Significance
CGG repeat expansion in the 5′ untranslated region of the NOTCH2NLC gene was reported to be associated with a neuromuscular/neurodegenerative disease, neuronal intranuclear inclusion disease (NIID). Here, we developed a fly model of CGG repeat expansion in NOTCH2NLC, recapitulated key pathological and clinical features of NIID, and characterized the mitochondrial dysfunction in these model organisms, human samples, and cellular models. Moreover, idebenone treatment restored mitochondrial function and alleviated neurodegenerative phenotypes in transgenic flies.
Keywords: neuronal intranuclear inclusion disease, polyG, transgenic Drosophila model, mitochondrial oxidative phosphorylation defects, idebenone treatment
Abstract
Neuronal intranuclear inclusion disease (NIID) is a neuromuscular/neurodegenerative disease caused by the expansion of CGG repeats in the 5′ untranslated region (UTR) of the NOTCH2NLC gene. These repeats can be translated into a polyglycine-containing protein, uN2CpolyG, which forms protein inclusions and is toxic in cell models, albeit through an unknown mechanism. Here, we established a transgenic Drosophila model expressing uN2CpolyG in multiple systems, which resulted in progressive neuronal cell loss, locomotor deficiency, and shortened lifespan. Interestingly, electron microscopy revealed mitochondrial swelling both in transgenic flies and in muscle biopsies of individuals with NIID. Immunofluorescence and immunoelectron microscopy showed colocalization of uN2CpolyG with mitochondria in cell and patient samples, while biochemical analysis revealed that uN2CpolyG interacted with a mitochondrial RNA binding protein, LRPPRC (leucine-rich pentatricopeptide repeat motif-containing protein). Furthermore, RNA sequencing (RNA-seq) analysis and functional assays showed down-regulated mitochondrial oxidative phosphorylation in uN2CpolyG-expressing flies and NIID muscle biopsies. Finally, idebenone treatment restored mitochondrial function and alleviated neurodegenerative phenotypes in transgenic flies. Overall, these results indicate that transgenic flies expressing uN2CpolyG recapitulate key features of NIID and that reversing mitochondrial dysfunction might provide a potential therapeutic approach for this disorder.
Microsatellites are short tracts of two to six DNA base pairs repeated multiple times in the human genome. However, expansion over a threshold limit of some microsatellite repeats is responsible for more than 50 human disorders (1, 2). When embedded in open reading frames (ORFs), these repeat expansions are consequently translated and lead to expression of mutant proteins containing a stretch of repeated amino acids, notably polyglutamine (polyQ), polyalanine (polyA), or polyglycine (polyG) (3, 4). PolyQ tracts encoded by (CAG)n repeat expansions are associated with various neurodegenerative diseases, such as Huntington’s disease (HD) and several spinocerebellar ataxias (5, 6). PolyA tracts encoded by (GCG)n repeat expansions are associated with eight congenital disorders and a neuromuscular disease, oculopharyngeal muscular dystrophy (7, 8). More recently, polyG tracts encoded by (CGG)n repeat expansions were found to be associated with two neurodegenerative diseases, fragile X-associated tremor/ataxia syndrome (FXTAS) and neuronal intranuclear inclusion disease (NIID) (9–11).
NIID is a progressive neuromuscular and/or neurodegenerative disease characterized by varied muscle and central or peripheral nervous system dysfunctions associated with the presence of eosinophilic hyaline ubiquitin- and p62-positive inclusion bodies (12, 13). Consequently, NIID was essentially diagnosed by its core clinical features associated with hyperintense signal at the corticomedullary junction on diffusion-weighted imaging of MRI and the presence of intranuclear inclusion body in skin biopsies (13, 14). In 2019, an expansion of CGG repeats in the 5′ untranslated region (UTR) of the NOTCH2 N-terminal like C gene (NOTCH2NLC, MIM: 618025) was identified as the genetic cause of NIID (15–18). Using genetic testing for NOTCH2NLC repeat expansion, recent studies have found that NIID comprises a much wider spectrum of clinical symptoms, including Alzheimer’s disease (AD) (16), frontotemporal dementia (19), parkinsonism-related disorders (16, 20–22), multiple system atrophy (23), essential tremor (ET) (24, 25), adult leukoencephalopathy (26), amyotrophic lateral sclerosis (27), and oculopharyngodistal myopathy (OPDM) (28, 29). Moreover, histopathological analyzes revealed that NIID pathology expanded to almost all systems (30). These results suggest that the terms systemic intranuclear inclusion disease and/or NOTCH2NLC-related repeat expansion disorders (NREDs) would more accurately reflect the diversity of clinical features associated with the NOTCH2NLC CGG repeat expansion (31–33).
Corollary to its recent genetic identification, research on the pathogenesis of NIID is just beginning (34). Recent studies indicate that expansions over 41 CGG repeats but below 300 to 500 repeats are considered pathogenic (16, 20, 22). Different repeat numbers may be related to different phenotypes. The largest CGG repeat expansion (118 to 517 repeats) is related to a muscle weakness-dominant phenotype, a shorter (91 to 268 repeats) expansion might result in a dementia-dominant phenotype, and a much shorter repeat expansion (66 to 102 repeats) might cause a parkinsonism-dominant phenotype (16, 33). However, there was no difference in the number of repeats between juveniles (139 and 172 repeats in two juvenile-onset patients) and adults (29, 33, 35). Importantly, neither NOTCH2NLC transcript levels nor methylation levels showed significant differences between NIID patients and normal controls (28). These observations exclude a promoter-silencing and loss-of-function mechanism in NIID, and suggest that the mutant NOTCH2NLC messenger RNA (mRNA) with a CGG repeat expansion is toxic through a gain-of-function mechanism at the RNA and/or protein level. Several studies have observed foci of RNA containing expanded CGG repeats that colocalize with specific RNA binding proteins in NIID cell models and/or patients’ muscle or skin samples, albeit with undetermined pathological consequences (35, 36). A second proposed mechanism is that NIID is caused by translation of the CGG expansion in a pathogenic protein. In this aspect, two recent studies found that this microsatellite is located in a small upstream open reading frame (uORF), located in front of the NOTCH2NLC main ORF, and translated by canonical initiation at an AUG start codon, resulting in expression of polyG-containing protein. So this protein was named upstream open reading frame of NOTCH2NLC (uN2CpolyG) or N2NLCpolyG (10, 11). Expression of uN2CpolyG in the mouse nervous system through an adeno-associated virus (AAV)-based approach leads to formation of ubiquitin- and p62-positive inclusions and locomotor alterations, neuronal cell loss, and premature death of the animals (10). Concomitantly, Zhong and collaborators (11) found that uN2CpolyG expression and aggregation impairs nuclear lamina and nucleocytoplasmic transport but does not necessarily cause acute neuronal cell death. Thus, the role of uN2CpolyG causing neuronal, muscular, and/or multisystemic degeneration is still unclear, and a transgenic animal model expressing uN2CpolyG to mimic NIID was missing.
In this study, we established a transgenic Drosophila model expressing uN2CpolyG in multiple systems. Interestingly, expression of uN2CpolyG recapitulates the major clinical and pathological features of NIID, notably the formation of protein inclusions and the progressive neuronal cell loss, locomotor deficiency, and shortened lifespan. Moreover, mitochondrial swelling was observed in both transgenic flies and NIID muscle biopsied samples, and uN2CpolyG protein was found to predominantly interact with the outer mitochondrial membrane (OMM) in NIID patient samples and the neuronal cell model. Remarkably, RNA sequencing (RNA-seq) analysis and functional assays showed mitochondrial oxidative phosphorylation (OXPHOS) defects, especially complex I impairment, in both transgenic fly and NIID patient samples. Importantly, restoring mitochondrial OXPHOS by idebenone (IDB) treatment rescued neurodegenerative phenotypes, mitochondrial dysfunction, and morphological defect in flies. Overall, these data indicate that uN2CpolyG transgenic flies can serve as a useful animal model to uncover the pathogenic mechanisms and investigate therapeutical approaches for NOTCH2NLC-related disorders, notably in regards to mitochondrial functions.
Results
Expression of uN2CpolyG in Fly Eyes Leads to Progressive Retinal Degeneration.
