Significance
Intestinal symbionts regulate host physiology by influencing the accumulation and activation of various immunologic cell types via secreted metabolites or contact-triggering of host receptors. From this study, we propose a model in which interaction of the host with the commensal Escherichia coli Nissle and downstream T cells activation can be regulated by the presence of a bacterial capsule. Mutants lacking the capsule are sequestered in the gut lumen in large intraluminal casts, involving immunoglobulin A and host cell exudates, impeding recognition and activation of T cells. Because un-capsulated E. coli Nissle exacerbated colitis, this physical barrier could be a way for the host to avoid prolonged exposure to potentially harmful microbes.
Keywords: gut, CD4+ T cell, genetic screen, capsule, IgA
Abstract
T cells that express the transcription factor RORγ, regulatory (Treg), or conventional (Th17) are strongly influenced by intestinal symbionts. In a genetic approach to identify mechanisms underlying this influence, we performed a screen for microbial genes implicated, in germfree mice monocolonized with Escherichia coli Nissle. The loss of capsule-synthesis genes impaired clonal expansion and differentiation of intestinal RORγ+ T cells. Mechanistic exploration revealed that the capsule-less mutants remained able to induce species-specific immunoglobulin A (IgA) and were highly IgA-coated. They could still trigger myeloid cells, and more effectively damaged epithelial cells in vitro. Unlike wild-type microbes, capsule-less mutants were mostly engulfed in intraluminal casts, large agglomerates composed of myeloid cells extravasated into the gut lumen. We speculate that sequestration in luminal casts of potentially harmful microbes, favored by IgA binding, reduces the immune system’s actual exposure, preserving host–microbe equilibrium. The variable immunostimulation by microbes that has been charted in recent years may not solely be conditioned by triggering molecules or metabolites but also by physical limits to immune system exposure.
The relationship between mammals and microbes that populate their digestive tract is multifaceted. Several bacterial species can induce pathology but most are innocuous and even beneficial to the host. The immune system has evolved an array of innate or adaptive detectors for distinct conserved structures on pathogens, but microbes have also learned to utilize these sensors to manipulate the host to their own advantage (1, 2). Commensal bacteria that reside in the intestinal lumen possess surface structures quite similar to those of pathogens, but they achieve a harmless coexistence with the local immune system. These immunostimulatory molecules include structures indispensable to bacterial function, such as flagellins, peptidoglycans, and lipopolysaccharides, which act as ligands for Toll-like-receptor (TLR) sensors. Conversely, bacteria employ stealth strategies to limit recognition by the immune system, such as hiding their stimulatory components behind a polysaccharidic capsule (3). Importantly, these capacities to manipulate the immune system are widely shared between different phyla, and there is more variation between strains of a given species than there is between genera or phyla, a variability that is likely important to ensure flexibility and adaptability to bacterial species (4, 5).
Among the well-recognized lymphoid populations elicited by commensal bacteria are the T cells that express the RORγ transcription factor. Earliest recognized were conventional CD4+ T helper (Th17) cells that express cytokines of the interleukin (IL)-17 family. These neutrophil-recruiting cytokines induce anti-microbial peptides and tight-junction proteins in intestinal epithelial cells, thereby buttressing gut barrier integrity and protecting against fungi and extracellular bacterial infections (6–10). On the other hand, Th17 cells can be detrimental to the host. They have been implicated in various inflammatory and autoimmune disorders (10, 11), and several studies have distinguished beneficial “homeostatic” types of Th17 cells needed for border control versus pathogenic types, typically marked by interferon gamma expression (12–14) although there is likely a continuum between these two entities (15, 16). The induction of Th17 cells by segmented filamentous bacteria (SFB) (17, 18) is the archetype of the control of T cell differentiation by microbes, but other commensals also have that ability (19, 20), including some used as probiotics (20). The other RORγ+ T cell population influenced by microbes is a subset of FoxP3+ T regulatory cells (Treg). Colonic Tregs are comprised of two major subsets, which can be distinguished operationally and phenotypically by the expression of specific transcription factors, RORγ (or c-Maf) vs. Helios (or Gata3) (21–27). The Helios+Gata3hi population, not dissimilar to Treg populations found in other tissues, is homeostatically controlled by IL-33, and expresses several transcripts linked to tissue repair (e.g., Areg) (22). The RORγ+ Maf+ pool, a major component of colonic lamina propria (LP) Tregs, is far less frequent in other body locations (23, 24). Several functions have been attributed to colonic Tregs, in particular the RORγ+ population (2, 28). They influence the intensity of generic gut inflammation, control immunoglobulin A (IgA) production, dampen Th1/2/17-biased conventional T cell (Tconv) activation, and control food allergy (29). RORγ+ Tregs are strikingly responsive to fluctuations in bacterial load and composition: germ-free (GF) condition or treatment with broad-spectrum antibiotics result in reduced RORγ+ Tregs (23, 24, 30). Single strains of commensal bacteria can induce RORγ+ Tregs upon colonization of GF mice (23, 31, 32); this property is widespread (31, 33), and is shared by members of diverse genera, such as Clostridiae, Bacteroides, or Staphylococcus.
The mechanisms and molecules underlying microbial control of RORγ+ T cells remain uncertain. Induction of Th17 cell by SFB involves a multicellular cascade initiated by tight adherence of the bacteria to epithelial cells (2), but the pathways used by other commensals are unclear. For Tregs, several microbe-controlled metabolites have been reported to influence their induction, like short-chain fatty acids [somewhat controversially (23, 34–37)], or bile acids (37–40). In addition, the enteric nervous system seems to modulate the induction of RORγ+ Tregs by specific microbes (41, 42).
As a means to elucidate these mechanisms, we leveraged intraspecies variation. A first comparison involving a panel of Bacteroides strains did not succeed in identifying genes or operons correlated with RORγ+ Tregs, perhaps because of overly broad genetic variation (33). We then turned to Escherichia coli Nissle (EcN), a human commensal that can be readily manipulated and can induce moderate Th17 and RORγ+ Treg responses (33). Comparison with a genetically close but ineffective E. coli strain revealed several candidate operons, informing an ablation screen that identified a mutant unable to induce intestinal RORγ+ T cell responses. While addressing the mechanism of this phenotype, we found a surprising result. Instead of the fine molecular identification we had sought, we discovered physical containment of the microbe in luminal casts, large structures composed of dead extravasated immunocytes and IgA-coated microbes, and we propose that this physical barrier constrained microbe exposure and limited T cell responses.
Results
A Bacterial Screen Identifies a Capsule Mutant Unable to Induce Intestinal T Cells.
