Abstract
The Toscana virus (family Bunyaviridae, genus Phlebovirus) is the only sandfly-transmitted virus that demonstrates neurotropic activity. Clinical cases ranging from aseptic meningitis to meningoencephalitis caused by Toscana virus are yearly observed in central Italy during the summer, and several cases have been reported among tourists returning from zones of endemicity (Italy, Portugal, Spain, and Cyprus). In Toscana virus patients, immunoglobulin M (IgM) antibodies, usually present at the onset of symptoms, can reveal elevated titers by enzyme-linked immunosorbent assay and can persist for at least 1 year. IgG antibodies can be absent at the onset of symptoms: titers rise in convalescent sera and persist for many years. At least five proteins have been identified in Toscana virus-infected cells: nucleoprotein N, glycoproteins G1 and G2, a large protein (L) assumed to be a component of the polymerase, and two nonstructural proteins, NSm and NSs. We report results of a study on the antibody response to individual viral proteins in patients with Toscana virus-associated acute neurologic disease. Immunoblotting and semiquantitative radioimmunoprecipitation assay (RIPA) allow identification of nucleoprotein N as the major antigen responsible for both IgM and IgG responses. Antibodies to proteins other than nucleoprotein N are detected only by RIPA. Antibodies to glycoproteins are detected in about one-third of patients, and whereas their presence always predicts neutralization, some serum samples with neutralizing activity have undetectable levels of antibodies to G1-G2. Antibodies to nonstructural proteins NSm and NSs are also identified. The results obtained raise some questions about antigenic variability and relevant neutralization epitopes of Toscana virus.
Toscana virus (family Bunyaviridae, genus Phlebovirus) was first isolated in 1971 from Phlebotomus perniciosus sandflies collected in central Italy (25). Since then, it has been repeatedly isolated from P. perniciosus and Phlebotomus perfiliewi sandflies collected in different foci in Italy and from the brain of a bat (Pipistrellus khuli) captured in an area where infected sandflies were present (26). By seroepidemiological studies, a high prevalence of antibodies to Toscana virus was demonstrated in healthy populations living in areas of endemicity (16). The virus was associated with clinical cases of acute central nervous system disease (meningitis and meningoencephalitis) occurring during the summer in foci of endemicity (17). Sporadic imported cases of central nervous system disease due to infection with Toscana virus have been reported; one occurred in a Swede visiting Portugal and others occurred in American and German tourists returning from central Italy (3, 5, 21).
Like other members of the Bunyaviridae, Toscana virus possesses a tripartite genome consisting of three single-stranded RNA segments: large (L; 6,404 nucleotides), medium (M; 4,215 nucleotides), and small (S; 1,869 nucleotides). The L segment codes for a large (L; >200-kDa) protein assumed to be a component of the polymerase (1). The M segment codes for two glycoproteins with the same molecular mass (G1 and G2; ∼65 kDa) and for a nonstructural protein (NSm; ∼30 kDa) (4). The S segment codes for the nucleocapsid protein (N; 27 kDa) and for a nonstructural protein (NSs; 37 kDa) (7).
At present, the serologic diagnosis of Toscana virus infection is performed by detecting specific immunoglobulin M (IgM) and IgG antibodies by using a number of different tests like hemagglutination inhibition (HI), plaque reduction neutralization (PRNT), enzyme-linked immunosorbent assay (ELISA), and indirect immunofluorescence assay (6, 17).
A limited study conducted by immunoblotting with nine patients indicated that N is the major virus antigen recognized by the humoral response in Toscana virus patients. However, these studies did not give any indication about the role of other viral proteins (22).
For a more complete understanding of the humoral response to Toscana virus, we designed immunoassays allowing studies of the antibody specificity against different structural and nonstructural proteins.
MATERIALS AND METHODS
Cells and media.
Vero (Cercopithecus aethiops kidney; ATCC CCL81) and BHK-21 (Mesocricetus auratus kidney; ATCC CCL10) cells were propagated in minimum essential medium with Earle’s salts supplemented with nonessential amino acids, 10% (Vero) or 5% (BHK-21) fetal calf serum, 100 IU of penicillin G per ml, and 100 IU of streptomycin per ml.
Virus.
Toscana virus prototype strain (ISS.Phl.3) at third passage in suckling mouse brain was plaque purified three times on Vero cells. Virus stocks were prepared on Vero cell monolayers.
Immunoblotting.
