Abstract
The continuous emergence of SARS-CoV-2 variants prolongs COVID-19 pandemic. Although SARS-CoV-2 vaccines and therapeutics are currently available, there is still a need for development of safe and effective drugs against SARS-CoV-2 and also for preparedness for the next pandemic. Here, we discover that astersaponin I (AI), a triterpenoid saponin in Aster koraiensis inhibits SARS-CoV-2 entry pathways at the plasma membrane and within the endosomal compartments mainly by increasing cholesterol content in the plasma membrane and interfering with the fusion of SARS-CoV-2 envelope with the host cell membrane. Moreover, we find that this functional property of AI as a fusion blocker enables it to inhibit the infection with SARS-CoV-2 variants including the Alpha, Beta, Delta, and Omicron with a similar efficacy, and the formation of syncytium, a multinucleated cells driven by SARS-CoV-2 spike protein-mediated cell-to-cell fusion. Finally, we claim that the triterpene backbone as well as the attached hydrophilic sugar moieties of AI are structurally important for its inhibitory activity against the membrane fusion event. Overall, this study demonstrates that AI is a natural viral fusion inhibitor and proposes that it can be a broad-spectrum antiviral agent against current COVID-19 pandemic and future outbreaks of novel viral pathogens.
Keywords: SARS-CoV-2 variants, COVID-19, Astersaponin I, Aster koraiensis, Membrane fusion, Syncytium
Abbreviations: AI, Astersaponin I; AK, Aster koraiensis; COVID-19, coronavirus disease 2019; ACE2, angiotensin converting enzyme 2; SARS-CoV-2, Severe acute respiratory syndrome coronavirus-2
1. Introduction
The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to an unprecedented global health problems and economic tragedy. The recent development of clinically effective vaccines and therapeutics against COVID-19 represents an important step toward ending this pandemic. But, multiple SARS-CoV-2 variants including Alpha (lineage B.1.1.7, designated by the Pango nomenclature system (Rambaut et al., 2020)), Beta (lineage B.1.351), Gamma (lineage P.1), Delta (lineage B.1.617.2), and Omicron (lineage B.1.1.529) have emerged around the world. Current global outbreak of Omicron and its sub-variants are of concern since they are highly transmissible and could compromise the effectiveness of the vaccines (Andrews et al., 2022; Halfmann et al., 2022). Most of these SARS-CoV-2 variants carry mutations in spike (S) protein, which is responsible for binding to host cell receptor angiotensin-converting enzyme 2 (ACE2) and fusion between virus and cell membranes during the viral entry (Hoffmann et al., 2020; Jackson et al., 2021). The mutations on S protein were shown to confer more efficient attachment of SARS-CoV-2 to ACE2, resulting in higher viral infection and increasing the possibility of transmission (Cai et al., 2021; Harvey et al., 2021). The Delta and Omicron variants are known to have 16 and 32 mutations to the S protein respectively, and thus a higher risk for re-infection and viral escape from neutralizing antibodies (Callaway and Ledford, 2021). In order to control the ongoing COVID-19 pandemic due to the continuous emergence of more infectious SARS-CoV-2 variants, therapeutic agents that are effective against SARS-CoV-2 variants regardless of S mutation status are needed.
Traditional medicinal plants and derived natural products have been used for a long time as important remedies for the prevention and treatment of various infectious diseases (Dvorkin-Camiel and Whelan, 2008; Li and Peng, 2013; Qi et al., 2013) and some of them have thus been suggested as potential drug candidates for COVID-19 with minimal adverse effects (Alam et al., 2021; Benarba and Pandiella, 2020; Chandramouli et al., 2021; Kuchta et al., 2021). But, natural agents that have been proven to be active against SARS-CoV-2 variants are limited. In this study, we evaluated a potential anti-SARS-CoV-2 activity of several plants containing saponins, including Asparagus cochinchinensis, Aster pseudoglehnii, Aster scabra, Aster koraiensis, and Polygala tenuifolia.
2. Materials and methods
2.1. Cell culture
H1299 from the Korean Cell Line Bank (KCLB, South Korea, 25803) were maintained in RPMI-1640 (Gibco, USA), and HEK293T cells (CRL-3216) and Vero cells (CCL-81) from the American Type Culture Collection (ATCC, USA) were maintained in Dulbecco's Modified Eagle's Medium (DMEM; Corning, USA) in a 5% CO2 incubator at 37 °C. All medium was supplemented with 10% fetal bovine serum (FBS) (Gibco, USA) and 1X Penicillin-Streptomycin solution (HyClone, USA). A549-ACE2-TMPRSS2 cells that stably express human ACE2 and TMPRSS2 were purchased from National Institute for Biological Standards and Control (NIBSC) and grown at 37 °C with 5% CO2 in Ham's F–12K (Kaighn's) medium, supplemented with 10% heat-inactivated FBS and 2 mg/ml Geneticin (G418), 200 μg/ml Hygromycin B and 1x Antibiotic-Antimycotic solution (Gibco/Thermo Fisher Scientific, Waltham, MA, USA).
2.2. Plant material
The aerial parts of Aster pseudoglehnii Y.Lim, J.O.Hyun & H.Shin (voucher specimen #: ASPS2-2021) and Aster scabra Thunb. (voucher specimen #: ASSC1-2021) were purchased from a food market, Dondaemun-gu, Seoul, Korea, in May 2021. The leaves of Aster koraiensis Nakai (voucher specimen #: ASKO2-2021) was collected at Maru Park, Gangnam-gu, Seoul Korea, in May 2021. The roots of Asparagus cochinchinensis (Loureio) Merrill (voucher specimen #: ASCO1-2017) were purchased from Nanuum pharmaceutical Co., Youngcheon-si, Kyungsangbukdo, Korea, in May 2017. The roots of Polygala tenuifolia Willd. (voucher specimen #: POTE1-2016) were obtained from Hyunjin pharmaceutical Co., Dongdaemun-gu, Seoul, Korea, in October 2016. The origins of the plant materials were authenticated by one of the authors D.S.J. and voucher specimens were deposited at the College of Pharmacy, Kyung Hee University, Korea.
2.3. Extraction and isolation
The parts of each plant used for food or medicine were used as experimental materials. The fresh leaves of A. pseudoglehnii (150 g), A. scabra (92 g), and A. koraiensis (320 g) were sliced and extracted three times in an ultrasonic bath at room temperature (RT) for 2 h with 2 L, 1.5 L, and 3 L of 70% EtOH, respectively. The extracts were filtered and evaporated in vacuo at 45 °C to give 70% EtOH extracts of A. pseudoglehnii (6.94 g, 4.63%), A. scaber (3.70 g, 4.02%), and A. koraiensis (17.21 g, 5.38%). The leaves of Aster koraiensis for isolation of triterpenoid saponins were collected at Pyeongchang, Gangwon-Do, Korea in 2017. The dried and ground leaves (5.0 kg) of A. koraiensis were extracted twice with 25 L of 95% EtOH at 70 °C for 3 h and extract solutions were removed using a rotary evaporator. The 95% EtOH extract (500 g) was chromatographed over Diaion HP−20 (φ 9.8 × 63.0 cm) eluting with an acetone-H2O gradient (from 0:1 to 1:0 v/v) to afford 28 fractions (C1 ∼ C28). Fraction C11 was separated into seven subfractions by column chromatography using Sephadex LH-20 (φ 3.6 × 65.0 cm), with 50% acetone to separate astersaponin I (96.5 mg). The dried roots of P. tenuifolia (400 g) was extracted twice with 4 L of 70% EtOH at 80 °C in water bath for 3 h and the solvent was evaporated in vacuo at 45 °C. The 70% EtOH extract (84 g) was chromatographed over Diaion HP-20 (φ 6.8 × 30.5 cm) with a MeOH–H2O gradient (from 6:4 → 8:2 → 1:0 v/v) to afford 14 fractions (Fr.1∼Fr.14). Fraction 9 (1.8 g) was fractionated further by using a flash chromatography system using Redi Sep-C18 (130 g, MeOH–H2O, 1:1 to 4:1 v/v) cartridge to obtain onjisaponin R (40.4 mg). Fraction 10 (5.2 g) was chromatographed over silica gel CC as stationary phase with EtOAc/BuOH/MeOH/H2O mixture (3/4/1.5/1.5 v/v) to isolate onjisaponins B (212.4 mg), Fg (25.8 mg), and J (33.2 mg) was purified from fraction 10-6 (600 mg) by a flash chromatography system using Redi Sep-C18 (40 g, MeOH–H2O, 25:75 to 9:1 v/v) cartridge. The dried roots of A. cochinchinensis (800 g) were extracted with distilled water (8 L) at 100 °C for 2 h, and the solvent was removed using rotary evaporator to give a hot water extract (300.0 g). The steroidal saponins tested in this study, protodioscin, methyl protodioscin, aspacochioside A, aspacochioside C, 15−hydroxypseudoprotodioscin, and chamaedroside E, were previously isolated from the hot water extract of A. cochinchinensis (Kim et al., 2021a). The purities of all isolates were determined as >95% by high-performance liquid chromatography (HPLC) and nuclear magnetic resonance (NMR) experiments.