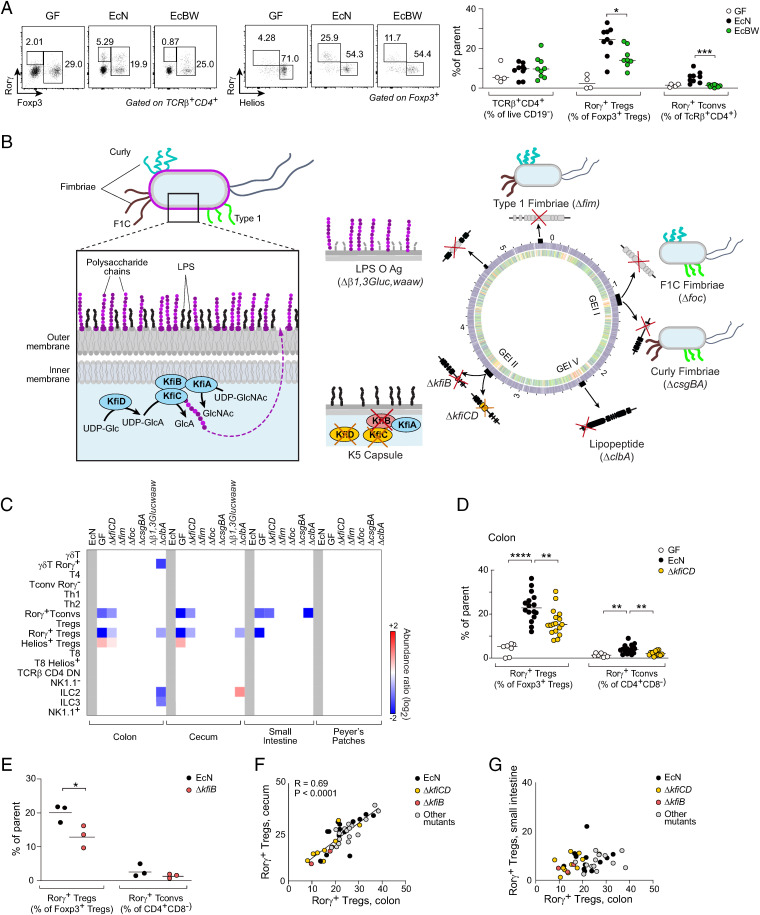
Closely related strains within a bacterial species can vary widely in their immunostimulatory capacity, as exemplified by EcN versus E. coli BW25113 (EcBW). Monocolonization of GF mice with EcN resulted in the induction of both RORγ+ Tconvs and RORγ+ Tregs in the colonic lamina propria (LP), while EcBW was unable to elicit these populations (Fig. 1A). The two strains were found at similar densities in the feces, indicating that overall levels of colonization did not account for the difference (SI Appendix, Fig. S1A), implying that genetic variation between the two strains affected loci that influence the accumulation of colonic T cells. We compared EcN and EcBW genomes using the RAST annotation tool (43) and identified variation in ∼1,000 genes (∼600 genes unique to EcN and ∼400 genes with low similarity, out of 4,832 total coding sequences in the E. coli genome) (Fig. 1B and Dataset S1). We filtered out mobile elements, phage, and replication machinery or hypothetical proteins, leaving 200 coding sequences. Many of these sequences clustered in genomic islands, regions that allow rapid evolution and segregation of pathogenicity within the E. coli species (44, 45). Some of the differential loci related to mobile elements, phage specificity or growth (iron transporters), which are unlikely to be related to immune system crosstalk, but others encoded cell wall and outer membrane structures that could plausibly influence immunocytes. To assess the relevance of some of these candidate differences, we devised a screen that inactivated the corresponding operons in EcN, either by large deletion or by silencing genes known to be essential for the operon’s function, as schematized in Fig. 1B. These mutants included ΔkfiCD for the K5 capsule, the semirough lipopolysaccharide (LPS) O-antigen (Δb1,3Glucwaaw), type 1 (Δfim), F1C (Δfoc), and Curli (ΔcsgBA) fimbriae and a recently described lipopeptide encoded in the colibactin cluster (ΔclbA) (listed in Dataset S2). Mutant bacteria were introduced into GF mice, the intestines of which they populated at similar levels, as judged by bacterial load in feces (SI Appendix, Fig. S1C); for some key mutants—see below—we also verified that the bacterial load in colonic or cecal content was unaffected (SI Appendix, Fig. S1D), as was the population of epithelial-adherent bacteria in the colon (SI Appendix, Fig. S1E). Detailed immunophenotyping on intestinal tissues from these mice (panels illustrated in SI Appendix, Fig. S2 A and B) revealed differences in lymphocyte populations (Fig. 1C). Mice monocolonized with the capsule mutant (ΔkfiCD) showed a clear reduction in EcN-induced RORγ+ T cells (both Tregs and Tconvs) in several tissues, almost reverting to GF levels (Fig. 1 C and D). The mutant lacking lipopeptide (ΔclbA) partially reproduced these effects in Tregs but not in Tconvs (46). Disruption of the LPS O-antigen or fimbriae did not affect immunocyte representation.
Fig. 1.
Screening of EcN mutants’ immunomodulatory activities. (A) Representative dot plots and proportions of CD4+ RORγ+ Tconv (Foxp3– RORγ+) and RORγ+ Tregs from colonic LP in germfree and mice monocolonized with EcN and EcBW (4 independent experiments, t test *P value <0.05, ***P value <0.001). (B) Left: schematic illustration of EcN outer membrane structure with capsule polysaccharides (purple) and Kfi components of their biosynthetic pathway. Right: schematic of EcN and EcBW genome comparison (SI Appendix, Fig. S1B), with the positions of the mutants created here and their impact on EcN outer membrane. (C) Average fold changes (at P < 0.01) among immunocytes of different organs of mice monocolonized with the mutants, relative to EcN. (D) Frequencies of RORγ+ Treg and Tconv from colonic LP in GF and mice monocolonized with EcN and capsule mutant ΔkfiCD (five independent experiments, t test **P < 0.01, ****P < 10–4). (E) As (D) for mutant ΔkfiB (*P < 0.05). (F, G) Frequencies of RORγ+ Tregs in colon, cecum, and small intestine LP.
We elected to focus on the capsule deficiency because it gave most robust differences. Importantly, neither ΔkfiCD, nor EcN induced any inflammatory response in the gut or systemic dissemination in the mice. We first confirmed the results by constructing an independent mutant (ΔkfiB) that also affected capsule biosynthesis and yielded similar results (Fig. 1E). When comparing results across the entire screen, we observed very similar responses in the colon and cecum of monocolonized mice, with a strong correlation across mice (Fig. 1F). This relationship did not extend to the small intestine (Fig. 1G), indicating specificity for the terminal digestive tract in keeping with E. coli’s preferential localization. The same patterns were detected in IELs (SI Appendix, Fig. S2C). In addition, Tconvs secreted IL-17 in EcN-monocolonized mice, less so in ΔkfiCD and ΔkfiB (SI Appendix, Fig. S2D). Thus, induction of RORγ+ T cells by EcN was strongly dependent on the presence of the capsule.
EcN Δkfi Capsule Mutants Are Highly Coated with Specific IgA.
Binding of secretory IgA is an important means for mammalian organisms to constrain intestinal microbes (47–49), and there are strong relationships between the production and secretion of IgA by intestinal B cells and the microbes that affect RORγ+ T cells. SFB, the prototypic Th17-inducer, activates IgA production (50, 51), as did several Th17 inducers in our pan-phylum survey (33). Conversely, our recent work on maternal control of Treg setpoints demonstrated a negative correlation between RORγ+ Tregs and IgA levels (52). E. coli strains are known to be particularly targeted by secretory IgA, and antigen-specific IgAs target a wide range of their membrane-associated antigens (49, 53, 54).
