Abstract
We have conducted a double-blind study to assess the possible involvement of the human herpesviruses (HHVs) HHV6, HHV7, Epstein-Barr virus (EBV), and cytomegalovirus in chronic fatigue syndrome (CFS) patients compared to age-, race-, and gender-matched controls. The CFS patient population was composed of rigorously screened civilian and Persian Gulf War veterans meeting the Centers for Disease Control and Prevention’s CFS case definition criteria. Healthy control civilian and veteran populations had no evidence of CFS or any other exclusionary medical or psychiatric condition. Patient peripheral blood mononuclear cells were analyzed by PCR for the presence of these HHVs. Using two-tailed Fisher’s exact test analyses, we were unable to ascertain any statistically significant differences between the CFS patient and control populations in terms of the detection of one or more of these viruses. This observation was upheld when the CFS populations were further stratified with regard to the presence or absence of major axis I psychopathology and patient self-reported gradual versus acute onset of disease. In tandem, we performed serological analyses of serum anti-EBV and anti-HHV6 antibody titers and found no significant differences between the CFS and control patients.
Chronic fatigue syndrome (CFS) is a poorly understood disease of unknown etiology. It is characterized by a profound state of debilitating fatigue that lasts more than 6 months and does not resolve with bed rest. CFS is accompanied by a plethora of associated symptoms, including fever, sore throat, myalgia, lymphadenopathy, sleep disturbances, neurocognitive difficulties, and depression (19). Frequently, patients report a sudden onset of symptoms following an acute flu-like illness (23). Since many CFS patients present with acute-onset, persistent symptoms reminiscent of a viral infection and have been reported to have elevated serum antiviral antibody titers, a viral component in the pathogenesis of this disease has been suspected. Among the variety of viruses evaluated to date, including enteroviruses (17), retroviruses (18), and human herpesviruses (HHVs) (1, 8, 15, 24), HHV6 has exhibited the most promise as a candidate for a CFS-associated virus (7, 11, 22, 27, 28, 31). Unfortunately, many of the studies addressing the possible involvement of HHV6 in CFS have produced ambiguous results (6, 7, 11, 22, 27, 28, 31). Some studies have relied heavily on HHV6 serological evidence (23, 27), which is known to be insufficient to provide ample evidence of active HHV6 infection (6, 20, 28). Others have examined relatively small CFS populations (11, 22, 31), lacked adequately matched controls (28), or failed to take into account the possible cross-reactivity of reagents utilized with the closely related virus HHV7.
Our goal was to ascertain whether there is any correlation between infection with one or more HHVs in CFS patients compared to age-, race-, and gender-matched controls. The control populations were composed of healthy civilian and Persian Gulf War veterans who exhibited no evidence of CFS or any other exclusionary medical or psychiatric condition. In this study, the focus was on the detection by PCR of HHV6, HHV7, Epstein-Barr virus (EBV), and human cytomegalovirus (HCMV) genomic DNA in circulating peripheral blood mononuclear cells (PBMCs). We also examined the serum anti-HHV6 and anti-EBV antibody titers of a substantial number of patients. The experiments were conducted by a double-blinded protocol.
MATERIALS AND METHODS
Patient selection.
Our patients were both civilians and veterans of the Persian Gulf War. Veterans from 10 states east of the Mississippi River were evaluated by a screening questionnaire designed to identify those with CFS and those who were in good health. After being identified, veterans came to the Veterans Administration Medical Center (VAMC) in East Orange, N.J., where they were evaluated by a physician in the Gulf War Research Center and divided into those fulfilling the 1994 Centers for Disease Control and Prevention CFS case definition (14) (n = 46) and those in good health (n = 32). Veterans with CFS also underwent a computerized psychiatric interview (3, 25) that was administered by trained personnel to identify those with (n = 33) and without (n = 13) concurrent major axis I psychopathology (e.g., major depression, posttraumatic stress disorder, or a generalized anxiety disorder).
Civilians either were self- or physician referred to the Chronic Fatigue Syndrome Research Center (VAMC, East Orange, N.J.) or were screened by the same means used for veterans. Potential research study participants were then evaluated by a neurologist or nurse practitioner and were grouped into those satisfying the 1994 CFS case definition criteria (n = 76) and those in good health (n = 73). Healthy civilians and those with CFS were further evaluated via the computerized psychiatric interview to provide an axis I diagnosis. None of the healthy civilians had an axis I diagnosis, and the CFS group was divided into those with a concurrent axis I diagnosis (n = 32) and those lacking such a diagnosis (n = 44). In both the veteran and civilian research populations, the CFS patients and healthy controls were carefully matched for age, gender, race, and educational level.
Patient demographics.
The Persian Gulf War veteran population was 83% male and 73% Caucasian and had a mean age of 35.3 years (range, 26 to 46). The civilian population was 84% female and 93.5% Caucasian and had a mean age of 35.2 years (range, 26 to 44). In addition, the mean educational levels for the veteran and civilian groups, ± the standard deviations, were 14 ± 1.95 and 15.2 ± 2.60 years, respectively.
Clinical specimens.
Heparinized-blood samples were collected from veterans and civilians at the Gulf War and CFS Research Centers at the VMAC in East Orange, N.J. PBMCs were isolated from the heparinized blood by using Ficoll-Hypaque gradients (Pharmacia, Uppsala, Sweden). Thereafter, PBMCs were aliquoted, pelleted, and immediately frozen at −70°C until required. When possible, a serum sample was also taken from the patient for analysis. All patient PBMC pellets and sera collected were numerically encrypted prior to shipment to our laboratory for analysis.
Avoidance of contamination.
Great care was taken to avoid specimen contamination during the course of this investigation. Sample preparation and analysis of amplified products were conducted in designated physically separated laboratory areas. All reagents were aliquoted and stored in distinct locations. Only dedicated PCR pipetting equipment and aerosol-resistant barrier pipette tips were used. Meticulous laboratory technique and adherence to standard PCR anticontamination procedures were the norm. A number of positive and negative controls were included in each PCR assay.
DNA extraction.
PBMC pellets were resuspended in 450 μl of autoclaved distilled-deionized water and subjected to three rounds of freezing and thawing. Fifty microliters of 10× proteinase K reaction buffer (50 mM KCl, 100 mM Tris-HCl, 15 mM MgCl2; pH 8.3) and 100 μg of proteinase K (Boehringer Mannheim, Indianapolis, Ind.) were added to the cell lysate. Digestion was performed for 90 min at 65°C; this was followed by heat inactivation at 98°C for 20 min. Alternatively, digestion was allowed to proceed overnight at 50°C with gentle shaking, prior heat inactivation. The resultant crude DNA extract was visually examined for the presence of cellular debris. If such debris was present, the extract was centrifuged at 1,000 × g for 5 min and the supernatant was used as a template DNA for subsequent PCR analysis. As a rule, mock DNA extracts, consisting of autoclaved deionized water or JJhan cells (a continuous human T-cell line [30]), were prepared concurrently with the patient PBMC samples to serve as internal negative controls.
