Graphical abstract
Keywords: Pediococcus pentosaceus, Bacteriocin, Pan-genome, Comparative genomics
Abstract
Bacteriocins are antimicrobial peptides produced by different species of bacteria, especially the Gram-positive lactic acid bacteria (LAB). Pediococcus pentosaceus is widely applied in the industry and stands out as Bacteriocin-Like Inhibitory Substances (BLIS) producer known to inhibit pathogens commonly considered a concern in the food industries. This study aimed to perform in silico comparisons of P. pentosaceus genomes available in the public GenBank database focusing on their pediocin-like bacteriocins repertoire. The pan-genome analysis evidenced a temporal signal in the pattern of gene gain and loss, supporting the hypothesis that the complete genetic repertoire of this group of bacteria is still uncovered. Thirteen bacteriocin genes from Class II and III were predicted in the accessory genome. Four pediocin-like bacteriocins (54% of the detected bacteriocin repertoire) and their accompanying immunity genes are highlighted; penocin A, coagulin A, pediocin PA-1, and plantaricin 423. Additionally, in silico, modeling of the pediocin-like bacteriocins revealed different configurations of the helix motif compared to other physically determined pediocin-like structures. Comparative and phylogenomic analyses support the hypothesis that a dynamic mechanism of bacteriocin acquisition and purging is not dependent on the bacterial isolation source origin. Synteny analysis revealed that while coagulin A, pediocin PA-1, and Plantaricin 423 loci are associated with insertion sequences mainly from the IS30 family and are likely of plasmid origin, penocin A lies in a conserved chromosomal locus. The results presented here provide insights into the unique pediocin-like bacteriocin peptide fold, genomic diversity, and the evolution of the bacteriocin genetic repertoire of P. pentosaceus, shedding new insights into the role of these biomolecules for application in inhibiting bacterial pathogens, and suggesting that prospecting and sequencing new strains is still an alternative to mining for new probiotic compounds.
1. Introduction
Over the years, the massive use of antibiotics has led to increased antimicrobial resistance, a current public health issue [1]. Alternative methods have been developed to avoid and reduce this increased resistance worldwide. The use of probiotics, which are microorganisms able to provide potentially therapeutic benefits, has been proved to be an effective food additive that brings several beneficial effects [2], [3]. As a promising trend in biotechnology [4], these organisms have been increasingly studied in the last decades and are widely used in the food industry and animal husbandry [5].
For instance, the Gram-positive lactic acid bacterium (LAB), Pediococcus pentosaceus (Firmicutes phylum, Bacilli class, and Lactobacillales order), are present in different environments, including food, plants, beverages, and feces [6], and has been extensively isolated and assessed for probiotic potential exhibiting positive results [7]. P. pentosaceus attracts substantial attention from the pharmaceutical, scientific, and food sectors because it produces antimicrobial peptides (AMP), known as bacteriocins that inhibit pathogens, such as Listeria monocytogenes Pseudomonas aeruginosa, Escherichia coli, and Clostridium perfringens [8], [9], [10].
The use of genomics to assess the potential of these probiotic species has revealed remarkable information regarding their main biological processes and physiological roles involved in inhibiting pathogens in different hosts [7]. Indeed, it is essential to investigate the production of bacteriocins – AMP produced by LAB with biopreservative ability and broad-spectrum inhibitory activity [11] – which have been “generally regarded as safe” [12]. Those are reported to be produced by many LAB and can be grouped into different classes depending on their function, structure, and mode of action [13].
Class I bacteriocins comprehend peptides that undergo post-translational modifications and contain lanthionine and/or methyllantionine residues, also called lantibiotics. Class II refers to small bacteriocins that do not suffer post-translational modification, categorized into different subclasses depending on their structures. Class III is a more recent class of large, heat-labile peptides, which are not classified as antibacterial [13].
Mainly, bacteriocins that are known to inhibit L. monocytogenes belong to the class IIA (pediocin-like) and are commonly arranged in operons, with well-known function and organization [14], containing the structural bacteriocin gene. The pediocin-like gene cluster encodes three primary genes; the prebacteriocin; an immunity protein; an ABC transporter, indispensable for secretion of the pediocin-like peptide; and a complementary gene of unknown function [11]. The prominent structural landmark of pediocin-like peptides is the presence of a YGNGV-motif, which gives them aS-shaped fold in their N-terminus, influencing their stability [13].
Although many studies have experimentally revealed the functional role of bacteriocin-like inhibitory substances (BLIS) produced by P. pentosaceus isolated from different sources [15], [16], [17], the employment of genomic analyses to perform a comparatively genomic assessment of these AMP can further evince particularities among strains and their correspondent roles associated to inhibitory function [6]. Therefore, this study aimed to perform an extensive in silico comparative analysis of P. pentosaceus, focusing on the genomic analysis across all available sequenced strains to assess their bacteriocin-producing abilities, as well as particularities of pediocin-like bacteriocin sequences, structures, and loci, relating these findings to their potential functional roles.
2. Material and methods
2.1. Pediococcus pentosaceus genomes retrieval and pan-genome analysis
After an initial search of the GenBank RefSeq database [18], 109 P. pentosaceus genomes were retrieved using the esearch script from the NCBI Edirect tool (https://dataguide.nlm.nih.gov/edirect/documentation.html). Furthermore, 67 other P. pentosaceus raw reads deposited on the SRA (Sequence Read Archive) database were retrieved using the fasterq-dump script from the SRA toolkit v3 (https://github.com/ncbi/sra-tools). The raw reads were trimmed with the fastp tool v0.20 [19] and further assembled with SPades 3.15.2 [20]. To normalize the analyses, all 176 P. pentosaceus genomes were annotated by Prokka [21]. The genome completeness was assessed by CheckM [22] for each P. pentosaceus fasta sequence, and only genomes showing completeness above 95 % and contamination below 5 % were considered for pan-genome analysis. All P. pentosaceus genomic metadata (e.g., isolation source, country of origin, number of contigs, assembly N50 and L50, number of genes, completeness, and others) were retrieved for the respective BioProject and BioSample and manually compiled in Table S1. The Panaroo pipeline [23] was employed to determine the P. pentosaceus pan- and core genome. The pan-genome graph-based visualization was determined with Cytoscape (https://cytoscape.org/) using the final_graph.gml file generated by the Panaroo pipeline. The pan-genome pre-processing was based on multidimensional scaling (MDS) projection of pairwise Mash [24] distances and gene and contig counts plots generated by the Panaroo pipeline. The pan-genome post-processing was performed with the panstripe tool (https://github.com/gtonkinhill/panstripe) to determine potential evidence of temporal signals in the gene gain and loss pattern. In addition, the accumulation curve was generated with the post-plot-runner.py script from the Panaroo pipeline. For this work, we used the Panaroo default values for pan-genome computation (e.g., the core genome was considered a group of genes shared by>98 % of strains; the soft-core genes were the ones shared by 95 %-98 % of genomes, and shell genes were shared by 15 %-95 % of the genomes. Finally, the cloud gene group refers to those shared by less than 15 % of analyzed genomes).