Confluent monolayers of BHK-21 cells were infected at 1 PFU/cell with Toscana virus. Forty-eight hours postinfection, the cells were scraped off from the culture dish and washed in Dulbecco’s phosphate-buffered saline (PBS), and the final pellet was dissolved in sample buffer (0.0625 M Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 1% 2-mercaptoethanol, 10% glycerol, 0.0025% bromophenol blue). Cells from one 10-cm-diameter petri dish were heated for 5 min at 95°C and subjected to SDS-polyacrylamide gel electrophoresis according to the method of Laemmli (12) on a 14-cm-wide 10 to 20% acrylamide gel in the presence of 0.5 M urea. After equilibration in transfer buffer (25 mM Tris [pH 8.3], 192 mM glycine, 20% [vol/vol] methanol), proteins were blotted onto nitrocellulose membrane (Hoefer; pore size, 220 nm) in a tank blot apparatus. Transfer efficiency was monitored by the use of color-labelled molecular weight markers (Sigma Color Markers Wide Range C 3437). Nitrocellulose sheets were saturated in 0.05 M Tris-HCl (pH 8)–0.15 M NaCl (Tris-buffered saline [TBS])–2% bovine serum albumin for 2 h at 39°C and then stored at +4°C until used.
The blotted membranes were cut into 0.4-cm strips and incubated overnight at room temperature with test serum samples diluted 1:50 in TBS–3% nonfat dry milk (Bio-Rad). The strips were washed with TBS–0.05% Tween 20, incubated for 1 h at room temperature with 1 μCi of 35S-protein A (Amersham) per ml, washed again, air dried, and exposed to X-ray film.
Concanavalin A extraction of glycoproteins.
Toscana virus-infected BHK-21 cells were treated as described by Smith and Wright (24). Briefly, monolayers of BHK-21 cells were infected at 1 PFU/cell with Toscana virus. Twenty-four hours postinfection, the cells were scraped off from the culture dish and washed once in PBS and the final pellet was dissolved in lysis buffer (10 mM Tris-acetate [pH 7.6], 0.5 mM Mg-acetate, 1 mM dithiothreitol, 0.5% sodium deoxycholate), homogenized, and centrifuged at 10,000 rpm in a Sorvall HB-4 rotor. Supernatant was incubated for 90 min with concanavalin A-Sepharose (Pharmacia) previously washed three times in buffer A (10 mM Tris-acetate [pH 7.6], 0.5 mM Mg-acetate, 1 mM dithiothreitol, 1 M NaCl). The resin was then washed twice in buffer A for 15 min and twice in 0.1% SDS for 15 min. All incubations were performed at room temperature in a shaker. Glycoproteins were then recovered from the resin by three 5-min treatments at 95°C with 8 M urea–0.5% SDS. Supernatants were pooled, electrophoresed, and blotted as described above.
Radioimmunoprecipitation assay (RIPA).
Confluent monolayers of BHK-21 cells were infected at 1 PFU/cell with Toscana virus. Thirty-six hours postinfection, the culture medium was replaced by Dulbecco’s modified minimum essential medium with Earle’s salts without methionine, cysteine, and fetal calf serum. Twelve hours later, 50 μCi of [35S]methionine per ml and 50 μCi of [35S]cysteine per ml were added and cells were reincubated for 2 h. Cells from a 10-cm-diameter petri dish were scraped off and washed in PBS, and the pellet was resuspended in 1 ml of TBS-RIPA buffer (0.05 M Tris-HCl [pH 8], 0.15 M NaCl, 1% Triton X-100, 0.1% bovine serum albumin)–500 kallikrein inhibitor units of aprotinin (Sigma A-6279) per ml (TBS-RIPA-AP buffer) and sonicated. Five microliters was precipitated by trichloroacetic acid and filtered onto a nitrocellulose disk (pore size, 450 nm) with a Millipore apparatus. Disks were transferred to scintillation vials. The radioactivity was measured in a scintillation counter (Packard TRI-CARB 1500) after adding scintillation fluid (Packard Filter Count).
Seventy microliters of agarose-linked anti-human IgG or IgM (Sigma A-3316 and A-9935) was incubated for 1 h at +4°C with 50 μl of lysate obtained from unlabelled uninfected BHK-21 cells treated as described above. After one washing in TBS-RIPA buffer, the resin was incubated with 25 μl of undiluted serum sample and 50 μl of TBS-RIPA-AP buffer for 1 h at +4°C. Samples were washed in TBS-RIPA buffer and incubated overnight at +4°C with 3 × 106 cpm of 35S-labelled Toscana virus-infected cell lysate (in 200 μl of TBS-RIPA-AP buffer). Samples were then washed five times in TBS-RIPA buffer and once in TBS, resuspended in 50 μl of sample buffer, heated for 5 min at 95°C, and electrophoresed on a 10 to 20% acrylamide gradient gel in the presence of 0.5 M urea. Gels were dried and exposed to X-ray film for 48 h at −80°C.
