Abstract
How the germ line achieves a clean transition from maternal to zygotic gene expression control is a fundamental problem in sexually reproducing organisms. Whereas several mechanisms terminate the maternal program in the soma, this combined molecular reset and handover are poorly understood for primordial germ cells (PGCs). Here, we show that GRIF-1, a TRIM32-related and presumed E3 ubiquitin ligase in Caenorhabditis elegans, eliminates the maternal cytoplasmic poly(A) polymerase (cytoPAP) complex by targeting the germline-specific intrinsically disordered region of its enzymatic subunit, GLD-2, for proteasome-mediated degradation. Interference with cytoPAP turnover in PGCs causes frequent transgenerational sterility and, eventually, germline mortality. Hence, positively acting maternal RNA regulators are cleared via the proteasome system to avoid likely interference between maternal and zygotic gene expression programs to maintain transgenerational fertility and acquire germline immortality. This strategy is likely used in all animals that preform their immortal germ line via maternally inherited germplasm determinants.
A maternal feed forward loop triggers selective protein turnover in primordial germ cells to ensure transgenerational fitness.
INTRODUCTION
Germ cells connect generations indefinitely, thereby establishing an immortal lineage. To perform this transgenerational task in animals, the germ line is typically set aside from somatic cell lineages during early embryogenesis, leading to the formation of primordial germ cells (PGCs) (1). As early embryogenesis primarily occurs under global transcriptional repression, posttranscriptional regulation of maternal transcripts and posttranslational regulation of maternal proteins predominate. Consequently, the developing embryo has to liberate itself from the instructive parental load of gene products until zygotic gene products take charge. This maternal-to-zygotic transition (MZT) is a molecular handover that removes maternal mRNA and protein products and prepares for zygotic genome activation (ZGA) (2). How this MZT is achieved in PGCs to promote germline immortality is currently poorly understood.
In particular, animals that set aside a germ line by preformation delay the MZT in the lineage that leads up to PGC specification (2, 3). Instrumental to preformation is selective inheritance of germ plasm from the oocytes, which organizes distinct liquid-liquid phase-separated organelles (germplasm granules) of stored mRNA transcripts and mRNA-associated regulatory proteins (1, 4). Consistent with an ability to maintain germ cell identity, germ plasm and its RNA granules are lost in somatic lineages but remain present throughout the germ lineage. Nonetheless, some of their maternal protein constituents, representing important posttranscriptional mRNA regulators, are eliminated from PGCs before or accompanying germline-specific ZGA in many systems (5–7), suggesting a potential incompatibility with subsequent zygotic gene expression programs. Whereas several protein clearance mechanisms have been identified that contribute to the MZT in the soma (5, 8), they are largely unknown in PGCs. Moreover, their impact on germline immortality remains to be determined for any organism.
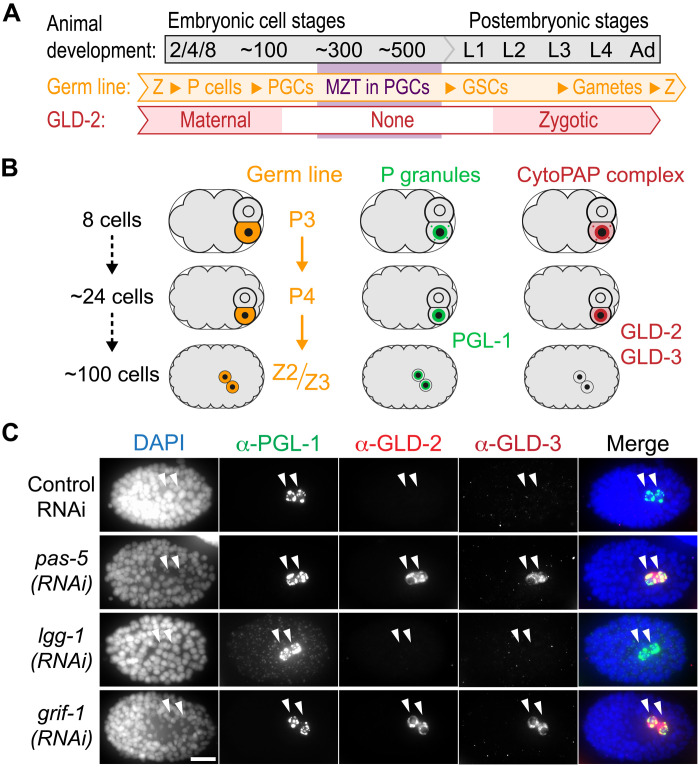
Embryonic germline formation in Caenorhabditis elegans constitutes an exemplary model system of preformation. A stereotypical cascade of four asymmetric cell divisions segregates germline precursors (P blastomeres) from soma (Fig. 1, A and B). After gastrulation has relocated P4 into the interior of the embryo, the two PGCs (Z2 and Z3) are born from a symmetric cell division at approximately the 100-cell stage (Fig. 1A). Its germ plasm is marked by species-specific proteins such as PGL (P granule abnormality)-1 (Fig. 1, A and B) and, in addition to conserved translational repressors such as PIE (pharynx and intestine in excess)-1 or NOS (Nanos related)-2 (9–11), also contains positively acting mRNA regulatory machinery such as GLD (defective in germline development)-2 and GLD-3 (Fig. 1, A and B). The latter form together a functional cytoplasmic polyadenylate [poly(A)] polymerase (cytoPAP) that promotes mRNA stability broadly and translation in a gene-specific manner (12–14). By maintaining and extending the poly(A) tails of mRNAs in the cytoplasm, GLD-2–like poly(A) polymerases counteract RNA turnover factors (i.e., deadenylases) and translational repressors in several animal models, including nematodes (15, 16). Contrary to PGL-1, a likely generic RNA binding protein that is maternally donated to the embryonic P lineage and permanently expressed in the germ plasm (17, 18), the aforementioned essential maternal translational regulators abruptly vanish from PGCs before early germline genome activation between the 100- and ~300-cell stages (14, 19) (Fig. 1, A and B). Given that zygotic reexpression of GLD-2 enzyme and its stimulatory subunit GLD-3 begins already in undifferentiated germ cells at early larval stages to promote postembryonic germ cell development (Fig. 1A) (14, 20), the cytoPAP complex serves as a paradigm to investigate the mechanisms underlying clearance of posttranscriptional germplasm RNA regulators and its impact on germline development when obstructed.
Fig. 1. Maternal GLD-2 and GLD-3 are degraded in PGCs.
(A and B) Starting from fertilization (Z, zygote), maternal GLD-2 protein is specifically maintained in the germline (P) lineage, marked by the permanent P granule component PGL-1. During later embryonic stages, GLD-2 and its activator, GLD-3, are absent during the ZGA stages of either PGC (Z2 and Z3). Germline development continues postembryonically, and PGCs become dividing germline stem cells (GSCs). Zygotic GLD-2 abundance gradually increases in later larval (L) stages and remains high through late female gametogenesis. (C) Fixed ~100-cell stage wild-type embryos, treated with control RNAi, or targeting 26S proteasome (pas-5), autophagosome (lgg-1), or a RING finger gene (grif-1), stained for DNA [4,6-diamidino-2-phenylindole (DAPI)] and indicated proteins, whose given color code corresponds to the respective fluorescent detection channel (see Materials and Methods for further clarification). CytoPAP subunits GLD-2 and GLD-3 are stabilized in PGCs upon pas-5 (96%, n = 118) or grif-1 (100%, n > 100) but not control (100%, n > 90) or lgg-1 (100%, n > 100) knockdown. Merge, all channels. Scale bar, 10 μm.
Here, we demonstrate that the proteasome, but not autophagy, regulates clearance of the maternal cytoplasmic polyadenylation GLD-2/GLD-3 machinery in PGCs. We identify the tripartite motif 32 (TRIM32)–like E3 ligase, GLD-2–interacting RING finger protein 1 (GRIF-1), as a germ lineage–specific turnover factor of GLD-2 enzyme and demonstrate its crucial role in transgenerational fitness. We find that maternal GRIF-1 expression itself depends on GLD-2 cytoPAP activity and that GRIF-1 associates with germplasm granules before GLD-2 turnover. We provide evidence that prolonged GLD-2 cytoPAP expression in PGCs critically interferes with germline immortality.
RESULTS
GLD-2 cytoPAP components are subject to GRIF-1–dependent proteasome degradation
Autophagy is a major degradation system that eliminates maternal proteins in mammals (21) or nongermplasm-localized PGL-1 in somatic lineages of C. elegans (22). Also, the ubiquitin proteasome system (UPS) contributes to the MZT in the soma via multisubunit ubiquitin E3 ligases (5, 23–25). To distinguish between a role of both prevalent protein turnover systems on GLD-2 cytoPAP expression in PGCs, we compromised either by RNA interference (RNAi) and monitored maternal expression by immunofluorescent analyses (Fig. 1 and fig. S1). As the proteasome is essential for early embryogenesis (26, 27), we devised a partial knockdown RNAi feeding regime against two of its core subunits, pas-5 and pbs-6, to harvest UPS-compromised embryos that still complete embryogenesis (fig. S1A). RNAi efficiency was gauged by scoring living F1 progeny (fig. S1B) and subsequent fertility at adulthood (fig. S1C), as well as by the appearance of multiple PGCs in late embryogenesis (fig. S1, D and E), a previously noted, but not further described, partial-loss-of-function phenotype of UPS-compromised embryos (22). To investigate an influence of autophagy, we knocked down lgg-1, an ortholog of the LC3/ATG8 protein family required for the clearance of the two constitutive P granule proteins, PGL-1 and PGL-3, in somatic sisters of the germ lineage (22).
Similar to wild-type embryos, 100-cell stage or older control RNAi embryos invariantly had two PGL-1–expressing PGCs but lacked both cytoPAP components (Fig. 1C and fig. S1D). By contrast, GLD-2 and GLD-3 proteins were both detected in most of the pas-5 and pbs-6 RNAi-compromised PGCs at similar and later stages of embryogenesis (Fig. 1C and fig. S1D). This prolonged expression of both cytoPAP components was also detected in embryos with more than two presumed PGCs (fig. S1, D to F). While RNAi-mediated knockdown of the autophagy factor and ATG (autophagy related) 8 homolog, lgg-1, did result in somatic accumulation of PGL-1 (22), no prolonged expression of GLD-2 and GLD-3 was detectable in PGCs of the very same embryos (Fig. 1C). These results suggest that UPS, rather than the somatic bulk protein clearance mechanisms of autophagy, promotes clearance of cytoPAP components in PGCs.
In search for a potential ubiquitin ligase that might regulate cytoPAP degradation, several full-length (FL) complementary DNA (cDNA) clones of Y51F10.2 were identified in a yeast two-hybrid (Y2H) screen with GLD-2(FL) as bait. This gene stood out as a prominent candidate for GLD-2 turnover as it encodes a RING finger domain (Fig. 2A and fig. S2, A and B), a hallmark of known E3 ubiquitin ligases, which also inspired its name, GRIF-1. Upon RNAi knockdown of grif-1, GLD-2 and GLD-3 expression was extended in PGCs (Fig. 1C), indicating that it is a likely candidate for mediating maternal cytoPAP clearance in PGCs. Unlike proteasome RNAi, no extra PGCs were detected in grif-1(RNAi) embryos, arguing that prolonged GLD-2 cytoPAP expression is a specific protein turnover defect, independent of PGC specification or division.
