Significance
Meiotic recombination is essential for producing genetically diversified haploid gametes and initiates from the programmed formation of double-strand breaks (DSBs), which usually take place in accessible chromatin with features of active transcription in multiple species. However, the relationship between the regulation of gene transcription and DSB hotspots is not well understood. Here, we demonstrate that the largest DNA polymerase epsilon subunit POL2A recruits SU(VAR)3-9 homologs SUVH2 to the meiotic DSB sites, thereby jointly repressing the transcription of meiotic DSB-associated genes via a nonclassical RdDM pathway. This study provides new insights into how gene transcription is suppressed at meiotic DSB sites in plants, which could be conserved in other organisms.
Keywords: meiosis, gene silencing, DNA polymerase epsilon, SUVH2
Abstract
Meiotic recombination is initiated by the SPORULATION 11 (SPO11)–triggered formation of double-strand breaks (DSBs) that usually occur in open chromatin with active transcriptional features in many eukaryotes. However, gene transcription at DSB sites appears to be detrimental for repair, but the regulatory mechanisms governing transcription at meiotic DSB sites are largely undefined in plants. Here, we demonstrate that the largest DNA polymerase epsilon subunit POL2A interacts with SU(VAR)3 to 9 homologs SUVH2 and SUVH9. N-SIM (structured illumination microscopy) observation shows that the colocalization of SUVH2 with the meiotic DSB marker γ-H2AX is dependent on POL2A. RNA-seq of male meiocytes demonstrates that POL2A and SUVH2 jointly repress the expression of 865 genes, which have several known characteristics associated with meiotic DSB sites. Bisulfite-seq and small RNA-seq of male meiocytes support the idea that the silencing of these genes by POL2A and SUVH2/9 is likely independent of CHH methylation or 24-nt siRNA accumulation. Moreover, pol2a suvh2 suvh9 triple mutants have more severe defects in meiotic recombination and fertility compared with either pol2a or suvh2 suvh9. Our results not only identify a epigenetic regulatory mechanism for gene silencing in male meiocytes but also reveal roles for DNA polymerase and SUVH2/9 beyond their classic functions in mitosis.
Meiosis is a specialized cell division, required for eukaryotic sexual reproduction, that produces haploid gametes from diploid progenitor cells. During meiotic prophase I, meiotic recombination is initiated from double-strand breaks (DSBs) generated by a conserved DNA topoisomerase-VI-like complex that includes SPO11 (SPORULATION 11) (1, 2). Meiotic DSBs can be repaired by several mechanisms, including those that result in exchanges of DNA between homologous chromosomes known as crossovers (COs) (3–6). COs, in addition to creating novel allelic combinations, are also required for proper chromosome segregation.
Studies in several organisms show that meiotic DSBs have a strikingly nonrandom distribution along chromosomes and preferentially cluster in small regions (∼1 to 2 kb) called hotspots (7, 8). Epigenetic and genomic features are important features of DSB hotspot location and activation (7–10). In budding yeast, the histone H3K4 methyltransferase Set1 complex tethers H3K4me3 on the chromosome axis to promote DSB formation (11, 12). In several mammalian species, including mice and humans, a SET domain protein called PR/SET DOMAIN 9 (PRDM9) binds specific DNA motifs and catalyzes both H3K4me3 and H3K36me3 to trigger DSBs in adjacent nucleosome-depleted regions (13–16). Although plants lack PRDM9 homologs, DSB and CO hotspots in Arabidopsis are also associated with H2A.Z, H3K4me3, and low nucleosome density (LND) and preferentially occur near transcriptional start sites (TSSs) (9, 17). Thus, DSB sites share features of active chromatin that are also associated with RNA polymerase II (RNA POL II) activity (17). However, in budding yeast, DSB sites are enriched near the TSSs of genes with a high density of H3K4me3, but RNA POL II cannot localize and facilitate transcription at DSB sites due to competition from the recombinase Mer2 (12, 18). Mammalian DSBs initiated by PRDM9 are preferentially located far from promoter and enhancer regions to avoid gene transcription and inefficient DSB repair (10, 19, 20). In human somatic cells, transient transcriptional silencing is triggered when DSBs occur near transcriptionally active genes, which is important for DNA repair and genomic stability (21). DNA-RNA hybrids induced by transcription have been shown to impede recombinational repair (22); thus, aberrant active transcription at meiotic DSB sites may also lead to genome instability. Meiotic DSBs in plants are also preferentially initiated in accessible chromatin (4, 9), but the transcriptional state and the molecular mechanism that prevents transcriptional activation at DSB sites are still unknown.
DNA replication guarantees both genetic and epigenetic inheritance during cell cycles. DNA polymerases, such as POL α, POL ε, and POL δ, play a role in the preservation of the epigenetic memory (23, 24). In Arabidopsis, both POL α and POL ε regulate the expression of flowering genes by affecting H3K27me3 (25–27). In fission yeast, POL ε binds a silencing complex to promote chromatin assembly via recruiting RNA interference (RNAi) machinery and histone methyltransferase of H3K9me2 (28, 29). In budding yeast, POL ε is required for the inheritance of silenced telomeres and ribosomal DNA (rDNA) after replication (30, 31). More recently, Arabidopsis POL ε was found to prevent DNA CHG (H denotes A, T, or G) hypermethylation of transposable elements (TEs) when mediating heterochromatin silencing in mitosis (32). Although Pol ε is highly conserved in eukaryotes, its roles in plant epigenetic maintenance and gene silencing remain unclear, especially in meiocytes.
The SU(VAR)3 to 9 homologs (SUVHs) have a role in catalyzing H3K9 methylation (33, 34). For example, SUVH4, SUVH5, and SUVH6 help mediate H3K9 dimethylation, which reinforces CHG methylation (35, 36). Highly homologous SUVH2 and SUVH9 lack a post-SET domain and do not have histone methyltransferase activity, but can bind methylated DNA through their N-terminal (NT) really interesting new gene (RING)-associated (SAR) domains to participate in RNA-directed DNA methylation (RdDM) (37, 38). SUVH2 and SUVH9 play an important role in the accumulation of RNA POL IV-dependent small interfering RNAs (siRNAs) and recruiting RNA POL V to promote CHH methylation at most RdDM loci, suggesting that SUVH2 and SUVH9 function redundantly (37, 39). Moreover, the two proteins also work with adenosine triphophatases (ATPases) MORC1/MORC6 and the chromatin-remodeling complex to mediate chromatin condensation, thus enhancing transcriptional gene silencing downstream of RdDM (40, 41).
Meiotic DSB repair involves DNA synthesis and chromatin remodeling, but the relationship between them is elusive. In this study, we found that the catalytic subunit of POL ε, POL2A interacts with SUVH2 and SUVH9 to regulate meiotic gene silencing. The genes suppressed by POL2A-SUVH2/9 have characteristics associated with DSB hotspots, and POL2A is required for the localization of SUVH2 on DSB sites in meiocytes. Although POL2A and SUVH2/9 have a genome wide effect on CHH methylation specifically in meiocytes, they likely suppress these genes independently of CHH methylation and 24-nt siRNAs. SUVH2/9 deficiency alone has relatively minor effects on meiotic gene silencing and DSB repair, but has a stronger phenotype when POL2A function is compromised. The pol2a suvh2 suvh9 triple mutants have severe defects in gene silencing at DSB sites and meiotic DSB repair. Taken together, our results provide an insight that the POL2A-SUVH2/9 module plays important roles in mediating DSB-associated transcriptional silencing and maintaining normal DSB repair in meiocytes.
Results
POL2A Interacts with SU(VAR)3 to 9 Homolog SUVH2 In Vitro and In Vivo.
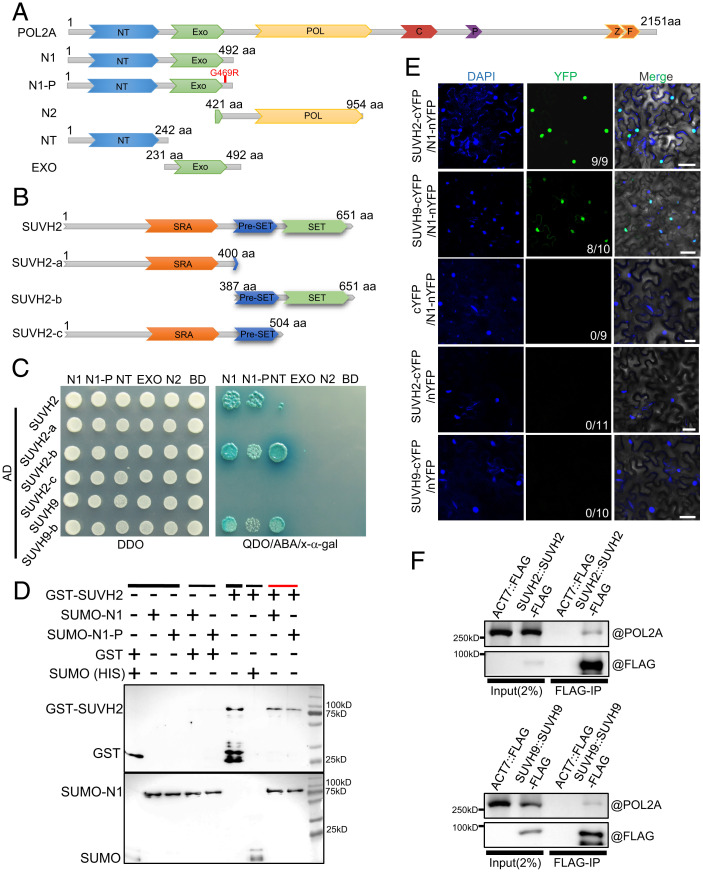
Arabidopsis POL2A is the catalytic subunit of Pol ε and functions not only in DNA replication but also in many other biological processes (24). Previously, by using two hypomorphic alleles, pol2a-1 and pol2a-2, we showed that AtPOL2A plays a role in regulating meiotic DSB repair (42). The mutations in these alleles are in their NT, so we used yeast two-hybrid (Y2H) screening to identify proteins that interact with the POL2A NT (N1: NT plus EXO; SI Appendix, Fig. S1A). We identified one candidate, the SU(VAR)3 to 9 homolog SUVH2 (38). We also used Y2H assays to demonstrate that SUVH2 interacts with the POL2A NT domain via its C-terminal Pre-SET-SET domain (Fig. 1 A–C). As SUVH2-c (SRA and pre-SET domains) is insufficient for the interaction with POL2A (Fig. 1 B and C), we speculate that the SET domain of SUVH2 is the direct interaction region for POL2A. Moreover, we found that the G469R mutation adjacent to the EXO domain in pol2a-2 attenuates its interaction with SUVH2 (Fig. 1 A, C, and D and SI Appendix, Fig. S1B). Previous studies showed that SUVH2 and its homolog SUVH9 function redundantly in CHH methylation in the RdDM pathway in somatic cells (37–39). However, POL2A only interacts with the C terminus of SUVH9 (SUVH9-b) rather than the full-length SUVH9 in the Y2H assay (Fig. 1C). We further confirmed that the full-length and C termini of both SUVH9 and SUVH2 interact with the POL2A NT using a bimolecular fluorescence complementation (BiFC) assay in tobacco (Fig. 1 B and E and SI Appendix, Fig. S1 C and D). To confirm their interaction in vivo, we raised and validated a polyclonal antibody against POL2A, and used coimmunoprecipitation (coIP) to show that POL2A coprecipitates with both SUVH2-FLAG and SUVH9-FLAG (Fig. 1F). These results suggest that POL2A can form a complex with SUVH2/9 in vivo.