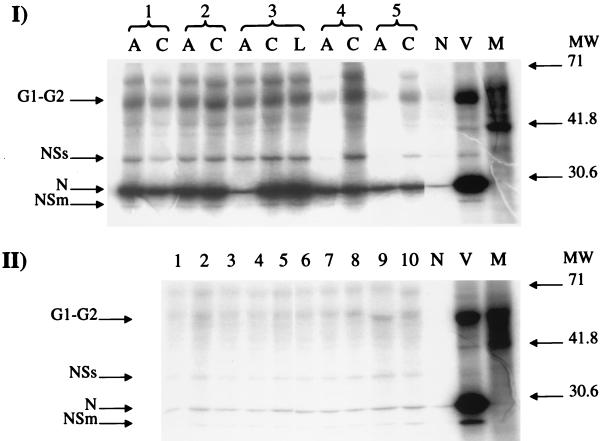
Figure 1 shows a typical pattern of proteins obtained in RIPA by patient sera (I) or by negative sera (II). Autoradiographs were read with an UltroScan XL laser densitometer (Pharmacia) and evaluated with GSXL software (Pharmacia). For each sample, areas corresponding to individual viral proteins were calculated. As described by Di Bonito et al. (4), the G1 and G2 proteins of Toscana virus comigrate in denaturing gels; moreover, G1 is extremely heat sensitive and is degraded at 95°C. As RIPA samples were heated before being loaded onto acrylamide gels, the areas corresponding to the G1 and G2 proteins were considered as a single area. In each gel, a blank consisting of agarose-linked anti-human IgG or IgM, treated as described above except for incubation with serum, was included. The values of areas obtained for this sample were considered as background.
FIG. 1.
Autoradiograph of a RIPA. (I) Toscana virus patients. 1 to 5, patient identification numbers; A, acute serum; C, convalescent serum; L, late convalescent serum; N, agarose with no serum; V, infected cell lysate; M, mock-infected cell lysate. MW, molecular mass markers (kilodaltons). (II) Negative sera. Lanes 1 to 10, patient identification numbers; N, agarose with no serum; V, infected cell lysate; M, mock-infected cell lysate. MW, molecular mass markers (kilodaltons). The film was scanned and elaborated with Corel Photo Paint program, version 7. The final image was computer elaborated with the Power Point 97 program.
Cutoff and titer calculations.
For each serum sample, values corresponding to individual viral proteins were calculated as follows: (area for a given protein [G1-G2, N, NSs, or NSm] in patient serum lane) − (area for the corresponding protein in agarose-without-serum lane). For each viral protein, mean area value plus 3 standard deviations obtained with 52 serum samples known to be negative for Toscana virus antibodies were utilized as cutoff values.
For patients’ sera, relative antibody titers to each viral protein were calculated as follows: (area for a given protein in patient serum lane − area for corresponding protein in agarose-without-serum lane) divided by the cutoff value for that given protein.
RESULTS
Immunoblot.
Sera from 86 patients were tested by immunoblotting: for 3 patients, one sample was available; for 37, two samples were available; for 35, three samples were available; and for 11, four samples were available, for a total of 226 serum samples. All patients had been diagnosed as being infected with Toscana virus by HI or N tests. All sera reacted to the N protein, with no differences in the intensity of bands. None of them reacted with other viral or cellular proteins.
To detect specific antibodies against viral glycoproteins, the test was repeated with the glycoproteic fraction of infected-cell lysates, obtained by treatment with concanavalin A-Sepharose before electrophoresis and blotting. No antibodies against any viral glycoprotein could be detected in any sera by such a test.
None of the 17 serum samples from nine patients diagnosed as non-Toscana virus infected by HI and N tests, utilized as negative controls, showed any reaction to viral proteins.
RIPA.
Sera from 30 patients diagnosed as Toscana virus infected were tested by this assay. For all patients, paired sera were collected at hospitalization (acute sera) and at discharge (usually 7 to 15 days after) (convalescent sera). For eight patients, followup sera were available, taken between 15 days and 18 months after the first sample. Figure 1 shows a typical pattern of proteins obtained by this test.
(i) IgM.
Table 1 shows the cumulative results of RIPA for IgM antibodies. For 25 of 30 patients, the acute serum was positive for antibodies against Toscana virus N protein, the mean value of antibody titers being 10.7 ± 7.4. In convalescent sera, antibody titers against N rose in 12 patients, decreased in 11 patients, and became negative in 3 patients. The mean titer and the median value for 23 positive serum samples were lower than those for acute sera. Antibodies to N at a lower titer than that in convalescent sera were present in three followup serum samples; a single patient became negative in the late sample.
TABLE 1.
Cumulative results of RIPA for IgM antibodies
| Serum (na) | Result | Value for viral protein:
|
|||
|---|---|---|---|---|---|
| N | G1-G2 | NSm | NSs | ||
| 1st (30) | No. of positives | 25 | 2 | 2 | 1 |
| Titer | |||||
| Mean ± SD | 10.7 ± 7.4 | 1.4 | |||
| Range | 1.0–24.3 | 1.0–1.5 | 1.6–2.7 | ||
| Median | 8.3 | ||||
| 2nd (30) | No. of positives | 23 | 3 | 1 | 2 |
| Titer | |||||
| Mean ± SD | 3.7 ± 6.5 | 1.5 ± 0.3 | 1.4 | ||
| Range | 1.0–28.3 | 1.2–1.9 | 2.0–2.9 | ||
| Median | 6.7 | 1.5 | |||
| 3rd (8) | No. of positives | 3 | 0 | 0 | 0 |
| Titer | |||||
| Mean ± SD | 4.7 ± 2.5 | ||||
| Range | 1.9–6.5 | ||||
| Median | 5.8 | ||||
n = number of tested serum samples.
