Abstract
A 120-amino-acid polypeptide selected from the transmembrane protein region (tTM) and the major capsid protein p26 of bovine immunodeficiency-like virus (BIV) were expressed as fusion proteins from recombinant baculoviruses. The antigenic reactivity of both recombinant fusion proteins was confirmed by Western blot with bovine and rabbit antisera to BIV. BIV-negative bovine sera and animal sera positive for bovine syncytial virus and bovine leukemia virus failed to recognize the recombinant fusion proteins, thereby showing the specificity of the BIV Western blot. One hundred and five bovine serum samples were tested for the presence of anti-BIV antibodies by the recombinant protein-based Western blot and a reference Western blot assay using cell culture-derived virions as test antigens. There was a 100% concordance when the p26 fusion protein was used in the Western blot. However, the Western blot using the tTM fusion protein as its test antigen identified four BIV-positive bovine sera which had tested negative in both the p26 recombinant-protein-based and the reference Western blot assays. This resulted in the lower concordance of 96.2% between the tTM-protein-based and reference Western blot assays. The results of this study showed that the recombinant p26 and tTM proteins can be used as test antigens for the serodetection of BIV-infection in animals.
Bovine immunodeficiency-like virus (BIV) is a lentivirus of the Retroviridae family which shares morphologic, genetic, and/or antigenic properties with human immunodeficiency virus (HIV) type 1 and other animal lentiviruses (14, 41). The BIV genome resembles that of other retroviruses with the typical 5′-to-3′ gag, pol, and env gene organization (15). In addition, it contains six nonstructural- and regulatory-protein-encoding genes between or overlapping the pol and env reading frames (15). The structural gag-encoded capsid protein (p26) and env-encoded surface (SU) and transmembrane (TM) proteins (gp110 and gp42, respectively) have been shown to be highly immunogenic (3, 4, 7, 18, 31, 39, 43).
BIV, like other retroviruses, establishes a permanent infection once the proviral DNA integrates into the host cell genome (15). BIV has been recently shown to infect a variety of cells, including peripheral lymphocytes, neurons, microglial cells, and endothelial cells of experimentally infected calves (47). Although clinical disease cannot yet be attributed to BIV, associations of BIV with immune system dysfunctions, central nervous system disorders, and progressive emaciation in naturally and experimentally BIV-infected cattle have been reported (6, 26, 41). Several secondary conditions, including mastitis, pododermitis, and other bacterial diseases, have been associated with BIV infection, suggesting a possible impact on dairy herd productivity and general health (24, 34). Serological data indicate the worldwide distribution of BIV (1, 10, 17, 19, 24, 29). In certain regions in the United States, the individual serological prevalence of BIV infection has been reported to be greater than 50% (15, 35). In Ontario, BIV seroprevalence was shown to be 5% in dairy cows (24).
Discrimination between infected and noninfected cattle is an essential step in controlling BIV infection by segregation and/or removal of virus-infected animals (34). To achieve this, convenient and reliable BIV-specific diagnostic tests are necessary. BIV isolation from peripheral blood buffy coat cells with cell culture cocultivation has been reported (41). However, this method is time-consuming and technically laborious. Moreover, this procedure may be associated with interference from other known bovine retroviruses (e.g., bovine syncytial virus [BSV] and bovine leukemia virus [BLV]), thus requiring further discrimination by immunological or molecular analyses (34). PCR has also been used to detect BIV in the DNA of peripheral blood mononuclear cells from BIV-infected animals and tissue culture cells infected with BIV (25, 39, 46). Serodetection techniques such as the immunofluorescence assay, Western blot assay, and enzyme-linked immunosorbent assay (ELISA) have been used to identify BIV-exposed animals by using native virion antigens (35, 43, 44).
Previous studies have demonstrated the reactivity of bovine serum antibody against the BIV major capsid protein p26, encoded by the gag gene (43, 44). The anti-p26 reactivity was associated with the presence of linear epitopes within the protein (3). In cattle experimentally infected with BIV, the immunological reactivity to BIV p26, as determined by Western blotting with a virion-derived antigenic preparation, was detected early (within 20 days) and persisted several months after BIV exposure (18, 39). On the other hand, the amino-terminal region of the TM envelope gp42 protein was also shown to contain at least one major linear epitope which is highly immunogenic in BIV-infected cattle (7). The immune reactivity against the gp42 protein, which appeared later than that observed for p26, was still readily detectable in the sera of cattle 3.5 years after experimental BIV infection, whereas antibodies to p26 were undetectable in the same animal sera (18).
In this study, we took advantage of recombinant techniques to produce recombinant fusion proteins expressed in the baculovirus system that target BIV TM envelope gp42 and capsid p26 proteins. The antigenic reactivity of the recombinant fusion proteins was analyzed in a Western blot assay using sera from rabbits and cattle experimentally infected with BIV. The Western blot was then used to test a panel of bovine field sera, and the results were compared with those obtained with a reference Western blot assay in which native virus proteins were used as test antigens (24, 44).
