Abstract
In order to maintain global food security, it will be necessary to increase yields of the cereal crops that provide most of the calories and protein for the world’s population, which includes common wheat (Triticum aestivum L.). An important wheat yield component is the number of grain-holding spikelets which form on the spike during inflorescence development. Characterizing the gene regulatory networks controlling the timing and rate of inflorescence development will facilitate the selection of natural and induced gene variants that contribute to increased spikelet number and yield. In the current study, co-expression and gene regulatory networks were assembled from a temporal wheat spike transcriptome dataset, revealing the dynamic expression profiles associated with the progression from vegetative meristem to terminal spikelet formation. Consensus co-expression networks revealed enrichment of several transcription factor families at specific developmental stages including the sequential activation of different classes of MIKC-MADS box genes. This gene regulatory network highlighted interactions among a small number of regulatory hub genes active during terminal spikelet formation. Finally, the CLAVATA and WUSCHEL gene families were investigated, revealing potential roles for TtCLE13, TtWOX2, and TtWOX7 in wheat meristem development. The hypotheses generated from these datasets and networks further our understanding of wheat inflorescence development.
Subject terms: Gene regulatory networks, Transcriptomics, Plant development, Shoot apical meristem
Introduction
The world population is expected to exceed 9 billion people by 2050, signaling that further increases in grain production will be required to ensure food security1. Because there remain few opportunities to expand arable land area, increasing the yield of major cereal crops through genetic improvement will be critical to meet this goal. In common wheat (Triticum aestivum L.) characterizing the genetic pathways regulating grain size and grain number will facilitate the rational combination of superior alleles in wheat breeding programs to help drive continued yield improvements2.
Grain number in wheat is determined to a large extent by inflorescence architecture and is influenced by the environment and genotype-environment interactions. By integrating photoperiod and temperature cues, the vegetative shoot apical meristem (SAM) transitions to the reproductive inflorescence meristem (IM), during which the developing spike passes through the characteristic double ridge (DR) stage, forming a lower leaf ridge and an upper spikelet ridge3. The lower leaf ridge is repressed by the MIKC-MADS box transcription factors (TFs) VRN1, FUL2 and FUL34, whereas the upper ridges develop glumes, lemmas, and floret primordia. As the IM elongates, spikelet meristems are added at the growing apex, while basal spikelets continue to develop. Wheat spikes are determinate structures and the addition of lateral spikelets ends when the terminal spikelet is formed. Therefore, spikelet number is determined by the timing and rate of meristem development preceding terminal spikelet formation. Each spikelet has the potential to form between 3 and 6 grains5 and spikelet number is correlated with grain number and yield6–8.
Shoot meristems are organized around the organizing center and stem cell maintenance is governed by the conserved CLAVATA-WUSCHEL negative feedback loop9. In Arabidopsis, the homeodomain TF WUS induces CLV3, which encodes a secreted peptide that forms receptor complexes repressing WUS10. Manipulation of this pathway confers variation in locule number in tomato (Solanum lycopersicum) and kernel row number in maize (Zea mays)11,12. The wheat genome contains 104 CLAVATA3/EMBRYO SURROUNDING REGION (CLE) peptides13 and 44 WUSCHEL RELATED HOMEOBOX (WOX) TFs14, but the specific ones regulating inflorescence meristem development in wheat are yet to be identified.
Inflorescence development is controlled by a complex regulatory network involving multiple classes of TFs which orchestrate rapid and dynamic changes in gene expression. The Type ΙΙ MIKC MADS-box TFs play critical roles in flower development across the angiosperms and can be divided into A, B, C, D and E-classes that interact mainly as tetrameric complexes in a spatially regulated manner to direct sepal (A- and E-), petal (A-, B-, E-), stamen (B-, C-, E-), and carpel development (C- and E-class genes)15,16. This family expanded during cereal evolution and the hexaploid wheat genome contains 201 MIKC MADS-box genes, classified into 15 phylogenetic subclades17.
The SHORT VEGETATIVE PHASE (SVP) subclade members SVP1, VRT2, and SVP3 promote the transition from the vegetative SAM to the IM, along with the AP1/SQUA subclade genes VRN1, FUL2 and FUL34,18. Subsequently, AP1/SQUA genes suppress the expression of SVP genes, which may be required to promote interactions between AP1/SQUA proteins and the E-class MIKC-MADS proteins SEPELLATA1 (SEP1) and SEP3, which are predominantly expressed in floral organogenesis during early reproductive growth18. The natural VRT2pol allele from Triticum polonicum exhibits ectopic expression and is associated with elongated glumes and increased grain length19. VRT2-overexpression lines show reduced transcript levels of B-class (PI and AP3) and C-class (AG1 and AG2) MIKC-MADS box genes, although the role of these latter subclades in wheat inflorescence development remains to be characterized18.
Although much has been learned about wheat inflorescence development from positional cloning, reverse genetics and comparative genetic approaches, we lack a full understanding of the regulatory networks controlling meristem determinacy and developmental transitions. Only a fraction of the hundreds of QTL for thousand kernel weight, kernel number per spike, and spikelet number have been cloned and validated to date, indicating that a large proportion of quantitative variation in these traits remains uncharacterized7.
Transcriptomics provides a complementary approach to characterize the regulatory networks underlying inflorescence development that is empowered by an expanding set of wheat genomic resources20,21. Co-expression and gene regulatory networks (GRNs) are powerful tools to interpret temporal correlation and causal relationships between genes, and to help identify critical hub genes that coordinate development22,23. Previous transcriptomic studies in wheat inflorescence tissues described the differential expression profiles of thousands of genes during vegetative and floral meristem development, including the stage-specific expression of different TFs and hormone biosynthesis and signaling genes24,25. A population-associative transcriptomic approach was used to identify regulators of wheat spike architecture, including CEN2, TaPAP2/SEP1-6, and TaVRS1/HOX2, which were validated in functional studies26.
In the current study, a series of co-expression and gene regulatory networks were assembled to characterize the predominant transcriptional profiles associated with the progression of wheat inflorescence development, revealing 2 consecutive regulatory shifts at the DR and TS stages. Core regulatory candidate genes were identified including both known TFs and novel candidates with potential roles in regulating spike architecture.
Results
Early wheat inflorescence development is defined by 2 major transcriptional shifts
To characterize the wheat transcriptome during inflorescence development, RNA was sequenced from tetraploid durum wheat meristem tissue at 5 developmental stages; vegetative meristem (W1), double ridge (W2), glume primordium (W3.0), lemma primordium (W3.25), and terminal spikelet (W3.5) (Fig. 1A)3. An average of 28.9 M reads per sample (79.6% of all reads) mapped uniquely to the A and B genomes, and unanchored scaffolds from the IWGSC RefSeqv1.0 assembly. Of the 190,391 gene models on these chromosomes, 82,019 (43.1%) were expressed (> 0 Transcripts per Million (TPM)) and 45,243 (23.8%) had a mean expression greater than 1 TPM in at least 1 timepoint (Supplementary Data 1). Of the 3861 gene models annotated as TFs (2.0% of gene models), 2874 (74.5%) were expressed (> 0TPM) and 1703 (44.1%) had a mean expression greater than 1 TPM in at least 1 timepoint (Supplementary Data 2).
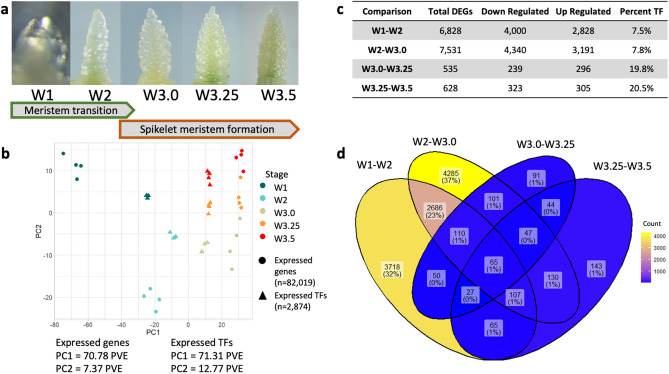
Figure 1.