2.4. Reagents
Platycodigenin (purity:≥98%) were purchased from Chemface (China).
2.5. Cloning and plasmids
The full length of ACE2 and SARS-CoV-2 S sequence were amplified by PCR from pCEP4-myc-ACE2 (a gift from Erik Procko, Addgene plasmid #141185) and SARS-CoV-2 spike plasmid (a gift from Fang Li, Addgene plasmid #145032) respectively and cloned into pHR-CMV or pHR-CMV_IRES-EmGFP lentiviral expression plasmid (a gift from A. Radu Aricescu, Addgene plasmid #113887 and #113888) via EcoRI and AgeI restriction sites. Firefly luciferase gene were cloned into pHR-CMV via EcoRI and AgeI restriction sites. TMPRSS2 lentiviral expression vector, RRL.sin.cPPT.SFFV/TMPRSS2 (variant 1).IRES-neo.WPRE (MT130) was gifted by Caroline Goujon (Addgene plasmid # 145843).
2.6. Generation of stable cell lines
H1299 cells stably expressing ACE2 (ACE2+) or ACE2 plus TMPRSS2 (ACE2/TMPRSS2+) cells, ACE2/TMPRSS2+ H1299 cells expressing mRuby2, and H1299 cells co-expressing S protein and GFP (Spike-H1299) were generated by lentiviral transduction. For lentiviral production, HEK293T cells were co-transfected with lentiviral expression plasmids, packaging plasmid psPAX2, and envelope plasmid pMD2.G using Lipofectamine 3000 transfection reagent (Thermo Fisher, USA). Supernatants containing the lentivirus were collected at 24 h and 48 h post-transfection and filtered through 0.45 μm-pore-size filters. For the generation of stable cell lines, viral supernatants supplemented with 4 μg/ml polybrene (Merck, Germany) were added into H1299 cultures that were ∼50% confluent. The next day, the viral supernatants were replaced with fresh culture medium and incubated for 2–3 more days before the experiments.
2.7. pSARS-CoV-2 entry assay
SARS-CoV-2 spike pseudotyped lentivirus (pSARS-CoV-2) was generated as described in our previous study (Kim et al., 2021b). Briefly, HEK293T cells that were ∼80% confluent were co-transfected with pHR-CMV-firefly luciferase plasmid, psPAX2, and SARS-CoV-2 S plasmid (Addgene #145032) using Lipofectamine 3000 transfection reagent (Invitrogen, USA). Viral supernatants were collected 24 h and 48 h after transfection and centrifuged at 500×g for 5 min to remove detached cells. ACE2+ or ACE2/TMPRSS2+ H1299 cells that were ∼80% confluent in a 48 well plate was treated with the collected supernatant containing pSARS-CoV-2 virus particles and plant extracts or compounds after 1 h pretreatment with plant extracts or compounds. Next day, pSARS-CoV-2 entry efficiency was quantified by measuring firefly luciferase activity in cell lysates using a luciferase assay kit (Promega, USA) with SpectraMax iD5 Multi-Mode Microplate Reader (Molecular Devices, USA). DMSO-treated groups were set as 100% and IC50 (half-maximal inhibitory concentration) values were calculated and plotted with Prism v.9.0.0 software (GraphPad, USA). For the experiment comparing the infectivity of WT and D614G mutant of SARS-CoV-2, D614G mutant construct were generated by PCR-based mutagenesis using Pfu DNA polymerase. The sense primer sequence for the point mutation is as follows: 5′-GTG GCC GTG CTG TAC CAG GGC GTG AAT TGC ACC GAG GTG -3′. Culture media containing WT and D614G pSARS-CoV2 viruses were prepared as described above and subjected to ultracentrifugation in a Beckman SW28 rotor at 28,000 rpm for 2 h at 4 °C. The virus pellet was suspended in PBS buffer and viral titers were determined using qPCR Lentivirus Titration Kit (LV900, ABM). Viral titers ranging from 0.6 to 2.7 × 10 IU (infectious unit)/ml were obtained for WT and D614G and equivalent viral titer were used for pSARS-CoV-2 entry assay.
2.8. Dose-response curve (DRC) analysis using authentic SARS-CoV-2
Five lineages of SARS-CoV-2 viruses (Ancestral virus, Alpha, Beta, Delta, Omicron) were obtained from Korea Disease Control and Prevention Agency (KDCA). All experiments using these infectious viruses were conducted in an enhanced biosafety level 3 (BSL-3) containment facility at Institut Pasteur Korea using BSL-3 procedures in laboratories as approved for such use by the KDCA. The viruses were propagated on Vero E6 cells and the viral titers were determined by plaque formation on Vero cells. For DRC analysis, Vero cells were seeded at 1.2 × 104 cells per well in black, 384-well, μClear plates (Greiner Bio-One, Austria). The following day, 2-fold serial dilution series of compounds prepared in dimethyl sulfoxide (DMSO) were added to each well and the cells were infected with the viruses at a multiplicity of infection (MOI) of 0.008, incubated for 24 h, and then fixed with 4% paraformaldehyde (PFA). For visualization of infected cells, the fixed cells were stained with an antibody against SARS-CoV-2 nucleocapsid (N) protein and DNA fluorochrome Hoechst 33342. The images were acquired using Operetta high-throughput imaging device (PerkinElmer) and analyzed using Columbus software (PerkinElmer, Inc. Waltham, MA) to quantify cell numbers and infection ratios. Antiviral activity was normalized to infection control (0.5% DMSO) in each assay plate. DRCs were generated using Prism7 software (GraphPad). IC50 values were measured in duplicates and calculated using nonlinear regression analysis.
2.9. SARS-CoV-2 nanoluciferase (NLuc) antiviral assay
A549-hACE2-TMPRSS2 cells (1.2 × 104 cells per well) were seeded in white 384-well μClear plates (Greiner Bio-One) and the next day treated with 2-fold serial dilution series of AI, followed by infection with SARS-CoV-2-NLuc (MOI 0.01) (Rihn et al., 2021). After 24 h incubation, nano luciferase substrates (Promega, USA) were added and luciferase signals were measured using a VICTOR3™ multilabel plate reader (PerkinElmer, USA). The relative luciferase signals of the AI-treated groups were normalized to that of non-infection control (set as 0%) and DMSO-treated groups (set as 100%). The plots for DRCs were generated using Prism7 software (GraphPad), and IC50 values were calculated using a nonlinear regression model.
2.10. Cell viability assay
H1299, Vero, and A549 cells were seeded in 96-well plate (5 × 103 cells/well) and the next day they were treated with the indicated concentrations of plant extracts or each compounds. After 24 h culture, WST-8 solution (Biomax, Korea) was added to each well and incubated for 2 h at 37 °C in CO2 incubator. The absorbance of each well was measured at 450 nm using SpectraMax iD5 Multi-Mode Microplate Reader (Molecular Devices, USA). The relative cell viability was calculated as the ratio between the mean absorbance values of the drug-treated samples to those of cells treated with DMSO.
2.11. SARS-CoV-2 S and ACE2 binding assay
To monitor the binding of SARS-CoV-2 S protein to ACE2, a recombinant protein S-RBD-GFP comprising receptor binding domain of SARS-CoV-2 spike protein (RBD) fused to GFP were produced in Expi293F cells as previously described (Kim et al., 2021b). ACE2+ H1299 cells were treated with DMSO or indicated concentrations of AK extracts or AI for 30 min, and medium containing S-RBD-GFP was added with the ratio of 10:1 and incubated for further 10 min at 37 °C in CO2 incubator. The mixture of ACE2+ H1299 cells and S-RBD-GFP were then washed with PBS containing 1% BSA and 1 × 104 cells analyzed for the binding of S-RBD-GFP to ACE2 on the surface of H1299 cells using LSRFortessa flow cytometer (BD Biosciences). The data was analyzed using FlowJo software (BD Life Sciences).
2.12. Cell fusion assay using time-lapse imaging
ACE2/TMPRSS2+ H1299-mRuby2 cells were plated in clear bottom 24 well plates (ibidi, #82426) (1 × 105 cells per well) and allowed to adhere overnight. The cell cultures were treated with DMSO or 10 μM AI for 1 h before 1 × 104 of GFP-H1299 or Spike-H1299 co-expressing GFP cells were added. Fluorescence microscopy images were acquired at 37 °C every 4 min for 2 h at x40 magnification under Leica DMi8 microscope (Leica Microsystems, Germany).
