Abstract
Killer-cell immunoglobulin-like receptors (KIR) are essential for acquiring natural killer (NK) cell effector function, which is modulated by a balance between the net input of signals derived from inhibitory and activating receptors through engagement by human leukocyte antigen (HLA) class I ligands. KIR and HLA loci are polygenic and polymorphic and exhibit substantial variation between individuals and populations. We attempted to investigate the contribution of KIR complex and HLA class I ligands to the genetic predisposition to lung cancer in the native population of southern Iran. We genotyped 16 KIR genes for a total of 232 patients with lung cancer and 448 healthy controls (HC), among which 85 patients and 178 HCs were taken into account for evaluating combined KIR-HLA associations. KIR2DL2 and 2DS2 were increased significantly in patients than in controls, individually (OR 1.63, and OR 1.42, respectively) and in combination with HLA-C1 ligands (OR 1.99, and OR 1.93, respectively). KIR3DS1 (OR 0.67) and 2DS1 (OR 0.69) were more likely presented in controls in the absence of their relative ligands. The incidence of CxTx subset was increased in lung cancer patients (OR 1.83), and disease risk strikingly increased by more than fivefold among genotype ID19 carriers (a CxTx genotype that carries 2DL2 in the absence of 2DS2, OR 5.92). We found that genotypes with iKIRs > aKIRs (OR 1.67) were more frequently presented in lung cancer patients. Additionally, patients with lung cancer were more likely to carry the combination of CxTx/2DS2 compared to controls (OR 2.04), and iKIRs > aKIRs genotypes in the presence of 2DL2 (OR 2.05) increased the likelihood of lung cancer development. Here we report new susceptibility factors and the contribution of KIR and HLA-I encoding genes to lung cancer risk, highlighting an array of genetic effects and disease setting which regulates NK cell responsiveness. Our results suggest that inherited KIR genes and HLA-I ligands specifying the educational state of NK cells can modify lung cancer risk.
Subject terms: Cancer, Immunology
Introduction
Lung cancer, the most common cause of cancer-related mortality worldwide is generally classified into main histological subtypes, including non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC)1,2. Lung mucosa is constantly exposed to inhaled environmental pollution, dust, smoke, and pathogens; hence a dynamic network of tissue-resident immune cells continually keeps monitoring the lung to maintain tissue homeostasis3. The immune system also has a vital role in cancer initiation and progression. Natural killer cells (NK cells) are part of innate immune cells that serve as a first-line of defense with the capacity to eliminate virally infected cells and neoplastic transformations without prior sensitization4. These cytotoxic cells mediate antitumor responses by exploiting death receptors, releasing pro-inflammatory cytokines and perforin/granzyme granules exocytosis5.
Interestingly, over 10% of resident lymphocytes in the lung are NK cells6. The association of NK cell dysfunction with tumor progression was revealed in KRAS-driven lung cancer, in which NK cells protect tumors at the early stage but cannot prevent tumor progression7. Also, it is shown that resident NK cells have a pivotal role in resistance to experimental lung metastasis by producing IFN-γ8. Although the infiltration of NK cells was indicated as a favorable prognostic factor in lung cancer9,10, the functional reactivity of intratumoral NK cells is crucial in addition to the degree of NK cell infiltration. It has been demonstrated that NK cell dysfunction at the late stage is caused by the suppressive tumor microenvironment (TME)7. NK cells isolated from NSCLC patients, which depict different expression patterns, have impaired interferon-γ (IFN-γ) production capability and lower cytotoxicity than non-tumoral NK cells11–13.
The activity of NK cells is modulated by a balance between the net input of signals derived from an array of inhibitory and activating receptors14. Among these germ-line encoded receptors, killer-cell immunoglobulin-like receptors (KIR) are essential for acquiring NK cell effector function15,16. KIRs are encoded by a cluster of polymorphic and homologous genes located at chromosomal region 19q13.417. KIR region comprises activating (2DS1-5, 3DS1) and inhibitory (2DL1-3, 2DL5A/B, 3DL1) genes, divided by framework KIRs into centromeric (3DL3 to 3DP1) and telomeric (2DL4 to 3DL2) intervals18. Although (5′) centromeric and (3′) telomeric regions contain various combinations of activating (aKIR) and inhibitory (iKIR) genes, certain adjacent KIRs tend to be strongly linked17,19. Considering strong linkage disequilibrium (LD) among KIR genes, it is difficult to isolate each KIR gene's effect on NK cell response20. Based on gene content within both intervals, the KIR locus segregates in distinct haplotypes, mainly known as A (fixed combination of genes, mostly iKIRs) and B (variable number of iKIRs/aKIRs)17,21. The KIR gene cluster displays tremendous variation due to gene-content diversity and haplotypic variety17. In addition to gene content diversity, allelic polymorphism extends KIR variations and accounts for differential expression levels of KIRs on the surface of NK cells22,23.
The classical HLA class I molecules (HLA-A, B, C) as major KIR ligands have been categorized into 4 types of KIR-binding epitopes (C1, C2, Bw4, A3/A11) according to amino acid sequences24. KIR2DL1 and KIR2DL2/3 mainly bind to a group of ligands encoded by HLA-C alleles that differed by Lys/ Asn dimorphism at position 80 (HLA-C2 and HLA-C1, respectively)25. KIR3DL1 was found to interact with HLA-Bw4, and KIR3DL2 recognizes HLA-A3/A1126,27. The Ile/Thr dimorphism at position 80 defines the affinity of KIR binding to the Bw4 motif, in which Ile80 exhibits a greater affinity for 3DL128. The growing understanding of activating KIR/ HLA interactions indicates that activating KIR3DS1, 2DS1, and 2DS2 bind to the same HLA class I with lower affinity than their homologous inhibitory counterparts29–32. However, other KIR/HLA pair interactions haven’t been distinctly demonstrated. It is shown that different iKIRs exhibit various binding affinities for HLA-I ligands. Interestingly the peptide presented by HLA subtypes seems to play a role in binding affinity alteration33. Signals derived from iKIRs interacting with self-HLA ligands set a threshold of activation for NK cells leading to a maturation process titled “education” or “licensing”34,35. Besides development and self-tolerance, education renders NK cells capacity to recognize diseased cells with downregulated or lacking HLA class I expression, referring to the “missing self” hypothesis34. Lacking a specific HLA-I ligand or any responding iKIR, yields hypo-responsive NK clones with high activation threshold30. It is particularly noteworthy that KIR-gene complex diversity influences surface expression, ligand specificities, ligand binding affinity, and subsequent signal transduction through KIR-HLA class I interaction36–38. Such an intense heterogeneity in KIR and HLA complex prominently affects NK cell responses and is associated with disease susceptibility39. Genome-wide association studies (GWAS) have identified 45 loci associated with lung cancer risk40. More specifically, SNPs from 5p15.33, 6p21.33, and 15q25.1 regions are strongly associated with lung cancer in Caucasians41. However, the composition of multiple highly-homogeneous gene content, intense polymorphism, strong linkage disequilibrium between multiple loci, and low/no coverage by GWAS reagents pose challenges to studying the polymorphism of KIR (19q13) and HLA (6p21) gene families by GWAS.
The present study aimed to investigate the contribution of KIR complex and HLA-I ligands to the genetic predisposition to lung cancer in the native population of Fars province, located in the southern part of Iran, and to disclose possible associations with the dysfunctional state of NK cells in the context of lung cancer (Table 1).
Table 1.