The early wheat inflorescence development transcriptome. (a) Sampling stages of Kronos apical meristems according to the Waddington development scale3; W1—vegetative meristem, W2—double ridge, W3.0—glume primordium, W3.25—lemma primordium, W3.5—terminal spikelet. (b) Whole transcriptome and transcription factor expression principal component analysis of samples, PC1 plotted on the x-axis and PC2 plotted on the y-axis. PVE-percent variance explained. (c) Differentially expressed genes (DEGs) in sequential pairwise comparisons (W1–W2, W2–W3.0, W3.0–W3.25, W3.25–W3.5). The total number of genes, the number up- and down-regulated and the proportion encoding transcription factors (TF) are described. (d) Venn diagram of DEGs in each consecutive pairwise comparison from (c). Each category is shaded according to the number of sequential DEGs shared among the 4 comparisons.
Comparison of the tetraploid ‘Kronos’ inflorescence development transcriptome with 2 whole-plant hexaploid wheat development transcriptome datasets27,28 revealed 3682 genes with spike-dominant expression profiles (τ > 0.9, where 0 means constitutive expression and 1indicates tissue-specific expression) (Supplementary Data 3). These genes were most strongly enriched for gene ontology (GO) terms relating to histone assembly and chromosome organization (Supplementary Data 4), but also included 286 genes (7.8%) encoding TFs, including both LEAFY homoeologs, 15 GROWTH REGULATING FACTOR (GRF) TFs (of 20 expressed during the time course), 7 SHI RELATED SEQUENCE (SRS) TFs (out of 10), 20 TCP TFs (out of 49) and 10 WOX TFs (out of 28, Supplementary Data 3). Despite their known roles in regulating inflorescence development, only 2 out of 130 MIKC-MADS box and 6 out of 41 SPL TFs exhibited spike-dominant expression profiles, suggesting they play more diverse roles across plant development. There were 86 spike-specific genes with 0 expression in all other stages of development (τ = 1), including putative bHLH and MYB TFs and TE-related orthologs of DAYSLEEPER family genes (Supplementary Data 3). It is interesting to note that 31.7% of spike-dominant and 75.6% of spike-specific genes were annotated as low-confidence gene models, indicating they may represent less-studied genes (Supplementary Data 3).
Principal component analysis (PCA) using the whole transcriptome grouped the 4 biological replicates of each growth stage closely together and revealed that the transition from the vegetative meristem to double ridge formation is associated with a major transcriptional shift (Fig. 1B). These changes are described by PC1, which accounted for 71.8 percent variation explained (PVE). The transition from W1 to W2 was associated with 6828 DEGs, 58.6% of which were downregulated (Fig. 1C, Supplementary Data 5) and most significantly enriched for GO terms relating to “cell wall organization” (GO:0071554), including at least 16 genes predicted to encode cellulose synthase enzymes, and lignin and hemicellulose metabolic processes (Supplementary Data 6). Surprisingly, the 2828 (41.4%) DEGs upregulated between W1 and W2 were most significantly enriched for GO terms relating to photosynthesis (GO:0015979) despite the transition from leaf to floral meristem development (Supplementary Data 5). For example, genes encoding Photosystem I reaction center subunit IV (TraesCS5B02G496000, TraesCS5A02G482800) were upregulated sharply at stage W2.
The transition from W2 to W3.0 was associated with 7,531 DEGs (57.6% downregulated, Supplementary Data 5, 6). The 3,191 DEGs upregulated between these timepoints were most significantly enriched for “meristem maintenance” (GO:0010073) and “flower development” (GO:0009908) GO terms, including two genes encoding helicase-related family proteins (TraesCS3A02G212600, TraesCS3B02G242700) (Supplementary Data 6), suggesting that multiple genes triggering floral meristem formation are first activated at this stage.
By contrast, the second transcriptomic shift from W3.0 to terminal spikelet formation was associated with 12.3-fold fewer DEGs than during the transition from the vegetative meristem to stage W3.0 (Fig. 1C). These changes were distributed across PC2, which accounts for just 7.4 PVE (Fig. 1B). Just 535 DEGs were found between W3.0 and W3.25 (55.3% upregulated) and 628 DEGs between W3.25 and W3.5 (48.6% upregulated) (Supplementary Data 5). Genes upregulated across these 3 timepoints were most significantly enriched for “floral organ identity” (GO:0048437) and included PISTILLA-like MADS-box genes (TraesCS1A02G264300, TraesCS1B02G275000) (Supplementary Data 6). There are fewer developmental changes between W3.25 and W3.5, relative to changes between W1 and W3.0, which may be due in part to basal and apical spikelets being at similar developmental stages between the latter timepoints29.
Of the 11,669 DEGs in at least 1 of the 4 consecutive pairwise comparisons, 899 (7.7%) encoded a TF, representing a 2.2-fold enrichment (hypergeometric P = 2.22e−62). This enrichment was strongest after DR through terminal spikelet formation (5.2-fold enrichment, P = 8.73e−73) where TFs accounted for 19.8% and 20.5% of all DEGs in pairwise comparisons (Fig. 1C). A PCA using only TF expression resulted in the same spatial arrangement of biological samples as in the whole-transcriptome PCA but with improved resolution between stages (Fig. 1B), and explained a greater proportion of variation for PC2 than when including the whole transcriptome (Fig. S1).
Taken together, these analyses show that less than half of the wheat transcriptome but nearly three-quarters of TFs are expressed during inflorescence development, including a set of genes which are spatially and temporally restricted to early inflorescence tissues. The transcriptomic shift that occurs during terminal spikelet formation is associated with comparatively less transcriptional variation relative to stages preceding W3.25 and the strong enrichment in TFs suggests they play critical roles during this stage.
Co-expression networks reveal predominant transcriptome profiles during inflorescence development
Co-expression networks were assembled to identify highly correlated modules of genes that define the major transcriptional profiles during early inflorescence development. To prioritize genes more likely to regulate inflorescence development, all networks were assembled using a set of 22,566 genes that were differentially expressed in at least one of the 10 possible pairwise combinations between timepoints (Fig. 1D) and that were also defined as significantly differentially expressed using Impulse DE2, a package used to analyze longitudinal transcriptomic datasets (Supplementary Data 5).
A consensus network constructed with repeated subsampling and randomized parameters with WGCNA (see “Materials and methods” section) assembled these genes into 21 modules with a mean connectivity score of 0.485 (Fig. 2A, Supplementary Data 7). A standard WGCNA network was also constructed using ‘best practices’ parameters but with no repeated subsampling and randomization and had a connectivity score of 0.327 which skewed to zero (Fig. 2A). In both networks, the majority of genes clustered into modules 1 and 2, which contained many of the same genes (Jaccard index > 0.86, Fig. S2). However, other modules exhibited dissimilar expression profiles between networks (Jaccard index < 0.5), indicating the consensus network clustered genes into a greater number of modules with distinct expression profiles not captured in the standard network. Based on the improved correlation of co-clustered genes within modules and the detection of distinct regulatory profiles, the consensus network was used in all subsequent analyses.
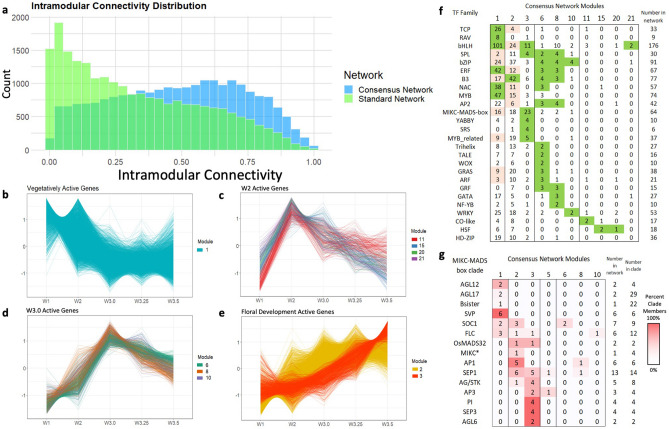
Figure 2.
Co-expression networks showing the dominant transcriptional profiles during wheat inflorescence development. (a) Histogram of intramodular connectivity scores for 22,566 genes clustered in consensus (blue) or standard (green) network. (b–e) Expression profiles during inflorescence development of discussed modules in the consensus network. Lines represent scaled time course expression of each gene in the module. Modules with similar expression profiles are grouped together for comparison. (f) Number of TF family members clustered in each discussed consensus module. Modules enriched (green) or depleted (pink) for TF families are highlighted (P < 0.01). (g) Number of MIKC-MADS box clade members clustered in each consensus modules. Co-expressed MIKC-MADS box groups are shaded relative to the total number of genes in the clade.