MATERIALS AND METHODS
Sources of sera.
Field bovine sera, serum samples from cattle that were experimentally infected with BIV, and bovine BSV-specific antisera were provided by Robert M. Jacobs, Department of Pathobiology, University of Guelph, Guelph, Ontario, Canada. Bovine serum samples positive for anti-BLV antibodies were generously provided by Diagnostics Biovet Inc., St-Hyacinthe, Québec, Canada. The equine serum specific to equine infectious anemia virus (EIAV) was provided by Alain M. P. Bouillant (Virology Section, Animal Diseases Research Institute, Canadian Food Inspection Agency, Nepean, Ontario, Canada). Rabbit serum samples reactive to BIV or BSV were obtained from animals experimentally inoculated by the intraperitoneal route with BIV-infected (28) or BSV-infected (2a) cells.
Viral RNA isolation and oligonucleotide primers.
Viral genomic RNA was extracted by the guanidium isothiocyanate method (8) from the supernatant of fetal bovine embryonic lung cells chronically infected with the R-29 isolate of BIV (41). The cells were propagated in minimal Eagle medium supplemented with 10% fetal bovine serum and antibiotics. The oligonucleotide primers for reverse transcription-PCR amplification of the nucleic acid sequences encoding the BIV p26 (nucleotides 700 to 1401) and a 120-amino-acid-long truncated form of the TM gp42 envelope protein (amino acids 31 to 150; nucleotides 7170 to 7529) (tTM) were selected according to the BIV genomic sequence (GenBank accession no. M32690) (13). The primers were synthesized by a commercial supplier (Gibco/BRL, Gaithersburg, Md.). The specific sense and antisense primers used to amplify the p26-encoding nucleic acid sequence were from nucleotide positions 700 to 715 and 1401 to 1384, respectively, while those used to amplify the tTM-encoding nucleic acid sequence were from nucleotide positions 7170 to 7190 and 7529 to 7505, respectively. In addition, all primers contained short 5′ extensions in which restriction endonuclease cleavage sites were present for cloning and subcloning purposes. The expected sizes of PCR products were 701 and 359 bp for the p26- and the tTM-encoding nucleic acid sequences, respectively.
Reverse transcription-PCR amplification, cloning, and sequencing.
The BIV genomic RNA was converted to cDNA by reverse transcription using random hexadeoxyribonucleotides [pd(N)6; Pharmacia Biotech] as previously described (36). The cDNA was then amplified with a programmable thermal cycler by 30 successive cycles of denaturation at 95°C for 1 min, primer annealing at 52°C (for the p26-encoding nucleic acid sequence) or 60°C (for the tTM-encoding nucleic acid sequence) for 1 min, and DNA chain extension at 72°C for 2 min. The amplified cDNA products were subsequently cloned into the pCR II TA vector according to the manufacturer’s instructions (Invitrogen, Palo Alto, Calif.) and sequenced by the chain termination method of Sanger et al. (33) to confirm the BIV-specific nature of the amplified products.
Construction of recombinant transfer plasmids.
The recombinant pCR II TA plasmids containing the p26- or tTM-protein-encoding nucleic acid sequences were digested with BamHI and EcoRI or with Xho and HindIII, respectively. The fragments of interest were then purified with a low-melting-temperature agarose gel (32) and ligated into the pBlue-bac His2 transfer plasmid (Invitrogen) digested with the corresponding enzymes. This enabled the amplified cDNA products encoding BIV p26 and tTM proteins to be cloned in frame with nucleic acid sequences which code for a stretch of six histidine residues and an enterokinase cleavage site, resulting in the expression of recombinant fusion proteins. After transformation in competent Escherichia coli DH5α cells, ampicillin-resistant bacterial colonies were screened for the presence of the appropriate recombinant transfer plasmids using standard procedures (32). The resulting cDNA clones were then sequenced as described above to confirm that the cloned sequences were in the correct reading frame.
Generation of recombinant baculovirus.
Recombinant transfer plasmid DNA and wild-type Autographa californica nuclear polyhedrosis virus (AcNPV) DNA were used to cotransfect Spodoptera frugiperda 9 (Sf9) cells grown in Grace medium supplemented with 10% fetal bovine serum, according to the supplier’s instructions (Invitrogen). Blue recombinant baculovirus plaques were then selected (42) and expanded once in Sf9 cells. After 72 h of incubation at 27°C, the cell culture medium was harvested, and total DNA was isolated from infected cells by the standard phenol-chloroform extraction method (32). The presence of the p26- and tTM-encoding nucleic acid sequences from the recombinant baculoviruses were then confirmed by PCR using the corresponding primers described above. Two or three rounds of plaque purification were then performed according to the supplier’s instructions. Plaque-purified recombinant viruses were thereafter propagated in Sf9 cells, and the cell culture supernatants were harvested to generate the recombinant baculoviruses BIV p26-rAcNPV and BIV ptTM-rAcNPV.
Expression and purification of recombinant fusion proteins.