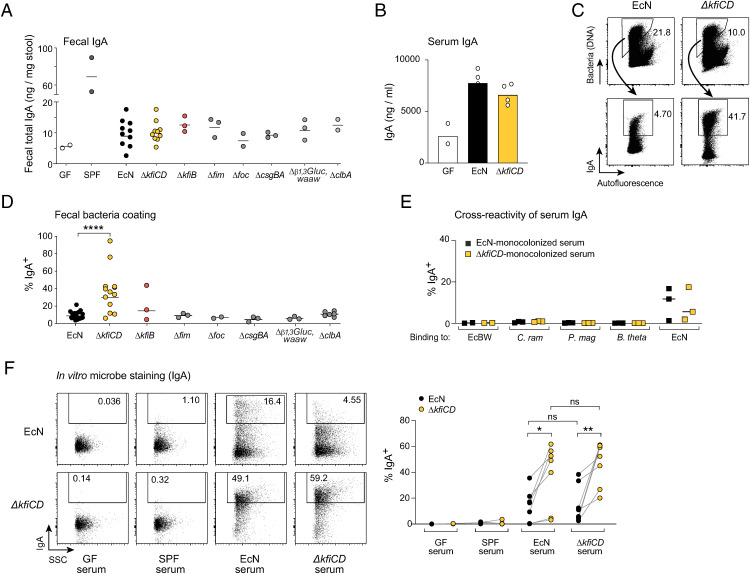
Thus, we asked whether the inability of the capsule mutants ΔkfiCD and ΔkfiB to induce RORγ+ T cells might affect their ability to induce IgA. Overall, the induction of IgA synthesis upon monocolonization of GF mice, assessed as total fecal IgA (Fig. 2A) or IgA in blood (Fig. 2B), was equivalent for EcN and the mutants. On the other hand, we noted a striking difference in coating by IgA of the microbes: ΔkfiCD and ΔkfiB cells were coated with significantly more IgA than were to wild-type (WT) cells and other mutants (Fig. 2 C and D). This enhanced binding did not involve IgG (SI Appendix, Fig. S3A). Microbe-reactive IgA can bind specific surface molecules or can bind to structurally unrelated bacterial antigens (polyreactive IgA) (48, 49). Binding of serum IgA from EcN- or ΔkfiCD-monocolonized mice was highly specific: there was no binding to other commensal bacteria, not even the closely related EcBW (Fig. 2E). Conversely, monocolonization with several microbes did not elicit EcN-reactive serum IgA (SI Appendix, Fig. S3B). However, serum IgA from either EcN or ΔkfiCD-monocolonized mice bound more effectively to cultured ΔkfiCD than to WT EcN (Fig. 2F). The same heightened binding was observed with serum immunoglobulin G (IgG) (SI Appendix, Fig. S3C). Together, these results demonstrate that, despite their reduced ability to induce RORγ+ T cells, the capsule-deficient mutants induced a strong IgA response and, in fact, exposed more epitopes for IgA binding, both in vivo and in vitro.
Fig. 2.
EcN capsule-less Δkfi mutant is highly coated with specific IgA. (A) Total IgA (from ELISA) in stool of germ-free and monocolonized mice. (B) Total serum IgA (from ELISA) in germ-free and monocolonized mice (day 14). (C) Representative cytometry profiles and quantification (D) of IgA-coated bacteria in EcN or ΔkfiCD monocolonized mice (t test ****P < 0.0001). (E) Binding of serum IgA from EcN or ΔkfiCD monocolonized mice, assessed by flow cytometry, against EcBW, Clostridium ramosum, Peptostreptococcus magnus, and Bacteroides thetaiotaomicron. (F) Representative cytometry plots and quantification of binding to EcN or ΔkfiCD bacteria of serum IgA from EcN or ΔkfiCD monocolonized mice, GF or SPF serum for reference (paired t test *P < 0.05, **P < 0.01).
Single-Cell RNA and T Cell Receptor Sequencing Revealed a General Impairment in Activation of CD4 T Cells.
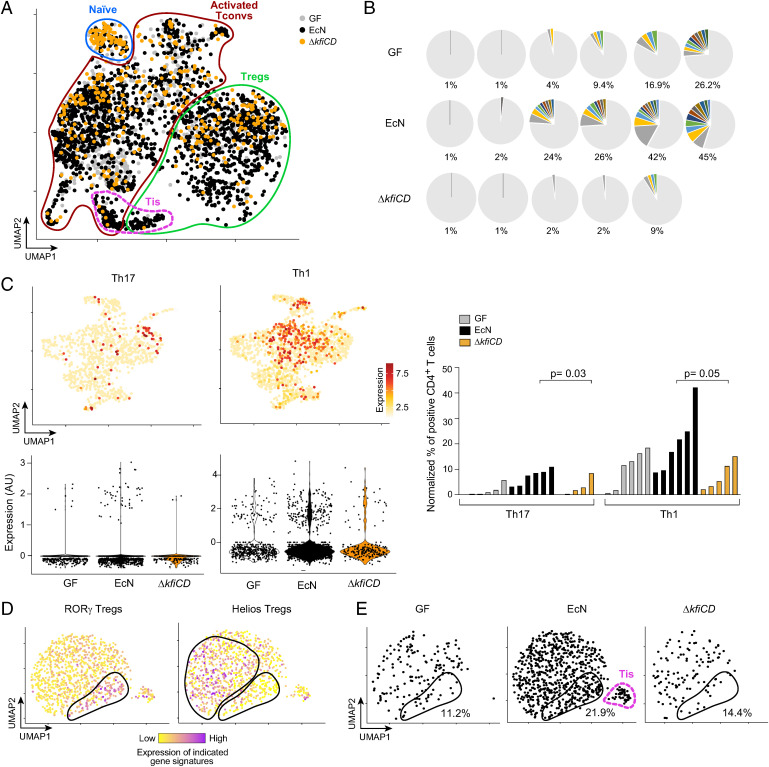
Thus, the Δkfi capsule deficiency yielded an apparently paradoxical outcome, with reduced ability to induce RORγ+ T cells, both Tconvs and Tregs, but a heightened binding of IgA. The capsule is generally thought to be an essential shield for pathogens to evade host defenses and confer resistance to phages (3, 55, 56). Capsule components exert immunomodulatory activity by themselves (57). As an initial step of mechanistic exploration, we performed single-cell RNA sequencing (scRNA-seq) on CD4+ T cells from GF mice monocolonized by WT EcN or the ΔkfiCD mutant, using hash-tagging (58) to group cells from the three different types of mice into the same encapsulation for optimal comparison between groups. Three independent experiments were performed with sorted colonic CD4+ T cells from 17 mice, obtaining QC-passing data for 29,182 cells altogether. Dimensionality reduction and visualization on a Uniform Manifold Approximation and Projection (UMAP) (representative experiment in Fig. 3A) showed that cells from all mice resolved into the same general cell types, identified with signature genes and modules: naive Tconvs, activated Tconv, and Tregs (SI Appendix, Fig. S4A). We also observed a unique cluster of cells expressing high levels of interferon signature genes, as previously reported [named “Tis” in (16)]. Here, Tis cells included both a Treg and a Tconv component (Fig. 3A). Cells from all mice populated the various clusters, with the expected preponderance of naive Tconvs from GF (also high with ΔkfiCD). Tis cells were almost exclusively found in EcN-colonized mice, not in the GF or ΔkfiCD groups.
Fig. 3.