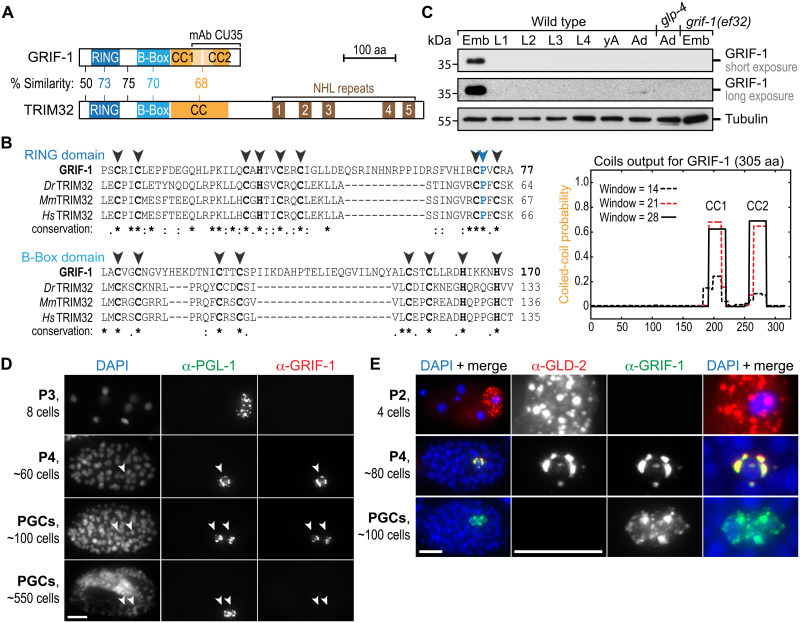
Fig. 2. GRIF-1 is a TRIM32-like protein abundant in PGCs.
(A and B) Across its primary sequence (top BLAST hit score of 1 × 10−6) and in its protein domain architecture, GRIF-1 is most similar to the E3 ubiquitin ligase TRIM32. However, its similarity is limited to the N-terminal half of TRIM32 because GRIF-1 lacks the additional RNA binding (NHL) domain. At its C terminus, GRIF-1 contains a noncontiguous CC domain (orange) predicted to be split into two parts (CC1 and CC2). (B) Multiple sequence alignments of the RING (blue) and single B-Box (cyan) domain. Despite several insertions, key amino acids [aa; cysteine (C) and histidine (H)] important for zinc finger formation (black arrowheads) or presumed E3 ligase activity [e.g., proline (P), blue arrowhead] are conserved: full stop (.), similar amino acid; colon (:), highly similar amino acid; asterisk (*), identical amino acid. Dark orange regions correspond to COILs (version 2.2) output, using strings of 14, 21, or 28 amino acids. (C to E) GRIF-1 expression is restricted to the embryonic germ line, temporarily overlapping with GLD-2 on P granules in P4 and early PGCs. (C) Western blots using mAb CU35 on extracts of wild-type larvae (L), adults without (yA) and with (Ad) embryos (Emb), and no germline (glp-4). (D and E) Fluorescent images of wild-type embryos (100%, n > 50 for each stage) stained with DAPI and antibodies against indicated proteins. Note that GRIF-1 expression always ceases in aged PGCs. Scale bars, 10 μm.
Upon closer inspection of GRIF-1’s polypeptide sequence, we found that the annotated N-terminal “cross-braced” zinc finger–type RING domain is followed by a type 2 B-Box domain and a large coiled-coil (CC) region at its C terminus (Fig. 2, A and B). This RBCC (RING, B-Box, and coil-coil) domain architecture is stereotypical for TRIM proteins (Fig. 2A), a large and diverse subgroup of E3 ligases that are independent of multisubunit ubiquitin-ligase complexes (28, 29). Consistent with these structural determinants, GRIF-1 is, across its entire sequence, most similar to the mammalian subfamily C-V and in particular to TRIM32 proteins (Fig. 2A), which, in addition to acting as protein turnover factors, also bind and regulate RNA (29, 30). Whereas functionally essential amino acids for E3 ligase activity are conserved across the RING and B-Box domains (Fig. 2B), GRIF-1 lacks the RNA binding NHL (NCL1, HT2A, and LIN-41) repeats at its C terminus (Fig. 2A). In addition, TRIM32’s extended CC domain appears to be split into two shorter CC domains in GRIF-1 (CC1 and CC2; Fig. 2, A and B, and fig. S2B), which is further corroborated by recent AlphaFoldv2.0 predictions (31). As GRIF-1 has diverged in the overall domain organization from other TRIM32 protein members but maintained its RBCC architecture with strong sequence similarity to TRIM32 proteins, we hypothesize that it might exclusively be an E3 ubiquitin ligase and only indirectly involved in posttranscriptional RNA regulation as a potential negative regulator of GLD-2 cytoPAP.
GRIF-1 expression is restricted to the late embryonic germ lineage
The unique developmental expression profile of GRIF-1 supports this hypothesis. A monoclonal antibody (mAb; CU35) that recognizes the C-terminal CC region of GRIF-1 (Fig. 2A and fig. S2, B and C), detected on Western blots of wild-type embryo extracts a single protein band of predicted molecular weight (Fig. 2C), which was absent in grif-1(0) mutants, such as grif-1(ef32) and grif-1(ok1610ef40), or of expected reduced molecular weight in grif-1(ok1610) animals (Fig. 2C and fig. S2, A to C). The latter allele represents a molecular deletion that removes parts encoding for the second CC domain (fig. S2A), leading to the production of a stable truncated GRIF-1(ok1610) protein isoform (fig. S2, B and C), as further verified with an N-terminal–specific mAb against GRIF-1 (fig. S2D). A barely detectable signal of GRIF-1 in wild-type adults is a likely consequence of fully developed gonads containing embryos, rather than expression derived from soma (Fig. 2C).
Consistent with mRNA expression data indicating that Y51F10.2 mRNA is maternally donated and potentially translationally repressed (11), we observed in immunostaining analyses that embryos before the 24-cell stage lack a GRIF-1–specific signal, further in line with reported epitope-tagged Y51F10.2 protein expression (11). Moreover, we observed that gastrulating embryos begin GRIF-1 expression exclusively in the late P lineage just before the birth of both PGCs, and GRIF-1 levels remain high in PGCs until approximately the 500-cell stage before dropping below detection levels at the threefold stage past the onset of germline ZGA (Fig. 2, D and E, and fig. S3A). While a signal was absent in grif-1(0) (fig. S3B), a much weaker signal than may be expected from immunoblots was detected in grif-1(ok1610) (figs. S2, C and D, and S3B), suggesting that the C-terminally truncated GRIF-1 protein of ok1610 is correctly expressed in PGCs, but its three-dimensional fold may evade full antibody detection as it lacks CC2.
GRIF-1 subcellular localization is dynamic and starts with an initial colocalization with GLD-2 on P granules (Fig. 2, D and E, and fig. S3A). Conspicuously, in ~150-cell stage embryos, when all GLD-2 signal is lost, we noticed that GRIF-1 gradually dissociates from P granules, becomes exclusively cytosolic, and enriches in small but distinct puncta in the cytosol (Fig. 2E and fig. S3A). Over time, as embryogenesis proceeds, the diffuse cytosolic signal is lost, and puncta begin to reduce in intensity. We suspect that this behavior of foci formation may manifest sites of GRIF-1 protein turnover, similar to other TRIM proteins that self-associate and autoubiquitinate once they lose their turnover targets, to inflict self-directed protein turnover (32–34). In summary, GRIF-1 expression peaks and enriches on P granules at the time just before GLD-2 elimination and dissociates from P granules when GLD-2 turnover has been completed.
GRIF-1 interacts with the germline-specific IDR of GLD-2
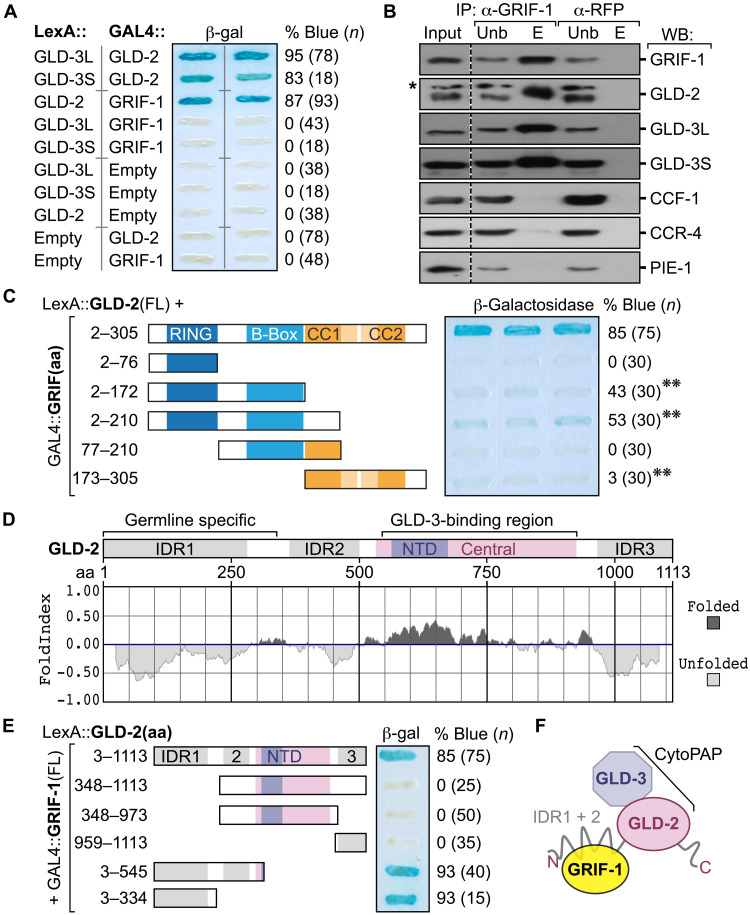
To assess protein interactions between GRIF-1 and particular cytoPAP subunits and to map sites important for physical interaction, we performed directed Y2H tests and coimmunoprecipitation (coIP) experiments from embryo extracts (Fig. 3). As GRIF-1(FL) fused to the DNA binding domain led to autoactivation in yeast, we fused GRIF-1 variants to the activation domain in all our experiments and surveyed fusion protein expression by Western blotting (fig. S4). The known interactions between GLD-2 and either of the embryo-expressed GLD-3 isoforms, GLD-3L or GLD-3S, are mediated by the central catalytic region of GLD-2 (12, 14) and served as positive controls.
Fig. 3. GRIF-1 interacts with the N-terminal IDR of GLD-2 cytoPAP.
(A) Beta-galactosidase (β-gal) activity (percentage of blue) of yeast colonies (n) coexpressing indicated fusion proteins; empty, no fusion. GRIF-1 interacts with GLD-2, but not with either isoform of GLD-3 (L or S), each binding to GLD-2. (B) Immunoprecipitated (IP) materials from embryonic extracts with antibodies directed against GRIF-1 or RFP that serves as a specificity control were probed in Western blots (WBs) for indicated proteins; 5% input (Inp) and unbound (Unb) materials and 50% of eluted (E) material loaded. Asterisk (*) shows unspecific background signal of anti–GLD-2 antibody. (C) Only FL GRIF-1 fused to GAL4 activation domain binds efficiently to GLD-2 in direct Y2H tests. Label as in (A); double asterisk (**) indicates colonies that became slightly blue after longer reaction times than in (A). (D) FoldIndex algorithm predicts three IDRs in GLD-2, including the entire N-terminal domain, encoded only in germline-specific transcripts of gld-2. The catalytical heart of GLD-2, the nucleotidyl-transferase domain (NTD), is embedded in a well-folded central domain that binds GLD-3 isoforms tightly for further enzymatic stimulation. (E) The germline-specific IDR of GLD-2 mediates the interaction with GRIF-1 in yeast. Label as in (A). (F) Working model of the interaction among GRIF-1 and GLD-2/3 cytoPAP subunits; GLD-3 represents either GLD-3L or GLD-3S.
