa1702900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis I.D., Martin A.J., Stockler M.R., Begbie S., Chi K.N., Chowdhury S., Coskinas X., Frydenberg M., Hague W.E., Horvath L.G., et al. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N. Engl. J. Med. 2019;381:121–131. doi: 10.1056/NEJMoa1903835. [DOI] [PubMed] [Google Scholar]
- 8.Chi K.N., Agarwal N., Bjartell A., Chung B.H., Gomes A.J.P.D.S., Given R., Soto Á.J., Merseburger A.S., Özgüroğlu M., Uemura H., et al. Apalutamide for Metastatic, Castration-Sensitive Prostate Cancer. N. Engl. J. Med. 2019;381:13–24. doi: 10.1056/NEJMoa1903307. [DOI] [PubMed] [Google Scholar]
- 9.Hamid A.A., Huang H.-C., Wang V., Chen Y.-H., Feng F., Den R., Attard G., Allen E.M.V., Tran P.T., Spratt D.E., et al. Transcriptional Profiling of Primary Prostate Tumor in Metastatic Hormone-Sensitive Prostate Cancer and Association with Clinical Outcomes: Correlative Analysis of the E3805 CHAARTED Trial. Ann. Oncol. 2021;32:1157–1166. doi: 10.1016/j.annonc.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson D., Van Allen E.M., Wu Y.-M., Schultz N., Lonigro R.J., Mosquera J.-M., Montgomery B., Taplin M.-E., Pritchard C.C., Attard G., et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aparicio A.M., Shen L., Tapia E.L.N., Lu J.-F., Chen H.-C., Zhang J., Wu G., Wang X., Troncoso P., Corn P., et al. Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers. Clin. Cancer Res. 2016;22:1520–1530. doi: 10.1158/1078-0432.CCR-15-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beltran H., Prandi D., Mosquera J.M., Benelli M., Puca L., Cyrta J., Marotz C., Giannopoulou E., Chakravarthi B.V.S.K., Varambally S., et al. Divergent Clonal Evolution of Castration-Resistant Neuroendocrine Prostate Cancer. Nat. Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marín-Aguilera M., Codony-Servat J., Reig Ò., Lozano J.J., Fernández P.L., Pereira M.V., Jiménez N., Donovan M., Puig P., Mengual L., et al. Epithelial-to-Mesenchymal Transition Mediates Docetaxel Resistance and High Risk of Relapse in Prostate Cancer. Mol. Cancer Ther. 2014;13:1270–1284. doi: 10.1158/1535-7163.MCT-13-0775. [DOI] [PubMed] [Google Scholar]
- 14.Jiménez N., Reig Ò., Montalbo R., Milà-Guasch M., Nadal-Dieste L., Castellano G., Lozano J.J., Victoria I., Font A., Rodriguez-Vida A., et al. Cell Plasticity-Related Phenotypes and Taxanes Resistance in Castration-Resistant Prostate Cancer. Front. Oncol. 2020;10:594023. doi: 10.3389/fonc.2020.594023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beltran H., Wyatt A.W., Chedgy E.C., Donoghue A., Annala M., Warner E.W., Beja K., Sigouros M., Mo F., Fazli L., et al. Impact of Therapy on Genomics and Transcriptomics in High-Risk Prostate Cancer Treated with Neoadjuvant Docetaxel and Androgen Deprivation Therapy. Clin. Cancer Res. 2017;23:6802–6811. doi: 10.1158/1078-0432.CCR-17-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research Network The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mak M.P., Tong P., Diao L., Cardnell R.J., Gibbons D.L., William W.N., Skoulidis F., Parra E.R., Rodriguez-Canales J., Wistuba I.I., et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin. Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George J.T., Jolly M.K., Xu S., Somarelli J.A., Levine H. Survival Outcomes in Cancer Patients Predicted by a Partial EMT Gene Expression Scoring Metric. Cancer Res. 2017;77:6415–6428. doi: 10.1158/0008-5472.CAN-16-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbons D.L., Creighton C.J. Pan-Cancer Survey of Epithelial-Mesenchymal Transition Markers across the Cancer Genome Atlas. Dev. Dyn. 2018;247:555–564. doi: 10.1002/dvdy.24485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris W.P., Mostaghel E.A., Nelson P.S., Montgomery B. Androgen Deprivation Therapy: Progress in Understanding Mechanisms of Resistance and Optimizing Androgen Depletion. Nat. Clin. Pract. Urol. 2009;6:76–85. doi: 10.1038/ncpuro1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang R.Y.-J., Kuay K.T., Tan T.Z., Asad M., Tang H.M., Ng A.H.C., Ye J., Chung V.Y., Thiery J.P. Functional Relevance of a Six Mesenchymal Gene Signature in Epithelial-Mesenchymal Transition (EMT) Reversal by the Triple Angiokinase Inhibitor, Nintedanib (BIBF1120) Oncotarget. 2015;6:22098–22113. doi: 10.18632/oncotarget.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marín-Aguilera M., Codony-Servat J., Kalko S.G., Fernández P.L., Bermudo R., Buxo E., Ribal M.J., Gascón P., Mellado B. Identification of Docetaxel Resistance Genes in Castration-Resistant Prostate Cancer. Mol. Cancer Ther. 2012;11:329–339. doi: 10.1158/1535-7163.MCT-11-0289. [DOI] [PubMed] [Google Scholar]
- 23.Marín-Aguilera M., Reig Ò., Milà-Guasch M., Font A., Domènech M., Rodríguez-Vida A., Carles J., Suárez C., Del Alba A.G., Jiménez N., et al. The Influence of Treatment Sequence in the Prognostic Value of TMPRSS2-ERG as Biomarker of Taxane Resistance in Castration-Resistant Prostate Cancer. Int. J. Cancer. 2019;145:1970–1981. doi: 10.1002/ijc.32238. [DOI] [PubMed] [Google Scholar]
- 24.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster Analysis and Display of Genome-Wide Expression Patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saldanha A.J. Java Treeview—Extensible Visualization of Microarray Data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 26.Scher H.I., Halabi S., Tannock I., Morris M., Sternberg C.N., Carducci M.A., Eisenberger M.A., Higano C., Bubley G.J., Dreicer R., et al. Design and End Points of Clinical Trials for Patients with Progressive Prostate Cancer and Castrate Levels of Testosterone: Recommendations of the Prostate Cancer Clinical Trials Working Group. J. Clin. Oncol. 2008;26:1148–1159. doi: 10.1200/JCO.2007.12.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C., et al. Systematic RNA Interference Reveals That Oncogenic KRAS-Driven Cancers Require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hänzelmann S., Castelo R., Guinney J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Development Core Team R. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2020. [Google Scholar]
- 30.Wu W., Maneix L., Insunza J., Nalvarte I., Antonson P., Kere J., Yu N.Y.-L., Tohonen V., Katayama S., Einarsdottir E., et al. Estrogen Receptor β, a Regulator of Androgen Receptor Signaling in the Mouse Ventral Prostate. Proc. Natl. Acad. Sci. USA. 2017;114:E3816–E3822. doi: 10.1073/pnas.1702211114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaurasiya S., Widmann S., Botero C., Lin C.-Y., Gustafsson J.