T cell activation and clonal expansion are impaired in mice monocolonized with the ΔkfiCD mutant. scRNA-seq and TCR-seq of GF or EcN and ΔkfiCD monocolonized mice (day14). (A) UMAP projection of transcriptomes from CD4+ T cells; cell populations distinguished by expression of signature gene and modules (per S4A). One representative experiment shown. (B) Proportion of αβ TCR clonotypes in individual mice, GF, or monocolonized. Nonexpanded clones in gray, colored clonotypes present in two or more cells (Dataset S3 for details). (C) Projection on a UMAP dimensionality reduction (activated Tconv cells only) of averaged Th1 and Th17 signatures (listed in Materials and Methods), with corresponding violin plots below. Right: quantification of activated T conv cells positive for Th1 and Th17 gene signatures (normalized to total CD4+ T cells). Each bar corresponds to one mouse (Mann-Whitney P value). (D) UMAP projection (Treg cells from all mice in one experiment), color-coded for expression of RORγ+ and Helios+ Treg gene signatures (genes in Materials and Methods and Dataset S4). (E) As (D), but split between hash tagged cells from different mice; proportion of RORγ+ aTregs and the position of Tis Tregs are shown.
The scRNA-seq data were also processed to identify the αβ T cell receptors (TCRs) expressed, revealing a range of clonality. Cells expressing the same αβ TCR alpha and beta chain clonotype (identity at the level of CDR3 nucleotides), and thus resulting from clonal amplification, were observed at low frequencies in colons of GF- and ΔkfiCD-monocolonized mice (9.6% and 3.0% on average, per 100 cells). Amplified clonotypes were more frequent in EcN-monocolonized mice (23.6% on average), some expressed by dozens of cells across the Tconv compartment (Fig. 3B and Dataset S3). These results indicate that EcN drove a stronger immune response than did the ΔkfiCD mutant.
Focusing the transcriptome analysis on the activated Tconv population (Fig. 3C), we asked whether the lesser clonal expansion seen in ΔkfiCD-colonized GF mice was also accompanied by lower Tconv differentiation toward effector phenotypes. We assessed the single-cell transcriptomes for signatures of Tconv differentiation. In accordance with Kiner et al. (16), no areas of the UMAP corresponded to discrete Th populations. Cells with high levels of the Th1 and Th17 signatures were observed after colonization with EcN, but markedly less frequent in ΔkfiCD-colonized mice (Fig. 3C). Accordingly, bulk RNA sequencing (RNA-seq) of sorted CD4+ T cells showed an over-representation of a curated gene signature of T cell activation (59) after WT vs. ΔkfiCD monocolonization (SI Appendix, Fig. S4B).
Thus, in terms of clonal expansion and effector differentiation, the capsule-deficient mutant was less potent at stimulating colonic Tconv cells, consistent with the flow cytometry data. We similarly interrogated the Treg compartment, parsing these cells into a specific UMAP and using curated gene signatures [(60) and Dataset S4] to visualize the main two subpopulations of colonic Tregs: RORγ+ and Helios+ (Fig. 3D). In this framework, colonization with neither WT nor mutant microbe brought forth a specific Treg subset not already present in GF hosts but only shifted their representation, with an increase in the RORγ+ component after EcN colonization (Fig. 3E). The exception was Treg Tis cells, which were only observed in EcN-colonized mice. Thus, these data further support the idea that when EcN lacks a capsule, it is unable to elicit a response promoting colonic CD4+ T conv activation and RORγ+ Treg accrual.
Cellular Relays?
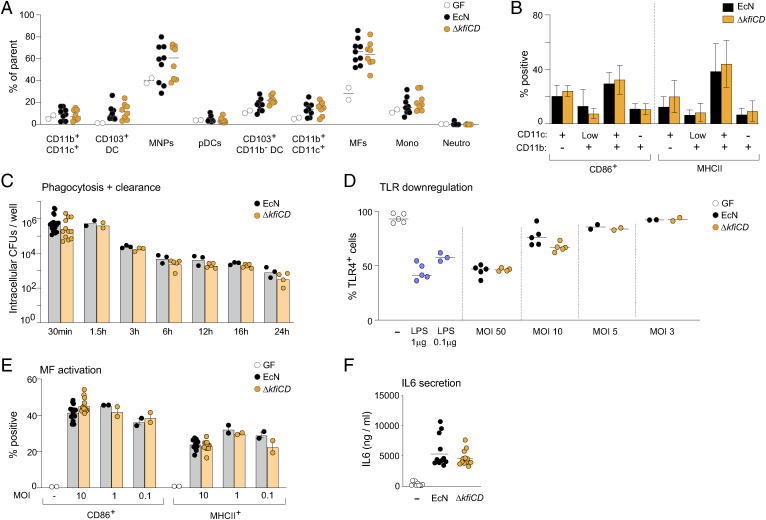
SFB activates Th17 cells through a multicellular cascade of events: epithelial cells are triggered by adhesion of SFB to release free radicals and serum amyloid A (SAA) proteins, which in turn activate myeloid cells (DCs) to produce IL-1b, IL-6 and IL-23, which then favor Tconv differentiation to RORγ+ Il17-producing cells (2). Our observations raised a similar question: since it seemed unlikely that capsule polysaccharides on EcN would directly activate T cells, what might be the relays that boost both RORγ+ Treg amplification and Th17 cell differentiation? Myeloid cells seemed a plausible candidate, since they can be directly activated by bacterial polysaccharides (32, 61), as demonstrated by experiments in vitro where treating DCs with Bifidobacterium polysaccharides enhanced iTreg generation in the same wells (32). The loss of capsule polysaccharides might be expected to unmask LPS. Immunophenotyping of myeloid cells in the colon of monocolonized mice (strategy in SI Appendix, Fig. S2 A and B) did not reveal changes in major myeloid populations, in their abundance (Fig. 4A) or in their activation state, as assessed by expression of CD86 and MHC-II molecules (Fig. 4B) (neither did other mutants in the panel) (SI Appendix, Fig. S5). In addition, gene-expression profiling of total colonic myeloid cells, performed 1 or 14 d after colonization with EcN or ΔkfiCD, did not show any reproducible differences in independent experiments. Given the possible unmasking of LPS in ΔkfiCD cells, we assessed the impact of the mutation in vitro by challenging bone-marrow-derived macrophages (BMDM) with EcN or ΔkfiCD. Both microbes were phagocytosed and then killed equally effectively by BMDM (Fig. 4C). They also induced similar endocytosis of the TLR4 receptor from the surface of BMDM (reflected as loss of surface staining; Fig. 4D). This observation suggested that loss of the capsule may not have exposed more LPS to sensing by macrophages in this instance. The two microbes also activated BMDM similarly, judging from CD86 and MHC-II induction (Fig. 4E) or IL-6 secretion (Fig. 4F). Altogether, these data suggest that the presence or absence of the E. coli K5 capsule did not alter the bacteria’s ability to trigger myeloid cells, and modified interaction with myeloid cells was not the relay between the ΔkfiCD mutation and its reduced ability to invoke T cell responses.
Fig. 4.
Myeloid cells are similarly activated by both EcN and its capsule-less mutant. (A) Myeloid cell proportions, measured by flow cytometry, in colonic LP of GF, or EcN and ΔkfiCD monocolonized mice (day 14; gating strategy from (SI Appendix, Fig. S1E)). (B) Activation of colonic LP myeloid cells assessed by expression of CD86 and MHCII expression 24 h after monocolonization with EcN or ΔkfiCD. (C) Phagocytosis and clearance by BMDM in culture at different times after infection with live EcN and ΔkfiCD at a multiplicity of infection (MOI) = 10, evaluated as cell-associated cfu. (D) TLR4 surface expression on immortalized BMDM after 20 min exposure to EcN or ΔkfiCD at different MOI (LPS used as a positive control). Different concentration of LPS was used as positive control. (E) BMDM activation 24 h after exposure to live EcN and ΔkfiCD in vitro, assessed as expression of CD86 and MHCII. (F) As (E), IL-6 concentration measured by ELISA from culture supernatants after 24 h.
