Abstract
Filopodia are actin-rich membrane protrusions essential for cell morphogenesis, motility, and cancer invasion. How cells control filopodium initiation on the plasma membrane remains elusive. We performed experiments in cellulo, in vitro, and in silico to unravel the mechanism of filopodium initiation driven by the membrane curvature sensor IRSp53 (insulin receptor substrate protein of 53 kDa). We showed that full-length IRSp53 self-assembles into clusters on membranes depending on PIP2. Using well-controlled in vitro reconstitution systems, we demonstrated that IRSp53 clusters recruit the actin polymerase VASP (vasodilator-stimulated phosphoprotein) to assemble actin filaments locally on membranes, leading to the generation of actin-filled membrane protrusions reminiscent of filopodia. By pulling membrane nanotubes from live cells, we observed that IRSp53 can only be enriched and trigger actin assembly in nanotubes at highly dynamic membrane regions. Our work supports a regulation mechanism of IRSp53 in its attributes of curvature sensation and partner recruitment to ensure a precise spatial-temporal control of filopodium initiation.
Curvature-sensing protein IRSp53 synergizes with actin elongator VASP to control actin-filled membrane protrusion formation.
INTRODUCTION
Plasma membrane shaping relies on a precisely controlled coupling of the plasma membrane and the actin cytoskeleton (1–3). A prominent example is filopodia: thin, finger-like membrane protrusions with a typical diameter of 100 to 300 nm, which can extend from cell edges with lengths of the order of 10 μm (4, 5). Filopodia are filled with parallel actin filaments bundled by fascin (typically 10 to 30 filaments) (6, 7). Cells use filopodia to explore and sense their environment (8). Thus, filopodia are critical in numerous cellular processes, including polarized cell migration and adhesion, and in the tissue environment, embryogenesis, cancer invasion, and cell-cell communication (2, 4, 5). Filopodium formation uses different mechanisms involving different sets of actin regulatory proteins (4, 5, 9). So far, two distinct mechanisms of filopodium generation have been proposed: the convergent elongation model relying on the reorganization of the preexisting Arp2/3 complex–mediated branched actin network in lamellipodia (10–12) and the tip nucleation model in which de novo actin assembly is triggered by formin family actin nucleators (4). The two mechanisms are not necessarily mutually exclusive and are most likely cell type dependent (4, 13–15). Moreover, it is likely that other distinct mechanisms remain to be unraveled (13). Notably, recent cell biology studies proposed another filopodium generation mechanism in which the membrane curvature–sensing protein IRSp53 (insulin receptor substrate protein of 53 kD) forms local clusters on the plasma membrane to recruit the actin polymerase VASP (vasodilator-stimulated phosphoprotein), which promotes actin filament elongation at the onset of filopodium initiation (16–18). However, it remains poorly understood how cells control precisely when and where to trigger actin assembly on the plasma membrane to initiate filopodia (4, 5, 9).
IRSp53 [also known as BAIAP2 (BAR/IMD domain containing adaptor protein 2)], an inverse Bin-Amphiphysin-Rvs161/167 (I-BAR) domain protein, is a crucial player in coordinating actin assembly and membrane dynamics in filopodium formation (16–23). IRSp53 features an N-terminal I-BAR domain [also known as IMD (IRSp53-missing-in-metastasis (MIM) homology domain)], followed by a CRIB-PR (Cdc42/Rac interactive binding–proline rich) domain, and a canonical SH3 (Src homology 3) domain. Purified I-BAR domains spontaneously assemble into crescent-shaped dimers (23) that can bind to negatively charged lipids such as PS (phosphatidylserine) and PI(4,5)P2 [hereafter termed PIP2 (phosphatidylinositol-4,5-bisphosphate)] (24, 25). The SH3 domain of IRSp53 allows it to interact with many actin regulators, such as VASP (16, 17), Eps8 (20), neural Wiskott-Aldrich syndrome protein (N-WASP) (17), WASP-family verprolin-homologous protein (WAVE) (26, 27), and mDia1 (27). It was shown that IRSp53 exhibits a closed, autoinhibited conformation due to the binding of its SH3 domain to its CRIB-PR motif (28). The most well-known activator of IRSp53 is Cdc42 (16, 17, 19, 28). In addition, PIP2 and cytoskeleton effectors, such as Eps8 and VASP, have been shown to synergize with Cdc42 in IRSp53 activation to its fully open conformation (19, 21, 28–30). It has been shown that 14-3-3 binds to phosphorylated IRSp53, keeping it in the inhibited state (21, 29, 30).
Biophysical studies in cellulo and in reconstituted systems have revealed that the I-BAR domain of IRSp53 can sense and generate similar negative membrane curvature that is found in filopodia (25, 31–33). Consistently, when overexpressing either the I-BAR domain or the full-length IRSp53 protein in cells, the generation of membrane protrusions reminiscent of filopodia was observed (16–22). Most of these I-BAR–induced protrusions contain actin filaments, albeit many of them have a low amount of actin (18). Notably, it was shown that full-length IRSp53 forms clusters on the plasma membrane that are followed by the accumulation of VASP for filopodium initiation (16). The tendency of IRSp53 to cluster was suggested by an in vitro study where, by pulling membrane tubes from giant unilamellar vesicles (GUVs) encapsulating the I-BAR domain, it was shown that at low concentrations of the I-BAR domain and low membrane curvature, the I-BAR domain phase separates along the tubes (32). By using mouse embryonic fibroblast (MEF) cells derived from IRSp53-null mice stably infected with vectors expressing IRSp53 to levels comparable to the endogenous protein, it was shown that IRSp53 is required for the recruitment and further clustering of VASP at the plasma membrane (16). Moreover, theoretical modeling has predicted how IRSp53 and actin cooperatively drive the formation of membrane protrusions (34). However, little is known about how IRSp53 by itself clusters on the plasma membrane and recruits VASP and actin. Moreover, we still lack a comprehensive mechanistic description of how IRSp53 and VASP cooperate with actin and its regulatory proteins to initiate filopodia.
VASP is a member of the Ena/VASP protein family that has been indicated to contribute to filopodium formation in cells (4, 5, 9, 10, 15, 35). VASP forms homotetramers that are weakly processive as actin polymerases (36). When forming oligomers or clusters or when actin filaments are bundled by fascin, VASP becomes more processive on the barbed ends of the actin filaments (37–40). Consistent with its actin polymerase activity, VASP clusters are part of the filopodial “tip complex,” where actin assembly occurs on the plasma membrane to elongate filopodia (10, 12, 41). Recent cell biology studies have identified protein partners involved in VASP clustering on the plasma membrane, notably IRSp53, lamellipodin, and myosin-X (16, 42, 43). While IRSp53-VASP clustering has been proposed to contribute to filopodium formation, lamellipodin- and myosin-X–dependent VASP clustering appear to be responsible for the formation of another distinct type of protrusion called microspikes (16, 19, 22, 38, 42–44). However, how VASP clusters facilitate filopodial initiation remains to be explored (4, 5).
To uncover how IRSp53, VASP, and actin cooperate to initiate filopodia, we performed experiments in cellulo, in vitro, and in silico. Using in vitro reconstitution systems, we demonstrated that on PIP2 membranes, purified full-length IRSp53 self-assembles into clusters that are crucial for the recruitment of VASP to locally trigger actin assembly, giving rise to filopodium-like membrane protrusions. Our coarse-grained (CG) simulations showed that PIP2 is key for IRSp53 clustering. Our in vitro assays revealed that fascin is not required for filopodium initiation; however, we observed that, in live cells, fascin enhances filopodium elongation and stability. Last, to unravel the regulation of IRSp53 activity in live cells, we performed an in cellulo biophysical assay in which membrane nanotubes having a membrane geometry reminiscent of cellular filopodia, but initially devoid of actin, are pulled from the cells using optical tweezers. This assay allows us to reveal the regulated activation of IRSp53 in cells by detecting two of its functions: its abilities to sense the negative membrane curvature of the pulled nanotubes and to trigger actin assembly inside the nanotubes. We found that IRSp53 is active only in membrane nanotubes that are pulled from regions of cell surfaces where the plasma membrane exhibits dynamic shape changes. Together, our results provide fundamental insights into how the curvature sensor protein IRSp53 synergizes with the actin polymerase VASP in filopodium initiation. Our findings further suggest that IRSp53 is tightly regulated in cells to have a rigorous control over filopodium initiation.
RESULTS
IRSp53 self-assembles into clusters that recruit VASP on PIP2 membranes
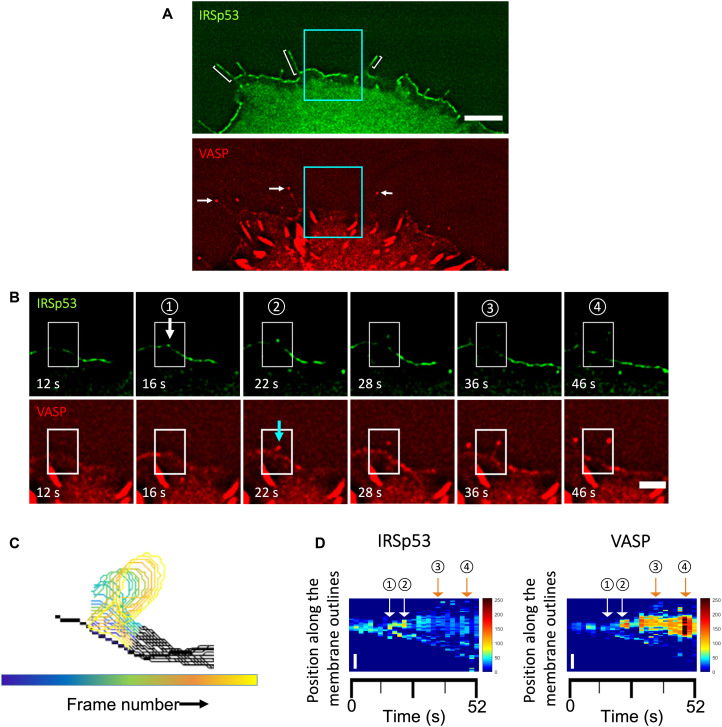
Earlier work on MEF cells derived from IRSp53-null mice provided evidence that IRSp53 is responsible for the formation of VASP clusters at the leading edge of migrating cells (16). It was shown that the accumulation of IRSp53 precedes the recruitment of VASP before the formation of filopodia (16). To further examine the roles of IRSp53 and VASP in filopodium initiation, we expressed IRSp53–enhanced green fluorescent protein (eGFP) and red fluorescent protein (RFP)–VASP in live Rat2 cells, which assemble frequent endogenous filopodia (45). By tracking their fluorescence signals at the plasma membrane, we found that IRSp53 is present along filopodia (Fig. 1A, brackets), while VASP tends to enrich at the filopodial tips (Fig. 1A, arrows). Before the appearance of filopodia, IRSp53 is enriched at the plasma membrane often as small clusters, while VASP displays quite a diffuse cytoplasmic localization with enrichment at focal adhesions (Fig. 1A). At the onset of a filopodium, the emergence of larger IRSp53 clusters occurs, and this is followed by the birth of VASP clusters (Fig. 1B and movie S1). To reveal the evolution of IRSp53 and VASP clusters at the plasma membrane before and after filopodium formation, we generated adaptive kymographs with a previously developed automated method (42). The adaptive kymographs follow the movement of the plasma membrane (Fig. 1C) and map the signals of IRSp53 and VASP into vertical lines to create space-time plots (Fig. 1D). The kymographs confirm that IRSp53 clustering at the plasma membrane precedes VASP accumulation and filopodium initiation (Fig. 1D). To quantify the colocalization of IRSp53-eGFP and RFP-VASP during filopodium formation, we used an intensity correlation analysis by following a well-established method (see Materials and Methods for details) (46). This method allows us to obtain, for each pixel, the normalized mean deviation product (nMDP) of florescent signals of IRSp53 and VASP, , termed the colocalization index, where I, , and Imax are the intensity of a corresponding pixel, the mean, and the maximum intensities of the image, respectively. For each pixel, the colocalization index indicates how the two protein signals are correlated in terms of their fluorescence intensities. The index varies from −1 to 1, with negative values for noncorrelated pixels while positive values for correlated ones. By using this method, we found that the colocalization indexes of IRSp53 and VASP were generally higher than 0, thus indicating their colocalization during filopodium formation (N = 28 filopodia, n = 2 experiments) (fig. S1, A and B). Moreover, in 46% of the tracked filopodia, we observed that the values of the IRSp53-VASP colocalization indexes globally increased during the onset of filopodium formation (fig. S1, C and D). Our results support the earlier work by Disanza et al. (16) and indicate the potential synergistic function of IRSp53 and VASP in filopodium initiation. Our data, together with earlier work (16–18), provide evidence supporting a previously proposed mechanism in which IRSp53 clustering at the plasma membrane induces VASP recruitment to locally polymerize actin on the membrane for driving protrusion formation (16–18). In this scenario, the synergistic functions of IRSp53, VASP, actin, and the filopodium-specific actin filament bundling protein, fascin, would be sufficient to drive protrusion formation.
Fig. 1. Dynamics of VASP clusters assembled from preexisting IRSp53 clusters on the plasma membrane in filopodium initiation.
(A) Wide-field fluorescence image of a representative Rat2 cell transfected with IRSp53-eGFP and RFP-VASP. Brackets indicate some filopodia where IRSp53 is present along them. White arrows indicate the same filopodia to demonstrate that VASP is enriched in their tips. Scale bar, 5 μm. (B) Time-lapse images of a filopodium formation. Images are magnifications of the indicated area (cyan boxes) in (A). The white arrow indicates the appearance of an IRSp53 cluster followed by a VASP cluster indicated by a cyan arrow at the onset of filopodium formation. White boxes indicate the selected area used to generate outlines of plasma membrane positions over time shown in (C). Scale bar, 2 μm. (C) Colored outlines of membrane positions in the region indicated by the white boxes shown in (B). Total of 27 frames and frame interval of 2 s. (D) Adaptive kymograph maps replot the detected membrane profiles in (C) in the y axis and the corresponding time points in the x axis to show the dynamics of IRSp53 (left) and VASP (right) on the plasma membrane over time. Y axis shows the membrane positions of the proteins, and the x axis shows the time (in seconds; total of 27 frames). Scale bars, 1 μm (y axis). Color maps: Low fluorescence intensity is in blue, and high fluorescence intensity is in red. Circled numbers correspond to the frames indicated in (B).
