Abstract
The strong immunogenicity of bacterial fimbriae results from their polymeric and proteinaceous nature, and the protective role of these immunogens in experimental or commercial vaccines is associated with their capacity to induce antiadhesive antibodies. Fimbria-mediated intestinal colonization by enteropathogens typically leads to similar antibody responses. The possibility of taking advantage of these properties was investigated by determining whether enteroadhesive fimbriae, like the 987P fimbriae of enterotoxigenic Escherichia coli, can serve as carriers for foreign antigens without losing their adhesive characteristics. Random linker insertion mutagenesis of the fasA gene encoding the major 987P subunit identified five different mutants expressing wild-type levels of fimbriation. The linker insertion sites of these mutants were used to introduce three continuous segments of viral surface glycoproteins known to be accessible to antibodies. These segments encode residues 11 to 19 or 272 to 279 of herpes simplex virus type 1 (HSV-1) glycoprotein D [gD(11–19) and gD(272–279), respectively] or residues 379 to 388 of the transmissible gastroenteritis virus (TGEV) spike protein [S(379–388)]. Studies of bacteria expressing fimbriae incorporating mutated FasA subunits alone or together with wild-type FasA subunits (hybrid fimbriae) indicated that foreign epitopes were best exported and displayed on assembled fimbriae when they were inserted near the amino terminus of FasA. Fimbriated bacteria expressing FasA subunits carrying the HSV gD(11–19) or the TGEV S(379–388) epitope inserted between the second and third residues of mature FasA elicited high levels of foreign epitope antibodies in all rabbits immunized parenterally. Antibodies against the HSV epitope were also shown to recognize the epitope in the context of the whole gD protein. Because the 987P adhesive subunit FasG was shown to be present on mutated fimbriae and to mediate bacterial attachment to porcine intestinal receptors, polymeric display of foreign epitopes on 987P offers new opportunities to test the potential beneficial effect of enteroadhesion for mucosal immunization and protection against various enteric pathogens.
Since the original studies documenting how critical the fimbriae of enterotoxigenic Escherichia coli (ETEC) are for enteral colonization and diarrhea in animals and humans were published (50, 58), fimbriae have been considered antigens for potential vaccine development. Fimbriae of ETEC are highly immunogenic proteins, inducing protective antibodies which inhibit bacterial adhesion and colonization (28, 34). For example, piglets of dams injected with purified 987P fimbriae are protected against experimental infections with 987P-fimbriated ETEC, and this protection correlates with the presence of specific antiadhesive anti-987P antibodies in the colostrum (27, 28, 42, 43). In the veterinary field, anti-ETEC vaccines consisting of the epidemiologically most important fimbriae have been used for many years and are considered both safe and effective (39, 40). Currently tested vaccines against ETEC infections in humans include fimbrial antigens (51).
Two major properties of fimbriae explain their high levels of immunogenicity. These are their proteinaceous composition and their quasi-homopolymeric structures as fimbriae consist typically of the multimeric assembly of one major type of subunit. The repetitive nature of the helically arranged subunits results in the presentation of the same epitopes 102 to 103 times on each fimbrial thread, or 105 to 106 times on each bacterial surface, rendering fimbriae major immunogens of fimbriated killed or live bacterial vaccines. Several investigators have proposed taking advantage of the strong immunogenic properties of fimbriae by using them as carriers of protective microbial foreign epitopes. Concentrating essentially on the feasibility of creating fimbrial chimeras, most studies noted that there appeared to be unpredictable structural constraints dictating the length or sequence of the genetically inserted foreign peptide (44). Some of these limitations may have resulted from the fimbrial locations used for insertion, the target sites having been based exclusively on comparative and predictive analysis of primary structure information. Only hypervariable domains (3, 5, 61, 62) or predicted surface-exposed domains of fimbrial proteins (25, 45) were considered potential permissive insertion sites, namely, sites which accept insertions without affecting fimbrial expression.
In this study, we have taken a new experimental approach, based on a random mutagenesis technique, allowing us to avoid the bias of theoretical predictions for localizing permissive insertion sites in the 987P major subunit FasA. An earlier version of this technique was used successfully to study the topography of the 987P outer membrane or usher protein FasD (53). Here, random mutagenesis was designed to specifically target only DNA encoding the mature portion of FasA, keeping the other 987P genes intact for complementing regulation and export functions (6, 17, 19, 53). Identification and characterization of the best permissive site in FasA for carrying a foreign epitope and for surface exposure was evaluated with two epitopes of glycoprotein D (gD) of herpes simplex virus (HSV) and one epitope of the spike protein of transmissible gastroenteritis virus (TGEV) (10, 11, 14, 29, 47). 987P fimbriae displaying foreign epitopes were shown to induce specific anti-foreign epitope antibodies in all immunized rabbits.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
E. coli strains used in this study are listed in Table 1. Cultures for colony isolations or plasmid purifications were grown in L medium (57) with appropriate antibiotics used at the following concentrations: ampicillin, 200 μg/ml; chloramphenicol, 30 μg/ml; tetracycline, 10 μg/ml. Media components were purchased from Difco (Detroit, Mich.), and, unless specified otherwise, reagents were purchased from Sigma Chemical Co. (St. Louis, Mo.). Restriction and modification enzymes were from New England Biolabs (Beverly, Mass.), the ApaI linker was from Pharmacia LKB Biotechnology (Piscataway, N.J.), and pancreatic DNase I was from GIBCO BRL (Gaithersburg, Md.). Oligonucleotides used as PCR primers or for inserting sequences encoding foreign epitopes (see Table 2) were synthesized on an Applied Biosystems model 380B DNA synthesizer and purified with a final trityl group.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristics | Reference(s) or source |
|---|---|---|
| E. coli strains | ||
| SE5000 | MC4100 recA56(Fim−) | 57 |
| MH6085 | dcm-6 dam-3 metB galK galT lacY tsx thi ton mtl? | M. Howe |
| Plasmids | ||
| pDMS158 | pACYC184 fas+ | 6, 53 |
| pDMS161 | pDPS158 fasA (deletion of 5′ end of fasA) | This study |
| pDMS167 | pBR322 fas+ | 53 |
| pDMS175 | pKS fasA+ | This study |
| pDMS183 | pGEM4z cat | This study |
| pRS205 | pUC19 fasA+ | This study |
| pRS206 | pKS fasA signal sequence | This study |
| pRS207 | pRS206 fasA | This study |
| pRS207A(2aG5) | pRS207 ApaI site at nucleotide 86 of fasA | This study |
| pRS207A(2bE11) | pRS207 ApaI site at nucleotide 289 of fasA | This study |
| pRS207A(5bC8-2) | pRS207 ApaI site at nucleotide 289 of fasA | This study |
| pRS207A(5bC8-3) | pRS207 ApaI site at nucleotide 424 of fasA | This study |
| pRS207A(2aF6) | pRS207 ApaI site at nucleotide 574 of fasA | This study |
| pRS207A(3bE12) | pRS207 ApaI site at nucleotide 578 of fasA | This study |
| pRS207A(N168P) | pRS207 ApaI site at nucleotide 499 of fasA | This study |
| pRS210 | pRS207A(2aG5) with insert for gD(272–279) | This study |
| pRS211 | pRS207A(2bE11) with insert for gD(272–279) | This study |
| pRS212 | pRS207A(5bC8-2) with insert for gD(272–279) | This study |
| pRS213 | pRS207A(2aF6) with stop linker at fasA 3′ end | This study |
| pRS214 | pRS207A(3bE12) with stop linker at fasA 3′ end | This study |
| pRS215 | pRS213 with insert for HSV gD(272–279) | This study |
| pRS216 | pRS207A(2bE11) with insert for TGEV S(379–388) | This study |
| pRS218 | pRS207A(2aG5) with insert for TGEV S(379–388) | This study |
| pRS221 | pRS213 with insert for TGEV S(379–388) | This study |
| pRS223 | pRS207A(5bC8-2) with insert for TGEV S(379–388) | This study |
| pRS225 | pRS207 ApaI site at nucleotide 76 of fasA | This study |
| pRS226 | pRS207A(2aG5) with insert for HSV gD(11–19) | This study |
| pRS227 | pRS225 with insert for HSV gD(272–279) | This study |
| pRS228 | pRS225 with insert for HSV gD(11–19) | This study |
| pRS229 | pRS207A(2aF6) with nucleotide 141 to stop codon of pRS207A(5bC8-3) (3′ end duplication) | This study |
| pRS230 | pDMS167 with fasA from pRS228 | This study |
| pRS233 | pRS225 with insert for TGEV S(379–388) | This study |
| pRS234 | pDMS167 with fasA from pRS233 | This study |
| pRS239 | pRS225 with insert for HSV gD(7–23) | This study |
TABLE 2.