Characteristics of the study population.
| Characteristics | Lung cancer patients (n = 232) | Healthy controls (n = 448) |
|---|---|---|
| Mean age ± SD | 64.4 ± 11.1 | 58.13 ± 12.66 |
| Gender | ||
| Female | 34 (14.7%) | 150 (33.5%) |
| Male | 198 (85.3%) | 298 (66.5%) |
| Smoking status | ||
| Smoker | 51 (21.98%) | 64 (14.3%) |
| Non-smoker | 0 (0.0%) | 3 (0.7%) |
| Unknown | 181 (78.02%) | 381 (85.0%) |
| Lung cancer subtypes | ||
| SCLC | 53 (22.8%) | |
| NSCLC | 179 (77.2%) | |
| Squamous cell carcinoma | 144 (62.1%) | |
| Adenocarcinoma | 35 (15.1%) | |
Results
Susceptibility/resistance influence of specific B haplotype-associated KIRs on lung cancer risk
The distribution of 16 KIR genes were determined in patients and HCs (Table 2). As observed, framework genes (3DL3, 3DP1, 2DL4, 3DL2) were presented in all subjects. Two adjacent B haplotype-associated genes 2DL2 (67.2% vs. 55.4%, p = 0.003, OR 1.63, CI 1.17–2.26) and 2DS2 (62.9% vs. 54.6%, p = 0.041, OR 1.42, CI 1.02–1.96) were significantly increased in patients in comparison with controls. Activating genes 2DS1 (50.2% vs. 41.4%, p = 0.029, OR 0.69, CI 0.5–0.95) and 3DS1 (47.3% vs. 37.5%, p = 0.015, OR 0.67, CI 0.48–0.92) were more frequently presented in controls than patients, conferring protection against the lung cancer. Table 2 also shows the results with further assessment of KIR genes and their associations with lung cancer subtypes (NSCLC, SCLC).
Table 2.
KIR gene frequencies among patients with lung cancer and healthy controls.
| KIR genes | Healthy controls | Lung cancer | Comparisons | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lung cancer | NSCLC | SCLC | Lung cancer versus HC | NSCLC versus HC | SCLC versus HC | |||||
| n = 448 | n = 232 | n = 179 | n = 53 | p value | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | |
| %F (N + /n) | %F (N + /n) | %F (N + /n) | %F (N + /n) | |||||||
| Group-A haplotype-associated KIR genes | ||||||||||
| 2DL1 | 98.0 (439/448) | 97.4 (226/232) | 97.2 (174/179) | 98.1 (52/53) | ||||||
| 2DL3 | 88.2 (395/448) | 85.3 (198/232) | 84.4 (151/179) | 88.6 (47/53) | ||||||
| 3DL1 | 92.6 (415/448) | 95.3 (221/232) | 96.6 (173/179) | 92.4 (49/53) | ||||||
| 2DS4 | 92.9 (416/448) | 94.8 (220/232) | 95.5 (171/179) | 92.4 (49/53) | ||||||
| 2DS4fl | 12.9 (31/242) | 10.4 (18/174) | 10.2 (14/137) | 10.8 (4/37) | ||||||
| 2DS4del | 73.5 (178/242) | 70.6 (123/174) | 70.8 (97/137) | 70.3 (26/37) | ||||||
| 2DS4fl,del | 13.6 (33/242) | 19.0 (33/174) | 19.0 (26/137) | 18.9 (7/37) | ||||||
| Group-B haplotype-associated KIR genes | ||||||||||
| 2DL2 | 55.4 (248/448) | 67.2 (156/232) | 65.9 (118/179) | 71.6 (38/53) | 0.003 | 1.63 (1.17–2.26) | 0.016 | 1.56 (1.087–2.24) | 0.027 | 2.04 (1.09–3.82) |
| 2DL5 | 66.1 (296/448) | 63.8 (148/232) | 64.2 (115/179) | 62.2 (33/53) | ||||||
| 3DS1 | 47.3 (212/448) | 37.5 (87/232) | 36.8 (66/179) | 39.6 (21/53) | 0.015 | 0.67 (0.48–0.92) | 0.02 | 0.65 (0.43–0.93) | ||
| 2DS1 | 50.2 (225/448) | 41.4 (96/232) | 41.3 (74/179) | 41.5 (22/53) | 0.029 | 0.69 (0.5–0.95) | ||||
| 2DS2 | 54.5 (244/448) | 62.9 (146/232) | 62.0 (111/179) | 66.0 (35/53) | 0.041 | 1.42 (1.02–1.96) | ||||
| 2DS3 | 41.5 (186/448) | 40.1 (93/232) | 39.6 (71/179) | 41.5 (22/53) | ||||||
| 2DS5 | 39.5 (177/448) | 32.8 (76/232) | 32.9 (59/179) | 32.0 (17/53) | ||||||
| Framework genes/pseudogenes | ||||||||||
| 2DL4 | 100 (448) | 100 (232) | 100 (179) | 100 (53) | ||||||
| 3DL2 | 100 (448) | 100 (232) | 100 (179) | 100 (53) | ||||||
| 3DL3 | 100 (448) | 100 (232) | 100 (179) | 100 (53) | ||||||
| 2DP1 | 97.8 (438) | 96.1 (223) | 94.9 (170) | 100 (53) | ||||||
| 3DP1 | 100 (448) | 100 (232) | 100 (179) | 100 (53) | ||||||
N+ number of individuals positive for the gene, n number of individuals tested for the gene, HC healthy control, OR odds ratio, CI confidence interval. p < 0.05: statistically significant; based on two-tailed Fisher’s exact test.
Risk-association of KIR-HLA combinations with lung cancer
To evaluate the contribution of KIR-HLA combinations to lung cancer risk, we analyzed the distribution of KIR genes and cognate HLA-I ligands within a group of 85 patients along with 178 HCs (Table 3). Similar to the results with individual KIR genes, coexistence of 2DL2/C1 (55.3% vs. 38.3%, p = 0.026, OR 1.99, CI 1.11–3.56) and 2DS2/C1 (50.8% vs. 34.7%, p = 0.036, OR 1.93, CI 1.08–3.46) were found to occur more frequently in lung cancer patients than controls. A significantly less frequent carriage of 3DL1-Bw4 combination was detected in patients with lung cancer than HCs (44.2% vs. 62%, p = 0. 014, OR 0.48, CI 0.25–0.91). However, no significant differences were observed between the two groups for the prevalence of 2DS1 and 3DS1 genes combined with respective HLA-C2 and HLA-Bw4 ligands, suggesting that particular educational states of NK cells may alter NK cell functionality in the lung cancer setting.
Table 3.
Frequency of KIR-HLA combinations, and HLA class-I ligands among patients with lung cancer and healthy controls.
| KIR/HLA | Healthy controls | Lung cancer | Lung cancer versus HC | |
|---|---|---|---|---|
| n = 448 | n = 232 | p value | OR (95% CI) | |
| %F (N + /n) | F% (N + /n) | |||
| KIR-binding motif | ||||
| HLA-C1 | 74.2 (124/167) | 81.5 (53/65) | ||
| HLA-C2 | 73.6 (123/167) | 72.3 (47.65) | ||
| HLA-Bw4 | 62.1 (95/153) | 48.1 (25/52) | ||
| Bw4T80 | 45.7 (70/153) | 44.2 (23/52) | ||
| Bw4I80 | 16.3 (25/153) | 3.8 (2/52) | 0.018 | 0.2 (0.047–0.9) |
| HLA-A3/A11 | 39.2 (31/79) | 38.1 (24/63) | ||
| HLA-A23/24/25/32 | 49.3 (39/79) | 44.4 (28/63) | ||
| KIR-HLA combination | ||||
| 3DL2 + A3/A11 | 39.2 (31/79) | 38.1 (24/63) | ||
| 2DL1 + C2 | 71.8 (120/167) | 72.3 (47/65) | ||
| 2DL3 + C1 | 68.8 (115/167) | 69.2 (45/65) | ||
| 2DL2 + C1 | 38.3 (64/167) | 55.3 (36/65) | 0.026 | 1.99 (1.11–3.56) |
| 3DL1 + Bw4 | 62.0 (95/153) | 44.2 (23/52) | 0.014 | 0.48 (0.25–0.91) |
| 2DS1 + C2 | 26.6 (45/169) | 27.7 (18/65) | ||
| 2DS2 + C1 | 34.7 (58/167) | 50.8 (33/65) | 0.036 | 1.93 (1.08–3.46) |
| 3DS1 + Bw4 | 20.2 (31/153) | 15.3 (8/52) | ||
N+ number of individuals positive for the gene, n number of individuals tested for the gene, HC healthy control, OR odds ratio, CI confidence interval. p < 0.05: statistically significant; based on two-tailed Fisher’s exact test.