To test this hypothesis in a better-controlled experimental environment than in live cells, we developed an in vitro reconstitution system composed of GUVs as model membranes and purified IRSp53, VASP, actin, and fascin, all in full-length forms. We first assessed whether IRSp53 can self-assemble into clusters on model membranes without the presence of other proteins. We purified full-length human IRSp53 that is a constitutive dimer (23) and labeled it with Alexa Fluor 488 (AX488) dyes (AX488-IRSp53) for visualization by fluorescent confocal microscopy. We generated GUVs containing 5 mole percent (mol %) of PIP2, given that it is the key phosphatidylinositol for an IRSp53-membrane interaction and thus necessary for filopodium generation (25). We used IRSp53 at 16 nM in its dimer form (i.e., 32 nM as a monomer) to be comparable to the cellular concentration of IRSp53 (29.7 to 453 nM, in which 29.7 nM is sufficient for filopodium generation) (17). By incubating AX488-IRSp53 with PIP2-GUVs, we observed clusters of IRSp53 on the GUV membranes [72%; N = 58 GUVs (technique replicates), n = 3 independent sample preparations (biological replicates) (47)] (Fig. 2A, arrows). These clusters are reminiscent of IRSp53 clusters observed in cells at the onset of filopodium formation. In addition, we observed that a small fraction of GUVs (usually less than 10% of the whole GUV population) has inward membrane protrusions, where IRSp53 is in the interior of the protrusions (Fig. 2A, arrowhead, and fig. S2). This observation demonstrates that the full-length IRSp53 is functional such that it can generate negative membrane curvature.
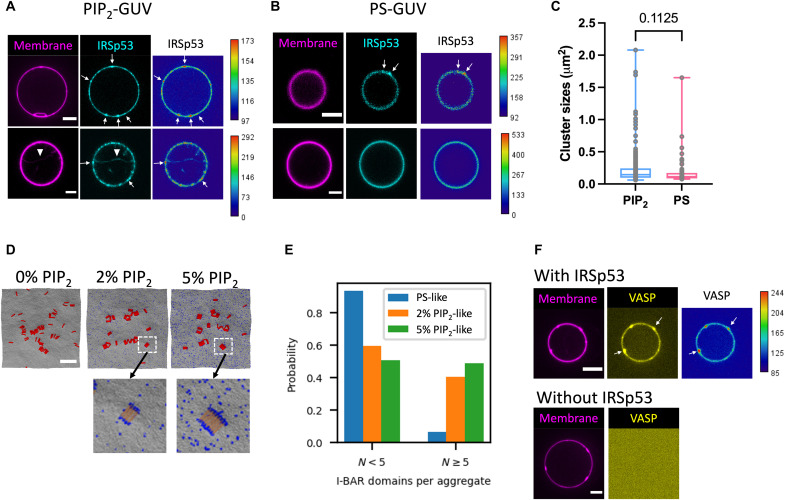
Fig. 2. IRSp53 self-assembles into clusters and recruits VASP on PIP2 membranes.
(A and B) Representative confocal images of GUVs incubated with AX488-labeled IRSp53 (16 nM). GUVs contain brain total lipid extract (TBX) with 0.5% TR-ceramide and either 5% PIP2 (PIP2-GUVs) in (A) or 25% 1,2-dioleoyl-sn-glycero-3-phospho-L-Serine (DOPS) (PS-GUVs) in (B). TR-ceramide in magenta and IRSp53 in cyan. Arrows indicate IRSp53 clusters on GUV membranes. Arrowhead indicates an inward membrane tube generated by IRSp53. Scale bar, 5 mm. (C) Sizes of IRSp53 clusters on PIP2-GUVs and on PS-GUVs. Each data point represents one cluster. PIP2-GUV: total of 225 clusters; N = 42 GUVs, three sample preparations. PS-GUV: total of 55 clusters; N = 12 GUVs, two sample preparations. Statistical analysis: two-tailed Mann-Whitney test, P = 0.1125. (D) Top: Representative snapshots of CG simulations from PS-like (0% PIP2), 2% PIP2-like, and 5% PIP2-like membranes. Membrane CG beads are in gray, PIP2-like CG beads are in blue, and I-BAR domains are in red. Scale bar, 50 nm. Bottom: Enlarged areas as indicated by the white dashed boxes. Only the central portion of the I-BAR domain is shown (yellow) to visualize PIP2 clusters (blue). (E) Probability of I-BAR domain aggregate size to be <5 or ≥5 molecules for membranes shown in (D). (F) Representative confocal images of GUVs incubated with AX488-labeled VASP (yellow) together with (top) or without (bottom) IRSp53 (unlabeled). Protein concentrations: 16 nM IRSp53 and 4 nM VASP. GUVs contain TBX with 0.5% TR-ceramide (magenta) and 5% PIP2. Heatmaps in (A) and (B) for IRSp53 signals and (F) for VASP signals; low fluorescence intensity is in blue, and high fluorescence intensity is in red. Scale bars, 5 μm.
To explore the role of PIP2 in IRSp53 clustering, we tested whether IRSp53 could form clusters upon interacting with another negatively charged lipid, PS, given that IRSp53-PIP2 binding is driven by electrostatic interactions (25). We replaced PIP2 with PS (25 mol %; PS has a net charge of −1) on GUVs while keeping the amount of negative charges comparable to that of PIP2-GUVs [5 mol % of PIP2; at pH 7, the charge of PIP2 is expected to be around −4 (48)]. We found that IRSp53 can form clusters also on PS-GUVs (Fig. 2B). The number of clusters per GUV are comparable on PIP2-GUVs and PS-GUVs (fig. S3). However, the clusters are larger on PIP2-GUVs as compared to PS-GUVs (Fig. 2C). To elucidate the mechanism underlying IRSp53 clustering on PIP2 membranes, we performed CG simulations. We generated PS-like and PIP2-like membrane sheets. The PS-like membrane is a quasi-monolayer of membrane beads that uniformly interacts with the I-BAR domain membrane binding surface. The PIP2-like membranes are nearly identical except a subset of membrane beads (2 or 5%) preferably interact with the ends of the I-BAR domains, as previously reported (25). In both membrane cases, the I-BAR domains have purely repulsive direct interactions with each other and are attracted to each other because of curvature coupling and Casimir-like forces mediated by the membrane as well as a membrane composition–mediated force that occurs only as PIP2-like membrane beads cluster to the ends of the I-BAR domains (25). Thus, the functional difference between the PS-like membrane (i.e., 0% PIP2-like membrane) and the PIP2-like membrane is the additional attraction to the ends of the I-BAR domain for only a small percentage of the membrane beads. Consistent with our observation on GUVs, we found that I-BAR domain clustering occurs on both PS-like (i.e., 0% of PIP2-like membrane) and PIP2-like membranes (2 and 5% of PIP2-like membranes; Fig. 2D, top). Moreover, we found that the addition of PIP2-like membrane beads increases the aggregation of I-BAR domains. Without PIP2-like membrane beads, we found a high probability of free or small aggregates containing less than five I-BAR domains. In the presence of PIP2-like membrane beads, there is a substantial decrease in free or small aggregates and a corresponding increase of larger aggregates with five or more I-BAR domains (Figs. 2D, top, and 2E, and movie S2). It has been shown that I-BAR domains can induce stable PIP2 microdomains upon membrane binding (31, 49). We found an enrichment of PIP2-like membrane beads near I-BAR domain aggregates (Fig. 2D, bottom, blue dots, and movie S3). In the 2% PIP2-like membranes, we found that 27% of the neighboring membrane beads are PIP2-like, which is an approximately 14 times enrichment of PIP2-like membrane beads around the I-BAR domain aggregates compared to the total 2% concentration of PIP2-like beads on the membrane. Similarly, in the 5% PIP2-like membranes, we found that 33% of the neighboring membrane beads are PIP2-like, which is a 6.7 times enrichment of PIP2-like membrane beads to the aggregates compared to the total 5% concentration of PIP2-like membrane beads. The enrichment of PIP2-like membrane beads around I-BAR domain aggregates depletes PIP2 in the bulk membrane that is not adjacent to an I-BAR domain aggregate. In other words, the PIP2 percentages in the membrane adjacent to the I-BAR domain aggregates are enriched to 27 and 33%, while the rest of the membrane is depleted to 0.7 and 3.4% of PIP2-like membrane beads for 2 and 5% PIP2-like membrane systems, respectively. Our results thus indicate a positive feedback of the assembly of I-BAR domain aggregates mediated by PIP2, and the enrichment of PIP2 around the aggregates facilitates further I-BAR domain recruitment. Together, our simulation and experimental reconstitution results indicate the key role of PIP2 in IRSp53 clustering on membranes.
It was reported that, in bulk, as well as on small vesicles containing PIP2, IRSp53 and VASP interact directly via their SH3 domain and PR domain, respectively (16). We thus examined this interaction in our reconstituted system. We purified full-length human VASP and labeled it with AX488 dyes (AX488-VASP). When incubating PIP2-GUVs with AX488-VASP (4 nM VASP tetramer) together with unlabeled full-length IRSp53 (16 nM IRSp53 dimer), we observed that VASP is recruited on GUV membranes (Fig. 2F, top, arrows), which is not the case in the absence of IRSp53 (Fig. 2F, bottom). Consistently, we observed VASP recruitment on GUV membranes when replacing full-length IRSp53 with a truncated version of IRSp53 that contains the I-BAR domain and the SH3 domain of IRSp53 (fig. S4A); in contrast, no VASP recruitment was observed when replacing full-length IRSp53 with the isolated I-BAR domain of IRSp53 (fig. S4B). These findings are consistent with the previous observations that IRSp53 and VASP interact directly in solution, on model membranes, and in filopodia (16, 17, 19). Notably, we found clusters of VASP on GUV membranes (Fig. 2F, arrows) that are reminiscent of what has been observed in live cells [Fig. 1B and (16)]. Furthermore, for GUVs having IRSp53-generated membrane tubules, we observed that VASP is recruited in these tubules (fig. S5). Our results indicate that IRSp53 can recruit VASP into clusters on the relatively flat GUV surfaces and to negatively curved membrane tubes, a characteristic of filopodial membranes.
Self-assembly of IRSp53, VASP, fascin, and actin on PIP2 membranes generates actin-filled membrane protrusions
We next assessed whether IRSp53, VASP, actin, and fascin can self-organize on PIP2 membranes to drive protrusion formation, as hypothesized above. We kept the bulk concentration of IRSp53 and VASP relatively low (16 nM IRSp53 dimer and 4 nM VASP tetramer) to allow the formation of IRSp53-VASP clusters on PIP2 membrane as seen in cells. To visualize actin, we used AX488-labeled globular actin (G-actin; ~10 to 27% AX488 labeled and total actin concentration of 0.5 μM). To ensure that actin polymerization occurs at the membrane only, as in cells and not in solution, we included capping protein (CP; 25 nM) and profilin (0.6 μM) in the protein mixture. CP binds to the barbed ends of filamentous actin (F-actin) with high affinity [dissociation constant (Kd) = 0.1 nM] and inhibits F-actin elongation in the bulk (1, 50). In cells, most G-actin is associated with profilin (Kd = 0.1 μM) (51, 52), which suppresses spontaneous actin nucleation in the bulk. In addition, it was shown that, at high ionic strengths, profilin is required for VASP to be more effective in actin polymerization (36). Last, to introduce actin filament bundling as in filopodia, we used fascin at 250 nM (11).
By performing pyrene actin polymerization assays, we verified that, in our experimental conditions, VASP increases actin polymerization in the presence of profilin, and CP suppresses F-actin elongation (fig. S6, solid lines). To verify the presence of actin filaments in our reconstitution systems, we performed some experiments using unlabeled G-actin and included AX488 phalloidin (0.66 μM) in the protein mixture, given that AX488 phalloidin binds to F-actin and its fluorescence is higher on F-actin than in the bulk (53). We verified by pyrene actin assays that the actin polymerization activity of VASP is preserved when phalloidin is present (by comparing the green dashed curve and the black dashed curve in fig. S6). In addition, the suppression of actin polymerization by CP is preserved in the presence of phalloidin, as indicated by the initial actin polymerization rates with 56 pM polymerized actin/s and 134 pM polymerized actin/s in the presence and absence of CP, respectively (by comparing the initial slopes of the dashed pink and dashed green curves in fig. S6; see Materials and Methods for details of the calculation). Last, the actin bundling activity of fascin was verified by performing two independent assays: an actin-fascin cosedimentation assay at low centrifugation speed, showing that most of the actin filaments were present in the pellet fraction only when fascin was present (fig. S7A), and an actin-fascin copolymerization assay observed by confocal microscopy, showing bundles of actin filaments only when fascin was present (fig. S7B).
By incubating PIP2-GUVs with IRSp53, VASP, actin, fascin, CP, and profilin, we observed inward membrane tubes filled with actin on the GUVs (Fig. 3A). The tubes are not static but move rapidly inside GUVs (movies S4 and S5). In addition to the tubes, there are actin signals on GUV membranes, indicating the formation of an actin shell on the membrane (Fig. 3A). On average, 32% of the GUVs had tubes, and 93% of these tube-positive GUVs had at least one tube filled with actin (N = 140 GUVs, n = 3 sample preparations; fig. S8A). By using AX488 phalloidin together with the unlabeled G-actin, we observed clear AX488 phalloidin signals inside the tubes and on GUV membranes, confirming the presence of F-actin (Fig. 3B and movies S6 and S7). In the presence of phalloidin, on average, 55% of the GUVs had at least one actin-filled tube (N = 366 GUVs, n = 7 sample preparations; fig. S8, B and C). Last, we verified the presence of IRSp53, VASP, and fascin in actin-filled membrane protrusions by using AX488-labeled IRSp53, VASP, and fascin. Regardless of phalloidin, we observed that VASP and fascin are present throughout the protrusions, while IRSp53 is either present throughout the protrusions or enriched at the tips of protrusions where the fluorescence signal of actin is somewhat weaker compared to the rest of the protrusion (fig. S9).