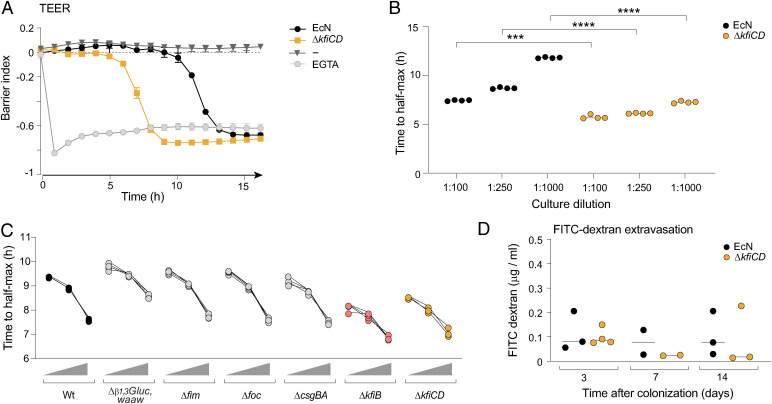
We next addressed the possibility of a differential interaction with other cell types. SFB and other Th17-inducing microbes are closely associated with intestinal epithelial cells (IECs), and this adhesion helps initiate the signaling cascade (17, 19). In addition, EcN’s capsule modulates bacterial interaction with IECs in vitro (62, 63). To investigate the ability of EcN and ΔkfiCD to affect IEC barrier function, we prepared cultures of Caco-2 cells in Maestro Edge plates, exposed them to the microbes, and measured epithelial integrity over time by trans epithelial electrical resistance (TEER). Both bacteria induced a drop in TEER, sharp when initiated, but the transition occurred markedly faster with ΔkfiCD than with EcN (Fig. 5A). The timing of the TEER drop was dose-dependent, implying a cumulative signal delivered by the microbes over time, but the faster response to ΔkfiCD was significant at all doses (Fig. 5B). To test whether this was a unique trait induced by lack of capsule, we performed similar experiments with our other EcN mutants. Only the ΔkfiCD and ΔkfiB mutations affected IEC barrier function at an accelerated rate (Fig. 5C). Interestingly, the LPS O-antigen Δb1,3Glucwaaw mutation slightly delayed the TEER drop, suggesting that in this context the semirough O-Ag had a detrimental effect on the integrity of IECs (Fig. 5C and SI Appendix, Fig. S6).
Fig. 5.
Lack of capsule increases damaging effects on epithelial cells in vitro. (A) Caco-2 cells were cultured in CytoView-Z 96 plates, and TEER was recorded over time after challenge (1:1,000) with EcN or ΔkfiCD (EGTA as a positive control). The barrier index was calculated by “axion impedance module” as the ratio between cellular resistance at low frequency (1 Hz) vs. high frequency (41 Hz). (B) As (A), the time of transition (50% drop in barrier index) was measured in cultures supplemented with EcN or ΔkfiCD at different dilutions. (t test, ***P < 0.001, ****P < 10–4); each point a quadruplicate within an experiment, representative of three experiments. (C) As (B), time to transition after exposure to various EcN mutants at three dilutions (1:1,000, 1:5,000, and 1:104). (D) Assessment of in vivo gut permeability as leakage of FITC dextran (4 kDa) administered by gavage and detected in serum 4 h after oral gavage at different times after monocolonization with EcN or ΔkfiCD (each point a different mouse).
These results suggest that the loss of capsule actually increased the ability of E. coli to damage the IEC barrier. For an in vivo correlate of these findings, we analyzed extravasation from the gut into the circulation of fluorescein isothiocyanate (FITC)-dextran after oral gavage, as a test for intestinal barrier function in GF mice monocolonized with EcN or ΔkfiCD. In contrast to the results obtained in vitro, no clear difference was noted between the two microbes at various times after colonization (Fig. 5D).
Luminal Casts Engulf the Capsule-Deficient Mutant.
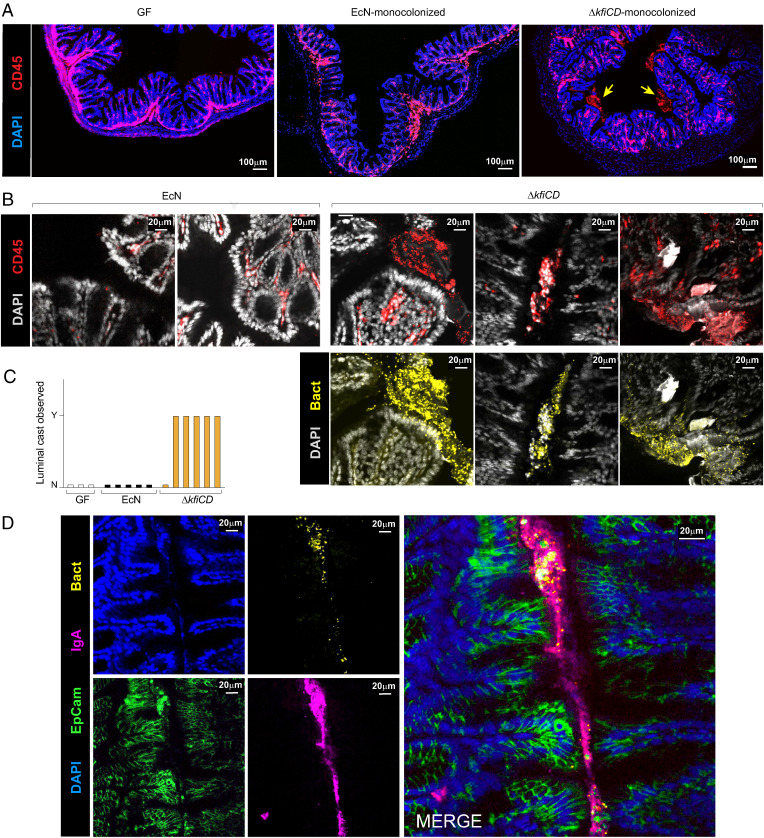
These results left us with a conundrum. The capsule-deficient mutants were able to perturb myeloid cells, induce IgA, and destabilize epithelial cells just as efficiently as the EcN parent strain (even more strongly for IECs): yet they seemed largely ignored by T cells. This paradox led us to hypothesize that, during monocolonization, ΔkfiCD and WT EcN might interact differentially with epithelial cells, engaging signaling pathways that lead to different T cell outcomes. To better understand the interaction of bacteria with the intestinal wall and immunocytes during GF monocolonization, we performed confocal imaging of colon tissue. Agglomerates of CD45-positive immunocytes were observed in the lumen of ΔkfiCD-monocolonized mice (Fig. 6A, higher magnification in Fig. 6B). These structures were absent in both GF and EcN-colonized mice (Fig. 6C). The intraluminal CD45+ structures were large (50–150 μm equivalent to 4–15 cell diameters), with visible nuclei in some, but not all, cases, usually located alongside the epithelial border (Fig. 6B and SI Appendix, Fig. S7A). They included high densities of ΔkfiCD bacteria (Fig. 6 B, Bottom) and also colocalized with IgA deposits (Fig. 6D and SI Appendix, Fig. S7B). These structures were very reminiscent of the “intraluminal casts” described in a recent study, which appear during Toxoplasma gondii infection and accompanying overgrowth of Proteobacteria (64). These casts result from granulocyte extravasation into the lumen, where they contribute to the encapsulation of bacteria, and were proposed to be a mechanism by which the host contains pathobiont outgrowth during adventitious inflammation (65, 66).