In some instances, the crude DNA extracts were further purified by phenol-chloroform extraction followed by ethanol precipitation. Briefly, the crude DNA extract was thoroughly mixed with 2 volumes of 25:24:1 (vol/vol/vol) Tris-buffered phenol-chloroform-isoamyl alcohol–0.1% (wt/vol) 8-hydroxyquinoline and centrifuged at 14,000 × g for 5 min at 4°C. The top, clear, aqueous phase was transferred to a new tube and combined with 2 μl of Pellet Paint coprecipitant (Novagen, Madison, Wis.) with 0.1 volume of 3 M sodium acetate, pH 5.2. Thereafter, 2 volumes of −20°C absolute ethanol were added. The DNA was precipitated by a 30-min incubation at −70°C. The DNA was then pelleted by a 4°C centrifugation at 14,000 × g for 15 min, the supernatant was removed, and the DNA pellet was washed with ice-cold 70% ethanol. The DNA pellet was allowed to air dry and then was resuspended in 200 μl of autoclaved deionized water.
HHV6 PCR protocol.
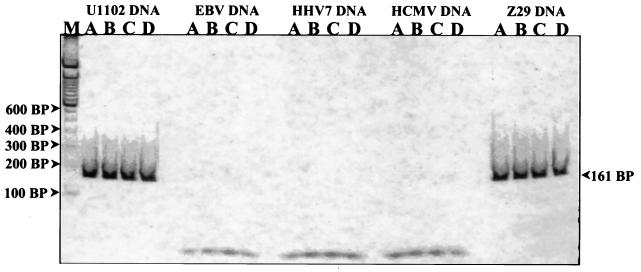
PCR was performed with HHV6 primers, denoted HHV6 A and HHV6 B (Table 1), that amplify a 161-bp region of a strain-conserved open reading frame (ORF), U30 (16, 20, 21). The ORF U30 gene exhibits the highest degree of sequence homology to the HCMV UL47 gene, which encodes a capsid assembly myosin (16). This set of primers is specific for both HHV6 strains and does not exhibit any cross-reaction with HHV7, EBV, or HCMV (Fig. 1).
TABLE 1.
Primers used in this study
| Primer pair | Refer-ence(s) | Sense | Sequence | GenBank locus (accession no.) | Bases spanned | PCR product size (bp) | Target gene |
|---|---|---|---|---|---|---|---|
| TCR-β #1 | 13 | Sense | 5′ GTGTTCCCACCCGAGGTCGCTGTGTTTGAGCC 3′ | C2CDNA | 16–47 | 124 | TCR-β |
| TCR-β #2 | Antisense | 5′ GTGCTGACCCCACTGTGCACCTCCTTCCCATT 3′ | 139–110 | ||||
| HHV6 A | 16, 20, 21 | Sense | 5′ TCTCACAGCCCAGGACAATGGATTATATA 3′ | HHV6AGNM (X83413) | 43155–43183 | 161 | ORF U30 (putative capsid myosin) |
| HHV6 B | Antisense | 5′ TGAGATCATTCTCCCGTTCTTCTTGAGGG 3′ | 43315–43295 | ||||
| HHV7 #1 | 5, 11 | Sense | 5′ CAGAAATGATAGACAGATGTTGG 3′ | HHU43400 (U43400) | 15678–15700 | 124 | ORF U10 |
| HHV7 #2 | Antisense | 5′ TAGATTTTTTGAAAAAGATTTAATAAC 3′ | 15801–15775 | ||||
| EBV #1 | 26 | Sense | 5′ AAGGAGGGTGGTTTGGAAAG 3′ | EBV (V01555) | 109331–109350 | 296 | EBNA1 |
| EBV #2 | Antisense | 5′ AGACAATGGACTCCCTTAGA 3′ | 109628–109609 | ||||
| HCMV #1 | 13 | Sense | 5′ CACGCAACTTTTGGCCGCCACACCTGTCAC 3′ | HS5PPBC (M15120) | 2081–2110 | 419 | Phosphorylated ma-trix protein pp65 |
| HCMV #2 | Antisense | 5′ CACCACGCAGCGGCCCTTGATGTTT 3′ | 2500–2476 |
FIG. 1.
HHV6 primer specificity control. Shown is a nondenaturing polyacrylamide gel stained with a 1:20,000 dilution of Sybr Green I nucleic acid stain (Molecular Probes). The first lane (M) represents a 100-bp DNA ladder (Gibco BRL Life Technologies). Serial twofold dilutions of the various HHV DNAs, ranging from 1,000 to 125 pg (lanes A to D), were amplified with HHV6 PCR primers A and B for a total of 40 cycles. A prominent 161-bp band can be seen in each of the HHV6 A strain U1102 and HHV6 B strain Z29 lanes. In contrast, no bands are apparent in the HHV7, EBV, or HCMV genomic-DNA dilution lanes. Therefore, we conclude that this primer set is specific for HHV6 alone and does not cross-react with its genetically proximate viral relatives.
Thirty microliters of the crude DNA extract was used as the template DNA for each PCR. A 50-μl PCR reaction mixture consisted of 0.25 μM each primer and 0.2 mM each of four deoxynucleoside triphosphates (Gibco BRL Life Technologies, Grand Island, N.Y.) in 10 mM Tris-HCl (pH 8.3)–50 mM KCl-1.5 mM MgCl2, and 1 U of Taq DNA polymerase (Gibco BRL Life Technologies). Before being added to the PCR reaction mixture, the Taq DNA polymerase was incubated for 5 min with Taq Start antibody (Clontech Laboratories, Palo Alto, Calif.). Taq Start inactivates Taq DNA polymerase until it is denatured at 70°C, reducing the amount of mispriming and increasing the chances of amplifying rare target DNAs. Reaction mixtures were initially denatured by heating to 94°C for 5 min and then subjected to 40 successive rounds of cycling as follows: 94°C for 1 min, 55°C for 1 min, and 72°C for 75 s. These cycling conditions were optimized for use with a DNA Thermo Cycler model 480 (Perkin-Elmer Cetus Corporation, Norwalk, Conn.). For each experiment, a number of positive and negative controls were included. The positive controls consisted of serial dilutions of HHV6 genomic DNA. The negative controls consisted of the mock and JJhan cell DNA extracts coupled with water samples that were interspersed within assays to ensure that no cross-contamination occurred during pipetting. Using this PCR protocol, we could routinely detect ≥25 fg of HHV6 genomic DNA. This level of sensitivity permits detection of less than one viral genome copy per cell.
HHV7 PCR protocol.