In yeast, GRIF-1 robustly interacted with GLD-2(FL) but neither of the GLD-3 isoforms (Fig. 3A and fig. S4A). Concomitant with an enrichment of GRIF-1 in coIPs from embryos, all three known cytoPAP subunits, GLD-2 and GLD-3L or GLD-3S, were specifically enriched, but not other maternal RNA regulators, such as the two deadenylase subunits of the CCR4-Not complex, CCF-1 and CCR-4 (35), or the P granule–associated RNA regulator and nuclear transcriptional repressor PIE-1 (9) (Fig. 3B). In delineating the potential interaction surfaces between GRIF-1 and GLD-2, we noticed that any perturbation of GRIF-1 protein leads to essentially negative results (Fig. 3C and fig. S4B). Only a weak positive result was obtained in this assay with GRIF-1 fragments that were truncated in their CC region, partially or in full (Fig. 3C and fig. S4B), suggesting that similar to other TRIM proteins, all GRIF-1 domains may cooperate as an integrated modular unit for an efficient and target-specific interaction. Its interaction with GLD-2 required the germ cell–specific intrinsically disordered region (IDR) of GLD-2’s N terminus (Fig. 3, D and E, and fig. S4C) (14), substantiating that GRIF-1 specifically associates with the GLD-2/3 cytoPAP complex, likely on P granules, by binding to the N-terminal IDR of GLD-2, using all of its protein domains for target recognition or stable association (Fig. 3F).
GRIF-1 promotes GLD-2 turnover
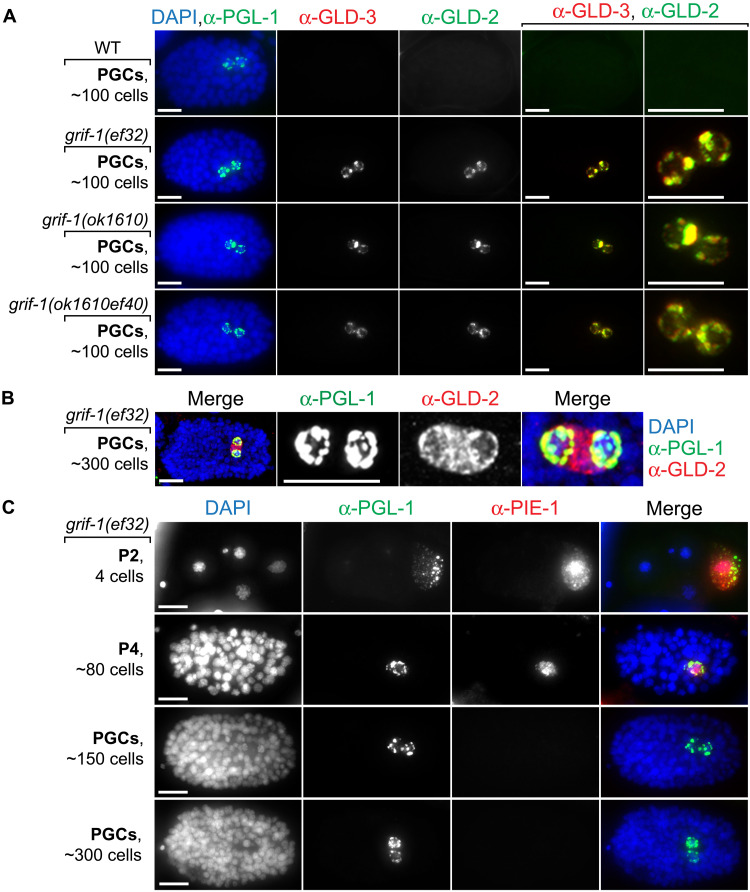
As a potential turnover factor of GLD-2 cytoPAP, loss of GRIF-1 may prolong cytoPAP’s presence, and ectopic expression of GRIF-1 may negatively affect GLD-2 abundance. Hence, we first compared wild-type to maternally depleted grif-1 embryos, derived either from RNAi-mediated knockdown experiments (Fig. 1C) or grif-1(−) homozygote mutant mothers (Fig. 4). Similar to wild type, neither PGL-1 nor PIE-1 localization or expression was overtly changed in both PGCs of grif-1 embryos (Fig. 4, A and C). In all grif-1(−) embryos (n > 100, per genotype), PGL-1 remained expressed in the embryonic P lineage and associated with P granules (Fig. 4). Compared to ~3% in wild type (n = 30), a slightly increased variability of the initiation of PIE-1 elimination up until the 150-cell stage was detected in ~6% of grif-1(−) embryos (n = 40, per genotype); PIE-1 was efficiently eliminated in 98% of all embryos of any genotype beyond the 300-cell stage (Fig. 4C, n > 100). On the contrary, GLD-2 and GLD-3 remained expressed from the 100-cell stage onward and localized to P granules in both PGCs in all analyzed grif-1(−) embryos (n > 100, per genotype) (Figs. 1C and 4A), suggesting that grif-1 activity does not generally turn over P granule–associated RNA regulators but specifically destabilizes the subunits of the entire GLD-2 cytoPAP complex. Intriguingly, stabilized GLD-2/3 cytoPAP components largely detached from P granules and remained exclusively cytosolic by the ~300- to 500-cell stages (Fig. 4B), suggesting a subcellular relocation upon the onset of germline ZGA potentially due to a change in mRNA targets.
Fig. 4. Expression profile changes in grif-1 mutant PGCs.
(A to C) Fluorescent images of embryos at different developmental embryonic stages stained for DNA (DAPI) and indicated proteins. Merge, all channels. Scale bars, 10 μm. (A and B) GLD-2 and GLD-3 protein expression endures and remains P granular in freshly born PGCs of various grif-1 mutant ~100-stage embryos (100%, n > 100, each genotype). (B) GLD-2 redistributes from P granules in younger embryos to become strongly cytosolic in older grif-1 mutant embryos (100%, n > 100) correlating with minor genome activation at the ~300-cell stage in wild-type PGCs. (C) The maternal expression of the cytoplasmic RNA regulator and nucleoplasmic transcriptional repressor PIE-1 is, at large, unperturbed in grif-1(0) embryos and becomes undetectable beyond the 150-cell stage (see main text for details).
In TRIM proteins, the RING finger confers catalytic E3 ligase activity and affinity to its target substrate (28, 36). To further strengthen a functional relationship between GRIF-1 and its in vivo target GLD-2, we expressed wild-type GRIF-1(WT) and a RING domain mutant GRIF-1(C43A) in postembryonic germ cells of otherwise wild-type animals (fig. S5). We chose this ectopic expression approach to gain enough material for subsequent coIP experiments and to probe for potential effects on animal fertility, as a postembryonic reduction of GLD-2 activity causes infertility (14, 37).
Although the achievable expression level of each transgene was low at 20°C, it was comparable among GRIF-1(WT) and GRIF-1(C43A) (fig. S5B, compare lanes 2 to 3 or 7 to 8). Nonetheless, we repeatedly and specifically pulled down less GRIF-1(C43A) compared to GRIF-1(WT) from whole-animal protein extracts with anti–GLD-2 antibodies (fig. S5B), suggesting that the RING domain of GRIF-1 contributes to stable GLD-2 association. At 25°C where transgene expression is typically stronger in C. elegans, a partial reduction of GLD-2 in whole-animal protein extracts of GRIF-1(WT) compared to GRIF-1(C43A) transgenic strains was evident (fig. S5C). Furthermore, only ectopically expressing GRIF-1(WT) animals displayed infrequent animal sterility that, upon inspection by Nomarksi microscopy, included, among other germline phenotypes, gld-2–like sterility defects, i.e., fully developed gonads with diplotene-arrested germ cells, proximal proliferation, or dead early embryos (fig. S5, D and E) (14, 37). While these defects may also be a consequence of off-target activity, together, our observations are consistent with an enzymatic and substrate-targeting role of GRIF-1’s RING domain, likely contributing to GLD-2 turnover.
Loss of grif-1 affects transgenerational fertility and germline immortality
Mutations in genes required for germ cell development are typically maintained as heterozygote (+/−) animals and analyzed in F1 (−/−) homozygote animals. grif-1(−) animals were fertile as F1 homozygotes, consistent with a maternal role of grif-1. However, when grif-1(−) animals were further propagated as homozygote lines lacking further maternal contributions, we noticed a reduction in overall fecundity, and sterile animals stochastically appeared in any generation from F2 onward at 20°C. In these affected populations, the frequency of steriles increased in subsequent generations until this line could not be maintained any longer. Moreover, at 25°C, the frequencies at which sterility appeared increased further, and the number of generations it took to reach full sterility shortened. These observations are hallmarks of a mortal germline phenotype (Mrt), which is a multigenerational defect in which a selfing lineage and population become progressively sterile (38–40).
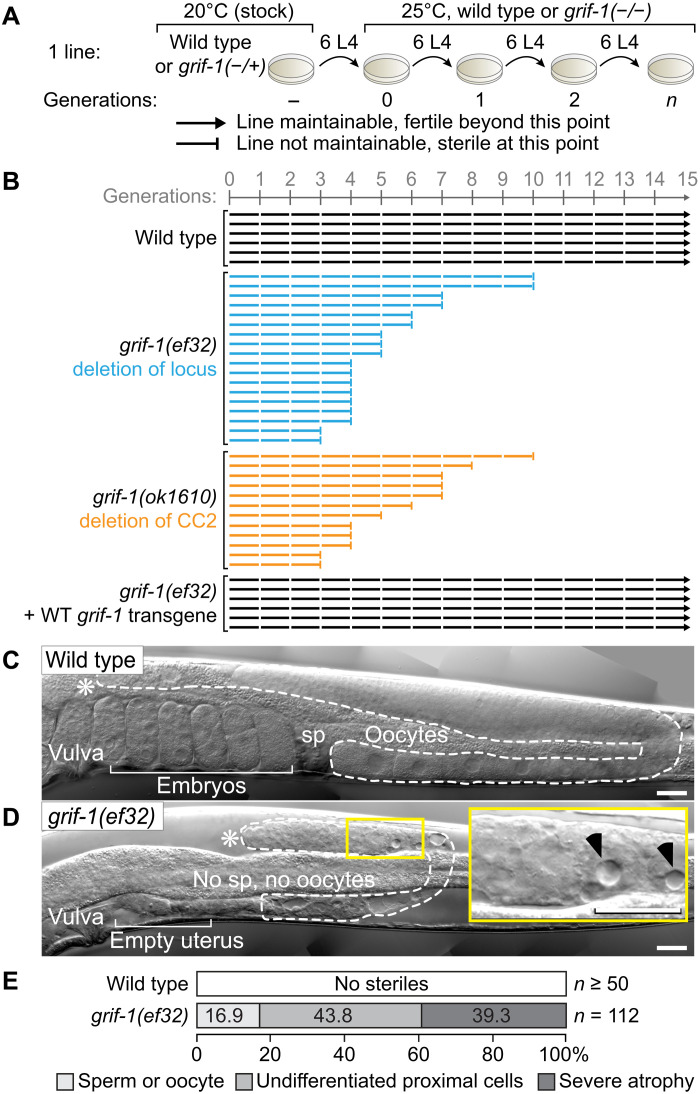
To characterize these phenotypes of grif-1 loss, we maintained several lines of different genotypes for 15 or 20 generations according to a standard passing regime (Fig. 5A). Despite the occasional sterile worm appearing in any population, all wild-type lines could be propagated normally (Fig. 5B), demonstrating that a population of wild type is indefinitely fertile. By contrast, populations of grif-1 mutant animals, irrespective of their genetic lesion, became completely sterile over varying generations, and no line could be maintained for more than 12 generations, (Fig. 5B and fig. S6C). This phenotype was less pronounced at 20°C, where it usually takes more than 20 generations for the entire population to become completely sterile. At any temperature, expression of a wild-type grif-1 transgene restored both fertility and germline immortality, at the level of an individuum or population, respectively (Fig. 5B and fig. S6, A to C), suggesting that embryonic expression of GRIF-1 is sufficient to prevent these defects, and, judging from grif-1(ok1610), its C-terminal CC2 region is critical for maintaining germline immortality.