Fig. 6.
Luminal IgA-immune cell casts engulf the capsule-less EcN mutant. Confocal imaging of colonic tissue was collected for imaging from GF, EcN, or ΔkfiCD monocolonized mice (day 14). (A) Confocal imaging of colonic tissue immunostained for pan-immunocytes (CD45) and cell nuclei (DAPI). Arrows point to intraluminal CD45+ structures. (B) As (A), with counterstaining for bacteria (anti-mCherry). (C) Quantification of intraluminal casts (scored as presence of discrete CD45+ structures, 100 μm or more) in the colon of GF, EcN, and ΔkfiCD monocolonized mice. Each bar represents an individual mouse. (D) As (B), staining for IgA, Epcam (epithelial cells), and bacteria (anti-mCherry).
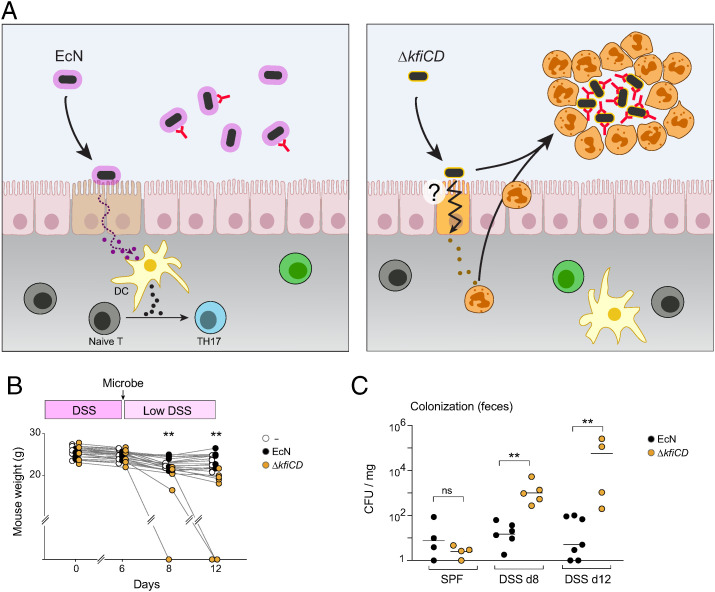
We thus propose that the same phenomenon may operate here, as modeled in Fig. 7A: the capsule-deficient mutant, coated with high amounts of IgA, provokes myeloid cell extravasation and cast formation, encapsulating the microbes to exclude them and arrest an otherwise stronger insult to the epithelial border. We attempted to identify the extravasated cells by flow cytometric analysis of the luminal content, but very few live cells could be identified (with marker profiles suggestive of monocytes). We note that Molloy et al. (64) detected extravasated cells by flow cytometry only during a narrow period after T. gondii infection, a time window that may not apply after bacterial colonization of GF mice.
Fig. 7.
Deleterious effects of the capsule-less mutant during DSS-induced colitis. (A) Schematic representation of proposed model, comparing the epithelial-initiated response to EcN or ΔkfiCD that leads either to RORγ+ cell activation or to myeloid cell extravasation. (B, C) Specific pathogen-free animals were fed 2.5% DSS for 6 d, then gavaged with 107 EcN or ΔkfiCD bacteria (DSS dose lowered to 1.5% for the remaining time). Weight was recorded and normalized to their initial weight. (C) As (B), measure of live bacteria (as cfu/mg of feces) at days 8 or 12 (t test, **P < 0.01).
In this model, physical containment within solid structures of potentially aggressive bacteria (here ΔkfiCD) is conditioned by the initial interactions by the epithelial layer, innocuous symbionts (here EcN) initiating a different response and remaining in the lumen. Encapsulation into casts may be an unstable solution, however. Thus, we asked how EcN and ΔkfiCD microbes would behave in a context of adventitious damage to the IEC layer, for instance after damage induced by dextran sulfate (DSS). Mice housed under normal (SPF) conditions were fed DSS for 5 d, inoculated with either EcN or ΔkfiCD by oral gavage (∼107 colony-forming unit [cfu]/mouse), and more then maintained on a lower dose of DSS to maintain inflammation. Administration of ΔkfiCD had a detrimental impact on the mice, as shown by increased weight loss and sudden death in three of eight mice (Fig. 7B), while EcN had no such effect. In normal SPF mice, EcN colonized more effectively than did ΔkfiCD, likely reflecting the protective role of the capsule, but the capsule-less mutant colonized more effectively under conditions of DSS-induced inflammation (Fig. 7C). Thus, we conclude that the containment offered by intraluminal casts can break down, revealing the more aggressive nature of the capsule-deficient mutant.
Discussion
Starting with a symbiotic strain of E. coli, one that amplifies both Treg and Th17 RORγ+ compartments, and one normally so innocuous that it is used as a probiotic, we initially aimed to identify molecules that induced RORγ+ T cells in the intestine by harnessing the power of microbial genetics. Instead, we ended up rediscovering a strategy through which the host walls off a potentially inflammatory variant of normally tolerated symbionts, previously described in a different context.
In GF mice monocolonized with the capsule-deficient mutant, the colon lumen contained abundant agglomerates that included IgA-coated ΔkfiCD bacteria and CD45+ immunocyte. This disposition resembled the intraluminal casts that engulf Proteobacteria expansion during T. gondii-induced inflammation (64). In the model proposed above (Fig. 7A), we hypothesize that WT EcN induces RORγ+ Th17 and Treg cells by initiating a signaling cascade involving IECs that modulates T cell activation by DCs—this by analogy to mechanisms downstream of SFB triggering the epithelium (67–69). In contrast, loss of the capsule in the ΔkfiCD mutant, which would expose immunoactive components of the outer membrane, like LPS, leads to a more damaging interaction with IECs, as revealed by the faster loss of integrity in TEER assays. This heightened interaction with IECs would initiate a different response: instead of RORγ+ Th17 and Treg recruitment, enhanced extravasation of neutrophils and monocytes, leading to cast formation (plausibly involving DNA release and “netosis”). We also propose that encapsulation of microbes into the cast structures is conditioned by high amounts of bound IgA, as for ΔkfiCD here, as a sorting mechanism for selective inactivation. In this model, the ΔkfiCD mutant was not incapable of immunologic activation: it elicited a robust production of highly specific IgA, possibly through a T-independent route, although the apparently high specificity and affinity of the IgA induced by ΔkfiCD suggests that it was able to activate T follicular helper cells (70). On the other hand, as evidenced by the flow cytometry and scRNA-seq results, the ΔkfiCD strain was unable to activate the pathways connecting to RORγ+ T cells. The shielding of microbes within established casts would prevent the adhesion and endocytosis events required for Th17 induction (19, 71). Prior to encapsulation, IEC activation by ΔkfiCD might be diverted in favor of myeloid cell extravasation instead of RORγ+ T cell inducing paths (e.g., not inducing IL-6), or elicit a dominant inhibitory response. Overall, the key distinction between a microbe that becomes physically excluded in casts vs. a commensal that elicits a mild “homeostatic” Th17 response (like EcN or Bifidobacterium) would be in the initial response of the EICs, working in tandem with the subsequent IgA response toward an effective exclusion of pathobionts.