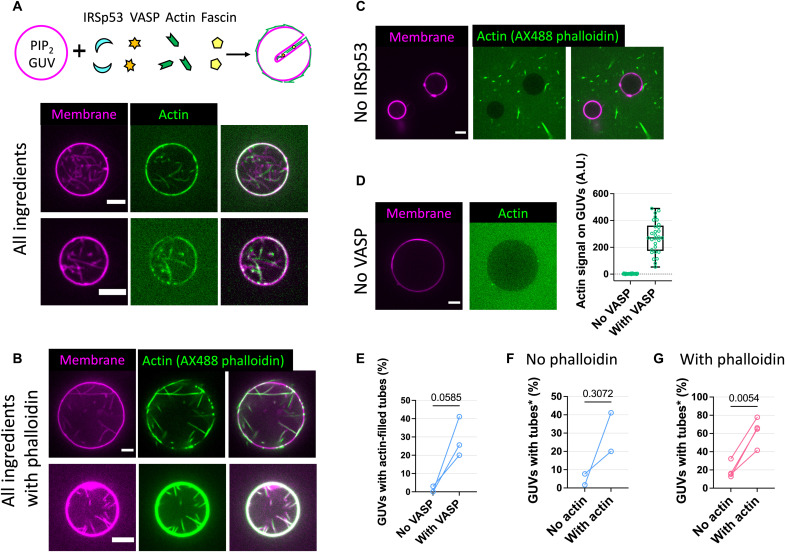
Fig. 3. IRSp53 and VASP synergistically drive the formation of actin-filled membrane protrusions.
(A to D) Representative GUVs (membranes, magenta; actin, green) incubated with (A) all proteins, (B) where actin was labeled instead with AX488 phalloidin, and where (C) IRSp53 and (D) VASP were excluded. Cartoon in (A) depicts GUVs incubated with all proteins (CP and profilin not shown). All proteins: IRSp53 (16 nM), VASP (4 nM), actin (0.5 μM; 10 to 27% AX488 labeled), fascin (250 nM), CP (25 nM), and profilin (0.6 μM). GUV composition: TBX, 0.5% TR-ceramide, and 5% PIP2. Scale bars, 5 μm. (D) Right: Pixel-averaged actin signals on GUVs. “No VASP,” N = 29 GUVs; “With VASP,” N = 29, one preparation (see fig. S12A for another two preparations). (E) Percentages of GUVs having actin-filled tubes with (“With VASP”) and without (“No VASP”) VASP. “With VASP,” N = 56, 39, and 45; “No VASP,” N = 42, 41, and 33; three preparations. Statistical analyses: chi-square test on pooled data, P < 0.0001; paired t test, P = 0.0585. (F and G) Percentages of tube-positive GUVs in the absence (“No actin”) and presence (“With actin”) of actin. “With actin” GUVs were counted only when having actin-filled tubes (*). (F) “No phalloidin” corresponds to AX488-labeled actin. “No actin,” N = 39 and 60; “With actin,” N = 45 and 56; two preparations. Statistical analyses: chi-square test on pooled data, P < 0.0001; paired t test, P = 0.3072. (G) In the presence of AX488 phalloidin and no AX488-labeled actin. “No actin,” N = 31, 31, 26, and 39; “With actin,” N = 54, 57, 41, and 56; four preparations. Statistical analyses: chi-square test on pooled data, P < 0.0001; paired t test, P = 0.0054.
Using the fluorescence signals of either AX488 actin monomers or AX488 phalloidin, we quantified that, on average, there are ~2 and ~11 actin filaments in the tubes, respectively (fig. S10A). Although the estimated numbers of filaments in tubes are lower than those in filopodia (typically 10 to 30 filaments) (6, 7), they are comparable to theoretical predictions in which 2 to 3 growing filaments are required to overcome membrane-restoring forces to generate extensions (54). Moreover, the membrane enclosing the actin bundles was predicted to stabilize the bundle against buckling in which the minimum number of filaments required for protrusion stability is four (55). In our reconstitution system, given the previously reported ability of the I-BAR domain in stabilizing membrane tubes (32), we anticipate that the presence of IRSp53 in the actin-filled tubes could contribute to their stability. We noted that the higher number of filaments in the presence of phalloidin could be due to the previously reported effect of phalloidin in promoting actin nucleation (56). Moreover, phalloidin was shown to increase the persistence length of actin filaments, thus effectively making them stiffer (57). We thus assessed the orientational properties of the actin-filled tubes by performing image analysis (see Materials and Methods for details). As shown in fig. S10, color-coded orientation maps showed the presence of dominant orientation directions of the tubes in the presence of phalloidin but not in its absence. Consistently, in the presence of phalloidin, higher coherency values, which indicate one dominant orientation of local structures, can be seen in places where the protrusions are, as readily visible by the white patches in the coherency map [indicated by arrows in fig. S10 (D and E)]. Thus, consistent with the higher number of actin filaments in the tubes and the abovementioned side effects of phalloidin, our results show that the actin-filled tubes are straighter in the presence of phalloidin.
Besides tubes, we observed membrane deformations on the GUV membranes independent of the presence of phalloidin (fig. S11, arrows). The deformed bulge shape of the GUV membranes is reminiscent of membrane deformation driven by the I-BAR domain of IRSp53 on GUVs (58) and indicates the presence of pushing forces acting on the GUV membranes toward the interior of the GUVs. Together, our results show that IRSp53, VASP, actin, and fascin can spontaneously organize locally on PIP2 membranes to generate actin-based membrane protrusions.
IRSp53 is indispensable for protrusion formation by recruiting VASP to facilitate actin polymerization in protrusions
To reveal the contribution of individual protein components in the generation of actin-filled membrane protrusions, we performed loss-of-function assays by removing one protein component at a time. To ensure that protein activities are identical in the loss-of-function assays, we performed paired experiments in which we used the same batches of GUVs and protein stocks in each independent sample preparation. Then, in each sample preparation, we compared the efficiencies of the generation of actin-filled tubes in the absence of a protein of interest and the reference condition where all the proteins are present. To this end, we counted the number of GUVs with and without tubes and if an actin signal was readily detected in at least one or more tubes of the GUVs.
When IRSp53 was absent, we found that none of the GUVs had tubes (N = 57 GUVs, n = 3 sample preparations, in the presence of AX488 phalloidin; Fig. 3C). Thus, in our experimental condition, IRSp53 is essential for the generation of actin-filled membrane tubes. In the absence of VASP, we observed a nearly complete lack of actin signal on the GUV membranes (Fig. 3D and fig. S12A), and nearly no GUV had actin-filled tubes (of a total of 116 GUVs, n = 3 sample preparations, only 8 GUVs were found with tubes, in which only 1 GUV had actin-filled tubes; Fig. 3E and fig. S12B). Our observations thus indicate the key role of VASP in the generation of membrane tubes, via recruiting actin on GUVs. Given that VASP is an actin polymerase, we assessed whether VASP facilitates actin polymerization in tubes by using AX488 phalloidin. We observed that because of the presence of VASP, there is an increase in the number of GUVs having actin-filled tubes (fig. S13, A and B) and an increase in the number of actin filaments in the tubes (fig. S13C). We note that in the presence of phalloidin, there are actin-filled tubes on GUVs even when VASP is absent (fig. S13D). This observation indicates that phalloidin aids actin polymerization even when CP and profilin are present. To understand how phalloidin influences actin polymerization in our system, we performed pyrene actin polymerization assays. We observed that phalloidin enhances VASP-mediated actin nucleation (fig. S6, compare the magenta solid and dashed curves), consistent with a previous report (56). It was shown in vitro that in the bulk, IRSp53 (after being activated by Eps8) and its I-BAR domain can interact with F-actin and induce actin bundle formation (20, 23). Thus, in the absence of VASP, phalloidin facilitates actin polymerization, and IRSp53 recruits F-actin to GUV membranes, resulting in the formation of actin-filled tubes. Consistently, we found that the percentages of GUVs having actin-filled tubes are higher in the presence of phalloidin compared to its absence (fig. S8C). Together, our results show that through IRSp53-driven recruitment to PIP2 membranes, VASP plays a key role in protrusion generation via its actin polymerization activity.
Biophysical studies using reconstitution systems and theoretical modeling have revealed how the interplay between the mechanical properties of membranes and actin bundles/networks determines the formation of actin-driven membrane protrusions (12, 59, 60). Given that actin’s role in the initiation of IRSp53-driven cellular protrusions is not fully understood (18), we assessed whether actin facilitates IRSp53-based protrusion formation using our reconstitution systems. In the absence of actin, we observed 2% to 34% of GUVs having tubes (regardless of phalloidin; Fig. 3, F and G). These tubes were generated by IRSp53 because when IRSp53 is absent, no GUV has tubes (Fig. 3C). Notably, we observed a global increase in the amount of GUVs having tubes because of the presence of actin (Fig. 3, F and G). This effect is more pronounced when phalloidin is present (N = 127 and 208 total GUVs, without and with actin, respectively, n = 4 sample preparations; Fig. 3G). In the absence of phalloidin, in a sample preparation where there was a relatively high amount of GUVs with tubes in the absence of actin (34.1%; N = 41 GUVs), the addition of actin did not aid tube generation (25.6%; N = 39 GUVs). However, in experimental sets where the amount of GUVs with tubes was low (less than 10%) in the absence of actin, the addition of actin increased the amount of GUVs with tubes (N = 99 and 101 total GUVs, without and with actin, respectively, n = 2 sample preparations; Fig. 3F). Together, these results indicate that actin contributes to IRSp53-based tube generation. Furthermore, given that phalloidin facilitates actin nucleation on membranes in our system, our results point out the essential role of actin nucleation to enhance actin’s function in protrusion formation in cells.
Fascin is not required for protrusion generation but enhances protrusion elongation and stability
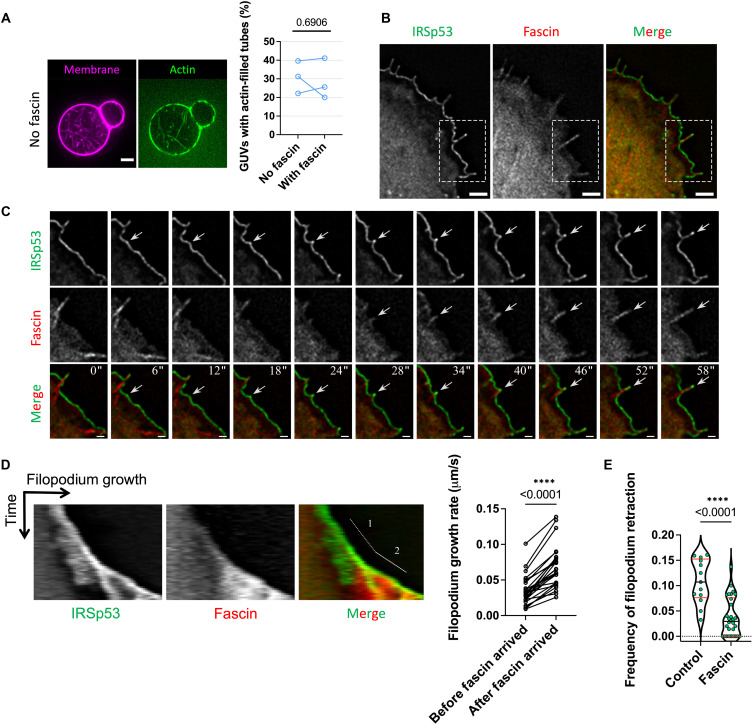
Given that fascin is the specific actin bundler in filopodia, we assessed its role in protrusion formation. In the presence and absence of fascin, we did not observe significant differences considering the number of GUVs having actin-filled tubes, regardless of the absence (Fig. 4A and fig. S14A) or presence of phalloidin (fig. S14, B and C). Furthermore, there is no significant difference in the amount of F-actin in tubes in the presence and absence of fascin (fig. S14D). Because of the rapid movement of tubes inside GUVs, we could not characterize the dynamics of tube generation and elongation. We thus assessed how fascin affects protrusion dynamics in live cells. To this end, we performed experiments using Rat2 cells that expressed eGFP-tagged IRSp53 and mCherry-tagged fascin (Fig. 4B and movie S8). We observed that the recruitment of fascin in IRSp53-based protrusions coincides with their elongation (Fig. 4, C and D). By tracking IRSp53-based protrusions, we also found that fascin significantly increases the growth rates of the protrusions (Fig. 4D) and that its overexpression decreases the frequency of their retraction (Fig. 4E). Thus, our in vitro reconstitution data demonstrate that actin polymerization facilitates the formation of IRSp53-dependent membrane protrusions, while the cell data suggest that fascin increases the growth rate and enhances the stability of these protrusions.
Fig. 4. Fascin facilitates filopodium growth and prevents filopodium retraction.
(A) Left: Representative images of GUVs incubated with all protein ingredients (IRSp53, VASP, actin, CP, and profilin) besides fascin. Right: Percentages of GUVs having actin-filled tubes in the absence (“No fascin”) and presence (“With fascin”) of fascin. GUV composition: TBX, 0.5% TR-ceramide, and 5% PIP2. “No fascin,” N = 53, 32, and 54 GUVs; “With fascin,” N = 56, 45, and 39; three sample preparations. Statistical analyses: chi-square test on pooled data, P = 0.8652; paired t test, P = 0.6906. (B) Wide-field fluorescence image of a Rat2 cell transfected with IRSp53-eGFP and mCherry-fascin. (C) Time-lapse images of the formation of a filopodium. Images are magnifications of the indicated area (white dashed boxes) in (B). White arrows indicate the appearance of IRSp53 clusters followed by fascin recruitment. Time is in seconds. (D) Left: Representative kymograph of IRSp53 and fascin fluorescence signals in a growing filopodium showing the growth before (number 1) and after (number 2) the presence of fascin. Right: Quantification of filopodial growth rate before (“Before fascin arrived”) and after (“After fascin arrived”) fascin recruitment. N = 26 filopodia. Statistical analysis: Mann-Whitney nonparametric test, P = 0.00001633. (E) Frequency of filopodium retractions in IRSp53-eGFP–expressing Rat2 cells transfected with either an empty mCherry plasmid (“Control”) or mCherry-fascin (“Fascin”). The frequency of each event was calculated for the period of filopodium growth as the number of retractions per second. “Control,” N = 13 filopodia; “Fascin,” N = 30. Statistical analysis: Mann-Whitney nonparametric test, P = 0.000083802. Scale bars, 5 μm (A) and 2 μm (B and C).