The HHV7 PCR assay followed the protocol for HHV6 with a few modifications. The primer annealing temperature was increased to 58°C and the optimum MgCl2 concentration was determined to be 4.5 mM. The HHV7 primers used, denoted HHV7 #1 and HHV7 #2 (Table 1), amplify a 124-bp region of a conserved gene named ORF U10 (5, 11). These primers are specific for HHV7 DNA alone and do not cross-react with EBV, HCMV, or either strain of HHV6 (data not shown). Again, this PCR protocol permitted the detection of ≥25 fg of HHV7 genomic DNA.
EBV PCR protocol.
The EBV PCR protocol was the same as that used for HHV7 except the optimum MgCl2 concentration was determined to be 2.5 mM. The EBV primers used, denoted EBV #1 and EBV #2 (Table 1), amplify a conserved region of the EBV nuclear antigen 1 (EBNA1)-encoding BKRF1 gene outside the gly-ala repeat that varies among wild-type EBV strains (26). The resultant PCR product is 296 bp in size. These primers are specific for EBV alone and do not cross-react with HHV7, HCMV, or either strain of HHV6 (data not shown). The sensitivity of this protocol is comparable to those for HHV6 and HHV7.
HCMV PCR protocol.
The HCMV primers used, denoted HCMV #1 and HCMV #2 (Table 1), specifically amplify a 419-bp region of the highly conserved CMV lower-matrix phosphoprotein pp56 gene (13). These primers were designed by Mary Jo Evans (Children’s Hospital, Buffalo, N.Y.), who graciously provided us with their sequences. Currently, these primer sequences are protected by patent applications (13a). The cycling parameters were essentially the same as for HHV6, but the cycle number was 35. These primers are specific for HCMV alone and do not cross-react with HHV7, EBV, or either strain of HHV6 (data not shown). Once more, we were able to routinely obtain sensitivity levels comparable to those of the other viral PCR assays.
TCR-β PCR protocol.
To verify that each patient PBMC DNA extract prepared did indeed contain cellular DNA and to show that inhibitors of PCR amplification were absent, we employed a PCR assay specific for human T-cell receptor beta chain (TCR-β). The human TCR-β gene locus is located on chromosome 7 and is present at a copy number of two per cell genome. The TCR-β primers were designed by Mary Jo Evans, who kindly provided us with their sequences. These primers, denoted TCR-β #1 and TCR-β #2 (Table 1), amplify a 124-bp portion of the TCR-β constant region (13). Each TCR-β primer was used at a final concentration of 0.2 μM. Amplification was carried out as previously described except that the cycling elongation time was shortened to 1 min and the cycle number was reduced to 35. A positive TCR-β result was considered proof that the DNA extraction procedure was successful and that overt PCR inhibitors were absent.
Control DNAs.
Positive-control HHV genomic DNAs, including HHV6 strain A (U1102), HHV6 strain B (Z29), HHV7 strain JI, EBV strain B95-8, and HCMV DNAs, were purchased from Advanced Biotechnologies Inc. (Columbia, Md.). The DNA concentrations of these viral DNA stocks were determined spectrophotometrically. The stocks were serially diluted in the presence of 10 ng of purified JJhan cell DNA/μl, and titrations of them near the limits of the PCR protocol’s detection capabilities were included in each PCR assay. These controls ensured that assay conditions were up to par and also allowed us to gauge the relative amount of PCR product amplified in positive patient PBMC samples. Negative controls consisted of autoclaved deionized water or JJhan cell DNA.
Fluorescence-based detection of amplified products.
During the course of this study, two slightly different methods were employed to evaluate the amplified PCR products. The first involved mixing 20 μl of the amplified PCR product with 5 μl of a gel loading buffer containing 25 μg of bromphenol blue/ml, 25 μg of xylene cyanol FF, 50% (wt/vol) glycerol, and 1:4,000-diluted Sybr Green I nucleic acid gel stain (Molecular Probes, Eugene, Oreg.) and loading it onto a nondenaturing 5% polyacrylamide gel. The products were electrophoresed for 2 h and 15 min at 125 V, with 0.5× TBE (44.5 mM Tris base, 44.5 mM boric acid, 1 mM EDTA; pH 8.0) as the running buffer. After electrophoresis, without the gel being removed from the glass plates, it was inserted into a Fluor Imager model 575 fluorimeter (Molecular Dynamics, Sunnyvale, Calif.) and scanned. The gel image was projected onto a computer screen, and amplified PCR product bands were directly quantitated by using Image QuanT software (Molecular Dynamics).
Alternatively, 20 μl of the PCR product was mixed with 4 μl of a gel loading buffer containing 20% (wt/vol) Ficoll 400 (Pharmacia Biotech, Piscataway, N.J.), 10 mM Na2 EDTA (pH 8.0), and 2.5 μg of xylene cyanol FF/ml and loaded into a 2% Ultra Pure Aqua Por low-electroendosmosis agarose gel (National Diagnostics, Atlanta, Ga.). Electrophoresis was allowed to proceed at 125 V for 2 h and 15 min, with 1× TBE as the running buffer. Afterward, the gel was stained for 20 min in a 1-μg/ml ethidium bromide solution. The gel was then destained for 5 min in distilled-deionized water. The PCR products were visualized by using Molecular Analyst Gel Doc 1000 (Bio-Rad Laboratories, Hercules, Calif.). The amplified PCR product bands were directly quantitated by using Molecular Analyst software (Bio-Rad Laboratories). Using either method, the relative amounts of PCR product from the patients’ PBMC samples were determined by linear-regression analysis. For each PCR assay, a standard curve was generated by using known concentrations of viral genomic DNA, with limiting DNA dilutions being run on each gel. Standard curves were deemed acceptable only if they were composed of at least four data points and had a correlation coefficient of at least 0.87. Each patient PBMC sample was tested for the presence of each virus in at least three separate experiments. Only patient samples yielding consistently positive results with both virus and TCR-β primers were considered positive.
Additional controls.
To test for the presence of subtler PCR inhibitors, we selected patient PBMC DNA extracts that had tested negative for each of the four HHV DNAs evaluated. We spiked PCR tubes containing 30 μl of these respective patient PBMC template DNAs with known concentrations of the DNAs. The relative amounts of amplified PCR product generated were compared to those generated using the same viral DNA concentrations in the absence of the patient template DNA. We found that the relative amounts of product generated were virtually identical (data not shown). This implies that the likelihood of a false-negative result due to the presence of a Taq DNA polymerase inhibitor in the extract is small.
We also randomly selected crude patient DNA extracts and further purified them as described above. The purified patient DNA extracts were then reevaluated for the presence or absence of viral genomic DNAs (data not shown). We observed similar results whether the sample tested was in the form of a crude or a purified DNA extract.
Statistical analysis.
The statistical significance of the results generated from this study was determined by a two-tailed Fisher’s exact test. A P value of ≤0.05 was considered statistically significant.
Virus culture procedure.