2.13. Cell fusion assay using flow cytometry
ACE2/TMPRSS2+ H1299-mRuby2 cells were plated in 12 well plate (2 × 105 cells/well) and allowed to adhere overnight. The cell cultures were treated with DMSO or 10 μM of each compounds as indicated for 30 min before 2 × 104 Spike-H1299 cells co-expressing GFP were added. After 1 h incubation, the co-cultures were harvested using trypsin-EDTA (Gibco, USA). 1 × 104 cells were analyzed by flow cytometry (LSRFortessa™, BD Biosciences) to determine the percentage of GFP and mRuby double-positive cells as a measure of cell fusion. The data was analyzed using FlowJo software (BD Life Sciences).
2.14. Split-GFP fusion assay
Two fragments of GFP, GFP1-10 (residues 1–214) and GFP11 (residues 215–230), were separately expressed in ACE2/TMPRSS2+ H1299 cells by lentiviral expression systems as described in a previous study (Buchrieser et al., 2020). Equal numbers of ACE2/TMPRSS2+ H1299-GFP1-10 and -GFP11 were seeded in clear bottom 96 well plates (ibidi, #89626), infected the next day with lentiviral particles expressing S protein, and cultured for 48 h until a substantial number of GFP-positive cells are formed. To investigate the effects of AI on the S-mediated syncytia formation, the cell cultures were treated with varying concentration of AI 6 h after S protein expression via lentiviral infection and further incubated for 42 h. The cells were then fixed with 4% PFA and stained with DAPI (Invitrogen, D1306) to visualize cell nuclei. The cell images were acquired at x20 magnification on an ImageXpress Pico automated cell imaging system (Molecular Devices) and were analyzed using cell scoring function (2 channel assay for scoring cells based on a nuclei stain and a GFP signal) of CellReporterXpress software. GFP-positive cells in the field of view were designated as multinucleated syncytia. Nuclei contained in GFP-positive cells and free nuclei were automatically pseudocolored in green and red, respectively and the percentage of fused cells was calculated as the ratio of green-to red-colored nuclei.
2.15. Filipin staining of intracellular cholesterol
ACE2+ were plated on glass coverslips and allowed to attach overnight. The cells were treated with indicated concentrations of compounds for 1 h and fixed with 4% PFA, followed by staining with 5 μg/ml filipin-III (Cayman, USA) for 2 h. The stained cells were examined using a LSM 700 confocal microscope (Carl Zeiss, Germany) and the fluorescence intensity in the plasma membrane was quantified using image J software (NIH, USA).
2.16. Protein extraction and immunoblot analysis
Protein lysates from 48 h co-cultures of GFP-H1299 and ACE2/TMPRSS2+, and Spike-H1299 and ACE2/TMPRSS2+ pretreated with DMSO or 10 μM AI for 1 h were prepared by dissolving the co-cultures directly in 2X SDS sample buffer, separated by SDS-PAGE, and transferred onto nitrocellulose membranes. The membranes were then blocked with 5% skim milk in TBST (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.05% Tween 20) for 1 h at RT, followed by incubation with primary antibodies in 1% bovine serum albumin (BSA) in TBST overnight at 4 °C. The primary antibodies used were as follows; rabbit anti-Cleaved Caspase-9 (Cell Signaling, #9505), rabbit anti-Cleaved PARP (Cell Signaling, #9541), and mouse anti-GAPDH (Abcam, ab8245). The membranes were incubated with the corresponding horseradish peroxidase-conjugated secondary antibodies (KPL, USA) for 1 h at RT. Immuno-reactive protein bands were detected using ECL Western Blotting Substrate (Thermo Fisher Scientific, USA).
2.17. Statistical analysis
The data shown in this study are representative of two or three independent experiments and presented as the mean ± SEM of triplicate samples. The results were analyzed using Student's t-test or one-way analysis of variance (ANOVA) followed by Tukey's post hoc test and a p value less than 0.05 was considered to indicate statistical significance (*P < 0.05; ****P < 0.0001; NS not significant). Prism v.9.0.0 software was used for statistical analyses.
3. Results
3.1. Astersaponin I from Aster koraiensis blocks two main infection routes of SARS-CoV-2
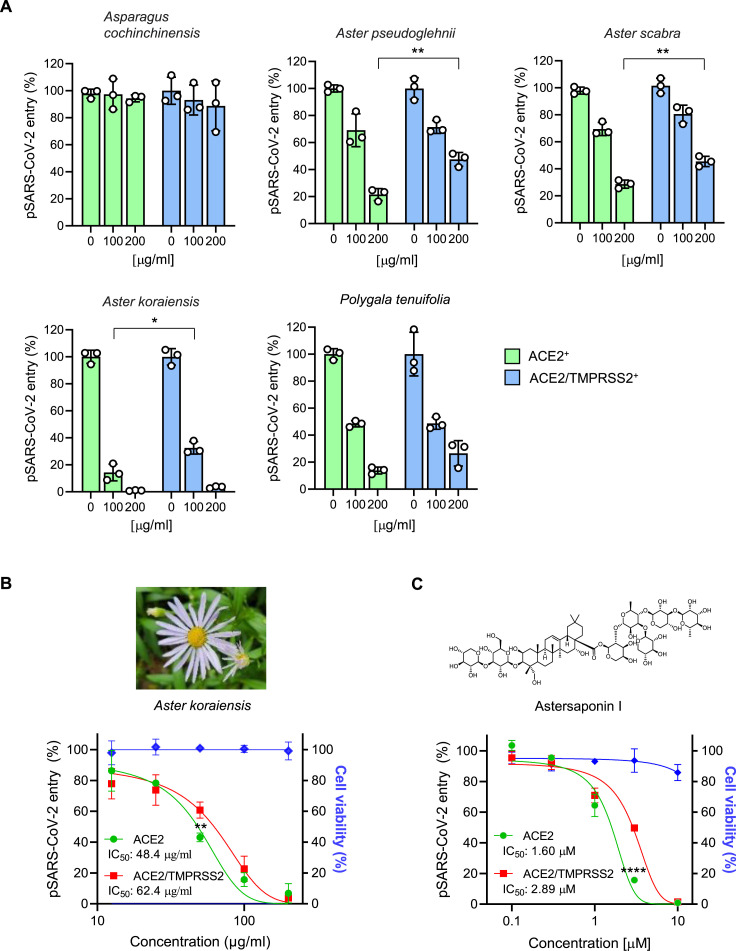
To test the antiviral potency of five different plant extracts against SARS-CoV-2, we constructed and utilized SARS-CoV-2 pseudovirus that carry SARS-CoV-2 S protein on HIV-based lentiviral particles (pSARS-CoV-2) as previously described (Kim et al., 2021b). The pseudovirus was then allowed to infect the model host cell lines that stably overexpress either the host cell receptor ACE2 (ACE2+) or ACE2 plus TMPRSS2 (ACE2/TMPRSS2+) in H1299 cells. These cell lines were used to determine the effects of plant extracts and compounds on two different SARS-CoV-2 infection routes of the endosomal and TMPRSS2-mediated entry pathways respectively. pSARS-CoV-2 entry assay demonstrated that four out of five plant extracts, besides the roots of A. cochinchinensis, exhibited potent inhibitory activity against both entry routes of SARS-CoV-2 (Fig. 1 A). Among them, the 70% EtOH extracts from the leaves of A. koraiensis (AK) exhibited the most potent anti-SARS-CoV-2 activity with IC50 values of 48.4 and 62.4 μg/ml in ACE2+ and ACE/TMPRSS2+ respectively, while it did not show any sign of cytotoxicity up to 200 μg/ml (Fig. 1B). Note that the inhibitory effect of plant extracts on ACE2/TMPRSS2+ cells was less efficient compared with ACE2+ cells, which might be due to the fact that TMPRSS-mediated SARS-CoV-2 entry pathway is much more efficient than the endosomal pathway (Ou et al., 2021). Next, in an attempt to identify the bioactive compound(s) present in AK, we isolated a major triterpenoid saponin from the AK extracts. By analyzing MS and 1D NMR spectroscopic data (Supplementary Figs. S1–3) and comparing with published literature, the chemical structure of the triterpenoid saponin was identified as astersaponin I (AI) (Kwon et al., 2019). AI consists of 30-carbon triterpene backbone with two sugars at position C3 and a hydrophilic oligosaccharide chain attached at position C28 (Fig. 1C). pSARS-CoV-2 entry assay showed that AI effectively blocked both SARS-CoV-2 entry pathways with IC50 values of 1.60 μM in ACE2+ and 2.89 μM in ACE2/TMPRSS2+ without obvious cytotoxicity (Fig. 1C), suggesting that AI is a bioactive compound exerting antiviral effects in AK extracts.