IRSp53 regulation in cells revealed by membrane nanotube pulling experiments
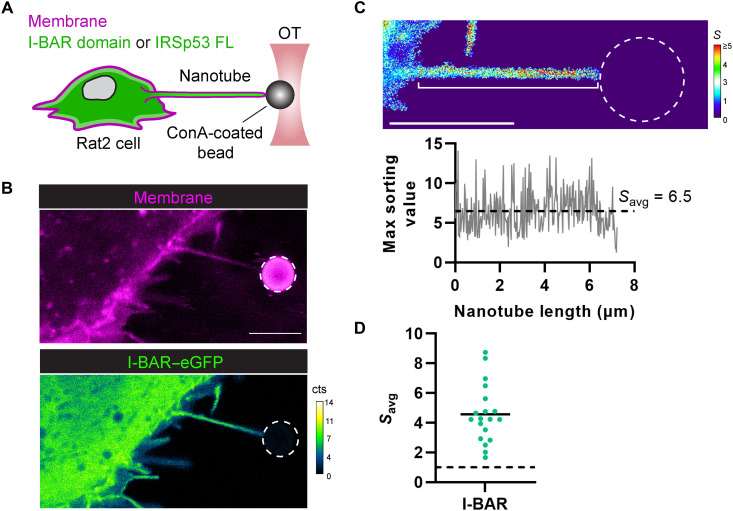
Our results obtained using the reconstituted system demonstrate the function of IRSp53 in the generation of actin-filled membrane protrusions and show that IRSp53 is primed to self-assemble into clusters that drive the association of downstream partners (e.g., VASP and fascin) for protrusion growth. However, it remains unclear how cells regulate the activity of IRSp53 to control, for instance, protrusion formation at specific membrane locations. With the abilities of IRSp53 to sense membrane curvature and promote actin assembly mediated by VASP, we expect that any local deformation of the plasma membrane could induce filopodial growth. To reveal IRSp53’s possible regulation directly in cellulo, we generated artificial protrusions having identical topologies to filopodia by pulling membrane nanotubes (i.e., tethers) from the plasma membrane of Rat2 fibroblasts using optically trapped polystyrene beads and micromanipulation (Fig. 5A). The pulled nanotubes serve as tractable models to directly assess the recruitment of IRSp53 into tubular geometries and the possible actin assembly inside these nanotubes as a consequence of IRSp53 enrichment. Cells were transfected with either IRSp53’s I-BAR domain (I-BAR–eGFP), its putative membrane-deforming and curvature-sensitive region, or the full-length IRSp53 protein (IRSp53-eGFP). The plasma membrane was exogenously labeled using the lipophilic CellMask Deep Red stain. Rat2 I-BAR–eGFP–expressing cells show strong I-BAR–eGFP fluorescence in pulled nanotubes (Fig. 5B) that was visible after nanotube formation (fig. S15 and movie S9), suggesting an innate preference of the I-BAR domain to sort into these nanotubes. Protein enrichment in the nanotube was quantified by determining the sorting parameter S, which corresponds to the ratio of the protein intensity in the nanotube (at a given pixel) relative to the protein’s average intensity measured in the plasma membrane (i.e., the bulk). This nanotube/membrane ratio for the protein is further normalized by the same ratio for a lipid marker (here, the CellMask Deep Red stain) to correct for cell-to-cell signal differences in the membrane and protein fluorescence channels (e.g., differences in expression levels and staining efficiencies). This calculation ultimately results in a color-coded image of S values (we term an S map) from which we then can determine a mean enrichment value for a given nanotube (Savg) (Fig. 5C; see Materials and Methods for further details). Note that Savg values greater than 1 indicate preferential protein enrichment for a given nanotube. We measured the Savg of the I-BAR domain for multiple nanotubes (N = 19 nanotubes) and determined an ensemble average of 4.5 (Fig. 5D). This value is similar to previously reported sorting values obtained from nanotubes pulled from GUVs encapsulating the purified I-BAR domain (32).
Fig. 5. IRSp53’s I-BAR domain is robustly recruited into pulled membrane nanotubes.
(A) Experimental setup for pulling membrane nanotubes using a concanavalin A (ConA)–coated bead trapped in an optical tweezer (OT). Rat2 fibroblasts, expressing eGFP fusions (green) of either IRSp53’s I-BAR domain or the full-length (FL) IRSp53 protein, were labeled with CellMask Deep Red plasma membrane stain (magenta); protein enrichment in the membrane nanotube was monitored by confocal fluorescence microscopy using single-photon avalanche detectors (cts, counts). (B) Representative confocal image of a pulled membrane nanotube from a Rat2 cell expressing IRSp53’s I-BAR domain showing high enrichment of the I-BAR domain. (C) Top: Calculated sorting map of the nanotube in (B) with low sorting (S) values in blue and high S values in red. Bottom: Plot of the maximum sorting value at each pixel position along the length of the nanotube (white bracket in the sorting map) and the mean sorting value for the protein (Savg). (D) Measured Savg values for IRSp53’s I-BAR domain in pulled nanotubes (N = 19 nanotubes). Savg > 1 (dashed black line) indicates protein enrichment. Black solid line, mean of the data points. Dashed white circles in the figure outline the trapped bead. Scale bars, 5 μm.
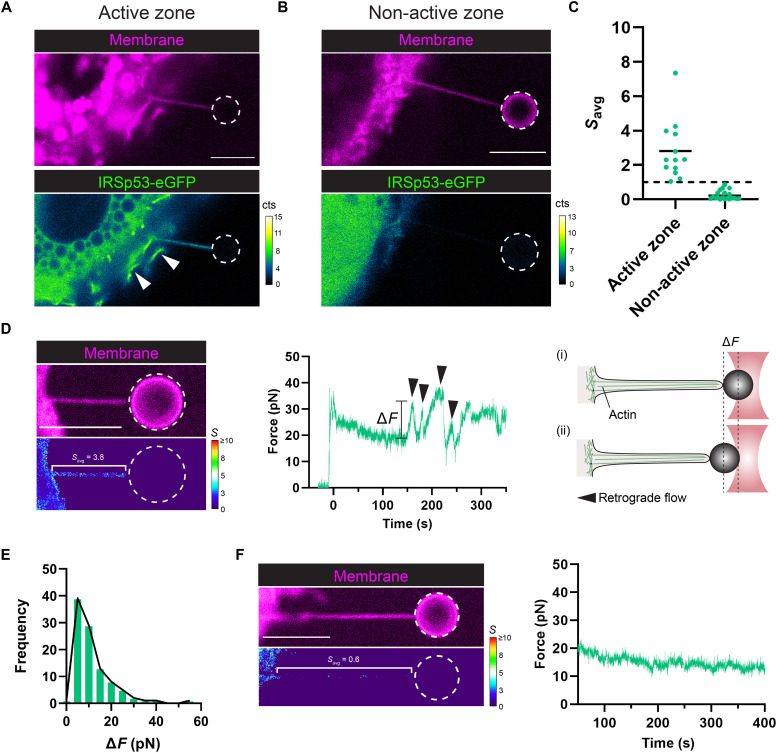
Contrary to the robust enrichment of the I-BAR domain, we observed a more complex behavior for the sorting of the full-length IRSp53 protein in nanotubes that is dependent on the local cellular membrane activity near the sites of the nanotubes. Pulled nanotubes from IRSp53-eGFP–expressing cells show strong IRSp53 signal only when the nanotube was pulled near zones exhibiting “active” processes of membrane remodeling (Fig. 6A), such as lamellipodia and membrane ruffles, which were frequently found at the cell leading edge. Similar to the fast arrival of the I-BAR domain in pulled nanotubes (fig. S15), we observed a rapid stabilization of IRSp53 intensity in pulled nanotubes within 30 s (fig. S16). However, no IRSp53-eGFP fluorescence was observed in nanotubes when pulled from “non-active” zones (Fig. 6B), such as those found near the cell trailing edge. Subsequent imaging over time showed that nanotubes pulled from active and non-active regions stably retained either the presence or absence of IRSp53, respectively, indicating that an equilibrium situation was reached in our measurements (fig. S17). As seen in Fig. 6C, nanotube Savg values for IRSp53 near active zones (N = 13 nanotubes) averaged 2.8 and were comparable to the I-BAR domain case (Fig. 5D). However, Savg values for IRSp53 near non-active zones (N = 20 nanotubes) averaged 0.2, indicating an exclusion of the protein from the nanotubes. The dichotomy of sorting behaviors for IRSp53 is completely opposite to the results that we observed for the I-BAR domain, where the I-BAR domain sorting is consistently stronger and did not depend on the cellular region where the nanotube pulling was performed. Together, our results highlight that IRSp53’s curvature-sensing ability is innately part of the I-BAR domain (32, 33) and further suggest that cellular regulation mechanisms are at play in modulating IRSp53’s activity.
Fig. 6. IRSp53 is recruited into membrane nanotubes pulled from highly active cellular zones and coincides with actin filament assembly.
(A and B) Representative confocal images of pulled membrane nanotubes from Rat2 cells expressing the full-length IRSp53 protein (IRSp53-eGFP). (A) Pulled nanotubes near cellular zones of active membrane remodeling, such as membrane ruffling (white arrowheads), exhibit recruitment of IRSp53 in the nanotube. (B) Pulled nanotubes near non-active zones instead show no recruitment of IRSp53. (C) Mean sorting values for a given nanotube, Savg, were determined from active and non-active zones for IRSp53-expressing cells. Dashed black line, Savg = 1. Black solid lines, mean of the data points. Active zone, N = 13 nanotubes; non-active zone, N = 20 nanotubes. (D) IRSp53 enrichment corresponds with eventual actin development within the pulled nanotube. Representative sorting (S) map (left) and the corresponding force plot (center) for a nanotube showing high IRSp53 sorting. Peaks in the force plot (black arrowheads) are signatures of actin in the tube and arise when retrograde flows outcompete actin polymerization (at the nanotube tip), causing bead displacement toward the cell body and hence a rise in the force (right; i and ii). (E) Distribution of the force peak magnitudes (ΔF). Sample size, 100 peaks. (F) Representative sorting map (left) and the corresponding force plot (right) for a nanotube showing no IRSp53 enrichment and hence no actin development. Color maps: Low S values are in blue, and high S values are in red. Dashed white circles in the figure outline the trapped bead. Scale bars, 5 μm.
We observed that the sorting of IRSp53 has a direct consequence on the assembly of F-actin within the nanotubes. Nanotubes with high IRSp53 sorting (Fig. 6D) exhibited characteristic signatures of F-actin in the measured force profile and behaviors mimicking those of true filopodia. The observed bead displacement within the optical trap (i.e., a rise in the force, ΔF) indicates that F-actin development has reached the end of the nanotube, allowing retrograde and contractile forces to transmit from the cell body to the bead. The magnitude of the force peaks generates traction forces of 5 to 10 pN (Fig. 6E) and is of the same order of magnitude as those reported for true filopodia (61, 62). In addition, events of nanotube curling (i.e., helical buckling) were also frequently observed (fig. S18), as previously shown for filopodia (62). In essence, IRSp53-positive nanotubes eventually mature into a “pseudo” filopodium. However, nanotubes with no IRSp53 enrichment (Fig. 6F) exhibit a constant force profile, and consequently, no F-actin development was observed. Consistent with our GUV results, in IRSp53-positive nanotubes, we also observed the recruitment of both VASP and fascin (fig. S19 and movies S10 and S11) in cells coexpressing the proteins of interest, confirming that bundled F-actin is indeed present within the core of the nanotube. Together, our tube pulling results show that although IRSp53 is curvature sensitive through its I-BAR domain, its activity is tightly controlled within cells to constrain local F-actin development for protrusion generation.
DISCUSSION
How cells control the formation of protrusions such as filopodia and microvilli at specific membrane locations remains largely unclear. Using in vitro reconstitution systems, we demonstrated that a minimum set of proteins composed of the membrane curvature sensor IRSp53, the actin polymerase VASP, and actin can spontaneously organize to generate actin-based membrane protrusions. By performing loss-of-function assays, we investigated the function of proteins in protrusion formation. We demonstrated that IRSp53 is the essential player in protrusion generation by recruiting VASP to promote local actin assembly. Thus, by using a precisely controlled reconstitution system, our study provides a strong support to the previously proposed mechanism of IRSp53-VASP–driven filopodium generation that was largely based on cell biology studies (16, 17).
A key finding of our in vitro assay is that purified full-length IRSp53 is active in the absence of its activators such as Cdc42. Our results indicate that once IRSp53 binds to the plasma membrane, it can self-assemble into clusters and readily recruit VASP to generate actin-filled protrusions. It was reported that when expressing a nonregulated constitutively active mutant of IRSp53 in cells, an aberrant explosive formation of filopodia was observed (21). Thus, to prevent generating unwanted protrusions, cells must have a tight regulation of IRSp53, i.e., how much, when, and where to activate IRSp53. To reveal the regulation of IRSp53 activity in cells, we generated filopodium-like membrane geometries in live cells by pulling membrane nanotubes from the plasma membrane. We found that IRSp53 can only be recruited to nanotubes pulled from active membrane regions of the cell (e.g., where the membrane exhibits nearby dynamic ruffling, lamellipodium extension, and filopodium formation) but not from regions where no membrane remodeling was observed, and thus, IRSp53 is presumed inactivated and unable to bind to the plasma membrane to exert its function. Moreover, actin polymerization extending throughout the IRSp53-enriched nanotube was detected, creating, in essence, a pseudo filopodium that was positive for VASP and fascin. Our finding is consistent with the location of IRSp53’s activator Cdc42 at the front edge of migratory cells where ruffling and protrusions develop (63, 64) and with IRSp53’s function in inducing Rac-dependent membrane ruffling (26, 29). Our results indicate that to finely regulate IRSp53 activity, it is probably even more important to keep IRSp53 inhibited such that its binding to membranes is prevented. It was shown that the binding of 14-3-3 to IRSp53 counteracts the activation by Cdc42 and other downstream cytoskeleton effectors (21). Notably, 14-3-3 binding to phosphorylated IRSp53 keeps it in an inhibited state, resulting in impaired filopodium formation and dynamics (21, 29, 30). Collectively, our results reflect a finely tuned regulatory mechanism where IRSp53 is kept in its inhibited state by phosphorylation and binding to 14-3-3 and activated by binding to activators such as Cdc42. Controlling IRSp53 by three different signaling pathways—phosphorylation, Cdc42 binding, and PIP2 binding—thus ensures its precise spatial-temporal regulation at the plasma membrane such that IRSp53 is activated only at specific regions of cells.