2.2. Pan-genome annotation and enrichment analysis
The core, soft core, shell, and cloud reference genes were extracted from the “gene_presence_absence.csv” and “gene_data.csv” files generated by the Panaroo pipeline. The reference pan-genome was annotated with the eggNOG-mapper v2 [25]. The enrichment analysis was carried out to test the overrepresentation of Gene Ontology terms in the different pan-genome gene lists (core, soft core, shell, and cloud) with the find_enrichment.py script from the GOATOOLS package [26], using the fisher's exact test, and multiple testing correction. Only results with p-values < 0.05 corrected using Benjamini-Hochberg multiple test correction were considered.
2.3. Phylogenomics analysis
The Pediococcus pentosaceus phylogenomics analysis was inferred using 402 single-copy core genes (397 being single-copy in >= 90.0 of the analyzed species), identified with BUSCO v5.4.2 [27] using the lactobacillales_odb10 database. The 402 single-copy genes were processed using the supermatrix approach with the BUSCO_Phylogenomics pipeline (https://github.com/jamiemcg/BUSCO_phylogenomics). The maximum-likelihood (ML) phylogenomic tree was built using IQ-Tree v2.1.3 [23] with the best-of-fit model WAG + F + R10 according to AIC criteria [28], with the software ModelFinder [29]with 1,000 fast bootstrap replicates. L. lactis strains G121 (NZ_CP053671.2), UC11 (NZ_CP015904.1), II1403 (AE005176.1), and KF147 (NC_013656.1) were used as out-group. The tree drawing was performed using iTOL v5 [30] using the collected genomic metadata and annotation as decoration features.
2.4. Identification and annotation of bacteriocins-encoding Areas of Interest (AOI) and synteny analysis with a focus on pediocin-like peptides
BAGEL4 tool [31] was employed to predict Areas Of Interest (AOI) in the genomes that contained Ribosomally synthesized and post-translationally modified peptides (RiPPs) and bacteriocins. The previously described pediocin-like motifs and conserved domains [32], [33] were identified through sequence alignment and visual inspection using the CLC Genomics Workbench 22.0 (QIAGEN). For the comparative analysis across the different P. pentosaceus AOI encoding pediocin-like peptides, we firstly re-annotated each AOI using prokka with the BAGEL4 proteins database and UniProt [34] set of plasmid-derived sequences (e.g., VirB, TraA, relaxase, and other plasmid derived sequences) using the --proteins parameter. The synteny was determined using the clinker & clustermap.js tool [35]. All figures were manually edited with Inkscape (https://inkscape.org).
2.5. In silico modeling of pediocin-like bacteriocins
The pediocin-like sequences had their leader peptides, including the double-glycine peptides that are cleaved during the protein secretion, identified and removed using SignalP v6.0 [36]. The mature bacteriocins sequences were compared against the available protein structures deposited in the Protein Data Bank (PDB) [37] and UniProt Databases. The in silico modeling of the pediocin-like structures found in P. pentosaceus genomes was performed using the ROBETTA platform [38]. The 3D models were further refined using the 3DRefine server [39], and the SAVES 6.0 pipeline was employed for the stereochemical quality analysis based on the Ramachandram plot using PROCHECK [40]. In addition, the physicochemical properties corresponding to sequences of pediocin-like mature bacteriocins were computed using the ProtParam tool [41].
3. Results
3.1. Pan-genome analysis revealed temporal signals in the pattern of gene gain and loss, supporting the hypothesis that the complete genetic repertoire of Pediococcus pentosaceus is still uncovered
The comparative analyses of the 176 P. pentosaceus genomes available in the GenBank database revealed that the majority of the strains are well assembled (average of 20 contigs/scaffolds), encoding in an average of 1,800 genes, except for P. pentosaceus strains isolated from metagenome projects (Metagenome-Assembled Genomes (MAGs)) exhibiting 1,200 to 1,400 genes and showing genome completeness between 75 and 98 % (Fig. 1 A and B, and Table S1). At least 92 % of P. pentosaceus strains present genome completeness above 98 % (Table S1). The main finding regarding the genome completeness analysis was related to the FHNFQ30 strain, isolated from human feces in China and exhibiting a higher contamination level of 23.14 %. Therefore, the FHNFQ30 strain was removed for the following pan-genome analysis. Interestingly, the SRCM100892 strain, primarily isolated from food material in South Korea, exhibits the most significant number of genes (Fig. 1 B). This strain presents 98.46 % and 0.31 % of genome completeness and contamination, respectively, supporting a well-assembled genome.
Fig. 1.
P. pentosaceus pan-genome pre-processing and post-processing analyses. A and B. The number of contigs and genes distributed across all available P. pentosaceus genomes. C. Multidimensional scaling projection of pairwise Mash distances, indicating that the strain LN-PT16 is potentially misclassified in the Pediococcus genus. D. plot of the phylogeny and the corresponding gene presence/absence of P. pentosaceus pan-genome (showing only genes that vary). E. plot of the fitted gene gain and loss events across P. pentosaceus phylogeny, indicating that there are still occurring gene gain and loss events at the tips of the tree. F. Pan-genome accumulation curve shows the accessory (acc) diversity in function of increasing the number of new genomes added (number of samples). G. Panstripe summary, showing a significant association with gene gain and loss. Core: indicates whether the branch lengths in the phylogeny are associated with gene gain and loss. Depth: indicates whether the rate of gene gain and loss changes significantly with the depth of a branch. Istip: indicates associations with genes observed on the tips of the phylogeny. p: the inferred index parameter of the underlying Tweedie distribution. Phi: the inferred dispersion parameter.