When HLA-I ligands were analyzed separately, HLA-Bw4 (Ile80) was less prevalent (3.8% vs. 16.3%, p = 0.018, OR 0.2, CI 0.47–0.9) in lung cancer patients than in controls (Table 3). This points to the possibility that the difference in the distribution of 3DL1-Bw4 combination could be driven by Bw4 (Ile80). The results of comparing the frequency of remaining HLA-I genes didn’t reach the level of statistical significance.
Subgroup analysis wasn’t accomplished regarding the association of KIR-HLA combinations and HLA-I ligands with different lung cancer subtypes owing to the inadequate sample size included in the HLA typing method.
Susceptibility/resistance influence of specific Bx genotype-associated gene clusters on lung cancer risk
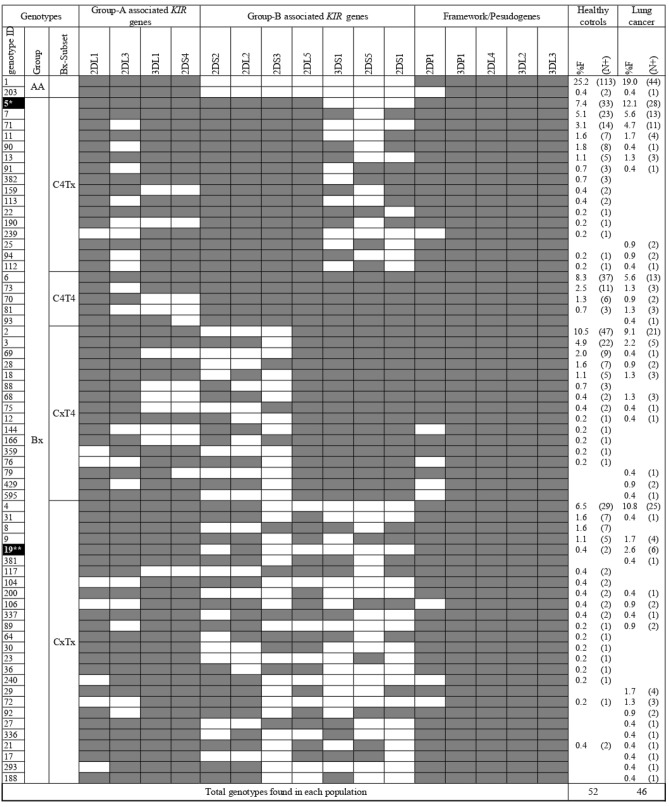
KIR genotype profiles of 232 lung cancer patients and 448 HCs are listed in Table 4. A set of 65 genotypes differentiated by KIR gene content were detected in a total of 680 study participants from southern Iran. Thirty-three genotypes occurred in both patients and controls, 13 genotypes occurred only in patients, and 19 genotypes occurred only in controls.Genotype ID5 (12.1% vs. 7.4%, p = 0.048, OR 1.73, CI 1.015–2.93) was significantly more frequent in patients in comparison with controls. Strikingly, the genotype ID19, which is a rare CxTx genotype carrying 2DL2 in the absence of 2DS2 was associated with a more than fivefold increase in lung cancer risk (2.4% vs. 0.4%, p = 0.021, OR 5.92, CI 1.18–29.58) (Fig. 1).
Table 4.
KIR genotype frequencies among patients with lung cancer and healthy controls.
| KIR | Healthy controls | Lung cancer | Comparisons | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lung cancer | NSCLC | SCLC | Lung cancer versus HC | NSCLC versus HC | SCLC versus HC | |||||
| n = 448 | n = 232 | n = 179 | n = 53 | p value | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | |
| %F (N+) | %F (N+) | %F (N+) | %F (N+) | |||||||
| KIR genotypes | ||||||||||
| AA | 25.7 (115) | 19.4 (45) | 19.5 (35) | 16.9 (9) | ||||||
| Bx | 74.3 (333) | 80.6 (187) | 80.4 (144) | 83.1 (44) | ||||||
| CxT4 | 22.7 (102) | 17.6 (41) | 18.4 (33) | 15.0 (8) | ||||||
| C4Tx | 23.4 (105) | 28.4 (66) | 29.6 (53) | 24.5 (13) | ||||||
| C4T4 | 12.7 (57) | 9.5 (22) | 8.37 (15) | 13.2 (7) | ||||||
| CxTx | 15.4 (69) | 25.0 (58) | 24.0 (43) | 30.2 (16) | 0.0035 | 1.83 (1.23–2.71) | 0.015 | 1.74 (1.13–2.66) | 0.011 | 2.37 (1.25–4.5) |
| C4 Linkage groups | 36.2 (162) | 37.9 (88) | 37.9 (68) | 37.7 (20) | ||||||
| T4 Linkage groups | 35.4 (159) | 27.1 (63) | 26.8 (48) | 28.3 (15) | 0.03 | 0.67 (0.47–0.97) | 0.0039 | 0.66 (0.45–0.97) | ||
N + number of individuals positive for the gene, n number of individuals tested for the gene, HC healthy control, OR odds ratio, CI confidence interval. p < 0.05: statistically significant; based on two-tailed Fisher’s exact test.
Figure 1.
KIR gene content diversity among patients with lung cancer and healthy controls. N+ : number of individuals positive for the gene; n: number of individuals tested for the gene; Gene content of 65 KIR genotypes are displayed by presence/shaded boxes or absence/white boxes of 16 KIR genes. Distribution of genotypes with ID5, and ID19 highlighted by dark boxes were found to be significantly different between lung cancer patients and controls. *p = 0.048, OR 1.73, 95% CI: (1.015–2.93); **p = 0.021, OR 5.92, 95% CI: (1.18– 29.58).
The distribution of main genotypes AA and Bx were comparable among the two groups. Significant differences in the frequencies of Bx genotype subsets categorized based on C4 and T4 gene clusters were observed, whereby T4 gene cluster (35.4% vs. 27.1%, p = 0.03, OR 0.67, CI 0.47–0.97) was found to be associated with reduced risk of lung cancer in our study population. In contrast, we noticed significantly more frequent CxTx subset (25% vs. 15.4%, p = 0.0035, OR 1.83, CI 1.23–2.71) in patients indicating an association between this subset and increased risk of lung cancer (Table 4). KIR genotype frequencies and their statistical associations with lung cancer subtypes (NSCLC, SCLC) are summarized in Table 4.
Risk-association of co-existing susceptibility factors with lung cancer
To explore further the possible associations of carrying gene contents varied in the number of inhibitory and activating KIR genes on susceptibility to lung cancer, we assessed comparisons with regard to different numbers of iKIRs and aKIRs (Table 5). Carriage of genotypes with iKIRs > 4 was more likely presented in patients (75% vs. 67%, p = 0.035, Pc = 0.14, OR 1.48, CI 1.036–2.11), on the contrary genotypes with aKIRs > 4 were more frequently found in HCs (50% vs. 40.1%, p = 0.015, Pc = 0.06, OR 0.66, CI 0.48–0.92). Lung cancer patients were more likely to carry genotypes with iKIRs > aKIRs (64.7% vs. 52.2%, p = 0.002, Pc = 0.008, OR 1.67, CI 1.2–2.32), and this difference remained significant after being corrected for multiple comparisons, suggesting the strong association of genotypes with iKIRs > aKIRs with susceptibility to lung cancer.
Table 5.