For protein expression, groups of 2 × 106 Sf9 log-phase cells were each infected with one of the plaque-purified recombinant baculoviruses at a multiplicity of infection of 4. Uninfected Sf9 cells and those infected with wild-type baculovirus were processed in parallel and used as controls. After 5-day incubation at 27°C, the cells were pelleted by centrifugation at 3,000 × g for 10 min. The cell pellets were then resuspended in 0.5 ml (for 106 cells) of 2% sodium dodecyl sulfate (SDS)–0.2 M NaCl–0.2 M Tris (pH 7.5)–1.5 mM MgCl2 and heated to 100°C for 3 min (16). These cell lysates and cell culture supernatants were then analyzed on an SDS–12% polyacrylamide gel (32).
For the standardization of the Western blot assay, the recombinant fusion proteins were purified by the electroelution of the protein that was previously cut out of an SDS-polyacrylamide gel (22). The concentration of purified proteins was estimated by densitometry after comparison with standard proteins on a Coomassie blue-stained SDS-polyacrylamide gel.
Western blot analysis of BIV recombinant fusion proteins.
Purified proteins were electrophoresed on SDS–12% polyacrylamide gels as described above and were then electrotransferred to nitrocellulose membranes. Nonspecific binding to nitrocellulose was blocked by using a solution of 1% enzyme immunoassay-grade gelatin (Bio-Rad, Palo Alto, Calif.) and 0.2% Tween 20 in 50 mM Tris hydrochloride-buffered saline solution (pH 7.5) (TBS). Bovine serum samples (used at a dilution of 1:50) and rabbit and horse sera (used at a dilution of 1:100) were allowed to react for 2 h at room temperature with the test antigen (approximately 200 ng of purified fusion protein). The membranes were then washed three times in TBS containing 0.05% Tween 20 before the appropriate peroxidase-labeled conjugate was added for 1 h at room temperature. The immune reactivity was revealed after the peroxidase substrate (TBS, H2O2, methanol, and 4-chloro-naphthol) was added 10 to 15 min (32).
BIV reference Western blot assay.
The BIV reference Western blot assay, using cell culture supernatant-derived semipurified virions as test antigens, was performed in accordance with the method described by Whetstone et al. (44), as adapted by McNab et al. (24).
RESULTS
Expression of BIV p26- and tTM-encoding nucleic acid sequences in Sf9 cells.
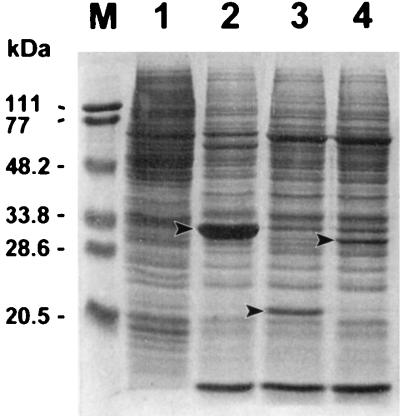
Segments of the BIV genome containing the BIV p26 or BIV tTM nucleic acid sequences were amplified and inserted into the pBlue-bac His2 transfer vector. Recombinant baculovirus virions (BIV p26-rAcNPV and BIV ptTM-rAcNPV) were then successfully obtained following the cotransfection of Sf9 cells with the respective plasmid transfer construct and wild-type AcNPV DNA. The recombinant viruses and wild-type baculovirus were then used to infect Sf9 cells. As shown in Fig. 1, the baculovirus polyhedrin protein (lane 2), the BIV tTM recombinant fusion protein (lane 3), and the BIV p26 recombinant fusion protein (lane 4) were readily observable on a Coomassie brilliant blue-stained SDS-polyacrylamide gel from the pellets of Sf9 cells infected with wild-type baculovirus and BIV ptTM-rAcNPV and BIV p26-rAcNPV recombinant baculoviruses, respectively. The recombinant fusion proteins were not present in any of the cell culture supernatants (data not shown). As expected, the BIV p26 recombinant fusion protein had a molecular size of approximately 29.6 kDa (Fig. 1, lane 4), due to the presence of the fusion partner containing an enterokinase cleavage site and six histidine residues at the NH2 terminus of the expressed fusion protein. However, the BIV tTM recombinant fusion protein, with a predicted molecular size of 18 kDa, migrated at approximately 20.6 kDa (Fig. 1, lane 3). Although no systematic experiments were conducted, this result may reflect the fact that the BIV tTM polypeptide is predicted to contain three glycosylation sites and, therefore, may be glycosylated in insect cells, as reported elsewhere (31).
FIG. 1.
Expression of BIV p26 and tTM proteins in Sf9 insect cells from recombinant baculoviruses. Proteins synthesized in Sf9 cells were analyzed by SDS-polyacrylamide gel electrophoresis and visualized by staining with Coomassie brilliant blue. Lane 1, extracts (10 μg) of control noninfected cell cultures; lanes 2 to 4, extracts (10 μg) of cell cultures infected with wild-type AcNPV baculovirus, recombinant BIV ptTM-rAcNPV, and recombinant BIV p26-rAcNPV, respectively. The polyhedrin protein expressed from wild-type baculovirus and BIV recombinant fusion proteins are indicated by arrowheads. Lane M, molecular mass standards.
