Abstract
Objectives
High‐magnitude CD8+ T cell responses are associated with mild COVID‐19 disease; however, the underlying characteristics that define CD8+ T cell‐mediated protection are not well understood. The antigenic breadth and the immunodominance hierarchies of epitope‐specific CD8+ T cells remain largely unexplored and are essential for the development of next‐generation broad‐protective vaccines. This study identified a broad spectrum of conserved SARS‐CoV‐2 CD8+ T cell epitopes and defined their respective immunodominance and phenotypic profiles following SARS‐CoV‐2 infection.
Methods
CD8+ T cells from 51 convalescent COVID‐19 donors were analysed for their ability to recognise 133 predicted and previously described SARS‐CoV‐2‐derived peptides restricted by 11 common HLA class I allotypes using heterotetramer combinatorial coding, which combined with phenotypic markers allowed in‐depth ex vivo profiling of CD8+ T cell responses at quantitative and phenotypic levels.
Results
A comprehensive panel of 49 mostly conserved SARS‐CoV‐2‐specific CD8+ T cell epitopes, including five newly identified low‐magnitude epitopes, was established. We confirmed the immunodominance of HLA‐A*01:01/ORF1ab1637–1646 and B*07:02/N105–113 and identified B*35:01/N325–333 as a third epitope with immunodominant features. The magnitude of subdominant epitope responses, including A*03:01/N361–369 and A*02:01/S269–277, depended on the donors' HLA‐I context. All epitopes expressed prevalent memory phenotypes, with the highest memory frequencies in severe COVID‐19 donors.
Conclusion
SARS‐CoV‐2 infection induces a predominant CD8+ T memory response directed against a broad spectrum of conserved SARS‐CoV‐2 epitopes, which likely contributes to long‐term protection against severe disease. The observed immunodominance hierarchy emphasises the importance of T cell epitopes derived from nonspike proteins to the overall protective and cross‐reactive immune response, which could aid future vaccine strategies.
Keywords: CD8+ T cells, convalescence, epitopes, immunodominance, infection, SARS‐CoV‐2
Fifty‐one convalescent COVID‐19 donors were analysed for their ability to recognise 133 predicted SARS‐CoV‐2‐derived peptides restricted by 11 common HLA‐I allotypes using heterotetramer combinatorial coding. Forty‐nine mostly conserved SARS‐CoV‐2‐specific CD8+ T cell epitopes, including five new, were identified. This study revealed three dominant epitopes (HLA‐A*01:01/ORF1ab1637–1646, B*07:02/N105–113 and B*35:01/N325–333). The magnitude of subdominant epitope responses, including HLA‐A*03:01/N361–369 and A*02:01/S269–277, largely depended on the donors' HLA context. All epitopes had a prevalent memory phenotype, which were significantly higher in severe COVID‐19 donors.
Introduction
Over the last 2 years, substantial progress has been made to improve our understanding of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)‐specific immunity. However, the associated coronavirus disease 2019 (COVID‐19) still results in substantial morbidity and mortality worldwide. 1 COVID‐19 disease severity ranges from asymptomatic and mild self‐limiting disease to critical illness with death as a possible outcome. 2
Multiple studies showed a key role for SARS‐CoV‐2‐specific CD8+ cytotoxic T cells in the modulation and resolution of COVID‐19 pathogenesis. 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 Robust SARS‐CoV‐2‐specific CD8+ T cell immunity is associated with a mild disease outcome. 3 , 11 CD8+ T cells impair viral replication through recognition of viral peptide human leukocyte antigen class I (pHLA‐I) complexes displayed on the cell surface of infected cells, by which they accomplish rapid viral clearance and thereby reduce disease severity. 11 Most individuals generate SARS‐CoV‐2‐specific CD8+ T memory cells, indicative for the development of long‐lasting immunity. 12 , 13 Furthermore, SARS‐CoV‐2‐specific CD8+ T cells can recognise conserved viral epitopes, suggesting that they can provide broad protection against variants of SARS‐CoV‐2. Indeed, it has been shown that CD8+ T cells can continue to recognise SARS‐CoV‐2 variants of concern (VOC), 14 , 15 , 16 which makes them an attractive target for the development of broadly protecting COVID‐19 vaccines. Current COVID‐19 vaccines are designed to induce antibodies directed against the highly variable receptor‐binding domain (RBD) of the spike protein (S). 17 , 18 However, recent data indicate that the neutralising antibodies induced following natural SARS‐CoV‐2 infection and/or vaccination are less effective against emerging SARS‐CoV‐2 VOC, including Delta and Omicron. 14 , 19 , 20 In addition, SARS‐CoV‐2 vaccines have been proven very successful in inducing T cell‐mediated immunity. 21 , 22 , 23 , 24 It is therefore of vital importance to gain knowledge on the dynamics of the CD8+ T cell response following SARS‐CoV‐2 infection and vaccination. More specifically, we need to identify key immunogenic regions, their respective immunodominance and longevity of the SARS‐CoV‐2‐specific CD8+ T cell response.
Understanding the longevity and level of cross‐reactivity of the SARS‐CoV‐2 CD8+ T cell response between ancestral SARS‐CoV‐2 and VOCs within the human population requires assessment of SARS‐CoV‐2‐derived peptide recognition across a wide range of HLA‐I allotypes. Previous studies have addressed the peptide‐specific T cell reactivity after infection and vaccination in individuals expressing diverse repertoires of HLA‐I allotypes. 18 , 21 , 22 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 A substantial number of SARS‐CoV‐2 epitopes of CD8+ T cells in a wide range of HLA allotypes have been identified. However, technical limitations have thus far prevented the parallel detection and analysis of a myriad of SARS‐CoV‐2 epitope‐specific CD8+ T cells, hampering in‐depth assessment of their immunodominance profiles. 23 , 25 , 32 , 33 Gangaev et al. 30 characterised virus‐specific CD8+ T cells ex vivo and reported the immunodominance of HLA‐A*01:01/ORF1ab1637–1646 during acute SARS‐CoV‐2 disease; however, the hierarchy in relation to many other SARS‐CoV‐2‐derived CD8+ T cell epitopes remains uncharted. Furthermore, comprehensive characterisation of the memory phenotypes across a large amount of SARS‐CoV‐2‐specific CD8+ T cell epitopes directly ex vivo has not yet been reported and is of great importance to understand the longevity and strength of the CD8+ cell response in the months and years following infection and vaccination.
In this study, we analysed samples from 51 convalescent COVID‐19 donors for CD8+ T cell recognition of 133 predicted and previously identified SARS‐CoV‐2 pHLA‐I complexes, restricted by 11 common HLAs using combinatorial encoded pHLA class I tetramers directly ex vivo. Combinatorial encoding allowed simultaneous screening of up to 30 SARS‐CoV‐2 pHLA‐I combinations per donor, resulting in the identification of 49 distinct SARS‐CoV‐2‐specific CD8+ T cell epitopes, including the identification of five novel pHLA‐I combinations. Unique to our study is the depth of the combinatorial encoding assay that permitted the parallel establishment of the immunodominance and phenotype of the SARS‐CoV‐2 epitope‐specific CD8+ T cells. Our results established the hierarchy and phenotype of CD8+ T cells recognising distinct SARS‐CoV‐2 epitopes in convalescent donors, with higher frequencies and immunodominant features observed for nonspike epitopes. Our study could have important implications for the selection of CD8+ T cell epitopes in next‐generation COVID‐19 vaccine design, which aim to provide broad CD8+ T cell‐driven protection against current and emerging SARS‐CoV‐2 variants.
Results
Characteristics of study participants
A cohort of 51 convalescent SARS‐CoV‐2 seropositive donors was recruited in the spring of 2020 (Figure 1a; Supplementary table 1). Blood samples were taken from recruited donors on average 93‐day postsymptom onset (21–353 days) (Figure 1b). The median age of the cohort was 41 years (range 22–66 years), 47% were female and 62.7% of the cohort recovered from mild disease, 21.6% had experienced a severe infection, 3.9% was critically ill and 11.8% did not disclose the severity of their illness, who were therefore labelled unknown (Figure 1c–e; Supplementary figure 1a, Supplementary table 1). Seroconversion status of the donors was used to confirm previous SARS‐CoV‐2 infection. RBD, S and nucleocapsid (N)‐specific IgG antibody levels were measured by enzyme‐linked immunosorbent assay (ELISA). Convalescent donors who experienced a severe SARS‐CoV‐2 infection had significantly higher RBD, S and N IgG antibody titres than donors, who experienced a mild infection (Figure 1f). To study SARS‐CoV‐2‐specific CD8+ T cell responses, classical HLA‐I typing was performed on all donors. HLA typing revealed 11 common HLA‐I allotypes of interest for which combinatorial encoded HLA class I tetramers were available in our cohort, namely HLA‐A*01:01, A*02:01, A*03:01, A*11:01, A*24:02, B*07:02, B*08:01, B*15:01, B*27:05, B*35:01 and B*40:01 (Figure 1g). The distribution of those HLA‐I allotypes in our cohort was representative of those found in the Dutch population (Allele Frequency Net Database). 34 Several donors simultaneously expressed 2 (n = 23), 3 (n = 15) or 4 (n = 2) HLA‐I allotypes of interest (Figure 1h). Thus, our cohort includes donors with distinct and representative HLA‐I profiles providing us with the means to determine the dynamics of the SARS‐CoV‐2‐specific CD8+ T cell response in a diverse HLA‐I context.