Carrier frequency of various susceptibility-related factor combinations among patients with lung cancer and healthy controls.
| KIR combination | Healthy controls | Lung cancer | Lung cancer versus HC | ||
|---|---|---|---|---|---|
| n = 448 | n = 232 | p value | Pc | OR (95% CI) | |
| %F (N + /n) | F% (N + /n) | ||||
| KIR genotypes | |||||
| iKIR > aKIR | 52.2 (234/448) | 64.7 (150/232) | 0.002 | 0.008* | 1.67 (1.2–2.32) |
| aKIR > iKIR | 19.4 (87/448) | 13.4 (31/232) | |||
| iKIR > 4 | 67.0 (300/448) | 75.0 (174/232) | 0.035 | 0.14* | 1.48 (1.036–2.11) |
| aKIR > 4 | 50.0 (224/448) | 40.1 (93/232) | 0.015 | 0.06* | 0.66 (0.48–0.92) |
| KIR combined genotypes | |||||
| CxTx/2DS2 | 11.1 (50/448) | 20.2 (47/232) | 0.0074 | 0.066** | 1.84 (1.18–2.88) |
| CxTx/2DL2 | 12.3 (55/448) | 23.7 (55/232) | 0.0012 | 0.011** | 2.04 (1.34–3.13) |
| iKIR > aKIR carriers subgroup | |||||
| 2DS2 presence | 45.2 (105/232) | 59.3 (89/150) | 0.0087 | 0.078** | 1.76 (1.16–2.67) |
| 2DL2 presence | 49.1 (114/232) | 66.6 (100/150) | 0.0008 | 0.0072** | 2.05 (1.35–3.17) |
N+ number of individuals positive for the gene, n number of individuals tested for the gene, HC healthy control, OR odds ratio, CI confidence interval. p < 0.05: statistically significant; based on two-tailed Fisher’s exact test; Pc: corrected p values, Pc*: correction factor = 4, Pc**: correction factor = 9.
We next performed a comparative analysis to explore whether simultaneous inheritance of disease risk-related factors influences disease susceptibility (Table 5). We found that patients with lung cancer were more likely to carry the combination of CxTx/2DS2 compared to controls (20.2% vs. 11.1%, p = 0.0074, Pc = 0.066, OR 1.84, CI 1.18–2.88). The likelihood of carrying CxTx/2DL2 combination was also significantly higher in patients (23.7% vs. 12.3%, p = 0.0012, Pc = 0.011, OR 2.04, CI 1.34–3.13). Likewise, disease susceptibility was conferred by the presence of 2DL2 within individuals carrying iKIRs > aKIRs (66.6% vs. 49.1%, p = 0.0008, Pc = 0.0072, OR 2.05, CI 1.35–3.17), this association was weakened in the presence of 2DS2 (59.3% vs. 45.2%, p = 0.0087, Pc = 0.078, OR 1.76, CI 1.16–2.67).
Discussion
In the present study, we assessed the contribution of KIR gene content and their corresponding HLA-I ligands to lung cancer development in the ethnically homogeneous population of southern Iran. Although previous studies have examined KIRs at genetic, transcriptional, and expression levels in lung cancer, to our knowledge, this is the first report that demonstrates individual KIR genes and certain genotypes seem to be associated with susceptibility to lung cancer. Given our sizeable dataset, suggestive interactions between KIR-HLA class I ligands can influence the dynamics of NK cell responses in the lung cancer setting.
Despite existing research addressing the KIR-HLA pair's role in lung cancer, their findings are less consistent. Most recently, in the Chinese Han population, studies conducted by Li et al.42 and Yu et al.43, found no association between KIRs and KIR-HLA combinations with metastatic NSCLC (mSCLC) and adenocarcinoma, respectively. Consisting with these findings, Wisniewski et al. couldn’t find a significant difference between KIR genes or combinations of KIR-HLA in 269 Polish Caucasians with NSCLC compared with 690 HCs44. However, Wisniewski et al. reported carriers of homozygous HLA-C1 and C2 were more frequent in NSCLC patients, which was not detected in our study44. Furthermore, Al Omar et al. observed significantly increased 2DL1/C2 and decreased 2DL3/C1 in NSCLC patients from England and Northern Ireland45. Decreased frequency of Ile80 allele in NSCLC patients positive for 3DL1/Bw4 and decreased Thr80 allele in SCLC patients positive for 3DS1/Bw4, which is observed by Al Omar et al.45, Conforms to our findings of less frequent 3DS1, 3DL1/Bw4 in lung cancer patients. In part, these inconsistent results may be elucidated by the small sample size, heterogeneity of the target population (study population), and divergent distribution of KIRs and HLAs in different ethnic groups. Importantly, cross-talk of NK cells and the unique microenvironment of each lung cancer subtype46 could be responsible for behavioral differences in NK cells, assessing distinct histologic subtypes including (mNSCLC, NSCLC, and SCLC) can presumably lead to conflicting results observed in mentioned studies.
Although we didn’t examine survival rate and response to treatment in lung cancer patients, previous studies obtained interesting results. Yu et al. noted that chemotherapy-treated mNSCLC patients with KIR2DS4del and HLA-Bw4 (Thr80) gene expression at the mRNA level exhibited poor overall survival (OS)43. Wisniewski et al. reported the striking association of 2DL2/2DS2/C1 combination with more prolonged survival and better response to therapy in Polish patients44, which is discordant with the predisposing effect of 2DL2/C1, 2DS2/C2 on lung cancer risk observed in our study. Given the cancer setting, it is crucial to consider the impact of chemotherapy agents on the sensitization of tumor cells to NK cell activity. As it has been shown that stress signals induced by chemotherapy and other treatment modalities can elevate the expression of NK cell-activating ligands47–49 or downregulate inhibitory ligands50,51, the transient deleterious effect of chemotherapeutic agents on NK cells has also been observed52,53.
Our findings primarily determined an association between the carriage of KIR2DL2 and its activating counterpart 2DS2 with an increased risk of lung cancer. We identified similar results when further analyzing 2DL2 and 2DS2 in the presence of their corresponding HLA-C1 allele. Consistent with our results, 2DL2 has been demonstrated to confer susceptibility to endometriosis54, leukemia55, and could be predisposing to lymph node metastasis (LNM) in HNSCC as well56. The carrier frequency of 2DL2/C1 in malignant melanoma patients with the advanced stage was significantly higher compared with lower-stage patients57. Similar results were reported by Naumova et al. showing the association of 2DL2/C1 with malignant melanoma58. Additionally, 2DL2 and 2DS2 have been shown to confer a predisposition to lymphatic invasion in ER + and PR + breast cancer cases59.
According to the “licensing” model, NK cell education via inhibitory receptors integrating with cognate HLA-I ligands endows NK cells with full effector functions and self-tolerance34, while NK cell licensing by iKIRs translates into effective sensing of missing HLA I targets, “missing-self” hypothesis, aKIR mediated licensing in the presence of its HLA I ligand induces hypo-responsiveness and renders NK cells impaired responsiveness60. Analysis of infiltration pattern and immune cell localization in NSCLC patients revealed that HLA-I negative tumors are predominantly TIL-free and encapsulated by stromal tissue, which consists of a dense structure of FAP + fibroblasts61. In addition, reduced TIL infiltration, bigger tumor size, and lymphatic spread have been observed among HLA-I−/PD-L1+ tumors62. To this extent, our finding implies that NK cells of 2DL2/C1 carriers are incapable of mounting an efficient response against lung cancer tumors sustaining HLA-I expression due to a defect in “missing-self” recognition. Stromal tissue surrounding tumor lesions, which restrains TILs, including NK cells61, could hypothetically represent another immune escape mechanism to avoid NK cell attack in lung cancer with total loss or downregulated HLA-I. It is shown that KIRs recognize altered peptides presented by cognate HLA-I ligands63, indicating that alterations in peptide repertoire mostly occurring in the process of tumorigenesis could be detected by KIRs64. As a result, stimulation of aKIRs with neoantigens and tumor-inducible ligands expressed on lung cancer cells may prompt cytokine release instead of cytolytic function. Supporting examples would be recent studies in which β2-microglobulin–independent ligand has been suggested to be recognized by 2DS265 and 2DS4 interacting with melanoma-derived non-class I MHC proteins66. The 2DS2-mediated education in the presence of C1 ligand could raise activation threshold and cause hypo-responsiveness in carriers of 2DS2/C1 combination, it also remains possible that upregulated “induced self” ligands mentioned above are unable to overcome such hypo-responsiveness. Due to the tight LD between 2DS2/2DL2, the co-carriage of this combination may exacerbate the detrimental impact attributed to individual genes. More investigation is needed to distinguish the predisposing effect of these two genes on lung cancer.