Further inspection of the SRCM100892 strain revealed that in addition to the circular main replicon, exhibiting the average P. pentosaceus genome size of 1.7 Mb, this genome also harbors three shorter and three larger plasmids showing 8, 14, 18, 52, 55, and 68 kb. Therefore, the unique plasmid content can explain the higher number of genes in the SRCM100892 strain. In contrast, the LN-PT16 strain isolated from fermentable vegetals in China appears as an outlier in the MDS projection (Fig. 1 C), indicating potential misclassification. Further analysis of the LN-PT16 strain revealed completeness of 99.38 % and contamination of 0 %. This genome is assembled in 2 Mb showing 36 contigs, with N50 of 341 kb, thus, indicating a high-quality assembly. Furthermore, our phylogenomic analyses also strongly support that this strain does not belong to the Pediococcus genus (see below), supporting the misclassification hypothesis. Therefore, the LN-PT16 was also not used in the further pan-genomic calculations.
After the pre-processing, 167 Pediococcus pentosaceus were considered for the pan-genome analyses. The reference pan-genome comprises 5,749 genes, and the core genome consist of 1,158 genes (representing ∼ 20 % of species coding set). Additionally, 245 genes that belonged to a group of soft-core genes were identified. A group of shell genes, composed of 712 genes, was shared by between 15 % and 95 % of genomes, and a final group of 3,691 cloud genes was shared by less than 15 % of the analyzed isolates (Fig. 1 D and E, and Figure S1). The pan-genome post-processing plots across P. pentosaceus phylogeny indicate that gene gain and loss events occur at the tree's tips. (Fig. 1 E). Furthermore, the accumulation curves and p-value of the core branch lengths corroborated a significant association between gene gain/loss, firmly supporting an open pan-genome scenario (Fig. 1 F and G).
3.2. Pediococcus pentosaceus pan-genome annotation reveals a variable repertoire of 13 bacteriocins in the accessory genome
The cluster of Orthologous Groups (COG) annotation of the reference pan-genome revealed that most of the genes were associated with unknown functions (S) (Fig. 2 A). The other prominent COGs are Replication, recombination, and repair (L), followed by Transcription (K), Carbohydrate transport and metabolism (G), and Cell wall/membrane/envelope biogenesis (M). In contrast, the lesser main COG categories are Cell motility (N), secondary metabolites biosynthesis, transport, and catabolism (Q).
Fig. 2.
A. Clusters of Orthologous Genes (COG) functional annotation of P. pentosaceus reference pan-genome. B. Predicted bacteriocin by automatic annotation using the BAGEL4 platform and manual curation. The pediocin-like peptides are shown with black dots, and the number of genomes carrying each bacteriocin is shown between parenthesis.
Gene Ontology enrichment analyses revealed that across the core genes, the cellular metabolic process (GO:0044237), primary metabolic process (GO:0044238), gene expression (GO:0010467), and many other GO terms related to the primary metabolism and housekeeping are over-represented. The soft-core genes exhibit enrichment of GO terms potentially involved with probiotics features, such as vitamin biosynthetic process (GO:0009110), water-soluble vitamin biosynthetic process (GO:0042364), organic substance metabolic process (GO:0071704), and small molecule biosynthetic process (GO:0044283). In contrast, across the shell genes, only the monosaccharide catabolic process (GO:0046365) is over-represented, and many GO terms related to primary metabolism and housekeeping are underrepresented. Furthermore, the cloud genes do not present any GO term enrichment, and as observed in the shell genes set, the primary metabolism and housekeeping-related GO terms are underrepresented (Table S2).
Among the analyzed P. pentosaceus genomes, 13 bacteriocins, mostly from class II and accounting for up to 286 genes, were identified in the accessory genome (Fig. 2 B and Table S3). A total of 47 (26 %) P. pentosaceus genomes do not encode bacteriocin. Conversely, in the bacteriocinegic strains, the gene content is highly variable, where each strain carries up to three Areas Of Interest (AOI) (mean of 1.78). These findings are compatible with the concept of dynamic genomes regarding bacteriocin content.
The pediocin-like bacteriocins: penocin A, coagulin A, pediocin PA-1, and plantaricin 423, corresponds to ∼ 54 % of the detected bacteriocins. These pediocin-like bacteriocins present the conserved pediocin-box, formed by an N-terminal consensus characteristic YGNGVxCxxxxCxVxWxxA motif commonly found in the class IIA bacteriocins (Figures S2, S3, S4, and S5). Interestingly, the immunity genes entA and pediocin ACH appears associated with the pediocin-like bacteriocins. Alignment analysis indicates three different EntA groups (Figure S6) were identified. In contrast, the ACH immunity proteins are much more conserved, showing slight variations (Figure S7).
The pediocin-like bacteriocins exhibit highly conserved amino acid composition, showing specific particularities. For instance, non-synonymous mutations were identified in some coagulin A members (Figure S2), in particular, across the pediocin box (V → L, T → Y, H → Y, and W → R). In addition, some strains' leader peptide regions of coagulin A and penocin A are not fully present (Figure S2).
The physicochemical profile of predicted pediocin-like bacteriocins indicates that all predicted sequences of mature bacteriocins peptides were positively charged, which relates mainly to their hydrophilic N-terminal regions (Table 1). For these bacteriocins, the net charge is essential for electrostatic interactions once pediocin-like bacteriocins are reported to have their mode of action interacting with negatively charged cell surfaces [11]. Furthermore, all evaluated pediocin-like bacteriocins had a similar molecular weight. Additionally, according to their instability indices, they are stable under normal conditions (should be less than 40) and can be potentially manipulated in test conditions.
Table 1.
Physicochemical properties of pediocin-like mature bacteriocins of P. pentosaceus.
| Characteristics | Bacteriocin | |||
| Penocin A | Coagulin A | Pediocin PA-1 | Plantaricin 423 | |
| Amino acid residues | 42 | 44 | 44 | 37 |
| Molecular weight (Da) | 4689.31 | 4615.19 | 4628.19 | 5719.54 |
| Theoretical pI | 9.45 | 8.85 | 8.85 | 8.93 |
| Extinction Coefficient | 21095 | 14230 | 14230 | 8605 |
| Instability index | -23.82 | -0.48 | -0.48 | 26.56 |
| Aliphatic index | 55.71 | 40.00 | 40.00 | 58.87 |
| Grand Average of Hydropathicity | -0.586 | -0.430 | -0.493 | -0.323 |
3.3. Molecular modeling of pediocin-like bacteriocin reveals similar but unique structural configurations in the helix motif
So far, except for pediocin PA-1, there are no accurate templates in the Protein Data Bank [37] showing higher similarity or close relation to the helix-motif pediocin-like bacteriocins identified in P. pentosaceus (Table 2). The molecular modeling revealed that 87.5 %, 91.2 %, 94.1 %, and 82.8 % of the amino acid residues of the penocin A, coagulin A, pediocin PA-1, and plantaricin 423 are located in the most favorable regions, respectively (Figure S8).