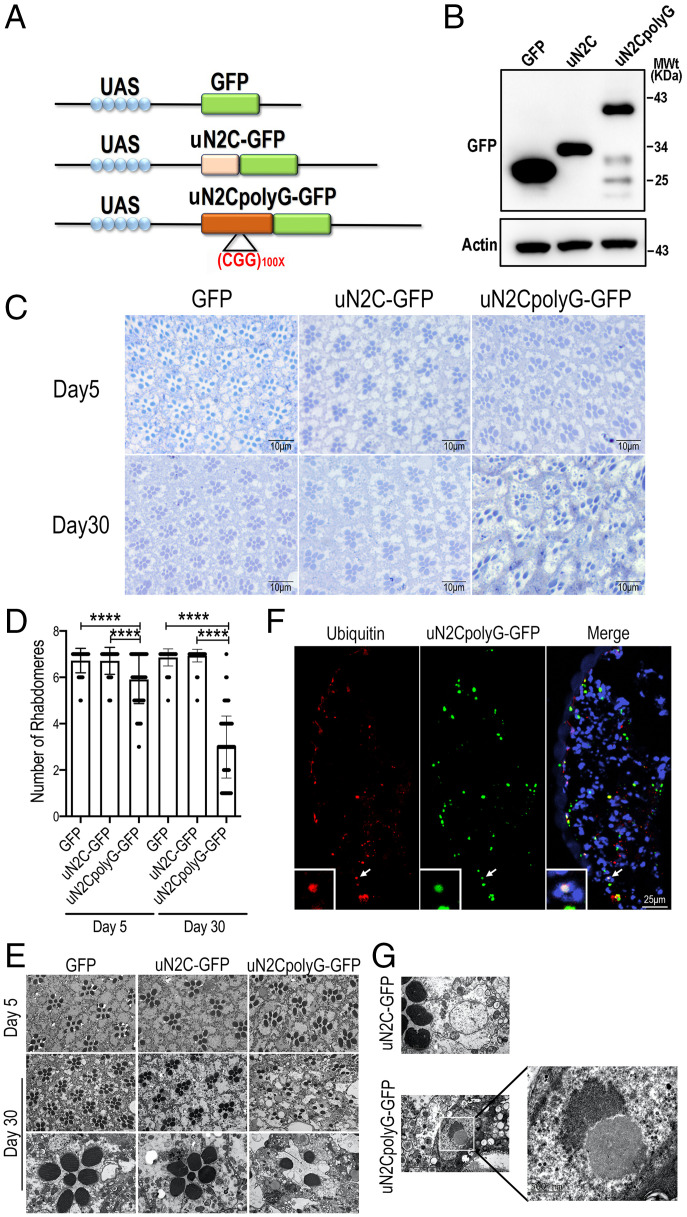
The NOTCH2NLC CGG repeat microsatellite sequence is embedded in a short AUG-initiated uORF encoding in control condition (nine CGG repeats in RefSeq) for a small, ∼6-kDa protein, which was named uN2C. Thus, expansion between ∼40 and ∼300 to 500 repeats of the CGG track in NIID individuals results in expression of a longer protein, where each GGC codon encodes for a glycine, that was consequently named uN2CpolyG (10, 11). To evaluate the pathogenic role of uN2CpolyG in a transgenic animal model, we used the upstream active sequence-GALactose-regulated upstream promoter element(UAS-Gal4) system to express green fluorescent protein (GFP), control uN2C-GFP, and uN2CpolyG-GFP in flies (Fig. 1A). The fly eye is an advantageous choice for neurodegeneration research because it is the most accessible part of the Drosophila nervous system (37). Therefore, we first used a glass multiple reporter-GALactose-regulated upstream promoter element (GMR-Gal4) driver to express uN2CpolyG in fly eyes. Equivalent expression levels of GFP, uN2C-GFP, and uN2CpolyG-GFP proteins were detected by western blot (Fig. 1B). Consistent with the previous finding that uN2CpolyG exerted moderate neuronal toxicity at an early time point (11), expression of uN2CpolyG did not cause obvious eye surface degeneration but did lead to mild rhabdomere loss at day 5, while fly eyes expressing GFP or uN2C reveal intact ommatidial structures with seven rhabdomeres (Fig. 1 C–E and SI Appendix, Fig. S1). Of interest, small GFP-positive and ubiquitin-positive inclusions were present in uN2CpolyG-GFP–expressing fly eyes as early as day 5, but no inclusions were observed in flies expressing GFP or uN2C-GFP (Fig. 1F and SI Appendix, Fig. S1). To investigate if expressing uN2CpolyG causes age-dependent neurodegeneration, we examined the ommatidial structures in fly eyes at day 30. Importantly, expression of uN2CpolyG caused severe ommatidial degeneration in 30-d-old flies, while flies expressing GFP or uN2C-GFP had an intact ommatidial structure. Quantification indicated that the average number of rhabdomeres in flies expressing uN2CpolyG at day 30 was three, as compared with six rhabdomeres at day 5 (Fig. 1 C–F). Remarkably, large intranuclear inclusions were found in flies expressing uN2CpolyG but not in flies expressing the control uN2C protein (Fig. 1G). To exclude the effect of GFP fusion protein, we expressed uN2C or uN2CpolyG lacking the GFP tag in flies. Consistently, ommatidial degeneration was observed in flies expressing uN2CpolyG but not in flies expressing uN2C (SI Appendix, Fig. S2). These data indicate that expression of the uN2CpolyG protein in Drosophila leads to progressive neurodegeneration and intranuclear inclusion formation, which recapitulates the major clinical and pathological features of NIID.
Fig. 1.
Expression of uN2CpolyG causes progressive neurodegeneration in the eyes of Drosophila. (A) Schematic diagram of pUAST-uN2C-(CGG)n-GFP constructs. Top: Schematic of UAS-GFP. Middle: Schematic of UAS-uN2C-GFP with the full NOTCH2NLC uORF fused to GFP. Bottom: Schematic of UAS-N2CpolyG-GFP with the full NOTCH2NLC uORF with 100 CGG repeats fused to GFP. (B) Western blot analysis of protein expression from the fly heads. Genotypes: GFP, GMR-Gal4 > UAS-GFP; uN2C, GMR-Gal4 > UAS-N2C-GFP; and uN2CpolyG, GMR-Gal4 > UAS-N2CpolyG-GFP. (C) Toluidine blue staining shows intact ommatidial structures with seven rhabdomeres in both 5-d-old and 30-d-old female adult flies expressing GFP and uN2C-GFP protein. But in the flies expressing uN2CpolyG-GFP protein, the rhabdomeres were lost when 5 d old and more severely damaged when 30 d old. (D) Quantification of rhabdomeres in three groups at day 5 and day 30. Data were analyzed using one-way ANOVA with Bonferroni post hoc test (****P < 0.0001). (E) EM images show intact ommatidial structures with seven rhabdomeres in 5-d-old and 30-d-old female adult flies expressing GFP and uN2C protein, whereas the rhabdomere structures are damaged or completely lost in the retinae of flies expressing uN2CpolyG protein. (F) Confocal micrographs of transverse retinal sections from GMR-Gal4 > UAS-uN2CpolyG-GFP flies at day 5 reveal nuclear and cytoplasmic inclusions that colocalize with ubiquitin (indicated by arrow). (G) EM images show intranuclear inclusion formation in flies expressing uN2CpolyG protein but not in flies expressing uN2C protein. Error bars correspond to the SEM. MWT, molecular weight.
Expression of uN2CpolyG in Fly Leads to Locomotor Impairment and Reduced Lifespan.
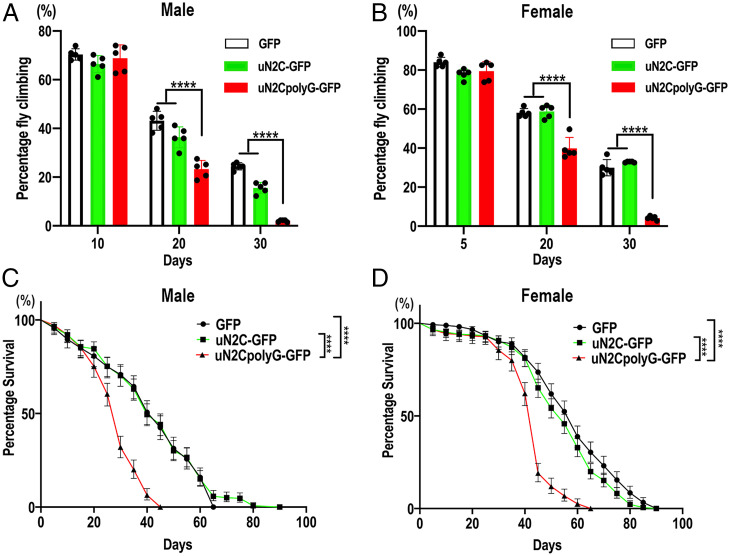
To investigate the systemic multisystem toxicity of uN2CpolyG in transgenic flies, GFP, uN2C-GFP, or uN2CpolyG-GFP were then ubiquitous expressed using an actin5C-Gal4 driver, and we examined the locomotor ability of the adult flies. Consistent with the skeletal muscle weakness and the neurological clinical features observed in NIID, the movements of flies expressing uN2CpolyG-GFP were significantly and progressively reduced in both male and female groups compared with the GFP or uN2C-GFP group (Fig. 2 A and B). In addition, the lifespan was significantly shortened in flies expressing uN2CpolyG-GFP, with medians of 40, 40, and 30 d in males and of 60, 55, and 45 d in females expressing GFP, uN2C-GFP, and uN2CpolyG-GFP, respectively (Fig. 2 C and D). In contrast to ubiquitous expression driven by actin5C-Gal4, flies with neuron-specific expression of uN2CpolyG driven by the Elav-Gal4 driver did not show motor deficits, whereas nearly all flies that multisystemically expressed uN2CpolyG at day 30 lost their locomotor ability (SI Appendix, Fig. S3). Since NIID is a multisystem proteinopathy, ubiquitous expression of uN2CpolyG could better mimic the phenotype of NIID.
Fig. 2.
