Abstract
Background
Although some of the regulatory genes, signaling pathways, and gene regulatory networks altered in medulloblastomas (MB) are known, the roles of non-coding RNAs, particularly long non-coding RNAs (lncRNAs), are poorly described. Here we report that the lncRNA SPRIGHTLY (SPRY4-IT1) gene is upregulated in group 4 medulloblastoma (G4 MB).
Methods
SPRIGHTLY expression was assessed in MB subgroup patient-derived xenografts, cell lines, and patient samples. The effect of SPRIGHTLY hemizygous deletion on proliferation, invasion, apoptosis, and colony formation were assessed in vitro and on tumor growth in vivo. dChIRP pull-down assays were used to assess SPRIGHTLY-binding partners, confirmed by immunoprecipitation. SMYD3 ΔE5 transcripts were examined in cell lines and publicly available RNA-seq data. Pathway analysis was performed by phospho-kinase profiling and RNA-seq.
Results
CRISPR/Cas9 deletion of SPRIGHTLY reduced cell viability and invasion and increased apoptosis in G4 MB cell lines in vitro. SPRIGHTLY hemizygous-deleted G4 MB cells injected into mouse cerebellums produced smaller tumors than those derived from parental cells expressing both copies of SPRIGHTLY. SPRIGHTLY lncRNA bound to the intronic region of the SMYD3 pre-mRNA transcript. SPRIGHTLY also interacted with PTPB1 protein to regulate SMYD3 exon skipping to produce an aberrant protein. SPRIGHTLY-driven SMYD3 regulation enhanced the expression of EGFR pathway genes in G4 MB cell lines and activated cell coagulation/hemostasis-related gene expression, suggesting a novel oncogenic role in G4 MB.
Conclusions
These results demonstrate the importance of SPRIGHTLY lncRNA as a promoter of G4 MB and the role of the SPRIGHTLY-SMYD3-PTPB1 axis as an important oncogenic regulator in MB.
Keywords: long non-coding RNA, medulloblastoma, PTPB1, SMYD3, SPRIGHTLY
Key Points.
The lncRNA SPRIGHTLY is highly expressed in group 4 medulloblastomas.
Together with its novel binding partner, PTPB1, SPRIGHTLY regulates SMYD3 exon skipping.
The downstream effects of these regulatory events modify oncogenic signaling and potentially the microenvironment in group 4 medulloblastomas.
Importance of the Study.
Long non-coding RNAs (lncRNAs) are emerging as important regulators of cancer growth and behavior, but few have been characterized in medulloblastomas. We now report that the lncRNA SPRIGHTLY is expressed in the group 4 subtype of medulloblastomas, for which the driver pathways are poorly described. We establish that SPRIGHTLY lncRNA binds to PTPB1 protein, and in doing so regulates the expression of SMYD3 transcript variants, leading to modulating the EGFR signaling pathways. These regulatory events governed by SPRIGHTLY affect group 4 medulloblastoma tumor growth and appears to modulate the microenvironment, providing new insights into the molecular biology of this poorly described subgroup of medulloblastoma.
Medulloblastoma (MB) is the most common malignant pediatric brain tumor.1 Molecular profiling studies have subcategorized MB into four main molecular subgroups: wingless pathway (WNT) activated, sonic hedgehog pathway (SHH) activated, group 3 (G3), and group 4 (G4).2,3 While the main common driver pathways for G3 and G4 MB have not been identified, these groups are characterized by c-Myc and MYCN signatures, respectively.3,4 Among the four subgroups, G3 is the most aggressive MB with a 45–60% 5-year survival rate,5 while other subgroups (WNT > 90%,6 SHH 60–80%,7 and group 4 75–80%)5 have more favorable prognoses.
The standard treatment for MB includes surgical resection, radiotherapy, and chemotherapy.8 Although craniospinal irradiation after surgery improves long-term outcomes, the development of radio-resistance hampers therapeutic efficacy and survival.9 Even with advanced molecular classification, robust candidate therapeutic targets for the different subgroups, especially G3 and G4 MB, have yet to be identified.10 Therefore, there is an urgent need to identify underlying molecular mechanisms in MB subgroups to discover new therapeutic targets.
MBs develop through various genetic, epigenetic, and non-coding RNA (ncRNA)-related mechanisms. However, the roles played by ncRNAs, particularly long non-coding RNAs (lncRNAs), in MB development remain poorly defined. Several lncRNAs associated with MB have been reported.11 The fusion of PVT1 oncogene, which encodes a lncRNA, with MYC and NDRG1 is frequently observed in G3 MB.12 Similarly, lnc-HLX-2-7 was highly upregulated in G3 MBs, and its knock-down reduced tumor growth both in vitro and in vivo.13 Varon et al.14 reported that the lncRNA TP73-AS1, an antisense transcript of p73 mRNA, is upregulated in the SHH subgroup; its silencing in the SHH MB human cell line DAOY reduced cellular proliferation, increased apoptosis, and reduced tumorigenicity in xenografts in mouse brains relative to parental DAOY cells. TP73-AS1 acts as an oncogene by regulating proliferation, migration, and invasion by sponging miR-493-3p.15 By contrast, a relative absence of understanding of the molecular pathogenesis of G4 MB makes it important to investigate this subclass of medulloblastomas.
We previously identified and reported that the SPRIGHTLY lncRNA regulates cell proliferation, invasion, and apoptosis in melanomas.16 Further studies showed that SPRIGHTLY serves as an intra-nuclear organizing hub for pre-mRNA molecules,17 but its specific role in mRNA splicing was speculative. Since skin and brain tissues are both derived from neural crest cells of the primary ectoderm of the embryo,18,19 and neural stem cells give rise to both neural cells and melanocytes,20 common molecular pathways are involved in the early differentiation and morphogenesis of these two organs. For example, Notch signaling pathways are important in the early morphogenesis of both tissues,21 and abnormalities in these pathways are associated both with melanomas22 and brain tumors, including medulloblastomas.23,24 These considerations led us to speculate whether there might be a more intimate molecular connection involving not only protein-mediated signaling cascades but also non-coding RNA circuitries between skin and brain tumors. More specifically, we hypothesized that as in melanoma, the lncRNA SPRIGHTLY, which controls the transcript levels of SMYD3, which is a direct target of HIF2α (a member of the SHH signaling pathway and a regulator of the EGFR signaling cascade,25 might similarly control EGFR pathways in medulloblastoma. This is especially interesting in light of the observation that simultaneous inhibition of Notch and EGFR pathways exhibit additive effects on medulloblastoma cellular properties,26 and, because there are several reports of cross-talks among the SHH, Notch, and EGFR pathways,27–29 especially in medulloblastomas.30
Consistent with the above speculation, here we report that SPRIGHTLY expression is highly upregulated in G4 MB patient RNAseq datasets, in cell lines and patient-derived xenografts (PDXs), and that SPRIGHTLY regulates EGFR and its downstream pathway gene transcription in these cells. Knocking down SPRIGHTLY in G4 MB cell lines reduced cellular proliferation rates, invasiveness, and colony forming ability. SPRIGHTLY-gene deleted G4 MB cells injected into mouse cerebellums produced smaller tumors than those injected with equivalent numbers of parental G4 MB cells. We discovered that SPRIGHTLY lncRNA binds to the intronic region of SMYD3 pre-mRNA molecules and that this binding leads to the skipping of exon 5 of SMYD3 pre-mRNA during splicing. PTBP1 protein, a member of the heterogeneous nuclear RNA (hnRNA) binding protein group, is associated with this exon skipping property of SPRIGHTLY on SMYD3 pre-mRNA. The expression patterns of SMYD3 and SMYD3 E5-deleted transcripts in patient RNA-seq data suggest that an altered ratio of the two transcripts is associated with G4 MB tumor development and/or maintenance.
Materials and Methods
Cell Culture
The group 4 MB CHLA-01-MED (hereafter CHLA01) cell line was purchased from the American Type Culture Collection (ATCC). Cells were cultured in DMEM/F-12 medium supplemented with B-27 supplement (Invitrogen, Carlsbad, CA, USA), 20 ng/ml EGF (R&D Systems, Minneapolis, MN, USA), and 20 ng/ml bFGF (R&D Systems, Minneapolis, MN, USA) in a humidified incubator at 37°C in 5% CO2.