Immunological reactivity of the baculovirus-expressed recombinant fusion proteins.
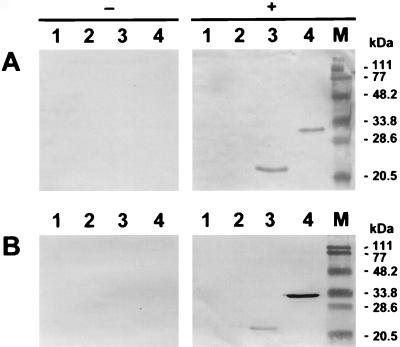
In order to analyze the BIV-specific immunological reactivity of the 29.6- and 20.6-kDa baculovirus-expressed recombinant fusion proteins, protein preparations from crude cell extracts were subjected to Western blotting with serum samples from a cow naturally infected with BIV and from a rabbit experimentally exposed to BIV. Both bovine (Fig. 2A) and rabbit (Fig. 2B) BIV-positive sera recognized tTM (lanes 3) and p26 (lanes 4) recombinant fusion proteins. No immune reactivity was observed when these BIV-positive sera were allowed to react with proteins prepared from noninfected Sf9 cells (Fig. 2, lanes 1) or Sf9 cells infected with wild-type baculovirus (lanes 2). No immune reactivity was observed when the Sf9-derived proteins and the recombinant fusion protein preparations were allowed to react with sera from BIV-negative cattle or with rabbit serum prior to BIV exposure (Fig. 2A and B, respectively).
FIG. 2.
Immunoreactivity of BIV p26 and tTM recombinant fusion proteins expressed in Sf9 cells from recombinant baculoviruses. Extracts (10 μg each) from uninfected cell cultures (lane 1) and cultures infected with wild-type AcNPV baculovirus (lane 2), recombinant BIV ptTm-rAcNPV (lane 3), and recombinant BIV p26-rAcNPV (lane 4) were electrophoresed, transblotted to nitrocellulose membranes, and exposed to a 1:50 dilution of bovine sera (A) or a 1:100 dilution of rabbit sera (B) positive (+) or negative (−) for BIV antibodies. Lane M, molecular mass standards.
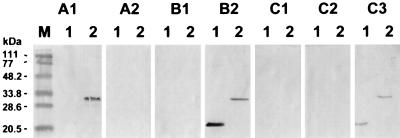
Immune cross-reactivity with proteins derived from the Sf9 cells was observed with certain bovine sera. To decrease background activity, BIV recombinant fusion proteins were gel purified. When these purified recombinant fusion proteins were exposed to rabbit or bovine BIV antisera, positive immune reactivity was detected with both proteins (Fig. 3, panels B2 and C3, respectively). Immune cross-reactivity was observed when the BIV p26 recombinant fusion protein was exposed to an equine serum containing anti-EIAV antibodies (Fig. 3, panel A1, lane 2). This was expected on the basis of prior studies in which such immune cross-reactivity between BIV p26 and EIAV-specific equine antisera was observed (3, 14). No immune cross-reactivity was observed when the EIAV horse antiserum was allowed to react with the BIV tTM recombinant fusion protein (Fig. 3, panel A1, lane 1). Finally, no cross-reactions were detected when the BIV recombinant fusion proteins were allowed to react with bovine antisera specific to BSV and BLV (Fig. 3, panels C1 and C2, respectively) or immune serum from a rabbit experimentally exposed to BSV (Fig. 3, panel B1).
FIG. 3.
Specificity analysis of purified BIV p26 and tTM fusion proteins expressed in Sf9 cells from recombinant baculoviruses. A total of 200 ng of purified tTM (lanes 1) and p26 (lanes 2) recombinant fusion proteins was electrophoresed, transblotted to nitrocellulose membranes, and exposed to a 1:100 dilution of horse sera positive (A1) or negative (A2) for EIAV antibodies, to a 1:100 dilution of rabbit sera positive for BSV (B1) or BIV (B2) antibodies, and to a 1:50 dilution of bovine sera positive for BSV (C1), BLV (C2), or BIV (C3) antibodies. Lane M, molecular mass standards.
Comparison of the BIV recombinant fusion protein-based Western blot with the BIV reference Western blot assay.
A total of 105 bovine serum samples were tested for antibodies to BIV by recombinant-fusion-protein-based Western blotting and the BIV reference Western blot assay, in which whole virus proteins were used as test antigens. For the recombinant-protein-based Western blot, serum samples were tested separately against each purified recombinant fusion protein. As shown in Table 1, we were able to detect antibodies in 22 serum samples when the BIV p26 recombinant fusion protein was used as the test antigen. When the BIV tTM recombinant fusion protein was used as the test antigen, four additional sera which were not reactive against the BIV p26 recombinant fusion protein or in the reference Western blot assay tested positive for BIV antibodies. To investigate whether those four positive blot samples were false positives, an inhibition assay was performed with sera which had been preincubated with an excess of tTM (1 μg) in an Eppendorf tube for 1 h at 37°C and then tested in the tTM-based Western blot assay. This procedure completely abolished the immune reactivity of these sera to the tTM in the Western blot assay (data not shown), thereby suggesting that these sera were likely to be true BIV-positive samples. Finally, when the results obtained from the Western blots with the p26 and tTM recombinant fusion proteins were compared with the reference Western blot assay results, concordances of 100 and 96.2% were observed, respectively.