Figure 1.
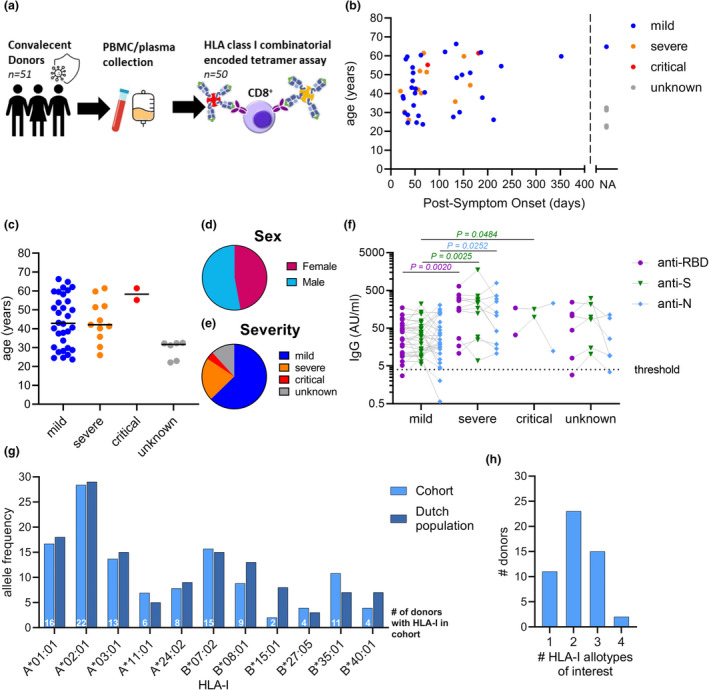
Convalescent COVID‐19 donor cohort and seroconversion. (a) Overview of convalescent donor cohort and study design. Distribution of days postsymptom onset (b), age (c), sex (d) and disease severity (e). Each dot represents an individual (n = 51 in total) (b, c). Donor with unspecified days postonset symptoms or severity was set as not available (NA; n = 7) (b) or unknown (n = 6) (c), respectively. (f) Plasma IgG titres to SARS‐CoV‐2 RBD, spike (S) and nucleocapsid (N) stratified per severity group; dotted line indicates the seroconversion threshold, and individual donors were connected by lines. Statistical significance between severity groups was determined with the unpaired Mann–Whitney U‐test, considering P‐values < 0.05 as significant. (g) Allelic frequency of 11 HLA‐I allotypes of interest in cohort (n = 51) compared with the general Dutch population obtained from the Allele Frequency Net Database (AFND; n = 1305). (h) Number of simultaneous expressed HLA‐I allotypes of interest across our cohort.
SARS‐CoV‐2 peptide selection
In addition to 47 previously identified peptides, 78 unique putative SARS‐CoV‐2 CD8+ T cell pHLA‐I combinations were selected based on the peptide prediction tools NetMHC‐4.0 and/or NetMHCpan‐4.1 and 8 peptides were selected based on their homology (> 75%) with SARS‐CoV‐1 and/or seasonal coronaviruses (Supplementary table 2). HLA‐I peptide binding with one or more HLA‐I allotypes of interest was validated using in vitro binding assays (Figure 2a). A total of 89 out of 133 predicted peptides had a > 50% binding efficiency to their respective HLA‐I; these were selected for further analysis. In addition, four peptides with lower binding affinities (ranging from 22.6% to 49.4%) were also selected based on epitope‐specific CD8+ T cell responses reported by others (Supplementary table 2). 9 , 15 , 26 , 27 The 93 selected pHLA‐I combinations covered 11 HLA allotypes (Figure 2b), including four peptides restricted to both HLA‐A*03:01 and A*11:01. The 93 peptide sequences were derived from SARS‐CoV‐2 structural proteins, namely S (n = 58), N (n = 20), membrane (M; n = 8) and envelope (E; n = 1) and nonstructural proteins including ORF1ab (n = 31), ORF3a (n = 3) and ORF6 (n = 1) (Figure 2b). The selected spike protein‐derived peptides are associated with the highest number of HLA‐I allotypes (n = 10), followed by the nucleocapsid protein (n = 9) and ORF1ab (n = 8) (Figure 2b). HLA‐A*02:01 binds the most diverse peptide repertoire, consisting of 30 peptides spanning seven viral proteins.
Figure 2.

Selection of 93 pHLA‐I complexes for heterotetramer combinatorial coding (HTCC). (a) Distribution of 133 predicted epitopes across HLA‐I and their binding avidity. Peptides with ≥ 50% binding to their respective HLA‐I allotype or previous confirmed as CD8+ T cell epitope were selected for CD8+ T cell analysis. (b) Distribution of selected peptides across viral proteins per HLA‐I allotype. (c) Overview of the HTCC approach. UV exposure cleaves the UV‐cleavable peptide in the HLA molecules and is exchanged for SARS‐CoV‐2‐specific peptides (red, green and blue). Next, pHLA‐I complexes were conjugated to two different fluorophores to generate the dual‐coded tetramers for each pHLA combination. Peripheral blood mononuclear cells were stained with the combinatorial encoded tetramers and analysed using flow cytometry. (d) Representative flow cytometry plots (donor D04) created by Boolean gating CD8+ T cell populations expressing tetramer fluorophores; associated gating strategy is provided in Supplementary figure 2. Each plot in the matrix represents a unique tetramer dual‐coding combination; cells that are double‐positive for both fluorophores are indicated in red, single‐positive cells are indicated in green, and cells negative for all tetramer fluorophores are in blue.
Identification of SARS‐CoV‐2‐derived CD8 + T cell epitopes
Heterotetramer combinatorial coding (HTCC)‐linked pHLA‐I complexes were used to establish which peptides could be recognised by SARS‐CoV‐2‐specific CD8+ T cells present in convalescent COVID‐19 donors (Figure 2c and d). Up to 30 unique SARS‐CoV‐2 pHLA‐I complexes were screened simultaneously in a single donor directly ex vivo (Figure 2d; Supplementary figure 2). Positive responses were identified as CD8+ T cells double‐positive for both fluorophores of a specific pHLA complex (Figure 2d). In total, 49 double‐positive pHLA‐I complexes were identified (Figure 3a), including two novel CD8+ T cell epitopes (A*03:01/N160–169 and A*03:01/ORF1ab807–816) and three previously identified peptides associated with novel HLA‐I allotypes (A*11:01/S529–537, B*07:02/S1014–1022 and B*35:01/N352–360) albeit at low frequencies (Figure 3a; Supplementary table 2). The remaining 44 epitopes were previously described (Figure 3a; Supplementary table 2). All peptides recognised by CD8+ T cells were linked to a single HLA‐I allotype, with exception of N361–369 which could be recognised in complex with HLA‐A*03:01 and HLA‐A*11:01 (Figure 3a). No epitopes were identified for HLA‐B*08:01 (Figure 3a). For 9 out of 11 included HLA‐I allotypes, at least one epitope was recognised by 90–100% of the donors expressing that specific HLA (Figure 3a). The most highly recognised epitopes were restricted to HLA‐A*01:01, with four epitopes recognised by > 90% of the HLA‐A*01:01+ donors (Figure 3a). Combined these nine HLA‐I allotypes are commonly expressed in 89% of the world population, as shown by the IEDB‐AR population coverage tool. 35 , 36 Recognised epitopes were derived from six SARS‐CoV‐2 proteins (Figure 3b). A total of 18 epitopes were found in ORF1ab, which were restricted to seven HLA‐I allotypes. Epitopes originating from spike (n = 10) and nucleocapsid (n = 15) proteins were both restricted to 8 HLA allotypes. Although only two ORF3a peptides restricted to two HLAs were identified, 100% of the donors expressing either HLA‐A*01:01 and/or HLA‐A*02:01 were able to recognise the respective ORF3a epitopes (Figure 3c). In contrast, ORF1ab‐specific CD8+ T cells were only identified in 55% of the donors with the potential to recognise ORF1ab epitopes (Figure 3c).
Figure 3.
