Abstract
One of the key features of any infectious disease is whether infection generates long-lasting immunity or whether repeated reinfection is common. In the former, the long-term dynamics are driven by the birth of susceptible individuals while in the latter the dynamics are governed by the speed of waning immunity. Between these two extremes a range of scenarios is possible. During the early waves of SARS-CoV-2, the underlying paradigm was for long-lasting immunity, but more recent data and in particular the 2022 Omicron waves have shown that reinfection can be relatively common. Here we investigate reported SARS-CoV-2 cases in England, partitioning the data into four main waves, and consider the temporal distribution of first and second reports of infection. We show that a simple low-dimensional statistical model of random (but scaled) reinfection captures much of the observed dynamics, with the value of this scaling, , providing information of underlying epidemiological patterns. We conclude that there is considerable heterogeneity in risk of reporting reinfection by wave, age-group and location. The high levels of reinfection in the Omicron wave (we estimate that 18% of all Omicron cases had been previously infected, although not necessarily previously reported infection) point to reinfection events dominating future COVID-19 dynamics. This manuscript was submitted as part of a theme issue on “Modelling COVID-19 and Preparedness for Future Pandemics”.
Keywords: COVID-19, Immunity, Waning, Cross-projection, Statistical model, Alpha, Delta, Omicron
1. Introduction
The pattern of SARS-CoV-2 cases in England and elsewhere can be conceptualised as a series of waves, often associated with the emergence of a new variant. In England, there have been four main waves: the first from January to July 2020 was due to the Wildtype variant, the second from August 2020 to May 2021 was due to the Wildtype variant followed by the Alpha variant, the third from May to December 2021 was attributable to the Delta variant, while the fourth wave has been driven by the Omicron variant (with two sub-waves due to sub-types BA.1 from December 2021 to March 2022, and BA.2 from March 2022 onwards). The scale of these waves is dependent on the characteristics of each variant (Dyson et al., 2021) (with each variant having a higher transmission potential than the last and by immune escape for the Omicron variant), population mixing (Gimma et al., 2022) (impacted by control measures and precautionary behaviour driven by perceived risk) and population immunity (Anderson et al., 2020), with the latter being the most relevant for the long-term dynamics.
From the start of the epidemic in January 2020 until 14th April 2022 (our chosen end date due to changes in national testing policy in England UK Health Security Agency (2022)), there were 18.3 million reported infections, equivalent to 33% of the English population. This is likely to be a considerable underestimate of the true number of infections, especially in the early waves when testing was far more limited. During the first wave (from the first reported cases in England in late January 2020 until 15th July 2020) there were around 250,000 reported cases — whereas the first REACT study estimated around 3.4 million people had been infected by this time (Ward et al., 2021a). In the second wave, testing became more wide-spread with national testing centres and postal tests available. From April 2021 to April 2022, covering much of the third and fourth waves, free Lateral Flow Device (LFD) tests were freely available in England, and many groups were encouraged to perform regular tests, hence during these later times we may expect the ratio of reported cases to infection to increase. The number of reported cases (from both regular tests and symptomatic testing) is likely to be influenced by age-dependent changes in testing behaviour, reduced symptoms associated with second infections and the reduction in symptoms due to vaccine protection. Despite these biases, the study of identified cases and in particular individuals who test positive on two (or more) occasions is extremely valuable.
During the first three waves (associated with Wildtype, Alpha and Delta variants) reported reinfections were rare; less that 1% of cases before December 2021 were in individuals that had previously reported infection. Reinfections increased dramatically during the Omicron waves, with around 10% of cases from January to April 2022 having previously reported infection.
2. Methods
England has had multiple methods of testing for SARS-CoV-2 infection operating in parallel. These can be split into: Pillar 1 testing — generally PCR-based in hospitals and care homes; and Pillar 2 testing — generally at home or in testing centres (Department of Health and Social Care, 2020). The nature of Pillar 2 testing has changed over the course of the pandemic, with such testing largely absent during the first wave. Subsequently, during the second and third waves of the pandemic PCR testing was advised for all symptomatic infections and as a follow-up to positive LFD tests, with PCR comprising 95% of all positive Pillar 2 tests in 2020 and 2021. From early 2022 onward LFD tests dominate. Here, we do not distinguish between the type of test, but use episode number and an anonymised unique identifier to link first and second infections — we combine reported cases from both Pillar 1 and Pillar 2 test, LFD and PCR positive tests without discriminating. It is worth noting that PCR (polymerase chain reaction) detects the presence of viral genetic material, it is highly sensitive and therefore often seen as the gold-standard but can continue to detect virus after the infectious period; by contract LFD (lateral flow devices) detect the presence of antigen, have slightly lower sensitivity and are generally considered to be a marker of infectiousness (Dinnes et al., 2021, Smith et al., 2021). We label to be the number of individual reported cases that first test positive on day and then subsequently on day (); we also define as the total number of all positive tests on day , irrespective of whether they are a first or subsequent infection. We note that there is a 90-day threshold imposed on the data we receive, such that a new episode is only defined if it is more the 90 days since the last positive test. This 90-day threshold ensures that long-duration infections with multiple positive tests are not counted as repeat infections (Harvey et al., 2021); however this threshold could hide some rapid reinfection or might not exclude all long-duration infections — we will see that secondary episodes at 90 days are relative common.