Cell Invasion Assay
Cell invasion assays were performed using a trans-well chamber (Corning, Tewksbury, MA, USA). DMEM/F-12 medium supplemented with B-27 supplement, EGF and bFGF was added to the bottom chambers, and cells were suspended in DMEM/F-12 medium and seeded into the top chamber coated with Matrigel. After 48 h incubation at 37°C, cells in the top chamber were removed with a cotton swab. The cells were fixed in 4% paraformaldehyde for 5 min, followed by staining with 0.5% crystal violet in 95% ethanol for 40 min. Cells were counted using a phase-contrast microscope and quantified with Image J software.31
Anchorage-Independent Colony Growth Assay
Anchorage-independent growth assays were performed using the soft agar colony formation assay.32 Briefly, CHLA01 cells were cultured in the growth medium that contained 0.6% agar at 1000 cells per 24-well plate. Cells were cultured in a humidified incubator at 37°C in 5% CO2. The complete growth medium on top of the agar was changed each 3 days. After 21 days incubation, the colonies were stained with 0.005% crystal violet in 95% ethanol for 30 min and all colonies were counted using Cytation 1 cell imaging reader (BioTek, Winooski, VT, USA). Colony number was determined using the ImageJ software.31
RNA Samples
Normal human cerebellum (CB) was purchased from BioChain (Newark, CA, USA). RNAs from various MB cell lines and PDXs were kindly provided by Dr. Wechsler-Reya (Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA).33–35
RNA-Fluorescence in Situ Hybridization (RNA-FISH)
RNA was visualized in paraffin-embedded tissue sections using the QuantiGene ViewRNA ISH Tissue Assay Kit (Affymetrix, Frederick, MD, USA). In brief, tissue sections were rehydrated and incubated with proteinase K. Subsequently, they were incubated with ViewRNA probe sets designed to target human SPRIGHTLY and MYC (Affymetrix, Santa Clara, CA, USA). Custom Type 1 primary probes targeting SPRIGHTLY and Type 6 primary probes targeting MYC were designed and synthesized by Affymetrix (Santa Clara, CA, USA; Supplementary File 1). Hybridization was performed according to the manufacturer’s instructions.
Immunofluorescence
CHLA01 cells were fixed with freshly made 4% paraformaldehyde and immuno-stained with anti-SMYD3 (ab187149, 1:250, Abcam, Cambridge, UK) and anti-MYC tag (ab32, 1:200, Abcam) antibodies. Primary antibody-antigen complexes were visualized using anti-mouse Alexa Fluor 488 and anti-rabbit Alexa Fluor 546 secondary antibodies (Molecular Probes, Eugene, OR, USA; 1:500). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Stained sections were imaged using a confocal laser-scanning microscope (Nikon C1 confocal system, Nikon Corp, Tokyo, Japan). The acquired images were processed using NIS (Nikon) and analyzed with Image J software (https://imagej.nih.gov/ij/).
Intracranial Medulloblastoma Xenografts
Mouse studies were approved and performed in accordance with the policies and regulations of the Animal Care and Use Committee of Johns Hopkins University. Intracranial MB xenografts were established by injecting CHLA01 cells or SPRIGHTLY-deleted CHLA01 cells into the cerebellums of NOD-SCID mice (Jackson Laboratory, Bar Harbor, ME, USA). Cerebellar coordinates were ‐2 mm from lambda, +1 mm laterally, and 1.5 mm deep. Tumor growth was evaluated by weekly bioluminescence imaging using an in vivo spectral imaging system (IVIS Lumina II, Xenogen, Alameda, CA, USA).
Quantitative Real-Time PCR (qRT-PCR)
RNA was isolated using the Direct-zol RNA miniprep kit (Zymo Research, Irvine, CA, USA) and cDNA was synthesized using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Waltham, MA, USA). Real-time PCR was performed using a Power SYBR green PCR master mix (Applied Biosystems) in the QuantaStudio 5 Real-Time PCR systems (Thermo Fisher Scientific). ACTB was used for normalization. Primer sequences are listed in Supplementary File 1.
Domain-Specific Chromatin Isolation by RNA Purification (dChIRP)
dChIRP was performed as previously described.17,36 Cells were cross-linked with 0.3% formaldehyde for 10 min. Sheared chromatin was diluted with hybridization buffer and probes were added. The mixture was incubated overnight at 37°C with rotation. Streptavidin-magnetic C1 beads (Thermo Fisher Scientific, Waltham, MA, USA) were added and incubated for another 2 h at 37°C. Beads were re-suspended in buffer containing proteinase K (Sigma-Aldrich, St. Louis, MO) and incubated at 50°C for 45 min followed by overnight incubation at 65°C for reverse crosslinking. RNAs were extracted by TRIzol:chloroform and precipitated with ethanol. The purified RNAs were treated with DNA-free water (Life Technologies, Carlsbad, CA, USA) according to manufacturer’s protocol. RNAs were quantified qRT-PCR. Primer and probe sequences are available in reference.17
SMYD3, SMYD3 ΔE5, SPRY4, and SPRIGHTLY Expression in Patient RNA-seq Data
Raw FASTQ files for the RNA-seq data were collected from the European Genome-Phenome Archive (EGA, http://www.ebi.ac.uk/ega/, accession number: EGAD00001003279) after obtaining permission from the ICGC Data Access Compliance Office (DACO). The data represented 175 medulloblastoma samples (n = 18 WNT, n = 46 SHH, n = 45 G3, and n = 66 G4). Libraries for these data had been prepared using a strand-specific method (TruSeq or a modified method) and were sequenced on the Illumina HiSeq2000 platform for 2 × 51 cycles. The ultra-fast FASTQ preprocessor package fastp37 was used for quality control, adapter trimming, and filtering low-quality FASTQ read data. Filtered FASTQ files were then mapped to SMYD3 and SMYD3 E5-deleted sequences separately using BWA read aligner. Samtools was used to count primary aligned reads to the SMYD3/SMYD3 E5-deleted genomic sequence.
Overexpression and siRNA-Mediated Knock-Down
The plasmid containing PTBP1 was obtained from Addgene (Watertown, MA, USA). One µg of plasmids for PTBP1 (Addgene plasmid #64925) or SPRIGHTLY17 were transfected for 48 h using FugeneHD (Promega, Madison, WI, USA). siRNAs targeting PTBP1 (catalog no. 4427038, ID: s11436) and negative control (siNEG, catalog no. AM4611) were purchased from Ambion (Foster City, CA, USA). siRNAs were transfected at 20 nM for 48 h using Lipofectamine RNAiMAX (Life Technologies, Carlsbad, CA, USA). The efficiency was determined by qRT-PCR.
SMYD3-Myc and SMYD3 ΔE5-Myc Overexpressing Cell Lines
SMYD3 cDNA was cloned into pcDNA4/TO/myc-HisB (Thermo Fisher Scientific) using an In-Fusion cloning system according to the manufacturer’s instructions (Takara Bio USA Inc., San Jose, CA, USA). SMYD3 ΔE5 gBlock was synthesized by Integrated DNA Technologies (IDT, Coralville, IA, USA) and cloned into pcDNA4/TO/myc-HisB. To move SMYD3-Myc and SMYD3 ΔE5-Myc fragments into lenti-plasmids, the dCas9-VP64 gene in pLenti-dCas9-VP64_blast (addgene #61425) was replaced with SMYD3-Myc or SMYD3 ΔE5-Myc fragments. Lentiviruses were generated using a Lenti-X packaging single shot (Takara Bio USA Inc.) and CHLA01 cells were transduced with lentiviruses containing the SMYD3-Myc or SMYD3 ΔE5-Myc gene. Primer sequences are available in Supplementary File 1.
Western Blotting
Cell lysates were fractionated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5% bovine serum albumin (Fisher Scientific, Hampton, NH, USA) in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 0.5% Tween 20) for 60 min, then incubated with antibodies targeting PTBP1 (Medical and Biological Laboratories (MBL), Nagoya, Aichi, Japan; catalog no. RN011P), lamin B1 (MBL; catalog no. PM064), phospho-ERK1/2 (Cell Signaling Technology, Danvers, MA, USA; catalog no. 4370), SMYD3 (Abcam; catalog no. ab183498), Myc (Abcam; catalog no. ab32), and tubulin (Abcam; catalog no. ab6046) at 4°C overnight. Membranes were washed and incubated with a 1:10,000 dilution of horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies for 1 h. Blots were washed and developed with the ECL system (Amersham Biosciences, Piscataway, NJ, USA) according to the manufacturer’s protocols.