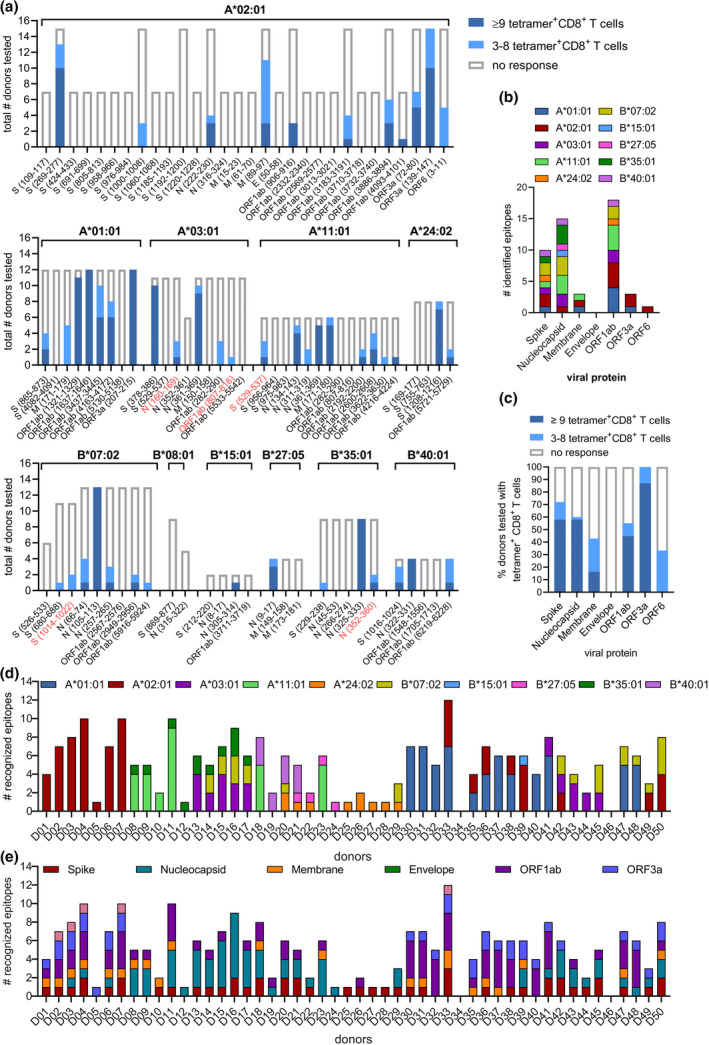
Identification of SARS‐CoV‐2‐specific CD8+ T cell epitopes. (a) Height of the bar indicates the number of donors with a specific HLA‐I allotype tested for the presence of SARS‐CoV‐2 pHLA‐specific responses. Tetramer+CD8+ T cells detected at ≥ 9 cells counted within the dual‐tetramer‐positive gate are indicated in dark blue, those with 3–8 dual‐positive events are indicated in light blue. (b) Distribution of identified epitopes across SARS‐CoV‐2 viral proteins and HLA‐I restrictions. (c) Frequency of donors able to recognise various SARS‐CoV‐2 peptides. (d) Number of SARS‐CoV‐2 epitopes recognised by individual donors and their distribution across their HLA profile. (e) Number of epitopes derived from various SARS‐CoV‐2 peptides recognised by individual donors.
Next, the distribution of SARS‐CoV‐2 epitope recognition across individual donors in the convalescent cohort was studied. 96% of the convalescent donors recognised one or more SARS‐CoV‐2 epitopes (Figure 3d). On average, five epitopes were recognised per donor, one donor recognised 12 epitopes, six donors recognised only a single epitope and two donors did not recognise any of the tested SARS‐CoV‐2 epitopes (Figure 3d; Supplementary figure 3a). The diversity of the tested HLA‐I profiles in an individual donor correlated with the amount of tested and recognised epitopes (Figure 3d; Supplementary figure 3b and c). 84% of the donors was capable of recognising epitopes derived from multiple viral proteins (Figure 3e). Combined the 50 convalescent donors recognised 49 different SARS‐CoV‐2 epitopes, which target a broad spectrum of the viral genome and spanning various HLA‐I allotypes.
Dynamic immunodominance landscapes of SARS‐CoV‐2‐specific CD8 + T cell epitopes in convalescent donors
To probe the magnitude of the individual SARS‐CoV‐2 epitope‐specific CD8+ T cell populations, we assessed the percentage of tetramer‐positive cells in the total CD8+ T cell population of a donor directly ex vivo. Epitopes were ordered based on the mean frequency of all HLA‐I allotype+ donors tested per allotype, which showed that each HLA‐allotype has a favorable SARS‐CoV‐2 epitope that displays a high frequency of CD8+ T cell recognition in all donors (Figure 4a). Epitopes recognised by all tested donors did not consistently have the highest mean frequency, as observed for HLA‐A*02:01 and A*11:01 (Figure 4a). Overall, epitope‐specific CD8+ T cell frequencies were relatively stable up to 228‐day postinfection (Supplementary figure 3d).
Figure 4.
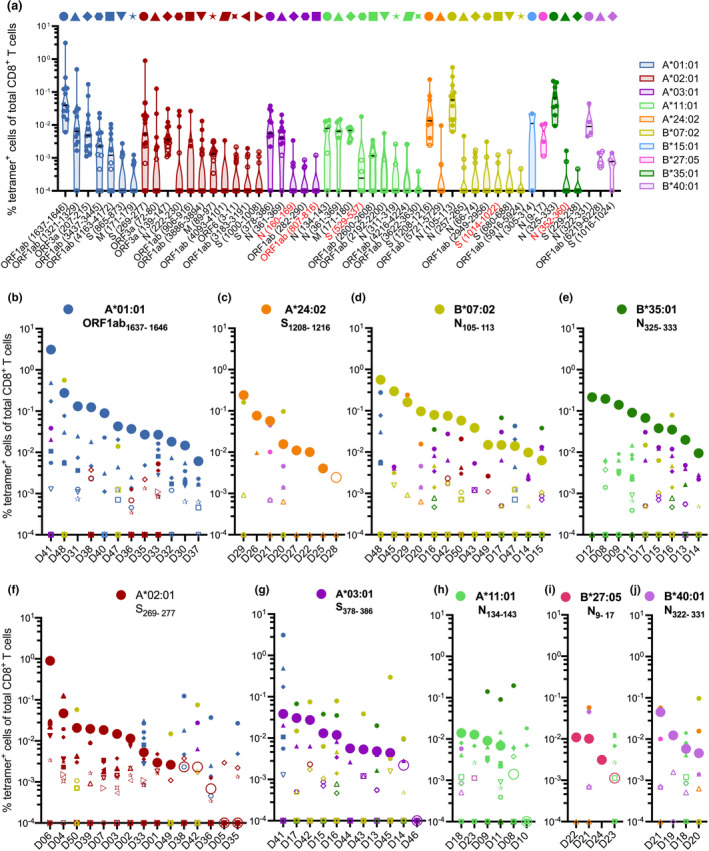
Frequency and immunodominance landscapes of SARS‐CoV‐2‐specific CD8+ T cell epitopes in convalescent donors. (a) Frequency of tetramer+CD8+ T cells for SARS‐CoV‐2 epitopes, each dot represents an individual donor, and epitopes are ordered based on mean frequency per HLA‐I. Colours correspond to different HLA‐I restrictions. Tetramer+CD8+ T cells detected at < 9 cells counted within a dual‐positive gate are indicated by open symbols (Supplementary figure 4) and were excluded from phenotypic analysis. Bar indicates median. (b–j) Immunodominance landscapes of single SARS‐CoV‐2 epitope‐specific CD8+ T cell populations in individual donors grouped based on their HLA‐I expression. Unique symbol/colour combinations are assigned for each individual epitope as indicated by the caption above (a). Graphs (b–j) are ordered based on the mean frequency of the epitope with the highest mean frequency for the respective HLA (large circles) identified in a.
A key benefit of combinatorial encoded pHLA‐I tetramers is that it allowed us to evaluate immunodominance hierarchies of up to 30 epitopes simultaneously within a donor directly ex vivo (Figure 4b–j; Supplementary figure 4a and b). We defined immunodominance based on how the sizes of SARS‐CoV‐2 epitope‐specific CD8+ T cell responses, as measured by tetramer binding, are impacted by the HLA context of an individual. Of all the epitopes that displayed the highest magnitude within a certain HLA, HLA‐A*01:01, B*07:02 and B*35:01 and possibly A*24:02 (Figure 4b–e) were more likely to be dominant over HLA‐A*02:01, A*03:01, A*11:01, B*27:05 and B*40:01 epitopes (Figure 4f–j). Hence, the frequency of epitope‐specific CD8+ T cells seems to be dependent on the HLA‐I context of a donor, as changes in immunodominance hierarchy can be observed between donors with different HLA‐I allotypes. Epitopes with seemingly lower immunodominance display a higher frequency of epitope‐specific CD8+ T cells when the ability to present more dominant epitopes on other HLA‐I allotypes is absent. For example, the frequency of HLA‐A*02:01/S269–277‐specific CD8+ T cells is significantly lower in donors, who also express the HLA‐A*01:01 allotype, suggesting that the HLA‐A*01:01 response is dominant over the A*02:01 response (Figure 4b and f; Supplementary figure 4c). A similar trend for lower frequency of HLA‐A*02:01/S269–277‐specific CD8+ T cells was also observed in HLA‐A*02+B*07:02+ donors when compared to A*02:01+ donors (Figure 4d and f; Supplementary figure 4d). In addition, interplay is detected within the epitopes with immunodominant features, for instance, HLA‐B*07:02/N105–113‐specific CD8+ T cell responses are generally immunodominant, but in some cases can be overtaken by HLA‐A*03:01, A*24:02 and B*35:01 epitope‐specific CD8+ T cell responses (Figure 4d). Because of HLA‐I diversity beyond the 11 tested HLA‐I allotypes, it remains difficult to grasp a complete picture. However, the data strongly suggest a prominent role of HLA‐A*01:01/ORF1ab1637–1646, B*35:01/N325–333 and B*07:02/N105–113 in CD8+ T cell immunity against SARS‐CoV‐2.