Expression of uN2CpolyG in flies leads to locomotive impairment and shortened lifespan. (A and B) The locomotor index was measured in adult flies at different time points. Expression of uN2CpolyG protein in adult flies led to progressive locomotor deficits in both male flies (A) and female flies (B) compared with flies expressing GFP or uN2C protein. (C and D) Lifespan analysis assay. Survival of adult male flies (C) and female flies (D). Fly genotypes: GFP, Actin-Gal4 > UAS-GFP; uN2C-GFP, Actin-Gal4 > UAS-uN2C-GFP; and uN2CpolyG-GFP, Actin-Gal4 > UAS-uN2CpolyG-GFP. Flies expressing uN2CpolyG exhibited a decrease in the median lifespan. The median lifespan was 30 d for uN2CpolyG-GFP–expressing male flies versus 40 d for GFP- or uN2C-GFP–expressing male flies (C) and 45 d for uN2CpolyG-GFP–expressing female flies versus 55 d for N2C-GFP–expressing female flies and 60 d for GFP-expressing female flies (D) (log-rank Mantel-Cox test; n = 235–356/genotype for female flies and n = 233–279/genotype for male flies; ****P < 0.0001). Error bars correspond to the SEM.
uN2CpolyG Causes Mitochondrial Swelling in NIID Patients and Transgenic Flies.
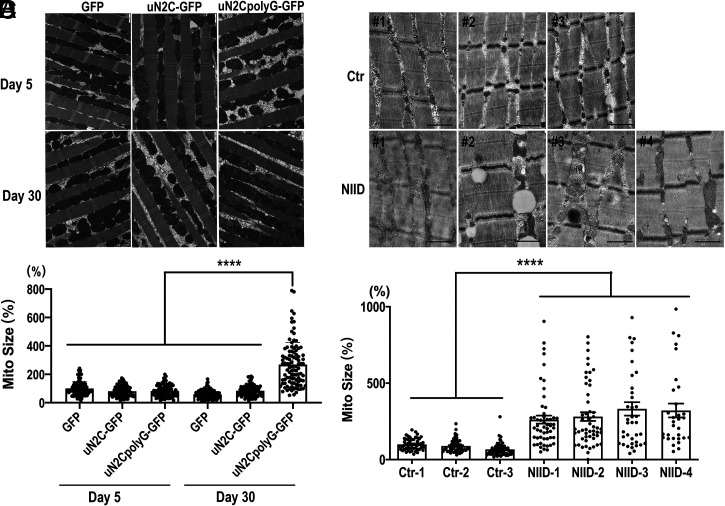
To explore further the pathological changes induced by uN2CpolyG expression in vivo, we examined muscle fibers of actin5C-Gal4-driven GFP, uN2C-GFP, or uN2CpolyG-GFP transgenic flies by transmission electron microscopy (TEM). Muscle fibers were intact in flies expressing uN2CpolyG at both day 5 and day 30, but a remarkable increase of mitochondrial size was observed in flies expressing uN2CpolyG compared with flies expressing GFP or uN2C-GFP at day 30. Quantification revealed that mitochondrial size was normal at day 5 but significantly increased in 30-d-old flies expressing uN2CpolyG (Fig. 3B), suggesting that uN2CpolyG expression causes progressive mitochondrial changes. To investigate if uN2CpolyG also induces some mitochondrial damage in NIID patients, we examined skeletal muscle samples from NIID patients and normal controls by TEM. Importantly, increased mitochondrial size was also observed in NIID patients compared with normal controls (Fig. 3 C and D). Taken together, these data suggest that uN2CpolyG expression leads to progressive mitochondrial dysfunctions.
Fig. 3.
EM analysis showing increased mitochondrial size in muscle samples from flies expressing uN2CpolyG and NIID patients. (A) Muscle fibers were intact in flies expressing GFP, uN2C-GFP, and uN2CpolyG-GFP at both day 5 and day 30. Normal mitochondrial size was observed in three groups expressing GFP, uN2C-GFP, and uN2CpolyG-GFP at day 5. Meanwhile, increased mitochondrial size was observed in flies expressing uN2CpolyG-GFP at day 30 compared with five other groups. (B) Statistical analysis of mitochondrial size in flies expressing GFP, uN2C-GFP, and uN2CpolyG-GFP at day 5 and day 30. A significant difference was observed in uN2CpolyG-GFP–expressing flies at day 30 compared with five other groups. (C) Increased mitochondrial size was observed in four NIID patients compared with three normal controls. Massive lipid droplets accumulated in muscle fibers of patient NIID#2. (D) Statistical analysis of mitochondrial size in NIID patients and normal controls. A significant difference was observed between NIID patients and normal controls. Scale bar, 1 μm (A and B); ****P < 0.0001. Error bars correspond to the SEM. Ctr, control; Mito, mitochondrial.
uN2CpolyG Partly Colocalizes with Mitochondria.
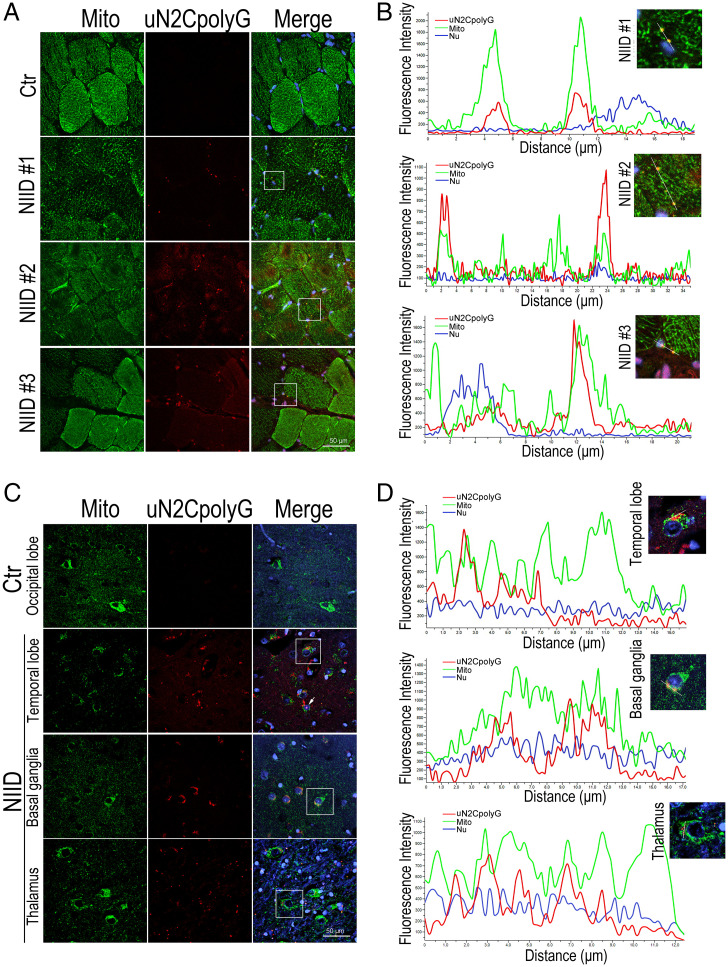
To examine if uN2CpolyG protein may alter mitochondria by direct interaction, we performed immunofluorescence on skeletal muscle and brain samples from NIID patients. Of interest, we observed that some uN2CpolyG partially colocalized with mitochondria in both muscle and brain sections of NIID patients but not in age-matched controls (Fig. 4 A and C). Line scan analysis confirmed uN2CpolyG colocalization with mitochondria in NIID patient samples, as indicated by overlapping fluorescence intensity (Fig. 4 B and D).
Fig. 4.
Colocalization of uN2CpolyG and mitochondria in skeletal muscle and brain from NIID patients. (A) Representative immunofluorescence microscopy images of skeletal muscle samples from three NIID patients and one normal control, which reveals the distribution of uN2CpolyG in muscle fibers. uN2CpolyG protein partially colocalized with mitochondria. (B) Line scan analysis of the merged images shows uN2CpolyG was colocalized with mitochondria in muscle fibers from NIID patients. (C) Representative immunofluorescence microscopy images of brain samples from one NIID patient and one normal control, which reveals the distribution of uN2CpolyG in brain samples. uN2CpolyG protein partially colocalized with mitochondrial. The arrow indicates uN2CpolyG-positive intranuclear inclusion. (D) Line scan analysis of the merged images shows uN2CpolyG was colocalized with mitochondria in three different brain regions (temporal lobe, basal ganglia, and thalamus) from the NIID patient. Scale bar, 50 μm (A and C).Nu, nucleus.