Moreover, the frequency of KIR2DS1 and 3DS1 in the HC group was higher than in patients. We couldn’t detect a significant association of 2DS1/C2, 3DS1/Bw4 combinations with lung cancer, though the HLA-Bw4 (Ile80) allele and KIR3DL1/Bw4 were strongly associated with protection against lung cancer. NK cells expressing 2DS1 exhibit an anergy state in individuals carrying HLA-C2/C2, but not in HLA-C1/Cx carriers67. Likewise, Bw4 (Ile80) could be recognized by 3DS1 positive NK cells derived from donors lacking Bw4 (Ile80), in contrast with NK cells from donors positive for Bw4 (Ile80)68. Referring to these studies, it can be suggested that NK cells generated from those 3DS1 and 2DS1 carriers lacking putative HLA-Bw4 and HLA-C2 ligands have lower activation threshold and might recognize different ligands associated with lung tumor transformation. Therefore, carrying 3DS1 and 2DS1 in the absence of cognate HLA class I ligands could confer better protection against lung cancer. Supporting this explanation, HLA-F open conformers (OCs) have been reported as high-affinity ligands for 3DS169, and Kiani et al. have shown that 3DS1 positive NK cells could be activated upon ligation with HLA-F in which stimulation with HLA-F results in increased antiviral function in NK cells70. Interestingly a highly expressed level of HLA-F has been detected in lung cancer71, suggesting that HLA-F might be a factor in the association of 3DS1/2DS1 with protection in lung cancer. Furthermore KIRs exhibit various degrees of peptide selectivity, implying that KIRs are sensitive to altered peptides, and this sensory mechanism is more sensitive than “missing self-detection” of lowered HLA-I expression on target cells72. Regarding iKIRs, several studies have reported changing peptide repertoire in tumor cells function as peptide antagonism, which down-modulates NK cell inhibition by reducing inhibitory ligands on tumor cells, caused by low-affinity interaction of KIR-HLA73,74. The protective effect conferred by 3DL1/Bw4 could result from 3DL1 sensitivity to subtle alterations in presented peptides leading to a reduction in inhibitory signals and triggering cytotoxicity in the absence of HLA downregulation.
Estimating the immunological genetic profile could preferably characterize the pathogenesis of lung cancer. Our findings with more frequent T4 gene cluster carriers in HCs, which is likely imparted by the presence of 3DS1/2DS1 combination in T4 gene cluster, are in contravention of CxT4 predisposing role in head and neck squamous cell carcinoma and colorectal adenocarcinoma reported in our previous studies of the same population75,76. The association of CxTx with lung cancer risk is in line with susceptibility to meningioma reported in CxTx carriers77. The results of comparing the different number of iKIR and aKIR genes, in addition to signifying the strong positive association with lung cancer risk in individuals carrying more inhibitory genes; displayed the influence of activating genes on protection against disease regardless of not being significant after the p value correction.
As we emphasized the tight LD between 2DL2 and 2DS2, it is difficult to dissect the contribution of individual 2DL2 and 2DS2 genes in susceptibility to the disease. Hence, assessing rare genotypes lacking either 2DL2 or 2DS2 is highly informative. Accordingly, we noticed a striking association of the genotype ID19, a rare CxTx genotype carrying 2DL2 in the absence of 2DS2, with increased lung cancer risk by more than fivefold, suggesting a predominant detrimental impact of 2DL2 over 2DS2. Regarding our analysis of co-existing susceptibility factors with lung cancer, simultaneous inheritance of CxTx/2DL2 was shown to predispose carriers to lung cancer. Although, the simultaneous presence of CxTx/2DS2 did not meet the significant level after p value correction and seems to confer a slightly lower risk of lung cancer compared to CxTx/2DL2. Superior adverse effect was noticeably related to individuals with iKIR > aKIR in the presence of 2DL2 rather than 2DS2. Our findings are strengthened by the report denoting the correlation of higher expression in inhibitory KIRs with poor prognosis in lung cancer patients. A higher proportion of NK cells expressing inhibitory KIRs was noticed in NSCLC patients in which lower cytotoxicity and reduced IFN-γ production were also shown78. Consistently, separate consideration of inhibitory and activating counterparts demonstrated the susceptibility of 2DL1+/S1− genotype carriers to cutaneous melanoma and the formation of sentinel lymph node metastasis within individuals with homozygous HLA-C279. A previous study by Momot et al. suggested that carrying 2DS2+/L2− combination is related to a higher risk of scleroderma disease80, and similar results were illustrated regarding systemic sclerosis81. However, this disagreement with our observations might be due to different mechanisms involved in tumorigenesis and autoimmune disorders. The fact that the difference of genotypes with iKIRs > aKIRs and CxTx combined with 2DL2 but not 2DS2 remained significant after correction of p values implies that these combinations are significantly more strongly associated with lung cancer development. It can also be presumed that the presence of 2DL2 intensifies disease associations.
It is important to highlight the suppressive effects on NK cells suggested to be driven by alveolar macrophages and epithelial lining fluid of the lower respiratory tract82. Despite comprising a well-differentiated phenotype, NK cells in lung tissue are exposed to a restricting microenvironment in homeostasis, causing a hypo-functional state of lung NK cells to stimuli in comparison with NK cells of peripheral blood83,84. Attenuated killing potency in TME owing to the presence of regulatory T cells (TREGs) and myeloid-derived suppressor cells (MDSCs), limitations of leukocyte infiltration, as well as immunosuppressive factors in lung TME such as adenosine and transforming growth factor β (TGF-β) are yet to be overcome4,85. Taken together, it can be speculated that carrying more inhibitory gene content may interfere with NK-mediated immunosurveillance, favoring tumor evasion in the existing suppressor context of lung tissue. On the other end, activating gene content is presumed to restore the functional competence of NK cells, especially in suppressive lung settings.
In conclusion, we report new susceptibility factors and the contribution of KIR and HLA-I encoding genes to lung cancer risk, highlighting an array of genetic effects and disease setting that regulates NK cell responsiveness. Our results suggest that inherited KIR genes and HLA-I ligands specifying the educational state of NK cells can modify lung cancer risk. In the current study, HLA-I ligands and co-associations of KIR-HLA were examined for a limited number of subjects; also an inadequate number of patients with different lung cancer subtypes hindered the evaluation of KIRs and HLA-I genes impact within subgroups. Thus, larger cohorts assessing the contribution of KIR-HLA combinations are needed to confirm these associations. Functional analysis might help extend associations to the potential therapeutic strategies against lung cancer. The unique microenvironment of each lung cancer subtype with a varied composition of immune cells is assumed to affect NK cell characteristics86. Further investigations on mechanisms involved in NK cell dysfunction in different subtypes might develop into NK-based immunotherapies in lung cancer.