Conservation of SARS‐CoV‐2‐specific CD8 + T cell epitopes in SARS‐CoV‐2 variants of concern
Next, the amino acid conservation of the identified epitopes in the five main VOC (Alpha, Beta, Gamma, Delta and Omicron) was established. 37 Of the 49 epitopes identified in this study, only two epitopes were mutated to a significant extent (> 50%), namely HLA‐B*07:02/S680–688 (P681H in Alpha and Omicron and P681R in Delta) and HLA‐B*27:05/N9–17 (P13L in Omicron) (Table 1). According to the above‐established immunodominance profiles, HLA‐B*07:02/S680–688 is considered a subdominant epitope and although HLA‐B*27:05/N9–17 is the only and highly prevalent epitope found for HLA‐B*27:05 its immunodominance potential relative to other HLA‐I epitopes is limited (Figure 4i). The remaining 17 mutations were found in < 10% of variant strains sequenced (Table 1). Interestingly, 9.2% of the Delta strains display a P1640L mutation in the immunodominant HLA‐A*01:01/ORF1ab1637–1646 epitope. Furthermore, previous studies have shown that a P272L mutation of the highly prevalent HLA‐A*02:01/S269–277 epitope is associated with reduced recognition by CD8+ T cells. 38 However, our sequence analysis revealed that this mutation was found in < 1% of current and past VOC sequences and thus this mutation is not included in Table 1. These data indicate that the SARS‐CoV‐2 CD8+ T cell epitopes are highly conserved in VOC.
Table 1.
Amino acid sequence identity of the viral peptides of the SARS‐CoV‐2 tetramers across variants of concern
| HLA‐I | Epitope | Peptide sequence | Mutation | B.1.1.7 | B.1.351 | P.1 | B.1.617.2 | B.1.1.529 | BA.2 | BA.5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alpha | Beta | Gamma | Delta | Omicron | Omicron | Omicron | |||||||||
| Sequence | % | Sequence | % | Sequence | % | Sequence | % | Sequence | % | Sequence | Sequence | ||||
| A01:01 | ORF1ab1637–1646 | TTDPSFLGRY | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 90.8 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐ | |
| P1640L | ND | ND | ND | ‐‐‐L‐‐‐‐‐‐ | 9.2 | ND | |||||||||
| A01:01 | ORF1ab1321–1329 | PTDNYITTY | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A01:01 | ORF3a207–215 | FTSDYYQLY | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A01:01 | ORF1ab3437–3445 | GTDLEGNFY | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A01:01 | ORF1ab4163–4172 | CTDDNALAYY | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐ | |
| A01:01 | S865–873 | LTDEMIAQY | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A01:01 | M171–179 | ATSRTLSYY | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A02:01 | S269–277 | YLQPRTFLL | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A02:01 | ORF3a72–80 | ALSKGVHFV | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 98.8 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 95.5 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A72S/H78Y | ND | S‐‐‐‐‐‐‐‐ | 1.2 | ND | ND | ‐‐‐‐‐‐Y‐‐ | 4.5 | ‐‐‐‐‐‐Y‐‐ | |||||||
| A02:01 | ORF3a139–147 | LLYDANYFL | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 98.9 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| L140F | ND | ND | ND | ‐F‐‐‐‐‐‐‐ | 1.1 | ND | ‐F‐‐‐‐‐‐‐ | ||||||||
| A02:01 | N222–230 | LLLDRLNQL | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A02:01 | ORF1ab906–916 | YLFDESGEFKL | ‐‐‐‐‐‐‐‐‐‐‐ | 98.9 | ‐‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐‐ | |
| E913D | ‐‐‐‐‐‐‐D‐‐‐ | 1.1 | ND | ND | ND | ND | |||||||||
| A02:01 | ORF1ab3886–3894 | KLWAQCVQL | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A02:01 | M89–97 | GLMWLSYFI | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A02:01 | ORF1ab4093–4101 | ALWEIQQVV | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A02:01 | ORF63–11 | HLVDFQVTI | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| D6G | ND | ND | ND | ND | ND | ‐‐‐G‐‐‐‐‐ | |||||||||
| A02:01 | ORF1ab3183–3191 | FLLNKEMYL | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A02:01 | S1000–1008 | RLQSLQTYV | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A03:01 | S378–386 | KCYGVSPTK | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 98.2 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| P384L | ND | ‐‐‐‐‐‐L‐‐ | 1.8 | ND | ND | ND | |||||||||
| A03:01 | N361–369 | KTFPPTEPK | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 92.2 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| T362I | ND | ‐I‐‐‐‐‐‐‐ | 7.8 | ND | ND | ND | |||||||||
| A03:01 | N160–169 | QLPQGTTLPK | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐ | |
| A03:01 | ORF1ab282–290 | KTIQPRVEK | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A03:01 | ORF1ab807–816 | MVTNNTFTLK | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐ | |
| A11:01 | N134–143 | ATEGALNTPK | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐ | |
| E136D | ND | ND | ND | ND | ND | ‐‐D‐‐‐‐‐‐‐ | |||||||||
| A11:01 | N361–369 | KTFPPTEPK | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 92.2 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| T362I | ND | ‐I‐‐‐‐‐‐‐ | 7.8 | ND | ND | ND | |||||||||
| A11:01 | M171–180 | ATSRTLSYYK | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐ | |
| A11:01 | S529–537 | KSTNLVKNK | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A11:01 | ORF1ab2600–2608 | STFNVPMEK | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A11:01 | ORF1ab2192–2200 | ASMPTTIAK | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A11:01 | N311–319 | ASAFFGMSR | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A11:01 | ORF1ab4216–4224 | VTDTPKGPK | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A11:01 | ORF1ab3622–3630 | SAFAMMFVK | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A24:02 | S1208–1216 | QYIKWPWYI | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| A24:02 | ORF1ab5721–5729 | VYIGDPAQL | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| B07:02 | N105–113 | SPRWYFYYL | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| B07:02 | N257–265 | KPRQKRTAT | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| B07:02 | N66–74 | FPRGQGVPI | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 98.6 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| B07:02 | ORF1ab2949–2956 | RPDTRYVL | ‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐ | |
| B07:02 | S1014–1022 | RAAEIRASA | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| B07:02 | S680–688 | SPRRARSVA | ‐‐‐‐‐‐‐‐‐ | 1.0 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 92.3 | ‐‐‐‐‐‐‐‐‐ | 0.9 | ‐‐‐‐‐‐‐‐‐ | 0.8 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| P681H/P681R | ‐H‐‐‐‐‐‐‐ | 99.0 | ND | ‐H‐‐‐‐‐‐‐ | 5.2 | ‐R‐‐‐‐‐‐‐ | 99.1 | ‐H‐‐‐‐‐‐‐ | 99.2 | ‐H‐‐‐‐‐‐‐ | ‐H‐‐‐‐‐‐‐ | ||||
| A688V | ND | ND | ‐‐‐‐‐‐‐‐V | 2.5 | ND | ND | |||||||||
| B07:02 | ORF1ab5916–5924 | IPRRNVATL | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| B15:01 | N305–314 | AQFAPSASAF | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐ | |
| B27:05 | N9–17 | QRNAPRITF | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 93.7 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 93 | ‐‐‐‐‐‐‐‐‐ | 2.3 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| Q9H/Q9L | ND | H‐‐‐‐‐‐‐‐ | 1.4 | ND | L‐‐‐‐‐‐‐‐ | 7 | ND | ||||||||
| P13L/P13S/P13F | ND | ‐‐‐‐S‐‐‐‐ | 4.9 | ND | ND | ‐‐‐‐L‐‐‐‐ | 97.7 | ‐‐‐‐L‐‐‐‐ | ‐‐‐‐L‐‐‐‐ ‐‐‐‐F‐‐‐‐ | ||||||
| B35:01 | N325–333 | TPSGTWLTY | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 98.3 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| S327L | ND | ND | ND | ‐‐L‐‐‐‐‐‐ | 1.7 | ND | |||||||||
| B35:01 | N352–360 | LLNKHIDAY | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
| B35:01 | S229–238 | LPIGINITRF | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐ | |
| B40:01 | N322–331 | MEVTPSGTWL | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 98.3 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐ | |
| S327L | ND | ND | ND | ‐‐‐‐‐L‐‐‐‐ | 1.7 | ND | |||||||||
| B40:01 | ORF1ab6219–6228 | IEYPIIGDEL | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐‐ | |
| B40:01 | S1016–1024 | AEIRASANL | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | 100 | ‐‐‐‐‐‐‐‐‐ | ‐‐‐‐‐‐‐‐‐ | |
Access date: 15/08/2022. % = frequency of all sequence on outbreak.info database threshold 1%, exact values BA.2 and BA.5 unknown; ND = mutation not detected in variant.
SARS‐CoV‐2‐specific CD8 + T cells display a memory phenotype
We established the phenotypic profile of the epitope‐specific CD8+ T cells to understand whether they were recruited during primary SARS‐CoV‐2 infection, resulting in immunological memory formation. Here, combinatorial encoded pHLA‐I tetramers were combined with CD27 and CD45RA staining, allowing characterisation of central memory (Tcm; CD27+CD45RA−), effector memory (Tem; CD27−CD45RA−), terminally differentiated effector memory CD45RA (Temra; CD27−CD45RA+) and naive‐like (Tnaive‐like; CD27+CD45RA+) SARS‐CoV‐2‐specific CD8+ T cells (Figure 5a; Supplementary figure 2). Phenotypic profiles for 38 epitopes with ≥ 9 tetramer‐positive events were established (Supplementary figure 4b), demonstrating a clear central memory response for 36 out of 38 epitopes (Figure 5b), with the exception of HLA‐A*02:01/ORF1ab4093–4101 (68.4% Tnaive‐like) and B*40:01/ORF1ab6219–6228 (55.56% Tnaive‐like) which were detected in a single donor (Figure 5b). After combining all epitopes per HLA‐I allotype, phenotypic profiles displayed significantly higher Tcm than Tnaive‐like frequencies in HLA‐A*01:01, A*02:01, A*03:01, A*11:01, and B*07:02 (Figure 5c). Similar trends were observed for HLA‐A*24:02, B*15:01, B*27:05, B*35:01 and B*40:01 allotypes (Figure 5c; Supplementary figure 5a). To assess the relation between disease severity and phenotype distribution, all responding peptides were combined per severity group (Figure 5d). Significantly higher frequencies of SARS‐CoV‐2‐specific CD8+ Tcm responses at the expense of Tnaive‐like responses were observed in convalescent donors, who experienced severe and/or critical COVID‐19 compared to those who experienced mild disease (Figure 5d). Individuals with severity‐related characteristics age (≥ 60 years) displayed significantly higher frequencies of Temra and lower frequencies of Tnaive‐like SARS‐CoV‐2 epitope‐specific CD8+ T cells (Supplementary figure 5b). Overall, phenotype frequencies were relatively stable up to 228‐day postinfection (Supplementary figure 5c–h). Together these results demonstrate that all SARS‐CoV‐2 convalescent donors generated a broad memory CD8+ T cell response across a large variety of CD8+ T cell epitopes.