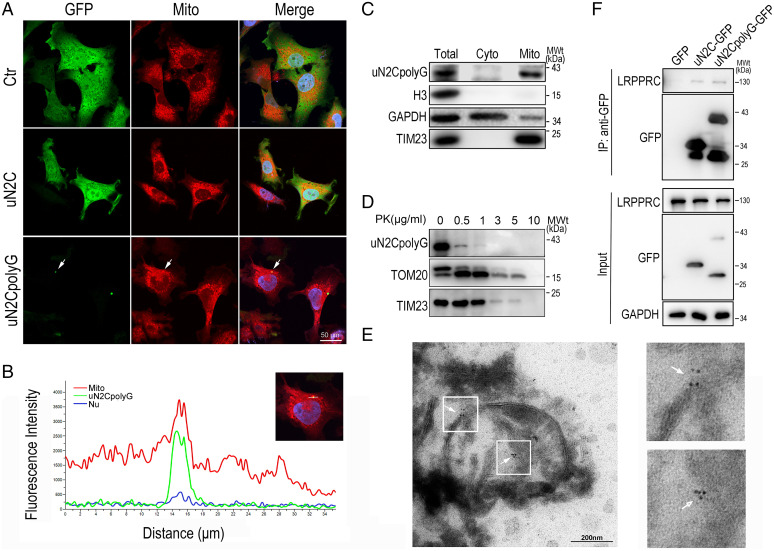
To further confirm colocalization of uN2CpolyG with mitochondria in vitro, GFP, uN2C-GFP, or uN2CpolyG-GFP was expressed in a neuronal cell line, SH-SY5Y. Consistent with NIID tissue analyzes, uN2CpolyG aggregates partially colocalized with mitochondria, while GFP or uN2C were diffusely distributed in the cytoplasm (Fig. 5 A and B). Then, we isolated mitochondria from SH-SY5Y cells expressing uN2CpolyG-GFP. Western blotting experiments showed that the mitochondrial samples were enriched for the mitochondrial marker TIM23 but lacked either the cytoplasmic marker GAPDH or the nuclear marker Histone H3. The uN2CpolyG protein was consistently detected in these mitochondrial preparations (Fig. 5C). To explore further uN2CpolyG localization within mitochondria, isolated mitochondria were subjected to proteinase K (PK) digestion. Western blot revealed that the OMM was broken by PK treatment (0.5 to 1 μg/mL PK) and that uN2CpolyG was sensitive to PK digestion, a pattern identical to that of the OMM protein marker TOM20, while the mitochondrial inner membrane marker TIM23 remained intact (Fig. 5D). These data indicated that uN2CpolyG was predominantly associated with the OMM, while a small amount of uN2CpolyG was translocated into the intermembrane space (IMS). To verify the submitochondrial location of uN2CpolyG in vivo, we performed immunoelectron microscopy (immuno-EM) analysis using specific anti-uN2CpolyG antibody on NIID brain tissues. Consistently, uN2CpolyG was found localized in both OMM and IMS (Fig. 5E), suggesting that endogenous uN2CpolyG is associated with mitochondria in the NIID brain.
Fig. 5.
Colocalization of uN2CpolyG and mitochondria in a SH-SY5Y cell model and NIID brain tissue. (A) Representative immunofluorescence microscopy images of SH-SY5Y cells expressing GFP, uN2C-GFP, and uN2CpolyG-GFP at 24 h posttransfection. Mitochondria were partially colocalized with uN2CpolyG (indicated by arrows). (B) Line scan analysis of the merged images showed uN2CpolyG was colocalized with mitochondria in SH-SY5Y cells expressing uN2CpolyG-GFP. (C) Isolated mitochondria were prepared from SH-SY5Y cells expressing uN2CpolyG-GFP. The mitochondrial purity was confirmed by the detection of mitochondrial TIM23 and the absence of the cytoplasmic proteins such as GAPDH or nuclear protein Histone H3. The uN2CpolyG protein was localized to mitochondria. (D) Isolated mitochondria from SH-SY5Y cells expressing uN2CpolyG-GFP were subjected to digestion with PK. The uN2CpolyG protein, the OMM protein, TOM20, and the inner mitochondrial membrane (IMM) protein TIM23 were detected. (E) Immuno-EM images of brain tissues from the NIID patient showed uN2CpolyG-immunostaining signals associated with mitochondria labeled with 6-nm immunogold particles (indicated by arrows; uN2CpolyG is labeled by a 4D12-specific antibody). (F) uN2CpolyG-LRPPRC interaction was detected by coIP assay. Western blot was performed using corresponding specific antibodies following immunoprecipitation of cell lysates with anti-GFP. Cyto, cytoplasm; IP, immunoprecipitation.
Of interest, a previous study based on uN2polyG immunoprecipitation followed by mass spectrometry analysis provided a list of proteins potentially interacting with uN2CpolyG (10), revealing that 17.4% of these putative interactants are related to mitochondria, which further supports the possibility that uN2CpolyG alters mitochondrial functions. Hence, we performed coimmunoprecipitation (coIP) assays and confirmed an interaction of a mitochondrial RNA binding protein LRPPRC (leucine-rich pentatricopeptide repeat motif-containing protein) with both uN2C and uN2CpolyG (Fig. 5F). As uN2CpolyG is prone to form protein inclusions, its interaction with LRPPRC might cause coaggregation and loss of function of LRPPRC associated with mitochondrial dysfunctions. Interestingly, overexpressing Bsf (a homolog of human LRPPRC) (38) in uN2CpolyG transgenic flies could partially rescue the retinal degeneration (SI Appendix, Fig. S5).
Mitochondrial Complex I Coding Genes Are Down-Regulated in NIID.
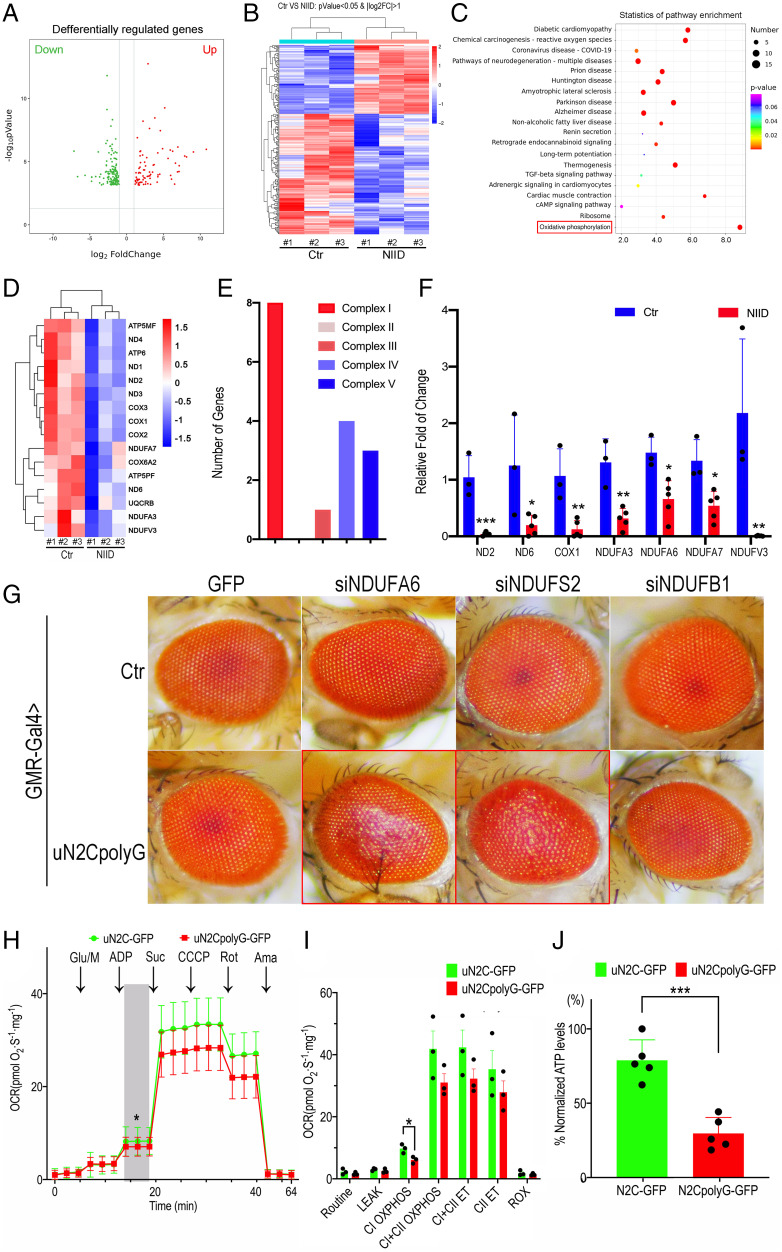
Since skeletal muscle samples from NIID exhibited substantial pathological changes, including rimmed vacuoles, lipid accumulation, and ubiquitin-positive uN2CpolyG deposits in the nucleus (SI Appendix, Fig. S4), we sought to explore further the mitochondrial and molecular alterations involved in NIID. Skeletal muscle biopsy samples of three NIID patients (N1 to N3) with muscle pathology and three age-matched controls (C1 to C3) were analyzed by RNA-seq. The volcano plot and heatmap of the genome-wide mRNA expression profiles revealed numerous gene expression changes between control and NIID samples (Fig. 6 A and B). Importantly, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed that the most significantly down-regulated expressed genes correspond to the OXPHOS pathway (Fig. 6C), with notable decreased expression of mitochondrial and nuclear genes encoding mitochondrial complexes I, III, IV, and V (Fig. 6 D and E). qRT-PCR confirmed transcriptional down-regulation of complex I-encoding genes, including ND2, ND6, NDUFA3, NDUFA6, NDUFA7, and NDUFV3 mRNAs in skeletal muscle samples of an additional five NIID patients compared with three control individuals (Fig. 6F).