Fig. 1.
N-terminal of POL2A interacts with SET-domain protein SUVH2. (A) Illustration of POL2A protein showing the full-length, point mutation, and different truncated forms. (B) Diagrams showing the full-length and truncated forms of SUVH2. (C) The N-terminal (N1) of POL2A interacts with the SET domain SUVH2 by Y2H assay. The point mutation near EXO domain (pol2a-2) of POL2A may attenuate the interaction between them. DDO refers to SD/-Leu/-Trp medium, and QDO refers to SD/-Ade/-His/-Leu/-Trp medium. (D) Pull-down assay showing the interaction between POL2A and SUVH2 in vitro. N1 of POL2A with SUMO-HIS and SUVH2 with GST were precipitated using Ni-NTA beads. Black lines indicate input (thick line) and negative control (thin line), and the red line indicates the experimental group. (E) The interaction between POL2A and SUVH2/SUVH9 as verified by BiFC assay. At least 9 leaves from 3 individual plants were used for infiltration. Numbers in the Bottom Right Corners indicate the number of positive interactions/all leaves for infiltration. (F) In vivo coIP of POL2A and SUVH2/SUVH9 using SUVH2/SUVH9-FLAG transgenic plant inflorescences by anti-FLAG antibody.
POL2A and SUVH2/9 Jointly Repress a Particular Class of Gene Transcription in Meiocytes.
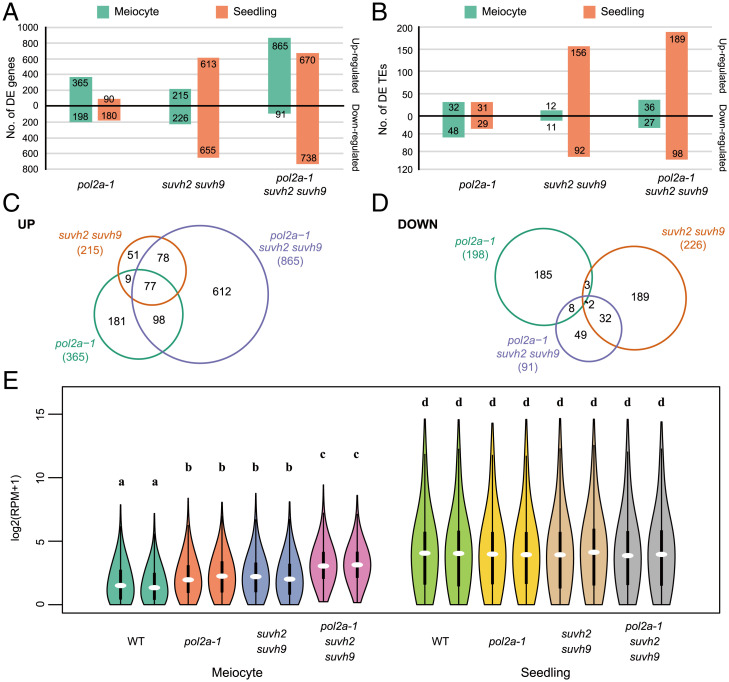
Because SUVH2/9 is required for TE and gene silencing (38, 39, 41), we used mRNA sequencing to examine the transcriptomes of meiocytes and seedlings in pol2a-1, suvh2 suvh9 and pol2a-1 suvh2 suvh9 (SI Appendix, Figs. S2 and S3 A and B). Compared with pol2a-1, suvh2 suvh9 has more differentially expressed genes (DEGs) and TEs in seedlings than in meiocytes (Fig. 2 A and B). The abundance of up-regulated TEs in suvh2 suvh9 seedlings is consistent with previous findings (38, 39, 41). Interestingly, DEGs/TEs in the pol2a-1 suvh2 suvh9 triple mutant seedlings resemble those in the suvh2 suvh9 double mutant more than the pol2a-1 single mutant, but the patterns differ in meiocytes (Fig. 2 A and B and SI Appendix, Fig. S3 C and D). In meiocytes, pol2a-1 suvh2 suvh9 has a pattern similar to pol2a-1 but has a dramatic increase in up-regulated genes (Fig. 2 A and B and Dataset S1). We observed that only a few TEs are up-regulated in the meiocytes of all mutants, including suvh2 suvh9 (Fig. 2B). These results suggest that POL2A and SUVH2/9 have distinct roles in regulating the expression of genes/TEs in meiocytes relative to seedlings. We then examined the up-regulated genes in pol2a-1 (365), suvh2 suvh9 (215), and pol2a-1 suvh2 suvh9 (865) meiocytes. As expected, the 865 genes de-repressed in pol2a-1 suvh2 suvh9 overlap significantly with those in pol2a-1 and suvh2 suvh9 (Fig. 2C). We also examined the down-regulated genes in these mutants, but did not find significant overlap, suggesting that POL2A and SUVH2/9 do not directly function in promoting gene expression (Fig. 2D). Moreover, the overall expression level of these 865 genes is heavily suppressed in meiocytes compared to seedlings in wild type (WT), and is increased in pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9 meiocytes, but not in seedlings (Fig. 2E and SI Appendix, Fig. S3E). We also examined the expression of 111 genes with known meiotic functions, and with the exception of POL2A, which is significantly down-regulated in the corresponding mutants, we did not observe any other significant changes in gene expression (Dataset S2), indicating that POL2A and SUVH2/9 do not have a specific effect on the expression of known meiotic genes. We then performed Gene Ontology (GO) analysis for the 865 genes up-regulated in pol2a-1 suvh2 suvh9 and found that the enriched terms are mostly related to abiotic/biotic stimulus (SI Appendix, Fig. S3F). These results support the hypothesis that POL2A and SUVH2/9 specifically suppress expression of a particular class of genes in meiocytes.
Fig. 2.
POL2A and SUVH2/9 mediate gene silencing in meiocytes. (A) Diagram showing the number of DEGs in meiocytes and seedlings of pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9 compared to WT. (B) Diagram showing the number of DE TEs in meiocytes and seedlings of pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9 compared to WT. (C) Venn diagram for up-regulated genes in meiocytes of each mutant compared to WT. (D) Venn diagram for down-regulated genes in meiocytes of each mutant compared to WT. (E) Violin plot for the expression level of the 865 up-regulated genes of triple mutants in meiocytes and seedlings of WT and each mutant. The letters above violins indicate different homogenous subsets based on the results of Tukey’s test (95% confidence level).
To validate our observations using the pol2a-1 allele, we also tested the transcriptomic changes in pol2a-2 meiocytes and seedlings (SI Appendix, Fig. S4). The transcriptional profile of pol2a-2 meiocytes resembles pol2a-1 with more up-regulated genes than down-regulated ones, but the number of affected genes is higher in pol2a-2 compared with pol2a-1 (SI Appendix, Fig. S4 C, E, and F), possibly because the former is a stronger allele. Specifically, although the up-regulated genes and TEs in pol2a-1 and pol2a-2 have substantial overlap in meiocytes and seedlings, they are primarily not shared between meiocytes and seedlings (SI Appendix, Fig. S4 E and G), supporting a divergent function for POL2A in seedlings and meiocytes. GO analysis of the combined 791 up-regulated genes from both pol2a mutants also revealed an enrichment of GO terms related to biotic/abiotic stimulus but not to meiosis (SI Appendix, Fig. S4I).
Genes Transcriptionally Suppressed by POL2A-SUVH2/9 Have Characteristics Associated with DSB Hotspots.