Inflorescence meristem development is associated with the down-regulation of RAV and TCP transcription factors
Module 1 was the largest in the network and grouped 10,102 genes defined by high transcript levels in the vegetative meristem and early meristem transition followed by down-regulation after DR and as the spike develops (Fig. 2B). Several TF families were enriched in this module, including 101 basic Helix-Loop-Helix (bHLH) TFs, 47 MYB TFs and 8 of the 9 differentially expressed RELATED TO ABI3 AND VP1 (RAV) TFs included in the network (Fig. 2F). Twenty-six of the 33 total TCP TFs clustered in this module, 9 of which were also spike-dominant expressed (Fig. 2F). Although at the whole family level MIKC-MADS TFs are significantly under-represented in module 1 (Fig. 2F, hypergeometric P = 8.6e−4), all 6 SVP genes (A and B homoeologs of SVP1, VRT2 and SVP3) cluster in this module, consistent with their specific role regulating early stages of inflorescence development. In addition, both AGL12 subclade genes, and 3 of the 6 FLC subclade genes clustered in this module (Fig. 2G).
A small number of genes are transiently expressed during double ridge formation
Genes which showed a peak at the double ridge stage (W2) followed by a decline in later stages were clustered in modules 11 (131 genes), 15 (104 genes), 20 (44 genes) and 21 (42 genes). These clusters share broadly similar expression profiles (Fig. 2C) and were enriched for genes with spike-dominant expression profiles (between 2.1 and 3.0-fold enrichment). Genes in modules 15 and 20 were significantly enriched for development functional terms including “shoot system development” and “carpel development” (Supplementary Data 8) including 3 TERMINAL FLOWER1-like genes CENTRORADIALIS2 (CEN2), CEN4, and CEN-5A (Supplementary Data 7). All 3 modules were enriched for the functional term “response to auxin” and included several auxin-responsive factors (ARF), indole-3 acetic acid (IAA), and SAUR-like protein family members, indicating that auxin signaling may promote double ridge formation.
Inflorescence transition and spike architecture genes are upregulated at W3.0
Modules 6 (267 genes), 8 (211 genes), and 10 (144 genes) share broadly similar profiles defined by maximum expression at stage W3.0 and subsequent downregulation (Fig. 2D). Each of these modules was significantly enriched (between 2.3 and 5.3-fold) for spike-dominant genes, indicating they likely play highly specific roles restricted to developing meristems and inflorescence initiation. Module 6 included 18 genes previously associated with variation in spikelet number and 5 orthologs of rice genes with roles in panicle development, including the ERF TF WHEAT FRIZZY PANICLE (WFZP) and KAN2, a MYB TF which functions in establishing lateral organ polarity in Arabidopsis30,31.
Inflorescence and spikelet meristem formation is associated with sequential activation of different classes of TFs
The 8971 genes in module 2 were defined by the inverse transcriptional profile to module 1, with low expression in the vegetative meristem followed by sustained upregulation from the double ridge stage onwards (Fig. 2E). Transcription factors were under-represented in this module, and only the B3 family (42 of 77 B3 TFs assembled in the co-expression network) was significantly enriched (Fig. 2F). There were 18 MIKC-MADS box TFs which were upregulated early in the transition to the inflorescence meristem including all genes in the AP1/SQUA subclade (with the exception of VRN-A1) and 6 of the 13 genes in the SEP1 subclade (Fig. 2G). Several genes with characterized roles in inflorescence development clustered in this module, including the durum wheat orthologs of FLOWERING LOCUS T2 (FT-A2), Q, and RAMOSA2 (RA-B2) (Supplementary Data 7)32,33.
The 708 genes clustered in module 3 exhibited a similar transcriptional profile to module 2, with a delayed upregulation and stronger peak at the terminal spikelet stage (Fig. 2E). These genes are significantly enriched for developmental functional terms including “specification of floral organ identity”, suggesting they include floral patterning and developmental genes that regulate spikelet meristem formation (Supplementary Data 8). This module was significantly enriched for both spike-dominant expressed genes (106 genes, P < 0.001) and for TFs (86 genes, 12.1%, P < 0.001), consistent with pairwise DE analysis between stages W3.0 and W3.5 (Fig. 1C). These included 4 members of the SRS TF family, 4 YABBY TFs, and the class Ι HD-ZIP TFs Grain Number Increase 1 (GNI1) and HOX2 (Supplementary Data 7). Transcript levels of GNI-A1 and HOX-A2 are highest at the terminal spikelet stage, consistent with their expression profiles in barley and hexaploid wheat inflorescences reported previously34,35. All members of the MIKC-MADS subclades PI, AGL6 and SEP3 were clustered in module 3, as well as 2 of the 3 AP3 subclade genes, 4 of the 5 AG/STK subclade genes and 5 SEP1 subclade genes (Fig. 2G).
Gene regulatory networks predict high-confidence interactions between transcription factors
To identify the most robust co-expression patterns, the consensus adjacency matrix used for previous co-expression analyses was filtered for genes which co-clustered with at least 1 gene every time they were co-sampled in 1000 networks assembled with variable, randomized parameters. The 18,174 genes that met this criterion were assembled into a conensus100 network consisting of 924 modules with a median size of 3 (Supplementary Data 7).
Module 9 of this network comprised 167 genes (including 32 TFs) which were most highly expressed at the terminal spike stage (Fig. S3) and significantly enriched for the GO terms “specification of floral organ identity” and “flower development” (Supplementary Data 9), suggesting it may represent a core regulatory network for wheat spikelet and/or floret development. The genes with the highest connectivity (Kw, a measure of each gene’s intramodular co-expression) in this module are SEP1-A2 and SEP1-B2, which may be related with the intermediate position of the SEP genes between the meristem identity SQUAMOSA MADS-BOX genes and the anther and carpel development MADS-box genes. This module also groups WAPO-A1, that influences spikelet number and stamen identity36 and a gene encoding an F-box protein that is a component of an SCF ubiquitin ligase that may be targeted by TB137 (Supplementary Data 7).
To predict interactions between TFs during inflorescence development, a de novo Causal Structure Inference (CSI) network was constructed using all 970 TFs from the consensus100 network. This gene regulatory network consisted of 704 genes (nodes) with 5604 predicted interactions (edges) with interaction strength (edge weight) > 0.001 (Supplementary Data 10). To prioritize the most important regulatory candidate genes, the network was screened for interactions with an edge weight ≥ 0.03, leaving 88 genes with 177 interactions. The majority of these genes were from consensus modules 1 (37 genes, 42.0%) and 3 (36 genes, 40.9%), with 27 of the latter genes clustered in consensus 100 module 9 (Fig. 3).
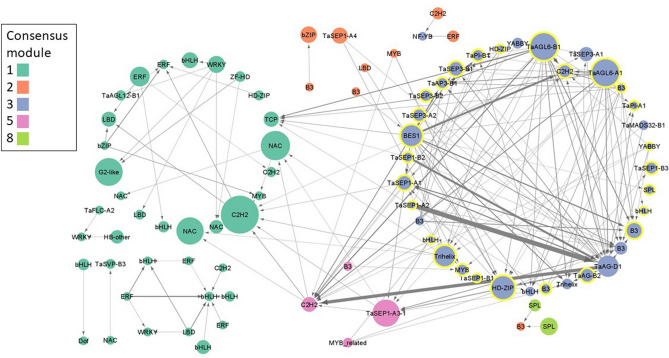
Figure 3.
Causal structural inference prediction of interacting transcription factors, filtered for edge weight ≥ 0.03. Nodes (genes) are colored by their consensus network modules, and consensus100 module 9 genes are highlighted with a yellow border. Node diameter is scaled to betweenness centrality to indicate its importance within the network. Directional interactions are indicated by arrows and width is scaled to predicted interaction strength.