Fig. 5. Transgenerational sterility and germline atrophy in grif-1 mutants.
(A) Experimental workflow to determine a mortal germline phenotype and legend to all data compilations as shown in (B). Six wild-type or grif-1(−/−) L4 animals, with or without expressing a wild-type (WT) grif-1 transgene, are passed across >15 generations at 25°C, starting from grif-1(+/−) heterozygotes, and maintained at 20°C. (B) A line (horizontal bar) is regarded as sterile (ending on a vertical bar) when mothers produce less than two progenies on average and further propagation became impossible. Wild-type or rescued grif-1 mutant animals continued to be fertile (ending on an arrowhead), and grif-1(−/−) mutant lines became completely sterile at early but varying generations. See fig. S6A for other grif-1 alleles and the rescuing transgene. (C and D) Nomarski images of a single gonadal arm (dotted line) in adults. Asterisks (*), distal end of gonad; self-fertile wild-type animals store sperm (sp) in their proximal spermatheca. Inset enlarges signatures of germline atrophy (black arrowheads); no differentiated gametes are present in this small gonad of a sterile grif-1 adult. Scale bars, 20 μm. (E) Sterile grif-1 animals display three major germ cell phenotypes in their gonads: defective gametes, no gametes but undifferentiated cells in the proximal region, and severe cell death and atrophy either proximally or throughout.
Nomarski microscopy revealed that all sterile grif-1 animals have much smaller gonads than wild type (Fig. 5, C and D, and fig. S6D). While a fraction of these contain gonads with either defective gametes or germ cells that failed to differentiate into gametes (Fig. 5E), we also observed a substantial fraction of grif-1 steriles that display germline tissue degeneration defects with partially filled gonads and strong vacuolization (Fig. 5, D and E, and fig. S6D). This phenotype ranges in severity from highly atrophic gonads with distal clusters of undifferentiated germ cells to empty gonads, lacking any recognizable germ cell nuclei (Fig. 5F); underproliferation and atrophy are already apparent in randomly selected younger larvae. As germ cell death occurred to a large fraction independently of the critical apoptotic pathway gene ced-4 (fig. S6E) (7, 20, 41), the pathway promoting germline atrophy remains unknown. We conclude that grif-1 promotes transgenerational fitness by promoting fertility and preventing the occurrence of pleiotropic germline defects across generations, presumably as a result of imprecise or delayed MZT in PGCs.
Ectopic persistence of GLD-2 induces transgenerational defects
The extended expression of GLD-2 in PGCs and transgenerational fertility defects in grif-1 animals might be at least in part due to prolonged GLD-2 cytoPAP activity during embryogenesis. We tested this idea by generating transgenic (TG+) animals that expressed FL or N-terminally truncated variants (Δaa) of GLD-2 in the germ line of gld-2(0) animals (Fig. 6A and fig. S7). As GLD-2 is essential for gametogenesis (37) and early embryogenesis (14), exclusively depending on its enzymatic activity (42), we first analyzed the expression levels and rescuing capabilities of each transgene.
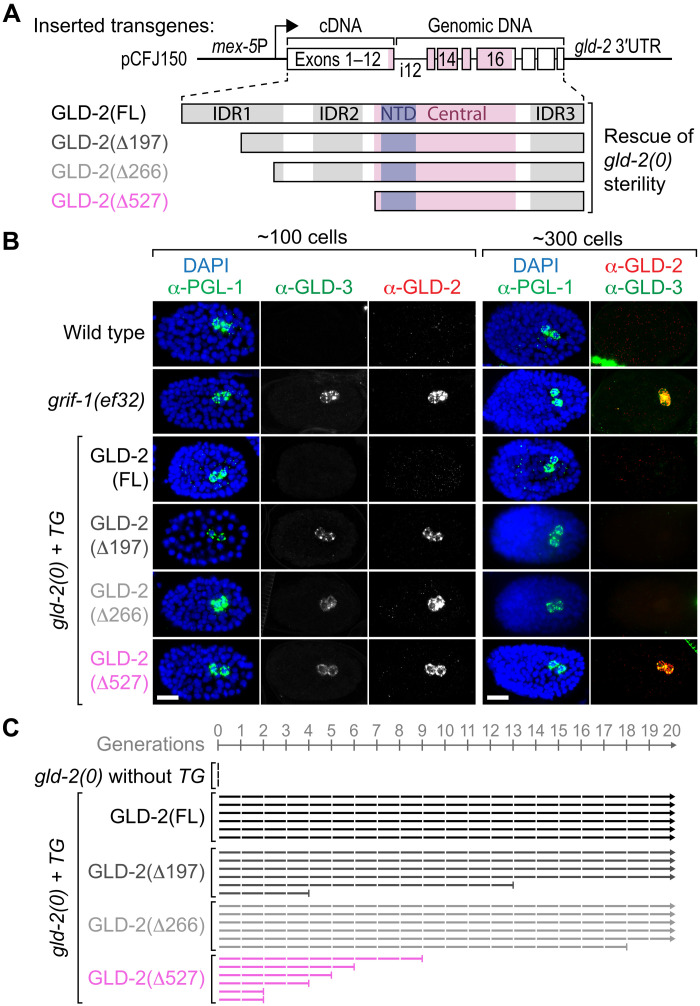
Fig. 6. Prolonged GLD-2 expression in PGCs affects transgenerational fitness at 25°C.
(A) Stick diagrams of all transgene-expressed N-terminally truncated (Δ) GLD-2 protein variants used to rescue postembryonic germline development defects and early embryonic arrests of gld-2(0) mutant F1 animals and F2 embryos, respectively; labels as in Fig. 3D. Partially intronized (i) chromosome II–integrated gld-2 transgenes under the control of germ cell–specific mex-5 promoter (arrow) and cognate 3′ untranslated region (UTR) sequences. (B) Unlike in wild-type or FL GLD-2–rescued gld-2(0) animals and similar to PGCs in grif-1(ef32), PGCs of any GLD-2(ΔN terminus)–rescued gld-2(0) embryos display prolonged cytoPAP expression. Note that extended GLD-2 and GLD-3 perdurance is only maintained in PGCs of older grif-1(ef32) and GLD-2(Δ527aa) embryos (for quantitation, see main text). Scale bars, 10 μm. (C) Transgenerational defects are most severe upon removal of the entire IDR in GLD-2(Δ527aa). gld-2(0) transgenic animals analyzed and displayed as in Fig. 5. See also fig. S7 for extended analyses.
Correlating with higher expression levels at 25°C when compared to 20°C, GLD-2(FL) transgenic lines showed a more robust brood size that is similar to that of wild type (fig. S7, A to C). All GLD-2(Δaa) transgenic lines were expressed similar to wild type already at 20°C (fig. S7A) and robustly rescued gld-2(0) F1 sterility at both temperatures (fig. S7, B and C). Although less penetrant for GLD-2(Δ527) than for the other two shorter truncations, GLD-2(Δ197) and GLD-2(Δ266), these rescuing data support the notion that the germ cell–specific N-terminal IDR of GLD-2 is largely dispensable for GLD-2’s postembryonic germ cell functions and essential roles in early embryogenesis. Instead, GLD-2’s core functions reside in its enzymatic domain that is stimulated by other cytoPAP subunits, such as GLD-3.
Next, we examined the contribution of the N-terminal IDR in GLD-2 stability by focusing on embryonic germ cells. Each IDR-truncated transgenic GLD-2 variant remained expressed in freshly minted PGCs (Fig. 6B), yet the extent of prolonged expression depended on IDR length: GLD-2(Δ197) and GLD-2(Δ266) were expressed up to the 150-cell stage in most embryos (72%, n = 68 and 69.5%, n = 59, respectively) and occasionally to the 300-cell stage (8.3%, n = 36 and 7.7%, n = 26, respectively). Only GLD-2(Δ527) expression was, similar to endogenous GLD-2 in grif-1(0) animals, persistently observed in PGCs throughout embryogenesis (100%, n > 50) (Fig. 6B), demonstrating that the entire N-terminal IDR is essential for efficient GLD-2 turnover. Intriguingly, GLD-3 expression was also extended in grif-1(0) or GLD-2(Δ527) animals (Fig. 6B), further suggesting that GLD-3 removal is linked to GLD-2 elimination and that a presumed enzymatically active GLD-2/3 cytoPAP complex remains present in PGCs.
To test a role for prolonged GLD-2 cytoPAP in transgenerational fertility, we assessed, at the individual level, the reproductive output of rescued GLD-2(TG+) animals at generations F1 and F3 and, at the population level, germline mortality across 20 generations at 25°C. The number of progeny sired by GLD-2(FL) mothers did not decline across generations (fig. S7C), and lines could be propagated without any signs of germline mortality (Fig. 6C and fig. S7D). By contrast, fertility of GLD-2(Δaa) animals decreased across generations, and transgenerational sterility increased in populations most severe for GLD-2(Δ527) (Fig. 6C and fig. S7, C and D). Together, we conclude that the entire N-terminal IDR is important for an efficient turnover of GLD-2 in PGCs and contributes to prevent germline mortality.
Maternal GLD-2 promotes embryonic GRIF-1 expression
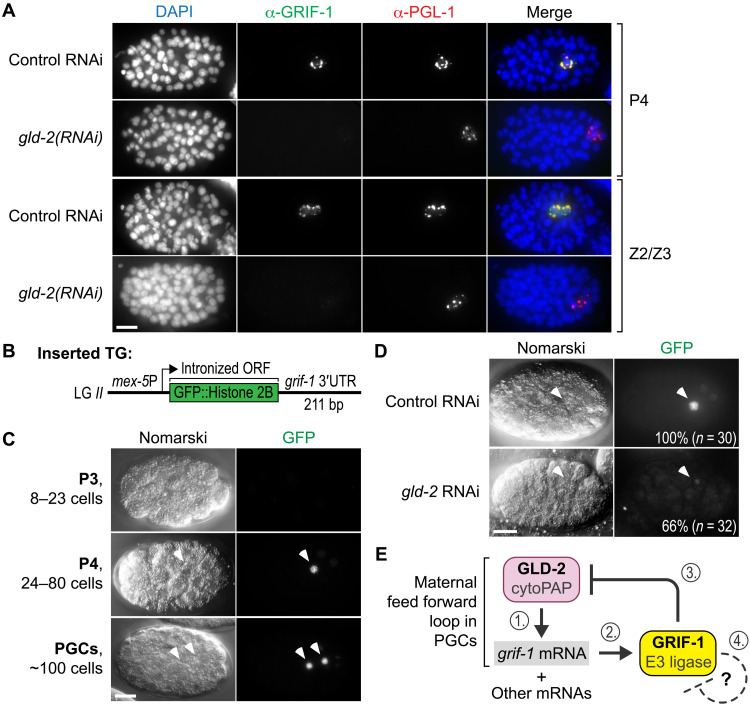
Before this study, GRIF-1 was characterized as a maternally donated transcript that is likely subjected to posttranscriptional repression and activation (11). As grif-1 mRNA was also reported in a global study to be a potential target of GLD-2–mediated mRNA stability or translatability (13), we tested the idea that GLD-2 might be important for maternal GRIF-1 protein expression in P4 and its PGC daughters.