TABLE 1.
Comparison of BIV p26 and tTM recombinant-fusion-protein-based Western blots with the reference Western assay for the serodetection of BIV
| Western blot assay | No. of serum samples
|
No. of samples identical to the reference assay | Concordancea to the reference assay (%) | |
|---|---|---|---|---|
| Positive | Negative | |||
| p26 based | 22 | 83 | 105 | 100 |
| tTM protein based | 26 | 79 | 101 | 96.2 |
| Referenceb | 22 | 83 | 105 | NAc |
Concordance: (number of serum samples identical to the reference Western blot/total number of sera tested) × 100.
Assay using whole virus proteins as test antigens.
NA, not applicable.
DISCUSSION
Although several laboratories have used various serological methods to detect BIV-infection in animals, diagnostic assays for BIV infection are not yet commercially available. Here, we have produced BIV p26 and tTM fusion proteins from recombinant baculoviruses and investigated their ability to detect serum anti-BIV antibodies by a Western blot procedure. The capsid (p26) and tTM proteins were targeted as potential test antigens because they have been shown to induce the production of specific serum antibodies to BIV (7, 18, 43, 44). In addition, as reported with BIV and other lentiviruses (9, 12, 37), both of these proteins are likely to be genetically and antigenically well conserved. This is an important issue in light of the emergence of new BIV isolates which may be genetically divergent from the R-29 reference strain, as has been recently reported in the United States (37). In fact, hypervariable regions in the SU envelope gene have been identified among several BIV isolates (38), thereby suggesting that the SU BIV protein is unlikely to be a reliable reagent for the serodiagnosis of BIV. In contrast, antigenic relationships on the basis of serological immune reactivity to p26 among various BIV isolates have been reported (37).
Similar to that reported with the HIV p24 capsid protein in the course of HIV infection in the human (11), immune reactivity associated with the major capsid protein p26 appears early in animals experimentally exposed to BIV (43, 44). In another study, the p26-specific antibodies were shown to decrease to undetectable levels within 2 to 3 years after experimental infection in cattle (18). In that study, infectious virus was recovered from peripheral blood mononuclear cells of each BIV-infected animal before and after the loss of p26-specific antibodies. In contrast, immune reactivity to the TM protein, which appears later in the course of BIV infection, was still detectable at the end of the experiment period (3.5 years postinfection). Under this circumstance, using the TM protein would allow the detection of BIV infection in cattle whose sera failed to recognize the p26 protein (18). On the basis of all the above observations, one may conclude that a diagnostic test for the serodetection of BIV infection should include both the capsid p26 and TM proteins as test antigens in an immunoassay.
As mentioned above, the presence of major linear epitopes has been reported in BIV p26 and TM env proteins (3, 7). In the study on the env proteins, the TM protein was shown to be much more antigenic than the extracellular SU protein (7), an observation which is similar to that reported for other lentiviruses (5, 27, 45). By using several cDNA mutant clones that expressed mutated fusion proteins in bacterial cells, Chen et al. (7) demonstrated that the strong immune reactivity of the BIV TM protein was confined to a 134-amino-acid polypeptide beginning in the amino-terminal region of the protein (i.e., amino acids 1 to 134). Based on this, and on the results from hydrophilicity and antigenicity index analyses of this peptide region with appropriate software programs (21, 23) (data not shown), the 120-amino-acid polypeptide to be expressed in this study was selected. This polypeptide contained most of the 134-amino-acid region described by Chen et al. (7), spanning amino acid residues 31 through 150 of the BIV TM env protein.
The recombinant baculovirus system for expression of proteins to be used as test antigens in ELISA and Western blot procedures for diagnostic purposes has been used in other viral systems (16, 40). Moreover, the baculovirus system has been used to successfully express the entire gag and env nucleic acid sequences of BIV (30, 31). The results of the present study showed that both the BIV p26 and tTM proteins were adequately expressed in Sf9 insect cells and could be successfully used in a Western blot procedure to detect BIV antibodies in sera from cattle naturally or experimentally infected with BIV. The results also showed that, due to the reactivity of insect proteins derived from the crude cell extract with some bovine sera, the gel-purified proteins should be the antigens of choice for the Western blot assay.