Low-titer IgM antibodies against G1-G2 were detected at the onset of symptoms in two patients. These two patients became negative in the second sample, whereas three patients, negative for G1-G2 antibodies in the acute phase, became positive later. No patient had antibodies against viral glycoproteins in the third sample.
One of the patients with antibodies against G1-G2 in the acute serum also had antibodies against NSm in the same sample. A second patient, with antibodies against NSm at hospitalization, did not react to G1-G2. In the second sample, only one patient was still positive for G1-G2 antibodies and became negative in the third sample.
Similar results were obtained for NSs antibodies: one patient was positive at the onset of the disease, with a titer increasing in the second sample, and one patient showed a seroconversion from negativity to positivity between the first and the second sample. No patient had antibodies to NSm in the third sample.
(ii) IgG.
All but one patient had antibodies against N at hospitalization (Table 2). Titers varied largely: the mean was 43.0 ± 34.5 (median, 37.6) in a range of 3.1 to 149.3. All 29 patients were still positive for N antibodies at discharge. Titers increased in 18 cases and decreased in 10 cases, and in one case no changes were observed. The mean and median values were higher than those in acute sera. All patients for whom a third sample was available were positive for N antibodies with an increased titer for two of them and a decreased titer for others.
TABLE 2.
Cumulative results of RIPA for IgG antibodies
| Serum (na) | Result | Value for viral protein:
|
|||
|---|---|---|---|---|---|
| G1-G2 | N | NSm | NSs | ||
| 1st (30) | No. of positives | 9 | 29 | 18 | 10 |
| Titer | |||||
| Mean ± SD | 6.0 ± 4.8 | 43.0 ± 34.5 | 4.8 ± 5.6 | 7.5 ± 9.4 | |
| Range | 1.5–14.5 | 3.1–149.3 | 1.0–25.2 | 1.0–28.4 | |
| Median | 4.2 | 37.6 | 3.0 | 3.1 | |
| 2nd (30) | No. of positives | 12 | 29 | 22 | 10 |
| Titer | |||||
| Mean ± SD | 4.7 ± 4.1 | 54.3 ± 35.0 | 4.8 ± 5.4 | 3.6 ± 2.2 | |
| Range | 1.0–14.9 | 2.0–142.0 | 1.0–20.8 | 1.1–7.9 | |
| Median | 3.6 | 51.0 | 2.9 | 3.3 | |
| 3rd (8) | No. of positives | 3 | 8 | 5 | 3 |
| Titer | |||||
| Mean ± SD | 2.3 ± 1.7 | 40.9 ± 34.0 | 3.9 ± 3.4 | 2.3 ± 0.8 | |
| Range | 1.2–4.3 | 5.1–106.2 | 1.2–9.7 | 1.6–3.1 | |
| Median | 1.4 | 39.7 | 2.9 | 2.1 | |
n = number of tested serum samples.
Nine patients had antibodies against G1-G2 in acute serum. Four patients had increased antibody titer in the convalescent serum, five had decreased titers, and three patients, negative at the onset of symptoms, became positive later. Only for four patients with antibodies against G1-G2 in the second sample was a third one available: one of them became negative, one had a lower titer, and two showed an increased titer.
Eighteen patients had antibodies against NSm in the first sample. In the second sample, one of them became negative, seven showed a lower titer, eight had increased titers, and five became positive: a total of 22 of 30 convalescent serum samples were positive for NSm antibodies. Six patients with NSm antibodies had a followup serum: in two cases they became negative, in one case antibody titer decreased, in two cases it increased, and for one patient no changes were observed. A single patient had detectable antibodies against NSm only in the third sample.
Antibodies to NSs were detected in acute serum of 10 patients. In five cases, antibodies were still present in convalescent serum (two with higher titer and three with lower titer). Five patients became positive in the second sample. Three patients with antibodies to NSs had a third serum sample and were still positive (one with a lower titer and two with a higher titer).
Antibody patterns.
Antibody patterns are shown in Table 3. As for IgM, the majority of patients had antibodies only against the N protein (22 in the first sample, 20 in the second, and 4 in the third). Only four had antibodies to the N and G1-G2 proteins in the first or in the second sample (in one case also for NSm and in two cases also for NSs). No patient had antibodies against all Toscana virus proteins in any sample of serum. Some patients showed no reaction at all in the acute (three), convalescent (six), or followup (four) serum. Positivity in one or more serum samples against one or more viral proteins was demonstrated for 27 of 30 patients.
TABLE 3.