Fig. 1.
Astersaponin I from Aster koraiensis inhibits SARS-CoV-2 infection. (A) The effects of various plant extracts on the entry of SARS-CoV-2 pseudovirus (pSARS-CoV-2) into ACE2+ and ACE2/TMPRSS2+ H1299 cells. (B, C) pSARS-CoV-2 entry and cell viability assays for the 70% EtOH extract of Aster koraiensis (B) and triterpenoid saponins isolated from A. koraiensis (C) in ACE2+ and ACE2/TMPRSS2+ H1299 cells. The data from pSARS-CoV-2 entry assay were representative of three independent experiments. The error bars indicate the SEM (n > 3). P values were determined by the unpaired, two-tailed Student's t-test. *P < 0.05; **P < 0.01, ****P < 0.0001.
3.2. Steroidal saponins and triterpenoid sapnoins with hydrophobic moiety exhibit a limited antiviral activity against SARS-CoV-2
Since AI showed a potent anti-SARS-CoV-2 activity, we then asked whether different saponins isolated from other plants might also show such antiviral effects. Interestingly, the results from pSARS-CoV-2 assay demonstrated that steroidal saponins isolated from the roots of A. cochinchinensis including protodioscin, methyl protodioscin, 15-hydroxyl pseudoprotodioscin, aspacochioside A, and chamaedroside E did not show any inhibitory activity against SARS-CoV-2 infection in ACE2+ until a concentration of 10 μM (Fig. 2 A). Moreover, onjisaponins, triterpenoid saponins possessing a hydrophobic phenyl group on sugar chain, isolated from the roots of P. tenuifolia were tested, and all exhibited no or limited antiviral effects until 10 μM (Fig. 2B). These results suggest that not all saponins exhibit anti-SARS-CoV-2 activity and that saponins with specific chemical features like AI, have a potent inhibitory activity against SARS-CoV-2 infection.
Fig. 2.
Steroidal saponins and triterpenoid saponins with a phenyl moiety have a limited inhibitory activity against SARS-CoV-2 infection. (A) Chemical structures and the effects of the steroidal saponins obtained from A. cochinchinensis on the entry of pSARS-CoV-2 into ACE2+ H1299 cells. (B) Chemical structures and the effects of onjisaponins obtained from P. tenuifolia on the entry of pSARS-CoV-2 into ACE2+ H1299 cells. The data from pSARS-CoV-2 entry assay were representative of two or three independent experiments. The error bars indicate the SEM (n > 3).
3.3. Astersaponin I inhibits SARS-CoV-2 S protein-mediated membrane fusion
To understand the mechanism by which AI blocks SARS-CoV-2 infection, we first investigated whether AI inhibits the binding of S protein to ACE2 using a recombinant protein containing receptor-binding domain (RBD) of the SARS-CoV2 S protein fused with GFP (RBD-GFP) (Fig. 3 A). The strong binding of RBD-GFP to ACE2 expressed on the surface of H1299 cells was confirmed by flow cytometry (Fig. 3B). Importantly, the pretreatment of AK extracts or AI did not lead to blockade of the interaction between RBD-GFP and ACE2 at concentrations up to 250 μg/ml and 10 μM respectively, but significant inhibitory effects were seen at 1000 μg/ml AK extracts and 20 μM AI (Fig. 3B), indicating that, despite they can interfere with the binding of S protein to ACE2 at high concentrations, the antiviral activity of AK extracts and AI observed in pSARS-CoV-2 entry assay (Fig. 1B and C) is not attributed to reduced S-ACE2 protein interactions. Next, we observed that removal of AI after 1 h pretreatment of ACE2+ with 1 or 5 μM AI did not change its antiviral activity, offering an important clue that AI affects host cells, but not SARS-CoV-2 (Fig. 3C). Previously, we proposed that platycodin D (PD), a triterpenoid saponin from Platycodon grandiflorum, can inhibit SARS-CoV-2 fusion with host cells by incorporating into host cell membrane (Kim et al., 2021b). Since AI has a similar chemical structure to PD, we hypothesized that AI might also incorporate into host cell membrane and interferes with viral envelope fusion with host cell membrane. To test this possibility, we established a SARS-CoV-2 S-driven cell fusion system in which H1299 cells overexpressing S protein (Spike-H1299, labeled with GFP) are added to a monolayer of ACE2/TRMPRSS2+ (labeled with mRuby) to monitor the cell-to-cell fusion between the two cell lines (Fig. 3D). Time-lapse microscopy showed that Spike-H1299 cells gradually moved close to and interacted with the ACE2/TMPRSS2+, followed by a rapid fusion between two cells within 10 min after adding cells (Fig. 3E, Movie 1). The fusion event causes the hybrid cells to display S proteins on their surface, which allows them to fuse continuously with neighboring ACE2/TMPRSS2+, giving rise to enlarged multinucleated cells (Fig. 3E). The cell-to-cell fusion is mediated by the interaction between S protein and ACE2, because this process was not observed in co-culture of control GFP-H1299 (no S protein) with ACE2/TMPRSS2+ (Fig. 3E, Movie 2). Strikingly, AI treatment almost completely blocked the fusion between the two cells, thus preventing the formation of multinucleated cells (Fig. 3E, Movie 3). Note that even though the fusion between Spike-H1299 and ACE2/TMPRSS2+ occasionally occurred, it did not lead to a further fusion with adjacent cells. To quantitatively measure these fusion events, we further performed a flow cytometry analysis after adding Spike-H1299 cells onto ACE2/TRMPRSS2+. The results showed that around 10% of total cells were participated in the fusion event, generating hybrid cells (both mRuby- and GFP-positive; Fig. 3F). Again, the fusion was not observed in the mixture of control GFP-H1299 cells (no S protein) with ACE2/TMPRSS2+ (Fig. 3F). Importantly, AI pretreatment effectively inhibited the cell-to-cell fusion, with less than 1% of cells showing cell fusion hybrids (Fig, 3F), which is consistent with the results shown in Fig. 3E. Next, we examined the effect of other natural saponins on the S-mediated cell fusion. Platycodigenin, a triterpenoid aglycone without hydrophilic sugar moieties, did not exhibit inhibitory effect on the cell-to-cell fusion (Fig. 3F). Moreover, steroidal saponin such as protodioscin (Fig. 2A) and onjisaponin Fg, a triterpenoid saponin from P. tenuifolia (Fig. 2B) did not show any anti-fusogenic activity (Fig. 3F). These results indicate that AI from AK, but not other saponins, can antagonize the S-mediated viral membrane fusion event, which leads to an inhibition of SARS-CoV-2 entry into host cells. Given that cholesterol is essential for membrane fusion and the penetration of enveloped viruses including SARS-CoV-2 into host cells (Sanders et al., 2021; Yang et al., 2016), we next investigated whether AI alters cholesterol content of the plasma membrane. After treatment with 5 μM AI for 1 h, ACE2+ were stained with filipin to visualize intracellular cholesterol. Confocal imaging showed that the amounts of free cholesterol in the plasma membrane were markedly increased upon AI treatment (3.3-fold increase above DMSO-treated control) (Fig. 3G and H). Meanwhile, platycodigenin, protodioscin, and onjisaponin Fg did not affect membrane cholesterol content even at higher concentration of 10 μM (Fig. 3G and H). Together, these results showed a correlation between changes in cholesterol levels in cell membranes and anti-fusogenic activity of saponins, suggesting that the increase in cholesterol at the plasma membrane by AI can contributes to the inhibition of the S-mediated membrane fusion and SARS-CoV-2 entry.
Fig. 3.