The results of testing a group of bovine sera by our Western blot with the recombinant BIV p26 as the test antigen were 100% in agreement with those obtained in the reference Western blot assay with whole virus as the test antigen. This was not surprising because it is well known that the protein preparation derived from cell culture-derived semipurified BIV virions contained p26 in a relatively large quantity (30). However, four bovine sera that tested negative by both of these assays readily tested positive by the Western blot assay with the BIV tTM recombinant fusion protein as the test antigen. Although false-positive test results cannot be completely ruled out despite the specificity of the reaction demonstrated in our inhibition studies, the discrepancy between these test results might be attributed, as mentioned above, to the fact that the level of antibodies to p26 decreased in the course of BIV infection while immune reactivity to the TM protein remains detectable (18). Another explanation that might account for the discrepancy between these assays is that the cell culture-derived virion preparations used as test antigens in the reference Western blot assay may contain negligible amounts of the env glycoproteins, such that immune reactivity of sera containing low levels of BIV antibody to these proteins may not be detected by this assay. In fact, protein virion preparations used for Western blotting may generate a weak signal to the env glycoproteins even with some control sera (18a).
In summary, the results of this study showed that both recombinant p26 and tTM virus proteins can be used as test antigens for the serodiagnosis of BIV-infected animals by Western blotting. This test appears to be highly sensitive and specific to BIV, since no immune cross-reactivity was shown with BLV and BSV. This point is important because multiple retrovirus infections in cattle are common (1, 19). The test also has several advantages, including a relatively short test time and, most importantly, an adequate and reproducible supply of recombinant proteins to be used as test antigens in the Western blot assay. Further research should investigate the immune reactivity of the recombinant proteins to a larger number of BIV isolates and the feasibility of using these proteins in an ELISA that might be more convenient, in terms of screening a larger number of sera at once.
ACKNOWLEDGMENTS
We are grateful to Carole Villeneuve for secretarial work and Claude Daniel for the photographs.
This work was partly supported by research grants from the National Sciences and Engineering Research Council of Canada, Agriculture Canada, the Conseil des recherches en pêche et en agroalimentaire du Québec (Entente Auxiliaire Canada-Québec), and Diagnostics Biovet Inc., St-Hyacinthe, Québec, Canada, to D. Archambault. Y. Abed holds a postdoctoral fellowship from the University of Québec in Montréal (Fondation UQAM). D. Archambault holds a research scholarship from the Fonds de la Recherche en Santé du Québec.
REFERENCES
- 1.Ambroski G F, Lo J L, Seger C L. Serological detection of multiple retroviral infections in cattle: bovine leukemia virus, bovine syncytial virus and bovine visna virus. Vet Microbiol. 1989;20:247–253. doi: 10.1016/0378-1135(89)90048-5. [DOI] [PubMed] [Google Scholar]
- 2.Archambault D, Wang Z-M, Lacal J C, Gazit A, Yaniv A, Dahlberg J E, Tronick S R. Development of an enzyme-linked immunosorbent assay for equine infectious anemia virus detection using recombinant Pr55gag. J Clin Microbiol. 1989;27:1167–1173. doi: 10.1128/jcm.27.6.1167-1173.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Archambault, D. Unpublished data.
- 3.Atkinson B, Liu Z Q, Wood C. Use of bacterial TrpE fusion vectors to express and characterize the bovine immunodeficiency-like virus core protein. J Virol Methods. 1992;36:35–49. doi: 10.1016/0166-0934(92)90155-7. [DOI] [PubMed] [Google Scholar]
- 4.Baron T, Betemps D, Mallet F, Cheynet V, Levy D, Belli P. Detection of bovine immunodeficiency-like virus infection in experimentally infected calves. Arch Virol. 1998;143:181–189. doi: 10.1007/s007050050278. [DOI] [PubMed] [Google Scholar]