HHV6 A strain U1102 (12) was propagated in a continuous human CD4+-T-cell line known as JJhan (30). Both the cells and virus were kindly donated by Frank J. Jenkins (Uniformed Services University of the Health Sciences, Bethesda, Md.). The cells were grown in RPMI 1640 medium (Gibco BRL Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum, 1 mM l-glutamine, 50 μg of gentamicin/ml, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Cultures were incubated at 37°C in a humidified 5% CO2 incubator. For routine propagation of virus, uninfected JJhan cells were cocultured with U1102-infected JJhan cells at a ratio of 5:1 (2). Infected cells were harvested at the peak of infection, usually between 5 and 7 days postinfection, for use in the subsequent experiments. The peak of infection was assessed by the appearance of large “ballooning” refractile cells in culture, indirect immunofluorescence assay (IFA), and HHV6-specific PCR.
Indirect IFA.
U1102-infected JJhan cells harvested at the peak of infection and uninfected JJhan cells cultured for the same time period were used as the antigen in indirect IFAs. After being collected, the cells were washed with sterile phosphate-buffered saline (PBS; pH 7.4), air dried onto 10-mm-diameter circular glass slides (Fluoroslides; Erie Scientific Company, Portsmouth, N.H.), and fixed in ice-cold 80% acetone for 10 min. The fixed cells were then washed three times in PBS and stored at −70°C until needed. The primary antibody in this assay was either a dilution (in sterile PBS) of a patient serum or mouse anti-HHV6 p41 monoclonal antibody (designated 9A5D12). This monoclonal antibody, which was used as a positive-control serum, was a gift from N. Balachandran of the University of Kansas Medical Center, Kansas City. It reacts with HHV6 41- and 110-kDa proteins (4). HHV6 P41 is a viral phosphoprotein that is expressed both early and late during infection (4, 9, 30).
Briefly, the fixed cells were incubated at room temperature for 1 h with a 1:100 dilution of antibody. Then the cells were washed three times with sterile PBS prior to incubation for 1 h at room temperature, in the dark, with either a 1:32 dilution of a fluorescein isothiocyanate (FITC)-labeled goat anti-human immunoglobulin G (IgG) (gamma-chain specific) F(ab′)2 fragment (Sigma Immunochemicals, St. Louis, Mo.) or, if the monoclonal antibody was used, a 1:320 dilution of FITC-labeled goat anti-mouse IgG (Fab specific) F(ab′)2 fragment (Sigma Immunochemicals). Subsequently, the cells were washed three more times with PBS and then counterstained for 20 s with a 1:100,000 dilution of Evans blue. This was followed by two washings with PBS. Finally, the stained cells were mounted, using Johnsons’ medium (0.1% [wt/vol] p-phenylenediamine, 10% PBS, 90% anhydrous glycerol; pH 8.0), and were observed under a UV microscope.
A positive reaction, indicating the presence of anti-HHV6 antibody, was defined as extensive fluorescent staining of the infected cells compared to that of uninfected JJhan cells. Initially, patient sera were screened for anti-HHV6 antibodies via indirect IFA, using a dilution of 1:100 in PBS. If a positive reaction was observed, the serum was serially diluted twofold to determine the fluorescence end point. If a sample exhibited fluorescent staining in control JJhan cells, it was adsorbed and reevaluated. Adsorption was performed by incubating a 1:100 dilution of the cross-reactive human serum with 3 × 106 JJhan cells overnight at 4°C. The next day, the cells were pelleted by centrifugation at 1,000 × g for 5 min and the adsorbed serum was harvested for subsequent use.
EBV serological analysis.
Serological analyses of selected civilian sera for EBV were commercially performed by SmithKline Beecham Laboratories (Philadelphia, Pa.). Titers to EBV viral capsid antigen (VCA) (both IgG and IgM), EBNA1, and immediate-early antigen (IE) were obtained by indirect IFA.
RESULTS
Evaluation of DNA extraction.
All 149 civilian and 78 veteran patient PBMC samples analyzed exhibited a positive TCR-β result (data not shown). These data provided assurance that the DNA extractions were indeed successful and free of any overt PCR inhibitors.
Detection of HHV6 and HHV7 DNAs in PBMCs from civilians.
Typical PCR gels for HHV6 and HHV7 are shown in Fig. 2 and 3, respectively. The civilian two-tailed Fisher’s exact test data are summarized in Table 2. These data can be subdivided into four categories, i.e., the detection of HHV6 DNA alone, HHV7 DNA alone, either HHV6 or HHV7 DNA, or both HHV6 and HHV7 DNAs. In Table 2, comparisons of frequencies of detection of HHV6 and HHV7 DNAs are made (i) for healthy adult controls versus the CFS patient population as a whole, (ii) with respect to the presence (axis I) or absence of concurrent psychological symptoms, and (iii) with regard to acute versus gradual onset of disease. Despite these multiple comparisons, we failed to find any statistically significant correlation between infection with either of these HHVs and CFS in patients compared to the healthy controls. The PCR-detected prevalences of HHV6 and HHV7 DNAs in the peripheral blood for the entire civilian population assayed were 31 and 78.4%, respectively.
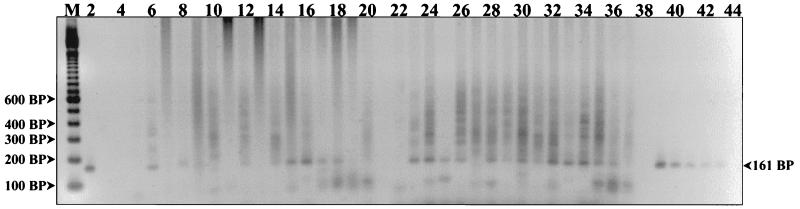
FIG. 2.
Detection of HHV6 DNA from human PBMCs. PCR products were resolved on a 2% agarose gel and stained with ethidium bromide. The first lane (M) represents a 100-bp DNA ladder (Gibco BRL Life Technologies). Lane 4 is the reagent control, and lanes 3, 21, 38, and 44 are empty. The HHV6 A strain U1102 DNA titration controls are in lanes 2 and 39 to 43 and represent template DNA quantities of 800, 400, 200, 100, 50, and 25 fg, respectively. Amplified products generated from various patient PBMC DNA extracts are in the remaining lanes. HHV6 DNA was detected in patient sample lanes 6, 8, 12, 14 to 18, 23 to 26, and 28 to 36.
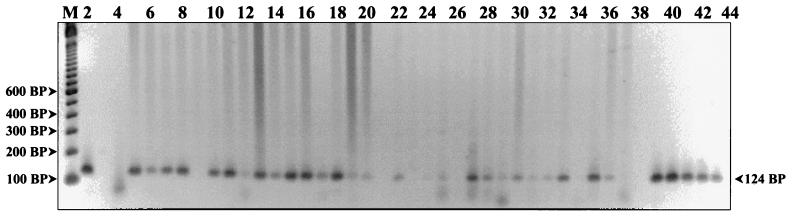
FIG. 3.