Fig. 6.
Down-regulation of mitochondrial complex I in NIID patient samples and in transgenic flies. (A) Volcano plots of significantly differentially expressed genes. The criteria of corrected P < 0.05 and |fold change| ≥ 2; red, up-regulated; green, down-regulated. (B) Heatmap showing hierarchical clustering of the differentially expressed mRNAs. (C) Significant enrichment items of KEGG analyses. KEGG pathway classification associated with dysregulated mRNAs in skeletal muscle biopsies between individuals with NIID and controls. The OXPHOS pathway, highlighted by a red square, is verified in this study. (D) Heatmap showing hierarchical clustering of the differentially expressed mRNAs. Part of the mitochondrial DNA and nuclear DNA encoding genes were down-regulated. (E) The number of down-regulated genes encoding complex I, complex III, complex IV, and complex V, among which complex I has the largest number of down-regulated genes. (F) Alteration of transcriptional levels of mitochondrial-encoded and nuclear-encoded genes in muscle samples of five NIID cases, determined by qRT-PCR in three biological replicate experiments. The relative mRNA levels of ND2, ND6, COX1, NDUFA3, NDUFA6, NDUFA7, and NDUFV3 were significantly down-regulated in NIID. (G) Representative microscopic images of fly eyes expressing GFP or uN2CpolyG-GFP in combination with small interfering RNA (siRNA)-mediated knockdown of mitochondrial complex I subunits. RNAi knockdown of NDUFA6 and NDUFS2 genes aggravated retinal degeneration in flies expressing uN2CpolyG protein. Fly genotypes: Control (Ctr): GMR-Gal4 > UAS-GFP, GMR-Gal4 > UAS-GFP/UAS-siNDUFA6, GMR-Gal4 > UAS-GFP/UAS-siNDUFS2, and GMR-Gal4 > UAS-GFP/UAS-siNDUFB1; uN2CpolyG: GMR-Gal4 > UAS-N2CpolyG-GFP, GMR-Gal4 > UAS-N2CpolyG-GFP/UAS-siNDUFA6, GMR-Gal4 > UAS-N2CpolyG-GFP/UAS-siNDUFS2, and GMR-Gal4 > UAS-N2CpolyG-GFP/UAS-siNDUFB1. (H and I) Average experimental oxygen consumption rate (OCR) measurements at baseline and after addition of glutamate/malate, adenosine diphosphate (ADP), succinate, carbonyl cyanide 3-chlorophenylhydrazone (CCCP), rotenone, and antimycin are shown. (J) Mitochondrial adenosine-triphosphate (ATP) synthesis was measured. ATP levels were significantly reduced in uN2CpolyG-GFP–expressing flies compared with uN2C-GFP–expressing flies (n = 3 for all conditions; *P < 0.05, ***P < 0.001). Fly genotypes: Actin-Gal4 > UAS-uN2C-GFP and Actin-Gal4 > UAS-uN2CpolyG-GFP. Error bars correspond to the SEM. VS, versus; log2FC, log2 fold change = mean(log2(certain gene transcription level of NIID patient)) - mean(log2(the gene transcription level of control)); Glu/M, glucose or malic acid; Suc, succinic acid; Rot, rotenone; Ama, antimycin A; CII, complex II; ROX, rates of oxidation.
To investigate further the role of mitochondrial complex I subunits in uN2CpolyG-induced pathogenesis, we examined whether down-regulation of mitochondrial complex I modified uN2CpolyG-induced toxicity. Interestingly, knockdown of mitochondrial complex I coding genes, including NDUFA6 and NDUFS2, exacerbated retinal degeneration in flies expressing the uN2CpolyG protein, whereas knockdown of these genes in control flies did not show any detectable effect (Fig. 6G). It suggested that exacerbation of the eye phenotype could be an independent event rather than a synergistic effect, in which susceptibility is increased with the uN2CpolyG expression, together with the OXPHOS deficit.
Finally, given mitochondrial morphological and molecular alterations in uN2CpolyG-expressing flies, we wondered whether these animals exhibited altered mitochondrial respiration. Using the Oroboro Oxygraph system, we observed a significantly reduced mitochondrial respiration capacity in flies expressing uN2CpolyG protein compared with flies expressing the control uN2C protein (Fig. 6H). Mitochondrial alterations were particularly evident for coupled respiration at complex I (CI OXPHOS) (Fig. 6I). Furthermore, we measured the levels of mitochondrial ATP synthesis in the whole body of these two groups of flies. Importantly, flies expressing the uN2CpolyG protein had significantly lower levels of ATP synthesis compared with flies expressing the control uN2C protein (Fig. 6J). These data suggest that changes in mitochondrial complex I could be a key pathogenic process in NIID, potentially unveiling a therapeutic target for this disease.
IDB Restores Motor Performance, Lifespan, ATP Synthesis, and Mitochondrial Morphology in Flies Expressing uN2CpolyG.
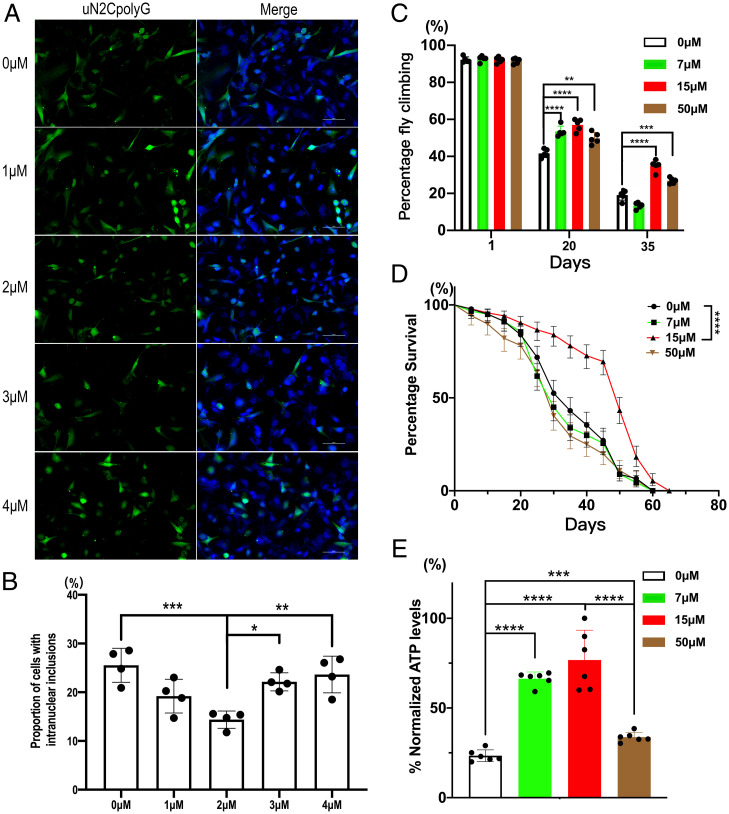
IDB is an analog of coenzyme Q that facilitates electron transfer along the respiratory chain and increases mitochondrial complex I activity (39). As our data suggest that complex I deficiency may be involved in uN2CpolyG pathogenesis, SH-SY5Y neuronal cells expressing uN2CpolyG-GFP were treated with increasing concentrations of IDB. Immunofluorescence showed that a concentration of 2 µM IDB significantly reduced uN2CpolyG aggregate formation (Fig. 7 A and B). Moreover, uN2CpolyG-expressing flies feed 20 d with a 7, 15, or 50 μM concentration of IDB showed significantly improved motor performance (P < 0.0001, P < 0.0001, and P < 0.01, respectively) (Fig. 7C). Importantly, treatment with 15 μM IDB significantly extended the lifespan of flies expressing uN2CpolyG protein (P < 0.0001) (Fig. 7D), although it did not fully restore the animals to normal lifespan. Finally, we measured whole-body mitochondrial ATP synthesis levels in 25-d-old uN2CpolyG-expressing flies treated with IDB. Treatment with 7, 15, or 50 μM IDB significant improved ATP synthesis compared with control flies (P < 0.0001, P < 0.0001, and P < 0.001, respectively), with an optimal dose of 15 μM (Fig. 7E). Notably, the 15 μM dose of IDB significantly mitigated mitochondrial swelling in uN2CpolyG-expressing flies at day 15 (SI Appendix, Fig. S6). These data suggested that IDB attenuated mitochondrial dysfunction and neurodegeneration in a transgenic Drosophila model of NIID, serving as a potential therapeutic agent for the disease.
Fig. 7.