Figure 5.
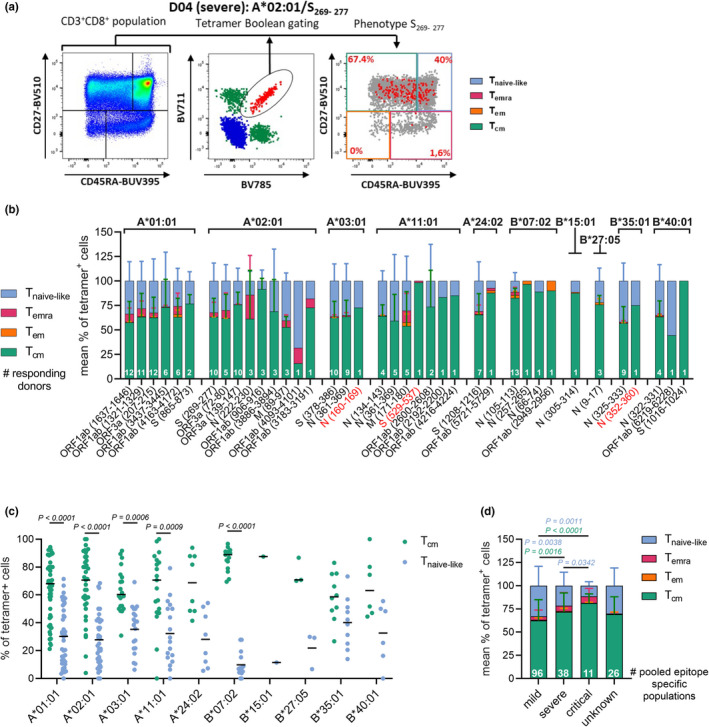
SARS‐CoV‐2 epitope‐specific CD8+ T cells display a clear memory phenotype. (a) Representative FACS panels indicate the gating strategy used to characterise the phenotype profile of SARS‐CoV‐2 epitope‐specific CD8+ T cells. CD27 and CD45RA were used to identify Tcm (CD27+CD45RA−), Tem (CD27−CD45RA−), Temra (CD27−CD45RA+) and Tnaive‐like (CD27+CD45RA+) cells. Gates were set based on the total CD8+ T cell population (left panel). The right panel displays a combination of total CD8+ T cells (grey dots) with dual‐tetramer‐positive cells (red dots) (b) Mean phenotypic frequencies of SARS‐CoV‐2 epitope‐specific CD8+ T cells pooled donors per epitope. (c) Tcm and Tnaive frequencies of tetramer+CD8+ T cells, populations of multiple donors and epitopes with the respective HLA‐I restriction were pooled. (d) Mean phenotypic frequency of tetramer+CD8+ T cells per severity group. Mean (c) and SD (b, d) are shown and statistical significance was determined using the Wilcoxon (c) or the Mann–Whitney U‐test, considering P‐values < 0.05 as significant (d).
Discussion
The 49 SARS‐CoV‐2 CD8+ T cell epitopes spanned 10 HLA‐I allotypes and included five novel pHLA‐I combinations. The simultaneous evaluation of this study confirmed the immunodominance of A*01:01/ORF1ab1637–1646, B*07:02/N105–113 and identified B*35:01/N325–333 as a third SARS‐CoV‐2 epitope with immunodominant features. The immunodominance hierarchies of some of the less dominant epitopes, such as A*02:01/S269–277, largely depended on the HLA‐I context in which the epitope was expressed. Despite the variable epitope‐specific CD8+ T cell frequencies, all epitope‐specific CD8+ T cell populations had a clear memory phenotype, with the highest frequency of CD8+ Tcm cells observed in the most severe COVID‐19 patients. Thus, SARS‐CoV‐2 infection initiates a broad CD8+ T cell response across multiple HLA‐I allotypes.
The process of peptide prediction and parallel pHLA screening using combinatorial encoded tetramers resulted in the identification of 49 SARS‐CoV‐2 CD8+ T cell epitopes, including five novel pHLA‐I combinations that have not been previously identified, namely HLA‐A*03:01/N160–169, A*03:01/ORF1ab807–816, A*11:01/S529–537, B*07:02/S1014–1022 and B*35:01/N352–360. The importance of the five novel pHLA‐I combinations after infection needs to be further analysed, as only a fraction of respective HLA‐positive donors recognised these epitopes and frequencies were relatively low. One of the limitations of our study is that due to the low frequency of the novel epitope‐specific CD8+ T cells, we were unable to verify the functionality of the novel epitope‐specific CD8+ T cell response in our donors. However, the CD8+ T cell response directed against these novel peptides may improve by repeated infection and/or vaccination. Furthermore, the HLA‐A*03:01/N160–169 epitope overlaps with a conserved immunodominant B cell epitope region in SARS‐CoV‐1. 39 The remaining 44 epitopes were confirmed in other studies, which used techniques ranging from in vitro stimulation with overlapping peptide pools to ex vivo pHLA‐I tetramer staining (Supplementary table 2). 4 , 9 , 19 , 23 , 26 , 27 , 28 , 29 , 30 , 39 , 40 , 41 , 42 For 42 selected peptides, no epitope‐specific CD8+ T cell populations could be detected using the combinatorial encoded tetramers. In addition, no ex vivo responses were found for A*01:01/S4082–4091 and A*01:01/ORF1ab5130–5138 that were identified as potential SARS‐CoV‐2 CD8+ T cell epitopes by others, following in vitro expansion. 26 , 27
The novelty of this study lies in the broad characterisation of immunodominance of CD8+ T cell epitopes of SARS‐CoV‐2, which has been initiated by earlier studies for a few epitopes. 19 , 30 A key advantage of the HTCC setup is that it permits the evaluation of immunodominance hierarchies of up to 30 epitopes simultaneously within a single donor through direct ex vivo analysis, which was performed in a cohort of 50 convalescent COVID‐19 donors. Previous immunodominance studies were limited by the number of HLA‐I allotypes that could be studied simultaneously in a single donor. The co‐expression of multiple HLA‐I of interest in our convalescent COVID‐19 cohort allowed us to assess immunodominance across 2 or 3 HLA‐I allotypes in 26 donors. Comparing immunodominance hierarchies of epitopes across HLA‐I profiles revealed that HLA‐A*01:01/ORF1ab1637–1646 epitope had the strongest immunodominant features followed by B*07:02/N105–113 and B*35:01/N325–333. HLA‐A*01:01/ORF1ab1637–1646, A*24:02/S1208–1216 and B*07:02/N105–113 epitopes were previously identified as potential immunodominant epitopes based on the high frequency of epitope‐specific CD8+ T cells. 23 , 25 , 30 , 31 Our study was able to extend these prior studies by establishing their immunodominance hierarchy across multiple epitopes and HLA‐I allotypes simultaneously. Furthermore, HLA‐B*35:01/N325–333 was identified as an immunodominant epitope over HLA‐A*11:01 and A*03:01 epitopes. Contradictory results have been found for HLA‐A*02:01/S269–277, a previous study described the epitope as immunodominant, 23 whereas another study categorised it as subdominant relative to other HLA‐I restricted epitopes. 31 The current study revealed that the immunodominance or subdominance of the A*02:01/S269–277 epitope largely depended on the HLA‐I context in which the epitope was expressed. This likely explains the different A*02:01/S269–277 immunodominance profiles that have been previously described. 23 , 31 This study indicates that the magnitude of the SARS‐CoV‐2‐specific CD8+ T cell response strongly depends on the HLA‐I context within an individual. Therefore, the HLA‐I allotype profile could be indicative for the level of CD8+ T cell protection in future SARS‐CoV‐2 infections. However, association studies correlating specific HLA‐I allotypes to COVID‐19 remain inconclusive. 43 Higher resolution may be required to establish whether the dynamic immunodominance profiles of HLA‐I epitopes are linked to disease progression and/or viral infectivity. Another interesting aspect that warrants further investigation is whether and how the immunodominance profiles will change following additional infections (exposure to total viral proteome) and/or vaccinations (exposure to spike protein only).