Astersaponin I blocks viral membrane fusion with the host cell membrane. (A) Schematic illustration of the binding of SARS-CoV-2 spike receptor binding domain (RBD) fused to GFP (RBD-GFP) to ACE2 protein overexpressed in H1299 cells. (B) Examination of the effect of AK extracts and AI on the interaction between RBD-GFP and ACE2 on the surface of H1299 cells by flow cytometry after treatment with indicated concentration of AK extracts and AI. The grey peaks indicate the control experiments without RBD-GFP addition. (C) ACE2+ H1299 cells were treated with 1 or 5 μM AI for 1 h and washed with fresh media (Wash) or maintained (No wash), followed by the addition of pSARS-CoV-2. The data from pSARS-CoV-2 entry assay were representative of two independent experiments. The error bars indicate the SEM (n > 3). P values were determined by the unpaired, two-tailed Student's t-test (NS, not significant). (D) Schematic illustration of the SARS-CoV-2 S protein-mediated cell fusion. Addition of the cell suspension of H1299 cells stably expressing S protein and GFP (Spike-H1299) to a monolayer of ACE2/TMPRSS2+ H1299 cells with mRuby fluorescence (ACE2/TMPRSS2-H1299) leads to cell-to-cell fusion. (E) Still images at different time points from time-lapse imaging of S-mediated cell fusion. (F) The fusion between Spike-H1299 and ACE2/TMPRSS2+ H1299 was determined by counting the number of cells double-positive for GFP and mRuby by flow cytometry. H1299 cells expressing only GFP (no spike) were used for control experiment. All compounds were used at the concentration of 10 μM. The data were representative of three independent experiments. (G, H) Filipin cholesterol staining of ACE2+ H1299 cells after treatment with DMSO, AI (5 μM), and other indicated compounds (10 μM) for 1 h (G) and quantification of the intensity of membrane filipin staining using Image J software (n = 20 for each group). Error bars in the graphs indicate the SEM. P values were determined by one-way ANOVA followed by Tukey's post hoc test. ****P < 0.0001; NS not significant (H).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.antiviral.2022.105428
The following is/are the supplementary data related to this article:
2
3
4
3.4. Astersaponin I blocks syncytia formation
SARS-CoV-2-infected cells undergo continuous cell fusion with neighboring cells, leading to enlarged multinucleated cells, i.e. a syncytium. Syncytia is often detected in the lung tissue from COVID-19 patients and regarded as a hallmark of COVID-19 (Bussani et al., 2020). To investigate the effect of AI on the syncytium formation, we employed a split-GFP technology, in which each of the two fragments of GFP are expressed in separate cell lines to produce a detectable GFP fluorescence upon a cell-to-cell fusion (Fig. 4 A). When S protein was overexpressed by lentiviral gene transfer in ACE2/TMPRSS2+ expressing each fragment of split-GFP's, GFP-positive multinucleated cells was formed, indicating that S protein promoted syncytium formation (Fig. 4B). Automated image analysis determined that the population of GFP-positive syncytia reached up to around 30% of the total number of plated cells, 48 h after expression of S protein (Fig. 4C). Importantly, this syncytia formation was dose-dependently and effectively reduced by treatment with AI, showing IC50 values of 3.9 μM (Fig. 4C and D).
Fig. 4.
Astersaponin I prevents SARS-CoV-2 S-induced syncytia formation. (A) Schematic illustration of Split-GFP assay. Ectopic expression of SARS-CoV-2 S protein into cultures of the mixture of ACE2/TMPRSS2+ H1299 cells expressing GFP1-10 or GFP11 leads to cell-to-cell fusion, generating GFP fluorescence. (B) Images of the S-mediated cell-to-cell fusion using Split-GFP. GFP and blue fluorescence indicate the cell-to-cell fusion and DAPI-stained nuclei, respectively. The nuclei are automatically pseudocolored in white and overlapped with GFP fluorescence by CellReporterXpress software. (C) Examination of effect of AI on the S-mediated cell-to-cell fusion using Split-GFP assay. Representative images of GFP-positive cell-to-cell fusion. The GFP signal and DAPI nuclei staining are automatically pseudocolored in green and red by CellReporterXpress software. (D) Quantitative evaluation for cell-to-cell fusion using Split-GFP assay. Images of GFP and nuclei stained with DAPI were obtained in five random fields per well. The percentage of fusion cells were calculated by dividing number of nuclei in GFP-positive cells by total number of nuclei. The data were representative of three independent experiments. The error bars indicate the SEM (n > 3). P values were determined by one-way ANOVA followed by Tukey's post hoc test. *P < 0.05; ****P < 0.0001; NS not significant. (E) Protein lysates from co-culture experiments were assessed by Western blot. GAPDH was used as a loading control.
Virus-induced syncytia formation generally leads to cell death by apoptosis or necrosis depending on cell types (Buchrieser et al., 2020; Hooper et al., 2001; Nardacci et al., 2015). We investigated whether the S-mediated syncytia formation promotes apoptosis in our model system. Co-culture of Spike-H1299 cells with ACE2/TMPRSS2+ for 48h induced apoptosis as evidenced by Western blot analysis of the proteolytic cleavage of caspase 9 and poly (ADP-ribose) polymerase (PARP) (Fig. 4E). Notably, these apoptotic markers were not detected either when ACE2/TMPRSS2+ were co-cultured with control GFP-H1299 (no spike) cells or when ACE2/TMPRSS2+ were pretreated with AI for 1 h before adding Spike-H1299 cells (Fig. 4E). Overall, these results indicate that AI blocks SARS-CoV-2 S-induced syncytia formation and consequent apoptotic cell death.
3.5. Astersaponin I effectively inhibits SARS-CoV-2 infection regardless of S mutation
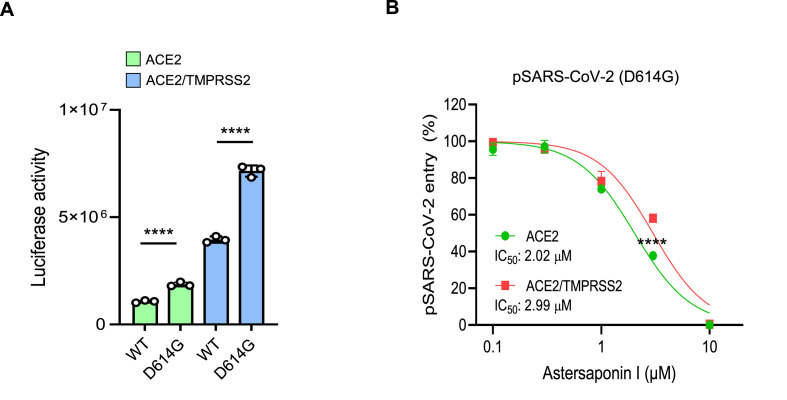
Most of the SARS-CoV-2 variants carry spontaneous mutations in genes encoding S protein such as D614G, N501Y, E484K, L452R, and K417N, which leads to an increase in SARS-CoV-2 infectivity by enhancing the binding affinity to ACE2 (Ozono et al., 2021; Plante et al., 2021). Consistently, pSARS-CoV-2 carrying the D614G mutation that found in B lineage SARS-CoV-2 variants showed a significant increase in infectivity compared to the wild type virus in both ACE+ and ACE2/TMPRSS2+ cells (Fig. 5 A). As the viral membrane fusion occurs after the attachment of S protein to ACE2, we considered that AI, as a viral membrane fusion blocker, could effectively inhibit the infection of SARS-CoV-2 variants despite their enhanced binding affinity to ACE2. As expected, AI inhibited the entry of the D614G mutant virus with IC50 of 2.02 and 2.99 μM in ACE2+ and ACE2/TMPRSS2+ respectively (Fig. 5B), which were comparable to those of the WT virus (Fig. 1C).
Fig. 5.
Astersaponin I equally inhibits the infection of SARS-CoV-2 WT and D614G mutant. (A) pSARS-CoV-2 entry assay in ACE2+ and ACE2/TMPRSS2+ H1299 cells. The data were representative of two independent experiments. The error bars indicate the SEM (n > 3). P values were determined by the unpaired, two-tailed Student's t-test (****P < 0.0001). (B) The effects of AI on the infection of WT and D614G mutant of pSARS-CoV-2 in ACE2+ and ACE2/TMPRSS2+ H1299 cells. The data were representative of three independent experiments. The error bars indicate the SEM (n > 3). P values were determined by the unpaired, two-tailed Student's t-test (****P < 0.0001).
3.6. Astersaponin I shows an inhibitory activity against authentic SARS-CoV-2 and its variants
We next tested whether AI exhibits an inhibitory activity against SARS-CoV-2 variants by measuring the infection of authentic SARS-CoV-2 into Vero cells that express abundant ACE2 but not TMPRSS2 (Kim et al., 2021b). Staining of SARS-CoV-2 nucleocapsid (N) protein demonstrated that AI impeded the infection of various SARS-CoV-2 variants including Alpha, Beta, Delta, and Omicron with a similar range of IC50 values of 1.91–2.04 μM, which was almost identical to that of ancestral virus (IC50: 1.94 μM) (Fig. 6 A and B). These results provide evidence that AI can effectively and equally hinder the infection of ancestral SARS-CoV-2 and their variants, regardless of the types of S-mutation. Meanwhile, chloroquine, which was used as a control drug, exhibited less effective and variable inhibitory activity toward ancestral SARS-CoV-2 (IC50: 10.64 μM) and their variants (IC50: 7.98–16.39 μM) compared to AI (Fig. 6C and D). Of note, chloroquine showed relatively poor inhibitory activity against Omicron, which was consistent with a previous report (Zhao et al., 2022). Finally, we verified the inhibitory activity of AI against TMPRSS2-dependent entry pathway by measuring the infection of replicable SARS-CoV-2 recombinant virus expressing nanoluciferase (Nluc) into A549 cells that overexpress ACE2 and TMPRSS2, and found that AI effectively inhibited the viral entry in these cells with an IC50 of 4.95 μM (Fig. 6E). This observation corroborates that AI restricts the TMPRSS2-mediated SARS-CoV-2 entry pathway as effectively as it inhibits the endosomal infection route.