As such is a matrix of values, where and convey information on the likely variants while the value of informs on the time-varying testing behaviour and the level of cross-protection. Examining all the positive tests in England from late January 2020 to 14 April 2022, we find that approximately 95% are first reports, 5% are second reports and only 0.064% (approximately 12,000 cases) are third or subsequent reports of infection. For this reason we restrict our attention to first and second reports only, which simplifies the interaction between variants and reporting interval that needs to be considered.
We partition the period January 2020 to April 2022 into four distinct waves and note the dominant variant(s): Wave 1, Wildtype (from the first case on 30th January–15th July 2020); Wave 2, Wildtype and Alpha (16th July 2020–30th April 2021); Wave 3, Delta (1st May–12th December 2021); and Wave 4, Omicron, both BA.1 and BA.2 (from 12th December 2021 until 12th April 2022 when testing dramatically declined). The boundaries between the waves are chosen to correspond to local troughs in reported cases separating the main peaks of infection; the exception is the transition to Wave 4 on 12th December 2021 which corresponds to the switch from predominantly Delta infections to predominantly Omicron. We compare the data on first and second reported cases () at times and , to a simple model () in which reinfection is a random process, that could have wave-specific scaling:
where is the population size (approximately 56 million for England), is the number of cases at time (we note that as refers to all cases, whereas are only individuals that report two or more infections) and refers to the wave number at a particular time point. Under this formulation, would be for a homogeneous infection obeying the SIS paradigm with no immunity and equal risk of reporting all cases, so that a proportion of the reported cases are expected to have reported previous infection at time . Immunity will act to reduce ; we expect where informs about degree of cross-protection afforded by previous infection (or alternatively is the relative risk of reinfection); however, heterogeneity in the risk of infection and reporting within the population will act to increase - as the effective population size experiencing and reporting infection will be smaller.
We consider two particular forms of , estimated such that is a good fit to , in terms of maximising the likelihood (assuming that reinfection events are Poisson distributed; alternative assumptions such as calculated the likelihood based on a Binomial distribution, such that some fraction of cases are reinfections, or simply matching the total number of reinfection events over the entire time period being considered, produce very similar estimates). Firstly, a homogeneous model where just two values of are used, an initial value for the first three waves when reinfection was rare, and a second higher value during the fourth Omicron wave when the degree of protection was lower and reinfection was more common:
Alternatively, we can compute a different for every pair of waves (noting that ).
To delve into the interactions within and between waves in more detail, we consider the different pairs of waves and the delay, , between first and second infections in waves and as captured by
where is an indicator function (it is is the input is true or otherwise) and is used to identify if a time period is associated with a particular wave. The simple model, (making either the homogeneous and variant-specific assumptions for ) can also be used to generate a similar function:
capturing the expected pattern of reported reinfection events over time.
We therefore use the estimated values of to understand the general patterns of reinfection between waves, noting that both greater risk of re-infection or greater heterogeneity can lead to an increased value of , such that we cannot uniquely attribute every change in to a direct epidemiological cause — although the relative level of reported cases provides some guidance. We use the comparison between and to explore the finer structure within each wave, and to assess longer-term trends.
3. Results
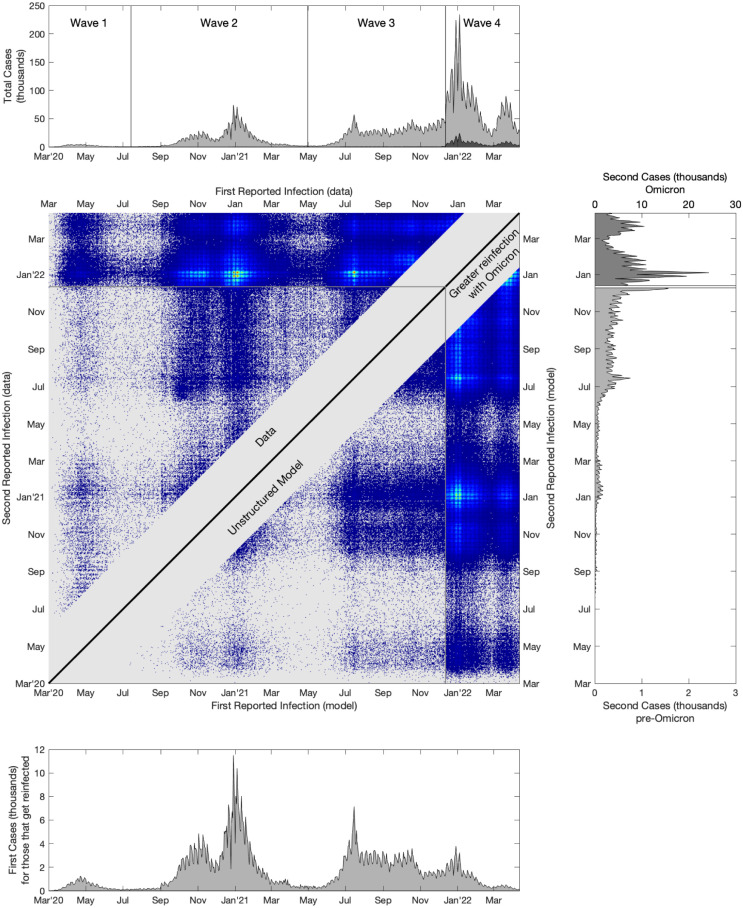