- 5.Benichou S, Venet A, Beyer G, Tiollais P, Madaule P. Characterization of B-cell epitopes in the envelope glycoproteins of simian immunodeficiency virus. Virology. 1993;194:870–874. doi: 10.1006/viro.1993.1333. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter S, Miller L D, Alexandersen S, Whetstone C A, VanDerMaaten M J, Viuff B, Wannemuehler Y, Miller J M, Roth J A. Characterization of early pathogenic effects after experimental infection of calves with bovine immunodeficiency-like virus. J Virol. 1992;66:1074–1083. doi: 10.1128/jvi.66.2.1074-1083.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P, Liu Z Q, Wood C. Use of TrpE fusion protein to identify antigenic domains within the BIV envelope protein. J Virol Methods. 1994;47:331–343. doi: 10.1016/0166-0934(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Fontenot J D, Hoover E A, Elder J H, Montelaro R C. Evaluation of feline immunodeficiency virus and feline leukemia virus transmembrane peptides for serological diagnosis. J Clin Microbiol. 1992;30:1885–1890. doi: 10.1128/jcm.30.7.1885-1890.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forman A J, Gibson C A, Rodwell B J. Serological evidence for the presence of bovine lentivirus infection in Australia. Aust Vet J. 1992;69:337. doi: 10.1111/j.1751-0813.1992.tb09919.x. [DOI] [PubMed] [Google Scholar]
- 11.Gaines H, von Sidow M, Sonnerborg A, Albert J, Czajkowski J, Pehrson P O, Chiodi F, Moberg L, Fenyo E M, Asjo B, Forsgren M. Antibody response in primary human immunodeficiency virus infection. Lancet. 1987;ii:1249–1253. doi: 10.1016/s0140-6736(87)92696-1. [DOI] [PubMed] [Google Scholar]
- 12.Gallaher W R, Ball J M, Garry R F, Griffen M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retrovir. 1989;5:431–439. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 13.Garvey K J, Oberste M S, Elser J E, Braun M J, Gonda M A. Nucleotide sequence and genomic organization of biologically active proviruses of bovine immunodeficiency-like virus. Virology. 1990;175:391–409. doi: 10.1016/0042-6822(90)90424-p. [DOI] [PubMed] [Google Scholar]
- 14.Gonda M A, Braun M J, Carter S G, Kost T A, Jr, Bess J W, Arthur L O, Van Der Maaten M J. Characterization and molecular cloning of a bovine lentivirus related to the human immunodeficiency virus. Nature (London) 1987;330:388–391. doi: 10.1038/330388a0. [DOI] [PubMed] [Google Scholar]
- 15.Gonda M A, Luther D G, Fong S E, Tobin G T. Bovine immunodeficiency virus: molecular biology and virus-host interactions. Virus Res. 1994;32:155–181. doi: 10.1016/0168-1702(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 16.He J, Tam A W, Yarbough P O, Reyes G R, Carl M. Expression and diagnostic utility of hepatitis E virus putative structural proteins expressed in insect cells. J Clin Microbiol. 1993;31:2167–2173. doi: 10.1128/jcm.31.8.2167-2173.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirai N, Kabeya H, Ohashi K, Sugimoto C, Onuma M. Detection of antibodies against bovine immunodeficiency-like virus in dairy cattle in Hokkaido. J Vet Med Sci. 1996;58:455–457. doi: 10.1292/jvms.58.455. [DOI] [PubMed] [Google Scholar]
- 18.Isaacson J A, Roth C, Wood C, Carpenter S. Loss of Gag-specific antibody reactivity in cattle experimentally infected with bovine immunodeficiency-like virus. Viral Immunol. 1995;8:27–36. doi: 10.1089/vim.1995.8.27. [DOI] [PubMed] [Google Scholar]
- 18a.Jacobs, R. Unpublished data.
- 19.Jacobs R M, Smith H E, Gregory B, Valli V E O, Whetstone C A. Detection of multiple retroviral infections in cattle and cross-reactivity of bovine immunodeficiency-like virus and human immunodeficiency virus type 1 proteins using bovine and human sera in a Western blot assay. Can J Vet Res. 1992;56:353–359. [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs R M, Pollari F L, McNab W B, Jefferson B. A serological survey of bovine syncytial virus in Ontario: associations with bovine leukemia and immunodeficiency-like viruses, production records, and management practices. Can J Vet Res. 1994;59:271–278. [PMC free article] [PubMed] [Google Scholar]
- 21.Jameson B A, Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci. 1988;4:181–186. doi: 10.1093/bioinformatics/4.1.181. [DOI] [PubMed] [Google Scholar]
- 22.Kheyar A, Martin S, St.-Laurent G, Timoney P J, McCollum W H, Archambault D. Expression cloning and humoral immune response to the nucleocapsid and membrane proteins of equine arteritis virus. Clin Diagn Lab Immunol. 1997;4:648–652. doi: 10.1128/cdli.4.6.648-652.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle R R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.McNab W B, Jacobs R M, Smith H E. A serological survey for bovine immunodeficiency-like virus in Ontario dairy cattle and association between test results, production records and management practices. Can J Vet Res. 1994;58:36–41. [PMC free article] [PubMed] [Google Scholar]
- 25.Nadin-Davis S A, Chang S C, Smith H, Jacobs R M. Detection of bovine immunodeficiency-like virus by polymerase chain reaction. J Virol Methods. 1993;42:323–336. doi: 10.1016/0166-0934(93)90043-q. [DOI] [PubMed] [Google Scholar]
- 26.Onuma M, Koomoto E, Furuyama H, Yasutomi Y, Taniyama H, Iwai H, Kawakami Y. Infection and dysfunction of monocytes induced by experimental inoculation of calves with bovine immunodeficiency-like virus. J Acquired Immune Defic Syndr. 1992;5:1009–1015. [PubMed] [Google Scholar]