Fig. 6.
Astersaponin I inhibits the infection of SARS-CoV-2 and its variants with a similar efficiency. (A, C) Dose-response curves for AI (A) and chloroquine (C) against ancestral SARS-CoV-2 (A, black circle) and SARS-CoV-2 variants Alpha (B.1.1.7, red square), Beta (B.1.351, orange triangle), Delta (B.1.617.2, pink inverted triangle), and Omicron (B.1.1.529, green diamond) in Vero cells. The blue diamond represents cell viability. The mean ± SEM was calculated from duplicate experiments. (B, D) Confocal images of SARS-CoV-2 N protein (green) and cell nuclei at concentrations near the IC50 of AI (B) and chloroquine (D) in Vero cells. (E) Dose-response curve for AI against replicable SARS-CoV-2 recombinant viruses expressing Nanoluciferase (Nluc) into A549 cells that overexpress ACE2 and TMPRSS2. The blue diamond represents cell viability.
4. Discussion
The advent of highly transmissible variants of SARS-CoV-2 has raised concerns that they could escape immune protection conferred by natural infection or vaccination. New members of the growing Omicron family of SARS-CoV-2 including BA.1, BA.2, BA.3, BA.4, and BA.5 have been reported to be highly resistant to neutralization by therapeutic antibodies and vaccine-elicited antibodies (Arora et al., 2022; Koyama et al., 2022; Wang et al., 2022a). Mechanistically, mutations in the RBD of SARS-CoV-2 S protein can impair neutralization and enhance viral infection with efficient engagement with ACE2 (Lupala et al., 2021; Wang et al., 2021, 2022b). Thus, at this point where the efficacy of current COVID-19 vaccines and antibody therapies are threatened, effective therapeutic agents against SARS-CoV-2 variants are desperately needed. We here found that extracts from annual leafy plant of AK have a potent inhibitory activity against infection of SARS-CoV-2. AK is a perennial herb native to Korea and has long been used as a food ingredient and herbal medicine for treating several diseases including diabetes, chronic bronchitis, pertussis, and pneumonia (Hyun et al., 2018; Kim et al., 2018). Al was recently isolated from AK as a new triterpenoid saponin and its autophagy regulating function was reported (Kwon et al., 2019). However, other biological activities including the antiviral effect of AI have never been explored elsewhere.
After the COVID-19 outbreak, multiple triterpenes and triterpenoid saponins have been suggested as potential therapeutic agents for SARS-CoV-2. Several in-silico studies predicted that the molecular targets of these compounds are diverse including many viral components such as main proteases, NSP15, and S protein as well as cellular receptor such as ACE2 (Diniz et al., 2021; Luo et al., 2020; Sinha et al., 2021). Recent experimental studies demonstrated that Esculentoside A, a triterpenoid saponin isolated from roots of Phytolacca esculenta, has an anti-SARS-CoV-2 activity in a concentration ranging from 5 to 20 μg/ml (6–24 μM) by binding to S protein of SARS-CoV-2 (Zeng et al., 2021). Yang et al. also tested several triterpenoid saponins from licorice including glycyrrhetinic acid, 3-O-β-D-glucuronosyl-glycyrrhetinic acid, licorice-saponin A3, betulinic acid, and ursolic acid against interaction between SARS-CoV-2 S RBD and ACE2 by ELISA assay and showed IC50 values ranging from 8.3 to 15.1 μM (Yi et al., 2022). Similarly, our data from the RBD-ACE2 binding assay demonstrates that AI inhibits the interaction between these two proteins at a concentration higher than 10 μM (Fig. 3B). But, these inhibitory effects were not detected in platycodigenin and four different onjisaponins until 20 μM (Supplementary Fig. S4). Overall, these data suggest that triterpenoid aglycones and triterpenoid saponins have a poor inhibitory activity against S protein and ACE2, and their interaction. Importantly, in keeping with these findings, our results showing the activity of AI to inhibit SARS-CoV-2 infection at substantially lower concentrations ranging from 1.69 to 2.04 μM (Fig. 1, Fig. 6A) suggest that the main target of AI may be other than S protein and ACE2.
Previously, we discovered that platycodin D (PD), a triterpenoid saponin from Platycodon grandiflorum, inhibits the endosomal and TMPRSS2 driven entry pathways with IC50 values of 1.19 and 4.79 μM, respectively (Kim et al., 2021b). PD has a similar chemical structure to AI, consisting of 30-carbon triterpene backbone with a linear oligosaccharide chain attached at position C28. Our molecular modeling experiments predicted that pentacyclic triterpene core of PD can incorporate into lipid bilayers because of its structural similarities to cholesterol and the hydrophilic sugar moieties could stretch out of the membrane, thus possibly hindering close proximity between viral and cellular membranes for fusion (Kim et al., 2021b). Because of the similar chemical structure of these two natural compounds, we reasoned that AI would function through the same or similar mechanisms of action with PD and aimed to test this possibility by establishing several cell-to-cell fusion assay systems. Based on the results from these assays, we can conclude that AI indeed hinders the S-mediated membrane fusion event, thereby inhibiting SARS-CoV-2 infection. To initiate viral membrane fusion, S protein consisting of S1 receptor-binding subunit and S2 fusion subunit (Xia et al., 2020), should be cleaved at S1/S2 site by cellular proteases, resulting in exposure and insertion of hydrophobic fusion peptide (FP) of S2 subunit into lipid bilayer of the cellular membrane (Huang et al., 2020). The fusion inhibitory effect of AI might be related to the hydrophilic properties of oligosaccharide of AI, which can hinder the insertion of hydrophobic FP into cellular membrane. This notion is supported by our results showing that onjisaponins, triterpenoid saponins possessing a hydrophobic phenyl group on the oligosaccharide chain did not exhibit significant inhibitory effects on viral entry and the cell-to-cell fusion (Fig. 2B and Fig. 3F). Furthermore, we found that steroidal saponins did not exhibit any antiviral effects (Fig. 2A and Fig. 3F). It is probably due to the inability of C-27 steroidal backbone, which has no structural resemblance to cholesterol, to intercalate into the cell membrane. Our results indicate that the triterpenoid saponins with a unique chemical structure containing hydrophilic oligosaccharide at position C28 like AI and PD can hinder viral membrane fusion. Furthermore, we observed that cholesterol levels at the plasma membrane were remarkably increased by AI, but not by other saponins tested. Cholesterol is an important constituent of cell membranes and crucial role in virus-induced membrane fusion during infection (Yang et al., 2016). Since cholesterol content directly affects the kinetics of membrane fusion through its effect on the biophysical properties of the cell membrane (Najafinobar et al., 2016), increasing cholesterol concentration at plasma membrane by AI can influence the S-mediated membrane fusion events. In line with this, it was recently reported that exogenously added 27-hydroxycholesterol accumulates in the plasma membrane lipid rafts, leading to inhibition of SARS-CoV-2 entry (Marcello et al., 2020). Taken together, we propose a model in which AI hinders viral membrane fusion, possibly by altering membrane cholesterol and through sugar moiety protrusion after incorporation into the plasma membrane, which leads to prevent SARS-CoV-2 infection.
As the viral envelope fusion to host cell membrane takes place after ACE2 binding during the viral entry, fusion blockers can inhibit the entry of SARS-CoV-2 variants with a similar efficiency to that of the ancestral virus despite their increased affinity for ACE2. Supporting this notion, we demonstrated that AI effectively inhibits the infection of the SARS-CoV-2 variants as well as their ancestral virus with similar IC50 values ranging from 1.91 to 2.04 μM. Another important advantage of fusion inhibitors over other viral entry blockers is that they can simultaneously block two major SARS-CoV-2 entry routes with comparable efficiency because the fusion process is shared in the two different entry pathways. In these respects, synthetic peptides and a cholesterol-conjugated peptides derived from a fusogenic domain of the S protein have been developed as fusion inhibitors and tested for their ability to inhibit SARS-CoV-2 infection in vitro and in vivo (de Vries et al., 2021; Duzgunes et al., 2021; Kandeel et al., 2021).