Antibody patterns and neutralization titers in Toscana virus patients
| Patient no. | Result for testa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| PRNTb
|
RIPA IgM
|
RIPA IgG
|
|||||||
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | |
| 1 | >80 | 320 | G1-G2, N, NSm | NSm | Neg | Neg | |||
| 2 | — | — | N, NSs | G1-G2, N, NSs | Neg | N | N | N | |
| 3 | — | 320 | N, NSm | N | N | N | |||
| 4 | — | 160 | N | N | N | N | |||
| 5 | 40 | 20 | G1-G2 | Neg | N | N | |||
| 6 | 20 | 20 | N | Neg | N | N | |||
| 7 | — | 80 | 80 | N | N | N | N | N, NSm | G1-G2, N, NSm |
| 8 | 10 | 10 | 10 | N | N | Neg | N | N, NSm, NSs | N, NSm, NSs |
| 9 | — | 40 | N | N | N | N, NSm, NSs | |||
| 10 | — | 10 | N | N | N | N, NSm, NSs | |||
| 11 | — | 160 | N | N | N | G1-G2, N, NSm, NSs | |||
| 12 | — | ND | 160 | N | N | N | N, NSs | N, NSs | G1-G2, N, NSm, NSs |
| 13 | 40 | 160 | N | N | N, NSm | G1-G2, N, NSm | |||
| 14 | 10 | 10 | N | N | N, NSm | G1-G2, N, NSm | |||
| 15 | — | 1,280 | N | N | N, NSm | G1-G2, N, NSm, NSs | |||
| 16 | — | ND | ND | N | N | N | N, NSm | N, NSm | N |
| 17 | — | — | 160 | Neg | Neg | Neg | N, NSm | N, NSm | N, NSm |
| 18 | — | — | Neg | N | N, NSm | N, NSm | |||
| 19 | — | 20 | N | N | N, NSm | N, NSm | |||
| 20 | — | — | N | N | N, NSm | N, NSm | |||
| 21 | — | 20 | N | N | N, NSm, NSs | N, NSm | |||
| 22 | ND | ND | N | N | N | G1-G2, N, NSm | G1-G2, N, NSm | N | |
| 23 | — | 20 | N | G1-G2, N | G1-G2, N, NSm, NSs | G1-G2, N, NSm | |||
| 24 | 20 | 256 | N | N | G1-G2, N, NSm, NSs | G1-G2, N, NSm | |||
| 25 | — | 128 | N | G1-G2, N, NSs | G1-G2, N, NSm, NSs | G1-G2, N, NSm | |||
| 26 | 128 | 320 | Neg | Neg | Neg | G1-G2, N, NSm, NSs | G1-G2, N, NSm, NSs | G1-G2, N, NSm, NSs | |
| 27 | 80 | 320 | N | N | G1-G2, N, NSm, NSs | G1-G2, N, NSm, NSs | |||
| 28 | 80 | 320 | N | Neg | G1-G2, N, NSm, NSs | G1-G2, N, NSm, NSs | |||
| 29 | 80 | 80 | Neg | Neg | G1-G2, N, NSm, NSs | G1-G2, N, NSm, NSs | |||
| 30 | 128 | 256 | N | N | G1-G2, N, NSm, NSs | N | |||
Serum: 1, acute serum; 2, convalescent serum; 3, late-convalescent or follow-up serum. Neg, negative.
Reciprocal of highest serum dilution showing 80% reduction. —, <10; ND, not done.
As for IgG, eight patients had antibodies against all viral proteins in the first sample, six had them in the second sample, and two had them in the third sample. Antibodies to N were present in 29 of 30 patients, in some cases alone (10 patients in the first sample, 6 in the second, and 3 in the third) and in some cases with antibodies to one or more other viral proteins. Only one patient was completely negative for all proteins in the first and second samples (the third one was not available): this patient had only IgM antibodies to Toscana virus proteins in his sera (against G1-G2, N, and NSm in the first sample and against only NSm in the second one).
DISCUSSION
Several serologic techniques have been utilized for the detection of specific antibodies to Toscana virus including indirect immunofluorescence assay, HI, PRNT, and ELISA (3, 5, 17, 21). None of these assays allowed the specific identification of antibodies against the individual viral proteins.
In other viral systems, it has been shown that antibodies directed against different viral proteins arise at different times after vaccination or after the onset of the disease (8, 9, 13).
The immunoblot is the easiest technique, allowing the identification of antibodies against a given protein. It has been extensively employed in studies with a number of different viruses including members of the Bunyaviridae (22, 28). However, in the case of Toscana virus, it seems to be of limited usefulness, as it permits only identification of antibodies against the N protein. As this could be due to technical problems caused by the nucleoprotein being heavily expressed in cell lysates utilized to prepare membranes, the technique has been improved with concanavalin A-Sepharose, which permits separation of the glycoprotein fraction. Unfortunately, this technique did not get the expected results. For other members of the Bunyaviridae, it has been demonstrated that antibody-glycoprotein reaction involves conformational epitopes that are very fragile (2, 13). On the other hand, Di Bonito et al. showed that at least the G1 glycoprotein of Toscana virus is extremely sensitive to SDS and heat (4). In the immunoblot technique, cell lysates are treated with detergents, like SDS, and denatured with urea and heated before contact with antibodies. Therefore, the lack of reaction of sera to glycoproteins could be a consequence of the denaturing treatment, which destroys conformational epitopes. The antibody response to the N protein is probably directed not only to conformational epitopes but also to linear epitopes, which are not affected by denaturing treatment, allowing identification of antigen-antibody reaction in the immunoblot test.