Figs. 1 and S1 graphically compare the data () to the simple models of reinfection (). Focusing on the homogeneous assumption for (Fig. 1) we estimate that (% CI ) and (% CI ) which taken together explain 51% and 85% of the variance in reinfection data in the pre-Omicron (waves 1–3) and Omicron periods (wave 4) respectively. (That is, is 0.51 for in the first three waves and 0.85 for in the fourth wave; with much of the remaining variance due to the relatively small and integer nature of , whereas is represents a continuous expectation.) The top panel of Fig. 1 is simply the number of reported cases over time (), with reinfections in dark grey; the bottom panel is the number of first cases of individuals who report a secondary infection (, plotted against ), while the right-hand panel is the number of secondary reported infections (, plotted against ). We note that due to the much larger number of reinfection events for Omicron (presumably caused by a lower value and less cross-protection from previous infection, reflected in ), we have been forced to use two different scales for the pre-Omicron and Omicron periods, as captured by the top and bottom axes. Finally, the middle panel shows the data on reinfections ( upper-left triangle) and the simple model ( lower-right triangular) — we note that the bottom and right panels are therefore the appropriate projections of the sum of the matrix of data (). From inspection, it is clear that reinfection is far more common in the Omicron waves (as exemplified by being at least three times larger than ), and that the simple model captures the bulk patterns of reinfection, even with this homogeneous set of assumptions for .
Fig. 1.
A comparison of reported first and second infections in England to the simple homogeneous assumption model. The central panel shows reinfection data ( upper-left triangle) and the corresponding model fit (in particular we show a Poisson sample of in the lower-right triangle, to allow a better visual comparison when is low), brighter colours correspond to more reported reinfections, while grey is zero reinfections in the data or model. The upper panel shows the total number of reported cases (pale grey) and the number of second reported cases (dark grey); the lower panel shows the number of reported first cases for those that report twice, while the right-hand panel shows the second reports — note that second reported cases during the Omicron wave (after 12th December 2021) are plotted on a different scale for clarity.
Figure S1 shows similar results for the model in which is wave specific, with the four main UK waves (associated with Wildtype, Wildtype/Alpha, Delta and Omicron). This greater heterogeneity in explains more of the variance in : 56% and 90% for the pre-Omicron and Omicron periods respectively. The relatively small improvement in the pre-Omicron results (compared to a single value) is because is dominated by the single combination of reinfections in wave 3 (Delta variant) after an initial infection in wave 2.
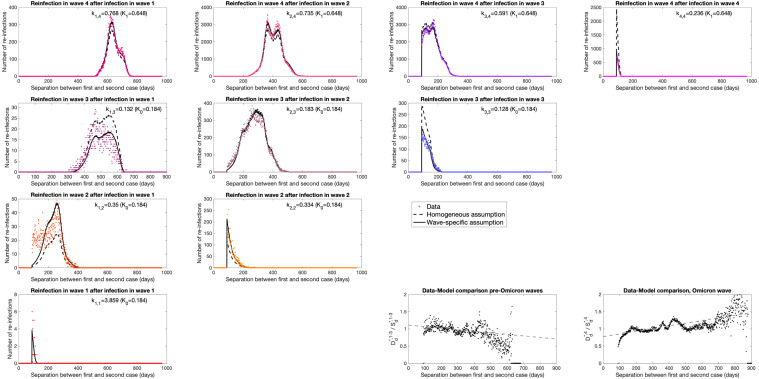
We now consider the data transformed to examine the separation between first and second cases for each wave (, Fig. 2), which in plotted in the same upper triangular form as the data in Fig. 1. To this data we add lines as produced from the simple model () with both the homogeneous (dashed line) and wave-specific (solid line) assumptions for .
Fig. 2.
Wave–wave interactions for reported reinfections. Each panel in the upper-left triangle shows the number of reported cases with a given delay between the first and second report (, coloured dots), separated by the associated waves. For each, we also show the same numbers based on the simple model with both the wave-specific (solid lines) and homogeneous (dashed lines) assumptions for . Note, to enable visual inspection of the empirical data compared to the simple model, we have used different -axis ranges across subplots. The two panels in the bottom right perform a comparison of data () and model (, using the homogeneous assumption) when the second cases are from the pre-Omicron () and Omicron () waves respectively, enabling us to detect temporal trends, shown as a linear fit to the points.