Detection of HHV7 DNA from human PBMCs. PCR products were resolved on a 2% agarose gel and stained with ethidium bromide. The first lane (M) represents a 100-bp DNA ladder (Gibco BRL Life Technologies). Lane 4 is the mock DNA extract control, and lanes 3, 38, and 44 are empty. The HHV7 strain JI genomic-DNA titration controls are in lanes 2 and 39 to 43 and represent template DNA quantities of 800, 400, 200, 100, 50, and 25 fg, respectively. Amplified products generated from various patient PBMC DNA extracts are in the remaining lanes. A prominent positive 124-bp band can be seen in patient lanes 5 to 8, 10, 11, 13 to 18, 22, 27, 28, 30, 33, 35, and 36. Faint positive bands can be seen in lanes 12, 19, 20, 25, 29, 31, and 32.
TABLE 2.
Prevalence of HHV6 and HHV7 in the civilian population: healthy adults versus the stratified CFS subsets
| PBMC donors | No. with virus/no. tested (%)
|
|||
|---|---|---|---|---|
| HHV6a | HHV7b | Either HHV6 or HHV7 | Both HHV6 and HHV7 | |
| Healthy adults | 19/71 (26.7%) | 51/73 (69.9%) | 56/73 (76.7%) | 14/71 (19.7%) |
| CFS patients | 26/74 (35.1%) | 58/75 (77.3%) | 61/76 (80.3%) | 23/73 (31.5%) |
| P valuec | 0.29 | 0.35 | 0.69 | 0.13 |
| CFS patients with axis I diagnosis | 11/32 (34.4%) | 24/31 (77.4%) | 27/32 (84.4%) | 8/31 (25.8%) |
| CFS patients without axis I diagnosis | 15/42 (35.7%) | 34/44 (77.3%) | 34/44 (77.3%) | 15/42 (35.7%) |
| P valuec | 0.55 | 0.65 | 0.70 | 0.17 |
| CFS patients reporting acute onset | 17/52 (32.7%) | 41/53 (77.4%) | 42/53 (79.3%) | 42/53 (79.3%) |
| CFS patients reporting gradual onset | 9/22 (40.9%) | 17/22 (77.3%) | 19/23 (82.6%) | 19/23 (82.6%) |
| P valuec | 0.43 | 0.60 | 0.89 | 0.26 |
Two CFS and healthy adult control donor samples were omitted from the HHV6 PCR analysis because a definitive determination as to the presence or absence of viral DNA could not be made.
One CFS patient sample was omitted from the HHV7 PCR analysis because a definitive determination as to the presence or absence of viral DNA could not be made.
Determined by Fisher’s exact test.
Detection of HHV6 and HHV7 DNAs in PBMCs from Persian Gulf War veterans.
Interestingly, HHV6 DNA was detected in only one healthy-veteran donor PBMC sample. Hence, statistical analyses were performed on data generated from the detection of HHV7 DNA alone. As evident in Table 3, the prevalences of HHV7 DNA in circulating PBMCs of veterans with CFS and their healthy comrades did not significantly differ (P = 0.82). When the Gulf War veteran CFS population was subdivided on the basis of the presence or absence of concurrent psychological symptoms and compared to the healthy controls (P = 0.19), as well as to each other; again no significant differences were observed (P = 0.48). Based on these data, we fail to find any correlative connection between detection by PCR of HHV7 genomic DNA in circulating PBMCs of Persian Gulf War veterans with CFS compared to their healthy comrades. Also, HHV7 genomic DNA was found in the peripheral blood of 46.2% of the Gulf War veteran population assayed, which is a lower prevalence than that observed in the civilian population.
TABLE 3.
Prevalence of HHV7 in the Persian Gulf War veteran population
| PBMC donor population | No. with HHV7/no. tested (%) |
|---|---|
| Healthy veteran controls | 14/32 (43.8%) |
| Veterans with CFS | 22/46 (47.8%) |
| P valuea | 0.82 |
| Veteran CFS patients with axis I diagnosis | 13/33 (39.4%) |
| Veteran CFS patients without axis I diagnosis | 9/13 (69.2%) |
| P valuea | 0.19 |
| Veteran CFS patients reporting acute onset | 1/5 (20.0%) |
| Veteran CFS patients reporting gradual onset | 21/41 (51.2%) |
| P valuea | 0.48 |
Determined by Fisher’s exact test.
Detection of EBV and HCMV DNAs in PBMCs from both civilian and veteran populations.
EBV DNA was detected in the PBMC from one civilian CFS patient and one veteran CFS patient. An example of a representative gel for EBV is shown in Fig. 4. HCMV DNA was not detected by PCR in the peripheral-blood lymphocyte samples taken from any civilian or veteran. A representative gel for HCMV is depicted in Fig. 5.
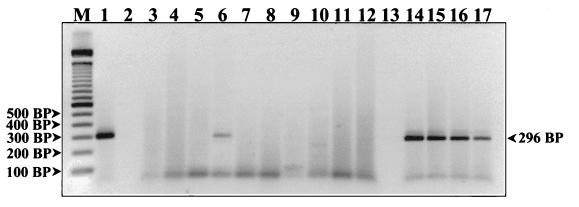
FIG. 4.
Detection of EBV DNA from human PBMCs. PCR products were resolved on a 2% agarose gel and stained with ethidium bromide. The first lane (M) represents a 100-bp DNA ladder (Gibco BRL Life Technologies). Lanes 2 and 13 are empty. The EBV strain B95-8 DNA titration controls are in lanes 1 and 14 to 17 and represent template DNA quantities of 800, 400, 200, 100, and 50 fg, respectively. Amplified products generated from various patient PBMC DNA extracts are in the remaining lanes. A prominent positive 296-bp band can be seen in patient lane 6. The remaining patient samples on this gel are negative.
FIG. 5.
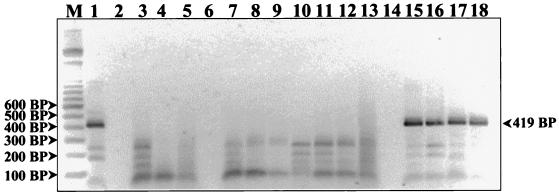
Detection of HCMV DNA from human PBMCs. PCR products were resolved on a 2% agarose gel and stained with ethidium bromide. The first lane (M) represents a 100-bp DNA ladder (Gibco BRL Life Technologies). Lanes 2, 6, and 14 are empty. The HCMV DNA titration controls are in lanes 1 and 15 to 18 and represent template DNA quantities of 800, 400, 200, 100, and 50 fg, respectively. Amplified products generated from various patient PBMC DNA extracts are in the remaining lanes. All of the patient DNA extracts represented on this gel are negative and fail to exhibit a band at 419 bp.