Despite the large variety of frequencies and immunodominance profiles, the ex vivo HTCC analysis revealed that SARS‐CoV‐2‐specific CD8+ T cells had a strong memory profile for 36 out of 39 epitopes. It is unlikely that the observed memory phenotypes were the result of previous seasonal coronavirus infections, due to the lack of amino acid sequence homology. Hence, these results therefore suggest that the epitope‐specific CD8+ T cells were successfully recruited during primary seasonal SARS‐CoV‐2 infection. However, it should be noted that potential homology with other pathogen‐derived peptides could not be excluded. Donors who recovered from severe/critical disease had significantly higher Tcm and lower Tnaive‐like populations than convalescent donors who experienced mild disease, which is in correspondence to other studies that found larger overall T cell responses in patients who recovered from severe disease. 44 Lower Tnaive‐like frequencies were also identified in elderly, which was expected as elderly are more susceptible to severe COVID‐19. 45 Although the memory phenotype of SARS‐CoV‐2‐specific CD8+ T cells identified in this study suggests persistence, to confirm their longevity requires study of epitope‐specific T cell responses over long periods of time.
The strength of the CD8+ T cell response against future SARS‐CoV‐2 infections also depends on their ability to recognise conserved epitopes in emerging viral variants. Overall, SARS‐CoV‐2 CD8+ T cell epitopes were highly conserved among all VOCs, with HLA‐B*07:02/S680–688 and B*27:05/N9–17 being the only exceptions. The impact of the mutations in the HLA‐B*07:02/S680–688 epitope on the overall CD8+ T cell response is probably limited as this epitope was subdominant and therefore unlikely to be the result of selective immune escape. Potentially, the N9–17 P13L mutation in Omicron is of higher concern, as it affects the only identified and highly prevalent HLA‐B*27:05 SARS‐CoV‐2 epitope. Although the P13L mutation is not predicted to affect peptide‐HLA‐I binding considerably (NetMHC‐4.0), T cell receptor (TCR) recognition was found to be altered. 46 Hence, although Omicron evades previously induced neutralising antibody responses, 47 CD8+ T cell responses to Omicron, and other VOCs, are hardly affected. 15 , 16 , 48 However, it is to be expected that over the years, due to repeated SARS‐CoV‐2 infections of people with existing SARS‐CoV‐2‐specific immunity, mutations arise in future VOCs that permit escape from existing CD8+ T cell‐driven immunity, as is also observed for other seasonal infections like influenza. 49 , 50
Vaccination is the most effective way to protect against (severe) COVID‐19. Current vaccines have been shown to induce highly effective CD8+ T cell responses, but generally only exploit the S protein. 51 This means that vaccine‐induced CD8+ T cells target a narrow range of spike‐derived epitopes. The current study identified 10 spike‐derived epitopes across eight HLA‐I allotypes, of which A*02:01/s269–277, A*03:01/S378–386 and A*24:02/S1208–1216 are the only epitopes that induced high CD8+ T cell frequencies in the majority of convalescent donors expressing the respective HLA‐I allotype. Spike‐derived epitopes were not identified for HLA‐B*08:01, B*15:01 and B*27:05, which may have been the result of insufficient peptide prediction for these HLA‐I allotypes. A recent study has identified HLA‐B*15:01 restricted spike‐specific CD8+ T cell epitopes, both the SARS‐CoV‐2 and seasonal coronavirus variant of the epitopes could be recognised by the same T cell receptors. 23 To the best of our knowledge, no spike‐specific epitopes have been identified for HLA‐B*08:01 and B*27:05, suggesting that vaccination may result in lower spike‐specific CD8+ T cell responses in individuals expressing one or a combination of these HLA‐I allotypes. CD8+ T cells recognising spike‐derived peptides restricted by HLA‐A*01:01, A*11:01, B*07:02, B*35:01 and B*40:01 were induced at low frequencies after infection‐induced immunity. However, these epitopes may play a central role after spike‐based vaccination, as in this setting the more immunodominant epitopes derived from other viral proteins will be absent. Indeed, a recent study by Minervina et al. 23 demonstrated that HLA‐A*01:01, A*02:01, A*24:02, B*15:01 and B*44:02 restricted spike epitope‐specific CD8+ T cells were preferentially boosted following vaccination of individuals with and without a history of previous SARS‐CoV‐2 infection. It will be of special interest to determine whether CD8+ T cells recognising the newly identified HLA‐A*11:01/S529–537 and B*07:02/S1014–1022 epitopes have a similar potential to become important in spike‐vaccine‐induced immunity, as these are the most prominent spike‐derived epitopes for their respective HLA‐I allotype. However, the high mutation rate observed in the spike protein, relative to the rest of the SARS‐CoV‐2 genome, 52 emphasises the importance of including other SARS‐CoV‐2 proteins in next‐generation vaccines that aim to provide broad CD8+ T cell‐driven protection against severe disease caused by current and emerging SARS‐CoV‐2 variants. In prospect, it would be interesting to study how spike epitopes develop in relation to other SARS‐CoV‐2‐derived epitopes over time, after reinfections and/or repeated vaccination.
To summarise, we reported immunodominance hierarchies and memory phenotype profiles of conserved SARS‐CoV‐2 CD8+ T cell epitopes simultaneously in 50 convalescent COVID‐19 donors. The ability of epitope‐specific CD8+ T cells to respond to a large variety of conserved SARS‐CoV‐2 epitopes and differentiate into a sustainable memory population is likely to contribute to long‐term protection against severe disease inflicted by future SARS‐CoV‐2 strains. Furthermore, this study underlines and extends the currently known HLA‐I epitope repertoire against SARS‐CoV‐2 and described the high level of conservation of CD8+ T cell epitopes. In addition, the immunodominance hierarchy emphasises the importance of CD8+ T cell epitopes derived from viral proteins, besides spike, to the overall protective and cross‐reactive immune response. Overall, these epitopes could play an essential role in developing broad‐protective SARS‐CoV‐2 vaccines, which aim to protect against severe disease resulting from current and emerging SARS‐CoV‐2 variants.
Methods
Study participants and sample collection
Fifty‐one convalescent SARS‐CoV‐2 donors infected in the spring of 2020 were recruited as part of the COVID‐19 Convalescent Plasma programme at Sanquin Blood Supply Foundation, Amsterdam, the Netherlands, between 30 March and 6 September 2020. SARS‐CoV‐2 infection was confirmed based on RBD, spike and/or N IgG serology. A questionnaire was used to determine the date of symptom onset, date of recovery and severity of the infection. Donors were categorised into the following categories: mild (stay at home, minimal symptoms), severe (hospitalised, ward) and critical (hospitalised, intensive care unit) or unknown (donors did not provide information on their disease progression). Demographics of all participants are listed in Supplementary table 1 and Supplementary figure 1. The study is in accordance with the declaration of Helsinki and according to Dutch regulations. Data and samples were collected only from voluntary, nonremunerated, adult donors who provided written informed consent as part of routine blood collection procedures of the Sanquin Blood Supply Foundation (Blood Bank). The study was approved by the Ethics Advisory Council of Sanquin Blood Supply Foundation.
Blood was collected at least 2 weeks after recovery. Peripheral blood mononuclear cells (PBMCs) were isolated from heparinised peripheral blood or buffy coats by Ficoll‐Paque separation, and plasma was collected for serology and granulocytes for HLA‐I typing.
Serology
Anti‐RBD, spike and nucleocapsid antibody titres were quantified by ELISA, as described previously. 53 , 54 , 55 In short, the viral antigens were coated overnight at 4°C on MaxiSorp microtitre plates (Thermo Fisher Scientific, Landsmeer, the Netherlands). Plasma samples were diluted 1200‐ and/or 3600‐fold in PBS supplemented with 0.1% polysorbate‐20 and 0.3% gelatin (PTG) and were subsequently incubated on the antigen‐coated plate for 1 h at room temperature (RT). After washing, 0.5 μg mL−1 of HRP‐conjugated anti‐human IgG (MH16‐1; Sanquin, Amsterdam, the Netherlands) was added in PTG and incubated for 30 min at RT, after which the plated was washed. The enzymatic conversion of TMB substrate was monitored, and absorbance was measured at 450 and 540 nm. The signals were quantified using a serially diluted calibrator consisting of a reference plasma pool of previously confirmed convalescent COVID‐19 patients that were included on each plate. This calibrator was arbitrarily assigned a value of 100 AU mL−1, and seroconversion threshold of IgG was set at 4 AU mL−1 levels as determined by using preoutbreak samples. 53 , 54 Since plasma samples of donors D09 and D29 were collected after PBS dilution and the Ficoll centrifugation, ELISA results were multiplied accordingly to the predilution of the sample.
HLA‐I‐typing and coverage
HLA class I genotyping was performed on genomic DNA extracted from the granulocytes using the QIAamp DNA mini‐Kit (Qiagen, Hilden, Germany) by the department of Immunogenetics Sanquin Diagnostiek B.V.
Allelic frequencies were calculated by dividing the total number of a specific allele of interest (HLA‐A*01:01, HLA‐A*02:01, HLA‐A*03:01, HLA‐A*11:01, HLA‐A*24:02, HLA‐B*07:02, HLA‐B*08:01, HLA‐B*15:01, HLA‐B*27:05, HLA‐B*35:01 and HLA‐B*40:01) observed in the 51 HLA‐typed donors by the total number of all allele copies of the associated genetic locus (alleles of interest/2n). The HLA‐I frequencies of our cohort were compared with the general Dutch populations (Allele Frequency Net Database, 34 Netherlands Leiden n = 1305, accessed on 3 December 2020).