For Wildtype–Wildtype reinfections in wave 1 (Fig. 2, bottom left), the value of for the variant-specific model is large (approximately 3.859), suggesting that reinfection with wildtype is much more common than expected from case numbers; we attribute this to the extreme heterogeneity in reporting leading to a much smaller effective population size, as only a small fraction of the population was likely to be tested (in general only those severely ill and requiring hospital treatment). (All values of for the variant-specific assumption together with 95% confidence intervals, are given in Table 1.) Reinfections in wave 2 show a similar pattern to our observations for wave 1 (Fig. 2, row 3), where the wave-specific model is larger (0.350 and 0.334) than in the homogeneous model (0.184), but far smaller than for wave 1 due to increased population-level testing. The model generally captures the timing of wave 2 reinfections but substantially underestimates rapid reinfection (days 90–150) in the transition between wave 1 and wave 2 (which would correspond to Wildtype–Wildtype reinfections), although the total number of such reinfections is relatively small.
Table 1.
The scaling parameter that generates the best fit between the simple model and the data in terms of maximising the likelihood (together with 95% confidence intervals) assuming that the data represent a Poisson sample with a mean given by the model prediction (). We maintain the same upper-triangular structure as seen in Fig. 1 and Fig. 2 for ease of comparison.
| Wave | Wave, first case |
|||
|---|---|---|---|---|
| second case | 1 | 2 | 3 | 4 |
| 4 | 0.768 (0.759 - 0.776) | 0.735 (0.733 - 0.737) | 0.591 (0.589 - 0.593) | 0.236 (0.231 - 0.240) |
| 3 | 0.132 (0.127 - 0.136) | 0.183 (0.181 - 0.184) | 0.128 (0.125 - 0.131) | – |
| 2 | 0.350 (0.341 - 0.359) | 0.334 (0.326 - 0.341) | – | – |
| 1 | 3.859 (2.918 - 5.101) | – | – | – |
Delta variant reinfections during wave 3 (Fig. 2, row 2) show far more agreement between the simple model and the data. Secondary cases with Delta after a primary case in wave 2 (Fig. 2, row 2 column 2) numerically dominate, and for such reports the wave-specific and homogeneous models generate similar values ( compared to ). The fact that is less than one is a reflection of the protection afforded by the primary case; however, it does not suggest that the degree of cross-protection, , is around 82% (), as due to heterogeneity in infection and reporting risk, the effective population size is likely to be far smaller than the true population size. We note a slightly reduced level of reinfection (for Delta reinfection after wave 1 cases, although the numbers are again small, and for Delta after Delta) in the data and variant-specific model compared to the homogeneous model, which may be a signal of greater cross-protection.
Lastly, for reinfections in the Omicron (fourth) wave, (Fig. 2, top row) the data and models are in relatively good agreement, with both models generating similar fits. The estimated values of are all relatively large (0.591 to 0.766) which we attribute to high rates of reinfections due to limited cross protection. The exception to this pattern is Omicron–Omicron pairs where the variant-specific is 0.236, suggesting that there is a higher degree of protection against the same variant. Considering the temporal pattern in more detail, we note that decreases from initial cases during wave 1 through to initial cases with Delta in wave 3, hinting a decline in cross-protection over time. In addition, within each subplot there is a tendency for the model to slightly overestimate the number of most recent reinfections or underestimate the number of longer duration reinfections — which again suggests a slight decline in cross-protection over time.
To explore this temporal aspect in more detail, we compare the data, , with the model, (by plotting against ). We contrast when the second infection, , occurs during any of the first three waves, with when reinfection occurs in the fourth Omicron wave (Fig. 2, bottom right). We perform this comparison using the homogeneous model ( pre-Omicron or for Omicron) as this removes the temporal component that is inherent within the wave-specific assumptions. In the pre-Omicron period we observe a slight but statistically significant () decline in with the separation between cases, ; whereas during Omicron the trend is stronger and in the opposite direction (). Therefore during the Omicron wave, the model (with homogeneous assumption for ) tends to underestimate the number of recent reported reinfections, and consequently overestimate the number of longer separation reinfections; the trend of more infections at longer separations may be a signal of waning immunity.
We can use the analysis of these reported reinfection patterns to generate estimates of the total level of reinfection. For the four waves considered here, we estimate a reporting rate of 7.3% (6.6–8.0%) in wave 1, 32% (28%–38%) in wave 2, 56% (46%–65%) in wave 3 and 31% (25%–40%) in wave 4, using the fitted Warwick model (Keeling et al., 2021a, Keeling et al., 2022a, Keeling et al., 2022c). This agrees with primary measurements: estimates from the REACT-2 study (Ward et al., 2021b) lead to a 7.5% (7.2–7.9%) reporting rate in wave 1 (based on measurements that 6.0% (5.8–6.1%) were infected in the first wave (Ward et al., 2021a)), while cumulative incidence estimates from the ONS study (Office for National Statistics, 2022) lead to a 38% (36%–41%) and 42% (39%–46%) reporting rate for waves 2 and 3 respectively (fig. S3). Using this level of reporting through time, and assuming that reported cases are representative of all infections, we can estimate the true level of reinfection (after 90-days) over the course of the pandemic. For wave 1, the amount of reinfection is low at around 0.6% (0.5–0.7%), for waves 2 and 3 this increases to 2.5% (2.2–2.8%) and 2.3% (2.0–2.5%) respectively, but for wave 4 and the Omicron variant this increases to 18% (16%–20%). We note that the pre-Omicron figures are lower that those generated by the SIREN study (Hall et al., 2022) in which around 7.1% of infections from December 2020 to September 2021 (during the Alpha and Delta waves) were reinfections; we attribute this to the far higher risks of infection for the health care workers that were recruited to the SIREN study. These results are strongly suggestive that the future dynamics of SARS-CoV-2 will be contingent on reinfections within the population, although the extent to which these are mild or severe infections will be driven by the characteristics of new variants that emerge.