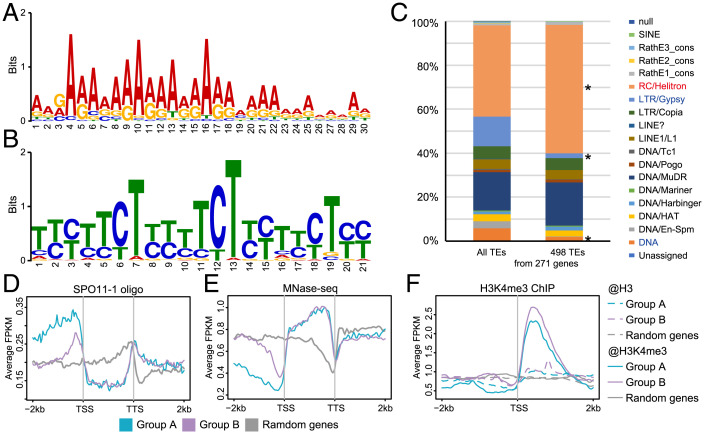
The functional enrichment analysis of up-regulated genes in pol2a and pol2a-1 suvh2 suvh9 shows GO terms similar to those associated with meiotic DSB hotspots according to a previous report (9). We then examined the relationship of the 865 genes with DSB hotspot-associated characteristics by analyzing the genomic features in the windows of gene bodies and 2-kb flanking sequences around these genes. We found that A-rich and CTT-repeat motifs known to be enriched near DSBs and CO hotspots (9, 17, 43–45) exhibit high enrichment in the promoters (852/865) and gene bodies (821/865), respectively (Fig. 3 A and B). Different TE families were also reported to be correlated and anticorrelated with SPO11-1-oligo hotspots (9), among which SPO11-1-oligo-associated RC/Helitron TEs are the most preferentially represented in the promoter region of the 865 genes, whereas SPO11-oligos anticorrelated LTR/Gypsy TEs are depleted (Fig. 3C). We further investigated the other DSB-associated features around these genes using published data (9, 46). As expected, SPO11-1 oligos are overrepresented in the promoters, especially near the TSSs (SI Appendix, Fig. S5A), and the nucleosome occupancy levels at TSSs are lower, compared to random genes (SI Appendix, Fig. S5B). Moreover, H3K4me3 occupancy of these genes is lower than the average level near the TSS with the depleted nucleosomes, but higher downstream from the TSS (SI Appendix, Fig. S5C). These features are consistent with the notion that the A-rich motif excludes nucleosomes and the CTT-repeat motif is associated with H3K4me3 (43). Taken together, these results provide strong evidences that TSSs of these 865 genes occupy obvious DSB hotspot characteristics (9).
Fig. 3.
The 865 genes regulated by POL2A-SUVH2/9 have features of meiotic DSB hotspots. (A) WebLogo plot for a 30-bp A-rich motif identified in promoter regions (2 kb upstream TSS) of the 865 up-regulated genes from pol2a-1 suvh2 suvh9 meiocytes. (B) WebLogo plot for a 21-bp CTT-repeat motif identified in gene bodies of the 865 up-regulated genes. (C) Proportion of TE superfamilies located in promoter regions of genes. Only 271 genes from the 865 genes have TEs in the promoters. RC/Helitron is significantly enriched, and LTR/Gypsy and DNA are significantly depleted among TEs located in promoters (2 kb) of the 865 up-regulated genes compared to all TEs (P < 0.001, Fisher's exact test). (D) Density of SPO11-1-oligos around the 865 up-regulated genes and random genes in WT. (E) Nucleosome occupancy around the 865 up-regulated genes and random genes in WT. (F) Distribution of H3K4me3 along the 865 up-regulated genes and random genes in WT. The 865 up-regulated genes are divided into genes with Helitron TEs in promoters as group A and others without Helitron TEs in promoters as group B.
We further divided the 865 genes into two groups according to the presence of RC/Helitron TEs in their promoters (Fig. 3C): group A includes 173 genes with RC/Helitron TEs in their promoters and group B includes the remaining 692 genes. Interestingly, group A promoters have more DSB hotspot features than group B, including a higher enrichment of SPO11-1-oligos (Fig. 3D), a lower density of nucleosomes (Fig. 3E), and a higher H3K4me3 at TSSs (Fig. 3F). Hence, among the 865 genes, the TSSs of genes with RC/Helitron TEs in their promoters may be peculiarly prone to DSBs.
SUVH2 Is Recruited to Meiotic Chromatin and DSB Sites in a POL2A-Dependent Manner.
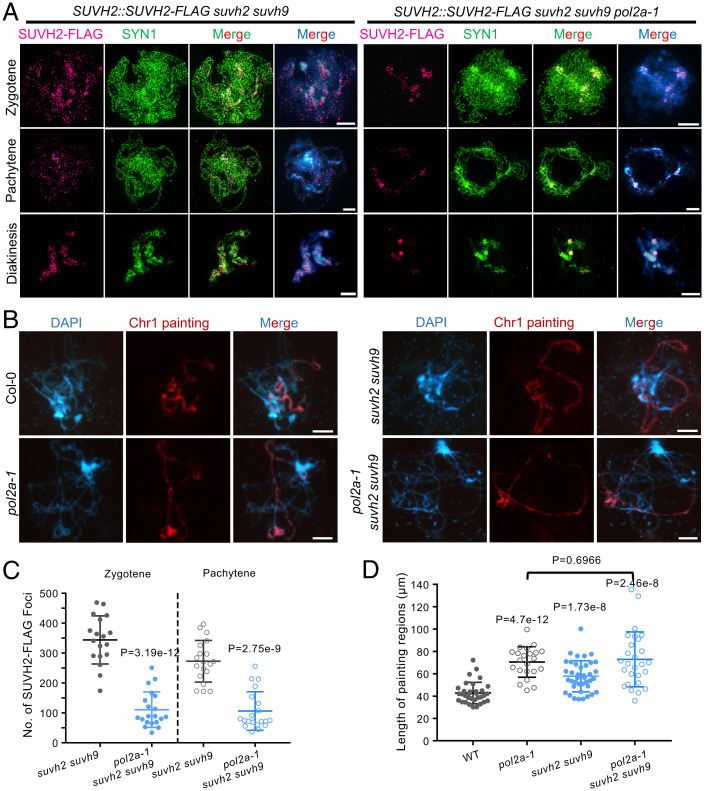
To examine how POL2A and SUVH2 function together in meiosis, we generated SUVH2pro::SUVH2-FLAG transgenic plants in suvh2 suvh9 and pol2a-1 suvh2 suvh9 backgrounds. We validated the expression of SUVH2-FLAG in these transgenic plants (SI Appendix, Fig. S6A), and found that POL2A expression decreases dramatically in pol2a-1 suvh2 suvh9, similar to our previous RT-PCR results in pol2a-1 (42). We then performed immunofluorescence (IF) in male meiocytes using anti-FLAG antibody with N-SIM and found punctate SUVH2-FLAG signals mainly distributed along chromosomes from zygotene to diakinesis in suvh2 suvh9 (Fig. 4A and SI Appendix, Fig. S6B). In contrast, SUVH2-FLAG signals are significantly reduced at all stages in pol2a-1 suvh2 suvh9 meiocytes (Fig. 4 A and C). Notably, the residual SUVH2-FLAG signal in some pol2a-1 suvh2 suvh9 meiocytes tends to associate with DAPI-bright heterochromatic regions (Fig. 4A). These results support the idea that POL2A is required for SUVH2 localization and distribution on meiotic chromosomes.
Fig. 4.
POL2A is required for SUVH2 localization on meiotic prophase I chromosome and functions in chromatin condensation. (A) Immunostaining detecting localization of SUVH2-FLAG at zygotene, pachytene, and diakinesis in suvh2 suvh9 and suvh2 suvh9 pol2a-1 backgrounds. Cohesion SYN1 is used as a control to infer the chromatin regions. In suvh2 suvh9, SUVH2-FLAG tends to form small bodies along chromosome regions stained by DAPI before diakinesis. In contrast, the SUVH2-FLAG is significantly reduced in suvh2 suvh9 pol2a-1. (B) Chr I painting at pachytene in WT, pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9. (C) Statistics of SUVH2-FLAG foci at zygotene and pachytene in suvh2 suvh9 and suvh2 suvh9 pol2a-1 backgrounds. P values were calculated using the two-tailed Student’s t test. (D) The average length of Chr I painted regions in WT, pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9. P values report the comparison between WT and mutants, unless otherwise specified, using the two-tailed Student’s t test. Scale bars: 5 µm.
During meiosis chromosomes undergo dramatic conformational changes from long threads to highly compacted bivalents. SUVH2/9 has been reported to associate with chromatin remodeling factors to promote chromatin condensation downstream of RdDM (40, 41). To evaluate the roles of POL2A and SUVH2/9 in chromatin compaction in meiocytes, we analyzed the length of chromosome I (Chr I) at pachytene in WT and different mutants using a chromosome-specific painting assay (Fig. 4B). Quantitative analysis shows that pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9 have longer Chr I signals compared to WT (WT< suvh2 suvh9 < pol2a-1 ∼ pol2a-1 suvh2 suvh9; Fig. 4D), indicating a less condensed state. Moreover, pol2a-1 and pol2a-1 suvh2 suvh9 have similar chromosome compaction phenotypes (Fig. 4D), which suggests that pol2a-1 may be epistatic to suvh2 suvh9 in regulating chromosome structure.