The interaction between S protein and ACE2 promotes not only fusion of virus with host cells but also fusion of infected cells with neighboring cells to form multinucleated syncytia. These syncytia were often detected in cell cultures and patient tissues infected with many enveloped viruses including respiratory syncytial virus (RSV), human immunodeficiency virus (HIV), SARS-CoV-1, and MERS-CoV (Frankel et al., 1996; Franks et al., 2003; Johnson et al., 2007; Qian et al., 2013). SARS-CoV-2 S-induced syncytia play important roles in virus protection against neutralizing antibodies and viral spreads through cell-to-cell transmission, and contributes to cytopathic effect (Rajah et al., 2021; Zeng et al., 2022). Moreover, recent studies reported that S-mediated syncytia formation eventually leads to cell death via apoptosis or pyroptosis, causing the release of virus to infect neighboring cells and/or triggering an inflammatory response (Ma et al., 2021; Santana et al., 2021). Here, we demonstrated that AI effectively inhibits the formation of S-mediated syncytia and abolishes syncytial apoptosis, indicating that AI could also block viral spread after infection and prevent viral cytopathic effects.
5. Conclusion
AI is a natural viral fusion blocker that effectively prevents SARS-CoV-2 and its variants from infecting host cells and syncytia formation by hindering the S-mediated membrane fusion. We propose that AI can be a broad-spectrum antiviral drug not only against SARS-CoV-2 but also against other novel enveloped viral pathogens that might arise in the future.
Author contributions
TYK, DSJ, and CJL conceived and designed the overall study. TYK performed the pSARS-CoV-2-related experiments. SJ performed the authentic SARS-CoV-2 experiments under the supervision of SK. JYK and HCK carried out the extraction and isolation of plant materials. SJL concentrated pSARS-CoV-2 virus for titration. HJ performed flow cytometry analysis. TYK analyzed the data and wrote the manuscript with input from coauthors.
Declaration of competing interest
The authors declare no conflict of interests.
Acknowledgement
We thank Ph. D. Taek Seung Kim, a senior engineer at the Research Solution Center (RSC) in IBS, for imaging cell-to-cell fusion using Leica DMi8 microscope. The pathogen resources (NCCP43326, NCCP43381, NCCP43382, NCCP43390 and NCCP43408) for this study were provided by the National Culture Collection for Pathogens. This work was supported by the Institute for Basic Science (IBS), Center for Cognition and Sociality (IBS-R001-D2) to C.J.L., National Research Foundation of Korea (NRF) with grants funded by the Korean government (MSIT) (NRF-2017M3A9G6068245 and NRF-2020M3E9A1041756), NRF funded by the MSIT, Republic of Korea (NRF-2019R1A2C1083945), and the Korea Institute of Science & Technology (KIST) Institutional Program (Project No. 2E30650-20-154).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105428.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Alam S., Sarker M.M.R., Afrin S., Richi F.T., Zhao C., Zhou J.R., Mohamed I.N. Traditional herbal medicines, bioactive metabolites, and plant products against COVID-19: update on clinical trials and mechanism of actions. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.671498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O'Connell A.M., Simons D., Blomquist P.B., Zaidi A., Nash S., Iwani Binti Abdul Aziz N., Thelwall S., Dabrera G., Myers R., Amirthalingam G., Gharbia S., Barrett J.C., Elson R., Ladhani S.N., Ferguson N., Zambon M., Campbell C.N.J., Brown K., Hopkins S., Chand M., Ramsay M., Lopez Bernal J. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P., Zhang L., Rocha C., Sidarovich A., Kempf A., Schulz S., Cossmann A., Manger B., Baier E., Tampe B., Moerer O., Dickel S., Dopfer-Jablonka A., Jack H.M., Behrens G.M.N., Winkler M.S., Pohlmann S., Hoffmann M. Comparable neutralisation evasion of SARS-CoV-2 omicron subvariants BA.1, BA.2, and BA.3. Lancet Infect. Dis. 2022;22(6):766–767. doi: 10.1016/S1473-3099(22)00224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarba B., Pandiella A. Medicinal plants as sources of active molecules against COVID-19. Front. Pharmacol. 2020;11:1189. doi: 10.3389/fphar.2020.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser J., Dufloo J., Hubert M., Monel B., Planas D., Rajah M.M., Planchais C., Porrot F., Guivel-Benhassine F., Van der Werf S., Casartelli N., Mouquet H., Bruel T., Schwartz O. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020;39 doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussani R., Schneider E., Zentilin L., Collesi C., Ali H., Braga L., Volpe M.C., Colliva A., Zanconati F., Berlot G., Silvestri F., Zacchigna S., Giacca M. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Lavine C.L., Rawson S., Peng H., Zhu H., Anand K., Tong P., Gautam A., Lu S., Sterling S.M., Walsh R.M., Jr., Rits-Volloch S., Lu J., Wesemann D.R., Yang W., Seaman M.S., Chen B. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. Science. 2021;373:642–648. doi: 10.1126/science.abi9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E., Ledford H. How bad is Omicron? What scientists know so far. Nature. 2021;600:197–199. doi: 10.1038/d41586-021-03614-z. [DOI] [PubMed] [Google Scholar]
- Chandramouli V., Niraj S.K., Nair K.G., Joseph J., Aruni W. Phytomolecules repurposed as covid-19 inhibitors: opportunity and challenges. Curr. Microbiol. 2021;78:3620–3633. doi: 10.1007/s00284-021-02639-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R.D., Schmitz K.S., Bovier F.T., Predella C., Khao J., Noack D., Haagmans B.L., Herfst S., Stearns K.N., Drew-Bear J., Biswas S., Rockx B., McGill G., Dorrello N.V., Gellman S.H., Alabi C.A., de Swart R.L., Moscona A., Porotto M. Intranasal fusion inhibitory lipopeptide prevents direct-contact SARS-CoV-2 transmission in ferrets. Science. 2021;371:1379–1382. doi: 10.1126/science.abf4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz L.R.L., Perez-Castillo Y., Elshabrawy H.A., Filho C., de Sousa D.P. Bioactive terpenes and their derivatives as potential SARS-CoV-2 proteases inhibitors from molecular modeling studies. Biomolecules. 2021;11 doi: 10.3390/biom11010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duzgunes N., Fernandez-Fuentes N., Konopka K. Inhibition of viral membrane fusion by peptides and approaches to peptide design. Pathogens. 2021;10 doi: 10.3390/pathogens10121599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorkin-Camiel L., Whelan J.S. Tropical American plants in the treatment of infectious diseases. J. Diet. Suppl. 2008;5:349–372. doi: 10.1080/19390210802519648. [DOI] [PubMed] [Google Scholar]
- Frankel S.S., Wenig B.M., Burke A.P., Mannan P., Thompson L.D., Abbondanzo S.L., Nelson A.M., Pope M., Steinman R.M. Replication of HIV-1 in dendritic cell-derived syncytia at the mucosal surface of the adenoid. Science. 1996;272:115–117. doi: 10.1126/science.272.5258.115. [DOI] [PubMed] [Google Scholar]
- Franks T.J., Chong P.Y., Chui P., Galvin J.R., Lourens R.M., Reid A.H., Selbs E., McEvoy C.P., Hayden C.D., Fukuoka J., Taubenberger J.K., Travis W.D. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 2003;34:743–748. doi: 10.1016/S0046-8177(03)00367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P.J., Kuroda M., Maemura T., Chiba S., Armbrust T., Wright R., Balaram A., Florek K.R., Bateman A.C., Kawaoka Y. Efficacy of vaccination and previous infection against the Omicron BA.1 variant in Syrian hamsters. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Consortium C.-G.U., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper P., Zaki S., Daniels P., Middleton D. Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microb. Infect. 2001;3:315–322. doi: 10.1016/s1286-4579(01)01385-5. [DOI] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S.W., Kim J., Jo K., Kim J.S., Kim C.S. Aster koraiensis extract improves impaired skin wound healing during hyperglycemia. Integr Med Res. 2018;7:351–357. doi: 10.1016/j.imr.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2021 doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.E., Gonzales R.A., Olson S.J., Wright P.F., Graham B.S. The histopathology of fatal untreated human respiratory syncytial virus infection. Mod. Pathol. 2007;20:108–119. doi: 10.1038/modpathol.3800725. [DOI] [PubMed] [Google Scholar]