Peptide and oligonucleotide sequences of foreign epitopes
| Source | Residues | Antibodya | Peptide and oligonucleotide sequences | Reference(s) |
|---|---|---|---|---|
| gD protein of HSV | 272–279 | MAb DL6 | Gly Pro Glu Asp Pro Glu Asp Ser Ala Leu Leu Gly Pro | 29 |
| CCG GAA GAC CCG GAA GAC TCC GCT CTG CTG GGG GCC | ||||
| C CGG GGC CTT CTG GGC CTT CTG AGG CGA GAC GAC CC | ||||
| C GAA GAC CCG GAA GAC TCC GCT CTG CTG GGG CC | ||||
| CC GGG CTT CTG GGC CTT CTG AGG CGA GAC GAC C | ||||
| gD protein of HSV | 11–19 | MAb 1D3 | Gly Pro Met Ala Asp Pro Asn Arg Phe Arg Gly Gly Pro | 14, 29 |
| CCG ATG GCT GAC CCG AAC CGT TTC CGT GGT GGG GCC | ||||
| C CGG GGC TAC CGA CTG GGC TTG GCA AAG GCA CCA CC | ||||
| C ATG GCT GAC CCG AAC CGT TTC CGT GGT GGG CC | ||||
| CC GGG TAC CGA CTG GGC TTG GCA AAG GCA CCA C | ||||
| Spike protein of TGEV | 379–388 | PAb anti-peptide S(379–388) | Gly Pro Ser Ser Phe Phe Ser Tyr Gly Glu Ile Pro Gly Pro | 10, 11 |
| CCG TCC TCC TTC TTC TCC TAC GGT GAA ATC CCG GGG GCC | ||||
| C CGG GGC AGG AGG AAG AAG AGG ATG CCA CTT TAG GGC CC | ||||
| C TCC TCC TTC TTC TCC TAC GGT GAA ATC CCG GGG CC | ||||
| CC GGG AGG AGG AAG AAG AGG ATG CCA CTT TAG GGC C |
PAb, polyclonal antibody.
Plasmid constructs.
Standard procedures (49) were used to construct the following plasmids. The 5′ end of fasA of plasmid pDMS158 was removed as a SalI-SpeI fragment to obtain pDMS161. This plasmid, which expresses the fasB to fasH genes, was used in complementation assays to study fimbrial expression by the various fasA mutants. Plasmid pDMS175 was constructed by cloning the fasA-containing HindIII-ClaI fragment from pDMS167 (53, 54) into phagemid pKS (Stratagene, La Jolla, Calif.) and by deleting a HindIII-EspI fragment upstream from fasA and several restriction sites in vector DNA (deletion of the EcoRV-SmaI fragment and trimming of the overhangs of the ApaI site with T4 DNA polymerase). Plasmid pDMS175 was engineered by PCR in two steps to contain unique restriction sites flanking the reading frame encoding only processed fasA. First, a 520-bp fragment encoding essentially only processed fasA was amplified on a thermal cycler (model 480; Perkin-Elmer Corp., Norwalk, Conn.) with pDMS175 used as a template, primers U453 (5′-GCTCTAGATGCTAGCTGCGCCCGCTGAAAC-3′) and L965 (5′-GCTCTAGATGTCGACTTACGGTGTACCTGCTGAAC-3′), and Taq DNA polymerase, as suggested by the manufacturer (Perkin-Elmer). PCR was carried out for 16 cycles, each consisting of 1 min at 94°C, 1 min at 65°C, and 1 min at 72°C. The 520-bp fragment was restricted with XbaI and cloned into pUC19 to give plasmid pRS205, resulting in the insertion of a silent NheI site just downstream of the DNA encoding the FasA signal sequence cleavage site and a SalI site following the fasA stop codon. In a second step, a vector containing only the signal sequence of fasA was prepared by inverse PCR. For this, primers U563 (5′-GGAATTCATACCGTCGACCTC-3′) and L4660 (5′-GGAATTCTGCTAGCGAGTAACCACTG-3′), with appropriately placed EcoRI and NheI or SalI sites, respectively, were used to amplify pDMS175 DNA as described above, with a 3-min extension time at 72°C. The resulting 3.1-kb fragment was digested with EcoRI and ligated to give plasmid pRS206. Finally, the 520-bp NheI-SalI fragment of pRS205 was subcloned into NheI-SalI-restricted pRS206 to generate pRS207. Plasmids pRS213 and pRS214 were constructed from two plasmids with ApaI linkers at the 3′ ends of fasA (see below) by replacing their ApaI-SalI fragments with a short linker encoding stop codons in the corresponding reading frames. Plasmid pRS225 was constructed by inverse PCR as described above with primers U467 (5′-GGGGGCCCGCTGAAAACAACAC-3′) and L455 (5′-GGGGGCCCTGGCGCTGCTAGCATCTAGA-3′) for insertion of an ApaI site after the 6th bp of the open reading frame for mature FasA in plasmid pRS207. The expected constructs were confirmed by DNA sequencing.
Mutagenesis.
The following approach was used to create a library of randomly inserted polylinkers in 987P DNA encoding only processed FasA. Plasmid pRS205 was CsCl purified (49) and was digested with limiting concentrations of DNase I in the presence of Mn2+, as described previously (53) with the following modifications. The linearized band was gel purified and ligated with T4 DNA ligase to a chloramphenicol resistance (Cmr) gene flanked by ApaI sites. This cassette is a SmaI restriction fragment of pDMS183, which contains a Cmr gene flanked by overlapping SmaI/ApaI restriction sites (18). The ligation mixture was used to transform competent SE5000 or MH6085 cells by electroporation (16), and the cells were plated on L agar with chloramphenicol. To select for random insertions in pRS205, approximately 10,000 Cmr colonies were pooled and grown to stationary phase, and plasmid DNA was isolated as described above. The CsCl-purified DNA was then restricted with the fasA flanking enzymes NheI and SalI to prepare a second library with the Cmr gene inserted exclusively in the fasA-containing fragment. The DNA fragment containing the fasA gene with randomly inserted Cmr cassette was isolated by agarose gel electrophoresis and recloned into the original vector (pRS206). More than 5,000 Cmr transformants were pooled to prepare a CsCl-purified plasmid library, as described above. This DNA was restricted with ApaI to excise the Cmr gene, and linearized DNA was isolated, ligated, and used to transform SE5000(pDMS161) for complementation studies. Transformants with a functional mutated FasA protein were screened for fimbriation by colony immunoblots and/or seroagglutination tests. Site-directed mutagenesis was performed to insert an ApaI site in fasA at one of the predicted surface-exposed sites of its product by using the Altered Sites II in vitro mutagenesis system (Promega Corp.) and mutagenic primer U889, 5′-ACCACCACAGGGCCCCCTGATACAAACGGT-3′, as described elsewhere (7). The resulting fasA allele-containing plasmid, designated pRS207A(N168P), encoded a proline residue substituted for Asn residue 145 of mature FasA.
Insertion of foreign epitopes.
Linker insertion sites in fasA were used as target sites for the addition of foreign DNA encoding antigenic epitopes. Only the four plasmids encoding mutated FasA proteins which behaved like wild-type FasA, based on the complementation assay for fimbriation, were studied. The plasmid constructs encoding foreign epitopes in FasA are derivatives of pRS207 having the foreign DNA inserted at an ApaI linker insertion site identified as a permissive site. Epitope insertions were created by digestion of these plasmids with ApaI and ligation with a 100-fold excess of phosphorylated double-stranded oligonucleotides specifying an epitope (Table 2). Pairs of hybridizing oligonucleotides were flanked by ApaI sites for in-frame insertion at the preexisting ApaI linker sites in fasA. The designs of all oligonucleotides encoding foreign epitopes were based on an E. coli codon usage table (48). Plasmids of transformants carrying the insertions were identified by restriction analysis. The orientation of each insertion was screened by PCR with appropriate primers and was further confirmed by nucleotide sequencing.
DNA sequencing.
DNA sequencing of fasA constructs was undertaken to determine or confirm linker insertion sites as well as the correct in-frame insertion of foreign DNA into fasA. All plasmids constructed or modified by PCR were sequenced by the chain termination method (49) with an Applied Biosystems model 373A sequencer, with double-stranded plasmid DNA as a template. Various appropriate primers were used, including forward and reverse lacZ primers and a specifically designed primer corresponding to DNA encoding the signal sequence of FasA.
Seroagglutination and bacterial aggregation by enterocyte brush borders.
Slide agglutinations were performed with preadsorbed rabbit anti-987P antiserum (52), with anti-FasG (6), or with foreign epitope-specific antibodies. Monoclonal antibodies (MAbs) DL6 and 1D3 recognize specific continuous epitopes of gD of HSV, as previously reported with synthetic peptide antigens (14, 29). The anti-TGEV epitope antibody was prepared as described below. Bacterial aggregation by enterocyte brush borders containing 987P receptors was determined as described previously (33, 55).
Colony immunoblotting.
For the selection of bacteria expressing modified 987P fimbriae on the bacterial surface after linker insertion into fasA, colonies were screened for fimbriation by immunoblotting with the quaternary structure-specific anti-987P MAb E11 (52), as described previously (53). Briefly, L broth with ampicillin and chloramphenicol in microtiter wells was inoculated with individual colonies, and bacteria were grown overnight and replica plated onto nitrocellulose discs placed on L agar dishes containing the appropriate antibiotics. Bacteria were grown for 10 to 18 h, and the blots were exposed to chloroform vapor for 15 min, transferred to a blocking buffer containing 3% bovine serum albumin (BSA) in TNT (0.01 M Tris [pH 7.3], 0.9% NaCl, 0.05% Tween 20) for 1 h, and washed with TNT (49). Rinsed blots were processed for 987P detection by antibody E11 (52), peroxidase-conjugated secondary anti-rabbit immunoglobulin G antibodies, and 3,3′-diaminobenzidine. To assess the surface accessibility of foreign epitopes inserted into FasA, intact bacterial cells were probed by the following blotting procedure. Bacteria grown overnight in L broth with the appropriate antibiotics were centrifuged, washed, and resuspended in phosphate-buffered saline. Ten microliters of three fourfold dilutions of the cells (1:4, 1:16, and 1:64) were applied to a Polymacron membrane (Kalyx Biosciences Inc., Nepean, Ontario, Canada) previously wetted in 20% ethanol, washed with water, and air dried. The spots were allowed to air dry, membranes were blocked with a modified 5× Denhardt solution (DS) (49) consisting of DS with 0.1% Nonidet P-40, 1.5% BSA, 1× borate-buffered saline (pH 8.2), 0.05% gelatin, and 0.04% NaN3 (8). The DL6 or 1D3 MAb recognizing HSV gD epitopes (20, 29) inserted into FasA was used at a 1:2,000 dilution, and the secondary antimouse antibodies conjugated to horseradish peroxidase were used at a 1:5,000 dilution in blocking buffer. The membranes were incubated with antibodies for 1 h, and all washings were done three times in a solution identical to the blocking solution, except that no BSA was added to the DS. The blots were developed by using the Renaissance enhanced chemiluminescence (ECL) detection system (DuPont NEN, Boston, Mass.).