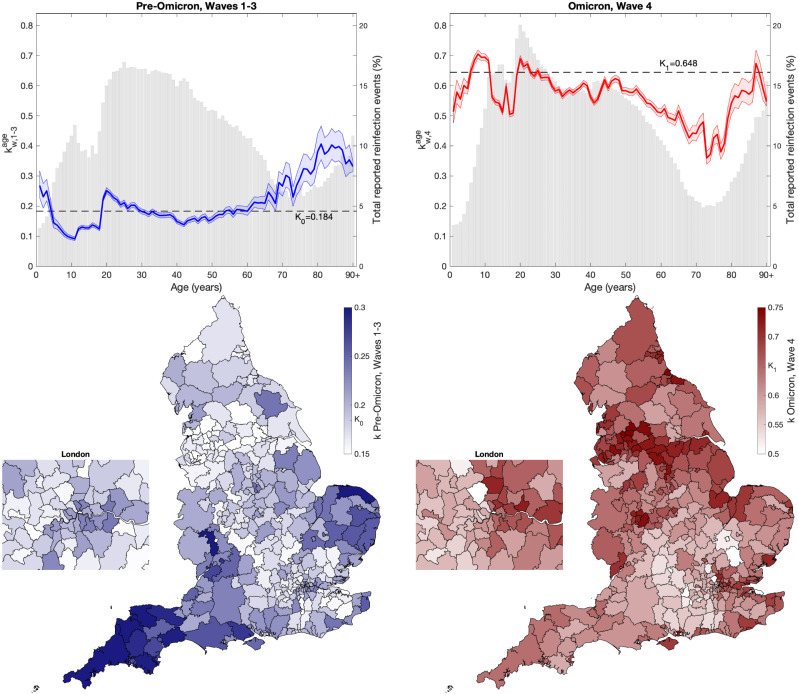
Finally, we perform the same analysis but on subsets of the population; in particular we partition the population by age and location (Fig. 3). We calculate in the pre-omicron waves (Fig. 3, blue) and Omicron wave (Fig. 3, red). Both show distinct deviation from the population average values ( and ), with patterns that differ between pre-Omicron and Omicron waves. Pre-Omicron (January 2020 to December 2021, Fig. 3 left) the scaling parameter is large for those over 60, those between 20 and 30, and those under 5 (there are more reported reinfections than expected from population averages); for the Omicron wave, it is only the 7–11 year olds and 20–22 year olds that are appreciably greater than , with the 70–80 year olds showing a substantially reduced level of reported reinfection compared to population-level expectations. Spatially, in the pre-Omicron waves, there is greater reporting reinfection than expected in Cornwall, Devon and East Anglia; whereas during the Omicron wave, we observe that it is predominantly Southern Central England that has lower than expected reinfection.
Fig. 3.
The values of the scalingwhen partitioning the data by age or spatial location, in comparison to the homogeneous assumptionor. Top row shows the mean value of in one-year age groups (from to ), together with 95% confidence intervals, the left-hand figure (blue) is for waves 1–3 pre-Omicron, while the right-hand figure (red) is for Omicron wave 4. The grey bars (and right-hand y-axis) show the percentage of all reported cases that are reinfections (). The lower panels are when the data is partitioned by Lower Tier Local Authority (LTLA) of which there are 317 in England; darker colours refer to higher values, with (pre-Omicron waves national average value for ) and (Omicron wave national average value for ) shown on the colorbar.
4. Discussion
We have passed the point in the pandemic where the overwhelming majority of people in England have already been infected, with estimates of 70.7% (90% credible interval: 66.0–75.6%) infected by 11th February 2022 (Office for National Statistics, 2022) — although many of these will have been mild or asymptomatic infections and not reported. The ability of SARS-CoV-2 to reinfect individuals is a key factor in both its persistence and control in the longer-term and the transition to endemicity (Keeling et al., 2021b, Rustom and Halloran, 2021). Here, using detailed individual-level case reports for England from the start of the pandemic (January 2020) to 14th April 2022, we consider the pattern of reported cases and the number of reported reinfection events. By far the most striking patterns are the massive increase in reported reinfection during the Omicron waves, and the general agreement between the data and a simple model of random reinfection.