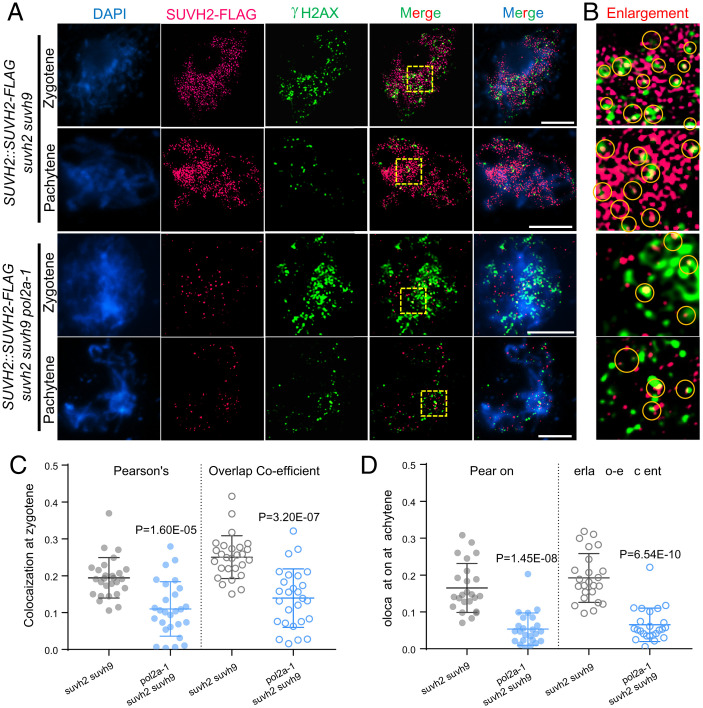
Meiotic DSBs are initiated by SPO11 during leptotene-zygotene transition (4) and are marked by γ-H2AX (phosphorylated histone H2AX) in multiple organisms (47). To explore whether SUVH2 is located at DSB sites at zygotene and pachytene, we examined the colocalization of SUVH2-FLAG with γ-H2AX using N-SIM (Fig. 5 A and B). At zygotene, we quantified the colocalization between SUVH2-FLAG and γ-H2AX signals using ImageJ (48). The overlap coefficient (0.25, n = 27) and Pearson’s correlation coefficient (r = 0.19, n = 27) show a certain degree of colocalization of SUVH2-FLAG and γ-H2AX (Fig. 5C). At pachytene, the number of γ-H2AX foci decreases sharply from 201 ± 26 (n = 27) to 78 ± 23 (n = 24) following DSB repair (Fig. 5A), whereas SUVH2-FLAG foci remain relatively persistent (Figs. 4 A and C and 5A). We were still able to determine some overlapping signals between the remaining γ-H2AX foci and SUVH2-FLAG foci with slightly decreased overlap coefficient (0.19, n = 24) and Pearson’s correlation coefficient (r = 0.17, n = 24) (Fig. 5 A, B, and D). In both zygotene and pachytene, some of the colocalized SUVH2-FLAG and γ-H2AX foci partially overlap (Fig. 5B), implying that the precise physical nature of the association is still unclear. In contrast, SUVH2-FLAG foci in pol2a-1 suvh2 suvh9 meiocytes are significantly reduced and have less colocalization with γ-H2AX in either zygotene (average r = 0.11, n = 27) or pachytene (average r = 0.05, n = 24) (Fig. 5 A–D). Based on these data, we hypothesize that POL2A not only functions in meiotic DSB-associated DNA synthesis (42) but also recruits SUVH2/9 to DSB sites to mediate nearby gene silencing.
Fig. 5.
POL2A is required for localization of SUVH2 at DSB sites in meiosis. (A) Immunostaining showing colocalization of SUVH2-FLAG (carmine) and γ-H2AX (green) at zygotene and pachytene in suvh2 suvh9 background and suvh2 suvh9 pol2a-1 background by N-SIM. Scale bars: 5 µm. (B) Enlarged images of the yellow squares in (A). Yellow cycles indicate the overlapped signals of SUVH2-FLAG and γ-H2AX. (C and D) Coefficient analysis of colocalization between SUVH2-FLAG and γ-H2AX at zygotene (C) and pachytene (D) in suvh2 suvh9 and suvh2 suvh9 pol2a-1. P values were calculated by the two-tailed Student’s t test.
POL2A and SUVH2/9 Likely Regulate Gene Silencing Independent of CHH Methylation.
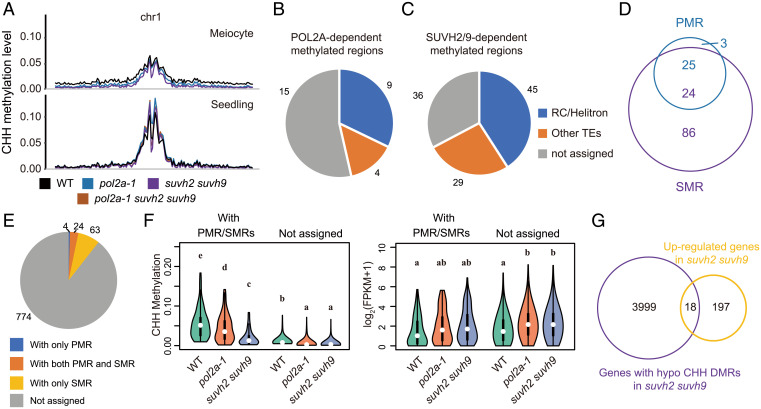
In somatic cells, SUVH2 and SUVH9 are required for the establishment of CHH methylation via the RdDM pathway (37–39). To investigate whether POL2A and SUHV2/9 regulate local DNA methylation, we assessed whole-genome DNA methylation of WT, suvh2 suvh9, and pol2a-1 meiocytes and seedlings (SI Appendix, Fig. S7 A–C) by bisulfite sequencing (BS-seq), as well as the published drm1 drm2 (designated as drm hereafter) methylome as a control (49). We did not analyze the pol2a-2 and the pol2a-1 suvh2 suvh9 triple mutants because they do not produce sufficient numbers of meiocytes. Consistent with previous observations (49), the CHH methylation of meiocytes is lower than that of seedlings in heterochromatin (Fig. 6A). In meiocytes, only CHH methylation is decreased in pol2a-1 on a genome-wide scale, not CG or CHG methylation, and this decrease is more severe in suvh2 suvh9 and drm (Fig. 6A and SI Appendix, Fig. S7 D and E). By contrast, CHH methylation in seedlings is only decreased in suvh2 suvh9 and pol2a-1 suvh2 suvh9, but not in pol2a-1 (Fig. 6A). To further characterize the role of POL2A and SUVH2/9 on DNA methylation in meiocytes, we analyzed the differentially methylated regions (DMRs) between the WT and mutants. We observed 1,327 and 5,843 hypo-CHH DMRs in the meiocytes of pol2a-1 and suvh2 suvh9, respectively (SI Appendix, Fig. S7F), in which 86.7% POL2A-dependent DMRs overlap with hypo-CHH DMRs in suvh2 suvh9 (SI Appendix, Fig. S7F), indicating that POL2A may interact with SUVH2/9 to facilitate CHH methylation in meiocytes. We then focused on the POL2A-SUVH2/9–mediated 865 genes and found that only a few POL2A- and SUVH2/9-dependent DMRs (28 POL2A-dependent methylated regions [PMRs] and 110 SUVH2/9-dependent methylated regions [SMRs]) are in the promoters of these genes (Fig. 6 B–D). When matching PMRs/SMRs to the genes, only 10.5% of the genes (91/865) have these hypomethylated regions in promoters (Fig. 6E). CHH methylation in promoters of the other 774 genes is also compromised in the 2 mutants, but the methylation level is very low (Fig. 6F and SI Appendix, Fig. S8A), and its contribution to gene silencing is difficult to determine. The changes in promoter CHH methylation and gene expression are not positively correlated, as demonstrated by low Pearson’s correlation coefficients (Fig. 6F and SI Appendix, Fig. S8 B and C). Consistent with these observations, genes known to be located in previously defined recombination hotspots such as 3a, 3b, RAC1, and 130x (17, 50–52) are all up-regulated in pol2a-1 suvh2 suvh9, but the adjacent genes are not (SI Appendix, Fig. S9A). Relatively low CHH DNA methylation around these genes is not significantly reduced in any of the mutants (SI Appendix, Fig. S9 B–E). In addition, we found that suvh2 suvh9 has very low CHH methylation levels (Fig. 6 A and F and SI Appendix, Fig. S8A), but has few up-regulated genes in meiocytes (Fig. 2C). Only 18 of 215 up-regulated genes have SMRs in promoters in suvh2 suvh9 (Fig. 6G). These results suggest that CHH methylation induced by POL2A-SUVH2/9 may contribute very little to gene silencing at DSB sites in meiocytes.
Fig. 6.
POL2A and SUVH2/9 likely suppress gene expression independently of CHH methylation. (A) The CHH methylation profiles of Chr 1 in meiocytes and seedlings of WT (black), pol2a-1 (blue), suvh2 suvh9 (purple), and pol2a-1 suvh2 suvh9 (brown). The global CHH methylation is less compromised in pol2a-1 meiocytes than that in suvh2 suvh9. (B and C) The genomic features of POL2A (B) and SUVH2/9-dependent (C) methylated regions (PMR and SMR, respectively) in the promoters of 865 up-regulated genes. More than half of these regions are located in TEs, the majority of which belong to the RC/Helitron superfamily. (D) Venn diagram showing the overlap between PMRs and SMRs. Most PMRs (25/28) overlap with SMRs, suggesting that POL2A may affect CHH methylation through the function of SUVH2/9. (E) A pie chart showing the distribution of PMRs and SMRs on 865 up-regulated genes in pol2a-1 suvh2 suvh9 compared to WT. Only 91 genes accommodate PMRs and SMRs. (F) Violin plots of the CHH methylation level (Left Panel) in promoters and expression level (Right Panel) of genes with or without PMR/SMRs shown in (E). Although the CHH methylation of these genes is compromised and the expression increased in both pol2a-1 and suvh2 suvh9, higher methylation and more changes in genes with PMR/SMRs do not promise more dramatic changes in gene expression. (G) Venn diagram showing the overlap between 215 up-regulated genes in suvh2 suvh9 and 4017 genes with SMRs in promoters. Most up-regulation in suvh2 suvh9 is not related to SMRs.