SDS-PAGE and Western blotting.
Cells producing fimbriae consisting of FasA and/or hybrid FasA subunits were cultured to the mid-exponential phase of growth. The fimbriae were then isolated by heat shock treatment and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting, essentially as described previously (33). After protein transfer from polyacrylamide gels to nitrocellulose sheets, the blots were processed as described above for intact bacterial cells by ECL.
Electron microscopy.
Immune electron microscopy of fimbriae or cells expressing 987P fimbria with foreign epitopes was performed as described previously (6) by using MAbs DL6 and 1D3 and 5-nm colloidal gold protein A (Auroprobe; Amersham, Arlington Heights, Ill.).
Preparation of fimbriae.
Fimbriae expressed on the bacterial surface were prepared by heat extraction (60°C for 30 min) essentially as described previously (33). Moreover, with preparations containing nonnegligible amounts of contaminating proteins, these proteins were first precipitated with 5% saturated ammonium sulfate for approximately 30 min at 4°C. Pellets were removed by a centrifugation step, and the supernatants were precipitated with 30% ammonium sulfate for overnight at 4°C. Following centrifugation and pellet resuspension in phosphate-buffered saline, the purity of the fimbrial preparations was judged to be >90% by SDS-PAGE. Fimbrial preparations were desalted by several concentration and dilution steps using ultrafiltration (Centricon 10; Amicon, Beverly, Mass.). The yields of pure fimbriae (36) were 2.7 to 9 mg per liter of culture.
Immunization and peptide antigen.
Adult New Zealand White rabbits were immunized by injecting subcutaneously 200 μg of fimbriae in complete Freund’s adjuvant, followed by three to four booster injections of 100 to 200 μg of fimbriae in incomplete Freund’s adjuvant at 1- to 3-week intervals. Sera were screened for reactivity by dot blot assay. A synthetic peptide corresponding to amino acid residues 379 to 388 of the TGEV spike protein [S(379–388)] with a cysteine added to the carboxy terminus (SFFSYGEIPC) was prepared and cross-linked to keyhole limpet hemocyanin (KLH) as described elsewhere (20). Antibodies against the TGEV epitope were induced by injecting the TGEV peptide coupled to KLH into rabbits by the protocol described above with the fimbrial antigens. Antibody titers of approximately 1/50,000 were determined by enzyme-linked immunosorbent assay, as described previously (52), using coated peptide antigen.
Dot blot assay.
The antigen spot test was used as described elsewhere (8), with the following modifications. Nitrocellulose strips were spotted with 3 μl of different dilutions of purified gD or TGEV peptides (750, 75, and 7.5 ng) and gD protein (150, 15, and 1.5 ng). All dilutions were made in TNS buffer (0.01 M Tris [pH 7.3], 0.15 M NaCl, 0.1% Nonidet P-40). The strips were air dried and blocked as described above for immunoblotting of intact cells. Incubations with dilutions of sera raised against mutated 987P fimbriae carrying the HSV epitope gD(11–19) (i.e., residues 11 to 19 of gD) or the TGEV epitope S(379–388) were done at room temperature. The blots were developed with goat antirabbit antibodies conjugated to horseradish peroxidase at a dilution of 1:1,500 and detected by ECL.
RESULTS
Identification of permissive linker insertion sites in FasA.
FasA is the major structural subunit of the 987P fimbrial polymer. Fimbrial subunit export and assembly on the bacterial surface results from the dynamic interaction of structural subunits with components of the 987P export apparatus. For this, sets of specific FasA domains are thought to interact sequentially with opposing surfaces of the periplasmic chaperone FasB and of the outer membrane usher protein FasD. In addition, consistent with current fimbrial biogenesis models, during and following export through the usher channel, distinct regions of FasA are thought to initiate and finalize the strong noncovalent intermolecular associations typifying the polymeric structure of fimbriae. The FasA subunit being a small molecule, it can be assumed that only few segments of its primary structure can be altered without interfering with bacterial fimbriation. To determine whether such permissive domains exist and to map their location, the fasA gene was subjected to random linker insertion mutagenesis. The search for nonessential domains of FasA was rendered more stringent by inserting a linker encoding in two reading frames proline and glycine, which act as breakers of secondary structures such as β-sheets or α-helices. Identified permissive sites should essentially represent structurally nonessential domains which are required neither for subunit export nor for assembly of fimbriae.
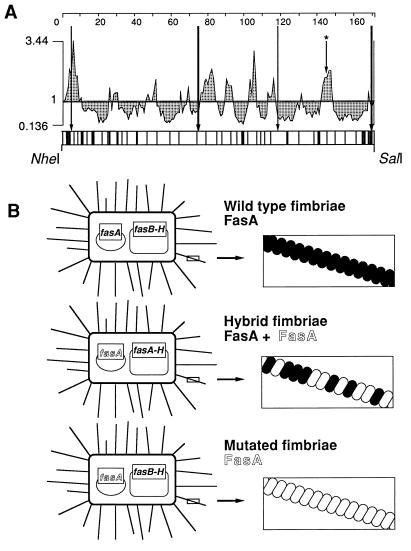
To carry out DNA manipulations only in fasA without affecting other fimbrial genes, a transcomplementation system was created by constructing two separate compatible plasmids. E. coli SE5000(pDS161), carrying all of the fimbrial genes with the exception of fasA, is not fimbriated. In contrast, this strain expresses 987P fimbriae after being transformed with pDMS175, which carries only the fasA gene, indicating that the two plasmids complement each other. To later select for mutations occurring exclusively in the mature FasA protein, pDMS175 was modified by introducing two unique restriction sites (NheI and SalI) flanking the DNA encoding processed FasA, to obtain pRS207. A Cmr cassette flanked by ApaI linkers was inserted randomly into pRS207 linearized previously with DNase I. A final library with the ApaI linker inserted only in the fasA region encoding the mature product was obtained by subcloning the NheI-SalI fragments containing the randomly inserted Cmr cassette and excising the cassette. This plasmid library was then used to transform strain SE5000(pDS161) for complementation analysis. Permissive sites in FasA were identified by colony immunoblot assay with MAbs specific for the quaternary structure of the 987P fimbriae, to exclude colonies expressing export- or assembly-deficient fasA products. Of 450 individual colonies, 22 reacted with the antibodies, and the isolates were confirmed to be fimbriated by seroagglutination. Restriction mapping revealed that linker insertion occurred randomly over the targeted fasA sequence and that some of the permissive sites were located near the amino or carboxy terminus of mature FasA (Fig. 1A). DNA sequencing of fasA from the fimbriated strains identified six different insertional mutants (Table 3), most of the 22 isolates being identical. Five of these mutants seroagglutinated like the wild-type strain containing the complementing nonmutated plasmids (Table 3). The two insertions at the 3′ end of fasA created frameshifts resulting in the addition of 13 or 15 amino acid residues at the carboxy terminus of FasA. That the resulting allelic FasA proteins were functional suggested that the carboxy terminus of FasA is permissive to the addition of extraneous stretches of amino acids. To prepare the 3′ end of fasA for the replacement of the frameshift-originated tails by foreign epitopes, a stop codon was engineered downstream from the ApaI site in the appropriate frames. Only the construction keeping the original carboxy-terminal proline residue still expressed fimbriae (Table 3), suggesting that this residue, in contrast to the expendable penultimate residue (threonine), is essential for the stability or function of FasA. Moreover, a FasA allelic protein (pRS229) with duplication of the 52 carboxy-terminal residues of FasA did not complement the other Fas proteins for 987P fimbriation, highlighting a limitation to the approach used for adding residues to this end of FasA (data not shown). Curiously, with the exception of the permissive site identified at the amino terminus, all of the other permissive sites identified by our random linker insertion approach did not localize in predicted surface-exposed domains (Fig. 1A). Moreover, introduction of an ApaI linker by site-directed mutagenesis into the last predicted surface-exposed domain of FasA [pRS207A(N168P)] inhibited fimbrial expression (data not shown). Taken together, the data suggested that our experimental approach was successful in identifying permissive sites which would not have been found with commonly used predictive algorithms.
FIG. 1.
(A) Surface probability profile of FasA and mapping of linker insertion sites in fasA. Horizontal axis values correspond to the amino acid sequence of mature FasA. Vertical axis values are probabilities of surface exposure (21). Approximately 60 linker insertion sites were mapped by restriction analysis (±25 bp), as indicated by vertical bars in the bottom box, which represents the NheI-SalI-flanked fasA DNA segment encoding the mature FasA protein. Permissive linker insertion sites, more precisely identified by sequence analysis, are indicated by long arrows. The arrow with the asterisk indicates a construct specifically targeted at a predicted surface-exposed domain, which was later characterized as a nonpermissive site. (B) Comparison of 987P fimbria display systems illustrating bacteria containing two compatible and complementing plasmids to express wild-type fimbriae incorporating only wild-type FasA subunits (shown in black), hybrid fimbriae incorporating both wild-type FasA and mutated FasA subunits (shown in white), or mutated fimbriae incorporating only mutated FasA.