The fact that the random model fits the data so well, even using the homogeneous assumption for (which involves the fitting of just two scaling quantities), strongly suggests that there are at most sparse immunological signals in this data. The data are well captured by a model in which reinfections occur randomly in a subset of the population (accounting for protection from past infection, heterogeneous risk of infection and heterogeneous likelihood of testing in a single parameter, ), with only a minimal impact of the temporal separation between infections. For second reported infections with Omicron during wave 4, there is a slight signal of waning immunity — reporting reinfection when the initial infection was over 18 months ago is 35% more likely than reporting reinfection when the initial infection was less than 6 months ago given the relative abundance of reported cases over these periods (Fig. 2, bottom-right). However, before Omicron this trend was reversed (although weaker) with shorter separations being relatively more likely (Fig. 2, bottom column 3).
The scaling parameter is seen to vary substantially between waves, and between ages and spatial locations. is a combination of two factors — the degree of protection offered by past infection (which reduces ) and the degree of population heterogeneity (which increases ); in general it is impossible to disentangle these two elements, from alone; however combining changes in with changes in reported cases and reinfections offers some insights. The large jump in the number of reported reinfections in the Omicron wave suggests that the associated increase in is primarily associated with a reduction in cross protection. In contrast, we believe that the high value in the first wave () reflects the narrow subset of the population that would be tested for SARS-CoV-2 infection, thereby reducing the effective population size. In particular, given the same pattern of infection, the value of will double if only half the population would report an infection, as the effective population size is halved. This basic concept can be extended, with heterogeneity in risk of infection or the chance of reporting leading to a increase in . Moreover, vaccination is likely to increase if both first and second case are post vaccination roll-out, as this changes the heterogeneity in risk across the population (those unvaccinated have a higher chance of infection).
The age data require more care to interpret. In the Omicron wave (Fig. 3, top-right), the shape of closely follows the pattern of reported reinfections, suggesting that the low in 70–80 years old is due to increased cross protection (or behavioural changes) following previous infection, leading to relatively fewer cases. In contrast, the high value and low reinfection probability in 7–11 year olds hints at highly heterogeneous levels of infection and reporting, potentially due to a subset of this age-group regularly self-testing. The patterns pre-Omicron (Fig. 3, top-left) show that the elderly age-groups have high and relatively low reported reinfections, suggestive of dynamics driven by population heterogeneity and again increased testing in a smaller subset of the population.
There are some important caveats to this work. Firstly, we are only dealing with reported cases; this either requires symptomatic illness followed by testing (and reporting) or periodic testing to detect mild or asymptomatic infection. This is important for four main reasons. The pattern of testing has changed throughout the pandemic as different forms of testing have become available, in the first wave testing was of severely ill patients only and using PCR, whereas from April 2021 to April 2022 free lateral flow device (LFD) tests were available nationally. There is also a strong age-bias in testing with younger individuals more likely to test regularly and therefore detect asymptomatic infections. We note that there is evidence that for individuals that are infected more than once, the most common pattern is for just one severe episode with other infections being mild or asymptomatic (Mensah et al., 2022, Office for National Statistics, 2021) — this means that reporting of multiple infections is likely to be an underestimate unless there is regular asymptomatic testing. In addition, the action of the vaccine reduces the amount of symptomatic illness, which again introduces a temporal and age-dependent bias. Finally, only positive tests separated by 90 days are recorded as a new reported episode, meaning that shorter separations between reinfection events are not included and long-duration persistent infections, although rare, may be recorded as multiple episodes.
Secondly, we have subdivided the timeline into four periods, roughly corresponding to the four main waves; while waves 1, 3 and 4 are dominated by Wildtype, Delta and Omicron variants respectively, wave 2 is a mixture of Wildtype and Alpha. An alternative characterisation, splitting wave 2 into Wildtype and Alpha dominated periods (fig. S2), does not greatly improve the fit; in particular there are problems capturing reported reinfections with Wildtype variants in wave 2 after initial infection in wave 1. Our definition of waves is not rigorous, but we generally transition at the point of minimum reported infections, such that the precise timing of the transition does not affect the bulk proprieties reported; the only exception to this rule is the transition between waves 3 and 4, which is aligned with the dominance of the Omicron variant. In addition, we do not split the two Omicron waves attributable to BA.1 (during December 2021, January and February 2022) and BA.2 (mainly during March and April 2022), this is due to the short time-scales involved — the 90 day minimum separation between reported infections means that we are left with relatively little discriminatory power and certainly no prospect of have recorded two infections within the BA.2 time period.
Finally, the pattern of reported reinfection events as a function of the time between them (Fig. 2) is largely driven by the timing of the infection waves. Thus while it is intuitively tempting to seek a immunological explanation of the observed pattern of declining reinfection events with separation for cases within the same wave (diagonal subplots in Fig. 2), the decline in reinfection events is a facet of the waves of infection and is echoed in the behaviour of the simple model. When infection has largely peaked in a single wave, the most common separation between any two infection events is short with long separations corresponding to rarer infection events at the start and end of the wave.