Table 2.
BLAST results show the best templates for homology modeling for each pediocin-like bacteriocin identified in P. pentosaceus genomes. The leader peptide sequence was removed for a more accurate query search.
| Description | Organism | Hits | Coverage | Similarity | PDB |
| Penocin_A |
Latilactobacillus sakei Pediococcus acidilactici Chemically synthesized Enterococcus faeciumM3K31 Carnobacterium maltaromaticum Leuconostoc gelidum Latilactobacillus curvatus |
Sakacin P Pediocin PA-1 Pediocin PA-1 M31L analog Enterocin HF Carnobacteriocin B2 Leucocin A Curvacin A |
88.0% 59.0% 59.0% 61.0% 45.0% 76.0% 83.0% |
62.0% 80.0% 80.0% 73.0% 73.0% 68.0% 68.0% |
1OG7_A 7VLY_A 5UKZ_A 2N4K_A 1CW5_A 1CW6_A 2A2B_A |
| Coagulin_A |
Pediococcus acidilactici Chemically synthesized Latilactobacillus sakei Enterococcus faecium M3K31 Carnobacterium maltaromaticum Leuconostoc gelidum Latilactobacillus curvatus |
Pediocin PA-1 Pediocin PA-1 M31L analog Sakacin P Enterocin HF Carnobacteriocin B2 Leucocin A Curvacin A |
100% 100% 100% 90.0% 44.0% 52.0% 81.0% |
97.0% 97.0% 77.0% 65.0% 84.0% 78.0% 55.0% |
7VLY_A 5UKZ_A 1OHM_A 2N4K_A 1CW5_A 1CW6_A 2A2B_A |
| Pediocin_PA-1 |
Pediococcus acidilactici Chemically synthesized Latilactobacillus sakei Enterococcus faecium M3K31 Leuconostoc gelidum Carnobacterium maltaromaticum Latilactobacillus curvatus |
Pediocin PA-1 Pediocin PA-1 M31L analog Sakacin P Enterocin HF Leucocin A Carnobacteriocin B2 Curvacin A |
100% 100% 100% 90.0% 52.0% 43.0% 81.0% |
100% 100% 77.0% 65.0% 78.0% 84.0% 55.0% |
7VLY_A 5UKZ_A 1OHM_A 2N4K_A 1CW6_A 1CW5_A 2A2B_A |
| Planaricin_423 |
Leuconostoc gelidum Pediococcus acidilactici Chemically synthesized Latilactobacillus sakei Carnobacterium maltaromaticum Enterococcus faecium M3K31 |
Leucocin A Pediocin PA-1 Pediocin PA-1 M31L analog Sakacin P Carnobacteriocin B2 Enterocin HF |
94.0% 81.0% 81.0% 64.0% 54.0% 59.0% |
74.0% 90.0% 90.0% 83.0% 85.0% 81.0% |
1CW6_A 7VLY_A 5UKZ_A 1OHM_A 1CW5_A 2N4K_A |
Except for coagulin A and pediocin PA-1, which present identical peptide structures and very similar sequences; all pediocin-like bacteriocins from P. pentosaceus are structural divergent to the closest related bacteriocins having physical structure determined (e.g., Leucocin A, Sakacin P, and Pediocin PA-1) (Fig. 3). The main differences are located at the helix motif, where alpha-helices present a different number of right-hand turns, and the predicted beta-sheet position slightly differs.
Fig. 3.
A. Molecular modeling of pediocin-like bacteriocins of P. pentosaceus, and comparisons against the peptide fold of other known and related bacteriocin available at PDB. The AxxxA and SxxxS motif in the Pediocin PA-1, Coagulin, and Plantaricin 423 is shown as a purple arrow.; B. Alignment of the primary amino acid sequences of P. pentosaceus coagulin A and pediocin PA-1 against Pediococcus acidilactici and the chemically synthesized Pediocin PA-1 available on the PDB database, showing the differences in the N terminal, AxxxA mofif, and M → L substitution.
As previously determined [42], the differences between coagulin A and pediocin PA-1 are related to substituting one threonine residue at position 44 in the place of asparagine (Fig. 3). Interestingly, both peptides present prominent differences from the Pediocin PA-1 (M31L 5UKZ) physically determined structure. The structural difference can be associated with methionine to conservative valine substitution (different amino acids showing similar biochemical properties) at position 34 (Fig. 3). These results support the idea of distinct functional implications of the P. pentosaceus pediocin-like bacteriocins.
3.4. Phylogenomics comparative analysis reveals that it is not possible to determine a correlation between the bacteriocigenic repertoire and strain isolation source or sampling region
Phylogenomics analyses revealed that P. pentosaceus strains form at least nine different clades. In general, no clear correlation between the bacteriocin repertoire, isolation source, and clades topology can be made, supporting the hypothesis that a dynamic mechanism of bacteriocin acquisition and purging is independent of the evolutive driven forces associated with each isolation source (Fig. 4). Moreover, the phylogenomics analysis strongly supports that the LN-PT16 strain does not belong to P. pentosaceus species. Indeed, the LN-PT16 strain is located between the Lactococcus and P. pentosaceus clades. In addition, the LN-PT16 strain does not encode bacteriocins or AOI.
Fig. 4.
Phylogenomics analyses and evolutionary relationships across available P. pentosaceus genomes associated to the bacteriocin gene content and source of isolation of each strain. The nine different clades are shown in roman numerals. The L. lactis G121, UC11, and KF147 genomes were used as an out-group.
3.5. The penocin A loci are conserved and located in the main chromosome, whereas the coagulin A and pediocin PA-1 loci exhibit variations, presenting association to Insertion Sequences and plasmid origin
Comparative analysis across the four pediocin-like bacteriocins AOI revealed distinct patterns. Each AOI generally exhibits some level of conserved synteny but specific features. For instance, the penocin A AOI is located in the main P. pentosaceus chromosome and has a conserved gene context. Most penocin A genes are next the immunity protein (EntA). Interestingly, except for one strain (DYNDL40), no transposase or other mobile genetic elements, such as plasmid maintenance genes, were identified in the penocin A AOI (Fig. 5). This finding supports the idea that penocin A AOI is currently little affected by P. pentosaceus mobilome.
Fig. 5.
Synteny of penocin A AOI identified in P. pentosaceus genomes. For brevity, the repeated AOI sharing > 95 % of identity and > 95 % of coverage were not shown.