Most predicted interactions were between genes in the same consensus module, with the majority occurring within module 3 and involving MIKC-MADS box TFs, suggesting a closely coordinated network during spikelet meristem and terminal spikelet formation (Fig. 3). Among the genes with the highest betweenness centrality, a measure of each gene’s importance in the overall network, were AGL6-A1 and AGL6-B1 which were predicted to interact with 31 other TFs in the network, including 13 MIKC-MADS genes such as PI-1, SEP3-1, AP3-1, SEP1-1 and AG1 (Fig. 3). Interaction strengths implicated a role for AG-D1 as a regulatory hub with strong incoming interactions from other MIKC-MADS-box genes from the SEP1, SEP3, AG, PI, and AP3 subclades, as well as outgoing interactions with genes such as the LOFSEP MIKC-MADS box TF SEP1-1 (Fig. 3). The BES1 TF BES1/BZR1 HOMOLOG 2-like had high betweenness centrality and was predicted to have outgoing interactions with MIKC-MADS and Trihelix TFs, as well as the class Ι HD-ZIP TFs GNI-A1 and HOX-A2 (Fig. 3).
Cross-module interactions included 16 outgoing edges from module 3 to module 1, including 6 outgoing interactions to a PCF-type TCP TF (Fig. 3). Although only 4 TFs from module 5 were assembled in the network, they included SEP1-A3 and a C2H2 TF with 10 incoming interactions from module 3 including AGL6-B1, BES1/BZR1 HOMOLOG 2-like and AG-D1 (Fig. 3).
Integrating transcriptomics to prioritize candidate genes underlying natural variation
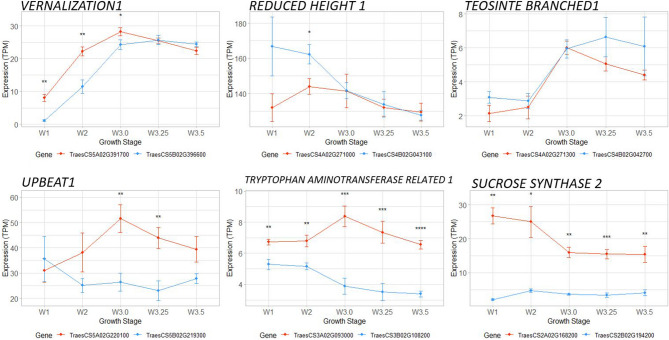
The consensus network includes 4637 high confidence homoeologous gene pairs, the majority of which (3636, 78.4%) clustered either in the same module, or in modules with highly similar expression profiles (Supplementary Data 7). We hypothesized that homoeologous genes clustering in different modules may have divergent expression profiles resulting from natural variation in 1 homoeolog. Of these 1001 divergently expressed gene pairs, 221 encoded TFs, including VRN1 (where ‘Kronos’ carries a dominant VRN-A1 spring allele with an intron 1 deletion that results in its expression at an earlier stage of inflorescence development compared to the wild-type VRN-B1 allele), RHT1 (where the Rht-B1b semi-dwarfing allele is more highly expressed in the vegetative meristem than RHT-A1), and TEOSINTE BRANCHED 1 (TB1, where TB-B1 expression is maintained at higher levels than TB-A1 during terminal spikelet formation, Fig. 4A).
Figure 4.
Divergent expression of homoeologous gene pairs during inflorescence development. Examples include the expression profiles of characterized domestication and adaptation alleles and genes close to QTL for spike architecture or grain size. Expression values are in TPM ± standard error. A-genome homoeologs are in orange, B-genome homoeologs are in blue. Paired t-tests were used to indicate differences between homoeolog expression at each time point, P values < 0.05 (*), 0.01 (**), 0.001 (***), 0.0001 (****).
Each of these 3 genes lies within 250 kb of a QTL for either grain number or grain size (Supplementary Data 7), so we hypothesized that other differentially expressed homoeologs located close to a yield-component QTL might point to natural regulatory sequence variation associated with yield traits in wheat. For example, UPBEAT-A1 is upregulated at the double ridge stage to a much greater degree than UPBEAT-B1 (Fig. 4), is close to a QTL for TKW, and encodes an ortholog of a bHLH TF that regulates cell proliferation in Arabidopsis38. Similarly, TRYPTOPHAN AMINOTRANSFERASE RELATED-A1 (TAR-A1) is also upregulated at the double ridge stage compared to TAR-B1 (Fig. 4) and is proximal to a QTL for grain yield (Supplementary Data 7). These genes encode enzymes in the IAA biosynthesis pathway and their overexpression has previously been shown to modify inflorescence development in wheat39. Co-expression networks and observations from meta-analysis are available for developing hypotheses on inflorescence development (Supplementary Data 7).
Identification of CLE/WOX genes expressed during wheat inflorescence development
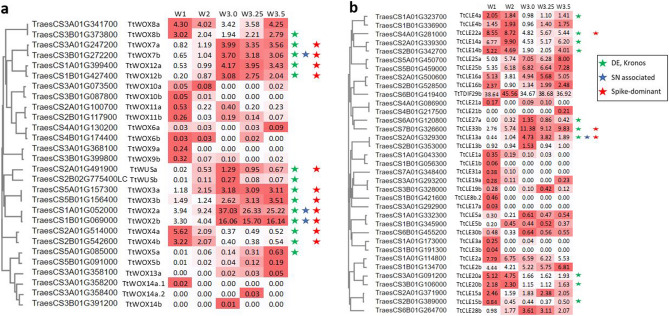
To identify members of the conserved CLAVATA-WUSCHEL pathway that may regulate stem cell maintenance in wheat spike meristems, the expression profiles of genes encoding WOX TFs and CLE peptides were analyzed. Of 29 WOX TFs, 28 were expressed during early inflorescence development and 11 were both significantly differentially expressed during the time course and exhibited a spike-specific expression profile (Fig. 5A). Two orthologs of OsWOX4 were co-expressed in module 1 with rapid down-regulation before transition to the inflorescence meristem, suggesting they may play a role in vegetative meristem maintenance but not in inflorescence development. Seven WOX genes clustered in module 2, characterized by rising expression during inflorescence development, including the orthologs of AtWUS (TtWUSa and b). The homoeologs TtWOX2a and 2b are both associated with variation in spikelet number and are clustered into separate co-expression modules (Supplementary Data 7).
Figure 5.
Expression profiles of WOX TFs (a) and CLE peptides (b) during wheat inflorescence development. Stars indicate additional evidence of a possible role in spike regulation (green = differential expression in ‘Kronos’ inflorescence, blue = associated with variation in spikelet number, red = spike-dominant expression profile). Heatmaps show expression (TPM) relative to each gene’s minimum and maximum expression. Only genes with TPM ≥ 0.01 are shown.
Of the 64 CLE genes, 35 were expressed during inflorescence development and just 9 were differentially expressed across the time course (Fig. 5B). Three wheat genes orthologous to OsFON2/4 (putatively TtCLV3, TraesCS2A02G329300 and TraesCS2B02G353000) exhibit spike-dominant DR-peaking expression profiles.
Discussion
Temporal transcriptomic datasets can help to characterize the regulatory networks controlling the development of complex organs such as the wheat inflorescence. One strategy to reduce spurious co-clustering of genes is to assemble a consensus co-expression network using a matrix of co-clustering frequencies from multiple independent networks, each assembled with randomized parameters and gene selection40–42. Co-expression networks have been successfully applied to unravel gene function in yeast (Saccharomyces cerevisiae), floral and fruit developmental pathways in strawberry (Fragaria vesca), and regulatory networks underlying leaf development in maize (Zea mays)41–43. In the current study, this approach generated a consensus network with a larger number of modules with improved intramodular connectivity compared to a standard WGCNA network (Fig. 2A). A further refinement to screen for genes co-clustering in every network assembly that they were both included revealed a consensus 100 module 9 of 167 genes that likely contribute to spikelet meristem and terminal spikelet formation (Fig. S3), indicating that consensus networks can help improve the accuracy of co-expression predictions and module assignment.
