Abstract
SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) is the key enzyme required for viral replication and mRNA synthesis. RdRp is one of the most conserved viral proteins and a promising target for antiviral drugs and inhibitors. At the same time, analysis of public databases reveals multiple variants of SARS-CoV-2 genomes with substitutions in the catalytic RdRp subunit nsp12. Structural mapping of these mutations suggests that some of them may affect the interactions of nsp12 with its cofactors nsp7/nsp8 as well as with RNA substrates. We have obtained several mutations of these types and demonstrated that some of them decrease specific activity of RdRp in vitro, possibly by changing RdRp assembly and/or its interactions with RNA. Therefore, natural polymorphisms in RdRp may potentially affect viral replication. Furthermore, we have synthesized a series of polyphenol and diketoacid derivatives based on previously studied inhibitors of hepatitis C virus RdRp and found that several of them can inhibit SARS-CoV-2 RdRp. Tested mutations in RdRp do not have strong effects on the efficiency of inhibition. Further development of more efficient non-nucleoside inhibitors of SARS-CoV-2 RdRp should take into account the existence of multiple polymorphic variants of RdRp.
Keywords: RNA-Dependent RNA polymerase, SARS-CoV-2, RdRp mutations, Viral replication, Non-nucleoside inhibitors
Abbreviations: RdRp, RNA-dependent RNA polymerase; PP, polyphenols; DA, diketoacids; DAA, analogs of diketoacids
1. Introduction
RdRp is the most conserved viral enzyme and a proved target for inhibition by antiviral compounds [[1], [2], [3], [4]]. Coronaviruses have unusually large RNA genomes, and efficient and accurate genome replication is essential for virus survival [[5], [6], [7]]. During replication and subgenomic RNA synthesis, RdRp is assisted by several cofactors that ensure processive RNA synthesis, capping and proofreading [2,8]. The replicase complex of coronaviruses includes the catalytic RdRp subunit nsp12 and its cofactors nsp7 and nsp8 required for interactions with RNA (Fig. 1 ) [[9], [10], [11], [12]], two molecules of the nsp13 helicase potentially involved in template switching and RNA proofreading [2,13], the proofreading exonuclease nsp14 with its cofactor nsp10 [14,15], as well as factors involved in RNA capping (nsp9, nsp14, nsp16) (reviewed in Ref. [2]). The nsp14 exonuclease is essential in SARS-CoV-2 [16] and can likely remove unnatural nucleotides and chain terminators from the nascent RNA 3′-end [14,15,17], thus explaining the resistance of SARS-CoV-2 to nucleoside inhibitors that can inhibit replication of other viruses (e.g. Ref. [18]).
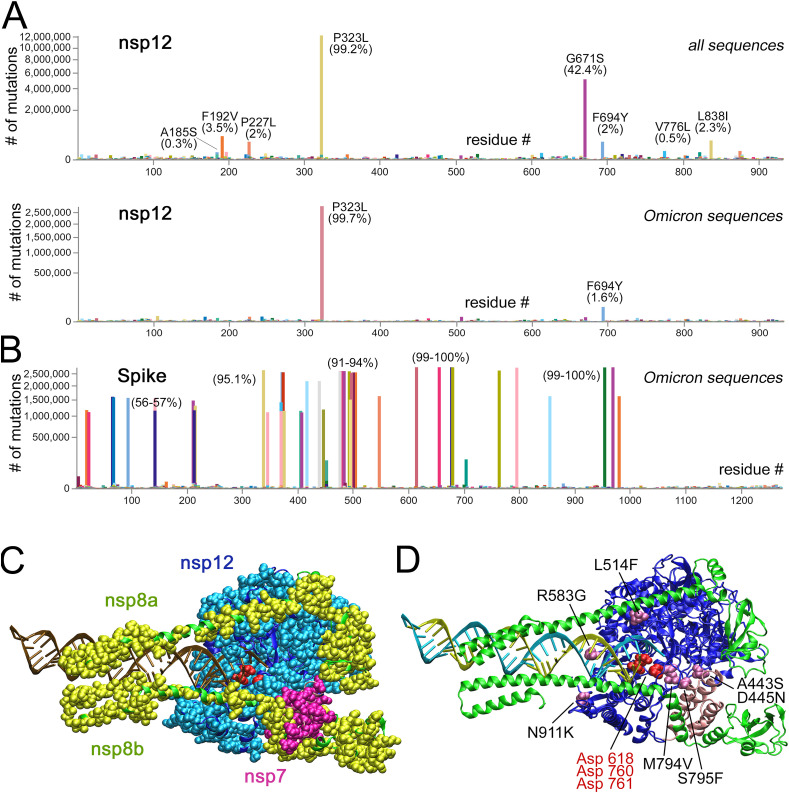
Fig. 1.
Naturally occurring amino acid substitutions in nsp12. (A) Frequencies of amino acid substitutions that emerged in nsp12 from December 15, 2019 (the start of pandemic) until May 25, 2022, for all sequenced SARS-CoV-2 genomes (top, 12,365,901 genomes) and for the Omicron lineage (bottom, 2,789,360 genomes). The data were retrieved from the GISAID database on September 21, 2022 [21]. The number of sequenced genomes containing substitutions at each nsp12 position is shown on the y-axis (log scale). Substitutions are shown relative to the reference strain WIV04. Most frequent substitutions and their percentage among all SARS-CoV-2 sequences are indicated. (B) Frequencies of amino acid substitutions in the spike protein in the Omicron lineage (sublineages BA.1, BA.1.1, BA.2, BA.2.12.1, BA.2.3, BA.4, BA.5, BA.5.2.1, BA.5.2) in comparison with the reference strain WIV04, counted for the same period. (C) Structure of the replication complex of SARS-CoV-2 RdRp (PDB ID: 6XEZ). Nsp12, nsp8 and nsp7 are shown as ribbon models in dark blue, pink and green, respectively. Natural substitutions in nsp12, nsp8 and nsp7 are shown as CPK models in light blue, magenta and yellow, respectively; the active site residues are red. (D) RdRp structure showing substitutions tested in this study (mauve). Primer RNA is yellow, template RNA is blue. The catalytic site aspartate residues are shown in red.