SARS‐CoV‐2 epitopes
SARS‐CoV‐2‐specific CD8+ T cell peptides, 8–11 amino acids in length, were selected based on the ancestral Wuhan‐Hu‐1 proteome of which sequences were obtained from National Center for Biotechnology Information (NCBI) database (Spike YP_009724390.1, Membrane YP_009724393.1, Nucleocapsid YP_009724397.2, Envelop YP_009724392.1, ORF1ab YP_009724389.1, ORF3a YP_009724391.1, ORF6 YP_009724394.1 and ORF7a protein YP_009724395.1). Epitope predictions were based on NetMHC‐4.0 and/or NetMHCpan‐4.1, using their default settings, and 126 strong (< 0.5) and weak binding (0.5–2) predicted epitopes were identified covering 120 unique peptides, 47 of which had already been confirmed by the scientific community at the start of this study. Another seven peptides were selected based on their high homology (> 75%) with SARS‐CoV‐1 and/or seasonal human coronaviruses (Supplementary table 2). All 127 peptides (JPT, Berlin, Germany) were tested on actual binding to their predicted HLA‐I allotype of interest, by in vitro binding assays. Peptides with ≥ 50% binding avidity relative to the HLA‐I allotype control peptide or previously confirmed as CD8+ T cell epitope in other studies with lower < 50% binding avidity were selected for HTCC.
Generation of combinatorial encoded pHLA‐I tetramers
HLA‐I complexes with UV‐cleavable peptides were generated in‐house by the Reagents department of Sanquin, as described previously. 56 In short, recombinant HLA‐A*01:01, A*02:01, A*03:01, A*11:01, A*24:02, B*07:02, B*08:01, B*15:01, B*27:05, B*35:01 and B*40:01 heavy chains and the B2M light chain were produced in Escherichia coli. pHLA‐I complexes were formed by combining heavy chain, light chain and UV‐cleavable peptide, 57 and purified by gel‐filtration high‐performance liquid chromatography (HPLC). After biotinylation, pHLA‐I complexes were stored at −20°C until use. UV‐mediated exchanges, by subjecting the pHLA complex to 366 nm UV light, created SARS‐CoV‐2‐specific pHLA‐I complexes. 56 HLA‐I tetramers were generated by conjugating 10 different fluorescent streptavidin conjugates [PE, PE‐Cy7, APC (Thermo Fisher), BUV661, BUV737, BV421, BV605, BV711, PE‐CF594 (BD bioscience, Vianen, the Netherlands) and BV785 (Biolegend, Amsterdam, the Netherlands)] to the SARS‐CoV‐2‐specific pHLA‐I complexes. 58 The UV‐exchange and combinatorial coding techniques are patent‐protected in Europe, the US and other countries WO 2010/060439 and WO 2006/080837. Due to high sequence similarity of some peptides, several pHLA tetramer combinations could not be tested simultaneously, in those cases peptides with higher in vitro binding avidity and/or those previously described as CD8+ T cell epitopes by us or others were preferentially selected. Those three peptides are identified in Supplementary table 2. Altogether, CD8+ T cell analysis was performed for 93 epitopes.
Flow cytometry assay
For flow cytometry analysis, donors with overlapping HLA‐I allotypes were grouped and one donor (D51) was excluded because of technical complications (Supplementary table 1). PBMCs were thawed in RPMI 1640 (Life Technologies; 21875‐034) supplemented with 10% FCS (Bondinco, Alkmaar, the Netherlands), 1% penicillin–streptomycin (Sigma, Zwijndrecht, the Netherlands), 1% l‐glutamine (Sigma) and 1:1000 DNase (Worthington Biochemical Corporation, Lakewood, USA; cat. LS002140, 10 mg mL−1). PBMCs were washed twice in MACS buffer (0.5% BSA and 2 mm EDTA in PBS), after which 4–8 million cells per donor were resuspended in FACS‐buffer [0.5% BSA (Sigma; cat. A7030) 0.1% NaN3 in PBS, 0.2 μm filtered (Whatman, Medemblik, the Netherlands)]. HLA‐I tetramer pools were generated to stain with up to 30 different peptide‐HLA‐I tetramers simultaneously, in the presence of Brilliant Staining Buffer Plus (BD Bioscience; cat. 566385) according to the manufacturer's instructions. After incubating cells with HLA‐I tetramers for 30 min on ice, the antibody mix was added, and cells were incubated for an additional 30 min on ice. Antibody mix contained: AF700 anti‐human CD3 (clone UCHT1; BD Bioscience; cat. 557943), FITC anti‐human CD8 (clone SK1; BD Bioscience; cat. 345772), BUV395 anti‐human CD45RA (clone HI1000; BD Bioscience; cat. 740298), BV510 anti‐human CD27 (clone O323; BD Bioscience; cat. 751672) and Near‐IR‐Dye (Invitrogen, Carlsbad, USA; cat. L10119). Subsequently, cells were washed twice and fixated with IntraStain (Agilent Dako, Santa Clara, USA; cat. K231111‐2) following the manufacturer's instructions. Next, cells were washed twice and resuspended in FACS‐buffer for acquisition on the BD FACSymphony™ A5 with FACSDiva software (BD Biosciences). Data were analysed using FlowJo (v10.8.1; Treestar, Ashland, USA). The threshold to identify tetramer+CD8+ T cells as antigen‐specific was ≥ 3 double‐tetramer‐positive cells. A threshold of ≥ 9 tetramer+CD8+ T cells was used for phenotypic characterisation.
Amino acid sequence identity
Amino acid sequence identity of the viral peptides of confirmed SARS‐CoV‐2 epitopes across the different VOCs, Alpha, Beta, Gamma, Delta and Omicron was determined using outbreak.info, 37 accessed on 15 August 2022. Mutations are reported in Table 1 when the mutation was detected in ≥ 1% of the total sequences in the database for one of the VOCs.
Statistics
All statistical analyses were performed on GraphPad Prism (v9.1.1). The assumed nonparametric datasets (two‐tailed) were tested for statistical significance with the Mann–Whitney U‐test (unpaired) or the Wilcoxon (paired), comparing two groups at a time. Two‐tailed simple linear regression analysis was performed with 95% confidence interval. Differences were considered significant if P ≤ 0.05.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Jet van den Dijssel: Formal analysis; investigation; methodology; validation; visualization; writing – original draft; writing – review and editing. Ruth R Hagen: Formal analysis; investigation; writing – review and editing. Rivka de Jongh: Data curation; writing – review and editing. Maurice Steenhuis: Formal analysis; investigation; methodology; writing – review and editing. Theo Rispens: Formal analysis; investigation; methodology; writing – review and editing. Dionne M Geerdes: Formal analysis; investigation; methodology; writing – review and editing. Juk Yee Mok: Formal analysis; investigation; methodology; writing – review and editing. Angela HM Kragten: Formal analysis; investigation; methodology; writing – review and editing. Mariël C Duurland: Investigation; writing – review and editing. Niels JM Verstegen: Formal analysis; validation; writing – review and editing. S Marieke van Ham: Data curation; writing – review and editing. Wim JE van Esch: Conceptualization; formal analysis; investigation; methodology; validation; writing – review and editing. Klaas PJM van Gisbergen: Validation; writing – original draft; writing – review and editing. Pleun Hombrink: Conceptualization; writing – review and editing. Anja ten Brinke: Data curation; writing – review and editing. Carolien E van de Sandt: Conceptualization; formal analysis; investigation; methodology; supervision; validation; visualization; writing – original draft; writing – review and editing.
Supporting information
Supplementary figures 1–5
Supplementary tables 1–2
Acknowledgments
We thank all donors who participated in the study, the Sanquin COVID‐19 cryo and biobank facility for processing of samples and the Sanquin Core Facility: Erik Mul, Simon Tol and Mark Hoogenboezem for providing technical assistance. CES has received funding from the European Union's Horizon 2020 research, innovation programme under the Marie Skłodowska‐Curie grant agreement (#792532).