To avoid early embryonic arrest, we devised an RNAi regime that partially depleted gld-2 activity and found in rare escapers that GLD-2 activity is required for endogenous GRIF-1 expression (Fig. 7A). Moreover, a transgenic germ cell–specific translational reporter, under the posttranscriptional control of the cognate 3′ untranslated region (3′UTR) sequence of grif-1 (Fig. 7B), recapitulates maternal GRIF-1 expression and depends on GLD-2 activity (Fig. 7, C and D), arguing that GLD-2 itself promotes GRIF-1 expression in the embryonic germ lineage.
Fig. 7. GRIF-1 expression depends on posttranscriptional positive regulation via GLD-2 cytoPAP.
(A) Fixed wild-type embryos of late P lineage stages, treated with low doses of RNAi targeting gld-2, and stained for DNA (DAPI) and indicated proteins. In rare escapers that also have mild gastrulation defects, endogenous GRIF-1 expression is compromised in PGL-1–positive P4 or PGC stages (100%, n = 20). Merge, all channels. Scale bar, 10 μm. (B) Cartoon display of strains harboring a genome-integrated green fluorescent protein (GFP) translational reporter transgene (TG) whose mRNA is transcribed from the germline-specific mex-5 promoter and posttranscriptionally regulated through the grif-1 3′UTR. The ORF of GFP is interrupted by three synthetic introns (not shown) and fused in frame to a histone 2B protein piece. Upon translation, the resulting fusion protein accumulates in the nucleus on chromatin. (C and D) Nomarski images of embryos at different developmental embryonic stages with corresponding autofluorescence of the translational reporter GFP signal (100%, n > 50). Scale bars, 10 μm. (C) Translational GFP reporter expression recapitulates developmental GRIF-1 expression profile. (D) RNAi knockdown compromises translational GFP reporter expression in many gld-2, but not in control, embryos. (E) Working model of GLD-2 cytoPAP–dependent grif-1 expression in P4 and early PGCs (steps 1 and 2). GRIF-1–mediated turnover of GLD-2 (step 3) stops this feed forward loop of maternal components also for other maternal—and potentially even zygotic—mRNA targets of GLD-2 cytoPAP. We hypothesize that a potential autoregulatory feedback loop of the presumed TRIM E3 ligase GRIF-1 may cause its elimination once its protein targets might be exhausted (step 4).
DISCUSSION
Maternal proteome clearance is a conserved feature of the MZT, but it was previously unclear how it is achieved in PGCs and what consequence it may have for the continuation of the germ line. Our study proposes a maternal germ cell–specific feed forward mechanism that eliminates the enzymatic component of the cytoplasmic poly(A) machinery, GLD-2 cytoPAP, from germplasm granules in PGCs (Fig. 7E). Initially, as a posttranscriptional activator of maternally donated grif-1 mRNA, GLD-2 cytoPAP directly contributes to the germ cell–specific expression of GRIF-1, a presumed TRIM-type E3 ubiquitin ligase. Via its germ cell–specific IDR (Fig. 3F), GLD-2 attracts GRIF-1 protein to promote its UPS-dependent degradation. At least in part by clearing PGCs of an omnipotent posttranscriptional activator complex, GRIF-1 activity assists the MZT in germ cells to ensure transgenerational fertility and germline immortality.
The UPS achieves its task by selectively removing ubiquitinated proteins once they were flagged by target-specific E3 ubiquitin ligases (43). To support the MZT by quickly clearing early zygotes from their maternal protein load after fertilization in animals, several multisubunit E3 ligases (i.e., Cullin-based complexes and the C-terminal to Lis Homology complex) were identified in invertebrates to eliminate cell cycle regulators and posttranscriptional RNA repressors from somatic lineages (5, 25, 44). Only in mice, an oocyte-donated single RING finger domain–containing protein (RNF114) is known to promote the MZT by directly supporting ZGA in early embryos, hereby ubiquitinating select chromatin and transcriptional regulators (45, 46). Our work on PGCs in C. elegans expands the repertoire of UPS-targeting devices in the MZT to TRIM proteins, one of the largest subfamilies of single RING E3 ligases (47, 48). In particular, we found GRIF-1, whose evolutionary sequence conservation identifies it as a relative of mammalian TRIM32, a human disease-linked gene (48).
Belonging to the subclass of TRIM-NHL, TRIM32 is endowed with dual functionality of ligating ubiquitin to protein substrates via its RING domain and binding RNA to influence RNA fates posttranscriptionally via its NHL domain. Conspicuously, this duality is not apparent from the domain architecture of GRIF-1, as it lacks the NHL domain, a versatile RNA binding platform consisting of NHL repeats. Conversely, some TRIM-NHL protein family members (C. elegans LIN-41 and Drosophila melanogaster Brat and Wech) lack functional RING domains (29, 30), arguing that this subclass of TRIM proteins is subject to evolutionary functionalization. However, by binding to GLD-2 cytoPAP, a broadly acting posttranscriptional mRNA activator in germ cells, GRIF-1 is still intimately connected to RNA regulation, albeit indirectly, further strengthening a possible ancient common role of this protein family. Moreover, by eliminating GLD-2 cytoPAP, GRIF-1 is expected to indirectly destabilize also maternal mRNAs, acting as an indirect posttranscriptional repressor, which is consistent with the documented posttranscriptional RNA repressor functions of RNA binding TRIM-NHL proteins. Hence, an ancient direct link to negative RNA regulation may have been eventually traded in favor of protein turnover specialization in the case of GRIF-1. Although highly speculative, this scenario may also offer a potential avenue of how this maternal-only function of GRIF-1 may have been selected for. A second avenue of selection pressure may have arisen from GRIF-1’s role in maintaining transgenerational fertility (see below).
In contrast to multidomain E3 ligases, the biochemical mechanisms of ubiquitin transfer appear more complex, diverse, and less well understood in TRIM proteins (28, 43). This may also be due to the intricate RBCC architecture and the different types of possible ubiquitylation outcomes carried out by TRIM proteins (28, 34, 49). One common denominator, however, is that TRIM proteins use their RING domains to catalyze ubiquitin ligation and oligomerize via their elongated CC domain; both contribute to substrate binding and ubiquitin transfer from E2-conjugating enzymes to targets (33, 36). Consistent with this notion, we find that a functional RING finger domain in GRIF-1 is important for a stable GLD-2 interaction. Furthermore, GRIF-1’s RING domain induces negative effects on germline development upon ectopic expression in postembryonic germ cells. While one reason for the partially penetrant sterility might have been the low expression levels of GRIF-1, it might also reflect on a limited availability of the cognate E2-conjugating enzyme in these cells or a physical inaccessibility to the N-terminal IDR of GLD-2. In either case, GRIF-1 requires its C-terminal CC domain to bind to GLD-2 and to mediate GLD-2 clearance in PGCs. Deletion of CC2, as evidenced by GRIF-1(ok1610), is detrimental to GLD-2 turnover and germline immortality. While these data are consistent with a potential tetrameric assembly of TRIM32 proteins via their CC domains promoting E3 ligase activity, it is also consistent with a contribution to substrate interaction (36). As our experiments cannot distinguish between these possibilities, further detailed biochemical studies will be required to provide mechanistic insight into the molecular mechanisms of GLD-2 turnover.
At the level of substrate specificity, we found that GRIF-1 binds to the N-terminal IDR of GLD-2. Intriguingly, only germ cell transcripts of GLD-2 encode this entire IDR in full; somatic transcripts encode a severely N-terminally truncated GLD-2 protein (14). Although the biological functions of somatic GLD-2 are unclear, amino acids 528 to 923 of GLD-2 are sufficient to polyadenylate RNAs in vitro (12). To execute its molecular function as a poly(A) polymerase efficiently, GLD-2 must be stimulated either by RNP-8, which is not maternally contributed, or GLD-3 isoforms, which are maternally contributed (20, 50), and either enzymatic stimulator of GLD-2 cytoPAP binds in a mutually exclusive manner to its central catalytic domain independent of GLD-2’s IDR (51). In full compliance with these previous findings, we provided evidence that distinct GLD-3 isoform–containing trimeric GRIF-1/GLD-2/GLD-3 complexes are formed, and all enzymatic roles of GLD-2 can be rescued with GLD-2 isoforms that lack IDR sequences, either partly or entirely. While we cannot exclude a potential influence on enzymatic activity in vivo, our combined experiments support the notion that the entire IDR of GLD-2 mediates a major regulatory function in timing cytoPAP destabilization during the MZT in PGCs, contributing to transgenerational fertility and germline immortality. Such a long IDR might be especially suited to efficiently attract cytosolic GRIF-1 as GLD-2 cytoPAP accumulates in germplasm RNA granules. On the other hand, enhanced accumulation of GRIF-1 in P granules might increase its local concentration, required for efficient E3 ligase activity. Furthermore, we demonstrated that upon GLD-2 elimination, GLD-3 also becomes degraded. Whether this is also mediated directly by GRIF-1 E3 ligase activity, due to lack of GLD-2, or as a consequence of other protein turnover pathways during the MZT needs to be investigated in the future.
The timing of GRIF-1 expression onset correlates with germline specification and may mark the molecular start of the MZT in the maternal germ lineage. We found that the beginning of GRIF-1 protein synthesis is restricted to the initial PGC P4 and dependent on GLD-2, via posttranscriptional regulatory information encoded in the 3′UTR of grif-1 mRNA. Consistent with this, other studies reported that grif-1 mRNA is maternally loaded into the embryo where it is likely subject to prior posttranscriptional repression as expression of epitope-tagged Y51F10.2 (aka GRIF-1) begins in P4 (11, 52). Additional large-scale studies are in support of GLD-2–mediated positive expression of GRIF-1 and reported that a longer poly(A) tail of grif-1 mRNA in the early P lineage depends on gld-2 activity (13, 53). This posttranscriptional regulation may also depend on POS (posterior segregation)-1, a P lineage–enriched maternal mRNA regulator (6, 11, 53). Whether GLD-2 cytoPAP or POS-1 prolongs RNA stability to eventually enrich grif-1 mRNA in P4 or promotes also derepression and translation of grif-1 mRNA must be clarified in further experiments. Irrespective of these mechanistic details, maternal expression of GRIF-1 is a consequence of de novo translation, likely depending on GLD-2 cytoPAP activity. As part of the MZT in PGCs, this sets a feed forward mechanism in motion that terminates the positively contributing regulatory potential of the maternally inherited cytoPAP machinery. However, other protein targets of GRIF-1 may also be likely, and an even broader effect on the MZT can be envisioned. The narrow window of GRIF-1 expression in PGCs argues that GRIF-1 is a particularly suited factor to promote the MZT and to provide a molecular reset for the developing germ line.