Beyond co-expression profiles, context-specific gene regulatory networks provide information on the centrality of each gene (a measure of its importance to the flow of information through a network), as well as the strength and directionality of interactions between individual genes44. This network predicts that the MIKC-MADS box TF AGL6 is a critical gene in inflorescence development regulatory networks, and functions together with MIKC-MADS TFs from the PI and SEP subclades (Fig. 3). This is consistent with its role in rice, where AGL6 functions as a cofactor with A, B, C, and D class proteins during floral development, as well as in wheat, where it interacts with ABCDE proteins, likely as a bridge in complex protein–protein interactions to regulate whorl development45–47. This network also revealed novel candidate genes for future characterization studies. For example, the BES1 TF BES1/BZR1 HOMOLOG 2-like is predicted to interact with several TFs, including the class Ι HD-ZIP TFs GNI-A1 and HOX-A2, suggesting a role for brassinosteroid signaling in wheat inflorescence development.
During the inflorescence development time course in tetraploid Kronos presented here, 43.1% of genes were expressed in at least 1 timepoint, comparable to the 40.2% and 42.5% of genes expressed in similar inflorescence development time courses in the hexaploid wheat genotypes ‘Chinese Spring’ and ‘Kenong 9204’ when these reads were reanalyzed using the same mapping parameters and reference genome24,25. Of these genes, 3,682 exhibited spike-dominant expression profiles, defined by a high proportion of expression in the ‘Kronos’ inflorescence compared to two whole-plant hexaploid transcriptome datasets (τ > 0.9). It is important to note that these genes might include those that are absent or poorly expressed in the genotypes used to generate these reference datasets. Decades of selection in bread wheat and durum wheat breeding programs may have contributed to differences in the gene regulatory networks underlying inflorescence development in these species. Among these genes were 7 of 10 SRS TFs, including the wheat ortholog of six-rowed spike 2 (HvVRS2) that modulates hormone activity in the developing barley spike48. Its expression profile in wheat, coupled with its association with spikelet number in an earlier study26, suggests it plays a conserved role in wheat inflorescence development. It would also be interesting to characterize the function of 4 other SRS TFs that exhibit spike-specific expression profiles peaking towards terminal spikelet formation (Supplementary Data 7). Ten of 15 GRF TFs were expressed predominantly in spike tissues, including the durum wheat ortholog of TaGRF4 which improves regeneration efficiency in tissue culture when co-expressed with GIF cofactors49. The broadly similar, spike-specific expression profiles of genes in this family suggest other members may also contribute to meristem differentiation and inflorescence development (Supplementary Data 7).
A subset of WOX TFs and CLE peptides exhibited dynamic and spike-dominant expression profiles across the time course, consistent with the differential regulation of OsWUS, OsWOX3, OsWOX4, and OsWOX12 during panicle development in rice50. The overexpression of TaWOX5 (named TtWOX9 in the current study) enhances wheat transformation and callus regeneration efficiency51. Several other WOX TFs are co-clustered with this gene and exhibit similar expression profiles in the wheat inflorescence (Fig. 5), suggesting they may also be candidates to enhance regeneration efficiency (Fig. 5). Among CLE peptides, TaCLV3 was negatively associated with spikelet number in a set of Chinese wheat landraces26, consistent with its proposed role as a negative regulator of SAM size and activity in rice and maize52,53.
Analyses of principal components and co-expression profiles indicate that the transition from the vegetative meristem to the double ridge stage is associated with major reprogramming of the wheat transcriptome (Fig. 1), consistent with an earlier study25. Several TF families were enriched in module 1, characterized by high expression in the vegetative meristem before rapid downregulation after the double ridge stage, including 8 of the 9 RAV TFs in the consensus network. In Arabidopsis, the RAV genes TEMPRANILLO1 (TEM1) and TEM2 repress FT to prevent precocious flowering54,55. In rice, the TEM orthologs OsRAV8 and OsRAV9 bind the promoters of OsMADS14 and Hd3a to suppress the floral transition, indicating this function is conserved in monocots56. The rapid downregulation of the wheat orthologs of these genes before double ridge formation, as well as homologs of OsRAV11 and OsRAV12 that act in reproductive patterning in rice56, suggests this family may act as local repressors of meristem identity genes in the developing wheat spike.
There were also 26 TCP TFs clustered in module 1, including TtTCP-A9 and TtTCP-B9, negative regulators of spikelet number and grain size in durum wheat57. It is likely that other members of the TCP TF family also play roles as negative regulators of grain development. For example, TtTCP-A17 and -B17 are both downregulated during inflorescence development, are within 250 kbp of QTL for grain size, and are orthologous to genes associated with spikelet number variation in rice (Supplementary Data 7). Eight TCP TFs clustered in different modules and were most highly expressed during spikelet meristem formation, including TEOSINTE BRANCHED 1, which integrates photoperiod signals to regulate spike architecture in a dosage-dependent manner58, and a paralogous copy on chromosome 5B, BRANCHED AND INDETERMINATE SPIKE, that regulates spike architecture in barley59. Four other uncharacterized TCP TFs with homology to RETARDED PALEA1 exhibit spike-dominant expression profiles and would be promising candidates to characterize their role in inflorescence development in wheat (Supplementary Data 7). Gene regulatory networks controlling inflorescence development may also influence male and female reproductive traits for hybrid wheat production. For example, variation in the rate and extent of anther extrusion, which impacts pollen shedding to facilitate out-crossing, might be impacted during inflorescence development to control lemma and anther physiology. It will be interesting to analyze the expression of candidate genes for hybrid wheat production within the inflorescence time course. Similarly, regulatory networks can be important resources to identify and characterize genes that underlie variation in fruiting efficiency, an important yield component60.
Although association and linkage mapping studies in wheat have described hundreds of QTL for agronomic traits, relatively few causative genes have been cloned and validated7. One example is the Mov-1 locus on chromosome 2D that confers variation in grains per spikelet in hexaploid wheat61. Transcriptomic data can help prioritize candidate gene selection within a mapping interval based on spatial or temporal expression profiles62. Furthermore, changes in transcription may indicate the presence of dominant or semi-dominant gain-of-function variants in cis-regulatory elements or of structural variation that confer changes in phenotype through modified expression profiles. Because of the functional redundancy of the polyploid wheat genome, such variants underlie the majority of cloned genes to date63, including domestication alleles of PPD1, VRN1 and RHT1, which clustered in different co-expression modules to their wild-type homoeologous allele (Fig. 4). Such divergent expression profiles, especially for those genes in close proximity to QTL for traits relating to grain number and grain size, might be strong candidates for allele mining to explore the extent of natural variation in wheat germplasm collections, and to engineer novel variation by targeted editing of cis-regulatory regions64. In order to apply this approach in crop breeding, it will be necessary to develop efficient methods to identify specific cis-regulatory elements in these highly variable regions of the genome. It may also be possible to engineer changes in inflorescence development by over-expressing candidate TFs identified in this study, although regulatory restrictions and consumer opposition may limit the commercial application of such tools in some parts of the world.
Conclusions
Consensus and gene regulatory networks provide the means to analyze temporal transcriptomic datasets as a complementary approach to characterize functional pathways underlying wheat inflorescence development. The incorporation of higher resolution datasets at both the spatial and temporal levels within meristem tissues will build on these findings29. Although reverse genetics will be required to validate the hypotheses generated from in silico network analyses, the integration of functional datasets from wheat and related species facilitates the identification of critical regulators65. Functional characterization studies focused on these predicted genes will improve our understanding of inflorescence development and help breeders identify and combine superior alleles to drive increased grain number.
Materials and methods
Plant materials and growth conditions
All experiments were performed in the tetraploid Triticum turgidum L. subsp. durum (Desf.) var. Kronos (genomes AABB). Kronos has a spring growth habit conferred by a VRN-A1 allele containing a deletion in intron 1 and carries the Ppd-A1a allele that confers reduced sensitivity to photoperiod66,67. Plants were grown in controlled conditions in PGR15 growth chambers (Conviron, Manitoba, Canada) under a long day photoperiod (16 h light/8 h dark) at 23 ℃ day/17 ℃ night temperatures and a light intensity of ~ 260 µM m−2 s−1. Developing apical meristems were harvested under a dissecting microscope using a sterile scalpel and placed immediately in liquid nitrogen. All samples were harvested within a 1 h period approximately 4 h after the lights were switched on (± 30 min) to account for possible differences in circadian regulation of gene expression. Approximately 20 apices were combined for each biological replicate of samples harvested at stages W1 (shoot apical meristem, SAM) and W2 (early double ridge, EDR) and approximately 12 apices for samples harvested at stages W3.0 (double ridge, DR), W3.25 (lemma primordia, LP) and W3.5 (terminal spikelet, TS)3. Four biological replicates were harvested at each timepoint.