Gene Silencing or CHH Methylation Induced by POL2A and SUVH2/9 Is Independent of 24-nt siRNA Accumulation.
siRNAs are a key component of the RdDM pathway and functions in gene silencing (34, 53). SUVH2 and SUVH9 are also involved in the accumulation of RNA POL IV-dependent siRNAs in somatic cells (37, 39). Unexpectedly, neither pol2a-1 nor suvh2 suvh9 have significant 24-nt siRNA production defects in meiocytes (SI Appendix, Fig. S10 A and B), while suvh2 suvh9 seedlings produce significantly fewer 24-nt siRNAs (SI Appendix, Fig. S10 C and D), consistent with previous reports (37, 39). We examined the differential siRNA regions (DSRs, see Materials and Methods for details) in each mutant and found that only 956 and 295 regions have fewer (hypo) 24 nt-siRNAs mapped to them in pol2a-1 and suvh2 suvh9 meiocytes, respectively (SI Appendix, Fig. S10 E and G). In contrast, 5,034 24-nt hypo DSRs are observed in suvh2 suvh9 seedlings (SI Appendix, Fig. S10F). We found that very few hypo DSRs overlap with the POL2A-SUVH2-associated suppressed genes or methylated regions, and >83% of these genes show no nearby siRNA accumulation (SI Appendix, Fig. S10 H and I). These results resemble reports that the siRNA-independent POL V pathway is sufficient for low-level DNA methylation, and POL IV-dependent siRNA biogenesis enhances local DNA methylation (54). These results indicate that the POL2A-SUVH2/9 dependent gene silencing and CHH methylation in meiocytes may be independent of 24-nt siRNAs.
Loss of SUVH2/9 in pol2a-1 Shows Additive Defects in Meiosis.
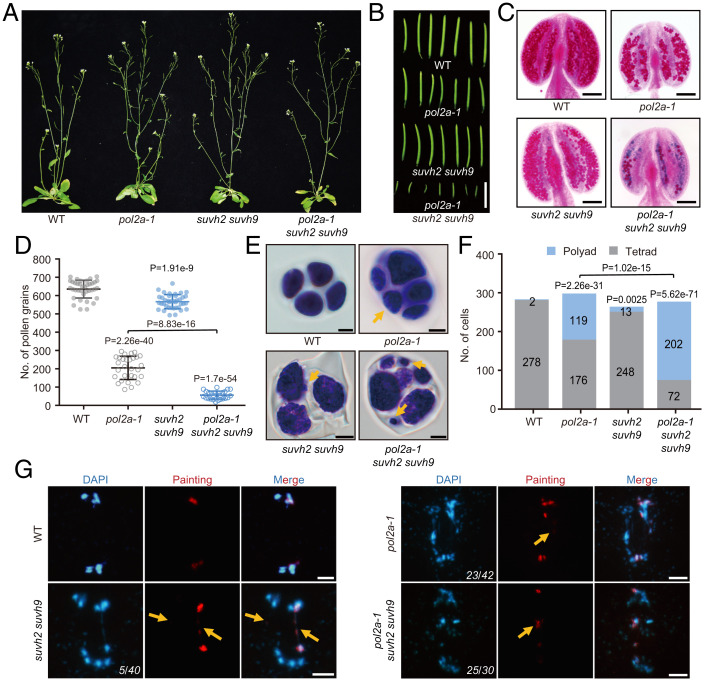
We previously showed that POL2A is required for fertility and meiotic recombination (42), but the effects of SUVH2/9 in meiotic DSB repair have not been investigated. To investigate the role of SUVH2/9 in meiosis, we examined the fertility of pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9 plants compared to WT. Consistent with previous results (42), pol2a-1 has shorter siliques, fewer viable pollen grains (205 ± 63, n = 29), and polyads of microspores rather than tetrads (119/295) (Fig. 7 A–F). In contrast, vegetative growth and silique length in suvh2 suvh9 are indistinguishable from WT (Fig. 7 A and B). Furthermore, suvh2 suvh9 has a slight reduction in viable pollen grains (567 ± 39, n = 39) compared to WT (636 ± 49, n = 37) (Fig. 7 C and D), and a small fraction of meiosis produced polyads with variably sized microspores (13/261) (Fig. 7 E and F). Unexpectedly, pol2a-1 suvh2 suvh9 plants have severe defects in silique length, number of viable pollen grains (56 ± 22, n = 26), and a higher frequency of polyad microspores (202/274) compared to pol2a-1 (Fig. 7 A–F). To check whether these phenotypes are reflected in meiotic chromosome behavior, we examined chromosome spreads stained with DAPI and observed no obvious difference before diakinesis among meiocytes from WT, pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9 plants (SI Appendix, Fig. S11 A–D). After diakinesis, a small fraction of suvh2 suvh9 meiocytes have abnormal chromosome morphologies, including chromosome bridges and fragments, compared with WT (Fig. 7G and SI Appendix, Fig. S11 A–E), potentially indicating recombination between nonhomologous chromosomes and/or unrepaired DSBs in the mutant. The triple mutant has more severe meiotic defects, including chromosome multivalents and chromosomal fragmentation from anaphase I to telophase II, compared to pol2a-1 (Fig. 7G and SI Appendix, Fig. S11 A–E). We also investigated another strong allele, pol2a-2, and pol2a-2 suvh2 suvh9 mutants. We found that the triple mutants have a little more severe meiotic defects than pol2a-2 (SI Appendix, Fig. S12), and both are more severe than pol2a-1 (Fig. 7). By comparing the transcription data (Fig. 2 and SI Appendix, Fig. S4), we found that more derepressed genes in meiocytes are accompanied by more serious DSB repair defects in these mutants. Since these genes are closely associated with DSB hotspots (Fig. 3), we speculate that the gene silencing induced by POL2A and SUVH2/9 at DSB sites is important for normal meiotic DSB repair in Arabidopsis.
Fig. 7.
POL2A and SUVH2 cooperatively function in meiotic progression and fertility. (A–C) Whole plants, the first 7 siliques, and Alexander red–stained anthers of WT, pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9. (D) The number of viable pollen grains of WT, pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9. Two-tailed Student’s t test. (E) Developing pollen at the tetrad stage of WT, pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9 by toluidine blue staining. Yellow arrows indicate small nuclear bodies. (F) Histogram of counts of microspore tetrads and polyads in WT and mutants. P values were calculated using the χ2 test. (G) Chromosome spreads with Chr I painting of WT, pol2a-1, suvh2 suvh9, and pol2a-1 suvh2 suvh9 at anaphase I. Yellow arrows indicate the chromosome fragmentation or chromosome bridges. Numbers in the Bottom Right Corners indicate the number of abnormal cells out of all of the cells counted. Scale bars: A, 3 cm; B, 1 cm; C, 1 mm; E, 5 μm, G, 5 μm. P values refer to the comparison between WT and mutants unless otherwise specified.
Discussion
Transcriptional Silencing at DSB Sites.
Meiotic recombination is initiated by the formation of SPO11-induced DSBs (3–6), which are preferentially localized in euchromatic regions with active transcription markers, including H3K4me3, H2A.Z, and LND (7–10). In Saccharomyces cerevisiae, DSBs are preferentially localized near TSSs with high levels of H3K4me3, but the activity of POL II and gene expression are inhibited (12, 18). In contrast, in many mammals, DSB hotspots are distributed far away from TSSs, relying on the function of PRDM9 (10, 19, 20). In prdm9, meiotic DSB formation is unaffected, but their distribution reverts to localization near TSSs with enriched H3K4me3 (19), resulting in the inefficient repair of DSBs and pachytene arrest (55). Mutant dnmt3L (required for de novo DNA methylation) mice have abnormally high numbers of meiotic DSBs and transcriptional activity at retrotransposons, which results in meiotic arrest at pachytene (56). In this study, we identified a mechanism involving POL2A and SUVH2/9 for regulating gene silencing at DSB sites in Arabidopsis. Unlike RdDM-dependent DNA CHH methylation associated with TE silencing in somatic cells (Fig. 2B) and previous reports (34, 53, 57), we found that POL2A recruits SUVH2 to meiotic DSB sites (Fig. 5) and represses a particular class of genes with characteristics associated with DSB hotspots (Figs. 2 and 3). Congruously, we observed the derepression of genes and an increase in meiotic DSB repair defects in pol2a, suvh2 suvh9, and triple mutant (Figs. 2 and 7), indicating the derepressed transcription at meiotic DSB sites may lead to impaired repair. Consistent with our results, a recent report showed that DNA-RNA hybrid accumulation at DNA breaks interferes with DSB repair (22). These data suggest that aberrant active transcription at meiotic DSB sites can impede the repair of DSBs, leading to meiotic defects.
How Do POL2A and SUVH2/9 Regulate Gene Transcription at DSB Sites?
As a DNA polymerase, POL ε is known to function mainly in DNA synthesis during DNA replication or repair (58, 59). POL ε itself may be incapable of affecting epigenetic states or regulating gene expression, but it is able to associate with different epigenetic regulators to mediate chromatin condensation and modification in some organisms (24, 28). For example, POL ε associates with the RNAi machinery, H3K9me2 methyltransferase, deacetylases, and chromatin remodelers to play a crucial role in the heterochromatin assembly and transcriptional silencing in fission yeast (28, 29). In plants, POL ε interacts with Polycomb repressive complex (PRC) proteins to silence some flowering genes by inducing H3K27me3 in seedlings (25). In meiocytes, our results demonstrate that the catalytic subunit of POL ε (POL2A) can recruit SUVH2/9 to promote DSB-associated gene silencing in meiocytes (Figs. 2 and 3). SUVH2/9 is known as an RdDM component in somatic cells (39). Although POL2A-SUVH2/9 has effects on CHH methylation in meiocytes, CHH methylation may not directly play a role in gene silencing in meiocytes, or only CHH methylation is not sufficient for gene silencing (Fig. 6 E–G and SI Appendix, Figs. S8 and S9). The majority of the up-regulated genes in mutants lack siRNA accumulation (SI Appendix, Fig. S10H). Our explanation of these results is that, in meiocytes, POL2A-SUVH2/9 may directly associate POL V and DRM1/2 for establishing siRNA-independent CHH methylation. This is consistent with previous reports that a POL V–dependent pathway is sufficient to trigger CHH methylation even in the absence of POL IV and siRNAs, whereas siRNA biogenesis can further potentiate targeted methylation (54). This is also supported by a recent report that male meiocytes likely import siRNAs from tapetal cells in a nonautonomous manner and POL IV–generated siRNAs and POL V–mediated CHH methylation are likely decoupled in meiocytes (60, 61). Except to promoting CHH methylation, SUVH2/9 can also interact with ATPases MORC1/MORC6 and chromatin remodeling factors to promote gene silencing and chromatin condensation downstream of RdDM (40, 41). Indeed, we found that POL2A and SUVH2/9 are required for chromatin condensation in meiocytes (Fig. 4D). Hence, POL2A may recruit SUVH2/9 to directly promote chromatin remodeling and condensation for gene silencing after DNA repair at meiotic DSB sites. In addition, CHH methylation is also required for SUVH2/9 residence (37) and chromatin remodeling downstream (41), which may be a by-product or footprint at meiotic DSB sites, especially on the 774 up-regulated genes with low CHH methylation (Fig. 6 E and F). A reasonable hypothesis is that POL2A facilitates the localization of SUVH2/9 to the newly synthesized DNA without DNA methylation at meiotic DSBs. SUVH2/9 then induces low CHH methylation without siRNA, and maintains its localization via SRA domain binding methylated DNA after DSB repair. These actions will corporately promote gene silencing and chromatin condensation at meiotic DSB sites.