IDB decreases uN2CpolyG aggregation in cells and restores motor performance, lifespan, and ATP synthesis in uN2CpolyG-GFP–expressing flies. (A) IDB treatments at concentrations of 0, 1, 2, 3, and 4 μM were applied to cells expressing uN2CpolyG. Representative immunofluorescence images showed 2 μM IDB decreased uN2CpolyG aggregate formation in SH-SY5Y cells. (B) The proportion of the uN2CpolyG aggregates at different IDB concentrations. The concentration of 2 μM IDB significantly decreased the number of aggregates. (C) Flies expressing uN2CpolyG were fed with 0, 7, 15, and 50 μM IDB. The locomotor index was measured in adult flies at different ages. IDB treatment significantly restored motor performance of flies expressing uN2CpolyG at day 20 and day 35. (D) Effect of IDB on the lifespan of uN2CpolyG-expressing flies. 15 μM IDB treatment significantly improved the survival rate of flies expressing uN2CpolyG. The median lifespan was 35, 30, 55, and 35 d for flies treated at concentrations of 0, 7, 15, and 50 μM IDB, respectively. (log-rank Mantel-Cox test; n = 233–279/genotype for flies; **P < 0.01, ***P < 0.001, ****P < 0.0001). (E) IDB significantly improved ATP levels in flies expressing uN2CpolyG protein. Fly genotype: Actin-Gal4 > UAS-uN2CpolyG-GFP. Error bars correspond to the SEM.
Discussion
NOTCH2NLC is one of the human-specific NOTCH2NL genes, which were thought to be involved in the cortical neurogenesis and the expansion of the human cortex (40, 41). Since the expansion of CGG repeats located in the 5′ UTR of the NOTCH2NLC gene has been identified as the causative mutation of NIID (15–18), numerous clinical studies have confirmed that NIID is a heterogeneous and systemic neurological disease with clinical symptoms and pathological changes involving almost all systems (30). Meanwhile, several studies have demonstrated that the abnormal expression of CGG repeat expansion not only was confined in neurons but also existed in multiple other cells and formed eosinophilic intranuclear inclusions. The intranuclear inclusions can be widely observed in 1) neurons and astrocytic glial cells of central nervous system, including cerebral cortex, basal ganglia, brainstem, hippocampus, and spinal cord (13); 2) Schwann cells of the peripheral nerve system; and 3) multiple cells of other organs, such as skin, blood vessels, colon, bladder, heart, stomach, kidney, and lung tissues (13, 30, 42). However, as a corollary to the recent identification of this mutation, studies on pathogenic mechanisms are just emerging. The National Center for Biotechnology Information (NCBI) Gene Database documented two mRNA isoforms of NOTCH2NLC: NM_001364012.2 (the canonical transcript variant, https://www.ncbi.nlm.nih.gov/nuccore/NM_001364012.2) and NM_001364013.2 (the transcript variant uses an alternative splice site that is expected to result in the use of an upstream start codon, https://www.ncbi.nlm.nih.gov/nuccore/NM_001364013.2) (11). Previous studies verified that NM_001364012.2 was the physiologically dominant transcript variant, and two recent studies were all based on this isoform. They revealed the NOTCH2NLC CGG repeat expansion located in a small uORF, leading to translation of these repeats into a toxic polyG-containing protein, uN2CpolyG, in the absence of the entire NOTCH2NLC gene (10, 11). This protein forms and accumulates in ubiquitin- and p62-positive inclusions in cell and animal models of NIID. Moreover, antibodies against uN2CpolyG stain the typical inclusions in tissue sections of individuals with NIID. Importantly, expression of uN2CpolyG is pathogenic in cell and mouse models, albeit through an unknow mechanism (10, 11). In vitro studies demonstrated that uN2CpolyG protein impairs nuclear lamina and nucleocytoplasmic transport but may not cause acute neuronal cell death (10, 11). Moreover, current animal models are based on transient uN2CpolyG expression in the mouse brain through AAV injection or intrauterine electroporation and thus cannot mimic the multisystem involvement observed in NIID patients.
Here, we established a transgenic Drosophila model expressing ubiquitously the uN2CpolyG protein from the dominant transcript through the UAS-Gal4 system. We report a transgenic animal model expressing uN2CpolyG protein and enabling study of systemic changes. We demonstrated that expression of uN2CpolyG alone is sufficient to induce formation of protein inclusions and age-dependent progressive degeneration, which are key features of NIID (working model in Fig. 8). Interestingly, formation of uN2CpolyG aggregates occurs at a very early stage and precedes massive neuronal cell loss. This is consistent with an observation of ubiquitin-positive inclusions in morphologically intact neurons and years before NIID symptom onset (43). Thus, our study confirms that detection of ubiquitin-positive uN2CpolyG deposits can be an early diagnosis biomarker for NOTCH2NLC-related diseases.
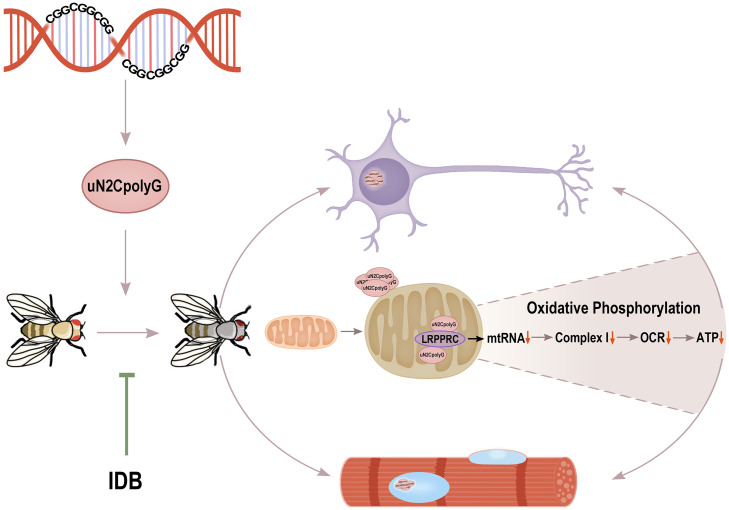
Fig. 8.
Working model for uN2CpolyG-induced mitochondrial dysfunction and progressive neurodegeneration. In this study, the CGG repeat expansion of NOTCH2NLC is translated into toxic uN2CpolyG protein, which leads to intranuclear inclusion formation and progressive neurodegeneration in multiple systems of a Drosophila model. The uN2CpolyG protein is associated with mitochondria and interacts with a mitochondrial RNA binding protein, LRPPRC, which may lead to mitochondrial RNA (mtRNA) instability and reducing mitochondrial OXPHOS complex-related RNA expression. Eventually, down-regulation of complex I activity, reduced OCRs, and decreased ATP synthesis contribute to neuronal and muscle cell degeneration. The use of IDB may reverse mitochondrial damage and alleviate neurodegenerative phenotypes in a transgenic fly model.
Importantly, we observed mitochondrial morphological and molecular alterations in uN2CpolyG-expressing flies. These results were confirmed in tissue sections of individuals with NIID, suggesting that the toxic uN2CpolyG protein may lead to progressive neurodegeneration by impairing mitochondrial functions. These data are reminiscent of another neurodegenerative disease, FXTAS, caused like NIID by a polyG-containing protein, FMRpolyG (9, 44), which also forms protein inclusions and alters mitochondrial functions (45). Furthermore, various mitochondrial alterations are reported in cell models or tissues of FXTAS (46–49). Considering that both NIID and mitochondrial disease are episodic diseases with high clinical heterogeneity, NIID may have pathogenesis similar to that of mitochondrial disease, and mitochondrial abnormalities may be a trigger event for these diseases (50). This is reminiscent of other toxic proteins translated from expanded trinucleotide repeats. Notably, an association between mutant huntingtin with a polyQ expansion and the TIM23 complex has been shown in HD, which leads to deficient mitochondrial protein import and subsequent neuronal cell death (51). Similarly, an abnormal accumulation of PABPN1 with an extended polyA tract has been observed on the inner membrane of mitochondria, resulting in alteration of the mitochondrial protein import complex TIM23 and reduced expression of OXPHOS complexes (52). Together, these findings suggest that mitochondrial dysfunctions caused by different toxic protein interactions can be a common pathogenesis in repeat expansion disorders. However, it remains to be determined by what precise molecular mechanisms the uN2CpolyG protein alters the mitochondrial metabolism in NIID. In that aspect, we identified an interaction between uN2CpolyG and LRPPRC. The LRPPRC protein is involved in posttranscriptional regulation of mitochondrial gene expression and is associated with defective complex I assembly and decreased steady-state levels of mitochondrial mRNAs (53, 54). These findings suggest that uN2CpolyG may impair LRPPRC functions, thereby reducing mitochondrial RNA stability and reducing RNA levels, such as down-regulation of the mitochondrial encoding gene for OXPHOS, as shown in uN2CpolyG-expressing flies and in NIID tissue samples (working model in Fig. 8). To support this hypothesis, expression of Bsf, a homolog of human LRPPRC, in uN2CpolyG transgenic fly eyes could partially restore retinal degeneration.
Of clinical interest, our identification of mitochondrial dysfunctions in NIID animal models and tissues may help to pinpoint a potential therapeutic pathway to target and cure these diseases. Notably, we found that IDB treatment improves uN2CpolyG-induced neurodegeneration in Drosophila. IDB is a coenzyme Q10 analog with better pharmacological properties and contains a redox-active moiety, so it is able to transport electrons to protect cells from oxidative damage and help to preserve mitochondrial respiratory capacity (55). IDB has been used to treat numerous neurodegenerative disorders, including AD (56), Friedreich ataxia (57), and dementia (58). Considering that 15 μM IDB could significantly restore lifespan and motor performance of uN2CpolyG-expressing flies, possibly by improving mitochondrial activities and ATP levels, as well as ameliorating mitochondrial morphological deficits, we propose that mitochondria can be an important therapeutic target, and IDB a potential therapeutic agent, for NREDs (working model in Fig. 8).