In the RIPA, antigen and antibody are allowed to react before strong denaturing treatment. For this reason probably, better results are obtained with Toscana virus proteins in this test, also permitting differentiation of IgG and IgM responses.
All patients had antibodies to N in their acute sera, in most cases both IgG and IgM (Tables 1 and 2). Antibody titers varied considerably, being in general higher for IgG than for IgM. In convalescent sera, IgM antibodies to N were still present in 23 cases, but titers were lower than those in acute sera with regard to mean and range (Table 1). IgG antibodies to nucleoprotein were present in almost all convalescent sera, and titers increased as mean or median, the range being similar to that obtained at the onset of the disease. In late sera, IgM antibodies to N were still present in three of eight patients: these data confirm previous reports in which the presence of IgM antibodies to Toscana virus in toto was demonstrated by ELISA a year after the onset of symptoms (17). IgG antibodies to N were always present in late sera: the mean titer was slightly lower than that in convalescent sera, with a similarly wide range. The results obtained are in accordance with those obtained with other members of the Bunyaviridae, demonstrating that the major antibody response in hantavirus patients is directed against the nucleoprotein (11, 13, 28).
Results for antibodies to other Toscana virus proteins, either structural or nonstructural, were less uniform. IgM antibodies were generally present only in a few serum samples, and values of mean titer and range were low. No IgM antibodies to any of these proteins could be detected in followup sera. Why IgM antibodies against different viral proteins could not be measured at the same extent at different stages of the infection is unclear. As hypothesized by Groen et al. (9) in the case of Puumala virus patients, the differences found in antibody levels against different proteins can reflect the later appearance of antibodies to glycoproteins due to the delay in their production or different sensitivities of the respective test systems.
IgG antibodies to glycoproteins were present in about one-third of patients, regardless of the phase of the disease (Table 2). They were always accompanied by antibodies to NSm, the nonstructural protein encoded by the M segment (the same segment coding for glycoproteins). In all but three patients, antibodies to glycoproteins appeared at the onset of the disease; only three patients showed a seroconversion from negativity to positivity for antibodies to G1-G2 between the first and the second samples, and in one case they appeared later. Antibodies to the NSm protein were present in a higher number of cases, but the time course was similar to that found for glycoproteins: they could usually be detected in acute serum, and in only a few cases could seroconversion be demonstrated. Antibodies to NSs were detected in about one-third of patients, but their presence did not seem to be related to antibodies to G1-G2 or to NSm (Table 3). Titers of antibodies to glycoproteins and nonstructural proteins were similar: the mean was lower than that for antibodies to N, and ranges were smaller.
Data obtained in this study show that, in patients with acute neurologic disease due to Toscana virus infection, both IgM and IgG antibody responses are present in sera at the onset of symptoms, indicating that the incubation period is relatively long. It has been reported that in the Toscana virus imported cases, the onset of symptoms was observed within 5 days after the patients left the zone of endemicity (3, 5, 21). The antibodies are long lasting: IgG antibodies to Toscana virus have been detected in sera taken 2 years after the onset of disease, and data based on serological surveys of healthy populations indicate that the antibodies may persist for life. However, as Toscana virus is endemic to the areas surveyed, it is possible that periodic reinfections act as a booster to immunity.
The major antibody response in Toscana virus patients is against the N protein: these data could be explained by the fact that N is the first protein to be expressed in Toscana virus-infected cells, and it is always present in infected cells at very high levels. Moreover, it is not exposed on the surface of the virus, and its role is to interact with the RNA to protect it from nucleases. Even if antibodies to N have no neutralizing activity, protection studies conducted in vivo demonstrated that monoclonal antibodies to the N protein partially protect mice from challenge with the virulent virus (18). Similar results have been obtained in hantavirus infections where antibodies to the N protein have been shown to protect mice, probably via antibody-dependent cell-mediated cytotoxicity and complement-mediated cytolysis (11, 14, 15, 27).
Unpublished studies on genomic sequences coding for the N protein in several Toscana virus strains, isolated from patients or sandflies, demonstrated that this protein is highly conserved: no more than one amino acid was changed in different strains (19). It is probable that changes in the amino acid sequence that make this protein less efficient in its interaction with viral nucleic acid are lethal for the virus and have no chance to be selected. Thus, by using the prototype strain as antigen it would be possible to evidence antibodies to N, even if the strain that infected the patient is a different one.