How does this model integrate with other mechanisms proposed for IgA function? IgA is generally considered to discriminate between noninvasive and invasive microbes, hence protecting the host from bacterial colonization and disease (48, 49). Several mechanisms have been proposed: microbe killing (e.g., recruiting complement), inactivation of important functions (e.g., of adhesion or motility by binding to fimbriae or flagella), or modulation of bacterial gene expression (49). In other instances, some symbionts seem to have co-opted binding of IgA to actually enhance their productive colonization (72). Such effects cannot be excluded here, but the colocalization of IgA and bacteria in casts suggests that IgA binding to microbes might enhance their interaction with intraluminal phagocytes and inclusion in casts.
Encapsulation into robust cast structures is an effective way to physically segregate harmful pathobionts, but this isolation strategy might also be a dangerously unstable one should containment fail. Indeed, the harmful potential of ΔkfiCD was revealed in the context of DSS-induced inflammation, where it induced more severe inflammation (and death of half the animals). There may be correlates in human pathology, such as the E. coli blooms that accompany inflammatory bowel disease.
In fairness, and although this model does account for the different facets of the peculiar interaction of ΔkfiCD with the immune system, we have not formally demonstrated the causal connection between cast formation and the mutant’s inability to trigger RORγ+ T cells. For instance, it is possible that the mutation perturbs the secretion of immunostimulatory molecules, or that it prevents components of the microbe to be routed into specific cellular compartments required to effectively trigger RORγ+ cells. We also cannot rule out that the high degree of IgA binding to mutant microbes might be an epiphenomenon not related to cast formation.
In conclusion, this study highlights the importance of commensals microbe capsules as a means to minimize insult to the host and favor symbiotic relationships. More generally, it implies that the diversity of outcomes that microbial species induce by the immune system may result from variations in physical barriers that they form or encounter in addition to the diversity of molecular agents they encode.
Limitations of Study.
Much of this work, albeit not all, relied on monocolonization of GF mice. This reductionist strategy rendered more tractable the identification of differences in immunomodulatory properties between individual bacterial strains but there is no doubt that it does represent a simplified approximation of the far more complex network of interactions in the microbiota of normal mammals. Because the mutation shunted the ΔkfiCD microbe along an unanticipated path, and the project with it, we did not uncover the actual mechanism leading to homeostatic Th17 and Treg activation by the WT EcN symbiont. Nor do we know how IgA responses were triggered equally well by EcN and mutants (little Tfh induction was detected with either). Finally, the model proposed here attempts to integrate a number of threads in the data, but several aspects remain speculative in the in vivo context.
Materials and Methods
Bacterial Strains and Generation of EcN Mutants.
Strains of the probiotic EcN 1917 wild-type and mutants (EcN; Mutaflor, DSM 6601) and EcBW utilized in this study are listed in Dataset S2. Mutants of EcN were constructed using the lambda red recombinase system (73), as previously described (74).
Generation of Monocolonized Mice.
For monocolonization, GF C57BL/6 mice at 4 wk of age were orally inoculated by gavage with LB-grown single bacteria (∼1 × 107 cfu/mL). Each group of mice was housed in autoclaved cages, with autoclaved food and water and kept under sterile conditions for 2 wk. Fecal material, colon content, and cecum content were collected and plated at 2 wk after bacterial inoculation to ensure monocolonization by a single bacterial strain. Colonization were all preformed and processed at the same time of the day (10 AM) to reduce diurnal variability.
Intestinal Immunocyte Analysis.
Intestinal tissues (colon, cecum, and small intestine) were collected at day 14 postmonocolonization in RPMI 2% fetal calf serum (FCS). Each intestinal section was then measured, cleaned, and treated with RPMI medium containing 1 mM DTT, 20 mM EDTA, 2% FCS at 37 °C for 15 min to remove epithelial cells, then dissociated in 1.5 mg/mL collagenase II (GIBCO), 0.5 mg/mL dispase, 1% FCS in RPMI with constant stirring at 37 °C for 40 min. Single-cell suspensions were then filtered and washed with 4% RPMI solution, stained with three constant panels of antibodies, and analyzed on a BD Symphony flow cytometer, with data analysis in FloJo. Fold-change values for population frequencies were calculated by dividing raw frequencies of the cytometry profiles of a given cell type for each mouse colonization by the average frequency obtained from control mice of the same experiment. For scRNA-seq and T cell receptor sequencing (three independent monocolonization experiments), CD4+ T cells from distal colons of GF or monocolonized mice were sorted (DAPI− CD19− CD4+ TCRβ+), and encapsulated together for scRNA-seq on the 10× Genomics 5′v2 platform after hashtagging with Biolegend TotalSeq-C reagents (10 different hashtags per experiment). For immunofluorescence staining of colon, mice were monocolonized with bacteria expressing mCherry. Tissues were collected 14 d postmonocolonization from distal colon and fixed in 4% PFA overnight. The following day, tissues were place in 30% sucrose for 12–24 h and then embedded in OCT and frozen at −80 °C.
Dynamics TEER Measurements.
Single colony of EcN and its related mutant strains were grown overnight in Lennox Luria-Bertani (LB; BD Difco) at 37 °C to concentration of ∼1 × 109 cfu/mL and diluted in Caco2 growth media without antibiotics. The diluted bacteria cocultured with the cells (seeded at 3 × 105 cells/well 4 d prior to coculture with bacteria) in the CytoView-Z 96 plate and TEER was recorded over time (at 37 °C and 5% CO2). The barrier index was calculated by Axio “Impedance” module (using AxIS Z software version 3.2.3/Axion BioSystems), as the ratio between cellular resistance at low frequency (1 Hz) vs. high frequency (41 Hz). The barrier index was measured at high temporal resolution of 1 min and was normalized to the reference time (t = 0). Additional correction was performed by normalizing to the barrier index of unstimulated cells.
Supplementary Material
Acknowledgments
We thank Drs. D. Yang, I. Chiu, M. Jost, M. McClelland, D. Kasper, and J. Kagan for insightful discussions, advice, and reagents, L. Yang, B. Vijaykumar, and N. Patel for help with computational analyses, T.B. Yanortsonag for mice, and P. Montero Llopis and O. Yaghi for confocal imaging advice and assistance. This work was supported by grants AI125603 and AI150686 from the National Institutes of Health (to C.B. and D.M.), grant 3114831 from the Israel Science Foundation (to N.Y.), grant 8165162 from the Broad-ISF exchange (to N.Y., C.B., and D.M.), and in part by a Sponsred Research Agreement from Evelo Biosciences. M.S.C. was a Cancer Research Irvington Fellow.
Footnotes
Reviewers: A.B., University of California-Davis; and Y.B., National Institutes of Health.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209624119/-/DCSupplemental.
Data, Materials, and Software Availability
The data reported in this paper have been deposited in the Gene Expression Omnibus database under accession numbers GSE211049 (75) and GSE213200 (76). Further details are available in SI Appendix, SI Materials and Methods.