TABLE 3.
Deduced sequences and functions of mutated FasA proteins
| Sitea | Mutationb | Wild-type sequencec | Mutated sequenced | Fimbriatione |
|---|---|---|---|---|
| 29 | 2I, 1S | 224 AAC AAC ACC 232 | 224 AAC AGG GCC CAC 235 | ++ |
| 28 Asn Asn Thr 30 | 28 Asn Arg Ala His 31 | |||
| 97 | 2I | 428 ATT GCT GGT AAT AAT 442 | 428 ATT GGG CCC GCT 439 | ++ |
| 96 Ile Ala Gly Asn Asn 100 | 96 Ile Gly Pro Ala 99 | |||
| 2I, 2D | 428 ATT GGG CCC AAT 439 | ++ | ||
| 96 Ile Gly Pro Asn 99 | ||||
| 141 | 2I | 563 GTA CCT GTG 571 | 563 GTA GGG CCC CCT 574 | + |
| 140 Val Pro Val 142 | 140 Val Gly Pro Pro 143 | |||
| 192 | 2I, 3D, 14A | 713 GCA GGT ACA CCG TAA 727 | 713 GCA GGG CCC GTA …f | ++ |
| 191 Ala Gly Thr Pro - 194 | 191 Ala Gly Pro Val … | |||
| 1D | 713 GCA GGG CCC TAA 724g | ++ | ||
| 191 Ala Gly Pro - 193 | ||||
| 193 | 2I, 2D, 15A | 716 GGT ACA CCG TAA 727 | 716 GGT AGG GCC CCC …h | ++ |
| 192 Gly Thr Pro - 194 | 192 Gly Arg Ala Pro … | |||
| 2D, 4A | 716 GGT AGG GCC CTA AGC TAA 733g | − | ||
| 192 Gly Arg Ala Leu Ser - 96 |
Insertion site in the FasA protein sequence, corresponding to the first altered residue in the sequence.
Data are the numbers of amino acid residues which were inserted (I), deleted (D), or substituted (S) in FasA or which were added (A) at the carboxy terminus of FasA because of a frameshift just upstream of the stop codon as a result of fasA mutagenesis.
fasA and FasA sequences. Numbers refer to the indicated first and last nucleotides or amino acid residues of the original sequences.
Linker insertion sites with resulting mutations in fasA and deduced amino acid changes. Numbers refer to the indicated first and last nucleotides or amino acid residues of the mutated sequences. Linker DNA (ApaI restriction site) and new amino acids are in italics.
Exported and assembled fimbriae quantitated by seroagglutination with anti-987P antibodies. ++, strong reaction during the first 10 s, equivalent to the reaction with bacteria expressing the wild-type FasA protein; +, weak reaction after 10 s; −, no reaction for 1 min.
Frameshift resulting in the substitution of 16 residues (not all shown) for the 3 residues at the carboxy terminus of wild-type FasA.
Mutants obtained by site-directed mutagenesis to remove the long residue tail at the carboxy terminus of mutated FasA.
Frameshift resulting in the substitution of 17 residues (not all shown) for the 2 residues at the carboxy terminus of wild-type FasA.
Fimbriation after the introduction of foreign epitopes into FasA.
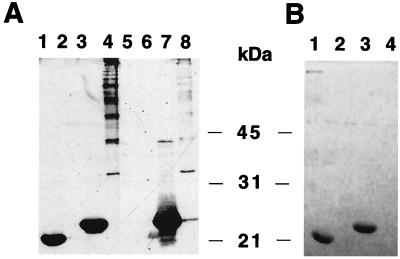
To determine whether the allelic FasA proteins with permissive linker insertion sites can be used as carriers of foreign antigenic determinants, two continuous epitopes, one of glycoprotein D [gD(272–279)] from HSV-1 and one of the spike protein [S(379–388)] from porcine TGEV, were introduced into the linker insertion sites which did not affect fimbriation (Table 3). These constructions included the linkers inserted at residue 29, the two linker inserts at residue 97, one designated 97 (2D) for the two deleted residues following the insert, and the linker insert at residue 192 with the stop codon in frame. The amino acid sequences of these antigenic determinants and the synthetic oligonucleotides used are listed in Table 2. Anticipating that insertion of foreign epitopes into FasA may have a deleterious effect on fimbriation, we determined whether the export and assembly of such subunits could be rescued in merodiploid bacteria also expressing the wild-type FasA protein. Therefore, in addition to the study of fimbrial expression mediated only by mutated FasA proteins, resulting in fimbriae designated “mutated fimbriae,” the potential assembly of “hybrid fimbriae,” consisting of mutated and wild-type FasA proteins, was evaluated. For this, plasmids encoding mutated subunits were complemented by plasmids expressing the FasB to FasH proteins or the FasA to FasH proteins to produce mutated or hybrid fimbriae, respectively (Fig. 1B). This approach was used to screen for the most consistent permissive site for foreign epitopes. Bacteria expressing hybrid fimbriae, including FasA subunits carrying either the HSV gD(272–279) or the TGEV S(379–388) epitope inserted after residue 5, or the TGEV epitope inserted at the carboxy terminus of FasA (position 192 of FasA [Table 3]), were agglutinated by the respective foreign epitope-specific antibodies, suggesting that these epitopes are displayed on hybrid fimbriae. Although no mutated fimbriae were detected by seroagglutination with anti-987P antibodies, small amounts of them carrying the gD(272–279) epitope of HSV inserted between residues 5 and 6 of mature FasA (position 29 of FasA [Table 3]) were isolated and detected by the more sensitive Western blot analysis (Fig. 2A, lane 3).
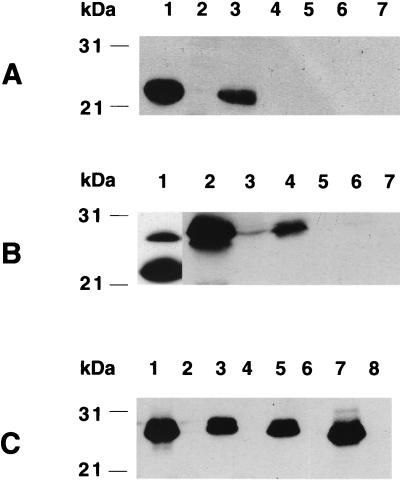
FIG. 2.
Western blot of fimbrial preparations from E. coli strains expressing HSV epitope gD(272–279) (A and B) or TGEV epitope S(379–388) (C) on mutated fimbriae (A) or on hybrid fimbriae (B and C). (A) Lanes 1 and 2, no epitope in FasA; lanes 3 and 4, gD epitope in position 29 of FasA; lanes 5 and 6, gD epitope in position 97 of FasA; lane 7, gD epitope in position 97(−2) of FasA; odd-numbered lanes, 100°C-treated fimbrial preparations. All lanes were probed with anti-987P fimbria antibody. (B) Lanes 1, 2, and 3, gD epitope in position 29 of FasA; lanes 4 and 5, gD epitope in position 97 of FasA; lanes 6 and 7, gD epitope in position 97(−2) of FasA; lanes 1, 2, 4, and 6, 100°C-treated fimbrial preparations. Lane 1 was probed with anti-987P fimbria antibody; both wild-type (bottom band) and mutated (top band) FasA are shown. Lanes 2 through 7 were probed with MAb DL6, and only mutated FasA is shown. (C) Lanes 1 and 2, TGEV epitope in position 29 of FasA; lanes 3 and 4, TGEV epitope in position 97 of FasA; lanes 5 and 6, TGEV epitope in position 97(−2) of FasA; lanes 7 and 8, TGEV epitope in position 192 of FasA; odd-numbered lanes, 100°C-treated fimbrial preparations. All lanes were probed with anti-TGEV epitope antibody; only mutated FasA is shown.
Incorporation of mutated and hybrid subunits into fimbriae.
To demonstrate the presence of both types of subunits in the fimbriae and to determine whether mutated subunits assembled like wild-type subunits, preparations of fimbrial proteins were submitted to Western blot analysis. Both subunits were detected in the fimbrial preparations, as shown with the strain expressing wild-type and mutated proteins carrying the gD(272–279) epitope in position 29 probed with anti-987P antibodies (Fig. 2B, lane 1). Densitometric comparisons of the two bands suggested that approximately 25% of the exported and assembled subunits were mutated subunits. Whereas lower levels of mutated subunits were exported when the gD(272–279) epitope was inserted into the other sites, 34 to 55% of the mutated subunits were exported and assembled when the TGEV S(379–388) epitope was inserted in one of the four studied sites (data not shown). The identities of the mutated FasA subunits carrying either the HSV epitopes (Fig. 2B, lanes 2, 4, and 7) or the TGEV epitopes (Fig. 2C, lanes 1, 3, 5, and 7) were confirmed by the corresponding epitope-specific antibodies. That these subunits were assembled as polymeric structures was confirmed by taking advantage of our earlier observation (33) that SDS-treated 987P fimbriae do not dissociate into their subunits unless they are simultaneously heated to 100°C (Fig. 2). That some of the hybrid fimbriae were identified in Western blots (Fig. 2B, lane 4, and Fig. 2C, lanes 3 and 5) but not by seroagglutination may result from differences in the sensitivities of the techniques. Alternatively, foreign epitopes in these sites may be accessible only after the denaturation steps used for Western blot analysis. Finally, that both mutated and wild-type FasA proteins assembled together in fimbrial filaments was suggested by the findings that most mutated subunits were not exported or assembled in the absence of wild-type FasA proteins. This concept was further supported by the different immunogold labeling patterns characterizing assembled hybrid and mutated fimbriae, as described below (see Fig. 6).