Identification of SPRIGHTLY-Binding Proteins
SPRIGHTLY-binding proteins were purified with the RiboTrap kit (MBL) according to the manufacturer’s protocol. Briefly, 1.0 × 107 cells were harvested and lysed in CE buffer with detergent solution. After removing the cytosol fraction, nuclei were re-suspended in NE buffer. The nuclear lysate was used for immunoprecipitation. BrU-labeled SPRIGHTLY was prepared with the MEGAstrip T7 system (Invitrogen). BrU-labeled RNA was mixed with the nuclear extracts and captured by Protein G agarose conjugated with an anti-BrdU antibody. Proteins were digested with trypsin for analysis by mass spectrometry. Mass spectrometry analysis was performed by the Johns Hopkins University School of Medicine Proteomics Core (https://www.hopkinsmedicine.org/research/labs/mass-spectrometry-core). Average peptide numbers between replicates were used for determining the specific binding partners. To measure the degree of variation between replicates, the coefficient of variation was calculated as the ratio of the standard deviation to the mean [= (standard deviation/mean) × 100].
Validation of PTBP1 Interaction with SPRIGHTLY by Co-immunoprecipitation
PTBP1 immunoprecipitation was performed using a Pierce classic IP kit (Thermo Fisher Scientific) according to the manufacturer’s protocols. Briefly, cell lysates (from 5.0 × 106 cells) were pre-cleared using control agarose resin and incubated with 1 μg of anti-PTBP1 antibody overnight. Immune complexes were captured with protein A/G agarose. After washing the captured immune complexes, half were used for protein elution with SDS sample buffer. The other half was used for RNA extraction by adding TRIzol and subjected to purification using a Direct-zol RNA Mini Prep kit (Zymo Research).
Cell Fractionation
To localize SPRIGHTLY or SMYD3 in the cell, cytosolic and nuclear fractions were prepared using the RiboTrap kit (MBL). The same amount of cytosol and nuclear extract was analyzed by western blotting using an anti-SMYD3 antibody. For SPRIGHTLY localization, RNAs were isolated from the cytosol and nuclear fractions using the Direct-zol RNA Mini Prep kit (Zymo Research), and SPRIGHTLY was detected by qRT-PCR.
Human Phospho-Protein Profiler Array
Lysates from DMSO- and EGF-treated cells were incubated with the Human Phospho-Kinase Array kit (ARY003C; R&D System, Minneapolis, MI, USAs) according to the manufacturer’s instructions. The protein signal was detected by chemiluminescent signal. The intensity of specific protein expression was quantified using ImageJ software.
Results
LncRNA SPRIGHTLY is Upregulated in G4 MB Cell Lines and in MB Patient-Derived Xenografts
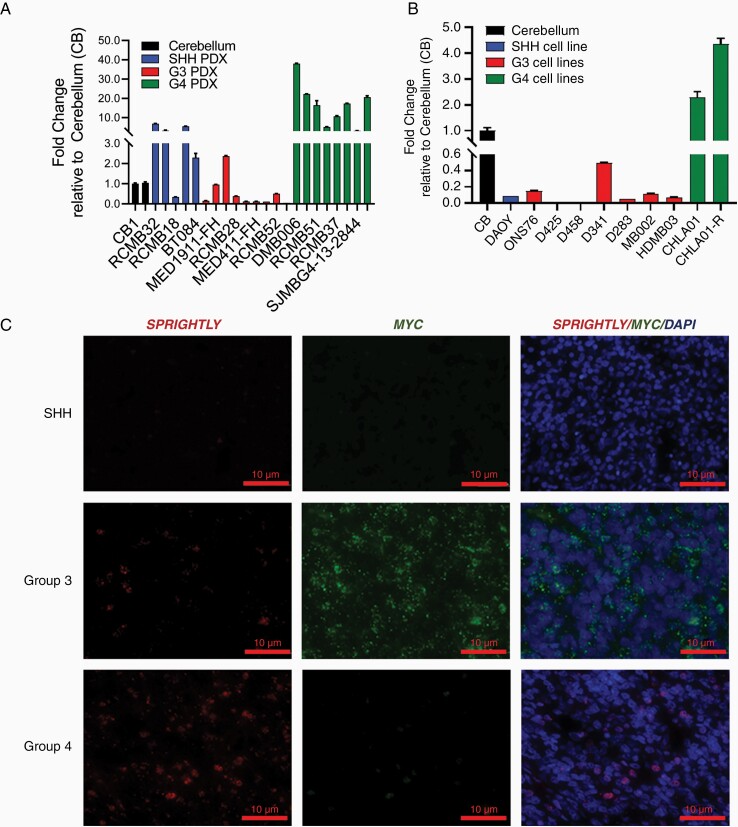
Since lncRNA expression is generally known to be more tissue-specific than that of mRNAs,38,39 we examined SPRIGHTLY expression in MB subgroup patient-derived xenografts (PDXs). SPRIGHTLY expression was highest in G4 PDXs (Figure 1A and Supplementary Figure S1A) relative to its expression in normal cerebellum. These observations were reflected in established MB cell lines: SPRIGHTLY expression was highest in the two G4 cell lines CHLA01 and CHLA01-R-MED (here after termed CHLA01-R; the drug resistant version of the CHLA01 cells from the same individual) compared with the other MB cell lines tested and normal cerebellum (Figure 1B and Supplementary Figure S1B). SPRIGHTLY-gene expression was further confirmed by RNA-FISH in patient tissue microarray samples and cell lines (Figure 1C and Supplementary Figure 1C). As shown in Figure 1C, SPRIGHTLY RNA-FISH signals were enriched in G4 patient tissues compared with the other groups. RNA-FISH further confirmed SPRIGHTLY transcript enrichment in G4 cell lines, with MYCN used as the G4 positive control4 (Supplementary Figure S1C). RNA-FISH analysis also showed SPRIGHTLY RNA localized to the nucleus, which was further confirmed by subcellular fractionation followed by qPCR analysis (Supplementary Figure S2). Lastly, we examined the expression level of SPRIGHTLY and SPRY4 (SPRIGHTLY host gene)16 expression levels reported in 175 patient-derived RNA-seq data.40SPRIGHTLY expression in G4 MB was higher than in other groups, especially relative to those in SHH MB. Interestingly, SPRIGHTLY expression pattern was quite different from that of its host gene SPRY4 in various subgroups of MB. SPRIGHTLY resides in the intron region of SPRY4 and the two RNA transcriptions were co-regulated in melanoma.41 By contrast to SPRIGHTLY expression pattern, the expression levels of SPRY4 mRNA in G4 MB samples were significantly lower than those in other groups (Supplementary Figure S3), possibly due to a different decay rate in MB cells.
Figure 1.
SPRIGHTLY expression in MB PDXs, cell lines, and patient samples. SPRIGHTLY expression was determined by qRT-PCR in various MB (A) PDX and (B) cell lines. (C) RNA-FISH analysis revealed that SPRIGHTLY is highly expressed primarily in Group 4 MBs compared with other groups. Student’s t-test. * ≤ .05, ** ≤ .01, **** ≤ .0001, ns, not significant.
SPRIGHTLY Knock-Down Decreases in Vitro and in Vivo Cellular Proliferation
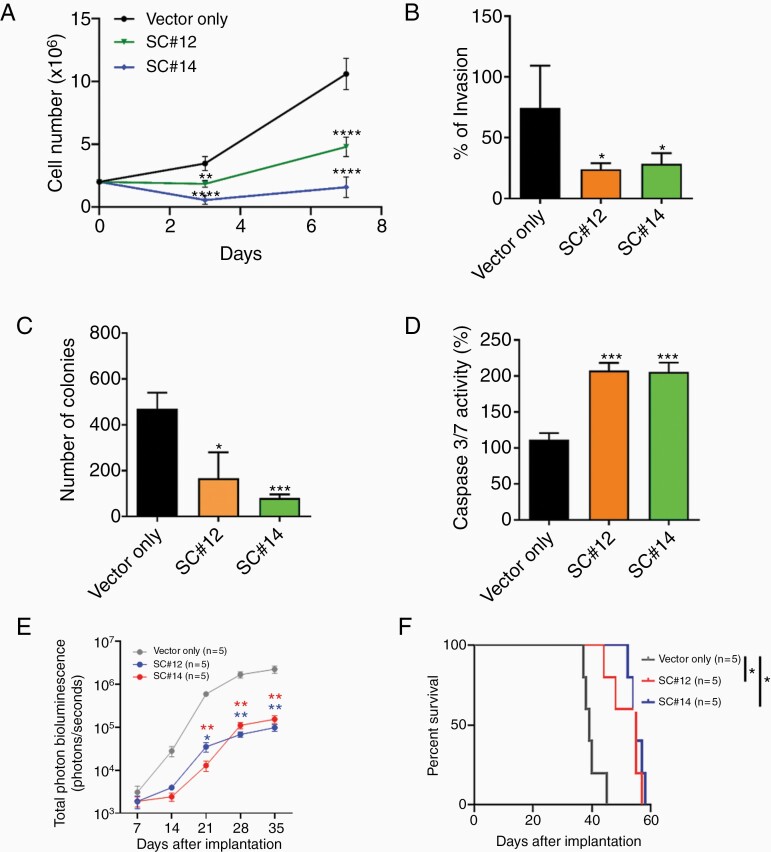
We next deleted the SPRIGHTLY in CHLA01 cells using CRISPR/Cas9 and isolated single clones that simultaneously exhibited PCR-determined hemizygous knockout of the SPRIGHTLY gene and at least 50% reduction in SPRIGHTLY expression (Supplementary Figure S4). Both of two independent single clones (SC#12 and SC#14) had reduced viability (Figure 2A), invasion, and colony formation in vitro (Figure 2B and C, Supplementary Figure S5). We postulated that the decreased viability arose from intrinsic apoptosis,42,43 which was reflected in the higher expression of caspases 3 and 7 in these cell lines relative to parental controls containing the “empty” lentiviral vector (Figure 2D).