It was shown previously that PIP2 binding by IRSp53’s I-BAR domain is required for the generation of plasma membrane protrusions (25). Our in vitro work and CG simulation results showed that PIP2 is key for the assembly of IRSp53 clusters: Higher PIP2 concentration results in larger I-BAR domain clusters. Our simulation results showed an at least sevenfold enrichment of PIP2 around the I-BAR domain clusters. Our finding is consistent with the previous reports of BAR and I-BAR domains inducing local and stable PIP2 clusters (31, 49, 65). Notably, it was proposed that these PIP2 clusters are transiently interacting with BAR domains such that PIP2 in the clusters is available for the recruitment of downstream partners having PIP2 binding motifs (31, 49, 65). We thus propose that, besides specific protein-protein interactions, IRSp53-induced PIP2 clusters could facilitate not only the further recruitment of IRSp53 but also other proteins such as actin nucleation–promoting factors, actin polymerases, and actin-binding proteins for protrusion formation and regulation (17, 24, 27, 66–68).
We observed clusters of IRSp53 preceding those of VASP on the plasma membrane at the onset of filopodium formation. Our observations are in agreement with what was reported by other studies using MEF cells and COS-7 cells (16, 21). VASP clustering has been shown to be an important factor to increase the processivity of VASP for actin filament elongation (37, 39). Consistently, using reconstitution systems, we showed that VASP clustering via IRSp53 efficiently polymerizes actin on PIP2 membranes and in protrusions. Moreover, we observed an increased number of actin filaments in protrusions when VASP is present. Notably, VASP clustering has also been attributed to the formation of another type of membrane protrusion called microspikes. As shown previously, despite containing actin bundles, microspikes differ from filopodia by being shorter protrusions, which are generated from and are largely embedded in the lamellipodial actin filament network. By focusing on the formation of microspikes, recent studies using B16F1 cells showed that VASP clustering is initiated by lamellipodin and myosin-X but not by IRSp53 (42). These studies and our work thus suggest that VASP requires specific membrane-interacting partners to bring it to membranes and induce its clustering to drive outward membrane deformation. The differences in the mechanisms of recruiting VASP on membranes may be also cell type dependent, i.e., cells may use different pathways to induce membrane deformation via VASP. Notably, in our current study, IRSp53 and VASP were overexpressed in cells, and the elevated expression levels could potentially influence the regulation of different molecular machineries involved in the generation of membrane protrusions. In the future, it will be important to study the roles of different proteins in VASP clustering during both filopodium and microspike formation in different cell types by expressing proteins of interest at levels comparable to their endogenous counterparts. This would allow to decipher how and under which physiological conditions cells use different VASP clustering pathways to generate distinct membrane protrusions.
In our in vitro GUV assays, we observed that IRSp53 is either present throughout the actin-filled protrusions or enriched at the tips of the protrusions. This observation is in agreement with the localization of full-length IRSp53 in cellular filopodia using super-resolution microscopy (69). Consistently, given that IRSp53 is responsible for the recruitment of VASP to GUV membranes, we observed that VASP is present throughout the protrusions. Along the same line, in our cell-tether pulling experiments, we observed that VASP is distributed throughout the tethers. These observations are different from the reported localization of VASP in filopodia, where VASP is mostly found at the tips of filopodia and, to a lesser extent, also along the shaft of filopodia. Notably, in contrast to the very simplified GUV systems, the formation of filopodia in cells involves an overwhelmingly large set of other proteins (70). We thus speculate that, in cells after IRSp53 initiates VASP-dependent actin elongation, other binding partners of VASP that are members of the filopodial tip complex could restrict the localization of VASP to be at the filopodial tip. Potential VASP partners could be myosin-X and formin mDia2, which were shown to cooperate with VASP in filopodium formation (43, 71, 72). Our in vitro work provides direct evidence that actin filament assembly facilitates IRSp53-based protrusion generation. This is consistent with what was previously proposed on the basis of electron microscopy and cell biology experiments (18). In our assay, actin assembly is driven by VASP, given that when VASP was eliminated, we observed nearly no actin signal on GUVs (Fig. 3D). We anticipate that other actin assembly factors can play similar roles as VASP. It was reported that IRSp53 interacts directly with formins, such as mDia1, in filopodium formation (27). Moreover, actin nucleation–promoting factors WAVE2 and N-WASP were shown to synergize with IRSp53 to generate filopodia (17, 27). In our in vitro system, fascin is not essential for protrusion initiation. However, by observing the dynamics of filopodia in Rat2 cells, we found that fascin enhances the elongation rate and stability of filopodia, consistent with the previous observation in cells that used RNA interference of fascin (6, 7). Our result thus supports the notion that fascin mechanically strengthens filopodia by bundling actin filaments together, thus facilitating filopodial extension and stabilization (4–7). Notably, Eps8, another actin bundler, has been reported to form a complex with IRSp53 during filopodium formation (20). Besides bundling, Eps8 can cap the barbed end of actin filaments. The dual function of Eps8 on actin filaments is fine-tuned in cells, allowing cells to generate filopodia via different signaling networks depending on cellular contexts (20). To understand how the abovementioned proteins regulate dynamic filopodium formation, elongation, retraction, and force generation, future work is required to elucidate the interplay between these proteins in the context of protrusion formation.
Together, our work demonstrates that IRSp53 is an efficient protrusion initiator: Once being activated, IRSp53 readily triggers the cascade of protrusion formation. Thus, to avoid the uncontrolled formation of protrusions, it is critical for cells to have a strict and precise control on when and where to activate or unlock IRSp53 from its inhibitory state.
MATERIALS AND METHODS
Pyrene actin polymerization assay
Polymerization assays were based on measuring the fluorescence change of pyrenyl-labeled G-actin (λexc = 365 nm, λem = 407 nm). Experiments were carried out on a Safas Xenius spectrofluorometer (Safas, Monaco). Polymerization assays were performed in buffer containing 60 mM NaCl, 1 mM MgCl2, 0.2 mM EGTA, 0.2 mM adenosine triphosphate (ATP), 10 mM dithiothreitol (DTT), 1 mM 1,4-Diazabicyclo[2.2.2]octane (DABCO), 5 mM tris (pH 7.5), and 0.01% NaN3 in the presence of actin (2 μM; 5% pyrenyl labeled), profilin (2.4 μM), VASP (15 nM in tetramer), CP (25 nM), and phalloidin (2 μM).
To reveal the upper limit of the fluorescence intensity for the pyrene actin polymerization, we performed positive control experiments of (i) actin and spectrin-actin seeds, (ii) actin alone, and (iii) actin and phalloidin. As shown in fig. S6, the sample of actin + spectrin seeds reaches completion with the fluorescence intensity of a value of 50 arbitrary units (A.U.) after around 1000 s.
To verify that, in the presence of VASP, phalloidin does not inhibit/prevent CP from suppressing F-actin elongation, we estimated the initial actin polymerization slopes of two conditions: (i) profilin actin + VASP + CP + phalloidin (dashed pink curve in fig. S6B) and (ii) profilin actin + VASP + phalloidin (dashed green curve in fig. S6B). We considered that the fluorescence reached the plateau value of 48.2 A.U. (obtained by using spectrin-actin seeds as shown in fig. S6A), corresponding to 1.9 μM actin, i.e., the total concentration of actin subtracts 0.1 μM (the critical concentration for actin polymerization). We obtained the following results: (i) profilin actin + VASP + CP + phalloidin (dashed pink curve): 56 pM polymerized actin/s, and (ii) profilin actin + VASP + phalloidin (dashed green curve): 134 pM polymerized actin/s.
GUV experiments
Reagents
Brain total lipid extract (TBX, 131101P), brain PIP2 (840046P), 18:1 PS [1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), 840035], and 1,2-dioleoyl-sn-glycero-3-phosphocholine-N-(Cyanine 5) (Cy5-PC) (850483) were purchased from Avanti Polar Lipids. BODIPY TR C5 ceramide (TR-ceramide, D7540) was purchased from Invitrogen. AX488- and AX594-tagged phalloidin (AX488 phalloidin and AX594 phalloidin) were purchased from Interchim. β-Casein from bovine milk (>98% pure; C6905) and other reagents were purchased from Sigma-Aldrich.
GUV preparation
The lipid mixture used contained TBX supplemented with 5 mol % of PIP2 at 0.5 mg/ml dissolved in chloroform. To visualize GUV membranes, the lipid mixture was supplemented with either 0.5 mol % of TR-ceramide or 0.5 mol % of Cy5-PC.
GUVs were prepared by using the polyvinyl alcohol (PVA) gel-assisted vesicle formation method as previously described (73). Briefly, a PVA gel solution [5% (w/w), dissolved in 280 mM sucrose and 20 mM tris (pH 7.5)] warmed up to 50°C was spread on clean coverslips (20 mm by 20 mm). The coverslips were cleaned by ethanol and then double-distilled water twice. The PVA-coated coverslips were incubated at 50°C for 30 min. Around 5 μl of the lipid mixture was spread on the PVA-coated coverslips and then dried under vacuum at room temperature (RT) for 30 min to remove the chloroform solvent. The coverslips were then placed in a petri dish, and around 500 μl of the inner buffer was pipetted on the top of the coverslips. The inner buffer contains 50 mM NaCl, 20 mM sucrose, and 20 mM tris (pH 7.5). The coverslips were kept at RT for at least 45 min, allowing GUVs to grow. Once done, we gently “tapped” the bottom of the petri dish to detach GUVs from the PVA gel. The GUVs were collected using a 1-ml pipette tip with its tip cut to prevent breaking the GUVs.
Protein purification and labeling
Muscle actin (UniProt, no. P68135) was purified from rabbit muscle and isolated in monomeric form in G-buffer [5 mM tris-Cl− (pH 7.8), 0.1 mM CaCl2, 0.2 mM ATP, 1 mM DTT, and 0.01% NaN3] as previously described (74). Actin was labeled with AX488 or AX594 succimidyl ester–N-hydroxysuccinimide (75).
The genes encoding full-length human IRSp53 (UniProt, no. Q9UQB8; Homo sapiens) and the I-BAR–SH3 domain of IRSp53 (UniProt, no. Q9UQB8; H. sapiens, amino acids 1 to 452) and the full-length human VASP (UniProt, no. P70460; H. sapiens) were provided by R. Dominguez (University of Pennsylvania) and J. Faix (Hannover Medical School), respectively, and subcloned into the pGEX-6P-1 vector (Cytiva). The expression plasmids were transformed into BL21 (DE3) competent cells. A single colony was resuspended in 10 ml of LB medium containing ampicillin (100 μg/ml) and was cultured at 37°C overnight. Then, the starter culture was inoculated into 1 liter of LB medium containing ampicillin (100 μg/ml) and cultured at 37°C until OD600 (optical density at 600 nm) reached 0.6. The protein expression was induced with 0.2 mM isopropyl-d-1-thiogalactopyranoside overnight at 15°C. After harvesting the cells, the pellets were resuspended and sonicated in a buffer containing 20 mM tris-HCl (pH 7.4), 500 mM NaCl, 1 mM EDTA, lysozyme (1 mg/ml), 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 1 mM DTT, followed by affinity purification with GSTrap FF (Cytiva). The glutathione S-transferase (GST)–tag was removed with PreScission protease (Cytiva). The proteins were concentrated to 50 to 100 μM using Amicon Ultra centrifugal filters (Merck). For protein labeling, Alexa Fluor maleimides (Thermo Fisher Scientific) were added to the protein solution, and the reaction was allowed to proceed at 4°C overnight in the dark to protect it from light. The labeled proteins were further purified and separated from free Alexa Fluor dye using a Superdex 75 10/300 GL gel filtration column (Cytiva) using either an ÄKTA protein purification system (Cytiva) or the NGC Chromatography System (Bio-Rad). The proteins were concentrated using Amicon ultra centrifugal filters (Merck) by replacing the buffer with 20 mM tris-HCl (pH 7.4), 150 mM NaCl, and 1 mM DTT, and then 0.1% (w/v) methylcellulose [M0512, Sigma-Aldrich; 4000 centipoise (cP)] was added. The purified proteins were frozen in liquid nitrogen and stored at −80°C before use. The labeling efficiency of AX488-IRSp53 (dimer) was 0.94, and that of AX488-VASP (tetramers) was 3.0. The purification of the isolated mouse I-BAR domain of IRSp53 (UniProt, no. Q8BKX1; mouse, amino acids 1 to 250) was performed as previously described (31).
Recombinant human profilin I was expressed in BL21 (DE3) Star competent bacterial cells and purified as described (76). The plasmid for the expression of mouse CP α1β2 [UniProt, no. P47753 (α1) and no. P47757 (β2); mouse] was cloned in a pRSFDuet-1 vector, providing a 6xHis tag at the N terminus of the α1 subunit. Mouse CP α1β2 was expressed in BL21 (DE3) and was purified as previously described (77).
The plasmid expressing GST-HsFascin1 was provided by D. Kovar (University of Chicago). The HsFascin1 full-length protein (UniProt, no. Q16658) was produced in BL21(DE3) competent bacterial cells from the pGEX-4T-3 vector, as described previously with some modifications (11). Briefly, the bacterial cells were grown for 24 hours at RT (around 20°C) in autoinduction media (AIMLB0210, Formedium, UK), inoculated with overnight growth starter culture. The cells were harvested by centrifugation and resuspended in phosphate-buffered saline (PBS) buffer supplemented with 1 mM DTT, 10 mM CaCl2, cOmplete protease inhibitor cocktail (no. 11697498001, Merck), and deoxyribonuclease I (Sigma-Aldrich, DN25-1G). The cells were disrupted by sonication and centrifuged for 1 hour at 18,000g, and the soluble fraction was then applied to a Glutathione Sepharose 4 Fast Flow purification resin (no. 17513201, Cytiva). After 2 hours of incubation at 4°C, the beads were extensively washed with the same buffer, and thrombin protease was added to the beads, thoroughly mixed, and incubated overnight at 4°C. The next day, the cleaved soluble fraction from the beads was collected and subjected to a HiLoad 16/600 Superdex 75 gel filtration chromatography column, preequilibrated with 20 mM Hepes (pH 8.0), 50 mM NaCl, and 0.01% NaN3. The elution fractions containing the protein were collected and concentrated with 30-kDa molecular weight cutoff of VivaSpin columns. Fascin was labeled with the AX488 C5 maleimide (Invitrogen) with a two times excess dye-to-protein molar ratio. Samples were left to react overnight at 4°C under agitation. Excess dye was removed using a PD MidiTrap G-25 column (Cytiva) in 20 mM Hepes (pH 8.0), 50 mM NaCl, and 0.01% NaN3. Aliquots were flash-frozen in liquid nitrogen and stored at −80°C until used for experiments.