Coronaviruses are rapidly evolving, and several highly transmissible SARS-CoV-2 strains have appeared during the pandemic, highlighting the importance of development of universal antiviral drugs and inhibitors targeting diverse virus variants [1,19,20]. While most previous studies of SARS-CoV-2 variants were focused on mutations in the spike protein affecting receptor recognition, cell entry and immune evasion, analysis of SARS-CoV-2 genome sequences reveals multiple substitutions in every coronavirus protein, including RdRp (Fig. 1) (covidcg.org, ref. [21]). Possible effects of these polymorphisms on the activity of SARS-CoV-2 replicase and the viral life cycle remain unknown. Intense research in the last two and a half years has allowed to discover several potent inhibitors of SARS-CoV-2, including nucleoside and non-nucleoside compounds [[22], [23], [24], [25], [26], [27], [28]], reviewed in Refs. [[2], [3], [4]]. Several nucleoside analogs have been clinically approved for COVID-19 treatment in various countries (see Discussion). Whether natural polymorphisms in SARS-CoV-2 RdRp may affect its sensitivity to various types of inhibitors remains to be investigated.
Here, we have analyzed several naturally occurring substitutions in SARS-CoV-2 RdRp and found that they can decrease RdRp activity in vitro. We have also tested several previously studied inhibitors of hepatitis C virus (HCV) RdRp and demonstrated that some of them have promise for further development of more efficient inhibitors of SARS CoV-2 RdRp.
2. Materials and methods
2.1. Analysis of natural substitutions in SARS-CoV-2 RdRp
The information on amino acids substitutions in the nsp7, nsp8 and nsp12 protein sequences relative to the reference strain WIV04 (MN996528.1) was downloaded from the COVID-19 CG site (covidcg.org [21]), for all geographical locations and the time period from the start of pandemic until May 25, 2021 or May 24, 2022 or September 21, 2022, as indicated in the figure legends. Substitutions were visualized on the replication complex structure (PDB ID: 6XEZ) with Visual Molecular Dynamics software [29].
2.2. Purification of SARS-CoV-2 RdRp
Wild-type codon-optimized nsp12 (with a C-terminal His8 tag) and fused nsp7/nsp8 genes (separated by a hexahistidine linker [30]) were cloned into pET-28 and expressed in E. coli BL21 (DE3) as previously described [31]. Mutatant variants of the nsp12 gene were obtained by PCR site-directed mutagenesis and expressed in the same way. The cells were disrupted with a high pressure homogenizer in lysis buffer containing 50 mM Tris-HCl, pH 7.9, 500 mM NaCl, 100 μg/ml PMSF, the lysate was cleared by centrifugation and loaded onto a 1 ml HiTrap TALON crude column (GE Healthcare) equilibrated in 40 mM Tris-HCl, pH 7.9, 500 mM NaCl. The column was washed with buffer solution containing 0, 30, and 100 mM imidazole at 1 ml/min, and RdRp was eluted with 300 mM imidazole. The samples were dialyzed against 50 mM Tris-HCl, pH 7.9, 50 mM NaCl, 5% glycerol, 0.1 mM DTT, 0.5 mM EDTA overnight and loaded onto a HiScreenQ HP column (GE Healthcare). The column was washed with the same buffer and the proteins were eluted with a NaCl gradient (50 mM–1 M) in the same buffer for 150 min at 0.5 ml/min. The fractions containing the nsp12 and nsp7-nsp8 complex were collected. Isolated nsp7-nsp8 proteins was purified in the same way. All protein samples were concentrated using Amicon Ultra-4 centrifugal filters, supplemented with NaCl, DTT and glycerol (250 mM, 1 mM and 50% final concentrations), aliquoted and stored at −70 °C.
2.3. Synthesis of prospective compounds for testing RdRp inhibition
Hydrazones PP1-PP11 were prepared from corresponding polyhydroxybenzaldehydes and substituted hydrazine derivatives as described in Refs. [32,33]. Diketo acids DA1-DA7 and compounds DAA3 and HQ1 (compound I-13e) were synthesized according to published methods [[34], [35], [36]]. The syntheses of heterocyclic N-hydroxy acids DAA1 and DAA2 and of hydroxyquinolines derivatives HQ2 and HQ3 were carried out according to original techniques that will be published elsewhere. The structures of the target compounds were confirmed by NMR spectroscopy (Supplementary Fig. S5).
2.4. Analysis of RdRp activity
RdRp activity was measured using a variant of RNA substrate described previously [37], which contained a 20 bp long RNA duplex followed by a 10 nt template segment (Fig. 2 A). Primer RNA was 5’-[32P]-labeled and annealed to template RNA in buffer containing 10 mM Tris-HCl, pH 7.9, 10 mM KCl, 2 mM MgCl2 and 1 mM DTT. The RNA substrate (25 or 50 nM final concentration) was mixed with RdRp (the nsp12-nsp7-nsp8 complex; 500 nM or 1 μM) in the same buffer and incubated for 15 min at 30 °C. When indicated, extra nsp7/nsp8 (1 μM) was added to the reaction mixture. Inhibitors were added to concentrations indicated in the figure legends (0.2 or 0.4 μl of stock solutions in DMSO per 10 μl sample). The same volume of DMSO was added to control reactions. The reaction was started by adding ATP (100 μM, unless otherwise indicated) and terminated after indicated time intervals by adding equal volume of formamide with 100 μg/ml heparin. The samples were heated for 4 min at 95 °C and separated by 15% (19:1) denaturing urea PAGE. The gels were scanned by a Typhoon 9500 scanner (GE Healthcare) and quantified with ImageQuant software (GE Healthcare).