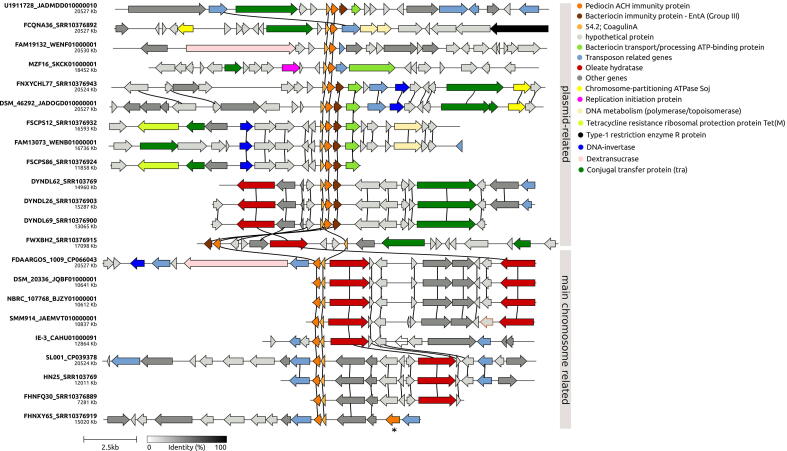
The coagulin A AOI presents a much more diverse genomic context, exhibiting up to two different immunity genes (EntA and pediocin ACH immunity protein) and several transposase genes. For instance, at least six AOI presents an IS30 transposase from the ISLpl1 element, flanking the Coagulin A gene (Fig. 6). Comparative annotation and analysis indicate that the coagulin A AOI are divided into the plasmid-derived and main chromosome groups (Fig. 6). The plasmid-derived group exhibits several genes related to plasmid mobilization, such as chromosome-partitioning ATPase Soj, conjugal transfer, DNA replication, and others. Additionally, the FSCPS12 and FSCPS86 strains carry a tetracycline resistance ribosomal protection protein Tet(M). The Tet(M) gene is commonly associated with plasmids in Lactobacillus species [43].
Fig. 6.
Synteny of coagulin A AOI identified in P. pentosaceus genomes. The asterisk denotes a different bacteriocin immunity gene.
In contrast, the FDAARGOS_1009, SL001, and the other strains located below in the Fig. 6 belong to the main chromosome group. The FDAARGOS_1009 and SL001 strains were fully sequenced and assembled in one circular contig, indicating that their corresponding coagulin A AOI does not present plasmid origin. The main features of the chromosomal AOI group are related to the presence of an oleate hydratase gene (EC 4.2.1.53). This enzyme belongs to the lyases family and is involved with the cleavage of carbon–oxygen bonds. Together, these findings support the hypothesis that the Coagulin A AOI was originated, acquired, or propagated to other compatible strains by conjugation and may also be integrated with the main chromosome.
The pediocin PA-1 AOI exhibits the immunity (pediocin ACH immunity protein), biosynthesis, and transport/processing of bacteriocin (LanT) genes close to the pediocin PA-1 gene (Fig. 7). These AOI are strongly associated with plasmid genes, showing a potential conjugation acquisition origin. Only two strains carry transposase-related genes in their respective AOI. For instance, the ET34 strain presents the IS30 transposase from the ISLpl1 element next to a bacteriocin immunity gene. Moreover, the FAM19080 strain AOI carries a transposase from the IS6 family showing 100 % similarity to the IS1216E element. Interestingly, the bacteriocin transport/processing ATP-binding (LanT) gene appears as a pseudogene in the FWXBH44 and FAM19080 strains.
Fig. 7.
Synteny of pediocin PA-1 AOI identified in P. pentosaceus genomes. The asterisk denotes a different bacteriocin immunity gene.
Finally, the plantaricin 423 AOI also presents the plantaricin 423 genes next to their respective immunity gene (e.g., EntA). Both genes are also next to a truncated/fragment of IS30 transposase (94 % of similarity and 27 % of coverage of the entire transposase) from the ISLpl1 element (Fig. 8). The other features of this AOI are the presence of a 5-deoxy-d-ribulose 1-phosphate aldolase, PTS system, HTH-type transcriptional repressor GlcR, MarR transcriptional regulator, and a cell surface protein. However, this AOI has no apparent and direct relationship to plasmid-derived genes.
Fig. 8.
Synteny of plantaricin 423 AOI identified in P. pentosaceus genomes.
4. Discussion
Due to its ability to produce antimicrobial peptides, the Gram-positive P. pentosaceus received during the last decade significant attention, especially in the food industry [8]. Previous comparative genomics analyses employing a fraction of the available P. pentosaceus genomes revealed distinct functional roles, which may or not be able to associate with strains able to produce bacteriocins [6]. In this study, we performed a more comprehensive P. pentosaceus pan-genome and in silico analyses focusing on the pediocin-like bacteriocins. Using all 176 publicly available P. pentosaceus genomes, we demonstrated that this LAB group encodes unique BLIS characteristics.
Using the graph-based method employed by the panaroo pipeline [23], we determined a more accurate picture of the P. pentosaceus pan-genome. As the first cut-off for pan-genome pre-processing, only high-quality genomes showing CheckM [22] completeness above 95 % and contamination below 5 % were used. This method was enough to determine that 92 % (160 out 176) of P. pentosaceus strains present genome completeness above 98 % and that not all strains derived from metagenomic projects (MAGs) are suitable for pan-genome analysis due to the low completeness and higher contamination levels. However, the genome completeness pre-processing was not adequate to detect other potential issues. For instance, the Mash MDS projection analyses revealed that the LN-PT16 strain appeared to be an outlier in the plot. The Mash is a sequence comparison algorithm that relies only on k-mers, providing accurate comparisons between genomes [24] and an exact estimation of differences that cannot be detected using genome completeness methods.
Furthermore, phylogenomics analyses confirmed these results. Therefore, the Mash distance and phylogenomics methods indicated that the deposited LN-PT16 sequences in the Genbank database were misclassified as P. pentosaceus. Indeed, one of the major concerns is related to the quality of the assembly and annotation of the deposited genomes and the impacts of contamination and low-quality genomes in the results of a pan-genome analysis [23]. The presented results reinforce that concern, pointing out that pre-processing steps using genome completeness, Mash MDS projection, and phylogenomics analyses must be considered before any pan-genome study to avoid inaccurate inferences.