To verify that nothing in the PCR protocol was amiss, we obtained some known HCMV-positive and -negative human bone marrow transplant patient lymphocyte samples from Mary Jo Evans. The results generated from these specimens, using our PCR protocol (data not shown), gave the positive results anticipated. In addition, we also spiked some of the crude patient DNA extracts with various concentrations of HCMV DNA and compared the results to those obtained by using the same HCMV DNA concentrations without the cellular extracts. The resultant amounts of an amplified HCMV 418-bp product obtained from the paired samples were virtually the same (data not shown). From this we conclude that the likelihood of a false-negative result due to the presence of a PCR inhibitor in the extract itself is remote. The HCMV and EBV data were not subjected to statistical analyses because the numbers involved were too small to be significant.
HHV6 serology.
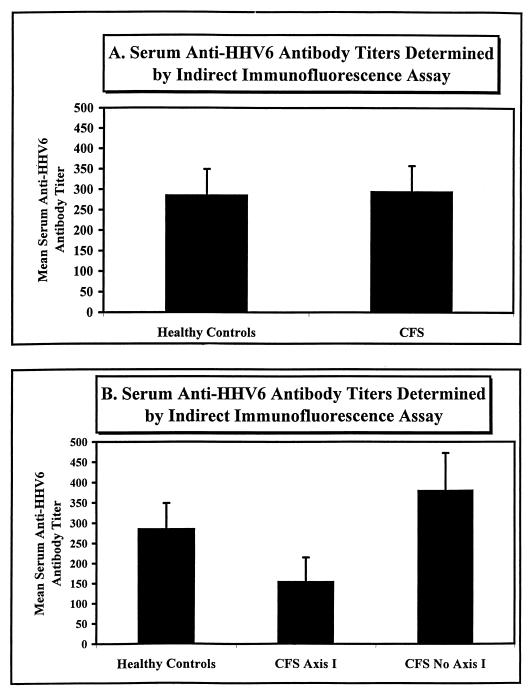
All of the patient sera tested by indirect IFA had anti-HHV6 antibody titers of between ≤100 and <3,200. We used a titer of 100 as an arbitrary threshold value. The indirect-IFA results generated from the sera of 51 healthy and 58 civilian CFS patients are graphically represented in Fig. 6. The serum anti-HHV6 antibody titers were essentially the same for both the healthy (mean ± standard deviation, 286 ± 62.9) and CFS (295 ± 62.6) populations. When the civilian CFS population was stratified on the basis of the presence (axis I; n = 22) or absence (no axis I; n = 36) of concurrent psychological symptoms (Fig. 6B), the latter had a higher mean anti-HHV6 antibody titer than the former. That is, the CFS group members lacking concurrent psychological symptoms had a titer of 381 ± 91.8, compared to the CFS axis I diagnosis group titer of 155 ± 59.8. However, this apparent difference was determined to lack statistical significance. It should be noted that this assay does not exclude the possibility of the presence of cross-reactive HHV7 antibodies in the patients’ sera. In addition, the assay is not totally objective, since it relies on making a subjective comparison between the intensities of fluorescent staining observed on virus-infected and on uninfected cells.
FIG. 6.
HHV6 serology determined by indirect IFA. (A) The indirect IFA results for 51 healthy civilians and 58 civilian CFS patients are graphically represented. The mean serum anti-HHV6 antibody titers ± the standard deviations were 286 ± 62.9 and 295 ± 62.6 for the healthy and CFS groups, respectively. (B) Shown are the same data as were represented in panel A but after stratification of the CFS patient population with regard to the presence (CFS Axis I) or absence (CFS No Axis I) of concurrent psychological symptoms. The CFS axis I diagnosis group (n = 22) had a mean serum anti-HHV6 antibody titer of 155 ± 59.8, compared to the CFS no-axis I diagnosis group (n = 36) mean titer of 381 ± 91.8.
EBV serology.
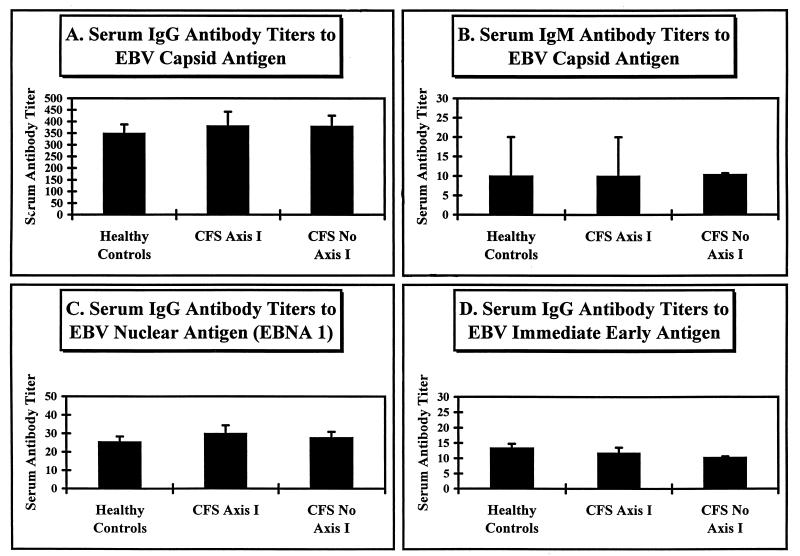
A representative portion of patient serum specimens were tested for the presence of serum antibodies against EBV VCA (both IgG and IgM), EBNA1, and EBV IE. The data are represented graphically in Fig. 7. In this assay, we examined the sera of 42 healthy civilian controls, 17 Axis I CFS civilian patients, and 30 CFS civilian patients lacking concurrent psychological symptoms. As can be seen, there were no significant differences among these groups in terms of VCA IgG, VCA IgM, EBNA1, or IE titers. Also, no correlations can be seen, using different serum anti-EBV antibody titers, between CFS patients as a whole (data not shown) or patients stratified on the basis of the presence or absence of psychological symptoms and the healthy control population.
FIG. 7.
EBV serology determined by indirect IFA. The indirect-IFA results generated from sera of 42 healthy civilians and 47 civilian CFS patients are graphically represented. The error bars indicate 1 standard deviation from the mean serum antibody titer. The CFS patient population shown was stratified by the presence (CFS Axis I; n = 17) or absence (CFS No Axis I; n = 30) of concurrent psychological symptoms. The mean serum anti-EBV VCA (both IgG [A] and IgM [B]), EBNA1 (C), and IE (D) antibody titers are depicted.