- 27.Pancino G, Chappey C, Saurin W, Sonigo P. B epitopes and selection pressures in feline immunodeficiency virus envelope glycoproteins. J Virol. 1993;67:664–672. doi: 10.1128/jvi.67.2.664-672.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pifat D Y, Ennis W H, Ward J M, Oberste M S, Gonda M A. Persistent infection of rabbits with bovine immunodeficiency-like virus. J Virol. 1992;66:4518–4524. doi: 10.1128/jvi.66.7.4518-4524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polack B, Schwartz I, Barthelemy M, Belloc C, Manet G, Vuillaume A, Baron T, Gonda M A, Levy D. Serologic evidence for bovine immunodeficiency virus infection in France. Vet Microbiol. 1996;48:165–173. doi: 10.1016/0378-1135(95)00138-7. [DOI] [PubMed] [Google Scholar]
- 30.Rasmussen L, Battles J K, Ennis W H, Nagashima K, Gonda M A. Characterization of virus-like particles produced by a recombinant baculovirus containing the gag gene of the bovine immunodeficiency-like virus. Virology. 1990;178:435–451. doi: 10.1016/0042-6822(90)90341-n. [DOI] [PubMed] [Google Scholar]
- 31.Rasmussen L, Greenwood J D, Gonda M A. Expression of bovine immunodeficiency-like virus envelope glycoproteins by a recombinant baculovirus in insect cells. Virology. 1992;186:551–561. doi: 10.1016/0042-6822(92)90021-g. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snider T G, Hoyt P G, Jenny B F, Coats K S, Luther D G, Storts R W, Battles J K, Gonda M A. Natural and experimental bovine immunodeficiency virus infection in cattle. Vet Clin N Am Food Anim Pract. 1997;13:151–176. doi: 10.1016/s0749-0720(15)30370-4. [DOI] [PubMed] [Google Scholar]
- 35.St-Cyr Coats K, Pruett S F, Nash J W, Cooper C R. Bovine immunodeficiency virus: incidence of infection in Mississippi dairy cattle. Vet Microbiol. 1994;42:181–189. doi: 10.1016/0378-1135(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 36.St-Laurent G, Morin G, Archambault D. Detection of equine arteritis virus following amplification of structural and nonstructural viral genes by reverse transcription-PCR. J Clin Microbiol. 1994;32:658–665. doi: 10.1128/jcm.32.3.658-665.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suarez D L, VanDerMaaten M J, Wood C, Whetstone C A. Isolation and characterization of new wild-type isolates of bovine lentivirus. J Virol. 1993;67:5051–5055. doi: 10.1128/jvi.67.8.5051-5055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suarez D L, Whetstone C A. Identification of hypervariable and conserved regions in the surface envelope gene in the bovine lentivirus. Virology. 1995;212:728–733. doi: 10.1006/viro.1995.1532. [DOI] [PubMed] [Google Scholar]
- 39.Suarez D L, Van Der Maaten M J, Whetstone C A. Improved early and long-term detection of bovine lentivirus by a nested polymerase chain reaction test in experimentally infected calves. Am J Vet Res. 1995;56:579–586. [PubMed] [Google Scholar]
- 40.Sullender W M, Britt W J. Antigenic and immunogenic analysis of group A and group B respiratory syncytial virus G proteins expressed from recombinant baculoviruses. J Gen Virol. 1996;77:641–648. doi: 10.1099/0022-1317-77-4-641. [DOI] [PubMed] [Google Scholar]
- 41.Van Der Maaten M J, Boothe A D, Seger C L. Isolation of a virus from cattle with persistent lymphocytosis. J Natl Cancer Inst. 1972;49:1649–1657. doi: 10.1093/jnci/49.6.1649. [DOI] [PubMed] [Google Scholar]
- 42.Webb N R, Summers M D. Expression of proteins using recombinant baculoviruses. Technique. 1990;2:172–188. [Google Scholar]
- 43.Whetstone C A, Van Der Maaten M J, Black J W. Humoral immune response to the bovine immunodeficiency-like virus in experimentally and naturally infected cattle. J Virol. 1990;64:3557–3561. doi: 10.1128/jvi.64.7.3557-3561.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whetstone C A, Van Der Maaten M J, Miller J M. A Western blot assay for the detection of antibodies to bovine immunodeficiency-like virus in experimentally inoculated cattle, sheep and goats. Arch Virol. 1991;116:119–131. doi: 10.1007/BF01319236. [DOI] [PubMed] [Google Scholar]
- 45.Windheuser M G, Wood C. Characterization of immunoreactive epitopes of the HIV-1 p41 envelope protein using fusion proteins synthesized in Escherichia coli. Gene. 1988;64:107–119. doi: 10.1016/0378-1119(88)90485-4. [DOI] [PubMed] [Google Scholar]
- 46.Zhang S, Xue W, Wood C, Chen Q, Kapil S, Minocha H C. Detection of bovine immunodeficiency virus antibodies in cattle by Western blot assay with recombinant gag protein. J Vet Diagn Investig. 1997;9:347–351. doi: 10.1177/104063879700900401. [DOI] [PubMed] [Google Scholar]
- 47.Zhang S, Troyer D L, Kapil S, Zheng L, Kennedy G, Weiss M, Xue W, Wood C, Minocha H C. Detection of proviral DNA of bovine immunodeficiency virus in bovine tissues by polymerase chain reaction (PCR) and PCR in situ hybridization. Virology. 1997;236:249–257. doi: 10.1006/viro.1997.8740. [DOI] [PubMed] [Google Scholar]