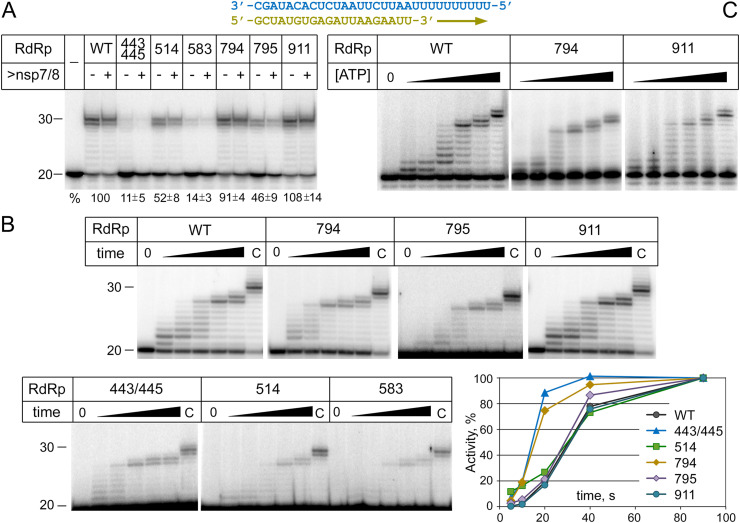
Fig. 2.
Catalytic activity of mutant RdRp variants. (A) RNA primer extension by the wild-type and mutant RdRp variants. The RNA substrate is shown on the top (primer, yellow; template, blue). The reactions were performed for 15 min at 30 °C with 100 μM ATP and 500 nM RdRp, either without or with addition of extra nsp7-nsp8 (1 μM). The activity of mutant RdRps is shown in percentage of wild-type RdRp (means and standard deviations from three independent experiments). (B) Kinetics of incorporation of ATP (1 μM) by the wild-type and mutant RdRps (1 μM) containing substitutions at indicated positions. The reactions were performed for 5, 10, 20, 40 or 90 s with 50 nM RNA substrate; chase reactions (‘C’) were performed for 7 min with 1 mM ATP. The plot shows quantification of the kinetics of full-length RNA synthesis, normalized by the maximum activity for each RdRp variant (except for R583G that had too low activity in this assay). (C) RNA extension by the WT, M794V and N911K RdRps at various ATP concentrations (100 nM, 250 nM, 1 μM, 2.5 μM, 10 μM, 1 mM). The reactions were performed for 10 s at 30 °C. Representative gels from two independent experiments are shown. Positions of the starting RNA primer (20 nt) and the extended RNA product (30 nt) are indicated.
3. Results
3.1. Polymorphisms in SARS-CoV-2 RdRp
Analysis of public databases reveals amino acid substitutions in almost every position of nsp12 in sequenced SARS-CoV-2 genomes collected from around the world, when compared with the original Wuhan strain (Fig. 1A; covidcg.org) [21]. In particular, substitutions can be found in 712 out of the 932 positions of nsp12 for genomes sequenced during the first wave of pandemic (from December 2019 until 25 May 2020) and in 915 positions during the two and a half years of pandemic (until 25 May 2022) (Fig. 1A, top). Visualization of these substitutions on the structure of the replication complex of SARS-CoV-2 RdRp shows that they cover almost the entire surface of nsp12 (Fig. 1C). Similarly, amino acid substitutions are found in various SARS-CoV-2 genomes at most positions of the nsp12 cofactors nsp7 and nsp8 (Fig. 1C; covidcg.org). Most of these substitutions are found in only a minor fraction of all genomes, however, some of them have become widespread during the pandemic. These include P323L found in 99.2% of all sequenced genomes, G671S found in 42.1% of genomes (mostly in genomes of the Delta lineage), and several others present in a percentage of all genomes (Fig. 1A, top).
The evolution of SARS-CoV-2 is in a large part driven by mutations in the spike protein, which affect receptor binding and virus entry, and change the transmission rates and virulence of newly emerging coronavirus strains [38,39]. In comparison with the original reference strain, the now dominant Omicron lineage of SARS-CoV-2 contains multiple substitutions in the S protein (Fig. 1B). To get insight into possible co-evolution of substitutions in RdRp and the S protein, we analyzed the frequencies of substitutions in nsp12 separately in the Omicron lineage. Strikingly, it was found that the frequencies of many substitutions found in the population are decreased in this lineage (except for P323L, which is also prevalent in other lineages) (Fig. 1A, bottom). Similarly, we found that the frequencies of amino acid substitutions in the exoribonuclease nsp14 are much decreased in the Omicron lineage in comparison with all sequenced genomes (Fig. S1). The only exception is an I24V substitution, which is found in most Omicron genomes. It can therefore be concluded that many polymorphic variants of nsp12 and nsp14 are being eliminated from the population during replacement of dominant SARS-CoV-2 lineages in the course of pandemic.
3.2. Natural substitutions in various parts of SARS-CoV-2 RdRp alter its activity in vitro
The effects of naturally occurring substitutions on the activity of SARS-CoV-2 RdRp have remained unknown. In this study, we analyzed several substitutions in nsp12 that could potentially alter its functional activity. When choosing substitutions, we excluded rare variants that could result from sequencing errors and focused on variants present in hundreds and thousands of sequenced genomes. The selected polymorphic variants of nsp12 included substitutions located close to the active site of nsp12 (M794V and S795F), to the interface of nsp12 with nsp7 (A443S/D445 N, which were combined together), with nsp8 (L514F), and with the primer-template RNA duplex (R583G and N911K) (Fig. 1D). In addition to the selected substitutions, other amino acid residues can also be found in these positions. The frequencies of all these substitutions in sequenced genomes of SARS-CoV-2 are shown in Supplementary Table S1.