DISCUSSION
To our knowledge, this is the first study to simultaneously test for the presence of HHV6, HHV7, EBV, and HCMV DNAs in PBMCs of CFS patients (both civilian and military) versus matched controls by PCR. Based on the PCR data generated, we failed to find any statistically significant correlation between the detection of one or more of these HHVs in the circulating PBMCs and CFS in patients compared to the healthy control population. This observation held true even when the CFS patient population was stratified by the presence or absence of concurrent psychological symptoms and by patient-reported sudden or gradual onset of disease. Data from both the civilian and veteran study populations confirmed this observation. In the literature, there are both positive (7, 8, 22, 31) and negative (1, 11, 24, 28) data with regard to correlation between the detection of HHV6 DNA in PBMCs of CFS patients versus healthy controls. Our present data clearly support the latter view, in that despite the use of well-characterized long-term CFS patients and an age-, race-, and gender-matched control population, there were no significant differences observed. Previous-study results contrary to our findings may be attributed to differences in the CFS patient population evaluated and the particular PCR protocols utilized.
We have gone further in our analyses than most previous studies, stratifying the CFS populations with the hope of uncovering subpopulations with particular virological markers. Even these detailed analyses, however, gave no obvious correlations. Nevertheless, our data do not conclusively rule out the possibility that one or more of these HHVs play a role in CFS pathogenesis in some cases, but at this stage it seems that routine screening for these viruses in CFS patients would have limited clinical value. In the present study, we did not differentiate between strains A and B of HHV6, but there are reports that CFS patients (11, 31) and those with lymphoproliferative disorders (10) harbor HHV6 strain A more frequently than they harbor strain B.
In keeping with the PCR data, we observed no significant differences in anti-HHV6 or anti-EBV serum antibody titers between the civilian CFS patients and healthy control populations. We also found that elevated serum anti-HHV6 antibody titers did not always correlate with the detection by PCR of HHV6 genomic DNA in circulating PBMCs. These results coincide with serological data published by others (6, 20, 27, 28). Viral serological analyses in and of themselves have proven to be of little diagnostic value in CFS. Patniak et al. (27) showed that both serum anti-HHV6 early-antigen (p41/38) IgM and IgG antibody titers were significantly higher in CFS patients than in healthy controls. The enzyme-linked immunosorbent assay technique utilized by them is more sensitive and quantitative than the indirect-IFA endpoint titration used in this study; however, Patnaik et al. failed to validate their serological data by detecting HHV6 DNA or by virus isolation. HHV6 serological evidence alone, as stated previously, is insufficient to confirm HHV6 infection or in this case most probably reactivation.
Finally, for an as-yet-undetermined reason, we observed a higher prevalence of both HHV6 and HHV7 genomic DNAs in the circulating PBMCs of the civilian population than in those of the veteran population. Both sets of civilian prevalence percentages are similar to those previously reported in the literature (11, 29), but we were surprised that the differences between relatively equally matched populations were so marked. This was particularly so with HHV6, whose detection level in the veteran population was close to zero. The primary difference between the veteran and civilian populations examined was that the veteran population was predominantly male, but we would not predict that this alone would be a determining factor. Perhaps the differences relate to some as-yet-undiscovered biological or environmental phenomenon.
ACKNOWLEDGMENTS
We thank Mary Jo Evans (Children’s Hospital, Buffalo, N.Y.) for the HCMV and TCR-β primer sequences and protocols. We also thank N. Balachandran (University of Kansas Medical Center, Kansas City) for the gift of the mouse anti-HHV6 p41 monoclonal antibody and to Neal E. Niesen and Donna Czechowski for technical and moral support.
This project was supported by grants from the U.S. Public Health Service (AI32670) and the Veterans Administration/Department of Defense Environmental Hazards Research Institute, E. Orange, N.J.
REFERENCES
- 1.Ablashi D V. Viral studies of chronic fatigue syndrome. Clin Infect Dis. 1994;18(Suppl. 1):S130–S133. doi: 10.1093/clinids/18.supplement_1.s130. . (Review.) [DOI] [PubMed] [Google Scholar]
- 2.Ablashi D V, Lusso P, Hung C L, Salahuddin S Z, Josephs S F, Llana T, Kramarsky B, Biberfeld P, Markham P D, Gallo R C. Utilization of human hematopoietic cell lines for the propagation and characterization of HBLV (human herpesvirus 6) Int J Cancer. 1988;42:787–791. doi: 10.1002/ijc.2910420526. [DOI] [PubMed] [Google Scholar]
- 3.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 3rd ed. Washington, D.C: American Psychiatric Association; 1987. [Google Scholar]
- 4.Balachandran N, Amelse R E, Zhou W W, Chang C K. Identification of proteins specific for human herpesvirus 6-infected human T cells. J Virol. 1989;63:2835–2840. doi: 10.1128/jvi.63.6.2835-2840.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berneman Z N, Ablashi D V, Li G, Eger-Fletcher M, Reitz M S, Hung C L, Brus I, Komaroff A L, Gallo R C. Human herpesvirus 7 is a T-lymphotrophic virus and is related to, but significantly different from, human herpesvirus 6 and human cytomegalovirus. Proc Natl Acad Sci USA. 1992;89:10552–10556. doi: 10.1073/pnas.89.21.10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchwald D, Ashley R L, Pearlman T, Kith P, Komaroff A L. Viral serologies in patients with chronic fatigue and chronic fatigue syndrome. J Med Virol. 1996;50:25–30. doi: 10.1002/(SICI)1096-9071(199609)50:1<25::AID-JMV6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 7.Buchwald D, Cheney P R, Peterson D L, Berch H, Wormsley S B, Geiger A, Ablashi D V, Salahuddin S Z, Saxinger C, Royce B, Kikinis R, Jolesz F A, Folks T, Balachandran N, Peter J B, Gallo R C, Komaroff A L. A chronic illness characterized by fatigue, neurologic and immunologic disorders, and active human herpesvirus type 6 infection. Ann Intern Med. 1992;116:103–113. doi: 10.7326/0003-4819-116-2-103. [DOI] [PubMed] [Google Scholar]
- 8.Buchwald D, Komaroff A L. Review of laboratory findings for patients with chronic fatigue syndrome. Rev Infect Dis. 1991;13:S12–S18. doi: 10.1093/clinids/13.supplement_1.s12. [DOI] [PubMed] [Google Scholar]
- 9.Chang C K, Balachandran N. Identification, characterization, and sequence analysis of a cDNA encoding a phosphoprotein of human herpesvirus 6. J Virol. 1991;65:2884–2894. doi: 10.1128/jvi.65.6.2884-2894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Luca D, Dolcetti R, Mirandola P, De Re V, Secchiero P, Carbone A, Boiocchi M, Cassai E. Human herpesvirus 6: a survey of presence and variant distribution in normal peripheral lymphocytes and lymphoproliferative disorders. J Infect Dis. 1994;170:211–215. doi: 10.1093/infdis/170.1.211. [DOI] [PubMed] [Google Scholar]
- 11.Di Luca D, Zorzenon M, Mirandola P, Colle R, Botta G A, Cassai E. Human herpesvirus 6 and human herpesvirus 7 in chronic fatigue syndrome. J Clin Microbiol. 1995;33:1660–1661. doi: 10.1128/jcm.33.6.1660-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Downing R G, Sewankambo N, Serwadda D, Honess R, Crawford D, Jarrett R, Griffin B E. Isolation of human lymphotrophic herpesviruses from Uganda. Lancet. 1987;ii:390. doi: 10.1016/s0140-6736(87)92403-2. [DOI] [PubMed] [Google Scholar]
- 13.Evans M, Edwards-Spring Y, Myers J, Wendt A, Povinelli D, Amsterdam A, Rittenhouse-Diakun K, Armstrong D, Murray B M, Greenberg S J, Riepenhoff-Talty M. Polymerase chain reaction assays for the detection of cytomegalovirus in organ and bone marrow transplant recipients. Immunol Investig. 1997;26:209–229. doi: 10.3109/08820139709048928. [DOI] [PubMed] [Google Scholar]
- 13a.Evans, M. J. U.S. patent applications 081/428,370 and 081/600,764.