All plant materials were sourced from the Dubcovsky lab at UC Davis, and all experiments were in compliance with institutional, national, and international guidelines and legislation.
RNA-seq library construction and sequencing
Tissues were ground into a fine powder in liquid nitrogen and total RNA was extracted using the Spectrum™ Plant Total RNA kit (Sigma-Aldrich, St. Louis, MO). Sequencing libraries were produced using the TruSeq RNA Sample Preparation kit v2 (Illumina, San Diego, CA), according to the manufacturer’s instructions. Library quality was determined using a high-sensitivity DNA chip run on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Libraries were barcoded to allow multiplexing and all samples were sequenced using the 100 bp single read module across 2lanes of a HiSeq3000 sequencer at the UC Davis Genome Center.
RNA-seq data processing
‘Kronos’ RNA-seq reads were trimmed and checked for quality Phred scores above 30 using Fastp v0.20.168. Trimmed reads were aligned to the IWGSC RefSeq v1.0 genome assembly consisting of A and B chromosome pseudomolecules and unanchored (U) scaffolds not assigned to any chromosome (ABU) using STAR 2.7.5 aligner (outFilterMismatchNoverReadLmax = 0.04, alignIntronMax = 10,000)20,69. Only uniquely mapped reads were retained for expression analysis. Transcript levels were quantified by featureCounts using 190,391 gene models from the ABU IWGSC RefSeq v1.1 annotations28,70 and converted to Transcripts Per Million (TPM) values using a custom python script available from https://github.com/cvanges/spike_development/ (Supplementary Data 1).
Raw RNA-seq reads for ‘Kenong9204’ and ‘Chinese Spring’ inflorescence development datasets were obtained from BioProjects PRJNA325489 and PRJNA38367724,25. RNA-seq reads were processed with Fastp as described above and aligned to the hexaploid ABDU RefSeq v1.0 genome assembly using the same methods and parameters. Transcript quantification and TPM were determined as above using the full ABDU IWGSC RefSeq v1.1 annotations.
RNA-seq reads and raw count data for each sample is available from NCBI Gene Expression Omnibus under the accession GSE193126 (https://www.ncbi.nlm.nih.gov/geo/).
Transcription factors
There were 3,838 ABU gene models annotated as transcription factors that were grouped into 65 TF families per IWGSC v1.1 annotations28. The following families were consolidated: “AP2” and “APETALA2”, “bHLH” and “HRT-like”, “MADS” and “MADS1”, “NFYB” and “NF-YB”, “NFYC” and “NF-YC”, and “SBP” and “SPL”, as well as “MADS2” and “MIKC”, which were consolidated into “MIKC-MADS”. After consolidation, there were 59 TF families. A previous study described the annotation of 201 MIKC-MADS box genes placed into 15 subclades17. There were 30 MIKC transcription factors on the A and B genomes absent from the IWGSC TF list, which were added to this family. Investigations of the CLE and WOX gene families were based on the naming reported in Li et al. (Ref.13) and Li et al. (Ref.14), with the addition of TtWUSb (TraesCS2B02G775400LC) to the WOX family, which was absent from these studies. In total, 3,861 TFs were included in this study (Supplementary Data 2).
Spike-dominant expression analysis
Expression data (TPMs) for 2 developmental studies were obtained from the Grassroots Data Repository (https://opendata.earlham.ac.uk/wheat/under_license/toronto/Ramirez-Gonzalez_etal_2018-06025-Transcriptome-Landscape/expvip/RefSeq_1.0/ByTranscript/)27,28. The first dataset, in ‘Chinese Spring’, included samples from 5 tissue types at 3 timepoints (mean of 2 biological replicates) for 15 total tissue/stages27. A second dataset from the variety ‘Azhurnaya’ comprised 209 unreplicated samples grouped into 22 “intermediate tissue” groups of various sizes28. Twelve samples overlapping with ‘Kronos’ spike samples were removed (tissue groups “coleoptile”, “stem axis”, and “shoot apical meristem”). For early spike tissue specificity analyses, the mean TPM expression of 15 ‘Chinese Spring’ tissues (n = 2) or the mean of 22 ‘Azhurnaya’ tissues (n ranging from 3 to 30) were compared to the ‘Kronos’ sampling stage with the highest mean expression (n = 4). Comparisons were made using the Tau (τ) tissue specificity metric where τ = 0 indicates ubiquitous expression and τ = 1 indicates tissue specific expression71,72. A custom R script was used to calculate tissue specificity and is available at github.com/cvanges/spike_development. Genes which were expressed predominantly in ‘Kronos’ inflorescence tissues (τ > 0.9) were defined ‘spike-dominant’ whereas genes only expressed in ‘Kronos’ inflorescence tissues (τ = 1) were defined ‘spike-specific’ (Supplementary Data 3).
Principal component analysis (PCA), differential expression, and GO enrichment
PCA was performed in R using prcomp in the r/stats package v2.6.2 including all replications for each time point. PCA plots were generated with ggplot2 v3.3.2. Whole transcriptome PCA used read counts from all expressed gene models (n = 82,019) and TF PCA used expression of 2874 expressed TFs. Randomized PCA distribution (Fig. S1) used independent random subsampling of 2874 expressed genes without replacement. Principle component percent variation explained and eigenvalues from prcomp were used for comparisons between whole transcriptome PCA and TF-only PCA.
Pairwise differential expression was determined using both EdgeR v3.24.3 and DESeq2 v1.22.2 for robustness73,74. Pairwise comparisons between consecutive timepoints were done using raw read counts for 4 biological replicates at each stage. Benjamin-Hochberg FDR adjusted P-values ≤ 0.01 was used as a stringent DE cut-off for both tools. Only genes DE using both tools were classified as pairwise DEGs (Supplementary Data 5). Differential expression of ‘Chinese Spring’ and ‘Kenong9204’ inflorescence development datasets was also determined with raw read counts and EdgeR and DESeq2 using the same method as for the ‘Kronos’ dataset. Adjustments to DE tests were made to compare all 4 timepoints (6 pairwise comparisons) with 2 biological replicates in ‘Chinese Spring’ as well as the 6 timepoints (15 pairwise comparisons) with 2 biological replicates in the ‘Kenong9204’ datasets. For network analyses, a second DE test was included which reinforced longitudinal DE determination, an impulse model (ImpulseDE2, https://github.com/YosefLab/ImpulseDE2) was used for ‘Kronos’ data75,76. Raw counts were used with default parameters and genes with Benjamin-Hochberg FDR adjusted P-values ≤ 0.05 considered differentially expressed. Functional annotation to generate GO terms for each high-confidence and low-confidence gene in the IWGSC RefSeq v1.1 genome was performed as described previously77.
Standard and consensus WGCNA network construction
Genes identified using pairwise differential expression (EdgeR and DESeq2) and ImpulseDE2 (22,566 genes total) were used for co-expression analyses. A standard co-expression network was built using the R package WGCNA v1.66 with the parameters: power = 20, networkType = signed, minimum module size = 30, and mergecutheight = 0.2578 (Supplementary Data 7). Parallel coordinate plots were produced in R by normalizing raw read counts and visualized with ggparacoord (scale = ‘globalminmax’) in GGally (version 1.5.0).
A consensus network was built using methods described in Shahan et al. (Ref.42). In brief, 1,000 WGCNA runs were performed with 80% of genes randomly subsampled without replacement and random parameters for power (1, 2, 4, 8, 12, 16, 20), minModSize (40, 60, 90, 120, 150, 180, 210), and mergeCutHeight (0.15, 0.2, 0.25, 0.3). The final consensus network was built using an adjacency matrix—adj = number of times gene i is clustered with gene j/number of times gene i is subsampled with gene j—with parameters power = 6 and minModuleSize = 30 (Supplementary Data 7). The consensus100 network was built by filtering the adjacency network for adj = 1 prior to network construction. Along with module assignments, we used the WGCNA package to find the connectivity of each gene with co-clustered genes (intramodularConnectivity.fromExpr()) and summarized module expression patterns (moduleEigengenes()). Python and R scripts for creating the adjacency matrix and consensus network are available at https://github.com/cvanges/spike_development. The Bioconductor package GeneOverlap was used to determine the overlap of module assignments between consensus and standard networks (http://shenlab-sinai.github.io/shenlab-sinai/)79.