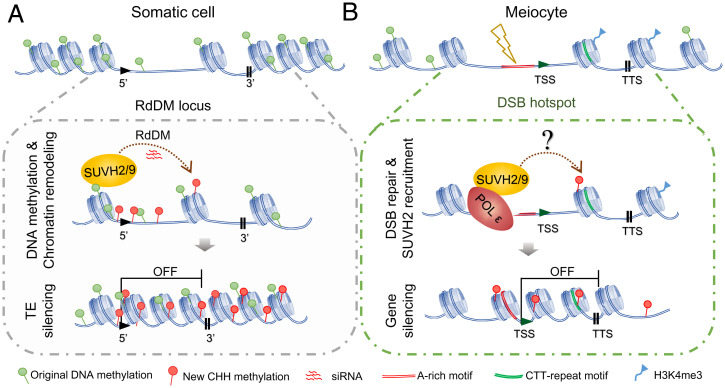
Based on these results, we present a model for comparing the mechanisms of SUVH2/9-dependent transcriptional silencing between somatic cells and meiocytes (Fig. 8). In somatic cells, SUVH2/9 involves two steps for transcriptional silencing at targeted loci, including siRNA-directed CHH methylation and chromosome condensation (Fig. 8A), in which SUVH2/9 can bind to methylated CG/CHG and trigger local DRM2-dependent CHH methylation (37, 38). Meanwhile, they interact with MORC proteins and chromatin remodeling complex to promote chromosome remodeling (40, 41). In contrast, in meiocytes, meiotic recombination initiates at DSBs that are enriched at gene TSSs within the “open” chromatin (Fig. 8B), which tend to be nucleosome-depleted, AT-rich, and seldom methylated in CG and CHG (9, 50, 62), with CTT-repeat motifs and enriched H3K4me3 in the neighboring gene bodies (9, 17, 43, 45) (Fig. 3 and SI Appendix, Fig. S5). Following DSB formation and procession, POL ε is recruited to the DSB sites to perform DNA synthesis during DSB repair (42). In the DSB hotspots, the DNA methylation level is quite low (9, 62), and SUVH2/9 may not be able to bind to these sites by its SRA domain. In this case, POL ε recruits SUVH2/9 using the NT domain of POL2A and directly promotes chromatin remodeling and condensation to suppress gene expression downstream of DSB sites (Fig. 8B). Meanwhile, male meiocytes seem to lack the canonical POL IV–dependent siRNA pathway (60). SUVH2 probably induces low levels of CHH methylation, depending on POL V and DRM2 at these sites (54). In pol2a-1 meiocytes the formation of meiotic DSBs appears unaffected (42), but the localization of SUVH2 and SUVH9 to DSB sites is compromised. As a consequence, RNA POL II is able to access the open chromatin to start gene transcription, which may be harmful to meiotic recombinational repair (22). In addition, POL2A may also function in gene silencing in a SUVH2-independent manner at DSB sites, because only limited significant up-regulated genes are found in the meiocytes of suvh2 suvh9 compared to those of the triple mutant (Fig. 2C). It is plausible that there is double insurance to suppress genes at DSB sites in meiocytes. For example, POL2A interacts with the PRC-dependent H3K27me3 pathway, similar to that in seedlings (25). Similarly, transcriptional silencing at DSB sites in mammalian cells is corporately induced by multilayered pathways, including catalyzing local H3K27me3, and DNA methylation is also found to increase in CpG islands at DNA damage sites (21). Future studies are warranted to identify the in-depth mechanisms that are involved in DSB-associated gene silencing in meiocytes.
Fig. 8.
A working model showing how POL2A-SUVH2/9 regulates meiotic DSB-associated gene silencing. (A) In somatic cells, the RdDM sites have highly methylated DNA. SUVH2/9 commonly recognize methylated CG/CHH to generate CHH methylation via the canonical RdDM pathway on heterochromatin. However, SUVH2/9 bind to methylated DNA and interact with MORCs-SWI/SNF complex to promote chromatin remodeling and condensation at RdDM sites. The outcome of RdDM mainly contributes to transcriptional silencing, especially for TEs, at target loci. (B) In meiocytes, the programmed formation of DSBs usually takes place around TSSs of genes within “open” chromatins, which tend to be lowly methylated, nucleosome depleted, and AT rich. Given that meiocytes seem to lack the cell-autonomous mechanism to produce siRNAs, SUVH2/9 may not target DSB sites as that in somatic cells and can be recruited by POL ε, which is required for meiotic DSB-associated DNA synthesis. The POL2A-SUVH2/9 likely directs DNA methylation and chromatin condensation around DSB sites independently of the classical RdDM pathway, and mediates the downstream gene silencing.
Materials and Methods
Plant Materials and Growth Conditions.
The pol2a-1 (SALK_096341) (42), pol2a-2 (til1-4) (42), suvh2 (SALK_079574) (38), suvh9 (SALK_048033) (38), and suvh2 suvh9 (38) were described previously. The SUVH2pro::SUVH2-FLAG and SUVH9pro::SUVH9-FLAG constructs were transformed into suvh2 suvh9. Then, SUVH2pro::SUVH2-FLAG suvh2 suvh9 plants were crossed with pol2a-1+/−suvh2−/− suvh9−/− to obtain transformants in pol2a-1 suvh2 suvh9 background. T2/T3 generations of them were used for investigation. All of the plants were grown in a greenhouse at 20 °C under a long-day cycle (16 h light/8 h dark, humidity 75%).
Y2H Assay.
The GAL4-based Matchmaker Gold Yeast Two-Hybrid System (Clontech) was used for the Y2H assay. N1 of POL2A was amplified from the complementary DNA (cDNA) of Arabidopsis and cloned into pGBKT7 vectors (Clontech) using EcoRI and SalI restriction enzyme sites. The construct was transformed into Y2H gold yeast strain using the LiAc/polyethylene glycol method (63). Then, the N1 yeast strain was used as bait to screen the Y187 strains containing the cDNA library of Arabidopsis inflorescence in pGADT7 (Clontech) following the manufacturer’s instructions.
The point mutation of N1 (N1-P) was constructed using the Fast Mutagenesis System (TransGen, FM201-01). N1-P, NT, EXO, and N2 of POL2A were also cloned into the pGBKT7 vector carried by the Y2H gold yeast strain. Whole coding regions and truncated forms of SUVH2 and SUVH9 were cloned into pGADT7 and were transformed into Y187 yeast strains. The truncated forms of SUVH2/9 were based on the study of Liu et al. (39). Positive transformed strains were mated on yeast peptone dextrose adenine medium for 24 h, and then transferred to SD/-Trp-Leu (DDO) plates and SD/-His-Ade-Trp-Leu (QDO) with X-α-Gal and aureobasidin A plates to test for positive interactions. The mating cells were grown using SD/-Trp/-Leu liquid medium until the optical density 600 reached 1.2; they were then diluted to different concentrations for gradient dilution assay. The primers are listed in SI Appendix, Table S1.
BiFC.
The POL2A N1 truncation was fused to pXY105 (cYFP-C), and SUVH2, SUVH9, and their truncated proteins were fused to pXY106 (nYFP-C) (the pXY series of plasmids from Yu et al. (64)). These positive constructs were transformed into Agrobacterium tumefaciens strain GV3101, and BiFC assays were performed as described previously (65). The primers are listed in SI Appendix, Table S1.
Affinity Purification Pull-Down Assay.
N1 and N1-P fragments were constructed using the pET28a-SUMO vector (Novagen) to fuse with a His-SUMO tag. SUVH2 was cloned into pGEX4T-1 (GE Healthcare) to fuse with a glutathione S-transferase (GST) tag. All of the constructs were transformed into Escherichia coli Rosseta (DE3) and proteins were expressed using isopropyl β-d-1-thiogalactopyranoside induction at 18 °C for 16 h. GST- and His-SUMO proteins were purified using GST-Bind Resin (Millipore, 70541) and Ni-NTA Resin (Millipore, 70666). Approximately 5 μg of each protein were mixed together in pull-down buffer (50 mM Tris⋅HCl pH 7.5, 2 mM EDTA, 200 mM NaCl, 0.2% Nonidet P-40) at 4 °C for 3 h. His binding resin at 15 μL was added and incubated with rotation for 1.5 h at 4 °C. Protein-bound beads were washed with pull-down buffer four times and incubated with boiling sodium dodecyl sulfate (SDS) loading buffer (EpiZyme, LT101) for 10 min. Eluted proteins were detected using western blots with anti-GST (Abmart, M20007) and anti-His (Abmart, M20001). The primers are listed in SI Appendix, Table S1.