These results indicate that reported reinfection events are very much driven by the contemporary and historic pattern of reported cases, with factors such as waning immunity playing a limited role. Heterogeneity is most profound in the difference between pre-Omicron (before December 2021) and Omicron (after December 2021) waves, and the differences between age-groups. Taken together, our results suggest that periodic reinfection is highly likely especially in an environment where new variants are constantly emerging. We predict that the Omicron wave (15th December 2021 to April 2022) was associated with 18% (16%–20%) of cases being reinfections. This pattern of reinfections aligns most closely with an SIRS-paradigm, such that we may expect seasonal waves of infection driven by waning immunity and virus evolution.
CRediT authorship contribution statement
Matt J. Keeling: Conceptualization, Methodology, Formal analysis, Writing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
I would like to thank Ed Hill, Louise Dyson and Mike Tildesley for their extremely helpful comments on the early versions of this work.
Funding
MJK was supported through the UKRI [JUNIPER modelling consortium; grant number MR/V038613/1] and the National Institute for Health Research (NIHR) [Policy Research Programme, Mathematical and Economic Modelling for Vaccination and Immunisation Evaluation, and Emergency Response; NIHR200411]. MJK is affiliated to the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections at University of Liverpool in partnership with UK Health Security Agency (UKHSA), in collaboration with University of Warwick. MJK is also affiliated to the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Genomics and Enabling Data at University of Warwick in partnership with UK Health Security Agency (UKHSA). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health and Social Care or UK Health Security Agency.
Ethical considerations
Data from the CHESS and SARI databases were supplied after anonymisation under strict data protection protocols agreed between the University of Warwick and Public Health England. The ethics of the use of these data for these purposes was agreed by Public Health England with the Government’s SPI-M(O)/SAGE committees.
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.jtbi.2022.111299.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
Spread sheet of the number of reported infections and reinfections in England from 1 January 2020 to 12 April 2022.
Additional figures showing further sub-division of the epidemic period, and comparison between model and data.
Data availability
Data on cases were obtained from the COVID-19 Hospitalisation in England Surveillance System (CHESS) data set that collects detailed data on patients infected with COVID-19. These data contain confidential information, with public data deposition non-permissible for socioeconomic reasons. The CHESS data resides with the National Health Service (www.nhs.gov.uk). The ethics of the use of these data for these purposes was agreed by Public Health England with the Governments SPI-M(O)/SAGE committees. More aggregate data is freely available from the UK Coronavirus dashboard: https://coronavirus.data.gov.uk/.
References
- Anderson R.M., Vegvari C., Truscott J., Collyer B.S. Challenges in creating herd immunity to SARS-CoV-2 infection by mass vaccination. Lancet. 2020;396(10263):1614–1616. doi: 10.1016/S0140-6736(20)32318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Health and Social Care R.M. 2020. Coronavirus (COVID-19): Scaling up testing programmes. URL https://www.gov.uk/government/publications/coronavirus-covid-19-scaling-up-testing-programmes. [Google Scholar]
- Dinnes J., Deeks J., Berhane S., Taylor M., Adriano A., Davenport C., Dittrich S., Emperador D., Takwoingi Y., Cunningham J., Beese S., Domen J., Dretzke J., Ferrante di Ruffano L., Harris I., Price M., Taylor-Phillips S., Hooft L., Leeflang M., McInnes M., Spijker R., Van den Bruel A. Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2021;(3) doi: 10.1002/14651858.CD013705.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson L., Hill E.M., Moore S., Curran-Sebastian J., Tildesley M.J., Lythgoe K.A., House T., Pellis L., Keeling M.J. Possible future waves of SARS-CoV-2 infection generated by variants of concern with a range of characteristics. Nature Commun. 2021;12:5730. doi: 10.1038/s41467-021-25915-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimma A., Munday J.D., Wong K.L., Coletti P., van Zandvoort K., Prem K., working group C.C.