Figure 2.
SPRIGHTLY knock-down affects cellular proliferation, invasion, soft agar colony formation, apoptosis, in vivo tumorigenicity, and mouse survival. Decreased SPRIGHTLY expression reduces (A) proliferation, (B) invasion, and (C) colony formation and increases (D) apoptosis. (E) Tumor volume Student’s t-test. * ≤ .05, ** ≤ .01, *** ≤ .001, **** ≤ .0001. (F) Kaplan-Meier survival curve of mice. The log-rank test was applied to assess the differences between groups. *P < .05, Mantel–Cox log-rank test.
Next, to study the in vivo effect of SPRIGHTLY knock-down, CHLA01 (transduced with empty lentiviral vector), SC#12, and SC#14 cells were separately injected intracranially into the cerebellums of NOD/SCID mice. SC#12 and SC#14 tumors were smaller than parental tumors at day 21 (P < .05). Signal intensity decreased by ~10-fold (at day 35) as judged by in vivo imaging of the orthotopic tumor (IVIS system; Figure 2E and Supplementary Figure S6A), representing a significant delay in tumor development in cells with hemizygous deletion of SPRIGHTLY. This result was further confirmed by H&E staining and Ki67 immunostaining showing reduced in vivo proliferation in SC#12 and SC#14 tumors compared with controls (Supplementary Figure S6B–D). In addition, according to the log-rank test, there was significant improvement of mice survival. Compared to mice implanted with parental G4 MB cells, the survival rates of mice injected with SC#12 and SC#14 cells were significantly higher (P < .05, Figure 2F). Together, these results demonstrate SPRIGHTLY expression promotes MB tumor growth in vivo.
SPRIGHTLY Binds to the Intronic Region of SMYD3 Pre-mRNA and Interacts with its Binding Partner PTBP1
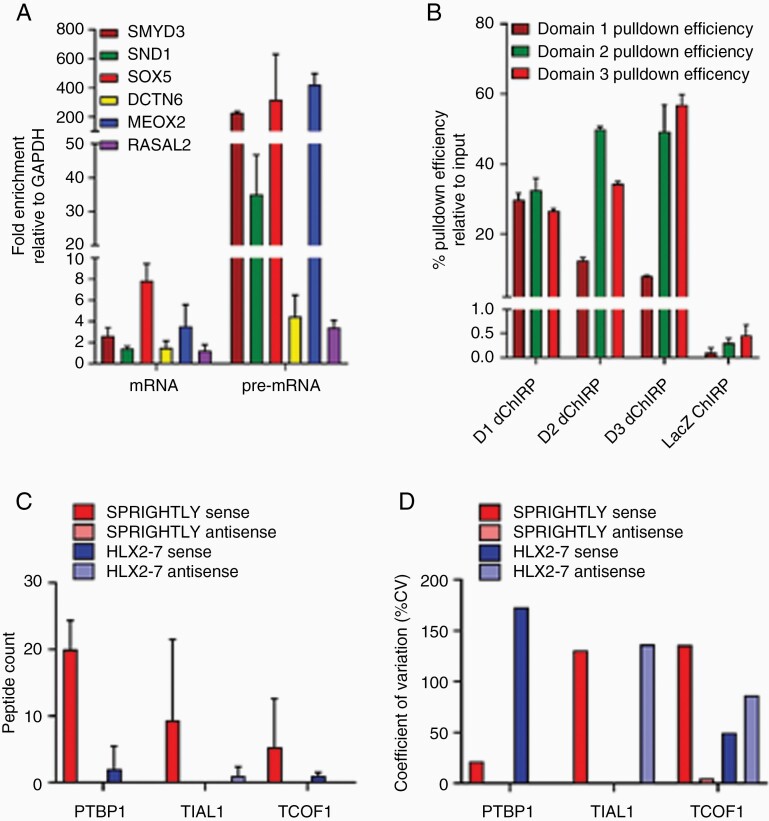
Since it is difficult to predict lncRNA function from its primary sequence, we sought to determine its binding partners, because the functions of its binding proteins might be informative. We previously identified a role for SPRIGHTLY in melanoma and reported its RNA-binding partners using domain-specific chromatin isolation by RNA purification (dChIRP).17 In melanoma cells, SPRIGHTLY binds to the intronic regions of six pre-mRNAs: SMYD3, SND1, SOX5, MEOX2, DCTN6, and RASAL2. We confirmed that the same six pre-mRNA partners still interacted with SPRIGHLY lncRNA in medulloblastoma cells by dChIPR-qPCR (Figure 3A). SPRIGHTLY has three distinguishable topological domains (D1–D3); three independent dChIRP pull-down assays were therefore carried out using the respective domain-specific biotinylated probes, and the pull-down efficiencies were determined by qPCR analysis using domain-specific qPCR primers17 (Figure 3B).
Figure 3.
SPRIGHTLY binds to its known RNA-binding partners and PTBP1 protein in the MB group 4 cell line CHLA01. dChIRP was performed with biotinylated probes, and SPRIGHTLY’s RNA-binding partners were determined by qRT-PCR. (A) dChIRP-qRT-PCR showing the co-purification of six RNA-binding partners with SPRIGHTLY. The relative fold change of co-purified RNAs are shown compared with the amount of non-specific GAPDH RNA. (B) Pull-down efficiency of SPRIGHTLY in each dChIRP. Ribo trap followed by mass spectrometry analysis of binding proteins shows that PTBP1 is a protein binding partner of SPRIGHTLY. (C) Top 3 protein binding partners. Identified peptide numbers are shown. (D) Coefficient of variation quantifying the variation between the three biological replicates.
The interaction between SPRIGHTLY and its six pre-mRNA binding partners is not mediated by sequence complementarity, suggesting that another factor, perhaps a protein, might be involved in this interaction.17 To identify a putative protein binding partner that might mediate such an interaction, we first labeled SPRIGHTLY transcripts with 5-bromo-UTP by in vitro transcription followed by incubation with nuclear extracts to form SPRIGHTLY-RNA-binding protein (RBP) complexes. The SPRIGHTLY-RBP complexes were then immunoaffinity-purified using anti-BrdU antibody. RBPs associated with SPRIGHTLY lncRNA were identified by mass spectrometry (Figure 3C and D). Antisense RNA of SPRIGHTLY (negative control for non-specific association), sense RNA of HLX-2-7 (positive control for a G3-specific lncRNA),13 and antisense RNA of HLX-2-7 (negative control for G3-specific lncRNA) were used in separate experiments to exclude proteins non-specifically pulled down with the SPRIGHTLY lncRNA. The total peptide count showed that polypyrimidine tract binding protein 1 (PTBP1) was the most enriched protein of those identified, followed by nucleolysin TIAR (TIAL1) and treacle ribosome biogenesis factor 1 (TCOF1) (Figure 3C). Non-specific binding proteins are expected to show less reproducibility in biological replicates. PTBP1 levels had the lowest coefficient of variation (CV) between replicates, suggesting that PTBP1 is most likely to be a specific binding partner protein of SPRIGHTLY (Figure 3D). To confirm the interaction between PTBP1 and SPRIGHTLY, we immunoprecipitated endogenous PTBP1 in CHLA01 cells and observed by qRT-PCR that SPRIGHTLY lncRNA had co-immunoprecipitated (Supplementary Figure S7). Of note, the primary sequence of SPRIGHTLY contains several PTBP1 consensus RNA-binding sequences44 (Supplementary Figure S8).