-Å., Strom A.M. Estrogen Receptor β Exerts Tumor Suppressive Effects in Prostate Cancer through Repression of Androgen Receptor Activity. PLoS ONE. 2020;15:e0226057. doi: 10.1371/journal.pone.0226057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyquist M.D., Corella A., Coleman I., De Sarkar N., Kaipainen A., Ha G., Gulati R., Ang L., Chatterjee P., Lucas J., et al. Combined TP53 and RB1 Loss Promotes Prostate Cancer Resistance to a Spectrum of Therapeutics and Confers Vulnerability to Replication Stress. Cell Rep. 2020;31:107669. doi: 10.1016/j.celrep.2020.107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abida W., Cyrta J., Heller G., Prandi D., Armenia J., Coleman I., Cieslik M., Benelli M., Robinson D., Allen E.M.V., et al. Genomic Correlates of Clinical Outcome in Advanced Prostate Cancer. Proc. Natl. Acad. Sci. USA. 2019;116:11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conteduca V., Wetterskog D., Sharabiani M.T.A., Grande E., Fernandez-Perez M.P., Jayaram A., Salvi S., Castellano D., Romanel A., Lolli C., et al. Androgen Receptor Gene Status in Plasma DNA Associates with Worse Outcome on Enzalutamide or Abiraterone for Castration-Resistant Prostate Cancer: A Multi-Institution Correlative Biomarker Study. Ann. Oncol. 2017;28:1508–1516. doi: 10.1093/annonc/mdx155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conteduca V., Jayaram A., Romero-Laorden N., Wetterskog D., Salvi S., Gurioli G., Scarpi E., Castro E., Marin-Aguilera M., Lolli C., et al. Plasma Androgen Receptor and Docetaxel for Metastatic Castration-Resistant Prostate Cancer. Eur. Urol. 2019;75:368–373. doi: 10.1016/j.eururo.2018.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graf R.P., Fisher V., Mateo J., Gjoerup O.V., Madison R.W., Raskina K., Tukachinsky H., Creeden J., Cunningham R., Huang R.S.P., et al. Predictive Genomic Biomarkers of Hormonal Therapy Versus Chemotherapy Benefit in Metastatic Castration-Resistant Prostate Cancer. Eur. Urol. 2022;81:37–47. doi: 10.1016/j.eururo.2021.09.030. [DOI] [PubMed] [Google Scholar]
- 37.Zhao S.G., Chang S.L., Erho N., Yu M., Lehrer J., Alshalalfa M., Speers C., Cooperberg M.R., Kim W., Ryan C.J., et al. Associations of Luminal and Basal Subtyping of Prostate Cancer With Prognosis and Response to Androgen Deprivation Therapy. JAMA Oncol. 2017;3:1663–1672. doi: 10.1001/jamaoncol.2017.0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cato L., de Tribolet-Hardy J., Lee I., Rottenberg J.T., Coleman I., Melchers D., Houtman R., Xiao T., Li W., Uo T., et al. ARv7 Represses Tumor-Suppressor Genes in Castration-Resistant Prostate Cancer. Cancer Cell. 2019;35:401–413.e6. doi: 10.1016/j.ccell.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Zhang Y., Li D., Ma X., Xu K., Ding B., Li H., Wang Z., Ouyang W., Long G., et al. Androgen Receptor Splice Variant 7 Predicts Shorter Response in Patients with Metastatic Hormone-Sensitive Prostate Cancer Receiving Androgen Deprivation Therapy. Eur. Urol. 2021;79:879–886. doi: 10.1016/j.eururo.2021.01.037. [DOI] [PubMed] [Google Scholar]
- 40.Bonkhoff H. Estrogen Receptor Signaling in Prostate Cancer: Implications for Carcinogenesis and Tumor Progression. Prostate. 2018;78:2–10. doi: 10.1002/pros.23446. [DOI] [PubMed] [Google Scholar]
- 41.Wu W., Wang L., Spetsieris N., Boukovala M., Efstathiou E., Brössner C., Warner M., Gustafsson J.-A. Estrogen Receptor β and Treatment with a Phytoestrogen Are Associated with Inhibition of Nuclear Translocation of EGFR in the Prostate. Proc. Natl. Acad. Sci. USA. 2021;118:e2011269118. doi: 10.1073/pnas.2011269118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gehrig J., Kaulfuß S., Jarry H., Bremmer F., Stettner M., Burfeind P., Thelen P. Prospects of Estrogen Receptor β Activation in the Treatment of Castration-Resistant Prostate Cancer. Oncotarget. 2017;8:34971–34979. doi: 10.18632/oncotarget.16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilson C., Ingleby F., Gilbert D.C., Parry M.A., Atako N.B., Ali A., Hoyle A., Clarke N.W., Gannon M., Wanstall C., et al. Genomic Profiles of De Novo High- and Low-Volume Metastatic Prostate Cancer: Results From a 2-Stage Feasibility and Prevalence Study in the STAMPEDE Trial. JCO Precis. Oncol. 2020;4:882–897. doi: 10.1200/PO.19.00388. [DOI] [PubMed] [Google Scholar]