GRIF-1 is essential for transgenerational fertility, and its activity prevents germline mortality. We provide evidence that prolonged cytoPAP activity contributes to this phenomenon, but it is also likely that other yet to be identified targets of GRIF-1 may play a role in germline immortality. This may include potential cross-talk with other RNA-related pathways that prevent germline mortality, such as nuclear RNAi (39), chromatin regulation via Piwi-interacting RNA machinery (54, 55), or endonuclease-mediated mRNA regulation (56), and will have to be investigated in future studies. As polyadenylation is the main function of cytoPAPs that typically stabilizes mRNAs and enhances their translational competency (42), our data suggest that the gene regulatory mechanisms of cytoplasmic polyadenylation must be terminated during MZT in PGCs. At least two, mutually not exclusive, scenarios for an extended capability of cytoplasmic polyadenylation can be envisioned: (i) Prolonged cytoPAP activity might either extend the half-life of the maternal transcriptome, contributing to germline sterility by overwhelming the in-parallel acting mRNA decay machinery (e.g., deadenylases), or promote continued translation of maternal regulators that may otherwise be targets of translational repression; (ii) cytoPAP perdurance may also interfere with posttranscriptional regulation of the zygotic transcriptome of PGCs. Although a mortal germline phenotype has not been reported for NOS-2, a known negative regulator of maternal mRNAs (10), double-mutant nos-2;grif-1 (a.k.a. Y51F10.2) mothers produced sterile offspring (11), implying that an extension of maternal mRNA expression may interfere with PGC specification. As little is still known about the early zygotic transcriptome of PGCs in C. elegans (10), further detailed studies will have to be conducted. In either case, our study argues that translational activators are incompatible with zygotic independence, and clearance should be completed before ZGA.
As many other animals use GLD-2–type enzymes to translationally regulate maternal mRNA gene expression programs (57–59), we envision cytoPAP clearance in PGCs as a potentially conserved mechanism to ensure transgenerational fitness in animals deploying germline preformation. In support of this speculation are several observations on a GLD-2 ortholog in Drosophila, documenting that maternal Wispy, which stabilizes germplasm mRNAs to enrich in the PGCs, is subject to proteasome-mediated turnover (5, 60, 61). Whether a TRIM-like turnover factor is involved must yet be clarified.
MATERIALS AND METHODS
Strain handling
All nematodes were handled according to standard procedures (62). Except otherwise noted, worms were grown and maintained at 20°C on nematode growth media (NGM) plates, seeded with Escherichia coli OP50. Phenotypes were analyzed 24 hours past larval stage mid-L4 at 20°C or 18 to 20 hours past mid-L4 at 25°C; RNAi-induced phenotypes and brood sizes were analyzed as stated below. Bristol N2 was used as wild type; all other strains and their genotypes are listed in table S1. Double and triple mutants, including those containing ced-4(n1162), were generated by standard crossing schemes. Genotyping primers are listed in table S2. gld-2(q497) null mutant animals are sterile and kept over the balancer hT2 [bli-4(e937) let-?(q782) qIs48] (I;III) [also known as hT2g(I;III)]. To avoid accumulation of transgenerational phenotypes, all grif-1 alleles were also maintained over hT2g(I;III), giving rise to nonmarked grif-1(−/−) segregants. glp-4(bn2ts) is temperature sensitive and was maintained at 16°C. Raised at restrictive temperature (25°C), glp-4 adults contain only a few germ cells and therefore were used in Western blot experiment to test for the somatic expression of GRIF-1 protein (Fig. 2C).
Mutants, genomic engineering, and transgenesis
The ethyl methanesulfonate (EMS)–induced grif-1(ok1610) allele was obtained from the C. elegans deletion consortium, immediately balanced, and backcrossed nine times. It lacks 1016 base pairs, affecting exon 4 and intron 4. cDNA analysis suggests that due to the deleted sequence in ok1610, a cryptic exon-intron junction gets used, and several splice variants of the transcript are produced; the predominant transcripts are depicted in the protein isoform given in fig. S2B. We also found evidence of a splice version that produces a C-terminally truncated GRIF-1 protein isoform, lacking amino acids 205 until 305, that contains 12 additional unrelated amino acids due to a frameshift. All other genomic mutations at the grif-1 locus were generated using the CRISPR-Cas9 RNP technique (63). Single guide RNAs were in vitro transcribed from polymerase chain reaction (PCR) products of pDR274-encoded sequences (64), using standard techniques; its sequences including their single-stranded DNA repair templates are listed in table S3. grif-1(ef32) contains a large out-of-frame deletion with additional base pairs, presumably added via the repair process, while grif-1(ef35) contains a premature stop codon almost immediately downstream of the cognate start codon; a stop codon was additionally inserted at the start codon of ok1610 to generate the null allele, grif-1(ok1610ef40). To generate transgenes (TGs) of choice, the Mos1-mediated Single Copy Insertion (MosSCI) genome engineering technique was used at linkage group (LG) II (65), and at least three independent lines were recovered; two randomly chosen ones were crossed into given mutant backgrounds (see also table S1). Constructs for MosSCI transgenesis were assembled using MultiSite Gateway Cloning (Invitrogen) of pCFJ150 (65). gld-2 open reading frame (ORF) was generated from reengineered gld-2 locus optimized for easier cloning and single-copy insertion transgenesis, using a cDNA sequence encoding for the N-terminal half of GLD-2 protein and fused with genomic sequence encoding for the C-terminal half of GLD-2 protein to generate a sequence that encodes for FL GLD-2 protein (graphically depicted in Fig. 6A). Shorter constructs deleting the N-terminal coding sequences were generated by PCR, using the FL sequence as template before cloning into pCFJ150 as described above. Similarly, FL grif-1 ORF was generated from reengineered grif-1 locus: Its first three exons were amplified from genomic sequence, and the last two exons were taken from cDNA; the two fragments were fused by PCR to generate partially intronized grif-1 TGs (graphically depicted in figs. S5A and S6A). The C43A mutation was introduced via site-directed mutagenesis PCR.
RNAi experiments
RNAi constructs targeting grif-1 and lgg-1 were generated from PCR-amplified cDNA with primers listed in table S2, cloned into pL4440 vector, and sequenced for verification. Feeding RNAi regimes to knock down grif-1 and lgg-1 were started at the L1 stage onward and performed according to (66). Their F1 embryos and those from in-parallel fed empty pL4440 vector control animals were extruded and used for immunostaining analyses. Effectiveness of grif-1 RNAi was always assessed by immunostaining, using GRIF-1–specific antibodies; effectiveness of lgg-1 was ascertained by monitoring known turnover defects of PGL-1 in somatic cells of lgg-1 RNAi embryos (22). Only batches of embryos that passed these quality control measures were further analyzed.
RNAi constructs targeting pas-5 and pbs-6 were described before (26). However, to ameliorate the embryonic lethal effect of pas-5 or pbs-6 RNAi treatment, a partial RNAi knockdown regime was devised that allowed a substantial number of embryos to complete embryogenesis. HT115(DE3) bacteria expressing double-stranded RNA to pas-5 and pbs-6 were diluted with equal amounts of control bacteria containing empty pL4440 plasmid. As RNAi exposure of mothers beyond 18 hours led to severe embryonic lethality phenotypes in their progeny, earlier time windows were selected for analysis. Embryos produced at 15 hours of feeding were used for immunostainings (as given in Fig. 1C and fig. S1D; quantitation in fig. S1, E and F), while those laid between 14 and 16 hours were analyzed for embryonic lethality and F1 fertility (as given in fig. S1, B and C). Once the experimental parameters were established, at least three consecutive repetitions were performed. The quantitation in fig. S1 (E and F) reflects these repetitions and their variations. For quantifying embryonic lethality and F1 fertility (fig. S1, B and C), a total of 100 mothers were individually moved to OP50 NGM plates at the 14-hour time point and allowed to lay embryos for 2 hours before all mothers were removed. Sired F1 embryos were analyzed after 24 hours. Hatched L1 larvae were counted as living progeny, while unhatched embryos were counted as dead embryos. Overall, we used both embryonic lethality and extra PGC phenotype as a positive control for the effectiveness of pas-5 and pbs-6 RNAi in early embryos (22) and as quality control criteria for analyzing embryos by immunofluorescence.
The pL4440 RNAi clone targeting gld-2 comprises the entire ORF of 3339 nucleotides. Similar to proteasome factors, gld-2 is important for worm fertility and early embryogenesis. Therefore, to circumvent these phenotypes, a short RNAi regime was performed: Adult worms were selected and exposed to either control or gld-2 RNAi feeding bacteria for 15 hours, and their embryos were analyzed within the 15- to 20-hour window.
Determination of brood size
For all brood size assessments, individual L4 hermaphrodites were transferred onto separate plates. Every 24 hours, they were moved to new plates. Before counting, the sired embryos from which the mother had just been removed were given another 24 hours to hatch and develop; unhatched embryos were counted as dead embryos, and hatched progenies were counted as living progenies. A sum of the two across 4 days was taken as the total brood size of the mother.
To analyze transgenerational fecundity of animals expressing gld-2 transgenes [gld-2(0)/hT2g I; gld-2 TG II] or grif-1 mutants with or without rescue transgene, F1 homozygote nongreen progenies of heterozygous animals were selected as L4 and shifted to 25°C. Brood sizes of these F1 mothers were determined as above. The F2 progenies of these F1 mothers were continuously maintained at 25°C until F3, and brood size determination was repeated as above for F3 mothers. Fecundity of F1 and F3 mothers was compared. Wild type was included as a control throughout these experiments. Box plot in fig. S7C is a combination of two independent lines generated for individual gld-2 transgenes, separately shown in fig. S7C to the left. Similarly, box plot in fig. S7E is a combination of two independent lines generated for individual grif-1 transgenes, separately shown in fig. S6B.
Sterility analysis
To determine the percentage sterility in animals ectopically expressing either wild-type or RING finger mutant GRIF-1 shown in fig. S5C, L4 worms (P0) were shifted and allowed to acclimatize for one generation at 25°C. Progenies were selected as L4 (F1) and shifted to new plates every 12 hours. The sired F2 progenies were then analyzed for sterility at adulthood. F2 adults containing no embryo in the uterus were scored as sterile, while those containing at least a single embryo in the uterus were scored as fertile.
Mrt phenotype analysis and Nomarski microscopy
To assess a mortal germline (Mrt) phenotype, the procedure described in (38) was performed with minor modifications. Six L4-staged homozygote grif-1 mutants, which were F1 progenies of heterozygotes, were randomly selected and shifted to 25°C. From this founder population, six L4 progenies were randomly selected and together passed onto a new plate at 25°C. This was repeated for successive generations until complete sterility. A line was regarded sterile at generation “n” when the total number of progenies sired by the six mothers was between 0 and 12. The original method passed six L1s at every successive generation (38); however, to avoid picking males that were occasionally produced by many mutant genotypes used in our analysis at a relatively higher rate than by wild type, L4 animals were randomly selected instead. Wild-type animals were treated the same way and never led to the termination of an analyzed line due to sterility. At 20°C, two L4 grif-1 mutants were maintained across generations compared to six maintained at 25°C.
To analyze gonadal defects associated with Mrt phenotype, sterile grif-1 adults (L4 + 20 hours) were analyzed from populations containing a substantial number of sterile animals. This means that during experiments to assay for Mrt, L4 larvae were picked from plates that were passed a generation or two before complete sterility. Since a prevalent number of mothers were sterile on these plates and fertile mothers laid a strongly reduced number of eggs, worms were preserved without starvation on a plate for up to a week later for analysis.