References
- 1. Johns Hookins University . COVID‐19 Dashboard. 2022. [cited March 31 2022]. Available from: https://coronavirus.jhu.edu/map.ht
- 2. Williamson EJ, Walker AJ, Bhaskaran K et al. Factors associated with COVID‐19‐related death using OpenSAFELY. Nature 2020; 584: 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rydyznski Moderbacher C, Ramirez SI, Dan JM et al. Antigen‐specific adaptive immunity to SARS‐CoV‐2 in acute COVID‐19 and associations with age and disease severity. Cell 2020; 183: 996–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weiskopf D, Immunol S, Weiskopf D et al. Phenotype and kinetics of SARS‐CoV‐2‐specific T cells in COVID‐19 patients with acute respiratory distress syndrome. Sci Immunol 2020; 5: eabd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schub D, Klemis V, Schneitler S et al. High levels of SARS‐CoV‐2–specific T cells with restricted functionality in severe courses of COVID‐19. JCI Insight 2020; 5: e142167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thevarajan I, Nguyen THO, Koutsakos M et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non‐severe COVID‐19. Nat Med 2020; 26: 453–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koutsakos M, Rowntree LC, Hensen L et al. Integrated immune dynamics define correlates of COVID‐19 severity and antibody responses. Cell Rep Med 2021; 2: 100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mathew D, Giles JR, Baxter AE et al. Deep immune profiling of COVID‐19 patients reveals distinct immunotypes with therapeutic implications. Science 2020; 369: eabc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sekine T, Perez‐Potti A, Rivera‐Ballesteros O et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID‐19. Cell 2020; 183: 158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu J, Yu J, Mcmahan K et al. CD8 T cells contribute to vaccine protection against SARS‐CoV‐2 in macaques. Sci Immunol 2022: eabq7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tan AT, Linster M, Tan CW et al. Early induction of functional SARS‐CoV‐2‐specific T cells associates with rapid viral clearance and mild disease in COVID‐19 patients. Cell Rep 2021; 34: 108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jung JH, Rha MS, Sa M et al. SARS‐CoV‐2‐specific T cell memory is sustained in COVID‐19 convalescent patients for 10 months with successful development of stem cell‐like memory T cells. Nat Commun 2021; 12: 4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adamo S, Michler J, Zurbuchen Y et al. Signature of long‐lived memory CD8+ T cells in acute SARS‐CoV‐2 infection. Nature 2021; 602: 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geers D, Shamier MC, Bogers S et al. SARS‐CoV‐2 variants of concern partially escape humoral but not T‐cell responses in COVID‐19 convalescent donors and vaccinees. Sci Immunol 2021; 6: eabj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tarke A, Sidney J, Methot N et al. Impact of SARS‐CoV‐2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med 2021; 2: 100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. GeurtsvanKessel CH, Geers D, Schmitz KS et al. Divergent SARS CoV‐2 Omicron‐reactive T‐ and B cell responses in COVID‐19 vaccine recipients. Sci Immunol 2022; 7: eabo2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaebler C, Wang Z, Lorenzi JCC et al. Evolution of antibody immunity to SARS‐CoV‐2. Nature 2021; 591: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma H, Zeng W, He H et al. Serum IgA, IgM, and IgG responses in COVID‐19. Cell Mol Immunol 2020; 17: 773–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tarke A, Sidney J, Kidd CK et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS‐CoV‐2 epitopes in COVID‐19 cases. Cell Rep Med 2021; 2: 100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keeton R, Tincho MB, Ngomti A et al. T cell responses to SARS‐CoV‐2 spike cross‐recognize Omicron. Nature 2022; 603: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Collier DA, Ferreira IATM, Kotagiri P et al. Age‐related immune response heterogeneity to SARS‐CoV‐2 vaccine BNT162b2. Nature 2021; 596: 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sahin U, Muik A, Vogler I et al. BNT162b2 vaccine induces neutralizing antibodies and poly‐specific T cells in humans. Nature 2021; 595: 572–577. [DOI] [PubMed] [Google Scholar]
- 23. Minervina AA, Pogorelyy MV, Kirk AM et al. SARS‐CoV‐2 antigen exposure history shapes phenotypes and specificity of memory CD8+ T cells. Nat Immunol 2022; 23: 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oberhardt V, Luxenburger H, Kemming J et al. Rapid and stable mobilization of CD8+ T cells by SARS‐CoV‐2 mRNA vaccine. Nature 2021; 597: 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rowntree LC, Petersen J, Juno JA et al. SARS‐CoV‐2‐specific CD8+ T‐cell responses and TCR signatures in the context of a prominent HLA‐A*24:02 allomorph. Immunol Cell Biol 2021; 99: 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schulien I, Kemming J, Oberhardt V et al. Characterization of pre‐existing and induced SARS‐CoV‐2‐specific CD8+ T cells. Nat Med 2020; 27: 78–85. [DOI] [PubMed] [Google Scholar]
- 27. Ferretti AP, Kula T, Wang Y et al. Unbiased screens show CD8+ T cells of COVID‐19 patients recognize shared epitopes in SARS‐CoV‐2 that largely reside outside the spike protein. Immunity 2020; 53: 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saini SK, Hersby DS, Tamhane T et al. SARS‐CoV‐2 genome‐wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID‐19 patients. Sci Immunol 2021; 6: eabf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kared H, Redd AD, Bloch EM et al. SARS‐CoV‐2‐specific CD8+ T cell responses in convalescent COVID‐19 individuals. J Clin Invest 2021; 131: e145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gangaev A, Ketelaars SLC, Isaeva OI et al. Identification and characterization of a SARS‐CoV‐2 specific CD8+ T cell response with immunodominant features. Nat Commun 2021; 12: 2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nguyen THO, Rowntree LC, Petersen J et al. CD8+ T cells specific for an immunodominant SARS‐CoV‐2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity. Immunity 2021; 54: 1066–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rowntree LC, Nguyen THO, Kedzierski L et al. SARS‐CoV‐2‐specific T cell memory with common TCRαβ motifs is established in unvaccinated children who seroconvert after infection. Immunity 2022; 55: 1299–12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Habel JR, Nguyen THO, van de Sandt CE et al. Suboptimal SARS‐CoV‐2−specific CD8+ T cell response associated with the prominent HLA‐A*02:01 phenotype. Proc Natl Acad Sci USA 2020; 117: 24384–24391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gonzalez‐Galarza FF, McCabe A, Dos Santos EJM et al. Allele frequency net database (AFND) 2020 update: gold‐standard data classification, open access genotype data and new query tools. Nucleic Acids Res 2020; 48: D783–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T‐cell epitope‐based diagnostics and vaccines. BMC Bioinformatics 2006; 7: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dhanda SK, Vita R, Ha B, Grifoni A, Peters B, Sette A. ImmunomeBrowser: a tool to aggregate and visualize complex and heterogeneous epitopes in reference proteins. Bioinformatics 2018; 34: 3931–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mullen JL, Tsueng G, Latif AA et al. Outbreak.info. 2020. [cited August 15 2022]. Available from: https://outbreak.info/
- 38. Dolton G, Rius C, Hasan MS et al. Emergence of immune escape at dominant SARS‐CoV‐2 killer T cell epitope. Cell 2022; 185: 2936–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS‐CoV‐2. Cell Host Microbe 2020; 27: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Agerer B, Koblischke M, Gudipati V et al. SARS‐CoV‐2 mutations in MHC‐I‐restricted epitopes evade CD8+ T cell responses. Sci Immunol 2021; 6: eabg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Francis JM, Leistritz‐Edwards D, Dunn A et al. Allelic variation in class I HLA determines CD8+ T cell repertoire shape and cross‐reactive memory responses to SARS‐CoV‐2. Sci Immunol 2022; 7: eabk3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nelde A, Bilich T, Heitmann JS et al. SARS‐CoV‐2‐derived peptides define heterologous and COVID‐19‐induced T cell recognition. Nat Immunol 2020; 22: 74–85. [DOI] [PubMed] [Google Scholar]
- 43. Deb P, Zannat K, Talukder S et al. Association of HLA gene polymorphism with susceptibility, severity, and mortality of COVID‐19: a systematic review. HLA 2022; 99: 281–312. [DOI] [PubMed] [Google Scholar]
- 44. Peng Y, Mentzer AJ, Liu G et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS‐CoV‐2 in UK convalescent individuals following COVID‐19. Nat Immunol 2020; 21: 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. CDC COVID‐19 Response Team . Severe outcomes among patients with coronavirus disease 2019 (COVID‐19) – United States, February 12‐March 16, 2020. MMWR Morb Mortal Wkly Rep 2020; 69: 343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Silva TI, Liu G, Lindsey BB et al. The impact of viral mutations on recognition by SARS‐CoV‐2 specific T cells. iScience 2021; 24: 103353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hoffmann M, Krüger N, Schulz S et al. The Omicron variant is highly resistant against antibody‐mediated neutralization: implications for control of the COVID‐19 pandemic. Cell 2022; 185: 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Naranbhai V, Nathan A, Kaseke C et al. T cell reactivity to the SARS‐CoV‐2 Omicron variant is preserved in most but not all individuals. Cell 2022; 185: 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van de Sandt CE, Kreijtz JHCM, Rimmelzwaan GF. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses 2012; 4: 1438–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van de Sandt CE, Kreijtz JHCM, Geelhoed‐Mieras MM et al. Differential recognition of influenza A viruses by M158‐66 epitope‐specific CD8+ T cells is determined by extraepitopic amino acid residues. J Virol 2015; 90: 1009–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pack SM, Peters PJ. SARS‐CoV‐2–specific vaccine candidates; the contribution of structural vaccinology. Vaccine 2022; 10: 236–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang S, Xu X, Wei C et al. Molecular evolutionary characteristics of SARS‐CoV‐2 emerging in the United States. J Med Virol 2022; 94: 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Steenhuis M, van Mierlo G, Derksen NIL et al. Dynamics of antibodies to SARS‐CoV‐2 in convalescent plasma donors. Clin Transl Immunology 2021; 10: e1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vogelzang EH, Loeff FC, Derksen NIL et al. Development of a SARS‐CoV‐2 total antibody assay and the dynamics of antibody response over time in hospitalized and nonhospitalized patients with COVID‐19. J Immunol 2020; 205: 3491–3499. [DOI] [PubMed] [Google Scholar]
- 55. Larsen MD, de Graaf EL, Sonneveld ME et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID‐19 severity. Science 2021; 371: eabc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rodenko B, Toebes M, Hadrup SR et al. Generation of peptide‐MHC class I complexes through UV‐mediated ligand exchange. Nat Protoc 2006; 1: 1120–1132. [DOI] [PubMed] [Google Scholar]
- 57. Garboczi DN, Hung DT, Wiley DC. HLA‐A2‐peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci USA 1992; 89: 3429–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hadrup SR, Bakker AH, Shu CJ et al. Parallel detection of antigen‐specific T‐cell responses by multidimensional encoding of MHC multimers. Nat Methods 2009; 6: 520–526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures 1–5
Supplementary tables 1–2