SPRIGHTLY lncRNA and PTBP1 Protein Regulate Exon 5 Skipping of SMYD3 Pre-mRNA in G4 MB Cells
PTBP1 is known to act as a repressor of alternative splicing, favoring exon skipping.45,46 Since SPRIGHTLY preferentially binds to at least six distinct pre-mRNA sequences (Figure 3A), we hypothesized that SPRIGHTLY lncRNA and PTBP1 protein cooperate to regulate RNA splicing in G4 MB cells. Among the six recognized targets of SPRIGHTLY lncRNA, we focused on SMYD3 because (a) it is a histone lysine-N-methyl transferase and thereby regulates epigenetic processes,47,48 (b) it is a direct target of HIF2α,25 which is controlled by the SHH signaling cascade, and (c), it is a regulator of EGFR signaling pathways as well direct regulatory of the telomerase gene.49 We therefore tested the hypothesis that SPRIGHTLY and PTBP1 together controls alternative splicing of SMYD3.
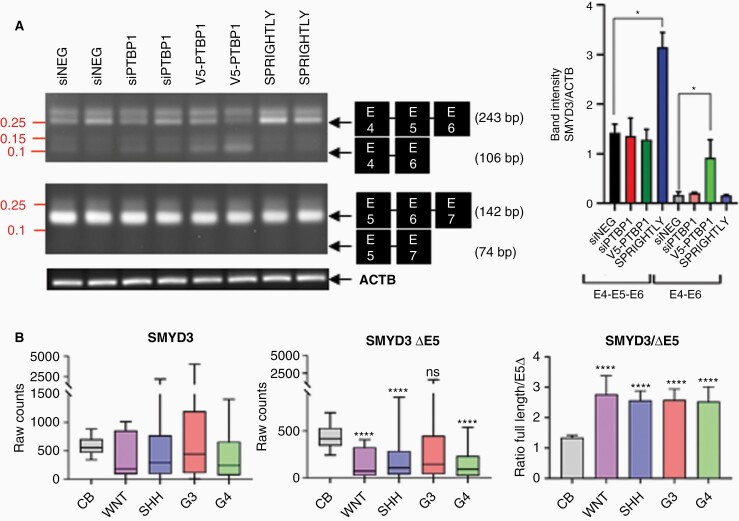
SPRIGHTLY binds to intron 5 of SMYD3 (NM_001167740.2),17 so if SPRIGHTLY and PTBP1 protein localize to intron 5 of SMYD3, there could be at least two possible alternative splicing scenarios: exon 5 skipping or exon 6 skipping (Supplementary Figure S9). We therefore designed PCR primers to detect the predicted exon-skipped and non-skipped transcripts and transfected cells with PTBP1 siRNA or overexpressed PTBP1 and SPRIGHTLY. As shown in Figure 4A, exon 5-skipped transcripts (SMYD3 ΔE5 transcript) were detected in the G4 cell line, CHLA01. Primers binding to exons 4 and 5 detected full-length (amplicon size 243 bp) and exon 5-skipped (amplicon size 106 bp) transcripts, indicating that both transcripts were transcribed together in CHLA01 cells. PTBP1 knock-down significantly reduced, and overexpression increased, SMYD3 ΔE5 transcripts, whereas the overexpression of SPRIGHTLY significantly increased the levels of full-length transcript (Figure 4A). Sanger sequencing further confirmed the predicted 243 bp and 106 bp PCR products of the skipped and non-skipped mRNAs: the 243 bp amplicon contained exons E4, E5, and E6, while the 106 bp amplicon contained only exons E4 and E6 (Supplementary Figure S10). Both full-length and ΔE5 transcripts were expressed in the G4 cell line CHLA01 (Figure 4A).
Figure 4.
The interaction of SPRIGHTLY with PTBP1 regulates SMYD3 exon skipping. The group 4 cell line was transfected with siPTBP1 or siNEG or PTBP1 or SPRIGHTLY was overexpressed to determine their effects on exon skipping. (A) The splicing variant lacking exon 5 was detected. Two independent experiments are shown. (B) The expression levels of full-length SMYD3 and SMYD3 E5 skipped (SMYD3 ΔE5) transcripts were determined in tissue RNA-seq data (left and middle panel). The ratio between SMYD3 ΔE5 and full length SMYD3 transcripts in each patient was determined (right panel). siNEG: siRNA for scrambled sequence, siPTBP1: siRNA for PTBP1, V5-PTBP1: V5 epitope-tagged PTBP1. Student’s t-test compared with normal cerebellum (CB). ns > .05, * ≤ .05, **** ≤ .0001.
To examine whether ΔE5 transcripts are expressed in patient samples, we retrieved RNA-seq data from twelve normal cerebellums50 and 175 MBs.4 The ratio of full-length to ΔE5 SMYD3 transcripts was indeed significantly higher in MBs than in normal patient samples, suggesting that the ΔE5 transcript is a characteristic of MB (Figure 4B, right panel bar graph). We suggest that the balance between the two transcript levels might affect MB development or maintenance.
The SMYD3/SPRIGHTLY Axis Stimulates the EGFR Pathway in G4 MBs
SMYD3 is a lysine methyltransferase with a variety of target proteins depending on the context and localization. Nuclear SMYD3 induces histone H3K4 di- or tri-methylation48 and histone H4K5 methylation,47 whereas cytosolic SMYD3 methylates MAP3K2 to stimulate the ERK1/2 pathway.51 Therefore, it is important to determine SMYD3 protein localization in G4 MB cells.
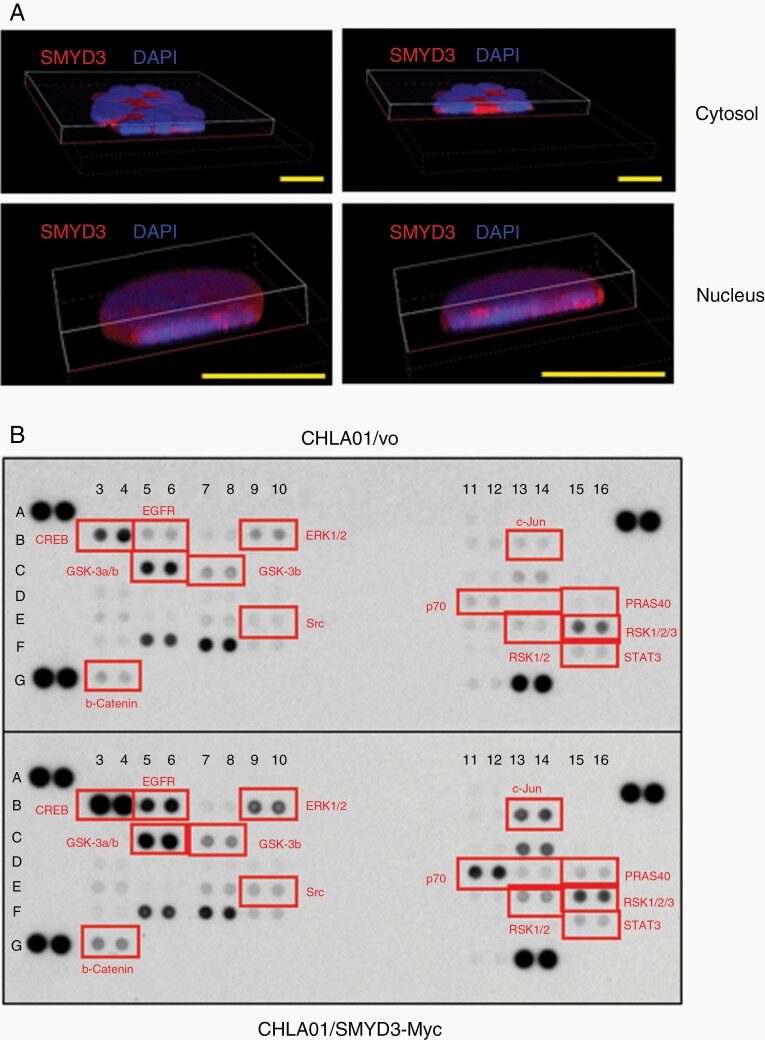
We performed immunofluorescence staining for endogenous SMYD3 to determine its cellular localization. Confocal immunofluorescence microscopy (Figure 5A) showed that most SMYD3 protein localizes to the cytoplasm, but 18% of cells showed nuclear SMYD3 (Figure 5A and Supplementary Figure S11A). Both localizations were confirmed by cell fractionation followed by western blot analysis with anti-SMYD3 antibodies (Supplementary Figure S11B). Interestingly, as shown in the middle and right panels of Supplementary Figure S11C, SMYD3 was enriched at cell–cell junctions. While CHLA01 cells tend to grow in suspension, they also form spheres of varying sizes. The localization of SMYD3 suggests that it might be involved in cell–cell contacts and cellular clumping. SMYD3 localization was further confirmed by stably overexpressing SMYD3-Myc fusion proteins in cell lines. Immunofluorescent confocal images with anti-Myc antibodies showed that SMYD3-Myc also localized to both the cytosol and nucleus, similar to endogenous SMYD3 protein (Supplementary Figure S12A). Stable clones of cells expressing SMYD3 ΔE5-Myc fusion proteins exhibited the same localization patterns (Supplementary Figure S12B).