The P. pentosaceus group encodes, on average, 1,800 genes. After thorough pre-processing, the pan-genome analyses revealed 5,749 genes comprising the set of all different genes found in this species. This is equivalent to ∼ 3x the average species coding set. Conversely, the core genome contains 1,158 genes (∼20 % of the species pan-genome set and ∼ 0.6x of the species coding set). However, the comparison of gene gain and loss rates between strains, as well as the pan-genome post-processing based on accumulation curves and panstripe pipeline, revealed that the complete genetic repertoire of the species is still uncovered. Therefore, we corroborated previous results, which used fewer strains (74 in total), indicating that the P. pentosaceus pan-genome is not yet fully closed but approaching closure [6]. However, in this study, we more than doubled the samples of P. pentosaceus, and the pan-genome is still open.
Further annotation and enrichment analysis of the reference pan-genome indicated an over-representation of many functions and processes related to the primary metabolism and housekeeping functions within the core gene set. This result is consistent with the role of core-genome with the bacterial cellular maintenance and survival, as observed in other bacterial genera, including P. pentosaceus [6], [44], [45]. Interestingly, potential probiotic functions, such as vitamin biosynthetic process (GO:0009110), water-soluble vitamin biosynthetic process (GO:0042364), and monosaccharide catabolic process (GO:0046365), are enriched in the soft-core and shell gene set. Conversely, the bacteriocins are features of the P. pentosaceus accessory genome, and contrary to a previous study that identified four different bacteriocin operons in P. pentosaceus [6] accessory genome, here, using much more samples (74 against 175) was possible to determine thirteen bacteriocin genes, from Class II and III.
We also showed the evolutionary distance across the different strains, corroborating previous studies that demonstrated that different P. pentosaceus strains were not correlated based on isolation source or sampling region [6] and also showing that it is impossible to determine a correlation between the bacteriocigenic repertoire with the strains phylogeny. Indeed, our phylogenomics analyses corroborate the hypothesis that the bacteriocin content is not a plesiomorphic characteristic; on the contrary, it may be independently acquired during the evolution and adaptation of each strain. This finding is supported by the fact that bacteriocin production is heavily influenced by its environment, with factors such as nutrition, temperature, pH, and aeration level playing a significant role [11]. In addition, the horizontal transfer of the bacteriocin gene has been demonstrated to instigate metabolic adaptation that increases cellular and adaptation fitness [46].
We also identified, annotated, and compared the P. pentosaceus bacteriocins arsenal, focusing on the pediocin-like bacteriocins, revealing the unique structural patterns of these peptides. The bacteriocin structures presented in this study were modeled using the ROBETTA server, which uses deep learning algorithms to predict protein models [47] accurately. This method can be considered the most reliable alternative when no protein crystals are available [37]. In this regard, all bacteriocins shown in Fig. 3 are of type IIa, demonstrating that pediocin PA-1 is very similar to Coagulin A, as shown in Table 2 (97 %). This is in agreement with what has been shown before [48]. Also, it was possible to observe the presence of the “GxxxG-like” AxxxA and SxxxS motif in the Pediocin PA-1 (AxxxA; 48–52 aa) and Coagulin (AxxxA; 48–52 aa) and in the Plantaricin 423 (SxxxS; 42–46 aa). These motifs are characteristic of type IIb bacteriocins [49] and are known to mediate helix-helix interactions in membrane proteins [50], [51]. Therefore, these motifs are essential. Since they were present in the helix (Fig. 3), it may suggest that these bacteriocins will act as two-peptide bacteriocins to form membrane-penetrating helix-helix structures [52]. Thus, the presented 3D models will provide a good baseline for further studies since none were previously available.
The synteny analyses revealed that penocin A is widespread across the species and located in a very conserved chromosomal genomic context, showing almost no relationship to mobile genetics elements, thereby corroborating previous studies [6], [53]. In contrast, the coagulin A locus is hugely diverse and is an excellent example to illustrate the plasticity of the bacterial genomes. For instance, some coagulin A loci are associated with transposases and plasmid-derived genes, including other antibiotic resistance genes. Conversely, another group of coagulin A locus is present in the chromosomal genomic context. The pediocin PA-1 and plantaricin 423 are more conserved. While the Pediocin PA1 locus is embedded in plasmid-derived genes, the plantaricin 423 exhibits transposase fragments. Interestingly, many pediocin-like AOIs present transposases from the IS30 family. This family is known to form compound transposon structures [54], thereby strongly indicating a potential role of elements from this family in shaping the penocin A, coagulin_A, pediocin_PA-1, and plantaricin_423 AOI evolution in P. pentosaceus.
Coagulin A has a very similar sequence and structure to Pediocin PA-1. It presents anti-Listeria inhibitory activity described from Bacillus coagulans [42], exhibiting high identity with other bacteriocins such as pediocin PA-1 and leucocin A [32]. However, comparing it to the sequence of Penocin A reveals more differences, both in the C- and N-terminal portions. The detected Plantaricin 423 has also been reported to have antilisterial activity when produced by the LAB Lactobacillus plantarum [55]; compared to Pediocin PA-1 and Coagulin A sequences, it is mostly similar. The production of pediocin-like bacteriocins requires the presence of complementary genes grouped in one or more clusters. The composition of these loci is essential for the function of the AMP [48], [56]. Class IIa bacteriocins are known for having components of the mannose phosphotransferase (Man-PTS) system involved in the target recognition, including Leucocin A and Pediocin PA-1 [57], [58]. These permeate the target cell membrane, leading to cell death by employing specific receptors on the target cell surface [59].
Also, the bacteriocin enterolysin is detected in many strains of this analysis and is known to degrade bacterial cell walls and be produced by bacteria of genera Enterococcus, Lactobacillus, Streptococcus, and Pediococcus, showing potential for inhibition of enterococci, pediococci, lactococci, and lactobacilli [60]. This bacteriocin belongs to class III, which are thermostable peptides with a broad inhibitory spectrum [61]. The enterolysin A has only been described recently on P. pentosaceus. Its presence has been associated with prophage genes and thus has been hypothesized that P. pentosaceus have seized its production ability from other species [6]. This is supported by the fact that the enterolysin has not been reportedly encoded by P. pentosaceus, and its operon is placed on mobile elements [6].
Taken together, the presented results suggest that probiotic features may be randomly spread over the different P. pentosaceus strains by different lateral gene transfer mechanisms (i.e., conjugation, transposition, transduction), supporting the hypothesis that new probiotics functions and antimicrobial peptides can eventually be found prospecting and sequencing new strains.