Materials and methods
Study subjects
A total of 232 patients with lung cancer (comprising NSCLC: squamous cell carcinoma and adenocarcinoma, SCLC subtypes) and 448 healthy controls (HC) from a homogenous population of the southern part of Iran (Fras province) were included in this case–control study. The enrolled group of 232 unrelated lung cancer patients was made up 85.3% of men and 14.7% of women with a mean age of 64.4 ± 11.1. Healthy controls were comprised 66.5% of men and 33.5% of women with a mean age of 58.13 ± 12.66. The pathologically confirmed lung cancer cases were recruited from Faghihi hospital, Shiraz University of medical sciences. Age and sex-matched HCs with no Family History of Cancer (FHC) were selected from the Motahari clinic. The demographic and clinical characteristics of lung cancer patients were gathered from medical records (Table 1). Due to the fact that smoking status for a very small portion of our lung cancer patients and HCs was available, stratification of the study population based on smoking status wasn’t carried out.
Informed consent from all research participants was obtained, and the study was carried out according to the declaration of Helsinki. This study was reviewed and ethically approved by the Medical Ethics Committee of Shiraz University of Medical Sciences (IR.SUMS.REC.1398.1110).
KIR genotyping
Genomic DNA extraction from whole blood samples was performed using QIAamp DNA Mini Kit (Qiagen, Germany) as detailed in the manufacturer’s instructions. The PCR-SSP method was used for genotyping 16 KIR genes, and KIR2DS4 variants (KIR2DS4fl: 2DS4 full variant, and KIR2DS4del: 2DS4 deleted variant) as previously described87. Detailed information on primer sequences, thermal conditions, and the mixture of each reaction are reported in our previous study59. Reference DNA samples from the UCLA KIR exchange program provided by Prof. Raja Rajalingam were applied to ensure typing accuracy. An alternative SSP-PCR method was used to confirm the unique and unusual KIR genotyping88.
The KIR-binding HLA-A, B, and C ligands of 85 lung cancer patients and 178 HCs were typed using the recently developed direct DNA sequencing method. The strategy includes PCR amplifying exons 2 and 3 using HLA-A, B, or C gene-specific primers and direct sequencing of the segment that encodes the KIR-binding region. The process was accomplished in accordance with the method described by Ashouri et al.89.
Data analysis and statistical methods
KIR genotypes of the study participants were assigned according to previous studies88,90. The genotype AA comprises fixed gene content (2DL3-2DL1-2DP1-3DL1-2DS4) surrounded by frameworks. Carriers of AA genotype-related genes were regarded as homozygous AA, and the remaining subjects were considered as Bx genotype carriers which can be heterozygous AB or homozygous BB. The KIR genotype ID was obtained using the allele frequency database (http://www.allelefrequencies.net/) for all participants.
Based on the linkage disequilibrium, two frequently occurring clusters that include distinct sets of B-haplotype-specific KIR genes have been identified91. C4 linkage group comprising KIR2DS2-2DL2-2DS3-2DL5B genes is located in the centromeric region of the KIR complex, while the T4 linkage group contains KIR3DS1-2DL5A-2DS5-2DS1 genes located at the telomeric region of the complex. Concerning the presence or absence of C4 and T4 linkage groups, The Bx genotype carriers were further divided into the following four subsets: C4Tx, CxT4, C4T4, and CxTx89. The frequency of C4 and T4 gene clusters were defined by subsequent formulas: C4 = nC4Tx + nC4T4, and T4 = nCxT4 + nC4T4 (n: number of individuals with a particular subset within each group).
The percentage of KIR genes in both study groups was indicated by direct counting (number of subjects positive for the gene divided by the number of subjects per population × 100). Differences in the distribution of each KIR gene, genotypes, KIR-binding HLA ligands, and KIR-HLA pairs between lung cancer patients and HCs were estimated by two-tailed Fisher Exact probability (p) test using SPSS (IBM, US) version 16.0 and Items with p < 0.05 were considered as statistically significant. Moreover, the Odds ratio (OR) and 95% Confidence Intervals (CI) were calculated to assess the magnitude of associations. The method expounded by Svejgaard and Ryde92 was applied to identify the combined effect of the lung cancer susceptibility factors CxTx+/2DL2+ and CxTx+/2DS2+. p values regarding the association of genotypes with certain number of genes (aKIRs > iKIRs, iKIRs > aKIRs, aKIRs > 4, iKIRs > 4) were corrected using Pn = 1 − (1 − P)n, where n represents the number of comparisons92.
Ethical approval
Ethical approval of the research was confirmed by the Medical Ethics Committee of Shiraz University of Medical Sciences [IR. SUMS.REC.1398.1110].
Acknowledgements
The study was financially supported by Shiraz University of Medical Sciences (Grant Number: 18267) and partly by Shiraz Institute for Cancer Research (Grant Number: ICR-100-508).
Author contributions
M.H.L. and E.A. performed K.I.R. and H.L.A. ligands typing. M.H.L. prepared the first draft of manuscript. S.B. contributed to the statistical analysis and edited the first draft of the manuscript. S.M.A.G. provided samples and clinical data. A.G. and R.R. designed the study, performed the interpretation, and edited the manuscript final version. The paper was reviewed and approved by all authors.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Abbas Ghaderi, Email: ghaderia@sums.ac.ir.
Raja Rajalingam, Email: Rajalingam.Raja@ucsf.edu.
References
- 1.Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: Epidemiology, etiology, and prevention. Clin. Chest Med. 2011;32:605–644. doi: 10.1016/j.ccm.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borish L. The immunology of asthma: Asthma phenotypes and their implications for personalized treatment. Ann. Allergy Asthma Immunol. 2016;117:108–114. doi: 10.1016/j.anai.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morvan MG, Lanier LL. NK cells and cancer: You can teach innate cells new tricks. Nat. Rev. Cancer. 2016;16:7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 5.Smyth MJ, Hayakawa Y, Takeda K, Yagita H. New aspects of natural-killer-cell surveillance and therapy of cancer. Nat. Rev. Cancer. 2002;2:850–861. doi: 10.1038/nrc928. [DOI] [PubMed] [Google Scholar]
- 6.Cong J, Wei H. Natural killer cells in the lungs. Front. Immunol. 2019;10:1416. doi: 10.3389/fimmu.2019.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong J, et al. Dysfunction of natural killer cells by FBP1-induced inhibition of glycolysis during lung cancer progression. Cell Metab. 2018;28:243–255.e5. doi: 10.1016/j.cmet.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 8.Takeda K, et al. IFN-γ production by lung NK cells is critical for the natural resistance to pulmonary metastasis of B16 melanoma in mice. J. Leukoc. Biol. 2011;90:777–785. doi: 10.1189/jlb.0411208. [DOI] [PubMed] [Google Scholar]
- 9.Takanami I, Takeuchi K, Giga M. The prognostic value of natural killer cell infiltration in resected pulmonary adenocarcinoma. J. Thorac. Cardiovasc. Surg. 2001;121:1058–1063. doi: 10.1067/mtc.2001.113026. [DOI] [PubMed] [Google Scholar]
- 10.Villegas FR, et al. Prognostic significance of tumor infiltrating natural killer cells subset CD57 in patients with squamous cell lung cancer. Lung Cancer. 2002;35:23–28. doi: 10.1016/s0169-5002(01)00292-6. [DOI] [PubMed] [Google Scholar]
- 11.Carrega P, et al. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(-) cells and display an impaired capability to kill tumor cells. Cancer. 2008;112:863–875. doi: 10.1002/cncr.23239. [DOI] [PubMed] [Google Scholar]
- 12.Platonova S, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011;71:5412–5422. doi: 10.1158/0008-5472.can-10-4179. [DOI] [PubMed] [Google Scholar]
- 13.Bruno A, et al. The proangiogenic phenotype of natural killer cells in patients with non-small cell lung cancer. Neoplasia. 2013;15:133–142. doi: 10.1593/neo.121758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djaoud Z, Parham P. HLAs, TCRs, and KIRs, a triumvirate of human cell-mediated immunity. Annu. Rev. Biochem. 2020;89:717–739. doi: 10.1146/annurev-biochem-011520-102754. [DOI] [PubMed] [Google Scholar]