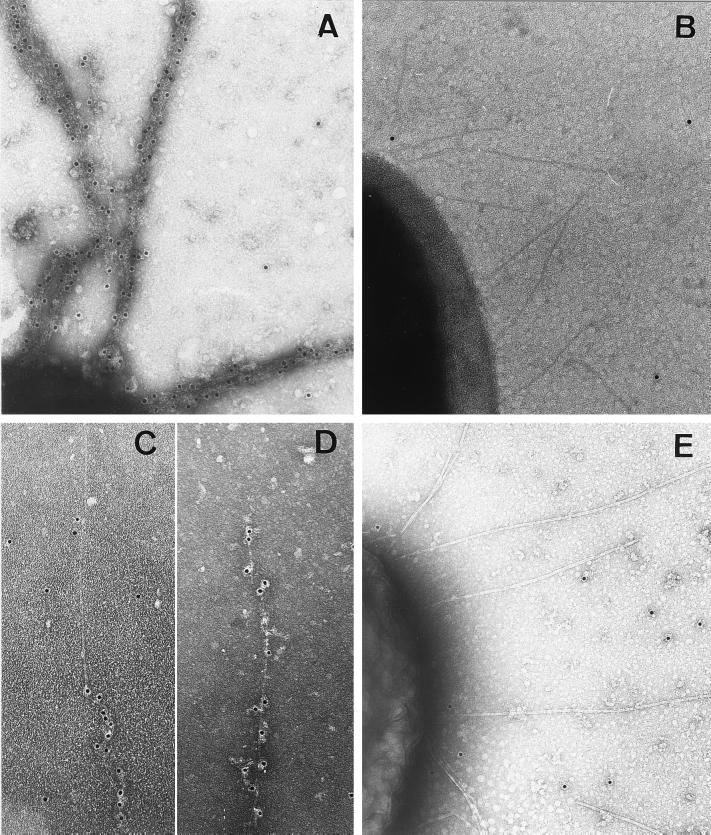
FIG. 6.
Electron micrographs of immunogold-labeled negatively stained mutated or hybrid fimbriae and fimbriated E. coli. (A) Strain SE5000(pRS228, pDMS161) displaying the HSV gD(11–19) epitope, bundled and labeled by MAb 1D3. (B) Negative labeling of strain SE5000(pRS228, pDMS161), with MAb DL6 as a control. (C and D) Hybrid fimbriae of strain SE5000(pRS227, pDMS158) displaying the HSV gD(272–279) epitope, labeled with MAb DL6. Individual fimbrial threads remain separate, with long continuous segments without gold particles. (E) Negative labeling of strain SE5000(pRS227, pDMS158), with MAb 1D3 as a control. Gold particles, 5 nm; magnifications, ×72,000.
Surface exposure of foreign immunodeterminants on 987P.
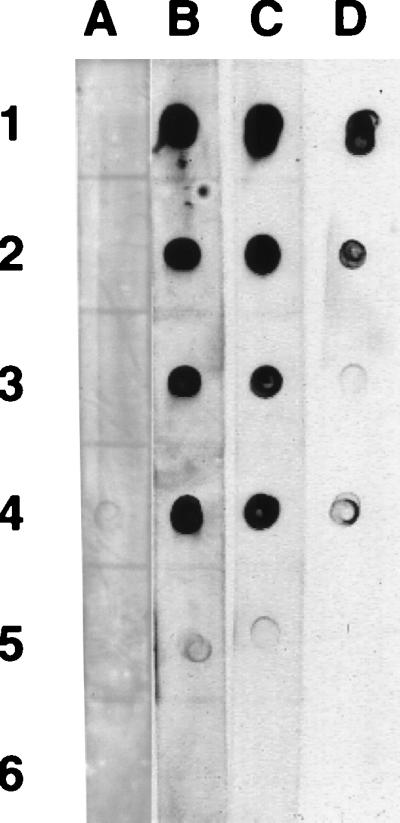
All bacteria with hybrid fimbriae that were detected by seroagglutination were also positive by native dot blot immunoassays with intact cells, confirming that the inserted foreign epitopes were accessible from the bacterial surface (Fig. 3). Epitope recognition in a previously undetected hybrid fimbria probably results from differences between the accessibility of antigen in solution and antigen fixed on a solid surface (Fig. 3A, fourth row). Most relevant for the identification of a permissive site for foreign epitopes, surface exposure of both the HSV and TGEV epitopes was detectable only with insertions at position 29, namely between residues 5 and 6 of mature FasA, suggesting that the amino terminus of FasA is more tolerant of modifications than are other domains.
FIG. 3.
Surface exposure of HSV epitope gD(272–279) (A) and TGEV epitope S(379–388) (B) on bacteria expressing hybrid fimbriae, by native dot blot immunoassay. (A) Row 1, HSV epitope expressed in the cell but not on fimbriae (negative control); row 2, no epitope in FasA; row 3, epitope in position 29 of FasA; row 4, epitope in position 97 of FasA; row 5, epitope in position 97(−2) of FasA; row 6, epitope in position 192 of FasA. Column a, 2.5 × 107 bacteria/dot; column b, 6.3 × 106 bacteria/dot. (B) Row 1, no TGEV epitope in FasA; row 2, TGEV epitope in position 26 of FasA; row 3, epitope in position 29 of FasA; row 4, epitope in position 97 of FasA; row 5, epitope in position 97(−2) of FasA; row 6, epitope in position 192 of FasA. Column a, 6.3 × 106 bacteria/dot; column b, 1.6 × 106 bacteria/dot.
Export and assembly of enteroadherent fimbriae in the absence of wild-type FasA subunits.
As the amino terminus of FasA seemed to be the most permissive to insertions for the expression of hybrid fimbriae, we determined whether shifting the insertion nearer to the signal sequence cleavage site would promote the production of fimbriae containing only mutated FasA subunits. Thus, a new ApaI site was introduced after amino acid residue 2 of mature FasA (position 26 [Table 1, pRS225]), a position which should not hinder FasA processing at the inner membrane. Mutated fimbriae were detected by seroagglutination after insertion of either of the two previously studied foreign epitopes into this new site, though bacteria expressed more mutated fimbriae with the TGEV S(379–388) epitope than with the HSV gD(272–279) epitope. Interestingly, introduction of the new epitope HSV gD(11–19) after residue 2 of mature FasA, but not after residue 5, also resulted in expression of mutated fimbriae, as detected by seroagglutination with the appropriate antiepitope or anti-987P antibodies. This epitope (Fig. 3B, row 2), like the two other epitopes at the same site (not shown), was expressed at high levels on hybrid fimbriae.
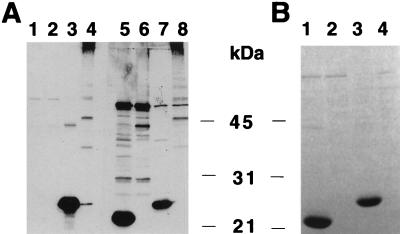
Since fimbrial expression is regulated by the fine-tuning effects of transcriptional and posttranscriptional mechanisms, changes in the relative numbers of fas gene products obtained by using two plasmids may not be optimal for fimbrial expression. To improve fimbrial expression, gene clusters encoding all of the Fas proteins on one plasmid were constructed and used for the bacterial production of mutated fimbriae carrying either the TGEV S(379–388) epitope or the HSV gD(11–19) epitope inserted after the second residue of mature FasA. Bacteria containing these plasmids strongly seroagglutinated with the anti-987P antibodies and with the corresponding foreign epitope antibodies. The presence of these epitopes in FasA was further demonstrated by SDS-PAGE and Western blot analysis (Fig. 4 and 5). Fimbrial depolymerization to subunits required 100°C treatments in 1% SDS, suggesting that the structure of assembled mutated fimbriae mimicked that of assembled wild-type fimbriae. Effective exposure of the epitope on the fimbriae was visualized by immune electron microscopy with foreign epitope-specific antibodies and protein-A labeled gold particles. Whereas mutated fimbriae were typically bundled by the antibodies and uniformly labeled (Fig. 6A), the antibody binding pattern on hybrid fimbriae was irregular and gold particles were less densely packed along fimbrial filaments (Fig. 6D and E). The labeling of some hybrid fimbriae appeared to be segmented, suggesting that the unlabeled and labeled regions may represent alternating domains consisting mainly of assembled wild-type or mutated subunits, respectively (Fig. 6C and D). Finally, the presence of the adhesive FasG subunits on the mutated fimbriae carrying either the TGEV S(379–388) epitope or the HSV gD(11–19) epitope was confirmed by seroagglutination with anti-FasG antibodies. Wild-type and both mutated fimbriae conferred similar levels of bacterial aggregation by enterocyte brush borders from piglets, suggesting that the FasG molecules present in the mutated fimbriae mediate bacterial enteroadhesion, as when they are assembled on wild-type 987P.
FIG. 4.
Western blot (A) and SDS-PAGE (B) of fimbrial preparations from E. coli strains expressing HSV gD(11–19) on mutated fimbriae. Lanes 1, 2, 5, and 6, no epitope in FasA; lanes 3, 4, 7, and 8, epitopes in position 26 of FasA; odd-numbered lanes, 100°C-treated fimbrial preparations. Lanes 1 through 4 were probed with anti-987P fimbria antibody; lanes 5 through 8 were probed with MAb 1D3.
FIG. 5.