The wild-type and mutant variants of SARS-CoV-2 RdRp (the nsp12-nsp7-nsp8 complex) were expressed and purified from E. coli using metal-affinity and anion-exchange chromatography (Fig. S2). The activity of all RdRp variants was tested using a primer-template RNA substrate containing an oligoU template region (Fig. 2A). Wild-type RdRp efficiently extended the primer until the end of the template RNA strand (Fig. 2A). The M794V and N911K RdRp variants had similar activities. In comparison, the L514F and S795F substitutions were ∼2-fold less active than wild-type RdRp, while the A443S/D445 N and R583G substitutions strongly decreased the RdRp activity (Fig. 2A). The addition of extra nsp7/nsp8 proteins did not increase the activity of mutant variants suggesting that their low activity was not due to insufficient nsp7/nsp8 binding (Fig. 2A).
We then measured the activity of mutant RdRp variants and their ability to interact with the RNA substrate at different RdRp concentrations. We found that RNA extension by the A443S/D445 N, L514F, R583G and S795F mutants is stimulated at high RdRp concentrations (Fig. S3A). Furthermore, it was shown that these mutant variants poorly bind the RNA substrate in an electrophoretic mobility shift assay (EMSA) (Fig. S3B). In particular, formation of the RdRp-RNA complex with the least active mutants (A443S/D445 N, L514F and R583G) could be detected only at the highest RdRp concentrations (Fig. S3B). We therefore conclude that the mutations may affect the assembly of the RdRp replicase complex and/or its interactions with the RNA substrate.
To reveal whether the mutations may also affect the catalytic properties of RdRp, we analyzed the kinetics of RNA synthesis at low ATP concentration, which allowed to visualize stepwise primer extension. It was found that all six mutant RdRp variants could extend RNA with similar kinetics, even though with different efficiencies of substrate utilization (Fig. 2B). The A443S/D445 N and M794V mutants were even somewhat faster than wild-type RdRp, since longer RNA products were observed for these mutants at short time points (Fig. 2B). We further compared RNA extension by wild-type RdRp and two of the mutants, M794V and N911K, at various concentrations of ATP. No strong differences in the nucleotide concentration dependence was observed between these enzymes (Fig. 2C), suggesting that these mutations do not affect nucleotide binding by RdRp. Furthermore, we explored the pattern of nucleotide misincorporation by the wild-type, L514F and N911K RdRp variants, by testing primer extension on the same RNA template in the presence of non-complementary NTPs. It was found that all three polymerases could misincorporate GTP opposite template U, when it was added at high concentrations (>100 μM), but did not misincorporate CTP or UTP (Fig. S4). Therefore, the mutations are unlikely to directly decrease the catalytic activity and fidelity of RdRp.
3.3. Synthesis and analysis of non-nucleoside inhibitors of RdRp
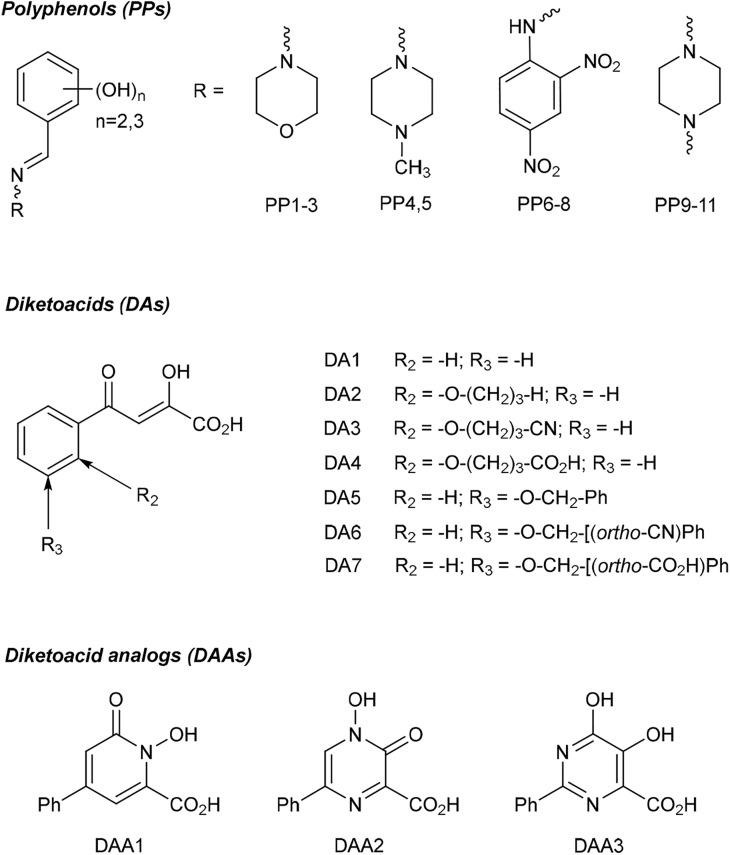
Repurposing of compounds known to inhibit RdRps from other viruses, such as influenza virus, Ebola virus or HCV, may aid quick development and approval of anticoronaviral drugs [3,4]. Based on structural homology between SARS-CoV-2 RdRp and HCV RdRp (NS5B) [40], we selected three classes of non-nucleoside compounds previously shown to inhibit HCV RdRp for testing their activity against SARS-CoV-2 RdRp (Fig. 3 , Fig. S5): (i) polyphenols (PP1-11) [33,41], (ii) derivatives of α,γ-diketoacids (DA1-7) [35,42], and (iii) their structural analogs, hydroxyl-containing heterocyclic acids (diketoacid analogs, DAA1-3) [34,43]. Previous in silico analysis predicted high affinity of diketoacids to the palm domain of SARS-CoV-2 RdRp [40].