- Kandeel M., Yamamoto M., Tani H., Kobayashi A., Gohda J., Kawaguchi Y., Park B.K., Kwon H.J., Inoue J.I., Alkattan A. Discovery of new fusion inhibitor peptides against SARS-CoV-2 by targeting the spike S2 subunit. Biomol Ther (Seoul) 2021;29:282–289. doi: 10.4062/biomolther.2020.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee Y.M., Jung W., Park S.B., Kim C.S., Kim J.S. Aster koraiensis extract and chlorogenic acid inhibit retinal angiogenesis in a mouse model of oxygen-induced retinopathy. Evid Based Complement Alternat Med. 2018;2018 doi: 10.1155/2018/6402650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Choi H.Y., Kim H.M., Choi J.H., Jang D.S. A novel cytotoxic steroidal saponin from the roots of Asparagus cochinchinensis. Plants. 2021;10 doi: 10.3390/plants10102067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.Y., Jeon S., Jang Y., Gotina L., Won J., Ju Y.H., Kim S., Jang M.W., Won W., Park M.G., Pae A.N., Han S., Kim S., Lee C.J. Platycodin D, a natural component of Platycodon grandiflorum, prevents both lysosome- and TMPRSS2-driven SARS-CoV-2 infection by hindering membrane fusion. Exp. Mol. Med. 2021;53:956–972. doi: 10.1038/s12276-021-00624-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Miyakawa K., Tokumasu R., S S.J., Kudo M., Ryo A. Evasion of vaccine-induced humoral immunity by emerging sub-variants of SARS-CoV-2. Future Microbiol. 2022;17:417–424. doi: 10.2217/fmb-2022-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta K., Cameron S., Lee M., Cai S.Q., Shoyama Y. Which East Asian herbal medicines can decrease viral infections? Phytochemistry Rev. 2021:1–19. doi: 10.1007/s11101-021-09756-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J., Ko K., Zhang L., Zhao D., Yang H.O., Kwon H.C. An autophagy inducing triterpene saponin derived from aster koraiensis. Molecules. 2019;24 doi: 10.3390/molecules24244489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Peng T. Traditional Chinese herbal medicine as a source of molecules with antiviral activity. Antivir. Res. 2013;97:1–9. doi: 10.1016/j.antiviral.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P., Liu D., Li J. Pharmacological perspective: glycyrrhizin may be an efficacious therapeutic agent for COVID-19. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupala C.S., Ye Y., Chen H., Su X.D., Liu H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun. 2021;590:34–41. doi: 10.1016/j.bbrc.2021.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Zhu Z., Lin H., Wang S., Zhang P., Li Y., Li L., Wang J., Zhao Y., Han J. Pyroptosis of syncytia formed by fusion of SARS-CoV-2 spike and ACE2-expressing cells. Cell Discov. 2021;7:73. doi: 10.1038/s41421-021-00310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello A., Civra A., Milan Bonotto R., Nascimento Alves L., Rajasekharan S., Giacobone C., Caccia C., Cavalli R., Adami M., Brambilla P., Lembo D., Poli G., Leoni V. The cholesterol metabolite 27-hydroxycholesterol inhibits SARS-CoV-2 and is markedly decreased in COVID-19 patients. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafinobar N., Mellander L.J., Kurczy M.E., Dunevall J., Angerer T.B., Fletcher J.S., Cans A.S. Cholesterol alters the dynamics of release in protein independent cell models for exocytosis. Sci. Rep. 2016;6 doi: 10.1038/srep33702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardacci R., Perfettini J.L., Grieco L., Thieffry D., Kroemer G., Piacentini M. Syncytial apoptosis signaling network induced by the HIV-1 envelope glycoprotein complex: an overview. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou T., Mou H., Zhang L., Ojha A., Choe H., Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T., Tokunaga K. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F.H., Wang Z.X., Cai P.P., Zhao L., Gao J.J., Kokudo N., Li A.Y., Han J.Q., Tang W. Traditional Chinese medicine and related active compounds: a review of their role on hepatitis B virus infection. Drug Discov Ther. 2013;7:212–224. doi: 10.5582/ddt.2013.v7.6.212. [DOI] [PubMed] [Google Scholar]
- Qian Z., Dominguez S.R., Holmes K.V. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajah M.M., Bernier A., Buchrieser J., Schwartz O. The mechanism and consequences of SARS-CoV-2 spike-mediated fusion and syncytia formation. J. Mol. Biol. 2021 doi: 10.1016/j.jmb.2021.167280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihn S.J., Merits A., Bakshi S., Turnbull M.L., Wickenhagen A., Alexander A.J.T., Baillie C., Brennan B., Brown F., Brunker K., Bryden S.R., Burness K.A., Carmichael S., Cole S.J., Cowton V.M., Davies P., Davis C., De Lorenzo G., Donald C.L., Dorward M., Dunlop J.I., Elliott M., Fares M., da Silva Filipe A., Freitas J.R., Furnon W., Gestuveo R.J., Geyer A., Giesel D., Goldfarb D.M., Goodman N., Gunson R., Hastie C.J., Herder V., Hughes J., Johnson C., Johnson N., Kohl A., Kerr K., Leech H., Lello L.S., Li K., Lieber G., Liu X., Lingala R., Loney C., Mair D., McElwee M.J., McFarlane S., Nichols J., Nomikou K., Orr A., Orton R.J., Palmarini M., Parr Y.A., Pinto R.M., Raggett S., Reid E., Robertson D.L., Royle J., Cameron-Ruiz N., Shepherd J.G., Smollett K., Stewart D.G., Stewart M., Sugrue E., Szemiel A.M., Taggart A., Thomson E.C., Tong L., Torrie L.S., Toth R., Varjak M., Wang S., Wilkinson S.G., Wyatt P.G., Zusinaite E., Alessi D.R., Patel A.H., Zaid A., Wilson S.J., Mahalingam S. A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D.W., Jumper C.C., Ackerman P.J., Bracha D., Donlic A., Kim H., Kenney D., Castello-Serrano I., Suzuki S., Tamura T., Tavares A.H., Saeed M., Holehouse A.S., Ploss A., Levental I., Douam F., Padera R.F., Levy B.D., Brangwynne C.P. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. Elife. 2021;10 doi: 10.7554/eLife.65962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana M.F., Pinto R.A.A., Marcon B.H., Medeiros L., Morais T., Dias L.C., Souza L.P., Melo G.C., Monteiro W.M., Lacerda M.V.G., Val F.A., Lalwani P.J., Ferreira L.C.L. Pathological findings and morphologic correlation of the lungs of autopsied patients with SARS-CoV-2 infection in the Brazilian Amazon using transmission electron microscopy. Rev. Soc. Bras. Med. Trop. 2021;54 doi: 10.1590/0037-8682-0850-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S.K., Shakya A., Prasad S.K., Singh S., Gurav N.S., Prasad R.S., Gurav S.S. An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J. Biomol. Struct. Dyn. 2021;39:3244–3255. doi: 10.1080/07391102.2020.1762741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., Shah J.G., Nguyen N., Chen Z., Meyers K., Yin M.T., Sobieszczyk M.E., Sheng Z., Huang Y., Liu L., Ho D.D. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature. 2022 doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Chen J., Wei G.W. Mechanisms of SARS-CoV-2 evolution revealing vaccine-resistant mutations in europe and America. J. Phys. Chem. Lett. 2021;12:11850–11857. doi: 10.1021/acs.jpclett.1c03380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang L., Li Q., Liang Z., Li T., Liu S., Cui Q., Nie J., Wu Q., Qu X., Huang W. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg. Microb. Infect. 2022;11:1–5. doi: 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.T., Kreutzberger A.J.B., Lee J., Kiessling V., Tamm L.K. The role of cholesterol in membrane fusion. Chem. Phys. Lipids. 2016;199:136–143. doi: 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y., Li J., Lai X., Zhang M., Kuang Y., Bao Y.O., Yu R., Hong W., Muturi E., Xue H., Wei H., Li T., Zhuang H., Qiao X., Xiang K., Yang H., Ye M. Natural triterpenoids from licorice potently inhibit SARS-CoV-2 infection. J. Adv. Res. 2022;36:201–210. doi: 10.1016/j.jare.2021.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Evans J.P., King T., Zheng Y.M., Oltz E.M., Whelan S.P.J., Saif L.J., Peeples M.E., Liu S.L. vol. 119. Proc Natl Acad Sci U S A; 2022. (SARS-CoV-2 Spreads through Cell-To-Cell Transmission). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M.S., Yu W.D., Wang H.X., Liu J.Y., Xu P.P. A potential antiviral activity of Esculentoside A against binding interactions of SARS-COV-2 spike protein and angiotensin converting enzyme 2 (ACE2) Int. J. Biol. Macromol. 2021;183:2248–2261. doi: 10.1016/j.ijbiomac.2021.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Lu L., Peng Z., Chen L.L., Meng X., Zhang C., Ip J.D., Chan W.M., Chu A.W., Chan K.H., Jin D.Y., Chen H., Yuen K.Y., To K.K. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microb. Infect. 2022;11:277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2
3
4
Data Availability Statement
Data will be made available on request.