Western blot (A) and SDS-PAGE (B) of fimbrial preparations from E. coli strains expressing TGEV epitope S(379–388) on mutated fimbriae. Lanes 1, 2, 5, and 6, no epitope in FasA; lanes 3, 4, 7, and 8, epitopes in position 26 of FasA; odd-numbered lanes, 100°C-treated fimbrial preparations. Lanes 1 through 4 were probed with anti-TGEV epitope S(379–388) antibody; lanes 5 through 8 were probed with anti-987P fimbria antibody.
Immunogenicity of foreign immunodeterminants in 987P.
Immunization studies were undertaken to determine whether the foreign epitopes exposed on 987P would be able to induce an epitope-specific immune response in hosts. Fimbrial antigens were prepared from the optimized one-plasmid system described above. To evaluate more stringently the immunogenic properties of mutated fimbriae, immunizations were undertaken in rabbits, which are notorious for displaying heterogeneous immune responses. All of the rabbits immunized with mutated fimbriae carrying the HSV gD(11–19) epitope or the TGEV S(379–388) epitope developed specific anti-foreign epitope antibodies, as well as anti-987P antibodies, as screened by seroagglutination. The mutated fimbriae carrying the HSV epitope elicited antibodies recognizing the gD(11–19) peptide in the three immunized rabbits. These antibodies also reacted with the gD protein spotted onto nitrocellulose, with titers reaching ≥10−3 at 5 weeks (Fig. 7). Similarly, all three rabbits immunized with mutated fimbriae carrying the TGEV S(379–388) epitope developed specific antibodies reacting with the TGEV S(379–388) peptide spotted onto nitrocellulose, with titers reaching ≥10−3 at 8 or 11 weeks (Fig. 8B). Similar titers were detected with the two rabbits inoculated with the TGEV S(379–388) peptide linked to KLH (Fig. 8A), although only one of the antibodies was seroagglutinating fimbriated E. coli displaying the TGEV S(379–388) epitope. Whereas sera from rabbits immunized with wild-type 987P fimbriae did not react with the viral antigens, the sera from rabbits immunized with mutated fimbriae contained both specific antiviral antigens and anti-987P antibodies (data not shown). Taken together, the data indicated that the 987P fimbrial carrier systems can induce specific anti-foreign epitope antibodies which are capable of recognizing foreign epitopes in the context of the full-length foreign protein.
FIG. 7.
Dot blot immunoassays with antibody from one representative rabbit immunized with mutated 987P fimbriae carrying HSV epitope gD(11–19) in position 26 of FasA. Column A, preimmunization control (diluted 1/20); columns B, C, and D, postimmunization (diluted 1/20, 1/100, and 1/1,000, respectively). Rows 1, 2, and 3, purified gD glycoprotein (150, 15, and 1.5 ng, respectively); rows 4, 5, and 6, gD(10–20) peptide (750, 75, and 7.5 ng, respectively).
FIG. 8.
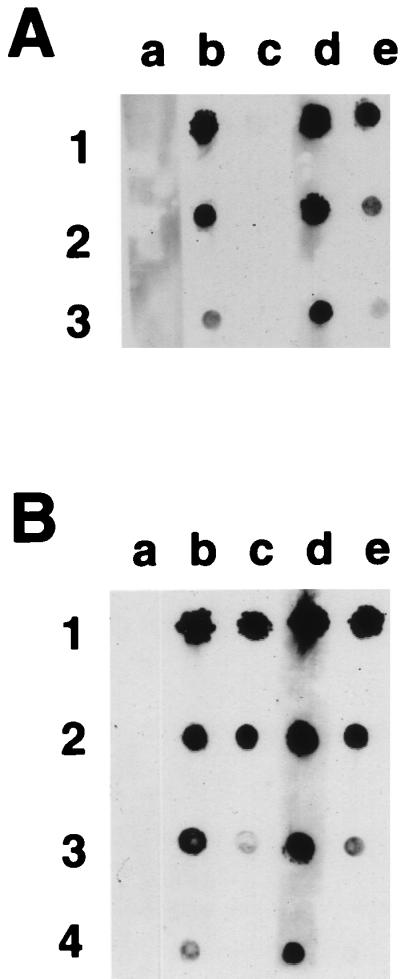
Dot blot immunoassays with peptide TGEV S(379–388) as antigen. (A) Antibody from one representative rabbit immunized with peptide TGEV S(379–388) linked to KLH. Column a, preimmunization controls diluted 1/100; columns b and c, 5 weeks postimmunization with serum diluted 1/100 and 1/1,000, respectively; columns d and e, 8 weeks postimmunization with serum diluted 1/100 and 1/1,000, respectively. Rows 1, 2, and 3 have 750, 75, and 7.5 ng of spotted peptide TGEV S(379–388), respectively. (B) Antibody from one representative rabbit immunized with mutated 987P fimbriae carrying TGEV epitope S(379–388) in position 26 of FasA. Column a, preimmunization controls diluted 1/100; columns b and c, 8 weeks postimmunization with serum diluted 1/100 and 1/1,000, respectively; columns d and e, 11 weeks postimmunization with serum diluted 1/100 and 1/1,000, respectively. Rows 1, 2, 3, and 4 have 750, 75, 7.5, and 0.75 ng of spotted peptide TGEV S(379–388), respectively.
DISCUSSION
In this study, stepwise approaches were taken to optimize the construction of a fimbrial expression system capable of integrating and displaying immunogenic epitopes on the bacterial surface. The first step was designed to randomly search for sites in the major subunit FasA of 987P accepting modifications that do not interfere with fimbriation. The use of these sites was further tested by studying hybrid and mutated fimbriae for surface expression of foreign epitopes. A comparative evaluation of the data suggested that, similarly to the pVIII protein of filamentous bacteriophages, the amino terminus of FasA may be an optimal location for epitope presentation. Three viral peptides known to be recognized by antivirus antibodies were introduced into the amino terminus of mature FasA, two residues distal from the signal sequence cleavage site and shown to be expressed on fimbria-like structures on the bacterial surface. These fimbriae, carrying an epitope either of the TGEV spike protein or of the HSV gD glycoprotein, were shown to be immunogenic and to induce antiepitope or antiviral glycoprotein-specific antibodies in all rabbits tested.
Although several types of fimbriae have been shown to be capable of displaying foreign epitopes (44), only a few of them have been studied for their immunogenicity. Moreover, some of the reported limitations to displaying foreign epitopes on fimbriae may have resulted from the difficulty of identifying optimal insertion sites in the subunits. Essentially two methods were applied to determine where insertions or replacements in the primary structure of the subunits do not interfere with fimbrial biogenesis. First, protein sequences of subunits of different antigenic variants were aligned to identify hypervariable and conserved domains. Hypervariable domains were proposed to be more expendable for fimbrial protein export and assembly than conserved ones and were targeted for insertion or replacement with antigenic viral epitopes (3, 5, 61, 62). Predictive algorithms were used for the second method to pinpoint potential surface-exposed domains of fimbrial proteins (25, 46, 59). Additions of foreign epitopes in such domains were proposed to be less deleterious to fimbrial protein export and assembly. Although both approaches gave results, several studied foreign epitopes could not be used without affecting fimbriation and/or losing antigenicity. Thus, instead of applying these methods, we preferred to randomly search for permissive sites in the 987P subunit FasA by linker insertion mutagenesis. This technique, previously employed to study the topography of the outer membrane protein FasD (53), identified five fasA mutants which accepted a 6-mer ApaI linker without affecting fimbriation. A similar approach has been used to identify permissive sites in the CS31A fimbrial subunit (13). To evaluate which of these sites would be the most permissive to the addition and display of foreign epitopes on the bacterial surface, we analyzed fimbriae expressed by E. coli containing only mutated fasA or both mutated and wild-type fasA. By using the second approach, we were able to show that the addition of wild-type subunits exerted a dominant phenotype resulting in fimbrial expression and, in some cases, suppressing deleterious effects of inserted epitopes on the export and assembly of mutated subunits. Suppression was most consistent with foreign epitopes in the permissive linker insertion site most proximal to the amino terminus of FasA, suggesting that this region was most receptive to modification.
Both the creation of hybrid fimbriae, as suggested by electron microscopic examination, and the identification of the amino terminus of processed FasA as an optimal location for foreign epitope exposure on 987P were reminiscent of the display of foreign peptides on coat protein pVIII of filamentous bacteriophages. Although biologically largely unexplained, hybrid phages consisting of mutated and wild-type pVIII proteins can be produced in specific cases. Both FasA and pVIII are the major subunits of helically arranged filamentous polymeric proteins. Similar to structural constrains observed for the export and assembly of major fimbrial subunits, increasing the length of the inserts in pVIII affects viral particle assembly, stability, and infectivity. Whereas pVIII usually accepts inserts of up to six amino acid residues (23), length restrictions for the 987P fimbriae and those previously found for other fimbriae seem to be less drastic (44, 59). For example, we recently found that 987P hybrid fimbriae can express the HSV epitope gD(7–23) on the bacterial surface (data not shown). A major advantage of fimbrial antigens being their polymeric nature, the use of hybrid fimbriae as immunogen requires that a significant proportion of the subunits are those carrying the foreign epitope. Therefore, mutated fimbriae including only carrier subunits are preferable over hybrid fimbriae for immunization. However, hybrid fimbriae have been demonstrated to be very useful for pinpointing potential permissive locations in fimbrial subunits. Moreover, hybrid fimbriae may be best suited for the development of ligand display systems, where affinity for receptors could be modulated by using insertion sites with reduced numbers of mutated subunits in each fimbria, permitting selection of ligands of higher affinities (35). That 987P can be modified to specifically recognize a new receptor was recently confirmed in preliminary studies with a displayed ligand which mediates 987P binding to complement component C3 (data not shown).