- 44.Mateo J., Seed G., Bertan C., Rescigno P., Dolling D., Figueiredo I., Miranda S., Rodrigues D.N., Gurel B., Clarke M., et al. Genomics of Lethal Prostate Cancer at Diagnosis and Castration Resistance. J. Clin. Investig. 2020;130:1743–1751. doi: 10.1172/JCI132031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamid A.A., Gray K.P., Shaw G., MacConaill L.E., Evan C., Bernard B., Loda M., Corcoran N.M., Van Allen E.M., Choudhury A.D., et al. Compound Genomic Alterations of TP53, PTEN, and RB1 Tumor Suppressors in Localized and Metastatic Prostate Cancer. Eur. Urol. 2019;76:89–97. doi: 10.1016/j.eururo.2018.11.045. [DOI] [PubMed] [Google Scholar]
- 46.Corn P.G., Heath E.I., Zurita A., Ramesh N., Xiao L., Sei E., Li-Ning-Tapia E., Tu S.-M., Subudhi S.K., Wang J., et al. Cabazitaxel plus Carboplatin for the Treatment of Men with Metastatic Castration-Resistant Prostate Cancers: A Randomised, Open-Label, Phase 1–2 Trial. Lancet Oncol. 2019;20:1432–1443. doi: 10.1016/S1470-2045(19)30408-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sweeney C., Bracarda S., Sternberg C.N., Chi K.N., Olmos D., Sandhu S., Massard C., Matsubara N., Alekseev B., Parnis F., et al. Ipatasertib plus Abiraterone and Prednisolone in Metastatic Castration-Resistant Prostate Cancer (IPATential150): A Multicentre, Randomised, Double-Blind, Phase 3 Trial. Lancet. 2021;398:131–142. doi: 10.1016/S0140-6736(21)00580-8. [DOI] [PubMed] [Google Scholar]
- 48.Crabb S.J., Griffiths G., Marwood E., Dunkley D., Downs N., Martin K., Light M., Northey J., Wilding S., Whitehead A., et al. Pan-AKT Inhibitor Capivasertib With Docetaxel and Prednisolone in Metastatic Castration-Resistant Prostate Cancer: A Randomized, Placebo-Controlled Phase II Trial (ProCAID) JCO. 2021;39:190–201. doi: 10.1200/JCO.20.01576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prat A., Saura C., Pascual T., Hernando C., Muñoz M., Paré L., González Farré B., Fernández P.L., Galván P., Chic N., et al. Ribociclib plus Letrozole versus Chemotherapy for Postmenopausal Women with Hormone Receptor-Positive, HER2-Negative, Luminal B Breast Cancer (CORALLEEN): An Open-Label, Multicentre, Randomised, Phase 2 Trial. Lancet Oncol. 2020;21:33–43. doi: 10.1016/S1470-2045(19)30786-7. [DOI] [PubMed] [Google Scholar]
- 50.Paré L., Pascual T., Seguí E., Teixidó C., Gonzalez-Cao M., Galván P., Rodríguez A., González B., Cuatrecasas M., Pineda E., et al. Association between PD1 MRNA and Response to Anti-PD1 Monotherapy across Multiple Cancer Types. Ann. Oncol. 2018;29:2121–2128. doi: 10.1093/annonc/mdy335. [DOI] [PubMed] [Google Scholar]
- 51.Narrandes S., Xu W. Gene Expression Detection Assay for Cancer Clinical Use. J. Cancer. 2018;9:2249–2265. doi: 10.7150/jca.24744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen T., Wallden B., Schaper C., Ferree S., Liu S., Gao D., Barry G., Dowidar N., Maysuria M., Storhoff J. Analytical Validation of the PAM50-Based Prosigna Breast Cancer Prognostic Gene Signature Assay and NCounter Analysis System Using Formalin-Fixed Paraffin-Embedded Breast Tumor Specimens. BMC Cancer. 2014;14:177. doi: 10.1186/1471-2407-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw counts from NanoString nCounter gene expression data generated in this study are available in Supplementary Table S2.