Actin cosedimentation assay
Nonmuscle actin (no. APHL-99, Cytoskeleton Inc.) at a final concentration of 5 μM was prepolymerized in G-buffer [2 mM tris (pH 7.5), 0.2 mM ATP, 0.5 mM β-mercaptoethanol, and 0.2 mM CaCl2] by adding 10× initiation mix [200 mM Hepes (pH 7.4), 1 M KCl, 1 mM EGTA, 10 mM MgCl2, and 2 mM ATP] and incubating for 30 min at RT. Then, recombinantly produced HsFascin was added at a final concentration of 1 μM for 10 min at RT. The mixture was then centrifuged at low speed (19,000 rpm; equivalent to around 14,000g), which sediments only actin filament bundles. As controls, HsFascin alone and F-actin alone were tested side by side. At the end of the centrifugation, the supernatant fractions were carefully separated from the pellets, Laemmli sample buffer was added, and fractions were resolved on SDS–polyacrylamide gel electrophoresis (no. 4561094, Bio-Rad).
GUV sample preparation and observation
For all experiments, coverslips were passivated with a β-casein solution at a concentration of 5 g liter−1 for at least 5 min at RT. Experimental chambers were assembled by placing a silicon open chamber (two-well culture inserts, Ibidi) on a coverslip.
Actin polymerization buffer (F-buffer) contained 60 mM NaCl, 1 mM MgCl2, 0.2 mM EGTA, 0.2 mM ATP, 10 mM DTT, 1 mM DABCO, and 5 mM tris (pH 7.5). Actin polymerization buffer without NaCl (F-buffer–no NaCl) contained 1 mM MgCl2, 0.2 mM EGTA, 0.2 mM ATP, 10 mM DTT, 1 mM DABCO, and 5 mM tris (pH 7.5).
GUVs were mixed sequentially with the ingredients to reach a total volume of 50 μl, if present, in the following order: NaCl (3 μl from 1 M stock to reach a final concentration of 60 mM), F-buffer–no NaCl, IRSp53, VASP, fascin, profilin, CP, phalloidin, GUVs (20 μl), and lastly actin. The GUV-protein mixture was then pipetted using a pipette tip with its tip cut into the experimental chamber, followed by incubating at RT for at least 15 min before observation. The final concentrations of proteins, if present, were as follows: 16 nM IRSp53, 4 nM VASP, 250 nM fascin, 0.6 μM profilin, 25 nM CP, and 0.5 μM actin. If needed, we diluted protein stocks in F-buffer so that the final concentrations of salt and ATP in the GUV-protein mixtures were held approximately constant. To visualize actin by confocal microscopy, depending on the experiments, we either used actin monomers having 10 to 27% fluorescently labeled with AX488 or AX594 or unlabeled actin that included AX488- or AX594-conjugated phalloidin (AX488 phalloidin or AX594 phalloidin) at a final concentration of 0.66 μM. Single actin filaments were prepared by mixing actin monomers with F-buffer to reach a final actin concentration of 0.5 μM.
Samples were observed using either spinning disk confocal microscopes or a laser scanning confocal microscope. The two spinning disk confocal microscopes used were as follows: (i) a Nikon Eclipse Ti-E microscope equipped with a Yokogawa CSU-X1 confocal head, a 100× CFI Plan Apo VC objective, and a complementary metal-oxide semiconductor (CMOS) camera (Prime 95B, Photometrics) and (ii) a Nikon eclipse Ti-E equipped with a 100×/1,4 OIL DIC N2 PL APO VC objective and an electron-multiplying charge-coupled device (CCD) camera (Evolve). The laser scanning confocal microscope was a Nikon TE2000 microscope equipped with a C1 confocal system and a 60× water immersion objective (Nikon, CFI Plan Apo IR 60XWI ON 1.27 DT 0.17).
Characterization of the orientational properties of actin-filled membrane tubes
To characterize the orientational properties of GUVs with actin-filled membrane tubes, we performed image analysis using the “OrientationJ” plugin in ImageJ/Fiji (78). This plugin performs orientation analysis and provides a visual representation of the orientation of the image and coherency of each pixel in the image that indicates whether the local structural feature is oriented or not. The coherency index C is between 0 and 1; C = 1 when the local structure has one dominant orientation and C = 0 when the image is locally isotropic.
Quantification of AX488 actin on GUV membranes
Membrane fluorescence signals of GUVs were used to detect the contours of the GUVs by using the “Fit Circle” function in ImageJ/Fiji. Then, a five-pixel-wide band centered on the GUV contours was used to obtain the actin intensity profile of the band where the x axis of the profile is the length of the band and the y axis is the averaged pixel intensity along the band width. Following background subtraction, the actin intensity was then obtained by calculating the mean value of the intensity values of the profile. To obtain the background intensity of AX488 fluorescence, we first manually drew a five-pixel-wide line perpendicularly across the GUV membranes (in which the x axis of the profile is the length of the line and the y axis is the averaged pixel intensity along the width of the line), and then the background intensity was obtained by calculating the mean value of the sum of the first 10 intensity values and the last 10 intensity values of the background intensity profile.
Estimation of the number of actin filaments in protrusions
To estimate the number of actin filaments inside protrusions, we extracted fluorescence signals of actin in protrusions and normalized them by the fluorescence signals of single actin filaments in the bulk prepared by using the same actin stock as those in the GUV-protein mixture. The microscope settings for image acquisition were identical for the GUV sample and for the corresponding single actin filaments in the bulk. We performed the following steps to extract actin signals in protrusions and in the bulk. We manually defined the region of interest (ROI), a line with a width of five pixels drawn perpendicularly across protrusions or single actin filaments. We then obtained the actin fluorescent intensity profile of the line where the x axis of the profile is the length of the line and the y axis is the averaged pixel intensity along the width of the line. The background intensity was obtained by calculating the mean value of the sum of the first five intensity values and the last five intensity values of the intensity profile. Last, the actin fluorescence intensities were obtained by subtracting the background intensity from the maximum intensity value in the intensity profile. This image process was performed using ImageJ/Fiji (79).
Characterization of IRSp53 clustering on GUVs
To define clusters of IRSp53 on a surface of a GUV, we segmented fluorescence images of the protein using a custom-made ImageJ/Fiji script based on the Rényi entropy algorithm (80). We first created a mask of the fluorescent image in the IRSp53 channel using a Rényi entropy threshold and then used the “Analyze Particles” function in ImageJ/Fiji to define clusters as a set of nonconnected areas that have nonzero values in the mask image, permitting quantification of the number of clusters and their areas.
CG model and clustering analysis
The CG model used here has been discussed in detail previously (81, 82). The low-resolution, phenomenological CG model contains a three-bead quasi-monolayer membrane model and a curved I-BAR domain model. The quasi-monolayer model is highly tunable and can accommodate significant remodeling, which makes it appealing for the application to I-BAR domains. The three beads interact internally with two harmonic bonds with a force constant of 25 kBT where kB is Boltzmann’s constant and T is temperature and an equilibrium distance of 0.9 nm and a harmonic angle potential with a force constant of 10 kBT and an equilibrium angle of 180°. The intermolecular forces were modeled using a soft pair potential, shown below, where A and B dictate the softness of the repulsion and the depth of the attraction and r0 and rc are the onset of repulsion cutoff and attraction cutoff, respectively
Both the membrane and the protein have an A value of 25 kBT to maintain an excluded volume and an rc value of 2 · r0, where r0 differs between the membrane and protein. Hence, r0 and B distinguish the species in the model. The membrane model is three linear beads: The two on the ends are smaller and only interact with other lipids while the central membrane bead is larger and attracted to other membrane beads and the membrane binding interface of the protein. The smaller beads have a B value of 0 (i.e., no attraction) and an r0 value of 1.125 nm. The central membrane beads have a membrane-membrane B value of 0.6 kBT and an r0 value of 1.5 nm. The PS-like and PIP2-like membrane beads interact the same with each other, the key difference being the protein-membrane interaction. With this parameter set, the quasi-monolayer is fluid with a bending modulus around 10 kBT. The protein model is made of three curved strings of beads to capture the curvature and size of the I-BAR domain of IRSp53 with an r0 value of 1.7 nm. There are two outer strings of beads (shown in red in Fig. 2D, top, and movie S2) that capture the shape with a B value of 0, while the central string is the membrane-binding interface. The middle beads of the central string represent a weakly binding surface that is attracted to the all-membrane beads with a B value of 0.235 kBT (shown in yellow in Fig. 2D, bottom, and movie S3), and the ends of the membrane-binding interface have a B value of 0.235 kBT to PS-like membrane beads and a B value of 0.705 kBT to PIP2-like membrane beads. The PIP2-like membrane beads are attracted to the ends of the I-BAR domain model to recapitulate the experimentally observed behavior of PIP2 clustering by I-BAR domains (31).
All systems were run using the large-scale atomic/molecular massively parallel simulator (LAMMPS) molecular dynamics engine with a time step of 200 fs (83). The initial positions of the system were a flat membrane of 188,031 membrane beads with 65 I-BAR proteins slightly above the membrane. The systems were equilibrated for 50 million time steps in the NPXYLZT ensemble at 0 surface tension using the Parrinello-Rahman barostat with a dampening constant of 200 ps (84). Temperature was maintained using the Langevin thermostat with a dampening constant of 20 ps (85). The simulations were then run for another 500 million time steps in the canonical ensemble.
Clustering analysis was performed by creating a network of neighbors, where I-BAR domains were neighbors if they were within 2 nm in the xy plane. Two I-BAR domains were part of the same cluster if and only if a path existed in the network of neighbors. Thus, any I-BAR domain without a neighbor was considered free and not part of any aggregate. The probability density was estimated using kernel density estimation from the histogram of neighbor sizes from the last 250 million time steps of the simulation and subsequently used to calculate the probability of I-BAR domain aggregates containing less than five I-BAR domains and five or more I-BAR domains. The PIP2-like membrane beads were separated into two groups: within 2 nm of an I-BAR domain in the xy plane (neighboring membrane beads) and not within 2 nm of an I-BAR domain (bulk membrane beads). The percent of PIP2-like membrane beads within each group was quantified to measure the enrichment of PIP2 near I-BAR domains. The CG model analysis was performed using numpy, scikit-learn, and freud python packages, and results were plotted using the matplotlib python package (86–89). All snapshots of the CG systems were created using visual molecular dynamics (VMD) 1.9.2 (90).
Live-cell imaging and membrane nanotube pulling experiments
Cell culture, plasmids, and transfection
Rat2 fibroblasts (CRL-1764, American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) GlutaMAX high-glucose media (Gibco) supplemented with 1 mM sodium pyruvate (Gibco), 10% fetal bovine serum (Eurobio), and 1% penicillin and streptomycin (Sigma-Aldrich). Cells were maintained in a humidified incubator with 5% CO2 at 37°C. Rat2 cells were routinely monitored for mycoplasma contamination and found to be negative. Common plasmids used for mammalian cell transfection include the following: (i) IRSp53 (human, full-length; UniProt, no. Q9UQB8) cloned into pmCherry-N1 and pEGFP-N1 vectors (Clontech), fusing the fluorescent proteins to the C-terminus of IRSp53 (IRSp53-mCherry and IRSp53-eGFP); (ii) IRSp53’s I-BAR domain (mouse, residues 1 to 250; UniProt, no. Q8BKX1) cloned into a pEGFP-N1 vector (I-BAR–eGFP); (iii) Fascin1 (human, full-length; UniProt, no. Q16658) cloned into pmCherry-C1 and pEGFP-C1 vectors (Clontech), fusing the fluorescent proteins to fascin’s N-terminus (mCherry-fascin and eGFP-fascin); (iv) pmTagRFP-VASP (mouse, full-length; UniProt, no. P70460), which was a gift from M. Davidson (Addgene, plasmid no. 58027); and (v) an empty mCherry vector.
For the live-cell imaging experiments (Figs. 1 and 4), cells were transiently transfected the day before imaging using the Xfect transfection reagent (Takara Bio) according to the manufacturer’s recommendations and replated on fibronectin-coated glass-bottom dishes (35 mm; no. 1.5H, Ibidi) 2 hours before imaging. Fibronectin (from human plasma, >95%; Sigma-Aldrich) coating was done by incubating the dishes 1 hour at RT (or overnight at 4°C) with a fibronectin solution (10 μg/ml) dissolved in PBS.
For the nanotube pulling experiments (Figs. 5 and 6), stable Rat2 cell lines expressing IRSp53-eGFP or I-BAR–eGFP were generated by antibiotic selection [geneticin (0.5 mg/ml) (G418), 7 days] after transfection (FuGENE HD, Promega) followed by cell sorting using an SH800 cell sorter (Sony Biotechnology) to collect a pool of fluorescent-positive cells. For fig. S19, wild-type Rat2 cells were transiently transfected using FuGENE HD (Promega) or Xfect (Takara Bio) reagents to achieve double-expressing IRSp53-eGFP/RFP-VASP or eGFP-Fascin/IRSp53-mCherry cells.
Live-cell imaging of filopodium formation
Time-lapse image series for double-transfected Rat2 cells expressing IRSp53-eGFP/RFP-VASP, IRSp53-eGFP/mCherry-fascin, or IRSp53-eGFP/mCherry were obtained using a GE DeltaVision Ultra wide-field microscope equipped with a pco.edge 4.2ge scientific CMOS camera (PCO GmbH), solid-state illuminators for fluorescence excitation, an incubation system set at 37°C and 5% CO2, and 63× or 100× oil objectives. Deconvolution of acquired time-lapse series was performed using the built-in microscopy software (softWoRx). Further analysis was conducted with the Microscopy Image Browser, a free MATLAB-based software developed by I. Belevich (University of Helsinki).
Filopodium quantification
Each analyzed filopodium was monitored from the initiation of membrane bending (cone formation) until the beginning of active retraction. Changes in spatial coordinates of the positions of base and tip with the times were used to calculate the length of growing filopodia (“Distance”). The rate of growth (μm/s) was calculated as the first derivative of filopodium length with respect to time (). Changes in the rate of growth were used to assess peculiarities of filopodium formation (stops, i.e., when = 0, and instantaneous retractions when < 0). The frequency of each event (s−1) was calculated as the number of stops or retractions per second registered for the period of active filopodium growth.