- 15.Olcese L, et al. Human and mouse killer-cell inhibitory receptors recruit PTP1C and PTP1D protein tyrosine phosphatases. J. Immunol. 1996;156:4531–4534. [PubMed] [Google Scholar]
- 16.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 17.Pyo CW, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE. 2010;5:e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norman PJ, et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ordóñez D, et al. Duplication, mutation and recombination of the human orphan gene KIR2DS3 contribute to the diversity of KIR haplotypes. Genes Immun. 2008;9:431–437. doi: 10.1038/gene.2008.34. [DOI] [PubMed] [Google Scholar]
- 20.Single RM, Martin MP, Meyer D, Gao X, Carrington M. Methods for assessing gene content diversity of KIR with examples from a global set of populations. Immunogenetics. 2008;60:711–725. doi: 10.1007/s00251-008-0331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guethlein LA, Norman PJ, Hilton HG, Parham P. Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol. Rev. 2015;267:259–282. doi: 10.1111/imr.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardiner CM, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J. Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 23.Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Variable NK cell receptors exemplified by human KIR3DL1/S1. J. Immunol. 2011;187:11–19. doi: 10.4049/jimmunol.0902332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajalingam R. Diversity of killer cell immunoglobulin-like receptors and disease. Clin. Lab. Med. 2018;38:637–653. doi: 10.1016/j.cll.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Dębska-Zielkowska J, et al. KIR receptors as key regulators of NK cells activity in health and disease. Cells. 2021;10:1777. doi: 10.3390/cells10071777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J. Exp. Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Döhring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J. Immunol. 1996;156:3098–3101. [PubMed] [Google Scholar]
- 28.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 30.Chewning JH, Gudme CN, Hsu KC, Selvakumar A, Dupont B. KIR2DS1-positive NK cells mediate alloresponse against the C2 HLA-KIR ligand group in vitro. J. Immunol. 2007;179:854–868. doi: 10.4049/jimmunol.179.2.854. [DOI] [PubMed] [Google Scholar]
- 31.Biassoni R, et al. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur. J. Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 32.Stewart CA, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubreuil L, Chevallier P, Retière C, Gagne K. Relevance of polymorphic KIR and HLA class I genes in NK-cell-based immunotherapies for adult leukemic patients. Cancers. 2021;13:3767. doi: 10.3390/cancers13153767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anfossi N, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 35.Kim S, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3053–3058. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J. Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez MI, et al. Maturation of paneth cells induces the refractory state of newborn mice to Shigella infection. J. Immunol. 2008;180:4924–4930. doi: 10.4049/jimmunol.180.7.4924. [DOI] [PubMed] [Google Scholar]
- 38.Bari R, et al. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. 2009;114:5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Béziat V, Hilton HG, Norman PJ, Traherne JA. Deciphering the killer-cell immunoglobulin-like receptor system at super-resolution for natural killer and T-cell biology. Immunology. 2017;150:248–264. doi: 10.1111/imm.12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bossé Y, Amos CI. A decade of GWAS results in lung cancer. Cancer. Epidemiol. Biomarkers. Prev. 2018;27:363–379. doi: 10.1158/1055-9965.EPI-16-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musolf AM, et al. Familial lung cancer: A brief history from the earliest work to the most recent studies. Genes. 2017;8:E36. doi: 10.3390/genes8010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, et al. The association of HLA/KIR genes with non-small cell lung cancer (adenocarcinoma) in a Han Chinese population. J. Cancer. 2019;10:4731–4738. doi: 10.7150/jca.33566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H, et al. Typing of killer-cell immunoglobulin-like receptors and their cognate human leukocyte antigen class I ligands predicts survival of Chinese Han patients with metastatic non-small-cell lung cancer. Mol. Clin. Oncol. 2017;6:279–285. doi: 10.3892/mco.2016.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiśniewski A, et al. KIR2DL2/S2 and HLA-C C1C1 genotype is associated with better response to treatment and prolonged survival of patients with non-small cell lung cancer in a Polish Caucasian population. Hum. Immunol. 2012;73:927–931. doi: 10.1016/j.humimm.2012.07.323. [DOI] [PubMed] [Google Scholar]
- 45.Al Omar S, et al. Associations between genes for killer immunoglobulin-like receptors and their ligands in patients with solid tumors. Hum. Immunol. 2010;71:976–981. doi: 10.1016/j.humimm.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 46.Seo JS, Kim A, Shin JY, Kim YT. Comprehensive analysis of the tumor immune micro-environment in non-small cell lung cancer for efficacy of checkpoint inhibitor. Sci. Rep. 2018;8:14576. doi: 10.1038/s41598-018-32855-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abruzzese MP, et al. Inhibition of bromodomain and extra-terminal (BET) proteins increases NKG2D ligand MICA expression and sensitivity to NK cell-mediated cytotoxicity in multiple myeloma cells: Role of cMYC-IRF4-miR-125b interplay. J. Hematol. Oncol. 2016;9:134. doi: 10.1186/s13045-016-0362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fionda C, et al. Inhibition of glycogen synthase kinase-3 increases NKG2D ligand MICA expression and sensitivity to NK cell-mediated cytotoxicity in multiple myeloma cells: Role of STAT3. J. Immunol. 2013;190:6662–6672. doi: 10.4049/jimmunol.1201426. [DOI] [PubMed] [Google Scholar]
- 49.Fionda C, et al. Heat shock protein-90 inhibitors increase MHC class I-related chain A and B ligand expression on multiple myeloma cells and their ability to trigger NK cell degranulation. J. Immunol. 2009;183:4385–4394. doi: 10.4049/jimmunol.0901797. [DOI] [PubMed] [Google Scholar]
- 50.Hogg SJ, et al. BET-bromodomain inhibitors engage the host immune system and regulate expression of the immune checkpoint ligand PD-L1. Cell Rep. 2017;18:2162–2174. doi: 10.1016/j.celrep.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi J, et al. Bortezomib down-regulates the cell-surface expression of HLA class I and enhances natural killer cell-mediated lysis of myeloma. Blood. 2008;111:1309–1317. doi: 10.1182/blood-2007-03-078535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markasz L, et al. Effect of frequently used chemotherapeutic drugs on the cytotoxic activity of human natural killer cells. Mol. Cancer Ther. 2007;6:644–654. doi: 10.1158/1535-7163.mct-06-0358. [DOI] [PubMed] [Google Scholar]
- 53.Sako T, et al. Cellular immune profile in patients with non-small cell lung cancer after weekly paclitaxel therapy. Acta Oncol. 2004;43:15–19. doi: 10.1080/02841860310016226. [DOI] [PubMed] [Google Scholar]
- 54.Marin MLC, et al. Inhibitory KIR2DL2 Gene: Risk for deep endometriosis in euro-descendants. Reprod. Sci. 2021;28:291–304. doi: 10.1007/s43032-020-00255-x. [DOI] [PubMed] [Google Scholar]
- 55.Verheyden S, Bernier M, Demanet C. Identification of natural killer cell receptor phenotypes associated with leukemia. Leukemia. 2004;18:2002–2007. doi: 10.1038/sj.leu.2403525. [DOI] [PubMed] [Google Scholar]
- 56.Barani S, Khademi B, Ghaderi A. KIR2DS4, KIR2DL2, and KIR2DS4del are linked with basaloid tumors, lymph node metastasis, advanced stage and metastatic risk in head and neck squamous cell carcinoma. Exp. Mol. Pathol. 2020;112:104345. doi: 10.1016/j.yexmp.2019.104345. [DOI] [PubMed] [Google Scholar]