Finally and most importantly, fimbrial preparations from E. coli expressing mutated fimbriae carrying the HSV gD(11–19) epitope or the TGEV S(379–388) epitope elicited high levels of foreign epitope antibodies in all rabbits immunized parenterally, with titers reaching ≥10−3. Antibodies against the HSV epitope were shown to recognize the epitope in the context of the whole gD protein. Recent studies with the CS31 fimbriae of certain bovine E. coli strains also identified the amino terminus of the major subunit as a useful target for foreign epitope insertion and display, where displayed TGEV S epitopes induced memory antibodies in outbred mice after boosting with TGEV (12, 38). Also, several studies have shown that peptides corresponding to the amino terminus of fimbrial subunits are usually good immunogens (1, 56), suggesting that this region is predisposed for immune recognition. In addition to the attractive immunogenic assets of fimbriae in general, fimbriae of enteric pathogens like ETEC have the advantage of inducing enteral immunity. Oral administration of 987P fimbriae is sufficient to induce mucosal antibodies (9), and killed or live vaccines based on nontoxigenic E. coli or attenuated Salmonella strains expressing cloned fimbriae have been used to immunize by the oral route and to successfully induce antiadhesive antibodies (2, 26, 41, 51, 60). The binding property, and not the toxic activity, of mucosal immunogens like the cholera and E. coli heat-labile enterotoxins was proposed to be sufficient for the adjuvant effect of these molecules (24, 63), suggesting that other enteroadherent immunogens like fimbriae could harbor similar properties. Unlike most ETEC fimbriae whose enteroadhesion is mediated by their major subunits (4, 15, 22, 30–32, 37), enteroadhesion of 987P is essentially mediated by a minor subunit, the FasG protein (33). Therefore, the advantage of the 987P carrier over other fimbrial systems relies on the ability to genetically engineer the major subunit FasA as the carrier molecule without affecting the enteroadhesive property of the fimbriae. The developed 987P carrier offers a unique opportunity for future testing of the potential adjuvant effects of a fimbrial carrier displaying epitopes of enteric viruses for mucosal immunization and protection.
ACKNOWLEDGMENTS
We thank R. J. Eisenberg and G. H. Cohen for herpes simplex virus antibodies, peptides, and glycoprotein D; W. M. Armstead for piglets; and W. C. Lawrence for critical reading of the manuscript.
Parts of this work were supported by a University of Pennsylvania Research Foundation grant, by a National Pork Producers Council (NPPC) research grant, and by Commonwealth of Pennsylvania Department of Agriculture grant ME 445129.
REFERENCES
- 1.Abraham S N, Beachey E H. Assembly of a chemically synthesized peptide of Escherichia coli type 1 fimbriae into fimbria-like antigenic structures. J Bacteriol. 1987;169:2460–2465. doi: 10.1128/jb.169.6.2460-2465.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attridge S, Hackett J, Morona R, Whyte P. Towards a live oral vaccine against enterotoxigenic Escherichia coli of swine. Vaccine. 1988;6:387–389. doi: 10.1016/0264-410x(88)90134-x. [DOI] [PubMed] [Google Scholar]
- 3.Bakker D, van Zijderveld F G, van der Veen S, Oudega B, de Graaf F K. K88 fimbriae as carriers of heterologous antigenic determinants. Microb Pathog. 1990;8:343–352. doi: 10.1016/0882-4010(90)90093-6. [DOI] [PubMed] [Google Scholar]
- 4.Bakker D, Willemsen P T J, Simons L H, van Zijderveld F G, de Graaf F K. Characterization of the antigenic and adhesive properties of FaeG, the major subunit of K88 fimbriae. Mol Microbiol. 1992;6:247–255. doi: 10.1111/j.1365-2958.1992.tb02006.x. [DOI] [PubMed] [Google Scholar]
- 5.Bousquet F, Martin C, Girardeau J P, Méchin M C, der Vartanian M, Laude H, Contrepois M. CS31A capsule-like antigen as an exposure vector for heterologous antigenic determinants. Infect Immun. 1994;62:2553–2561. doi: 10.1128/iai.62.6.2553-2561.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao J, Khan A S, Bayer M E, Schifferli D M. Ordered translocation of 987P fimbrial subunits through the outer membrane of Escherichia coli. J Bacteriol. 1995;177:3704–3713. doi: 10.1128/jb.177.13.3704-3713.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, B.-K., and D. M. Schifferli. The 987P fimbrial adhesin FasG harbors separate binding domains for its epithelial glycolipid and glycoprotein receptors. Submitted for publication.
- 8.Cohen J P, Dietzschold B, Ponce de Leon M, Long D, Golub E, Varrichio A, Pereira L, Eisenberg R J. Localization and synthesis of an antigenic determinant of herpes simplex virus glycoprotein D that stimulates the production of neutralizing antibody. J Virol. 1984;49:102–108. doi: 10.1128/jvi.49.1.102-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Aizpurua H J, Russell-Jones G J. Oral vaccination, identification of classes of proteins that provoke an immune response upon oral feeding. J Exp Med. 1988;167:440–451. doi: 10.1084/jem.167.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delmas B, Gelfi J, Laude H. Antigenic structure of transmissible gastroenteritis virus. II. Domains of the peplomer glycoprotein. J Gen Virol. 1986;67:1405–1418. doi: 10.1099/0022-1317-67-7-1405. [DOI] [PubMed] [Google Scholar]
- 11.Delmas B, Rasschaert D, Godet M, Gelfi J, Laude H. Four major antigenic sites of the coronavirus transmissible gastroenteritis virus are located on the amino-terminal half of spike glycoprotein S. J Gen Virol. 1990;71:1313–1323. doi: 10.1099/0022-1317-71-6-1313. [DOI] [PubMed] [Google Scholar]
- 12.Der Vartanian M, Girardeau J-P, Martin C, Rousset E, Chavarot M, Laude H, Contrepois M. An Escherichia coli CS31A fimbrillum chimera capable of inducing memory antibodies in outbred mice following booster immunization with the entero-pathogenic coronavirus gastroenteritis virus. Vaccine. 1997;15:111–120. doi: 10.1016/S0264-410X(96)00172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Der Vartanian M, Mechin M C, Jaffeux B, Bertin Y, Felix I, Gaillard-Martinie B. Permissible peptide insertions surrounding the signal peptide-mature protein junction of the ClpG prepilin: CS31A fimbriae of Escherichia coli as carriers of foreign sequences. Gene. 1994;148:23–32. doi: 10.1016/0378-1119(94)90229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietzschold B, Eisenberg R J, Ponce de Leon M, Golub E, Hudecz F, Varrichio A, Cohen G H. Fine structure analysis of type-specific and type-common antigenic sites of herpes simplex virus glycoprotein D. J Virol. 1984;52:431–435. doi: 10.1128/jvi.52.2.431-435.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Martino P, Girardeau J-P, Der Vartanian M, Joly B, Darfeuille-Michaud A. The central variable V2 region of the CS31A major subunit is involved in the receptor-binding domain. Infect Immun. 1997;65:609–616. doi: 10.1128/iai.65.2.609-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards R A, Cao J, Schifferli D M. Identification of major and minor chaperone proteins involved in the export of 987P fimbriae. J Bacteriol. 1996;178:3426–3433. doi: 10.1128/jb.178.12.3426-3433.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards R A, Keller L H, Schifferli D M. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207:149–157. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 19.Edwards R A, Schifferli D M. Differential regulation of fasA and fasH expression of Escherichia coli 987P fimbriae by environmental cues. Mol Microbiol. 1997;25:797–809. doi: 10.1046/j.1365-2958.1997.5161875.x. [DOI] [PubMed] [Google Scholar]
- 20.Eisenberg R J, Cerini C P, Heilman C J, Joseph A D, Dietzschold B, Golub E, Long D, Ponce de Leon M, Cohen J P. Synthetic glycoprotein D-related peptides protect mice against herpes simplex virus challenge. J Virol. 1985;56:1014–1017. doi: 10.1128/jvi.56.3.1014-1017.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emini E A, Hughes J V, Perlow D S, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froehlich B J, Karakashian A, Sakellaris H, Scott J R. Genes for CS2 pili of enterotoxigenic Escherichia coli and their interchangeability with those for CS1 pili. Infect Immun. 1995;63:4849–4856. doi: 10.1128/iai.63.12.4849-4856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenwood J, Willis A E, Perham R N. Multiple display of foreign peptides on a filamentous bacteriophage. Peptides from Plasmodium falciparum circumsporozoite protein as antigens. J Mol Biol. 1991;220:821–827. doi: 10.1016/0022-2836(91)90354-9. [DOI] [PubMed] [Google Scholar]
- 24.Guidry J J, Cárdenas L, Cheng E, Clements J D. Role of receptor binding in toxicity, immunogenicity, and adjuvanticity of Escherichia coli heat-labile enterotoxin. Infect Immun. 1997;65:4943–4950. doi: 10.1128/iai.65.12.4943-4950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedegaard L, Klemm P. Type 1 fimbriae of Escherichia coli as carriers of heterologous antigenic sequences. Gene. 1989;85:115–124. doi: 10.1016/0378-1119(89)90471-x. [DOI] [PubMed] [Google Scholar]
- 26.Hone D, Attridge S, van den Bosch L, Hackett J. A chromosomal integration system for stabilization of heterologous gene in Salmonella based vaccine strains. Microb Pathog. 1988;5:407–418. doi: 10.1016/0882-4010(88)90002-2. [DOI] [PubMed] [Google Scholar]