Causal structure inference network
Expression data (TPM) for 970 transcription factors retained in the consensus100 network was used to build a gene regulatory network using the Causal Structure Inference algorithm42. Network construction used CSI in Cyverse with default parameters42.
Conversion of wheat, rice, and barley gene IDs
Genes associated with wheat and rice spikelet number described in Wang et al., were identified from a previous set of annotated wheat gene models (ftp://ftp.ensemblgenomes.org/pub/plants/release-28/). To identify the corresponding IWGSC RefSeq v1.1 gene ID, each gene model coding sequence was extracted and used as a query in BLASTn searches against the IWGSCv1.1 ABU genome. Homologous gene pairs with > 99% identity to each query were considered spikelet number associated genes. Two previous studies reported genes DE during H. vulgare inflorescence development using IBSC_v2 annotations80,81. Each barley gene model coding sequence was extracted and used as a query in BLASTn searches against the IWGSCv1.1 ABU wheat genome. Genes with percent identity > 90% were retained and considered orthologs of barley DEGs (HvDE).
Enrichment analysis
Enrichment and depletion of genes among modules or DEG lists was determined using the cumulative distribution function of the hypergeometric distribution (http://systems.crump.ucla.edu/hypergeometric/).
QTL proximity and definition of homoeologous pairs
Using a previously published meta-analysis of yield component QTL studies, we searched the IWGSCv1.1 genome for expressed genes in our timecourse within 500 kbp of 428 loci associated with yield component traits (kernel number per spike, thousand kernel weight, spikelet number)7. Homoeologous gene pairs reported from Ramírez-González et al.28 were used to determine co-expressed homoeologs.
Supplementary Information
Acknowledgements
The authors thank Dr. Luis de Haro for assistance in RNA-seq library preparation and Dr. Cristobal Uauy for helpful input and discussion on project design and analysis. Work in this project was funded by the International Wheat Yield Partnership Grant IWYP76.
Author contributions
S.P. and J.D. conceived of and designed the project. C.V., J.H. and F.T. analyzed RNA-seq data. C.V., S.P. and J.D. wrote the manuscript.
Data availability
All RNA-seq data have been deposited with the Gene Expression Omnibus (GEO) database under record number GSE193126. Processed expression and gene annotation information are provided as Supplementary Files. The consensus expression network is provided as Supplementary File 10 and can be viewed in the open-source Cytoscape platform.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-21571-z.
References
- 1.Ray DK, Mueller ND, West PC, Foley JA. Yield trends are insufficient to double global crop production by 2050. PLoS ONE. 2013;8:e66428. doi: 10.1371/journal.pone.0066428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinton J, Uauy C. A reductionist approach to dissecting grain weight and yield in wheat. J. Integr. Plant Biol. 2019;61:337–358. doi: 10.1111/jipb.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waddington SR, Cartwright PM, Wall PC. A quantitative scale of spike initial and pistil development in barley and wheat. Ann. Bot. 1983;51:119–130. doi: 10.1093/oxfordjournals.aob.a086434. [DOI] [Google Scholar]
- 4.Li C, et al. Wheat VRN1, FUL2 and FUL3 play critical and redundant roles in spikelet development and spike determinacy. Development. 2019;146:175398. doi: 10.1242/dev.175398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnett OT. Inflorescences of Maize, Wheat, Rye, Barley, and Oats: Their Initiation and Development. University of Illinois; 1966. [Google Scholar]
- 6.Rawson HM. Spikelet number, its control and relation to yield per ear in wheat. Aust. J. Biol. Sci. 1970;23:1–15. doi: 10.1071/BI9700001. [DOI] [Google Scholar]
- 7.Cao S, Xu D, Hanif M, Xia X, He Z. Genetic architecture underpinning yield component traits in wheat. Theor. Appl. Genet. 2020;133:1811–1823. doi: 10.1007/s00122-020-03562-8. [DOI] [PubMed] [Google Scholar]
- 8.Würschum T, Leiser WL, Langer SM, Tucker MR, Longin CFH. Phenotypic and genetic analysis of spike and kernel characteristics in wheat reveals long-term genetic trends of grain yield components. Theor. Appl. Genet. 2018;131:2071–2084. doi: 10.1007/s00122-018-3133-3. [DOI] [PubMed] [Google Scholar]
- 9.Somssich M, Je BI, Simon R, Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016;143:3238–3248. doi: 10.1242/dev.133645. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher JC. The CLV-WUS stem cell signaling pathway: A roadmap to crop yield optimization. Plants. 2018;7:87. doi: 10.3390/plants7040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171:470–480. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, et al. Structural variation at the maize WUSCHEL1 locus alters stem cell organization in inflorescences. Nat. Commun. 2021;12:1–12. doi: 10.1038/s41467-021-22699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, et al. Identification and functional analysis of the CLAVATA3/embryo surrounding region (CLE) gene family in wheat. Int. J. Mol. Sci. 2019;20:4319. doi: 10.3390/ijms20174319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, et al. Identification of the WUSCHEL-related homeobox (WOX) gene family, and interaction and functional analysis of TaWOX9 and TaWUS in wheat. Int. J. Mol. Sci. 2020;21:1581. doi: 10.3390/ijms21051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 16.Theißen G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001;4:75–85. doi: 10.1016/S1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- 17.Schilling S, Kennedy A, Pan S, Jermiin LS, Melzer R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020;225:511–529. doi: 10.1111/nph.16122. [DOI] [PubMed] [Google Scholar]
- 18.Li K, et al. Interactions between SQUAMOSA and SHORT VEGETATIVE PHASE MADS-box proteins regulate meristem transitions during wheat spike development. Plant Cell. 2021;33:3621–3644. doi: 10.1093/plcell/koab243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamski NM, et al. Ectopic expression of Triticum polonicum VRT-A2 underlies elongated glumes and grains in hexaploid wheat in a dosage-dependent manner. Plant Cell. 2021;33:2296–2319. doi: 10.1093/plcell/koab119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The International Wheat Genome Sequencing Consortium (IWGSC) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2019;361:7191. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- 21.Walkowiak S, et al. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020;588:277–283. doi: 10.1038/s41586-020-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao X, Dixon RA. Co-expression networks for plant biology: Why and how. Acta Biochim. Biophys. Sin. (Shanghai) 2019;51:981–988. doi: 10.1093/abbs/gmz080. [DOI] [PubMed] [Google Scholar]
- 23.van den Broeck L, Gordon M, Inzé D, Williams C, Sozzani R. Gene regulatory network inference: Connecting plant biology and mathematical modeling. Front. Genet. 2020;11:457. doi: 10.3389/fgene.2020.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng N, et al. Transcriptome profiling of wheat inflorescence development from spikelet initiation to floral patterning identified stage-specific regulatory genes. Plant Phys. 2017;174:1779–1794. doi: 10.1104/pp.17.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, et al. A genome-wide view of transcriptome dynamics during early spike development in bread wheat. Sci. Rep. 2018;8:1–16. doi: 10.1038/s41598-018-33718-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Transcriptome association identifies regulators of wheat spike architecture. Plant Phys. 2017;175:746–757. doi: 10.1104/pp.17.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choulet F, et al. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345:1249721. doi: 10.1126/science.1249721. [DOI] [PubMed] [Google Scholar]
- 28.Ramírez-González RH, et al. The transcriptional landscape of polyploid wheat. Science. 2018;361:6089. doi: 10.1126/science.aar6089. [DOI] [PubMed] [Google Scholar]
- 29.Backhaus AE, et al. High expression of the MADS-box gene VRT2 increases the number of rudimentary basal spikelets in wheat. Plant Phys. 2022;189:1536–1552. doi: 10.1093/plphys/kiac156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emery JF, et al. Radial patterning of Arabidopsis shoots by Class III HD-ZIP and KANADI genes. Curr. Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 31.Du D, et al. Frizzy panicle defines a regulatory hub for simultaneously controlling spikelet formation and awn elongation in bread wheat. New Phytol. 2021;231:814–833. doi: 10.1111/nph.17388. [DOI] [PubMed] [Google Scholar]