- 14.Fukuda K, Straus S E, Hickie I, Sharpe M C, Dobbins J G, Komaroff A L, Schluederberg A, Jones J F, Lloyd A R, Wessely S, Gantz N M, Holmes G P, Steele L, Reyes M, Abbey S, Rest J, Jolson H, Peterson D L, Vercoulen J H M M, Tirelli U, Evengard B, Natelson B, Reeves W C. Chronic fatigue syndrome: a comprehensive approach to its definition and study. Ann Intern Med. 1994;121:953–959. doi: 10.7326/0003-4819-121-12-199412150-00009. [DOI] [PubMed] [Google Scholar]
- 15.Gold D, Bowden R J, Sixbey R, Riggs R, Katon W J, Ashley R, Obrigewitch R, Corey L. Chronic fatigue: a perspective clinical and virologic study. JAMA. 1990;264:48–53. doi: 10.1001/jama.264.1.48. [DOI] [PubMed] [Google Scholar]
- 16.Gomples U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus 6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 17.Gow J W, Behan W M H, Simpson K, McGarry F, Keir S, Behan P O. Studies on enteroviruses in patients with chronic fatigue syndrome. Clin Infect Dis. 1994;18:S126–S129. doi: 10.1093/clinids/18.supplement_1.s126. [DOI] [PubMed] [Google Scholar]
- 18.Heneine W, Woods T C, Sinha S D, Khan A S, Chapman L E, Schonberger L B, Folks T M. Lack of evidence for infection with human and animal retroviruses in patients with chronic fatigue syndrome. Clin Infect Dis. 1994;18:S121–S125. doi: 10.1093/clinids/18.supplement_1.s121. [DOI] [PubMed] [Google Scholar]
- 19.Holmes G P, Kaplan J E, Gantz N M, Komaroff A L, Schonberger L B, Straus S E, Jones J F, Dubois R E, Cunningham-Rundles C, Pahwa S, Tosato G, Zegans L S, Partilo D T, Brown N, Schooley R T, Brus I. Chronic fatigue syndrome: a working case definition. Ann Intern Med. 1988;108:387–389. doi: 10.7326/0003-4819-108-3-387. [DOI] [PubMed] [Google Scholar]
- 20.Jarrett R F, Clark D A, Josephs S F, Onions D E. Detection of human herpesvirus-6 DNA in peripheral blood and saliva. J Med Virol. 1990;32:73–76. doi: 10.1002/jmv.1890320113. [DOI] [PubMed] [Google Scholar]
- 21.Josephs S F, Ablashi D V, Salahuddin S Z, Jagodzinski L L, Wong-Staal F, Gallo R C. Identification of the human herpesvirus 6 glycoprotein H and putative large tegument protein genes. J Virol. 1991;65:5597–5604. doi: 10.1128/jvi.65.10.5597-5604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Josephs S F, Henry B, Balachandran N, Strayer D, Peterson D, Komaroff A L, Ablashi D V. HHV6 reactivation in chronic fatigue syndrome. Lancet. 1991;337:1346–1347. doi: 10.1016/0140-6736(91)93018-5. [DOI] [PubMed] [Google Scholar]
- 23.Kormaroff A L, Buchwald D. Symptoms and signs of chronic fatigue syndrome. Rev Infect Dis. 1991;13:S8–S11. doi: 10.1093/clinids/13.supplement_1.s8. [DOI] [PubMed] [Google Scholar]
- 24.Levy J A. Viral studies of chronic fatigue syndrome. Part III. Clin Infect Dis. 1994;18:S117–S120. doi: 10.1093/clinids/18.supplement_1.s117. [DOI] [PubMed] [Google Scholar]
- 25.Marcus S, Robins L S, Bucholz K. Quick diagnostic interview schedule III R, version 1. St. Louis, Mo: Department of Psychiatry, Washington University School of Medicine; 1990. [Google Scholar]
- 26.Oosterveer M A P, Markusse H M, Lennette E T, Zou J Z, Bolhuis R L H, Gratama J W. Characterization of Epstein-Barr viral strains in parotid gland saliva and peripheral blood of patients with primary Sjögren’s syndrome and healthy EBV carriers. J Med Virol. 1993;41:261–269. doi: 10.1002/jmv.1890410402. [DOI] [PubMed] [Google Scholar]
- 27.Patniak M, Kormaroff A L, Conley E, Ojo-Amaize E A, Peter J B. Prevalence of IgM antibodies to human herpesvirus 6 early antigen (p41/38) in patients with chronic fatigue syndrome. J Infect Dis. 1995;172:1364–1367. doi: 10.1093/infdis/172.5.1364. [DOI] [PubMed] [Google Scholar]
- 28.Wagner M, Krueger G R F, Ablashi D V, Whitman J E. Chronic fatigue syndrome (CFS): a critical evaluation of testing for active human herpesvirus-6 (HHV6) infection: review of data of 107 cases. J Chronic Fatigue Syndr. 1996;2:3–16. [Google Scholar]
- 29.Wiborn F, Schmidt C A, Lorenz F, Peng R, Gelderbolm H, Huhn D, Siegert W. Human herpesvirus type 7 in blood donors: detection by polymerase chain reaction. J Med Virol. 1995;47:65–69. doi: 10.1002/jmv.1890470113. [DOI] [PubMed] [Google Scholar]
- 30.Wyatt L S, Balachandran N, Frenkel N. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J Infect Dis. 1990;162:852–857. doi: 10.1093/infdis/162.4.852. [DOI] [PubMed] [Google Scholar]
- 31.Yalcin S, Kuratsune H, Yamaguchi K, Kitani T, Yamanishi K. Prevalence of human herpesvirus 6 variants A and B in patients with chronic fatigue syndrome. Microbiol Immunol. 1994;38:587–590. doi: 10.1111/j.1348-0421.1994.tb01827.x. [DOI] [PubMed] [Google Scholar]