Fig. 3.
Schematic presentation of molecular structures of studied compounds. Polyphenols (PP1-11), α,γ-diketoacids (DA1-7), and diketoacid analogs (DAA1-3).
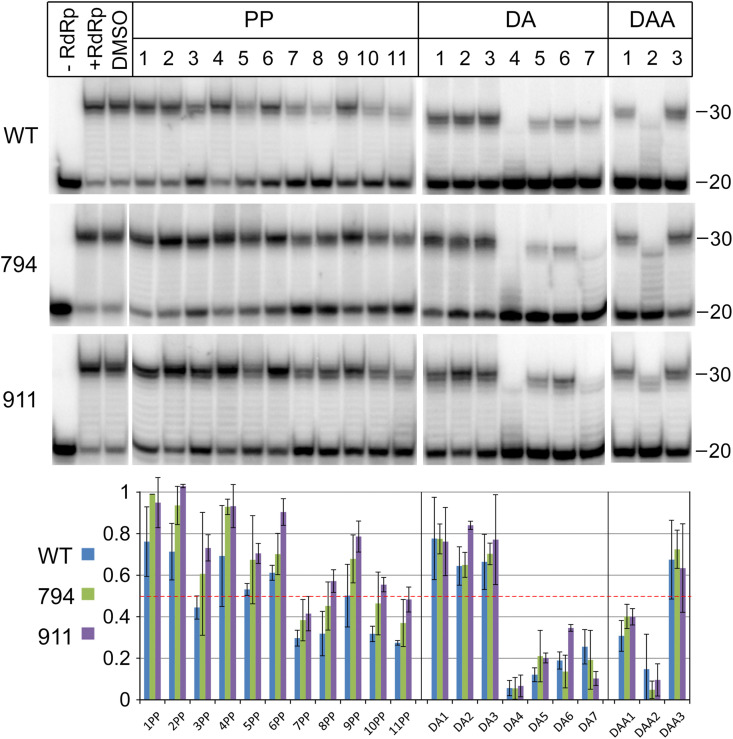
It was found that several of these compounds at high concentrations could indeed inhibit the activity of SARS-CoV-2 RdRp. These included PP7, PP8, PP10, PP11, DA4, DA5, DA6, DA7 and DAA2, which decreased the activity of RdRp 2-fold or more in the primer extension assay (Fig. 4 ). The strongest effects were observed for the DA4 and DAA2 compounds (Fig. 4). Titration of several selected compounds demonstrated that they are low affinity inhibitors, with their effects observed only at submillimolar or millimolar concentrations (PP7,8,10,11 in Fig. S6; DA4 and DAA2 in Fig. S7A). To rule out the possibility that the inhibitory effects of DA and DAA compounds may simply result from chelating of magnesium ions required for catalysis from the reaction solution, we performed similar experiments in a reaction buffer containing increased concentration of MgCl2. Although RdRp was less active in this buffer in comparison with standard conditions, the efficiency of inhibition of RdRp by DA4 and DAA2 was the same at both magnesium concentrations (compare Fig. S7A and Fig. S7B).
Fig. 4.
Inhibition of RdRp activity by various classes of tested compounds. The reactions were performed with wild-type, M794V and N911K RdRps (1 μM). Preformed replication complex was incubated with indicated compounds (PP1-11, DA1-7, DAA1-3, or DMSO in control reactions), ATP was added (100 μM) and the reactions were performed for 20 min at 30 °C. The plot at the bottom shows the activity of tested RdRp variants (measured by the full-length RNA synthesis) in the presence of indicated compounds relative to control reactions performed in the absence of inhibitors (means and standard deviations from three independent experiments). The red dotted line indicates a 2-fold decrease in the efficiency of RNA synthesis.
We then performed similar experiments with two most active mutant variants of RdRp, M794V and N911K. We observed that the overall pattern of inhibition of these mutants by different types of compounds was similar to the wild-type RdRp (Fig. 4, Fig. S7A). Polyphenols were somewhat less efficient in inhibiting mutant RdRps in comparison with the wild-type polymerase, while DA7 was more efficient, although these differences were not statistically significant (Fig. 4).
Previously, another class of non-nucleoside compounds, hydroxyquinoline derivatives were shown to inhibit the activity of SARS-CoV-2 RdRp in a cell-based screening assay [36]. The most potent of these inhibitors was compound I-13e (HQ1 in Fig. S5), which inhibited reporter gene expression with a micromolar 50% effective concentration (EC50) [36]. However, its activity with purified RdRp samples has not been tested. We tested the ability of compound I-13e and two related hydroxyquinoline derivatives (HQ2 and HQ3, Fig. S5) to inhibit SARS-CoV-2 RdRp in the in vitro assay. For all three compounds, considerable inhibition of the RdRp activity was observed only in submillimolar range of concentrations (Fig. S8), which was similar to other compounds tested in our study. Therefore, the much more potent inhibition observed with hydroxyquinolines in the in vivo assay [36] was not due to their direct interaction with RdRp but might have resulted from their indirect effects on cellular metabolism and mRNA synthesis. This result emphasizes the importance of in vitro studies of various types of RdRp inhibitors.