- 32.Shaw LM, et al. Flowering locus T2 regulates spike development and fertility in temperate cereals. J. Exp. Bot. 2019;70:193–204. doi: 10.1093/jxb/ery350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Debernardi JM, Greenwood JR, JeanFinnegan E, Jernstedt J, Dubcovsky J. APETALA 2-like genes AP2L2 and Q specify lemma identity and axillary floral meristem development in wheat. Plant J. 2020;101:171–187. doi: 10.1111/tpj.14528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakuma S, et al. Unleashing floret fertility in wheat through the mutation of a homeobox gene. Proc. Natl. Acad. Sci. U.S.A. 2019;116:5182–5187. doi: 10.1073/pnas.1815465116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gauley A, Boden S. Stepwise increases in FT1 expression regulate seasonal progression of flowering in wheat (Triticum aestivum) New Phytol. 2021;229:1163–1176. doi: 10.1111/nph.16910. [DOI] [PubMed] [Google Scholar]
- 36.Kuzay S, et al. WAPO-A1 is the causal gene of the 7AL QTL for spikelet number per spike in wheat. PLoS Genet. 2022;18:e1009747. doi: 10.1371/journal.pgen.1009747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong Z, et al. Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E8656–E8664. doi: 10.1073/pnas.1714960114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 39.Shao A, et al. The auxin biosynthetic tryptophan aminotransferase related TaTAR2.1-3A increases grain yield of wheat. Plant Phys. 2017;174:2274–2288. doi: 10.1104/pp.17.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monti S, et al. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 2003;52:91–118. doi: 10.1023/A:1023949509487. [DOI] [Google Scholar]
- 41.Wu LF, et al. Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat. Genet. 2002;31:255–265. doi: 10.1038/ng906. [DOI] [PubMed] [Google Scholar]
- 42.Shahan R, et al. Consensus coexpression network analysis identifies key regulators of flower and fruit development in wild strawberry. Plant Phys. 2018;178:202–216. doi: 10.1104/pp.18.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miculan M, et al. A forward genetics approach integrating genome-wide association study and expression quantitative trait locus mapping to dissect leaf development in maize (Zea mays) Plant J. 2021;107:1056–1071. doi: 10.1111/tpj.15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Penfold CA, Wild DL. How to infer gene networks from expression profiles, revisited. Interface Focus. 2011;1:857–870. doi: 10.1098/rsfs.2011.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su Y, et al. Wheat AGAMOUS like 6 transcription factors function in stamen development by regulating the expression of TaAPETALA3. Development. 2019;146:177527. doi: 10.1242/dev.177527. [DOI] [PubMed] [Google Scholar]
- 46.Li H, et al. Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell. 2011;23:2536–2552. doi: 10.1105/tpc.111.087262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong X, et al. The wheat AGL6-like MADS-box gene is a master regulator for floral organ identity and a target for spikelet meristem development manipulation. Plant Biotechnol. J. 2022;20:75–88. doi: 10.1111/pbi.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youssef HM, et al. VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 2016;49:157–161. doi: 10.1038/ng.3717. [DOI] [PubMed] [Google Scholar]
- 49.Debernardi JM, et al. A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020;38:1274–1279. doi: 10.1038/s41587-020-0703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng S, Huang Y, Zhu N, Zhao Y. The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene. 2014;549:266–274. doi: 10.1016/j.gene.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Wang K, et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants. 2022;8:110–117. doi: 10.1038/s41477-021-01085-8. [DOI] [PubMed] [Google Scholar]
- 52.Chu H, et al. The FLORAL ORGAN NUMBER4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Phys. 2006;142:1039–1052. doi: 10.1104/pp.106.086736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bommert P, Je BI, Goldshmidt A, Jackson D. The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature. 2013;502:555–558. doi: 10.1038/nature12583. [DOI] [PubMed] [Google Scholar]
- 54.Hu H, et al. TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2021;118:e2103895118. doi: 10.1073/pnas.2103895118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- 56.Osnato M, Matias-Hernandez L, Aguilar-Jaramillo AE, Kater MM, Pelaza S. Genes of the RAV family control heading date and carpel development in rice. Plant Phys. 2020;183:1663–1680. doi: 10.1104/pp.20.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao J, et al. Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.) Front. Plant Sci. 2018;9:1282. doi: 10.3389/fpls.2018.01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dixon LE, et al. TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum) Plant Cell. 2018;30:563–581. doi: 10.1105/tpc.17.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shang Y, et al. A CYC/TB1-type TCP transcription factor controls spikelet meristem identity in barley. J. Exp. Bot. 2020;71:7118–7131. doi: 10.1093/jxb/eraa416. [DOI] [PubMed] [Google Scholar]
- 60.Pretini N, et al. The physiology and genetics behind fruiting efficiency: A promising spike trait to improve wheat yield potential. J. Exp. Bot. 2021;72:3987–4004. doi: 10.1093/jxb/erab080. [DOI] [PubMed] [Google Scholar]
- 61.Mahlandt A, et al. High-resolution mapping of the Mov-1 locus in wheat by combining radiation hybrid (RH) and recombination-based mapping approaches. Theor. Appl. Genet. 2021;134:2303–2314. doi: 10.1007/s00122-021-03827-w. [DOI] [PubMed] [Google Scholar]
- 62.Yang Y, et al. Large-scale integration of meta-QTL and genome-wide association study discovers the genomic regions and candidate genes for yield and yield-related traits in bread wheat. Theor. Appl. Genet. 2021;134:3083–3109. doi: 10.1007/s00122-021-03881-4. [DOI] [PubMed] [Google Scholar]
- 63.Gaurav K, et al. Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol. 2022;40:422–431. doi: 10.1038/s41587-021-01058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swinnen G, Goossens A, Pauwels L. Lessons from domestication: Targeting cis-regulatory elements for crop improvement. Trends Plant Sci. 2016;21:506–515. doi: 10.1016/j.tplants.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 65.Uauy C, Wulff BBH, Dubcovsky J. Combining traditional mutagenesis with new high-throughput sequencing and genome editing to reveal hidden variation in polyploid wheat. Ann. Rev. Genet. 2017;51:435–454. doi: 10.1146/annurev-genet-120116-024533. [DOI] [PubMed] [Google Scholar]
- 66.Wilhelm EP, Turner AS, Laurie DA. Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.) Theor. Appl. Genet. 2008;118:285–294. doi: 10.1007/s00122-008-0898-9. [DOI] [PubMed] [Google Scholar]
- 67.Fu D, et al. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genom. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- 68.Chen S, Zhou Y, Chen Y, Gu J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:884–890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 71.Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- 72.Kryuchkova-Mostacci N, Robinson-Rechavi M. A benchmark of gene expression tissue-specificity metrics. Brief. Bioinform. 2017;18:205–214. doi: 10.1093/bib/bbw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fischer DS, Theis FJ, Yosef N. Impulse model-based differential expression analysis of time course sequencing data. Nucleic Acids Res. 2018;46:e119. doi: 10.1093/nar/gky675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spies D, Renz PF, Beyer TA, Ciaudo C. Comparative analysis of differential gene expression tools for RNA sequencing time course data. Brief. Bioinform. 2019;20:228–298. doi: 10.1093/bib/bbx115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pearce S, Kippes N, Chen A, Debernardi JM, Dubcovsky J. RNA-seq studies using wheat Phytochrome B and Phytochrome C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways. BMC Plant Biol. 2016;16:141. doi: 10.1186/s12870-016-0831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:1–13. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen, L. GeneOverlap: Test and visualize gene overlaps. Preprint at http://shenlab-sinai.github.io/shenlab-sinai/ (2021).
- 80.Digel B, Pankin A, von Korff M. Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. Plant Cell. 2015;27:2318–2334. doi: 10.1105/tpc.15.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu H, et al. Transcriptome profiling reveals phase-specific gene expression in the developing barley inflorescence. Crop J. 2020;8:71–86. doi: 10.1016/j.cj.2019.04.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data have been deposited with the Gene Expression Omnibus (GEO) database under record number GSE193126. Processed expression and gene annotation information are provided as Supplementary Files. The consensus expression network is provided as Supplementary File 10 and can be viewed in the open-source Cytoscape platform.