Interestingly, both NIID and FXTAS are caused by similar CGG repeat expansions translated into polyG-containing proteins (9–11). In addition to FMR1 and NOTCH2NLC, three other genes, LRP12, GIPC1, and RILPL1, have been recently reported to contain similar 5′ UTR-embedded CGG repeat expansions causing a neuromuscular disorder, OPDM, which presents some striking overlapping clinical symptoms and histopathological features with FXTAS and NIID (9). These findings suggest the existence of a class of disorders called the polyG diseases (15, 59–61). It remains to test whether mitochondrial dysfunctions may be a common pathogenesis of these diseases and whether reversing/blocking mitochondrial dysfunction may be a potential therapeutic approach to treat polyG diseases.
There are limitations in this study. We only used the dominant transcript variant (NM_001364012.2) of NOTCH2NLC to generate the transgenic fly model in accordance with previous studies (10, 11). Although the expression level of another transcript variant (NM_001364013.2) of this gene was much lower than that of the dominant transcript, its pathogenic role in NIID is still required to be elucidated. Moreover, the antisense transcripts of NOTCH2NLC appeared to be generated in fibroblasts of individuals with NIID (17). Previous studies showed that repeat-associated non-AUG (RAN) translation also occurred in an antisense transcript containing expanded C4G2 repeats of C9orf72 and expanded CCG repeats of FMR1 to generate toxic proteins (62, 63). Additional studies are needed to decipher whether toxic peptides could be translated from the antisense transcripts of NOTCH2NLC by RAN translation machinery.
In conclusion, our study indicates that transgenic Drosophila expressing the uN2CpolyG protein recapitulates key pathological and clinical features of NREDs. This powerful animal model will be a key asset for studying pathogenic mechanisms of these diseases and can be used for research on genetic modifiers or therapeutic agents in the future.
Materials and Methods
Transgenic Constructs and Fly Strains.
The uN2C and uN2CpolyG reported in the previous study (10) were subcloned into the attB-pUAST vector, which contains a UAS sequence in the promoter region (Fig. 1A) (64): uN2C with 9× glycine as wild-type uN2C and uN2CpolyG with 100× glycine as mutant uN2C. Both uN2C and uN2CpolyG protein fused at its carboxyl terminus to GFP. After the sequence of complementary DNA (cDNA) was verified, the constructs with or without GFP fusion protein were inserted into attP2 site of phiC31 stocks via standard microinjection, as in our previous study (65). Transgenic flies expressing GFP vector control, uN2C-GFP, or uN2CpolyG-GFP were generated. The RNA interference (RNAi) stocks of mitochondrial complex I were a gift from Dr. Jane Wu’s laboratory and UAS-Bsf stock was provided by Dr. Tao Wang’s laboratory (38). All Gal4 lines were obtained from the Bloomington Drosophila Stock Center. All fly strains were maintained on standard cornmeal agar medium at 25 °C with a 12-h light-dark cycle.
Climbing Assay and Lifespan Assay.
Sex-specific climbing assays and lifespan assays were performed using vials containing 20 to 30 flies of the same nature. All flies were separated within 1 d of eclosion and transferred to new vials every 4 d throughout the assay period. For climbing assay, flies were lightly tapped down to the bottom of the vial. Then, we recorded the number of flies climbing above 5 cm within 15 s. Each climbing trial was repeated five times, and the mean and SE were calculated. The lifespan assay conducted between three gender-matched and age-matched groups as described above. The climbing assay and lifespan assay were recorded every 5 d.
Extraction of Mitochondrial Fraction.
Mitochondria extraction of Drosophila tissues was based on the previous protocol (66). In brief, 50 alive flies were transferred into a glass-Teflon Dounce homogenizer filled with 500 µL of cold isolation buffer (220 mM mannitol, 70 mM sucrose, 20 mM HEPES, and 1 mM EDTA) and were homogenized on ice for 20 strokes. The homogenate was transferred to a 1.5-mL tube for centrifugation at 300 × g for 5 min at 4 °C. The supernatant was then centrifuged at 6,000 × g for 10 min at 4 °C to enrich for mitochondria. The mitochondrial pellet was washed in 1 mL of wash buffer (250 mM sucrose, 50 mM HEPES, and 1 mM EDTA, pH 7.4) and was then resuspended in the same buffer and stored at −80 °C before use.
PK Digestion Assay.
The fresh isolated mitochondrial samples were incubated with the different indicated concentrations of PK on ice for 10 min. Then, the reaction was terminated by phenylmethylsulfonyl fluoride at a final concentration of 2 mM, followed by 95 °C heating for 10 min. Mitochondrial proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were detected by western blot analysis.
qRT-PCR.
We isolated the total RNA from patients’ skeletal muscle samples using TRIzol reagent (Invitrogen). We performed the cDNA synthesis and qRT-PCR as described (61). Fold changes of RNA levels were calculated using the 2−ΔΔCT method and analyzed by Student’s t test. The primer pair sequences used for qPCR reactions are listed in SI Appendix, Table S1. ACTB was used as a reference gene.
Administration of IDB.
IDB was obtained from Qilu Pharmaceutical in the form of powder, stored at 4 °C, and dissolved in dimethyl sulfoxide (DMSO) at a storage concentration of 1 mM and stored at −20 °C. The drug was diluted to the working concentration with cornmeal agar medium of Drosophila or a fresh medium of cells, and the drug was freshly prepared before each experiment.
IDB was administered at the egg stage, and adults were transferred to fresh vials with the drug three times per week. IDB was dissolved in 0.025% DMSO [safe dietary concentration of DMSO in feeding medium for flies (67)] to final concentrations of 0, 7, 15, and 50 μM. These concentrations of IDB were used according to previously reported methods (68, 69).
SH-SY5Y cells expressing uN2CpolyG were treated with different concentrations (0, 1, 2, 3, and 4 μM) of IDB for 24 h. Then, immunofluorescence was performed as described above.
Full details regarding the electron microscopy (EM), western blot analysis, patients, muscle biopsies and brain autopsy, cell culture and transfection, immunofluorescence, immuno-EM, immunoprecipitation, RNA-seq, oxygen consumption rate, and ATP assay are provided in SI Appendix.
Statistical Analysis.
Data were shown as mean ± SEM for the number of observations. Comparisons of two groups were performed using Student’s unpaired t test for repeated measurements to determine the levels of significance for each group. The one-way ANOVA test is used to calculate the degree of significance wherever there were more than two groups (Dunnett’s multiple comparison test). Each experiment was repeated a minimum of two times independently, and probability values of P < 0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
This project was financially supported by the National Natural Science Foundation of China (Grants 81460199, 81571219, 82071409, 82171846, 82160252, and U20A20356), the Natural Science Foundation of Jiangxi province (20202BAB206029), the Double Thousand Talents program of Jiangxi province (jxsq2019101021), and Capital's Funds for Health Improvement and Research (2022-4-40716). We are indebted to the cooperation of the individuals and their families. We are grateful to Mr. Lijun Chai (Peking University First Hospital) for his help in taking EM images and to Ms. Jing Liu and Ms. Qingqing Wang (Peking University First Hospital) for their work in preparations for pathological sections. We are grateful for Jinfa Ma, Lu Song, and Dr. Li Zhu from Dr. Jane Wu’s laboratory (Institute of Biophysics) for providing RNAi stocks of mitochondrial complex I. We are grateful for Dr. Tao Wang and Ms. Yuting Zhang from the National Institute of Biological Sciences for providing UAS-Bsf stock. We thank Li Wang from the Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Science, for help with immunoelectron microscopy sample preparation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208649119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SIAppendix. The data is available upon reasonable request.