For live imaging, animals were mounted on 2% agar pads in a drop of M9 solution containing 10 to 25 mM sodium azide, covered by a glass coverslip, and observed with an Imager M1 microscope (Zeiss), equipped with a plan-apo 63×/1.40 oil iris objective. Individual images were acquired with a MRm camera (Zeiss) controlled by AxioVision Software (Zeiss) and stitched together with Fiji (ImageJ). Assembled images were processed in Photoshop CS6 (Adobe) and labeled in Illustrator CS6 (Adobe).
Y2H work
Using FL GLD-2 as bait, a plate-based Y2H screen was performed as described before (67), transforming yeast with a oligo(dT)-primed cDNA library from whole adults (Dualsystems Biotech), and plated on synthetic dropout media containing 5 mM 3-amino-1,2,4-triazole. For plate-based beta-galactosidase testing (20), FL proteins or truncations thereof were cloned into vectors (Clontech) containing either LexA DNA binding domain (pBTM116 derivatives; pLexKn2) or GAL4 activation domain (pACT2 AD derivatives). All fusion proteins were analyzed by Western blotting to verify their expression after generating protein extracts according to (20). Given colonies are representatives of several independently transformed yeast L40 batches to ensure reproducibility.
Antibody generation
Monoclonal α-GRIF-1 hybridoma supernatants were raised with the help of the antibody facility of the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG; Dresden). Mice were immunized with heterologously expressed and affinity-purified FL GRIF-1 fusion proteins, containing 6xHis::SUMO::GRIF-1::GFP, originally produced in BL21 bacteria and purified to homogeneity under denaturing conditions using nickel-affinity columns. Specific immunoreactivity was established in Western blotting and immunostaining experiments using yeast-expressed proteins and worm material. The anti–GLD-2 affinity-purified serum rt317 was generated by immunizing rats with a GLD-2(aa959–1113) glutathione S-transferase–fusion protein expressed and purified as described in (42).
Immunostaining analysis of embryos
To isolate embryos for immunofluorescence experiments, typically 35 gravid mothers per slide were cut open around the vulva to release their embryos on a coverslip into M9 buffer. Once absorbed to a poly-lysine–coated glass slide, they were immediately placed onto a dry ice–cooled metal plate for a minimum of 15 min to freeze-crack the egg shell (68, 69). After coverslip removal, they were fixed for 10 min in dry ice–cold 100% methanol and permeabilized for 5 min in dry ice–cold 100% acetone. Samples were rehydrated in 1× phosphate-buffered saline (PBS) for 5 min and blocked with PBSB (PBS + 0.5% bovine serum albumin) for 30 min at room temperature (RT) or longer at 4°C in a humidified chamber. Primary antibodies were applied overnight at 4°C and secondary antibodies for 2 hours at RT in PBSB. Samples were washed in between and after the two antibody incubation steps three times; last, they were mounted in a drop of Vectashield antifade medium (Vector Labs), dried in the dark for about 10 min, and sealed on three sides with transparent nail polish.
4,6-Diamidino-2-phenylindole was always included in the final washing steps to reveal the nuclear compartment and assess the developmental stages of embryos. In most immunofluorescent experiments, PGL-1 intensity was used as a positive control for correct tissue penetration and cellular organization. Wide-field images were acquired and processed as described for Nomarski microscopy. A maximum intensity projection of several planes around the given cells in question was chosen for display purposes of all fluorescent images. Laser scanning confocal microscopy was performed on a Zeiss LSM 700 at MPI-CBG (CZ5) for the wild-type embryos given in fig. S3B.
The following primary antibodies were used: α-GRIF-1 (supernatant mAb CU35) at 1:10 (this study), α-GLD-2 (rt317) at 1:100 (this study), α-GLD-2 (rb184) at 1:100 (42), α-GLD-3 (rt236) at 1:100 (20), α-PIE-1 (moP45G2) at 1:100 (9), and α-PGL-1 at 1:100 (17). Secondary antibodies (Jackson ImmunoResearch) were supplied by Dianova (BIOZOL) and used at a final dilution of 1:2000: Cy5 (magenta label)–, Cy3 (red label)–, or fluorescein isothiocyanate (green label)–conjugated AffiniPure donkey anti-rabbit, anti-mouse, or anti–guinea pig immunoglobulin G antibodies. All immunostainings were typically performed >3 times on >>100 mothers per independent biological material, and only quality-controlled embryos were included in the analysis (see also explanations given for the RNAi experiments); in case of less than 100% penetrance, quantitations are explicitly given.
Western blotting and coIP experiments
For protein extracts of adult worms, individual animals were collected by hand, boiled in 2× SDS sample buffer before loading, and separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), followed by Western blotting. Individual larval stages were generated by washing of synchronized animals in bulk from NGM plates. To generate embryo extracts, embryos were boiled and dissolved in 2× SDS sample buffer, after alkaline hypochlorite treatment of gravid adults. To detect GRIF-1 in these samples, approximately 8000 to 10,000 embryos were loaded in a single pocket/lane of an SDS-PAGE gel. Extracts of yeast were made according to (20).The following primary antibodies were used in Western blotting experiments: α-GRIF-1 (typically supernatant mAb CU35 or, where noted, mAb CV44) at 1:10 (this study), α-CCF-1 (G25-1) at 1:1000 (35), α-PIE-1 (cN19) at 1:1000 (Santa Cruz Biotechnology, sc-9245), α-GLD-2 (A4-4) at 1:1000 (70), α-GLD-3 (rat236) at 1:1000 (20), α-LexA (2-12) at 1:3000 (Santa Cruz Biotechnology, sc-7544), α-alpha-tubulin (Merck, B-5-1-2) at the range of 1:50,000 to 1:200,000 (Sigma-Aldrich, T5168), and α-DYN-1 at 1:1000 (DSHB, University of Iowa). Except for α-GRIF-1 (mAb CU35), which was diluted in 5% milk for primary antibody incubation to reduce background signals, all other antibodies were diluted in 0.5% milk in PBST (PBS+0.1% Tween 20). Horseradish peroxidase–conjugated anti-mouse, anti-rabbit, or anti–guinea pig secondary antibodies (Jackson ImmunoResearch) were applied at a dilution of either 1:20,000 or 1:40,000 in 0.5% milk in PBST and detected with enhanced chemiluminescence substrate (Promega, W1001).
For coIP experiments, embryo extracts were generated according to (71), with slight modifications. Gravid mothers were dissolved in alkaline hypochlorite solution to release embryos, which were shock-frozen in liquid nitrogen and pulverized at 30 Hz to embryo powder using a MR301 bead mill (Retsch). For the coIP procedure, a minimum of 2 million embryos must be used to generate a concentrated extract of approximately 600 to 800 μl divided as input for a monoclonal α-GRIF-1 antibody mix (mAbs CU33, CU35, CV31, and CV44) and anti–red fluorescent protein (RFP; 125B42-4), a gift from the antibody facility of MPI-CBG, coupled to Protein G Dynabeads (Invitrogen). Immunoprecipitated materials were resuspended in SDS loading buffer before boiling (=eluate) and analyzed in Western blotting experiments as described above. A similar procedure was performed for GLD-2 coIP experiments. Young adults were frozen in liquid nitrogen without alkaline hypochlorite treatment (71). GLD-2 pulldown was carried out with monoclonal α-GLD-2 (mAb A4-4) (70) and α-HA (12CA5), a gift from the antibody facility of MPI-CBG, as a background control. All coIPs were reproduced at least two times with different biological material (>2 experimental setups).
Protein sequence analysis of grif-1 and gld-2 gene products
At the primary sequence level, the TRIM32 protein is GRIF-1’s best hit in BLAST searches directed against the human genome. TRIM2 and TRIM3, two other TRIM32 subfamily members, have lower sequence similarity to GRIF-1 than TRIM32. Therefore, the amino acid sequence of GRIF-1 and its orthologs were retrieved from UniProt to generate multiple sequence alignments: GRIF-1 (UniProt entry Q8WTJ8), TRIM32 from zebrafish (Dr, UniProt entry A8WGA9), mouse (Mm, UniProt entry Q8CH72), and human (Hs, UniProt entry Q13049). Multiple sequence alignment of the entire sequences of the proteins was carried out with the Clustal Omega program (www.ebi.ac.uk/Tools/msa/clustalo/). The alignment identified both RING and B-box domains of GRIF-1 and is displayed in Fig. 2B.
To predict the presence of a potential CC domain in GRIF-1, its amino acid sequence was used as an input for the program COILs (version 2.2). COILs identified two putative CC domains in the C terminus of GRIF-1, displayed in Fig. 2A and fig. S2B. To predict the extent of intrinsic disorder within GLD-2 protein, FoldIndex software (https://fold.proteopedia.org/cgi-bin/findex) was fed with GLD-2 amino acid sequence, and its output is displayed in Fig. 3D (72).
Statistical analysis
Statistical data are represented with a box plot to display percentile distribution or a column plot to display mean. Statistical difference between datasets was calculated with two-tailed Student’s t test with unequal variance; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; n.s., not significant.
Acknowledgments
We thank A. Bhargava for initial protein interaction screen in yeast and S. Vogt for the help with bacterial protein expression constructs, M. Rockstroh and K. Patsias for contribution to worm strain generation, and the antibody facility of MPI-CBG (Dresden) with help to generate antibodies. We thank D. Rudel, E. Wahle, and members of the laboratory for providing input to the manuscript. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding: This work was funded by the German Research Foundation grant EC369/2-3 (to C.R.E.), German Research Foundation grant EC369/6-1 (to C.R.E.), German Research Foundation grant RTG1591 (to C.R.E.), German Research Foundation grant RTG2467 391498659 (to C.R.E.), and German Research Foundation Instrumentation grant INST-271/401-1FUGG, cofinanced by the state of Sachsen-Anhalt (to C.R.E.).