CRediT authorship contribution statement
Iago Rodrigues Blanco: Methodology, Investigation, Formal analysis, Visualization, Writing – original draft. Lucas José Luduverio Pizauro: Methodology, Investigation, Formal analysis, Writing – review & editing. João Victor dos Anjos Almeida: Methodology, Investigation, Formal analysis. Carlos Miguel Nóbrega Mendonça: Methodology, Formal analysis. Alessandro de Mello Varani: Conceptualization, Supervision, Methodology, Writing – review & editing. Ricardo Pinheiro de Souza Oliveira: Supervision, Project administration, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank the São Paulo Research Foundation (FAPESP, Brazil) for financial support under grant 2018/25511-1. The authors also thank the National Council for Scientific and Technological Development (CNPq, Brazil) for financial support through grants 312923/2020-1, 408783/2021-4, and 303061/2019-7, as well as the Coordination for the Improvement of Higher Education Personnel (CAPES, Brazil), Finance Code 001.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.09.041.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Marston H.D., Dixon D.M., Knisely J.M., Palmore T.N., Fauci A.S. Antimicrobial Resistance. JAMA. 2016;316(11):1193–1204. doi: 10.1001/jama.2016.11764. [DOI] [PubMed] [Google Scholar]
- 2.Sanders M.E., Marco M.L. Food formats for effective delivery of probiotics. Annu Rev Food Sci Technol. 2010;1(1):65–85. doi: 10.1146/annurev.food.080708.100743. [DOI] [PubMed] [Google Scholar]
- 3.Nader-Macías M.E.F., Juárez Tomás M.S. Profiles and technological requirements of urogenital probiotics. Adv Drug Deliv Rev. 2015;92:84–104. doi: 10.1016/j.addr.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Strack L., Carli R.C., da Silva R.V., Sartor K.B., Colla L.M., et al. Food biopreservation using antimicrobials produced by lactic acid bacteria. Res, Soc Dev. 2020;9(8) doi: 10.33448/rsd-v9i8.6666. e998986666–e998986666. [DOI] [Google Scholar]
- 5.Reid G., Gadir A.A., Dhir R. Probiotics: Reiterating What They Are and What They Are Not. Front Microbiol. 2019;10:424. doi: 10.3389/fmicb.2019.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang J., Yang B., Ross R.P., Stanton C., Zhao J., et al. Comparative Genomics of Pediococcus pentosaceus Isolated From Different Niches Reveals Genetic Diversity in Carbohydrate Metabolism and Immune System. Front Microbiol. 2020;11:253. doi: 10.3389/fmicb.2020.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang S., Cai L., Lv L., Li L. Pediococcus pentosaceus, a future additive or probiotic candidate. Microb Cell Fact. 2021;20(1):45. doi: 10.1186/s12934-021-01537-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porto M.C.W., Kuniyoshi T.M., Azevedo P.O.S., Vitolo M., Oliveira R.P.S. Pediococcus spp.: An important genus of lactic acid bacteria and pediocin producers. Biotechnol Adv. 2017;35(3):361–374. doi: 10.1016/j.biotechadv.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh B., Sukumar G., Ghosh A.R. Purification and characterization of pediocin from probiotic Pediococcus pentosaceus GS4, MTCC 12683. Folia Microbiol (Praha) 2019;64(6):765–778. doi: 10.1007/s12223-019-00689-0. [DOI] [PubMed] [Google Scholar]
- 10.Grilli E., Messina M.R., Catelli E., Morlacchini M., Piva A. Pediocin A improves growth performance of broilers challenged with Clostridium perfringens. Poult Sci. 2009;88(10):2152–2183. doi: 10.3382/ps.2009-00160. [DOI] [PubMed] [Google Scholar]
- 11.Papagianni M., Pediocins A.S. The bacteriocins of Pediococci. Sources, production, properties and applications. Microb Cell Fact. 2009;8(3) doi: 10.1186/1475-2859-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.] Sabo S da S, Mendes MA, Araújo E da S, Muradian LB de A, Makiyama EN, et al. Bioprospecting of probiotics with antimicrobial activities against Salmonella Heidelberg and that produce B-complex vitamins as potential supplements in poultry nutrition. Sci Rep 2020;10(1):7235. doi:10.1038/s41598-020-64038-9. [DOI] [PMC free article] [PubMed]
- 13.Acedo J.Z., Chiorean S., Vederas J.C., van Belkum M.J. The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol Rev. 2018;42(6):805–828. doi: 10.1093/femsre/fuy033. [DOI] [PubMed] [Google Scholar]
- 14.Drider D., Fimland G., Hechard Y., McMullen L.M., et al. The Continuing Story of Class IIa Bacteriocins. Microbiol Mol Biol Rev. 2006;70(2):564–582. doi: 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Azevedo PO de S, Converti A, Gierus M, Oliveira RP de S. Antimicrobial activity of bacteriocin-like inhibitory substance produced by Pediococcus pentosaceus: from shake flasks to bioreactor. Mol Biol Rep 2019;46(1):461–9. doi:10.1007/s11033-018-4495-y. [DOI] [PubMed]
- 16.de Souza de Azevedo PO, Mendonça CMN, Moreno ACR, Bueno AVI, de Almeida SRY, et al. Antibacterial and antifungal activity of crude and freeze-dried bacteriocin-like inhibitory substance produced by Pediococcus pentosaceus. Sci Rep 2020;10(1):12291. doi:10.1038/s41598-020-68922-2. [DOI] [PMC free article] [PubMed]
- 17.Todorov S.D., Dicks L.M. Bacteriocin production by Pediococcus pentosaceus isolated from marula (Scerocarya birrea) Int J Food Microbiol. 2009;132(2–3):117–126. doi: 10.1016/j.ijfoodmicro.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., et al. GenBank. Nucleic Acids Res. 2020;48(D1):D84–D86. doi: 10.1093/nar/gkz956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 22.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tonkin-Hill G., MacAlasdair N., Ruis C., Weimann A., Horesh G., et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21(1):180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ondov B.D., Treangen T.J., Melsted P., Mallonee A.B., Bergman N.H., et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17(1):132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantalapiedra C.P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol Biol Evol. 2021;38(12):5825–5829. doi: 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klopfenstein D.V., Zhang L., Pedersen B.S., Ramírez F., Warwick Vesztrocy A., et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci Rep. 2018;8(1):10872. doi: 10.1038/s41598-018-28948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manni M., Berkeley M.R., Seppey M., Zdobnov E.M. BUSCO: Assessing Genomic Data Quality and Beyond. Curr Protoc. 2021;1(12):e323. doi: 10.1002/cpz1.323. [DOI] [PubMed] [Google Scholar]
- 28.Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19(6):716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 29.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242-W245. doi:10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed]