-., Klepac P., Rubin G.J., Funk S., et al. Changes in social contacts in England during the COVID-19 pandemic between March 2020 and March 2021 as measured by the CoMix survey: A repeated cross-sectional study. PLoS Med. 2022;19(3) doi: 10.1371/journal.pmed.1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall V., Foulkes S., Insalata F., Kirwan P., Saei A., Atti A., Wellington E., Khawam J., Munro K., Cole M., Tranquillini C., Taylor-Kerr A., Hettiarachchi N., Calbraith D., Sajedi N., Milligan I., Themistocleous Y., Corrigan D., Cromey L., Price L., Stewart S., de Lacy E., Norman C., Linley E., Otter A.D., Semper A., Hewson J., D’Arcangelo S., Chand M., Brown C.S., Brooks T., Islam J., Charlett A., Hopkins S. Protection against SARS-CoV-2 after COVID-19 vaccination and previous infection. N. Engl. J. Med. 2022;386(13):1207–1220. doi: 10.1056/NEJMoa2118691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R.A., Rassen J.A., Kabelac C.A., Turenne W., Leonard S., Klesh R., Meyer W.A., Kaufman H.W., Anderson S., Cohen O., et al. Association of SARS-CoV-2 seropositive antibody test with risk of future infection. JAMA Internal Med. 2021;181(5):672–679. doi: 10.1001/jamainternmed.2021.0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J., Dyson L., Guyver-Fletcher G., Holmes A., Semple M.G., Tildesley M.J., Hill E.M. Fitting to the UK COVID-19 outbreak, short-term forecasts and estimating the reproductive number. Stat. Methods Med. Res. 2022;31:1716–1737. doi: 10.1177/09622802211070257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J., Dyson L., Tildesley M.J., Hill E.M., Moore S. Comparison of the 2021 COVID-19 roadmap projections against public health data. Nature Commun. 2022;113:4924. doi: 10.1038/s41467-022-31991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J., Hill E.M., Gorsich E.E., Penman B., Guyver-Fletcher G., Holmes A., Leng T., McKimm H., Tamborrino M., Dyson L., Tildesley M.J. Predictions of COVID-19 dynamics in the UK: Short-term forecasting and analysis of potential exit strategies. Flegg J.A., editor. PLOS Comput. Biol. 2021;17(1) doi: 10.1371/journal.pcbi.1008619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J., Thomas A., Hill E.M., Thompson R.N., Dyson L., Tildesley M.J., Moore S. 2021. Waning, boosting and a path to endemicity for SARS-CoV-2. MedRxiv, Cold Spring Harbor Laboratory Press. [DOI] [Google Scholar]
- Mensah A.A., Lacy J., Stowe J., Seghezzo G., Sachdeva R., Simmons R., Bukasa A., O’Boyle S., Andrews N., Ramsay M., Campbell H., Brown K. Disease severity during SARS-COV-2 reinfection: A nationwide study. J. Infect. 2022;84(4):542–550. doi: 10.1016/j.jinf.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Office for National Statistics A.A. 2021. Coronavirus (COVID-19) Infection Survey technical article: Analysis of reinfections of COVID-19: June 2021. URL https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/articles/coronaviruscovid19infectionsurveytechnicalarticleanalysisofreinfectionsofcovid19/june2021. [Google Scholar]
- Office for National Statistics A.A. 2022. Coronavirus (COVID-19) Infection Survey technical article: Cumulative incidence of the number of people who have tested positive for COVID-19, UK: 22 April 2022. https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/articles/coronaviruscovid19infectionsurveytechnicalarticlecumulativeincidenceofthenumberofpeoplewhohavetestedpositiveforcovid19uk/22april2022. [Google Scholar]
- Rustom A., Halloran M.E. Transition to endemicity: Understanding COVID-19. Immunity. 2021;54(10):2172–2176. doi: 10.1016/j.immuni.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.L., Gibson L.L., Martinez P.P., Ke R., Mirza A., Conte M., Gallagher N., Conte A., Wang L., Fredrickson R., Edmonson D.C., Baughman M.E., Chiu K.K., Choi H., Jensen T.W., Scardina K.R., Bradley S., Gloss S.L., Reinhart C., Yedetore J., Owens A.N., Broach J., Barton B., Lazar P., Henness D., Young T., Dunnett A., Robinson M.L., Mostafa H.H., Pekosz A., Manabe Y.C., Heetderks W.J., McManus D.D., Brooke C.B. Longitudinal assessment of diagnostic test performance over the course of acute SARS-CoV-2 infection. J. Infect. Dis. 2021;224(6):976–982. doi: 10.1093/infdis/jiab337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK Health Security Agency R.L. 2022. Changes to COVID-19 testing in England from 1 April. URL https://www.gov.uk/government/news/changes-to-covid-19-testing-in-england-from-1-april. [Google Scholar]
- Ward H., Atchison C., Whitaker M., Ainslie K.E., Elliott J., Okell L., Redd R., Ashby D., Donnelly C.A., Barclay W., et al. SARS-CoV-2 antibody prevalence in England following the first peak of the pandemic. Nature Commun. 2021;12(1):1–8. doi: 10.1038/s41467-021-21237-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H., Cooke G.S., Atchison C., Whitaker M., Elliott J., Moshe M., Brown J.C., Flower B., Daunt A., Ainslie K., et al. Prevalence of antibody positivity to SARS-CoV-2 following the first peak of infection in England: Serial cross-sectional studies of 365,000 adults. Lancet Reg. Health-Eur. 2021;4 doi: 10.1016/j.lanepe.2021.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spread sheet of the number of reported infections and reinfections in England from 1 January 2020 to 12 April 2022.
Additional figures showing further sub-division of the epidemic period, and comparison between model and data.
Data Availability Statement
Data on cases were obtained from the COVID-19 Hospitalisation in England Surveillance System (CHESS) data set that collects detailed data on patients infected with COVID-19. These data contain confidential information, with public data deposition non-permissible for socioeconomic reasons. The CHESS data resides with the National Health Service (www.nhs.gov.uk). The ethics of the use of these data for these purposes was agreed by Public Health England with the Governments SPI-M(O)/SAGE committees. More aggregate data is freely available from the UK Coronavirus dashboard: https://coronavirus.data.gov.uk/.