4. Discussion
Beginning from the start of pandemic, more than 12 millions of SARS-CoV-2 genomes have been sequenced, resulting in detection of multiple polymorphic virus variants with amino acid substitutions in key proteins involved in viral replication. Most common substitutions found in RdRp (Fig. 1) do not directly affect the interface of nsp12 with nsp7/nsp8 or RNA, suggesting that they may not strongly change the RdRp activity. One of these substitutions, P323L, has become dominant during the pandemic, in association with the D614G variant of the spike protein [44]. At the same time, most polymorphic variants are found in only a small fraction of all sequenced genomes,. In particular, substitutions analyzed in this study are found with frequencies from 7.49 × 10−6 to 4.75 × 10−4 (Table S1). No natural polymorphic variants of RdRp have been previously studied and their possible effects on the RdRp activity and the virus life cycle have remained unknown.
Our analysis of the six mutations in SARS-CoV-2 RdRp revealed that even single amino acid variations detected in natural SARS-CoV-2 isolates can have strong effects on the RdRp activity in vitro. Two of the tested mutants, M794V and N911K, which contain substitutions of amino acids near the active site and the upstream RNA duplex, retain high levels of activity. The other four mutants (A443S/D445 N, L514F, R583G and S795F) have much lower specific activity, and much higher concentrations of these RdRp variants are required to achieve considerable levels of RNA substrate utilization. Furthermore, RNA binding by these mutants is apparently weaker in comparison with wild-type RdRp. At the same time, while significantly less active than the wild-type RdRp, these mutant variants do not decrease the rate of RNA extension or fidelity of nucleotide incorporation (Fig. 2 and Fig. S4). We therefore propose that the mutations may primarily affect the assembly of the holoenzyme RdRp complex and/or in its interactions with the RNA substrate, rather than the catalytic performance of RdRp. In particular, the S795F and A443S/D445 N substitutions may affect the interactions of nsp12 with nsp7, while L514F and R583G may change nsp12 interactions with nsp8 and RNA (Fig. 1D). Since SARS-CoV-2 strains containing these RdRp substitutions have been isolated from humans and are likely viable, these defects may be suppressed by interactions of RdRp with other subunits of the replication complex. Further analysis is needed to reveal their possible effects on virus replication in vivo. It also remains to be established whether other substitutions in RdRp may modulate its activity in vitro and in vivo.
Remarkably, we have found that the frequencies of substitutions at many positions of the catalytic subunit of RdRp, as well as in the exoribonuclease subunit, are decreased in the now dominant Omicron lineage of SARS-CoV-2, in comparison with the complete set of sequenced genomes (Fig. 1A, Fig. S1). These include most of the substitutions selected for in vitro analysis in this study (except for N911K, Table S1). In Omicron strains, only one substitution in addition to P323L (which is dominant in all lineages) can be found in >1% of sequences (F694Y, Fig. 1A). This corresponds to the replacement of earlier SARS-CoV-2 lineages, containing more frequent substitutions in RdRp, with Omicron. However, it remains to be explored whether this replacement might be connected to possible unfavorable effects of these substitutions on virus replication. Overall, the frequency of mutations in the catalytic subunit nsp12 or in the proofreading exoribonuclease nsp14 is dramatically lower than in the spike protein (Fig. 1 and Fig. S1), indicating that the conservation of the replication machinery is critical for virus reproduction.
Viral RdRps are promising therapeutic targets for development of antiviral compounds [1]. Current efforts are focused on finding both nucleoside and non-nucleoside inhibitors of RdRp, using de novo screening of individual compounds or large chemical libraries, either by molecular docking or by direct testing of their effect on the RdRp activity in vitro and in cell-based assays. Another approach is repurposing of known drugs acting on polymerases from other viruses [3,4,18,45]. Three clinically approved SARS-CoV-2 RdRp inhibitors, remdesivir, favipiravir and molnupiravir, are nucleoside compounds that were originally developed for inhibiting other viral RdRps [[46], [47], [48]]. These nucleoside analogs are efficiently incorporated into nascent RNA, escape the proofreading activity of nsp14, and either cause delayed chain termination (remdesivir), or induce template-dependent mutagenesis (favipiravir and molnupiravir) and RdRp stalling (remdesivir) [[22], [23], [24], [25], [26], [27], [28]].
Here, we have tested the effects of different classes of non-nucleoside compounds previously shown to inhibit HCV RdRp [[33], [34], [35],[41], [42], [43]] on the activity of SARS-CoV-2 RdRp in vitro. It was found that they are less active against SARS-CoV-2 RdRp and must be present at high concentrations to inhibit its activity. Moreover, some inhibitors of SARS-CoV-2 RdRp that apparently decrease its activity in a cell-based screening in vivo [36], have only weak effects on its activity in vitro (Fig. S8), suggesting that their in vivo effects may be indirect. While drug repurposing is a straightforward approach for finding novel antivirals, many known antivirals are inactive against SARS-CoV-2 RdRp, such as sofosbuvir that is likely removed from RNA through the proofreading activity of nsp14 [18]. Another example is nucleoside inhibitors of HIV-1 reverse transcriptase that do not inhibit SARS-CoV-2 RdRp because the latter polymerase cannot efficiently incorporate them into nascent RNA [49]. Therefore, direct testing of prospective compounds originally selected for inhibition of other polymerases on the activity of SARS-CoV-2 RdRp in vitro is essential for finding potent inhibitors.
Importantly, some of the compounds tested in our study can almost fully inhibit RdRp activity at high concentrations, despite the long reaction time and high nucleotide concentrations used in our assays. In our previous studies of the inhibition mechanism of HCV RdRp by polyphenol and diketoacid compounds, we proposed that they may bind near the active site and inhibit phosphoryl transfer, in particular, through changes in chelation of catalytic Mg2+ ions in the catalytic center of polymerase [33,42]. SARS-CoV-2 RdRp may be inhibited through a similar mechanism, which needs to be further investigated. We have demonstrated that mutant variants of SARS-CoV-2 RdRp do not differ significantly from the wild-type polymerase in their sensitivity to the tested compounds (Fig. 4, Fig. S7 and our unpublished observations). Recent screening for RdRp mutations conferring resistance to remdesivir identified several substitutions around the active site, including V792I, located close to the substitutions analyzed in our study [50]. It is therefore possible that naturally occurring polymorphisms in SARS-CoV-2 RdRp may also change RdRp sensitivity to various types of nucleoside inhibitors. Derivatives of polyphenols and diketoacids analyzed in this study may be used as starting compounds for further development of more efficient RdRp inhibitors acting on both wild-type and mutant RdRp variants.