- 31.van Heel A.J., de Jong A., Song C., Viel J.H., Kok J., et al. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46(W1):W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fimland G., Johnsen L., Dalhus B., Nissen-Meyer J. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J Pept Sci. 2005;11(11):688–696. doi: 10.1002/psc.699. [DOI] [PubMed] [Google Scholar]
- 33.Nissen-Meyer J., Rogne P., Oppegård C., Haugen H.S., Kristiansen P.E. Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by gram-positive bacteria. Curr Pharm Biotechnol. 2009;10(1):19–37. doi: 10.2174/138920109787048661. [DOI] [PubMed] [Google Scholar]
- 34.UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2018;46(5):2699. doi:10.1093/nar/gky092. [DOI] [PMC free article] [PubMed]
- 35.Gilchrist C.L.M., Clinker C.-H. & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics. 2021;37:2473–2475. doi: 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 36.Teufel F., Almagro Armenteros J.J., Johansen A.R., Gíslason M.H., Pihl S.I., et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 2022;40(7):1023–1025. doi: 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 2004;32(Web Server issue):W526-531. doi:10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed]
- 39.Bhattacharya D., Nowotny J., Cao R., Cheng J. 3Drefine: an interactive web server for efficient protein structure refinement. Nucleic Acids Res. 2016;44(W1):W406–W409. doi: 10.1093/nar/gkw336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 41.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Marrec C., Hyronimus B., Bressollier P., Verneuil B., Urdaci M.C. Biochemical and Genetic Characterization of Coagulin, a New Antilisterial Bacteriocin in the Pediocin Family of Bacteriocins, Produced by Bacillus coagulans I4. Appl Environ Microbiol. 2000;66(12):5213–5220. doi: 10.1128/AEM.66.12.5213-5220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danielsen M. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid. 2002;48(2):98–103. doi: 10.1016/S0147-619X(02)00118-X. [DOI] [PubMed] [Google Scholar]
- 44.Mathee K., Narasimhan G., Valdes C., Qiu X., Matewish J.M., et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A. 2008;105(8):3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rooney A.P., Swezey J.L., Friedman R., Hecht D.W., Maddox C.W. Analysis of core housekeeping and virulence genes reveals cryptic lineages of Clostridium perfringens that are associated with distinct disease presentations. Genetics. 2006;172(4):2081–2092. doi: 10.1534/genetics.105.054601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krauss S, Harbig TA, Rapp J, Schaefle T, Franz-Wachtel M, et al. Horizontal transfer of bacteriocin biosynthesis genes requires metabolic adaptation to improve compound production and cellular fitness. bioRxiv, 2022;2022.07.28.501952. 10.1101/2022.07.28.501952. [DOI] [PMC free article] [PubMed]
- 47.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373(6557):871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui Y., Zhang C., Wang Y., Shi J., Zhang L., et al. Class IIa Bacteriocins: Diversity and New Developments. Int J Mol Sci. 2012;13(12):16668–16707. doi: 10.3390/ijms131216668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acedo J.Z., Towle K.M., Lohans C.T., Miskolzie M., McKay R.T., et al. Identification and three-dimensional structure of carnobacteriocin XY, a class IIb bacteriocin produced by Carnobacteria. FEBS Lett. 2017;591(10):1349–1359. doi: 10.1002/1873-3468.12648. [DOI] [PubMed] [Google Scholar]
- 50.Senes A., Engel D.E., DeGrado W.F. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14(4):465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Senes A., Ubarretxena-Belandia I., Engelman D.M. The Cα—H⋅⋅⋅O hydrogen bond: A determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci U S A. 2001;98(16):9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nissen-Meyer J., Oppegård C., Rogne P., Haugen H.S., Kristiansen P.E. Structure and Mode-of-Action of the Two-Peptide (Class-IIb) Bacteriocins. Probiotics Antimicrob Proteins. 2010;2(1):52–60. doi: 10.1007/s12602-009-9021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martino M.E., Maifreni M., Marino M., Bartolomeoli I., Carraro L., et al. Genotypic and phenotypic diversity of Pediococcus pentosaceus strains isolated from food matrices and characterisation of the penocin operon. Antonie Van Leeuwenhoek. 2013;103(3):1149–1163. doi: 10.1007/s10482-013-9897-1. [DOI] [PubMed] [Google Scholar]
- 54.Siguier P., Gourbeyre E., Varani A., Ton-Hoang B., Chandler M. Everyman’s Guide to Bacterial Insertion Sequences. Microbiol Spectr. 2015;3(2) doi: 10.1128/microbiolspec.MDNA3-0030-2014. MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 55.Vermeulen R.R., Van Staden A.D.P., Dicks L. Heterologous Expression of the Class IIa Bacteriocins, Plantaricin 423 and Mundticin ST4SA, in Escherichia coli Using Green Fluorescent Protein as a Fusion Partner. Front Microbiol. 2020;11:1634. doi: 10.3389/fmicb.2020.01634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mesa-Pereira B., O'Connor P.M., Rea M.C., Cotter P.D., Hill C., et al. Controlled functional expression of the bacteriocins pediocin PA-1 and bactofencin A in Escherichia coli. Sci Rep. 2017;7(1):3069. doi: 10.1038/s41598-017-02868-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diep D.B., Skaugen M., Salehian Z., Holo H., Nes I.F. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci U S A. 2007;104(7):2384–2389. doi: 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bédard F., Hammami R., Zirah S., Rebuffat S., Fliss I., et al. Synthesis, antimicrobial activity and conformational analysis of the class IIa bacteriocin pediocin PA-1 and analogs thereof. Sci Rep. 2018;8(1):9029. doi: 10.1038/s41598-018-27225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kjos M., Nes I.F., Diep D.B. Mechanisms of Resistance to Bacteriocins Targeting the Mannose Phosphotransferase System. Appl Environ Microbiol. 2011;77(10):3335–3342. doi: 10.1128/AEM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Surachat K., Kantachote D., Deachamag P., Wonglapsuwan M. Genomic Insight into Pediococcus acidilactici HN9, a Potential Probiotic Strain Isolated from the Traditional Thai-Style Fermented Beef Nhang. Microorganisms. 2020;9(1):50. doi: 10.3390/microorganisms9010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nilsen T., Nes I.F., Holo H. Enterolysin A, a Cell Wall-Degrading Bacteriocin from Enterococcus faecalis LMG 2333. Appl Environ Microbiol. 2003;69(5):2975–2984. doi: 10.1128/AEM.69.5.2975-2984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.