Figure 5.
SMYD3 overexpression activates the EGFR pathway in CHLA01 cells. (A) Localization of endogenous SMYD3. Representative ×60 Z-stack confocal images showing cytosolic and nuclear localization. Scale bar 10 μm (B) Human phospho-kinase array showing the activation of the EGFR pathway upon SMYD3 overexpression. The phosphorylation level of each protein in duplicate was determined by western blot analysis. The dot intensity was quantified using an Image J software and shown in Supplementary Figure S14. SMYD3-Myc overexpression cells showed the increased the phosphorylation of EGFR pathway proteins. Red indicates elevated phosphoproteins in the EGFR pathway.
Since SMYD3 protein localizes to the cytosol, it may function as a lysine methyltransferase to enhance MAPK pathway activity, as observed previously.51 To identify which pathways are affected by SMYD3, we first induced the EGFR receptor tyrosine kinase pathway in cells overexpressing SMYD3 (and in a negative control line containing the empty vector) by stimulating with EGF and basic FGF (bFGF). EGF treatment increased ERK1/2 phosphorylation, whereas bFGF did not (Supplementary Figure S13). To compare the downstream signaling profiles between CHLA01/vo and CHLA01/SMYD3-Myc cells, a human phospho-kinase antibody array analysis was carried out on whole cell lysates. As expected, SMYD3 overexpression enhanced EGFR pathway signaling in CHLA01 cells (Figure 5B and Supplementary Figure S14). This suggests that SMYD3 methylates MAP3K2 in CHLA01 cells to activate the EGFR pathway.
SMYD3 and SPRIGHTLY Together Regulate Tumor-Associated Hypercoagulation Pathways
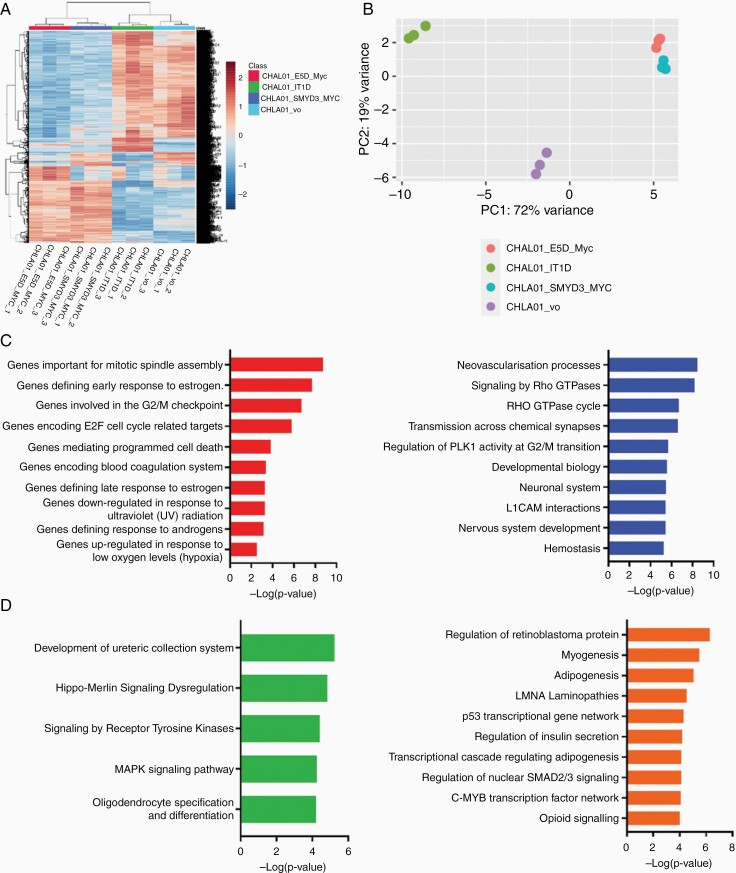
To understand the impact of SMYD3 and SPRIGHTLY in G4 MB, we subjected CHLA01/vo, CHLA01/SMYD3-Myc, CHLA01/SMYD3 ΔE5-Myc, and CHLA01/SPRIGHTLY-deleted (SC#14) cells to RNA-seq analysis. Differential gene expression analysis showed a clear distinctive gene expression pattern on SMYD3-Myc or SMYD3 ΔE5-Myc overexpression or SPRIGHTLY hemizygous deletion (Figure 6). Both unsupervised clustering (Figure 6A) and principal component analysis (Figure 6B) showed that the gene expression in SPRIGHTLY-deleted cells is distinct (clustering and PCA) from parental cells expressing both copies of SPRIGHTLY and the SMYD3 or SMYD3 ΔE5-overexpressing groups.
Figure 6.
RNA sequencing shows genes regulated by SMYD3 and SPRIGHTLY in CHLA01 cells. (A) Heatmap and (B) principal component analysis upon SMYD3 or SMYD3 ΔE5 overexpression or SPRIGHTLY depletion (CHLA01_IT1D). (C) Gene set enrichment analysis of common genes upregulated in SMYD3-overexpressing and downregulated in SPRIGHTLY-depleted cells. (left: hallmark gene set, right: canonical gene set). (D) Canonical gene sets of upregulated genes (left) or downregulated genes (right) in both SMYD3-overexpressing and SMYD3 ΔE5-overexpressing cells.
As predicted, SPRIGHTLY overexpression forced the splicing of SMYD3 towards the full-length SMYD3 transcript (Figure 4A), and, conversely, its deletion reduced full-length SMYD3 mRNA expression (Supplementary Figure S15). This suggests that both SPRIGHTLY and SMYD3 should regulate genes downstream of SMYD3 in the same direction. To test this, we subjected SMYD3-overexpressing and SPRIGHTLY-depleted RNA to RNA-seq analysis (Supplementary File 2). Gene set enrichment analysis (GSEA)52 showed that genes involved in mitotic spindle assembly were the major hallmark pathway genes in SMYD3-overexpressing cells (FDR 9.45 × 10-8; Figure 6C left panel). In addition, cell cycle regulation pathways (G2/M checkpoint genes and E2F target genes, FDR 3.25 × 10-6 and 2.09 × 10 -5, respectively) were regulated by both SMYD3 and SPRIGHTLY in CHLA01 cells.
Interestingly, “coagulation” and “hemostasis”-related gene sets were upregulated in SMYD3-overexpressing and downregulated in SPRIGHTLY-deleted cells (Figure 6C left and right panels). Cancer patients commonly exhibit a subclinical hypercoagulable state (an increased tendency to develop blood clots). Indeed, hypercoagulation is the second leading cause of cancer-related deaths53 and is known to promote tumor growth and progression by stimulating intracellular signaling pathways.54 For example, tumor cells secrete procoagulant and fibrinolytic molecules that are known to cause hypercoagulability in cancer patients.55 Tumor cells synthesize an activator of the blood coagulation called tissue factor (TF), thereby activating hemostasis. TF promotes thrombin generation and facilitates fibrin deposition, thus creating a hypercoagulation microenvironment that may protect tumor cells from immune system attack.56
As shown in Figure 6A and B, cells with endogenous SMYD3 or SMYD3 ΔE5 overexpression clustered together, suggesting that both proteins similarly affect downstream gene expression. In GSEA analysis (Figure 6D left and right panels), consistent with the human phospho-kinase array results (Figure 5B), receptor tyrosine kinase pathway (FDR 3.7 E-2) and MAPK kinase pathway genes (FDR 3.7 E-2) were upregulated in both SMYD3- and SMYD3 ΔE5-overexpressing cells (Figure 6D left panel), whereas, tumor suppressor gene pathways such as p53 signaling and the retinoblastoma protein regulation pathway were downregulated in both cell types (Figure 6D right panel).