Author contributions
AK, DE and SK conceived and supervised research, NM obtained RdRp mutations and analyzed inhibitors, MK designed and synthesized inhibitors, IP, DP and DE developed RdRp assays, IP analyzed the activity of RdRp mutants, MP performed bioinformatic analysis, NM, MP and AK prepared the figures, AK wrote the manuscript with contribution from all the authors.
Data availability statement
The authors declare that the data supporting the findings of this study are available within the article. Any additional raw data supporting the conclusions of this article will be made available by the authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Russian Foundation for Basic Research grant 20-04-60571.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biochi.2022.10.007.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lu L., Su S., Yang H., Jiang S. Antivirals with common targets against highly pathogenic viruses. Cell. 2021;184:1604–1620. doi: 10.1016/j.cell.2021.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Malone B., Urakova N., Snijder E.J., Campbell E.A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022;23:21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domling A., Gao L. Chemistry and biology of SARS-CoV-2. Chem. 2020;6:1283–1295. doi: 10.1016/j.chempr.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vicenti I., Zazzi M., Saladini F. SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19. Expert Opin. Ther. Pat. 2021;31:325–337. doi: 10.1080/13543776.2021.1880568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulyaeva A.A., Gorbalenya A.E. A nidovirus perspective on SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021;538:24–34. doi: 10.1016/j.bbrc.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020;295:12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillen H.S. Structure and function of SARS-CoV-2 polymerase. Curr Opin Virol. 2021;48:82–90. doi: 10.1016/j.coviro.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang X., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baddock H.T., Brolih S., Yosaatmadja Y., Ratnaweera M., Bielinski M., Swift L.P., Cruz-Migoni A., Fan H., Keown J.R., Walker A.P., Morris G.M., Grimes J.M., Fodor E., Schofield C.J., Gileadi O., McHugh P.J. Characterization of the SARS-CoV-2 ExoN (nsp14ExoN-nsp10) complex: implications for its role in viral genome stability and inhibitor identification. Nucleic Acids Res. 2022;50:1484–1500. doi: 10.1093/nar/gkab1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moeller N.H., Shi K., Demir O., Belica C., Banerjee S., Yin L., Durfee C., Amaro R.E., Aihara H. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2106379119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogando N.S., Zevenhoven-Dobbe J.C., van der Meer Y., Bredenbeek P.J., Posthuma C.C., Snijder E.J. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020;94 doi: 10.1128/JVI.01246-20. e01246-01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zandi K., Amblard F., Musall K., Downs-Bowen J., Kleinbard R., Oo A., Cao D., Liang B., Russell O.O., McBrayer T., Bassit L., Kim B., Schinazi R.F. Repurposing nucleoside analogs for human coronaviruses. Antimicrob. Agents Chemother. 2020;65 doi: 10.1128/AAC.01652-20. e01652-01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telenti A., Hodcroft E.B., Robertson D.L. The evolution and biology of SARS-CoV-2 variants. Cold Spring Harb Perspect Med. 2022;12 doi: 10.1101/cshperspect.a041390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz-Aravena M., McKee C., Gamble A., Lunn T., Morris A., Snedden C.E., Yinda C.K., Port J.R., Buchholz D.W., Yeo Y.Y., Faust C., Jax E., Dee L., Jones D.N., Kessler M.K., Falvo C., Crowley D., Bharti N., Brook C.E., Aguilar H.C., Peel A.J., Restif O., Schountz T., Parrish C.R., Gurley E.S., Lloyd-Smith J.O., Hudson P.J., Munster V.J., Plowright R.K. Ecology, evolution and spillover of coronaviruses from bats. Nat. Rev. Microbiol. 2022;20:299–314. doi: 10.1038/s41579-021-00652-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen A.T., Altschuler K., Zhan S.H., Chan Y.A., Deverman B.E. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. Elife. 2021;10 doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Hobartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon A., Le N.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon C.J., Tchesnokov E.P., Schinazi R.F., Gotte M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon A., Selisko B., Le N.T., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C., Coutard B., Peersen O., Canard B. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11:4682. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon C.J., Lee H.W., Tchesnokov E.P., Perry J.K., Feng J.Y., Bilello J.P., Porter D.P., Gotte M. Efficient incorporation and template-dependent polymerase inhibition are major determinants for the broad-spectrum antiviral activity of remdesivir. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2021.101529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tchesnokov E.P., Gordon C.J., Woolner E., Kocinkova D., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020;295:16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 30.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petushkov I., Esyunina D., Kulbachinskiy A. Effects of natural RNA modifications on the activity of SARS-CoV-2 RNA-dependent RNA polymerase. FEBS J. 2022 doi: 10.1111/febs.16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurskaya G.V., Zavodnik V.E., Zhukhlistova N.E., Kozlov M.V. Molecular structures of antiviral agents, 2,3-dihydroxybenzaldehyde 2,4-dinitrophenylhydrazone and 4-[(4-methylpiperazin-1-yl)imino]methyl-1,2-benzodiol. Crystallogr. Rep. 2008;53:626–630. [Google Scholar]