References
- 1.Depienne C., Mandel J. L., 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges? Am. J. Hum. Genet. 108, 764–785 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malik I., Kelley C. P., Wang E. T., Todd P. K., Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 22, 589–607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Spada A. R., Taylor J. P., Repeat expansion disease: Progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 11, 247–258 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieberman A. P., Shakkottai V. G., Albin R. L., Polyglutamine repeats in neurodegenerative diseases. Annu. Rev. Pathol. 14, 1–27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trottier Y., et al. , Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature 378, 403–406 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Bates G., Expanded glutamines and neurodegeneration--A gain of insight. BioEssays 18, 175–178 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Fan X., Dion P., Laganiere J., Brais B., Rouleau G. A., Oligomerization of polyalanine expanded PABPN1 facilitates nuclear protein aggregation that is associated with cell death. Hum. Mol. Genet. 10, 2341–2351 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Albrecht A., Mundlos S., The other trinucleotide repeat: Polyalanine expansion disorders. Curr. Opin. Genet. Dev. 15, 285–293 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Sellier C., et al. , Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron 93, 331–347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boivin M., et al. , Translation of GGC repeat expansions into a toxic polyglycine protein in NIID defines a novel class of human genetic disorders: The polyG diseases. Neuron 109, 1825–1835 e1825 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong S., et al. , Upstream open reading frame with NOTCH2NLC GGC expansion generates polyglycine aggregates and disrupts nucleocytoplasmic transport: Implications for polyglycine diseases. Acta Neuropathol. 142, 1003–1023 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Sung J. H., Ramirez-Lassepas M., Mastri A. R., Larkin S. M., An unusual degenerative disorder of neurons associated with a novel intranuclear hyaline inclusion (neuronal intranuclear hyaline inclusion disease). A clinicopathological study of a case. J. Neuropathol. Exp. Neurol. 39, 107–130 (1980). [DOI] [PubMed] [Google Scholar]
- 13.Sone J., et al. , Clinicopathological features of adult-onset neuronal intranuclear inclusion disease. Brain 139, 3170–3186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sone J., et al. , Neuronal intranuclear inclusion disease cases with leukoencephalopathy diagnosed via skin biopsy. J. Neurol. Neurosurg. Psychiatry 85, 354–356 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Ishiura H., et al. , Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat. Genet. 51, 1222–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Tian Y., et al. , Expansion of human-specific GGC repeat in neuronal intranuclear inclusion disease-related disorders. Am. J. Hum. Genet. 105, 166–176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sone J., et al. , Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat. Genet. 51, 1215–1221 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Deng J., et al. , Long-read sequencing identified repeat expansions in the 5'UTR of the NOTCH2NLC gene from Chinese patients with neuronal intranuclear inclusion disease. J. Med. Genet. 56, 758–764 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Jiao B., et al. , Identification of expanded repeats in NOTCH2NLC in neurodegenerative dementias. Neurobiol. Aging 89, 142 e141–142 e147 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Shi C. H., et al. , NOTCH2NLC Intermediate-Length Repeat Expansions Are Associated with Parkinson Disease. Ann. Neurol. 89, 182–187 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Yau W. Y., et al. ; Genomics England Research Consortium, Low prevalence of NOTCH2NLC GGC repeat expansion in white patients with movement disorders. Mov. Disord. 36, 251–255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma D., et al. , Association of NOTCH2NLC repeat expansions with Parkinson disease. JAMA Neurol. 77, 1559–1563 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang P., et al. , Repeat expansion scanning of the NOTCH2NLC gene in patients with multiple system atrophy. Ann. Clin. Transl. Neurol. 7, 517–526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Q. Y., et al. , Expansion of GGC repeat in the human-specific NOTCH2NLC gene is associated with essential tremor. Brain 143, 222–233 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Ng A. S. L., et al. , NOTCH2NLC GGC repeat expansions are associated with sporadic essential tremor: Variable disease expressivity on long-term follow-up. Ann. Neurol. 88, 614–618 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Okubo M., et al. , GGC repeat expansion of NOTCH2NLC in adult patients with leukoencephalopathy. Ann. Neurol. 86, 962–968 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Yuan Y., et al. , Identification of GGC repeat expansion in the NOTCH2NLC gene in amyotrophic lateral sclerosis. Neurology 95, e3394–e3405 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Yu J., et al. , The GGC repeat expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy type 3. Brain 144, 1819–1832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogasawara M., et al. , CGG expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy with neurological manifestations. Acta Neuropathol. Commun. 8, 204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H., et al. , Re-defining the clinicopathological spectrum of neuronal intranuclear inclusion disease. Ann. Clin. Transl. Neurol. 7, 1930–1941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao L., Yan Y., Zhao G., NOTCH2NLC-related repeat expansion disorders: An expanding group of neurodegenerative disorders. Neurol. Sci. 42, 4055–4062 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Huang X. R., Tang B. S., Jin P., Guo J. F., The phenotypes and mechanisms of NOTCH2NLC-related GGC repeat expansion disorders: A comprehensive review. Mol. Neurobiol. 59, 523–534 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Fan Y., Xu Y., Shi C., NOTCH2NLC-related disorders: The widening spectrum and genotype-phenotype correlation. J. Med. Genet. 59, 1–9 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Boivin M., Charlet-Berguerand N., Trinucleotide CGG repeat diseases: An expanding field of polyglycine proteins? Front. Genet. 13, 843014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng J., et al. , Genetic origin of sporadic cases and RNA toxicity in neuronal intranuclear inclusion disease. J. Med. Genet. 59, 462–469 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Fukuda H., et al. , Father-to-offspring transmission of extremely long NOTCH2NLC repeat expansions with contractions: Genetic and epigenetic profiling with long-read sequencing. Clin. Epigenetics 13, 204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y., et al. , Neddylation activity modulates the neurodegeneration associated with fragile X associated tremor/ataxia syndrome (FXTAS) through regulating Sima. Neurobiol. Dis. 143, 105013 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Jaiswal M., et al. , Impaired mitochondrial energy production causes light-induced photoreceptor degeneration independent of oxidative stress. PLoS Biol. 13, e1002197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angebault C., et al. , Idebenone increases mitochondrial complex I activity in fibroblasts from LHON patients while producing contradictory effects on respiration. BMC Res. Notes 4, 557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki I. K., et al. , Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell 173, 1370–1384.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiddes I. T., et al. , Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356–1369.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujita M., et al. , Case report: Adult-onset neuronal intranuclear inclusion disease with an amyotrophic lateral sclerosis phenotype. Front. Neurosci. 16, 960680 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motoki M., et al. , Neuronal intranuclear inclusion disease showing intranuclear inclusions in renal biopsy 12 years earlier. Neurology 91, 884–886 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Todd P. K., et al. , CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gohel D., et al. , FMRpolyG alters mitochondrial transcripts level and respiratory chain complex assembly in Fragile X associated tremor/ataxia syndrome [FXTAS]. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 1379–1388 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Ross-Inta C., et al. , Evidence of mitochondrial dysfunction in fragile X-associated tremor/ataxia syndrome. Biochem. J. 429, 545–552 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hukema R. K., et al. , Induced expression of expanded CGG RNA causes mitochondrial dysfunction in vivo. Cell Cycle 13, 2600–2608 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giulivi C., Napoli E., Tassone F., Halmai J., Hagerman R., Plasma metabolic profile delineates roles for neurodegeneration, pro-inflammatory damage and mitochondrial dysfunction in the FMR1 premutation. Biochem. J. 473, 3871–3888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez-Mora M. I., et al. , Impaired mitochondrial function and dynamics in the pathogenesis of FXTAS. Mol. Neurobiol. 54, 6896–6902 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Liang H., et al. , Clinical and pathological features in adult-onset NIID patients with cortical enhancement. J. Neurol. 267, 3187–3198 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Yano H., et al. , Inhibition of mitochondrial protein import by mutant huntingtin. Nat. Neurosci. 17, 822–831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doki T., et al. , Mitochondrial localization of PABPN1 in oculopharyngeal muscular dystrophy. Lab. Invest. 99, 1728–1740 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Oláhová M., et al. , LRPPRC mutations cause early-onset multisystem mitochondrial disease outside of the French-Canadian population. Brain 138, 3503–3519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siira S. J., et al. , LRPPRC-mediated folding of the mitochondrial transcriptome. Nat. Commun. 8, 1532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gueven N., Woolley K., Smith J., Border between natural product and drug: Comparison of the related benzoquinones idebenone and coenzyme Q10. Redox Biol. 4, 289–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar A., Singh A., A review on mitochondrial restorative mechanism of antioxidants in Alzheimer’s disease and other neurological conditions. Front. Pharmacol. 6, 206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clay A., Hearle P., Schadt K., Lynch D. R., New developments in pharmacotherapy for Friedreich ataxia. Expert Opin. Pharmacother. 20, 1855–1867 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Gillis J. C., Benefield P., McTavish D., Idebenone. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic use in age-related cognitive disorders. Drugs Aging 5, 133–152 (1994). [DOI] [PubMed] [Google Scholar]
- 59.Xi J., et al. , 5′ UTR CGG repeat expansion in GIPC1 is associated with oculopharyngodistal myopathy. Brain 144, 601–614 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Yu J., et al. , The CGG repeat expansion in RILPL1 is associated with oculopharyngodistal myopathy type 4. Am. J. Hum. Genet. 109, 533–541 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng J., et al. , Expansion of GGC repeat in GIPC1 is associated with oculopharyngodistal myopathy. Am. J. Hum. Genet. 106, 793–804 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mori K., et al. , Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Ladd P. D., et al. , An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 16, 3174–3187 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Brand A. H., Perrimon N., Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 65.Deng J., et al. , Novel and recurrent mutations in a cohort of Chinese patients with young-onset amyotrophic lateral sclerosis. Front. Neurosci. 13, 1289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng J., et al. , FUS interacts with ATP synthase beta subunit and induces mitochondrial unfolded protein response in cellular and animal models. Proc. Natl. Acad. Sci. U.S.A. 115, E9678–E9686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cvetković V., et al. , Toxicity of dimethyl sulfoxide against Drosophila melanogaster. Biol. Nyssana 6, 37–41 (2015). [Google Scholar]
- 68.Cohen A. R., New advances in iron chelation therapy. Hematology (Am. Soc. Hematol. Educ. Program) 2006, 42–47 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Soriano S., et al. , Deferiprone and idebenone rescue frataxin depletion phenotypes in a Drosophila model of Friedreich’s ataxia. Gene 521, 274–281 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SIAppendix. The data is available upon reasonable request.