Discussion and Conclusion
Here we have shown that the lncRNA SPRIGHTLY, expressed at high levels in G4 MB cells, prevents exon 5 skipping of SMYD3 precursor mRNA. This enables a higher expression level of the full-length SMYD3 transcript relative to that of the exon-skipped version. The expression of SPRIGHTLY and full-length SMYD3 mRNA was associated with increased expression of cell cycle, spindle apparatus, and hemostasis/coagulation genes and increased activity of EGFR pathway components accompanied by hyper-phosphorylation of ERK and downstream signaling proteins in G4 MB cell lines. Consistently, we demonstrate that a reduction in SPRIGHTLY levels through hemizygous deletion by CRISPR/Cas9 in G4 MB cells is accompanied by a reduction in the full-length SMYD3 mRNA, a corresponding increase in the exon 5-skipped version of the SMYD3 mRNA, and delayed G4 MB development in mice. Based on these evidence, we suggest that the abnormal expression (e.g., due to gene amplification) of SPRIGHTLY augments G4 MB through an over-production of the full-length SMYD3 protein. Our results establish a strong direct correlation between the ratio of the full-length to the exon-skipped shortened version of SMYD3 as an important parameter for G4 MB aggressiveness in a murine model xenografted with human MB cells. Since the SMYD3 gene maps at 1q44, the observed correlation is consistent with the frequent observation of chromosome 1 gain (24% of observed samples) in childhood medulloblastomas,57 and a case of cerebellar medulloblastoma with a chromosome 1q trisomy.58 This is because a gain in SMYD3 gene (located on 1q44) copies, and their increased levels of transcription to produce the precursor RNA, might titrate SPRIGHTLY/PTBP1 complexes such that the exon-skipped version of SMYD3 mRNA might be underproduced relative to that of full-length version, thus contributing to MB development.
The full-length SMYD3 protein has 428 amino acids and contains an MYND-type zinc finger domain and a SET domain at its N-terminal region.59 However, the SMYD3 ΔE5 transcript encodes a smaller protein of 140 amino acids due to the introduction of an early stop codon in exon 6 (Supplementary Figure S16A). Compared to full-length SMYD3, SMYD3 ΔE5 has lost most of the SET domain (N-lysine methyltransferase domain) but retains the MYND-type zinc finger domain, which might be involved in protein-protein interactions60 (Supplementary Figure S16B). Therefore, the ΔE5 version might help protein-protein interactions of the full-length protein or encode a dominant-negative protein that interferes with the normal SMYD3-encoded RNA-dependent methyltransferase. We therefore hypothesize that the expression of the full-length SMYD3 protein in G4 MB cells is related to its tumorigenic properties, and that in normal precursor stem cells—the progenitor of G4 MB cells—the ratio of the two versions of SMYD3 proteins is important for maintaining their stem cell characteristics. More specifically, the shortened protein from the exon 5-skipped SMYD3 mRNA is associated with a protective function whereas the full-length version of the SMYD3 protein promotes G4 MB. A salient prediction of this model, that the shortened protein from the exon 5-skipped version of SMYD3 mRNA is a dominant-negative regulator of the full-length SMYD3 protein, will be tested in future experiments. These results and the model of SPRIGHTLY action in G4 MB cells are summarized in Supplementary Figure S17.
Recent studies have noted that tumorigenesis induced by oncogene and tumor suppressor gene mutations can activate clotting pathways.56,61–63 For example, oncogenic mutation of KRAS and loss of function of p53 in colorectal cancer are associated with high TF expression.64 In a similar manner, glioblastomas, which often harbor oncogenic EGFR mutations, express high levels of TF.65 In MB, TF overexpression has been reported in the SHH subgroup66 but data are lacking for the G4 MB subgroup. Based on the results of GSEA analysis conducted on RNAseq data obtained from genetically modified G4 MB cells, we speculate that SPRIGHTLY and SMYD3, together with SMYD3-ΔE5 regulate hemostasis pathway gene expression patterns to influence clinical manifestations of G4 MB during later stages of tumor development.
Supplementary Material
Acknowledgments
We acknowledge JHU proteomics core for Mass Spec analysis to identify SPRIGHTLY-binding partners and Ms. Tiffany Casey for administrative support and manuscript submission.
Contributor Information
Bongyong Lee, Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, School of Medicine, Johns Hopkins University, 1650 Orleans St., Baltimore, MD 21231, USA; Johns Hopkins All Children’s Hospital, 600 5th St. South, St. Petersburg, FL 33701, USA.
Keisuke Katsushima, Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, School of Medicine, Johns Hopkins University, 1650 Orleans St., Baltimore, MD 21231, USA; Johns Hopkins All Children’s Hospital, 600 5th St. South, St. Petersburg, FL 33701, USA.
Rudramani Pokhrel, Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, School of Medicine, Johns Hopkins University, 1650 Orleans St., Baltimore, MD 21231, USA; Johns Hopkins All Children’s Hospital, 600 5th St. South, St. Petersburg, FL 33701, USA.
Menglang Yuan, Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, School of Medicine, Johns Hopkins University, 1650 Orleans St., Baltimore, MD 21231, USA; Johns Hopkins All Children’s Hospital, 600 5th St. South, St. Petersburg, FL 33701, USA.
Stacie Stapleton, Johns Hopkins All Children’s Hospital, 600 5th St. South, St. Petersburg, FL 33701, USA.
George Jallo, Johns Hopkins All Children’s Hospital, 600 5th St. South, St. Petersburg, FL 33701, USA.
Robert J Wechsler-Reya, Sanford Burnham Prebys Medical Discovery Institute, 10901 N. Torrey Pines Road, La Jolla, CA 92037, USA.
Charles G Eberhart, Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, School of Medicine, Johns Hopkins University, 1650 Orleans St., Baltimore, MD 21231, USA; Department of Pathology, Johns Hopkins University School of Medicine, 720 Rutland Ave – Ross Bldg 558, Baltimore, MD 21205, USA.
Animesh Ray, Riggs School of Applied Life Sciences, Keck Graduate Institute, Claremont CA, 91711, USA; Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA, USA.
Ranjan J Perera, Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, School of Medicine, Johns Hopkins University, 1650 Orleans St., Baltimore, MD 21231, USA; Johns Hopkins All Children’s Hospital, 600 5th St. South, St. Petersburg, FL 33701, USA.
Funding
This work was supported by the Schamroth Project funded by Ian’s Friends Foundation, Hough Family Foundation, and Susan and Robb Hough to Ranjan J. Perera and George Jallo, and NCI grant 1R37CA230400 to Ranjan J. Perera and Charles Eberhart.
Conflict of Interest Statement
The authors declare that they have no competing interests.
Authorship Statement
Conceptualization: RJP, BL, AR: data curation: BL, RP, KK; bioinformatics analysis: BL, RP; project administration: RJP; resources: CGE, RJW; supervision: RJP; validation: BL, KK, MY: visualization: BL, MY, KK, RP; writing—original draft preparation: BL, RJP; writing—review and editing: RJP, AR.