- 33.Kozlov M.V., Polyakov K.M., Ivanov A.V., Filippova S.E., Kuzyakin A.O., Tunitskaya V.L., Kochetkov S.N. Hepatitis C virus RNA-dependent RNA polymerase: study on the inhibition mechanism by pyrogallol derivatives. Biochemistry (Mosc.) 2006;71:1021–1026. doi: 10.1134/s0006297906090112. [DOI] [PubMed] [Google Scholar]
- 34.Summa V., Petrocchi A., Matassa V.G., Taliani M., Laufer R., De Francesco R., Altamura S., Pace P. HCV NS5b RNA-dependent RNA polymerase inhibitors: from alpha,gamma-diketoacids to 4,5-dihydroxypyrimidine- or 3-methyl-5-hydroxypyrimidinonecarboxylic acids. Design and synthesis. J. Med. Chem. 2004;47:5336–5339. doi: 10.1021/jm0494669. [DOI] [PubMed] [Google Scholar]
- 35.Summa V., Petrocchi A., Pace P., Matassa V.G., De Francesco R., Altamura S., Tomei L., Koch U., Neuner P. Discovery of alpha,gamma-diketo acids as potent selective and reversible inhibitors of hepatitis C virus NS5b RNA-dependent RNA polymerase. J. Med. Chem. 2004;47:14–17. doi: 10.1021/jm0342109. [DOI] [PubMed] [Google Scholar]
- 36.Zhao J., Zhang Y., Wang M., Liu Q., Lei X., Wu M., Guo S., Yi D., Li Q., Ma L., Liu Z., Guo F., Wang J., Li X., Wang Y., Cen S. Quinoline and quinazoline derivatives inhibit viral RNA synthesis by SARS-CoV-2 RdRp. ACS Infect. Dis. 2021;7:1535–1544. doi: 10.1021/acsinfecdis.1c00083. [DOI] [PubMed] [Google Scholar]
- 37.te Velthuis A.J., Arnold J.J., Cameron C.E., van den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Badua C.L.D.C., Baldo K.A.T., Medina P.M.B. Genomic and proteomic mutation landscapes of SARS-CoV-2. J. Med. Virol. 2021;93:1702–1721. doi: 10.1002/jmv.26548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin D.P., Lytras S., Lucaci A.G., et al. Selection analysis identifies clusters of unusual mutational changes in Omicron lineage BA.1 that likely impact spike function. Mol. Biol. Evol. 2022;39 doi: 10.1093/molbev/msac061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elghoneimy L.K., Ismail M.I., Boeckler F.M., Azzazy H.M.E., Ibrahim T.M. Facilitating SARS CoV-2 RNA-Dependent RNA polymerase (RdRp) drug discovery by the aid of HCV NS5B palm subdomain binders: in silico approaches and benchmarking. Comput. Biol. Med. 2021;134 doi: 10.1016/j.compbiomed.2021.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov A.V., Kozlov M.V., Kuzyakin A.O., Kostyuk D.A., Tunitskaya V.L., Kochetkov S.N. New non-nucleoside inhibitors of hepatitis C virus RNA-dependent RNA polymerase. Biochemistry (Mosc.) 2004;69:782–788. doi: 10.1023/b:biry.0000040204.82885.f1. [DOI] [PubMed] [Google Scholar]
- 42.Kozlov M.V., Polyakov K.M., Filippova S.E., Evstifeev V.V., Lyudva G.S., Kochetkov S.N. RNA-dependent RNA polymerase of hepatitis C virus: study on inhibition by alpha,gamma-diketo acid derivatives. Biochemistry (Mosc.) 2009;74:834–841. doi: 10.1134/s0006297909080033. [DOI] [PubMed] [Google Scholar]
- 43.Powdrill M.H., Deval J., Narjes F., De Francesco R., Gotte M. Mechanism of hepatitis C virus RNA polymerase inhibition with dihydroxypyrimidines. Antimicrob. Agents Chemother. 2010;54:977–983. doi: 10.1128/AAC.01216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ilmjarv S., Abdul F., Acosta-Gutierrez S., Estarellas C., Galdadas I., Casimir M., Alessandrini M., Gervasio F.L., Krause K.H. Concurrent mutations in RNA-dependent RNA polymerase and spike protein emerged as the epidemiologically most successful SARS-CoV-2 variant. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-91662-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian L., Qiang T., Liang C., Ren X., Jia M., Zhang J., Li J., Wan M., YuWen X., Li H., Cao W., Liu H. RNA-dependent RNA polymerase (RdRp) inhibitors: the current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021;213 doi: 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bravo J.P.K., Dangerfield T.L., Taylor D.W., Johnson K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell. 2021;81:1548–1552. doi: 10.1016/j.molcel.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naydenova K., Muir K.W., Wu L.F., Zhang Z., Coscia F., Peet M.J., Castro-Hartmann P., Qian P., Sader K., Dent K., Kimanius D., Sutherland J.D., Lowe J., Barford D., Russo C.J. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng J.Y., Du Pont V., Babusis D., Gordon C.J., Tchesnokov E.P., Perry J.K., Duong V., Vijjapurapu A., Zhao X., Chan J., Cohen C., Juneja K., Cihlar T., Gotte M., Bilello J.P. The nucleoside/nucleotide analogs tenofovir and emtricitabine are inactive against SARS-CoV-2. Molecules. 2022;27:4212. doi: 10.3390/molecules27134212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevens L.J., Pruijssers A.J., Lee H.W., Gordon C.J., Tchesnokov E.P., Gribble J., George A.S., Hughes T.M., Lu X., Li J., Perry J.K., Porter D.P., Cihlar T., Sheahan T.P., Baric R.S., Gotte M., Denison M.R. Mutations in the SARS-CoV-2 RNA dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms. Sci. Transl. Med. 2022 doi: 10.1126/scitranslmed.abo0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article. Any additional raw data supporting the conclusions of this article will be made available by the authors.