To assess the effect of fascin accumulation in filopodia on their elongation rates, separate kymographs of growing filopodia (time-space plots) for IRSp53-eGFP and mCherry-fascin channels were generated using the “Multi-Kymograph” plugin (https://biii.eu/multi-kymograph) in ImageJ/Fiji. Velocities were measured in ImageJ/Fiji for selected linear intervals of the kymographs (about 10 to 20 s) before and after fascin appearance using the “Velocity Measurement Tool” macro (https://dev.mri.cnrs.fr/projects/imagej-macros/wiki/Velocity_Measurement_Tool).
We followed the colocalization analysis as published previously (46) using the “Colocalization colormap” (91) plugin implemented in ImageJ/Fiji. For each pixel in an image, one calculates the deviation of the fluorescent intensity from the mean intensity of the whole image. Then, one calculates the mean deviation product of the two protein signals, , where I and are the pixel intensity and the mean intensity of the image, respectively. Last, to compare series of images, one calculates the nMDP, , where Imax is the maximum intensity of the corresponding image. A spatial representation of the correlation of the two protein signals can be obtained. We manually tracked filopodia, and for each time point, we averaged the nMDP values of four pixels located at the filopodial structures. We overlapped the nMDP values, termed the colocalization index, during filopodium formation.
Bead functionalization
An aliquot of streptavidin-coated polystyrene beads [0.5% (w/v); SVP-30-5, Spherotech] having a nominal diameter of 3 μm was washed three times in a 10× volume of PBS. Between washing steps, beads were pelleted using a centrifuge at 12,000 rpm for 5 min. The final pellet was resuspended in PBS to a concentration of 0.05% (w/v). Next, a volume of biotin-conjugated concanavalin A (ConA; C2272, Sigma-Aldrich), having a stock concentration of 1 mg ml−1 in PBS, was added to the bead suspension assuming a binding capacity of 10 μg of protein per milligram of solid particles. The mixture was incubated overnight at 4°C on a tabletop shaker set to 1500 rpm. The ConA-coated beads were rinsed three times according to the steps above and lastly resuspended in PBS to a concentration of 0.5% (w/v). ConA beads were stored at 4°C and generally usable up to 1 month.
Nanotube pulling experiments
A custom-built optical tweezer setup coupled to an inverted Nikon C1 Plus laser scanning confocal microscope, as previously described in (61), was used for pulling plasma membrane nanotubes, force measurements, and simultaneous fluorescence imaging. Briefly, a 1064-nm continuous wave ytterbium fiber laser (IPG Photonics) set to a 3-W input power was modulated to 400 mW (measured at the back aperture of the objective) using a polarizing beam splitter (Thorlabs), expanded through a telescope consisting of two plano-convex lenses with focal lengths of 100 and 150 mm (Thorlabs), and directed toward the back aperture of a Nikon CFI Plan Apochromat Lambda 100× 1.45 numerical aperture oil immersion objective. The trap stiffness κ was determined using the viscous drag method, including Faxen’s correction for calibration near surfaces (92) and averaged 60 pN μm−1. Displacements of a trapped bead from the fixed trap center were recorded using a Marlin F-046B CCD camera (Allied-Vision) at a frame rate of 20 frames/s and later analyzed by a custom MATLAB (MathWorks) script using the imfindcircles function to output the center location of the tracked bead (in micrometers). Forces were calculated from the determined bead positions according to the equation, F = κ ∙ (x − xo), where κ is the trap stiffness, x is the displaced bead position, and xo is the equilibrium reference position of the trapped bead. As the optical trap itself was stationary, all relative movements were performed using a piezo-driven stage (Nano-LP100, Mad City Labs). Atop the stage, a temperature- and CO2-controllable stage-top incubator (STXG-WELSX, Tokai Hit) was attached, allowing cells to be maintained at 37°C in a humidified, 5% CO2 atmosphere during experimentation.
The day before, Rat2 cells expressing I-BAR–eGFP or IRSp53-eGFP were plated on fibronectin-coated (35 μg ml−1) glass-bottom dishes (35 mm; no. 1.5, MatTek) at a density of ~30,000 cells cm−2. Thirty to 60 min before experimentation, the phenol-containing culture medium was removed, cells were rinsed with PBS, and phenol-free DMEM containing the ProLong Live Antifade Reagent (Invitrogen) at a 1:75 dilution, and β-casein (2 mg ml−1) (>98% pure, from bovine milk; C6905, Sigma-Aldrich) for surface passivation, was applied. The cells were taken to the optical tweezer setup and labeled with the CellMask Deep Red plasma membrane stain (Invitrogen) at a 1:2000 dilution for 10 min, and ConA-coated beads were added (1:50 to 1:100 dilution). Using a custom LabVIEW (National Instruments) program to control the piezo stage, membrane nanotubes were pulled by trapping an isolated floating bead, bringing it into contact with the cell for a short period of time (<10 s), and then moving the cell away from the bead in the x direction.
Confocal images were acquired using solid-state excitation lasers: 488 nm (Coherent), 561 nm (Coherent), and 642 nm (Melles Griot). The detection pathway consisted of a T560lpxr dichroic beam splitter (Chroma), an ET525/50 band-pass filter (Chroma), an ET665 long-pass filter (Chroma), and two PicoQuant tau single-photon avalanche diodes (τ-SPAD). The τ-SPADs were controlled by the SymPhoTime 64 software (PicoQuant). Images encompassing the nanotube and some of the cell body (typically 1024 pixels by 512 pixels, ×5 zoom) were gathered 5 min after the nanotube was pulled for protein sorting analysis; identical acquisition parameters were used when gathering the membrane and protein channel data for an individual nanotube.
Sorting analysis
Image analysis was performed using custom-written macros in the ImageJ/Fiji software. To quantify protein enrichment in the nanotube (t) relative to the bulk plasma membrane of the cell (ref), the sorting parameter (S) is defined as , where the ratio of the green protein fluorescence (IP) in the nanotube and in the cell is normalized by the same ratio of the red lipid fluorescence (IL). This is to correct for cell-to-cell differences in both the protein and membrane signal intensity (e.g., protein expression levels, different acquisition parameters between cells, and efficiency of membrane staining). Sorting values of S > 1 signify that the protein is enriched in the nanotube, while S < 1 signify that the protein is excluded from the nanotube, with respect to its average density in the cell plasma membrane. Given that Rat2 fibroblasts are quite flat, we focused on the ventral side of the cell to obtain reference images of the plasma membrane for both the protein and membrane channels. Images were background-subtracted, and common regions of interest (~20,000 pixels in size) were then manually drawn to encompass homogeneous areas of the plasma membrane (near the site of the nanotube and excluding areas where vesicular puncta were observed) to calculate average protein and lipid reference values, and , respectively. The protein and membrane channels of the in-focus nanotube were also subjected to background subtraction and then normalized by their respective reference values to generate a heatmap of S values. The resulting S map was filtered by a 3 × 3 adaptive median filter (https://weisongzhao.github.io/AdaptiveMedian.imagej/) to remove spurious pixels in the background; this processing was done given that the pixel values of the raw images are discrete values (photon counts) and not continuous values. The width of the nanotube was fit to a Gaussian, and a rectangular ROI (size of ±2σ of the Gaussian profile) was defined along the length of the nanotube in the S map. Orthogonal cross sections were iteratively generated pixel by pixel along the length of the nanotube within the ROI. The maximum S value for each cross section was determined and then averaged to report the mean sorting value of the protein in the nanotube (Savg).
Statistical analysis
All graphs and statistical analyses were performed using the GraphPad Prism software (versions 9.3.1 or 9.4.0 for Mac or Windows, respectively). P values of <0.05 were considered statistically significant, and all P values are indicated in the figures. The specific statistical tests performed, the number of independent experiments, and the total number of samples analyzed are indicated in the figure captions.
Acknowledgments
The computations were supported by the University of Chicago Research Computing Center (RCC). We thank E. Coudrier and C. Simon for insightful discussions. We also thank F. Di Federico for handling plasmids, F. Tabarin-Cayrac for cell sorting, and A.-S. Mace for ImageJ programming assistance. F.-C.T., C.L.C., and P.B. are members of the CNRS consortium AQV. F.-C.T. and P.B. are members of the Labex Cell(n)Scale (ANR-11-LABX0038) and Paris Sciences et Lettres (ANR-10-IDEX-0001-02). We acknowledge the Cell and Tissue Imaging Core facility (PICT IBiSA), Institut Curie, member of the French National Research Infrastructure France-BioImaging (ANR10-INBS-04).
Funding: This work was supported by Human Frontier Science Program (HFSP) grant RGP0005/2016 (to F.-C.T., J.M.H., G.A.V., P.L., and P.B.), Institut Curie and the Centre National de la Recherche Scientifique (CNRS) (to F.-C.T., J.M.H., and P.B.), Marie Curie actions H2020-MSCA-IF-2014 (to F.-C.T.), EMBO Long-Term fellowship ALTF 1527-2014 (to F.-C.T.), Pasteur Foundation Fellowship (to J.M.H.), Agence Nationale pour la Recherche ANR-20-CE13-0032 (to J.M.H. and P.B.) and ANR-20-CE11-0010-01 (to F.-C.T), Université Paris Sciences et Lettres-QLife Institute ANR-17-CONV-0005 Q-LIFE (to P.B.), FY 2015 Researcher Exchange Program between the Japan Society for the Promotion of Science and Academy of Finland (to Y.S.), the Takeda Science Foundation (to Y.S.), the Wesco Scientific Promotion Foundation (to Y.S.), Agence Nationale pour la Recherche ANR-18-CE13-0026-01 and ANR-21-CE13-0010-03 (to C.L.C.), Cancer Society Finland 4705949 (to P.L.), and U.S. National Institutes of Health (NIH) Institute of General Medical Sciences (NIGMS) grant R01-GM063796 (to G.A.V. and Z.J.).
Author contributions: G.A.V., P.L., and P.B. designed the initial project. F.-C.T., J.M.H., Z.J., E.K., Y.S., J.P., O.M., and J.M. performed experiments and analyzed results under the supervision of C.L.C., G.A.V., P.L., and P.B. F.-C.T. and J.P. developed GUV experiments, and F.-C.T. performed and analyzed data from the GUV experiments. J.M.H., Z.J., E.K., and J.P. performed and analyzed data from nanotube pulling experiments, computer simulations, live-cell imaging experiments, and actin pyrene assays, respectively. Y.S., J.P., and J.M. purified proteins. O.M. analyzed protein clustering on GUVs. F.-C.T. wrote the original draft with inputs and revisions from J.M.H. and P.B. Conceptualization: F.-C.T., J.M.H., G.A.V., P.L., and P.B. Methodology: F.-C.T., J.M.H., Z.J., and J.P. Investigation: F.-C.T., J.M.H., Z.J., E.K., and J.P. Resources: Y.S., J.P., and J.M. Visualization: F.-C.T., J.M.H., Z.J., and E.K. Supervision: C.L.C., G.A.V., P.L., and P.B. Writing—original draft: F.-C.T., J.M.H., and P.B. Writing—review and editing: F.-C.T., J.M.H., Z.J., E.K., Y.S., J.P., O.M., J.M., C.L.C., G.A.V., P.L., and P.B. Funding acquisition: G.A.V., P.L., and P.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S19
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S11
REFERENCES AND NOTES
- 1.Blanchoin L., Boujemaa-Paterski R., Sykes C., Plastino J., Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94, 235–263 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Rottner K., Schaks M., Assembling actin filaments for protrusion. Curr. Opin. Cell Biol. 56, 53–63 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Pollard T. D., Cooper J. A., Actin, a central player in cell shape and movement. Science 326, 1208–1212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faix J., Breitsprecher D., Stradal T. E., Rottner K., Filopodia: Complex models for simple rods. Int. J. Biochem. Cell Biol. 41, 1656–1664 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Mattila P. K., Lappalainen P., Filopodia: Molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9, 446–454 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Vignjevic D., Kojima S., Aratyn Y., Danciu O., Svitkina T., Borisy G. G., Role of fascin in filopodial protrusion. J. Cell Biol. 174, 863–875 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansen S., Collins A., Yang C., Rebowski G., Svitkina T., Dominguez R., Mechanism of actin filament bundling by fascin. J. Biol. Chem. 286, 30087–30096 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steketee M. B., Tosney K. W., Three functionally distinct adhesions in filopodia: Shaft adhesions control lamellar extension. J. Neurosci. 22, 8071–8083 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C., Svitkina T., Filopodia initiation: Focus on the Arp2/3 complex and formins. Cell Adh. Migr. 5, 402–408 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svitkina T. M., Bulanova E. A., Chaga O. Y., Vignjevic D. M., Kojima S., Vasiliev J. M., Borisy G. G., Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409–421 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vignjevic D., Yarar D., Welch M. D., Peloquin J., Svitkina T., Borisy G. G., Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 160, 951–962 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mogilner A., Rubinstein B., The physics of filopodial protrusion. Biophys. J. 89, 782–795 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young L. E., Heimsath E. G., Higgs H. N., Cell type–dependent mechanisms for formin-mediated assembly of filopodia. Mol. Biol. Cell 26, 4646–4659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steffen A., Faix J., Resch G. P., Linkner J., Wehland J., Small J. V., Rottner K., Stradal T. E., Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol. Biol. Cell 17, 2581–2591 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damiano-Guercio J., Kurzawa L., Mueller J., Dimchev G., Schaks M., Nemethova M., Pokrant T., Brühmann S., Linkner J., Blanchoin L., Sixt M., Rottner K., Faix J., Loss of Ena/VASP interferes with lamellipodium architecture, motility and integrin-dependent adhesion. eLife 9, e55351 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Disanza A., Bisi S., Winterhoff M., Milanesi F., Ushakov D. S., Kast D., Marighetti P., Romet-Lemonne G., Müller H.-M., Nickel W., Linkner J., Waterschoot D., Ampè C., Cortellino S., Palamidessi A., Dominguez R., Carlier M.-F., Faix J., Scita G., CDC42 switches IRSp53 from inhibition of actin growth to elongation by clustering of VASP. EMBO J. 32, 2735–2750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim K. B., Bu W., Goh W. I., Koh E., Ong S. H., Pawson T., Sudhaharan T., Ahmed S., The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J. Biol. Chem. 283, 20454–20472 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Yang C., Hoelzle M., Disanza A., Scita G., Svitkina T., Coordination of membrane and actin cytoskeleton dynamics during filopodia protrusion. PLOS ONE 4, e5678 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krugmann S., Jordens I., Gevaert K., Driessens M., Vandekerckhove J., Hall A., Cdc42 induces filopodia by promoting the formation of an IRSp53: Mena complex. Curr. Biol. 11, 1645–1655 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Disanza A., Mantoani S., Hertzog M., Gerboth S., Frittoli E., Steffen A., Berhoerster K., Kreienkamp H.