References
- 1. Louis DN, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6): 803– 820. [DOI] [PubMed] [Google Scholar]
- 2. Cavalli FMG, et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell. 2017;31(6): 737– 754 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Northcott PA, et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123(4): 615– 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Northcott PA, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663): 311– 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shih DJ, et al. Cytogenetic prognostication within medulloblastoma subgroups. J Clin Oncol. 2014;32(9): 886– 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramaswamy V, et al. Medulloblastoma subgroup-specific outcomes in irradiated children: who are the true high-risk patients? Neuro Oncol. 2016;18(2): 291– 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Northcott PA, et al. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012;8(6): 340– 351. [DOI] [PubMed] [Google Scholar]
- 8. Grassiot B, et al. Surgical management of posterior fossa medulloblastoma in children: The Lyon experience. Neurochirurgie. 2021;67(1): 52– 60. [DOI] [PubMed] [Google Scholar]
- 9. Grill J, et al. New research directions in medulloblastoma. Neurochirurgie. 2021;67(1): 87– 89. [DOI] [PubMed] [Google Scholar]
- 10. Perreault S, et al. MRI surrogates for molecular subgroups of medulloblastoma. AJNR Am J Neuroradiol. 2014;35(7): 1263– 1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katsushima K, et al. Long non-coding RNAs in brain tumors. NAR Cancer. 2021;3(1). zcaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Northcott PA, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488(7409): 49– 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katsushima K, et al. The long noncoding RNA lnc-HLX-2-7 is oncogenic in Group 3 medulloblastomas. Neuro Oncol. 2021;23(4): 572– 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Varon M, et al. The long noncoding RNA TP73-AS1 promotes tumorigenicity of medulloblastoma cells. Int J Cancer. 2019;145(12): 3402– 3413. [DOI] [PubMed] [Google Scholar]
- 15. Li B, et al. Long Noncoding RNA TP73-AS1 Modulates Medulloblastoma Progression In Vitro And In Vivo By Sponging miR-494-3p And Targeting EIF5A2. Onco Targets Ther. 2019;12:9873– 9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khaitan D, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71(11): 3852– 3862. [DOI] [PubMed] [Google Scholar]
- 17. Lee B, et al. The long noncoding RNA SPRIGHTLY acts as an intranuclear organizing hub for pre-mRNA molecules. Sci Adv. 2017;3(5): e1602505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richardson MK. and Sieber-Blum M.,. Pluripotent neural crest cells in the developing skin of the quail embryo. Dev Biol. 1993;157(2): 348– 358. [DOI] [PubMed] [Google Scholar]
- 19. Sieber-Blum M, et al. Distribution of pluripotent neural crest cells in the embryo and the role of brain-derived neurotrophic factor in the commitment to the primary sensory neuron lineage. J Neurobiol. 1993;24(2): 173– 184. [DOI] [PubMed] [Google Scholar]
- 20. Murphy M, et al. Neural stem cells. J Investig Dermatol Symp Proc. 1997;2(1): 8– 13. [DOI] [PubMed] [Google Scholar]
- 21. Cornell RA. and Eisen JS.,. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin Cell Dev Biol. 2005;16(6):663–672. [DOI] [PubMed] [Google Scholar]
- 22. Su S, et al. PLK1 and NOTCH Positively Correlate in Melanoma and Their Combined Inhibition Results in Synergistic Modulations of Key Melanoma Pathways. Mol Cancer Ther. 2021;20(1):161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bazzoni R. and Bentivegna A.,. Role of Notch Signaling Pathway in Glioblastoma Pathogenesis. Cancers. 2019;11(3):292. doi: 10.3390/cancers11030292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liang KH, et al. Notch signaling and natural killer cell infiltration in tumor tissues underlie medulloblastoma prognosis. Sci Rep. 2021;11(1) https://doi.org/ 10.1038/s41598-021-02651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu C, et al. VHL-HIF-2alpha axis-induced SMYD3 upregulation drives renal cell carcinoma progression via direct trans-activation of EGFR. Oncogene. 2020;39(21): 4286– 4298. [DOI] [PubMed] [Google Scholar]
- 26. Staberg M, et al. Combined EGFR- and notch inhibition display additive inhibitory effect on glioblastoma cell viability and glioblastoma-induced endothelial cell sprouting in vitro. Cancer Cell Int. 2016;26;16:34. doi: 10.1186/s12935-016-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aguirre A, Rubio ME., and Gallo V.,. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature. 2010;467(7313):323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma J, et al. Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J Clin Invest. 2010;120(1): 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frohlich H, et al. Dynamic Bayesian Network Modeling of the Interplay between EGFR and Hedgehog Signaling. PLoS One. 2015;16;10(11):e0142646. doi: 10.1371/journal.pone.0142646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gotschel F, et al. Synergism between Hedgehog-GLI and EGFR signaling in Hedgehog-responsive human medulloblastoma cells induces downregulation of canonical Hedgehog-target genes and stabilized expression of GLI1. PLoS One. 2013;8(6):e65403. doi: 10.1371/journal.pone.0065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schneider CA, Rasband WS., and Eliceiri KW,. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7): 671– 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Borowicz S., et al. The soft agar colony formation assay. J Vis Exp. 2014(92) (92):e51998. doi: 10.3791/51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rusert JM, et al. Functional Precision Medicine Identifies New Therapeutic Candidates for Medulloblastoma. Cancer Res. 2020;80(23):5393–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brabetz S, et al. A biobank of patient-derived pediatric brain tumor models. Nat Med. 2018;24(11):1752–1761. [DOI] [PubMed] [Google Scholar]
- 35. Smith KS, et al. Patient-derived orthotopic xenografts of pediatric brain tumors: a St. Jude resource. Acta Neuropathol. 2020;140(2):209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quinn JJ, et al. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32(9): 933– 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen S, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17): i884– i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jiang C, et al. Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget. 2016;7(6): 7120– 7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen X and Sun Z,. Novel lincRNA Discovery and Tissue-Specific Gene Expression across 30 Normal Human Tissues. Genes. 2021;21;12(5):614. doi: 10.3390/genes12050614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409): 100– 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mazar J, et al. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5(19): 8959– 8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fulda S. and Debatin KM,. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25(34): 4798– 4811. [DOI] [PubMed] [Google Scholar]
- 43. Pistritto G, et al. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging. 2016;8(4): 603– 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han A, et al. De novo prediction of PTBP1 binding and splicing targets reveals unexpected features of its RNA recognition and function. PLoS Comput Biol. 2014;10(1). doi: 10.1371/journal.pcbi.1003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spellman R and Smith CW,. Novel modes of splicing repression by PTB. Trends Biochem Sci. 2006;31(2): 73– 76. [DOI] [PubMed] [Google Scholar]
- 46. McCutcheon IE, et al. Expression of the splicing regulator polypyrimidine tract-binding protein in normal and neoplastic brain. Neuro Oncol. 2004;6(1): 9– 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hamamoto R, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6(8): 731– 740. [DOI] [PubMed] [Google Scholar]
- 48. Van Aller GS, et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics. 2012;7(4): 340– 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu C, et al. The telomerase reverse transcriptase (hTERT) gene is a direct target of the histone methyltransferase SMYD3. Cancer Res. 2007;67(6): 2626– 2631. [DOI] [PubMed] [Google Scholar]
- 50. Psych EC, Revealing the brain's molecular architecture. Science. 2018;362(6420): 1262– 1263. [DOI] [PubMed] [Google Scholar]
- 51. Mazur PK, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510(7504): 283– 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43): 15545– 1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khorana AA, et al. Thromboembolism is a leading cause of death in cancer patients receiving outpatient chemotherapy. J Thromb Haemost. 2007;5(3): 632– 634. [DOI] [PubMed] [Google Scholar]
- 54. Mandoj C, Tomao L, and Conti L,. Coagulation in Brain Tumors: Biological Basis and Clinical Implications. Front Neurol. 2019;10:181. doi: 10.3389/fneur.2019.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nasser NJ, Fox J, and Agbarya A,. Potential Mechanisms of Cancer–Related Hypercoagulability. Cancers. 2020;12(3): 566. doi: 10.3390/cancers12030566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Falanga A, Marchetti M, and Russo L,. The mechanisms of cancer-associated thrombosis. Thromb Res. 2015;135Suppl 1:S8– S11. [DOI] [PubMed] [Google Scholar]
- 57. Shlomit R, et al. Gains and losses of DNA sequences in childhood brain tumors analyzed by comparative genomic hybridization. Cancer Genet Cytogenet. 2000;121(1): 67– 72. [DOI] [PubMed] [Google Scholar]
- 58. Stratton MR, et al. A case of cerebellar medulloblastoma with a single chromosome abnormality. Cancer Genet Cytogenet. 1991;53(1): 101– 103. [DOI] [PubMed] [Google Scholar]
- 59. Fu W, et al. Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase. J Biol Chem. 2016;291(17): 9173– 9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Y, Li C, and Yang Z,. Is MYND Domain-Mediated Assembly of SMYD3 Complexes Involved in Calcium Dependent Signaling? Front Mol Biosci. 2019;6:121. doi: 10.3389/fmolb.2019.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Monteiro RQ, et al. Hypoxia regulates the expression of tissue factor pathway signaling elements in a rat glioma model. Oncol Lett. 2016;12(1): 315– 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. D'Asti E and Rak J,. Biological basis of personalized anticoagulation in cancer: oncogene and oncomir networks as putative regulators of coagulopathy. Thromb Res. 2016;140Suppl 1:S37– S43. [DOI] [PubMed] [Google Scholar]
- 63. Magnus N, et al. Oncogenes and the coagulation system--forces that modulate dormant and aggressive states in cancer. Thromb Res. 2014;133Suppl 2:S1– S9. [DOI] [PubMed] [Google Scholar]
- 64. Yu JL, et al. Oncogenic events regulate tissue factor expression in colorectal cancer cells: implications for tumor progression and angiogenesis. Blood. 2005;105(4): 1734– 1741. [DOI] [PubMed] [Google Scholar]
- 65. Rong Y, et al. PTEN and hypoxia regulate tissue factor expression and plasma coagulation by glioblastoma. Cancer Res. 2005;65(4): 1406– 1413. [DOI] [PubMed] [Google Scholar]
- 66. D'Asti E, Fang Y, and Rak J,. Brain neoplasms and coagulation-lessons from heterogeneity. Rambam Maimonides Med J. 2014;5 (4):e0030. doi: 10.5041/RMMJ.10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.