The low reactivity of patients to proteins other than nucleoprotein can be explained in several ways. It is possible that glycoprotein epitopes are so fragile that even mild conditions, such as those utilized in our RIPA, denature the majority of them, and that the reaction can be observed only in sera with high levels of antibodies. To evaluate this hypothesis, we compared patterns of antibody response in sera of Toscana virus patients with titers obtained by HI and PRNT tests (Table 3). With few exceptions, when glycoprotein-precipitating antibodies were present in acute serum, the patient had or would develop neutralizing titers higher than 1:80; when glycoprotein-precipitating antibodies were present in the convalescent serum, the patient had neutralizing antibodies higher than 1:80. This was a one-way relationship: some sera with high neutralizing titers failed to precipitate glycoproteins.
Virus neutralization is a complex phenomenon involving a variable number of epitopes. For Rift Valley fever virus, Besselaar and Blackburn found that at least seven epitopes important for neutralization are present on the G1 protein and four are present on G2 (2). The antibodies that they elicit vary noticeably in their biological activities. Moreover, neutralizing epitopes are not necessarily involved in immunoprecipitation; in most cases they depend on tertiary structure of proteins, and even a single amino acid substitution may be important for the biological activity (2, 10, 20, 23). No variability studies have been done on glycoproteins of Toscana virus strains. However, glycoproteins are present on the surface of virions and are exposed to the selective pressure of the host. It is probable that these proteins vary noticeably from one strain to another, particularly for those epitopes that are important for reaction to antibodies: in this case the antigen-antibody reaction is detectable only if the strain utilized as antigen in serologic tests is very similar to the one that infected the patient.
In conclusion, our study showed that the major response in Toscana virus neurologic disease is directed against the nucleoprotein N; however, RIPA might be valuable for assessing the importance of antibodies to Toscana virus antigens other than nucleoprotein N. Some questions arise from our results: future studies will be focused on the role of neutralizing antibodies in the outcome of the Toscana virus infection, as well as on the possibility that between different strains of Toscana virus that circulate in the same area, as demonstrated by ecoepidemiological studies (26), and infect humans, only a few are the cause of severe disease, whereas the majority of strains induce antibody response with no or minor symptoms of illness.
ACKNOWLEDGMENT
We thank Daniel W. Gleason for linguistic revision of the manuscript.
REFERENCES
- 1.Accardi L, Grò M C, Di Bonito P, Giorgi C. Toscana virus L genomic segment: molecular cloning, coding strategy and amino acid sequence in comparison with other negative strand RNA viruses. Virus Res. 1993;27:119–131. doi: 10.1016/0168-1702(93)90076-y. [DOI] [PubMed] [Google Scholar]
- 2.Besselaar T G, Blackburn N K. Topological mapping of antigenic sites on the Rift Valley fever virus envelope glycoproteins using monoclonal antibodies. Arch Virol. 1991;121:111–124. doi: 10.1007/BF01316748. [DOI] [PubMed] [Google Scholar]
- 3.Calisher C H, Winberg A N, Muth D J, Laznick J S. Toscana virus infection in a United States citizen returning from Italy. Lancet. 1987;i:165–166. doi: 10.1016/s0140-6736(87)92005-8. [DOI] [PubMed] [Google Scholar]
- 4.Di Bonito P, Mochi S, Grò M C, Fortini D, Giorgi C. Organization of the M genomic segment of Toscana phlebovirus. J Gen Virol. 1997;78:77–81. doi: 10.1099/0022-1317-78-1-77. [DOI] [PubMed] [Google Scholar]
- 5.Ehrnst A, Peters C J, Niklasson B, Svedmyr A, Holmgren B. Neurovirulent Toscana virus (a sandfly fever virus) in Swedish man after a visit to Portugal. Lancet. 1985;i:1212–1213. doi: 10.1016/s0140-6736(85)92886-7. [DOI] [PubMed] [Google Scholar]
- 6.Eitrem R, Niklasson B, Weiland O. Sandfly fever among Swedish tourists. Scand J Infect Dis. 1991;23:451–457. doi: 10.3109/00365549109075093. [DOI] [PubMed] [Google Scholar]
- 7.Giorgi C, Accardi L, Nicoletti L, Grò M C, Takehara K, Hildtch C, Bishop D H L. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian sandfly fever and Uukuniemi viruses. Virology. 1991;180:738–753. doi: 10.1016/0042-6822(91)90087-r. [DOI] [PubMed] [Google Scholar]