CoIP.
To detect POL2A interaction with SUVH2 in vivo, we raised a POL2A polyclonal antibody using the services of the Shanghai Ango Biotechnology Company. N1 protein of POL2A was purified and used to inject rabbits. The antibody was validated by western blot analysis using proteins extracted from the inflorescences of WT and mutants of pol2a-1 background (SI Appendix, Fig. S6A). We also generated transgenic plants expressing SUVH2-FLAG from its native promoter using the pCAMBIA2300 vector (primers listed in SI Appendix, Table S1). For coIP of POL2A with SUVH2-FLAG, proteins were extracted from ∼2 g of early inflorescences (stages 1 to 14) from transgenic plants with protein extraction buffer (50 mM Tris⋅HCl pH 8.0, 100 mM NaCl, 1.5 mM MgCl2, 5 mM β-mercaptoethanol, 10% glycerol, 0.5% Nonidet P-40, and protease inhibitor mixture (Bimake, B14001), and incubated for 1 h at 4 °C. The supernatant with extracted proteins was transferred to a new tube, and mixed with anti-FLAG affinity gel (Yeasen, 20585ES01) at 4 °C overnight. After washing four times with protein extraction buffer, the immunoprecipitated beads were eluted by SDS loading buffer and detected using western blots with anti-FLAG (GNI, GNI4110-FG) and anti-POL2A antibodies.
Isolation of Meiocytes and Extraction of Nucleic Acid.
The Arabidopsis meiocytes were isolated as described previously, with minor modifications (66, 67). Briefly, ∼30 anthers from stages 9 to 10 flower buds were placed in the cavity of double shallow depression slides with collection buffer (1× Dulbecco’s phosphate-buffered saline, Gibco, 14190-144). The anthers were squeezed by using a pair of forceps to release the meiocyte masses, which were collected in the microchamber by a micromanipulator system and transferred to a ZR BashingBead lysis tube (S6012) in liquid nitrogen.
For the observation of chromosome morphology, ∼80 clusters of meiocytes from stages 9 to 10 flower buds were digested (2.5% wt/vol) with cytohelicase (Sigma, C8274) + 1.5% cellulose (Yakult, F0250) + 1.5% macerozyme (Yakult, L0021) in 10 mM citrate buffer (pH 4.5) on a slide with one cavity for 5 min. Then, the digested meiocytes were carefully collected using the microchamber by a micromanipulator system and transferred to a slide with 20 μL ddH2O. This step was repeated three times to wash away the digestion buffer. The clusters of meiocytes were transferred to a slide and moved to a heat block at 45 °C to dry. Slides were observed with a Zeiss Axio Scope A1 (Zeiss) after DAPI staining.
For RNA sequencing (RNA-seq), recombinant RNase inhibitor (final concentration with 1 U/μL, Takara, 2313A) was added to the collection buffer. A total of 300 to 400 meiocyte masses were used to extract ∼400 to 1,000 ng RNA for a biological replicate. The RNA was extracted using TRIzol (ThermoFisher, 15596026), as described previously (67). For BS-seq, >1,500 meiocytes were used to extract ∼30 to 100 ng DNA for each biological replicate using the ChargeSwitch gDNA Micro Tissue Kit (ThermoFisher, CS11203).
mRNA-Seq and Analysis.
RNA-seq libraries were constructed with the TruSeq RNA Library Prep Kit version 2 (Illumina) following the manufacturer’s instructions. Paired-end sequencing was performed using Illumina Hiseq 2000/3000 with at least 20 million reads for each sample. pRNASeqTools (https://github.com/grubbybio/pRNASeqTools) was used for data analysis. Raw reads were trimmed with Cutadapt and mapped to TAIR10 using STAR version 2.7.9a (68) with parameters “–alignIntronMax 5000 –outSAMmultNmax 1 –outFilterMultimapNmax 50 –outFilterMismatchNoverLmax 0.1.” DE genes and TEs (fold change ≥2 and P < 0.01) were identified by the R package DESeq2 version 1.30.1 (69) based on the gene expression matrix quantified by featureCounts version 2.0.0 (70). For plotting, gene expression levels are shown using fragments per kilobase per million mapped reads.
BS-Seq and Analysis.
The genomic DNA was fragmented by sonication to 200 to 300 bp and treated with bisulfite using the EZ DNA Methylation-Gold Kit (Zymo Research). Libraries were constructed from two biological replicates of meiocytes and seedlings (20-d-old plants) of Col-0 (WT), suvh2 suvh9, and pol2a-1, and two biological replicates of pol2a-1 suvh2 suvh9 seedlings. We displayed only one replicate of each plant in all of the metaplots for easy observation because these two replicates showed good reproducibility.
Sequencing with 150-bp paired ends was done by Novogene Corporation using the Illumina platform. Published BS-seq data from meiocytes of drm1 drm2 (49) were also used.
FASTQ reads were analyzed using the plant sequencing analysis pipeline pRNASeqTools. Briefly, raw reads were treated using Cutadapt version 3.4 (71) to remove the adapters; the remaining reads were mapped to the genome, deduplicated, and quantified using Bismark version 0.22.3 (72) with Arabidopsis genome assembly TAIR10 (https://www.arabidopsis.org) and the modified parameter “–cutoff 4.” DMRs were called using an R package DMRcaller version 1.22.0 (73) with the “bins” mode and threshold 0.4/0.2/0.1 for CG/CHG/CHH, respectively. The Arabidopsis genome was divided into euchromatin and heterochromatin following the original genome annotation in the somatic cells, because euchromatin and heterochromatin regions have not been mapped in male meiocytes (74).
sRNA-seq and Analysis.
Small RNA libraries were constructed following the TruSeq small RNA library prep reference guide (Illumina), and single-end sequencing was performed on the Illumina HiSeq 3000 system. Raw reads were trimmed by Cutadapt and masked for rRNA, transfer RNA, and other noncoding RNAs. The remaining reads were mapped to TAIR10 using ShortStack version 3.8.5 (75) with parameters “–align_only –bowtie_m 1000 –ranmax 50 –mismatches 0 –nohp.” For calculating and comparing sRNA abundance in the WT and mutant libraries, the Arabidopsis genome was tiled into 100-bp bins, and reads were assigned to bins based on their 5′ nucleotide. Normalization was conducted by calculating the reads per million mapped reads value for each bin, and comparison was performed for each bin using DESeq2.
Analysis of the Features of DSB Hotspots.
SPO11-1 oligo sequencing and MNase-seq data in WT were downloaded from ENA and mapped to the TAIR10 genome with Bowtie2 following the report of Choi et al. (9). DNA motifs with at least 12 nucleotides were searched by the MEME module in the MEME suite (https://meme-suite.org/meme/) from the gene body and promoter (2 kb upstream) of up-regulated genes. Resulting motifs were plotted using Weblogo 3 (https://weblogo.threeplusone.com).
Morphological and Cytological Analysis.
Whole plants and siliques of WT and mutants were imaged by a Canon digital camera SX20 IS. Alexander red staining was used to test pollen viability from >5 individual plants (76) using a Zeiss Axio Scope A1. The tetrad-stage microspores were analyzed using toluidine blue, as described previously (77).
Chromosome spreading and centromere fluorescence in situ hybridization (FISH) were described previously (66). For chromosome painting, 27,000 oligo probes corresponding to Chr I of Arabidopsis were designed as described previously (78), except pericentromeric and centromeric regions. The oligo libraries were synthesized by Arbor Biosciences. The oligo libraries were processed into digoxigenin-labeled single-stranded oligos, which were used for FISH following a published protocol (78). The cytological images were photographed using a Zeiss Axio Scope A1.
The IF for anti-FLAG (1:100, GNI, GNI4110-FG) and anti-γ-H2AX (1:200) was as used previously (77). Secondary antibodies of Alexa Fluor 488 Goat Anti-Mouse IgG (H+L) (Invitrogen, A-11001) and Alexa Fluor 555 Goat Anti-Rabbit IgG (H+L) (Invitrogen, A-21428) were used with 1:500 and 1:1,000 dilutions, respectively. The images were photographed using the N-SIM (Nikon). Immunostaining of SUVH2-FLAG and SYN1 on chromosome spreading slides was as described previously, with modifications (79). Briefly, slides with chromosome spreads were denatured in the 10-mM citrate buffer (pH 6.0) at 95 °C for 1 min, and then transferred to the PBST buffer (0.1% [vol/vol] Tween 20) for 10 min at room temperature (RT). Slides were blocked in a moist chamber with goat serum (Bosterbio, AR0009) at RT for 1 h. The antibodies were diluted with goat serum and incubated for 36 h. The following steps are similar to IF, as described previously (66). The images were photographed using an N-SIM (Nikon). Images were handled using Adobe Photoshop, and any adjustments were applied globally. Inflorescences from ∼6 to 8 individual plants were used for IF or FISH assay to collect enough cells for statistics, and at least 10 plants were used with the pol2a-1 background because it produces fewer meiocytes. The statistics for meiotic phases of different mutants used the cells from stages 9 to 12 flower buds at different times after lights on.
Fluorescence colocalization and chromosome painting-based length measurements were analyzed using ImageJ (version 1.52). For colocalization analysis, we merged the anti-FLAG and anti-γ-H2AX immunostaining images. The merged images were split into two different color channels using ImageJ, the “threshold” tool was used for excluding the noise or diffused signals, and the overlap coefficient and Pearson’s correlation were measured by the Colocalization Finder plugin (version 1.3). The overlap coefficient indicates an actual overlap of the signals and represents the true degree of colocalization regardless of signal intensity; Pearson’s correlation describes the correlation of the intensity distribution between channels. For painting-based length measurements, the segmented lines tool was used to trace the painted regions of Chr I, and the traced lines were measured by the Measure tool.