Author contributions: Experimental design: T.D.O. and C.R.E. Laboratory work: T.D.O. Data analysis: T.D.O. and C.R.E. Manuscript preparation: T.D.O. and C.R.E.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S3
REFERENCES AND NOTES
- 1.Strome S., Updike D., Specifying and protecting germ cell fate. Nat. Rev. Mol. Cell Biol. 16, 406–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vastenhouw N. L., Cao W. X., Lipshitz H. D., The maternal-to-zygotic transition revisited. Development 146, dev161471 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Siddiqui N. U., Li X., Luo H., Karaiskakis A., Hou H., Kislinger T., Westwood J. T., Morris Q., Lipshitz H. D., Genome-wide analysis of the maternal-to-zygotic transition in Drosophila primordial germ cells. Genome Biol. 13, R11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voronina E., Seydoux G., Sassone-Corsi P., Nagamori I., RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 3, a002774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao W. X., Kabelitz S., Gupta M., Yeung E., Lin S., Rammelt C., Ihling C., Pekovic F., Low T. C. H., Siddiqui N. U., Cheng M. H. K., Angers S., Smibert C. A., Wühr M., Wahle E., Lipshitz H. D., Precise temporal regulation of post-transcriptional repressors is required for an orderly drosophila maternal-to-zygotic transition. Cell Rep. 31, 107783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabara H., Hill R. J., Mello C. C., Priess J. R., Kohara Y., Pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126, 1–11 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Subramaniam K., Seydoux G., Nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126, 4861–4871 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Rouget C., Papin C., Boureux A., Meunier A.-C., Franco B., Robine N., Lai E. C., Pelisson A., Simonelig M., Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467, 1128–1132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mello C. C., Schubert C., Draper B., Zhang W., Lobel R., Priess J. R., The PIE-1 protein and germline specification in C. elegans embryos. Nature 382, 710–712 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Lee C.-Y. S., Lu T., Seydoux G., Nanos promotes epigenetic reprograming of the germline by down-regulation of the THAP transcription factor LIN-15B. eLife 6, e30201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C.-Y. S., Putnam A., Lu T., He S. X., Ouyang J. P. T., Seydoux G., Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. eLife 9, e52896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakel K., Bonneau F., Eckmann C. R., Conti E., Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3. Proc. Natl. Acad. Sci. U.S.A. 112, 8614–8619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nousch M., Yeroslaviz A., Habermann B., Eckmann C. R., The cytoplasmic poly(A) polymerases GLD-2 and GLD-4 promote general gene expression via distinct mechanisms. Nucleic Acids Res. 42, 11622–11633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., Eckmann C. R., Kadyk L. C., Wickens M., Kimble J., A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419, 312–316 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Weill L., Belloc E., Bava F. A., Mendez R., Translational control by changes in poly(A) tail length: Recycling mRNAs. Nat. Struct. Mol. Biol. 19, 577–585 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Nousch M., Yeroslaviz A., Eckmann C. R., Stage-specific combinations of opposing poly(A) modifying enzymes guide gene expression during early oogenesis. Nucleic Acids Res. 47, 10881–10893 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawasaki I., Shim Y. H., Kirchner J., Kaminker J., Wood W. B., Strome S., PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Saha S., Weber C. A., Nousch M., Adame-Arana O., Hoege C., Hein M. Y., Osborne-Nishimura E., Mahamid J., Jahnel M., Jawerth L., Pozniakovski A., Eckmann C. R., Jülicher F., Hyman A. A., Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell 166, 1572–1584.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spencer W. C., Zeller G., Watson J. D., Henz S. R., Watkins K. L., McWhirter R. D., Petersen S., Sreedharan V. T., Widmer C., Jo J., Reinke V., Petrella L., Strome S., Von Stetina S. E., Katz M., Shaham S., Rätsch G., Miller D. M. III, A spatial and temporal map of C. elegans gene expression. Genome Res. 21, 325–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckmann C. R., Kraemer B., Wickens M., Kimble J., GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev. Cell 3, 697–710 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Tsukamoto S., Kuma A., Murakami M., Kishi C., Yamamoto A., Mizushima N., Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Yan L., Zhou Z., Yang P., Tian E., Zhang K., Zhao Y., Li Z., Song B., Han J., Miao L., Zhang H., SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 136, 308–321 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Higuchi C., Shimizu N., Shin S.-W., Morita K., Nagai K., Anzai M., Kato H., Mitani T., Yamagata K., Hosoi Y., Miyamoto K., Matsumoto K., Ubiquitin-proteasome system modulates zygotic genome activation in early mouse embryos and influences full-term development. J. Reprod. Dev. 64, 65–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeRenzo C., Reese K. J., Seydoux G., Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424, 685–689 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zavortink M., Rutt L. N., Dzitoyeva S., Henriksen J. C., Barrington C., Bilodeau D. Y., Wang M., Chen X. X. L., Rissland O. S., The E2 Marie Kondo and the CTLH E3 ligase clear deposited RNA binding proteins during the maternal-to-zygotic transition. eLife 9, e53889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kisielnicka E., Minasaki R., Eckmann C. R., MAPK signaling couples SCF-mediated degradation of translational regulators to oocyte meiotic progression. Proc. Natl. Acad. Sci. U.S.A. 115, E2772–E2781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi M., Iwasaki H., Inoue H., Takahashi K., Reverse genetic analysis of the Caenorhabditis elegans 26S proteasome subunits by RNA interference. Biol. Chem. 383, 1263–1266 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Wang H. T., Hur S., Substrate recognition by TRIM and TRIM-like proteins in innate immunity. Semin. Cell Dev. Biol. 111, 76–85 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tocchini C., Ciosk R., TRIM-NHL proteins in development and disease. Semin. Cell Dev. Biol. 47–48, 52–59 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Connacher R. P., Goldstrohm A. C., Molecular and biological functions of TRIM-NHL RNA-binding proteins. Wiley Interdiscip. Rev. RNA 12, e1620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., Žídek A., Green T., Tunyasuvunakool K., Petersen S., Jumper J., Clancy E., Green R., Vora A., Lutfi M., Figurnov M., Cowie A., Hobbs N., Kohli P., Kleywegt G., Birney E., Hassabis D., Velankar S., AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichimura T., Taoka M., Shoji I., Kato H., Sato T., Hatakeyama S., Isobe T., Hachiya N., 14-3-3 proteins sequester a pool of soluble TRIM32 ubiquitin ligase to repress autoubiquitylation and cytoplasmic body formation. J. Cell Sci. 126, 2014–2026 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Fiorentini F., Esposito D., Rittinger K., Does it take two to tango? RING domain self-association and activity in TRIM E3 ubiquitin ligases. Biochem. Soc. Trans. 48, 2615–2624 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiss L., Clift D., Renner N., Neuhaus D., James L. C., RING domains act as both substrate and enzyme in a catalytic arrangement to drive self-anchored ubiquitination. Nat. Commun. 12, 1220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nousch M., Techritz N., Hampel D., Millonigg S., Eckmann C. R., The Ccr4-Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. J. Cell Sci. 126, 4274–4285 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Koliopoulos M. G., Esposito D., Christodoulou E., Taylor I. A., Rittinger K., Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 35, 1204–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadyk L. C., Kimble J., Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125, 1803–1813 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Ahmed S., Hodgkin J., MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403, 159–164 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Buckley B. A., Burkhart K. B., Gu S. G., Spracklin G., Kershner A., Fritz H., Kimble J., Fire A., Kennedy S., A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barucci G., Cornes E., Singh M., Li B., Ugolini M., Samolygo A., Didier C., Dingli F., Loew D., Quarato P., Cecere G., Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat. Cell Biol. 22, 235–245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rybarska A., Harterink M., Jedamzik B., Kupinski A. P., Schmid M., Eckmann C. R., GLS-1, a novel P granule component, modulates a network of conserved RNA regulators to influence germ cell fate decisions. PLOS Genet. 5, e1000494 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nousch M., Minasaki R., Eckmann C. R., Polyadenylation is the key aspect of GLD-2 function in C. elegans. RNA 23, 1180–1187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng N., Shabek N., Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Verlhac M.-H., Terret M.-E., Pintard L., Control of the oocyte-to-embryo transition by the ubiquitin-proteolytic system in mouse and C. elegans. Curr. Opin. Cell Biol. 22, 758–763 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Zhou S., Guo Y., Sun H., Liu L., Yao L., Liu C., He Y., Cao S., Zhou C., Li M., Cao Y., Wang C., Lu Q., Li W., Guo X., Huo R., Maternal RNF114-mediated target substrate degradation regulates zygotic genome activation in mouse embryos. Development 148, dev199426 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Yang Y., Zhou C., Wang Y., Liu W., Liu C., Wang L., Liu Y., Shang Y., Li M., Zhou S., Wang Y., Zeng W., Zhou J., Huo R., Li W., The E3 ubiquitin ligase RNF114 and TAB1 degradation are required for maternal-to-zygotic transition. EMBO Rep. 18, 205–216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esposito D., Koliopoulos M. G., Rittinger K., Structural determinants of TRIM protein function. Biochem. Soc. Trans. 45, 183–191 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Hatakeyama S., TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci. 42, 297–311 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Kiss L., Zeng J., Dickson C. F., Mallery D. L., Yang J.-C., McLaughlin S. H., Boland A., Neuhaus D., James L. C., A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases. Nat. Commun. 10, 4502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim K. W., Wilson T. L., Kimble J., GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc. Natl. Acad. Sci. U.S.A. 107, 17445–17450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakel K., Bonneau F., Basquin C., Habermann B., Eckmann C. R., Conti E., Structural basis for the antagonistic roles of RNP-8 and GLD-3 in GLD-2 poly(A)-polymerase activity. RNA 22, 1139–1145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tintori S. C., Osborne Nishimura E., Golden P., Lieb J. D., Goldstein B., A transcriptional lineage of the early C. elegans embryo. Dev. Cell 38, 430–444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elewa A., Shirayama M., Kaymak E., Harrison P. F., Powell D. R., Du Z., Chute C. D., Woolf H., Yi D., Ishidate T., Srinivasan J., Bao Z., Beilharz T. H., Ryder S. P., Mello C. C., POS-1 promotes endo-mesoderm development by inhibiting the cytoplasmic polyadenylation of neg-1 mRNA. Dev. Cell 34, 108–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashe A., Sapetschnig A., Weick E.-M., Mitchell J., Bagijn M. P., Cording A. C., Doebley A.-L., Goldstein L. D., Lehrbach N. J., Pen J. L., Pintacuda G., Sakaguchi A., Sarkies P., Ahmed S., Miska E. A., piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heestand B., Simon M., Frenk S., Titov D., Ahmed S., Transgenerational sterility of piwi mutants represents a dynamic form of adult reproductive diapause. Cell Rep. 23, 156–171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qi W., Gromoff E. D. V., Xu F., Zhao Q., Yang W., Pfeifer D., Maier W., Long L., Baumeister R., The secreted endoribonuclease ENDU-2 from the soma protects germline immortality in C. elegans. Nat. Commun. 12, 1262 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winata C. L., Łapiński M., Pryszcz L., Vaz C., Ismail M. H. B., Nama S., Hajan H. S., Lee S. G. P., Korzh V., Sampath P., Tanavde V., Mathavan S., Cytoplasmic polyadenylation-mediated translational control of maternal mRNAs directs maternal-to-zygotic transition. Development 145, dev159566 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Lim J., Lee M., Son A., Chang H., Kim V. N., mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev. 30, 1671–1682 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui J., Sartain C. V., Pleiss J. A., Wolfner M. F., Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Dev. Biol. 383, 121–131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dufourt J., Bontonou G., Chartier A., Jahan C., Meunier A.-C., Pierson S., Harrison P. F., Papin C., Beilharz T. H., Simonelig M., piRNAs and Aubergine cooperate with Wispy poly(A) polymerase to stabilize mRNAs in the germ plasm. Nat. Commun. 8, 1305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sysoev V. O., Fischer B., Frese C. K., Gupta I., Krijgsveld J., Hentze M. W., Castello A., Ephrussi A., Global changes of the RNA-bound proteome during the maternal-to-zygotic transition in Drosophila. Nat. Commun. 7, 12128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S., Fire A. Z., Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeiser E., Frokjaer-Jensen C., Jorgensen E., Ahringer J., MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLOS ONE 6, e20082 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J., Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kraemer B., Crittenden S., Gallegos M., Moulder G., Barstead R., Kimble J., Wickens M., NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9, 1009–1018 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Strome S., Wood W. B., Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 79, 1558–1562 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duerr J. S., Antibody staining in C. elegans using “freeze-cracking”. J. Vis. Exp. , 50664 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Millonigg S., Minasaki R., Nousch M., Novak J., Eckmann C. R., GLD-4-mediated translational activation regulates the size of the proliferative germ cell pool in the adult C. elegans germ line. PLOS Genet. 10, e1004647 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jedamzik B., Eckmann C. R., Analysis of in vivo protein complexes by coimmunoprecipitation from Caenorhabditis elegans. Cold Spring Harb. Protoc. 2009, pdb prot5299 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Prilusky J., Felder C. E., Zeev-Ben-Mordehai T., Rydberg E. H., Man O., Beckmann J. S., Silman I., Sussman J. L., FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21, 3435–3438 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S3