- 57.Kandilarova SM, et al. The influence of HLA and KIR genes on malignant melanoma development and progression. Arch. Immunol. Ther. Exp. (Warsz) 2016;64:73–81. doi: 10.1007/s00005-016-0437-3. [DOI] [PubMed] [Google Scholar]
- 58.Naumova E, et al. Genetic polymorphism of NK receptors and their ligands in melanoma patients: Prevalence of inhibitory over activating signals. Cancer Immunol. Immunother. 2005;54:172–178. doi: 10.1007/s00262-004-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hematian Larki M, Barani S, Talei AR, Ghaderi A. Diversity of KIRs in invasive breast cancer patients and healthy controls along with the clinical significance in ER/PR/HER2+ patients. Genes Immun. 2020;21:380–389. doi: 10.1038/s41435-020-00117-1. [DOI] [PubMed] [Google Scholar]
- 60.He Y, Tian Z. NK cell education via nonclassical MHC and non-MHC ligands. Cell Mol. Immunol. 2017;14:321–330. doi: 10.1038/cmi.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perea F, et al. The absence of HLA class I expression in non-small cell lung cancer correlates with the tumor tissue structure and the pattern of T cell infiltration. Int. J. Cancer. 2017;140:888–899. doi: 10.1002/ijc.30489. [DOI] [PubMed] [Google Scholar]
- 62.Perea F, et al. HLA class I loss and PD-L1 expression in lung cancer: Impact on T-cell infiltration and immune escape. Oncotarget. 2018;9:4120–4133. doi: 10.18632/oncotarget.23469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajagopalan S, Long EO. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J. Exp. Med. 1997;185:1523–1528. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carrillo-Bustamante P, de Boer RJ, Keşmir C. Specificity of inhibitory KIRs enables NK cells to detect changes in an altered peptide environment. Immunogenetics. 2018;70:87–97. doi: 10.1007/s00251-017-1019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thiruchelvam-Kyle L, et al. The activating human NK cell receptor KIR2DS2 recognizes a β2-microglobulin-independent ligand on cancer cells. J. Immunol. 2017;198:2556–2567. doi: 10.4049/jimmunol.1600930. [DOI] [PubMed] [Google Scholar]
- 66.Katz G, et al. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J. Immunol. 2004;173:1819–1825. doi: 10.4049/jimmunol.173.3.1819. [DOI] [PubMed] [Google Scholar]
- 67.Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaëlsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood. 2010;115:1166–1174. doi: 10.1182/blood-2009-09-245746. [DOI] [PubMed] [Google Scholar]
- 68.Carlomagno S, et al. KIR3DS1-mediated recognition of HLA-*B51: Modulation of KIR3DS1 responsiveness by self HLA-B allotypes and effect on NK cell licensing. Front. Immunol. 2017;8:581. doi: 10.3389/fimmu.2017.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia-Beltran WF, et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat. Immunol. 2016;17:1067–1074. doi: 10.1038/ni.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kiani Z, et al. HLA-F on HLA-Null 721.221 cells activates primary NK cells expressing the activating killer Ig-like receptor KIR3DS1. J. Immunol. 2018;201:113–123. doi: 10.4049/jimmunol.1701370. [DOI] [PubMed] [Google Scholar]
- 71.Lin A, et al. HLA-F expression is a prognostic factor in patients with non-small-cell lung cancer. Lung Cancer. 2011;74:504–509. doi: 10.1016/j.lungcan.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 72.Moesta AK, Parham P. Diverse functionality among human NK cell receptors for the C1 epitope of HLA-C: KIR2DS2, KIR2DL2, and KIR2DL3. Front. Immunol. 2012;3:336. doi: 10.3389/fimmu.2012.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cassidy S, et al. Peptide selectivity discriminates NK cells from KIR2DL2- and KIR2DL3-positive individuals. Eur. J. Immunol. 2015;45:492–500. doi: 10.1002/eji.201444613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fadda L, et al. Peptide antagonism as a mechanism for NK cell activation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10160–10165. doi: 10.1073/pnas.0913745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barani S, Khademi B, Ashouri E, Ghaderi A. KIR2DS1, 2DS5, 3DS1 and KIR2DL5 are associated with the risk of head and neck squamous cell carcinoma in Iranians. Hum. Immunol. 2018;79:218–223. doi: 10.1016/j.humimm.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 76.Barani S, Hosseini SV, Ghaderi A. Activating and inhibitory killer cell immunoglobulin like receptors (KIR) genes are involved in an increased susceptibility to colorectal adenocarcinoma and protection against invasion and metastasis. Immunobiology. 2019;224:681–686. doi: 10.1016/j.imbio.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 77.Barani S, Taghipour M, Ghaderi A. Positive association of Bx genotype, KIR2L5, KIR2DS5 and full-length KIR2DS4 with the risk of meningioma. Immunobiology. 2020;225:151900. doi: 10.1016/j.imbio.2019.151900. [DOI] [PubMed] [Google Scholar]
- 78.Al Omar SY, Marshall E, Middleton D, Christmas SE. Increased killer immunoglobulin-like receptor expression and functional defects in natural killer cells in lung cancer. Immunology. 2011;133:94–104. doi: 10.1111/j.1365-2567.2011.03415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campillo JA, et al. KIR gene variability in cutaneous malignant melanoma: Influence of KIR2D/HLA-C pairings on disease susceptibility and prognosis. Immunogenetics. 2013;65:333–343. doi: 10.1007/s00251-013-0682-0. [DOI] [PubMed] [Google Scholar]
- 80.Momot T, et al. Association of killer cell immunoglobulin-like receptors with scleroderma. Arthritis Rheum. 2004;50:1561–1565. doi: 10.1002/art.20216. [DOI] [PubMed] [Google Scholar]
- 81.Salim PH, et al. Killer cell immunoglobulin-like receptor (KIR) genes in systemic sclerosis. Clin. Exp. Immunol. 2010;160:325–330. doi: 10.1111/j.1365-2249.2010.04095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robinson BW, Pinkston P, Crystal RG. Natural killer cells are present in the normal human lung but are functionally impotent. J. Clin. Invest. 1984;74:942–950. doi: 10.1172/JCI111513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marquardt N, et al. Human lung natural killer cells are predominantly comprised of highly differentiated hypofunctional CD69-CD56dim cells. J. Allergy. Clin. Immunol. 2017;139:1321–1330.e4. doi: 10.1016/j.jaci.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 84.Wang J, et al. Lung natural killer cells in mice: Phenotype and response to respiratory infection. Immunology. 2012;137:37–47. doi: 10.1111/j.1365-2567.2012.03607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chiossone L, Dumas PY, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 2018;18:671–688. doi: 10.1038/s41577-018-0061-z. [DOI] [PubMed] [Google Scholar]
- 86.Björkström NK, Ljunggren HG, Michaëlsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat. Rev. Immunol. 2016;16:310–320. doi: 10.1038/nri.2016.34. [DOI] [PubMed] [Google Scholar]
- 87.Vilches C, Castaño J, Gómez-Lozano N, Estefanía E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70:415–422. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 88.Du Z, Gjertson DW, Reed EF, Rajalingam R. Receptor-ligand analyses define minimal killer cell Ig-like receptor (KIR) in humans. Immunogenetics. 2007;59:1–15. doi: 10.1007/s00251-006-0168-4. [DOI] [PubMed] [Google Scholar]
- 89.Ashouri E, et al. Coexistence of inhibitory and activating killer-cell immunoglobulin-like receptors to the same cognate HLA-C2 and Bw4 ligands confer breast cancer risk. Sci. Rep. 2021;11:7932. doi: 10.1038/s41598-021-86964-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ashouri E, Farjadian S, Reed EF, Ghaderi A, Rajalingam R. KIR gene content diversity in four Iranian populations. Immunogenetics. 2009;61:483–492. doi: 10.1007/s00251-009-0378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Du Z, Sharma SK, Spellman S, Reed EF, Rajalingam R. KIR2DL5 alleles mark certain combination of activating KIR genes. Genes Immun. 2008;9:470–480. doi: 10.1038/gene.2008.39. [DOI] [PubMed] [Google Scholar]
- 92.Svejgaard A, Ryder LP. HLA and disease associations: Detecting the strongest association. Tissue Antigens. 1994;43:18–27. doi: 10.1111/j.1399-0039.1994.tb02291.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.