- 8.Grassi M, Wandele A I, Peterhaus E. Enzyme-linked immunosorbent assay for determination of antibodies to the envelope glycoprotein in rabies virus. J Clin Microbiol. 1989;27:899–902. doi: 10.1128/jcm.27.5.899-902.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groen J, Dalrymple J, Fisher-Hoch S, Jordans J G M, Clement J P, Osterhaus A D M E. Serum antibodies to structural proteins of hantavirus arise at different times after infection. J Med Virol. 1992;37:283–287. doi: 10.1002/jmv.1890370409. [DOI] [PubMed] [Google Scholar]
- 10.Hörling J, Lundkvist A. Single amino acid substitution in Puumala virus envelope glycoproteins G1 and G2 eliminates important neutralization epitopes. Virus Res. 1997;48:89–100. doi: 10.1016/s0168-1702(97)01436-6. [DOI] [PubMed] [Google Scholar]
- 11.Jenison S, Yamada T, Morris C, Anderson B, Torrez-Martinez N, Keller N, Hjelle B. Characterization of human antibody response to Four Corners hantavirus infections among patients with hantavirus pulmonary syndrome. J Virol. 1994;68:3000–3006. doi: 10.1128/jvi.68.5.3000-3006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Lundkvist A, Hörling J, Niklasson B. The humoral response to Puumala virus infection (nephropathia epidemica) investigated by viral protein specific immunoassays. Arch Virol. 1993;130:121–130. doi: 10.1007/BF01319001. [DOI] [PubMed] [Google Scholar]
- 14.Lundkvist A, Kallio-Kokko H, Brus-Sjölander K, Lankinen H, Niklasson B, Vaheri A, Vapalahti O. Characterization of Puumala virus nucleocapsid protein: identification of B-cell epitopes and domains involved in protective immunity. Virology. 1996;216:397–406. doi: 10.1006/viro.1996.0075. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura T, Yanagihara R, Gibbs C J, Gajdusek D C. Immune spleen cell-mediated protection against fatal Hantaan virus infection in infant mice. J Infect Dis. 1985;151:691–697. doi: 10.1093/infdis/151.4.691. [DOI] [PubMed] [Google Scholar]
- 16.Nicoletti L, Verani P, Lopes M C, Ciufolini M G, Zampetti P. Studies on Phlebotomus-transmitted viruses in Italy. II. Serological status of human beings. Zentbl Bakteriol Suppl. 1980;9:203–208. [Google Scholar]
- 17.Nicoletti L, Verani P, Caciolli S, Ciufolini M G, Renzi A, Bartolozzi D, Paci P, Leoncini F, Padovani P, Traini E, Baldereschi M, Balducci M. Central nervous system involvement during infection by Phlebovirus Toscana of residents in natural foci in Central Italy (1977-1988) Am J Trop Med Hyg. 1991;45:429–434. doi: 10.4269/ajtmh.1991.45.429. [DOI] [PubMed] [Google Scholar]
- 18.Nicoletti, L., and M. G. Ciufolini. Unpublished data.
- 19.Nicoletti, L., C. Giorgi, S. Mochi, A. Marchi, and M. G. Ciufolini. Unpublished data.
- 20.Schmaljohn C S, Che Y K, Schmaljohn A, Dalrymple J M. Antigenic subunits of Hantaan virus expressed by baculovirus and vaccinia virus recombinant. J Virol. 1990;64:3162–3170. doi: 10.1128/jvi.64.7.3162-3170.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz T F, Gilch S, Jager G. Travel-related Toscana virus infection. Lancet. 1993;342:803–804. doi: 10.1016/0140-6736(93)91568-7. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz T F, Gilch S, Pauli C, Jäger G. Immunoblot detection of antibodies to Toscana virus. J Med Virol. 1996;49:83–86. doi: 10.1002/(SICI)1096-9071(199606)49:2<83::AID-JMV2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 23.Sheshberadaran H, Niklasson B, Tkachenko E A. Antigenic relationship between Hantaviruses analyzed by immunoprecipitation. J Gen Virol. 1988;69:2645–2651. doi: 10.1099/0022-1317-69-10-2645. [DOI] [PubMed] [Google Scholar]
- 24.Smith G W, Wright P J. Synthesis of proteins and glycoproteins in Dengue type 2 virus-infected Vero and Aedes albopictus cells. J Gen Virol. 1985;66:559–571. doi: 10.1099/0022-1317-66-3-559. [DOI] [PubMed] [Google Scholar]
- 25.Verani P, Lopes M C, Nicoletti L, Balducci M. Studies on Phlebotomus-transmitted viruses in Italy. I. Isolation and characterization of a sandfly fever Naples-like virus. Zentbl Bakteriol Suppl. 1980;9:195–201. [Google Scholar]
- 26.Verani P, Ciufolini M G, Caciolli S, Renzi A, Nicoletti L, Sabatinelli G, Bartolozzi D, Volpi G, Amaducci L, Coluzzi M, Paci P, Balducci M. Ecology of viruses isolated from sand flies in Italy and characterization of a new Phlebovirus (Arbia virus) Am J Trop Med Hyg. 1988;38:433–439. doi: 10.4269/ajtmh.1988.38.433. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimatsu K, Yoo Y-C, Yoshida R, Isshihara C, Azuma I, Arikawa J. Protective immunity of Hantaan virus nucleocapsid and envelope protein studied using baculovirus-expressed proteins. Arch Virol. 1993;130:365–376. doi: 10.1007/BF01309667. [DOI] [PubMed] [Google Scholar]
- 28.Zöller L, Scholz J, Stohwasser R, Giebel L B, Sethi K K, Bautz E K F, Darai G. Immunoblot analysis of the serological response in Hantavirus infections. J Med Virol. 1989;27:231–237. doi: 10.1002/jmv.1890270309. [DOI] [PubMed] [Google Scholar]