Supplementary Material
Acknowledgments
We thank Dr. Xinjian He (National Institute of Biological Sciences, Beijing, China) for sharing the plant materials of SUVH2/9 and for his comments on the manuscript. We thank Dr. Hongbo Gao (Joint Center for Single Cell Biology, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China) for the advice on DNA extraction from meiocytes. This research was supported by the National Natural Science Foundation of China (31925005, 31870293, 32000246, 31900445, and 32170334), the State Key Laboratory of Genetic Engineering, the China Postdoctoral Science Foundation (2019M661343), the National Postdoctoral Program for Innovative Talents, and the Guangdong Laboratory for Lingnan Modern Agriculture. J.H. and G.P.C. were supported by a grant from the US NSF (IOS-1844264).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208441119/-/DCSupplemental.
Data, Materials, and Software Availability
The raw sequence data generated during this study were deposited into the NCBI Gene Expression Omnibus (GEO) database, accession number GSE203328 (80).
References
- 1.Keeney S., Giroux C. N., Kleckner N., Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Grelon M., Vezon D., Gendrot G., Pelletier G., AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M., The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66, 297–327 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Copenhaver G. P., Meiotic recombination: Mixing it up in plants. Annu. Rev. Plant Biol. 69, 577–609 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Hunter N., Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma H., A molecular portrait of Arabidopsis meiosis. Arabidopsis Book 4, e0095 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tock A. J., Henderson I. R., Hotspots for initiation of meiotic recombination. Front. Genet. 9, 521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petes T. D., Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2, 360–369 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Choi K., et al. , Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 28, 532–546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baudat F., Imai Y., de Massy B., Meiotic recombination in mammals: Localization and regulation. Nat. Rev. Genet. 14, 794–806 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Sommermeyer V., Béneut C., Chaplais E., Serrentino M. E., Borde V., Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol. Cell 49, 43–54 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Borde V., et al. , Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28, 99–111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parvanov E. D., Petkov P. M., Paigen K., Prdm9 controls activation of mammalian recombination hotspots. Science 327, 835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myers S., et al. , Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327, 876–879 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange J., et al. , The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell 167, 695–708.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baudat F., et al. , PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi K., et al. , Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45, 1327–1336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borde V., de Massy B., Programmed induction of DNA double strand breaks during meiosis: Setting up communication between DNA and the chromosome structure. Curr. Opin. Genet. Dev. 23, 147–155 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Brick K., Smagulova F., Khil P., Camerini-Otero R. D., Petukhova G. V., Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smagulova F., et al. , Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472, 375–378 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machour F. E., Ayoub N., Transcriptional regulation at DSBs: Mechanisms and consequences. Trends Genet. 36, 981–997 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Ortega P., Mérida-Cerro J. A., Rondón A. G., Gómez-González B., Aguilera A., DNA-RNA hybrids at DSBs interfere with repair by homologous recombination. eLife 10, e69881 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutierrez C., Desvoyes B., Vergara Z., Otero S., Sequeira-Mendes J., Links of genome replication, transcriptional silencing and chromatin dynamics. Curr. Opin. Plant Biol. 34, 92–99 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Pedroza-Garcia J. A., De Veylder L., Raynaud C., Plant DNA polymerases. Int. J. Mol. Sci. 20, 4814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del Olmo I., et al. , Arabidopsis DNA polymerase ϵ recruits components of Polycomb repressor complex to mediate epigenetic gene silencing. Nucleic Acids Res. 44, 5597–5614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.del Olmo I., et al. , EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J. 61, 623–636 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Hyun Y., et al. , The catalytic subunit of Arabidopsis DNA polymerase α ensures stable maintenance of histone modification. Development 140, 156–166 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Li F., Martienssen R., Cande W. Z., Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature 475, 244–248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He H., et al. , Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex. Proc. Natl. Acad. Sci. U.S.A. 114, 12524–12529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith J. S., Caputo E., Boeke J. D., A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19, 3184–3197 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iida T., Araki H., Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 217–227 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourguet P., et al. , DNA polymerase epsilon is required for heterochromatin maintenance in Arabidopsis. Genome Biol. 21, 283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimanelli D., Ingouff M., DNA methylation readers in plants. J. Mol. Biol., 10.1016/j.jmb.2019.12.043 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Law J. A., Jacobsen S. E., Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson L. M., et al. , The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 17, 379–384 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson J. P., Lindroth A. M., Cao X., Jacobsen S. E., Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Johnson L. M., et al. , SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507, 124–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson L. M., Law J. A., Khattar A., Henderson I. R., Jacobsen S. E., SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 4, e1000280 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z. W., et al. , The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 10, e1003948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z. W., et al. , Two components of the RNA-directed DNA methylation pathway associate with MORC6 and silence loci targeted by MORC6 in Arabidopsis. PLoS Genet. 12, e1006026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jing Y., et al. , SUVH2 and SUVH9 couple two essential steps for transcriptional gene silencing in Arabidopsis. Mol. Plant 9, 1156–1167 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Huang J., et al. , Formation of interference-sensitive meiotic cross-overs requires sufficient DNA leading-strand elongation. Proc. Natl. Acad. Sci. U.S.A. 112, 12534–12539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shilo S., Melamed-Bessudo C., Dorone Y., Barkai N., Levy A. A., DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in Arabidopsis. Plant Cell 27, 2427–2436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wijnker E., et al. , The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2, e01426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horton M. W., et al. , Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44, 212–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baerenfaller K., et al. , Diurnal changes in the histone H3 signature H3K9ac|H3K27ac|H3S28p are associated with diurnal gene expression in Arabidopsis. Plant Cell Environ. 39, 2557–2569 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Lowndes N. F., Toh G. W., DNA repair: The importance of phosphorylating histone H2AX. Curr. Biol. 15, R99–R102 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Manders E. M. M., Verbeek F. J., Aten J. A., Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382 (1993). [DOI] [PubMed] [Google Scholar]
- 49.Walker J., et al. , Sexual-lineage-specific DNA methylation regulates meiosis in Arabidopsis. Nat. Genet. 50, 130–137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yelina N. E., et al. , DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 29, 2183–2202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi K., et al. , Recombination rate heterogeneity within Arabidopsis disease resistance genes. PLoS Genet. 12, e1006179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drouaud J., et al. , Contrasted patterns of crossover and non-crossover at Arabidopsis thaliana meiotic recombination hotspots. PLoS Genet. 9, e1003922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matzke M. A., Kanno T., Matzke A. J., RNA-directed DNA methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66, 243–267 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Gallego-Bartolomé J., et al. , Co-targeting RNA polymerases IV and V promotes efficient de novo DNA methylation in Arabidopsis. Cell 176, 1068–1082.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayashi K., Yoshida K., Matsui Y., A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438, 374–378 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Zamudio N., et al. , DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 29, 1256–1270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H., Lang Z., Zhu J. K., Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506 (2018). [DOI] [PubMed] [Google Scholar]
- 58.McVey M., Khodaverdian V. Y., Meyer D., Cerqueira P. G., Heyer W. D., Eukaryotic DNA polymerases in homologous recombination. Annu. Rev. Genet. 50, 393–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pavlov Y. I., Shcherbakova P. V., DNA polymerases at the eukaryotic fork-20 years later. Mutat. Res. 685, 45–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Long J., et al. , Nurse cell-derived small RNAs define paternal epigenetic inheritance in Arabidopsis. Science 373, eabh0556 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Zhou X., et al. , 24-nt phasiRNAs move from tapetal to meiotic cells in maize anthers. New Phytol. 235, 488–501 (2022). [DOI] [PubMed] [Google Scholar]
- 62.Underwood C. J., et al. , Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 28, 519–531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jorrin-Novo J. V., Plant proteomics methods and protocols. Methods Mol. Biol. 1072, 3–13 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Yu X., et al. , Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 7618–7623 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cui J., et al. , Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28, 1078–1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y., Cheng Z., Lu P., Timofejeva L., Ma H., Molecular cell biology of male meiotic chromosomes and isolation of male meiocytes in Arabidopsis thaliana. Methods Mol. Biol. 1110, 217–230 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Huang J., et al. , Meiocyte-specific and AtSPO11-1-dependent small RNAs and their association with meiotic gene expression and recombination. Plant Cell 31, 444–464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10 (2011). [Google Scholar]
- 72.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Catoni M., Tsang J. M., Greco A. P., Zabet N. R., DMRcaller: A versatile R/Bioconductor package for detection and visualization of differentially methylated regions in CpG and non-CpG contexts. Nucleic Acids Res. 46, e114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arabidopsis Genome Initiative, Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000). [DOI] [PubMed] [Google Scholar]
- 75.Johnson N. R., Yeoh J. M., Coruh C., Axtell M. J., Improved placement of multi-mapping small RNAs. G3 (Bethesda) 6, 2103–2111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alexander M. P., Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122 (1969). [DOI] [PubMed] [Google Scholar]
- 77.Wang C., et al. , The largest subunit of DNA polymerase delta is required for normal formation of meiotic type I crossovers. Plant Physiol. 179, 446–459 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han Y., Zhang T., Thammapichai P., Weng Y., Jiang J., Chromosome-specific painting in cucumis species using bulked oligonucleotides. Genetics 200, 771–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chelysheva L. A., Grandont L., Grelon M., Immunolocalization of meiotic proteins in Brassicaceae: Method 1. Methods Mol. Biol. 990, 93–101 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Wang C., et al. , DNA polymerase epsilon interacts with SUVH2/9 to repress the expression of genes associated with meiotic DSB hotspot in Arabidopsis. NCBI: GEO. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203328. Deposited 18 May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data generated during this study were deposited into the NCBI Gene Expression Omnibus (GEO) database, accession number GSE203328 (80).