References
- 1.Chu H., Mazmanian S. K., Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivanov I. I., Tuganbaev T., Skelly A. N., Honda K., T cell responses to the microbiota. Annu. Rev. Immunol. 40, 559–587 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cress B. F., et al. , Masquerading microbial pathogens: Capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol. Rev. 38, 660–697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansonetti P. J., Di Santo J. P., Debugging how bacteria manipulate the immune response. Immunity 26, 149–161 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Diacovich L., Gorvel J. P., Bacterial manipulation of innate immunity to promote infection. Nat. Rev. Microbiol. 8, 117–128 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Kumar P., et al. , Intestinal interleukin-17 receptor signaling mediates reciprocal control of the gut microbiota and autoimmune inflammation. Immunity 44, 659–671 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maxwell J. R., et al. , Differential roles for interleukin-23 and interleukin-17 in intestinal immunoregulation. Immunity 43, 739–750 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Lee J. S., et al. , Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 43, 727–738 (2015). Correction in: Immunity 43, 1022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirota K., Ahlfors H., Duarte J. H., Stockinger B., Regulation and function of innate and adaptive interleukin-17-producing cells. EMBO Rep. 13, 113–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaffen S. L., Jain R., Garg A. V., Cua D. J., The IL-23-IL-17 immune axis: From mechanisms to therapeutic testing. Nat. Rev. Immunol. 14, 585–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanai T., Mikami Y., Sujino T., Hisamatsu T., Hibi T., RORγt-dependent IL-17A-producing cells in the pathogenesis of intestinal inflammation. Mucosal Immunol. 5, 240–247 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Lee Y., et al. , Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omenetti S., et al. , The intestine harbors functionally distinct homeostatic tissue-resident and inflammatory Th17 cells. Immunity 51, 77–89.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L., et al. , Niche-selective inhibition of pathogenic Th17 cells by targeting metabolic redundancy. Cell 182, 641–654.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaublomme J. T., et al. , Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell 163, 1400–1412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiner E., et al. , Gut CD4+ T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat. Immunol. 22, 216–228 (2021). Correction in: Nat. Immunol. 22, 666–668 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov I. I., et al. , Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaboriau-Routhiau V., et al. , The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Atarashi K., et al. , Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan T. G., et al. , Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. U.S.A. 113, E8141–E8150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josefowicz S. Z., et al. , Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482, 395–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schiering C., et al. , The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sefik E., et al. , Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohnmacht C., et al. , The microbiota regulates type 2 immunity through RORγ+ T cells. Science 349, 989–993 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Xu M., et al. , c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiSpirito J. R., et al. , Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci. Immunol. 3, eaat5861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell C., et al. , Extrathymically generated regulatory T cells establish a niche for intestinal border-dwelling bacteria and affect physiologic metabolite balance. Immunity 48, 1245–1257.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russler-Germain E. V., Rengarajan S., Hsieh C. S., Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol. 10, 1375–1386 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosovanu C., Neumann C., The many functions of Foxp3+ regulatory T cells in the intestine. Front. Immunol. 11, 600973 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chai J. N., et al. , Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci. Immunol. 2, eaal5068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faith J. J., Ahern P. P., Ridaura V. K., Cheng J., Gordon J. I., Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci. Transl. Med. 6, 220ra11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verma R., et al. , Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3+ regulatory T cells. Sci. Immunol. 3, eaat6975 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Geva-Zatorsky N., et al. , Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arpaia N., et al. , Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furusawa Y., et al. , Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Smith P. M., et al. , The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song X., et al. , Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hang S., et al. , Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W., et al. , A bacterial bile acid metabolite modulates Treg activity through the nuclear hormone receptor NR4A1. Cell Host Microbe 29, 1366–1377.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell C., et al. , Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yissachar N., et al. , An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell 168, 1135–1148.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Y., et al. , Interleukin-6 produced by enteric neurons regulates the number and phenotype of microbe-responsive regulatory T cells in the gut. Immunity 54, 499–513.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aziz R. K., et al. , The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juhas M., et al. , Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grozdanov L., et al. , Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186, 5432–5441 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pérez-Berezo T., et al. , Identification of an analgesic lipopeptide produced by the probiotic Escherichia coli strain Nissle 1917. Nat. Commun. 8, 1314 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pabst O., Slack E., IgA and the intestinal microbiota: The importance of being specific. Mucosal Immunol. 13, 12–21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y., Palm N. W., Immunoglobulin A and the microbiome. Curr. Opin. Microbiol. 56, 89–96 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Bunker J. J., Bendelac A., IgA responses to microbiota. Immunity 49, 211–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lécuyer E., et al. , Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 40, 608–620 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Talham G. L., Jiang H. Q., Bos N. A., Cebra J. J., Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect. Immun. 67, 1992–2000 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramanan D., et al. , An immunologic mode of multigenerational transmission governs a gut Treg setpoint. Cell 181, 1276–1290.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hapfelmeier S., et al. , Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rollenske T., et al. , Parallelism of intestinal secretory IgA shapes functional microbial fitness. Nature 598, 657–661 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Porter N. T., et al. , Phase-variable capsular polysaccharides and lipoproteins modify bacteriophage susceptibility in Bacteroides thetaiotaomicron. Nat. Microbiol. 5, 1170–1181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mues N., Chu H. W., Out-smarting the host: Bacteria maneuvering the immune response to favor their survival. Front. Immunol. 11, 819 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsieh S. A., Allen P. M., Immunomodulatory roles of polysaccharide capsules in the intestine. Front. Immunol. 11, 690 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stoeckius M., et al. , Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levine A. G., Arvey A., Jin W., Rudensky A. Y., Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15, 1070–1078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pratama A., Schnell A., Mathis D., Benoist C., Developmental and cellular age direct conversion of CD4+ T cells into RORγ+ or Helios+ colon Treg cells. J. Exp. Med. 217, e20190428 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Danne C., et al. , A large polysaccharide produced by helicobacter hepaticus induces an anti-inflammatory gene signature in macrophages. Cell Host Microbe 22, 733–745.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nzakizwanayo J., et al. , Disruption of Escherichia coli Nissle 1917 K5 capsule biosynthesis, through loss of distinct kfi genes, modulates interaction with intestinal epithelial cells and impact on cell health. PLoS One 10, e0120430 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hafez M., et al. , The K5 capsule of Escherichia coli strain Nissle 1917 is important in mediating interactions with intestinal epithelial cells and chemokine induction. Infect. Immun. 77, 2995–3003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Molloy M. J., et al. , Intraluminal containment of commensal outgrowth in the gut during infection-induced dysbiosis. Cell Host Microbe 14, 318–328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sumagin R., Robin A. Z., Nusrat A., Parkos C. A., Transmigrated neutrophils in the intestinal lumen engage ICAM-1 to regulate the epithelial barrier and neutrophil recruitment. Mucosal Immunol. 7, 905–915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fournier B. M., Parkos C. A., The role of neutrophils during intestinal inflammation. Mucosal Immunol. 5, 354–366 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Panea C., et al. , Intestinal monocyte-derived macrophages control commensal-specific Th17 responses. Cell Rep. 12, 1314–1324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goto Y., et al. , Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40, 594–607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sano T., et al. , An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell 163, 381–393 (2015). Correction in: Cell 164, 324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fagarasan S., Kawamoto S., Kanagawa O., Suzuki K., Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu. Rev. Immunol. 28, 243–273 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Ladinsky M. S., et al. , Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science 363, eaat4042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Donaldson G. P., et al. , Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sassone-Corsi M., et al. , Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.M. Sassone-Corsi et al., Sequestration of gut pathobionts in intraluminal casts, a mechanism to avoid dysregulated T cell activation by pathobionts. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE211049. Deposited 11 August 2022. [DOI] [PMC free article] [PubMed]
- 76.M. Sassone-Corsi et al., Sequestration of gut pathobionts in intraluminal casts, a mechanism to avoid dysregulated T cell activation by pathobionts. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213200. Deposited 12 September 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus database under accession numbers GSE211049 (75) and GSE213200 (76). Further details are available in SI Appendix, SI Materials and Methods.