-J., Milanesi F., Di Fiore P. P., Ciliberto A., Stradal T. E. B., Scita G., Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8–IRSp53 complex. Nat. Cell Biol. 8, 1337–1347 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Kast D. J., Dominguez R., IRSp53 coordinates AMPK and 14-3-3 signaling to regulate filopodia dynamics and directed cell migration. Mol. Biol. Cell 30, 1285–1297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govind S., Kozma R., Monfries C., Lim L., Ahmed S., Cdc42Hs facilitates cytoskeletal reorganization and neurite outgrowth by localizing the 58-kD insulin receptor substrate to filamentous actin. J. Cell Biol. 152, 579–594 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Millard T. H., Bompard G., Heung M. Y., Dafforn T. R., Scott D. J., Machesky L. M., Fütterer K., Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 24, 240–250 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suetsugu S., Kurisu S., Takenawa T., Dynamic shaping of cellular membranes by phospholipids and membrane-deforming proteins. Physiol. Rev. 94, 1219–1248 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Mattila P. K., Pykalainen A., Saarikangas J., Paavilainen V. O., Vihinen H., Jokitalo E., Lappalainen P., Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 176, 953–964 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miki H., Yamaguchi H., Suetsugu S., Takenawa T., IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408, 732–735 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Goh W. I., Lim K. B., Sudhaharan T., Sem K. P., Bu W., Chou A. M., Ahmed S., mDia1 and WAVE2 proteins interact directly with IRSp53 in filopodia and are involved in filopodium formation. J. Biol. Chem. 287, 4702–4714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kast D. J., Yang C., Disanza A., Boczkowska M., Madasu Y., Scita G., Svitkina T., Dominguez R., Mechanism of IRSp53 inhibition and combinatorial activation by Cdc42 and downstream effectors. Nat. Struct. Mol. Biol. 21, 413–422 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robens J. M., Yeow-Fong L., Ng E., Hall C., Manser E., Regulation of IRSp53-dependent filopodial dynamics by antagonism between 14-3-3 binding and SH3-mediated localization. Mol. Cell. Biol. 30, 829–844 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kast D. J., Dominguez R., Mechanism of IRSp53 inhibition by 14-3-3. Nat. Commun. 10, 483 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saarikangas J., Zhao H., Pykäläinen A., Laurinmäki P., Mattila P. K., Kinnunen P. K., Butcher S. J., Lappalainen P., Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 19, 95–107 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Prevost C., Zhao H., Manzi J., Lemichez E., Lappalainen P., Callan-Jones A., Bassereau P., IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nat. Commun. 6, 8529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breuer A., Lauritsen L., Bertseva E., Vonkova I., Stamou D., Quantitative investigation of negative membrane curvature sensing and generation by I-BARs in filopodia of living cells. Soft Matter 15, 9829–9839 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Gov N., Guided by curvature: Shaping cells by coupling curved membrane proteins and cytoskeletal forces. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20170115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young L. E., Latario C. J., Higgs H. N., Roles for Ena/VASP proteins in FMNL3-mediated filopodial assembly. J. Cell Sci. 131, jcs220814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen S. D., Mullins R. D., VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 191, 571–584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breitsprecher D., Kiesewetter A. K., Linkner J., Urbanke C., Resch G. P., Small J. V., Faix J., Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 27, 2943–2954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen S. D., Mullins R. D., Lamellipodin promotes actin assembly by clustering Ena/VASP proteins and tethering them to actin filaments. eLife 4, e06585 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brühmann S., Ushakov D. S., Winterhoff M., Dickinson R. B., Curth U., Faix J., Distinct VASP tetramers synergize in the processive elongation of individual actin filaments from clustered arrays. Proc. Natl. Acad. Sci. U.S.A. 114, E5815–E5824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harker A. J., Katkar H. H., Bidone T. C., Aydin F., Voth G. A., Applewhite D. A., Kovar D. R., Ena/VASP processive elongation is modulated by avidity on actin filaments bundled by the filopodia cross-linker fascin. Mol. Biol. Cell 30, 851–862 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rottner K., Behrendt B., Small J. V., Wehland J., VASP dynamics during lamellipodia protrusion. Nat. Cell Biol. 1, 321–322 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Cheng K. W., Mullins R. D., Initiation and disassembly of filopodia tip complexes containing VASP and lamellipodin. Mol. Biol. Cell 31, 2021–2034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.T. Pokrant, J. I. Hein, S. Körber, A. Disanza, A. Pich, G. Scita, K. Rottner, J. Faix, Ena/VASP clustering at microspike tips involves Lamellipodin but not I-BAR proteins, and absolutely requires unconventional Myosin-X. bioRxiv 10.1101/2022.05.12.491613 [Preprint]. 12 May 2022. 10.1101/2022.05.12.491613. [DOI] [PMC free article] [PubMed]
- 44.Faix J., Rottner K., Ena/VASP proteins in cell edge protrusion, migration and adhesion. J. Cell Sci. 135, jcs259226 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Jacquemet G., Stubb A., Saup R., Miihkinen M., Kremneva E., Hamidi H., Ivaska J., Filopodome mapping identifies p130Cas as a mechanosensitive regulator of filopodia stability. Curr. Biol. 29, 202–216.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaskolski F., Mulle C., Manzoni O. J., An automated method to quantify and visualize colocalized fluorescent signals. J. Neurosci. Methods 146, 42–49 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Pollard D. A., Pollard T. D., Pollard K. S., Empowering statistical methods for cellular and molecular biologists. Mol. Biol. Cell 30, 1359–1368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLaughlin S., Wang J., Gambhir A., Murray D., PIP2 and proteins: Interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 31, 151–175 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Zhao H., Michelot A., Koskela E. V., Tkach V., Stamou D., Drubin D. G., Lappalainen P., Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep. 4, 1213–1223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shekhar S., Pernier J., Carlier M.-F., Regulators of actin filament barbed ends at a glance. J. Cell Sci. 129, 1085–1091 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Kaiser D. A., Vinson V. K., Murphy D. B., Pollard T. D., Profilin is predominantly associated with monomeric actin in Acanthamoeba. J. Cell Sci. 112, 3779–3790 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Kinosian H. J., Selden L. A., Gershman L. C., Estes J. E., Interdependence of profilin, cation, and nucleotide binding to vertebrate non-muscle actin. Biochemistry 39, 13176–13188 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Huang Z., Haugland R. P., You W., Haugland R. P., Phallotoxin and actin binding assay by fluorescence enhancement. Anal. Biochem. 200, 199–204 (1992). [DOI] [PubMed] [Google Scholar]
- 54.Atilgan E., Wirtz D., Sun S. X., Mechanics and dynamics of actin-driven thin membrane protrusions. Biophys. J. 90, 65–76 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniels D. R., Turner M. S., Islands of conformational stability for filopodia. PLOS ONE 8, e59010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sampath P., Pollard T. D., Effects of cytochalasin, phalloidin and pH on the elongation of actin filaments. Biochemistry 30, 1973–1980 (1991). [DOI] [PubMed] [Google Scholar]
- 57.Isambert H., Venier P., Maggs A. C., Fattoum A., Kassab R., Pantaloni D., Carlier M.-F., Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 270, 11437–11444 (1995). [DOI] [PubMed] [Google Scholar]
- 58.Jarin Z., Tsai F.-C., Davtyan A., Pak A. J., Bassereau P., Voth G. A., Unusual organization of I-BAR proteins on tubular and vesicular membranes. Biophys. J. 117, 553–562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu A. P., Richmond D. L., Maibaum L., Pronk S., Geissler P. L., Fletcher D. A., Membrane-induced bundling of actin filaments. Nat. Phys. 4, 789–793 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simon C., Kusters R., Caorsi V., Allard A., Abou-Ghali M., Manzi J., Di Cicco A., Lévy D., Lenz M., Joanny J.-F., Campillo C., Plastino J., Sens P., Sykes C., Actin dynamics drive cell-like membrane deformation. Nat. Phys. 15, 602–609 (2019). [Google Scholar]
- 61.Bornschlögl T., Romero S., Vestergaard C. L., Joanny J.-F., Van Nhieu G. T., Bassereau P., Filopodial retraction force is generated by cortical actin dynamics and controlled by reversible tethering at the tip. Proc. Natl. Acad. Sci. U.S.A. 110, 18928–18933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leijnse N., Oddershede L. B., Bendix P. M., Helical buckling of actin inside filopodia generates traction. Proc. Natl. Acad. Sci. U.S.A. 112, 136–141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Etienne-Manneville S., Hall A., Rho GTPases in cell biology. Nature 420, 629–635 (2002). [DOI] [PubMed] [Google Scholar]
- 64.de Beco S., Vaidžiulytė K., Manzi J., Dalier F., Di Federico F., Cornilleau G., Dahan M., Coppey M., Optogenetic dissection of Rac1 and Cdc42 gradient shaping. Nat. Commun. 9, 4816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Picas L., Viaud J., Schauer K., Vanni S., Hnia K., Fraisier V., Roux A., Bassereau P., Gaits-Iacovoni F., Payrastre B., Laporte J., Manneville J. B., Goud B., BIN1/M-Amphiphysin2 induces clustering of phosphoinositides to recruit its downstream partner dynamin. Nat. Commun. 5, 5647 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Senju Y., Lappalainen P., Regulation of actin dynamics by PI (4, 5) P2 in cell migration and endocytosis. Curr. Opin. Cell Biol. 56, 7–13 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Senju Y., Kalimeri M., Koskela E. V., Somerharju P., Zhao H., Vattulainen I., Lappalainen P., Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc. Natl. Acad. Sci. U.S.A. 114, E8977–E8986 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsai F.-C., Bertin A., Bousquet H., Manzi J., Senju Y., Tsai M.-C., Picas L., Miserey-Lenkei S., Lappalainen P., Lemichez E., Coudrier E., Bassereau P., Ezrin enrichment on curved membranes requires a specific conformation or interaction with a curvature-sensitive partner. eLife 7, e37262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sudhaharan T., Hariharan S., Lim J. S. Y., Liu J. Z., Koon Y. L., Wright G. D., Chiam K. H., Ahmed S., Superresolution microscopy reveals distinct localisation of full length IRSp53 and its I-BAR domain protein within filopodia. Sci. Rep. 9, 2524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dobramysl U., Jarsch I. K., Inoue Y., Shimo H., Richier B., Gadsby J. R., Mason J., Szałapak A., Ioannou P. S., Correia G. P., Walrant A., Butler R., Hannezo E., Simons B. D., Gallop J. L., Stochastic combinations of actin regulatory proteins are sufficient to drive filopodia formation. J. Cell Biol. 220, e202003052 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tokuo H., Ikebe M., Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 319, 214–220 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Barzik M., McClain L. M., Gupton S. L., Gertler F. B., Ena/VASP regulates mDia2-initiated filopodial length, dynamics, and function. Mol. Biol. Cell 25, 2604–2619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinberger A., Tsai F.-C., Koenderink G. H., Schmidt T. F., Itri R., Meier W., Schmatko T., Schröder A., Marques C., Gel-assisted formation of giant unilamellar vesicles. Biophys. J. 105, 154–164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spudich J. A., Watt S., The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 (1971). [PubMed] [Google Scholar]
- 75.Ciobanasu C., Faivre B., Le Clainche C., Reconstituting actomyosin-dependent mechanosensitive protein complexes in vitro. Nat. Protoc. 10, 75–89 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Gieselmann R., Kwiatkowski D. J., Janmey P. A., Witke W., Distinct biochemical characteristics of the two human profilin isoforms. Eur. J. Biochem. 229, 621–628 (1995). [DOI] [PubMed] [Google Scholar]
- 77.Wioland H., Guichard B., Senju Y., Myram S., Lappalainen P., Jégou A., Romet-Lemonne G., ADF/cofilin accelerates actin dynamics by severing filaments and promoting their depolymerization at both ends. Curr. Biol. 27, 1956–1967.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Püspöki Z., Storath M., Sage D., Unser M., Transforms and operators for directional bioimage analysis: A survey. Adv. Anat. Embryol. Cell Biol. 219, 69–93 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sahoo P., Wilkins C., Yeager J., Threshold selection using Renyi’s entropy. Pattern Recognit. 30, 71–84 (1997). [Google Scholar]
- 81.J. Grime, J. J. Madsen, Efficient simulation of tunable lipid assemblies across scales and resolutions. arXiv:1910.05362 [physics.chem-ph] (11 October 2019).
- 82.Jarin Z., Pak A. J., Bassereau P., Voth G. A., Lipid-composition-mediated forces can stabilize tubular assemblies of i-bar proteins. Biophys. J. 120, 46–54 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Plimpton S., Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 117, 1–19 (1995). [Google Scholar]
- 84.Parrinello M., Rahman A., Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 85.Schneider T., Stoll E., Molecular-dynamics study of a three-dimensional one-component model for distortive phase transitions. Phys. Rev. B 17, 1302–1322 (1978). [Google Scholar]
- 86.Harris C. R., Millman K. J., Van Der Walt S. J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N. J., Kern R., Picus M., Hoyer S., van Kerkwijk M. H., Brett M., Haldane A., Del Río J. F., Wiebe M., Peterson P., Gérard-Marchant P., Sheppard K., Reddy T., Weckesser W., Abbasi H., Gohlke C., Oliphant T. E., Array programming with NumPy. Nature 585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay É., Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 88.Ramasubramani V., Dice B. D., Harper E. S., Spellings M. P., Anderson J. A., Glotzer S. C., freud: A software suite for high throughput analysis of particle simulation data. Comp. Phys. Commun. 254, 107275 (2020). [Google Scholar]
- 89.Hunter J. D., Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007). [Google Scholar]
- 90.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 91.Gorlewicz A., Krawczyk K., Szczepankiewicz A. A., Trzaskoma P., Mulle C., Wilczynski G. M., Colocalization colormap—An ImageJ plugin for the quantification and visualization of colocalized signals. Neuroinformatics 18, 661–664 (2020). [DOI] [PubMed] [Google Scholar]
- 92.Neuman K., Block S., Optical trapping. Rev. Sci. Instrum. 75, 2787–2809 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S19
Movies S1 to S11