- 27.Isaacson R E, Dean E A, Morgan R L, Moon H W. Immunization of suckling pigs against enterotoxigenic Escherichia coli-induced diarrheal disease by vaccinating dams with purified K99 and 987P pili: antibody production in response to vaccination. Infect Immun. 1980;29:824–826. doi: 10.1128/iai.29.2.824-826.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isaacson R E, Fusco P C, Brinton C C, Moon H W. In vitro adhesion of Escherichia coli to porcine small intestinal epithelial cells: pili as adhesive factors. Infect Immun. 1978;21:392–397. doi: 10.1128/iai.21.2.392-397.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isola V J, Eisenberg R J, Siebert G R, Heilman C J, Wilcox W C, Cohen G H. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J Virol. 1989;63:2325–2334. doi: 10.1128/jvi.63.5.2325-2334.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs A A C, Roosendaal B, van Breemen J F L, de Graaf F K. Role of phenylalanine 150 in the receptor-binding domain of the K88 fibrillar subunit. Infect Immun. 1987;169:4907–4911. doi: 10.1128/jb.169.11.4907-4911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs A A C, Simons B H, de Graaf F K. The role of lysine-132 and arginine-136 in the receptor-binding domain of the K99 fibrillar subunit. EMBO J. 1987;6:1805–1808. doi: 10.1002/j.1460-2075.1987.tb02434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobs A A C, Venema J, Leeven J, van Pelt-Heerschap H, de Graaf F K. Inhibition of adhesive activity of K88 fibrillae by peptides derived from the K88 adhesion. J Bacteriol. 1987;169:735–741. doi: 10.1128/jb.169.2.735-741.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan A S, Schifferli D M. A minor 987P protein different from the structural fimbrial subunit is the adhesin. Infect Immun. 1994;62:4233–4243. doi: 10.1128/iai.62.10.4233-4243.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine M M, Girón J O, Noriega F R. Fimbrial vaccines. In: Klemm P, editor. Fimbriae. Adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 255–270. [Google Scholar]
- 35.Lowman H B. Bacteriophage display and discovery of peptide leads for drug development. Annu Rev Biomol Struct. 1997;26:401–424. doi: 10.1146/annurev.biophys.26.1.401. [DOI] [PubMed] [Google Scholar]
- 36.Markwell M A, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 37.Marron M B, Smyth C J. Molecular analysis of the cso operon of enterotoxigenic Escherichia coli reveals that CsoA is the adhesin of CS1 fimbriae and that the accessory genes are interchangeable with those of the cfa operon. Microbiology. 1995;141:2849–2859. doi: 10.1099/13500872-141-11-2849. [DOI] [PubMed] [Google Scholar]
- 38.Méchin M-C, Der Vartanian M, Martin C. The major subunit ClpG of Escherichia coli CS31A fibrillae as an expression vector for different combinations of two TGEV coronavirus epitopes. Gene. 1996;179:211–218. doi: 10.1016/S0378-1119(96)00348-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moon H W. Colonization factor antigens of enterotoxigenic Escherichia coli in animals. Curr Top Microbiol Immunol. 1990;151:147–165. doi: 10.1007/978-3-642-74703-8_8. [DOI] [PubMed] [Google Scholar]
- 40.Moon H W, Bunn T O. Vaccines for preventing enterotoxigenic Escherichia coli infections in farm animals. Vaccine. 1993;11:200–213. doi: 10.1016/0264-410X(93)90020-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moon H W, Rogers D G, Rose R. Effects of an orally administered live Escherichia coli pilus vaccine on duration of lacteal immunity to enterotoxigenic Escherichia coli in swine. Am J Vet Res. 1988;49:2068–2071. [PubMed] [Google Scholar]
- 42.Morgan R L, Isaacson R E, Moon H W, Brinton C C, To C-C. Immunization of suckling pigs against enterotoxigenic Escherichia coli-induced diarrheal disease by vaccinating dams with purified 987 or K99 pili: protection correlates with pilus homology of vaccine and challenge. Infect Immun. 1978;22:771–777. doi: 10.1128/iai.22.3.771-777.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagy B, Moon H W, Isaacson R E, To C-C, Brinton C C. Immunization of suckling pigs against enteric enterotoxigenic Escherichia coli infection by vaccinating dams with purified pili. Infect Immun. 1978;21:269–274. doi: 10.1128/iai.21.1.269-274.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pallesen L, Klemm P. Chimeric fimbrial vaccines. In: Klemm P, editor. Fimbriae. Adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 271–276. [Google Scholar]
- 45.Pedersen P A, Andersen L N. Deletion and duplication of specific sequences in the K88ab fimbrial subunit protein from porcine enterotoxigenic Escherichia coli. Mol Gen Genet. 1991;229:285–291. doi: 10.1007/BF00272168. [DOI] [PubMed] [Google Scholar]
- 46.Pederson P A. Structure-function analysis of the K88ab fimbrial subunit protein from porcine enterotoxigenic Escherichia coli. Mol Microbiol. 1991;5:1073–1080. doi: 10.1111/j.1365-2958.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 47.Posthumus W P, Lenstra J A, Schaaper W M, van Nieuwstadt A P, Enjuanes L, Meloen R H. Analysis and simulation of a neutralizing epitope of transmissible gastroenteritis virus. J Virol. 1990;64:3304–3309. doi: 10.1128/jvi.64.7.3304-3309.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice P M, Elliston K, Gribskow M. DNA. In: Gribskow M, Devereux J, editors. Sequence analysis primer. W. H. New York, N.Y: Freeman and Co.; 1992. pp. 1–59. [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 50.Satterwhite T K, Evans D G, DuPont H L, Evans D J., Jr Role of Escherichia coli colonization factor antigen in acute diarrhoea. Lancet. 1978;ii:181–184. doi: 10.1016/s0140-6736(78)91921-9. [DOI] [PubMed] [Google Scholar]
- 51.Savarino S J, Brown F M, Hall E, Bassily S, Youssef F, Wierzba T, Peruski L, El-Masry N A, Safwat M, Rao M, Jertborn M, Svennerholm A M, Lee Y J, Clemens J D. Safety and immunogenicity of an oral, killed enterotoxigenic Escherichia coli-cholera toxin B subunit vaccine in Egyptian adults. J Infect Dis. 1998;177:796–799. doi: 10.1086/517812. [DOI] [PubMed] [Google Scholar]
- 52.Schifferli D M, Abraham S N, Beachey E H. Use of monoclonal antibodies to probe subunit- and polymer-specific epitopes of 987P fimbriae of Escherichia coli. Infect Immun. 1987;55:923–930. doi: 10.1128/iai.55.4.923-930.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schifferli D M, Alrutz M. Permissive linker insertion sites in the outer membrane protein of 987P fimbriae of Escherichia coli. J Bacteriol. 1994;176:1099–1110. doi: 10.1128/jb.176.4.1099-1110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schifferli D M, Beachey E H, Taylor R K. 987P fimbrial gene identification and protein characterization by T7 RNA polymerase induced transcription and TnphoA mutagenesis. Mol Microbiol. 1991;5:61–70. doi: 10.1111/j.1365-2958.1991.tb01826.x. [DOI] [PubMed] [Google Scholar]
- 55.Schifferli D M, Beachey E H, Taylor R K. Genetic analysis of 987P adhesion and fimbriation of Escherichia coli: the fas genes link both phenotypes. J Bacteriol. 1991;173:1230–1240. doi: 10.1128/jb.173.3.1230-1240.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt M A, O’Hanley P, Lark D, Schoolnik G K. Synthetic peptides corresponding to protective epitopes of Escherichia coli digalactoside-binding pilin prevent infection in a murine pyelonephritis model. Proc Natl Acad Sci USA. 1988;85:1247–1251. doi: 10.1073/pnas.85.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusion. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 58.Smith H W, Linggood M L. Observations on the pathogenic properties of the K88, Hly and Ent plasmids of Escherichia coli with particular reference to porcine diarrhoea. J Med Microbiol. 1971;4:467–485. doi: 10.1099/00222615-4-4-467. [DOI] [PubMed] [Google Scholar]
- 59.Stentebjerg-Olesen B, Pallesen L, Jensen L B, Christiansen G, Klemm P. Authentic display of a cholera toxin epitope by chimeric type 1 fimbria: effects of insert position and host background. Microbiology. 1997;143:2027–2038. doi: 10.1099/00221287-143-6-2027. [DOI] [PubMed] [Google Scholar]
- 60.Stevenson G, Manning P A. Galactose epimeraseless (galE) mutant G30 of Salmonella typhimurium is a good potential live oral vaccine carrier for fimbrial antigen. FEMS Microbiol Lett. 1985;28:317–321. [Google Scholar]
- 61.Thiry G, Clippe A, Scarcez T, Petre J. Cloning of DNA sequences encoding foreign peptides and their expression in the K88 pili. Appl Environ Microbiol. 1989;55:984–993. doi: 10.1128/aem.55.4.984-993.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Die I, van Oosterhout J, van Megen I, Bergmans H, Hoekstra W, Enger-Valk B, Barteling S, Mooi F. Expression of foreign epitopes in P-fimbriae of Escherichia coli. Mol Gen Genet. 1990;222:297–303. doi: 10.1007/BF00633832. [DOI] [PubMed] [Google Scholar]
- 63.Yamamoto S, Takeda Y, Yamamoto M, Kurazono H, Imaoka K, Yamamoto M, Fujihashi K, Noda M, Kiyono H, McGhee J R. Mutants in the ADP-ribosyltransferase cleft of cholera toxin lack diarrheagenicity but retain adjuvanticity. J Exp Med. 1997;185:1203–1210. doi: 10.1084/jem.185.7.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]